Abstract
Defining responses of the structural and immune cells in biologic systems is critically important to understanding disease states and responses to injury. This requires accurate and sensitive methods to define cell types in organ systems. The principal method to delineate the cell populations involved in these processes is flow cytometry. Although researchers increasingly use flow cytometry, technical challenges can affect its accuracy and reproducibility, thus significantly limiting scientific advancements. This challenge is particularly critical to lung immunology, as the lung is readily accessible and therefore used in preclinical and clinical studies to define potential therapeutics. Given the importance of flow cytometry in pulmonary research, the American Thoracic Society convened a working group to highlight issues and technical challenges to the performance of high-quality pulmonary flow cytometry, with a goal of improving its quality and reproducibility.
Keywords: flow cytometry, lung biology, reproducibility, cells
Contents
Overview
Introduction
Methods
Technology and Instrumentation
Recommendation
Other Technology
Recommendation
Sample Processing and Staining
Recommendation
Analysis and Data Presentation
Compensation and Gating
Autofluorescence
High-Content and Automated Data Analysis
Recommendation
Consensus Cellular Markers
Pulmonary Lymphocytes
Pulmonary Myeloid Cells
Pulmonary Structural Cells (Epithelial Cells, Endothelial Cells, and Stromal Cells)
Recommendation
Cell Sorting
Immunomagnetic Cell Sorting
Fluorescence-activated Cell Sorting
Collection Media
Recommendation
Components for Reporting Flow Cytometry Data in Publications
Conclusions
Overview
In this workshop report, we summarize key issues that exist while performing high-quality flow cytometry. Although these issues are common across flow cytometry applications, we focus on lung-specific concerns. The goal of the report is to improve the rigor and reproducibility of flow cytometry experiments and support potential users of this highly useful technology. The key findings of this workshop are as follows:
-
•
Flow cytometers use a combination of fluidics and lasers/detectors to identify surface and intracellular characteristics of single cells to define individual cells in complex tissues or biological fluids.
-
•
Performing high-quality flow cytometry experiments requires design based on the specific investigator’s research question. On the basis of this question, carefully considered decisions will need to be made on the type of cytometer, the tissue or biologic fluid to be used, the methods of digestion or sample preparation, the design of the flow cytometry panel, flow cytometry performance with attention to items such as compensation and autofluorescence, the analysis of data, and finally the method of reporting flow cytometry data for publication. All of these items require consideration and troubleshooting and can lead to faulty data if not well designed.
-
•
Lung tissue digestion methods require optimization for the desired cell type(s) and their viability. Poor tissue digestion either leads to ineffective liberalization of cells from lung tissue or excessive cell death. Furthermore, investigators should also consider the different pulmonary regions (airway, parenchyma, etc.) and if the tissue is normal or diseased, as these factors may alter the digestion.
-
•
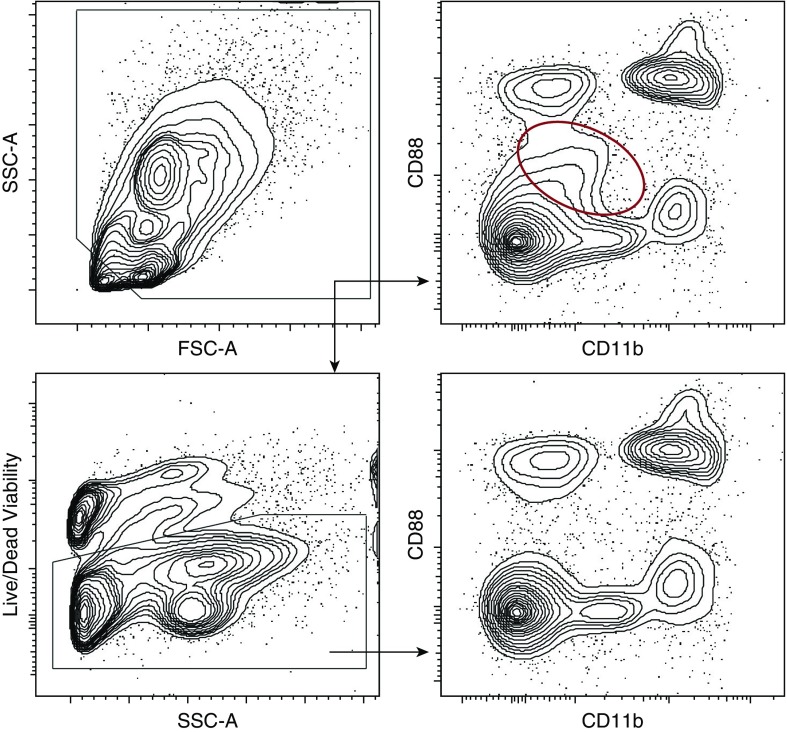
All flow cytometry samples should include an appropriate viability dye in their panel design. Flow cytometry data analysis then should be restricted to viable cells, as nonviable cells exhibit nonspecific fluorescence (Figure 1). It is not sufficient to use the light scatter properties of cells (forward scatter [FSC] vs. side scatter [SSC]) to define cell viability.
-
•
When defining individual cell populations, investigators should generally focus on defining cell types using standard markers and then describing how conditions alter surface expression (activation markers, etc.) or the frequency of cell populations. This is important so that the research community has common terminology, enabling better reproducibility of results across laboratories. To support this effort, cell surface markers are provided for standard pulmonary cell populations.
-
•
Reports of flow cytometry data for publication should include information on the antibodies (clone, commercial source, conjugated fluorophores, and dilution), the cytometer, and a clear gating strategy for how cells are defined. Placement of flow cytometry data on repositories is encouraged as a part of publication.
Figure 1.
Effect of apoptotic/necrotic cells and cell debris on flow cytometry plots with and without use of a viability dye. The red circle highlights a population of nonviable cells that are identified when a viability dye is not used in the staining conditions. Representative sample of a murine lung tissue digestion. FSC = forward scatter; SSC = side scatter.
Introduction
Increasingly, complex interactions of cells in organ systems are recognized to be drivers of injury and repair programs and crucial to organogenesis and maintenance of homeostasis. Unraveling complex cellular functions and interactions has tremendous potential to improve understanding of disease states and aid in the development of targeted therapeutics. This is particularly important to lung disease, as the lung is a principal immunologic organ, with complex interactions between immune and structural cells. These studies require careful identification and characterization of individual cell populations, principally performed by flow cytometry. Flow cytometry is the vital, mainstream tool for understanding the cellular basis of pathological processes and is the centerpiece of cellular phenotyping.
Cell phenotyping can provide a detailed snapshot of immune and structural function, which can serve to clarify responses in animal models, as a biomarker of disease state/activity, and potentially to direct the implementation of targeted therapeutics. In addition, flow cytometry is increasingly linked to novel -omics approaches (gene expression profiling, epigenomics, proteomics, metabolomics, etc.) and cellular and single-cell biology. However, despite its widespread and ever-increasing use, there are important technical and methodological concerns critical to the performance of high-quality flow cytometry (1). These challenges affect the reproducibility and generalizability of the results and can significantly hinder the advancement of research if performed incorrectly. A lack of standardized protocols and tools renders these issues particularly relevant.
Methods
To address these concerns with flow cytometry in the lung, the American Thoracic Society convened a panel of flow cytometry experts. The workshop co-chairs (R.M.T., A.V.M., and C.V.J.) identified international and U.S.-based participants on the basis of their flow cytometry expertise. All participants submitted conflict-of-interest statements before the workshop. The workshop considered the following general topic areas: sample processing, preparation, and cytometers; defining lung-specific reference panels; aspects of working with human lung samples; and methods of data analysis and reporting. The workshop convened on May 18, 2018 at the International Conference of the American Thoracic Society in San Diego, California. Individual workshop participants presented specific topics under the general topic sections. After those presentations, participants discussed the topics, and any key questions and needs were reviewed. Disagreements were resolved with discussion and consensus during the workshop and by subsequent discussions over e-mail. After the workshop, a writing committee was formed to be responsible for drafting the report (R.M.T., A.V.M., C.V.J., Y.-R.Y., B.D.S., and E.F.R.). After the generation of a draft document, all members of the workshop reviewed and revised the document before submission.
Technology and Instrumentation
Flow cytometers use fluidics to organize cell suspensions into a single-cell stream. Single cells, previously stained with antibodies conjugated to fluorochromes, are then subjected to defined light wavelengths to excite the individual fluorochromes. The emission spectra of these fluorochromes are then measured by specific detectors and converted to digital signals for analysis. The summative responses define individual cells by the unique expression patterns of these specific antibodies. Although in concept this process is relatively straightforward, flow cytometers vary considerably in terms of their configurations.
The components critical to individual cytometers include the laser source, mirrors, filters, and detectors. These features need to be considered both when designing experiments and when comparing results between different sites and instruments. A cytometer’s configuration should be reported in all flow cytometry publications—at a minimum, the cytometer make and model (2). Low-end analyzers have one to three lasers, whereas high-end analyzers are available with more than five lasers. These lasers can be set up in a colinear or a spatially separate arrangement. Fluorochromes emit light of different wavelengths, which are separated by a series of mirrors into individual wavelengths and then directed to detectors. Importantly, some fluorochromes are excited by different lasers on different instruments, which can change their emission characteristics. Before arriving at detectors, the wavelengths pass through filters to narrow the wavelengths directed to the detector. Long-pass filters allow all light above a specific wavelength to pass, short-pass filters allow light below a specific wavelength to pass, and band-pass filters allow only a specific range of wavelengths to pass. Therefore, it is important to make sure that the emission peak of individual antibody–fluorochrome combinations is appropriate for the associated mirrors and filters. Finally, the wavelengths reach the detectors, which are commonly photomultiplier tubes, although photodiodes are used in some instruments. The voltage can be set for each of the photomultiplier tubes, to maximize fluorescence resolution and dynamic range in the channels used. One generally accepted protocol for performing photomultiplier tube voltage optimization is the stain index voltration method (3). Given the complexity of lasers, mirrors, filters, and detectors, it is important to interact with individuals with flow cytometry expertise to ensure cytometry panels are designed appropriately for a given instrument’s optical configuration.
Recommendation
Identify if the configuration of the flow cytometer is optimized for the flow cytometry panel and the experimental design. We recommend consulting with a flow cytometry core staff or an individual with the expertise to assist with panel design to ensure optimal pairing of fluorochromes/antibodies to the instrument’s optical configuration.
Other Technology
Although traditional flow cytometry remains the main tool for single-cell analysis, several new instruments have been developed that can be useful for the pulmonary research community. These include spectral flow cytometers, imaging cytometers, and mass cytometers.
Spectral flow cytometers are similar to traditional flow cytometers, but instead of using only a portion of emitted signal (determined by the optical configuration, i.e., filters and mirrors), they capture the entire emitted spectrum, independent of the markers or fluorescent dyes. Spectral unmixing algorithms, similar to those used in fluorescence microscopy, are then used to deconvolute the data and unmask signal from fluorochromes with overlapping emission spectra. Unlike conventional fluorescence cytometers that dedicate one detector per fluorophore, spectral cytometers can resolve many more fluorophores simultaneously, regardless of the number of detectors.
Imaging flow cytometry bridges the gap between fluorescent microscopy (low throughput, low dimensionality, spatial context) and conventional flow cytometry (high throughput, high dimensionality, no spatial context). Current commercially available imaging flow cytometry instruments combine the design of a traditional flow cytometer and a microscope; they can capture images in up to 10 fluorescence channels and at different magnifications. This approach provides information about the cell size and shape and the spatial distribution of its fluorescent signal. Thus, imaging cytometry can provide researchers with high-dimensional data, which come at the expense of slow acquisition, large file size (>0.5 GB for 10,000 cells), and delayed analysis time, making it less suitable for the analysis of rare events.
Mass cytometry, also known as cytometry of time of flight, is an alternative technique that offers increased dimensionality and data yield in single-cell experiments. It uses stable isotopes of rare earth metals instead of fluorochromes. By design, mass cytometry is free from the issues related to spectral overlap/compensation and autofluorescence. In theory, more than 60 channels can be detected simultaneously with minimal signal overlap. However, care should be taken to understand the possibility of metal isotope contaminants in the sample. For example, reports have indicated that the use of medicinal iodine can interfere with mass cytometry analysis in lung samples (4). Although the potential gain of information from a well-designed and well-executed mass cytometry experiment is high, this technique requires significant upfront investment in reagents and optimization, as well as reliable infrastructure and experienced personnel. Most importantly, similar to any high-content data, analysis of mass cytometry data requires substantial computational infrastructure and expertise.
Recommendation
The choice of technology should be appropriate to answer the experimental question and match the researcher’s expertise and availability of resources.
Sample Processing and Staining
Sample processing and staining are a critical component in performing high-quality flow cytometry. The consensus among the workshop group is that poor handling and nonstandardized sample processing is the step most likely to result in poor flow cytometry performance and the one that most frequently leads to the presence of “false-positive” populations. This is because individual methods of tissue digestion and cell processing can alter the expression of cell surface markers and intracellular proteins (5–7). In addition, debris and apoptotic or necrotic cells accrued as a result of sample processing can lead to spurious cellular signals (Figure 1). Therefore, before initiating any study, a standardized methodology is required to limit issues with flow cytometry performance and interpretation.
The lung has several unique tissue compartments that can be sampled (airspace, vascular, and tissue/interstitium) and which require different considerations to obtain viable cells. BAL fluid is obtained by instillation and return of fluid through a bronchoscope (humans) or tracheal cannula (rodents). The type of lavage fluid and the method of sampling can vary widely in individual protocols and can affect the cell count (8). It is less clear if methodological differences affect the proportion of cell types. Standard methods should be used in human studies, including the type of lavage fluid used, the anatomic location of the lavage, the method of aspiration, and discarding or separately processing the return of the first-instilled aliquot (9, 10). After BAL, the lavage fluid should be maintained on ice and processed expeditiously to preserve the viability of the cells.
Lung tissue digestion has fewer standard processes and wide interlaboratory variability. The major concern with tissue digestion is maximizing cell recovery while limiting debris and cellular apoptosis/necrosis. In addition, it is clear that digestion protocols need to be tailored to the cell type, as some protocols favor structural cells over immune cells and vice versa. For example, digestion methods have been designed specifically for the isolation of epithelial cells (7, 11), whereas others have designed methods for myeloid cells (12, 13). Some labs use purely mechanical disaggregation methodologies without any enzymes to release viable hematopoietic cell types (14). Another consideration is the route of tissue digestion mixture administration, which can affect the recovery of specific cell types (7). In addition, some cells are more sensitive to digestion enzymes than others, and some digestion enzymes can cleave surface markers, thus preventing detection of the cells of interest (6, 7, 15). Finally, digestion protocols may require further optimization, depending on the nature of the disease and tissue injury. Therefore, before initiation of experiments, consideration and testing of the digestion protocol are crucial to the ultimate quality and reproducibility of flow cytometry experiments.
A specific lung tissue consideration is the tissue location of immune and structural cells. As the lung is highly vascularized, immune cells can be located within the vasculature. Investigators have traditionally used tissue perfusion to remove intravascular immune cells, including heparin administration before this perfusion (16). However, detailed studies demonstrate that intravascular immune cells remain despite perfusion (17). To address this concern, Desch and colleagues designed a protocol for intravascular administration of fluorescent dyes to segregate intravascular from intraparenchymal immune cells (18). This technique may be difficult to perform or not required in all experimental settings but should be considered in settings where careful compartment separation is required. An additional compartment consideration is that individual immune and structural cell populations will vary depending on the type of tissue. For example, tissue from central airways will increase the yield of airway epithelial cells and dendritic cells, whereas distal lung tissue will be enriched for alveolar epithelial cells and alveolar macrophages. This must be considered, particularly when attempting to quantify specific immune or structural cell populations.
Sample preservation is a variable in the performance of flow cytometry. There is limited consensus on this issue, particularly in the lung. Cryopreservation has been used with success in peripheral blood immune cells (19) but has not been explicitly studied in BAL cells or lung tissue. It is typically recommended to perform flow cytometry on fresh tissue or BAL cells. However, some investigators have fixed cells and then stained them (20). In these experiments, the cells were stained with a fixation-resistant viability dye before fixation. The samples then can be maintained for 2 to 3 weeks before performing staining. Alternatively, a design that was effective in a multicenter trial was to stain cells at clinical sites, fix them before shipping, and analyze them on a single flow cytometer (21). An initial consideration is that fixatives can degrade some organic and tandem dyes. Therefore, the recommendation is to either have short periods of fixation where the fixative is removed before staining or to account for the use of fixatives when these fluorochromes are used in panel design. Again, it is important to test these conditions between fresh and cryopreserved or fixed samples to confirm that preservation does not affect staining conditions.
Once cells are in a single-cell suspension, the next consideration is the panel design. This choice should be considered with care and in consultation with institutional expertise. To assist with design, the following section (Consensus Cellular Markers) suggests cell surface markers to define individual cell populations. Beyond the decision of individual cell-surface markers, other considerations are required. These include the antibody clone (which can affect staining characteristics), fluorochrome/antibody combination, and capabilities of the cytometer (reviewed in Reference 3). Panels should be tested and adjusted before performance of experiments to ensure they clearly define the cells and markers of interest. All panels should include a viability dye, particularly when using digested tissues. The use of light-scattering properties (FSC vs. SCC) as a sole method to assess cell viability is not sufficient and is associated with spurious flow cytometry results. In situations where excessive debris accumulation cannot be mitigated, a fluorescent DNA-binding dye compatible with the fluorescence panel design can be used on fixed samples to differentiate between cells and nonnucleated debris (22, 23).
During staining, antibodies should be used in a master mix cocktail to ensure that variations in staining do not result from differences in antibody concentrations between samples. Such antibody cocktails are stable for weeks and can reduce variability during the longitudinal studies. To this point, some commercial vendors offer lyophilized premade antibody cocktails. Preference should be made for use of monoclonal antibodies that are directly conjugated, which reduces the number of steps involved in staining and increases staining specificity. Particularly with phagocytic cells, Fc receptors should be blocked unless required for a specific cellular signal (24, 25). To reduce nonspecific staining, normal rat and mouse serum (in rodent flow cytometry) or normal human serum (in human flow cytometry) should be used in staining conditions. The majority of protocols perform incubation with blocking mixtures (typically Fc receptor block and normal serum) for a period before staining (16, 26). Staining should then occur in the presence of the blocking mixture. Furthermore, use of BSA in the staining buffer may prevent nonspecific binding of antibodies due to non-Fc receptor interactions. Antibody concentration should be titrated to work with a set number of cells to prevent over/under staining. After staining, typically at 4°C, washing steps should be performed to remove unbound antibodies. These conditions are important to limit the amount of nonspecific staining.
Recommendation
Sample processing and staining are critical components of performing reproducible, high-quality flow cytometry. Defining the samples collected, the methods of collection, the tissue processing/digestion, and cryopreservation/fixation needs to be carefully considered and tested. In addition, the conditions of staining need to include appropriate steps to limit nonspecific staining. Failure to do this will lead to spurious staining and poor ability to accurately and reproducibly define cell populations and activation markers.
Analysis and Data Presentation
Compensation and Gating
Proper compensation and gating are necessary to correctly visualize and interpret flow cytometry data (27). New users often perceive compensation as an overly complicated and mysterious procedure, leading them to avoid designing and setting up multicolor experiments in lieu of simple three- or four-color panels. Some experienced users who were trained during the time when modern reagents and algorithms for setting up compensation were lacking may recommend or even insist on using manual compensation. However, in reality, compensation is a simple and logical procedure, and a variety of detailed step-by-step guides are available (28, 29); for new and less-experienced investigators it produces better and more reproducible results. Most popular software packages include tools for calculating and applying automated compensation. Thus, users should avoid manual (i.e., nonautomated) compensation, as it is not nearly as accurate as automated compensation. Generally, the compensation matrix depends on a combination of fluorochromes used for staining, rather than on the cell type (see section below on autofluorescence).
To define truly positive populations in multicolor experiments, use of fluorescence-minus-one controls, where a sample is stained with all antibodies in a panel except for one, is highly recommended (29), although it may be difficult to implement when sample size is limited. An optimized panel containing fluorescence-minus-one controls is particularly important for flow sorting experiments to increase cell purity in a sorted population. Polystyrene antibody capture beads or amine reactive beads can be used to set up reliable compensation controls for fluorochrome-conjugated antibodies and amino-reactive fixable live-dead dyes, correspondingly. FSC (cell size) and SSC (cell granularity) can provide valuable information and assist identification of the cell type of interest (for example, high SSC of granulocytes or alveolar epithelial type II cells). However, these parameters should be used only in conjunction with specific cell markers and not on their own.
Autofluorescence
All cell types inherently possess autofluorescence due to differing amounts of natural fluorochromes, including nicotinamide adenine dinucleotide phosphate (NAD(P)H), flavins, porphyrin, lipofuscin, and others (30). Each of these endogenous fluorophores has distinct excitation and emission characteristics. However, autofluorescence is more pronounced in some cell types. In the lung, alveolar type II cells and alveolar macrophages—cells producing and metabolizing surfactants, respectively—have the highest autofluorescence (31). Various factors, such as smoking or environmental exposures, can increase cellular autofluorescence. Generally, autofluorescence is greatest in the violet and green wavelengths and less, though still present, in the red and far-red wavelengths (30, 32). Proper panel design and fluorochrome assignment can mitigate autofluorescence-related issues, or autofluorescence can be used to assist with cellular separation. It is important to recognize autofluorescence and distinguish it from undercompensated samples (33).
High-Content and Automated Data Analysis
Historically, analysis of flow cytometry data was performed by setting user-defined thresholds (gates), typically on two-dimensional plots. In the case of the complex panels, each gate can be further subsampled and reassessed using a different set of parameters, a practice known as sequential gating. Although this approach performs well for simple assays with well-defined markers, the increased number of markers that can be detected in cytometric assays necessitated the introduction of novel tools for analysis. Various dimensionality reduction, visualization, and clustering techniques have been adopted for identification of the specific cellular populations of flow cytometry data, including self-organizing maps, t-distributed stochastic neighbor embedding (tSNE), uniform manifold approximation and projection (UMAP), and Phenograph (34–40).
Several tools for supervised and unsupervised automated flow cytometry data analysis have been developed and have been shown to outperform expert-driven analysis, making them particularly attractive for analysis of data from large multicenter trials (41–45). Importantly, these packages support automated data preprocessing (cleaning) and can be organized into multipackage pipelines. Use of these tools and deposition of the raw data into public data repositories (such as FlowRepository) directly address the need for increased rigor, reproducibility, and transparency in flow cytometric studies (46, 47). Importantly, almost all of these powerful tools are available at no cost.
Recommendation
Instrument settings, experimental protocols, gating strategies, and reagents in accordance with appropriate guidelines should always be reported or referenced in publications. Use of open source tools for reproducible analysis and providing access to raw data is highly desirable.
Consensus Cellular Markers
A significant consideration in the development and utility of flow cytometry is panel design. Panel design should be focused on the individual investigator’s experimental question; therefore, there are unlikely to be single comprehensive panels for every study. However, the workshop members agreed that it is important to use common markers and terminology to define cells in the lung and BAL fluid. The importance of such standardization has been expressed in other research communities to harmonize terminology and identification of individual cell types (48, 49). Of note, some of these markers exhibit redundancy but are offered to be inclusive. Markers should be used to define the cell population of interest while also using specific markers to exclude other cell populations. Therefore, combinations are required to accurately identify specific cell populations. The workshop group recommended that investigators focus on defining cell types using standard markers and then describing how conditions alter surface expression (activation markers, etc.) or the frequency of cell populations. This is important so that the research community has common terminology, enabling better reproducibility of results across laboratories. To this end, the workshop panel focused on defining some consensus markers of pulmonary cell types. These panels are not meant to be definitive but rather to provide a guide to the pulmonary research community.
Pulmonary Lymphocytes
Most markers of lung and alveolar T-cell populations and subpopulations stem from observations in blood, secondary lymphoid organs, and other mucosal tissues (Table 1). Innate lymphoid cells, mucosal-associated invariant T cells, and natural killer (T) cells are not discussed in this section (50–52). In mice and humans, lung and alveolar T cells are generally identified by their low level of SSC and expression of a T-cell receptor complex component—CD3ε and specific T-cell receptor subtypes αβ or γδ. CD4+ and CD8+ subsets, which are nearly mutually exclusive in the lung, can be further defined once the T-cell receptor bearing, low-SSC T-cell population is identified (7). A critical CD4+ T-cell subtype required for maintenance of immune homeostasis, the Foxp3+ regulatory T (Treg) cell, is identified by staining for the Foxp3 transcription factor itself or a transgenic fluorochrome reporter knocked into the Foxp3 locus. In humans, low expression of CD127 (the IL-7 receptor) and high expression of CD25 (the IL-2 receptor α subunit) on the cell surface identify Treg cells (10, 53), although numerous other surface markers have been proposed (54). Murine effector T cells are identified by expression of CD44 and low levels of CD62L or CCR7; naive T cells display the opposite pattern (55, 56). Human CD4+ and CD8+ non-Treg T-cell populations include effector-memory (Tem), central memory (Tcm), and naive (Tn), in addition to Tem cells that express CD45RA (TemRA) (57–60). These four subsets can be defined based on expression of CCR7 (or CD62L) and CD45RA as follows: Tem, CCR7loCD45RAlo; Tcm, CCR7hiCD45RAlo; Tn, CCR7hiCD45RAhi; and TemRA, CCR7loCD45RAhi. T-helper subtypes are generally defined based on intracellular staining for canonical transcription factors and cytokines after ex vivo stimulation (e.g., T-box transcription factor 21 and IFN-γ for T-helper cell type 1 [Th1], Gata3 and IL-4 for Th2, retinoic acid–related orphan receptor γT and IL-17A for Th17, etc.). In addition, tissue-resident memory T cells (Trm) appear in the lung after complex signaling interactions (61). CD11a+CD69+ status identifies CD4+ Trm cells, and CD103+CD69+ status identifies CD8+ Trm cells (62–64). Finally, other surface markers often used in T cell panels, particularly for human samples, include human leukocyte antigen–DR isotype (HLA-DR) and CD38 (activation) and CD27 and CD45RO (naive/memory) (65).
Table 1.
Lymphocytes
| Population | Murine Markers | Human Markers | References |
|---|---|---|---|
| CD4+ T cell | Low SSC, CD3ε+ or TCRαβ or γδ+, CD4+ | — | |
| CD8+ T cell | Low SSC, CD3ε+ or TCRαβ or γδ+, CD8+ | — | |
| CD4+ Treg cell | CD4+ T cell expressing Foxp3-transgenic fluorochrome, CD25hi | CD4+ T cell, CD127lo, CD25hi; intra-nuclear staining for FOXP3 | 10, 53, 54 |
| Effector T cell | CD4+ or CD8+ T cell, CD62LloCD44hi, CD69hi | See below | 55, 56 |
| Naive T cell | CD4+ or CD8+ T cell, CD62LhiCD44lo, CD69lo | CD4+ or CD8+ T cell, CCR7hiCD45RAhi | 55–60 |
| Effector-memory T cell | — | CCR7loCD45RAlo | |
| Central-memory T cell | — | CCR7hiCD45RAlo | |
| T helper subset | Generally defined by transcription factor and cytokine staining (see text for examples) | — | |
| CD4+ tissue-resident memory T cells | CD4+ T cell, CD11a+CD69+ | — | 61–64 |
| CD8+ tissue-resident memory T cells | CD8+ T cell, CD103+CD69+ | — |
Definition of abbreviations: SSC = side scatter; Treg = regulatory T.
Pulmonary Myeloid Cells
Myeloid cells are cells that derive from a common myeloid progenitor, specifically a myeloblast. Their origin can be either tissue derived (seeded during embryonic development and locally maintained) or bone marrow derived (derived from circulating intermediates via bone marrow production). In the lung, myeloid compartment cells are a mix of tissue-derived (alveolar macrophages, subsets of interstitial macrophages, and monocytes) and bone marrow–derived (subsets of interstitial macrophages and monocytes, neutrophils, eosinophils, and basophils) cells. This origin definition has been worked out in rodents but has not been directly proven in humans. In the recent time frame, several panels have been developed to define pulmonary myeloid cells in rodents (16, 26, 66) and humans (18, 20, 67). The consensus of individual markers is summarized in Table 2.
Table 2.
Myeloid Cells
| Cell Group | Population | Murine Markers | Human Markers | References |
|---|---|---|---|---|
| Monocytes | Classical monocytes | CD11b+, CD115+, CCR2+, F4/80, CD43lo, Ly6Chi | CD64+, CD14hi, CD16lo/- | 16, 18, 20, 26, 67 |
| Nonclassical monocytes | CD11b+, CD115+, F4/80, CD43hi, CX3CR1hi, Ly6Clo | CD64+, CD14lo, CD16hi | 16, 18, 20, 26 | |
| CD14+ CD16+ monocytes | CD64+, CD14+, CD16+ | 18, 20 | ||
| Macrophages | Alveolar macrophages | CD11b−, CD11c+, CD64+, CD206+, CD169+, MerTK+, Siglec-F+ | CD11b+, CD11c+, CD64+, CD206+, CD169+, CD14lo/− | 16, 18, 20, 26 |
| Interstitial macrophages | CD11b+, CD11clo/+, CD64+, CD206+, CD169+/−, CX3CR1+, MerTK+, CCR2+/− | CD11b+, CD11c+, CD64+, CD206+, CD169lo, CD14hi | 76, 77 | |
| Monocyte-derived cells | Monocyte-derived macrophages (acute lung injury) | CD11b+, CD64+, CX3CR1+, Ly6C+/−, CCR2+/− | — | 16, 75, 78, 79 |
| Murine classical DC (cDC) | CD103+ CD11b− cDC | CD11c+, MHCII+, MerTK−, CD64−, CD24+, CD26+, CD11b−, CD103+, XCR1+ | — | 16 |
| CD103− CD11b+ cDC | CD11c+, MHCII+, CD64−, MerTK−, CD24+, CD26+, CD301−, CD11b+, SIRPα+ | — | ||
| Human DC | Pulmonary DC | — | CD11c+, MHCII+, CD64−, CD1c+, CD141−, CD206−, CD169−, CD14−, CD1a+ | 20 |
| CD1ahi DC | — | CD11c+, MHCII+, CD64−, CD1c+, CD141−, CD206+, CD169−, CD14lo/−, CD1a+ | ||
| CD1alo DC | — | CD11c+, MHCII+, CD64−, CD1c+, CD141−, CD206+, CD169−, CD14+, CD1a− | ||
| Granulocytes | Neutrophils | SSChi, CD24+, Ly6G+ | SSChi, CD24+, CD16+, CD15+, CD66bhi | 16, 20 |
| Eosinophils | SSChi, CD24+, Siglec-Flo, CD11c−, F4/80lo | SSChi, CD24+, CD16−, CD66blo, Siglec-8+ | 80 | |
| Basophils | SSClo, CD3−, CD19−, CD49b+, FceRI+, CD117−, IgE+ | SSClo, CD14−, HLA-DR, CD123+ | 81 |
Definition of abbreviations: DC = dendritic cell; HLA-DR = human leukocyte antigen–DR isotype; MerTK = tyrosine-protein kinase Mer; MHCII = major histocompatibility complex class II; Siglec = sialic acid–binding immunoglobulin-type lectins; SIRPα = signal regulatory protein α; SSC = side scatter; XCR = X-C motif chemokine receptors. “+/−” signify variable expression depending on the timing after acute lung injury.
Pulmonary Structural Cells (Epithelial Cells, Endothelial Cells, and Stromal Cells)
The development of appropriate markers for nonimmunologic cells is less mature than other pulmonary cell types. The panels rely on a few cell-specific markers for positive selection along with lineage markers used for negative selection. As a reminder, it is important to consider the digestion technique required to successfully liberate structural cells from tissues. These digestion techniques vary significantly for the isolation of different epithelial cell populations (68–70), fibroblasts (68), and endothelial cells (7) and can be different than what is used for immune cell–focused protocols. In addition, stromal cell identification leans heavily on the use of lineage labeling with genetic reporter mice (identified by italics) in addition to the use of specific fluorescently conjugated antibodies (71). These are defined in Table 3.
Table 3.
Pulmonary Structural Cells
| Cell Group | Population | Murine Markers | Human Markers | References |
|---|---|---|---|---|
| Epithelial cells | Epithelial cells (general) | EpCAM/CD326+, CD45−, CD31− | EpCAM/CD326+, CD45−, CD31− | |
| Basal cells | NGFR/CD271+, Krt5+ | NGFR/CD271+, ITGA+ | 68 | |
| Club cells | CD24+, Scgb1a1 | CD24+ | 82 | |
| Alveolar epithelial (type I) cells | RAGE (Ager), Hopx, T1α (Gp38/Podoplanin) | RAGE (Ager), T1α (Gp38/Podoplanin) | 69, 83 | |
| Alveolar epithelial (type II) cells | CD24−, Sftpc, sca 1−, Integrin B4−, lysotracker | HT2-280+, SPC+, CD24−, Integrin B4−, HLA-DR+, Lysotracker+ | 84–88 | |
| Alveolar epithelial progenitor cells | Axin2-Tdt, EpCAM+, CD31−, CD45− | TM4SF1, HT2-280 | 89 | |
| Bronchial epithelial cells | EpCAM/CD326high, CD24high, Integrin B4+ | — | 90 | |
| Endothelial cells | Endothelial cells (general) | CD31+, EpCAM−, CD45−, Thrombomodulin, ICAM-2, Tie2 | CD31+ EpCAM−, CD45−, Thrombomodulin, ICAM-2 | 91, 92 |
| Stromal cells | Fibroblasts | CD45−, CD31−, EpCAM− PDGFRα (CD140a), PDGFRβ (CD140b), Sca1, CD90, CD49e (Integrin α5), αSMA, lipidtox | CD45−, CD31−, EpCAM− PDGFRα (CD140a), PDGFRβ (CD140b), CD90, CD49e (Integrin α5), αSMA, lipidtox | 93–96 |
Definition of abbreviations: αSMA = α–smooth muscle actin; EpCAM = epithelial cellular adhesion molecule; ICAM = intercellular adhesion molecule; ITGA = integrin alpha; NGFR = nerve growth factor receptor; PDGFR = platelet-derived growth factor receptor; RAGE = receptor for advanced glycation endproducts; Sftpc = surfactant protein C.
Recommendation
Flow cytometry panels should be designed to address specific research questions. However, to improve the impact of observations and the reproducibility of the research across laboratories, flow panels should be designed with consideration of standard cell definitions and markers.
Cell Sorting
Cell sorting is an essential tool for studying lung biology and disease. It feeds numerous downstream applications: genotyping and transcriptional profiling, proteomics, and in vivo (adoptive transfers), and in vitro assays.
Immunomagnetic Cell Sorting
Cells can be labeled with antibodies conjugated to paramagnetic particles and thus can be retained in the presence of a strong magnetic field, while unlabeled cells can be washed away. Unlabeled cells can be incubated with a new set of antibodies, and the procedure can be repeated several times to collect fractions of interest (72). Various systems and reagent sets are available on the market. It is important to notice, however, that this procedure rarely achieves 100% purity and is generally used to enrich single-cell suspensions for cell types of interest before fluorescence-activated cell sorting.
Fluorescence-activated Cell Sorting
Cell sorters allow both simultaneous identification of cells and the collection of specific populations of interest. Most modern sorters create a stable stream of droplets, where each droplet contains a single cell. High-speed electronics allow modern instruments to precisely track the position of each cell and then, by applying an electric charge to the exact droplet of PBS carrying the target cell, an electric field created by the deflection plates can pull the single droplet into a collection vessel. Depending on the instrument, up to six different populations can be collected at the same time. Although each cell type and experimental question may have a unique setup, several general aspects, listed below, should be considered.
To create a stable stream of droplets, the sheath stream is vibrated at a specific frequency and amplitude by way of a transducer or piezo. The cells, in their sample fluid, are introduced into the carrier sheath fluid, where they are focused for laser interrogation and exit the nozzle distributed to one of the droplets. All this is done under significant shear stress and pressure; therefore, sorted-cell viability can be a concern. A small nozzle size generally operates at high pressure, which results in smaller droplets and allows faster sorts. Large nozzles operate at lower pressures, generate big droplets, and sort at a slower speed. Even though the largest cell types in the lung are much smaller than the smallest nozzle, users must keep in mind that cells are still being subjected to dramatic shearing forces as they travel through the nozzle at the high speed. Thus, the nozzle size should be selected appropriately. Although small cells (for example T and B cells) can be safely sorted at high speed on small nozzles, large or fragile cells, such as alveolar macrophages, alveolar epithelial cells fibroblasts, or endothelial cells, should be sorted using large nozzles.
Collection Media
Generally, capture media should be isotonic, buffered to maintain neutral pH, and contain some protein (FBS or BSA). However, when sorting for proteomic analysis, cells should be stained, kept, and sorted into protein-free buffers to avoid contamination with ambient proteins. For downstream RNA- or DNA-based assays, rare or fragile cells can be sorted directly into a lysis buffer (10, 73). However, because densities of the lysis buffer and sorting solution are different and they do not readily mix, users should remember to pause the sort every 3 to 5 minutes and flick the collection tube, to ensure proper lysis.
Recommendation
Modern cell sorters are easier to operate than their predecessors, and a relatively experienced user should be able to perform sorts independent from the flow core staff once the cytometer has been properly set up and quality-control steps performed. Users are encouraged to do optimization experiments and be fully in charge of all experimental steps, rather than delegating these tasks to the core facility staff.
Components for Reporting Flow Cytometry Data in Publications
To improve the quality and reproducibility of flow cytometry, it is important that specific flow cytometry components are reported in publications. These components were discussed by the workshop group and supported by prior publications (2, 74). The Methods sections of manuscripts should include all of the aspects required to reproduce an individual experiment. These aspects should include clear methods of tissue digestion, sample processing, and staining conditions. Information should be provided on the flow cytometer, including the machine type and configuration (2). Antibody panels should be clearly defined, including the source, clone, fluorophore, and the concentration of the antibody in the staining condition. Examples of cytometer configuration and antibody panels are provided in publications (16, 20, 26, 75). Data analysis should include a clear gating strategy used in the experiments. To allow the reader to understand how individual cells are defined, representative individual gates should be displayed in an overview figure. Preference should be made for biexponential display, as this allows for visualization of cells on the axis (27). Overlay analysis of cells can also be used to support clear separation of cell types and define how the expression of a specific marker varies across cell types. The method of data analysis needs to be clearly defined, including mean or median fluorescence intensity for the histograms used to define enhanced cell surface expression of an individual marker. Cell frequency should be reported and clearly defined in the text, preferably on the basis of either total cell numbers or, when indicated, as a percentages of total cells or of the parent gate. Finally, to facilitate use of datasets by other investigators and to permit secondary analysis of data, increased use of data repositories for published flow cytometry data should be considered.
Conclusions
Flow cytometry is a powerful tool to define immune and structural cells in complex biologic fluids and tissues. As the immunologic underpinnings of diseases increase, flow cytometry facilitates definition of cellular states to drive diagnosis and treatment of disease. However, generating accurate and reproducible flow cytometry data requires a clear understanding of the instrument, the methods of processing and preservation, staining conditions, and panel design and robust data analysis. We hope the issues and the discussion outlined as a part of this workshop group will assist the pulmonary research community considering the use of flow cytometry in their experimental designs.
Acknowledgments
This workshop report was prepared by an ad hoc task force of the ATS Assembly on Allergy, Immunology, and Inflammation.
Members of the task force are as follows:
Robert M. Tighe, M.D.1 (Co-Chair)
Alexander V. Misharin, M.D., Ph.D.2 (Co-Chair)
Claudia V. Jakubzick, Ph.D.3 (Co-Chair)
Ryan Brinkman, Ph.D.4,5*
Jeffrey L. Curtis, M.D.6,7†
Ryan Duggan, B.S.8*
Christine M. Freeman, Ph.D.6,7*
Susanne Herold, M.D., Ph.D.9†
William Janssen, M.D.10*
Hideki Nakano, Ph.D.11*
Elizabeth F. Redente, Ph.D.12,13‡
Benjamin D. Singer, M.D.2‡
Anne I. Sperling, Ph.D.14†
Suchitra Swaminathan, Ph.D.2*
Yen-Rei Yu, M.D., Ph.D.1‡
William J. Zacharias, M.D., Ph.D.15,16,17*
1Division of Pulmonary, Allergy, and Critical Care Medicine, Duke University Medical Center, Durham, North Carolina; 2Division of Pulmonary and Critical Care Medicine, Department of Medicine, Feinberg School of Medicine, Northwestern University, Chicago, Illinois; 3Department of Microbiology and Immunology, Geisel School of Medicine, Dartmouth College, Hanover, New Hampshire; 4Department of Medical Genetics, University of British Columbia, Vancouver, British Columbia, Canada; 5Terry Fox Laboratory, British Columbia Cancer Agency, Vancouver, British Columbia, Canada; 6Division of Pulmonary and Critical Care Medicine, University of Michigan, Ann Arbor, Michigan; 7VA Ann Arbor Healthcare System, Ann Arbor, Michigan; 8Immuno-Oncology Discovery, AbbVie, Inc., North Chicago, Illinois; 9Department of Medicine II, Pulmonary and Critical Care Medicine and Infectious Diseases, Universities of Giessen and Marburg Lung Center, member of the German Center for Lung Research (DZL), and Excellence Cluster Cardio-Pulmonary Institute, Giessen, Germany; 10Department of Medicine and 12Department of Pediatrics, National Jewish Health, Denver, Colorado; 11Immunity, Inflammation, and Disease Laboratory, National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina; 13Department of Research, Veterans Affairs Eastern Colorado Health Care System, Aurora, Colorado; 14Section of Pulmonary and Critical Care Medicine, University of Chicago, Chicago, Illinois; 15Department of Medicine and 16Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio; and 17Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio.
*Workshop speaker.
†Workshop participant.
‡Member of the writing subcommittee.
Footnotes
Supported by National Institutes of Health grants K08HL128867 and U19AI135964 (B.D.S.); Department of Veterans Affairs grant 1IK2BX002401 (E.F.R.); Department of Veterans Affairs grant I01 CX000911 (J.L.C.); National Institutes of Health/National Heart, Lung, and Blood Institute grant U01HL137880 (J.L.C.); German Research Foundation grant KFO309 project 284237345, SFB1021 project 197785619, and EXC2026 project 390649896 (S.H.); National Institutes of Health grants HL135124, AG049665, AI135964, and Department of Defense grant PR141319 (A.V.M.); National Institutes of Health grants R01ES027574, R01ES028829, and K08HL105537 (R.M.T.); and National Institutes of Health grants R35HL140039 and R01HL130938 (W.J.J.).
This official Clinical Practice Guideline of the American Thoracic Society was approved May 2019
Author Disclosures: R.M.T. received research support from Boehringer Ingelheim. R.B. has an ownership interest in Cytapex Bioinformatics. R.D. has patent 9663818 issued to The University of Chicago. B.D.S. has a pending U.S. patent application for 15/542,380, “Compositions and Methods to Accelerate Resolution of Acute Lung Inflammation.” W.J.Z. served as a speaker for Kyorin Pharmaceuticals. A.V.M., C.V.J., J.L.C., C.M.F., S.H., W.J., H.N., E.F.R., A.I.S., S.S., and Y.-R.U. report no relationships with relevant commercial interests.
Contributor Information
Collaborators: on behalf of the American Thoracic Society Assembly on Allergy, Immunology, and Inflammation
References
- 1.Cossarizza A, Chang HD, Radbruch A, Akdis M, Andra I, Annunziato F, et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur J Immunol. 2017;47:1584–1797. doi: 10.1002/eji.201646632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JA, Spidlen J, Boyce K, Cai J, Crosbie N, Dalphin M, et al. International Society for Advancement of Cytometry Data Standards Task Force. MIFlowCyt: the minimum information about a Flow Cytometry Experiment. Cytometry A. 2008;73:926–930. doi: 10.1002/cyto.a.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maciorowski Z, Chattopadhyay PK, Jain P. Basic multicolor flow cytometry. Curr Protoc Immunol. 2017;117:5.4.1–5.4.38. doi: 10.1002/cpim.26. [DOI] [PubMed] [Google Scholar]
- 4.Keller BC, Presti RM, Byers DE, Atkinson JJ. Significant interference in mass cytometry from medicinal iodine in human lung. Am J Respir Cell Mol Biol. 2016;55:150–151. doi: 10.1165/rcmb.2015-0403LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ford AL, Foulcher E, Goodsall AL, Sedgwick JD. Tissue digestion with dispase substantially reduces lymphocyte and macrophage cell-surface antigen expression. J Immunol Methods. 1996;194:71–75. doi: 10.1016/0022-1759(96)00067-1. [DOI] [PubMed] [Google Scholar]
- 6.Autengruber A, Gereke M, Hansen G, Hennig C, Bruder D. Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. Eur J Microbiol Immunol (Bp) 2012;2:112–120. doi: 10.1556/EuJMI.2.2012.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singer BD, Mock JR, D’Alessio FR, Aggarwal NR, Mandke P, Johnston L, et al. Flow-cytometric method for simultaneous analysis of mouse lung epithelial, endothelial, and hematopoietic lineage cells. Am J Physiol Lung Cell Mol Physiol. 2016;310:L796–L801. doi: 10.1152/ajplung.00334.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tighe RM, Birukova A, Yaeger MJ, Reece SW, Gowdy KM. Euthanasia- and lavage-mediated effects on bronchoalveolar measures of lung injury and inflammation. Am J Respir Cell Mol Biol. 2018;59:257–266. doi: 10.1165/rcmb.2017-0357OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins AM, Rylance J, Wootton DG, Wright AD, Wright AK, Fullerton DG, et al. Bronchoalveolar lavage (BAL) for research; obtaining adequate sample yield. J Vis Exp. 2014;(85):e4345. doi: 10.3791/4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter JM, Helmin KA, Abdala-Valencia H, Wunderink RG, Singer BD. Multidimensional assessment of alveolar T cells in critically ill patients. JCI Insight. 2018;3:e123287. doi: 10.1172/jci.insight.123287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zacharias W, Morrisey E. Isolation and culture of human alveolar epithelial progenitor cells. Protocol Exchange. 2018 doi: 10.1038/protex.2018.015. [accessed on 2019 Mar 24]. Available from: [DOI] [Google Scholar]
- 12.Yu YA, Tighe RM. Isolation and characterization of human lung myeloid cells. Methods Mol Biol. 2018;1809:111–119. doi: 10.1007/978-1-4939-8570-8_9. [DOI] [PubMed] [Google Scholar]
- 13.Gibbings SL, Jakubzick CV. A consistent method to identify and isolate mononuclear phagocytes from human lung and lymph nodes. Methods Mol Biol. 2018;1799:381–395. doi: 10.1007/978-1-4939-7896-0_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finch DK, Stolberg VR, Ferguson J, Alikaj H, Kady MR, Richmond BW, et al. Lung dendritic cells drive natural killer cytotoxicity in chronic obstructive pulmonary disease via IL-15Rα. Am J Respir Crit Care Med. 2018;198:1140–1150. doi: 10.1164/rccm.201712-2513OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leelatian N, Doxie DB, Greenplate AR, Mobley BC, Lehman JM, Sinnaeve J, et al. Single cell analysis of human tissues and solid tumors with mass cytometry. Cytometry B Clin Cytom. 2017;92:68–78. doi: 10.1002/cyto.b.21481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu YR, O’Koren EG, Hotten DF, Kan MJ, Kopin D, Nelson ER, et al. A protocol for the comprehensive flow cytometric analysis of immune cells in normal and inflamed murine non-lymphoid tissues. PLoS One. 2016;11:e0150606. doi: 10.1371/journal.pone.0150606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Z, Chiu S, Akbarpour M, Sun H, Reyfman PA, Anekalla KR, et al. Donor pulmonary intravascular nonclassical monocytes recruit recipient neutrophils and mediate primary lung allograft dysfunction. Sci Transl Med. 2017;9:eaal4508. doi: 10.1126/scitranslmed.aal4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desch AN, Gibbings SL, Goyal R, Kolde R, Bednarek J, Bruno T, et al. Flow cytometric analysis of mononuclear phagocytes in nondiseased human lung and lung-draining lymph nodes. Am J Respir Crit Care Med. 2016;193:614–626. doi: 10.1164/rccm.201507-1376OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barcelo H, Faul J, Crimmins E, Thyagarajan B. A practical cryopreservation and staining protocol for immunophenotyping in population studies. Curr Protoc Cytom. 2018;84:e35. doi: 10.1002/cpcy.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu YR, Hotten DF, Malakhau Y, Volker E, Ghio AJ, Noble PW, et al. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol. 2016;54:13–24. doi: 10.1165/rcmb.2015-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman CM, Crudgington S, Stolberg VR, Brown JP, Sonstein J, Alexis NE, et al. Design of a multi-center immunophenotyping analysis of peripheral blood, sputum and bronchoalveolar lavage fluid in the Subpopulations and Intermediate Outcome Measures in COPD Study (SPIROMICS) J Transl Med. 2015;13:19. doi: 10.1186/s12967-014-0374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adan A, Alizada G, Kiraz Y, Baran Y, Nalbant A. Flow cytometry: basic principles and applications. Crit Rev Biotechnol. 2017;37:163–176. doi: 10.3109/07388551.2015.1128876. [DOI] [PubMed] [Google Scholar]
- 23.Terstappen LWMM, Shah VO, Conrad MP, Recktenwald D, Loken MR. Discriminating between damaged and intact cells in fixed flow cytometric samples. Cytometry. 1988;9:477–484. doi: 10.1002/cyto.990090512. [DOI] [PubMed] [Google Scholar]
- 24.Andersen MN, Al-Karradi SN, Kragstrup TW, Hokland M. Elimination of erroneous results in flow cytometry caused by antibody binding to Fc receptors on human monocytes and macrophages. Cytometry A. 2016;89:1001–1009. doi: 10.1002/cyto.a.22995. [DOI] [PubMed] [Google Scholar]
- 25.Kuonen F, Touvrey C, Laurent J, Ruegg C. Fc block treatment, dead cells exclusion, and cell aggregates discrimination concur to prevent phenotypical artifacts in the analysis of subpopulations of tumor-infiltrating CD11b(+) myelomonocytic cells. Cytometry A. 2010;77:1082–1090. doi: 10.1002/cyto.a.20969. [DOI] [PubMed] [Google Scholar]
- 26.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 28.Roederer M. Compensation in flow cytometry. Curr Protoc Cytom. 2002;Chapter 1:Unit 1.14. doi: 10.1002/0471142956.cy0114s22. [DOI] [PubMed] [Google Scholar]
- 29.Roederer M. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry. 2001;45:194–205. doi: 10.1002/1097-0320(20011101)45:3<194::aid-cyto1163>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell AJ, Pradel LC, Chasson L, Van Rooijen N, Grau GE, Hunt NH, et al. Technical advance: autofluorescence as a tool for myeloid cell analysis. J Leukoc Biol. 2010;88:597–603. doi: 10.1189/jlb.0310184. [DOI] [PubMed] [Google Scholar]
- 31.Maus U, Rosseau S, Seeger W, Lohmeyer J. Separation of human alveolar macrophages by flow cytometry. Am J Physiol. 1997;272:L566–L571. doi: 10.1152/ajplung.1997.272.3.L566. [DOI] [PubMed] [Google Scholar]
- 32.Duan M, Li WC, Vlahos R, Maxwell MJ, Anderson GP, Hibbs ML. Distinct macrophage subpopulations characterize acute infection and chronic inflammatory lung disease. J Immunol. 2012;189:946–955. doi: 10.4049/jimmunol.1200660. [DOI] [PubMed] [Google Scholar]
- 33.Roederer M. Distributions of autofluorescence after compensation: be panglossian, fret not. Cytometry A. 2016;89:398–402. doi: 10.1002/cyto.a.22820. [DOI] [PubMed] [Google Scholar]
- 34.Amir el-AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir AD, Tadmor MD, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. doi: 10.1038/nbt.4314. [online ahead of print] 3 Dec 2018; DOI: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 37.Belkina AC, Ciccolella CO, Anno R, Spidlen J, Halpert R, Snyder-Cappione J. Automated optimal parameters for T-distributed stochastic neighbor embedding improve visualization and allow analysis of large datasets [preprint] bioRxiv. 2018:451690. doi: 10.1038/s41467-019-13055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 39.Saeys Y, Van Gassen S, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol. 2016;16:449–462. doi: 10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
- 40.McGrath-Morrow SA, Ndeh R, Helmin KA, Chen SY, Anekalla KR, Abdala-Valencia H, et al. DNA methylation regulates the neonatal CD4+ T-cell response to pneumonia in mice. J Biol Chem. 2018;293:11772–11783. doi: 10.1074/jbc.RA118.003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahim A, Meskas J, Drissler S, Yue A, Lorenc A, Laing A, et al. High throughput automated analysis of big flow cytometry data. Methods. 2018;134-135:164–176. doi: 10.1016/j.ymeth.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lux M, Brinkman RR, Chauve C, Laing A, Lorenc A, Abeler-Dörner L, et al. flowLearn: fast and precise identification and quality checking of cell populations in flow cytometry. Bioinformatics. 2018;34:2245–2253. doi: 10.1093/bioinformatics/bty082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fletez-Brant K, Špidlen J, Brinkman RR, Roederer M, Chattopadhyay PK. flowClean: automated identification and removal of fluorescence anomalies in flow cytometry data. Cytometry A. 2016;89:461–471. doi: 10.1002/cyto.a.22837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aghaeepour N, Chattopadhyay P, Chikina M, Dhaene T, Van Gassen S, Kursa M, et al. A benchmark for evaluation of algorithms for identification of cellular correlates of clinical outcomes. Cytometry A. 2016;89:16–21. doi: 10.1002/cyto.a.22732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahne F, LeMeur N, Brinkman RR, Ellis B, Haaland P, Sarkar D, et al. flowCore: a bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spidlen J, Breuer K, Rosenberg C, Kotecha N, Brinkman RR. FlowRepository: a resource of annotated flow cytometry datasets associated with peer-reviewed publications. Cytometry A. 2012;81:727–731. doi: 10.1002/cyto.a.22106. [DOI] [PubMed] [Google Scholar]
- 47.O’Neill K, Brinkman RR. Publishing code is essential for reproducible flow cytometry bioinformatics. Cytometry A. 2016;89:10–11. doi: 10.1002/cyto.a.22805. [DOI] [PubMed] [Google Scholar]
- 48.Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duerr CU, Fritz JH. Isolation of group 2 innate lymphoid cells from mouse lungs. Methods Mol Biol. 2017;1656:253–261. doi: 10.1007/978-1-4939-7237-1_16. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Li F, Zheng M, Sun R, Wei H, Tian Z. Lung natural killer cells in mice: phenotype and response to respiratory infection. Immunology. 2012;137:37–47. doi: 10.1111/j.1365-2567.2012.03607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malka-Ruimy C, Ben Youssef G, Lambert M, Tourret M, Ghazarian L, Faye A, et al. Mucosal-associated invariant T cell levels are reduced in the peripheral blood and lungs of children with active pulmonary tuberculosis. Front Immunol. 2019;10:206. doi: 10.3389/fimmu.2019.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singer BD, Mock JR, Aggarwal NR, Garibaldi BT, Sidhaye VK, Florez MA, et al. Regulatory T cell DNA methyltransferase inhibition accelerates resolution of lung inflammation. Am J Respir Cell Mol Biol. 2015;52:641–652. doi: 10.1165/rcmb.2014-0327OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singer BD, King LS, D’Alessio FR. Regulatory T cells as immunotherapy. Front Immunol. 2014;5:46. doi: 10.3389/fimmu.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rainbow DB, Yang X, Burren O, Pekalski ML, Smyth DJ, Klarqvist MD, et al. Epigenetic analysis of regulatory T cells using multiplex bisulfite sequencing. Eur J Immunol. 2015;45:3200–3203. doi: 10.1002/eji.201545646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bingaman AW, Patke DS, Mane VR, Ahmadzadeh M, Ndejembi M, Bartlett ST, et al. Novel phenotypes and migratory properties distinguish memory CD4 T cell subsets in lymphoid and lung tissue. Eur J Immunol. 2005;35:3173–3186. doi: 10.1002/eji.200526004. [DOI] [PubMed] [Google Scholar]
- 57.Tian Y, Babor M, Lane J, Schulten V, Patil VS, Seumois G, et al. Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA. Nat Commun. 2017;8:1473. doi: 10.1038/s41467-017-01728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheng SY, Gu Y, Lu CG, Zou JY, Hong H, Wang R. The distribution and function of human memory T cell subsets in lung cancer. Immunol Res. 2017;65:639–650. doi: 10.1007/s12026-016-8882-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song K, Rabin RL, Hill BJ, De Rosa SC, Perfetto SP, Zhang HH, et al. Characterization of subsets of CD4+ memory T cells reveals early branched pathways of T cell differentiation in humans. Proc Natl Acad Sci USA. 2005;102:7916–7921. doi: 10.1073/pnas.0409720102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romero P, Zippelius A, Kurth I, Pittet MJ, Touvrey C, Iancu EM, et al. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J Immunol. 2007;178:4112–4119. doi: 10.4049/jimmunol.178.7.4112. [DOI] [PubMed] [Google Scholar]
- 61.Shane HL, Klonowski KD. Every breath you take: the impact of environment on resident memory CD8 T cells in the lung. Front Immunol. 2014;5:320. doi: 10.3389/fimmu.2014.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zens KD, Chen JK, Guyer RS, Wu FL, Cvetkovski F, Miron M, et al. Reduced generation of lung tissue-resident memory T cells during infancy. J Exp Med. 2017;214:2915–2932. doi: 10.1084/jem.20170521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teijaro JR, Turner D, Pham Q, Wherry EJ, Lefrançois L, Farber DL. Cutting edge: tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J Immunol. 2011;187:5510–5514. doi: 10.4049/jimmunol.1102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turner DL, Bickham KL, Thome JJ, Kim CY, D’Ovidio F, Wherry EJ, et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 2014;7:501–510. doi: 10.1038/mi.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahlers JD, Belyakov IM. Memories that last forever: strategies for optimizing vaccine T-cell memory. Blood. 2010;115:1678–1689. doi: 10.1182/blood-2009-06-227546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zaynagetdinov R, Sherrill TP, Kendall PL, Segal BH, Weller KP, Tighe RM, et al. Identification of myeloid cell subsets in murine lungs using flow cytometry. Am J Respir Cell Mol Biol. 2013;49:180–189. doi: 10.1165/rcmb.2012-0366MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bharat A, Bhorade SM, Morales-Nebreda L, McQuattie-Pimentel AC, Soberanes S, Ridge K, et al. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol. 2016;54:147–149. doi: 10.1165/rcmb.2015-0147LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gonzalez RF, Dobbs LG. Isolation and culture of alveolar epithelial type i and type ii cells from rat lungs. Methods Mol Biol. 2013;945:145–159. doi: 10.1007/978-1-62703-125-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jansing NL, McClendon J, Kage H, Sunohara M, Alvarez JR, Borok Z, et al. Isolation of rat and mouse alveolar type II epithelial cells. Methods Mol Biol. 2018;1809:69–82. doi: 10.1007/978-1-4939-8570-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bantikassegn A, Song X, Politi K. Isolation of epithelial, endothelial, and immune cells from lungs of transgenic mice with oncogene-induced lung adenocarcinomas. Am J Respir Cell Mol Biol. 2015;52:409–417. doi: 10.1165/rcmb.2014-0312MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tighe S, Held MA. Isolation of total RNA from transgenic mouse melanoma subsets using fluorescence-activated cell sorting. Methods Mol Biol. 2010;632:27–44. doi: 10.1007/978-1-60761-663-4_2. [DOI] [PubMed] [Google Scholar]
- 74.Alvarez DF, Helm K, Degregori J, Roederer M, Majka S. Publishing flow cytometry data. Am J Physiol Lung Cell Mol Physiol. 2010;298:L127–L130. doi: 10.1152/ajplung.00313.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gibbings SL, Thomas SM, Atif SM, McCubbrey AL, Desch AN, Danhorn T, et al. Three unique interstitial macrophages in the murine lung at steady state. Am J Respir Cell Mol Biol. 2017;57:66–76. doi: 10.1165/rcmb.2016-0361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sabatel C, Radermecker C, Fievez L, Paulissen G, Chakarov S, Fernandes C, et al. Exposure to bacterial CpG DNA protects from airway allergic inflammation by expanding regulatory lung interstitial macrophages. Immunity. 2017;46:457–473. doi: 10.1016/j.immuni.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 78.Nakano H, Moran TP, Nakano K, Gerrish KE, Bortner CD, Cook DN. Complement receptor C5aR1/CD88 and dipeptidyl peptidase-4/CD26 define distinct hematopoietic lineages of dendritic cells. J Immunol. 2015;194:3808–3819. doi: 10.4049/jimmunol.1402195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larson SR, Atif SM, Gibbings SL, Thomas SM, Prabagar MG, Danhorn T, et al. Ly6C(+) monocyte efferocytosis and cross-presentation of cell-associated antigens. Cell Death Differ. 2016;23:997–1003. doi: 10.1038/cdd.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rose CE, Jr, Lannigan JA, Kim P, Lee JJ, Fu SM, Sung SS. Murine lung eosinophil activation and chemokine production in allergic airway inflammation. Cell Mol Immunol. 2010;7:361–374. doi: 10.1038/cmi.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schwartz C, Voehringer D. Identification of murine basophils by flow cytometry and histology. Methods Mol Biol. 2014;1192:229–237. doi: 10.1007/978-1-4939-1173-8_17. [DOI] [PubMed] [Google Scholar]
- 82.Zhou B, Flodby P, Luo J, Castillo DR, Liu Y, Yu FX, et al. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J Clin Invest. 2018;128:970–984. doi: 10.1172/JCI90429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Y, Tang Z, Huang H, Li J, Wang Z, Yu Y, et al. Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. Proc Natl Acad Sci USA. 2018;115:2407–2412. doi: 10.1073/pnas.1719474115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1:e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Corbière V, Dirix V, Norrenberg S, Cappello M, Remmelink M, Mascart F. Phenotypic characteristics of human type II alveolar epithelial cells suitable for antigen presentation to T lymphocytes. Respir Res. 2011;12:15. doi: 10.1186/1465-9921-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van der Velden JL, Bertoncello I, McQualter JL. LysoTracker is a marker of differentiated alveolar type II cells. Respir Res. 2013;14:123. doi: 10.1186/1465-9921-14-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Messier EM, Mason RJ, Kosmider B. Efficient and rapid isolation and purification of mouse alveolar type II epithelial cells. Exp Lung Res. 2012;38:363–373. doi: 10.3109/01902148.2012.713077. [DOI] [PubMed] [Google Scholar]
- 88.Chen H, Matsumoto K, Brockway BL, Rackley CR, Liang J, Lee JH, et al. Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury. Stem Cells. 2012;30:1948–1960. doi: 10.1002/stem.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Quantius J, Schmoldt C, Vazquez-Armendariz AI, Becker C, El Agha E, Wilhelm J, et al. Influenza virus infects epithelial stem/progenitor cells of the distal lung: impact on Fgfr2b-driven epithelial repair. PLoS Pathog. 2016;12:e1005544. doi: 10.1371/journal.ppat.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yunt ZX, Mohning MP, Barthel L, Kearns MT, Tuder RM, Hyde DM, et al. Kinetics of the angiogenic response in lung endothelium following acute inflammatory injury with bleomycin. Exp Lung Res. 2014;40:415–425. doi: 10.3109/01902148.2014.938202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fehrenbach ML, Cao G, Williams JT, Finklestein JM, Delisser HM. Isolation of murine lung endothelial cells. Am J Physiol Lung Cell Mol Physiol. 2009;296:L1096–L1103. doi: 10.1152/ajplung.90613.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Green J, Endale M, Auer H, Perl AK. Diversity of interstitial lung fibroblasts is regulated by platelet-derived growth factor receptor α kinase activity. Am J Respir Cell Mol Biol. 2016;54:532–545. doi: 10.1165/rcmb.2015-0095OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heinzelmann K, Noskovičová N, Merl-Pham J, Preissler G, Winter H, Lindner M, et al. Surface proteome analysis identifies platelet derived growth factor receptor-alpha as a critical mediator of transforming growth factor-beta-induced collagen secretion. Int J Biochem Cell Biol. 2016;74:44–59. doi: 10.1016/j.biocel.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 95.Akamatsu T, Arai Y, Kosugi I, Kawasaki H, Meguro S, Sakao M, et al. Direct isolation of myofibroblasts and fibroblasts from bleomycin-injured lungs reveals their functional similarities and differences. Fibrogenesis Tissue Repair. 2013;6:15. doi: 10.1186/1755-1536-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, et al. Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell. 2017;20:261–273. doi: 10.1016/j.stem.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]