Catabolism of flavonols contributes to S. sclerotiorum virulence during infection of Arabidopsis.
Abstract
Flavonols are widely distributed plant metabolites that inhibit microbial growth. Yet many pathogens cause disease in flavonol-containing plant tissues. We investigated how Sclerotinia sclerotiorum, a necrotrophic fungal pathogen that causes disease in a range of economically important crop species, is able to successfully infect flavonol-rich tissues of Arabidopsis (Arabidopsis thaliana). Infection of rosette stage Arabidopsis with a virulent S. sclerotiorum strain led to the selective hydrolysis of flavonol glycosidic linkages and the inducible degradation of flavonol aglycones to phloroglucinol carboxylic and phenolic acids. By chemical analysis of fungal biotransformation products and a search of the S. sclerotiorum genome sequence, we identified a quercetin dioxygenase gene (QDO) and characterized the encoded protein, which catalyzed cleavage of the flavonol carbon skeleton. QDO deletion lines degraded flavonols with much lower efficiency and were less pathogenic on Arabidopsis leaves than wild-type S. sclerotiorum, indicating the importance of flavonol degradation in fungal virulence. In the absence of QDO, flavonols exhibited toxicity toward S. sclerotiorum, demonstrating the potential roles of these phenolic compounds in protecting plants against pathogens.
Flavonoids are multifunctional phenolic natural products that are ubiquitous in plants (Williams and Grayer, 2004; Buer et al., 2010). With a C6-C3-C6 core structure, they are synthesized through the phenylpropanoid pathway and are divided into subclasses depending on their structure, including flavonols, flavanones, isoflavones, flavones, flavan-3-ols, and anthocyanins (Winkel-Shirley, 2001; Pereira et al., 2009). Flavonoids function in attraction of pollinators, protection against UV radiation, and regulation of auxin transport (Li et al., 1993; Mol et al., 1998; Peer and Murphy, 2007). Flavonoids are also much discussed as nutraceuticals because of their antioxidant and anticancer activity (Rice-Evans et al., 1995; Cook and Samman, 1996). Another frequently cited role for flavonoids in plants is as protection against pathogenic microbes. There are many reports of flavonoid antimicrobial activity, such as the strong inhibition of Fusurium culmorum growth in barley (Hordeum vulgare) by dihydroquercetin (Skadhauge et al., 1997). Moreover, pathogen infection often triggers local accumulation of flavonoids, such as the elevated levels of isoflavones, daidzein, and genistein, in soybean (Glycine max) after Sclerotinia sclerotiorum infection (Wegulo et al., 2005). However, it is unclear why many pathogens appear to grow well in flavonoid-rich plant tissues.
One of the most aggressive and widespread necrotrophic plant pathogens is S. sclerotiorum, which has a host range comprising more than 400 species, including many agriculturally important crops on which it causes substantial economic losses (Boland and Hall, 1994). Owing to the virulence of S. sclerotiorum and its long-term survival ability under harsh environmental conditions (Riou et al., 1991; Bolton et al., 2006), there are currently no sustainable strategies for controlling this fungus. S. sclerotiorum is known to overcome host plant defenses by producing pectinolytic and cellulolytic enzymes to break down the host cell wall and oxalic acid to suppress host defense responses, such as the oxidative burst and the hypersensitive response (Morrall et al., 1972; Riou et al., 1991; Cessna et al., 2000). In addition, S. sclerotiorum transforms plant defense compounds. For example, the indole-sulfur phytoalexins of the Brassicaceae, such as camalexin, brassinin, cyclobrassinin, and brassilexin, are detoxified by the fungus by glycosylation (Pedras and Ahiahonu, 2005; Pedras and Hossain, 2006). However, the molecular mechanisms underlying these detoxification reactions are poorly studied, and the effort to understand their significance for plant-pathogen interactions is still in its infancy.
In addition to indole-sulfur compounds, the Brassicaceae produce ample quantities of flavonols, a common group of flavonoids with a 3-OH function, which may also serve in antimicrobial defense. Flavonols such as quercetin, kaempferol, and isorhamnetin are mainly present in leaves as glycosides (Onyilagha et al., 2003; Cartea et al., 2010). However, it is not known if these compounds play a role in defense against pathogen infection and whether pathogens can circumvent them. In this study, we investigated the role of flavonols in the interaction between S. sclerotiorum and the Brassicaceae species Arabidopsis, which is very susceptible to this fungal pathogen. Leaves infected by S. sclerotiorum had reduced levels of flavonol glycosides due to fungal catabolism. Analysis of S. sclerotiorum flavonol degradation products resulted in the identification of a key enzyme in the breakdown of the flavonol ring system, a quercetin 2,3-dixogygenase. Characterization of this enzyme in S. sclerotiorum using molecular as well as biochemical methods confirmed its importance in flavonol degradation and in fungal virulence on Arabidopsis.
RESULTS
Flavonol Glycosides Are Metabolized by S. sclerotiorum In Planta and in Culture
To investigate the flavonoid defenses of the host plant against S. sclerotiorum infection, the levels of flavonol glycosides (K3(R2'G)7R, kaempferol 3-O-rhamnosyl 2'-glucoside-7-O-rhamnoside; Q3(R2'G)7R, quercetin 3-O-rhamnosyl 2'-glucoside-7-O-rhamnoside; K3G7R, kaempferol 3-O-glucoside-7-O-rhamnoside; Q3G7R, quercetin 3-O-glucoside-7-O-rhamnoside; K3R7R, kaempferol 3-O-rhamnoside-7-O-rhamnoside; Q3R7R, quercetin 3-O-rhamnoside-7-O-rhamnoside) in Arabidopsis leaves were analyzed after inoculation with the fungus. Flavonol glycoside concentrations decreased significantly by about 50% in S. sclerotiorum-inoculated Arabidopsis leaves (P < 0.05, Student’s t test) when compared with mock-inoculated plants 3 d postinoculation (Fig. 1A).
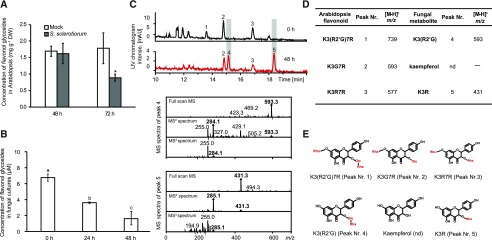
Figure 1.
S. sclerotiorum degrades Arabidopsis flavonol glycosides. A, Total flavonol glycoside levels in Arabidopsis 3 d postinoculation with S. sclerotiorum wild-type UF-70. Flavonol glycosides were analyzed using LC-UV (330 nm) and quantified using an external K3R7R calibration curve. Total flavonol glycosides represent the summed quantities of all the flavonol glycosides detected. Asterisk represents significant difference (P < 0.05, Student’s t test). B, Degradation of the major flavonol glycosides in Arabidopsis extracts by S. sclerotiorum in artificial medium over a time course of 48 h. Samples were analyzed by LC-UV (330 nm). Different letters above the bars indicate significant differences in residual flavonol glycoside content over time (P < 0.001, one-way ANOVA followed by Tukey’s post-hoc test). C, Representative HPLC chromatograms with UV detection (280 nm) from liquid cultures of S. sclerotiorum 0 and 48 h after addition of Arabidopsis flavonol glycoside extract. Peaks 1, 2, and 3 are the major flavonol glycosides in Arabidopsis. Cocultivation with the fungus for 48 h resulted in the appearance of peaks 4 and 5 (structures shown in E, ESI-MS2 and MS3 spectra in negative ionization mode shown below the chromatograms, are comparable to spectra for authentic standards shown in Supplemental Fig. S2). D, Negative molecular ions (m/z) of major Arabidopsis (Arabidopsis thaliana) flavonol glycosides and their corresponding S. sclerotiorum deglycosylated derivatives as identified by liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS). E, Structures of major flavonol glycosides in Arabidopsis and suggested structures of the products produced by S. sclerotiorum in culture. All data represent means ± se (n = 3). K3(R2'G)7R, kaempferol 3-O-rhamnosyl 2'-glucoside-7-O-rhamnoside; K3G7R, kaempferol 3-O-glucoside-7-O-rhamnoside; K3R7R, kaempferol 3-O-rhamnoside-7-O-rhamnoside; K3(R2'G), kaempferol 3-O-rhamnosyl 2'-glucoside; K3R, kaempferol 3-O-rhamnoside. DW, dry weight.
To investigate if the fungus can also reduce the levels of flavonol glycosides in vitro, a liquid culture of S. sclerotiorum was supplemented with a flavonol glycoside extract from Arabidopsis. Analysis of the medium collected at 0, 24, and 48 h after addition of the flavonol glycosides showed that the concentrations of these compounds were reduced significantly, approximately 70% in 2 d after inoculation (ANOVA, P < 0.001; Fig. 1B), suggesting that the fungus had rapidly degraded these metabolites. Liquid medium that contained the same amount of flavonol glycoside extract but no fungus was used as a negative control. The flavonol glycosides did not degrade at all, showing that flavonol glycosides are not degraded spontaneously in vitro (Supplemental Fig. S1).
The degradation products of the major flavonol glycosides from Arabidopsis accumulating during incubation with S. sclerotiorum were identified using LC-ESI-MS operated in the alternating ionization mode. Arabidopsis (Col-0) produces mainly K3(R2'G)7R ([M-H]− = 739]), K3G7R ([M-H]− = 593), and K3R7R ([M-H]− = 577; Veit and Pauli, 1999; Kerhoas et al., 2006; Saito et al., 2013). After 48 h incubation in medium with S. sclerotiorum, two new peaks appeared in the chromatogram with an [M-H]− of 593 and 431, respectively (peaks 4 and 5; Fig. 1C). According to mass spectral collision induced dissociation fragmentation data (MS2), peak 4 appeared to be K3(R2'G), derived from K3(R2'G)7R by hydrolysis of the 7-O-rhamnose residue (Fig. 1D). Although no standard of K3(R2'G) is commercially available, an extract containing this compound from Clitoria ternatea, where it is reported to be the major flavonol glycoside (Kazuma et al., 2003), gave a peak with the same retention time and mass spectrum as that produced by the S. sclerotiorum culture, confirming its identity (Supplemental Fig. S2A). Peak 5 was identified as K3R by comparison with a pure standard and is produced by the fungus from K3R7R (Fig. 1D; Supplemental Fig. S2B). These results suggested that S. sclerotiorum can readily cleave a 7-O-rhamnose or 7-O-glucose substituent. However, kaempferol 3-O-glucoside (K3G), which would be produced from K3G7R, was not detected in the medium. When we amended the S. sclerotiorum culture with K3G and K3R, only K3G was depleted in the medium after 15 h incubation, and no traces of kaempferol were detected (Supplemental Fig. S3A).
Flavonol Aglycones Are Also Metabolized by S. sclerotiorum in an Inducible Manner
The lack of flavonol aglycone accumulation suggested that these compounds were also degraded by S. sclerotiorum. To confirm this supposition, quercetin and kaempferol were added to the fungal culture and were shown to be degraded using LC-ESI-MS over a time course of 48 h. Approximately 25% of the initial quercetin was degraded in 24 h by the fungus, while the contents of kaempferol decreased by up to 90% during the same time interval, indicating that kaempferol is degraded more rapidly than quercetin (Fig. 2A).
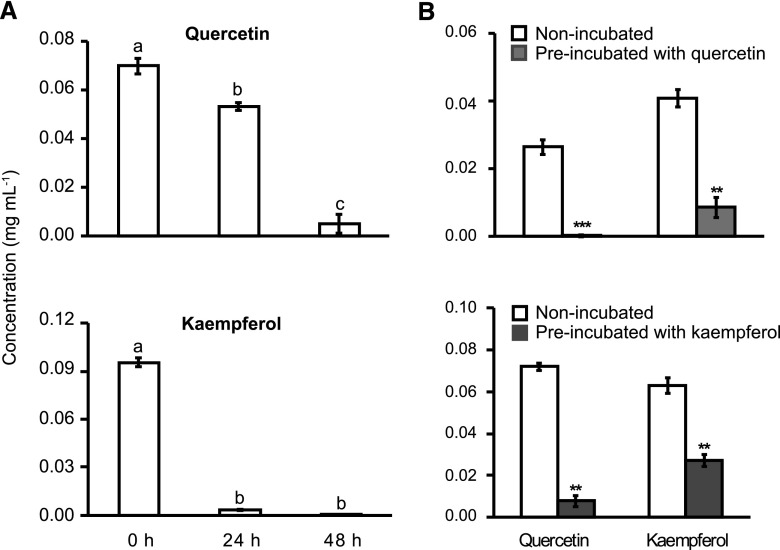
Figure 2.
S. sclerotiorum degrades flavonol aglycones in an inducible manner. A, Quercetin and kaempferol levels after treatment with S. sclerotiorum for 48 h in artificial medium. Different letters above the bars indicate significant differences (P < 0.001, one-way ANOVA followed by Tukey’s post-hoc test). B, Degradation of both quercetin and kaempferol by total protein from S. sclerotiorum cultures preincubated with either quercetin or kaempferol or non-preincubated. Asterisks represent significant differences between protein extracts of preincubated and non-preincubated fungi (Student’s t test, **P < 0.01; ***P < 0.001). All data represent means ± se (n = 3) and were acquired by LC-ESI-MS analysis.
We next conducted an in vitro enzyme assay using total protein extracts from S. sclerotiorum that had been cultured either with or without flavonol supplementation. Total protein was extracted from mycelium that was pretreated with 0.01 mg mL−1 quercetin or kaempferol for 2 h. This protein extract was then used in an in vitro enzyme assay with the substrates quercetin and kaempferol. Pretreatment with quercetin or kaempferol demonstrated that the activity for degradation of these flavonols increased from 2- to over 10-fold compared to protein extracts from nonpretreated fungus (Student’s t test; P < 0.01; Fig. 2B).
Identification and Heterologous Expression of a S. sclerotiorum Flavonol Degradation Enzyme, Quercetin 2,3-Dioxygenase
Enzyme-mediated quercetin degradation was previously shown to be the key step in the catabolism of rutin (quercetin 3-O-rhamnosyl-6′-glucoside) in some fungi and bacteria (Schaab et al., 2006; Tranchimand et al., 2010). In this pathway, the disaccharide moiety, rutinose, is hydrolyzed from rutin, and the resulting quercetin moiety is then cleaved by quercetin 2,3-dioxygenase (QDO) at the 2,3-position to form 2-protocatechuoyl phloroglucinol carboxylic acid. This degradation product of quercetin can be further metabolized to protocatechuic acid and phloroglucinol carboxylic acid by an esterase (Fig. 3A). We hypothesized that S. sclerotiorum degrades quercetin through the same pathway as we detected protocatechuic acid in the culture medium of S. sclerotiorum supplemented with quercetin (Supplemental Fig. S3B).
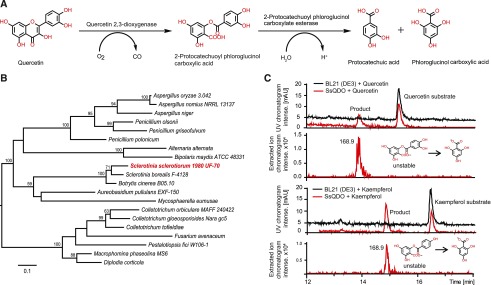
Figure 3.
Identification and heterologous expression of S. sclerotiorum quercetin 2,3-dioxygenase (SsQDO). A, The catabolism of quercetin by QDO. B, Phylogenetic analysis of QDO from S. sclerotiorum and other fungi. The maximum-likelihood tree was constructed using PhyML-3.1 employing the amino acid substitution model LG (Le and Gascuel, 2008). Nonparametric bootstrap analysis was performed with 1000 replicates, and values next to each branch point indicate the branch support percentages (values >50 are shown). The scale bar indicates amino acid substitution per site. C, S. sclerotiorum SsQDO heterologously expressed in Escherichia coli cleaves both quercetin and kaempferol at the 2,3 position. Shown are the LC-UV chromatograms at 280 nm and the extracted ion traces of m/z 169 in negative ionization mode generated by a LC-ESI-IonTrap-MS (= in-source fragment of oxidation products with m/z 305 and m/z 289, respectively). Products were further identified by UHPLC-ESI-TOF-MS (see Supplemental Fig. S4).
We used the annotated QDO from Penicillium olsonii to search the National Center for Biotechnology Information (NCBI)/GenBank databases and found a very similar gene in S. sclerotiorum designated SsQDO (accession number, MK992913). SsQDO has a 1113-bp open reading frame (ORF) that encodes a predicted protein of 371 amino acids. The ORF was interrupted by a 63-bp intron. We constructed a maximum-likelihood phylogenetic tree with protein sequences of similar annotated QDO genes and nonannotated genes from different fungal species (Fig. 3B). QDO enzymes are present in the Sordariomycetes, Dothideomycetes, Leotimycetes, and Eurotiomycetes. The putative QDO protein from S. sclerotiorum grouped closest to orthologs from Sclerotinia borealis and Botrytis cinerea with 86% and 84% similarity, respectively. Both S. sclerotiorum and B. cinerea belong to the class Leotiomycetes. QDO amino acid sequences from the Eurotiomycetes and Sordariomycetes were on average 53% and 46% similar, respectively, to SsQDO.
The coding region of the SsQDO gene was cloned and expressed in E. coli BL 21 (DE 3) using the Gateway expression vector pDest24. The crude protein extract of recombinant E. coli cleaved both quercetin and kaempferol (Fig. 3C). The quercetin oxidation product showed a molecular ion peak of mass-to-charge ratio (m/z) 305.0286 ([M-H]−), which was consistent with the molecular formula C14H10O8 (C14H10O8 − H)−, the calculated monoisotopic mass m/z 305.029194 (Supplemental Fig. S4). This corresponds to the molecular formula of 2-protocatechuoyl phloroglucinol carboxylic acid. Kaempferol oxidation resulted in a molecular ion peak of m/z 289.0336 ([M-H]−), which was consistent with the molecular formula C14H10O7 (C14H10O7 − H)−calculated monoisotopic mass m/z 289.034279 (Supplemental Fig. S4). This corresponds to the molecular formula of 2,4-dihydroxy-6-[(4-hydroxybenzoyl)oxy]benzoic acid. Both enzyme assay products showed strong insource fragmentation in negative mode ESI-MS to m/z 169. MS2 spectra of this in-source fragment ion matched the MS2 spectrum of an authentic standard of phloroglucinol carboxylic acid (Tronto Research Chemicals; Supplemental Fig. S4). However, the extracted ion of m/z = 169 from the SsQDO assay showed a different retention time compared to the authentic phloroglucinol carboxylic acid standard, suggesting that phloroglucinol carboxylic acid was not the direct product in the assay, but that m/z 169 is an in-source fragment of a larger molecule (Supplemental Fig. S4, A and C). We also tested the activity of SsQDO with other flavonol substrates, including fisetin, galangin, and an isoflavonol daidzein, but SsQDO did not show activity toward any of them (Supplemental Fig. S5). To compare the affinity of SsQDO to quercetin and kaempferol, we analyzed enzyme kinetics with heterologously expressed SsQDO, revealing that the enzyme has a higher affinity for kaempferol (Km = 0.12 mm) than for quercetin (Km = 0.55 mm; Supplemental Fig. S6). This result is consistent with our observation that S. sclerotiorum degrades kaempferol more rapidly than quercetin in vitro (Supplemental Fig. S2A).
We also analyzed the expression of SsQDO in cultures grown with or without quercetin or kaempferol. Addition of flavonols for 2 h led to significant increases in SsQDO transcript levels compared to those in the nontreated mycelium, further supporting its role in flavonol degradation (ANOVA; P < 0.001; Supplemental Fig. S7).
QDO Knockout Mutants of S. sclerotiorum (ΔSsQDO) Are Deficient in Flavonol Degradation Activity
To study the function of SsQDO in vivo and its role in the pathogenicity of the fungus, we generated SsQDO knockout mutants of S. sclerotiorum by polyethylene glycol (PEG)-mediated transformation and homologous recombination. A replacement vector containing a hygromycin cassette with upstream and downstream sequences flanking SsQDO was used to transform S. sclerotiorum protoplasts (Fig. 4A). PCR verification of S. sclerotiorum transformants showed that SsQDO was replaced by the hygromycin gene in three transformed lines (Fig. 4A). The other transformants were heterozygotes that contained both the SsQDO and the hygromycin gene, owing to the fact that S. sclerotiorum cells are multinuclear. The SsQDO knockout mutants were further verified by enzyme assay. After cocultivation with quercetin for 2 h, protein extracts from knockout mutants (ΔSsQDO) showed no activity for degradation of flavonols, as no product was detected in the assay (Supplemental Fig. S8), while the enzyme extracts from the wild-type fungus degraded most of the substrates (ANOVA; P < 0.001; Fig. 4B).
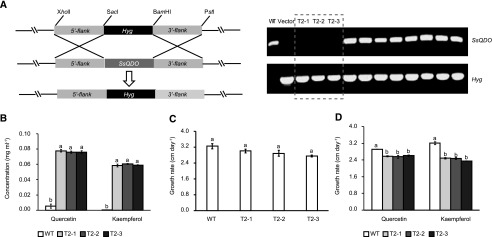
Figure 4.
Deletion of SsQDO and phenotypic characterization of the SsQDO replacement mutants (ΔSsQDO). A, Replacement of SsQDO with a hygromycin gene (Hyg) cassette by homologous recombination and PCR verification of transformants. The agarose gels depict (in order): wild type (WT), vector only, three transformed homozygotic lines, and eight heterozygotic lines. B, Flavonol degradation activity in ΔSsQDO and wild-type lines. Shown is 0.01 mg mL−1 quercetin and kaempferol after 2 h incubation with protein from preinduced wild-type and ΔSsQDO mycelium. Different letters above the bars indicate significant differences (P < 0.001, one-way ANOVA followed by Tukey’s post-hoc test). C, Growth rate of wild-type fungus and ΔSsQDO mutants on PDA medium. There were no significant differences in growth (P = 0.067; one-way ANOVA). D, Growth rate of wild-type fungus and ΔSsQDO mutants on PDA medium supplemented with 0.01 mg mL−1 quercetin or kaempferol. Different letters above the bars indicate significant differences (P < 0.01, one-way ANOVA followed by Tukey’s post-hoc test). All data represent means ± se (n = 3). T2-1, T2-2, and T2-3, ΔSsQDO mutants.
We then measured growth rates of the wild-type and ΔSsQDO S. sclerotiorum on PDA plates, and there was no significant difference in the growth rate between the wild-type and the SsQDO deletion mutants (ANOVA; P = 0.067; Fig. 4C). However, when flavonols were added to the medium, ΔSsQDO strains grew slower compared to the wild-type fungus (ANOVA, P < 0.01; Fig. 4D). These results indicate that SsQDO is important for flavonol degradation, and its loss of function is deleterious in a flavonol-amended medium.
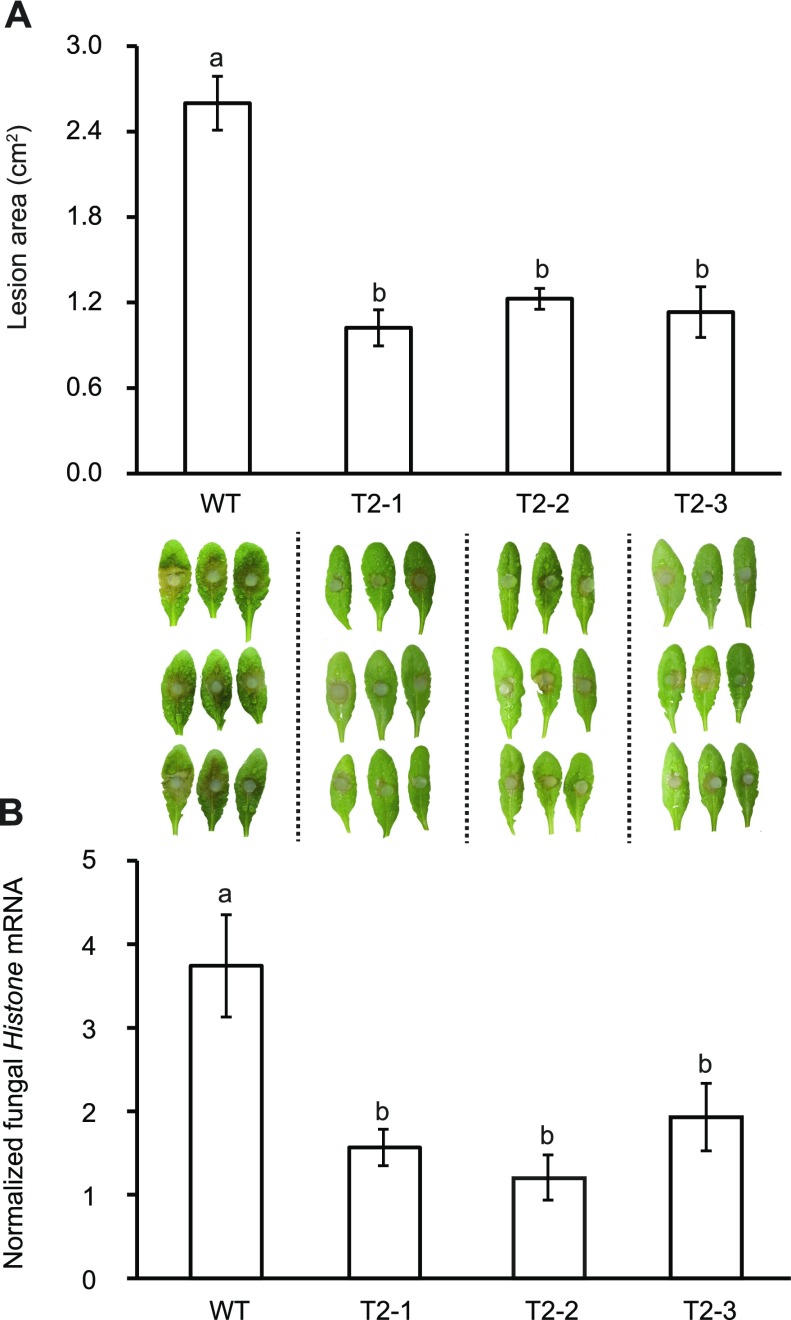
QDO Knockout Lines of S. sclerotiorum Caused Fewer and Smaller Necrotic Lesions in Arabidopsis Leaves
To test whether S. sclerotiorum SsQDO mutants with impaired abilities to degrade flavonols have an altered virulence, both wild-type and ΔSsQDO were used to inoculate both detached Arabidopsis leaves and intact plants. Lesion areas on detached leaves calculated at 24 h postinoculation showed that the ΔSsQDO lines formed significantly smaller lesions, less than 50% as large as wild-type lines (ANOVA; P < 0.01; Fig. 5A). After inoculation of intact plants, the ratio of the fungal Histone H3-encoding gene and Arabidopsis ACTIN gene transcript levels used to quantify fungal colonization in Arabidopsis at 24 h revealed that ΔSsQDO lines were less than half as pathogenic as the wild-type strain (ANOVA; P < 0.01; Fig. 5B).
Figure 5.
SsQDO deletion mutants showed less pathogenicity than wild-type (WT) S. sclerotiorum. A, Lesion area in ΔSsQDO and wild-type S. sclerotiorum lines 24 h after in vitro inoculation of detached Arabidopsis leaves. Leaf images have been digitally abstracted and made into a composite for comparison. B, Relative quantification of fungal Histone mRNA normalized to plant housekeeping gene ACTIN 24 h after in vivo inoculation of Arabidopsis leaves as determined by RT-qPCR. Different letters above the bars indicate significant differences (P < 0.01, one-way ANOVA followed by Tukey’s post-hoc test). Data for the in vitro inoculation experiment represent means of nine biological replicates. Data represent means ± se (n = 9 for in vitro inoculation experiment, n = 4 for in vivo inoculation). T2-1, T2-2, and T2-3, ΔSsQDO mutants.
Degradation of Flavonol Glycosides In Planta and in Culture Is Less Efficient in QDO Knockout Lines Than in Wild-Type S. sclerotiorum
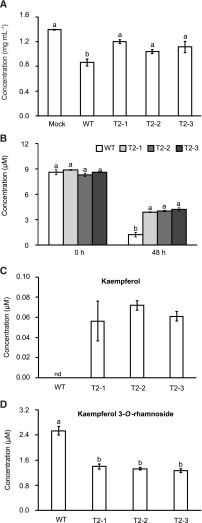
To determine how SsQDO deletion affected flavonol glycoside levels during infection, the flavonol glycoside concentration in Arabidopsis leaves was measured 72 h postinoculation both with wild-type and ΔSsQDO strains. Consistent with previous results, a significant decrease in flavonol glycoside content was observed in Arabidopsis leaves inoculated with the wild-type fungus compared to mock-treated leaves (ANOVA; P < 0.05; Fig. 6A). On the other hand, leaves inoculated with the SsQDO deletion mutants accumulated significantly higher levels of flavonol glycosides than leaves inoculated with wild-type S. sclerotiorum (ANOVA; P < 0.05; Fig. 6A). However, leaves inoculated with ΔSsQDO did show a decrease in flavonol glycoside levels at 72 h postinoculation compared to the mock treatment (even if not significantly different), suggesting that deletion of the SsQDO gene did not disable all possible mechanisms of flavonol glycoside degradation in this fungus.
Figure 6.
Deletion of SsQDO results in lower flavonoids catabolism in S. sclerotiorum. A, Flavonol glycosides in Arabidopsis (Col-0) inoculated with the wild-type (WT) fungus, in mock-inoculated control plants or in plants inoculated with ΔSsQDO mutants at 72 h. Different letters above the bars indicate significant differences (P < 0.001, one-way ANOVA followed by Tukey’s post-hoc test). B to D, Major flavonoid levels in cultures of wild-type and ΔSsQDO strains to which a concentrated flavonol extract from Arabidopsis was added. B, Catabolism of major flavonol glycosides in cultures of ΔSsQDO and wild type. Different letters above the bars indicate significant differences (P < 0.001, one-way ANOVA followed by Tukey’s post-hoc test). C, Kaempferol accumulation in ΔSsQDO and wild-type cultures. D, Kaempferol-3-O-rhamnoside production in cultures of ΔSsQDO and wild-type fungus. Different letters above the bars indicate significant differences (P < 0.001, one-way ANOVA followed by Tukey’s post-hoc test). All data were acquired by LC-ESI-MS and represent means ± se (n = 3). nd, not detected. T2-1, T2-2, and T2-3, ΔSsQDO mutants.
We next added flavonol glycoside extract from Arabidopsis to S. sclerotiorum cultures growing on artificial medium to validate the results of our in planta experiments. Media from both wild-type and ΔSsQDO cultures were analyzed at 0 and 48 h after flavonol glycoside extract addition. Higher levels of the major flavonol glycoside were recovered from medium colonized by the ΔSsQDO fungus compared to medium colonized by wild-type at 48 h (ANOVA; P < 0.001; Fig. 6B). We also quantified the biotransformed flavonols produced by the fungus from the compounds present in the extract. The kaempferol aglycone only appeared in the SsQDO mutant cultures after 48 h (Fig. 6C), confirming that further metabolism of this compound is blocked by the loss of QDO. Therefore, kaempferol aglycones are normally released by S. sclerotiorum from K3G7R, even though these were not detected in the wild-type culture. The reduction of K3R (peak 5; Fig. 1C) in the ΔSsQDO mutants compared to wild-type fungus (Fig. 6D), suggests that deletion of SsQDO not only disrupts cleavage of aglycones but also decreases the actual upstream reactions in this catabolic pathway. Taken together, these data show that deletion of SsQDO results in reduced both flavonol glycoside and flavonol aglycone metabolism by the fungus.
DISCUSSION
Flavonols, a subclass of flavonoids, are thought to play important roles in plant defense against pathogens because of their antimicrobial activity and local accumulation after fungal infection (Snyder and Nicholson, 1990; Snyder et al., 1991; Padmavati et al., 1997; Skadhauge et al., 1997; McNally et al., 2003; Hammerbacher et al., 2014; Ullah et al., 2017). Thus, we investigated the role of flavonols during fungal infection using an economically important plant pathogen, S. sclerotiorum, and the model plant, Arabidopsis, which produces high levels of flavonol glycosides, including kaempferol and quercetin glycosides (Graham, 1998; Veit and Pauli, 1999).
We unexpectedly found a significant decrease in flavonol glycoside levels in Arabidopsis 3 d after inoculation with S. sclerotiorum. Incubation of S. sclerotiorum with the major flavonol glycosides produced by Arabidopsis revealed that the fungus cleaves both 7-O-glucose and 7-O-rhamnose moieties to produce and accumulate K3(R'2G) and K3R. We further amended S. sclerotiorum cultures with K3R and K3G. We found that K3G was completely consumed by S. sclerotiorum, whereas K3R was not transformed by this fungus. In contrast, a previous study reported that Aspergillus flavus cleaves 3-O-rhamnose linkages of rutin (Krishnamurty and Simpson, 1970). Due to the fact that both K3R and K3(R'2G) cannot be further transformed by S. sclerotiorum, we hypothesize that the specificity of this fungal glycosyltransferase depends on both sugar linkage and position of flavonol glycosides. Moreover, K3G was completely consumed by S. sclerotiorum after 15 h in the liquid medium without leaving any traces of kaempferol, suggesting that S. sclerotiorum selectively cleaved glucoside moieties of flavonol glycosides and degraded the aglycones subsequently.
Plants also contain numerous glycoside hydrolases, which cleave flavonol glycosides during plant recovery from synergistic abiotic stresses, such as nitrogen deficiency and low temperature (Xu et al., 2004; Roepke and Bozzo, 2015; Roepke et al., 2017). The β-glucosidase BGLU15 characterized from Arabidopsis cleaves the 3-O-β-glucosides from K3G7R to produce kaempferol 7-O-rhamnoside (Roepke et al., 2017). Kaempferol 7-O-rhamnoside was, however, not found among the fungal degradation products of flavonol glycoside extracts in our study, suggesting a different hydrolysis pathway of glycosidic linkages in fungi. Whether plants regulate their flavonol glycoside metabolism in response to pathogen infections is also not known.
The degradation of glycosidic moieties from plant flavonol glycosides might be a carbohydrate assimilation process for S. sclerotiorum and beneficial for its expansion in host plants (Jobic et al., 2007), but the released flavonol aglycones are known to be more active antimicrobial compounds than the corresponding glycosylated derivatives (Liu et al., 2010). It was hypothesized that the basic structure of flavonol aglycones allows entry to microbial cells, and so individual functional groups may then target different components or functions of the cell (Cushnie and Lamb, 2005). For instance, quercetin can bind to the GyrB subunit of DNA gyrase and inhibits the enzyme’s ATPase activity in E. coli (Plaper et al., 2003). Many studies have shown the direct toxicity of flavonols toward various microbes. The growth of the rice (Oryza sativa) pathogen Xanthomonas oryzae causing leaf blight was inhibited by naringenin (Padmavati et al., 1997). Dihydroquercetin strongly reduced Fusarium growth and macrospore formation (Skadhauge et al., 1997). Moreover, naringenin and tangeretin, two constitutive flavonoids in Citrus aurantium, showed antifungal activity against Penicillium digitatum (Arcas et al., 2000). Many studies have also been conducted on how flavonoids are involved in plant-microbial interactions in the rhizosphere (Shaw et al., 2006). For example, naringenin, quercetin, and kaempferol accumulated significantly in root galls of Arabidopsis after Plasmodiophora brassicae infection and modulated auxin efflux from root galls (Päsold et al., 2010). In legumes, the role of flavonols is known as a key signal in regulating nodulation-related genes and rhizobial nodulation (Cooper, 2004). Interestingly, flavonols stimulate arbuscular mycorrhizal fungal growth, despite the fact that these compounds have antimicrobial activity toward a broad spectrum of pathogens in vitro (Gianinazzipearson et al., 1989; Padmavati et al., 1997).
Accordingly, we confirmed that the pathogenic fungus S. sclerotiorum is able to degrade flavonols, including kaempferol and quercetin. The degradation of flavonol aglycones in S. sclerotiorum is an inducible process, and the activity of enzymes involved in this process was significantly higher after exposure to quercetin or kaempferol. This suggests that the ability to degrade flavonols is not essential for development but for coping with specialized metabolites produced by the host plant. QDO was shown in previous studies to be responsible for flavonol degradation in numerous pathogenic fungi, such as A. flavus, Aspergillus niger, and Penicillium olsonii (Westlake and Simpson, 1961; Hund et al., 1999; Tranchimand et al., 2008). In our study, we found a homologous QDO gene in S. sclerotiorum as well as in its close relatives, such as Botrytis cinerea, suggesting that flavonol degradation might be a conserved function in fungal pathogens. The heterologously expressed SsQDO only showed activity toward quercetin and kaempferol but not toward other flavonoids. We analyzed the reaction products by LC-MS, whereas previous studies demonstrated enzyme activity only through detection of carbon monoxide released during catalysis using PdCl2 (Tranchimand et al., 2008). Flavonoids can also be oxidized by plant enzymes, such as polyphenol oxidases (PPOs). These enzymes are involved in the formation of brown pigments when internal plant tissues are exposed to oxygen (Pourcel et al., 2007). For example, aurone synthase is a catechol oxidase that belongs to the PPO enzyme family. This enzyme catalyzes the conversion of chalcones to aurones and is responsible for yellow coloration of Antirrhinum majus (Ono et al., 2006). However, a PPO enzyme specifically involved in quercetin oxidation in planta has not been reported yet, and therefore it is unlikely that the flavonol oxidation we observed after fungal inoculation is related to PPO activity (Jiménez and García-Carmona, 1999).
The role of flavonol degradation in fungal virulence was unknown and raised our interest. We generated three independent SsQDO deletion mutants, ΔSsQDO, and confirmed that these mutant strains could not degrade flavonols added to artificial medium. The ΔSsQDO mutants grew at similar rates as the wild-type fungus on medium in the absence of flavonols. However, the mutants grew significantly slower than the wild-type on medium amended with flavonols as well as on Arabidopsis leaves. These phenotypes combined with the inducibility of protein activity in response to flavonols confirmed our hypothesis that flavonol degradation is an important detoxification response for S. sclerotiorum and contributes to its virulence.
The effects of ΔSsQDO on flavonol glycoside metabolism was also monitored in planta and in culture. Arabidopsis leaves inoculated with ΔSsQDO contained more flavonol glycosides compared to leaves inoculated with the wild-type. In culture, flavonol glycosides were consumed by ΔSsQDO at a slower rate than by wild-type strains. Additionally, when we measured individual compounds, the wild-type accumulated more K3R than the mutants, but the kaempferol aglycone was only detected in ΔSsQDO cultures and not in wild-type cultures. This shows that the SsQDO gene deletion reduced flavonol glycoside metabolism in addition to the degradation of flavonol aglycones, although flavonol glycoside cleavage is not directly inhibited by knockout of QDO activity. We speculate that the flavonol, which is released after assimilation of the sugar moiety, inhibits the growth of S. sclerotiorum and inhibits the metabolism of flavonol glycosides via a negative feedback loop. The reduced virulence of ΔSsQDO may thus be attributed not only to the toxicity of flavonol aglycones, but might also be due to the reduced availability of Glc and other catabolites derived from flavonol glycosides as carbon and energy sources. This observation might also explain the slower growth of ΔSsQDO in planta compared to growth in artificial medium, which was amended only with flavonols.
Evidence to support that flavonols can serve as phytoalexins or phytoanticipins in planta is still lacking. Therefore, the characterization of SsQDO mutants in our study not only demonstrates the toxicity of flavonols to the fungus in culture, but also reveals that these phenolics cause a significant decline in fungal pathogenicity. This result shows that flavonols serve as antifungal defenses in plants unless a pathogen possesses specific adaptations to circumvent the toxicity of these compounds.
CONCLUSION
Among the flavonoids, isoflavones and flavan-3-ols are often reported as antifungal defense compounds (Naim et al., 1974; Harborne et al., 1976; Hammerbacher et al., 2014; Ullah et al., 2017). In contrast, information on the defensive roles of flavonols against fungal infection is sparse, although these compounds commonly occur in plants. For this reason, we hypothesize that plant pathogens may have evolved mechanisms to circumvent the toxicity of these compounds. We have shown here that S. sclerotiorum minimizes the potential toxicity of flavonols in Arabidopsis by degrading these metabolites. This fungus can cleave glycosidic linkages and degrade the resulting aglycones by employing an inducible dioxygenase that attacks the flavonol’s C ring. Future studies should focus on understanding how widespread this and other flavonol detoxification mechanisms are in the fungal kingdom. Given the broad distribution of flavonols in plants, the ability to detoxify these compounds or circumvent them in other ways could be an important determinant of fungal pathogenesis.
MATERIALS AND METHODS
Biological Materials
For inoculation experiments, the genome sequenced strain Sclerotinia sclerotiorum UF-70 (Amselem et al., 2011) was maintained on potato dextrose agar (PDA) at 25°C for 2 to 3 d. The fungus was cultured in potato dextrose broth (PDB) at 25°C, shaking at 150 rpm for 2 d for experiments in artificial medium.
The seeds of Arabidopsis (Arabidopsis thaliana) wild-type ecotype Columbia (Col-0) from the Arabidopsis Stock Centre (Nottingham, United Kingdom) were germinated on Murashige and Skoog medium. One-week-old seedlings were transferred to soil and grown under short-day conditions (10 h light/14 h dark cycle with fluorescent light at 120 to 180 μmol m−2s−1, 21°C, humidity 60%) for 4 to 5 weeks.
Plant Inoculation
Agar plugs (0.5 cm diameter) with actively growing mycelium were placed on five fully expanded leaves of 4- to 5-week-old Arabidopsis (Col-0) plants. Agar plugs without mycelium were used as controls (mock inoculation). Inoculation experiments were performed under the short-day conditions described above, and each treatment had three biological replicates unless otherwise stated. Both challenged and unchallenged Arabidopsis leaves were harvested 48 h and 72 h after inoculation and flash frozen.
Extraction and Analysis of Flavonoids of Arabidopsis
Inoculated plant samples were lyophilized using an Alpha 1-4 LD Plus freeze dryer (Martin Christ) for 2 d. Lyophilized leaves were ground with zirconium oxide beads in a shaking ball mill. Extraction was performed on 20 mg of tissue in 1 mL 80% (v v−1) methanol agitated for 10 min on a horizontal shaker at room temperature. After centrifugation at 18,000g for 10 min, supernatant (800 μL) was transferred onto a DEAE-Sephadex A-25 (Sigma-Aldrich) column conditioned with 800 μL distilled water and then with 500 μL 80% (v v−1) methanol in a 96-well filter plate. An aliquot of the flow-through (100 μL) collected in 96-well plates was diluted with 300 μL distilled water and used for analysis of flavonoids. The flavonol glycosides were identified as the major flavonol glycosides previously reported in Arabidopsis leaves (Veit and Pauli, 1999; Kerhoas et al., 2006; Saito et al., 2013). Analysis of flavonol glycosides was achieved on an HPLC 1100 (Agilent) with a Nucleodur Sphinx RP C-18 column (250 × 4.6 mm, 5 μm, Macherey-Nagel). Formic acid (0.2%) in water (solvent A) and acetonitrile (solvent B) were used as mobile phases for separation at a flow rate of 1 mL min−1 using a gradient as follows: 0 to 5 min, 100% A; 5 to 15 min, 0% to 45% B; 16 to 17 min, 100% B; 17 to 21 min, re-equilibration with 0% B. Flavonol glycosides were detected using a UV detector with wavelength 330 nm. K3R7R (High-Purity Compound Standard) was used as an external standard for quantification of flavonol glycosides. Quantification of flavonol glycosides was based on the UV (330 nm) absorption peak with the assumption that K3(R2'G)7R, Q3(R2'G)7R, K3G7R, Q3G7R, and Q3R7R had the same molar response as K3R7R.
Bioassays Using Crude Flavonol Glycoside Extract from Arabidopsis
Crude flavonol glycoside extracts from Arabidopsis were prepared for addition to experiments in artificial medium by extraction of lyophilized Arabidopsis leaf tissues (1 g) with 20 mL 80% (v v−1) methanol and agitation for 20 min on a horizontal shaker. The contents were centrifuged at 50,000g for 15 min (Avanti J25, Beckman). The supernatant was loaded onto a DEAE-Sephadex column (to eliminate glucosinolates from the raw extract), and flow-through was collected and concentrated to approximately 4 mL by using a rotatory evaporator. S. sclerotiorum 2-d-old cultures were supplied with the concentrated crude flavonol glycoside extracts using 1 volume of extract per 1000 volumes of medium. Liquid media without fungus supplied with the same amount of flavonol glycosides were used as controls. A 200-µL quantity of medium was collected 0, 24, and 48 h after addition to the fungal culture, and liquid medium was mixed with the same amount of methanol. Samples were centrifuged at 14,000g for 10 min, and supernatants were analyzed by LC-ESI-IonTrap-MS.
The flavonol glycosides in the extracts and their biotransformed products were identified via analysis on an Agilent HPLC1200 (Agilent Technologies) coupled to a Bruker Esquire 6000 ESI-IonTrap-MS system. Compounds were separated on a Nucleodur Sphinx RP C-18 column (250 × 4.6 mm, 5 mm, Macherey-Nagel) at a flow rate of 1 mL min−1. Formic acid (0.2%) in water (solvent A) and acetonitrile (solvent B) were used as mobile phases with the following gradient: 0 to 25 min, 95% to 45% A; 25 to 27 min, 100% B; 27 to 31 min, 95% A. An Esquire 6000 ESI-IonTrap mass spectrometer (Bruker Daltonik) was used to detect these compounds by scanning an m/z between 60 and 1100 with an optimal target mass of m/z 405 in alternating mode. The mass spectrometer parameters were set as follows: Skimmer voltage, −40 eV; capillary exit voltage, −121 eV; capillary voltage, ±3000 V; nebulizer pressure, 35 psi; drying gas, 11 L min−1; gas temperature, 330°C. The AutoMS function of Bruker Esquire Control software was used for MS2 analysis of enzyme assay products. Standard compounds K3G and K3R were bought from TransMIT PlantMetaChem (Giessen).
In Vitro Flavonol Biotransformation by S. sclerotiorum
The fungus S. sclerotiorum was cultured in PDB medium containing quercetin or kaempferol at a concentration of 0.1 mg mL−1 under the conditions described above. Medium (200 μL) was collected from each treatment at 0, 24, and 48 h for LC-MS analysis. Chromatography of flavonols was performed on the same equipment described for flavonol glycoside analysis with the following elution profile: 0 to 1 min, 100% A; 1 to 15 min, 0% to 65% B in A; 15 to 18 min, 100% B, 18.1 to 22 min, 100% A. Detection and quantification of flavonols was achieved using LC-ESI-IonTrap-MS in negative ionization mode scanning a range from m/z 100 to 1600 with an optimal target mass of m/z 405. The mass spectrometer parameters were set as follows: capillary voltage, ±3000 V; nebulizer pressure, 35 psi; drying gas, 11 L min−1 and gas temperature, 330°C. Capillary exit potential was kept at −121 V. The molecular ion peaks [M-H]− of the analytes (kaempferol m/z 285; quercetin m/z 301) were monitored in negative mode (extracted ion chromatograms), and the authentic external kaempferol (Carl Roth) and quercetin (Sigma-Aldrich) standards were used for quantification.
Total Protein Extraction from S. sclerotiorum and Enzyme Assays
S. sclerotiorum was cultured in liquid medium under the conditions described above. Mycelium was harvested 2 h after induction with 0.01 mg mL−1 quercetin or kaempferol, lyophilized for 1 d, and homogenized with steel beads in a shaking ball mill. After addition of 1 mL HEPES buffer (1 m sorbitol, 10 mm HEPES, 50 mm EDTA, 0.1 m KCl, 0.2% (v v−1) Triton X-100, pH 7) to 10 mg of the homogenate, the solution was incubated on ice for 30 min. Following centrifugation at 18,000g for 10 min at 4°C, the supernatant was collected for enzyme assays. Protein concentration was determined with a 2-d Quant Kit according to the manufacturer's instructions (GE Healthcare).
Enzyme assays were carried out as follows: 90 μL total protein (1 μg μL−1) was added to 10 μL of a 1 mg mL−1 quercetin or kaempferol solution. The reaction was incubated at 25°C for 30 min. The reactions were terminated by adding 100 µL methanol. A 10-μL portion from each reaction was used for LC-MS analysis described above for in vitro flavonol biotransformation assays.
Sequence and Phylogenetic Analysis of SsQDO
The protein sequence of an identified quercetin 2,3-dixogenase from Penicillium olsonii (accession number, ABV24349) was used to search the S. sclerotiorum genome using BLASTp on NCBI (Tranchimand et al., 2008). Only one protein (SS1G_11412 hypothetical protein) with 53% identity was found in S. sclerotiorum, which was later named SsQDO. Full-length homologous amino acid sequences of putative QDO enzymes from different fungal species were retrieved from the NCBI/GenBank databases. The sequences were aligned using a multiple sequence alignment program MAFFT v. 7 (Katoh and Standley, 2013) by employing the highly accurate method l-INS-I. The aligned sequences were then verified and manually adjusted using Mesquite 3.04 (http://mesquiteproject.org). The maximum likelihood tree was constructed with the aligned sequences (20) using the software PhyML v. 3.0 (Guindon et al., 2010). The LG amino acid substitution model was used (Le and Gascuel, 2008). The best optimized random starting tree was obtained out of five random trees using tree topology search BEST, which estimates the phylogeny using both NNI (nearest neighbor interchange) and SPR (subtree pruning and regrafting). To estimate branch support, a nonparametric bootstrap analysis was carried out (n = 1000). The tree with the highest log likelihood (−8862.81266) was viewed using Figtree (http://tree.bio.ed.ac.uk/software/figtree/) and rooted at the midpoint. The tree readability was improved using Adobe Illustrator CS5. Accession numbers of all protein sequences used in this phylogenetic analysis are provided at the end of the “Materials and Methods” section.
Cloning, Expression, and Enzyme Characterization of SsQDO in Escherichia coli
For heterologously expressing the ORF of SsQDO, the 1113-bp protein coding region was cloned using Gateway recombination technology (Invitrogen, Life Technologies Corporation). SsQDO was PCR amplified with Gateway compatible primers (Supplemental Table S1) from S. sclerotiorum cDNA using high-fidelity Phusion taq (New England BioLabs) and cloned into the pDONR 201 vector using BP clonase II (Invitrogen) to generate an entry clone. After sequencing, the protein-coding gene was transferred to a destination expression vector pDEST24, which enables the production of a recombinant protein with a C-terminal GST tag using LR clonase II (Invitrogen). This plasmid was transformed into BL 21 (DE 3) E. coli for protein expression. A single colony of the expression clone was cultured in Luria Bertani broth and incubated at 37°C. After an OD600 of 0.3 to 0.5 was reached, protein expression was induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside at 37 °C for 5 h. Bacteria were harvested by centrifugation and resuspended in 1× PBS buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mM KH2PO4) containing 10% (v v−1) glycerol, sonicated (2× 10% cycle, 60% power, 2 min), and centrifuged at 75,000g for 30 min at 4°C. The supernatant was used for enzyme assays. Total protein concentrations were determined according to the method described above.
Enzyme activity was performed as follows: 100 μm substrate, 0.5 mm phosphate buffer (pH 7.0), 50 μg crude protein, and deionized water to make a final volume 100 μL. Reaction time was decided by a linearity test with quercetin that showed that product formation exhibited a linear increase within 12 h. The reaction was incubated at 25°C for 2 h and terminated by adding 100 µL methanol. LC-MS analysis was conducted using the same equipment, gradient, and parameters described above for in vitro flavonol biotransformation assays. Protein from an untransformed BL21 (DE3) strain was used as a negative control.
The determination of the exact mass of enzyme assay products was performed on an ultra-HPLC-electrospray ionization-time-of-flight spectrometer (UHPLC-ESI-TOF-MS) system with a Dionex UltiMate 3000 rapid separation LC system (Thermo Fisher) and a timsTOF MS system (Bruker Daltonik). A Zorbax Eclipse XDB-C18 column (50 × 4.6 mm, 1.8 µm, Agilent) was used, and separation was accomplished using a mobile phase consisting of 0.1% (v v−1) formic acid in ultrapure water as solvent A and acetonitrile as solvent B with a flow rate of 300 μL min−1 at 25 °C. The gradient was as follows: 5% to 60% (v v−1) B (6 min), 100% B (1 min), 100% to 5% B (0.1 min), 5% B (2.4 min). ESI source parameters were set to 3500 V for capillary voltage, dry temperature 330°C, and a dry gas flow of 11 L min−1. Samples were measured in negative ionization mode at a mass range of m/z 50 to 1400. Instrument control, data acquisition, and reprocessing were performed using HyStar 4.1 (Bruker Daltonik).
Recombinant protein was further used for assays with quercetin and kaempferol in serial concentrations to compare the affinity of SsQDO. Each concentration was tested with three replicates. Enzyme reactions were set up as described above, and Km values were calculated using SigmaPlot (Systat Software).
Construction of SsQDO Replacement Vector
To delete the SsQDO gene in S. sclerotiorum, genomic DNA of this fungus was isolated with the Stratec Plant DNA Kit (Birkenfeld, Germany) following the manufacturer’s protocol. For cloning the flanking regions of SsQDO, a 1087-bp fragment (5′ flank-SsQDO) upstream of SsQDO was amplified with primers 5′ flank-SsQDO-forward (XhoI) and 5′ flank-SsQDO-reverse (SacI) using Phusion taq (New England BioLabs), following the manufacturer’s instructions. A 1346-bp downstream fragment (3′ flank-SsQDO) was amplified with primers 3′ flank- SsQDO-forward (BamHI) and 3′ flank-SsQDO-reverse (PstI). The purified upstream PCR product and the pXEH vector with a hygromycin phosphotransferase (hph) cassette containing a trpC promoter (Wang et al., 2016) were digested using XhoI and SacI and ligated using the Quick Ligation Kit (New England Biolabs). The final replacement vector pXEH-5′ flank-SsQDO-3′ flank was obtained by digesting the recombinant vector pXEH-5′ flank-SsQDO and the downstream flanking region with BamHI and PstI and ligating vector and fragments. Primers are listed in Supplemental Table S1.
Preparation of Protoplasts from S. sclerotiorum Mycelium
Protoplasts from S. sclerotiorum were prepared according to the protocol described by Rollins (2003). In short, wild-type S. sclerotiorum 1980 mycelium grown for 2 d in PDB medium was harvested by centrifugation and digested using a lysing enzyme from Trichoderma harzianum (Sigma). The lysing enzyme (0.4 mg) was first dissolved in 6 mL Novozyme buffer (1 m sorbitol, 50 mm sodium citrate, pH 5.8) and then added into 34 mL protoplast buffer (0.8 m MgSO4 • 7H2O, 0.2 m sodium citrate • 2H2O; pH 5.5). Mycelium was digested with the enzyme mixture for 3 h at 28°C. Protoplasts were washed with 20 mL 0.6 m KCl and then with 10 mL STC (1 m sorbitol, 50 mm Tris-HCl, pH 8, 50 mm CaCl2•2H2O) twice. The final concentration was adjusted to 1 × 108 protoplasts per mL STC. One milliliter protoplast stock was stored with 125 μL 60% PEG4000 (v v−1), 125 μL KTC (1.8 m KCl, 150 mm Tris-HCl, pH 8.0, 150 mm CaCl2), 1% DMSO, and 0.3 mg heparin at −80°C.
PEG-Mediated Protoplast Transformation and Analysis of Transformants
Approximately 1.5 μg chilled plasmid of the replacement vector pXEH-5′ flank-SsQDO-3′ flank was mixed with 100 μL (1 × 107 mL−1) protoplast stock solution and incubated on ice for 30 min. One milliliter PEG solution (two parts KTC, one part 60% PEG4000) was added to the protoplast-DNA suspension and mixed gently. The mixture was incubated at room temperature for 20 min and evenly spread on the surface of 20 mL bottom agar (239.6 g of Suc, 0.5 g of yeast extract, 15 g of agar per liter), which did not contain the selective agent. After overnight (15–20 h) regeneration of protoplasts in the dark at room temperature, 10 mL of top agar (239.6 g of Suc, 0.5 g of yeast extract, 8 g of agar per liter) containing 3000 μg hygromycin was overlaid on the plate (to make a final concentration at 100 μg mL−1). Transformants carrying the hygromycin gene were allowed to grow through the top agar for 5 to 7 d and then transferred at least three times to PDA medium containing 100 μg mL−1 hygromycin (Rollins, 2003).
PCR and enzyme assays were performed for verification of transformants. Genomic DNA of transformants was isolated, and PCR reactions with both SsQDO primers and hygromycin resistance gene primers (Supplemental Table S1) were performed. The transformants with the hygromycin gene but without the SsQDO gene were further verified by assaying the QDO activity of both wild-type and transformants after induction with quercetin.
Growth Rate Assay
An agar plug with actively growing mycelium was inoculated on PDA at the center of a 9-cm petri dish. Growth measurements were taken at 24 and 48 h postinoculation in two vertical directions on each plate. The average results from three replicates for each strain were calculated. The growth rate of both wild-type and mutants on PDA amended with 0.01 mg mL−1 quercetin or kaempferol was measured using the same method.
Pathogenicity Assay with Detached Leaves
Arabidopsis (Col-0) plants were cultivated under the conditions described above. Detached leaves with similar ages were placed on water-saturated filter paper in a 9-cm petri dish and inoculated with a 5-mm agar plug of actively growing mycelium from both wild-type S. sclerotiorum and the SsQDO knockout mutants. Petri dishes were maintained at 22°C to 25°C in the laboratory. Nine replicates were used for each fungal strain, and the experiment was repeated twice. Lesion areas were determined at 24 h after inoculation by photographing each leaf together with a 2 × 2 cm reference square. Image analysis was conducted with Photoshop CS5, using the magic wand tool to select symptomatic regions and reference regions. The pixel values of the symptomatic regions and the 4 cm2 reference region were used to calculate the lesion areas.
RT-qPCR
S. sclerotiorum was cultured in liquid medium under the conditions described above. Mycelium was harvested 2 h after induction either with 0.01 mg mL−1 quercetin or kaempferol. RNA was isolated from both non-pretreated and flavonol-pretreated mycelium. Fungal mycelium was ground in liquid nitrogen to a fine powder. Total RNA was isolated with the Stratec Plant RNA Mini Kit following the manufacturer’s protocol, and DNA was eliminated with DNase (Qiagen) using the modifications described by Wadke et al. (2016). Approximately 1 µg DNA-free RNA was used for cDNA synthesis with the Superscript II kit (Invitrogen) following the protocols of the manufacturer. Two-step RT-qPCR was performed using Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies) and a Bio-Rad CFX Connect Real-Time System with the following cycling conditions: 95°C for 5 min, 40 cycles at 95°C for 15 s, and extension at 60°C for 30 s. Primers used for RT-qPCR of SsQDO are listed in Supplemental Table S1. Primer efficiency and calibration curves were calculated with serially diluted templates using the same cycling (R2 > 0.99). Primer specificity was tested with cycling melting curve analysis from 65°C to 95°C at 0.5°C s−1 melting rate.
Relative quantification of wild-type and ΔSsQDO pathogenicity on Arabidopsis was also achieved through RT-qPCR of the fungal Histone gene (Li et al., 2018) against the Arabidopsis ACTIN gene (Czechowski et al., 2005; Supplemental Table S1). Plant inoculation, RNA isolation, cDNA synthesis, and RT-qPCR were conducted following the protocol described above.
Statistical Analysis
Data were analyzed using R version 3.4.0. Data normality and variances were verified using the Shapiro-Wilk and Levene’s test, respectively. To meet the assumptions of parametric tests, some data were subjected to square root or log transformation. Data were then analyzed either by Student’s t test or one-way ANOVA, depending on overall number of populations.
Accession Numbers
Accession numbers are as follows: EIT80042.1 (Aspergillus oryzae 3.042), XP_015403930.1 (Aspergillus nomius NRRL 13137), GAQ33184.1 (Aspergillus niger), ABV24349.1 (Penicillium olsonii), KXG50735.1 (Penicillium griseofulvum), OQD69664.1 (Penicillium polonicum), XP_018385410.1 (Alternaria alternata), XP_014075683.1 (Bipolaris maydis ATCC 48331), XP_001587420.1 now fully annotated MK992913 (S. sclerotiorum 1980 UF-70), ESZ96362.1 (Sclerotinia borealis F-4128), XP_001552378.1 (Botrytis cinerea B05.10), KEQ89945.1 (Aureobasidium pullulans EXF-150), KXS95020.1 (Mycosphaerella eumusae), ENH80076.1 (Colletotrichum orbiculare MAFF 240422), XP_007273525.1 (Colletotrichum gloeosporioides Nara gc5), KZL76304.1 (Colletotrichum tofieldiae), KIL83777.1 (Fusarium avenaceum), XP_007826906.1 (Pestalotiopsis fici W106-1), EKG13902.1 (Macrophomina phaseolina MS6), XP_020132085.1 (Diplodia corticola).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Major flavonol glycoside extracts were not degraded spontaneously in liquid medium within 48 h.
Supplemental Figure S2. Identification of S. sclerotiorum degradation products of Arabidopsis flavonol glycosides formed in artificial medium.
Supplemental Figure S3. Flavonol glycosides and aglycones were degraded by S. sclerotiorum.
Supplemental Figure S4. Identification of SsQDO degradation products of flavonols by UHPLC-ESI-TOF-MS in negative ionization mode.
Supplemental Figure S5. Enzyme assays of SsQDO with other flavonoids as substrates including daidzein, fisetin, and galangin.
Supplemental Figure S6. Michaelis-Menten constants (Km) of SsQDO with flavonols as substrates.
Supplemental Figure S7. Expression of SsQDO in S. sclerotiorum induced with quercetin and kaempferol.
Supplemental Figure S8. Quercetin degradation product protocatechuic acid was only detected in assay with protein from wild type, but not the ΔSsQDO mutant.
Supplemental Table S1. List of primers used in this study.
Acknowledgments
The authors thank Bettina Raguschke for all sequencing and for her assistance in the laboratory and all members of the greenhouse team, especially Andreas Weber and Elke Goschala, for growing Arabidopsis for this study. The pXEH vector was kindly provided by Prof. Hongyu Pan (Jilin University, Changchun, Jilin).
Footnotes
This work was supported by the China Scholarship Council (CSC; 201406170041) and the Max Planck Society (MPG).
Articles can be viewed without a subscription.
References
- Amselem J, Cuomo CA, van Kan JA, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, et al. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcas MC, Botía JM, Ortuño AM, Del Río JA (2000) UV irradiation alters the levels of flavonoids involved in the defence mechanism of Citrus aurantium fruits against Penicillium digitatum. Eur J Plant Pathol 106: 617–622 [Google Scholar]
- Boland GJ, Hall R (1994) Index of plant hosts of Sclerotinia sclerotiorum. Can J Plant Pathol 16: 93–108 [Google Scholar]
- Bolton MD, Thomma BP, Nelson BD (2006) Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol 7: 1–16 [DOI] [PubMed] [Google Scholar]
- Buer CS, Imin N, Djordjevic MA (2010) Flavonoids: New roles for old molecules. J Integr Plant Biol 52: 98–111 [DOI] [PubMed] [Google Scholar]
- Cartea ME, Francisco M, Soengas P, Velasco P (2010) Phenolic compounds in Brassica vegetables. Molecules 16: 251–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cessna SG, Sears VE, Dickman MB, Low PS (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12: 2191–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook NC, Samman S (1996) Flavonoids—chemistry, metabolism, cardioprotective effects, and dietary sources. J Nutr Biochem 7: 66–76 [Google Scholar]
- Cooper JE. (2004) Multiple responses of rhizobia to flavonoids during legume root infection. Adv Bot Res 41: 1–62 [Google Scholar]
- Cushnie TP, Lamb AJ (2005) Antimicrobial activity of flavonoids. Int J Antimicrob Agents 26: 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzipearson V, Branzanti B, Gianinazzi S (1989) In vitro enhancement of spore germination and early hyphal growth of a vesicular-arbuscular mycorrhizal fungus by host root exudates and plant flavonoids. Symbiosis 7: 243–255 [Google Scholar]
- Graham TL. (1998) Flavonoid and flavonol glycoside metabolism in Arabidopsis. Plant Physiol Biochem 36: 135–144 [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hammerbacher A, Paetz C, Wright LP, Fischer TC, Bohlmann J, Davis AJ, Fenning TM, Gershenzon J, Schmidt A (2014) Flavan-3-ols in Norway spruce: Biosynthesis, accumulation, and function in response to attack by the bark beetle-associated fungus Ceratocystis polonica. Plant Physiol 164: 2107–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB, Ingham JL, King L, Payne M (1976) Isopentenyl isoflavone luteone as a pre-infectional antifungal agent in genus Lupinus. Phytochemistry 15: 1485–1487 [Google Scholar]
- Hund HK, Breuer J, Lingens F, Hüttermann J, Kappl R, Fetzner S (1999) Flavonol 2,4-dioxygenase from Aspergillus niger DSM 821, a type 2 CuII-containing glycoprotein. Eur J Biochem 263: 871–878 [DOI] [PubMed] [Google Scholar]
- Jiménez M, García-Carmona F (1999) Oxidation of the flavonol quercetin by polyphenol oxidase. J Agric Food Chem 47: 56–60 [DOI] [PubMed] [Google Scholar]
- Jobic C, Boisson AM, Gout E, Rascle C, Fèvre M, Cotton P, Bligny R (2007) Metabolic processes and carbon nutrient exchanges between host and pathogen sustain the disease development during sunflower infection by Sclerotinia sclerotiorum. Planta 226: 251–265 [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuma K, Noda N, Suzuki M (2003) Malonylated flavonol glycosides from the petals of Clitoria ternatea. Phytochemistry 62: 229–237 [DOI] [PubMed] [Google Scholar]
- Kerhoas L, Aouak D, Cingöz A, Routaboul JM, Lepiniec L, Einhorn J, Birlirakis N (2006) Structural characterization of the major flavonoid glycosides from Arabidopsis thaliana seeds. J Agric Food Chem 54: 6603–6612 [DOI] [PubMed] [Google Scholar]
- Krishnamurty HG, Simpson FJ (1970) Degradation of rutin by Aspergillus flavus. Studies with oxygen 18 on the action of a dioxygenase on quercetin. J Biol Chem 245: 1467–1471 [PubMed] [Google Scholar]
- Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mu W, Veluchamy S, Liu Y, Zhang Y, Pan H, Rollins JA (2018) The GATA-type IVb zinc-finger transcription factor SsNsd1 regulates asexual-sexual development and appressoria formation in Sclerotinia sclerotiorum. Mol Plant Pathol 19: 1679–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Mou Y, Zhao J, Wang J, Zhou L, Wang M, Wang D, Han J, Yu Z, Yang F (2010) Flavonoids from Halostachys caspica and their antimicrobial and antioxidant activities. Molecules 15: 7933–7945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally DJ, Wurms KV, Labbe C, Belanger RR (2003) Synthesis of C-glycosyl flavonoid phytoalexins as a site-specific response to fungal penetration in cucumber. Physiol Mol Plant Pathol 63: 293–303 [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217 [Google Scholar]
- Morrall RAA, Duczek LJ, Sheard JW (1972) Variations and correlations within and between morphology, pathogenicity, and pectolytic enzyme activity in Sclerotinia from Saskatchewan. Can J Bot 50: 767–786 [Google Scholar]
- Naim M, Gestetner B, Zilkah S, Birk Y, Bondi A (1974) Soybean isoflavones. Characterization, determination, and antifungal activity. J Agric Food Chem 22: 806–810 [DOI] [PubMed] [Google Scholar]
- Ono E, Hatayama M, Isono Y, Sato T, Watanabe R, Yonekura-Sakakibara K, Fukuchi-Mizutani M, Tanaka Y, Kusumi T, Nishino T, et al. (2006) Localization of a flavonoid biosynthetic polyphenol oxidase in vacuoles. Plant J 45: 133–143 [DOI] [PubMed] [Google Scholar]
- Onyilagha J, Bala A, Hallett R, Gruber M, Soroka J, Westcott N (2003) Leaf flavonoids of the cruciferous species, Camelina sativa, Crambe spp., Thlaspi arvense and several other genera of the family Brassicaceae. Biochem Syst Ecol 31: 1309–1322 [Google Scholar]
- Padmavati M, Sakthivel N, Thara KV, Reddy AR (1997) Differential sensitivity of rice pathogens to growth inhibition by flavonoids. Phytochemistry 46: 499–502 [Google Scholar]
- Päsold S, Siegel I, Seidel C, Ludwig-Müller J (2010) Flavonoid accumulation in Arabidopsis thaliana root galls caused by the obligate biotrophic pathogen Plasmodiophora brassicae. Mol Plant Pathol 11: 545–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedras MS, Ahiahonu PW (2005) Metabolism and detoxification of phytoalexins and analogs by phytopathogenic fungi. Phytochemistry 66: 391–411 [DOI] [PubMed] [Google Scholar]
- Pedras MS, Hossain M (2006) Metabolism of crucifer phytoalexins in Sclerotinia sclerotiorum: Detoxification of strongly antifungal compounds involves glucosylation. Org Biomol Chem 4: 2581–2590 [DOI] [PubMed] [Google Scholar]
- Peer WA, Murphy AS (2007) Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Pereira DM, Valentao P, Pereira JA, Andrade PB (2009) Phenolics: From chemistry to biology. Molecules 14: 2202–2211 [Google Scholar]
- Plaper A, Golob M, Hafner I, Oblak M, Solmajer T, Jerala R (2003) Characterization of quercetin binding site on DNA gyrase. Biochem Biophys Res Commun 306: 530–536 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I (2007) Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci 12: 29–36 [DOI] [PubMed] [Google Scholar]
- Rice-Evans CA, Miller NJ, Bolwell PG, Bramley PM, Pridham JB (1995) The relative antioxidant activities of plant-derived polyphenolic flavonoids. Free Radic Res 22: 375–383 [DOI] [PubMed] [Google Scholar]
- Riou C, Freyssinet G, Fevre M (1991) Production of cell wall-degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum. Appl Environ Microbiol 57: 1478–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepke J, Bozzo GG (2015) Arabidopsis thaliana β-glucosidase BGLU15 attacks flavonol 3-O-β-glucoside-7-O-α-rhamnosides. Phytochemistry 109: 14–24 [DOI] [PubMed] [Google Scholar]
- Roepke J, Gordon HOW, Neil KJA, Gidda S, Mullen RT, Freixas Coutin JA, Bray-Stone D, Bozzo GG (2017) An apoplastic β-glucosidase is essential for the degradation of flavonol 3-O-β-glucoside-7-O-α-rhamnosides in Arabidopsis. Plant Cell Physiol 58: 1030–1047 [DOI] [PubMed] [Google Scholar]
- Rollins JA. (2003) The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol Plant Microbe Interact 16: 785–795 [DOI] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol Biochem 72: 21–34 [DOI] [PubMed] [Google Scholar]
- Schaab MR, Barney BM, Francisco WA (2006) Kinetic and spectroscopic studies on the quercetin 2,3-dioxygenase from Bacillus subtilis. Biochemistry 45: 1009–1016 [DOI] [PubMed] [Google Scholar]
- Shaw LJ, Morris P, Hooker JE (2006) Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ Microbiol 8: 1867–1880 [DOI] [PubMed] [Google Scholar]
- Skadhauge B, Thomsen KK, von Wettstein D (1997) The role of the barley testa layer and its flavonoid content in resistance to Fusarium infections. Hereditas 126: 147–160 [Google Scholar]
- Snyder BA, Nicholson RL (1990) Synthesis of phytoalexins in sorghum as a site-specific response to fungal ingress. Science 248: 1637–1639 [DOI] [PubMed] [Google Scholar]
- Snyder BA, Leite B, Hipskind J, Butler LG, Nicholson RL (1991) Accumulation of sorghum phytoalexins induced by Colletotrichum graminicola at the infection site. Physiol Mol Plant Pathol 39: 463–470 [Google Scholar]
- Tranchimand S, Ertel G, Gaydou V, Gaudin C, Tron T, Iacazio G (2008) Biochemical and molecular characterization of a quercetinase from Penicillium olsonii. Biochimie 90: 781–789 [DOI] [PubMed] [Google Scholar]
- Tranchimand S, Brouant P, Iacazio G (2010) The rutin catabolic pathway with special emphasis on quercetinase. Biodegradation 21: 833–859 [DOI] [PubMed] [Google Scholar]
- Ullah C, Unsicker SB, Fellenberg C, Constabel CP, Schmidt A, Gershenzon J, Hammerbacher A (2017) Flavan-3-ols are an effective chemical defense against rust infection. Plant Physiol 175: 1560–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M, Pauli GF (1999) Major flavonoids from Arabidopsis thaliana leaves. J Nat Prod 62: 1301–1303 [DOI] [PubMed] [Google Scholar]
- Wadke N, Kandasamy D, Vogel H, Lah L, Wingfield BD, Paetz C, Wright LP, Gershenzon J, Hammerbacher A (2016) The bark-beetle-associated fungus, Endoconidiophora polonica, utilizes the phenolic defense compounds of its host as a carbon source. Plant Physiol 171: 914–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu Y, Liu J, Zhang Y, Zhang X, Pan H (2016) The Sclerotinia sclerotiorum FoxE2 gene is required for apothecial development. Phytopathology 106: 484–490 [DOI] [PubMed] [Google Scholar]
- Wegulo SN, Yang XB, Martinson CA, Murphy PA (2005) Effects of wounding and inoculation with Sclerotinia sclerotiorum on isoflavone concentrations in soybean. Can J Plant Sci 85: 749–760 [Google Scholar]
- Westlake DW, Simpson FJ (1961) Degradation of rutin by Aspergillus flavus. Factors affecting production of the enzyme system. Can J Microbiol 7: 33–44 [DOI] [PubMed] [Google Scholar]
- Williams CA, Grayer RJ (2004) Anthocyanins and other flavonoids. Nat Prod Rep 21: 539–573 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Escamilla-Treviño L, Zeng L, Lalgondar M, Bevan D, Winkel B, Mohamed A, Cheng CL, Shih MC, Poulton J, et al. (2004) Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol Biol 55: 343–367 [DOI] [PubMed] [Google Scholar]