Both AtERO1 and AtERO2 play essential roles in oxidative protein folding in the endoplasmic reticulum, but AtERO1 may serve as the primary sulfhydryl oxidase relative to AtERO2
Abstract
Disulfide bonds are essential for the folding of the eukaryotic secretory and membrane proteins in the endoplasmic reticulum (ER), and ER oxidoreductin-1 (Ero1) and its homologs are the major disulfide donors that supply oxidizing equivalents in the ER. Although Ero1 homologs in yeast (Saccharomyces cerevisiae) and mammals have been extensively studied, the mechanisms of plant Ero1 functions are far less understood. Here, we found that both Arabidopsis (Arabidopsis thaliana) ERO1 and its homolog AtERO2 are required for oxidative protein folding in the ER. The outer active site, the inner active site, and a long-range noncatalytic disulfide bond are required for AtERO1’s function. Interestingly, AtERO1 and AtERO2 also exhibit significant differences. The ero1 plants are more sensitive to reductive stress than the ero2 plants. In vivo, both AtERO1 and AtERO2 have two distinct oxidized isoforms (Ox1 and Ox2), which are determined by the formation or breakage of the putative regulatory disulfide. AtERO1 is mainly present in the Ox1 redox state, while more AtERO2 exists in the Ox2 state. Furthermore, AtERO1 showed much stronger oxidative protein-folding activity than AtERO2 in vitro. Taken together, both AtERO1 and AtERO2 are required to regulate efficient and faithful oxidative protein folding in the ER, but AtERO1 may serves as the primary sulfhydryl oxidase relative to AtERO2.
The endoplasmic reticulum (ER) is a subcellular compartment where the eukaryotic secretory and membrane proteins are folded (Delaunay-Moisan and Appenzeller-Herzog, 2015). When protein folding in the ER is disturbed, or when loading of the secretory and membrane proteins exceeds the folding capacity of the ER, accumulation of unfolded proteins will cause ER stress (Meusser et al., 2005; Vitale and Boston, 2008). To recover from ER stress, the cell has evolved a sophisticated mechanism, called the unfolded protein response (UPR), to up-regulate ER chaperone or other gene expression (Deng et al., 2013a; Howell, 2013; Lindholm et al., 2017; Qian et al., 2018), thereby promoting the folding capacity of the ER. UPR in plants can be induced by the application of chemical reagents, such as DTT and tunicamycin (Tm; Liu et al., 2007; Iwata et al., 2008; Lu and Christopher, 2008; Fan et al., 2018; Ruberti et al., 2018), or by biotic stresses (Ye et al., 2011; Moreno et al., 2012; Kørner et al., 2015) and abiotic stresses (Liu and Howell, 2010; Deng et al., 2016; Zhang et al., 2017). There are at least two pathways mediating UPR signaling in Arabidopsis (Arabidopsis thaliana). One is the Inositol Required Protein1 (IRE1)/basic Leucine Zipper60 (bZIP60) pathway. Arabidopsis IRE1 splices bZIP60 mRNA in response to ER stress, resulting in the production of the spliced mRNA encoding the nucleus-targeted bZIP60. Another pathway is through ER membrane-associated transcription factors, such as bZIP28, which is cleaved by the protease Site-1 Protease and Site-2 Protease in the Golgi and migrates to the nucleus. Both bZIP60 and bZIP28 can up-regulate UPR gene expression (Liu et al., 2007; Iwata et al., 2008; Deng et al., 2013b; Liu and Howell, 2016).
Disulfide bonds play important roles in the folding, structural integrity, stability, localization, and functioning of secretory and membrane proteins (Bulleid and Ellgaard, 2011; Meyer et al., 2018). Disulfide bond formation is catalyzed by the ER resident protein disulfide isomerase (PDI; EC 5.3.4.1) in mammals (Hatahet and Ruddock, 2009; Bulleid and Ellgaard, 2011). Only properly folded proteins can exit the ER and reach their target destination. Unfolded proteins are either refolded again or retained in the ER and degraded via ER-associated degradation (Hong et al., 2008; Smith et al., 2011).
In yeast (Saccharomyces cerevisiae) and mammals, ER oxidoreductin-1 (Ero1) and its homologs are the major disulfide donors that supply oxidizing equivalents to the active centers of PDIs in the ER (Sevier and Kaiser, 2008; Meyer et al., 2019). Yeast has one Ero1, Ero1p, while mammals have two Ero1s, Ero1α (Cabibbo et al., 2000) and Ero1β (Pagani et al., 2000). Ero1p possesses two functional regions: the core region and a flexible shuttle arm, also called a flexible loop, outside the core region. The core region contains a redox-active di-Cys active site, termed the inner active site, which is oxidized by the proximally bound FAD cofactor. The disulfide bond is then transferred from the inner active site to the outer active site in the flexible shuttle arm of Ero1p, which, in turn, shuttles the disulfide to one active center of PDI (Gross et al., 2004, 2006; Sevier et al., 2007). Similarly, mammalian Ero1α uses the buried inner active site C394XXC397 associated with the FAD moiety to reduce molecular oxygen, generating peroxide in this process (Bertoli et al., 2004; Inaba et al., 2010). Then, the C394XXC397 site transfers a disulfide bond to the outer active site C94X4C99 located in the flexible loop of Ero1α, which enables the oxidation of the active site in the a′ domain of PDI, and the a′ domain of PDI oxidizes its a domain (Wang et al., 2009; Araki and Nagata, 2011).
Yeast and human (Homo sapiens) Ero1s also have regulatory disulfides formed between catalytic and noncatalytic Cys residues, which regulate the activities of Ero1s. For Ero1α, a regulatory disulfide Cys-94-Cys-131 was formed in the inactive oxidized state (Ox2), which inhibits the disulfide transfer from the inner active site to PDI via the outer active site. During the activation of Ero1α, Cys-94-Cys-131 is reduced, the outer active site is liberated, and Ero1α is shifted from Ox2 to Ox1, the enzymatically active state (Appenzeller-Herzog et al., 2008; Baker et al., 2008; Zhang et al., 2014).
In plants, rice (Oryza sativa) has one Ero1p ortholog, OsEro1, that is an N-glycosylated protein localized to the ER membrane in the endosperm subaleurone cells. OsEro1 is required for the formation of native disulfide bonds in the seed storage protein proglutelin, as demonstrated by RNA interference knockdown of OsEro1 (Onda et al., 2009). Soybean (Glycine max) has two Ero1p orthologs, GmERO1a and GmERO1b. GmERO1a is a type I membrane-bound glycoprotein that is localized to the ER. GmERO1a can oxidize multiple soybean PDIs (Matsusaki et al., 2016). Arabidopsis also has two Ero1 orthologs: AtERO1 and AtERO2 (Dixon et al., 2003). Relative to yeast and mammals, the mechanisms of plant Ero1 functions are far less understood.
In this work, we find that both AtERO1 and AtERO2 are required for oxidative protein folding in the ER, but they also exhibit differences in functioning as sulfhydryl oxidases. The outer active site, the inner active site, and a long-range noncatalytic disulfide bond are required for AtERO1’s function. Our work will advance our understanding of how the protein oxidative folding machinery in the ER operates to ensure efficient and faithful oxidative protein folding in plants.
RESULTS
Sequence Analysis of AtERO1/2 and Other Ero1 Homologs from Various Species
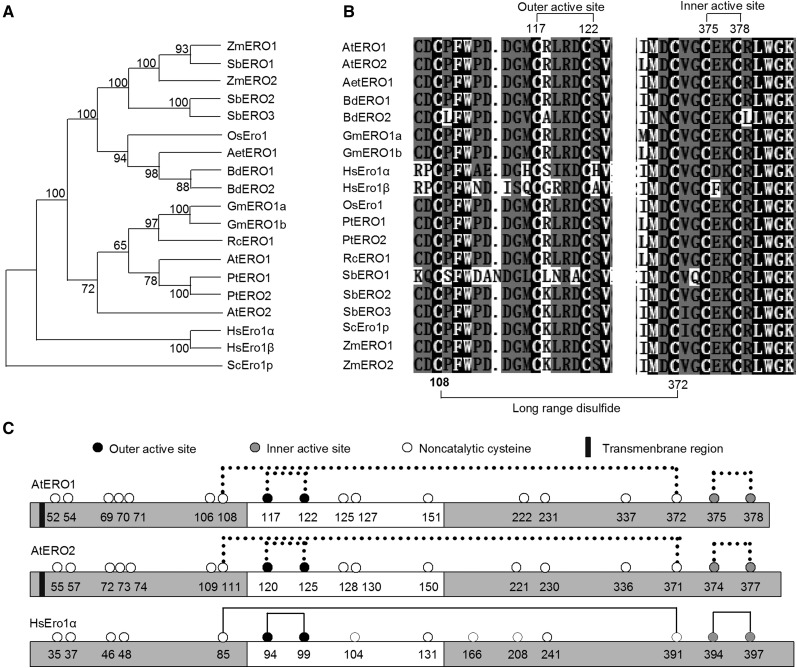
There are two Ero1 orthologs in Arabidopsis, AtERO1 and AtERO2 (Dixon et al., 2003). We performed phylogenetic analysis of Ero1 homologs from various species using yeast Ero1p as an outgroup (Supplemental Table S1). As shown in Figure 1A, all plant Ero1s form a clade, and human Ero1s form another clade. Furthermore, plant Ero1s fall into two subclasses: those of monocotyledons, such as Tausch’s goatgrass (Aegilops tauschii), Brachypodium distachyon, rice, sorghum (Sorghum bicolor), and maize (Zea mays), and Ero1s from dicotyledons, such as Arabidopsis, soybean, black cottonwood (Populus trichocarpa), and castor (Ricinus communis). Notably, in the subclass of dicotyledon Ero1s, AtERO2 shows large divergence from other Ero1s, including AtERO1. In addition, the genome of higher plant species analyzed here encodes two Ero1 homologs, except that castor, goatgrass, and rice all have one Ero1 homolog and sorghum has three Ero1 homologs. The Ero1 paralogs of most species are very similar to each other: BdERO1 and BdERO2 share 90.5% amino acid sequence identity, GmERO1a and GmERO1b share 97.8%, PtERO1 and PtERO2 share 86.3%, SbERO1, SbERO2, and SbERO3 share 96.3%, and ZmERO1 and ZmERO2 share 91.7%. Arabidopsis and human Ero1s are less similar: AtERO1 and AtERO2 share 63% amino acid sequence identity, while HsEro1α and HsEro1β share 60.3%. Furthermore, amino acid sequence alignment demonstrated that both AtERO1 and AtERO2 have the putative outer active site (Cys-117-Cys-122 for AtERO1 and Cys-120-Cys-125 for AtERO2), the putative inner active site (Cys-375-Cys-378 for AtERO1 and Cys-374-Cys-377 for AtERO2), a putative regulatory disulfide (Cys-117-Cys-151 for AtERO1 and Cys-120-Cys-150 for AtERO2), and a long-range disulfide (Cys-108-Cys-372 for AtERO1 and Cys-111-Cys-371 for AtERO2), which are conserved in Ero1s of various eukaryotic species from yeast, plants, and animals (Fig. 1, B and C).
Figure 1.
Analysis of Ero1s from different species. A, Phylogenetic analysis of Ero1s from different species. A neighbor-joining phylogenetic tree was constructed using MEGA 7.0. software based on putative amino acid sequences of various Ero1 homologs. Bootstrap values are indicated at each tree root. The amino acid sequence of yeast Ero1p was used as an outgroup. Aet, Aegilops tauschii; At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Gm, Glycine max; Hs, Homo sapiens; Os, Oryza sativa; Pt, Populus trichocarpa; Rc, Ricinus communis; Sb, Sorghum bicolor; Sc, Saccharomyces cerevisiae; Zm, Zea mays. B, Amino acid sequence alignment of Ero1 regions containing the outer/inner active sites from various species. Amino acid sequences of Ero1 proteins from various species were aligned using the ClustalW program. White-on-black letters indicate amino acid residues conserved in all sequences analyzed. Long-range disulfide bonds and active sites in Arabidopsis Ero1s are indicated. C, Schematic representation of AtERO1, AtERO2, and HsEro1α. Lines indicate disulfide bonds. The flexible loop region is represented by the white bars. Dotted lines indicate the putative disulfide bonds based on the results of amino acid sequence alignment of Ero1 homologs.
AtERO1/2 Are Required for the Growth of Plants under Reducing Conditions
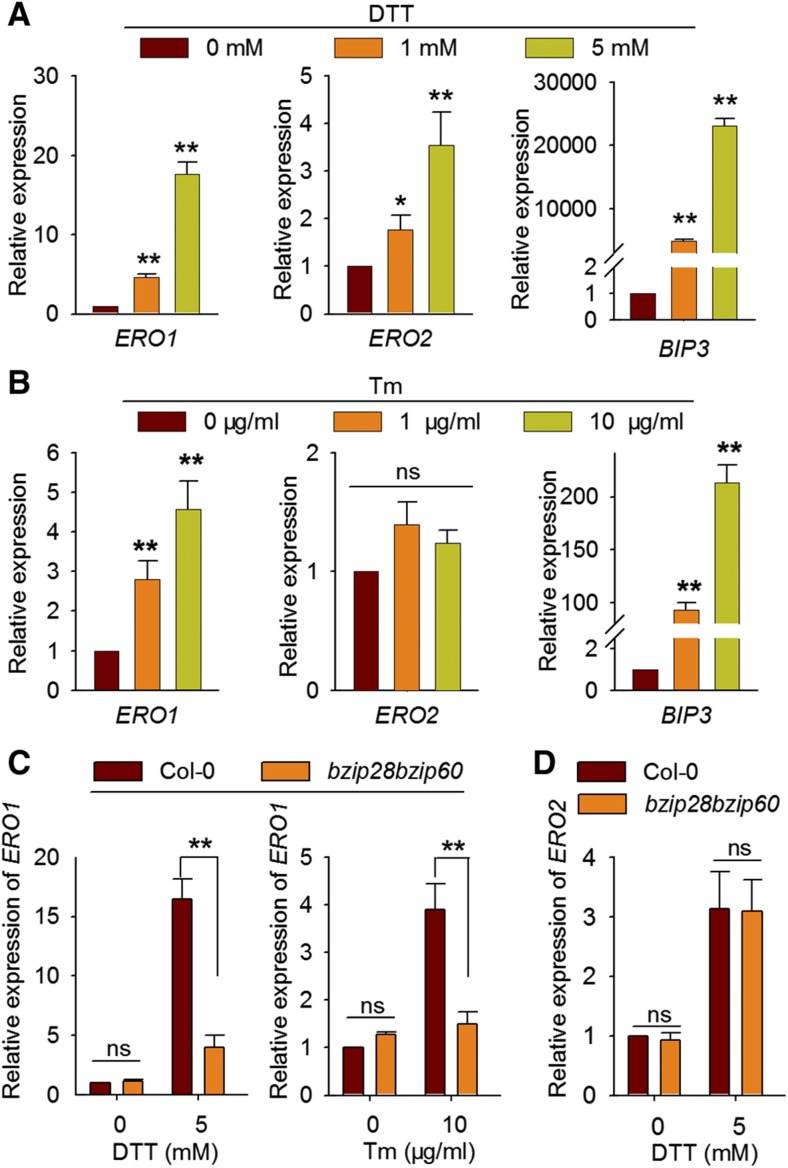
The reducing agent DTT and other pharmacological disruption of protein folding in plants trigger UPR, which is induced by the accumulation of unfolded proteins in the ER. Genes encoding chaperones and folding enzymes are induced by UPR to boost the protein-folding capacity in the ER (Martínez and Chrispeels, 2003; Lu and Christopher, 2008). We found that AtERO1/2 transcript levels were significantly increased upon DTT treatment (Fig. 2A). In addition, the expression of AtERO1, but not AtERO2, was also induced by Tm treatment (Fig. 2B), which triggers UPR by inhibiting N-linked glycosylation, a prerequisite for the folding of glycoproteins (Takatsuki et al., 1971). To examine whether the up-regulation of AtERO1/2 by DTT and AtERO1 by Tm treatment were plant responses to ER stress, we analyzed their expression in the background of the bzip28bzip60 UPR mutant (Deng et al., 2013b). We found that the induction of AtERO1 by DTT or Tm in bzip28bzip60 was significantly reduced compared with that in the wild-type (Fig. 2C), whereas the up-regulation of AtERO2 by DTT was not (Fig. 2D). These results suggest that the induction of AtERO1 by DTT or Tm treatment is caused by plant UPR and that AtERO1 is involved in protein folding in the ER. However, AtERO2 expression may not be induced by ER stress but is up-regulated by a reducing environment brought about by the DTT treatment, which results in the up-regulation of genes encoding enzymes catalyzing oxidative protein folding in the ER.
Figure 2.
Expression analysis of AtERO1 and AtERO2 upon DTT or Tm treatment. A and B, Seven-day-old Columbia-0 (Col-0) seedlings were treated with DTT (A) or Tm (B) at the indicated concentrations. The expression of AtERO1, AtERO2, and BIP3 was analyzed by reverse transcription quantitative PCR (RT-qPCR). The expression levels of AtERO1, AtERO2, or BIP3 were normalized to those of GAPC. Data are shown as means ± se (n = 3). Statistical significance compared with the control was analyzed using one-way ANOVA: *, P < 0.05; **, P < 0.01; ns, nonsignificant. C and D, Expression analysis of AtERO1 (C) and AtERO2 (D) in Col-0 and bzip28bzip60 plants treated with DTT or Tm by RT-qPCR. The expression levels of AtERO1 and AtERO2 were normalized to those of GAPC. Data are shown as means ± se (n = 3). Statistical significance compared with Col-0 was analyzed using one-way ANOVA: **, P < 0.01; ns, nonsignificant.
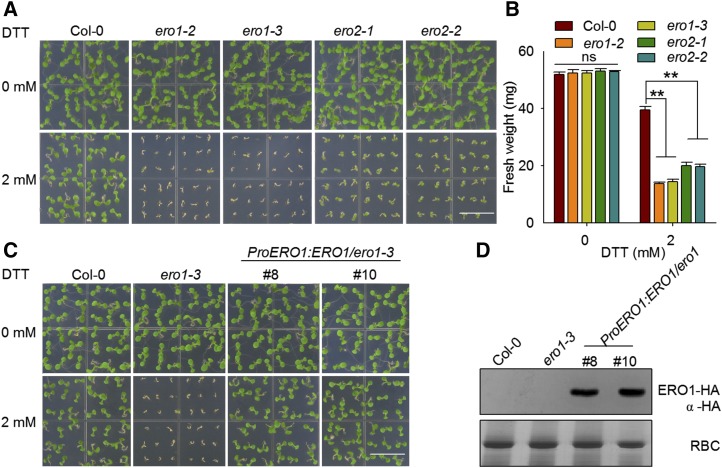
To examine the function of AtERO1/2 genetically, we used their mutant lines with T-DNAs inserted at different locations in AtERO1/2 (Supplemental Fig. S1). There was no obvious difference between mutant and Col-0 plants under normal conditions. However, when the plants were grown on Murashige and Skoog (MS) medium containing 2 mm DTT, the growth of ero1-2, ero1-3, ero2-1, and ero2-2 was dramatically inhibited (Fig. 3, A and B). The genetic complementation lines of ero1-3 transformed with the AtERO1 gene driven by its native promoter displayed a similar growth phenotype to wild-type plants upon DTT treatment (Fig. 3, C and D; Supplemental Fig. S2). These results suggest that AtERO1/2 are required for plants to survive under reducing conditions and that AtERO1/2 are involved in oxidative protein folding in plants. In addition, the cross-complementation lines of the ero2-1 mutant transformed with AtERO1 under the control of its native promoter also displayed a similar growth phenotype to wild-type plants when grown on medium containing 2 mm DTT (Supplemental Fig. S3), further suggesting that AtERO1 and AtERO2 have functional redundancy.
Figure 3.
AtERO1 and AtERO2 are required for plant growth under reducing conditions. A, Growth of AtERO1/2 mutant plants on medium containing DTT. Col-0, ero1-2, ero1-3, ero2-1, and ero2-2 plants were grown on one-half-strength MS medium with or without 2 mm DTT. Bar = 1 cm. B, Quantification of the fresh weight of 7-d-old Col-0, ero1-2, ero1-3, ero2-1, and ero2-2 plants. Data are shown as means ± se (n = 30). Statistical significance compared with Col-0 plants was analyzed using one-way ANOVA: **, P < 0.01; ns, nonsignificant. C, AtERO1 can complement the ero1-3 mutant. Col-0, ero1-3, and ProERO1:ERO1-HA/ero1-3 plants were grown on medium with or without 2 mm DTT for 7 d. Bar = 1 cm. D, Expression of AtERO1 proteins in ProERO1:ERO1-HA/ero1-3 transgenic plants. AtERO1-HA was detected by immunoblotting with an anti-HA antibody. Rubisco (RBC) stained with Coomassie Blue was used as a loading control.
We attempted to generate ero1ero2 double mutants by crossing ero1 and ero2 single mutants. However, no ero1ero2 homozygotes were obtained. We only obtained trans-heterozygous plants: ero1(+/−)ero2(−/−) and ero1(−/−)ero2(+/−). We dissected and observed the siliques from wild-type, ero1, ero2, ero1(+/−)ero2(−/−), and ero1(−/−)ero2(+/−) plants. ero1 and ero2 plants showed normal seed set. However, the seed set of ero1(+/−)ero2(−/−) and ero1(−/−)ero2(+/−) plants was significantly decreased compared with that of the wild-type. In addition, the number of seeds per silique of ero1(+/−)ero2(−/−) or ero1(−/−)ero2(+/−) plants was much less than that of the wild-type, ero1, or ero2 (Supplemental Fig. S4). Furthermore, the F1 progeny segregation ratios of selfed ero1(+/−)ero2(−/−) and ero1(−/−)ero2(+/−) dramatically deviated from the expected Mendelian segregation ratio (Supplemental Table S2). These data imply that the gametophytic transmission of the ero1ero2 double mutation was dramatically compromised.
AtERO1 and AtERO2 Are Present in Distinct Redox States in Vivo
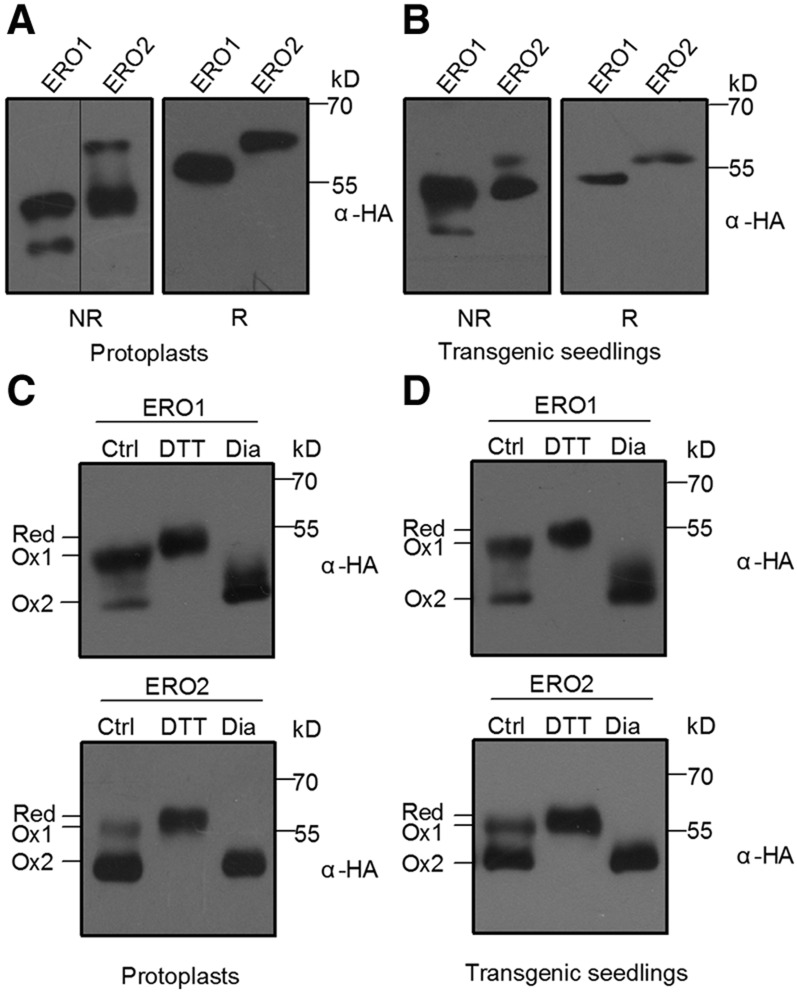
To understand the mechanism of AtERO1/2 functioning, we examined their redox states in vivo. We first transiently expressed AtERO1/2 fused to an HA tag (YPYDVPDYA) in Arabidopsis protoplasts. The free thiols in the transiently expressed AtERO1/2 were blocked by N-ethylmaleimide (NEM), and then the AtERO1/2 proteins were separated electrophoretically under nonreducing conditions, which allow disulfide bonds to be maintained. AtERO1/2 were present in two redox states on nonreducing gels, unlike their patterns under reducing conditions (Fig. 4A). The results were confirmed in transgenic plants overexpressing AtERO1/2-HA (Fig. 4B).
Figure 4.
AtERO1 and AtERO2 are present in two redox states in vivo. A, AtERO1 and AtERO2 are present in two redox states in Arabidopsis protoplasts. AtERO1-HA and AtERO2-HA were transiently expressed in Arabidopsis protoplasts. Free thiols were blocked with 20 mm NEM. Proteins were separated by 10% nonreducing or reducing SDS-PAGE followed by immunoblotting analysis with an anti-HA antibody. NR, Nonreducing gel; R, reducing gel. B, AtERO1 and AtERO2 are present in two redox states in transgenic plants. Proteins isolated from Pro-35S:ERO1/2-HA transgenic plants were separated with nonreducing or reducing SDS-PAGE. C, AtERO1 and AtERO2 are present in the putative Ox1 and Ox2 states in Arabidopsis protoplasts. AtERO1-HA and AtERO2-HA were expressed in Arabidopsis protoplasts. Transiently expressed proteins were treated with 5 mm DTT or 5 mm diamide for 10 min. Free thiols were blocked with 20 mm NEM. Proteins were separated with nonreducing SDS-PAGE and followed by immunoblot analysis. Red, Reduced form. Ox1 and Ox2 refer to different oxidized forms. D, AtERO1 and AtERO2 are present in the putative Ox1 and Ox2 states in transgenic plants. Proteins isolated from Pro-35S:ERO1/2-HA transgenic plants were separated with nonreducing SDS-PAGE.
To determine the nature of the redox states of AtERO1/2, we used DTT and diamide (a membrane-permeable oxidizing reagent) to pretreat the transiently expressed AtERO1/2 proteins, respectively. On nonreducing gels, one AtERO1/2 redox isoform migrated in a way identical to the diamide-treated AtERO1/2, suggesting that this isoform is fully oxidized, like the Ox2 isoform of human Ero1α. Another AtERO1/2 redox isoform migrated between the putative Ox2 and the DTT-treated species (Red), very likely reminiscent of a partially oxidized form of mammalian Ero1s, Ox1 (Benham et al., 2000; Fig. 4C). Notably, AtERO1 was present mainly in the putative Ox1 state in vivo, while AtERO2 was present mainly in the putative Ox2 state, and similar patterns of redox isoform distributions were also observed in the AtERO1/2-HA transgenic plants (Fig. 4D). For human Ero1α, its Ox2 isoform represents an inactive reservoir, while the Ox1 species is enzymatically active (Baker et al., 2008; Zhang et al., 2014). During the activation of mammalian Ero1α, the regulatory disulfide Cys-94-Cys-131 is reduced, resulting in the mobility shift of Ero1α proteins from Ox2 to Ox1 under nonreducing conditions (Zhang et al., 2014). The corresponding regulatory disulfide was also found in AtERO1/2 (Cys-117-Cys-151 for AtERO1 and Cys-120-Cys-150 for AtERO2; Fig. 1C). To examine whether the redox states of AtERO1/2 are affected by the formation or breakage of this putative regulatory disulfide, the Cys-to-Ala mutant proteins AtERO1C151A and AtERO2C150A were expressed transiently in the protoplasts and separated electrophoretically under nonreducing conditions. Consistent with the observation in Ero1α (Zhang et al., 2014), we found that the breakage of the regulatory disulfide in AtERO1/2 results in the shift of redox state from Ox2 to Ox1 (Supplemental Fig. S5).
AtERO1 Has Stronger Oxidase Activity Than AtERO2 In Vitro
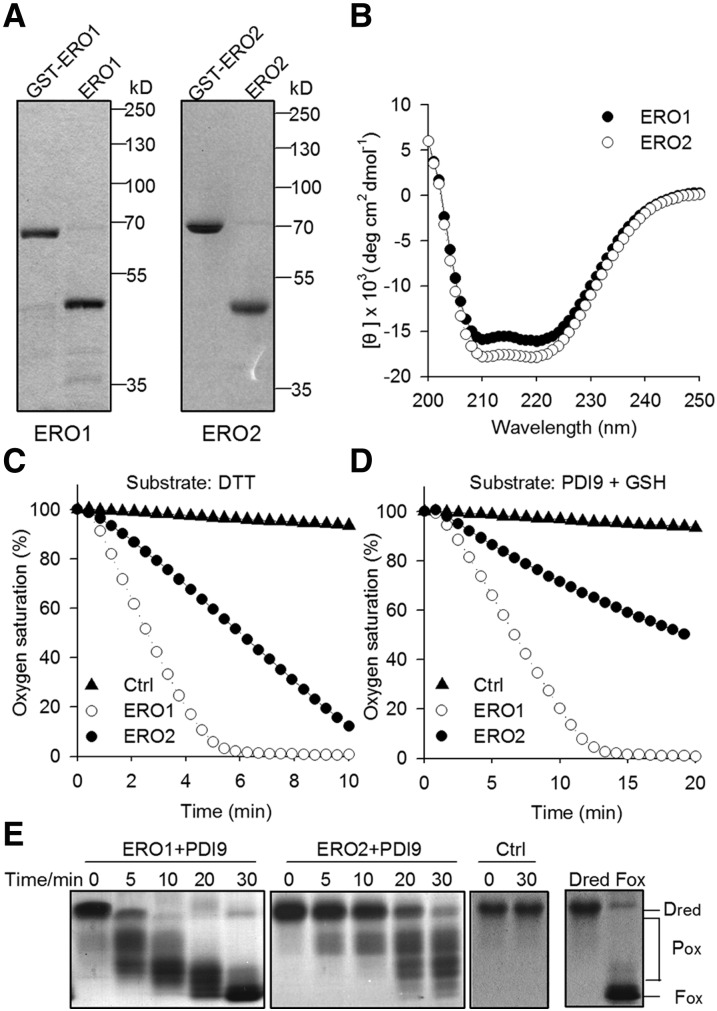
To analyze the oxidase activity of AtERO1/2, we purified the recombinant GST-fused AtERO1/2 protein from Escherichia coli, and the GST tag was subsequently removed (Fig. 5A). AtERO1/2 proteins showed a far-UV circular dichroism (CD) spectrum typical of well-folded proteins (Fig. 5B). We first employed DTT as a substrate to monitor the oxygen consumption catalyzed by AtERO1/2 using an oxygen electrode. As shown in Supplemental Figure S6, FAD was required for the functioning of AtERO1/2 as an oxidase. This is consistent with observations for other Ero1 homologs (Wang et al., 2011; Matsusaki et al., 2016), suggesting that FAD serves as a cofactor for AtERO1/2 to reduce molecular oxygen. We also tested the oxidase activity of AtERO1/2 in the presence of different amounts of exogenous FAD in the oxidation of DTT. The oxygen consumption rate of both AtERO1 and AtERO2 nearly reached the maximum when a 10-fold excess of exogenous FAD was added (Supplemental Fig. S7). Thus, we monitored the oxidase activity of AtERO1/2 at a 10:1 molar ratio of FAD to ERO1/2 in the following assays. The oxidase activity of AtERO1 was significantly higher than that of AtERO2 when DTT was used as a substrate (Fig. 5C; Supplemental Fig. S7). Similar results were also observed when AtPDI9 served as the substrate of AtERO1/2 in the presence of reduced glutathione (GSH; Fig. 5D). In addition, the dose-response curve relating AtERO1/2 concentration to their effects on the rate of oxygen consumption was determined using DTT or AtPDI9 as a substrate. The oxidase activity of AtERO1 was significantly higher than that of AtERO2 at every concentration of AtERO1/2 tested (Supplemental Fig. S8).
Figure 5.
Analysis of oxidative activities of AtERO1 and AtERO2 in vitro. A, Purification of AtERO1 and AtERO2. The GST tag was removed by digestion of recombinant GST-AtERO1/2 with PreScission Protease. AtERO1/2 lacks the signal peptide and N-terminal transmembrane domain. B, Far-UV CD spectrum of AtERO1 and AtERO2. C, Oxygen consumption assay of AtERO1/2 using DTT as a substrate. The reaction contains AtERO1/2, FAD, and DTT. Ctrl, Reaction without AtERO1/2. D, Oxygen consumption of AtERO1/2 using GSH as a substrate in the AtERO1/2-AtPDI9 system. The reaction contains AtERO1/2, FAD, AtPDI9, and GSH. E, Analysis of AtERO1/2 activities by gel-based RNase A reoxidation assay. AtERO1/2 and AtPDI9 were added to the denatured and reduced RNase A and FAD. Free thiols were blocked by the addition of nonreducing sample buffer containing AMS at the indicated time points. RNase A was separated by 15% nonreducing SDS-PAGE followed by Coomassie Blue staining. Dred RNase A and Fox RNase A are shown at far right.
We also reconstituted the AtERO1/2-AtPDI9 oxidative protein-folding system with reduced and denatured (Dred) RNase A as a substrate. The free thiol groups of RNase A were modified with 4-acetamido-4′-maleimidylstilbene -2,2′-disulfonic acid (AMS), which resulted in a 0.5-kD increase in the molecular mass of proteins when separated by nonreducing SDS-PAGE (Kobayashi et al., 1997). Dred RNase A and fully oxidized (Fox) RNase A are shown at far right in Figure 5E. AtERO1-AtPDI9 converted most of the Dred RNase A molecules into partially oxidized intermediates (Pox) within 5 min. The intermediates were converted into Fox forms in the next 25 min. Consistently, the conversion of Dred RNase A into Pox and Fox states by AtERO2-AtPDI9 was much slower than that by AtERO1-AtPDI9 (Fig. 5E). These results suggest that AtERO1 is more active than AtERO2 as a sulfhydryl oxidase.
The Outer Active Site and the Inner Active Site Are Required for AtERO1 Function
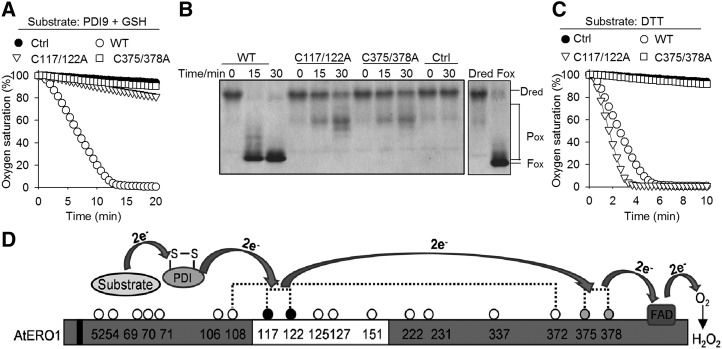
We then sought to investigate the mechanisms of how AtERO1 functions as an oxidase. Based on sequence alignment, the outer active site of AtERO1 is C117X4C122, while its inner active site is C375XXC378 (Fig. 1, B and C). To examine the functions of the potential inner/outer active sites of AtERO1, we generated a series of AtERO1 mutants with the Cys in the inner/outer active site mutated to Ala. We first found that these mutant proteins showed far-UV CD spectra typical of well-folded proteins (Supplemental Fig. S9). Then, we checked the effects of these mutations on their oxidase activities. In the AtERO1-AtPDI9-GSH in vitro system, when two Cys residues in the outer active site (AtERO1C117/122A) or the inner active site (AtERO1C375/378A) were mutated, the oxidase activity was drastically decreased, as determined by oxygen consumption assays (Fig. 6A). Similar results were observed in the AtERO1-AtPDI9-RNase A system, as demonstrated by the conversion of Dred RNase A into Pox and Fox states (Fig. 6B).
Figure 6.
Outer active site and inner active site are required for AtERO1 function. A, Oxygen consumption assay of AtERO1 mutants using GSH as a substrate in the AtERO1-AtPDI9 system. The reaction contains AtERO1/2, FAD, AtPDI9, and GSH. Ctrl, Reaction without AtERO1; WT, wild type. B, Analysis of activities of AtERO1 mutants using gel-based RNase A reoxidation assay in the presence of AtPDI9. Dred RNase A and Fox RNase A are shown at far right. C, Analysis of activities of AtERO1 mutants using DTT as a substrate. The reaction contains AtERO1/2, FAD, and DTT. D, Schematic representation of the electron transfer from the substrate via AtERO1 to oxygen in the presence of PDI. The electrons are transferred from AtPDIs to the inner active site of AtERO1 via the outer active site. The buried inner active site is oxidized by FAD, which reduces molecular oxygen, generating peroxide. The white box indicates the putative flexible loop region. Dotted lines indicate the putative disulfide bonds in AtERO1 based on the amino acid sequence alignment of Ero1 homologs. The Cys residues are represented by circles with numbering. The gray and black circles indicate the inner active site and the outer active site, respectively. The black box indicates the transmembrane region of AtERO1.
We also analyzed the activity of these AtERO1 mutants using DTT as a substrate. The activity of AtERO1C375/378A was almost completely impaired, whereas AtERO1C117/122A displayed even higher oxidase activity compared with wild-type AtERO1 (Fig. 6C). This could be explained on the assumption that AtERO1C117/122A has an intact inner active site, which can directly and more efficiently oxidize DTT and reduce oxygen. By contrast, in wild-type AtERO1, the inner active site first oxidizes the outer active site, which in turn oxidizes DTT. As for AtERO1C375/378A, its intact outer active site cannot transfer electrons to oxygen, and thereby loses its activity. Taken together, these results suggest that, in Arabidopsis, AtPDIs are oxidized by the outer active site in AtERO1, which, in turn, is reoxidized by its inner active site (Fig. 6D).
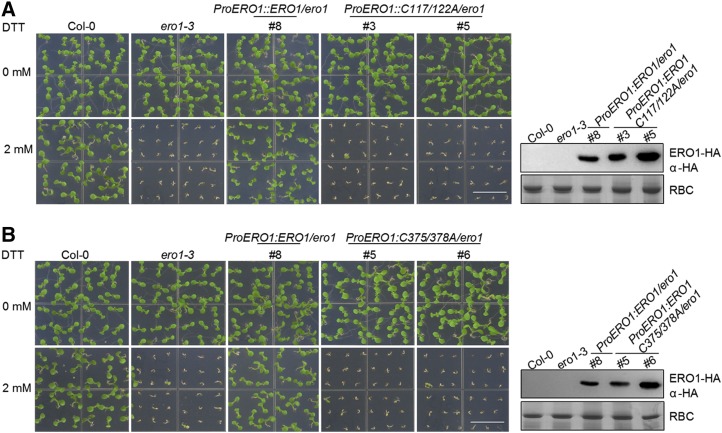
To confirm these results genetically, we transformed ero1-3 mutant plants with a construct containing wild-type AtERO1, AtERO1C117/122A, or AtERO1C375/C378A under the control of the native AtERO1 promoter. Expression of wild-type AtERO1 completely rescued the ero1-3 mutant, while expression of AtERO1C117/122A (Fig. 7A; Supplemental Fig. S10A) or AtERO1C375/C378A (Fig. 7B; Supplemental Fig. S10B) failed to rescue the ero1-3 mutant, suggesting that both the inner active site and the outer active site are required for AtERO1’s function in vivo.
Figure 7.
Outer active site and inner active site are required for AtERO1 function as demonstrated by genetic complementation analysis. The growth of Col-0, ero1-3, ProERO1:ERO1C117/122A-HA/ero1-3 (A), and ProERO1:ERO1C375/378A-HA/ero1-3 (B) transgenic plants on medium with or without 2 mm DTT is shown. AtERO1-HA was detected by immunoblotting with an anti-HA antibody. Rubisco (RBC) stained with Coomassie Blue was used as a loading control. Bars = 1 cm.
A Long-Range Disulfide Is Required for AtERO1’s Function
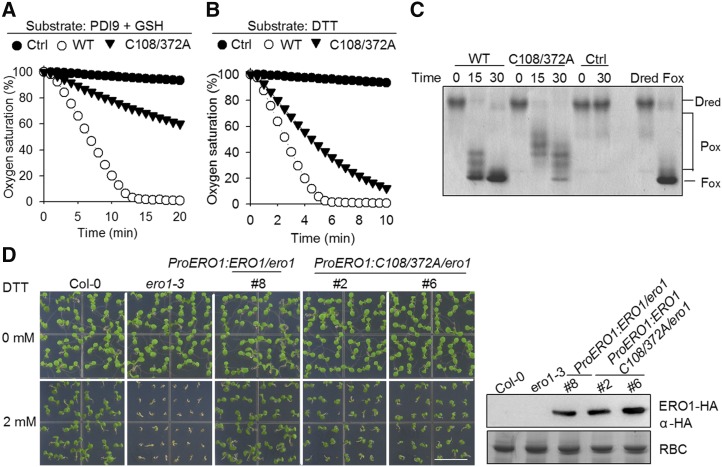
Based on sequence alignment, AtERO1 contains a conserved long-range disulfide, Cys-108-Cys-372 (Fig. 1, B and C; Supplemental Fig. S9). We found that AtERO1C108/372A has much weaker activity than wild-type AtERO1 (Fig. 8, A–C). Furthermore, the expression of AtERO1C108/372A only partially rescued the ero1-3 mutant (Fig. 8D; Supplemental Fig. S11). These results suggest that this long-range disulfide in AtERO1 is required for the function of AtERO1.
Figure 8.
The long-range disulfide Cys-108-Cys-372 is required for AtERO1 function. A, Oxygen consumption assay of AtERO1C108/372A using GSH as a substrate in the AtERO1-AtPDI9 system. The reaction contains AtERO1, FAD, AtPDI9, and GSH. Ctrl, Reaction without AtERO1; WT, wild type. B, Analysis of activities of AtERO1C108/372A using DTT as a substrate. The reaction contains AtERO1, FAD, and DTT. C, Analysis of activities of AtERO1C108/372A using gel-based RNase A reoxidation assay in the presence of AtPDI9. The two columns at far right show Dred RNase A and Fox RNase A. D, Genetic complementation analysis of the long-range disulfide mutant AtERO1C108/372A. The growth of Col-0, ero1-3, and ProERO1:ERO1C108/372A-HA/ero1-3 transgenic plants on medium with or without 2 mm DTT is shown. AtERO1-HA was detected by immunoblotting with an anti-HA antibody. Rubisco (RBC) stained with Coomassie Blue was used as a loading control. Bar = 1 cm.
DISCUSSION
The mechanisms underlying the functioning of yeast Ero1p and human Ero1s have been extensively studied. However, those of plant Ero1 functioning are far less understood. Here, using functional genomics, genetic, and biochemical approaches, we found that both AtERO1 and AtERO2 are required for oxidative protein folding in the ER. The outer active site and the inner active site are required for AtERO1’s function. AtPDIs (AtPDI9) are oxidized by the outer active site in AtERO1, which, in turn, is reoxidized by its inner active site. Based on sequence alignment, AtERO1 also contains a conserved long-range disulfide, Cys-108-Cys-372 (Fig. 1, B and C). However, there is a debate on the role of this long-range disulfide in human Ero1α (Cys-85-Cys-391): either negatively regulating Ero1α activity (Appenzeller-Herzog et al., 2008; Baker et al., 2008) or playing a positive structural role (Araki and Nagata, 2011; Zhang et al., 2014). As shown in Figure 8, A to C, AtERO1C108/372A has much weaker activity than wild-type AtERO1. Furthermore, the expression of AtERO1C108/372A only partially rescues the ero1-3 mutant (Fig. 8D; Supplemental Fig. S11). These results suggest that this long-range disulfide in AtERO1 is required for the function of AtERO1, further supporting its positive structural role.
Yeast has only one Ero1 gene, Ero1p, while humans have two Ero1s: Ero1α and Ero1β. Ero1p is essential for yeast viability, but mice (Mus musculus) deficient in both Ero1α and Ero1β are viable (Zito et al., 2010a). Mice may have supplementary pathways of disulfide bond formation in the ER (Bulleid and Ellgaard, 2011; Kakihana et al., 2012), such as peroxiredoxin IV (Tavender et al., 2010; Zito et al., 2010b), glutathione peroxidases (Nguyen et al., 2011; Wang et al., 2014), and vitamin K epoxide reductase (Schulman et al., 2010), all of which can serve as a disulfide bond donor to PDI. In addition, another thiol oxidase, Quiescin Sulfhydryl Oxidase (QSOX), can directly oxidize some secretory proteins (Alon et al., 2012). In plants, rice has one Ero1p homolog, and its knockdown mutant plants exhibit deficiency in the formation of native disulfide bonds in proglutelins, which leads to aggregation of proglutelins in the ER due to the nonnative intermolecular disulfide bonds formed. There are no other visible phenotypes in this RNA interference knockdown mutant, probably because it is not a null mutant of OsEro1 (Onda et al., 2009). Soybean has two Ero1p homologs; however, their null mutants have not been investigated yet (Matsusaki et al., 2016). In our work, we show that AtERO1/2 are required for plant growth under reducing conditions, underlining the importance of AtERO1/2 in oxidative protein folding. Interestingly, we failed to obtain the double mutant of AtERO1 and AtERO2, suggesting that they may also play redundant but essential roles in reproduction (Supplemental Fig. S4; Supplemental Table S2). Therefore, Arabidopsis may have no supplementary pathways for disulfide bond formation in the ER, although its genome encodes two QSOX homologs: QSOX1 and QSOX2 (Alejandro et al., 2007). However, this needs to be investigated in detail in future studies.
In humans, Ero1α is constitutively expressed while Ero1β, with higher oxidase activity, is abundantly expressed in specialized secretory tissues (e.g. in the pancreas; Zito et al., 2010a; Wang et al., 2011). The transcription of Ero1α is induced under hypoxia and that of Ero1β is induced by UPR (Pagani et al., 2000; Gess et al., 2003). In our work, we found that AtERO1 and AtERO2 exhibit distinct characteristics. For example, AtERO1 was induced by both DTT and Tm treatments, whereas AtERO2 was only induced by DTT. Human Ero1α is present in a reduced form and two partially oxidized forms, Ox1 and Ox2, in living cells (Benham et al., 2000). Ox1 is the active form of Ero1α, while Ox2 represents an inactive reservoir with a regulatory disulfide bond formed between C94 and C131 (Appenzeller-Herzog et al., 2008; Baker et al., 2008). In this work, we showed that plant Ero1 has two redox states in vivo. The fully reduced form of AtERO1/2 was not detected either in a transient expression system or in AtERO1/2 transgenic plants, but two oxidized forms were present in both cases. AtERO1 was mainly present in the Ox1 redox state, while more AtERO2 existed in the Ox2 state (Fig. 4, A and B). Furthermore, Cys-117-Cys-151 and Cys-120-Cys-150 are regulatory disulfides that modulate the redox states of AtERO1 and AtERO2, respectively (Supplemental Fig. S5). However, the significance of these redox forms of AtERO1/2 must still be determined. Interestingly, the ero1 plants are more sensitive to reductive stress than the ero2 plants (Fig. 3A), and AtERO1 showed much stronger oxidative protein-folding activity than AtERO2 in vitro (Fig. 5, C–E). These results suggest that AtERO1 and AtERO2 exhibit differences in catalyzing oxidative protein folding in the ER, and AtERO1 may function as the primary sulfhydryl oxidase in the ER relative to AtERO2.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild type (Col-0), AtERO1 mutants ero1-2 (SALK_003845) and ero1-3 (SALK_096805), AtERO2 mutants ero2-1 (SALK_201119C) and ero2-2 (SALK_074861), bzip28 (SALK_132285C; Liu et al., 2007), and bzip60 (SAIL_283_B03; Deng et al., 2011) were grown on soil in a growth chamber at 22°C with 60% relative humidity and a 12/12-h light/dark photoperiod. For seedlings grown on medium, seeds were surface sterilized with 50% (v/v) bleach for 5 min, washed several times with sterilized distilled, deionized water, and germinated on plates with one-half-strength MS medium containing 1% (w/v) Suc, 0.8% (w/v) agar, and 2.5 mm MES at pH 5.7, with or without DTT.
RNA Extraction, cDNA Synthesis, and RT-qPCR
Seven-day-old Arabidopsis seedlings were treated with 1 or 5 mm DTT for 12 h, 1 or 10 μg mL−1 Tm for 5 h, and the total RNA was isolated using TRIzol (Invitrogen) following the manufacturer’s instructions. The cDNA was synthesized from 1 μg of DNase I-treated total RNA in a 20-μL reaction system by RT (TaKaRa). qPCR amplification was performed using SYBR Green reagent (TaKaRa) on a Bio-Rad CFX Manager 3.1. The primer sequences are listed in Supplemental Table S3.
Plasmid Construction and Generation of Transgenic Plants
For protoplast transfection, the full-length coding sequences of AtERO1 and AtERO2 were amplified by RT-PCR from Col-0 RNA and cloned into a plant expression vector fused to an HA tag. The various mutants were generated by site-directed mutagenesis.
To generate transgenic plants, a 2-kb AtERO1 promoter was amplified by PCR from Col-0 genomic DNA. The ProERO1:ERO1-HA, Pro-35S:ERO1-HA, and Pro-35S:ERO2-HA transgenic plants were generated by Agrobacterium tumefaciens-mediated transformation in the Col-0, ero1-3, or ero2-1 background, and the transformants were selected by Basta resistance and confirmed by immunoblotting with an anti-HA antibody (Sigma).
For recombinant protein expression, full-length AtPDI9 was subcloned into pET-28a with a His and a FLAG in the N terminus. For AtERO1/2, the coding sequence fragments lacking the sequences encoding the N terminus containing the transmembrane domain were placed in the pGEX-6P-1 vector. The AtERO1 code sequence without the signal peptide sequence (without amino acids 1–38 for AtERO1 and amino acids 1–42 for AtERO2) was inserted into the pGEX-6P-1 vector at the BamHI and XhoI sites. The recombinant vector had a GST linked to the N terminus. The primers for plasmid construction and genotyping are listed in Supplemental Table S4.
Phylogenetic Analysis and Sequence Alignment
The bootstrapped phylogenetic trees of full-length Ero1 protein sequences from various species were constructed using the neighbor-joining method by MEGA 7.0. Bootstrap was set as 1,000, and bootstrap values are indicated at each tree root. The yeast (Saccharomyces cerevisiae) Ero1p was used as an outgroup. The amino acid sequences of Ero1 proteins from various species were aligned using the ClustalW program.
Recombinant Protein Expression and Purification
The expression and purification of the GST-ERO1 and GST-ERO2 fusion proteins were performed as described (Wang et al., 2009). The expression of recombinant PDI9 in Escherichia coli BL21 cells was induced by the addition of 0.5 mm isopropyl thiogalactoside at 25°C for 10 h. The recombinant PDI9 protein was purified using an Ni-NTA agarose purification kit (Qiagen) following the manufacturer’s instructions. The concentration of each protein was determined using the Bradford method, and aliquots were stored in buffer A (50 mm Tris-HCl and 150 mm NaCl, pH 7.6) at −80°C.
Transient Gene Expression in Arabidopsis Protoplasts
Arabidopsis protoplasts were isolated and transfected with plant expression vectors as previously described (Yoo et al., 2007).
Far-UV CD Spectra
The CD spectra in the far-UV region of 2 μm AtERO1 proteins were recorded in 20 mm sodium phosphate, pH 7.6, and 50 mm NaCl.
Oxygen Consumption Assay
The oxygen consumption assay was performed as described previously (Wang et al., 2009). Oxygen consumption was measured with an Oxygraph Clark-type oxygen electrode (Hansatech Instruments). When DTT was used as a substrate, 10 mm DTT and 10 μm FAD were freshly mixed in a total volume of 0.5 mL, and the reaction was initiated by injection of 1 μm AtERO1 or AtERO2 into the reaction vessel of the oxygen electrode. When GSH served as a substrate, 20 μm AtPDI9, 10 mm GSH, and 20 μm FAD were freshly mixed in a total volume of 0.5 mL, and the reaction was initiated by injection of 2 μm AtERO1 or AtERO2 into the reaction vessel of the oxygen electrode. All experiments were performed in buffer B (100 mm Tris-HAc, 50 mm NaCl, and 1 mm EDTA, pH 8).
Gel-Based RNase A Reoxidation Analyses
Gel-based RNase A reoxidation analyses were performed by incubating 3 μm AtPDI9, 3 μm AtERO1 or AtERO2, 100 μm FAD, and 8 μm denatured and reduced RNase A in buffer B at 25°C. At various time points, the reactions were terminated by the addition of 5× SDS loading buffer containing 10 mm AMS to block the free thiols. The samples were resolved by nonreducing 15% (w/v) SDS-PAGE, and the proteins were stained with Coomassie Blue.
Protein Redox State Determination
The redox state analyses for transiently expressed proteins were performed as described (Fan et al., 2018). For proteins isolated from transgenic plants, 7-d-old seedlings were ground in liquid nitrogen, and the samples were dissolved by the addition of 5× SDS loading buffer containing 20 mm NEM to block free thiols. Samples were incubated for 10 min at 25°C and resolved by nonreducing 10% (w/v) SDS-PAGE. The proteins were detected by immunoblotting with an anti-HA antibody.
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under the following accession numbers: AtERO1, At1G72280; AtERO2, At2G38960; AtPDI9, At2G32920; BIP3, At1G14540; bZIP28, At3G10800; and bZIP60, At1G42990. Mutants used in this article can be obtained from the Arabidopsis Biological Resource Center under the following accession numbers: ero1-2 (SALK_003845), ero1-3 (SALK_096805), ero2-1 (SALK_201119C), ero2-2 (SALK_074861), bzip28 (SALK_132285C), and bzip60 (SAIL_283_B03).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of AtERO1/2 mutants.
Supplemental Figure S2. AtERO1 can complement the ero1-3 mutant.
Supplemental Figure S3. AtERO1 can complement the ero2-1 mutant.
Supplemental Figure S4. Seed set in ero1(−/−)ero2(+/−) and ero1(+/−)ero2(−/−) mutants.
Supplemental Figure S5. Breakage of the regulatory disulfide in AtERO1/2 results in their redox state shift.
Supplemental Figure S6. FAD is required for AERO1 and AtERO2 activity in vitro.
Supplemental Figure S7. Oxygen consumption of AtERO1/2 in the presence of different amounts of exogenous FAD.
Supplemental Figure S8. Dose-response curve relating AtERO1/2 concentration to their effect on the rate of oxygen consumption.
Supplemental Figure S9. Purification of wild-type AtERO1 and its mutant proteins.
Supplemental Figure S10. AtERO1C117/122A and AtERO1C375/378A cannot complement ero1-3.
Supplemental Figure S11. AtERO1C108/372A can partially complement ero1-3.
Supplemental Table S1. Species and accession numbers of Ero1s used for phylogenetic analysis.
Supplemental Table S2. Segregation analysis of ero1(+/−)ero2(−/−) and ero1(−/−)ero2(+/−) mutants.
Supplemental Table S3. Primers used for RT-qPCR.
Supplemental Table S4. Primers used for construction and genotyping.
Acknowledgments
We thank Drs. Yi Guo and Rui Li from Hebei Normal University for assistance on observing the siliques of mutant plants and Drs. Yanmin Zou and Yongfeng Han from IGDB, Chinese Academy of Sciences for insightful discussions on this work.
Footnotes
This work was supported by the Chinese Ministry of Science and Technology (2015CB910200 and 2017YFA0504000), the National Natural Science Foundation of China (31322009 and 31571163), the State Key Laboratory of Plant Genomics of China, and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (to L.W.).
Articles can be viewed without a subscription.
References
- Alejandro S, Rodríguez PL, Bellés JM, Yenush L, García-Sanchez MJ, Fernández JA, Serrano R (2007) An Arabidopsis quiescin-sulfhydryl oxidase regulates cation homeostasis at the root symplast-xylem interface. EMBO J 26: 3203–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon A, Grossman I, Gat Y, Kodali VK, DiMaio F, Mehlman T, Haran G, Baker D, Thorpe C, Fass D (2012) The dynamic disulphide relay of quiescin sulphydryl oxidase. Nature 488: 414–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Riemer J, Christensen B, Sørensen ES, Ellgaard L (2008) A novel disulphide switch mechanism in Ero1alpha balances ER oxidation in human cells. EMBO J 27: 2977–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Nagata K (2011) Functional in vitro analysis of the ERO1 protein and protein-disulfide isomerase pathway. J Biol Chem 286: 32705–32712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KM, Chakravarthi S, Langton KP, Sheppard AM, Lu H, Bulleid NJ (2008) Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation. EMBO J 27: 2988–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benham AM, Cabibbo A, Fassio A, Bulleid N, Sitia R, Braakman I (2000) The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lalpha. EMBO J 19: 4493–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli G, Simmen T, Anelli T, Molteni SN, Fesce R, Sitia R (2004) Two conserved cysteine triads in human Ero1alpha cooperate for efficient disulfide bond formation in the endoplasmic reticulum. J Biol Chem 279: 30047–30052 [DOI] [PubMed] [Google Scholar]
- Bulleid NJ, Ellgaard L (2011) Multiple ways to make disulfides. Trends Biochem Sci 36: 485–492 [DOI] [PubMed] [Google Scholar]
- Cabibbo A, Pagani M, Fabbri M, Rocchi M, Farmery MR, Bulleid NJ, Sitia R (2000) ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem 275: 4827–4833 [DOI] [PubMed] [Google Scholar]
- Delaunay-Moisan A, Appenzeller-Herzog C (2015) The antioxidant machinery of the endoplasmic reticulum: Protection and signaling. Free Radic Biol Med 83: 341–351 [DOI] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA 108: 7247–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH (2013a) Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int J Mol Sci 14: 8188–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH (2013b) Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci USA 110: 19633–19638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Quilichini TD, Dong H, Bao Y, Horner HT, Howell SH (2016) IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J 88: 193–204 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Van Lith M, Edwards R, Benham A (2003) Cloning and initial characterization of the Arabidopsis thaliana endoplasmic reticulum oxidoreductins. Antioxid Redox Signal 5: 389–396 [DOI] [PubMed] [Google Scholar]
- Fan F, Zhang Y, Wang S, Han Y, Wang L, Lu D (2018) Characterization of the oxidative protein folding activity of a unique plant oxidoreductase, Arabidopsis protein disulfide isomerase-11. Biochem Biophys Res Commun 495: 1041–1047 [DOI] [PubMed] [Google Scholar]
- Gess B, Hofbauer KH, Wenger RH, Lohaus C, Meyer HE, Kurtz A (2003) The cellular oxygen tension regulates expression of the endoplasmic oxidoreductase ERO1-Lalpha. Eur J Biochem 270: 2228–2235 [DOI] [PubMed] [Google Scholar]
- Gross E, Kastner DB, Kaiser CA, Fass D (2004) Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell 117: 601–610 [DOI] [PubMed] [Google Scholar]
- Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, Thorpe C, Fass D (2006) Generating disulfides enzymatically: Reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci USA 103: 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW (2009) Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal 11: 2807–2850 [DOI] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH. (2013) Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64: 477–499 [DOI] [PubMed] [Google Scholar]
- Inaba K, Masui S, Iida H, Vavassori S, Sitia R, Suzuki M (2010) Crystal structures of human Ero1α reveal the mechanisms of regulated and targeted oxidation of PDI. EMBO J 29: 3330–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakihana T, Nagata K, Sitia R (2012) Peroxides and peroxidases in the endoplasmic reticulum: Integrating redox homeostasis and oxidative folding. Antioxid Redox Signal 16: 763–771 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K (1997) Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc Natl Acad Sci USA 94: 11857–11862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kørner CJ, Du X, Vollmer ME, Pajerowska-Mukhtar KM (2015) Endoplasmic reticulum stress signaling in plant immunity: At the crossroad of life and death. Int J Mol Sci 16: 26582–26598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D, Korhonen L, Eriksson O, Kõks S (2017) Recent insights into the role of unfolded protein response in ER stress in health and disease. Front Cell Dev Biol 5: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2010) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2016) Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol 211: 418–428 [DOI] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH (2007) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DP, Christopher DA (2008) Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol Genet Genomics 280: 199–210 [DOI] [PubMed] [Google Scholar]
- Martínez IM, Chrispeels MJ (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusaki M, Okuda A, Masuda T, Koishihara K, Mita R, Iwasaki K, Hara K, Naruo Y, Hirose A, Tsuchi Y, et al. (2016) Cooperative protein folding by two protein thiol disulfide oxidoreductases and ERO1 in soybean. Plant Physiol 170: 774–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T (2005) ERAD: The long road to destruction. Nat Cell Biol 7: 766–772 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Riemer J, Rouhier N (2019) Oxidative protein folding: State-of-the-art and current avenues of research in plants. New Phytol 221: 1230–1246 [DOI] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X, Orellana A, et al. (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 7: e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VD, Saaranen MJ, Karala AR, Lappi AK, Wang L, Raykhel IB, Alanen HI, Salo KE, Wang CC, Ruddock LW (2011) Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J Mol Biol 406: 503–515 [DOI] [PubMed] [Google Scholar]
- Onda Y, Kumamaru T, Kawagoe Y (2009) ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc Natl Acad Sci USA 106: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, Cabibbo A, Sitia R (2000) Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J Biol Chem 275: 23685–23692 [DOI] [PubMed] [Google Scholar]
- Qian D, Chen G, Tian L, Qu LQ (2018) OsDER1 is an ER-associated protein degradation factor that responds to ER stress. Plant Physiol 178: 402–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti C, Lai Y, Brandizzi F (2018) Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60, as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP28. Plant J 93: 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman S, Wang B, Li W, Rapoport TA (2010) Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci USA 107: 15027–15032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA (2008) Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta 1783: 549–556 [DOI] [PubMed] [Google Scholar]
- Sevier CS, Qu H, Heldman N, Gross E, Fass D, Kaiser CA (2007) Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell 129: 333–344 [DOI] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS (2011) Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science 334: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuki A, Arima K, Tamura G (1971) Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J Antibiot (Tokyo) 24: 215–223 [DOI] [PubMed] [Google Scholar]
- Tavender TJ, Springate JJ, Bulleid NJ (2010) Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J 29: 4185–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Boston RS (2008) Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 9: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Wang L, Li SJ, Sidhu A, Zhu L, Liang Y, Freedman RB, Wang CC (2009) Reconstitution of human Ero1-Lalpha/protein-disulfide isomerase oxidative folding pathway in vitro: Position-dependent differences in role between the a and a′ domains of protein-disulfide isomerase. J Biol Chem 284: 199–206 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhu L, Wang CC (2011) The endoplasmic reticulum sulfhydryl oxidase Ero1β drives efficient oxidative protein folding with loose regulation. Biochem J 434: 113–121 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang L, Niu Y, Sitia R, Wang CC (2014) Glutathione peroxidase 7 utilizes hydrogen peroxide generated by Ero1α to promote oxidative protein folding. Antioxid Redox Signal 20: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Dickman MB, Whitham SA, Payton M, Verchot J (2011) The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol 156: 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang L, Niu Y, Zhu L, Fang J, Wang X, Wang L, Wang CC (2014) Different interaction modes for protein-disulfide isomerase (PDI) as an efficient regulator and a specific substrate of endoplasmic reticulum oxidoreductin-1α (Ero1α). J Biol Chem 289: 31188–31199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SS, Yang H, Ding L, Song ZT, Ma H, Chang F, Liu JX (2017) Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. Plant Cell 29: 1007–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E, Chin KT, Blais J, Harding HP, Ron D (2010a) ERO1-beta, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J Cell Biol 188: 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E, Melo EP, Yang Y, Wahlander Å, Neubert TA, Ron D (2010b) Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol Cell 40: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]