Translational and posttranslational regulation play a crucial role in the plastid gene expression pathway of retrograde signaling and supports a function of GUN1 in plastid proteostasis.
Abstract
Retrograde signals emanate from the DNA-containing cell organelles (plastids and mitochondria) and control the expression of a large number of nuclear genes in response to environmental and developmental cues. Previous studies on retrograde signaling have mainly analyzed the regulation of nuclear gene expression at the transcript level. To determine the contribution of translational and posttranslational regulation to plastid retrograde signaling, we combined label-free proteomics with transcriptomic analysis of Arabidopsis (Arabidopsis thaliana) seedlings and studied their response to interference with the plastid gene expression pathway of retrograde signaling. By comparing the proteomes of the genomes uncoupled1 (gun1) and gun5 mutants with the wild type, we show that GUN1 is critical in the maintenance of plastid protein homeostasis (proteostasis) when plastid translation is blocked. Combining transcriptomic and proteomic analyses of the wild type and gun1, we identified 181 highly translationally or posttranslationally regulated (HiToP) genes. We demonstrate that HiToP photosynthesis-associated nuclear genes (PhANGs) are largely regulated by translational repression, while HiToP ribosomal protein genes are regulated posttranslationally, likely at the level of protein stability without the involvement of GUN1. Our findings suggest distinct posttranscriptional control mechanisms of nuclear gene expression in response to plastid-derived retrograde signals. They also reveal a role for GUN1 in the translational regulation of several PhANGs and highlight extensive posttranslational regulation that does not necessitate GUN1. This study advances our understanding of the molecular mechanisms underlying intracellular communication and provides new insight into cellular responses to impaired plastid protein biosynthesis.
Intracellular communication between different cell compartments is essential to optimize gene expression for differentiation, development, and responses to environmental challenges (Parikh et al., 1987; Cottage et al., 2010; Estavillo et al., 2011; Xiao et al., 2012; Esteves et al., 2014). Most protein complexes of plastids and mitochondria are mosaics of organelle-encoded and nucleus-encoded subunits. Consequently, tight coordination of gene expression from both genomes is crucial to cellular homeostasis and fitness of the whole organism (Liu and Butow, 2006; Jarvis and López-Juez, 2013; Chan et al., 2016). Already nearly four decades ago, it was realized that defects in plastid protein synthesis can repress the accumulation of nucleus-encoded plastid proteins (Bradbeer et al., 1979). It was proposed that signals emanating from plastids and relayed to the nucleus (later dubbed retrograde signals) control the expression of a large number of nuclear genes and, in this way, help to harmonize the functional state of the plastids with that of the nucleus (Woodson and Chory, 2008; Chan et al., 2016). Over the past decades, substantial progress has been made with identifying signaling components and distinct pathways of retrograde signaling that regulate chloroplast biogenesis (biogenetic control) or control plastid homeostasis in response to environmental stimuli (operational control; Susek et al., 1993; Wagner et al., 2004; Koussevitzky et al., 2007; Pogson et al., 2008; Estavillo et al., 2011; Woodson et al., 2011; Ramel et al., 2012; Xiao et al., 2012). Various potential signaling molecules have been identified, including intermediates of tetrapyrrole biosynthesis (TPB; Strand et al., 2003; Woodson et al., 2011), carotenoid oxidation products (Ramel et al., 2012; Shumbe et al., 2014), isoprenoid precursors (Xiao et al., 2012), phosphoadenosines (Estavillo et al., 2011), hydrogen peroxide (Balazadeh et al., 2012; Maruta et al., 2012), and carbohydrate metabolites (Heinrichs et al., 2012; Vogel et al., 2014).
The chloroplast translation inhibitor lincomycin (Lin; Mulo et al., 2003) and the carotenoid biosynthesis inhibitor norflurazon (NF; Oelmüller, 1989) have been widely used to inhibit chloroplast biogenesis and demonstrate the biogenic control exerted by plastid retrograde signaling. Six GENOMES UNCOUPLED (GUN) loci, whose inactivation uncouples nuclear gene expression from proper chloroplast function, have been identified through genetic screens (Susek et al., 1993; Woodson et al., 2011). Five of the six GUN genes (GUN2, GUN3, GUN4, GUN5, and GUN6) encode enzymes or regulators of the TPB pathway, underscoring the essential role of TPB-derived retrograde signals in chloroplast biogenesis (Mochizuki et al., 2001; Larkin et al., 2003; von Gromoff et al., 2008; Woodson et al., 2011). Inhibition of plastid gene expression (PGE) represses the expression of a number of photosynthesis-associated nuclear genes (PhANGs). Distinct from the other GUN genes, GUN1 is the only known GUN gene that, when defective, results in derepression of the expression of PhANGs in the presence of both Lin (which inhibits PGE by blocking chloroplast translation) and NF (which affects TPB signaling; Koussevitzky et al., 2007; Wu et al., 2018, 2019). Interestingly, the GUN1 mutation was shown to restore PhANG expression in plastid sigma factor mutants (Woodson et al., 2013), the prors1 mutant (defective in prolyl-tRNA synthetase in both chloroplasts and mitochondria; Tadini et al., 2016), and the prin2 mutant (affecting a locus required for full expression of genes transcribed by the plastid-encoded RNA polymerase; Kindgren et al., 2012a; Díaz et al., 2018). Together, these findings indicate a central role of GUN1 in the PGE pathway of retrograde signaling.
The vast majority of studies on retrograde signaling demonstrated regulation of nuclear genes by organellar signals at the transcript level (i.e. mRNA abundance and alternative splicing; Petrillo et al., 2014). Ferredoxin I (Fed-1) transcript abundance was shown to be modulated, in a light-dependent manner, posttranscriptionally at the level of mRNA stability (Elliott et al., 1989; Petracek et al., 1997, 1998). Loading of Fed-1 mRNA with ribosomes in the light likely results in protection from nucleolytic degradation (Dickey et al., 1994, 1998). This observation indicates a potential link between retrograde regulation of nuclear gene expression and cytosolic translation. The possible role of translational and posttranslational regulation in retrograde signaling is only beginning to draw more attention. For example, the translational response of Arabidopsis (Arabidopsis thaliana) to a shift from low light to high light (which induces operational control of retrograde signaling; Oelze et al., 2014) has been studied, the relationship between the accumulation of light-harvesting complex (LHC) proteins and their transcripts upon impaired plastid translation has been analyzed (Krupinska et al., 2019), and an important role of posttranslational modifications in mitochondrial retrograde regulation is emerging (Hartl and Finkemeier, 2012). The GOLDEN2-LIKE (GLK) transcriptional activator positively regulates the expression of a large number of PhANGs in plants (Yasumura et al., 2005; Waters et al., 2009). It was demonstrated that the expression of GLK1 is controlled by GUN1-dependent retrograde signals (Kakizaki et al., 2009). GLK1 protein accumulation under Lin and NF treatments is regulated posttranslationally by protein degradation through the cytosolic ubiquitin proteasome system (UPS; Tokumaru et al., 2017), indicating a role of posttranslational regulation in plastid retrograde signaling. GUN1 was also shown to regulate the accumulation of plastid ribosomal protein PRPS1 independent of PRPS1 mRNA abundance, although the underlying mechanism remains to be investigated (Tadini et al., 2016). Overall, the contribution of translational and posttranslational regulation to retrograde signaling and to retrograde regulation upon defective transcriptional regulation (e.g. in gun mutants) is still largely unknown.
To systematically investigate the contribution of translational and posttranslational mechanisms to plastid retrograde regulation, we investigated the translational and posttranslational regulation emanating from the PGE pathway in the wild type and the gun1 mutant. Our results demonstrated that, although PhANG expression is derepressed in gun1 at the transcript level, inhibition of plastid translation triggers similar changes in the proteomes of gun1 and the wild type, indicating strong translational and posttranslational components in retrograde responses in the PGE pathway. We identified 181 highly translationally or posttranslationally regulated (HiToP) genes in the wild type and classified them into two classes that differ in their patterns of gene regulation. We further show that HiToP PhANGs are largely regulated by translational repression, while HiToP ribosomal protein-encoding genes are regulated posttranslationally, likely through protein degradation. Our results suggest that the translational repression of PhANGs is dependent on GUN1, while the posttranslational regulation of ribosomal protein-encoding genes does not require the function of GUN1.
RESULTS
GUN1 Regulates Plastid Proteostasis upon Interference with Retrograde Signaling
In order to understand the regulation of nuclear genes by chloroplast retrograde signals at the protein level, we performed proteomic analyses of the wild type and the gun1 mutant. To this end, plants were grown under control conditions or treated with Lin to interfere with the PGE pathway of retrograde signaling (Fig. 1; Supplemental Data Set S1). Since the PGE pathway is mainly active during the first few days after germination when massive chloroplast biogenesis is occurring (Oelmüller et al., 1986), we germinated and grew the seedlings in the dark for 5 d followed by 2 d in continuous light to mimic the chloroplast biogenesis events occurring during the first days after germination. Also, using this experimental setup, enough material for proteomic analyses could be obtained. We included the gun5 mutant as a control, since GUN5 only derepresses PhANG expression under NF treatment but not under Lin treatment (Mochizuki et al., 2001; Hernández-Verdeja and Strand, 2018).
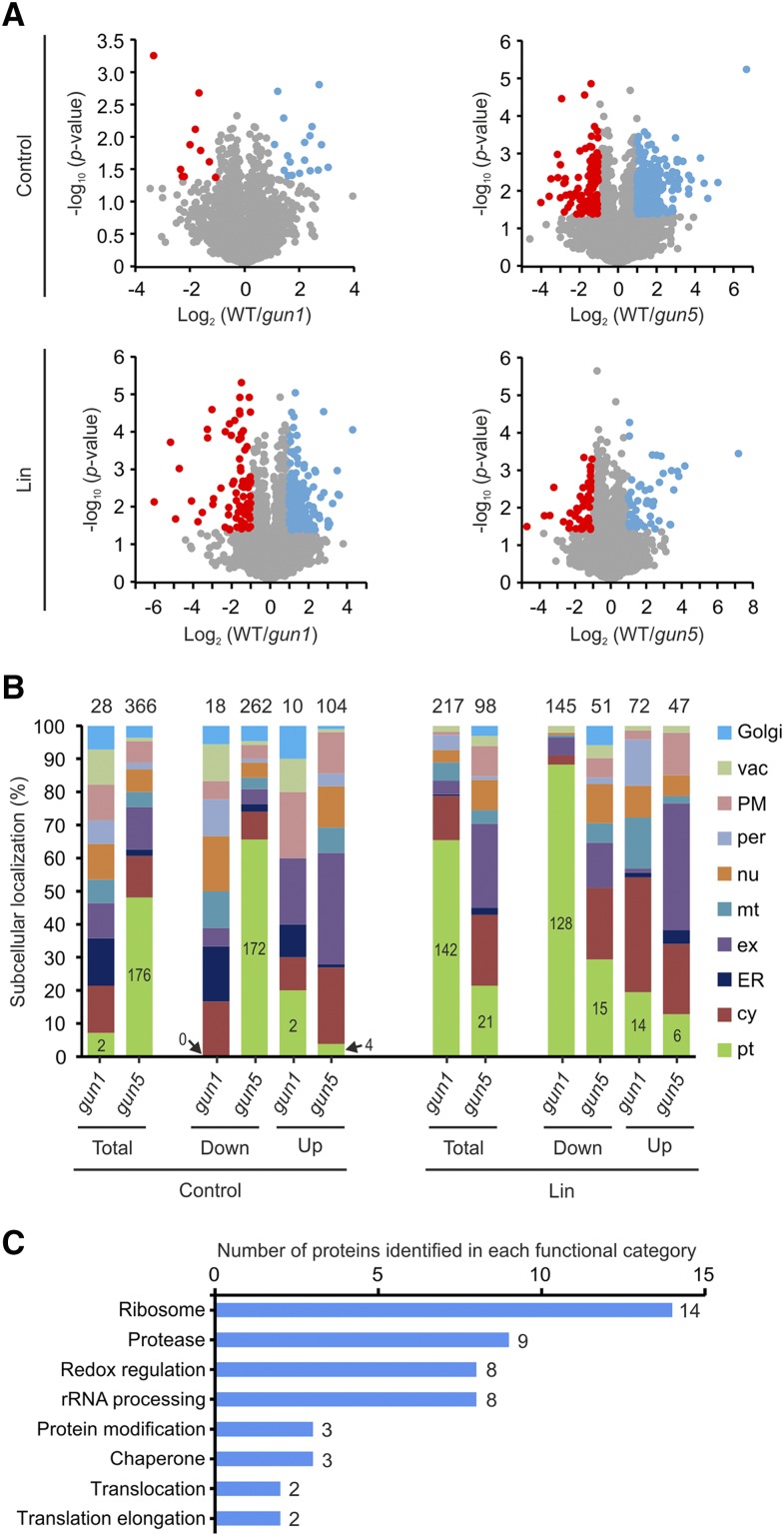
Figure 1.
GUN1 regulates plastid proteostasis in the PGE pathway of retrograde signaling. A, Volcano plots depicting the protein accumulation in gun1 and gun5 compared with the wild type (WT) in control conditions and upon Lin treatment. Proteins with P < 0.05 (Student’s t test) and a log2 value of the ratio wild type/mutant > 1 or < −1 are shown in blue and red, respectively. B, Histograms showing the subcellular localization of proteins that are differentially expressed in gun1 or gun5 compared with the wild type in control conditions and upon Lin treatment. The numbers above the bars indicate the number of proteins identified as being differentially expressed between mutant and the wild type. The number of plastid-localized proteins is indicated in the green part of each bar. cy, Cytosol; ER, endoplasmic reticulum; ex, extracellular; Golgi, Golgi apparatus; mt, mitochondrion; nu, nucleus; per, peroxisome; PM; plasma membrane; pt, plastid; vac, vacuole. C, Histogram illustrating the functional categories of chloroplast proteins that are potentially involved in plastid proteostasis and accumulate less in gun1 compared with the wild type upon Lin treatment.
Plastid-encoded proteins account for ∼18% of the total cellular protein under control conditions. The large subunit of Rubisco accounts for approximately half of this amount (Supplemental Fig. S1A). Upon Lin treatment, plastid-encoded proteins account for only ∼0.17% of the total cellular protein. By contrast, mitochondrially encoded proteins show no significant change in their abundance (Supplemental Fig. S1B), confirming the specific inhibition of plastid translation by Lin (Mulo et al., 2003).
While the gun1 mutant does not show a visible phenotype under normal growth conditions, the gun5 mutant displays a pale green phenotype (Mochizuki et al., 2001; Koussevitzky et al., 2007). Consistent with the absence of a visual mutant phenotype, the proteome of gun1 does not show many differences from the wild type under control conditions. We only identified 28 proteins that are differentially expressed between the wild type and gun1 (Supplemental Table S1). This set of proteins did not show any particular enrichment in functional classification or cellular compartment (Fig. 1A; Supplemental Table S1). By contrast, the proteome of gun5 shows large differences compared with that of the wild type. The number of proteins found to be differentially expressed in gun5 compared with the wild type was 366 (Fig. 1B; Supplemental Data Set S1). Among these, 262 proteins are down-regulated and 104 proteins are up-regulated in gun5. In agreement with the pale green phenotype, 172 of the 262 down-regulated proteins are localized in the chloroplast (according to SUBA4; http://suba.live/), including subunits of different photosynthetic complexes and proteins involved in chloroplast biogenesis. By contrast, only four chloroplast-localized proteins were identified among the 104 up-regulated proteins (Fig. 1B; Supplemental Data Set S1): ADP-Glc pyrophosphorylase (catalyzes the first and rate-limiting step in starch biosynthesis), aldehyde reductase, Gln phosphoribosylpyrophosphate amidotransferase1 (a key enzyme in the pathway of purine nucleotide biosynthesis), and ferretin1 (ferric iron binding).
When the seedlings were grown in the presence of Lin to affect the PGE pathway of retrograde signaling, the differences between the wild type and gun5 became smaller, with only 98 proteins showing differential expression (Fig. 1, A and B). Moreover, the plastid-localized proteins (21 out of 98) were no longer as strongly overrepresented as in the control conditions. This can potentially be explained by the plastids of the wild type and gun5 both remaining undifferentiated upon Lin treatment. In contrast to the increased similarity between the proteomes of the wild type and gun5, the proteome of gun1 shows strongly increased differences from that of the wild type (Fig. 1A). Compared with the control conditions, now 217 proteins are identified as differentially expressed between the wild type and gun1 (Fig. 1B; Supplemental Data Set S1). Among these, 145 proteins are down-regulated and 72 are up-regulated. Interestingly, more than 88% (128 out of 145) of the down-regulated proteins are plastid localized (Fig. 1B). By contrast, only ∼19% of the proteins up-regulated in gun1 are localized in the plastids, and only ∼29% (15 out of 51) of the proteins down-regulated in gun5 are plastid localized. The large-scale down-regulation of plastid proteins in gun1 upon Lin treatment indicates an important role of GUN1 in the maintenance of plastid protein homeostasis (proteostasis) when the PGE pathway of retrograde signaling is altered (Lin treatment). We further investigated the 128 down-regulated plastid proteins. Interestingly, when compared within the same genotype in the presence versus absence of Lin, only half (64 out of 128) of these proteins show more than 2-fold repression by Lin in the wild type (Supplemental Data Set S2). This indicates that (1) proteins that are repressed by Lin in the wild type are even more strongly repressed in gun1 and (2) proteins that are unchanged or even up-regulated in the wild type are down-regulated in gun1. Many of the 128 proteins are directly or indirectly involved in proteostasis, including a large number of ribosomal proteins (14), proteases (nine), rRNA processing factors (eight), and proteins involved in redox regulation (eight; Fig. 1C; Supplemental Data Set S2). This observation provides further evidence for plastid proteostasis in gun1 differing substantially from that in the wild type upon Lin treatment. The proteomes of gun1 and the wild type do not show many differences under control conditions, suggesting that GUN1 exerts its role in the regulation of plastid proteostasis only (or predominantly) under conditions when retrograde signaling is altered.
Strong Translational and Posttranslational Regulation Occurs When Retrograde Signaling Is Affected
The defective repression of PhANGs in gun1 upon Lin treatment is expected to result in higher PhANG protein accumulation compared with the wild type. However, our proteomic analyses of the wild type and the gun1 mutant revealed that this is not the case (Fig. 1; Supplemental Data Set S1). We identified only 14 plastid-localized proteins that overaccumulate in gun1 relative to the wild type (Supplemental Table S2). Ten out of these 14 proteins are typical PhANGs, including three subunits of LHC, two subunits of PSII, one subunit each of PSI and the NAD(P)H dehydrogenase (NDH) complex, carbonic anhydrases (CA1 and CA2), and Fru-bisphosphate aldolase5.
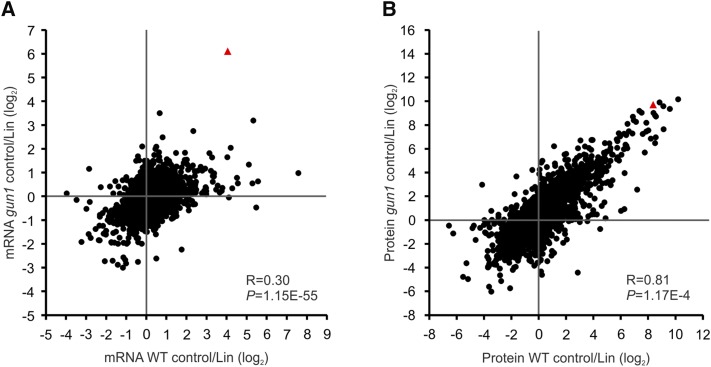
To resolve this seeming contradiction, we analyzed gene expression at the RNA level by whole-genome microarrays to enable side-by-side analysis of gene regulation at the transcript accumulation versus protein accumulation levels. From the proteomic data sets, all proteins that were detected in at least two biological replicates of at least one condition (genotype or treatment) were selected for further analysis. Since oligo(dT) primer was used for first-strand cDNA synthesis for the microarray experiments (and organellar transcripts are not normally polyadenylated), we excluded genes encoded in the plastid and mitochondrial genomes. Applying these criteria, 5,449 genes with transcript and protein expression data could be used for transcript-protein coanalysis (Fig. 2; Supplemental Data Set S3). When we compared transcript accumulation in the wild type and the gun1 mutant in the control conditions with that upon Lin treatment, the correlation of expression changes in the wild type and gun1 was found to be very low (Pearson correlation coefficient, 0.3; Fig. 2A). We identified only 26 genes that show differential expression at the transcript level between the wild type and gun1 under control conditions (Supplemental Table S3). Interestingly, the cytosolic chaperones involved in protein quality control, including two small heat shock proteins (HSP17.6A and HSP17.6II), HSP90.1, HSP70-2 and HSP70-4, and the transcription factor HSFA2 (involved in the responses to heat stress and accumulation of misfolded proteins in the cytosol), were up-regulated in gun1. The low correlation in response to Lin treatment indicates a different response of gun1 compared with the wild type. The differences are not just caused by the derepression of PhANGs in gun1, since a large number of non-PhANGs are differentially expressed between the two genotypes upon Lin treatment (Supplemental Data Set S4). Interestingly, when we compared the changes at the protein level, the wild type and the gun1 mutant responded surprisingly similarly to the Lin treatment (Pearson correlation coefficient, 0.81; Fig. 2B). This indicates that, although the response of gun1 at the transcript level is very different from that of the wild type, strong translational and posttranslational regulation overrides many of the changes in transcript abundance, thus making the response at the protein level more similar. In agreement with this, although 773 genes show differential expression (false discovery rate [FDR] < 0.05, fold change > 2) at the transcript level between the wild type and gun1 under Lin treatment (of which 254 can be detected by MS at the protein level), only 56 proteins show more than twofold changes in the same direction as their transcripts (i.e. both go up or down; Supplemental Data Set S4).
Figure 2.
Transcript and protein coanalysis reveals extensive translational and posttranslational regulation in retrograde signaling. Scatterplots show the high correlation of gene regulation at the protein level (B) compared with the transcript (mRNA) level (A) between the gun1 mutant and the wild type (WT) in response to Lin treatment. All nucleus-encoded genes whose protein products can be detected by mass spectrometry (MS; in a total of 5,449 genes; for details, see “Materials and Methods”) were included in the analysis. The fold changes in the Lin treatment relative to the untreated control (log2; averaged from three biological replicates) were compared between gun1 and the wild type to calculate the Pearson correlation coefficient (R) and the P value (P). Red triangles indicate the LOX2 gene.
Type I and Type II HiToP Genes
To further understand how gene expression is regulated by retrograde signaling, we performed a correlation analysis of transcripts and proteins in control conditions compared with Lin treatment for each genotype (Fig. 3, A and B). In the wild type, some PhANGs, such as LHCB1.4 and CA1, show a good correlation between mRNA and protein, in that both are repressed in the presence of Lin. For other PhANGs, for example, the small subunit of Rubisco (RBCS1B and RBCS3B), subunits of PSI (PsaG, PsaL, and PsaD), and the calcium-sensing receptor (CaSR), the down-regulation at the protein level is much stronger than that at the transcript level (Fig. 3A; Supplemental Data Set S3). This observation suggests that, also in the wild type, strong translational and/or posttranslational regulation is exerted in response to the altered PGE pathway of retrograde signaling (Lin treatment). However, compared with the wild type, the regulation is much stronger in gun1 (Fig. 3B). For example, the transcripts of all genes mentioned above did not show a substantial repression by Lin treatment, but all proteins, except CA1, were strongly down-regulated (Fig. 3B; Supplemental Data Set S3). By contrast, the expression of the non-PhANG lipoxygenase2 (LOX2) gene is repressed at the mRNA and protein levels both in the wild type and gun1 (Fig. 2; Supplemental Fig. S2).
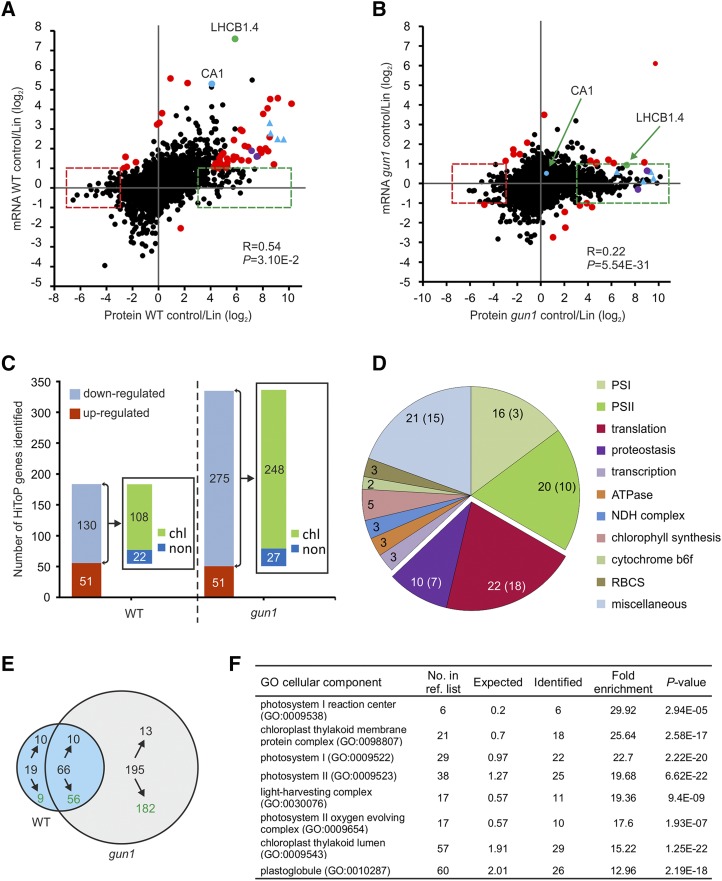
Figure 3.
HiToP genes identified. A and B, Scatterplots to compare the correlation of regulation between transcripts and proteins in control conditions versus Lin treatment in the wild type (WT; A) and the gun1 mutant (B). The HiToP genes were classified into two types: type I, log2 protein control/Lin > 3 or < −3 and −1 < log2 mRNA control/Lin < 1 (green and red dashed boxes); type II, log2 mRNA control/Lin > 1 or < −1 but log2 superratio of protein fold change to mRNA fold change of control to Lin treatment > 3 or < −3 (highlighted as red dots, purple dots, and blue triangles). Selected genes of special interest are marked: green dots, LHCB1.4; blue dots, CA1; purple dots, RBCS1B and RBCS3B (also identified as type II HiToP genes in the wild type); blue triangles, other examples of genes that were identified as type II HiToP genes in the wild type but as type I HiToP genes in gun1 (PsaG, PsaL, PsaD, and CaSR). See text for details. C, Up- and down-regulated HiToP genes identified in the wild type and the gun1 mutant. The numbers in the bars give the numbers of genes identified in each group. The insets show the number of HiToP genes that are down-regulated at the protein level upon Lin treatment and whose products are localized in chloroplasts (chl) or outside of chloroplasts (non). D, Pie chart illustrating the functional categories of the 108 plastid-localized HiToP proteins that are down-regulated in the wild type at the protein level upon Lin treatment. The numbers give the total number of genes identified in each functional category, while the numbers in parentheses denote the number of type I HiToP genes. The genes related to translation and proteostasis are separated out. E, Venn diagram illustrating the overlap of down-regulated type I genes (green boxes in A and B) between the wild type and the gun1 mutant. Green numbers indicate the number of genes whose protein products localize to plastids. F, GO enrichment of cellular component analysis of the 182 plastid-localized down-regulated type I HiToP genes in gun1. The 5,449 proteins identified by MS were used as the reference list for GO terms of enrichment analysis (http://www.pantherdb.org/). The fold enrichment was calculated by binomial test using the Bonferroni correction for multiple testing. The column “No. in ref. list” indicates the number of genes in each GO term among the 5,449 reference genes. The columns “Expected” and “Identified” indicate the number of genes that were expected or identified for each GO term within the 182 plastid-localized down-regulated type I HiToP genes.
Our observation that there is strong translational and posttranslational regulation also in the wild type suggests that this type of regulation is a general control mechanism in response to Lin treatment (altering the PGE pathway). To explore the extent of translational and posttranslational regulation more systematically, we analyzed transcript and protein accumulation in the wild type to identify HiToP genes. We classified HiToP genes into two types: type I, comprising genes whose expression changes at the protein level more than eightfold but less than twofold at the transcript level (log2 protein control/Lin > 3 or < −3, but −1 < log2 transcript control/Lin < 1); and type II, comprising genes that, although their expression changes at the transcript level more than twofold, the superratio of protein change to transcript change (reflecting the correlation of the transcript with its protein product, with a large superratio indicating low correlation and, thus, strong translational or posttranslational regulation) was higher than 8 (log2 transcript control/Lin > 1 or < −1, but log2 superratio > 3 or < −3). By these criteria, we identified 181 HiToP genes in the wild type, with 130 of them being down-regulated and 51 being up-regulated upon Lin treatment (Fig. 3, A and C; Supplemental Data Set S5). Among the 181 HiToP genes, 127 genes were identified as type I and 54 as type II genes. Importantly, most of the down-regulated HiToP genes (108 out of 130) encode plastid-localized proteins, thus underscoring the specific action of Lin treatment on plastid retrograde signaling.
We then further analyzed the functional annotation of the 108 down-regulated HiToP genes whose protein products localize to plastids to determine if they are enriched in specific pathways or macromolecular complexes (Fig. 3D; Supplemental Data Set S5). Interestingly, genes for PSI and PSII subunits were mainly among the type II HiToP genes, indicating that they are down-regulated also at the transcript level. By contrast, most of the genes for ribosomal subunits and genes involved in proteostasis were identified as type I. Of the 24 type I HiToP genes with a more than 16-fold down-regulation at the protein level (log2 protein control/Lin > 4), seven encode ribosomal proteins (Table 1).
Table 1. Type I HiToP genes encoding plastid-localized proteins.
Genes identified in the wild type as log2 control/Lin (C/L) > 4 at the protein level but −1 < log2 control/Lin < 1 at the mRNA level are listed. The superratio (SR) was calculated as protein fold change divided by mRNA fold change. The corresponding values in the gun1 mutant are included for comparison. The data represent averages from three biological replicates. Chl syn, Chlorophyll synthesis; –, unknown.
| Locus | Gene | Function | mRNA | Protein | Log2 SR | |||
|---|---|---|---|---|---|---|---|---|
| Log2 C/L | Log2 C/L | |||||||
| Wild Type | gun1 | Wild Type | gun1 | Wild Type | gun1 | |||
| AT4G32260 | ATPase, F0 complex, subunit B/B′ | ATPase | 0.95 | 0.10 | 7.38 | 9.17 | 6.43 | 9.07 |
| AT5G14320 | Ribosomal protein S13/S18 family | Translation | 0.65 | −0.15 | 6.50 | 8.07 | 5.85 | 8.22 |
| AT4G25080 | CHLM | Chl syn | 0.82 | −0.08 | 6.49 | 6.03 | 5.67 | 6.10 |
| AT3G56910 | PSRP5 | Translation | 0.53 | −0.19 | 6.42 | 7.14 | 5.89 | 7.33 |
| AT1G67090 | RBCS1A | RBCS | 0.27 | −0.05 | 6.27 | 7.26 | 6.00 | 7.31 |
| AT1G79040 | PsbR | PSII | 0.74 | 0.09 | 6.24 | 5.96 | 5.51 | 5.87 |
| AT3G55330 | PsbP-like protein1 (PPL1) | PSII | 0.93 | 0.22 | 6.07 | 5.49 | 5.14 | 5.27 |
| AT5G54190 | PORA | Chl syn | 0.30 | 0.19 | 6.04 | 0.75 | 5.73 | 0.56 |
| AT2G05620 | PGR5 | PSI | 0.92 | −0.49 | 5.75 | 6.07 | 4.84 | 6.56 |
| AT3G27160 | RPS21/GHS1 | Translation | 0.82 | −0.08 | 5.42 | 5.48 | 4.60 | 5.56 |
| AT5G17870 | PSRP6 | Translation | 0.79 | −0.13 | 5.41 | 3.33 | 4.62 | 3.46 |
| AT2G21960 | Metalloendopeptidase | Proteostasis | 0.30 | 0.09 | 5.38 | 5.49 | 5.08 | 5.40 |
| AT1G64770 | NDH45 | NDH | 0.63 | 0.39 | 5.25 | 4.96 | 4.61 | 4.57 |
| AT3G63540 | PsbP family protein | PSII | 0.34 | −0.06 | 5.23 | 5.84 | 4.89 | 5.89 |
| AT5G48220 | Aldolase-type TIM barrel family | – | 0.55 | 0.24 | 4.97 | 4.10 | 4.43 | 3.86 |
| AT3G54210 | RPL17 | Translation | 1.00 | −0.40 | 4.90 | 5.09 | 3.90 | 5.49 |
| AT5G40950 | RPL27 | Translation | 0.90 | −0.17 | 4.85 | 6.45 | 3.95 | 6.62 |
| AT3G15190 | RPS20 | Translation | 0.87 | 0.18 | 4.83 | 5.53 | 3.96 | 5.34 |
| AT1G16720 | HCF173 | Translation | 0.05 | −0.31 | 4.83 | 4.91 | 4.79 | 5.22 |
| AT2G26340 | Hypothetical protein | – | 0.65 | −0.56 | 4.73 | 4.03 | 4.09 | 4.59 |
| AT5G02830 | TPR superfamily protein | – | 0.41 | 0.29 | 4.30 | 4.34 | 3.89 | 4.05 |
| AT4G23890 | NDHS/CRR31 | NDH | 0.84 | 0.01 | 4.28 | 4.02 | 3.45 | 4.01 |
| AT4G18370 | DEG5 | Proteostasis | 0.74 | 0.17 | 4.24 | 3.28 | 3.50 | 3.12 |
| AT5G11450 | PsbP domain protein (PPD5) | PSII | 0.82 | −0.21 | 4.20 | 3.13 | 3.38 | 3.33 |
For comparison, we also analyzed HiToP genes in gun1, where we identified 326 HiToP genes in total (Fig. 3, B and C; Supplemental Data Set S5). While the numbers of up-regulated genes and down-regulated genes that encode non-plastid-localized proteins in gun1 are similar to those in the wild type, the number of down-regulated chloroplast-localized proteins is substantially larger in the gun1 mutant (Fig. 3C). Interestingly, among the 326 HiToP genes in gun1, only 21 genes were identified as type II genes (Fig. 3B; Supplemental Data Set S5). By contrast, more than 93% (305 out of 326) HiToP genes in gun1 were identified as type I genes (less than twofold change at the transcript level), further indicating that most of the genes are regulated posttranscriptionally.
To further analyze the regulation in gun1, we focused on the type I genes that are down-regulated at the protein level (Fig. 3, A and B, green boxes). We identified 85 and 261 down-regulated type I genes in the wild type and gun1, respectively (Fig. 3E). Sixty-six genes overlap between the two sets. Of the 195 genes only identified in gun1, 182 encode plastid-localized proteins (Fig. 3E). Gene Ontology (GO) analysis shows that they are enriched in functions related to PSI, PSII, and plastoglobules (Fig. 3F). These results indicate that the expression of many PhANGs is repressed in gun1 at the level of protein accumulation by translational or posttranslational mechanisms.
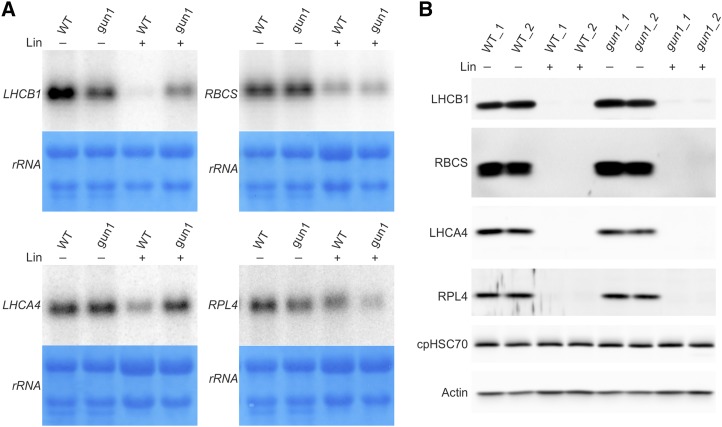
To confirm the pattern of posttranscriptional regulation seen in our microarray and MS studies, we selected several genes for northern-blot and western-blot analyses (Fig. 4). Based on antibody availability, we chose LHCA4, LHCB1, and RBCS as PhANGs and RPL4 as an example of a plastid ribosomal protein. While the degree of repression of transcript accumulation varied between the four genes (with LHCB1 mRNA accumulation being most strongly down-regulated; Fig. 4A), the proteins were all below the detection limit of the immunoblot (Fig. 4B). This finding lends further support to the idea that, as gun1 is unable to inhibit nuclear gene expression at the transcript level, the repression of plastid-localized proteins in gun1 relies largely on down-regulation at the translational and/or posttranslational levels, to ultimately reach a similarly low protein level to that in the wild type.
Figure 4.
Northern-blot and western-blot analyses to reveal the regulation of different types of HiToP genes. A, Northern-blot analysis of selected HiToP genes. Unlike LHCB1 (the probe used does not distinguish between different isoforms of LHCB1), whose transcript accumulation is largely repressed by Lin treatment in the wild type (WT), the transcripts of LHCA4 and RBCS (the probe used does not distinguish between different isoforms of RBCS) are less repressed and the RPL4 transcripts are nearly unchanged. Seeds from the wild type and gun1 were germinated and grown for 5 d in the dark followed by 2 d in continuous light in the absence (−) or presence (+) of 0.5 mm Lin. Methylene Blue staining of rRNAs after blotting was performed as a loading control. B, At the protein level, LHCA4, RBCS, and RPL4 are all below the detection limit of immunoblots both in the wild type and gun1 under Lin treatment. For LHCB1 in Lin treatment, faint bands can be detected in gun1 but not in the wild type. Actin and the chloroplast chaperone protein cpHSC70 were included as loading controls.
Repression of LHCB Accumulation in gun1 by Translational Inhibition
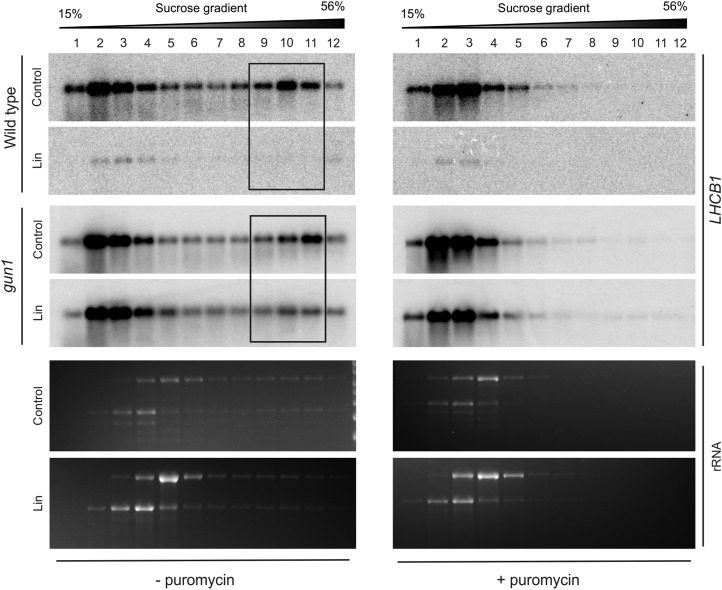
To distinguish between translational and posttranslational regulation, polysome profiles were analyzed for several mRNAs. We first examined genes whose transcripts are largely repressed in the wild type, but not in gun1, to determine how failure of accumulation of the corresponding proteins continues to occur in spite of the loss of GUN1. In comparison with LHCA4 and RBCS, mRNA accumulation of LHCB1 was very strongly repressed by Lin treatment in the wild type but not in gun1 (Fig. 4A; Supplemental Fig. S3). LHCB transcripts were also found to be dramatically repressed in deetiolating pea (Pisum sativum) seedlings in the presence of NF compared with the mild repression of RBCS and Fed-1 transcripts (Sagar et al., 1988). When we analyzed the polysome profiles of LHCB1 in both genotypes (Fig. 5), the transcripts in the wild type were found to be repressed to nearly undetectable levels, consistent with the microarray data. By contrast, LHCB1 continues to be expressed in gun1 in the presence of Lin (Figs. 4A and 5). Remarkably, while a large proportion of the mRNA is present in the polysome-containing fractions 9 to 11 under control conditions, polysome association is repressed by Lin treatment (Fig. 5). The redistribution of LHCB1 transcripts from polysome fractions in control conditions to the free mRNA pool upon Lin treatment suggests translational repression of LHCB1 in gun1 in response to Lin treatment.
Figure 5.
Polysome profiles demonstrate that translation of LHCB is repressed in gun1 upon Lin treatment. In the wild type, transcript accumulation of LHCB1 (the probe used does not distinguish between different isoforms of LHCB1) is dramatically reduced under Lin treatment (see Fig. 4A). In gun1, translation of LHCB1 is down-regulated upon Lin treatment, as evidenced by the distribution of LHCB1 transcripts in polysome fractions 9 to 11 (boxed). Puromycin-treated control samples were included to identify the polysome-containing gradient fractions. Puromycin releases ribosomes from the mRNA. The wild type and gun1-101 were grown in the presence (Lin) or absence (Control) of Lin in the dark for 5 d prior to 2 d in continuous light. The wedges above the blots indicate the increasing Suc concentration across the gradient. The ethidium bromide-stained rRNAs on the gels prior to blotting are also shown.
PhANGs and Ribosomal Proteins Are Regulated by Translational and Posttranslational Repression, Respectively, in Response to Chloroplast Translation Inhibition
We have identified 108 chloroplast-localized HiToP genes that are down-regulated in response to Lin treatment in the wild type (Fig. 3C). To shed light on the underlying mechanisms of posttranscriptional regulation, we analyzed polysome profiles for a set of PhANGs (mainly identified as type II HiToP genes) and non-PhANGs (e.g. nuclear genes encoding plastid ribosome proteins that mainly had been identified as type I HiToP genes; Fig. 3D). As examples of PhANGs, we analyzed RBCS and LHCA4, two classical genes regulated by retrograde signals (Woodson et al., 2011, 2013; Kindgren et al., 2012b). Different from LHCB1 genes, the transcripts of LHCA4 and RBCS still accumulate under Lin treatment (Fig. 4A), but the proteins are below the detection limit of immunoblots (Fig. 4B). As examples of chloroplast ribosomal proteins, we chose PSRP5 and RPL4 (Supplemental Data Set S5), because (1) PSRP5 was ranked as the strongest posttranscriptionally regulated ribosomal protein gene (Table 1) and (2) an antibody against RPL4 is available, thus allowing us to examine protein accumulation by immunoblotting (Fig. 4B).
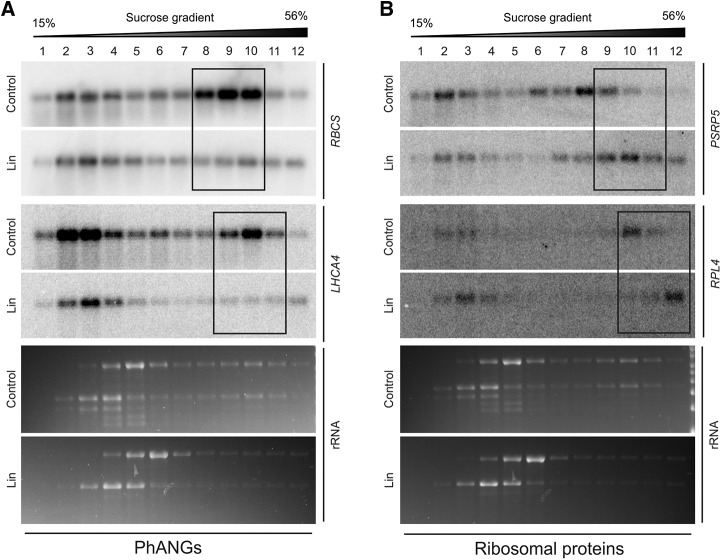
Analysis of polysome profiles of RBCS and LHCA4 mRNAs demonstrated that translation of both transcripts was strongly repressed, as revealed by strong reduction of mRNA abundance in polysome-containing fractions (fractions 8–10 for RBCS and 9–11 for LHCA4, respectively; Fig. 6A; Supplemental Fig. S4A). This observation suggests that expression of these HiToP PhANGs is largely controlled by translational repression.
Figure 6.
Polysome profiles uncover different modes of regulation of PhANGs and ribosomal protein-encoding HiToP genes. A, Comparison of polysome profiles under Lin treatment with those of the untreated control demonstrates the strong translation inhibition of RBCS and LHCA4 (HiToP PhANGs) in response to plastid translation inhibition in the wild type. The probe used does not distinguish between different isoforms of RBCS. The three major polysome-containing fractions are boxed. The wedges above the blots indicate the increasing Suc concentration across the gradient. B, Polysome profiles for nuclear genes encoding plastid ribosomal proteins (PSRP5 and RPL4) indicate that their translation is up-regulated in response to Lin treatment. The three major polysome-containing fractions are boxed for both mRNAs. For puromycin controls, see Supplemental Figure S4. The ethidium bromide-stained rRNAs on the gels prior to blotting are also shown.
Interestingly, the ribosomal protein-coding genes we tested show the opposite behavior. Unlike RBCS and LHCA4, both PSRP5 and RPL4 show increased translation in response to Lin treatment (Fig. 6B; Supplemental Fig. S4B). The PSRP5 mRNA accumulated in polysome fractions 7 to 9 in the untreated control but shifted to fractions 9 to 11 under Lin treatment, indicating denser loading with ribosomes. Similarly, the RPL4 mRNA was mainly present in polysome fraction 10 and shifted to fraction 12 in response to Lin treatment. Considering the undetectably low accumulation of these ribosomal proteins under Lin treatment (Fig. 4B), the repression of their expression obviously occurs at the posttranslational level, likely by regulation of protein stability/degradation.
GUN1-Dependent Plastid Retrograde Signals from the PGE Pathway Regulate Cytosolic Translation
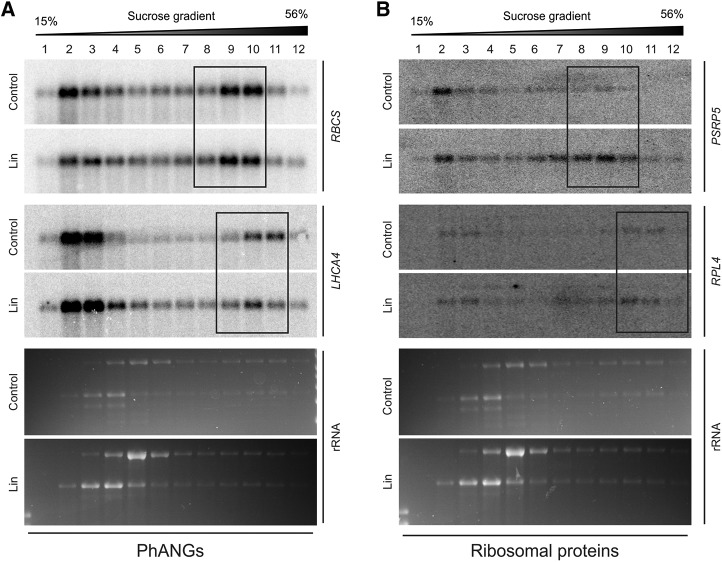
To examine if the translational regulation discovered in this study is regulated by GUN1-dependent retrograde signals, we analyzed the polysome profiles of PhANGs and ribosomal protein-coding genes in the gun1 mutant (Fig. 7; Supplemental Fig. S5). Interestingly, the translational repression of RBCS and LHCA4 and, to a lesser extent, the translational up-regulation of ribosomal protein-coding genes (PSRP5 and RPL4) observed in the wild type are compromised in gun1. These findings suggest that the cytosolic translational readjustment in response to altered PGE-derived retrograde signals requires the function of GUN1. In order to overcome the defective transcriptional and translational regulation (e.g. of RBCS and LHCA4), posttranslational control mechanisms set in when GUN1 is absent. Therefore, this posttranslational control is likely independent of GUN1.
Figure 7.
Translational regulation in the PGE pathway of retrograde signaling needs the action of GUN1. A, Comparison of the polysome profiles upon Lin treatment with those of the untreated controls for RBCS and LHCA4 demonstrates that translational inhibition of HiToP PhANGs in gun1 in response to Lin treatment is compromised. The probe used does not distinguish between different isoforms of RBCS. B, Comparison of polysome profiles upon Lin treatment with those of the untreated controls for PSRP5 and RPL4 demonstrates that up-regulation of their translation in response to Lin treatment is compromised in gun1 (in that the shift to heavier gradient fractions does not occur; compare with Fig. 6). In both A and B, the three major polysome-containing fractions are boxed. The wedges above the blots indicate the increasing Suc concentration across the gradient. The ethidium bromide-stained rRNAs on the gels prior to blotting are also shown. For puromycin controls, see Supplemental Figure S5.
DISCUSSION
Signals emanating from organelles have been shown to coordinate nuclear gene expression with organelle function (Jia et al., 1997; Butow and Avadhani, 2004; Chi et al., 2013; Hernández-Verdeja and Strand, 2018). Previous studies that established the concept of retrograde regulation largely focused on transcriptional control and transcript accumulation (Mochizuki et al., 2001; Strand et al., 2003; Koussevitzky et al., 2007; Lee et al., 2007; Estavillo et al., 2011; Xiao et al., 2012). To explore the possible role and the extent of translational and posttranslational regulation, we here analyzed gene expression in the PGE pathway of plastid retrograde signaling by comparing transcriptomes and proteomes of wild-type plants with those of knockout mutants for the proposed central regulator of retrograde signaling, GUN1, upon Lin treatment (Koussevitzky et al., 2007; Wu et al., 2019).
Interestingly, although the transcriptomes of the wild type and gun1 in response to Lin treatment differ very strongly, the proteomes of the two genotypes are much closer to each other (Fig. 2). GO analysis performed with the 182 down-regulated type I HiToP genes whose protein products localize to plastids shows that the GO terms for photosynthesis-related functions were significantly overrepresented (Fig. 3F). Many of the PhANG-encoded proteins accumulate to undetectably low levels in both the wild type and gun1 (Fig. 4B). The repression of PhANG expression in gun1 predominantly does not occur at the translational level (only repression of LHCB translation was observed). In fact, the translation inhibition of LHCA4 and RBCS genes in gun1 is even weaker than in the wild type (Figs. 6 and 7). Our results suggest that the posttranscriptional repression of PhANG expression in gun1 happens largely posttranslationally, at the level of protein degradation.
When we compared the transcriptomes and proteomes of the wild type, it became evident that translational and posttranslational regulation is crucial also in wild-type plants. We have identified 181 HiToP genes in the wild type. By comparing the polysome profiles of the different types of HiToP genes, we observed that the translation of two classes of HiToP PhANGs tested (RBCS genes and LHCA4) was strongly repressed. This observation suggests that translational inhibition acts on these genes to posttranscriptionally regulate their expression (Fig. 6A). Inhibited polysome loading of PhANGs (LHCB and Fed-1) was also observed in 3-(3,4-dichlorophenyl)-1,1-dimethylurea-treated dark-adapted tobacco (Nicotiana tabacum) plants during reillumination or exposure to high light (Petracek et al., 1997), treatments that interfere with the operational control of retrograde signaling. Taken together, these results and our findings indicate that translational regulation of PhANG expression is employed by both the biogenic control (PGE) and the operational control (redox and reactive oxygen species) exerted by retrograde signaling.
Compared with the repressed translation of RBCS genes and LHCA4, the translation of the two ribosomal protein-encoding genes tested (PSRP5 and RPL4) was even enhanced under Lin treatment (Fig. 6B). Lin represses plastid translation (Mulo et al., 2003), including the synthesis of all plastid-encoded subunits of the ribosome. One might have expected that, in order to achieve coordination with the plastid-encoded subunits, cytosolic translation of the nucleus-encoded ribosomal subunits would also be repressed. The plastid-encoded proteins account for ∼0.17% of the total cellular protein upon Lin treatment (Supplemental Fig. S1A), indicating that a basal level of translation still occurs. The observed up-regulated translation of nucleus-encoded ribosomal subunits simply could be a compensatory reaction that occurs in response to the deficiency in plastid-encoded subunits, or alternatively, it may prepare plastids that are blocked in their development by Lin (mimicking an early stage of plastid development) for subsequent enhancement of translation upon illumination.
A number of studies have suggested that translation and mRNA turnover are intimately connected. For example, the slow turnover of polysome-loaded Fed-1 mRNA in the light was hypothesized to be due to the destabilizing CAUU motif being hidden by translating ribosomes (Dickey et al., 1994). A possible explanation for the less repressed transcript accumulation of HiToP ribosomal protein genes could be that the up-regulated loading of the transcripts with ribosomes prevents them from being degraded. Irrespective of the mechanism of transcript stabilization, the encoded ribosomal proteins do not accumulate (Table 1) and cannot be detected by immunoblot analysis (Fig. 4B), indicating that they are epistatically regulated at the posttranslational level by protein degradation. Translational control provides more rapid changes in cellular concentrations of proteins compared with transcriptional regulation and, therefore, may be particularly important to mediate fast responses to environmental cues. Translational regulation mostly occurs at the initiation stage of translation (Sonenberg and Hinnebusch, 2009). How the cytosolic ribosome distinguishes between the different types of genes (PhANGs versus ribosomal protein-encoding genes) to differentially regulate their translation is currently unknown. It seems reasonable to speculate that translational activator and/or repressor proteins are involved that recognize specific features in the 5′ untranslated regions of the mRNAs.
GUN1 was suggested to regulate chloroplast proteostasis (Colombo et al., 2016; Tadini et al., 2016; Llamas et al., 2017). By conducting a proteomic analysis, we identified 128 down-regulated and 14 overaccumulating chloroplast proteins in gun1 in comparison with the wild type upon Lin treatment (Supplemental Table S2; Supplemental Data Set S2). By contrast, in the gun5 mutant, only 15 chloroplast proteins were identified as down-regulated and six as overaccumulated. Furthermore, among the 128 chloroplast proteins that were down-regulated in gun1, a large proportion functions in different aspects of protein metabolism (Fig. 1C). The reduced accumulation of these proteins in gun1 indicates that they are possible targets of GUN1 action and supports an important role of GUN1 in the control of plastid proteostasis. GUN1 was reported to restore the accumulation of the plastid ribosomal protein PRPS1 in gun1 prps1 and gun1 prps21 double mutants, regardless of unchanged PRPS1 transcript abundance (Tadini et al., 2016). We did not observe an overaccumulation of PRPS1 in gun1, neither under control conditions nor upon treatment with Lin (Supplemental Table S2; Supplemental Data Set S1). We recently demonstrated that GUN1 controls plastid protein import when retrograde signaling is affected either by genetic perturbations or by Lin or NF treatment (Wu et al., 2019). Although the exact function of GUN1 in protein import remains to be determined, its interaction with the cpHSC70-1 chaperone suggests that GUN1 may control the fate of newly imported proteins, in that it promotes either folding into a functional protein or degradation by envelope-associated Clp protease (Flores-Pérez et al., 2016).
The cytosolic UPS was demonstrated to degrade unimported chloroplast and mitochondrial precursor proteins (preproteins; Lee et al., 2009; Wrobel et al., 2015; Shanmugabalaji et al., 2018). The transcription factor GLK1 controls the expression of a large number of PhANGs and participates in GUN1-mediated retrograde communication (Kakizaki et al., 2009; Waters et al., 2009). It was shown that GLK1 is degraded by the UPS upon exposure to Lin or NF (Tokumaru et al., 2017), suggesting involvement of the cytosolic UPS in the posttranslational regulation of plastid proteostasis in retrograde signaling. GUN1 is required for efficient plastid protein import under conditions that affect retrograde signaling (Wu et al., 2019). Together, reduced protein import and compromised inhibition of translation would result in overaccumulation of preproteins in the cytosol of gun1 and activation of the cytosolic UPS (Wu et al., 2019). In agreement with this model, the cytosolic chaperones HSP90.1, HSP70-2, and HSP70-4 are strongly overaccumulated at the protein level in the gun1 clpc1 double mutant and the gun1 single mutant upon Lin or NF treatment (Wu et al., 2019). Interestingly, transcriptional up-regulation of these three HSPs was observed in gun1 also under control conditions (Supplemental Table S3), while they do not overaccumulate at the protein level (Supplemental Table S1; Wu et al., 2019). This finding suggests that the expression of these cytosolic chaperones is controlled mainly by posttranscriptional mechanisms.
In summary, the data presented here demonstrate extensive posttranscriptional regulation in the PGE pathway of plastid retrograde signaling and diverse cytosolic responses to plastid translation inhibition. Our work has determined the extent of two posttranscriptional layers of regulation in retrograde signaling: translational regulation (which is at least in part dependent on GUN1) and regulation of protein stability (which is independent of GUN1). It will be interesting to analyze other retrograde signaling pathways (e.g. the TPB pathway) to determine if these regulatory mechanisms are conserved across all pathways.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All experiments were performed with Arabidopsis (Arabidopsis thaliana) accession Columbia. The gun1-101 mutant used in this study was described previously (Wu et al., 2018). The gun5-1 mutant (Mochizuki et al., 2001) was kindly provided by Dr. Bernhard Grimm (Humboldt University, Berlin). For plant growth under aseptic conditions, seeds were surface sterilized with 1.2% (v/v) NaOCl for 10 min, washed five times with sterile water, and sown on 0.5× Murashige and Skoog medium (Murashige and Skoog, 1962) containing 1% (w/v) Suc in petri dishes. The seeds were stratified for 2 d in the dark at 4°C prior to germination. The seeds were first incubated for 8 h in the light to promote germination and then grown in the dark for 5 d followed by 2 d in continuous light. For Lin treatment, the Lin concentration in the medium was 0.5 mm.
RNA Extraction and RNA Gel-Blot Analyses
Total RNA was extracted with the NucleoSpin RNA Plant kit (Macherey-Nagel) according to the manufacturer’s instructions. To completely eliminate contaminating genomic DNA, a second DNase digestion was performed with the TURBO DNA-free kit (Invitrogen). For northern-blot analysis, total RNA samples were separated on 1% (w/v) denaturing agarose gels (containing formaldehyde), blotted onto Hybond-XL nylon membranes (GE Healthcare), and then cross-linked by UV light. Hybridization probes were generated by amplifying cDNA sequences with gene-specific primers, followed by labeling with [α-32P]dCTP with the Megaprime DNA-labeling system (GE Healthcare). Hybridizations were performed at 65°C overnight in Church buffer (Church and Gilbert, 1984). The experiments were repeated two times with similar results. Primers used for the generation of hybridization probes are listed in Supplemental Table S4.
MS and Label-Free Protein Quantification
For proteomic analysis of the wild type, gun1, and gun5 in control conditions or upon treatment with Lin, seedlings were grown for 5 d in the dark followed by 2 d in continuous light without (control) or with 0.5 mm Lin. Protein extraction and on-column digestion with trypsin and MS analyses were conducted essentially as described previously (Wu et al., 2018). In brief, peptides were separated by EASY-nLC 1000 nanoflow HPLC (Proxeon Biosystems) using a C18 LC column (Acclaim PepMap 100, 2 µm, 250 mm; Thermo Fisher Scientific) with a 230-min linear gradient (4.25%–51%, v/v) of acetonitrile and a final peptide elution step for 10 min with 76.5% (v/v) acetonitrile. A Q-Exactive Plus (Thermo Fisher Scientific) high-resolution Orbitrap hybrid mass spectrometer was used and run in positive ion mode. For full MS scans, the following settings were used: resolution, 70,000; automatic gain control target, 3E6; maximum injection time, 100 ms; scan range, 200 to 2,000 mass-to-charge ratio (m/z). For data-dependent MS2, the following settings were used: resolution, 175,000; automatic gain control target, 1E5; maximum injection time, 50 ms; loop count, 15; isolation window, 4 m/z; normalized collision energy, 30. The following data-dependent settings were used: underfill ratio, 1%; apex trigger, off; charge exclusion, unassigned, 1, 5, 5 to 8, >8; peptide match, preferred; exclude isotypes, on; dynamic exclusion, 20 s. Protein identification and quantification was done with the MaxQuant software (version 1.5.8.3), and unique peptides were used for quantification. Postdata analysis and statistical analysis were done with the Perseus software (Tyanova et al., 2016). For statistical analysis, proteins that could be detected in at least two biological replicates of one genotype in at least one condition were kept for further analysis. This resulted in 5,558 proteins used for downstream analysis, and the missing values (i.e. proteins that could not be detected in some samples/replicates) were imputed within the sample with a width of 0.3 (the Gaussian distribution relative to the SD of measured values) and downshift of 1.8 (units of the SD of the valid data) using the Perseus software to enable statistical analysis. Proteins with Student’s t test P < 0.05 and fold change > 2 were considered as differentially expressed between genotypes or treatments. For mRNA (transcript) and protein coanalysis, the organelle-encoded genes were excluded, because oligo(dT) primer had been used for reverse transcription in microarray sample preparation. For data visualization by scatterplots (Figs. 2 and 3), averaged protein expression values were determined as follows. If there were valid values (from detection by MS), the averaged protein expression values come from all valid values. If there were no valid values (i.e. the protein was not detected in all three replicates in the given condition or genotype), the protein expression values come from imputation.
Microarray Analysis
For whole-genome microarray analysis, the wild type and gun1 were grown in control conditions or treated with Lin as described above. After treatment with DNase, total RNA preparations were further purified with the peqGOLD Optipure regent (PEQLAB) to remove polysaccharide contaminations. The purified RNA samples were subjected to a quality check by denaturing RNA gel electrophoresis and assayed in the Agilent 2100 bioanalyzer with the Agilent RNA 6000 Nano Kit (Agilent). Antisense cRNA synthesis and labeling were carried out according to the manufacturer’s instructions (Affymetrix). Labeled antisense cRNA samples were hybridized to Affymetrix Arabidopsis Gene 1.0 ST Arrays (Affymetrix). Hybridization, washing, and scanning were conducted as described in the Affymetrix technical manual. The raw data were analyzed and normalized at the gene level with the RMA algorithm of the Expression Console v1.3 (Affymetrix). Two-way ANOVA was used to identify differentially expressed genes. The Benjamini and Hochberg algorithm was used to control the FDR of multiple tests in R (http://www.r-project.org/). Subsequently, pairwise comparisons between genotypes and conditions were conducted by Tukey’s honestly significant difference tests (using the TukeyHSD function in R). Genes with an FDR-adjusted P < 0.05 and fold change > 2 were identified as differentially expressed genes.
Protein Extraction and Western-Blot Analyses
Total cellular protein was extracted with a phenol-based method (Cahoon et al., 1992) and quantified with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Protein samples were separated by SDS-PAGE (15% [w/v]) and blotted onto polyvinylidene difluoride membranes. Immunoblot detection was performed using the ECL system (GE Healthcare) with specific antibodies. Signals were detected with the G:box Chemi Imaging System (Syngene). Antibodies were obtained from commercial suppliers (actin from Sigma-Aldrich; LHCA4, LHCB1, RBCS, RPL4, and cpHSC70 from Agrisera). The experiments were repeated two times with similar results.
Polysome Analyses
For isolation of polysomes, plants were germinated and grown under control conditions or treated with Lin exactly as described for the microarray and proteomic experiments. Polysome isolation and control treatments with puromycin were performed essentially as described (Barkan, 1998; Wu et al., 2018). Aliquots of 5 µL per fraction were analyzed by northern blotting as described above. The experiments were repeated two times with similar results.
Statistical Analysis
To identify genes with significantly altered expression in microarray experiments, two-way ANOVA was performed with the Benjamini and Hochberg algorithm to control the FDR of multiple tests. The post pairwise comparisons between genotypes and conditions were done by Tukey’s honestly significant difference tests. For proteomic analysis to identify differentially accumulated proteins, two-tailed unpaired Student’s t test was used when a pair of conditions/genotypes was compared.
Data Availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011759. The microarray data have been deposited in the Gene Expression Omnibus under the accession number GSE122667.
Accession Numbers
The sequence data from this article can be found in The Arabidopsis Information Resource or GenBank/EMBL database under the following accession numbers: GUN1 (AT2G31400), GUN5 (AT5G13630), PSRP5 (AT3G56910), RPL4 (AT1G07320), CA1 (AT3G01500), LOX2 (AT3G45140), PsaG (AT1G55670), PsaL (AT4G12800), PsaD (AT4G02770), CaSR (AT5G23060), rbcL (ATCG00490), RBCS1A (AT1G67090), RBCS1B (AT5G38430), RBCS2B (AT5G38420), RBCS3B (AT5G38410), LHCA1 (AT3G54890), LHCA2 (AT3G61470), LHCA3 (AT1G61520), LHCA4 (AT3G47470), LHCA5 (AT1G45474), LHCA6 (AT1G19150), LHCB1.1 (AT1G29920), LHCB1.2 (AT1G29910), LHCB1.3 (AT1G29930), LHCB1.4 (AT2G34430), LHCB1.5 (AT2G34420), LHCB2.1 (AT2G05100), LHCB2.2 (AT2G05070), LHCB2.3 (AT3G27690), LHCB3 (AT5G54270), LHCB4.1 (AT5G01530), LHCB4.2 (AT3G08940), LHCB4.3 (AT2G40100), LHCB5 (AT4G10340), LHCB6 (AT1G15820), NAD9 (ATMG00070), NAD5B (ATMG00665), NAD1A (ATMG01275), NAD2B (ATMG01320), NAD7 (ATMG00510), COX2 (ATMG00160), ORF25 (ATMG00640), COB (ATMG00220), and RPS3 (ATMG00090).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Lin specifically represses plastid protein synthesis.
Supplemental Figure S2. The expression of Lipoxygenase 2 (LOX2) is repressed in both the wild type and gun1 upon Lin treatment.
Supplemental Figure S3. Expression of LHCA, LHCB, and RBCS genes in response to Lin treatment in the wild type and the gun1-101 mutant.
Supplemental Figure S4. Puromycin-treated controls for the polysome profiles of LHCA4, RBCS, PSRP5, and RPL4 in the wild type shown in Figure 6.
Supplemental Figure S5. Puromycin-treated controls for the polysome profiles of LHCA4, RBCS, PSRP5, and RPL4 in gun1 shown in Figure 7.
Supplemental Table S1. Differentially expressed proteins in gun1 compared with the wild type under control conditions.
Supplemental Table S2. Plastid-localized proteins that overaccumulate in gun1 relative to the wild type upon Lin treatment.
Supplemental Table S3. Differentially expressed genes identified by microarray analysis in gun1 compared with the wild type under control conditions.
Supplemental Table S4. List of oligonucleotides used in this study.
Supplemental Data Set S1. MS data of proteomic analyses of the wild type, the gun1 mutant, and the gun5 mutant under control conditions and under Lin treatment, and significantly differentially expressed proteins identified between the wild type and the mutants.
Supplemental Data Set S2. The 128 plastid-localized proteins that accumulate to lower levels in gun1 compared with the wild type under Lin treatment and their response to Lin treatment.
Supplemental Data Set S3. Data used for scatterplots in Figures 2 and 3.
Supplemental Data Set S4. Differentially expressed genes between the wild type and the gun1 mutant under Lin treatment identified by microarray analysis.
Supplemental Data Set S5. HiToP genes identified in the wild type and gun1.
Acknowledgments
We thank Dr. Bernhard Grimm (Humboldt University Berlin) for gun5-1 mutant seeds.
Footnotes
This work was supported by the Max Planck Society and a grant from the Deutsche Forschungsgemeinschaft (SFB TR175 to R.B.).
Articles can be viewed without a subscription.
References
- Balazadeh S, Jaspert N, Arif M, Mueller-Roeber B, Maurino VG (2012) Expression of ROS-responsive genes and transcription factors after metabolic formation of H2O2 in chloroplasts. Front Plant Sci 3: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1998) Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol 297: 38–57 [Google Scholar]
- Bradbeer JW, Atkinson YE, Borner T, Hagemann R (1979) Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesized RNA. Nature 279: 816–817 [Google Scholar]
- Butow RA, Avadhani NG (2004) Mitochondrial signaling: The retrograde response. Mol Cell 14: 1–15 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Shanklin J, Ohlrogge JB (1992) Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc Natl Acad Sci USA 89: 11184–11188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L (2013) Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64: 559–582 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Tadini L, Peracchio C, Ferrari R, Pesaresi P (2016) GUN1, a jack-of-all-trades in chloroplast protein homeostasis and signaling. Front Plant Sci 7: 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottage A, Mott EK, Kempster JA, Gray JC (2010) The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J Exp Bot 61: 3773–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz MG, Hernández-Verdeja T, Kremnev D, Crawford T, Dubreuil C, Strand Å (2018) Redox regulation of PEP activity during seedling establishment in Arabidopsis thaliana. Nat Commun 9: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey LF, Nguyen TT, Allen GC, Thompson WF (1994) Light modulation of ferredoxin mRNA abundance requires an open reading frame. Plant Cell 6: 1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey LF, Petracek ME, Nguyen TT, Hansen ER, Thompson WF (1998) Light regulation of Fed-1 mRNA requires an element in the 5′ untranslated region and correlates with differential polyribosome association. Plant Cell 10: 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Dickey LF, White MJ, Thompson WF (1989) Cis-acting elements for light regulation of Pea Ferredoxin I gene expression are located within transcribed sequences. Plant Cell 1: 691–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves P, Pecqueur C, Ransy C, Esnous C, Lenoir V, Bouillaud F, Bulteau AL, Lombès A, Prip-Buus C, Ricquier D, et al. (2014) Mitochondrial retrograde signaling mediated by UCP2 inhibits cancer cell proliferation and tumorigenesis. Cancer Res 74: 3971–3982 [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú, Bédard J, Tanabe N, Lymperopoulos P, Clarke AK, Jarvis P (2016) Functional analysis of the Hsp93/ClpC chaperone at the chloroplast envelope. Plant Physiol 170: 147–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M, Finkemeier I (2012) Plant mitochondrial retrograde signaling: Post-translational modifications enter the stage. Front Plant Sci 3: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs L, Schmitz J, Flügge UI, Häusler RE (2012) The mysterious rescue of adg1-1/tpt-2—an Arabidopsis thaliana double mutant impaired in acclimation to high light—by exogenously supplied sugars. Front Plant Sci 3: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Verdeja T, Strand Å (2018) Retrograde signals navigate the path to chloroplast development. Plant Physiol 176: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA (1997) A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol 17: 1110–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Che FS, Terauchi R, Inaba T (2009) Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol 151: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Kremnev D, Blanco NE, de Dios Barajas López J, Fernández AP, Tellgren-Roth C, Kleine T, Small I, Strand A (2012a) The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J 70: 279–291 [DOI] [PubMed] [Google Scholar]
- Kindgren P, Norén L, López JdeD, Shaikhali J, Strand A (2012b) Interplay between Heat Shock Protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol Plant 5: 901–913 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Krupinska K, Braun S, Nia MS, Schäfer A, Hensel G, Bilger W (2019) The nucleoid-associated protein WHIRLY1 is required for the coordinate assembly of plastid and nucleus-encoded proteins during chloroplast development. Planta 249: 1337–1347 [DOI] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, Lee S, Jürgens G, Hwang I (2009) Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21: 3984–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA (2006) Mitochondrial retrograde signaling. Annu Rev Genet 40: 159–185 [DOI] [PubMed] [Google Scholar]
- Llamas E, Pulido P, Rodriguez-Concepcion M (2017) Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genet 13: e1007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, Noshi M, Tanouchi A, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S (2012) H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J Biol Chem 287: 11717–11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P, Pursiheimo S, Hou CX, Tyystjarvi T, Aro EM (2003) Multiple effects of antibiotics on chloroplast and nuclear gene expression. Funct Plant Biol 30: 1097–1103 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Oelmüller R. (1989) Photooxidative destruction of chloroplasts and its effect on nuclear gene-expression and extraplastidic enzyme levels. Photochem Photobiol 49: 229–239 [Google Scholar]
- Oelmüller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H (1986) Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168: 482–492 [DOI] [PubMed] [Google Scholar]
- Oelze ML, Muthuramalingam M, Vogel MO, Dietz KJ (2014) The link between transcript regulation and de novo protein synthesis in the retrograde high light acclimation response of Arabidopsis thaliana. BMC Genomics 15: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA (1987) The mitochondrial genotype can influence nuclear gene expression in yeast. Science 235: 576–580 [DOI] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Huber SC, Thompson WF (1997) Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell 9: 2291–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Nguyen TT, Gatz C, Sowinski DA, Allen GC, Thompson WF (1998) Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proc Natl Acad Sci USA 95: 9009–9013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo E, Godoy Herz MA, Fuchs A, Reifer D, Fuller J, Yanovsky MJ, Simpson C, Brown JW, Barta A, Kalyna M, et al. (2014) A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar AD, Horwitz BA, Elliott RC, Thompson WF, Briggs WR (1988) Light effects on several chloroplast components in norflurazon-treated pea seedlings. Plant Physiol 88: 340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugabalaji V, Chahtane H, Accossato S, Rahire M, Gouzerh G, Lopez-Molina L, Kessler F (2018) Chloroplast biogenesis controlled by DELLA-TOC159 interaction in early plant development. Curr Biol 28: 2616–2623.e5 [DOI] [PubMed] [Google Scholar]
- Shumbe L, Bott R, Havaux M (2014) Dihydroactinidiolide, a high light-induced β-carotene derivative that can regulate gene expression and photoacclimation in Arabidopsis. Mol Plant 7: 1248–1251 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Mühlhaus T, Schroda M, Masiero S, Pribil M, et al. (2016) GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170: 1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumaru M, Adachi F, Toda M, Ito-Inaba Y, Yazu F, Hirosawa Y, Sakakibara Y, Suiko M, Kakizaki T, Inaba T (2017) Ubiquitin-proteasome dependent regulation of the GOLDEN2-LIKE 1 transcription factor in response to plastid signals. Plant Physiol 173: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740 [DOI] [PubMed] [Google Scholar]
- Vogel MO, Moore M, König K, Pecher P, Alsharafa K, Lee J, Dietz KJ (2014) Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 26: 1151–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF (2008) Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell 20: 552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, Op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E, et al. (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. (2015) Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524: 485–488 [DOI] [PubMed] [Google Scholar]
- Wu GZ, Chalvin C, Hoelscher M, Meyer EH, Wu XN, Bock R (2018) Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol 176: 2472–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GZ, Meyer EH, Richter AS, Schuster M, Ling Q, Schöttler MA, Walther D, Zoschke R, Grimm B, Jarvis RP, et al. (2019) Control of retrograde signalling by protein import and cytosolic folding stress. Nat Plants 5: 525–538 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EEK, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Yasumura Y, Moylan EC, Langdale JA (2005) A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell 17: 1894–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011759. The microarray data have been deposited in the Gene Expression Omnibus under the accession number GSE122667.