CRISPR-based assessment reveals nonredundant functions and diversified evolution of enzymes in the gibberellin metabolic pathway in rice.
Abstract
Gibberellin (GA) functions as an essential natural regulator of growth and development in plants. For each step of the GA metabolic pathway, different copy numbers can be found in different species, as is the case with the 13 genes across four enzymatic steps in rice (Oryza sativa). A common view is that such gene duplication creates homologs that buffer organisms against loss-of-function (LOF) mutations. Therefore, knockouts of any single homolog might be expected to have little effect. To test this question, we generated clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) knockouts for these homologs and measured effects on growth and reproduction. Surprisingly, we report here that there is consistently one or more essential gene at each enzymatic step, for which LOF mutation induces death or sterility—suggesting that the GA pathway does not have a redundancy route and that each gene family is essential for GA metabolism. In most of these genes from the same gene family, we observed defects in plant height and infertility, suggesting that the duplicated members retain functions related to GA synthesis or degradation. We identified both subfunctionalization of the three recently diversified homologs OsKO1, OsKO2, and OsKO5 and neofunctionalization in OsKO3 and OsKO4. Thus, although the function of each step is conserved, the evolution of duplicates in that step is diversified. Interestingly, the CRISPR/Cas9 lines at the SD1 locus were typically sterile, whereas the natural sd1 mutants, related to the “Green Revolution” in rice, show normal setting rates. Collectively, our results identify candidates for control of GA production and provide insight into the evolution of four critical gene families in plants.
Gibberellin (GA) is a major phytohormone that regulates growth and developmental processes in plants, including seed germination, internode elongation, and the formation of flowers and seeds (Davies, 1995). GA-deficient mutants of crops have been widely used because dwarfism improves lodging resistance and increases yields. The miracle rice (Oryza sativa) cultivar IR8, which enabled dramatic yield increases and helped to avert food shortages in the “Green Revolution,” has a short height because of a mutation in the SD1 (GA20ox2) gene (Monna et al., 2002; Sasaki et al., 2002; Spielmeyer et al., 2002). The rice cultivar Tan-Ginbozu, which was considered the most productive cultivar in Japan, has a single nucleotide substitution in the d35 (KO2) gene (Itoh et al., 2004). These profound effects of single genes naturally lead to the question as to what roles are played by the rest of the specific genes in the GA metabolic pathway.
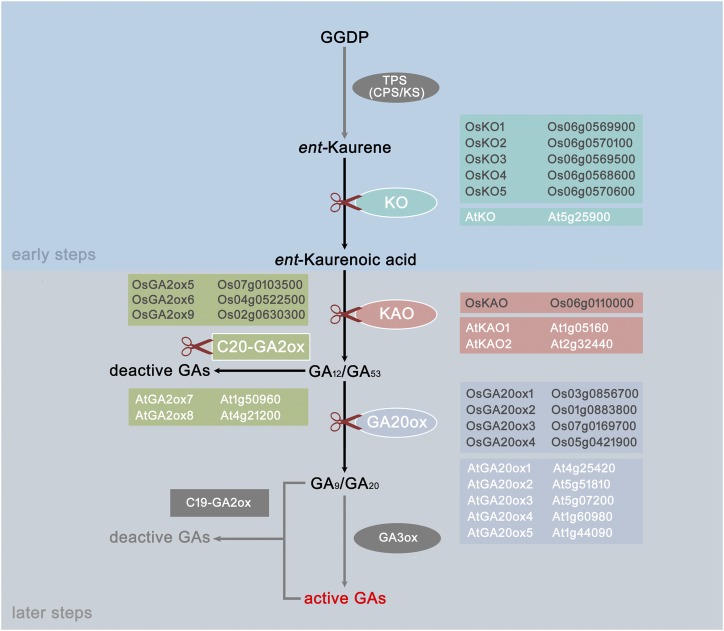
In angiosperms plants, GA synthesis starts from geranylgeranyl diphosphate, which is converted to ent-kaurene acid byent-copalyl diphosphate synthase, ent-kaurene synthase, and ent-kaurene oxidase (KO; Fig. 1). In addition to generating the precursor of GA, these early steps also provide substrates for production of phytoalexin compounds, such as brassinosteroids. In later steps, ent-kaurene acid is converted to bioactive GAs sequentially by ent-kaurenoic acid oxidase (KAO), GA20ox, and GA3ox (Fig. 1). Deactivation of GA is mostly catalyzed by GA2ox. C20-GA2ox uses C20-GAs as substrates and converts them to inactive forms (Fig. 1).
Figure 1.
The GA metabolism pathways in rice. Ellipses represent biosynthesis enzymes; rectangles represent breakdown enzymes. Scissors mark the families mentioned in this article with corresponding genes in rice.
In Arabidopsis, systemic analysis of the gene families of enzymes at particular steps in GA metabolism have been achieved through a comprehensive collection of T-DNA insertion loss-of-function (LOF) mutants (Schomburg et al., 2003; Rieu et al., 2008; Plackett et al., 2012; Regnault et al., 2014). However, in rice and other crop species, with their larger genomes and greater technical challenges, mutants are available for fewer genes and much less is known. For rice specifically, previous studies have included spontaneous mutants (Sasaki et al., 2002), overexpression lines (Oikawa et al., 2004; Qin et al., 2013; Shan et al., 2014a), and TOS 17 retrotransposon knockouts (Sakamoto et al., 2004). Although the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9)system has great potential to provide systemic assessment of metabolic pathway steps in rice and other plants, to our knowledge, this has not been accomplished previously.
The basic steps of the GA synthesis pathway are highly conserved across angiosperms. However, there are remarkable differences even between closely-related species in the number of homologous genes present at each enzymatic step. In rice, for example, there are up to five genes at one step in the GA synthesis pathway (Fig. 1). Such diversification raises questions about the functions of these new homologs. Do these new genes act in the same roles as the ancestral forms or have they undergone subfunctionalization and neofunctionalization?
A common view is that such gene duplication increases the robustness of organisms against LOF mutations in single genes. Many important enzymes in plants are represented by two or more genes, or homologs, in the genome. In many of these cases, knocking out one or the other of the homolog does not affect the plant phenotype, whereas knocking out both homologs results in a profound defect, such as in the case of fatty acid desaturases 3, 7, and 8 (Routaboul et al., 2000) and KAO1 and 2 in Arabidopsis (Regnault et al., 2014). Such cases are indicative of substantial redundancy in gene function. Theoretical models suggest that full redundancy of genes cannot persist over evolutionary time (Moore and Purugganan, 2005). However, comparative assessment of such recent homologs has been difficult before the advent of the CRISPR/Cas9 system.
In this study, to better understand the dynamics among genes at steps in GA synthesis, we employ CRISPR/Cas9 (Jinek et al., 2012) to create knockout mutants of each gene at four steps in the GA metabolic pathway and measure phenotypic alternations under common garden conditions to systematically reveal the functions of the homologs in these multigene families. Specifically, we ask the following questions: (1) Does rice have more than one pathway in GA metabolism, and which of these GA metabolic genes are essential for normal development? (2) Do all members of a gene family perform a similar function, and does the retention of duplicated genes fit the patterns of redundancy, subfunctionalization, or neofunctionalization? (3) Does the retention mode of the same gene family follow the same rules in rice compared to other species? (4) Is the phenotype of a mutant an accurate reflection of gene function? And, finally: (5) Do such analyses of GA have potential benefit in agricultural application?
RESULTS
We first identified 13 putative genes from four families in the GA metabolic pathway (Fig. 1) and obtained CRISPR/Cas9 full knockout lines of 11 of these genes (Table 1), including four out of five genes from the KO family (which catalyzes the sequential oxidation on C-19 to produce ent-kaurenoic acid), the sole gene from the KAO family (which converts ent-kaurenoic acid to GA12), three out of four genes from the GA20ox family (converting GA12 to GA4), and three out of three genes from the C20-GA2ox family. For nine genes, we phenotyped T1 generations and for three additional genes we phenotyped the T0 generation (Table 1). For the two genes, OsKO2 and OsGA20ox1, that could not be knocked out after three rounds of transformation, this was consistent with possible essential roles in the differentiation of callus. During each repeat of the transformation experiments, both the sequences of targeted loci and the phenotypes of all the transformants obtained were identical to wild type. We also confirmed the right spacers in those T0 lines, so we speculate that either heterozygous or homozygous, the successful knockout of any one of the two genes might induce failure in transformation.
Table 1. Genotypes of full knockout lines (homozygous for CRISPR allele).
| Gene Symbol | Locus Name | Background | Genotype | Generation |
|---|---|---|---|---|
| OsKO1 | Os06g0569900 | Kasalath Wuyungeng24 | Insufficient material for sampling, unable to obtain | T0 |
| OsKO2 | Os06g0570100 | Kasalath Wuyungeng24 | Unable to obtain | |
| OsKO3 | Os06g0569500 | Wuyungeng24 | Full knockout | T1 |
| OsKO4 | Os06g0568600 | Kasalath | Full knockout | T1 |
| OsKO5 | Os06g0570600 | Kasalath | Full knockout | T1 |
| OsKAO | Os06g0110000 | Kasalath Wuyungeng24 | Full knockout | T0 |
| OsGA20ox1 | Os03g0856700 | Kasalath Wuyungeng24 | Unable to obtain | |
| OsGA20ox2 | Os01g0883800 | Wuyungeng24 | Full knockout | T1 |
| OsGA20ox3 | Os07g0169700 | Kasalath | Full knockout | T1 |
| OsGA20ox4 | Os05g0421900 | Wuyungeng24 | Full knockout | T1 |
| OsGA2ox5 | Os07g0103500 | Kasalath Wuyungeng24 | Full knockout | T1 |
| OsGA2ox6 | Os04g0522500 | Kasalath Wuyungeng24 | Full knockout | T1 |
| OsGA2ox9 | Os02g0630300 | Kasalath | Full knockout | T0 |
Assessment of KO Homologs
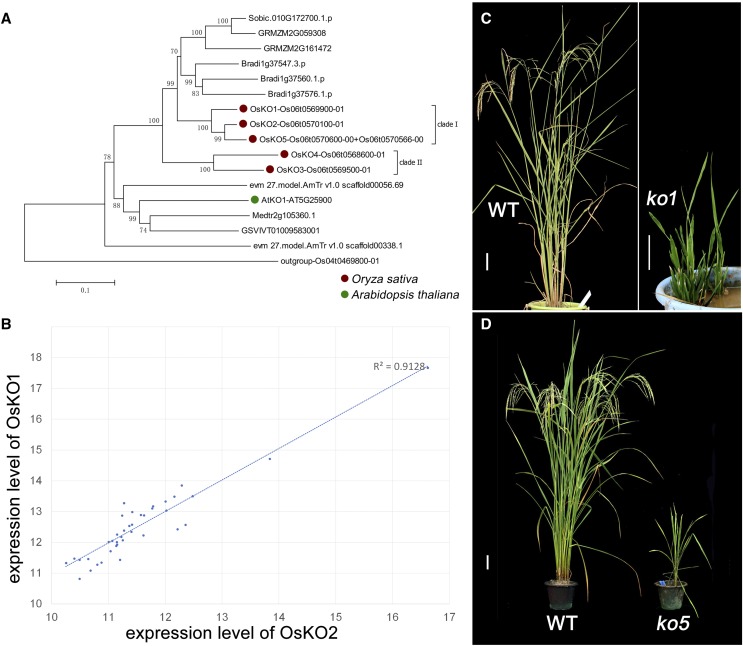
We began with the step performed by KO, the enzyme that converts ent-kaurene to kaurenoic acid (Fig. 1). As reported in Wang et al. (2012) and Mafu et al. (2016), the KO gene family in rice has two clades, with Clade I consisting of OsKO1, OsKO2, OsKO5 and Clade II consisting of OsKO3 and OsKO4 (Fig. 2A). Our data show that rice has two more KO homologs than is typical for plants in the Poaceae, four more KO homologs than monocots outside of the Poaceae, and three more KO homologs than the average dicot (Fig. 2A; Supplemental Table S1). In the monocots, especially in Oryzeae, there have been multiple independent duplication events (Fig. 2A; Supplemental Fig. S1). Our comparison of these clades across the angiosperm phylogeny suggest different evolutionary patterns of KO homologs in monocots and dicots, in that KO shows a tendency toward the single-copy state in dicots, such as in the case of Arabidopsis, whereas duplication events have occurred with some frequency in monocots.
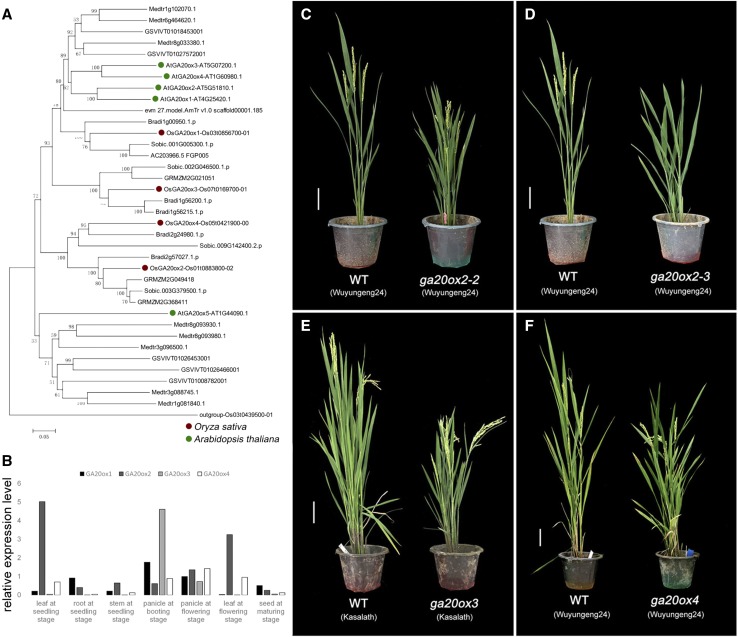
Figure 2.
Phylogenetic analysis, knockout phenotypes, and expression patterns of OsKO genes. Expression levels of KO homologs in rice. A, Phylogenetic analysis of protein sequences of KO encompassing paralogs from Amborella trichopoda, Z. mays, Sorghum bicolor, Brachypodium distachyon, O. sativa, Vitis vinifera, Medicago truncatula, and Arabidopsis. Scale = 0.1 amino acid substitutions per site. The “Clade I” bracket include three genes of Clade I OsKO. B, Significant coexpression (r2 = 0.9128) of OsKO1 (y axis) and OsKO2 (x axis) in 39 tissues of rice from Genevestigator database. C, Phenotype of 5-month–old wild-type (WT) lines and severely dwarfed osko1. Scale bar = 10 cm. D, Dwarfed phenotype of 4-month–old osko5. Scale bar = 10 cm.
To assess the relative roles of these genes, we used CRISPR/Cas9 to modify the sequences (Table 1). Our motivating question, based on the AtKO1 T-DNA insert mutant in Arabidopsis that exhibits severe dwarfism (Table 2), is whether single or multiple homologs among the five rice KO genes perform the same function as AtKO1. In other words, is there functional redundancy or functional differentiation? The one gene that would not yield mutants after three rounds of transformation was OsKO2 (CYP701A6) of Clade I, which suggests the essentiality of OsKO2 and is consistent with the report of the natural mutant d35, in which only a single nucleotide substitution induces semidwarfism (Itoh et al., 2004) and a previous reported OsKO2 mutant, where an insertion in exon 4 induces severe dwarfism and death (Sakamoto et al., 2004). Our spacer target in the second exon of OsKO2 may have induced severe deficiency that led to failure to obtain the transformant. In contrast, osko1 and osko5, which are also of Clade I, generally exhibited different extents of dwarfism. Knockout in OsKO1 was more severe (Fig. 2C), whereas knockout of OsKO5 (Fig. 2D) was less severe. Notably, even the osko5 mutants died before flowering (Table 2; Fig. 2B).
Table 2. An overview of KO, KAO, GA20ox, and C20-GA2ox mutants in O. sativa and Arabidopsis.
| Mutant | O. sativa | Arabidopsis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | Id | Mutation Type | Phenotype | Reference | Symbol | Id | Mutation Type | Phenotype | Reference | |
| KO | OsKO1 | Os06g0569900 | Cas9 knockout | Severe dwarf, sterile | This result | AtKO | AT5G25900 | 35S overexpression | Similar to wild type | (Swain et al., 2005) |
| OsKO2 | Os06g0570100 | Cas9 knockout | N/A | This result | ||||||
| Spontaneous d35: a single nucleotide substitution | Semidwarf | (Itoh et al., 2004) | ||||||||
| Tos17 retrotransposon: insertion into exon 4 | Severe dwarf, sterile | (Sakamoto et al., 2004) | ||||||||
| OsKO3 | Os06g0569500 | Cas9 knockout | Infected leaves, low setting rate | This result | A single nucleotide substitution introduces a stop codon | Severe dwarf | (Helliwell et al., 1998) | |||
| OsKO4 | Os06g0568600 | Cas9 knockout | Low setting rate | This result | ||||||
| OsKO5 | Os06g0570600a | Cas9 knockout | Severe dwarf, sterile | This result | ||||||
| KAO | OsKAO | Os06g0110000 | Cas9 knockout | Severe dwarf, sterile | This result | AtKAO1 | AT1G05160 | T-DNA insertion knockout | Similar to wild type (severe dwarf double knockout) | (Regnault et al., 2014) |
| Tos17 retrotransposon: insertion into exon 5 or 6 | Severe dwarf, sterile | (Sakamoto et al., 2004) | AtKAO2 | AT2G32440 | T-DNA insertion knockout | Similar to wild type (severe dwarf double knockout) | (Regnault et al., 2014) | |||
| GA20ox | OsGA20ox1 | Os03g0856700 | Cas9 knockout | N/A | This result | AtGA20ox1 | AT4G25420 | Spontaneous: a single nucleotide deletion | Alpine dwarf | (Luo et al., 2015) |
| 35S overexpression/ RNAi | Tall, low setting rate/ semidwarf | (Oikawa et al., 2004) | T-DNA insertion knockout | Semidwarf | (Rieu et al., 2008; Plackett et al., 2012) | |||||
| OsGA20ox2 | Os01g0883800 | Cas9 knockout | Semidwarf, low setting rate | This result | AtGA20ox2 | AT5G51810 | T-DNA insertion knockout | Delayed flowering, shorter siliques | (Rieu et al., 2008; Plackett et al., 2012) | |
| Spontaneous sd1: 383-bp deletion | Semidwarf | (Sasaki et al., 2002) | AtGA20ox3 | AT5G07200 | Ethyl-methane sulfonate Mutagenesis truncated protein and amino acid substitution | Little amplification on phenotype of ga20ox1/2 | (Plackett et al., 2012) | |||
| OsGA20ox3 | Os07g0169700 | Cas9 knockout | Semidwarf, low setting rate | This result | ||||||
| 35S overexpression/ RNAi | Tall, susceptible/ semidwarf, low setting rate | (Qin et al., 2013) | AtGA20ox4 | AT1G60980 | Natural 1-bp deletion | No altered phenotype | (Plackett et al., 2012) | |||
| OsGA20ox4 | Os05g0421900 | Cas9 knockout | Semidwarf, low setting rate | This result | AtGA20ox5 | AT1G44090 | T-DNA insertion knockout | No altered phenotype | (Plackett et al., 2012) | |
| C20-GA2ox | OsGA2ox5 | Os07g0103500 | Cas9 knockout | Large and chalky seeds, low setting rate | This result | AtGA2ox7 | AT1G50960 | T-DNA insertion knockout | GA excessive | (Schomburg et al., 2003) |
| 35S overexpression | Severe dwarf, irregularly shaped and small seeds | (Shan et al., 2014a) | 35S overexpression | Dwarf | (Schomburg et al., 2003) | |||||
| OsGA2ox6 | Os04g0522500 | Cas9 knockout | Large and chalky seeds | This result | ||||||
| 35S overexpression | Dwarf, normal flowering and seed production | (Huang et al., 2010) | AtGA2ox8 | AT4G21200 | T-DNA insertion knockout | GA excessive | (Schomburg et al., 2003) | |||
| OsGA2ox9 | Os02g0630300 | Cas9 knockout | Low setting rate | This result | 35S overexpression | Dwarf | (Schomburg et al., 2003) | |||
| 35S overexpression | Semidwarf, slightly decreased fertility | (Lo et al., 2008) | ||||||||
Note that Os06g0570600 and Os06g0570566 have been considered to be “OsKOL1” by Itoh et al. (2004). Here, we call it “OsKO5.”
Together, these results suggest that OsKO1, OsKO2, and OsKO5 are all essential for synthesis of GA and they seem to act together to provide precursors for GA synthesis, similar to AtKO1 in Arabidopsis (Table 2). To assess relative interactions among these KO proteins, we assessed expression data from GENEVESTIGATOR (https://genevestigator.com/gv/). From those data, we found a strong correlation (r2 = 0.91) between expression levels of OsKO1 and OsKO2 in 39 anatomical parts (Pearson correlation coefficient = 0.93, Fig. 2D). Combining phenotypes and expression data together, we propose two hypotheses: (1) OsKO1, OsKO2, and OsKO5 inherit different functional parts of the ancient KO separately, but all of the subfunctions are important and that is why loss of any one of these three KO genes induces severe dwarfism or death. (2) The three OsKO genes, which share similar expression patterns, might form a heterotrimer and act together, such that loss of any one of these three KO induces breakdown of the heterotrimer and leads to final loss of KO function. These two hypotheses provide direction for further assessment of gene function.
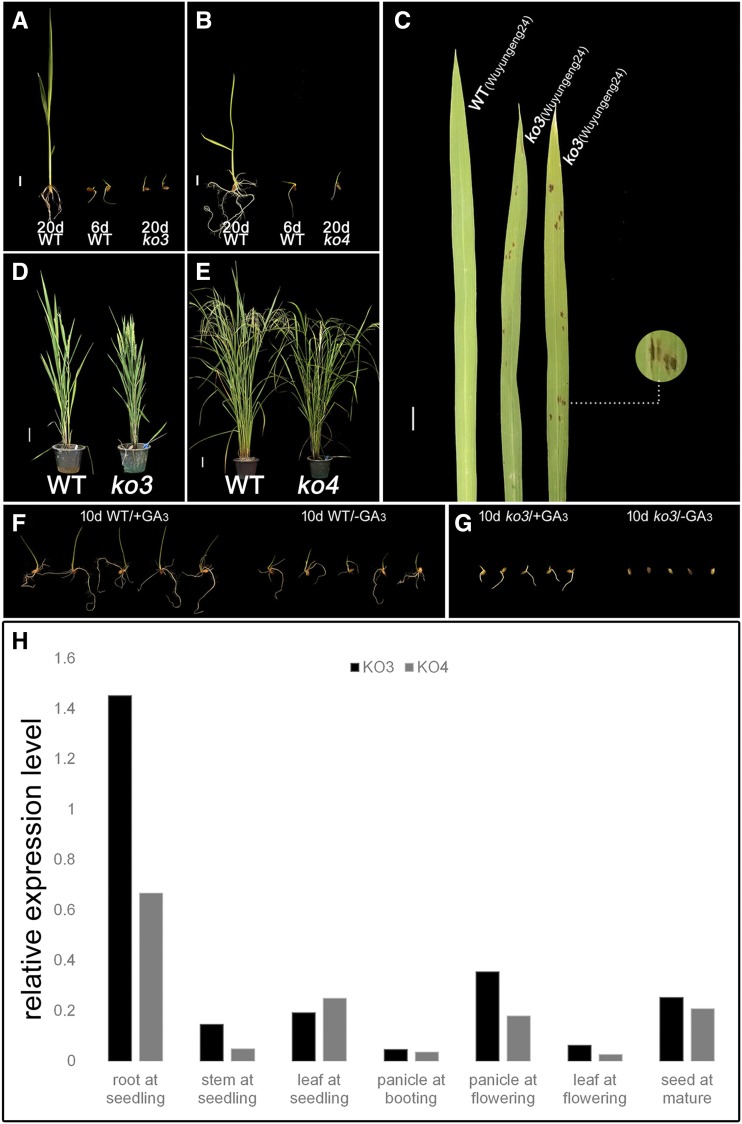
In contrast, osko3 and osko4 in Clade II did not exhibit plant height-related phenotypes of GA deficiency (Fig. 3, D and E), but both of them exhibited delayed germination and the osko3 knockout exhibited slower growth of the seminal root (Fig. 3, A and B). These results are consistent with our reverse transcription quantitative PCR (RT-qPCR) results that both of the KO genes in Clade II are mainly expressed in the root at the seedling stage (Fig. 3H) and suggest that the functions of these two genes contribute to seed germination and early root development in rice. Surprisingly, the delayed growth of these mutants was temporary and was later compensated such that they achieved normal flowering time despite the delayed germination. To assess whether these knockouts could be rescued by exogenous GA, we applied GA3 to seeds of the osko3 and osko4 lines and found that it partially rescued the root growth defect (Fig. 3, F and G), which demonstrated that the defect in osko3 and osko4 is related to the GA pathway.
Figure 3.
Analysis of KO lines from Clade II. A, Delayed germination of osko3 and −4. Scale bar = 1 cm. B, Delayed germination of osko3 and −4. Scale bar = 1 cm. C, Infection scabs on leaves of osko3. Scale bar = 1 cm. D and E, Similar phenotypes of 4-month–old (D) osko3 and (E) osko4 compared to wild type. Scale bar = 10 cm. F and G, 150 mg/L GA3 application complementation experiment of (F) wild type and (G) osko3. H, Expression of Clade II KO genes in different tissues. The data are means of three replicates.
Interestingly, the leaf lesion phenotype was observed on all the osko3 mutants (Fig. 3C), which supports the hypothesis of Itoh et al. (2004) that OsKO3 and OsKO4 may take part in phytoalexin biosynthesis because their expression was promoted by UV irradiation and/or elicitor treatment, but introduction of OsKO3 into the semidwarfed OsKO2 spontaneous mutant cannot rescue its dwarf phenotype. Taken together, these phenotypes of osko3 demonstrate that the neofunctionalization of disease resistance has arisen in Clade II. In other words, Clade II OsKOs have not only retained part of the function of GA-synthesis (especially for seed germination and early root development that has a substantially functional differentiation from Clade I), but have also evolved novel functions related to immune function at the same time.
Assessment of KAO
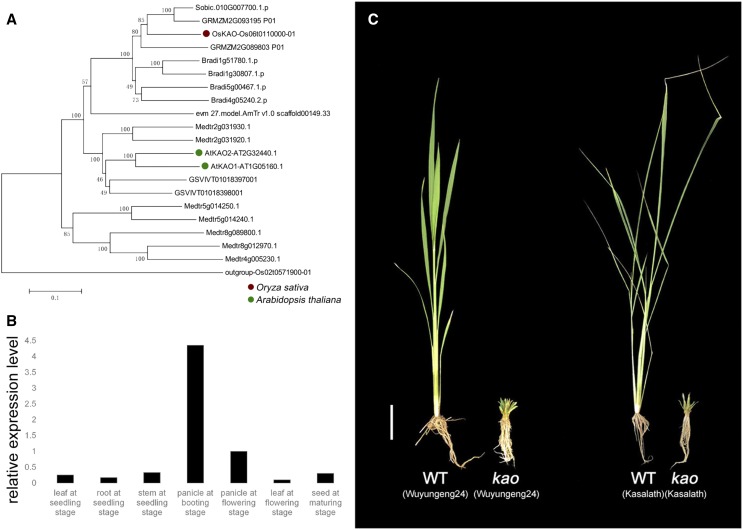
There is one KAO gene in rice, whereas Arabidopsis (Arabidopsis thaliana) has two homologs (Table 2). In Arabidopsis, atkao1− and atkao2− single gene knockouts are similar to wild type, while atkao1 and atkao2 double mutants exhibit typical GA-deficient severe dwarfism. There are overlapping roles of AtKAO1 and AtKAO2 throughout Arabidopsis development, suggesting functional redundancy of these two members (Regnault et al., 2014). For the sole copy of OsKAO in rice, we constructed mutants with two different knockout genotypes and both of them exhibited severe dwarfism and sterility (Fig. 4C), which is consistent with double mutants of atkao1–atkao2 in Arabidopsis (Table 2). Our RT-qPCR analysis revealed that OsKAO is expressed in most plant tissues, especially in panicles at the booting stage (Fig. 4B).
Figure 4.
Phylogenetic analysis, knockout phenotypes, and expression patterns of OsKAO genes. A, Phylogenetic analysis of protein sequences of KAO encompassing paralogs from Amborella trichopoda, Z. mays, Sorghum bicolor, Brachypodium distachyon, O. sativa, Vitis vinifera, Medicago truncatula, and Arabidopsis. Scale = 0.1 amino acid substitutions per site. B, Expression pattern of OsKAO genes. The data are means of three replicates. C, Severely dwarfed phenotypes of 40-d–old oskao. Scale bar = 5 cm.
Assessment of GA20ox Homologs
There are five and four homologs of GA20ox in Arabidopsis (AtGA20ox1-5) and rice (OsGA20ox1-4), respectively (Fig. 5). In the phylogenic tree (Fig. 4A), two distinct clades, Clade I and Clade II, were found. Clade I is unique to dicots and missing in monocots, and includes one member, AtGA20ox5, in Arabidopsis. In Clade II, AtGA20ox1-4 were grouped together, whereas the four members from rice have greater divergence, suggesting frequent copy-number variations in different species in this family (Fig. 5A). In Arabidopsis GA20ox genes, the T-DNA insertion mutant atga20ox1 is semidwarfed (Rieu et al., 2008; Plackett et al., 2012); atga20ox2 has delayed flowering time and only a slight decrease in plant height (Rieu et al., 2008); and atga20ox3 has no obvious phenotypic changes. Interestingly, the triple mutant atga20ox1/2/3 exhibits extreme dwarfism, strongly delayed flowering time and nearly complete infertility, which is similar to a completely GA-deficient phenotype (Plackett et al., 2012), indicating partial functional redundancy among these genes. In contrast, the deficient mutants of other two genes, AtGA20ox4 and AtGA20ox5, have no observed phenotypes (Plackett et al., 2012).
Figure 5.
Phylogenetic analysis, knockout phenotypes, and expression patterns of GA20ox genes. A, Phylogenetic analysis of protein sequences of GA20ox encompassing paralogs from Amborella trichopoda, Z. mays, Sorghum bicolor, Brachypodium distachyon, O. sativa, Vitis vinifera, Medicago truncatula, and Arabidopsis. Scale = 0.05 amino acid substitutions per site. B, Expression pattern of OsGA20ox genes in different tissues. The data are means of three replicates. C and D, Semidwarfed phenotypes of 3-month–old osga20ox2 and delayed flowering time of osga20ox2-3. Scale bars = 10 cm. E and F, Semidwarfed phenotypes of 3-month–old osga20ox3 and −4. Scale bars = 10 cm.
In the rice genome, we successfully knocked out three of the OsGA20ox genes, OsGA20ox2–4, but failed to knock out OsGA20ox1 even after three attempts. There is no report of a destructive mutant in the coding region of OsGA20ox1. Overexpression of OsGA20ox1 can increase plant height, but reduces fertility, whereas knockdown with RNA interference (RNAi) reduced plant height (Oikawa et al., 2004). Together, these results suggest that OsGA20ox1 may be an essential gene for rice.
Different natural mutants of OsGA20ox2, including sd1, have been studied and are generally semidwarfed and fertile (Asano et al., 2011). In contrast, our CRISPR/Cas9 knockouts were typically sterile, with pleiotropic differences depending upon the background. The knockout mutant in the indica cv Kasalath background, which has a complete OsGA20ox2, was severely dwarfed and could not flower normally. In contrast, knockout mutants in the japonica cv Wuyungeng background, which carries an allele that encodes an enzyme with less activity (Asano et al., 2011), showed semidwarfism and were partly infertile (∼60% setting rate). Moreover, there was a difference in flowering time between the two kinds of Wuyungeng mutants. Specifically, the homozygous 13-bp deletion (osga20ox2-3) mutant had delayed flowering time (Fig. 5D), while in the heterogeneous 13-bp/4-bp deletion (osga20ox2-2) mutant, flowering time was the same as wild type (Fig. 5C), which suggests that much is left to learn about the function of SD1. OsGA20ox2 has been shown to act in flower regulation (Dai and Xue, 2010). Here we report that OsGA20ox2 might affect flowering time and fertility.
According to a previous study, transgenic plants overexpressing OsGA20ox3 have increased stature and greater susceptibility to pathogens, whereas RNAi knockdown lines exhibited semidwarfism and greater resistance to rice blast and bacterial blight (Qin et al., 2013). The function of OsGA20ox4 has not been reported. In our experiments, knockout mutants of OsGA20ox3 and OsGA20ox4 both exhibit semidwarfism and partial infertility (∼30% setting rate, Fig. 5, D and E). Through RT-qPCR, we found that all members of GA20ox family have at least medium expression in panicle, but exhibit distinct expression patterns otherwise. OsGA20ox1 is expressed mainly in root and panicle, while both OsGA20ox2 and OsGA20ox4 share similar expression that is primarily in leaf and panicle, which is consistent with their closer genetic distance in the phylogenic tree. OsGA20ox3 is highly expressed specifically in panicle, which explains the sterility of osga20ox3 mutants (Fig. 5B).
Assessment of C20-GA2ox Homologs
In the GA2ox family, which inactivate GAs through 2β-hydroxylation, there is a larger class of C19-GA2oxs and a smaller class of C20-GA2oxs, based on the substrate used (Lo et al., 2008). There are two C20-GA2ox genes in Arabidopsis (AtGA2ox7, 8) and three in rice (OsGA2ox5, 6, 9; Fig. 6A; Table 1). None of these genes have natural mutants or T-DNA/Tos17 insert mutants. Previous knowledge of these genes has only relied on overexpression experiments. In Arabidopsis, overexpression of either of the two genes causes dwarfism. In rice, overexpression of OsGA2ox5 causes severe dwarfism and irregularly small seeds, whereas OsGA2ox6 overexpression plants exhibited dwarfism but normal seeds (Table 2).
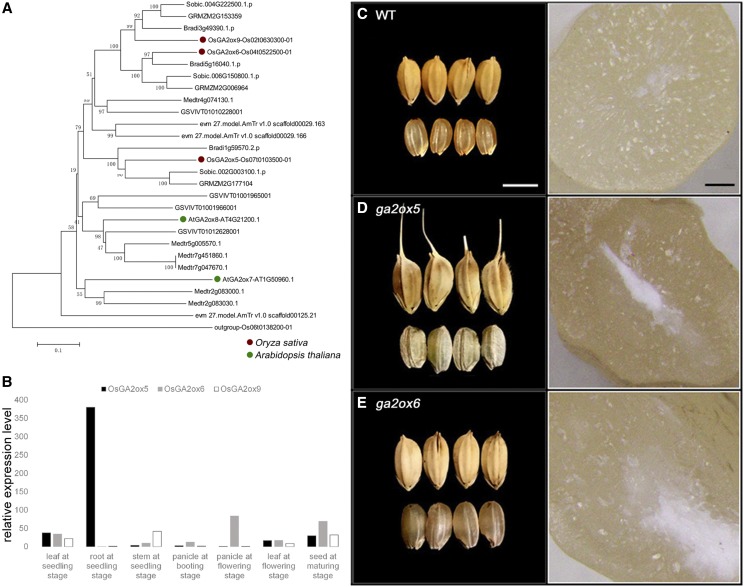
Figure 6.
Phylogenetic analysis, knockout phenotypes, and expression patterns of C20-OsGA2ox genes. A, Phylogenetic analysis of protein sequences of C20-GA2ox encompassing paralogs from Amborella trichopoda, Z. mays, Sorghum bicolor, Brachypodium distachyon, O. sativa, Vitis vinifera, Medicago truncatula, and Arabidopsis. Scale = 0.1 amino acid substitutions per site. B, Expression pattern of C20-OsGA2ox genes. The data are means of three replicates. C–E, Comparison of grain size (scale bar = 5 mm) and transection (scale bar = 50 μm) of wild type and C20-osga2oxs. C, Grains of wild type (left) and transection of wild-type grain (right). D, Grains of osga2ox5 (left) and transection of osga2ox5 grain (right). E, Grains of osga2ox6 (left) and transection of osga2ox6 grain (right).
Overexpression of OsGA2ox9 causes semidwarfism and slightly decreased fertility (Lo et al., 2008). However, in our knockouts of the three rice C20-GA2ox genes, no mutants exhibited alteration in plant height. This may prove that the natural function of C20-GA2ox is not in controlling plant height, and that the dwarfism phenotype may just be an overdose effect. In our RT-qPCR analysis, we found that the three members of C20-GA2ox family are moderately expressed in maturing seeds. OsGA2ox5 is mostly expressed in root, OsGA2ox6 is mostly expressed in panicle and maturing seed, and OsGA2ox9 is mostly expressed in stem (Fig. 6B). Both OsGA2ox5 and OsGA2ox6 mutants have large and chalky seeds. Moreover, mutants of OsGA2ox5 have partial infertility and reversion to the ancestral phenotype of an enclosing awn, a coarse layer outside the seeds. In contrast, mutants of OsGA2ox6 have large normal seeds displaying the chalky characteristic and are completely fertile (Fig. 6, C–E). Seed size and quality are also important artificial selection traits. Strong artificial selection signals also indicate the importance of this gene. A population genetics comparison (Table 3) also suggests there has been a selective sweep in OsGA2ox6 (nucleotide diversity [pi]-wild/pi-cultivated = 9.7). These results suggest that C20-OsGA2ox genes contribute to seed size and quality.
Table 3. Population genetics analyses of C20-GA2ox genes in indica, japonica, cultivated, and wild rice.
| Pi | Gene Id | Indica | Japonica | Cultivated | Wild | Ind–Jap | Cultivated–Wild |
|---|---|---|---|---|---|---|---|
| GA2ox5 | Os07g0103500 | 0.0004452 | 0.0010203 | 0.0009132 | 0.0013064 | 0.0010891 | 0.001249866 |
| GA2ox6 | Os04g0522500 | 0.000113 | 0.00017 | 0.0001428 | 0.001384 | 0.0001438 | 0.001622979 |
| GA2ox9 | Os02g0630300 | 0.0019105 | 0.001834 | 0.0020736 | 0.0009692 | 0.0022742 | 0.001652598 |
DISCUSSION
CRISPR/Cas9 Provides Us with a Convenient Tool to Study All Members of a Pathway
As one of the major phytohormones, GA functions as natural regulators of a variety of growth and developmental processes in plants, including seed germination, internode elongation, and flower and seed development (Davies, 1995). Previous studies of GA pathways have generally focused on the most canonical member of a family, such as OsKO2, which was reported to be the primary KO gene involved in GA synthesis (Yamaguchi, 2008). In fact, the study of the easily overlooked, noncanonical members is key to revealing the function of homologous genes, gene families, and metabolic pathways. Moreover, previous studies were carried out using natural mutants, T-DNA, and/or tos17 insertion mutants to reveal the function of each gene in the GA metabolic pathway, which has the following disadvantages: (1) Those approaches of mutant construction have been idiosyncratic, in that it is hard to obtain enough of the mutants of all genes in the pathway and thus, systematic study of gene functions between different copies could not be achieved at the same time. (2) Those approaches for obtaining mutants do not target the specific genes, so only nonlethal mutants with substantially phenotypic changes were identified. Indeed, there may be some genes that are not mutated after these random-chance methods, some genes that are essential, for which mutations are lethal to the mutant plant itself, and some genes that induce unapparent phenotypic changes after mutation. To study the function of those genes, the highly specific CRISPR/Cas9 system, which allows specific targeting of each gene in the pathway, has allowed us to compare mutations in nearly all genes in each step.
Among the 13 genes from four key steps in the GA metabolic pathway, only three were studied through spontaneous mutation or T-DNA insertion in previous studies. In this study, we use CRISPR/ Cas9 to edit each of these 13 genes individually and successfully obtained full knockout mutants for 11 of them. We found that two of the 13 genes (OsKO2 and OsGA20ox1) were incapable of producing knockouts. This is consistent with previous research, where editing in the posterior exon of OsKO2 induced severe dwarfism and no knockout mutants of OsGA20ox1 were reported (Oikawa et al., 2004). Our work represents a comprehensive vision for studying each key step in a pathway through mutation of each homolog. Our data suggest that the phenotypic defects in these knockout lines were not due to off-target effects of CRISPR/Cas9. This is due to the fact that our whole genome sequencing of the mixed sample of three Osga20ox6 mutants did not yield any mutations in potential off-target sites in a way that could explain the phenotypes observed. On that basis, we are confident that our phenotypes are related to the attributed genes.
In addition, our data suggest that GA metabolic genes have great potential for agricultural applications. There are lots of GA-deficient dwarf mutants found and applied to agriculture production because dwarfism improves lodging resistance and increases yields. However, even though it is one of the most valuable traits in agriculture, GA-related seed alteration has not been reported yet. In this study, we found that the C20-GA2ox gene family participates in seed development regulating size, shape, and quality of seeds, important to rice yields and flavor. Therefore, there is still tremendous potential for GA-metabolic genes and the study of these genes can provide a genetically ideal seed resource for future rice breeding.
The Evolutionary Dynamics of Functions of Duplicated Genes in the Rice GA Biosynthetic Pathway
In each step of GA metabolism, different gene copy numbers can be found in different species. Naturally, we wanted to know whether these duplicated genes provide functional redundancy or acquired new functions (Moore and Purugganan, 2005), because mechanisms that do not require evolution of new functions may play an important role in initial retention of duplicate genes (Panchy et al., 2016). As expected, whether in rice or Arabidopsis, most duplicated members in each step of the GA pathway have some similar functions in terms of the similar knockout outcomes (Table 2). That said, there is functional differentiation observed in the OsKO family in the case of the neofunctionalization of KO3 and KO4. In general, most phenotypes of these mutants involve lethality, plant height (dwarf/semidwarf), or infertility (or near-infertility), which is consistent with the phenotypes of GA variations, suggesting that these duplicated members still retain some functions of GA synthesis or degradation. In contrast, there are fewer homologous genes performing unrelated functions in the same families, which also indicates the importance and conservatism of this pathway. Below, we discuss subfunctionalization (e.g. three recently diversified homologs of OsKO1, OsKO2, and OsKO5) and neofunctionalization (e.g. OsKO3 and OsKO4) as they relate to these duplications. Essential genes play an important role in various aspects of plant growth and development; LOF of essential genes induces death or sterility. Many theories have predicted that duplication of genes can alleviate the selection pressure of these necessary genes, thereby improving the fitness of the organism. However, it is interesting that there is always at least one essential gene in each GA step, regardless of whether this step has a single gene or multiple genes (Table 1). For example, in the KO family, we found three essential genes, KO1, KO2, and KO5, all of which were unable to form knockout mutants or were dead soon after transformation. In the KAO family, LOF of the only member, OsKAO, induced dwarfism and death. In the GA20ox family, we did not obtain a GA20ox1 mutant after three repeated experiments, while GA20ox3 full knockout lines were sterile. In the C20-GA2ox family, GA2ox5 full knockout lines were sterile. Collectively, these results indicated that the GA pathway does not have a redundancy route for synthesis in rice. This contrasts with other important hormones, such as salicylic acid, which can be synthesized in most plants through both the isochorismate-dependent and Phe lyase-dependent pathways (Dempsey and Klessig, 2017).
On the other hand, although some of the duplicates may have similar or related functions, they also exhibit obvious and rapid differentiation. For instance, in the OsKO family that arose from tandem duplication, loss of the Clade I KO genes induces severe dwarfism or death; loss of the Clade II KO genes induces tardive root development and delayed germination. That these mutants can be partly rescued by exogenous GA application proves that the phenotypic changes are caused by GA-deficiency and both Clade I and Clade II KO genes are involved in the synthesis of GA. However, introduction of Clade II KO into Clade I KO mutants cannot rescue the GA-deficient dwarfism (Itoh et al., 2004), which suggests that the GA-related functions of Clade II KO are different from Clade I KO, which focus only on the germination and early development of roots. It is noteworthy that Clade II KO retained the original GA-synthesis function and developed the novel function of disease resistance at the same time. In consideration of the abundant transposable elements in this region, which might play an important role in the rapid functional differentiation, the discovery may provide details for deeper understanding of the pathway.
The four gene families studied in this article are present in all angiosperms. They are defined as core angiosperm gene families that show a strong preference toward the single-copy state according to Li et al. (2016), so it is interesting to study the retention pattern of these four families. However, we detected different patterns of duplicate retention. There are five KO members in rice, including Clade II (OsKO3, 4), which have undergone neofunctionalization, while in Arabidopsis, KO remains a single-copy gene. There is only one KAO in rice, while the two KAO members in Arabidopsis exhibit functional redundancy (Regnault et al., 2014). The gene balance hypothesis suggests that duplicates are retained because they are in balance with each other and selection against deletion of one member of a pair would prevent their rapid loss (Birchler et al., 2005). As we observed in the KO family in rice, loss of any one gene from KO Clade I could break the molecular complexes and induce death of the whole plant. The allocation in dosage keeps the duplicates in the original balance state and prevents them from rapid loss, which implies different species have chosen diverse evolutionary patterns even in gene families with similar function, which might be attributed to different environmental pressure. For the multigene families in both rice and Arabidopsis, GA20ox and C20-GA2ox, different expression patterns are observed in the rice knockout mutants. Gene expression plays an important role in the genotype-phenotype relationship, and changes in transcriptional regulation constitute a major component of the genetic basis for phenotypic evolution (Wray et al., 2003). The essential genes in the GA20ox and C20-GA2ox family, OsGA20ox1 and OsGA2ox5, show expression patterns different from other members from the same family, and thus it is possible that the diversified expression patterns contribute to their functional evolution.
As a matter of fact, gene loss is also an engine of evolutionary change (Olson, 1999). Just like the diurnal melatonin synthesis that plays a central role in the regulation of seasonal reproduction that was preserved in wild strains of Mus musculus although lost in laboratory mice (Ebihara et al., 1986; Goto et al., 1994), organisms can gain conditional benefits by discarding the functionality of widely conserved genes. In this study, as part of the agronomical selection for larger grains, loss of function of OsGA2ox6 is consistent with the less-is-more hypothesis, which might be selected in the future. These evolutionary patterns of diversity of gene families with critical functions provide a new path for us to explore the true nature of a certain type of genes.
Different Mutant Genotypes Have Different Phenotypes at Some Loci
Interestingly, CRISPR/Cas9 knockouts may yield different outcomes relative to T-DNA insertion knockouts and both may yield different results from natural allelic variants. Is the phenotype of a mutant that we observe the true reflection of a gene? Different mutation lines sometimes induce distinct phenotypes, such as sd1. In our research, most of our sd1 mutants, which possess strong alleles induced by knockout, are sterile, whereas the natural sd1 mutants are not (Monna et al., 2002; Sasaki et al., 2002; Spielmeyer et al., 2002; Asano et al., 2011). Among these natural mutants, weak alleles such as sd1-c resulting from substitutions in exon 2, sd1-j resulting from substitutions in exon 1, sd1-r resulting from substitutions in exon 3, and sd1-EQ resulting from substitutions in exon 1 and 3, along with sd1-d, which is resulted from 278-bp deletion in exons 1 and 2, merely exhibit semidwarfism with normal setting rate. The deletion in sd1-d might form a specific weak allele rather than be a reflection of the gene function. In previous research, gene function was mainly studied by regulating expression, such as in the C20-GA2ox family. In the literature, overexpression of C20-GA2oxs induces semidwarfism/dwarfism (Lo et al., 2008; Huang et al., 2010), but we did not observe alteration in plant height after C20-GA2oxs was knocked out in our experiment. Whether these phenotypic differences are due to the different genetic backgrounds or experimental approaches is still unclear. In another respect, some severe defects caused by mutation may have already been eliminated, and the phenotypes we observed in the corresponding mutants nowadays may have been the outcome of selection during artificial breeding. These differences suggest that research on the true functions of genes can be further improved with more mutation types, phenotypes, and genetic backgrounds being taken into consideration. Collectively, our results identify candidates for future control of GA production and provide insight into the evolution of four critical gene families in plants.
MATERIALS AND METHODS
Identification and Classification of Genes Related to GA Metabolism
Gene sequences for nine rice (Oryza sativa) relatives and seven additional angiosperms were obtained from the JGI Phytozome database v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html; Goodstein et al., 2012), NCBI GenBank, and individual genome project websites. The protein sequences of reported rice genes related to GA metabolism (Yamaguchi, 2008) were used as seeds to search for genes of all selected plants through the algorithm BLASTn (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch; Altschul et al., 1990) with an E-value cutoff set to 0.01. We set the criteria that only BLAST hits with identity ≥40% and coverage ≥50% were defined as GA-metabolic genes. The protein sequences of all identified genes were aligned with the tool ClustalW (https://www.genome.jp/tools-bin/clustalw) with default parameters. Phylogenetic trees were constructed using maximum likelihood with 5,000 replicates of bootstrap test through the software MEGA 6 (Tamura et al., 2013).
SNP Calling and Population Analyses
The resequencing data of japonica, indica, and wild samples were obtained from Yuan et al. (2017), and the cultivated samples were the sum of japonica and indica samples. The methods and algorithm used in population analyses were adapted from Yuan et al. (2017), and nucleotide diversity (pi) was calculated based on the coding sequence of C20-GA2ox genes in rice.
CRISPR/cas9-Mediated Gene Knockout and Rice Transformation
To screen spacers for highest specificity (Supplemental Table S2; Supplemental Fig. S2) for each of the 13 GA-metabolic genes, we used a standard method for all lines (Miao et al., 2013), with adjustments for Kasalath as suggested in Shan et al. (2014b). The complementary oligonucleotides of each spacer were inserted into the BsaI restriction site of the plasmid of single-guide RNAs (sgRNAs). Then sgRNA was incorporated into the Cas9 vector, which contained Cas9 driven by the maize (Zea mays) ubiquitin promoter with Gateway recombination method (Katzen, 2007). The vectors were then transformed into two rice cultivars O. sativa ssp Indica cultivar Kasalath and O. sativa ssp Japonica cultivar Wuyungeng24 using Agrobacterium strain EHA105 (Hiei et al., 1994). Kasalath is a traditional indica variety used extensively in rice functional characterization and is represented by a well-annotated genome sequence (Sakai et al., 2014). Wuyungeng24 is a representative cultivar of japonica rice that is known for reliable transformation. Our CRISPR/cas9 efforts yielded a total of 60 individuals from 26 full knockout lines for 10 genes and 33 individuals from 13 partial knockout lines for seven genes (including T0 and T1). Here, we used the full knockout lines (Supplemental Table S3) to describe the phenotypes induced by mutation for their statistical accuracy and hereditary stability.
Confirming Genotypes with PCR
Genomic DNA of transgenic lines was extracted using standard protocols (Monna et al., 2002). To identify mutations in regenerated plants, genomic DNA surrounding the targeted regions of sgRNAs was amplified by PCR and sequenced (Sanger et al., 1977; Supplemental Dataset). Plant lines with mutated alleles as well as wild-type lines were then used for phenotypic analyses.
Measurement of Phenotypes
GA-related mutants often have deficiency in flowering and seed setting (Sakamoto et al., 2004). To assess the setting rate, we calculated the percentage of plump grains in a representative subset of 20 randomly-selected grains. Mutants with plump percentage ≤80% were defined as having decreased fertility and ≤10% were defined as being sterile. Cross-sections of grains were determined by examination with a model no. SZX7 microscope (Olympus).
RT-qPCR Analyses
Total RNA was isolated from leaf, stem, and root at seedling stage, panicle at booting stage, panicle and leaf at flowering stage, and seeds at mature stage using the MiniBEST Universal RNA Extraction Kit (Takara) followed by reverse transcription using PrimeScript RT Master Mix (Takara) according to the manufacturer’s instructions. RT-qPCR was carried out by using a StepOnePlus (Applied Biosystems) real-time PCR system. Gene-specific primers (Supplemental Table S4) were designed with the software Oligo 6 (https://www.oligo.net/downloads.html). All RT-qPCR experiments were carried out in triplicate. RT-qPCR was performed with the following cycling parameters: 95°C for 30 s, 40 cycles of 95°C for 5 s and 60°C for 34 s, 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. Gene expression was quantified by the 2−ΔΔCT method in comparison with the endogenous control (G3PDH) gene (Schmittgen and Livak, 2008).
Assessment of Off-Target Effects
To quantify possible off-target effects in our CRISPR/Cas9 knockout approach, we performed whole genome sequencing of one mutant, Osga2ox6, using a mixed sample of three independent biological replicates. Upon obtaining the whole-genome sequencing data, we assessed the small indels and single-nucleotide variants (SNVs). After searching for the small indels and SNVs exhibiting putative moderate/high impact in coding sequences except the indels in the target gene, we detected 20 SNVs and 1 indel which are most likely to be true positives (Supplemental Table S5). None of these mutation sites were in genes that had known association with seed properties. As predicted by sequence similarity, none of these variations were in sequences within 100 bp of a potential off-target site .
Statistical Analyses
Gene expression was presented by relative gene expression. Relative gene expression level = 2−ΔΔCT.
CT was calculated by mean value of three replicates. Members from the same gene family were calculated relative to OsKAO, OsGA20ox1, and OsGA2ox5, respectively. Panicle at flowering stage was set as the reference sample in this study. No other statistical analyses were used in this study.
Accession Numbers
Sequence data in this article can be found in the GenBank data libraries under accession numbers: OsKO1(BAS98308.1), OsKO2(BAS98310.1), OsKO3(BAS98305.1), OsKO4(BAS98302.1), OsKO5(BAS98313.1,BAS98312.1), OsKAO(BAS95781.1), OsGA20ox1(BAS87451.1), OsGA20ox2(BAS75582.1), OsGA20ox3(BAT00232.1), OsGA20ox4(BAS94085.1), OsGA2ox5(BAS99697.1), OsGA2ox6(BAS90133.1), OsGA2ox9(BAS79897.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. KO gene tree including 10 Oryza species.
Supplemental Figure S2. Location of spacers for GA-related genes.
Supplemental Table S1. KO homologs in other groups compared relative to rice.
Supplemental Table S2. Spacers used in the study for GA-related genes.
Supplemental Table S3. Generations and genotypes of mutant lines used in this study.
Supplemental Table S4. The gene-specific primers used for RT-qPCR.
Supplemental Table S5. Assessment of off-target loci.
Supplemental Dataset. Sanger sequencing results of mutants obtained.
Footnotes
This work was supported by the National Science Foundation of China (grant nos. 31870205, 31870204, and 91731308), the National Major Special Project on New Varieties Cultivation for Transgenic Organisms (grant no. 2016ZX08009001-003), and Jiangsu Collaborative Innovation Center for Modern Crop Production.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Asano K, Yamasaki M, Takuno S, Miura K, Katagiri S, Ito T, Doi K, Wu J, Ebana K, Matsumoto T, et al. (2011) Artificial selection for a green revolution gene during japonica rice domestication. Proc Natl Acad Sci USA 108: 11034–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA (2005) Dosage balance in gene regulation: Biological implications. Trends Genet 21: 219–226 [DOI] [PubMed] [Google Scholar]
- Dai C, Xue H-W (2010) Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J 29: 1916–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ, ed (1995) Plant Hormones: Physiology, Biochemistry and Molecular Biology. Springer, Dordrecht, Netherlands [Google Scholar]
- Dempsey DA, Klessig DF (2017) How does the multifaceted plant hormone salicylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol 15: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara S, Marks T, Hudson DJ, Menaker M (1986) Genetic control of melatonin synthesis in the pineal gland of the mouse. Science 231: 491–493 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Oshima I, Hasegawa M, Ebihara S (1994) The locus controlling pineal serotonin n-acetyltransferase activity (Nat-2) is located on mouse chromosome 11. Brain Res Mol Brain Res 21: 349–354 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JA, Peacock WJ, Dennis ES (1998) Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA 95: 9019–9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Huang J, Tang D, Shen Y, Qin B, Hong L, You A, Li M, Wang X, Yu H, Gu M, Cheng Z (2010) Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J Genet Genomics 37: 23–36 [DOI] [PubMed] [Google Scholar]
- Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M (2004) A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol 54: 533–547 [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F. (2007) Gateway recombinational cloning: A biological operating system. Expert Opin Drug Discov 2: 571–589 [DOI] [PubMed] [Google Scholar]
- Li Z, Defoort J, Tasdighian S, Maere S, Van de Peer Y, De Smet R (2016) Gene duplicability of core genes is highly consistent across all angiosperms. Plant Cell 28: 326–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S-F, Yang S-Y, Chen K-T, Hsing Y-I, Zeevaart JAD, Chen L-J, Yu S-M (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Dong X, Yu T, Shi X, Li Z, Yang W, Widmer A, Karrenberg S (2015) A single nucleotide deletion in gibberellin20-oxidase1 causes alpine dwarfism in Arabidopsis. Plant Physiol 168: 930–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafu S, Jia M, Zi J, Morrone D, Wu Y, Xu M, Hillwig ML, Peters RJ (2016) Probing the promiscuity of ent-kaurene oxidases via combinatorial biosynthesis. Proc Natl Acad Sci USA 113: 2526–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu L-J (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23: 1233–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y (2002) Positional cloning of rice semidwarfing gene, sd-1: Rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res 9: 11–17 [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Oikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M (2004) A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol Biol 55: 687–700 [DOI] [PubMed] [Google Scholar]
- Olson MV. (1999) When less is more: Gene loss as an engine of evolutionary change. Am J Hum Genet 64: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu S-H (2016) Evolution of gene duplication in plants. Plant Physiol 171: 2294–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett ARG, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, Jikumaru Y, Benlloch R, Nilsson O, Ruiz-Rivero O, et al. (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24: 941–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Liu JH, Zhao WS, Chen XJ, Guo ZJ, Peng YL (2013) Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice. Mol Plant Microbe Interact 26: 227–239 [DOI] [PubMed] [Google Scholar]
- Regnault T, Davière J-M, Heintz D, Lange T, Achard P (2014) The gibberellin biosynthetic genes AtKAO1 and AtKAO2 have overlapping roles throughout Arabidopsis development. Plant J 80: 462–474 [DOI] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, et al. (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53: 488–504 [DOI] [PubMed] [Google Scholar]
- Routaboul J-M, Fischer SF, Browse J (2000) Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol 124: 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Kanamori H, Arai-Kichise Y, Shibata-Hatta M, Ebana K, Oono Y, Kurita K, Fujisawa H, Katagiri S, Mukai Y, et al. (2014) Construction of pseudomolecule sequences of the aus rice cultivar Kasalath for comparative genomics of Asian cultivated rice. DNA Res 21: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Mei Z, Duan J, Chen H, Feng H, Cai W (2014a) OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One 9: e87110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Gao C (2014b) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9: 2395–2410 [DOI] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA 99: 9043–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Singh DP, Helliwell CA, Poole AT (2005) Plants with increased expression of ent-kaurene oxidase are resistant to chemical inhibitors of this gibberellin biosynthesis enzyme. Plant Cell Physiol 46: 284–291 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Wu Y, Peters RJ (2012) CYP701A8: A rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol 158: 1418–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20: 1377–1419 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhang Q, Zeng S, Gu L, Si W, Zhang X, Tian D, Yang S, Wang L (2017) Selective sweep with significant positive selection serves as the driving force for the differentiation of japonica and indica rice cultivars. BMC Genomics 18: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]