Accumulation of photodamaged PSII core proteins enables retrograde signaling via the stress hormone salicylic acid and its signaling components, EDS1 and PAD4.
Abstract
Photodamage of the PSII reaction center (RC) is an inevitable process in an oxygen-rich environment. The damaged PSII RC proteins (Dam-PSII) undergo degradation via the thylakoid membrane-bound FtsH metalloprotease, followed by posttranslational assembly of PSII. While the effect of Dam-PSII on gene regulation is described for cyanobacteria, its role in land plants is largely unknown. In this study, we reveal an intriguing retrograde signaling pathway by using the Arabidopsis (Arabidopsis thaliana) yellow variegated2-9 mutant, which expresses a mutated FtsH2 (FtsH2G267D) metalloprotease, specifically impairing its substrate-unfolding activity. This lesion leads to the perturbation of PSII protein homeostasis (proteostasis) and the accumulation of Dam-PSII. Subsequently, this results in an up-regulation of salicylic acid (SA)-responsive genes, which is abrogated by inactivation of either an SA transporter in the chloroplast envelope membrane or extraplastidic SA signaling components as well as by removal of SA. These results suggest that the stress hormone SA, which is mainly synthesized via the chloroplast isochorismate pathway in response to the impaired PSII proteostasis, mediates the retrograde signaling. These findings reinforce the emerging view of chloroplast function toward plant stress responses and suggest SA as a potential plastid factor mediating retrograde signaling.
Chloroplast-generated reactive oxygen species (ROS) damage primarily the photosynthetic apparatus, thereby deleteriously affecting plant development. Among ROS, singlet oxygen (1O2), mainly generated by PSII, has been considered as a prime cause of PSII damage because of its proximity to PSII (Triantaphylidès et al., 2008; Kato and Sakamoto, 2009). Since 1O2 is a known by-product of photosynthesis, PSII reaction center (PSII RC) proteins undergo constant damage in a light-dependent manner (Ohad et al., 1984; Mishra et al., 1994). The damaged PSII (Dam-PSII) becomes repaired via a series of steps: (1) D1 oxidation and subsequent phosphorylation; (2) migration of the Dam-PSII from the grana core (appressed membranes) to the grana margin (nonappressed membranes); (3) dephosphorylation of D1; (4) degradation of the damaged D1 protein by the membrane-bound FtsH metalloprotease; (5) de novo synthesis of D1; and (6) reassembly and subsequent remigration to the grana core (Yamamoto et al., 2013; Yoshioka-Nishimura and Yamamoto, 2014). Accordingly, the impaired PSII repair in Arabidopsis (Arabidopsis thaliana) mutants lacking the membrane-bound FtsH protease leads to the failure of acclimation to a sublethal intensity of light, wherein wild-type plants rapidly acclimate without considerable damage to the photosystem apparatus (Khatoon et al., 2009; Kim et al., 2012). Interestingly, it was shown in cyanobacteria, but not yet in land plants, that degradation products of the D1 protein directly involve the expression of psbA encoding D1 (Stelljes and Koenig, 2007).
Apart from being toxic, 1O2 triggers two distinct retrograde signaling (RS) pathways in Arabidopsis. The first is mediated by β-carotene, a well-known 1O2 scavenger present in the PSII RC. β-Carotene acts as a 1O2 sensor and releases, upon interaction with 1O2, its volatile oxidative products such as β-cyclocitral (β-CC) under high-light stress, which in turn induce transcriptional reprogramming in the nucleus (Ramel et al., 2013a, 2013b; Havaux, 2014). The second RS is mediated by the nucleus-encoded plastid protein EXECUTER1 (EX1), which plays a critical role in the 1O2-triggered RS in the conditional fluorescent (flu) mutant that generates 1O2 in chloroplasts upon a dark-to-light shift (Goslings et al., 2004; Lee et al., 2007; Kim et al., 2008, 2012). Recent studies demonstrated that EX1-mediated 1O2 signaling is coordinated by the FtsH metalloprotease (Wang et al., 2016a; Dogra et al., 2017; Kato and Sakamoto, 2018). Upon 1O2 burst, EX1 proteins undergo degradation by the FtsH metalloprotease, which turns out to be an integral part of EX1-mediated 1O2 signaling (Wang et al., 2016a). Unlike β-carotene, which becomes oxidized in the grana core where PSII generates 1O2 under photoinhibitory conditions, the EX1-FtsH-mediated 1O2 signaling arises at the grana margin where the PSII repair cycle occurs. Hence, the newly synthesized chlorophyll, or its precursors that are required for the PSII reassembly, has been proposed as an alternative source of 1O2 generation in the grana margin (Wang et al., 2016a; Dogra et al., 2018). In addition to these two distinct signaling pathways, 1O2-driven oxygenated unsaturated fatty acid derivatives, also known as reactive electrophile species, are also involved in signaling (Sattler et al., 2006; Mueller et al., 2008; Fischer et al., 2012).
Remarkably, a recent study also demonstrated that β-CC stimulates salicylic acid (SA) synthesis in Arabidopsis wild-type plants upon exposure to high light (Lv et al., 2015). β-CC-primed SA accumulation and its related nuclear gene expression changes confer plant tolerance to a lethal dose of light intensity in Arabidopsis wild-type plants. Moreover, during the β-CC-primed acclimation process, vital immune components such as ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1) are found to positively regulate SA synthesis and SA signaling, respectively (Lv et al., 2015). SA appears to be mainly synthesized via the plastid isochorismate (ICS) pathway rather than through the cytosolic phenylalanine ammonium lyase (PAL) pathway (Wildermuth et al., 2001). Also, flu mutant plants up-regulate SA and SA-responsive genes, including EDS1, PATHOGENESIS RELATED1 (PR1), and PR5, upon release of 1O2 (Ochsenbein et al., 2006). Furthermore, the inactivation of EDS1 in flu mutant plants attenuates 1O2-triggerd stress responses, such as cell death and growth inhibition (Ochsenbein et al., 2006). In addition, a chloroplast-localized CALCIUM-SENSING PROTEIN (CAS) is involved in the plant immune signaling as an upstream component of SA synthesis and 1O2-triggered RS in Arabidopsis (Nomura et al., 2012). In the green alga Chlamydomonas reinhardtii, CAS regulates a subset of genes related to a CO2-concentating mechanism via RS, suggesting its role in regulating photosynthesis and immune responses (Wang et al., 2016b). These previous reports indicate a probable interconnection between the chloroplast-generated ROS and the SA-mediated signaling. Despite the importance of chloroplast-generated ROS and SA in plant stress responses, the molecular and genetic cross talk between them and a possible role of SA as a plastid factor functioning in RS remain unclear.
In this study, we reveal a plausible interconnection between the impaired PSII proteostasis and the induction of SA-responsive genes. In the Arabidopsis yellow variegated2-9 (var2-9) mutant, lacking a functional FtsH2 protease, SA-responsive genes were dramatically up-regulated in the absence of external stimuli. The stress hormone SA, synthesized via the chloroplast-established ICS pathway, was found to play a key role in the transcriptional reprogramming of the genes. More importantly, the SA accumulation via the ICS pathway is likely to be independent of transcriptional regulation of genes involved in SA synthesis, including ISOCHORISMATE SYNTHASE1 (ICS1; Wildermuth et al., 2001). This result suggests that the dysfunctional chloroplasts of the var2-9 mutant may induce the accumulation of SA via the ICS pathway, implying that the chloroplast-driven SA acts as a plastid factor mediating chloroplast-to-nucleus retrograde signaling.
RESULTS
FtsH2G267D Lacks Its Activity toward PSII Repair
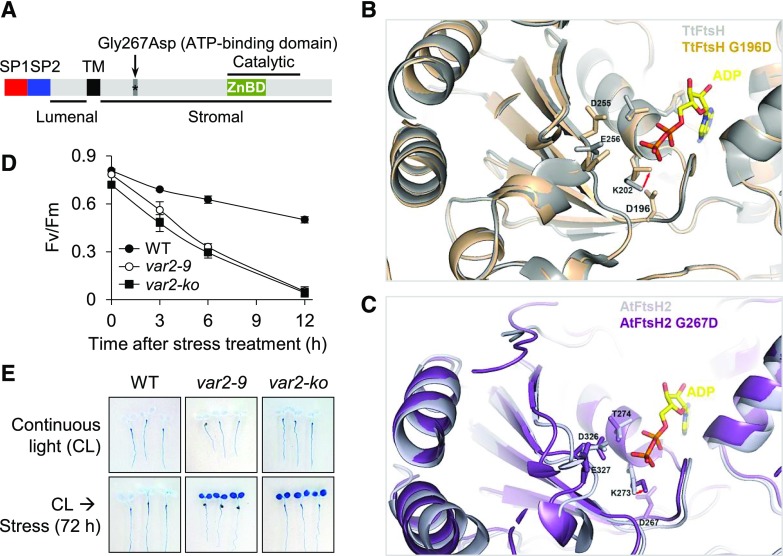
To explore whether the impaired PSII proteostasis triggers RS, we used the Arabidopsis var2-9 mutant that expresses a mutant form of the FtsH2 (FtsH2G267D) metalloprotease (Fig. 1A; Sakamoto et al., 2004; Liu et al., 2010; Zhang et al., 2010). The var2 null alleles, as well as var2-9, were initially isolated through an unbiased forward genetic screen for a leaf variegation phenotype (Sakamoto et al., 2004). Gly-267 (G267) is the first amino acid in the ATP-binding domain (Fig. 1A) and is highly conserved in FtsH proteases across a wide range of bacteria and plant species (Sakamoto et al., 2004). Arabidopsis FtsH2 (AtFtsH2) shares 51% sequence identity and 69% sequence similarity with the bacterial Thermus thermophilus FtsH (TtFtsH). Therefore, the known structure of TtFtsH (Protein Data Bank identifier 2DHR; Bieniossek et al., 2009) was used to predict the potential consequence of G267D in AtFtsH2, especially in the ATP-binding pocket. Four important residues involved in the interaction with ATP were identified in the structure of TtFtsH (Fig. 1B) and AtFtsH2 (Fig. 1C). The G267D mutation in AtFtsH2 and the corresponding G196D mutation in TtFtsH induce a conformational change of the ATP-interacting Lys (Lys-202 in TtFtsH and Lys-273 in AtFtsH2, respectively). The modeling result suggests a spatial shift of ATP-interacting Lys toward the ATP-binding site. In the analyzed mutated FtsH2 G-to-D proteins, the side chain of Asp enters the ATP-binding pocket, which may disturb the position of the γ-phosphate of ATP and interfere with ATP binding (Fig. 1, B and C).
Figure 1.
var2-9 and var2-ko seedlings are susceptible to photooxidative stress. A, Schematic illustration of FtsH2 and the missense mutation in var2-9. The mutant form of FtsH2 in the var2-9 allele contains a missense mutation (Gly-267-Asp) in the ATP-binding domain. SP1 and SP2 are the two signal peptides that are responsible for targeting to the chloroplast and the thylakoid membrane, respectively. TM marks the transmembrane domain, and Catalytic indicates the catalytic domain, which contains a Zn-binding domain. Lumenal and Stromal indicate different sides of the FtsH2 topology. B, The possible effect of G267D on ATP binding was analyzed using the known structure of TtFtsH (Protein Data Bank identifier 4EIW) as a template. The G267 in AtFtsH2 is equivalent to the G196 in TtFtsH. The residues K202, T203, D255, and E256 are known to be involved in the interaction with ATP. The TtFtsH and TtFtsHG196D structures are colored in gray and orange, respectively. The G196D mutation induces a conformational change of K202 because of steric hindrance, indicated by the red arrow. C, The AtFtsH2 structure is modeled using that of TtFtsH as a template. The residues K273, T274, D326, and E327 are likely to be involved in ATP binding in AtFtsH2. The AtFtsH2 and AtFtsH2G267D structures are colored in light purple and purple, respectively. AtFtsH2G267D may interfere with the ATP binding similar to that in TtFtsHG196D, indicated by the red arrow. Protein structures were visualized with PyMol (pymol.org). D, Time-course analysis of the rate of PSII damage. Five-day-old var2-9, var2-ko, and wild-type (WT) seedlings initially grown under CL (40 μmol m−2 s−1 at 22°C ± 2°C) were subjected to photoinhibitory (300 μmol m−2 s−1 at 10°C ± 2°C) conditions. The PSII activity (Fv/Fm) was determined at the indicated time points. At least 20 seedlings per genotype were used for each measurement. Error bars indicate sd. E, After 72 h of light stress treatment, the degree of cell death in the different genotypes was visualized by staining whole seedlings with Trypan Blue. Seedlings that were grown under nonstressful light (CL) were used as a control (top row).
Next, we determined whether the var2-9 mutant shows comparable susceptibility to photoinhibitory conditions as the var2 null (var2-ko) mutant (Kim et al., 2012). To analyze this, the var2-9 and var2-ko mutants and wild-type seedlings grown initially under continuous light (CL; 40 μmol m−2 s−1 at 22°C ± 2°C) for 5 d were examined under photoinhibitory conditions (300 μmol m−2 s−1 at 10°C ± 2°C). Lowering the temperature at the light intensity of 300 μmol m−2 s−1 was found to effectively inhibit PSII activity in Arabidopsis seedlings grown initially under low light and normal temperature conditions (Kim et al., 2012). The levels of photoinhibition were determined by measuring PSII activity (maximum photochemical efficiency of PSII [Fv/Fm]), and cell death was examined by using Trypan Blue that selectively dyes dead cells. Despite the lack of variegation in the cotyledons (Supplemental Fig. S1), both var2-9 and var2-ko mutant seedlings showed a drastic decline of PSII activity and a significant cell death response compared with the wild type (Fig. 1, D and E), indicating that the G267D mutation sufficiently compromises FtsH2 function.
Accumulation of Damaged PSII Core Proteins and Impaired Proteostasis in the var2-9 Mutant
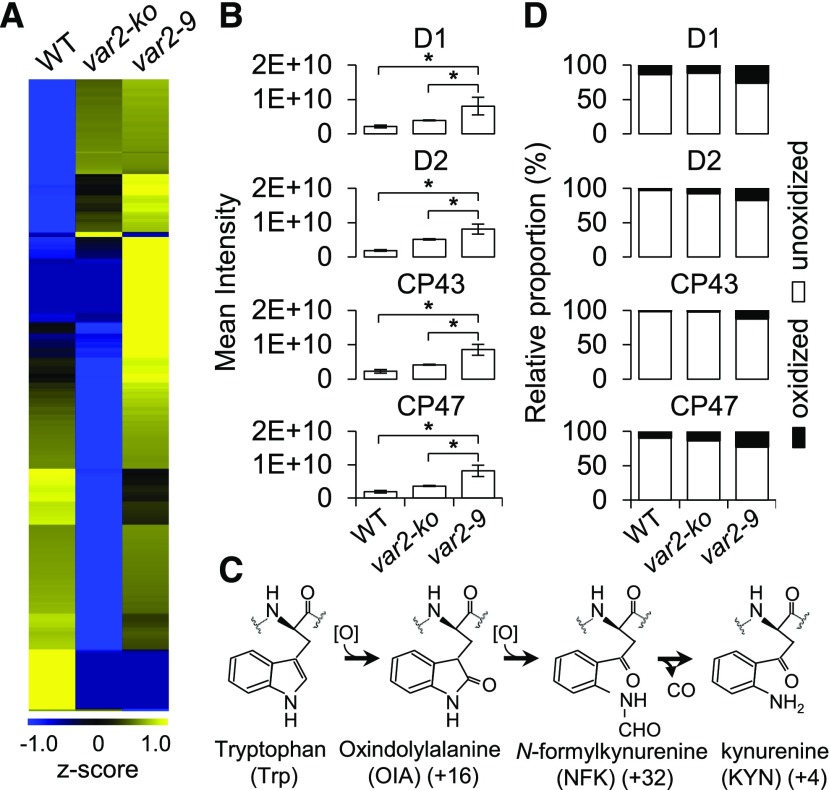
If the FtsH2 metalloprotease function is impaired in the var2-9 mutant, this should result in the accumulation of Dam-PSII proteins. To examine whether FtsH2G267D results in a differential accumulation of chloroplast proteins including its known substrate D1, intact green chloroplasts were isolated through a Percoll gradient step from var2-9, var2-ko, and wild-type plants grown under normal light conditions. The subsequent label-free quantitation using mass spectrometry disclosed differentially accumulated chloroplast proteins (Fig. 2A; Supplemental Table S1). Among a total of 1,594 proteins detected, 352 proteins were found to be at least twofold more abundant in the var2-9 mutant as compared with wild-type and var2-ko plants (Fig. 2A; Supplemental Table S1). As anticipated, the PSII core proteins, such as D1, D2, CP43, and CP47, were highly accumulated in the var2-9 mutant (Fig. 2B). A less significant accumulation of these proteins was observed in the var2-ko mutant followed by the wild type (Fig. 2B; Supplemental Table S1). Considering the higher content of various chloroplast proteins, including PSII core proteins in the var2-9 mutant, we assumed that this phenotype is due to the decreased content of Rubisco, which represents a large proportion of the total chloroplast proteins. If the Rubisco portion decreases, presumably the relative proportion of other chloroplast proteins would tend to increase. However, we found that the amount of Rubisco remained unchanged in the chloroplasts of the var2-9 mutant as compared with the wild type (Supplemental Table S1).
Figure 2.
Photodamaged PSII core proteins are highly accumulated in the var2-9 mutant. A total of 1 µg of chloroplast proteins was analyzed with mass spectrometric analysis. All samples were analyzed in triplicate. A, Heat map of the relative accumulation of chloroplast proteins in wild-type (WT), var2-ko, and var2-9 plants (Supplemental Table S1). B, PSII core proteins including D1, D2, CP43, and CP47 exhibited a higher accumulation in var2-9 compared with wild-type and var2-ko plants. Data shown are means ± se (n = 3 replicates). Asterisks indicate statistically significant differences between the var2-9 mutant and other genotypes (Student’s t test, P < 0.05). C, Trp oxidation pathway. D, Oxidative posttranslational modification analysis revealed the substantial accumulation of Trp-oxidized peptides of the PSII core proteins in the var2-9 mutant (Supplemental Table S2). Bar charts represent the proportions of oxidized and unoxidized protein content for the PSII proteins in the corresponding genotypes.
In plants, the membrane-bound FtsH protease is present as a hexameric heterocomplex composed of four FtsH isomers, such as FtsH1, FtsH2, FtsH5, and FtsH8 (Yu et al., 2005; Zaltsman et al., 2005). In comparison with the wild type, FtsH2 was completely absent in the var2-ko mutant with a significant reduction of other isomers, which is in line with an earlier report (Zhang et al., 2010), whereas the FtsH isomer levels (except for FtsH2G267D that appeared to be less accumulated) in the var2-9 mutant were similar to those in the wild type (Supplemental Fig. S2). The higher accumulation of PSII core proteins in the var2-9 mutant compared with the var2-ko mutant indicates that FtsH2G267D remains a component of the FtsH hexameric heterocomplex, impairing its function and leading to the accumulation of its substrates. When introducing the corresponding mutation into Escherichia coli FtsH (i.e. G195D), this EcFtsHG195D failed to rescue the phenotype of an E. coli mutant lacking the FtsH, indicating that the substitution results in a loss of FtsH activity (Zhang et al., 2010).
In the var2-ko mutant, the FtsH2-deficient FtsH hexameric heterocomplex composed of FtsH1, FtsH5, and FtsH8 is likely to be able to unfold and process its substrates for proteolysis, but with less efficacy compared with the intact FtsH in the wild type under nonphotoinhibitory conditions. Nonetheless, the difference between FtsH2G267D-containing and FtsH2-deficient hexameric FtsH proteases with respect to the accumulation of Dam-PSII and other chloroplast proteins seemed not apparent under the analyzed very mild photoinhibitory conditions, as both mutant seedlings failed to survive while wild-type seedlings rapidly acclimated (Fig. 1, D and E).
It was previously suggested that the oxidative posttranslational modification of PSII core proteins might be directly linked to their turnover (Anderson et al., 2002; Dreaden et al., 2011): certain Trp amino acid residues were found to be oxidized upon exposure to high light, resulting in the formation of various oxidized forms of Trp, namely oxindolyl-Ala, N-formylkynurenine, and kynurenine, with the corresponding mass shifts of +16, +32, and +4 D, respectively (Fig. 2C). Considering that the PSII repair cycle is a default pathway regardless of the light intensity and that 1O2 generated by PSII is a by-product of photosynthesis, Trp oxidation may also occur under normal light intensity. Due to the mutation in FtsH2, the proteolysis of the oxidized PSII RC proteins is expected to be compromised in the var2-9 mutant. In agreement with this assumption, the var2-9 mutant shows a comparatively higher portion of Trp oxidation in the PSII core proteins than in var2-ko and wild-type plants (Fig. 2D; Supplemental Table S2). Collectively, these results indicate that the var2-9 mutant can be utilized as a tool to study the effect of accumulated Dam-PSII in the context of RS in a noninvasive manner.
Impaired PSII Turnover in var2-9 Seedlings
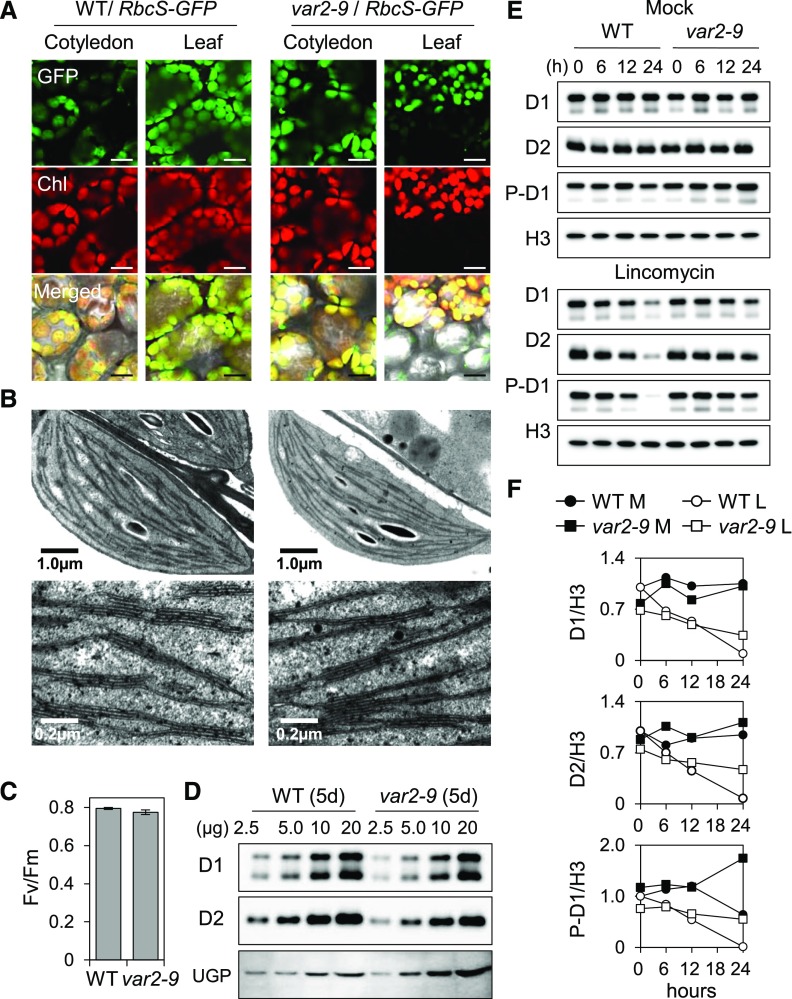
Next, we were interested in determining whether the mutation in var2-9 would lead to changes in the turnover of Dam-PSII proteins in cotyledons of young seedlings grown under nonstressful light conditions. As shown in Figure 3, the chloroplast size and the configuration of the thylakoid membrane, the intensity of chlorophyll fluorescence, the PSII activity (Fv/Fm), and the steady-state levels of PSII core proteins in the var2-9 mutant were similar to those in the wild type (Fig. 3, A–D). Despite this similarity, the var2-9 seedlings showed an impaired PSII proteostasis, as is evident from the reduced turnover rate of the PSII core proteins in the presence of lincomycin, a prokaryotic-type translation inhibitor (Fig. 3, E and F). It should be noted that lincomycin inhibits the de novo synthesis of plastid-encoded proteins, including D1 and D2. This result therefore suggests that G267D substitution in FtsH2 compromises its function toward PSII turnover. The relatively higher rate of D1 phosphorylation, which usually primes photodamaged D1 turnover (Fig. 3, E and F; Kato and Sakamoto, 2014; Yoshioka-Nishimura and Yamamoto, 2014), was also observed in the var2-9 seedlings compared with the wild type prior to the lincomycin treatment. In the presence of lincomycin, the amount of phosphorylated D1 remains relatively stable in var2-9 seedlings in comparison with the wild type, probably due to the impaired substrate-unfolding activity of FtsH2G267D.
Figure 3.
Impaired PSII proteostasis in cotyledons of var2-9 seedlings. Seedlings grown on Murashige and Skoog agar medium under CL (40 μmol m−2 s−1 at 22°C ± 2°C) were used. A, To compare the size and morphology of chloroplasts, 10-d-old transgenic wild-type (WT) and var2-9 seedlings expressing the Rubisco small subunit (RbcS) fused with a GFP tag under the control of the cauliflower mosaic virus (CaMV) 35S promoter were monitored by confocal microscopy. The selected area of the detached true leaf of the var2-9 mutant shows discrete signals of both the chlorophyll fluorescence and GFP because of the leaf variegation. The green fluorescence of GFP and red fluorescence of chlorophyll (Chl) were monitored separately. Bars = 25 μm. B, Characteristic ultrastructures of chloroplasts observed in the var2-9 cotyledons (left) and in the wild type (right). Top and bottom images show intact chloroplasts and the thylakoid membrane system in a granum, respectively. C, PSII activity (Fv/Fm) in 5-d-old wild-type and var2-9 seedlings. Results represent means from three independent measurements. For each measurement, at least 20 seedlings were analyzed. Error bars indicate sd. D, The steady-state levels of the D1 and D2 proteins in 5-d-old var2-9 and wild-type seedlings were determined by immunoblot analysis. The numbers on the top indicate the amount of proteins loaded on each lane. UGPase (UGP) was used as a loading control. E, Relative degradation rates of D1, D2, and phosphorylated D1 (P-D1) in 5-d-old wild-type and var2 seedlings in the absence (top) or presence (bottom) of 5 mm lincomycin. At the various indicated time points, total protein was extracted and the relative levels of D1 and D2 were probed by immunoblot analysis. Histone H3 (H3) was used as a loading control. F, The immunoblot signals in E from three independent biological replicates were quantified using ImageJ and normalized by the signals of H3. L, Lincomycin; M, mock.
Up-Regulation of SA-Responsive Genes in the var2-9 Mutant after the Onset of Photosynthesis
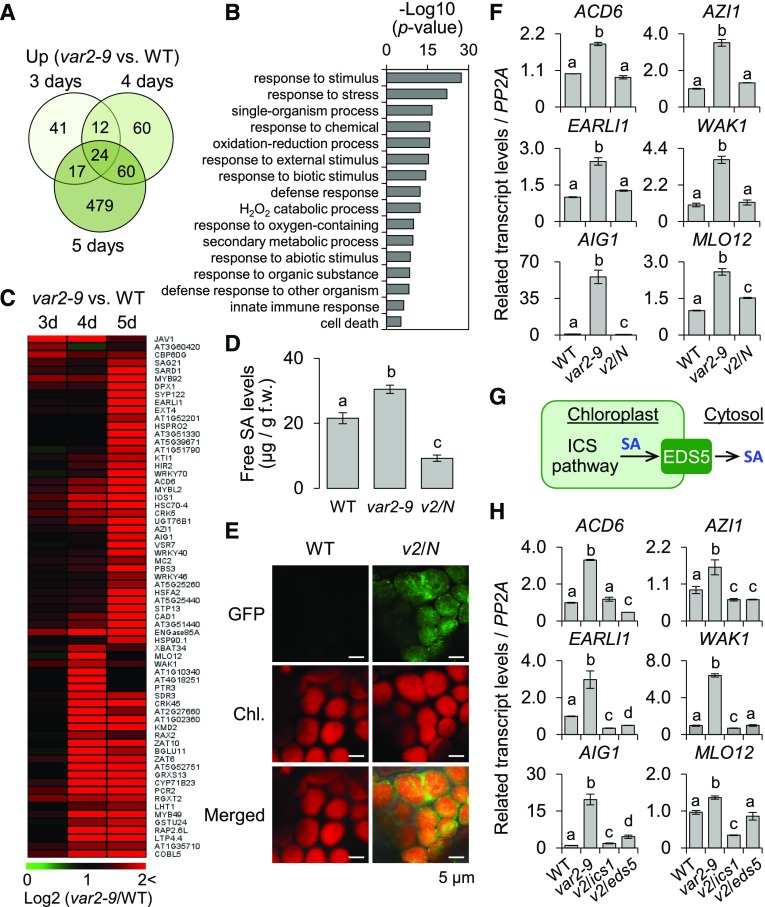
Accurate and prompt biogenesis of the photosynthetic apparatus is critical for the effective transition from heterotrophic to phototrophic growth. Therefore, PSII repair needs to be highly competent during the transition. Indeed, rapid turnover of PSII core proteins, such as D1 and D2, was observed in the wild-type seedlings after lincomycin treatment (Fig. 3, E and F). Hence, we hypothesized that the dysfunctional chloroplasts in the var2-9 mutant may transmit its functional status to the nucleus via retrograde signaling to readjust cellular homeostasis after the onset of photosynthesis. To test this hypothesis, we performed RNA sequencing (RNA-seq) analysis in 3-, 4-, and 5-d-old var2-9 and wild-type seedlings grown under CL (40 μmol m−2 s−1 at 22°C ± 2°C) conditions. The comparative transcriptome analysis revealed that a total of 693 genes were up-regulated in the var2-9 seedlings by at least twofold (false discovery rate [FDR] < 0.05) compared with the wild type (Fig. 4A; Supplemental Table S3). Gene Ontology (GO) term enrichment analyses (P < 0.05) for biological processes with these 693 genes revealed 16 overrepresented groups (Fig. 4B). Among them, response to stimulus (P = 5.05E-28) and response to stress (P = 9.44E-23) represented the most significantly enriched terms. Nearly 40% of the genes (275) were found to be involved in response to stimulus, including 189 genes involved in stress, defense, and immune responses (Fig. 4B; Supplemental Table S4).
Figure 4.
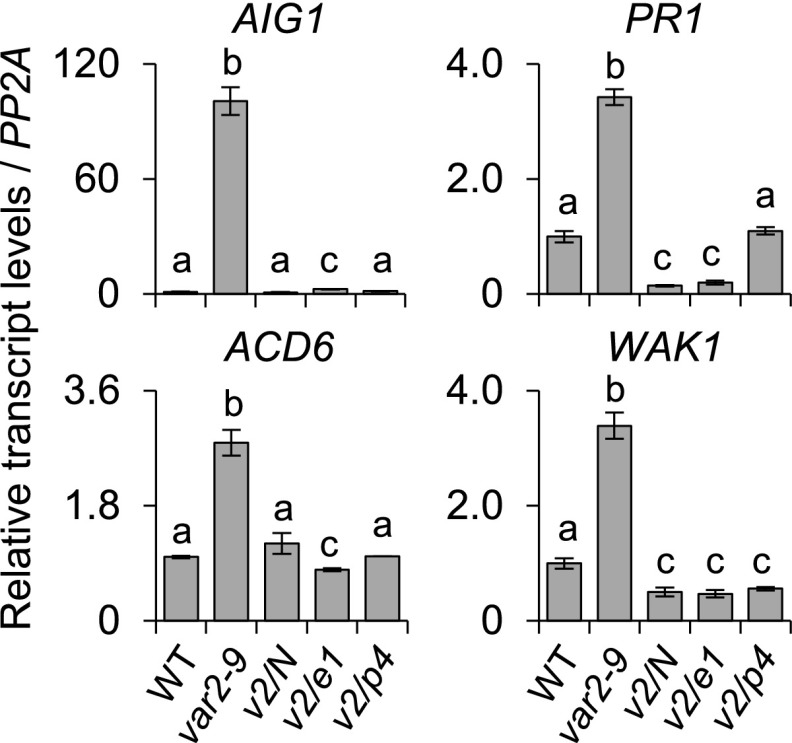
SA synthesized via the chloroplast ICS pathway primes the expression of SA-responsive genes in the var2-9 mutant. For the genome-wide transcriptome analysis, 3-, 4-, and 5-d-old wild-type (WT) and var2-9 seedlings grown under CL (40 μmol m−2 s−1 at 22°C ± 2°C) were used. A, Up-regulation of immune-related genes in var2-9 seedlings after the onset of photosynthesis. The Venn diagram shows the number of genes with at least a twofold up-regulation in var2-9 compared with the wild type. B, Enriched GO terms in the biological process category indicate that stress-, immune-, and defense-related genes were significantly overrepresented in the up-regulated genes (A) in the var2-9 mutant. C, Heat map showing the expression levels of the SRGs in the var2-9 seedlings compared with the wild type. D, The endogenous levels of free SA in wild-type, var2-9, and var2-9 cpNahG (v2/N) seedlings were determined. The data represent means of three independent biological replicates for each genotype. Error bars indicate sd. f.w., Fresh weight. E, Localization of cpNahG-GFP in a young leaf cell of the var2-9 mutant (right). Confocal images taken from the young leaf cell of wild-type seedlings are shown as a control (left). Chl., Chlorophyll. F, The transcript abundances of selected SRGs, including Avrrpt2-induced gene1 (AIG1), Azelaic acid induced1 (AZI1), early Arabidopsis aluminum induced1 (EARLI1; a paralog of AZI1), Accelerated cell death6 (ACD6), Mildew resistance locus O12 (MLO12), and Wall associated kinase1 (WAK1), in 5-d-old wild type, var2-9, and v2/N seedlings were examined using reverse transcription quantitative PCR (RT-qPCR). G, Chloroplast ICS pathway and SA export to the cytosol via the EDS5 transporter. H, Five-day-old wild-type, var2-9, var2-9 ics1 (v2/ics1), and var2-9 eds5 (v2/eds5) seedlings were collected to analyze the transcript levels of SRGs using RT-qPCR. Gene expression levels shown in F and H are means ± sd of three independent biological samples. PROTEIN PHOSPHATASE2A (PP2A) was used as an internal control. Lowercase letters in D, F, and H indicate statistically significant differences between mean values (P < 0.05, one-way ANOVA with posthoc Tukey’s honestly significant difference [HSD] test).
Interestingly, in-depth analysis of the 189 immune/defense-related genes (Supplemental Table S4) up-regulated in the var2-9 mutant revealed a suite of SA-responsive genes (SRGs). Of the 189 genes, 66 genes were previously identified as SRGs (Fig. 4C; Supplemental Table S5; Zhou et al., 2015). In agreement with this, an increased level of free SA in the var2-9 mutant, ∼1.5-fold higher than in wild-type seedlings, was observed (Fig. 4D). To explore whether the subtle increase of free SA in combination with the impaired chloroplast proteostasis is sufficient to prime the expression of SRGs, SA-depleted var2-9 transgenic lines were generated. The transgenic wild-type plants expressing the bacterial SA-hydrolyzing enzyme NahG fused with the signal peptide of small subunit of Rubisco and GFP at the N-terminal and C-terminal ends (hereafter cpNahG), respectively, driven by the constitutive Cauliflower mosaic virus (CaMV) 35S promoter (Fragnière et al., 2011) were crossed with var2-9 plants. Confocal imaging corroborated an exclusive localization of the cpNahG-GFP fusion proteins in the chloroplasts of the var2-9 cpNahG transgenic seedlings (Fig. 4E). As expected, this cpNahG construct reduced the SA levels (Fig. 4D). Comparison of the expression of the SRGs in var2-9 and var2-9 cpNahG plants suggests that the subtle increase of SA in the var2-9 mutant is sufficient to induce the SRGs, as evidenced by the significant repression of their expression in var2-9 cpNahG plants (Fig. 4F). These results also provide an important clue that not the cytosolic PAL pathway but the chloroplast-established ICS pathway is essential for the SA synthesis in the var2-9 mutant. ICS1 is a nucleus-encoded chloroplast enzyme, catalyzing the conversion of chorismate to ICS in the ICS pathway (Strawn et al., 2007). Transcriptional up-regulation of ICS1 in response to microbial pathogens was found to be a rate-limiting step in accumulating SA (Wildermuth et al., 2001). This chloroplast-synthesized SA is exported to the cytosol via ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5), a member of the MATE-transporter family localized in the chloroplast envelope (Fig. 4G; Nawrath et al., 2002; Serrano et al., 2013). If ICS-synthesized SA regulates the SRG expression upon its export to the cytosol in the var2-9 mutant, the SRG expression should be compromised in both double mutants var2-9 ics1 and var2-9 eds5. Indeed, the resulting data of the expression of several examined SRGs in the var2-9 ics1 and var2-9 eds5 mutant seedlings grown under normal light conditions further corroborated that the chloroplast-synthesized SA plays an essential role in var2-9 plants (Fig. 4H). Importantly, after lincomycin treatment, the degradation rates of the PSII core proteins in var2-9 ics1 and var2-9 eds5 seedlings were almost the same as those in var2-9 seedlings, suggesting that the accumulation of SA and the subsequent activation of SA-mediated retrograde signaling are downstream events of the impaired PSII repair in var2-9 plants (Supplemental Fig. S3). The observed FtsH2G267D-mediated phenotypes were abrogated by expressing the wild-type FtsH2 fused with the GFP tag and driven by its native promoter, confirming the causative role of FtsH2G267D in the instigation of the retrograde signaling (Supplemental Fig. S4, A–C).
Even though chloroplasts in both mature leaves and cotyledons showed impaired turnover of PSII core proteins (Figs. 2B and 3E), we examined the expression levels of SRGs in mature leaves to ensure that this retrograde signaling also occurs in the green sectors of mature leaves. To address this problem, using wild-type and var2-9 mature plants grown on soil, foliar discs were carefully collected from green and white sectors and total RNA was extracted. The resulting reverse transcription quantitative PCR (RT-qPCR) revealed that the expression patterns of SRGs in the green sectors (containing chloroplasts but deficient in PSII repair) of mature leaves were similar to those in the cotyledons (Supplemental Fig. S5), indicating that photosynthetically competent chloroplasts lacking PSII repair were indispensable in mediating this retrograde signaling in the var2-9 mutant.
The genes down-regulated in the var2-9 mutant in comparison with wild-type seedlings were also analyzed (Supplemental Fig. S6A). A total of 224 genes were down-regulated in the var2-9 mutant by at least twofold (FDR < 0.05; Supplemental Fig. S6B; Supplemental Table S6), and GO analysis revealed a significant enrichment of genes belonging to response to starvation (P = 3.74E-09), followed by response to extracellular stimulus (P = 1.69E-08; Supplemental Fig. S6C). As anticipated, genes involved in photosynthesis (P = 1.82E-05) were down-regulated in the var2-9 mutant, indicating a coupled expression of photosynthesis-associated genes to the dysfunctional chloroplasts in the var2-9 mutant. In addition, a subset of genes related to jasmonic acid, an antagonist of SA, was down-regulated. These genes include LIPOXYGENASE2, JASMONIC ACID CARBOXYL METHYLTRANSFERASE, and JASMONIC ACID RESPONSIVE2 (Supplemental Fig. S6D).
We also noticed that only 11 proteins among those differentially accumulated chloroplast proteins in the var2-9 mutant versus the wild type were found to coincide with their transcript abundance (Supplemental Table S7). This suggests that the var2-9-conferred chloroplast proteome mostly resulted from the imbalanced proteolysis and that the retrograde signaling instigated via the impaired proteostasis is mainly involved in extraplastidic stress responses.
1O2-Triggered Transcriptomes Are Distinct from the var2-9-Induced Transcriptome
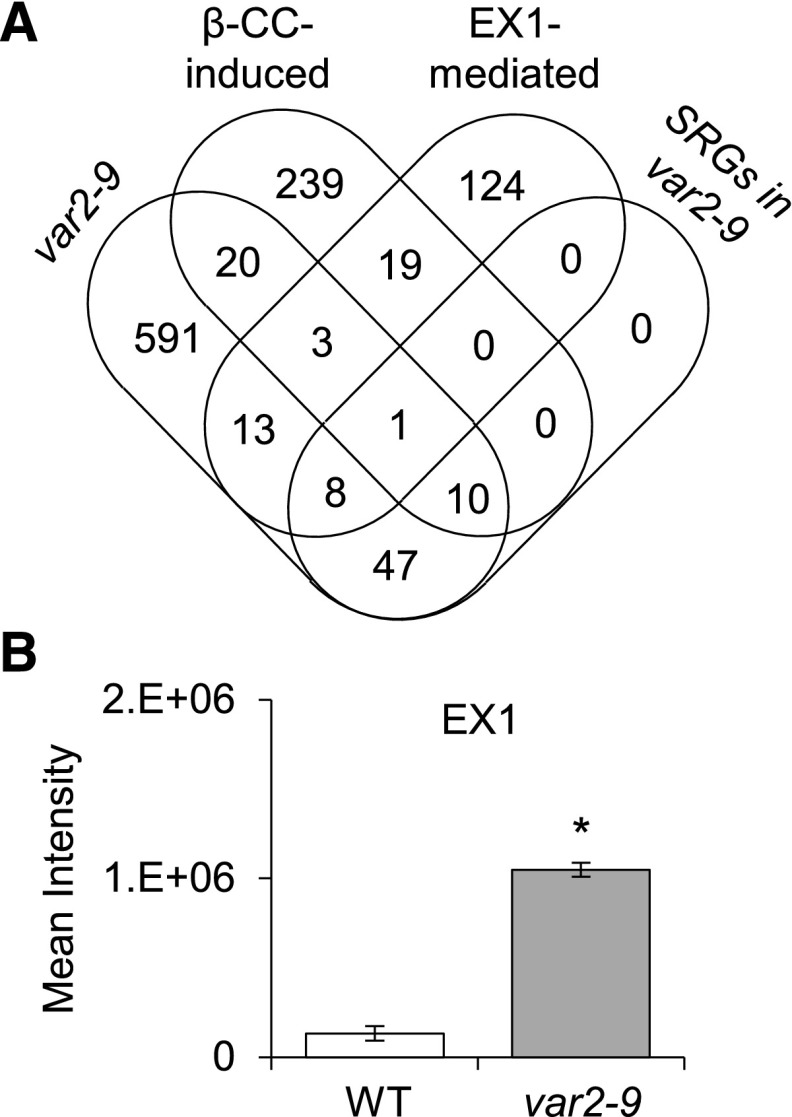
Given that 1O2 is a by-product of photosynthesis and that the photodamage of PSII proteins occurs in the var2-9 seedlings grown under nonstressful light conditions (Fig. 3E), it is possible that 1O2-triggered retrograde signaling may contribute to the var2-9-conferred transcriptome. Two putative 1O2 sensors, EX1 and β-carotene, were shown to mediate distinct 1O2 signaling pathways under nonphotoinhibitory and photoinhibitory conditions, respectively (Wagner et al., 2004; Ramel et al., 2012; Dogra et al., 2017, 2018). To determine whether EX1 or β-carotene contributes to the changes in nuclear gene expression in the var2-9 mutant, the EX1-mediated and β-CC-induced transcriptomes (Ramel et al., 2012; Dogra et al., 2017) were compared with the transcriptome caused by FtsH2G267D (Fig. 5A). The results indicated that the 1O2-triggered transcriptomes are largely distinct from that of the var2-9 mutant. Of the 693 genes up-regulated in the var2-9 mutant, only 34 and 25 genes were shared with β-CC-induced (292) and EX1-mediated (168) genes, respectively (Fig. 5A). Interestingly, a significant portion of these shared genes (11 of the 34 genes and nine of the 25 genes) are also SA responsive. None of the down-regulated genes in the var2-9 mutant were found in the β-CC-induced or EX1-mediated transcriptome (Supplemental Table S6). These results indicate that both EX1- and β-carotene-mediated signaling pathways are distinct from the one caused by FtsH2G267D. We also showed previously that FtsH2 coordinates EX1-mediated 1O2 signaling by promoting EX1 degradation (Wang et al., 2016a), suggesting that EX1 could be a substrate of the membrane-bound FtsH protease. Indeed, the chloroplast proteome data showed that EX1 was highly accumulated in the var2-9 mutant (Fig. 5B), confirming that the EX1-mediated 1O2 signaling was impaired in the var2-9 mutant. It is noteworthy that EX1 was less accumulated in var2-ko relative to var2-9 plants, indicating that the FtsH2-deficient FtsH hexameric complexes can degrade EX1 (like D1 and D2 shown in Fig. 2B) under normal light conditions.
Figure 5.
Impaired proteostasis-triggered retrograde signaling in the var2-9 mutant is distinct from that of 1O2-triggered retrograde signaling. A, The genes up-regulated in the var2-9 mutant are largely distinct from those of β-CC- and EX1-dependent SRGs. Among the 693 genes up-regulated in the var2-9 seedlings (Fig. 4A), 34 and 25 genes were shared with β-CC-induced (292) and EX1-mediated (168) genes, respectively. Of the 34 genes also induced by β-CC, 11 genes are SRGs that are also up-regulated in the var2-9 mutant. Of the 25 shared genes also up-regulated via EX1 in response to 1O2, nine genes are SRGs. B, Label-free protein quantification revealed higher accumulation of EX1 in var2-9 than in wild-type (WT) chloroplasts. Error bars indicate sd. The asterisk indicates a statistically significant difference (P < 0.01) determined by Student’s t test relative to the wild type.
No Transcriptional Regulation of Genes Involved in SA Accumulation Was Found in the var2-9 Mutant
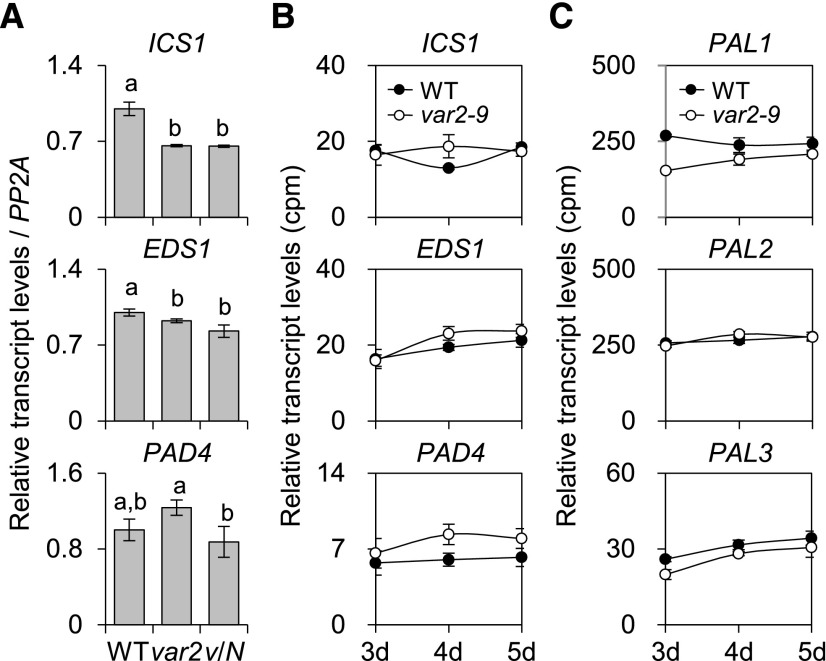
Earlier studies demonstrated that both EDS1 and PHYTOALEXIN DEFICIENT4 (PAD4), encoding lipase-like proteins functioning in basal plant disease resistance, are required for the accumulation of SA (Zhou et al., 1998; Feys et al., 2001; Wiermer et al., 2005). EDS1 and PAD4 are shown to form a heterodimer, which might be required in the amplification of defense responses against pathogens (Feys et al., 2001). To determine whether the accumulation of SA in the var2-9 mutant is mediated by the transcriptional regulation of genes involved in SA synthesis, the transcript levels of ICS1, EDS1, and PAD4 were examined by RT-qPCR. Interestingly, no obvious differences in their transcript levels were observed between var2-9 and wild-type seedlings (Fig. 6A), coinciding with the RNA-seq results (Fig. 6B). In addition, the expression levels of the selected nuclear genes encoding enzymes involved in the cytosolic PAL pathway remain unchanged compared with those in the wild type (Fig. 6C). These results indicate that SA levels were increased in the var2-9 mutant without transcriptional reprogramming of genes involved in the biosynthesis and accumulation of SA.
Figure 6.
The var2-9 mutant may accumulate SA in the absence of transcriptional reprogramming of genes involved in SA accumulation. A and B, Results of RT-qPCR (A) and RNA-seq (B) analyses show no noticeable changes of the transcript levels of ICS1, EDS1, and PAD4 in the var2-9 seedlings compared with the wild type (WT) and var2-9 cpNahG (v/N). For the RT-qPCR, PP2A was used as an internal control, and the data represent means of three independent biological replicates. Error bars indicate sd. Lowercase letters in A indicate statistically significant differences between mean values (P < 0.05, one-way ANOVA with posthoc Tukey’s HSD test). C, Transcript levels of three genes (PAL1, PAL2, and PAL3) encoding PAL in wild-type and var2-9 plants were obtained by RNA-seq analysis. Error bars indicate sd (n = 3).
Up-Regulation of SRGs in the var2-ko Mutant upon Exposure to Photooxidative Stress
The accumulation of Dam-PSII and the disrupted chloroplast proteostasis (Fig. 2A) caused by the impaired substrate-unfolding activity of FtsH2G267D play a causative role in triggering the retrograde signaling via SA. This assumption prompted us to examine the transcript levels of SRGs in the var2-ko seedlings upon exposure to photoinhibitory conditions, in which chloroplasts highly demand PSII repair and thus may lead to a similar accumulation of Dam-PSII and the disrupted chloroplast proteostasis as in the var2-9 seedlings grown under normal light conditions. In agreement with this assumption, the selected SRGs were highly up-regulated in var2-ko plants upon exposure to the photoinhibitory light conditions compared with the wild type (Supplemental Fig. S7).
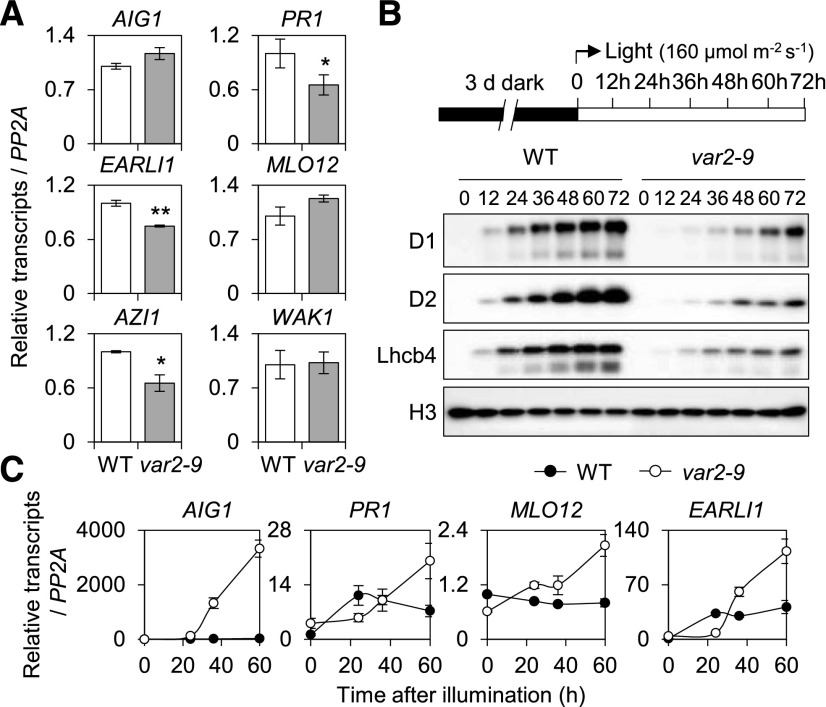
Light Is Indispensable for SRG Expression in the var2-9 Mutant
To explore whether the var2-9-induced SRGs are directly associated with the photosynthetic activity, which requires efficient PSII repair, the transcript levels of the SRGs were determined in dark-grown var2-9 and wild-type etiolated seedlings using RT-qPCR (Fig. 7A). The results showed no significant changes in their abundance in var2-9 as compared with wild-type plants. During deetiolation, however, a gradual and significant up-regulation of the SRGs was observed only in var2-9 but not in wild-type seedlings with the concurrent emergence of the PSII core proteins (Fig. 7, B and C). It is notable that the accumulation of D1, D2, and the light-harvesting protein Lhcb4 was remarkably delayed in var2-9 compared with wild-type plants (Fig. 7B), which supports the previous findings that FtsH2 functions not only in PSII repair but also in thylakoid biogenesis (Bailey et al., 2002; Sakamoto et al., 2002).
Figure 7.
Light is essential for the expression of SRGs in the var2-9 mutant. A, The transcript levels of selected SRGs (Fig. 4F) were determined in 5-d-old var2-9 and wild-type (WT) etiolated seedlings by RT-qPCR. The data represent means of three independent biological replicates. Error bars indicate sd. Asterisks indicate statistically significant differences (*, P < 0.05; **, P < 0.01) determined by Student’s t test relative to the wild type. B and C, Three-day-old etiolated seedlings were illuminated at the light intensity of 160 μmol m−2 s−1 at 22°C ± 2°C. Total RNA and protein were extracted at different time points as indicated. Accumulation of PSII proteins, such as D1, D2, and Lhcb4, during deetiolation was analyzed by immunoblot analysis (B), and the relative transcript levels of SRGs were determined by RT-qPCR (C). Gene expression levels are means ± sd of three independent biological samples. PP2A was used as an internal control. For immunoblot analysis, histone (H3) was used as a loading control.
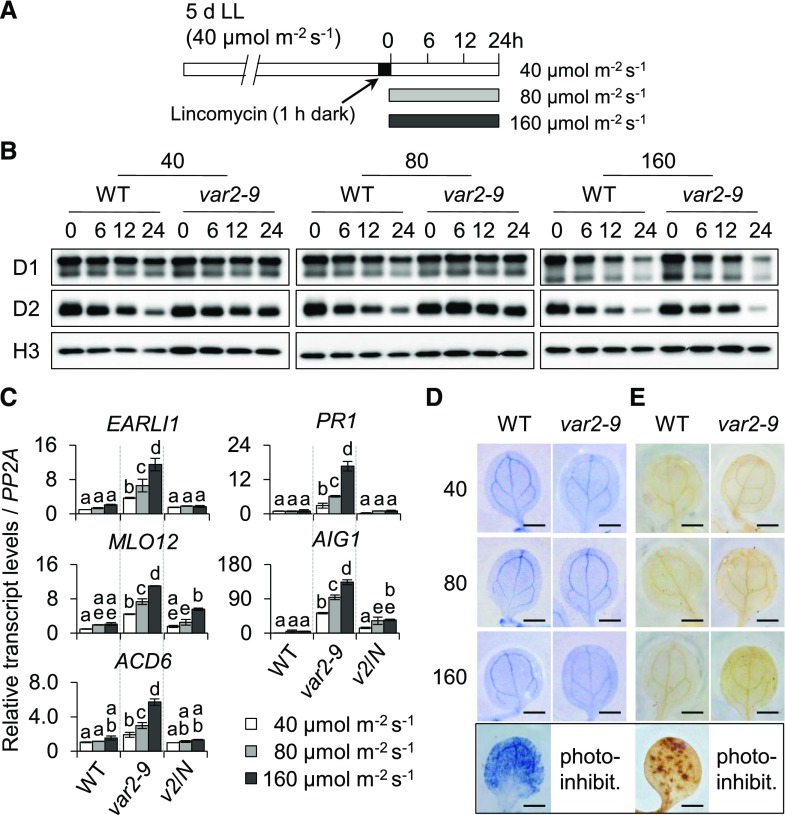
In addition, the effect of light intensity on the expression of SRGs in the var2-9 mutant was examined. The 5-d-old var2-9 and wild-type seedlings that were initially grown under CL for 5 d were subjected to different light intensities as indicated (Fig. 8A). The degradation rates of D1 and D2 in both wild-type and var2-9 seedlings treated with lincomycin were augmented in accordance with the light intensity, but it was less obvious in var2-9 compared with wild-type plants (Fig. 8B). The transcript levels of SRGs were determined under the same experimental conditions, except for the lincomycin treatment, to see the effect of light dose on the SRG expression. To further provide direct evidence that SA mediates the correlation between light dose and SRG expression, the expression levels of SRGs were also examined in var2-9 cpNahG transgenic seedlings under the same experimental conditions. It was clear that the transcript abundances of SRGs are proportional to the light intensity in the var2-9 mutant (Fig. 8C), suggesting that the expression of SRGs tends to enhance with the increased rate of photodamage. The RT-qPCR result also demonstrated a causal impact of SA in SRG expression, as is evident from the repression of the SRGs in the var2-9 cpNahG seedlings (Fig. 8C). Moreover, we determined foliar ROS levels to observe whether gradually increasing light intensity leads to gradual accumulation of ROS in the var2-9 mutant, which may act to increase cellular SA contents. As the oxygen signaling pathways were dormant in var2-9 plants under the examined growth conditions (Fig. 5), cellular superoxide anion (O2−) and hydrogen peroxide (H2O2) levels were determined. The wild-type seedlings exposed to photoinhibitory conditions were used as positive controls, since this condition leads to the accumulation of foliar ROS. In contrast to control seedlings, an accumulation of O2− and H2O2 was not observed in the var2-9 mutant (Fig. 8, D and E), indicating that ROS were not the cause of SRG expression.
Figure 8.
Increased photodamage in PSII core proteins reinforces the expression of SRGs in the var2-9 mutant. A, Schematic diagram showing the conditions used to examine the relative degradation rate of PSII proteins. Five-day-old var2-9 and wild-type seedlings initially grown under CL (40 μmol m−2 s−1 at 22°C ± 2°C) conditions were treated with 5 mm lincomycin for 1 h in the dark to block the de novo synthesis of chloroplast proteins and were then placed under different light intensities as indicated. B, The degradation rates of D1 and D2 upon exposure to different light intensities were analyzed by immunoblot analysis at the indicated time points. Histone (H3) was used as a loading control. C to E, With the exception of lincomycin treatment, under the same experimental conditions, wild-type (WT), var2-9, and var2-9 cpNahG (v2/N) seedlings were treated with different light intensities for 24 h. C, The transcript levels of SRGs were examined by RT-qPCR. Gene expression levels are means ± sd (n = 3 independent biological samples), normalized to that of PP2A. Lowercase letters indicate significant differences between genetic backgrounds (P < 0.05, one-way ANOVA with posthoc Tukey’s HSD test). D and E, O2− (D) and H2O2 (E) were detected with nitroblue tetrazolium (NBT) and 3,3′-diaminobenzidine (DAB) staining, respectively. Wild-type seedlings subjected to photoinhibitory conditions (photo-inhibit.; 300 μmol m−2 s−1 at 10°C ± 2°C) were used as positive controls producing O2− and H2O2 (bottom row). Representative images are shown at the same scale. Bars = 1 cm.
Key SA Signaling Components EDS1 and PAD4 Mediate Retrograde Signaling
Upon translocation to the cytosol through the SA transporter EDS5 located in the chloroplast envelope membrane, several SA-signaling components, such as NPR1, EDS1, and PAD4, were shown to relay SA signaling to the nucleus (Nawrath et al., 2002). To explore whether they are involved in the var2-9-conferred SRG expression, var2-9 eds1 and var2-9 pad4 double mutants were generated and the transcript levels of SRGs were determined using RT-qPCR. The inactivation of either of these key SA-signaling components in var2-9 plants resulted in a significant repression of the SRGs (Fig. 9). These results therefore establish a linear retrograde signaling pathway from the PSII proteostasis to the nuclear gene expression changes, wherein chloroplast-synthesized SA, EDS5, and EDS1-PAD4 are successively entailed, reminiscent of the pathway activated during the plant-pathogen interaction.
Figure 9.
SA, EDS1, and PAD4 are required for the expression of SRGs in the var2-9 mutant. Five-day-old wild-type (WT), var2-9, var2-9 cpNahG (v2/N), var2-9 eds1 (v2/e1), and var2-9 pad4 (v2/p4) seedlings grown under CL (40 μmol m−2 s−1 at 22°C ± 2°C) were collected to analyze the transcript levels of SRGs using RT-qPCR. Values are means ± sd of three independent biological samples. PP2A was used as an internal control. Lowercase letters indicate statistically significant differences between mean values (P < 0.05, one-way ANOVA with posthoc Tukey’s HSD test).
To investigate whether this chloroplast-mediated retrograde signaling potentiates plant immunity, 3-week-old wild-type, var2-ko, and var2-9 plants were surface inoculated with the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Pst DC3000) and the resistance was assessed by determining bacterial multiplication in plant tissue. Remarkably, the result showed an essential role of FtsH2 (or PSII repair) against bacterial pathogens: compared with the wild type, both var2-9 and var2-ko mutant plants were more susceptible to Pst DC3000 (Supplemental Fig. S8), which is in line with a previous report showing that chloroplasts rapidly accumulate ROS upon the perception of pathogens (de Torres Zabala et al., 2015). Presumably, the chloroplast-generated ROS damage the PSII, which needs to be repaired to maintain PSII homeostasis during plant-pathogen interaction. This might be one of the reasons why the var2-9 mutant also shows susceptibility to Pst DC3000 despite its primed defense responses. However, the enhanced resistance of var2-9 plants to Pst DC3000 in comparison with var2-ko could be directly associated with the increased levels of SA and SRGs.
DISCUSSION
Besides being clearly linked to signaling, 1O2 is a prime cause of photodamage of PSII. Interestingly, in cyanobacteria, the Dam-PSII core protein D1 regulates gene expression after being cleaved by proteases (Tyystjärvi et al., 1996). However, to date, the role of the photodamaged PSII proteins and/or its impaired proteostasis in the intracellular signaling network has not been elucidated in land plants. One major challenge to deciphering this problem is the difficulty in dissecting the 1O2-triggered signaling pathway from the photodamage-induced one. In this study, we have tackled this problem by using Arabidopsis var2-9 mutant plants whose PSII proteostasis is largely impaired because of the missense mutation in the ATP-binding domain of the FtsH2 protease (Fig. 1). As anticipated, var2-9 mutant plants accumulate photodamaged PSII core proteins after the onset of photosynthesis (Fig. 2, B and D). Therefore, the var2-9 mutant provides a noninvasive system for investigating the intracellular signaling network including RS resulting from the impaired PSII proteostasis. By utilizing the var2-9 mutant as a biological tool, we reveal that an impaired PSII proteostasis leads to the activation of retrograde signaling via the stress hormone SA after the onset of photosynthesis (Fig. 4, C and D).
Considering that 1O2 is a by-product of photosynthesis and that FtsH2G267D results in the accumulation of Dam-PSII, there is a possibility that the var2-9-conferred transcriptome is rather a result from the 1O2-triggered and EX1-dependent RS (Wagner et al., 2004; Lee et al., 2007; Dogra et al., 2017). However, we may rule out this possibility because the transcript levels of the majority of 1O2-responsive genes whose expression is mediated by EX1 via RS remain unchanged in var2-9 plants (Fig. 5A). On the other hand, one can also suppose that β-carotene, another putative 1O2 sensor, may direct RS in the var2-9 mutant. However, this is unlikely, since the accumulation of its volatile compounds that act as signaling molecules is only observed under extreme light stress conditions (Ramel et al., 2012, 2013a). Accordingly, the majority of the nuclear genes usually up-regulated via β-carotene-mediated 1O2 signaling remain unaffected in var2-9 plants (Fig. 5A).
It is widely accepted that plants inoculated with biotrophic bacterial pathogens accumulate SA mainly through the chloroplast ICS pathway. The rapid up-regulation of ICS1, encoding a key enzyme in the ICS pathway, has been associated with the transient rapid accumulation of SA during plant-pathogen interactions (Lee et al., 1995; Wildermuth et al., 2001). In contrast to this, the accumulation of subtle amounts of SA in var2-9 seedlings grown under CL conditions is not closely coupled to the transcriptional control of ICS1 (Fig. 6, A and B), suggesting that the dysfunctional chloroplasts in the var2-9 mutant may directly stimulate the ICS pathway. Intriguingly, a chorismate synthase, which catalyzes the transformation of 5-enolpyruvylshikimate 3-phosphate to chorismate, a precursor of SA, was previously found to be regulated by the redox status of the cofactor FMN, suggesting a possible posttranslational regulation of the ICS pathway (Macheroux et al., 1999). Notably, the uncoupled accumulation of SA to the transcript levels of ICS1 also suggests that SA may directly mediate retrograde signaling in the var2-9 mutant. Given that chloroplast-driven signaling molecules, also called plastid factors, directly emerge from chloroplasts in response to environmental changes (Chan et al., 2016; Kleine and Leister, 2016), SA accumulated in var2-9 plants without transcriptional control meets the aforementioned criteria to be considered as a genuine plastid factor. At present, we cannot exclude the possibility that SA O-β-glucoside, a vacuolar storage form of SA, might contribute to the increased levels of SA in the var2-9 mutant. Unlike SA, SA O-β-glucoside is biologically inactive and thus has no function in inducing the expression of SRGs. However, it can be rapidly converted into active SA by an SA β-glucosidase (Hennig et al., 1993; Seo et al., 1995; Dean et al., 2005; Song et al., 2008).
The susceptibility of the var2-9 mutant to Pst DC3000, in spite of the up-regulated SRGs (Supplemental Fig. S8), suggests a crucial role of PSII repair (or photoprotection) toward bacterial invasion. Therefore, we propose that during plant-pathogen interaction, the PSII-generated ROS (de Torres Zabala et al., 2015) might damage the PSII, which needs to be reassembled following the removal of the damaged proteins by FtsH protease. However, once chloroplast photodamage exceeds the repair capacity, the augmented photodamage and accumulation of damaged proteins may concurrently stimulate the ICS pathway to synthesize SA, which may contribute to the defense response via SA-mediated retrograde signaling. In this scenario, the impaired PSII repair would impact the establishment of the interaction with the pathogen, causing an enhanced susceptibility despite the subsequent production of SA. Interestingly, it was demonstrated that Pst DC3000 secretes a type III effector protein that attenuates the PSII-mediated ROS burst upon its translocation into the chloroplasts (Rodríguez-Herva et al., 2012). In conjunction with our findings, this previous report implies that PSII damage and the concurrent SA accumulation are entailed in plant immunity and that the chloroplast-targeted effector protein counteracts this response. The susceptibility of var2-ko and var2-9 mutants to both Pst DC3000 and light stress (Fig. 1E) also agrees with the existence of cross talk between the light-induced acclimation signaling pathway and immune responses (Muhlenbock et al., 2008; Trotta et al., 2014).
Our findings also raise questions that need to be answered to gain insight into this retrograde signaling pathway. First, how is the chloroplast proteostasis system intimately connected with the expression of SRGs? Second, does biotic stress lead to the impaired PSII proteostasis and the accumulation of photodamaged proteins, which eventually result in the instigation of retrograde signaling via SA? Answering these questions would enhance our current understanding regarding the role of chloroplasts in plant stress responses.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All the Arabidopsis (Arabidopsis thaliana) seeds were derived from the Columbia ecotype and were harvested on the same day from plants grown under CL (40 µmol m−2 s−1, 22°C ± 2°C). Arabidopsis mutant seeds used in this study were previously described elsewhere, including var2-9 (Sakamoto et al., 2004), eds1-2 (Bartsch et al., 2006), and eds5-1 (Nawrath et al., 2002). The cpNahG transgenic line overexpressing bacterial salicylate hydroxylase in chloroplast was reported previously (Fragnière et al., 2011). Arabidopsis T-DNA insertional mutant seeds of var2-ko (SAIL_253_A03), ics1 (SALK_042603), and pad4 (SALK_206548) were obtained from the Nottingham Arabidopsis Stock Centre. Homozygous double mutants and transgenic lines in the var2-9 background, including var2-9 eds1-2, var2-9 ics1, var2-9 eds5-1, var2-9 pad4, and var2-9 cpNahG, were obtained by crossing the two genotypes. Seeds were sterilized and plated on Murashige and Skoog medium (Duchefa Biochemie) with 0.8% (w/v) agar, supplemented with 0.5% (w/v) Suc. Seeds were stratified for 3 d at 4°C in darkness and then placed under CL (40 µmol m−2 s−1) at 22°C unless otherwise stated.
Plasmid Construction and Complementation Assay
The stop codon-less genomic VAR2 DNA containing the 2-kb promoter region was amplified by PCR using the primer pair 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTTAACTTTCACAACAATTATAGCCT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTATTAGACAGCAGCTGGTGTTGGT-3′ and cloned into a pDONR221 Gateway vector (Invitrogen) through the Gateway BP reaction (Invitrogen). Subsequently, the cloned gene was destined into the Gateway-compatible plant binary vector pGWB650 (Nakagawa et al., 2007) for C-terminal fusion with G3GFP through the Gateway LR reaction (Invitrogen). The resulting vector was transformed by electroporation into Agrobacterium tumefaciens strain GV3101. Transgenic plants in the var2-9 background were generated using A. tumefaciens-mediated transformation using the floral dip method (Clough and Bent, 1998), and homozygous transgenic plants were selected on Murashige and Skoog medium containing 12.5 mg L−1 Basta (Sigma-Aldrich). More than three independent transgenic lines were generated and used for the complementation assays.
RNA Extraction and RT-qPCR
Total RNA (1 µg) extracted using a Spectrum Plant Total RNA Kit (Sigma-Aldrich) was treated with RQ1 RNase-free DNase I (Promega). First-strand cDNA was synthesized by utilizing oligo(dT)15 primer (Promega) and Improm II reverse transcriptase (Promega) according to the kit’s protocol. The RT-qPCR was carried out using the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) and iTaq Universal SYBR Green PCR master mix (Bio-Rad). The relative transcript level of each gene was determined by using the comparative ΔCt method and normalized to the transcript level of PP2A (At1g13320). Primer sequences used in this study can be found in Supplemental Table S8.
Protein Extraction and Western-Blot Analyses
Total proteins were extracted from 5-d-old wild-type and var2-9 seedlings treated with or without lincomycin using the homogenization buffer (0.0625 m Tris-HCl [pH 6.8], 1% [w/v] SDS, 10% [v/v] glycerin, and 0.01% [v/v] 2-mercaptoethanol) and then quantified with the Pierce BCA protein assay kit (Thermo Scientific). For lincomycin treatment, seedlings were submerged in a solution containing 5 mm lincomycin hydrochloride (Sigma-Aldrich) and 0.002% (v/v) Silwet L-77 (GE Healthcare) for 30 min at room temperature. After treatment, the seedlings were further incubated under CL and collected at the indicated time points. The total proteins were separated by 10% SDS-PAGE gels and blotted onto an Immun-Blot polyvinylidene difluoride membrane (Bio-Rad). D1, phosphorylated D1, and D2 proteins were immunochemically detected using rabbit anti-D1 (1:10,000 dilution; Agrisera), rabbit anti-phosphorylated D1 (1:10,000; Agrisera), and rabbit anti-D2 (1:10,000; Agrisera) antibodies, respectively. Rabbit anti-histone H3 (1:3,000; Agrisera) and rabbit anti-UGPase (1:3,000; Agrisera) antibodies were used as loading controls.
RNA-Seq Library Construction and Data Analysis
Total RNA was extracted from three independent biological replicates of 3-, 4-, and 5-d-old wild-type and var2-9 seedlings grown under CL conditions (40 µmol m−2 s−1). Total RNA extracted using the RNeasy Plant Mini Kit (Qiagen) was subjected to on-column DNase digestion with RNase-free DNase Set (Qiagen) according to the manufacturer’s instruction. The purity of RNA was verified by a Nano Photometer spectrophotometer (IMPLEN). The Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies) was used to measure RNA concentration, and the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies) was used to evaluate RNA integrity. Only RNA samples that passed the quality control were used for RNA-seq analyses. RNA-seq libraries were constructed using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer’s instructions. RNA-seq libraries were sequenced on an Illumina HiSeq 2500 platform to generate 100-bp paired-end reads.
Bioinformatics Analysis
The sequencing data were filtered by SolexaQA (v2.2) to remove low-quality reads and to extract pair reads. Clean reads were then mapped to the Arabidopsis genome (TAIR10) using TopHat (Trapnell et al., 2009). After mapping, raw counts of annotated genes were obtained by the Python software HTseq-count. Lowly expressed genes were removed, and genes with an expression level of at least one transcript per million in at least one of the three replicates were selected for differential expression analysis. The R package edgeR, which uses counts per gene in different samples from the HTseq-count as input and performs data normalization using the trimmed mean of M values method, was used to identify differentially expressed genes (Robinson et al., 2010). The genes with at least twofold change in expression and FDR < 0.05 were considered to be differentially expressed. Gene expression was normalized to transcript per million according to the total number of mapped clean reads in each of library. The log2 values of normalized expression were used to build an expression matrix, and subsequent clustering and visualization were done using Multi-Experiment Viewer (MeV 4.9.0). GO enrichment analysis of differentially expressed genes was carried out using the Generic GO Term Finder tool (http://go.princeton.edu/cgi-bin/GOTermFinder) to determine the significantly enriched GO terms in the category of biological processes (Katari et al., 2010) with a significance of P < 0.05.
Confocal Laser-Scanning Microscopy
Cotyledons and leaves from 10-d-old transgenic wild-type and var2 seedlings expressing the plastid-localized marker protein RbcS-GFP under the control of a CaMV 35S constitutive promoter (Kim and Apel, 2004) were used for confocal laser-scanning microscopy analysis using a Leica TCS SP8 (Leica Microsystems).
Label-Free Quantitation
The intact chloroplasts were isolated from the 3-week-old plants of the wild type, var2-9, and var2-ko grown under CL conditions. The chloroplasts corresponding to equal amounts of chlorophyll were lysed to extract proteins. For normalization, protein contents from each sample were correlated with corresponding chloroplast numbers and chlorophyll amounts. Based on an obvious correlation between protein amount and chloroplast number (and chlorophyll amounts), equal amounts of chloroplast protein (2 µg µL−1) from independent biological samples were separated and analyzed by nanoAquity ultra-performance liquid chromatography coupled with the Q Exactive mass spectrometer.
Raw data files were processed and analyzed using MaxQuant software (version 1.5.8.3), and the intensity-based absolute quantification (iBAQ) algorithm was enabled as described previously (Luber et al., 2010; Schwanhäusser et al., 2011). Parent ion and MS2 spectra were searched against the FASTA database (http://www.arabidopsis.org/). The precursor ion tolerance was set at 7 ppm with an allowed fragment mass deviation of 20 ppm. Carbamidomethylation of Cys was set as a fixed modification, while N-terminal acetylation and oxidation of Met and Trp were defined as variable modifications. Peptides of a minimum of six amino acids and a maximum of two missed cleavages were allowed. The FDR was set to 0.01 for both peptide and protein identification. The iBAQ values were used to calculate the protein expression and abundance. Proteins were chosen if iBAQ values were detected in at least two of the three replicates. The expression matrix of the proteins was represented as a heat map prepared using Multi-Experiment Viewer (MeV 4.9.0). After log2 transformation of iBAQ values and data imputation (replacing missing values by normal distribution), proteins exhibiting at least twofold accumulation with P < 0.05 (Student’s t test) were considered as differentially accumulated in var2-9 in comparison with var2-ko or wild-type plants. The proportions of proteins undergoing oxidation were calculated using intensity values of the protein and the corresponding oxidized peptides.
Determinations of Photochemical Efficiency
Fv/Fm was determined with FlourCAM FC800-C/1010GFP (Photon Systems Instruments) according to the instrument manufacturer’s instructions.
Transmission Electron Microscopy
Cotyledons from 5-d-old wild-type and var2-9 seedlings were excised and prefixed in 0.1 m phosphate-buffered saline (PBS; pH 7.2) containing 5% (w/v) glutaraldehyde for 2 h at room temperature. After washing with 0.1 m PBS buffer three times, postfixation was performed in 1% (w/v) osmium tetroxide dissolved in 0.1 m PBS for 2 h. The samples were dehydrated serially in ethanol (seven steps from 30% to 100%) and embedded in Spurr Epoxy Resin (Electron Microscopy Sciences). Ultrathin sections were cut at 70 nm on a Leica UC7 Ultramicrotome (Leica Microsystems) using an ultra diamond knife (Diatome), then stained with uranyl acetate and lead citrate. Transmission electron microscopy analysis was performed on a Hittach HT7700 transmission electron microscope (Hitachi). At least 10 plants were examined for each genotype.
SA Measurements
The endogenous levels of free SA in wild-type, var2-9, and var2-9 cpNahG seedlings grown on agar plates were analyzed by an ACQUITY ultra-performance liquid chromatography system (Waters) coupled with a triple-TOF 5600+ mass spectrometer (AB Sciex). Briefly, the seedling was homogenized to a fine powder in liquid nitrogen with a mortar and pestle. Approximately 25 mg of the fine powder was mixed with 2 ng of d4-SA (internal standard; Sigma-Aldrich) and 500 µL of extraction solvent (acetone:50 mm citric acid [7:3, v/v]). After 2 h of shaking (1,000 rpm) in the dark at 4°C, the homogenate was centrifuged at 13,000 rpm for 5 min. The supernatant was transferred to a fresh tube and dried under a vacuum. The dried sample was dissolved in 300 µL of diethyl ether (Sigma-Aldrich) and then centrifuged at 5,000 rpm for 5 min. The upper organic phase was collected and dried under a vacuum. Subsequently, the dried samples were dissolved in 200 µL of 50% (v/v) acetonitrile (Sigma-Aldrich). After centrifuging at 10,000 rpm for 5 min, the supernatant was used for ultra-performance liquid chromatography-time of flight-mass spectrometry analysis.
ROS Determination
O2− and H2O2 were detected with NBT and DAB staining, respectively. Briefly, 5-d-old wild-type, var2-9, and var2-9 cpNahG seedlings initially grown under CL (40 µmol m−2 s−1) were treated with three different light intensities (40, 80, and 160 µmol m−2 s−1) for 24 h. The seedlings were then submerged and vacuum infiltrated for 10 min with NBT staining solution (1 mg mL−1 NBT, 10 mm sodium phosphate buffer, and 10 mm NaN3, pH 7.4) and DAB staining solution (1 mg mL−1 DAB, pH 3.8), respectively. After 2 h of incubation at room temperature, stained seedlings were boiled in bleaching solution (ethanol:acetic acid:glycerol [3:1:1, v/v/v]) for 10 min and then stored in 95% (v/v) ethanol for taking images.
Bacterial Pathogen Infections and Growth Assays
Pseudomonas syringae pv tomato DC3000 was grown overnight in Luria-Bertani medium at 28°C, washed, and resuspended in 10 mm Mg2Cl containing 0.01% (v/v) Silwet L-77. Fifty milliliters of the bacterial suspension with an OD600 = 0.1 (5 × 107 colony-forming units [cfu] mL−1) was dispensed onto plates containing 3-week-old Arabidopsis seedlings. The plates were incubated for 3 min at room temperature. The bacterial suspension was removed by decantation. Plates containing inoculated plants were incubated at 22°C ± 2°C under CL (80 µmol m−2 s−1). For each biological replicate, four seedlings from each genotype were ground from one plate, using three plates. Two days postinoculation, the weight of the aerial part from inoculated seedlings was measured and the tissue was ground and homogenized in 10 mm Mg2Cl. Diluted samples were plated onto Luria-Bertani medium, and cfu were counted after 2 d. The cfu count was normalized as cfu mg−1 using the total weight of inoculated seedlings. Bacterial populations were evaluated in three independent experiments. The results were statistically analyzed using Student’s t test.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis TAIR database (https://www.arabidopsis.org) under the following accession numbers: ACD6 (AT4G14400), AZI1 (AT4G12470), EARLI1 (AT4G12480), WAK1 (AT1G21250), AIG1 (AT1G33960), MLO12 (AT2G39200), ICS1 (AT1G74710), EDS1 (AT3G48090); PAD4 (AT3G52430), PR1 (AT2G14610), FTSH1 (AT1G50250), FTSH2 (AT2G30950), FTSH5 (AT5G42270), FTSH8 (AT1G06430), and EDS5 (AT4G39030).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The phenotypes of var2-9 and var2-ko seedlings.
Supplemental Figure S2. The relative abundance of FtsH isomers constituting the membrane-bound FtsH hexameric heterocomplex.
Supplemental Figure S3. The degradation rates of PSII core proteins in the var2-9 ics1 and var2-9 eds5 mutants.
Supplemental Figure S4. FtsH2 complements the var2-9-conferred phenotype.
Supplemental Figure S5. The expression of SRGs in the green versus white sectors in the rosette leaves of the var2-9 mutant.
Supplemental Figure S6. Genes down-regulated in the var2-9 mutant.
Supplemental Figure S7. The var2-ko mutant shows increased SRG expression under photoinhibitory conditions.
Supplemental Figure S8. Important role of FtsH2 against bacterial pathogens.
Supplemental Table S1. List of differentially accumulated chloroplast proteins in the var2-9 mutant relative to var2-ko and wild-type plants.
Supplemental Table S2. List of PSII RC proteins exhibiting oxidative modifications in var2-9, var2-ko, and wild-type plants.
Supplemental Table S3. List of genes that were up-regulated in var2-9 as compared with wild-type seedlings.
Supplemental Table S4. List of stress, defense, and immune-related genes up-regulated in var2-9 as compared with wild-type seedlings.
Supplemental Table S5. List of SA-responsive genes up-regulated in var2-9 as compared with wild-type seedlings.
Supplemental Table S6. List of genes down-regulated in var2-9 as compared with wild-type seedlings.
Supplemental Table S7. List of genes exhibiting coabundance of proteins and cognate transcripts in the var2-9 mutant.
Supplemental Table S8. List of qPCR primer sets used in this study.
Acknowledgments
We thank the Core Facilities of Genomics, Proteomics, and Cell Biology in the Shanghai Center for Plant Stress Biology for carrying out RNA sequencing, proteome profiling, and transmission electron microscopy, respectively. We thank Junghee Lee for critical reading of the article.
Footnotes
This work was supported by the Strategic Priority Research Program from the Chinese Academy of Sciences (XDB27040102), the 100-Talent Program of the Chinese Academy of Sciences, and the National Natural Science Foundation of China (31871397 to C.K.), as well as by the Research Fund for International Young Scientists Program of the National Natural Science Foundation of China (31850410478) and a President’s International Fellowship Initiative postdoctoral fellowship from the Chinese Academy of Sciences (2019PB0066 to V.D.).
References
- Anderson LB, Maderia M, Ouellette AJ, Putnam-Evans C, Higgins L, Krick T, MacCoss MJ, Lim H, Yates JR III, Barry BA (2002) Posttranslational modifications in the CP43 subunit of photosystem II. Proc Natl Acad Sci USA 99: 14676–14681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Thompson E, Nixon PJ, Horton P, Mullineaux CW, Robinson C, Mann NH (2002) A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J Biol Chem 277: 2006–2011 [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Niederhauser B, Baumann UM (2009) The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation. Proc Natl Acad Sci USA 106: 21579–21584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dean JV, Mohammed LA, Fitzpatrick T (2005) The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 221: 287–296 [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M, Littlejohn G, Jayaraman S, Studholme D, Bailey T, Lawson T, Tillich M, Licht D, Bölter B, Delfino L, et al. (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat Plants 1: 15074. [DOI] [PubMed] [Google Scholar]
- Dogra V, Duan J, Lee KP, Lv S, Liu R, Kim C (2017) FtsH2-dependent proteolysis of EXECUTER1 is essential in mediating singlet oxygen-triggered retrograde signaling in Arabidopsis thaliana. Front Plant Sci 8: 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra V, Rochaix JD, Kim C (2018) Singlet oxygen-triggered chloroplast-to-nucleus retrograde signalling pathways: An emerging perspective. Plant Cell Environ 41: 1727–1738 [DOI] [PubMed] [Google Scholar]
- Dreaden TM, Chen J, Rexroth S, Barry BA (2011) N-Formylkynurenine as a marker of high light stress in photosynthesis. J Biol Chem 286: 22632–22641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer BB, Ledford HK, Wakao S, Huang SG, Casero D, Pellegrini M, Merchant SS, Koller A, Eggen RI, Niyogi KK (2012) SINGLET OXYGEN RESISTANT 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 109: E1302–E1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragnière C, Serrano M, Abou-Mansour E, Métraux JP, L’Haridon F (2011) Salicylic acid and its location in response to biotic and abiotic stress. FEBS Lett 585: 1847–1852 [DOI] [PubMed] [Google Scholar]
- Goslings D, Meskauskiene R, Kim C, Lee KP, Nater M, Apel K (2004) Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J 40: 957–967 [DOI] [PubMed] [Google Scholar]
- Havaux M. (2014) Carotenoid oxidation products as stress signals in plants. Plant J 79: 597–606 [DOI] [PubMed] [Google Scholar]
- Hennig J, Malamy J, Grynkiewicz G, Indulski J, Klessig DF (1993) Interconversion of the salicylic acid signal and its glucoside in tobacco. Plant J 4: 593–600 [DOI] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, et al. (2010) VirtualPlant: A software platform to support systems biology research. Plant Physiol 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2009) Protein quality control in chloroplasts: A current model of D1 protein degradation in the photosystem II repair cycle. J Biochem 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2014) Phosphorylation of photosystem II core proteins prevents undesirable cleavage of D1 and contributes to the fine-tuned repair of photosystem II. Plant J 79: 312–321 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W (2018) FtsH protease in the thylakoid membrane: Physiological functions and the regulation of protease activity. Front Plant Sci 9: 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatoon M, Inagawa K, Pospísil P, Yamashita A, Yoshioka M, Lundin B, Horie J, Morita N, Jajoo A, Yamamoto Y, et al. (2009) Quality control of photosystem II: Thylakoid unstacking is necessary to avoid further damage to the D1 protein and to facilitate D1 degradation under light stress in spinach thylakoids. J Biol Chem 284: 25343–25352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Apel K (2004) Substrate-dependent and organ-specific chloroplast protein import in planta. Plant Cell 16: 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Apel K, Laloi C (2008) No single way to understand singlet oxygen signalling in plants. EMBO Rep 9: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Zhang S, Lee KP, Lakshmanan Ashok M, Blajecka K, Herrfurth C, Feussner I, Apel K (2012) Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Leister D (2016) Retrograde signaling: Organelles go networking. Biochim Biophys Acta 1857: 1313–1325 [DOI] [PubMed] [Google Scholar]
- Lee HI, León J, Raskin I (1995) Biosynthesis and metabolism of salicylic acid. Proc Natl Acad Sci USA 92: 4076–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S (2010) Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: A review. J Integr Plant Biol 52: 750–761 [DOI] [PubMed] [Google Scholar]
- Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O’Keeffe M, et al. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32: 279–289 [DOI] [PubMed] [Google Scholar]
- Lv F, Zhou J, Zeng L, Xing D (2015) β-Cyclocitral upregulates salicylic acid signalling to enhance excess light acclimation in Arabidopsis. J Exp Bot 66: 4719–4732 [DOI] [PubMed] [Google Scholar]
- Macheroux P, Schmid J, Amrhein N, Schaller A (1999) A unique reaction in a common pathway: Mechanism and function of chorismate synthase in the shikimate pathway. Planta 207: 325–334 [DOI] [PubMed] [Google Scholar]
- Mishra NP, Francke C, Vangorkom HJ, Ghanotakis DF (1994) Destructive role of singlet oxygen during aerobic illumination of the photosystem-II core complex. Biochim Biophys Acta 1186: 81–90 [Google Scholar]
- Mueller S, Hilbert B, Dueckershoff K, Roitsch T, Krischke M, Mueller MJ, Berger S (2008) General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20: 768–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S (2008) Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis (vol 20, pg 2339, 2008). Plant Cell 20: 3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura H, Komori T, Uemura S, Kanda Y, Shimotani K, Nakai K, Furuichi T, Takebayashi K, Sugimoto T, Sano S, et al. (2012) Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat Commun 3: 926. [DOI] [PubMed] [Google Scholar]
- Ochsenbein C, Przybyla D, Danon A, Landgraf F, Göbel C, Imboden A, Feussner I, Apel K (2006) The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. Plant J 47: 445–456 [DOI] [PubMed] [Google Scholar]
- Ohad I, Kyle DJ, Arntzen CJ (1984) Membrane protein damage and repair: Removal and replacement of inactivated 32-kilodalton polypeptides in chloroplast membranes. J Cell Biol 99: 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Ksas B, Akkari E, Mialoundama AS, Monnet F, Krieger-Liszkay A, Ravanat JL, Mueller MJ, Bouvier F, Havaux M (2013a) Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25: 1445–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Mialoundama AS, Havaux M (2013b) Nonenzymic carotenoid oxidation and photooxidative stress signalling in plants. J Exp Bot 64: 799–805 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Herva JJ, González-Melendi P, Cuartas-Lanza R, Antúnez-Lamas M, Río-Alvarez I, Li Z, López-Torrejón G, Díaz I, Del Pozo JC, Chakravarthy S, et al. (2012) A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell Microbiol 14: 669–681 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Tamura T, Hanba-Tomita Y, Murata M, Sodmergen (2002) The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7: 769–780 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Miura E, Kaji Y, Okuno T, Nishizono M, Ogura T (2004) Allelic characterization of the leaf-variegated mutation var2 identifies the conserved amino acid residues of FtsH that are important for ATP hydrolysis and proteolysis. Plant Mol Biol 56: 705–716 [DOI] [PubMed] [Google Scholar]
- Sattler SE, Mène-Saffrané L, Farmer EE, Krischke M, Mueller MJ, DellaPenna D (2006) Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell 18: 3706–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Seo S, Okamoto M, Seto H, Ishizuka K, Sano H, Ohashi Y (1995) Tobacco MAP kinase: A possible mediator in wound signal transduction pathways. Science 270: 1988–1992 [DOI] [PubMed] [Google Scholar]
- Serrano M, Wang B, Aryal B, Garcion C, Abou-Mansour E, Heck S, Geisler M, Mauch F, Nawrath C, Métraux JP (2013) Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol 162: 1815–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JT, Koo YJ, Seo HS, Kim MC, Choi YD, Kim JH (2008) Overexpression of AtSGT1, an Arabidopsis salicylic acid glucosyltransferase, leads to increased susceptibility to Pseudomonas syringae. Phytochemistry 69: 1128–1134 [DOI] [PubMed] [Google Scholar]
- Stelljes C, Koenig F (2007) Specific binding of D1 protein degradation products to the psbAI promoter in Synechococcus sp. strain PCC 7942. J Bacteriol 189: 1722–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn MA, Marr SK, Inoue K, Inada N, Zubieta C, Wildermuth MC (2007) Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J Biol Chem 282: 5919–5933 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M, Van Breusegem F, Mueller MJ (2008) Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol 148: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta A, Rahikainen M, Konert G, Finazzi G, Kangasjärvi S (2014) Signalling crosstalk in light stress and immune reactions in plants. Philos Trans R Soc Lond B Biol Sci 369: 20130235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjärvi T, Mulo P, Mäenpää P, Aro EM (1996) D1 polypeptide degradation may regulate psbA gene expression at transcriptional and translational levels in Synechocystis sp. PCC 6803. Photosynth Res 47: 111–120 [DOI] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, Op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E, et al. (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Wang L, Kim C, Xu X, Piskurewicz U, Dogra V, Singh S, Mahler H, Apel K (2016a) Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc Natl Acad Sci USA 113: E3792–E3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yamano T, Takane S, Niikawa Y, Toyokawa C, Ozawa SI, Tokutsu R, Takahashi Y, Minagawa J, Kanesaki Y, et al. (2016b) Chloroplast-mediated regulation of CO2-concentrating mechanism by Ca2+-binding protein CAS in the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 113: 12586–12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE (2005) Plant immunity: The EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Hori H, Kai S, Ishikawa T, Ohnishi A, Tsumura N, Morita N (2013) Quality control of photosystem II: Reversible and irreversible protein aggregation decides the fate of photosystem II under excessive illumination. Front Plant Sci 4: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka-Nishimura M, Yamamoto Y (2014) Quality control of photosystem II: The molecular basis for the action of FtsH protease and the dynamics of the thylakoid membranes. J Photochem Photobiol B 137: 100–106 [DOI] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR (2005) Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol 138: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A, Feder A, Adam Z (2005) Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts: Implications for thylakoid formation and photosystem II maintenance. Plant J 42: 609–617 [DOI] [PubMed] [Google Scholar]
- Zhang D, Kato Y, Zhang L, Fujimoto M, Tsutsumi N, Sodmergen, Sakamoto W (2010) The FtsH protease heterocomplex in Arabidopsis: Dispensability of type-B protease activity for proper chloroplast development. Plant Cell 22: 3710–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wang W, Karapetyan S, Mwimba M, Marqués J, Buchler NE, Dong X (2015) Redox rhythm reinforces the circadian clock to gate immune response. Nature 523: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]