PuHSFA4a, which activates the antioxidant program and root development-related genes, directly targets PuGSTU17 and PuPLA2, positively regulating excess Zn tolerance in Populus ussuriensis roots.
Abstract
Zinc (Zn) is an essential micronutrient but in excess is highly toxic to plants. Plants regulate Zn homeostasis and withstand excess Zn through various pathways; these pathways are generally tightly regulated by a specific set of genes. However, the transcription factors involved in excess Zn tolerance have yet to be identified. Here, we characterized a Populus ussuriensis heat shock transcription factor A4a (PuHSFA4a) that acts as a positive regulator of excess Zn tolerance in P. ussuriensis. We used overexpression (PuHSFA4a-OE) and chimeric dominant repressor (PuHSFA4a-SRDX) lines to identify the targets of PuHSFA4a. PuHSFA4a transcription is specifically induced in roots by high Zn. Overexpression of PuHSFA4a conferred excess Zn tolerance and a dominant repressor version of PuHSFA4a increased excess Zn sensitivity in P. ussuriensis by regulating the antioxidant system in roots. PuHSFA4a coordinately activates genes related to abiotic stress responses and root development and directly binds to the promoter regions of glutathione-s-transferase U17 (PuGSTU17) and phospholipase A2 (PuPLA2). PuGSTU17 overexpression significantly increased GST activity and reduced reactive oxygen species levels in roots while PuGSTU17-RNA interference lines exhibited the opposite phenotype. Furthermore, PuPLA2 overexpression promoted root growth under high Zn stress. Taken together, we provide evidence that PuHSFA4a coordinately activates the antioxidant system and root development-related genes and directly targets PuGSTU17 and PuPLA, thereby promoting excess Zn tolerance in P. ussuriensis roots.
Zinc (Zn) is an essential micronutrient but its excess in soil is highly toxic to plants (Broadley et al., 2007; Wang et al., 2009). To cope with Zn toxicity, plants have evolved complex adaptive responses to allow them to tolerate, acclimate to, or avoid excess Zn stress (Lin and Aarts, 2012; Schvartzman et al., 2018). Numerous studies have shown that, after exposure to stress due to the presence of excess Zn, the expression levels of many genes significantly increase or decrease, indicating that transcriptional reprogramming plays an important role in adaptive responses to Zn stress (Lin and Aarts, 2012; Luo et al., 2016).
At the cellular level, high Zn accumulation induces oxidative stress by producing reactive oxygen species (ROS). Elevated ROS levels create a strongly oxidizing environment, causing membrane lipid peroxidation and activating signaling pathways that lead to cell death (Lin and Aarts, 2012). Plants respond to such stresses by activating antioxidant enzymes (Wang et al., 2009). In tomato (Solanum lycopersicum), increased ascorbate peroxidase (APX) activity eliminates H2O2 caused by Zn accumulation in the roots (Xu et al., 2008). Another study in green bean (Phaseolus vulgaris) found that enhanced glutathione reductase activity in leaves may be a response to Zn-induced oxidative stress (Chaoui et al., 1997). Similarly, the enzymes glutathione-s-transferase (GST) and glutathione peroxidase use glutathione as a substrate to detoxify H2O2 and sequester Zn by acting as a precursor for synthesis of phytochelatins (Garg and Kaur, 2013). In addition, several genes have been shown to be involved in excess Zn detoxification. These include: Arabidopsis (Arabidopsis thaliana) AtHMA4 (Verret et al., 2004) and AtMTP1 (Desbrosses-Fonrouge et al., 2005), rice (Oryza sativa) OsHMA2 (Takahashi et al., 2012) and OZT1 (Lan et al., 2013), Saccharomyces cerevisiae ZRC1 (Miyabe et al., 2001), Populus alba AQUA1 (Ariani et al., 2016), and cyanobacteria (Synechococcus PCC 7942) SmtA (Xu et al., 2010). Moreover, in both Drosophila and humans (Homo sapiens), the Zn finger transcription factor (TF) metal-responsive TF-1 appears to confer excess Zn tolerance (Laity and Andrews, 2007). Although these studies have identified many genes involved in Zn detoxification and tolerance, the TFs that regulate the mechanisms of Zn detoxification have not yet been characterized in plants.
Heat shock transcription factors (HSFs) regulate the transcription of genes related to heat and other abiotic stresses (von Koskull-Döring et al., 2007). HSFs are divided into three categories—A, B, and C—based on their structures. HSFA4, a class A HSF, is found in a variety of plant species and confers tolerance to certain environmental stresses. For example, estradiol-dependent induction of Arabidopsis AtHSFA4a confers enhanced tolerance to salt and oxidative stress by interacting with MPK3/MPK6 signaling (Pérez-Salamó et al., 2014). Overexpression of CmHSFA4 enhances salt tolerance by regulating Na+/K+ ion and ROS homeostasis in Chrysanthemum (Li et al., 2018). Ectopic overexpression of Brassica napus BnHSFA4a in Arabidopsis improved desiccation tolerance (Lang et al., 2017). Overexpression of wheat TaHSFA4a conferred cadmium (Cd) tolerance and rice OsHSFA4a knockouts showed Cd hypersensitivity (Shim et al., 2009). However, to date no studies have assessed whether HSF TFs are involved in regulating tolerance to excess Zn stress.
The genus Populus (Populus spp) comprises deciduous hardwood trees that are mainly distributed in the northern hemisphere. Populus has been recognized as a suitable tree for phytoremediation due to its rapid growth, high biomass, extensive root system, and ease of propagation (Luo et al., 2016). Some studies have performed transcriptomic analysis and genetic manipulation to examine the molecular mechanisms responsible for Cd, copper (Cu), and Zn remediation in Populus species, including Populus euphratica (Han et al., 2014), Populus trichocarpa × deltoides (Kohler et al., 2004), Populus canescens (Luo et al., 2016), and Populus nigra (Bittsánszky et al., 2005). Populus spp have been reported to uptake several inorganic pollutants including Zn, but their capacity for heavy metal tolerance may be capable of further improvement (Bittsánszky et al., 2005).
The forested areas in the northeast of China are rich in Zn mineral resources. Years of mining activities have caused the destruction of surface vegetation near Zn mines in these regions (Hu et al., 2013). Populus ussuriensis, belonging to Populus sect. Tacamahaca in the Salicaceae, is a unique tree species native to the forested regions of northeastern China. It is one of the fastest growing tree species—especially in the young forest stage of forest succession—and is thus one of the main tree species used for forest renewal in parts of northeastern China (Su et al., 2001). Thus, P. ussuriensis is an excellent candidate for forest restoration in former Zn mining areas in northeastern China. In this study, we characterized PuHSFA4a, a P. ussuriensis TF belonging to the Class A4a subgroup of the HSF family. We demonstrate that PuHSFA4a specifically confers tolerance to excess Zn in transgenic P. ussuriensis. We also provide evidence that PuHSFA4a affects antioxidant scavenging and root development-related gene expression by directly targeting PuGSTU17 to eliminate ROS in roots and PuPLA2 (phospholipase A) to promote root growth under excess Zn. This study improves our understanding of the physiological and molecular responses to Zn exposure in Populus roots.
RESULTS
PuHSFA4a Is Specifically Induced in Roots by Excess Zn
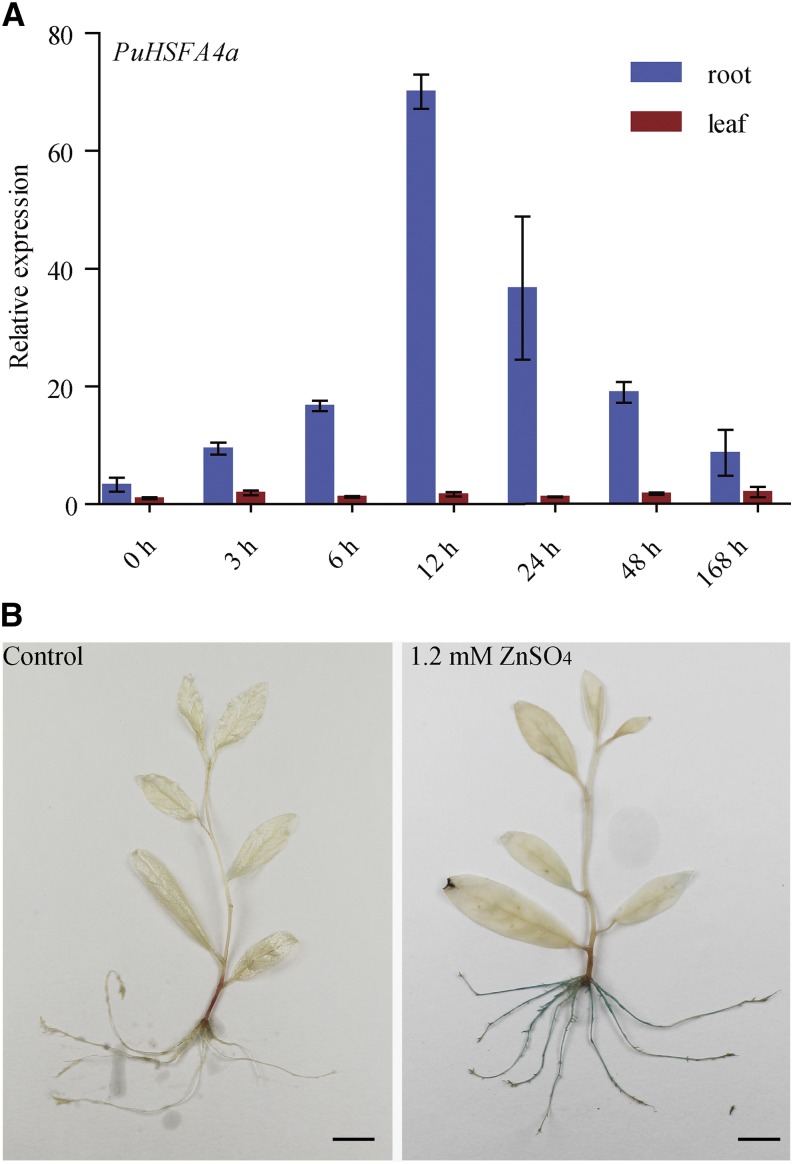
The expression profiles of P. ussuriensis Class A HSF genes in response to abiotic stress were investigated using reverse transcription quantitative PCR (RT-qPCR). Among the Class A PuHSF genes assessed, only PuHSFA4a was substantially upregulated specifically in roots in response to high Zn stress (Fig. 1A; Supplemental Fig. S1A). The expression of PuHSFA4a was higher in roots than in leaves under control conditions (Fig. 1A). The expression level of PuHSFA4a in roots did not significantly increase in response to Cd, Cu, iron (Fe), PEG6000, abscisic acid (ABA), and NaCl stresses. (Supplemental Fig. S1B).
Figure 1.
Relative expression of PuHSFA4a in roots and leaves of P. ussuriensis and histochemical analysis of the expression of the PuHSFA4a promoter in P. ussuriensis tissues as visualized by glucuronidase staining under excess Zn. A, Relative expression of PuHSFA4a in roots and leaves under excess Zn. Two-week–old plants were grown in media supplemented with 1.2 mm of ZnSO4 for 0–48 h and 1 week. Data are presented as means of three biological replicates, and error bars = sd. B, GUS activity staining of ProPuHSFA4a::GUS transgenic plants that were grown on half strength Murashige and Skoog (MS) medium supplemented with or without 1.2 mm of ZnSO4 for 1 week. Scale bars = 1 cm.
Next, the 2,203-bp PuHSFA4a promoter was fused to the GUS (uidA) gene, and the ProPuHSFA4a::GUS construct was transformed into P. ussuriensis. No GUS activity was observed under normal culture conditions (Fig. 1B). However, under high Zn stress, strong GUS activity was detected in whole roots but not in shoots (Fig. 1B). These observations suggest that PuHSFA4a functions specifically in root in response to high Zn stress.
Cloning and Sequence Characterization of PuHSFA4a
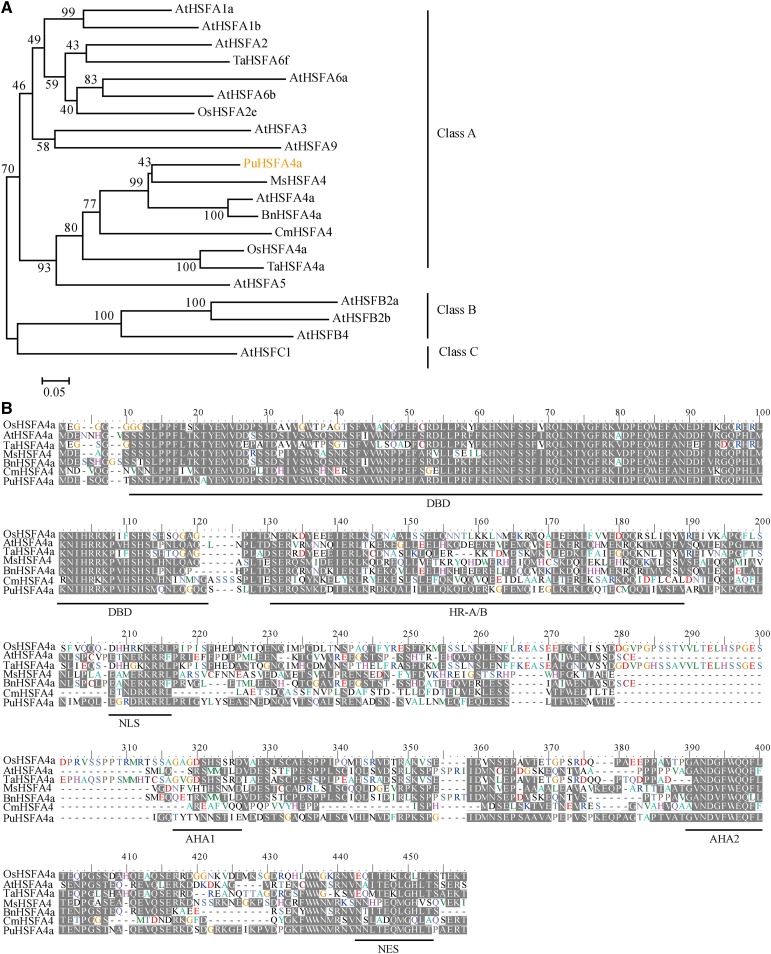
The full-length PuHSFA4a gene was obtained from P. ussuriensis. Phylogenetic analysis was performed on PuHSFA4a and similar HSFA4a proteins found in other plant species. The results of these comparisons showed that PuHSFA4a belongs to the HSF ClassA4a subgroup, where it clustered with AtHSFA4a, BnHSFA4a, MsHSFA4a, CmHSFA4a, OsHSFA4a, and TaHSFA4a (Fig. 2A). Multiple alignment results revealed that PuHSFA4a shares ∼40% to 56% of its amino acid sequence identity with homologous proteins in other plants species (Fig. 2B). Structure analysis showed that PuHSFA4a is a typical class A HSF and contains motifs typical of that class of proteins, including a predicted nuclear localization signal (NLS) domain and a nuclear export signal (NES) that is responsible for nuclear and cytoplasmic localization (Lyck et al., 1997; Li et al., 2018). However, the conserved DNA-binding domain (DBD) of PuHSFA4a does not contain the amino acids Ala-31 and Leu-42, two changes linked to Cd tolerance in TaHSFA4a (Shim et al., 2009).
Figure 2.
Phylogenetic tree and multiple sequence alignments of HSF proteins. A, Phylogenetic tree of HSF proteins. The number next to the branches is the percentage of trees that relate to taxa clustered together. B, Multiple sequence alignment of the HSFA4a amino acid sequence in P. ussuriensis (PuHSFA4a), Arabidopsis (AtHSFA4a), Triticum aestivum (TaHSFA4a), rice (OsHSFA4a), B. napus (BnHSFA4a), Medicago sativa (MsHSFA4a), and Chrysanthemum (CmHSFA4a). PuHSFA4a contains six domains: the DBD, the hydrophobic heptad repeat region for oligomerization (HR-A/B), the NLS, the transcriptional activators AHA1 and AHA2, and the Leu-rich NES domains (underlined in black).
PuHSFA4a Is Localized in the Nucleus and the Cytoplasm and Functions as a Transcriptional Activator
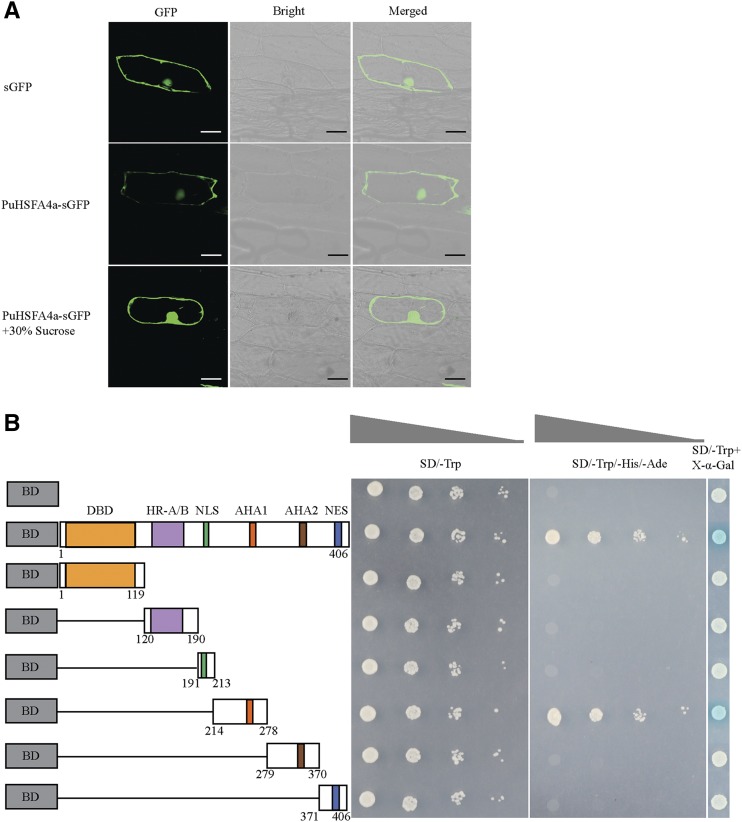
We generated a 35S::PuHSFA4a-GFP plasmid by inserting a GFP tag at the 3′end of the PuHSFA4a gene. The fusion genes 35S::PuHSFA4a-GFP and 35S::GFP were transiently transformed into P. ussuriensis leaves, after which we performed an immunoblot analysis using an anti-GFP antibody (Supplemental Fig. S2). The results of this experiment revealed two bands distinguishing the PuHSFA4a-GFP and GFP proteins, thereby confirming that PuHSFA4a-GFP was a fusion protein (Supplemental Fig. S2). Subcellular localization of the PuHSFA4a protein was examined by expressing a GFP fusion construct that was introduced into onion (Allium cepa) epidermal cells. The results showed that the control (i.e. construct only containing GFP) was distributed throughout the cell. In contrast, the PuHSFA4a-GFP fusion protein was observed only in the nucleus and the cytoplasm (Fig. 3A). These results are similar to LlHSFA1, which is localized primarily in the nucleus, but is also present in the cytoplasm (Gong et al., 2014). PuHSFA4a has six domains. These include an N-terminal DBD, a hydrophobic heptad repeat region for oligomerization, a nuclear import signal (NLS), a Leu-rich NES, and the transcriptional activator domains activator of Hsp90 ATPase 1 (AHA1) and AHA2. Correspondingly, we divided PuHSFA4a into six parts, with each part containing a structural domain (Fig. 3B). To measure the transactivation activity of PuHSFA4a, various versions of PuHSFA4a (Fig. 3B) were constructed and inserted into pGBKT7 vectors before transformation into AH109 yeast strains. The transactivation activity of PuHSFA4a was examined by assessing yeast transformant growth on a selection medium (SD/-Trp-His-Ade) or determined by assessing β-galactosidase (β-gal) activity. Expression of the PuHSFA4a full-length fusion protein in yeast resulted in high expression of the reporter gene, demonstrating that PuHSFA4a had strong activity as a transcription activator. Deletion of the region between amino acids 214 and 278 decreased the transactivation activity of PuHSFA4a, while deletion of other domains did not alter this activity. β-gal activity assays further confirmed these results. Taken together, these findings reveal both that PuHSFA4a acts as a transcriptional activator and that the region between amino acids 214 and 278—which contains the AHA1 domain—was required for transcription activation (Fig. 3B).
Figure 3.
Subcellular localization and transactivation assay of PuHSFA4a. A, Subcellular localization of GFP fusion PuHSFA4a protein in onion epidermal cells. Images were obtained in a dark field to detect green fluorescence, and again in bright light to observe the morphological characteristics of the cells. Bars = 50 μm. B, Transactivation assay of PuHSFA4a in yeast cells. The full-length construct and several partial deletion constructs of PuHSFA4a were fused to GAL4 DBD and expressed in the yeast strain AH109 Gold. Transformed yeast was grown in either SD/-Trp or SD/-Trp/-His/-Ade media. pGBKT7 was used as a negative control. LacZ activity was observed in SD/-Trp medium containing X-α-Gal.
PuHSFA4a Positively Regulates Root Growth under Excess Zn in P. ussuriensis
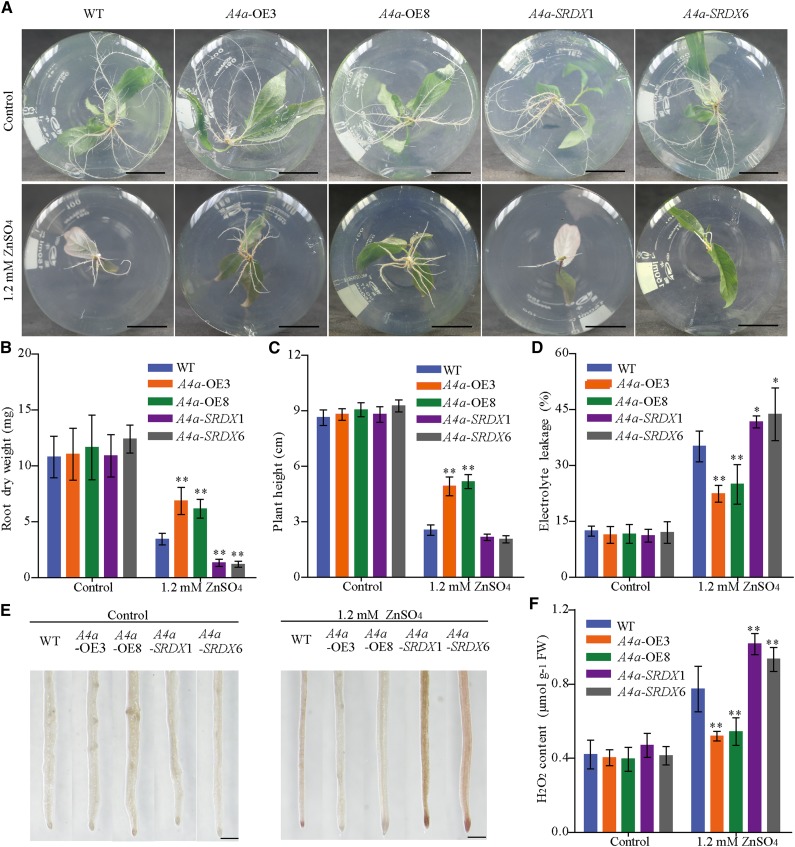
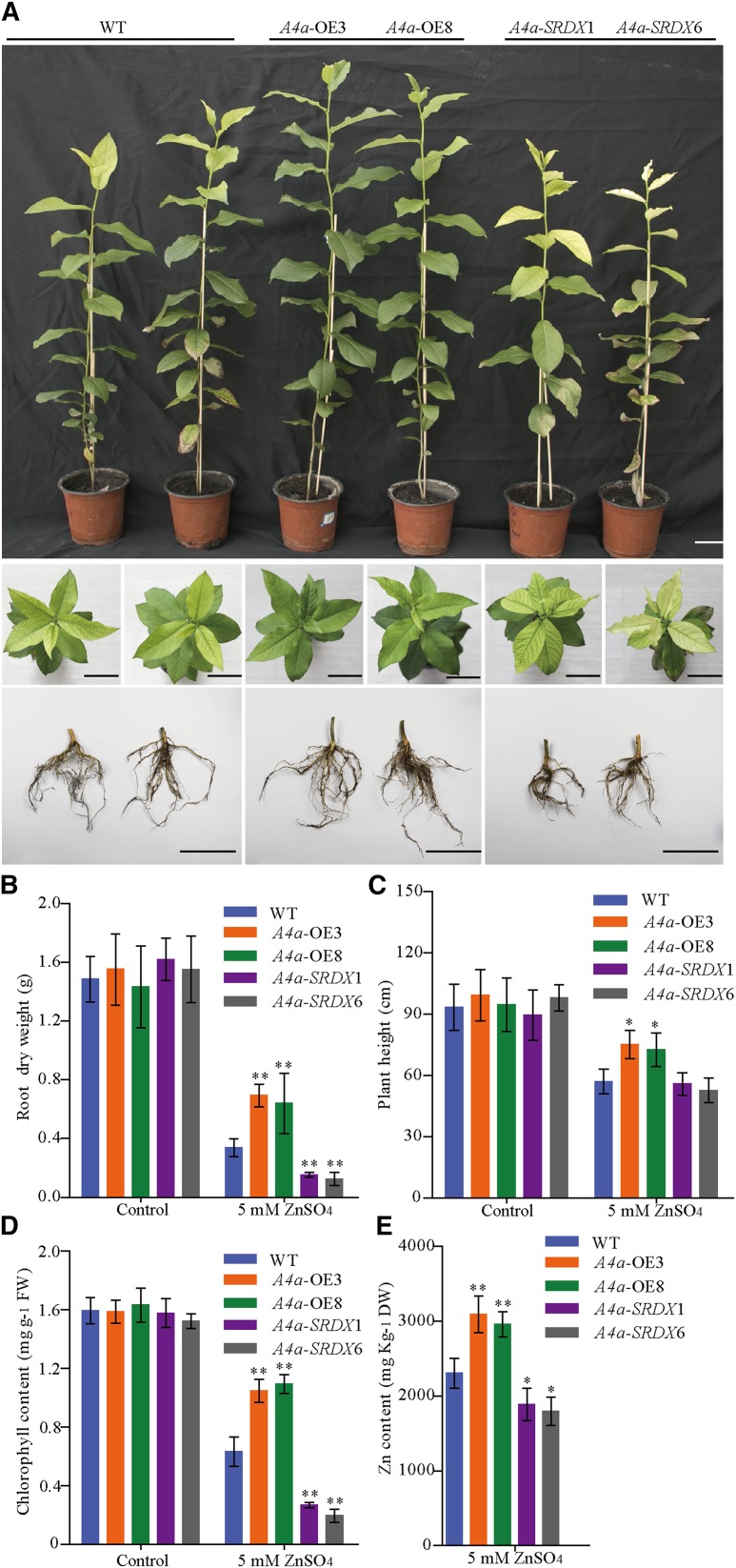
To investigate the function of PuHSFA4a, we generated overexpression PuHSFA4a transgenic lines of P. ussuriensis (PuHSFA4a-OE). We also generated PuHSFA4a dominant repressor lines (PuHSFA4a-SRDX) of P. ussuriensis using Chimeric Repressor Silencing Technology by fusing the full-length PuHSFA4a gene in frame with the SRDX repression domain (for SUPERMAN repression domain; Chai et al., 2014). The elevated expression of PuHSFA4a was confirmed in independent PuHSFA4a-OE lines and the elevated expression of PuHSFA4a-SRDX fusion gene was confirmed in independent PuHSFA4a-SRDX lines by RT-qPCR (Supplemental Fig. S3, A and B). Under normal conditions, both PuHSFA4a-OE and PuHSFA4a-SRDX transgenic plants showed the same phenotype relative to the wild type (Fig. 4; Supplemental Fig. S3C). After exposure to high Zn stress, the PuHSFA4a-OE lines had higher root dry weight and plant height than wild-type plants (Fig. 4, A–C; Supplemental Fig. S3C), while PuHSFA4a-SRDX lines displayed significantly inhibited root growth (Fig. 4, A–C).
Figure 4.
Phenotypes of wild-type, PuHSFA4a-overexpression (A4a-OE3 and A4a-OE8), and PuHSFA4a-SRDX dominant repressor (A4a-SRDX1 and A4a-SRDX6) P. ussuriensis plants. A, Phenotypes of A4a-OE3, A4a-OE8, A4a-SRDX1, A4a-SRDX6, and wild-type (WT) plant roots grown with or without 1.2 mm of ZnSO4 for two weeks. Scale bars = 2 cm. B and C, Root dry weight (B) and plant height (C) of A4a-OE3, A4a-OE8, A4a-SRDX1, A4a-SRDX6, and wild-type plants with or without 1.2 mm of ZnSO4. Each value represents the mean of 30 plants, and error bars = sd. D, Measurement of EL in A4a-OE3, A4a-OE8, A4a-SRDX1, A4a-SRDX6, and wild-type plants with or without 1.2 mm of ZnSO4. Data are presented as means of six biological replicates, and error bars = sd. E and F, In situ accumulation of H2O2 in wild-type and transgenic plants, as revealed by histochemical staining with DAB with or without 1.2 mm of ZnSO4 (E). Scale bars = 500 µm. Measurement of the H2O2 content of roots in wild-type and transgenic plants by luminol assay as described in the “Materials and Methods” (F). Data are presented as means of three biological replicates, and error bars = sd. In (B), (C), (D), and (F), Asterisks indicate significant differences as determined by a Student’s t test, *P < 0.05 and **P < 0.01.
After two months of growth in normal soil followed by four weeks of growth while being irrigated with 5 mm of ZnSO4 solution (Fig. 5A), the root dry weight, plant height, and chlorophyll content of leaves from PuHSFA4a-OE line plants were all significantly higher than those of wild-type plants (Fig. 5, A–D). In contrast, the root dry weight and chlorophyll content of PuHSFA4a-SRDX plants were significantly lower than in wild-type plants (Fig. 5, A–D). The root Zn concentration was significantly higher in PuHSFA4a-OE lines and lower in PuHSFA4a-SRDX lines relative to wild-type roots when irrigated with 5 mm of ZnSO4 solution (Fig. 5E). By contrast, after two weeks, above-ground and root growth were similar in PuHSFA4a-OE, PuHSFA4a-SRDX, and wild-type plants exposed to high Cu, Fe, Cd, ABA, NaCl, and PEG6000 stress (Supplemental Fig. S4).
Figure 5.
Phenotypes of PuHSFA4a-OE, PuHSFA4a-SRDX dominant repressor, and wild-type (WT) P. ussuriensis plants after transplantation in soil under excess Zn. A, Phenotypic comparison of PuHSFA4a transgenic and wild-type plants with respect to above-ground parts, leaves, and roots after irrigation with 5.0 mm of ZnSO4 for four weeks. Scale bars = 5 cm. B and C, Root dry weight (B) and plant height (C) of A4a-OE3, A4a-OE8, A4a-SRDX1, A4a-SRDX6, and wild-type P. ussuriensis plants in response to excess Zn. Each value represents the mean of 40 plants, and error bars = sd. D, Chlorophyll content of leaves of PuHSFA4a transgenic and wild-type plants with or without excess Zn treatment. E, Zn content of PuHSFA4a transgenic and wild-type plants roots under excess Zn treatment. Two-month–old plants were treated with 5.0 mm of ZnSO4 for four weeks. Data are presented as means of three biological replicates, and error bars = sd. Asterisks indicate significant differences as determined by a Student’s t test, *P < 0.05 and **P < 0.01.
PuHSFA4a Overexpression Decreased ROS Production in Roots under Excess Zn
Electrolyte leakage (EL) and malondialdehyde (MDA) are two widely used indicators of damage caused by abiotic stress. Under normal growth conditions, EL and MDA content was similar in transgenic lines and wild-type roots. However, under excess Zn, PuHSFA4a-OE lines showed decreases of ∼37% in EL and ∼35% in MDA content in roots compared to wild-type plants (Fig. 4D; Supplemental Fig. S3D). In addition, PuHSFA4a-SRDX plants displayed a higher EL and MDA than the wild type in roots (Fig. 4D; Supplemental Fig. S3D). Next, hydrogen peroxide (H2O2) levels were compared in roots among PuHSFA4a-OE, PuHSFA4a-SRDX, and wild-type plants under excess Zn (Fig. 4, E and F). No visible differences in H2O2 levels were detected between transgenic lines and wild-type roots under control conditions. In the presence of excess Zn, PuHSFA4a-OE lines were stained lighter compared with wild type, while PuHSFA4a-SRDX lines were stained darker than wild type (Fig. 4E). These results were further confirmed using quantitative measurements (Fig. 4F), implying that the roots of PuHSFA4a-OE plants had significantly less H2O2 content compared with wild-type plants, whereas PuHSFA4a-SRDX plants had significantly higher H2O2 content than wild-type plants (Fig. 4, E and F).
PuHSFA4a Regulates Genes Involved in Abiotic Stress Responses and Root Development
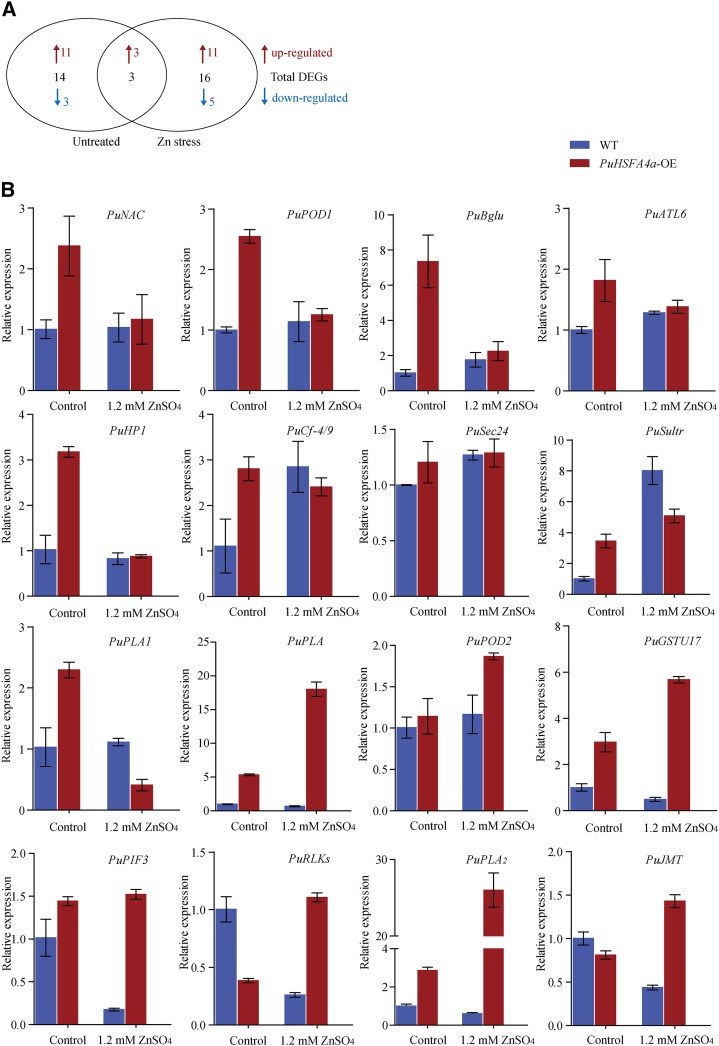
To identify genes regulated by PuHSFA4a in response to Zn-induced stress in roots, we performed RNA sequencing (RNA-seq) experiments using PuHSFA4a-OE and wild-type plant roots that were either treated with 1.2 mm of ZnSO4 for two weeks or were mock-treated as a control. We found 733 upregulated and 708 downregulated genes when comparing control and excess Zn-stressed wild-type plants (false discovery rate [FDR]-adjusted P value < 0.01), and 485 upregulated and 479 downregulated genes when making the same comparison of PuHSFA4a-OE plants (FDR-adjusted P value < 0.01; Supplemental Tables S1 and S2). Pairwise comparisons of relative expression data revealed that 33 genes showed substantially differences in expression between PuHSFA4a-OE and wild-type plants including three common genes (FDR-adjusted P value < 0.01). These genes included 17 genes under normal conditions and 19 genes under excess Zn treatment (Fig. 6A; Supplemental Tables S3 and S4). Gene Ontology term enrichment analysis was performed on the 33 differentially expressed genes (DEGs) between PuHSFA4a-OE and wild-type roots. Our results showed enrichment of genes in pathways related to: cellular responses to hypoxia, monocarboxylic acid biosynthetic processes, and lateral root formation. In general, these pathways are involved in defense and root growth (Supplemental Fig. S5). We found that overexpressing PuHSFA4a resulted in the upregulation of 16 abiotic stress-related genes. We validated our RNA-seq results via RT-qPCR analysis. The results of this validation for the expression levels of these 16 upregulated stress-response DEGs are shown in Figure 6B. All selected genes showed similar expression patterns in our RT-qPCR and RNA-seq analyses (Supplemental Tables S3 and S4).
Figure 6.
Transcriptome analyses of PuHSFA4a overexpression and wild-type (WT) P. ussuriensis plants under high Zn stress. A, Venn diagrams comparing the DEGs among the PuHSFA4a-OE and wild-type P. ussuriensis lines with or without 1.2 mm of ZnSO4. “Untreated” represents the DEGs between wild-type and PuHSFA4a-OE under control conditions. “Zn stress” represents the DEGs between wild-type and PuHSFA4a-OE under high Zn stress. Red and blue colors indicate upregulated and downregulated genes, respectively. B, RT-qPCR analysis of the transcript levels of PuHSFA4a target genes with or without 1.2 mm of ZnSO4. Data are presented as means of three biological replicates, and error bars = sd.
PuHSFA4 Binds To the Promoter Regions of PuGSTU17 and PuPLA2
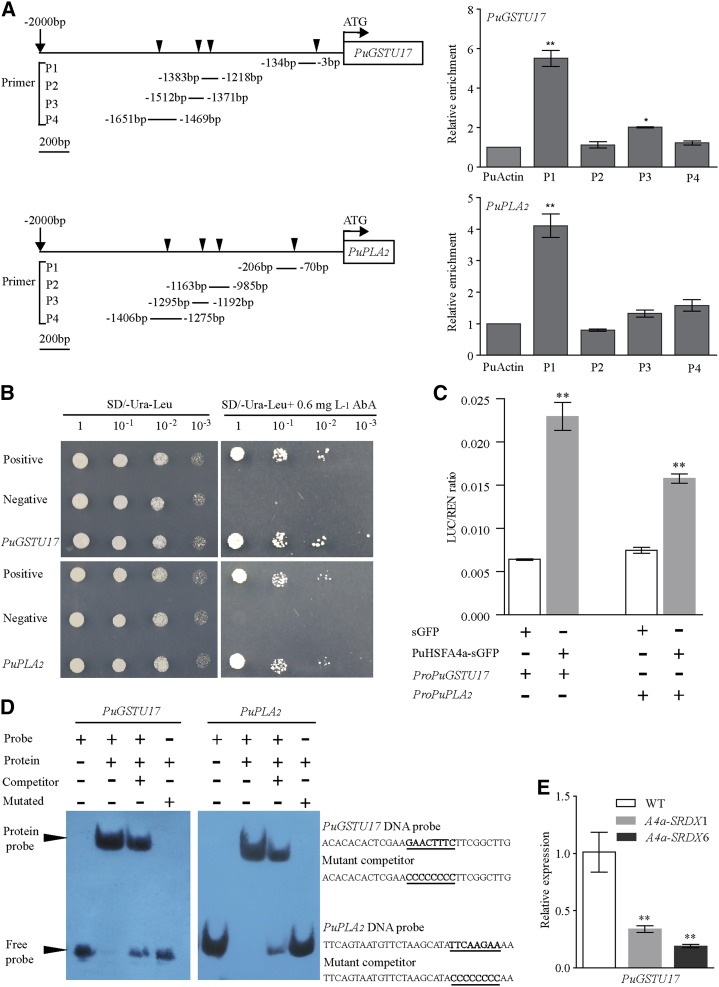
We used the PlantCARE and Plant PAN databases to search for putative heat shock elements (HSEs) in the ∼2-kb promoter regions of each of the 16 selected genes that were upregulated in PuHSFA4a-OE plants. From these searches, we found 13 genes that contained enriched HSE elements in their promoter regions (primers shown in Supplemental Table S5). Chromatin immunoprecipitation (ChIP)-PCR assays with anti-GFP from 35S::PuHSFA4a-GFP plants were used to test the capacity of PuHSFA4a to bind to the promoters of these 13 predicted target genes. ChIP-PCR showed robust enrichment of PuHSFA4a-GFP in the promoter regions of PuGSTU17 and PuPLA2, but not in the promoters of the other 11 genes (Supplemental Fig. S6). Another ChIP-qPCR analysis with anti-GFP antibody verified that the PuGSTU17 and PuPLA2 promoters showed significantly higher PuHSFA4a binding enrichment at binding sites relative to a control (Fig. 7A). Yeast one-hybrid (Y1H) assays showed that when fused PuGSTU17-AbAi or PuPLA2-AbAi was coexpressed with PuHSFA4a-AD in yeast, the two strains could grow on SD/-Ura/-Leu/AbA plates (Fig. 7B). This result revealed that PuHSFA4a bound specifically to the promoters of the PuGSTU17 and PuPLA2 genes in vivo. A luciferase reporter (LUC) assay showed that coexpression of 35S::PuHSFA4a with ProPuGSTU17::Luc or ProPuPLA2::Luc led to an obvious increase in luminescence intensity (Fig. 7C), thus confirming that PuHSFA4a interacts with the promoters of PuGSTU17 and PuPLA2. Additionally, an electrophoretic mobility shift assay (EMSA) showed that PuHSFA4a bound to a DNA probe containing a typical HSE motif (GAACTTTC or TTCAAGAA) was out-competed by an unlabeled DNA probe. In contrast, PuHSFA4a failed to bind to a mutant probe (Fig. 7D). A further RT-qPCR experiment showed that the expression of PuGSTU17 was significantly reduced under Zn stress in the roots of PuHSFA4a-SRDX lines relative to wild-type plants (Fig. 7E). Taken together, these results demonstrate that PuHSFA4a positively regulates PuGSTU17 and PuPLA2 expression in response to high Zn stress by binding directly to their promoter regions.
Figure 7.
Verification of the PuHSFA4a target genes PuGSTU17 and PuPLA2. A, ChIP-qPCR analysis of PuHSFA4a binding to PuGSTU17 and PuPLA2 promoter fragments using anti-GFP tag antibody. Schematic diagrams showing the PuHSFA4a binding sites in the regions 2,000 bp upstream of the transcriptional start (ATG) sites of the PuGSTU17 and PuPLA2 genes. Triangular arrowheads indicate PuHSFA4a binding sites (P1, P2, P3, and P4). B, Y1H assay of PuHSFA4a binding to PuGSTU17 and PuPLA2 promoter fragments. C, Binding of PuHSFA4a to the promoters of PuGSTU17 and PuPLA2 as assayed using the dual luciferase system. D, EMSA analyses showing the binding of PuHSFA4a to the DNA probes of the PuGSTU17 and PuPLA2 promoters in vitro. The free and bound DNAs (arrows) were separated by acrylamide gel electrophoresis. The unlabeled probes were used as competitors, and mutated probes were produced by replacing the HSE motifs with GGGGGGGG. E, Relative expression levels of PuGSTU17 in PuHSFA4a-SRDX and wild-type plants assessed under excess Zn conditions and normalized to PuActin (MH644084). In (A), (C), and (E), data are presented as means of three biological replicates, and error bars = sd. Asterisks indicate significant differences as determined by a Student’s t test, *P < 0.05 and **P < 0.01.
PuGSTU17 Positively Regulates Zn Tolerance and ROS Scavenging Capacity in Roots
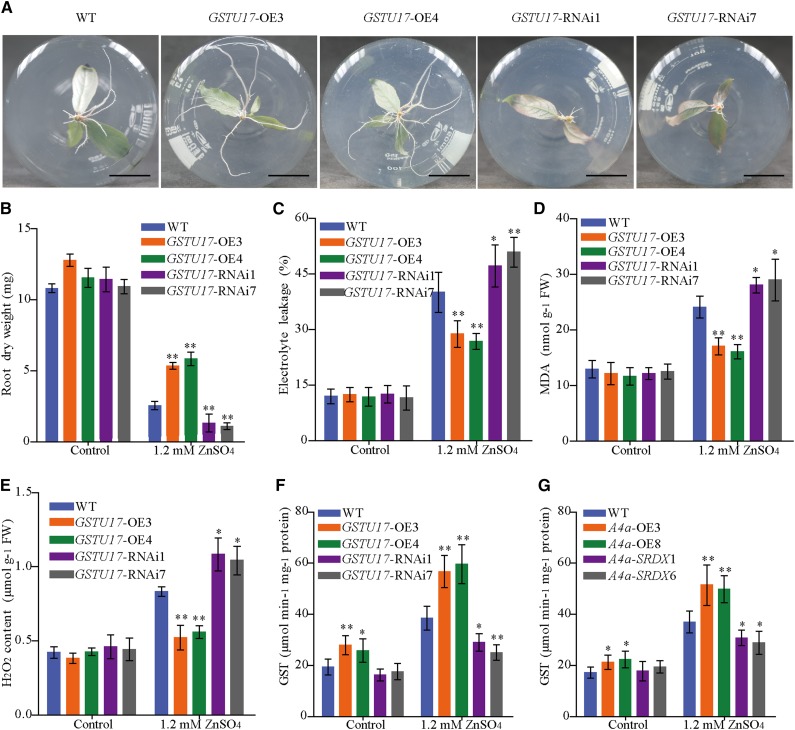
To determine whether PuGSTU17 confers Zn tolerance, we generated 35S::PuGSTU17 (PuGSTU17-OE) lines of P. ussuriensis, and also generated PuGSTU17-RNA interference (RNAi) lines of P. ussuriensis (PuGSTU17-RNAi). The expression of PuGSTU17 was confirmed in transgenic lines by RT-qPCR (Supplemental Fig. S7). Under normal culture conditions, PuGSTU17-OE and PuGSTU17-RNAi plants showed the same phenotype as wild-type plants (Supplemental Fig. S7). When plants were cultured on media containing 1.2 mm of ZnSO4 for two weeks, plant heights were not significantly different between PuGSTU17 transgenic lines and wild-type plants. However, the root dry weight of PuGSTU17-OE plants was significantly higher than that of wild-type plants, while PuGSTU17-RNAi lines displayed significantly inhibited root growth (Fig. 8, A and B). Moreover, PuGSTU17-OE plant roots had lowered EL and MDA content than wild-type plants when exposed to high Zn stress while PuGSTU17-RNAi plants displayed a higher EL and MDA than the wild type in roots (Fig. 8, C and D). Our results showed that PuGSTU17-OE plants suffered less cell membrane damage than wild-type and PuGSTU17-RNAi plants. In addition, quantitated H2O2 content data indicated that roots of PuGSTU17-OE plants displayed lower, and PuGSTU17-RNAi plants had higher, ROS levels than wild type under Zn stress (Fig. 8E).
Figure 8.
Phenotype of PuGSTU17-overexpression (GSTU17-OE3 and GSTU17-OE4), PuGSTU17-RNAi (GSTU17-RNAi1 and GSTU17-RNAi7), and wild-type (WT) P. ussuriensis plants under excess Zn. A, Phenotypic comparison of the roots of PuGSTU17-OE, PuGSTU17-RNAi, and wild-type plants in response to excess Zn. Scale bars = 2 cm. B, Root dry weight of PuGSTU17-OE, PuGSTU17-RNAi, and wild-type plants with or without Zn treatment. Each value represents the mean of 30 plants, and error bars = sd. C, Measurement of EL in PuGSTU17-OE, PuGSTU17-RNAi, and wild-type plants with or without Zn treatment. D, Measurement of MDA in PuGSTU17-OE, PuGSTU17-RNAi, and wild-type plants with or without Zn treatment. E, Measurement of the H2O2 content in PuGSTU17-OE, PuGSTU17-RNAi, and wild-type plants with or without Zn treatment. F, Measurement of GST activity in PuGSTU17-OE, PuGSTU17-RNAi, and wild-type plants with or without Zn treatment. G, Measurement of GST activity in PuHSFA4a-OE, PuHSFA4a-SRDX, and wild-type plants with or without Zn treatment. In (C) to (G), data are presented as means of three biological replicates, and error bars = sd. Asterisks indicate significant differences as determined by a Student’s t test, *P < 0.05 and **P < 0.01.
We quantified the GST activity of roots of PuGSTU17-OE, PuGSTU17-RNAi, and wild-type plants. Under excess Zn stress, the GST activity of the roots of PuGSTU17-OE plants was higher than the activity of the roots of wild-type plants (Fig. 8F). After inhibiting the expression of PuGSTU17, the GST activity in roots was lower compared with wild type (Fig. 8F). In addition, we assessed GST activity in the roots of PuHSFA4a-OE, PuHSFA4a-SRDX, and wild-type plants. The results showed PuHSFA4a-OE lines displayed significantly higher, and PuHSFA4a-SRDX lines had lower, root GST activity than wild type under high Zn stress (Fig. 8G). The root Zn concentration was higher in PuGSTU17-OE lines and lower in PuGSTU17-RNAi lines relative to wild-type roots when irrigated with a 1.2-mm ZnSO4 solution (Supplemental Fig. S7D). Next, we compared plant tolerance to CdCl2, PEG6000, and NaCl stresses in PuGSTU17-OE and wild-type plants. These comparisons revealed no phenotypic differences between PuGSTU17-OE and wild-type plants in response to these stresses for 2 weeks (Supplemental Fig. S8, A–D).
PuPLA2 Overexpression Increased Root Growth under Excess Zn
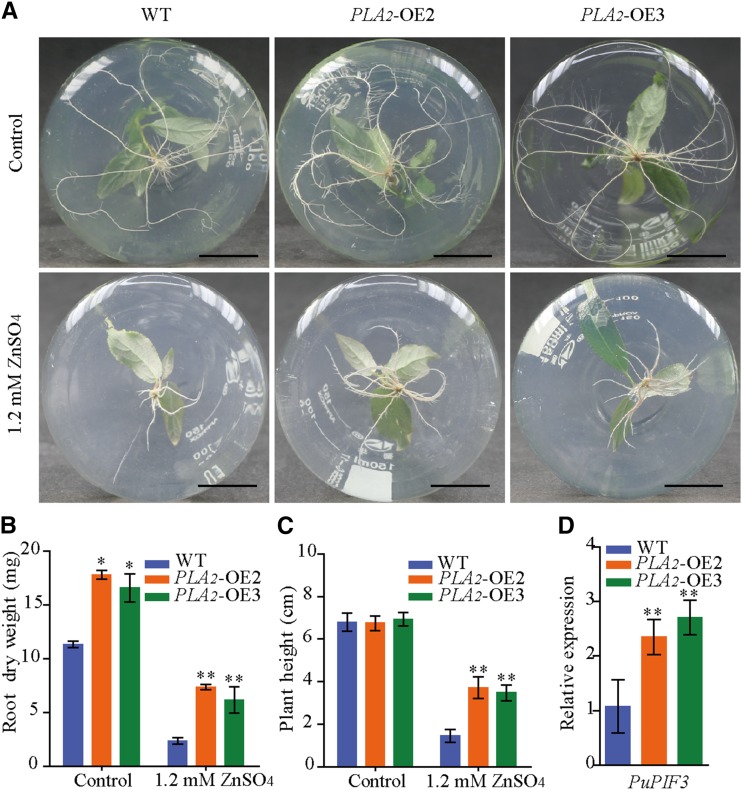
We then used the plant expression vector 35S::PuPLA2 to overexpress PuPLA2 (PuPLA2-OE) in P. ussuriensis. The expression levels of PuPLA2 were significantly increased in PuPLA2 overexpression (PLA2-OE2, PLA2-OE3) lines than in wild-type plants (Supplemental Fig. S9A). Under normal culture conditions in vitro, PuPLA2-OE plants had larger root systems than wild-type plants (Fig. 9, A and B). In contrast, plant height did not significantly differ between PuPLA2-OE and wild-type plants (Fig. 9C). When cultured on media containing 1.2 mm of ZnSO4 for 2 weeks, root growth and plant height of both transgenic and wild-type plants were suppressed. However, under high Zn stress, PuPLA2-OE lines showed higher root dry weight and plant height than wild-type plants (Fig. 9, A–C; Supplemental Fig. S9B). To further determine the role played by PuPLA2 in the Zn stress response, we assessed the expression of the auxin signaling-related gene Phytochrome-Interacting Factor 3 (PuPIF3), which was upregulated in PuHSFA4a-OE plants under Zn stress (Supplemental Table S4). We then found that, in response to excess Zn, PuPIF3 showed increased expression in PuPLA2-OE lines relative to wild-type plants (Fig. 9D).
Figure 9.
Phenotype of PuPLA2-OE and wild-type (WT) P. ussuriensis plants with or without Zn treatment. A, Phenotypic comparison of the roots of PuPLA2 transgenic and wild-type plants with or without excess Zn. Scale bars = 2 cm. B and C, Root dry weight (B) and plant height (C) of PuPLA2-OE and wild-type plants under excess Zn stress. Each value represents the mean of 30 plants, and error bars = sd. D, The relative expression levels of PuPIF3 in PuPLA2-OE and wild-type plants assessed under excess Zn conditions, and a comparison of the relative expression of PuPIF3 in PuPLA2-OE lines compared to wild-type plants. All values are normalized to PuActin (MH644084). Data are presented as means of three biological replicates, and error bars = sd. For (B)–(D), Asterisks indicate significant differences as determined by a Student’s t test, *P < 0.05 and **P < 0.01.
DISCUSSION
Plant HSFs are known to be involved in abiotic stress responses, including to heat, salt, oxidative, drought, Cd, and starvation stresses (Panchuk et al., 2002; von Koskull-Döring et al., 2007; Shim et al., 2009; Pérez-Salamó et al., 2014). However, to date there are no reports of plant HSFs that are specifically involved in the Zn stress response. In this study, we characterized PuHSFA4a, an HSF found in P. ussuriensis that acts as a positive regulator of excess Zn tolerance in roots. PuHSFA4a is specifically induced in root tissues by excess Zn. When exposed to excess Zn, a dominant repressor version of the PuHSFA4a lines (PuHSFA4a-SRDX) decreased Zn tolerance and root growth, while PuHSFA4a overexpression (PuHSFA4a-OE) significantly increased Zn tolerance and root growth. Moreover, we also found that PuHSFA4a contributed to excess Zn tolerance by directly regulating PuGSTU17 to control ROS levels in roots. PuHSFA4a overexpression was also found to enhance Zn tolerance by largely upregulating target PuPLA2 transcript levels, and thus promoting root growth when exposed to high levels of Zn. These results demonstrate that PuHSFA4a functions positively in regulating excess Zn response.
Because roots are the first tissue confronted by excess heavy metal content in the soil, toxic symptoms of excess exposure to heavy metals are more common in roots than in other tissues (Chang et al., 2005). The primary response to Zn toxicity is growth inhibition, often characterized by reduced root growth, increased root thickening, and impaired cell division/elongation (Barceló and Poschenrieder, 1990; Vaillant et al., 2005). For instance, overexpression of the ACS gene in Arabidopsis improves tolerance to excess Fe by protecting growing roots (Li et al., 2015). Moreover, in response to stress caused by exposure to excess Cd, suppression of GPL4 in Arabidopsis results in increased root biomass and tolerance to Cd (Khare et al., 2017). Overexpression of MAN3 in Arabidopsis in response to excess Cd also results in larger roots (i.e. roots with heavier root fresh weights) and enhanced Cd tolerance (Chen et al., 2015). In this study, under excess Zn, PuHSFA4a-OE transgenic line plants showed lower root ROS, MDA, and EL levels and stronger root systems than wild-type plants, whereas PuHSFA4a-SRDX plants had higher root ROS, MDA, and EL levels and weaker roots than wild-type plants. Previous studies have reported mechanisms by which plants have evolved to cope with excess ROS induced by Zn stress, mainly by activating antioxidant enzymes and by producing nonenzymatic compounds (Chaoui et al., 1997; Wang et al., 2009). For example, overexpression of HvAPX1 in barley alleviated Zn toxicity-induced damage in Arabidopsis by limiting oxidative damage (Xu et al., 2008). Based on these findings, we speculate that PuHSFA4 confers tolerance to excess Zn by regulating ROS levels and membrane lipid peroxidation to ensure root cell integrity.
Transcriptomic analysis of PuHSFA4a-OE and wild-type Populus roots identified many genes whose expression was substantially regulated by PuHSFA4a overexpression. Consistent with the observed phenotype changes, expression of genes related to abiotic stress response and root growth were increased substantially by high Zn stress in roots. In the last decade, many studies have improved our understanding of the molecular mechanisms that regulate excess Zn tolerance. For example, MTP1 from P. trichocarpa × P. deltoides (Blaudez et al., 2003), AtHMA1 (Kim et al., 2009), AtHMA3 (Morel et al., 2009), AtMTP1 (Desbrosses-Fonrouge et al., 2005), AtMTP3 (Arrivault et al., 2006), AQUA1 (Ariani et al., 2016), OZT1 (Lan et al., 2013), the cyanobacterial metallothionein gene SmtA (Xu et al., 2010), and HvAPX1 (Xu et al., 2008) have all been found to contribute to the detoxification of excess Zn in diverse species. In this study, transcriptomic analysis revealed that overexpression of PuHSFA4a did not affect the expression of any previously identified Zn stress response genes. In addition, studies of various other plant species have identified target genes regulated by HSFA4as, including the Arabidopsis HSP17.6A (Pérez-Salamó et al., 2014) and yeast MT (Shim et al., 2009) genes. However, our RNA-seq data did not reveal any known target genes that were affected by PuHSFA4a expression. Collectively, our results suggest that those genes that are differently expressed in response to PuHSFA4a expression are independent of both previously defined Zn stress response genes and known HSFA4a target genes found in P. ussuriensis roots. Further functional study of downstream genes will help to clarify this issue.
The antioxidative enzyme GST was reported to play an important role in abiotic stress resistance by reducing ROS toxicity (Jiang and Yan, 2018). In Juglans regia, overexpression of JrGSTTau1 confers chilling stress tolerance by decreasing ROS accumulation and thereby reducing cell damage (Yang et al., 2016). Ectopic expression of rice OsGSTU4 improves tolerance to salinity and oxidative stresses in Arabidopsis by enhancing GST activity, thereby suppressing ROS accumulation (Sharma et al., 2014). In our study, a tau class PuGST (PuGSTU17), homologous to Arabidopsis AtGSTU17, was identified as a direct target of PuHSFA4a by RT-qPCR, ChIP-qPCR, Y1H, and LUC assays, and by EMSA. Furthermore, ROS accumulation was significantly decreased in PuGSTU17 overexpressing lines but obviously increased in the PuGSTU17-RNAi lines compared with wild type under excess Zn. Accordingly, overexpression of PuHSFA4a significantly increased GST activity in roots under excess Zn, while a dominant repressor version of PuHSFA4a decreased GST activity. Taken together, our results demonstrate that PuHSFA4a can activate the target gene PuGSTU17 to promote GST activity to eliminating excess Zn-induced ROS in roots and thereby improve Zn tolerance. This finding agrees with previous studies showing that overexpression of tau GSTs from Pyrus pyrifolia increased tolerance to Cd (Liu et al., 2013), tau GSTs from Gly soja and Salicornia brachiata enhanced salt tolerance, and tau GSTs from G. soja improved drought tolerance in tobacco (Nicotiana benthamiana; Ji et al., 2010; Jha et al., 2011). However, we found that both PuHSFA4a and PuGSTU17 overexpression specifically improves plant resistant to excess Zn but does not confer tolerance to some other heavy metal stresses. In this regard, the relationship between PuHSFA4a and PuGSTU17 under Zn stress remains to be clarified.
The developmental plasticity of root shape is important for root function and plant survival after exposure to toxic heavy metals (Lin and Aarts, 2012). PLA enzymatic hydrolysate free fatty acids were reported to be second messengers that are involved in auxin functions such as activation of elongation and cell division (Holk and Scherer, 1998). In previous studies, PLA was identified as a regulator of plant growth and development (Wang et al., 2000; Holk et al., 2002; La Camera et al., 2005; Schvartzman et al., 2018). For instance, Arabidopsis AtPLAIVA-null mutants have reduced lateral root development by reduced auxin signaling (Schvartzman et al., 2018). In this study, ChIP-qPCR, Y1H, and LUC assays and EMSA confirmed that PuPLA2 was also a direct target gene of PuHSFA4a (Fig. 7; Supplemental Fig. S6). Under excess Zn, both the PuPLA2-OE and PuHSFA4a-OE lines displayed similar phenotypes with respect to root growth compared to wild-type plants. In addition, PuPIF3 expression was upregulated by both PuHSFA4a overexpression and PuPLA2 overexpression under excess Zn conditions. Previous studies reported that PIFs are involved in the regulation of auxin signaling and auxin biosynthesis in plants. For instance, Arabidopsis PIF3 lies downstream of PHYB and RGL3, and has been shown to reduce the inhibitory effect of nitric oxide on root growth in Arabidopsis (Bai et al., 2014). Arabidopsis PIF4 also regulates hypocotyl growth by directly targeting the auxin biosynthesis gene YUCCA8 and the auxin signaling gene IAA29 (Sun et al., 2012, 2013), and regulates root growth by modulating the expression of the auxin biosynthesis genes TAA1 and CYP79B2 in response to high temperatures (Franklin et al., 2011). These results suggest that PuHSFA4a might confer excess Zn tolerance partly by regulating PuPLA2 to promote the expression of PIF3 and thereby influencing the auxin signaling pathway, thus contributing to root growth. Further elucidation of the mechanisms underlying PuHSFA4a regulation of PuPLA2 and PuPIF3 expression under Zn stress should be a focus of future work.
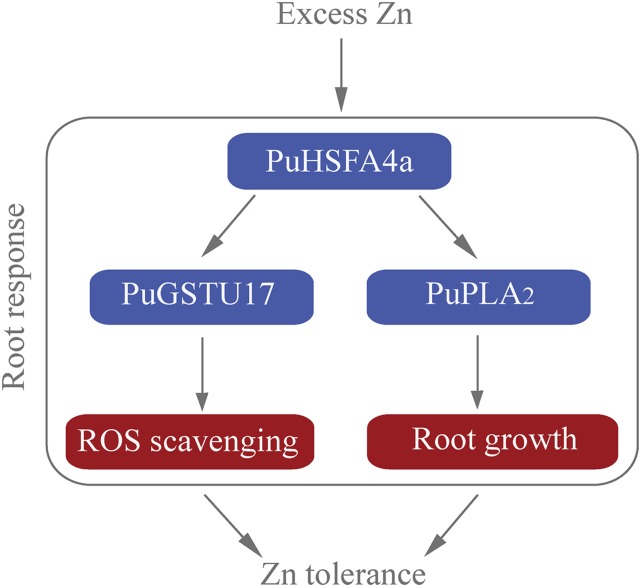
Based on our findings, we propose a model of action for PuHSFA4a in response to excess Zn in roots (Fig. 10). In this article, we have provided evidence that the root-specific–induced gene PuHSFA4a positively regulates high Zn tolerance in roots of P. ussuriensis by coordinately activating the antioxidant system and root development genes and that it directly targets PuGSTU17 and PuPLA2. This study thus identifies PuHSFA4a as a positive and specific regulator of excess Zn tolerance in P. ussuriensis and provides insight into the mechanisms of excess Zn tolerance in Populus roots.
Figure 10.
A proposed model illustrating excess Zn tolerance regulated by the PuHSFA4a. PuHSFA4a is induced by high Zn content in P. ussuriensis roots. Under excess Zn stress, PuHSFA4a activates PuGSTU17 expression by binding with its promoters, followed by enhancing GST activity, which resulted in enhancing tolerance by reducing ROS. Meanwhile, overexpression of PuHSFA4a promotes root growth by directly binding with the PuPLA2 promoter to activate its transcription under Zn stress. Thus, PuHSFA4a functions as a positive regulator of excess Zn tolerance by activating the antioxidant system and promoting root growth in P. ussuriensis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Populus ussuriensis clone Donglin plants were grown in vitro under 16-h–light/8-h–dark cycles at 46 μmol photons m−2 s−1 irradiation at 25°C. Plants were grown on half-strength MS plates with 0.6% (w/v) agar and 2% (w/v) Suc for two weeks followed by culturing on a medium supplemented with 1.2 mm of ZnSO4 for 0, 3, 6, 12, 24, 48 h, and 1 week, respectively. The plants were also exposed to 80 μm of CdCl2, 100 μm of CuSO4, 1.0 mm of FeSO4, 7% (w/v) PEG6000, 30 μm of ABA, and 150 mm of NaCl treatments for 0, 12, and 24 h, respectively. At the end of the experimental period, leaf and root tissues were collected, frozen in liquid nitrogen, and stored at −80°C. For gene expression and physiology analysis, plants were grown in a growth chamber at 25°C, in a 16-h–light,/8-h–dark photoperiod with a light intensity of 46 μmol photons m−2 s−1.
RNA Extraction and RT-qPCR Analysis
Total RNA was extracted using the cetyltrimethylammonium bromide method (Zhang et al., 2015), and reverse-transcription was performed using the PrimeScript RT First Strand cDNA Synthesis Kit (Toyobo). First-strand cDNA synthesized from 0.5 μg of purified RNA was reverse-transcribed using a Reverse Transcriptase Kit (TaKaRa). RT-qPCR was performed with primers corresponding to sequences from the UniGene database developed from a nonparametric RNA-seq of P. ussuriensis. RT-qPCR was carried out using a TransStart Top Green qPCR SuperMix (TransGen Biotech) with PCR conditions following the manufacturer’s instructions. We also used a qTOWER 3G Cycler and qPCR software (Analytik) to perform RT-qPCR and analyze data. The P. ussuriensis Actin (PuActin) gene was used as an internal control. The 2−ΔΔCt method was used to determine the relative abundance of transcripts. All primers used in this study are listed in Supplemental Table S5.
Phylogenetic Analysis and Protein Domain Identification
The HSF proteins were searched against the NCBI database (http://www.ncbi.nlm.nih.gov/). A phylogenetic tree was constructed based on HSF amino acid sequences using the neighbor-joining method in the software MEGA v5.0 (Tamura et al., 2011) and a bootstrap test with 1,000 iterations. Multiple sequence alignment of the full-length protein sequences was performed using the software BioEdit v7.1 (Hall, 1999).
Generation of Transgenic Plants
A 2,203-bp genomic promoter sequence upstream of the coding region of PuHSFA4a was cloned into a pCAMBIA1301 binary vector instead of the cauliflower mosaic virus 35S promoter. The full-length coding sequences (CDSs) of PuHSFA4a and PuGSTU17 were amplified by PCR and cloned into the pBI121 vector containing the cauliflower mosaic virus 35S promoter to generate the 35S::PuHSFA4a and 35S::PuGSTU17 constructs, respectively. A 27-bp DNA sequence encoding the SRDX repression domain (Chai et al., 2014) was fused in-frame to the 3′ end of the PuHSFA4a coding regions and then cloned into the pBI121 vector to generate the 35S::PuHSFA4a-SRDX constructs, respectively. The coding region of the PuPLA2 gene was cloned into a pCAMBIA1300 vector. For the PuGSTU17 RNAi construct, a 179-bp specific fragment beginning with an ATG starting at base 507 and ending at base 685 from PuGSTU17 was PCR-amplified using primers PuGSTU17-RNAi-F and PuGSTU17-RNAi-R. The resulting PCR product was cloned into a pENTR vector (Invitrogen) and was subsequently transferred into the RNAi destination vector pH7GWIWG2(II); Karimi et al., 2005). The vectors constructed included ProPuHSFA4a::GUS, 35S::PuHSFA4a-GFP, 35S::PuHSFA4a-SRDX-GFP, 35S::PuGSTU17-GFP, PuGSTU17-RNAi, and 35S::PuPLA2-GFP. These were introduced into Agrobacterium strain EHA105 before transformation by leaves. Transformants were selected on media containing 40 mg L−1 kanamycin except ProPuHSFA4a::GUS, PuGSTU17-RNAi, and PuPLA2-OE plants, which were selected on media containing 4.0 mg L−1 hygromycin. All transgenic plants were confirmed by genomic PCR and by RT-qPCR. At least 10 transgenic lines were used for phenotypic observations. From these lines, data were obtained from four or more transgenic lines that showed a common and stable phenotype. All primer sequences used in this study are listed in Supplemental Table S5.
Abiotic Stress and Zinc Tolerance Assays
Seven-day–old in vitro plants (wild type, PuHSFA4a-OE, PuHSFA4a-SRDX, PuGSTU17-OE, PuGSTU17-RNAi, and PuPLA2-OE) were transferred to half-strength MS medium containing 1.2 mm of ZnSO4, 70 μM or 80 μM of CdCl2, 100 μM of CuSO4, 1.0 mm of FeSO4, 7% (w/v) PEG6000, 30 μM of ABA, and 150 mm of NaCl for 2 weeks, respectively, unless otherwise specified. In addition, 2-month–old soil grown plants were irrigated with 5.0 mm of ZnSO4 for four weeks under the same light regime in soil. In a preliminary experiment, we evaluated the effect of different concentrations of ZnSO4 on in vitro and soil grown P. ussuriensis. The results showed that 1.2-mm and 5-mm Zn concentrations could reduce but not abolish in vitro and soil grown P. ussuriensis, respectively. Thus, 1.2-mm and 5-mm Zn concentrations were used as moderate excess Zn stress treatment for in vitro and soil grown plants in this study, respectively.
Measurement of EL, MDA Content, ROS Accumulation, and GST Activity
Seven-day–old in vitro plants were cultured on half-strength MS medium with or without 1.2 mm of ZnSO4, respectively. After 2 weeks of culture, the roots were collected and used for analysis.
MDA content was quantified as described by Metwally et al. (2003). About 50 mg of roots were ground in liquid nitrogen and homogenized in 1 mL of 0.1% (w/vTCA for 10 min and centrifuged at 10,000g for 15 min at 4°C. A quantity of 0.2 mL of the supernatant was reacted with 0.8 mL of 20% (w/v) TCA containing 0.5% (w/v) thiobarbituric acid. The mixture was boiled for 20 min and cooled immediately, then centrifuged at 10,000g for 5 min at 4°C. The absorbance of the supernatant was measured in 532 and 600 nm, respectively. The concentration of MDA was calculated using the extinction coefficient of 155 mmol−1 L−1 cm−1 and expressed as nmol g−1 fresh weight (Hossain et al., 2017).
Measurements of EL were performed as described in Duan et al. (2017) with some modifications. Briefly, three entire roots were immersed in 150-mL tubes containing 80 mL of deionized water. After 15 min of vacuum treatment and shaking at 140 rpm for 2 h at room temperature, the conductivities (C1) were determined with an electrical conductivity meter (DDS-307A; Hinotek). The samples were then boiled for 15 min. After cooling to 25°C, the conductivities of the roots (C2) and deionized water (C0) were determined again. The relative ion leakage% = (C1-C0) / (C2-C0)*100% was subsequently calculated.
For histochemical detection of H2O2 accumulation, the roots were treated with 3, 3′-diaminobenzidine (DAB) solutions (1.0 mg mL−1 DAB-HCl at pH 3.8; Sigma-Aldrich; Zhang et al., 2014). The stained roots were bleached in acetic acid-ethanol (1:3, v/v) solution at 25°C for 3 h, and then stored in a glycerol-ethanol (1:4, v/v) solution. Pictures were taken with SZ Stereo Microscopes (Olympus).
H2O2 was quantified by chemiluminescence (CL) with luminol as described by König et al. (2014). About 100 mg of roots were pulverized with a mortar and pestle in liquid nitrogen, and then mixed with 0.5 mL of 5% (w/v) TCA at 4°C. The samples were placed in the dark for 40 min and centrifugated at 10,000g for 10 min at 4°C. After dilution with 0.1-m sodium carbonate buffer, 50-μL aliquots were incubated with 100 U of catalase (bovine liver; Sigma-Aldrich) or with the same volume of water for 30 min at 30°C as a control. H2O2 was determined by CL with luminol. The sample (5 μL) was added to 100 µL of reagent solution. The emitted photons were counted over 7 s with a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek). The difference between catalase-treated and untreated samples (ΔCL) was considered as H2O2-specific CL. A standard curve was generated using appropriate dilutions of 30% (w/v) H2O2 (Farooq et al., 2016).
For detection of GST activity, frozen roots were ground in a liquid-nitrogen prechilled mortar containing 50 mm of phosphate-buffered saline at pH 7.2, 1 mm of EDTA, 8% (w/v) polyvinylpolypyrrolidone, 2 mm of DTT, and 0.01% (v/v) Triton 100-X. The extract was passed through Miracloth and then centrifuged at 23,000g for 30 min at 4°C (Davoine et al., 2006). The supernatants were used for GST activity analysis by using a GST (glutathione-ST) Assay Kit (Nanjing Jiancheng Bioengineering Institute) according to the manufacturer’s instructions. The total proteins in the samples were quantified by using a Bradford Protein Assay Kit (Solarbio) according to the manufacturer’s instructions.
Chlorophyll and Zn Content Analyses
Two-month-old soil grown plants were irrigated with or without 5.0 mm of ZnSO4 for 4 weeks.
The third leaf was collected for measurements of chlorophyll and the whole roots were collected for measurements of Zn content, respectively.
For total chlorophyll quantification, ∼20–30 mg leaves were ground in liquid nitrogen and thoroughly mixed with 2.5 mL of 80% (v/v) acetone by vigorous vortexing. The samples were then incubated in darkness overnight at 4°C. The mixtures were centrifuged at 10,000g for 20 min at 4°C. The absorbance of supernatant was measured at 663 and 645 nm (Porra, 2002). Total chlorophyll content was calculated and expressed as mg g−1 fresh weight.
The concentration of Zn in roots was analyzed as described by Cho et al. (2003) with some modifications. The roots were harvested, briefly washed using deionized water, and dried at 60°C until their weights remained constant. About 500 mg of dry homogeneous sample was digested with 5-mL 65% (v/v) HNO3 at 150°C until complete solubilization of the sample occurred. The cleared extracts were then diluted with ultra-pure water to 25 mL and analyzed by inductively coupled plasma optical emission spectrometry (Agilent Technologies).
Histochemical GUS Staining
Histochemical GUS-staining assays were performed as described in Jefferson et al. (1987) with minor modifications. Two-week–old ProPuHSFA4a::GUS plants were treated with or without ZnSO4 (1.2 mm) for 7 d. After this treatment all plants were incubated in a solution for ∼16 h at 37°C in 100 mm of sodium P buffer (pH 7.0) containing 0.6 g of 5-bromo-4-chloro-3-indolyl-β-d-GlcA cyclohexylammonium salt, 5 mm of potassium ferrocyanide, 5 mm of potassium ferricyanide, 0.1 m of EDTA disodium salt, and 0.1% (v/v) Triton X-100. Tissues were fixed with fixative solution (glacial acetic acid-absolute ethanol, 1:3 [v/v]) and then immersed in transparent solution (glycerin-absolute ethanol, 1:4). GUS activity was imaged using at least three replicates.
Subcellular Localization and Transcriptional Activation of PuHSFA4a
Subcellular localization was performed by expressing 35S::GFP and 35S::PuHSFA4a-GFP in onion (Allium cepa) tissue epidermal cells via a biolistic bombardment method (Lee et al., 2008). GFP fluorescent signals for the fusion protein were examined with a LSM 710 Meta confocal microscope (Zeiss) at an excitation wavelength of 488 nm and an emission wavelength of 509 nm. Plasma membrane localization was confirmed by plasmolysis after onion epidermal cells were treated with 30% (w/v) Suc solution for 20 min. To estimate PuHSFA4a transcriptional activation, full-length and six partial CDSs of the PuHSFA4a gene were cloned into the NcoI and SalI sites of the pGBKT7 vector to fuse with the GAL4 DBD (Aoyama and Chua, 1997). The vector was then transformed into the yeast strain AH109 (Clontech). The transformed yeast was spread on SD/-Trp, SD/-Trp/X-α-Gal, and SD/-Trp/-His/-Ade media (Coolaber) and incubated at 30°C for 3–5 d. The negative control, a pGBKT7 empty vector in AH109, was treated the same way. The primer sequences for vector construction and six partial CDSs are shown in Supplemental Table S5.
Protein Extraction from Transgenic Lines and Immunoblot Analysis
35S::GFP and 35S::PuHSFA4a-GFP reporter constructs were transiently expressed in P. ussuriensis leaves as described in Takata and Eriksson (2012). Total proteins were extracted from infected leaves expressing either GFP or PuHSFA4a fused with GFP using protein extraction buffer (10 mm of Tris-Cl at pH 7.5; 150 mm of NaCl; 0.5 mm of EDTA; 1 mm of phenylmethanesulfonyl fluoride; 0.5% [w/v] Nonidet P-40). We detected GFP and PuHSFA4a-GFP protein abundance by using a rabbit anti–GFP antibody (Abcam) and a goat anti-rabbit IgG H&L (HRP; ab205718; Abcam) secondary antibody. Immunoreactive polypeptides were visualized by using the western ECL blotting substrate BeyoECL Moon (Beyotime). Imaging was performed using a model no. LAS-4000 Imaging System (Fijifilm).
Transcriptome Analysis
To assess PuHSFA4a-mediated excess Zn tolerance in P. ussuriensis using RNA-seq experiments, in vitro cuttings of PuHSFA4a-OE and wild type were grown for 7 d in half strength MS medium, and were then transferred to half strength MS medium supplemented with 1.2 mm of ZnSO4 for two weeks. A separate cohort of plants was grown in half strength MS medium for all three weeks as a control. Total RNA isolation, library construction, and sequencing were performed by Anoroad Technologies. In total, 2 μg RNA was prepared for library construction. Sequencing was performed on a HiSeq 4000 (Illumina). Obtained reads were mapped to the P. trichocarpa genome (http://phytozome.jgi.doe.gov) using the software TopHat2 (http://ccb.jhu.edu/software/tophat/index.shtml; Kim et al., 2013), differential expression analysis was conducted using the programs Cuffdiff (http://cole-trapnell-lab.github.io/cufflinks/) and DEseq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html; Love et al., 2014). Heatmaps were generated using the software GENE (https://software.broadinstitute.org/GENE-E/). Ontology enrichment was analyzed using the software BiNGO (https://www.psb.ugent.be/cbd/papers/BiNGO/Home.html). Finally, our RNA-seq results were verified by RT-qPCR and all primers used are shown in Supplemental Table S5.
ChIP Assays
In vitro cuttings of PuHSFA4a-OE and wild type were grown for 7 d in half strength MS medium, and were then transferred to 1/2 MS medium supplemented with or without 1.2 mm of ZnSO4 for 2 weeks. Then, the roots of PuHSFA4a-OE transgenic plants were harvested for analysis using ChIP-PCR (Li et al., 2014) and ChIP-qPCR (Huang et al., 2017). GFP antibodies (Beyotime) bound to Protein-A agarose beads (Sigma-Aldrich) were used for the ChIP assays. A rabbit anti-hemagglutinin antibody was used as a negative control (Mock), and PuActin was used as a control. The degree of enrichment of promoter fragments over 13 genomic fragments were determined by PCR and RT-qPCR. The primers used for these analyses are listed in Supplemental Table S5.
Y1H Assays
The promoters of PuGSTU17 (1,654-bp) and PuPLA2 (1,673-bp) were PCR-amplified and inserted into a pAbAi-BR yeast integrating vector (bait–reporter; Clontech) and the PuHSFA4a gene was cloned into the pGADT7-rec vector (Clontech) in frame with the GAL4 activation domain. The pGADT7-PuHSFA4a and pAbAi-ProPuGSTU17 or pGADT7-PuHSFA4a and pAbAi-ProPuPLA2 vectors were cotransformed into yeast Y1HGold strain using a Matchmaker One-Hybrid Library Construction and Screening Kit (Clontech). Cotransformed yeast cells were selected on SD/-Ura-Leu (a synthetic dropout medium lacking Ura and Leu) with or without AbA (0.6 mg L−1; Sigma-Aldrich), and were then incubated for 3–5 d at 30°C. The Y1H assay was conducted as described in Wang et al. (2014). Both positive (pGAD-p53+p53-pAbAi) and negative (pGADT7-AD+ProPuGSTU17-pAbAi or ProPuPLA2-pAbAi) controls were carried out in the same manner. The primers used for these analyses are listed in Supplemental Table S5.
Dual LUC Assay
The promoters of PuGSTU17 (1,654 bp) and PuPLA2 (1,673 bp) were cloned into pGreenII0800-LUC vectors as reporters. Full-length open reading frames of PuHSFA4a were cloned into pBI121-GFP vectors as the effector. The effector and reporter vectors were cotransformed into leaves of Nicotiana benthamiana as previously described by He et al. (2013). Detection of dual LUC activity was also carried out according to He et al. (2013). A Synergy H1 Microplate Reader (Bio-Tek) and a Dual-Luciferase Assay Kit (Promega) were used to detect the firefly LUC and Renilla luciferase activities according to the manufacturer’s instructions. The primers used for these analyses are listed in Supplemental Table S5.
EMSA
The full-length CDS of PuHSFA4a was inserted into vector pET32a (Novagen) for production of recombinant PuHSFA4a protein in Escherichia coli BL21. Transformed E. coli were then grown in liquid media to an OD600 nm of 0.5, treated with 0.6 mm of isopropyl-b-d-thiogalactopyranoside for 7 h at 30°C, and then collected by centrifugation. The His-PuHSFA4a fusion protein was purified using PureCube Ni-NTA Agarose (Cube Biotech). The probes were labeled with biotin using an EMSA Probe Biotin Labeling Kit (Beyotime) with all procedures performed as per the manufacturer’s instructions, and an unlabeled probe was used as the competitor. EMSA was conducted using a Chemiluminescent EMSA Kit (Beyotime). Reactions were incubated at 25°C for 30 min after the addition of 2 µg of protein from the His purification eluate. Four probes were generated for PuGSTU17 and PuPLA2 promoters using the same oligonucleotides described in Supplemental Table S5.
Statistical Analysis
Stress treatments were repeated at least three times. The statistical significance between two means (except for microarray data) was determined using ANOVAs followed by posthoc t tests to examine pairwise differences. All tests were reported such that asterisks indicated a significance level of P < 0.05 and ** indicated P < 0.01.
Accession Numbers
Nucleic acid sequences of PuHSFA4a, PuGSTU17, and PuPLA2 from P. ussuriensis have been deposited into GenBank with accession numbers of MH668277, MH668278, and MH668279. The raw sequence data of RNA-seq experiments reported in this study have been deposited in the Gene Expression Omnibus with accession numbers of GSE117778.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-qPCR analysis of Class A of HSFs expression under Zn stress and PuHSFA4a and PuGSTU17 expression under abiotic stresses.
Supplemental Figure S2. Immunoblot analysis used anti-GFP antibody to verify PuHSFA4a-GFP fusion protein expression.
Supplemental Figure S3. Phenotypes and RT-qPCR confirmation of PuHSFA4a-overexpression (A4a-OE3 and A4a-OE8) and a dominant repressor version of PuHSFA4a (A4a-SRDX1 and A4a-SRDX6) P. ussuriensis plants.
Supplemental Figure S4. Phenotypes of wild type, PuHSFA4a-overexpression (PuHSFA4a-OE), and PuHSFA4a-suppression (PuHSFA4a-SRDX) transgenic P. ussuriensis plants under various abiotic stresses.
Supplemental Figure S5. Gene Ontology categorization of PuHSFA4a-binding genes under excess Zn stress.
Supplemental Figure S6. ChIP-PCR analysis of PuHSFA4a binding to PuGSTU17 and PuPLA2 gene promoters.
Supplemental Figure S7. Phenotypes and RT-qPCR confirmation of PuGSTU17-overexpression (PuGSTU17-OE) and PuGSTU17 suppression (PuGSTU17-RNAi) in P. ussuriensis.
Supplemental Figure S8. Phenotypes of wild-type and PuGSTU17-overexpression (PuGSTU17-OE) transgenic P. ussuriensis plants under different abiotic stresses.
Supplemental Figure S9. RT-qPCR analysis to confirm PuPLA2-overexpression transgenic lines and phenotypic observation of plants.
Supplemental Table S1. DEGs in wild-type P. ussuriensis plants with or without excess Zn stress (log2 Fold Change > 1, P value < 0.01).
Supplemental Table S2. DEGs in PuHSFA4a-overexpression (PuHSFA4a-OE) transgenic P. ussuriensis plants with or without excess Zn stress (log2 Fold Change > 1, P value < 0.01).
Supplemental Table S3. DEGs between wild-type and PuHSFA4a-overexpression (PuHSFA4a-OE) transgenic P. ussuriensis plants under control conditions (log2 Fold Change > 1, P value < 0.01).
Supplemental Table S4. DEGs between wild-type and PuHSFA4a-overexpression (PuHSFA4a-OE) transgenic P. ussuriensis plants under excess Zn conditions (log2 Fold Change > 1, P value < 0.01).
Supplemental Table S5. Primers used in this study.
Acknowledgments
We acknowledge TopEdit English Editing LLC for the linguistic editing and proofreading during the preparation of this article.
Footnotes
This work was sup\ported by the Fundamental Research Funds for the Central Universities of China (2572018CL04 to C.L.), the 111 Project (B16010 to C.L.), Fundamental Research Funds for the Central Universities of China (2572015AA20 to H.Z.), and the National Natural Science Foundation of China (31400573 to J.Y.).
Articles can be viewed without a subscription.
References
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Ariani A, Francini A, Andreucci A, Sebastiani L (2016) Over-expression of AQUA1 in Populus alba Villafranca clone increases relative growth rate and water use efficiency, under Zn excess condition. Plant Cell Rep 35: 289–301 [DOI] [PubMed] [Google Scholar]
- Arrivault S, Senger T, Krämer U (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J 46: 861–879 [DOI] [PubMed] [Google Scholar]
- Bai S, Yao T, Li M, Guo X, Zhang Y, Zhu S, He Y (2014) PIF3 is involved in the primary root growth inhibition of Arabidopsis induced by nitric oxide in the light. Mol Plant 7: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceló J, Poschenrieder C (1990) Plant water relations as affected by heavy metal stress: A review. J Plant Nutr 13: 1–37 [Google Scholar]
- Bittsánszky A, Kömives T, Gullner G, Gyulai G, Kiss J, Heszky L, Radimszky L, Rennenberg H (2005) Ability of transgenic poplars with elevated glutathione content to tolerate zinc(2+) stress. Environ Int 31: 251–254 [DOI] [PubMed] [Google Scholar]
- Blaudez D, Kohler A, Martin F, Sanders D, Chalot M (2003) Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 15: 2911–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley MR, White PJ, Hammond JP, Zelko I, Lux A (2007) Zinc in plants. New Phytol 173: 677–702 [DOI] [PubMed] [Google Scholar]
- Chai G, Qi G, Cao Y, Wang Z, Yu L, Tang X, Yu Y, Wang D, Kong Y, Zhou G (2014) Poplar PdC3H17 and PdC3H18 are direct targets of PdMYB3 and PdMYB21, and positively regulate secondary wall formation in Arabidopsis and poplar. New Phytol 203: 520–534 [DOI] [PubMed] [Google Scholar]
- Chang H, Lin C, Huang H (2005) Zinc-induced cell death in rice (Oryza sativa L.) roots. Plant Growth Regul 46: 261–266 [Google Scholar]
- Chaoui A, Mazhoudi S, Ghorbal MH, El Ferjani E (1997) Cadmium and zinc induction of lipid peroxidation and effects on antioxidant enzyme activities in bean (Phaseolus vulgaris L). Plant Sci 127: 139–147 [Google Scholar]
- Chen J, Yang L, Gu J, Bai X, Ren Y, Fan T, Han Y, Jiang L, Xiao F, Liu Y, et al. (2015) MAN3 gene regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis thaliana. New Phytol 205: 570–582 [DOI] [PubMed] [Google Scholar]
- Cho M, Chardonnens AN, Dietz KJ (2003) Differential heavy metal tolerance of Arabidopsis halleri and Arabidopsis thaliana: A leaf slice test. New Phytol 158: 287–293 [Google Scholar]
- Davoine C, Falletti O, Douki T, Iacazio G, Ennar N, Montillet JL, Triantaphylidès C (2006) Adducts of oxylipin electrophiles to glutathione reflect a 13 specificity of the downstream lipoxygenase pathway in the tobacco hypersensitive response. Plant Physiol 140: 1484–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses-Fonrouge AG, Voigt K, Schröder A, Arrivault S, Thomine S, Krämer U (2005) Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett 579: 4165–4174 [DOI] [PubMed] [Google Scholar]
- Duan M, Zhang R, Zhu F, Zhang Z, Gou L, Wen J, Dong J, Wang T (2017) A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 29: 1748–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq MA, Detterbeck A, Clemens S, Dietz KJ (2016) Silicon-induced reversibility of cadmium toxicity in rice. J Exp Bot 67: 3573–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N, Kaur H (2013) Response of antioxidant enzymes, phytochelatins and glutathione production towards Cd and Zn stresses in Cajanus cajan (L.) Millsp. genotypes colonized by arbuscular mycorrhizal fungi. J Agron Crop Sci 199: 118–133 [Google Scholar]
- Gong B, Yi J, Wu J, Sui J, Khan MA, Wu Z, Zhong X, Seng S, He J, Yi M (2014) LlHSFA1, a novel heat stress transcription factor in lily (Lilium longiflorum), can interact with LlHSFA2 and enhance the thermotolerance of transgenic Arabidopsis thaliana. Plant Cell Rep 33: 1519–1533 [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Han Y, Sa G, Sun J, Shen Z, Zhao R, Ding M, Deng S, Lu Y, Zhang Y, Shen X (2014) Overexpression of Populus euphratica xyloglucan endo transglucosylase/hydrolase gene confers enhanced cadmium tolerance by the restriction of root cadmium uptake in transgenic tobacco. Environ Exp Bot 100: 74–83 [Google Scholar]
- He J, Li H, Luo J, Ma C, Li S, Qu L, Gai Y, Jiang X, Janz D, Polle A, et al. (2013) A transcriptomic network underlies microstructural and physiological responses to cadmium in Populus × canescens. Plant Physiol 162: 424–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holk RUPA, Scherer GF (1998) Fatty acids and lysophospholipids as potential second messengers in auxin action. Rapid activation of phospholipase A2 activity by auxin in suspension‐cultured parsley and soybean cells. Plant J 16: 601–611 [Google Scholar]
- Holk A, Rietz S, Zahn M, Quader H, Scherer GF (2002) Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol 130: 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, ElSayed AI, Moore M, Dietz KJ (2017) Redox and reactive oxygen species network in acclimation for salinity tolerance in sugar beet. J Exp Bot 68: 1283–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Wang XC, Yang JY (2013) [Interannual variation patterns of heavy metals concentrations in tree rings of Larix gmelinii near Xilin lead–zinc mine, Yichun of Northeast China]. Ying Yong Sheng Tai Xue Bao 24: 1536–1544 [PubMed] [Google Scholar]
- Huang X, Zhang X, Gong Z, Yang S, Shi Y (2017) ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J 89: 354–365 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha B, Sharma A, Mishra A (2011) Expression of SbGSTU (tau class glutathione s-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance. Mol Biol Rep 38: 4823–4832 [DOI] [PubMed] [Google Scholar]
- Ji W, Zhu Y, Li Y, Yang L, Zhao X, Cai H, Bai X (2010) Over-expression of a glutathione s-transferase gene, GsGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnol Lett 32: 1173–1179 [DOI] [PubMed] [Google Scholar]
- Jiang D, Yan S (2018) Effects of Cd, Zn, or Pb stress in Populus alba berolinensis on the antioxidant, detoxifying, and digestive enzymes of Lymantria dispar. Environ Entomol 47: 1323–1328 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning and expression of tagged fluorescent protein in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Khare D, Mitsuda N, Lee S, Song WY, Hwang D, Ohme-Takagi M, Martinoia E, Lee Y, Hwang JU (2017) Root avoidance of toxic metals requires the GeBP-LIKE 4 transcription factor in Arabidopsis thaliana. New Phytol 213: 1257–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YY, Choi H, Segami S, Cho HT, Martinoia E, Maeshima M, Lee Y (2009) AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J 58: 737–753 [DOI] [PubMed] [Google Scholar]
- Kohler A, Blaudez D, Chalot M, Martin F (2004) Cloning and expression of multiple metallothioneins from hybrid poplar. New Phytol 164: 83–93 [DOI] [PubMed] [Google Scholar]
- König K, Galliardt H, Moore M, Treffon P, Seidel T, Dietz KJ (2014) Assessing redox state and reactive oxygen species in circadian rhythmicity. Methods Mol Biol 1158: 239–271 [DOI] [PubMed] [Google Scholar]
- La Camera S, Geoffroy P, Samaha H, Ndiaye A, Rahim G, Legrand M, Heitz T (2005) A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in Arabidopsis. Plant J 44: 810–825 [DOI] [PubMed] [Google Scholar]
- Laity JH, Andrews GK (2007) Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1). Arch Biochem Biophys 463: 201–210 [DOI] [PubMed] [Google Scholar]
- Lan HX, Wang ZF, Wang QH, Wang MM, Bao YM, Huang J, Zhang HS (2013) Characterization of a vacuolar zinc transporter OZT1 in rice (Oryza sativa L.). Mol Biol Rep 40: 1201–1210 [DOI] [PubMed] [Google Scholar]
- Lang S, Liu X, Xue H, Li X, Wang X (2017) Functional characterization of BnHSFA4a as a heat shock transcription factor in controlling the re-establishment of desiccation tolerance in seeds. J Exp Bot 68: 2361–2375 [DOI] [PubMed] [Google Scholar]
- Lee MO, Cho K, Kim SH, Jeong SH, Kim JA, Jung YH, Shim J, Shibato J, Rakwal R, Tamogami S, et al. (2008) Novel rice OsSIPK is a multiple stress responsive MAPK family member showing rhythmic expression at mRNA level. Planta 227: 981–990 [DOI] [PubMed] [Google Scholar]
- Li F, Zhang H, Zhao H, Gao T, Song A, Jiang J, Chen F, Chen S (2018) Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol J 16: 1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Xu W, Kronzucker HJ, Shi W (2015) Ethylene is critical to the maintenance of primary root growth and Fe homeostasis under Fe stress in Arabidopsis. J Exp Bot 66: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lin YC, Li Q, Shi R, Lin CY, Chen H, Chuang L, Qu GZ, Sederoff RR, Chiang VL (2014) A robust chromatin immunoprecipitation protocol for studying transcription factor-DNA interactions and histone modifications in wood-forming tissue. Nat Protoc 9: 2180–2193 [DOI] [PubMed] [Google Scholar]
- Lin YF, Aarts MG (2012) The molecular mechanism of zinc and cadmium stress response in plants. Cell Mol Life Sci 69: 3187–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Liu Y, Rao J, Wang G, Li H, Ge F, Chen C (2013) [Overexpression of the glutathione s-transferase gene from Pyrus pyrifolia fruit improves tolerance to abiotic stress in transgenic tobacco plants]. Mol Biol (Mosk) 47: 591–601 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZB, He J, Polle A, Rennenberg H (2016) Heavy metal accumulation and signal transduction in herbaceous and woody plants: Paving the way for enhancing phytoremediation efficiency. Biotechnol Adv 34: 1131–1148 [DOI] [PubMed] [Google Scholar]
- Lyck R, Harmening U, Höhfeld I, Treuter E, Scharf KD, Nover L (1997) Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta 202: 117–125 [DOI] [PubMed] [Google Scholar]
- Metwally A, Finkemeier I, Georgi M, Dietz KJ (2003) Salicylic acid alleviates the cadmium toxicity in barley seedlings. Plant Physiol 132: 272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe S, Izawa S, Inoue Y (2001) The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem Biophys Res Commun 282: 79–83 [DOI] [PubMed] [Google Scholar]
- Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P (2009) AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol 149: 894–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schöffl F (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Salamó I, Papdi C, Gábor R, Zsigmond L, Vilela B, Lumbreras V, Nagy I, Horváth B, Domoki M, Darula Z (2014) The heat shock factor HSFA4A confers salt tolerance and is regulated by oxidative stress and the MAP kinases, MPK3 and MPK6. Plant Physiol 114: 237891–237933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ. (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Schvartzman MS, Corso M, Fataftah N, Scheepers M, Nouet C, Bosman B, Carnol M, Motte P, Verbruggen N, Hanikenne M (2018) Adaptation to high zinc depends on distinct mechanisms in metallicolous populations of Arabidopsis halleri. New Phytol 218: 269–282 [DOI] [PubMed] [Google Scholar]
- Sharma R, Sahoo A, Devendran R, Jain M (2014) Over-expression of a rice tau class glutathione s-transferase gene improves tolerance to salinity and oxidative stresses in Arabidopsis. PLoS One 9: e92900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim D, Hwang JU, Lee J, Lee S, Choi Y, An G, Martinoia E, Lee Y (2009) Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21: 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Huang Q, Zhang X (2001) Study on gene resources in Populus ussuriensis. For Res 14: 472–478 [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Zhai Q, Li C (2013) PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25: 2102–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T, Nishizawa NK, Nakanishi H (2012) The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ 35: 1948–1957 [DOI] [PubMed] [Google Scholar]
- Takata N, Eriksson ME (2012) A simple and efficient transient transformation for hybrid aspen (Populus tremula × P. tremuloides). Plant Methods 8: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant N, Monnet F, Hitmi A, Sallanon H, Coudret A (2005) Comparative study of responses in four Datura species to a zinc stress. Chemosphere 59: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P (2004) Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett 576: 306–312 [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Wang C, Zien CA, Afitlhile M, Welti R, Hildebrand DF, Wang X (2000) Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 12: 2237–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhang SH, Wang PF, Qian J, Hou J, Zhang WJ, Lu J (2009) Excess Zn alters the nutrient uptake and induces the antioxidative responses in submerged plant Hydrilla verticillata (L.f.) Royle. Chemosphere 76: 938–945 [DOI] [PubMed] [Google Scholar]
- Wang Y, Peng W, Zhou X, Huang F, Shao L, Luo M (2014) The putative Agrobacterium transcriptional activator-like virulence protein VirD5 may target T-complex to prevent the degradation of coat proteins in the plant cell nucleus. New Phytol 203: 1266–1281 [DOI] [PubMed] [Google Scholar]
- Xu J, Tian YS, Peng RH, Xiong AS, Zhu B, Hou XL, Yao QH (2010) Cyanobacteria MT gene SmtA enhance zinc tolerance in Arabidopsis. Mol Biol Rep 37: 1105–1110 [DOI] [PubMed] [Google Scholar]
- Xu W, Shi W, Liu F, Ueda A, Takabe T (2008) Enhanced zinc and cadmium tolerance and accumulation in transgenic Arabidopsis plants constitutively overexpressing a barley gene (HvAPX1) that encodes a peroxisomal ascorbate peroxidase. Botany 86: 567–575 [Google Scholar]
- Yang G, Xu Z, Peng S, Sun Y, Jia C, Zhai M (2016) In planta characterization of a tau class glutathione s-transferase gene from Juglans regia (JrGSTTau1) involved in chilling tolerance. Plant Cell Rep 35: 681–692 [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang J, Wang W, Li D, Hu X, Wang H, Wei M, Liu Q, Wang Z, Li C (2015) Genome-wide identification and expression profiling of the copper transporter gene family in Populus trichocarpa. Plant Physiol Biochem 97: 451–460 [DOI] [PubMed] [Google Scholar]
- Zhang L, Ma H, Chen T, Pen J, Yu S, Zhao X (2014) Morphological and physiological responses of cotton (Gossypium hirsutum L.) plants to salinity. PLoS One 9: e112807 [DOI] [PMC free article] [PubMed] [Google Scholar]