The plant-specific protein GL6 determines grain length and spikelet number in rice by affecting cell proliferation through gene expression regulation via the RNAPIII transcription machinery
Abstract
Grain size is one of the key determinants of grain yield. Although a number of genes that control grain size in rice (Oryza sativa) have been identified, the overall regulatory networks behind this process remain poorly understood. Here, we report the map-based cloning and functional characterization of the quantitative trait locus GL6, which encodes a plant-specific plant AT-rich sequence- and zinc-binding transcription factor that regulates rice grain length and spikelet number. GL6 positively controls grain length by promoting cell proliferation in young panicles and grains. The null gl6 mutant possesses short grains, whereas overexpression of GL6 results in large grains and decreased grain number per panicle. We demonstrate that GL6 participates in RNA polymerase III transcription machinery by interacting with RNA polymerase III subunit C53 and transcription factor class C1 to regulate the expression of genes involved in rice grain development. Our findings reveal a further player involved in the regulation of rice grain size that may be exploited in future rice breeding.
Rice (Oryza sativa) is one of the three major cereal crops in the world, and the most important staple food in Asia. Rice has served as a model monocot plant for molecular genetic dissection since its reference genome sequence was generated in 2005 (International Rice Genome Sequencing Project, 2005). The exploitation of rice genetics to increase grain yield and improve plant architecture are the focus of current rice-breeding programs (Huang et al., 2009).
Grain size, one of the most important determinate factors of grain yield, is specified by grain length, width, and thickness. In recent years, a number of genes and quantitative trait loci (QTLs) that control grain size have been identified and functionally characterized in rice. These grain size genes have been found to be involved in signaling pathways mediated by guanine nucleotide-binding proteins (G-proteins), proteasomal degradation, phytohormones, protein kinases, and transcriptional factors that control cell division and/or cell expansion during seed development (Zuo and Li, 2014). GS3, a major gene controlling grain length and weight, encodes a putative Gγ protein that functions in G-protein signaling (Fan et al., 2006; Mao et al., 2010). GS3 suppresses the interaction between Gβ protein RGB1 (G protein β subunit 1) and two other Gγ proteins, namelyDEP1 (dense and erect panicle1) and GGC2 (G protein γ subunit type C2), to antagonistically regulate grain size (Sun et al., 2018). These Gβγ subunits are also colocalized in the nucleus and interact directly with the downstream effector OsMADS1 (MADS‐box gene 1) to regulate rice grain length and yield (Liu et al., 2018). GW2 (grain width 2) encodes a RING-type E3 ubiquitin ligase that functions in protein degradation via the ubiquitin-proteasome pathway and negatively regulates cell division of the spikelet hull, thus affecting grain width (Song et al., 2007). GLW7 (grain length and weight on chromosome 7), a target gene of OsmiR156 (microRNA 156), encodes the plant-specific transcription factor OsSPL13 that positively regulates grain length and yield. The large-grain allele of GLW7 in tropical japonica varieties was identified to be introgressed from indica varieties under artificial selection (Si et al., 2016). Copy number variation at the GL7 locus affects the expression of two linked genes to regulate grain length (Wang et al., 2015b). GW8, encoding the further SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-domain transcription factor OsSPL16, positively regulates grain width and directly binds to the GW7/GL7 promoter to repress its expression (Wang et al., 2012, 2015a). Some genes involved in auxin response and brassinosteroid signaling, such as TGW6 and GW5, also influence the grain size and yield in rice (Ishimaru et al., 2013; Liu et al., 2017). Most of these genes regulate grain size by increasing or decreasing cell number (Qi et al., 2012; Zhang et al., 2012; Liu et al., 2015; Song et al., 2015; Yu et al., 2018; Zhao et al., 2018a), and a few genes control grain length by regulating cell size or length (Wang et al., 2015b; Si et al., 2016). In the case of GS2 and GS5, they affect grain shape by controlling both cell number and size (Li et al., 2011; Che et al., 2015; Hu et al., 2015).
These findings have enriched our knowledge of the regulatory mechanisms behind grain size in rice. However, how these genes are integrated into signaling pathways, as well as into the regulatory networks behind grain development, and the cross talk between them remain poorly understood. Therefore, identification and molecular characterization of new QTLs/genes involved in grain size will help to comprehensively describe regulatory networks and serve for future improvement of rice yield (Duan et al., 2015).
Here, we report the identification and characterization of the rice grain length QTL GL6. GL6 encodes a plant AT-rich sequence- and zinc-binding (PLATZ) protein and is preferentially expressed in young panicles. A mutant version of GL6 that carries a premature stop codon results in short grains via impaired RNA polymerase III (RNAPIII)-mediated transcription. We demonstrate that GL6 functions in a further molecular mechanism that modulates grain length and grain weight.
RESULTS
Map-Based Cloning of GL6
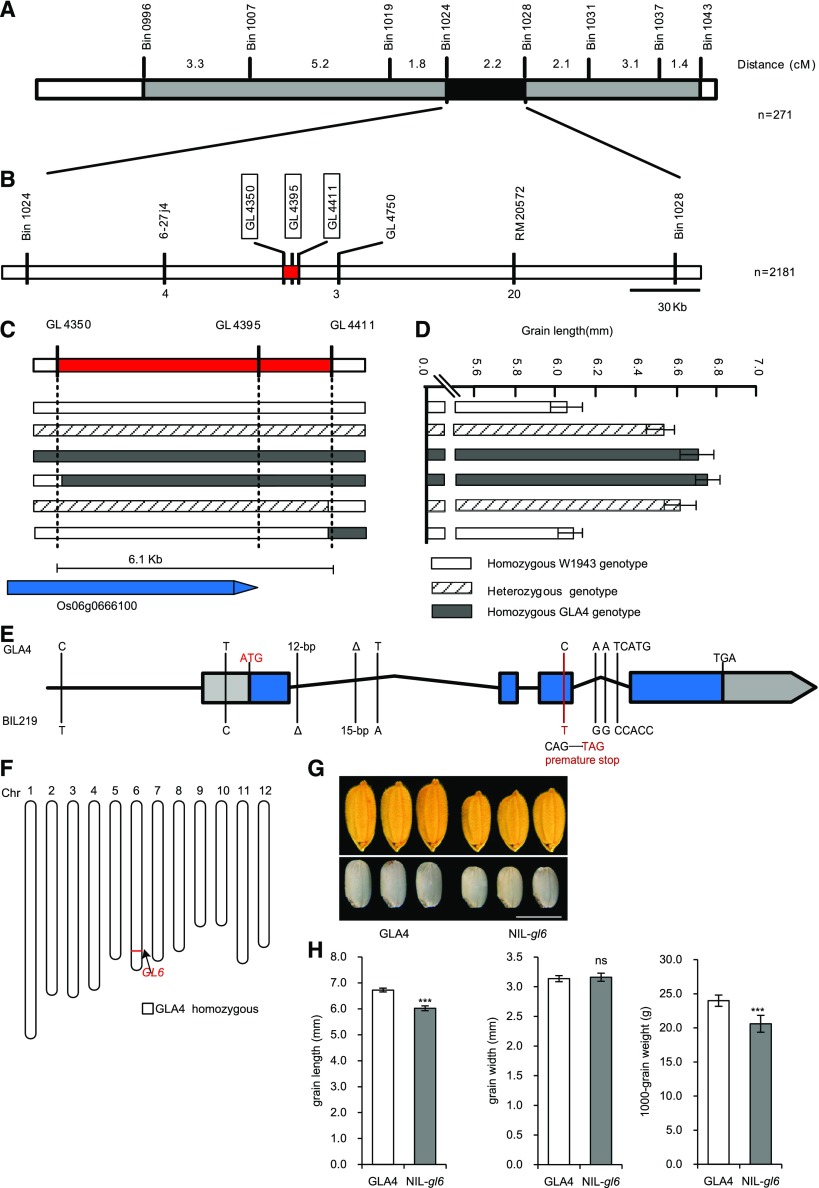
In our previous work, we mapped a major QTL, GL6, which explained 20.5% of the phenotypic variation (R2) in grain length within a set of 271 lines derived from a cross between a cultivated rice variety, O. sativa ssp. indica Guangluai4 (GLA4) and a wild rice accession, Oryza rufipogon W1943 (W1943; Huang et al., 2012). The GL6 locus was initially mapped to the region between the recombination bins Bin_1024 (53.3 cM) and Bin_1028 (55.5 cM) on chromosome 6.
To fine map this locus, one backcross inbred line, BIL219, with shorter grain length, harboring the GL6 locus and carrying several other segments from W1943 (Supplemental Fig. S1), was selected and backcrossed to GLA4. We then carried out high-resolution mapping using 2,181 BC1F5 individuals, and the locus was finally delimited to a 6.1-kb region between the two markers GL4350 and GL4411 (Fig. 1, B–D).
Figure 1.
Map-based cloning of GL6. A, Location of GL6 on rice chromosome 6, with 271 chromosome segment substitution lines. B, High-resolution mapping of the GL6 region was performed with 2,181 BC1F5 lines. The number of recombinants between molecular markers is indicated below the linkage map. C, GL6 was narrowed down to a 6.1-kb genomic region. D, Grain length is shown for each recombinant line. Values are given as the mean ± sd. E, Gene structure and allelic variations of GL6 between GLA4 and BIL219. Colored boxes and black lines represent exons and introns, respectively. The start and stop codons are indicated above the gene, and the coding region is blue. F, Chromosome maps of NIL-gl6. NIL-gl6 contained the T-type W1943 allele at GL6 in the 17-kb region on chromosome 6, shown as a red bar. G, Mature paddy grain (upper) and brown rice (lower) morphology of GLA4 and NIL-gl6. Scale bar = 5 mm. H, Comparison of grain length, width, and 1,000-grain weight between GLA4 and NIL-gl6. Values are given as the mean ± sd. ***P < 0.001, significant difference determined by Student’s t test.
According to the Rice Annotation Project annotation (http://rapdb.dna.affrc.go.jp/), this region contains only one candidate gene, Os06g0666100, which encodes a PLATZ protein. Compared to GLA4, the coding region of GL6 from BIL219 contained a C-to-T substitution in the third exon at nucleotide 352 that introduced a stop codon resulting in premature termination of translation (Fig. 1E; Supplemental Fig. S2). Interestingly, the original donor parent W1943 represented a T/C heterozygote at the premature stop codon allele; however, all other single-nucleotide polymorphisms (SNPs) were identical to those in BIL219. Thus, we considered that BIL219 inherited the T allele from the generation of progeny. In addition, we isolated three genotypes of GL6 in a filial generation by self-pollinating the heterozygous W1943, and found that the grain size of W1943 was related to different GL6 genotypes (Supplemental Fig. S3). Thus, we hypothesized that this premature stop codon might cause reduction or loss of function of GL6, thus affecting rice grain length.
Characterization and Validation of GL6 Function in Regulating Grain Length
To further investigate the function of GL6, we developed a near isogenic line (NIL), NIL-gl6, containing a 17-kb “T-type” W1943 chromosomal region at the GL6 locus in the GLA4 genetic background (Fig. 1F). Compared with GLA4, NIL-gl6 showed shorter grains (a 10.4% decrease) and lower 1,000-grain weight (a 14% decrease). However, the width of the grains was not affected (Fig. 1, G and H).
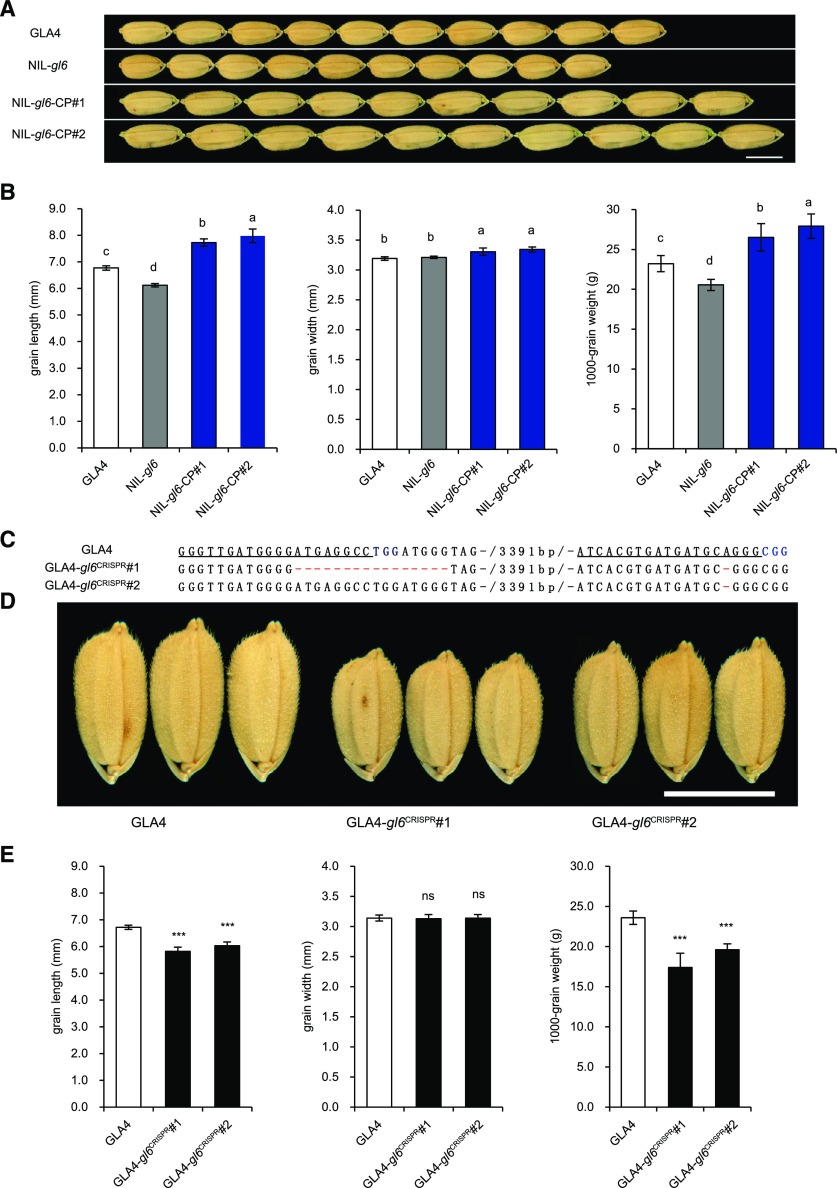
In order to verify the function of the candidate gene GL6, we generated a genetic complementation construct in which the GL6 locus from GLA4, containing the entire gene region, the 9277-bp promoter region, and the 728-bp downstream sequence, was introduced into NIL-gl6. In comparison with NIL-gl6, the transgenic genetic complementation lines NIL-gl6-CP1 and NIL-gl6-CP2 showed obvious increases in grain length and weight, even greater than in GLA4, alongside a slight increase in grain width (Fig. 2, A and B; Supplemental Fig. S4). In addition, we carried out gene mutation of GL6 by using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome editing system (Ma et al., 2015) in GLA4 (Fig. 2C) and a japonica variety, Nipponbare (Supplemental Fig. S5A). In GLA4, the resulting GL6 loss-of-function CRISPR-Cas9-derived mutants, GLA4-gl6CRISPR1 and GLA4-gl6CRISPR2, displayed reduced grain length and weight when compared with the wild-type plants (Fig. 2, D and E; Supplemental Fig. S5, B–E). Altogether, these results demonstrate that Os06g0666100 is the causative gene for the QTL GL6 and functions in the positive regulation of rice grain length and weight.
Figure 2.
Validation that GL6 controls grain length. A, Grain morphology of GLA4, NIL-gl6, and two independent complemented transgenic lines (NIL-gl6-CP1 and NIL-gl6-CP2), Scale bar = 5 mm. B, Comparisons of grain length, width, and 1,000-grain weight among the lines shown in A. Lowercase letters indicate significant differences (P < 0.05) as determined by Duncan’s multiple range test. C, Sequence of the CRISPR mutant alleles. The wild-type sequence is shown at the top, with the target sites underlined in black and the protospacer adjacent motif sequence highlighted in blue. Deletions are shown as red dashes. D, Grain phenotype of GLA4 and two independent CRISPR transgenic lines (GLA4-gl6CRISPR1 and GLA4-gl6CRISPR2). Scale bar = 5 mm. E, Comparisons of grain length, width, and 1,000-grain weight among the lines shown in D. Values are given as the mean ± sd. *** Significant difference (P <0.001, Student's t test).
GL6 Influences Cell Proliferation to Regulate Grain Length
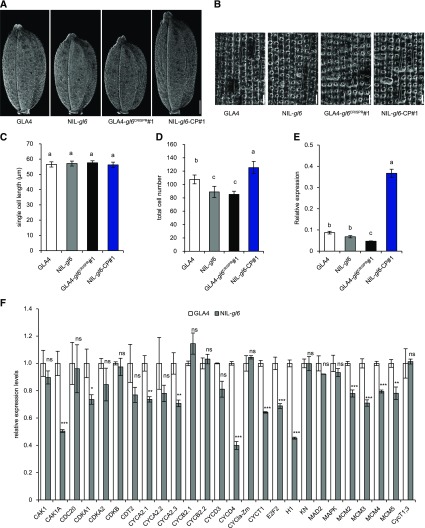
Since cell division and cell expansion are responsible for altering grain size and grain size is restricted by the size of the spikelet hull, we compared the epidermal cells of GLA4 and NIL-gl6 by scanning electron microscope. Observations of the outer glume surface showed that total cell number along the longitudinal axis in NIL-gl6 was reduced by 17.38% compared with that in GLA4, without any significant difference in single cell length (Fig. 3, A–D). Similarly, total cell number of the outer epidermal cells was also decreased by 20.72% in GLA4-gl6CRISPR1 and increased by 16.54% in NIL-gl6-CP1 (Fig. 3, A–D). The CRISPR/Cas9-derived gl6 mutant in Nipponbare, NIP-gl6CRISPR1, also showed a reduction in cell number (Supplemental Fig. S5, F–I). Consistent with this, comparative measurement of GL6 expression level among these plants showed that NIL-gl6-CP1 exhibited elevated GL6 transcripts, whereas decreased GL6 expression levels observed in GLA4-gl6CRISPR1 and NIP-gl6CRISPR1 (Fig. 3E; Supplemental Fig. S5J). Together, these results show that higher expression level of GL6 contributes to increased cell numbers for the spikelet hull, thus resulting in large grains. As expected, expression levels of 12 cell cycle-related genes, including CAK1A, CYCD4, CYCT1, E2F2, and H1, were significantly down-regulated in NIL-gl6 (Fig. 3F), indicating that the reduced cell number in NIL-gl6 might result from decreased expression of genes that promote cell proliferation. Hence, these results suggest that GL6 positively regulates grain length by altering cell division instead of cell expansion to control cell number of the glume during spikelet development.
Figure 3.
The effect of GL6 on cell number contributes to grain length. A and B, Scanning electron micrographs of the whole grain (A) and outer glume surfaces (B) in GLA4, NIL-gl6, GLA4-gl6CRISPR1, and NIL-gl6-CP1. Scale bars = 1 mm (A) and 100 μm (B). C and D, Comparisons of single cell length (C) and total cell number (D) among the lines shown in A. E, Comparisons of relative expression levels of GL6 among the lines shown in A. Values are given as the mean ± sd. Lowercase letters indicate significant differences (P < 0.05) as determined by Duncan’s multiple-range test. F, Relative expression levels of 26 cell cycle-related genes in young panicles (0–1 cm) of GLA4 and NIL-gl6. OsUBQ5 was used as the control and the values of expression levels in GLA4 were set to 1. Values are given as the mean ± sd. Student’s t test significant difference: *P < 0.05, **P < 0.01 and *** P < 0.001; ns, not significant.
GL6 Negatively Regulates Grain Number per Panicle
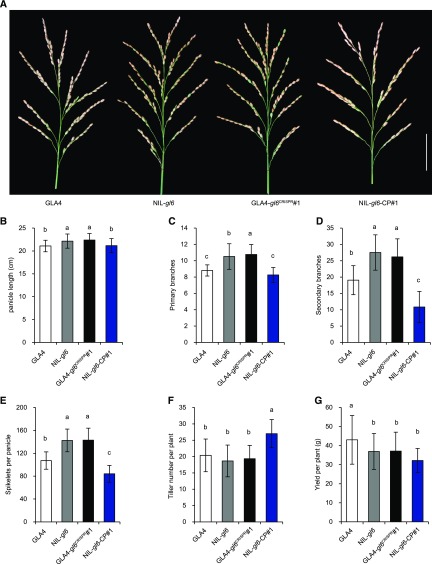
In addition to the grain size and weight, we also compared other agronomic traits among GLA4, NIL-gl6, and transgenic lines in field trials. We found that larger seed had fewer grains per panicle (Fig. 4A). Compared to that in GLA4 (107.25 ± 15.16), the number of grains per panicle was increased by 32.9% and 33.4% in NIL-gl6 (141.53 ± 19.89) and GLA4-gl6CRISPR1 (143.10 ± 20.82), respectively. However, the complemented transgenic line NIL-gl6-CP1 showed a dramatically reduced grain number (84.16 ± 14.74; Fig. 4E). We then measured the panicle length, primary branches, and secondary branches in these lines (Fig. 4, B–D). NIL-gl6 and GLA4-gl6CRISPR1 exhibited more secondary branches, whereas NIL-gl6-CP1 had fewer secondary branches, compared to GLA4 (Fig. 4D). The increased grain number was mainly attributed to the increased number of secondary branches. Other panicle phenotypes, such as panicle length and primary branches, showed only a slight effect on grain number. Moreover, we observed that the complemented line showed significantly increased tiller number per plant compared to other lines (Fig. 4F). Taking all these effects into account, the grain yield per plant was lower in NIL-gl6, GLA4-gl6CRISPR1, and NIL-gl6-CP1 than in GLA4 (Fig. 4G). These results reveal that GL6 influences both panicle and spikelet development and that a balance between grain number and grain size may exist that determines grain yield.
Figure 4.
GL6 affects panicle morphology. A, Panicles of GLA4, NIL-gl6, GLA4-gl6CRISPR1, and NIL-gl6-CP1. Scale bar = 5 cm. B to G, Comparisons of panicle length (B), primary branches (C), secondary branches (D), spikelets per panicle (E), tiller number per plant (F), and grain yield per plant (G), among the lines shown in A. Values are given as the mean ± sd. Lowercase letters indicate significant differences (P < 0.05) as determined by Duncan’s multiple-range test.
Spatial Expression Pattern of GL6
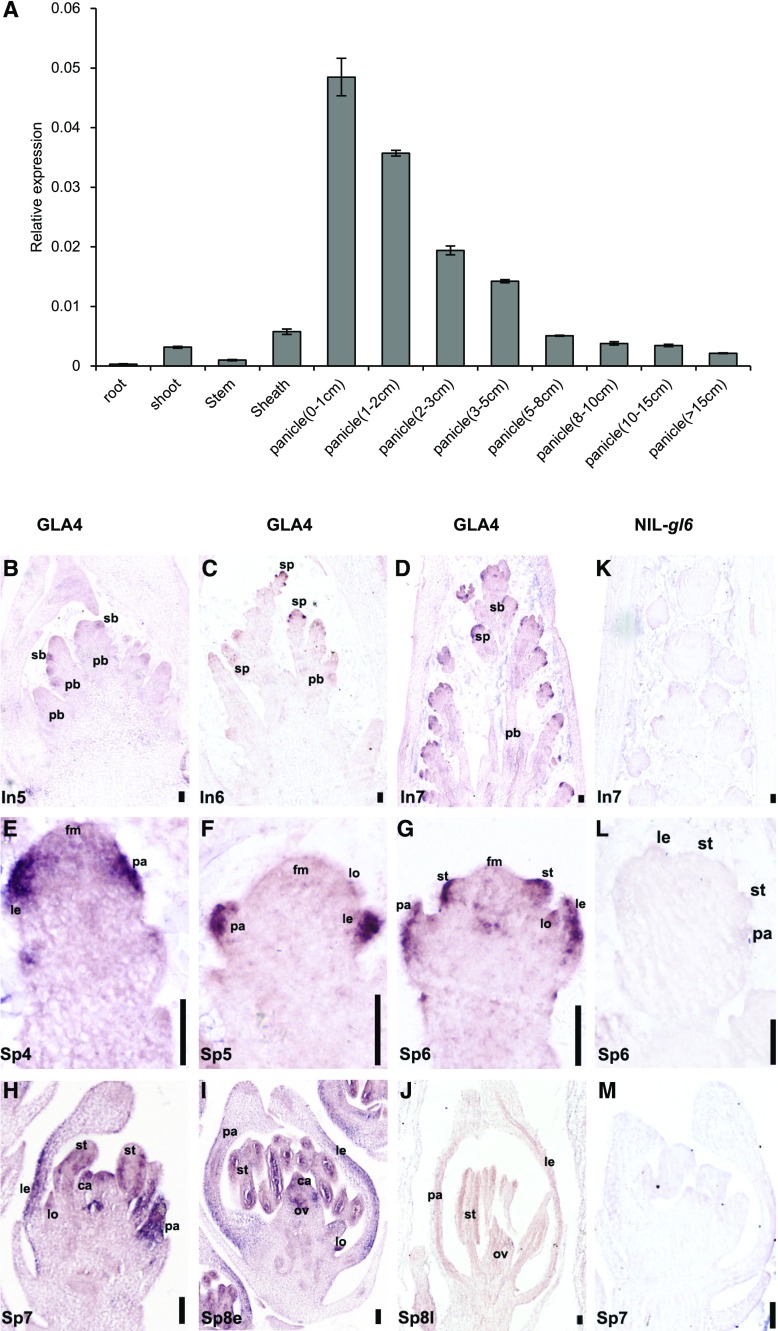
The expression pattern of GL6 was detected by reverse transcription quantitative PCR (RT-qPCR) analysis. GL6 expression was found in all organs and tissues examined; higher expression levels were observed in young panicles (<1 cm), but these gradually decreased during panicle development (Fig. 5A). Furthermore, we investigated the specific temporal and spatial expression pattern of GL6 by RNA in situ hybridization (Fig. 5, B–M). GL6 transcripts were initially detected when the secondary branch primordia were formed (Fig. 5B). During subsequent growth, GL6 transcripts were observed in abundance in both the lemma and palea primordia (Fig. 5, C, E, and F). With the development of floral organs, lemma, palea, and stamen primordia all exhibited a strong expression signal of GL6 at stage spikelet development 6 (Sp6; Fig. 5G). Subsequently, the expression of GL6 was restricted to the stamen and carpel primordia, and gradually decreased in the primordia of lemma and palea (Fig. 5, H and I). Finally, after floral organ differentiation, GL6 expression signals disappeared during late-stage Sp8 (Fig. 5J). No signals were detected with the sense probe (Supplemental Fig. S6A). By contrast, compared to expression of the positive control HISTONE H4, we could barely detect GL6 expression in NIL-gl6 at any stage (Fig. 5, K–M; Supplemental Fig. S6B).
Figure 5.
The spatial expression pattern analysis of GL6 in rice. A, Relative expression levels of GL6 mRNA in vegetative tissues and developmental panicles. The abundance of GL6 transcripts was normalized to that of OsUBQ5 (ubiquitin 5). Values are given as the mean ± sd. B to J, In situ analysis of GL6 in GLA4 during different inflorescence stages and flower development processes. GL6 transcripts at stages In5 (B), In6 (C), In7 (D), Sp4 (E), Sp5 (F), Sp6 (G), Sp7 (H), Sp8e (I), and Sp8e (J). K to M, In situ analysis of GL6 in NIL-gl6 at stages In7 (K), Sp6 (L), and Sp7 (M). ca, carpel; fm, floral meristem; le, lemma; lo, lodicule; ov, ovule; pa, palea; pb, primary branch; sb, secondary branch; sp, spikelet; st, stamen; Sp8e, early-stage Sp8; Sp8l, late-stage Sp8. Scale bars = 50 μm.
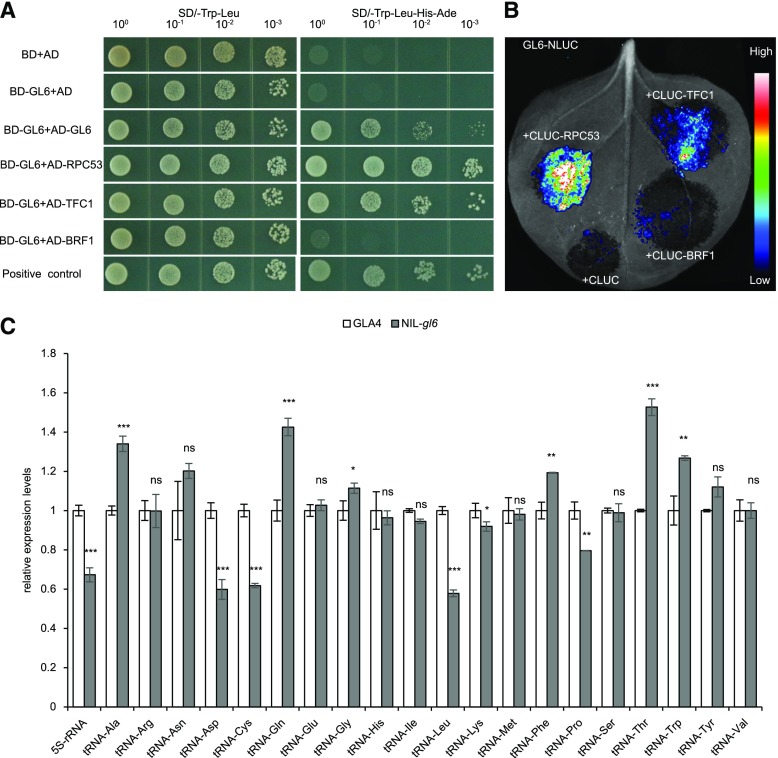
GL6 Interacts with OsRPC53 and OsTFC1
RNAPIII is a multisubunit complex eukaryotic RNA polymerase that transcribes tRNA genes, 5S ribosomal RNA (5S rRNA), RNase P, and other noncoding RNAs to regulate RNA and protein synthesis for multiple cellular developmental processes (Abascal-Palacios et al., 2018). The maize PLATZ protein FL3 was revealed to interact with RPC53 (RNA polymerase III subunit C53) and TFC1 (transcription factor class C1) of the RNAPIII complex to modulate the RNAPIII transcription machinery (Li et al., 2017). Amino acid sequence analysis showed that GL6 shared high sequence similarity with other PLATZ proteins, and phylogenetic analysis revealed that GL6 belonged to the PLATZ family (Supplemental Figs. S7 and S8). Considering the conserved PLATZ domain and potential functional conservation among the PLATZ family, we wondered whether GL6 could affect the function of RNAPIII in rice. We tested the interaction between GL6 and three rice homologs of RNAPIII transcription machinery, namely OsRPC53, OsBRF1 (TFIIIB B-related factor 1), and OsTFC1, respectively. Yeast two-hybrid assays indicated that GL6 interacts with OsRPC53 and OsTFC1, but not OsBRF1 (Fig. 6A). Further in vivo interactions were validated using a bimolecular luciferase complementation (BiLC) assay, and we found that the interaction between GL6 and OsRPC53 was much stronger than that between GL6 and OsTFC1 (Fig. 6B). Additionally, RNAPIII-dependent transcripts of tRNAs and 5S rRNA were decreased in NIL-gl6 compared with GLA4. These results suggest that GL6 might participate in mediating RNAPIII to coordinate ribosome biogenesis (Fig. 6C).
Figure 6.
GL6 interacts with RPC53 and TFC1 to regulate tRNA transcripts. A, Yeast two-hybrid assay, showing how GL6 interacts with OsRPC53, OsTFC1, and itself. AD, activation domain; BD, binding domain; 10-fold serial dilutions show the gradients. B, BiLC assay showing that GL6 interacts with OsRPC53 and OsTFC1 in vivo. C, Relative expression levels of tRNAs and 5S rRNA in young panicles (0–1 cm) of GLA4 and NIL-gl6. OsUBQ5 was used as the control and the values of expression levels in GLA4 were set to 1. Values are given as the mean ± sd. Student’s t test significant difference,*P < 0.05, **P < 0.01 and ***P < 0.001; ns, not significant.
RNA-Seq Analysis of the GL6 Downstream Regulatory Network
To further explore the regulatory mechanism of GL6, we carried out RNA sequencing (RNA-seq) analysis of young panicles from GLA4 and NIL-gl6 to investigate the downstream gene regulatory networks. Principle-components analysis of the gene expression data showed close clustering of biological replicates and clear differentiation of separate samples (Supplemental Fig. S9A). A total of 2,711 differentially expressed genes (DEGs) were detected (P < 0.05), of which 52.6% (1,426) up-regulated genes and 47.4% (1,285) down-regulated genes were found in NIL-gl6 relative to GAL4 (Supplemental Table S1). Gene Ontology (GO) analysis of these DEGs showed significant enrichment (false discovery rate < 0.05) of the biological process associated with transcription, transport, translation, and hormone stimulus (Supplemental Fig. S9B). Similarly, molecular functional categories associated with DNA binding, transferase activity, transcription regulator activity, protein binding, and signal transducer activity were also highly enriched (Supplemental Fig. S9, C and D). In addition, we found that OsMADS1 (a known gene controlling grain size; Liu et al., 2018), eIF1 (a protein translation factor), and TFIIS (a transcription elongation factor) were down-regulated in NIL-gl6, and further RT-qPCR analyses verified the differential expression of these genes (Supplemental Fig. S9E). These results support the hypothesis that GL6 functions as a transcription factor to regulate downstream gene expression.
Intriguingly, we found that a series of genes including TAW1, OsMADS22, OsMADS47, and OsMADS55, which function in regulatory pathways that control inflorescence architecture by delaying the developmental meristem phase transition from inflorescence meristem to spikelet meristem (Yoshida et al., 2013), were significantly up-regulated in NIL-gl6, and these expression changes were confirmed by RT-qPCR (Supplemental Fig. S9F). High expression of these five genes resulted in more secondary branches and spikelets, which was consistent with the increased grain number in NIL-gl6 compared with GLA4. These results imply that the null gl6 mutation might affect grain number by extending the activity of the inflorescence meristem to produce more seeds in NIL-gl6.
Natural Variation in the GL6 Gene
To investigate the variation of GL6 in diverse rice germplasms, 67 accessions from the rice pan-genome dataset (Zhao et al., 2018b) were selected alongside additional GL6 sequences of 52 wild rice (O. rufipogon) accessions (Huang et al., 2012). The resulting 119 rice accessions consisted of 54 cultivated rice and 65 wild rice accessions (Supplemental Table S2). Although the null gl6 allele was from wild rice W1943, comparisons of nucleotide diversity and neutrality testing between cultivars and wild rice revealed that GL6 was not a locus targeted by human selection during domestication (Supplemental Table S3).
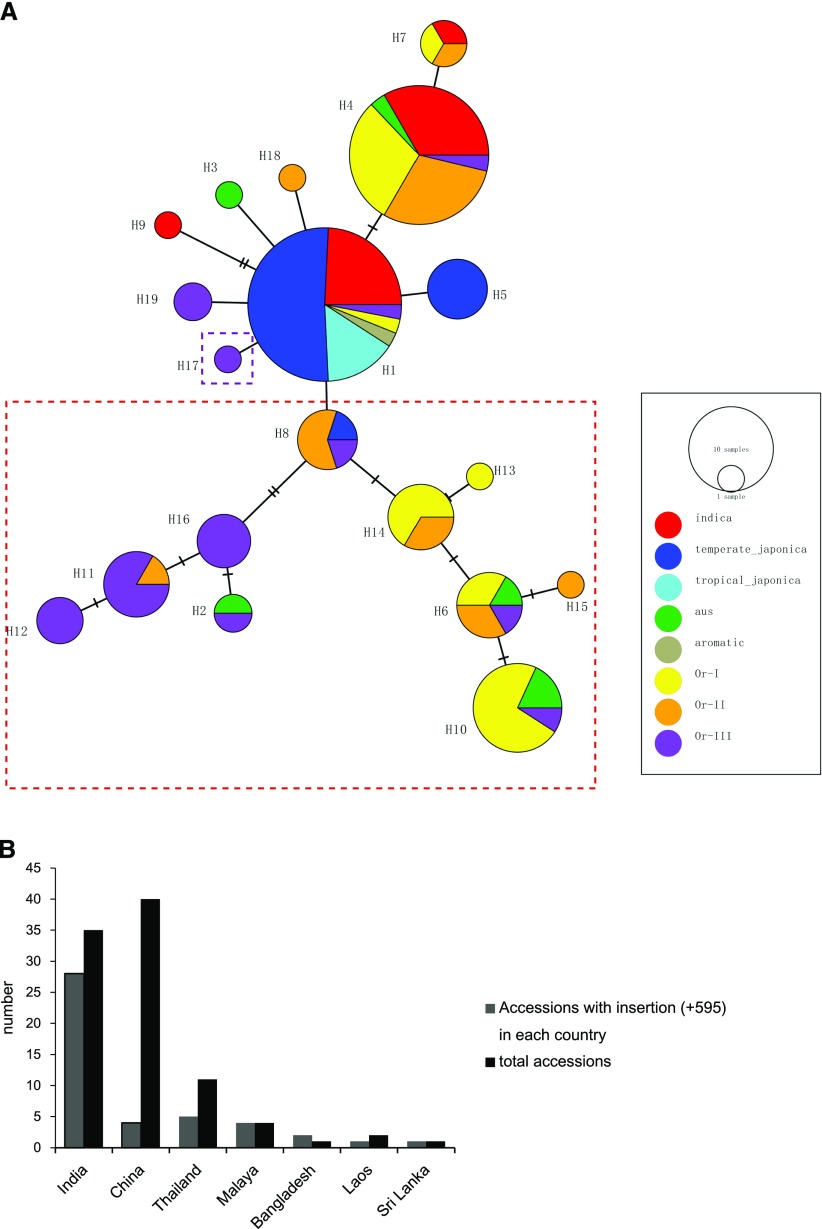
We then analyzed the GL6 open reading frame (ORF) to mine other natural variations in the GL6 gene, and a total of 20 polymorphic sites were observed, including 16 SNPs and four insertions or deletions (InDels). Among these variations, 13 SNPs were synonymous mutations, whereas no other variety except W1943 carried the identical SNP (premature stop codon), T allele, or T/C allele. The remaining four InDels and two missense mutation SNPs were located in the last exon of GL6. A total of 19 haplotypes based on these variations were generated and named H1 to H19 (Fig. 7A; Supplemental Table S4). Most of the japonica varieties showed no differences from the Nipponbare genome, belonging to the H1 haplotype, whereas the indica varieties were distributed among several haplotypes. Interestingly, 60% of the wild rice and most aus accessions, such Kasalath, shared one identical insertion (+595) in the last GL6 exon, which resulted in an insertion of three to six histidines. The geological distribution of these accessions originated mainly from India, indicating that this insertion was fixed in this region (Fig. 7B). However, whether the Kasalath allele and other InDels have effects on the determination of grain size needs more investigation in the future.
Figure 7.
Natural variation in the GL6 ORF. A, Haplotype network of GL6 ORF among 119 rice accessions. Each circle represented one of the 19 different haplotypes and circle size was proportional to haplotype frequency. Different colors refer to different rice subpopulations. The red dashed square encloses rice lines with the same insertion (+595), and the purple dashed square refers to W1943. B, Comparisons of distribution among accessions with the insertion (+595).
DISCUSSION
PLATZ family proteins are a class of plant-specific transcription factors with widespread distribution in dicots, monocots, mosses, and algae. All PLATZ family members share a conserved PLATZ domain that is ∼82 amino acids in length and consists of two noncanonical zinc finger domains (Nagano et al., 2001; Wang et al., 2018). The first PLATZ gene was isolated from pea (Pisum sativum), which binds to the A/T-rich sequence to negatively regulate the enhancer element of the pea plastocyanin gene (petE; Nagano et al., 2001). At present, only a few PLATZ genes have been isolated. Arabidopsis (Arabidopsis thaliana) AtPLATZ1 and AtPLATZ2 were confirmed to enhance seed desiccation tolerance (González-Morales et al., 2016). The maize (Zea mays) PLATZ protein FL3 interacts with RNA polymerase III for biogenesis of tRNA and 5S rRNA to regulate endosperm storage filling (Li et al., 2017). Moreover, AtORE15, an Arabidopsis ortholog of GL6, was identified recently as being involved in the regulation of leaf growth and suppression of senescence via cooperation with the growth regulating factor/growth regulating factor-interacting factor regulatory pathway (Kim et al., 2018).
There are 15 PLATZ genes in rice, yet there has been little functional characterization of rice PLATZ genes. In this study, we first identified the rice PLATZ gene GL6, which affects grain length and yield in rice. GL6 positively regulates cell division to increase cell numbers of the spikelet hull, resulting in larger grains.
RNAPIII synthesizes various small noncoding RNAs that are essential for general biological activities. Dysregulation of the RNAPIII machinery results in multiple reported instances of reduced cellular and organismal growth (Dauwerse et al., 2011; Soprano et al., 2013, 2017; Borck et al., 2015; Johnson et al., 2016). However, the regulation of RNAPIII transcriptional activity remains poorly understood in plants. Our data show that GL6 interacts with two critical RNAPIII subunits, RPC53 and TFC1, to regulate tRNAs and 5S rRNA, indicating that rice GL6 might have a similar function as that of the maize PLATZ protein FL3.
Seed size is crucial for evolutionary fitness in plants. Considering rice yield potential, there is a balance between panicle architecture and grain size. Larger grains are often associated with reduced grain number, and smaller grain size indicates that more seeds are produced (Guo et al., 2018). For example, the rare allele gw2 increases grain size and weight but also reduces grain number per panicle (Song et al., 2007). Similarly, in our study, we observed that NIL-gl6 and GLA4-gl6CRISPR lines that had shorter grains showed increased grain number per panicle compared to GLA4, consistent with the observed dramatic reduction in grain number per panicle in NIL-CP lines that had large grains. These phenotypes demonstrate the negative correlation between grain size and grain number due to a trade-off between inflorescence and spikelet development.
In conclusion, we identified a new QTL, GL6, which functions in the RNAPIII transcription machinery to affect rice grain length and number. Although the detailed mechanism by which GL6 regulates expression of tRNAs and 5S rRNA is still unclear, our data provided here indeed contribute toward understanding this process. The extended regulatory pathway surrounding GL6 remains to be investigated in further detail. Moreover, the exploitation of GL6 may be a potential approach to manipulate the molecular balance between grain size and number for the development of elite rice varieties with improved grain productivity.
MATERIALS AND METHODS
Plant Materials and Trait Measurement
BIL219 (small grain) and GLA4 (large grain) were used as two parents for QTL mapping, and Nipponbare was used for transgenic confirmation. All rice plants were cultivated under field conditions with transplant spacing of 20 × 20 cm in Shanghai and Hainan, China. The measurement of grain yield-related field agronomic traits was conducted with edge lines excluded. Plant height, panicle morphology, and grain number were obtained from the main culm. Grain yield per plant and tiller number were measured from the whole plant. Fully filled dry grains were used for measuring grain length, width, and weight by image analysis method provided with SC-E software (Hangzhou Wanshen Detection Technology Co., Ltd.). All trait measurements were repeated at least 3 times.
Fine Mapping of GL6
The backcross inbred line BIL219 was selected to carry out the backcross with GLA4 to generate the BC1F2 population, which contained 288 lines. Then we performed marker assisted selection on each substitution segment to purify the genetic background. Further fine mapping using 2,181 BC1F5 lines narrowed the GL6 locus down to a 6.1-kb region between markers GL4350 and GL4411.
Primers
All primers used in this study are listed in Supplemental Table S5.
Transgene Constructs and Plant Transformation
The entire 14365-bp GL6 genomic region was digested with SacI from GLA4 bacterial artificial chromosome clone osigba0159g02 and then inserted into the binary vector pCAMBIA1301 to generate the complementation construct. The gene editing constructs of GL6 via CRISPR/Cas9 were designed as previously described (Ma et al., 2015). All these constructs were introduced into Agrobacterium tumefaciens strain EHA105 and subsequently transferred into Nipponbare, GLA4, and NIL-gl6 by Agrobacterium-mediated transformation. More than 10 independent transgenic lines were generated, respectively. All analyzed phenotypes were measured in the T2 generation of transgenic plants.
Scanning Electron Microscopy
Mature seeds were first cleaned ultrasonically several times to remove epidermal hairs and dust. The samples were then dried in a critical point drier and coated with gold sputter. For glume cell observation, the outer surfaces of the spikelet glumes were observed by scanning electron microscope (Hitachi). Cell size and cell number were calculated along the longitudinal axis.
Phylogenetic Analysis
Protein sequences of PLATZ family members in rice and other organisms were obtained by BLAST from the National Center for Biotechnology Information database. Multiple sequence alignments of protein were performed using the ClustalW program. The phylogenetic tree of aligned sequence was constructed by MEGA7 using a neighbor-joining tree with 1000 bootstrapped replicates.
Neutrality Test
Multiple sequences of GL6 genomic DNA were aligned with ClustalW. Nucleotide diversity and Tajima’s D test were calculated and performed using DnaSP version 6.12.03 (Rozas et al., 2017).
Haplotype Network
Multiple sequences of GL6 ORF were aligned with ClustalW. Haplotype-frequency data were processed with DnaSP version 6.12.03 and visualized Median-joining networks were generated by PopART with some modifications (each continuous InDel was considered as one site; Leigh et al., 2015).
Yeast Two-Hybrid Assays
For the two-hybrid assay, the full-length coding region of GL6 was amplified and fused in frame with the GAL4 DNA-binding domain via cloning into pGBKT7 DNA-BD vector as the bait plasmid (Clontech). The entire coding regions of GL6, OsRPC53, OsBRF1, and OsTFC1 were introduced into the pGADT7 AD vector (Clontech) prey vector. The resulting constructs were then transformed into yeast strain AH109. The cotransformants were diluted (1, 1/10, 1/100, 1/1000) and spotted on control medium (SD/-Trp/-Leu) and selective medium (SD/-Trp/-Leu/-His/-Ade) and incubated at 30°C for 3 d.
BiLC Assay
The coding-region sequence of GL6 was cloned to the N-terminal luciferase (Luc) fusion vector JW771-NLUC, and OsRPC53, OsBRF1, and OsTFC1 were cloned respectively to the C-terminal Luc fusion vector JW772-CLUC. The BiLC assay procedure was performed as previously described (Gou et al., 2011). A. tumefaciens [strain GV3101 (pSoup-p19)] transformants containing the testing split LUC fusion constructs were cotransfected into Nicotiana benthamiana leaves via infiltration, and LUC activity was captured using a cooled CCD-image system (Tanon 5200) with injecting 0.94 mm luciferin (cat. no. 122799; PerkinElmer) after growing for 48 h under 16 h light/d.
RNA Extraction and RT-qPCR Analysis
Total RNA was extracted using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Then 500 ng of total RNA was used to synthesize first-strand cDNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo). Real-time PCR was performed on the Applied Biosystems QuantStudio 5 PCR system with diluted cDNA was used as a template using THUNDERBIRD SYBR qPCR Master Mix (Toyobo). Rice gene UBQ5 (Os01g0328400) was used as the internal control to normalize all data. Each set of experiments was repeated three times.
RNA In Situ Hybridization
Fresh young panicles of GLA4 and NIL-gl6 were collected and fixed in formaldehyde-alcohol-acetic acid solution at 4°C overnight, dehydrated by series ethanol procedures, and embedded in paraplast. The tissues were sliced into 8-μm sections with a microtome (Leica). The gene-specific region of GL6 was then amplified from FL-cDNA and used to generate digoxigenin-labeled RNA probes (Roche). In situ hybridization was performed as described (Luo et al., 1996).
RNA-Seq Analysis
Total RNA was extracted from young GLA4 and NIL-gl6 inflorescences (<1 cm) with two biological replicates using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. After treatment with DNase, the mRNA was purified using the NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490; New England Biolabs). Libraries were synthesized using the NEBNext Ultra II RNA Library Prep Kit for Illumina (E7770; New England Biolabs) and sequenced on an Illumina HiSeq2500. A total of 269 million 150-bp paired-end reads were generated, yielding 40.34 Gb raw reads. After trimming of Illumina adaptors and low-quality reads, the filtered clean reads were aligned to the MSU version 7 genome assembly (http://rice.plantbiology.msu.edu) using HISAT2 (Kim et al., 2015). The aligned read files were sorted and indexed by SAMtools (Li et al., 2009) and reads of each sample were then used to calculate raw counts for each gene and transcript using the function SummarizeOverlaps within the GenomicAlignments package. The sample-to-sample distances were presented by principal-components analysis, which was based on the transcript count of each sample. DESeq2 software packages (bioconductor.org/) were used to detect DEGs with the threshold of genes with Benjamini-Hochberg-adjusted p-values <0.05 and absolute log2 fold change >0.6. GO enrichment analyses were performed using agriGO V2.0 (Tian et al., 2017).
Statistical Analysis
Statistical analyses were carried out using Excel 2010 with two-tailed Student’s t test for comparison of two groups and R package “agricolae” with Duncan’s multiple-range tests for multiple mean comparisons.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers MK959365 (GL6 cDNA from W1943), XM_015781518.2 (OsRPC53), XM_015766861.2 (OsTFC1), and XM_015785297.2 (OsBRF1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Graphical genotype and phenotype of BIL219.
Supplemental Figure S2. Sequence alignment of GL6 in GLA4 and NIL-gl6.
Supplemental Figure S3. Relationship between different GL6-type and grain length in W1943.
Supplemental Figure S4. GL6 positively regulates grain length.
Supplemental Figure S5. Suppression of GL6 in Nipponbare results in smaller grains.
Supplemental Figure S6. RNA in situ hybridization analysis.
Supplemental Figure S7. Amino acid sequence alignments of GL6 and its homologs in various species.
Supplemental Figure S8. Phylogenetic analysis of GL6 and other related PLATZ proteins.
Supplemental Figure S9. GO enrichment analysis of RNA-seq DEGs.
Supplemental Table S1. The DEGs and GO enrichment in GLA4 and NIL-gl6.
Supplemental Table S2. The list of 119 rice accessions in the collection.
Supplemental Table S3. Nucleotide diversity and Tajima's D test.
Supplemental Table S4. Sequence variation and distribution of GL6 ORF haplotypes among 119 rice germplasms.
Supplemental Table S5.List of primers used in this study.
Acknowledgments
We thank the China National Rice Research Institute for providing the cultivated rice germplasm and the Chinese wild rice accessions. We thank Jiawei Wang (Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for providing the JW771 and JW772 plasmids and Yaoguang Liu (South China Agricultural University) for the CRISPR/Cas9 vector. We thank Jiqin Li, Xiaoyan Gao, and Xiaoshu Gao (Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for technical support.
Footnotes
This work was supported by the Ministry of Science and Technology of China (2016YFD0100902), the National Natural Science Foundation of China (31630055 and 31788103), and the Chinese Academy of Sciences (XDB27010301).
Articles can be viewed without a subscription.
References
- Abascal-Palacios G, Ramsay EP, Beuron F, Morris E, Vannini A (2018) Structural basis of RNA polymerase III transcription initiation. Nature 553: 301–306 [DOI] [PubMed] [Google Scholar]
- Borck G, Hög F, Dentici ML, Tan PL, Sowada N, Medeira A, Gueneau L, Thiele H, Kousi M, Lepri F, et al. (2015) BRF1 mutations alter RNA polymerase III-dependent transcription and cause neurodevelopmental anomalies. Genome Res 25: 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, et al. (2015) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants 2: 15195. [DOI] [PubMed] [Google Scholar]
- Dauwerse JG, Dixon J, Seland S, Ruivenkamp CAL, van Haeringen A, Hoefsloot LH, Peters DJM, Boers AC, Daumer-Haas C, Maiwald R, et al. (2011) Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet 43: 20–22 [DOI] [PubMed] [Google Scholar]
- Duan P, Ni S, Wang J, Zhang B, Xu R, Wang Y, Chen H, Zhu X, Li Y (2015) Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat Plants 2: 15203. [DOI] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112: 1164–1171 [DOI] [PubMed] [Google Scholar]
- González-Morales SI, Chávez-Montes RA, Hayano-Kanashiro C, Alejo-Jacuinde G, Rico-Cambron TY, de Folter S, Herrera-Estrella L (2016) Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 113: E5232–E5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Chen K, Dong NQ, Shi CL, Ye WW, Gao JP, Shan JX, Lin HX (2018) GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30: 871–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, et al. (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8: 1455–1465 [DOI] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41: 494–497 [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, et al. (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, et al. (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45: 707–711 [DOI] [PubMed] [Google Scholar]
- Johnson KC, Yu Y, Gao L, Eng RC, Wasteneys GO, Chen X, Li X (2016) A partial loss-of-function mutation in an Arabidopsis RNA polymerase III subunit leads to pleiotropic defects. J Exp Bot 67: 2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim J, Jun SE, Park S, Timilsina R, Kwon DS, Kim Y, Park SJ, Hwang JY, Nam HG, et al. (2018) ORESARA15, a PLATZ transcription factor, mediates leaf growth and senescence in Arabidopsis. New Phytol 220: 609–623 [DOI] [PubMed] [Google Scholar]
- Leigh JW, Bryant D, Nakagawa S (2015) popart: Full-feature software for haplotype network construction. Methods Ecol Evol 6: 1110–1116 [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang J, Ye J, Zheng X, Xiang X, Li C, Fu M, Wang Q, Zhang Z, Wu Y (2017) The maize imprinted gene Floury3 encodes a PLATZ protein required for tRNA and 5S rRNA transcription through interaction with RNA polymerase III. Plant Cell 29: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, et al. (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43: 1266–1269 [DOI] [PubMed] [Google Scholar]
- Liu J, Chen J, Zheng X, Wu F, Lin Q, Heng Y, Tian P, Cheng Z, Yu X, Zhou K, et al. (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants 3: 17043. [DOI] [PubMed] [Google Scholar]
- Liu Q, Han R, Wu K, Zhang J, Ye Y, Wang S, Chen J, Pan Y, Li Q, Xu X, et al. (2018) G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat Commun 9: 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Hua L, Dong S, Chen H, Zhu X, Jiang J, Zhang F, Li Y, Fang X, Chen F (2015) OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J 84: 672–681 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A Robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci USA 107: 19579–19584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano Y, Furuhashi H, Inaba T, Sasaki Y (2001) A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences. Nucleic Acids Res 29: 4097–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang L, Gao JP, Lin HX (2012) The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res 22: 1666–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34: 3299–3302 [DOI] [PubMed] [Google Scholar]
- Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y, et al. (2016) OsSPL13 controls grain size in cultivated rice. Nat Genet 48: 447–456 [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39: 623–630 [DOI] [PubMed] [Google Scholar]
- Song XJ, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, et al. (2015) Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc Natl Acad Sci USA 112: 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano AS, Abe VY, Smetana JH, Benedetti CE (2013) Citrus MAF1, a repressor of RNA polymerase III, binds the Xanthomonas citri canker elicitor PthA4 and suppresses citrus canker development. Plant Physiol 163: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano AS, Giuseppe PO, Shimo HM, Lima TB, Batista FAH, Righetto GL, Pereira JGC, Granato DC, Nascimento AFZ, Gozzo FC, et al. (2017) Crystal structure and regulation of the citrus Pol III repressor MAF1 by auxin and phosphorylation. Structure 25: 1360–1370.e4 [DOI] [PubMed] [Google Scholar]
- Sun S, Wang L, Mao H, Shao L, Li X, Xiao J, Ouyang Y, Zhang Q (2018) A G-protein pathway determines grain size in rice. Nat Commun 9: 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45(W1): W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ji C, Li Q, Zhou Y, Wu Y (2018) Genome-wide analysis of the plant-specific PLATZ proteins in maize and identification of their general role in interaction with RNA polymerase III complex. BMC Plant Biol 18: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44: 950–954 [DOI] [PubMed] [Google Scholar]
- Wang S, Li S, Liu Q, Wu K, Zhang J, Wang S, Wang Y, Chen X, Zhang Y, Gao C, et al. (2015a) The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet 47: 949–954 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Xu J, et al. (2015b) Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet 47: 944–948 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen R, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, et al. (2013) TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci USA 110: 767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Miao J, Zhang Z, Xiong H, Zhu X, Sun X, Pan Y, Liang Y, Zhang Q, Abdul Rehman RM, et al. (2018) Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol J 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang J, Huang J, Lan H, Wang C, Yin C, Wu Y, Tang H, Qian Q, Li J, et al. (2012) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA 109: 21534–21539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DS, Li QF, Zhang CQ, Zhang C, Yang QQ, Pan LX, Ren XY, Lu J, Gu MH, Liu QQ (2018a) GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat Commun 9: 1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, Zhan Q, Lu Y, Zhang L, Huang T, et al. (2018b) Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet 50: 278–284 [DOI] [PubMed] [Google Scholar]
- Zuo J, Li J (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet 48: 99–118 [DOI] [PubMed] [Google Scholar]