Abstract
Background
Low temperature restricts the planting range of all crops, but cold acclimation induces adaption to cold stress in many plants. Camellia sinensis, a perennial evergreen tree that is the source of tea, is mainly grown in warm areas. Camellia sinensis var. sinensis (CSS) has greater cold tolerance than Camellia sinensis var. assamica (CSA). To gain deep insight into the molecular mechanisms underlying cold adaptation, we investigated the physiological responses and transcriptome profiles by RNA-Seq in two tea varieties, cold resistant SCZ (classified as CSS) and cold susceptible YH9 (classified as CSA), during cold acclimation.
Results
Under freezing stress, lower relative electrical conductivity and higher chlorophyll fluorescence (Fv/Fm) values were detected in SCZ than in YH9 when subjected to freezing acclimation. During cold treatment, 6072 and 7749 DEGs were observed for SCZ and YH9, respectively. A total of 978 DEGs were common for both SCZ and YH9 during the entire cold acclimation process. DEGs were enriched in pathways of photosynthesis, hormone signal transduction, and transcriptional regulation of plant-pathogen interactions. Further analyses indicated that decreased expression of Lhca2 and higher expression of SnRK2.8 are correlated with cold tolerance in SCZ.
Conclusions
Compared with CSA, CSS was significantly more resistant to freezing after cold acclimation, and this increased resistance was associated with an earlier expression of cold-induced genes. Because the greater transcriptional differentiation during cold acclimation in SCZ may contribute to its greater cold tolerance, our studies identify specific genes involved in photoinhibition, ABA signal conduction, and plant immunity that should be studied for understanding the processes involved in cold tolerance. Marker-assisted breeding focused on the allelic variation at these loci provides an avenue for the possible generation of CSA cultivars that have CSS-level cold tolerance.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5988-3) contains supplementary material, which is available to authorized users.
Keywords: Assamica tea, Camellia sinensis, Cold acclimation, Gene expression profiling, Low temperature response
Background
Low temperature, as an abiotic stress, restricts the growth, geographic distribution, and productivity of crops. Plant cold injury yields various symptoms, including stunted bud, wilting/yellowing of leaves, and tissue death [1]. Perennial plants have developed strategies to tolerate the cold stress in seasonal cycles. However, a sudden drop to freezing temperatures can cause severe damage or death even in cold-tolerant plants. Cold acclimation can help avoid this cold damage and has been the subject of experiments for many decades [2–5]. Cold acclimation is an induced adaptive process that increases freezing tolerance and is achieved by exposing the plant to various low temperatures above 0 °C for various periods of time, depending on the plant. Cold acclimation activates a systemic response to cold stress, and can be integrated with control of seed germination by the process called vernalization [6], which blocks premature germination during autumn warm spells. Hence, the study of cold acclimation is of wide interest in plant biology and agriculture.
Mechanisms of cold acclimation have been investigated especially well in cereal crops such as wheat and in model plants like Arabidopsis [3]. These studies identified biochemical changes associated with cold acclimation, including an increased generation of reactive oxygen species (ROS), an up-regulation of antioxidant enzymes, alteration in plant hormone levels, increases in metabolites such as γ-aminobutyric acid and soluble sugar, and accumulation of amino acids with osmotic effects, such as proline [3]. Cold acclimation and cold tolerance are associated with multiple biological processes and complicated regulatory networks [2, 7]. For instance, lipid membranes change in response to cold, including their glycoprotein content, lipid profile, membrane fluidity and structural organization [8–10]. Plant hormones, including abscisic acid (ABA) and gibberellin (GA), have long been known to be involved in cold response [11, 12]. Current cold responsive genes mostly originate from ancient cold responsive genes that have been conserved during the diversification of land plants, and have been especially vital in plant lineages that have undergone long-term radiation from tropical regions to temperate regions [13].
Investigations in many plant species have shown that numerous genes are up- or down-regulated during cold treatment, and these studies have been particularly informative in Arabidopsis [2, 11, 14, 15]. These studies show that gene regulation in cold acclimation and freezing stress acts at multiple levels. For instance, removal of repressive histone modifications on chromatin by HOS15, a WD40-repeat protein, has been shown to increase transcription of cold-regulated (COR) genes [16]. Epigenetic regulation of VERNALIZATION INSENSITIVE3 and FLOWERING LOCUS C is also involved, and requires both cold acclimation and the absence of daily warm temperature (15 °C or higher) [17]. C-repeat-binding factors 1–3 (CBF1–3) are transcription factors that integrate cold signals with altered gene expressions, including such COR genes as mitogen-activated protein kinases (MAPKs) [11, 18].
Compared with cereal crops and Arabidopsis, studies on cold acclimation in trees are relatively few. The tea plant (Camellia sinensis (L.) O. Kuntze) is widely planted to produce leaves for making tea. It is a perennial and evergreen tree that is restricted to temperate, subtropical and tropical production zones because tea plants are vulnerable to cold injury, especially during freezing winters. Low temperature appears to cause damage in the thylakoid membrane of cold-susceptible tea plants [19], as has been seen in other cold-susceptible species [20, 21]. The C. sinensis variety (var.) sinensis (CSS) and C. sinensis var. assamica (CSA) are two widely grown categories of germplasm, but CSS exhibits better cold resistance than CSA. Several publications have reported physiological responses, including gene expression, in response to cold stress in the tea plant [22, 23]. Early low temperatures (4–16 °C) may increase the cold resistance of tea plants to a later freezing winter [24]. A 4–5 °C acclimation has been reported to increase the activity of enzymes such as GR, SOD, and APX, and to the accumulation of high levels of sucrose, all proposed to be processes that could increase cold resistance [25, 26]. The contents of numerous metabolites in leaves were affected by low temperature, including products derived from sucrose hydrolysis that increase under cold treatment [27]. Changes in some small RNAs were associated with responses to low temperature (4 °C) after 1–48 h in the tea plant [28]. The transcriptome profiles in leaves of tea variety Camellia sinensis (L.) O. Kuntze cv. Longjing 43 were investigated under field conditions during overwintering [24]. All of these studies are limited to one or a few varieties under a few environments, so they lack any power to identify global responses that are conserved across environments and germplasm variation. Hence, our understanding of the intricacies of gene expression and regulation mechanisms underlying cold adaptation are still highly deficient for C. sinensis.
In this study, we comprehensively profiled dynamic transcriptomes via high through-put RNA-Seq analysis of tea leaves across controlled cold acclimation to investigate gene expression. The treatments were a 6 h short-term cold stimulus (CS), a 7-day long-term chilling acclimation (CA), a 7-day long-term freezing acclimation (FA), and a 7-day long-term de-acclimation (DA). To discover the gene expression responsible for higher cold resistance, we compared the transcriptomes in two tea varieties, CSS cv. Shuchazao (SCZ) with a high cold tolerance and CSA cv. Yinghong9 (YH9) with weak tolerance. Our results reveal the key regulated genes and their gene ontologies. We identify the key transcriptional pathways for the cold stimulus response, for chilling acclimation and for freezing acclimation in the tea plant. Interestingly, many of the genes identified are involved in photosynthesis, hormone signaling and plant immunity. Comparisons identified some key gene expression patterns associated with the capability for greater cold tolerance. Thus, our study provides novel insights into gene regulation from cold acclimation to cold adaptation in tea plants, and we discuss how this relates to similar investigations in other plant species.
Results
Different cold tolerance in two tea varieties during cold acclimation
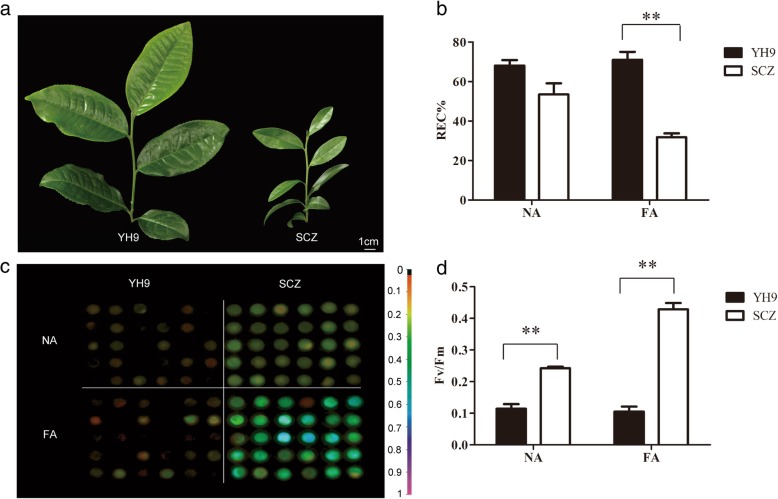
To investigate cold adaption in CSS and CSA, we assayed two cultivars with very different cold tolerances [26]. CSS tea variety SCZ is commonly planted in temperate and sub-tropical areas while cv. YH9, a CSA tea variety, is mainly planted in sub-tropical areas (Fig. 1a). Hence, SCZ is expected to have a higher cold tolerance than YH9, in agreement with the geographic difference in planting areas [26]. We treated both cultivars with − 5 °C freezing stress after FA treatment to confirm differences in cold tolerance. Relative electrical conductivity (REC), an indicator of cell leakage under freezing damage, was increased to more than 50% in both cultivars if there was no acclimation (NA), indicating that the freezing stress caused serious cell damage. However, REC was significantly lower in SCZ than YH9 if treated with freezing acclimation (FA), indicating a highly increased freezing tolerance in SCZ (Fig. 1b). The chlorophyll fluorescence (Fv/Fm) value was higher in SCZ than YH9 after treatment of FA (Fig. 1c, d). Together, this confirmed that cold acclimation can induce a higher freezing tolerance in cold resistant SCZ than in cold susceptible YH9.
Fig. 1.
Different cold responses of two Camellia sinensis cultivars. The two tea varieties (a) were exposed to cold acclimation for two weeks followed by freezing stress. The relative electrical conductivity (REC) (b), chlorophyll fluorescence by Fv/Fm image (c), and Fv/Fm value (d) were examined. NA and FA represent no cold acclimation and treatment with freezing acclimation followed by − 5 °C for 12 h. Statistically significant differences between SCZ and YH9 were calculated using one-way ANOVA test using Statistical Package for the Social Sciences (SPSS) 16.0. ** p < 0.01
C. sinensis transcriptome profiles during cold acclimation
To characterize the transcriptomes in tea leaves in response to cold acclimation, we transferred one-year-old tea plants propagated from cuttings of each cultivar into a growth chamber for 4 weeks at 25 °C during the day and 20 °C at night to synchronize growth. We first treated the tea plants with CS for a short time (6 h), followed by a seven-day cyclic CA treatment at 10 °C during the day and 4 °C at night, and by subsequent seven-day FA treatment at 4 °C during the day and 0 °C at night. The tea plants were then grown at 25 /20 °C, the same as before the CS-CA-FA treatments, for 7 days to provide the DA treatment. The control group was the plants with NA, which were received no CS, CA, FA or DA treatments.
To understand gene expression during cold acclimation and de-acclimation, we examined the transcriptome profiles in tea leaves by using RNA-Seq analysis [24]. The mRNA of each sample was sequenced on an Illumina HiSeq X 10 platform. More than 41 million 150-bp paired-end reads were generated for each sample, totaling 479,445,976 and 466,081,620 clean reads from SCZ and YH9, respectively. We aligned the RNA-Seq reads to the C. sinensis draft genomic assembly [29] with tool HiSAT2 and StringTie to assemble and compare transcripts [30]. Approximately 81% and ~ 84% of reads were mapped onto the genome assembly for SCZ and YH9, respectively (Additional file 1: Table S1). A slightly higher mapping ratio was found in YH9, probably because YH9 is a C. sinensis var. assamica, as is the draft genome assembly utilized [29]. We obtained 85,949 and 87,842 transcripts, from 46,223 and 46,736 gene loci in SCZ and YH9, respectively (Additional file 1: Table S1). Of these candidate genes, 9272 from SCZ (20%) and 9785 from YH9 (21%) were not predicted as genes in the draft genome assembly [29]. The RNA-Seq data and transcript assemblies have been deposited and are publicly available in SRA and TSA at NCBI (https://www.ncbi.nlm.nih.gov/) under bio-project accession PRJNA387105.
Overview of differential gene expression during cold acclimation
To understand gene expression in response to cold acclimation, we measured transcript levels in terms of fragments per kilo-base exon per million reads (FPKM) based on RNA-Seq data. Genes were called Differentially Expressed Genes (DEGs) if they exhibited an at least 2-fold change in transcript abundance (Wald test, p < 0.05) using R package DESeq2 [31]. First, we investigated DEGs specific to each cold treatment by comparing with the stage immediately before the corresponding treatment. The CS group was treated with a short-term cold stimulus, from which 879 and 1589 DEGs were discovered in SCZ and YH9, respectively, compared with the control NA, indicating genes that are regulated for a rapid response to cold stimulus (Fig. 2a). The CA group was treated with 7d chilling acclimation, which generated a 2383 and 381 DEGs in SCZ and YH9, respectively (Fig. 2a), suggesting chilling acclimation genes. The FA group was treated with subsequent 7d freezing acclimation, which led to 1684 and 297 DEGs not observed in the CA group in SCZ and YH9 (Fig. 2a), respectively, suggesting responsive genes to freezing and cold resistance. The DA group, experiencing potential de-acclimation, yielded 1857 and 5968 DEGs that were not observed at the previous stage FA in SCZ and YH9, respectively (Fig. 2a). Fewer DEGs were found in the short-term and recovering treatment, while more DEGs in CA and FA treatment were observed in SCZ than YH9 (Fig. 2a).
Fig. 2.
Differential expression of genes during chilling stimulus, chilling acclimation, freezing acclimation and de-acclimation. Image (a) shows the number of differentially expressed genes (DEGs) compared with the stage just before the corresponding treatment. Image (b) shows the number of DEGs in each stage compared with stage NA. Venn diagram images (c and d) show the number of DEGs across treatments. Image (e) shows the expression level of DEGs. Image f shows the clusters of expression patterns of DEGs. The change of expression level is shown after log10 conversion
In total, 6072 and 7749 DEGs were identified in cultivar SCZ and YH9 in response to one or more of the four treatments. Of these DEGs, 978 common DEGs were present in both cultivars (Fig. 2e). Their expression patterns fell into four major clusters (Fig. 2f). A total of 583 DEGs in cluster 1 (338 genes) and cluster 2 (245 genes) showed different patterns between the two tea cultivars in response to cold acclimation (Fig. 2f), indicating that these shared common DEGs responded differently and thus are candidate contributors to the difference in cold resistance between the two cultivars. The 394 genes in cluster 3 and 4 showed similar patterns in each cultivar (Fig. 2f).
We analyzed the DEGs after sequential CS, CA, and FA treatments using the starting stage NA as a control. Results revealed that the CA treatment led to the most DEGs in SCZ while the FA treatment led to the most DEGs in YH9 (Fig. 2b), suggesting an earlier peak time of most DEGs was associated with greater cold resistance in SCZ compared to YH9. Regarding the DEGs after the DA treatment, we used the FA treatment as the control, suggesting that these DEGs were responsible for recovering from cold acclimation. Comparison showed that 25 DEGs were shared by both CS and CA treatments while the number was reduced to 7 DEGs after further FA treatment in SCZ. Hence, these 25 common DEGs could be crucial for long-term CA or FA. The 7 common DEGs didn’t exhibit differential expression in the DA treatment, further suggesting these genes were responsible for cold response only (Fig. 2c). In YH9, we discovered 384 common DEGs after CS and CA treatments and this number was reduced to 291 DEGs after FA treatment. Of the 291 DEGs, 225 DEGs were cold responsive only and not present after DA recovery treatment (Fig. 2d).
Functional comparison of cold responsive DEGs
To gain insights into the functions of discovered DEGs, we first assigned gene ontology (GO) term(s) to each DEG with the GO database (version 2016.04, http://geneontology.org) by similarity search using Blastx in the NCBI Blast+ package [32]. We then conducted an enrichment analysis using the statistical tool GOseq (version 1.18.0) [33]. DEGs relative to the NA stage were significantly (Wallenius test, p < 0.05) enriched for 317 and 700 GO terms in SCZ and YH9, respectively. The top 10 GO terms in each treatment, ranked by the number of DEGs, varied with treatment stages, suggesting altered functional responses at each stage.
GO terms of shared DEGs between the two cultivars could reflect common responses to cold acclimation. Thus, we assigned GO terminology to the shared 978 DEGs, and found that they belonged to 317 known GO terms. Of these GO terms, more were classified into the “biological process” category than the “cellular component” or the “molecular function” category. The top 10 enriched (Wallenius test, p < 0.05) GOs of common DEGs exhibited a very similar rank, although the DEG numbers in each GO term differed between cultivars (Fig. 3a). The shared top GO terms included “regulation of transcription”, “flower development”, “abscisic acid-activated signaling pathway”, and “photosynthesis” in the biological process category. The top enriched GOs in the cellular component category included “membrane”, “apoplast”, and “chloroplast thylakoid membrane”. The top enriched GOs in the molecular function category included “transcription factor activity”, “hydrolase activity”, “zinc ion binding” and “oxidoreductase activity” (Fig. 3a). We found that other GOs, besides the top enriched terms, also contributed to the responses to cold treatment. For instance, we found that GOs of two DEGs from the comparison CA_vs_ CS in SCZ were defined as cellular response to freezing (GO:0071497). GO terms for a DEG from the comparison CA_vs_NA in YH9 were also assigned to cellular response to freezing. The differences in “cellular response to freezing” may also reflect the difference in cold tolerances between cultivars.
Fig. 3.
Comparison of top GO terms and pathways of DEGs in tea during cold stress, chilling acclimation, freezing acclimation and de-acclimation. The numbers of DEGs in each GO term are shown in three categories defined in the GO database (a). The numbers of DEGs in each pathway are shown for each treatment comparison (b). The comparison is described as A_vs_B, which means the DEG was found in the A treatment relative to the B treatment. NA, CS, CA, FA and DA indicate the no acclimation control, cold stimulus, chilling acclimation, freezing acclimation, and de-acclimation, respectively
Enriched pathways associated with DEGs in response to cold acclimation
To understand the metabolic or signal pathways involved in cold treatment responses, we first mapped all DEGs to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to retrieve involved pathways in each cultivar. In total, 22 and 44 significantly (p < 0.05) enriched pathways were identified for all DEGs in SCZ and YH9, respectively. Then, we searched for the pathways shared by both tea cultivars and found 18 common enriched pathways (Fig. 3b), of which “Plant-pathogen interaction” had the most abundant DEGs enriched, while “Plant hormone signal transduction” and “Ribosome” were also ranked high. We found different numbers of DEGs in the same pathway in the two cultivars (Fig. 3b). Among the top five pathways, the most abundant DEGs appeared in the comparison CA_vs_CS, which was after the CA treatment, in SCZ while they appeared at a later stage in the comparison FA_vs_CA after the FA treatment in YH9, suggesting earlier altered gene expression in the shared pathways in highly cold-tolerant SCZ.
We also compared the pathways associated with DEGs relative to the starting stage NA. DEGs from the short-term CS treatment were mapped into 66 and 108 KEGG pathways in SCZ and YH9, respectively, but only eight pathways were significantly (p < 0.05) enriched in SCZ. We found the enriched pathways were different from the shared pathways mentioned above (Fig. 3b). The top three pathways ranked by the number of DEGs were “plant pathogen interaction (22 DEGs)”, “plant hormone signal transduction (15 DEGs)” and “photosynthesis (5 DEGs)” in SCZ. However, 13 significantly (p < 0.05) enriched pathways were found in YH9, and the top three pathways with the most DEGs were “Carbon metabolism (24 DEGs)”, “Glycerolipid metabolism (12 DEGs)” and “Phagosome (12 DEGs)”. This suggested fewer and more pathways were affected in the high and low cold-tolerant cultivar after the short-term CS, respectively. Moreover, the cold-sensitive cultivar appeared to be mostly changing expression of general metabolism genes, while the cold-tolerant cultivar was modifying general resistance/stress pathways.
After the long-term CA treatment, the top five of nine enriched (p < 0.05) pathways were “Plant hormone signal transduction” (42 DEGs), “Pyruvate metabolism” (18 DEGs), “Phagosome” (18 DEGs), “Glycolysis / Gluconeogenesis” (16 DEGs) and “Carbon fixation in photosynthetic organisms” (16 DEGs) in SC. In contrast, in YH9, the top five of nine enriched (p < 0.05) pathways were “Ribosome” (11 DEGs), “Carbon metabolism” (21 DEGs), “Phagosome” (17 DEGs), “Glyoxylate and dicarboxylate metabolism” (13 DEGs) and “Ribosome biogenesis in eukaryotes” (10 DEGs). After the subsequent long-term FA treatment, only two pathways termed “RNA degradation” (11 DEGs) and “Photosynthesis” (4 DEGs) were enriched in SCZ; however, 12 enriched pathways were found in YH9, including the top five pathways termed as “Carbon metabolism” (47 DEGs), “Phagosome” (24 DEGs), “Purine metabolism” (23 DEGs), “Pyruvate metabolism” (21 DEGs), and “Carbon fixation in photosynthetic organisms” (20 DEGs). This suggested that the final effects after chilling acclimation and freezing acclimation induced less change in pathways in SCZ, which matches the observed higher adaptation to cold and stable physiological responses in SCZ (Fig. 1).
Key DEGs in the photosynthesis pathway
As the photosynthesis pathway was enriched in DEGs, we further characterized functional annotation of these DEGs. The proteins encoded by the DEGs were mainly located in the chloroplast, and annotated to function as antenna proteins (Fig. 4a), photosystems I and II components, cytochromes, and electron transport (KEGG map 00062, Fig. 4b, Additional file 2: Table S2). Comparison of these DEGs’ expression between the two cultivars revealed 17 genes that had similar patterns of expression. In contrast, light-harvesting chlorophyll gene 2 (Lhca2) had quite different expression patterns in the two cultivars (Fig. 4c). Lhca2 encodes a protein in the membrane complex of photosynthesis system I (Fig. 4a). Lhca2 was highly expressed in YH9 but much less expressed in SCZ in response to cold treatments. During recovery from cold to warm temperature, Lhca2 expression showed a reversed pattern, increasing in SCZ and decreasing in YH9 (Fig. 4c and d). We validated the Lhca2 expression level using RT-qPCR (Fig. 4d, more validation in Additional file 3: Figure S1). Hence, low expression of Lhca2 was negatively correlated with cold-tolerant capability.
Fig. 4.
Differentially expressed genes involved in photosynthesis and chloroplast. Images (a and b) show differentially expressed genes shaded in a green box in the photosynthesis pathways depicted in KEGG (map 00195). Image (c) shows a comparison of gene expression patterns in SCZ and YH9. The heatmap was generated from the log2FPKM mean value calculated from two replicates of RNA-Seq data. The asterisk before a gene product name indicates the most different patterns between two cultivars. Image d shows the relative expression level of gene Lhca2 validated from RT-qPCR
Regulation of key DEGs in plant-pathogen related immunity
We further characterized the identified DEGs involved in plant-pathogen related immunity in the KEGG database to locate the proposed regulatory pathway. We also used the UniProt database for additional functional annotation based on protein similarity. Ten common DEGs in this category were found in both SCZ and YH9 (Table 1); however, 70 and 56 DEGs were present only in SCZ and YH9, respectively, indicating different regulation in pathways in the two tea varieties. Many of the predicted functions of these DEGs are defense-related kinases, transcription factors, or heat shock proteins (Table 1). Multiple members of gene families BAK1, HSP, and PBS1 were expressed, but only a subset of members was differentially expressed (Table 1), indicating functional diversity of these families in cold response. Among these genes, FLS2 exhibited very different expression patterns between SCZ and YH9 (Fig. 5a, c).
Table 1.
DEGs related to plant immunity shared between SCZ and YH9
| Gene name | Pathway entry | Manually annotated function |
|---|---|---|
| CSA015695 | K13416 | brassinosteroid insensitive 1-associated receptor kinase 1 (BAK1.10) |
| CSA001565 | K13420 | LRR receptor-like serine/threonine-protein kinase (FLS2) |
| CSA030713 | K04079 | heat shock protein 83-like (HSP.2) |
| CSA017980 | K13430 | serine/threonine-protein kinase PBS1 (PBS1.6) |
| CSA007045 | K13430 | serine/threonine-protein kinase PBS1 (PBS1.10) |
| CSA024279 | K13430 | serine/threonine-protein kinase PBS1 (PBS1.12) |
| CSA025610 | K13449 | basic form of pathogenesis-related protein 1 (PR1) |
| CSA004479 | K13426 | WRKY transcription factor 29 (WRKY29) |
| CSA000715 | NA | unknown |
| CSA009117 | NA | unknown |
Fig. 5.
Expression patterns of DEGs associated with biotic defense in the tea plant. The heatmap (a) represents the gene expression patterns generated from the log2FPKM of the mean value of replicates from RNA-Seq data. The asterisk before a gene product name listed in the right side of each row indicates the most different patterns in RNA-Seq data from the two cultivars. The relative expression level of gene PBS1.10, PBS1.12 (b) and gene FLS2.11 (c). NA, CS, CA, FA and DA represent the cold treatments
The column “Gene name” represents the gene ID in the publicly available draft tea genome assembly. Pathway entry represents the ID in the database KEGG.
Regulation of key DEGs in plant hormone signal transduction
We identified 115 DEGs involved in hormone signal transduction from both cultivars of SCZ and YH9 in response to cold treatments. Of those, 15 DEGs were shared in the two tea cultivars, while 59 and 41 DEGs were only present in the respective SCZ and YH9, indicating both common and cultivar-specific regulation under cold treatments (Table 2). The DEGs were mapped into hormone signal transduction pathways of auxin, cytokinin, abscisic acid, gibberellin, ethylene, brassinosteroid, jasmonic acid and salicylic acid in Arabidopsis and Populus (Table 2, Fig. 6). Most of the known genes in auxin, cytokinin, abscisic acid and ethylene signal pathways were identified as DEGs in this study (Fig. 6a). Different expression patterns of common DEGs were found between the two tea cultivars in response to cold treatments (Fig. 6b), indicating regulation of these common DEGs was also associated with different cold tolerances. The expression of SnRK2.8, functioning in ABA signal transduction, osmotic stress and glycol-metabolism [34], was significantly higher in SCZ than YH9 in response to cold acclimation (Fig. 6b).
Table 2.
DEGs involved in plant hormone signal transduction
| Gene name | KEGG # | Manually annotated function | Shared DEG |
|---|---|---|---|
| CSA026554 | K14492 | two-component response regulator ARR-A family (A-ARR) | |
| CSA025614 | K21568 | auxin-responsive protein IAA5-like (IAA5) | Yes |
| CSA003423 | K14432 | ABA responsive element binding factor (ABF) | Yes |
| CSA025810 | K14490 | histidine-containing phosphotransfer protein (AHP) | |
| CSA014553 | K14490 | histidine-containing phosphotransfer protein (AHP) | Yes |
| CSA002625 | K14486 | auxin response factor (ARF) | |
| CSA014562 | K14484 | auxin-responsive protein IAA (AUX/IAA.1) | |
| CSA019710 | K14484 | auxin-responsive protein IAA (AUX/IAA.2) | |
| CSA023080 | K14484 | auxin-responsive protein IAA (AUX/IAA.3) | |
| CSA027576 | K14491 | two-component response regulator ARR-B family (B-ARR.1) | |
| CSA036208 | K14491 | two-component response regulator ARR-B family (B-ARR.2) | |
| CSA036403 | K14491 | two-component response regulator ARR-B family (B-ARR.3) | |
| CSA015695 | K14502 | brassinosteroid insensitive 1-associated receptor kinase 1-like (BIN_like) | Yes |
| CSA001186 | K14502 | protein brassinosteroid insensitive 2 (BIN2) | |
| CSA007850 | K14500 | BR-signaling kinase (BSK.1) | |
| CSA032457 | K14500 | BR-signaling kinase (BSK.2) | |
| CSA022935 | K14503 | brassinosteroid resistant 1/2 (BZR1/2) | |
| CSA016248 | K14489 | cytokinin receptor (CRE1) | Yes |
| CSA000728 | NA | Unknown | Yes |
| CSA006127 | NA | hypothetical protein | Yes |
| CSA009443 | NA | Unknown | Yes |
| CSA022307 | NA | Unknown | Yes |
| CSA029041 | K14515 | EIN3-binding F-box protein (EBF1/2) | Yes |
| CSA008678 | K14513 | mitogen-activated protein kinase kinase 4/5 (EIN2) | |
| CSA031389 | K14509 | ethylene receptor (ETR.1) | Yes |
| CSA024687 | K14509 | ethylene receptor (ETR.2) | |
| CSA004165 | K14509 | ethylene receptor (ETR.3) | |
| CSA033915 | K14487 | auxin responsive GH3 gene family (GH3) | |
| CSA012511 | K14493 | gibberellin receptor GID1 (GID1.1) | Yes |
| CSA020494 | K14493 | gibberellin receptor GID1 (GID1.2) | |
| CSA020665 | K14493 | gibberellin receptor GID1 (GID1.3) | |
| CSA021414 | K14493 | gibberellin receptor GID1 (GID1.4) | |
| CSA009988 | K14506 | jasmonic acid-amino synthetase (JAR1) | |
| CSA010349 | K14497 | protein phosphatase 2C (PP2C) | |
| CSA028964 | K14497 | Highly ABA-induced PP2C gene 2 (PP2C2) | Yes |
| CSA025610 | K13449 | pathogenesis-related protein 1 (PR-1) | Yes |
| CSA027144 | K14496 | abscisic acid receptor PYR/PYL family (PYR/PYL) | |
| CSA006311 | K14498 | serine/threonine-protein kinase SRK2 (SnRK2.8) | |
| CSA007707 | NA | systemin receptor SR160 | Yes |
| CSA036164 | K14485 | transport inhibitor response 1 (TIR1) | |
| CSA022307 | K13415 | protein brassinosteroid insensitive 1 (BRI1) |
Fig. 6.
DEGs in hormone signaling pathways in response to cold treatment. a Green boxes show the mapped DEGs in plant hormone signaling pathways, as modified from a KEGG map (ID 04075). b The expression patterns are shown as a heatmap. The most different patterns of expression between the two tea cultivars are marked by asterisks. The shared DEGs in the cultivars are tagged by the suffix .com. NA, CS, CA, FA and DA denote the cold treatments
The k number in the KEGG column is the pathway entry in the KEGG database. The abbreviation of the gene in parenthesis was added with a dot and digital suffix if multiple copies of DEGs were annotated as the same function.
Discussion
Identifying the genes responsible for high cold resistance could help the breeding of new cultivar to extend tea plantations into cooler zones. To date, the expression of several genes [26] or miRNAs [28] were reported in response to cold stress in tea plant, but these studies did not take acclimation or cultivar differences into account. The transcriptome profile under natural temperature changes was reported in a tea variety between winter and early spring [24]. Here, the systematic transcriptome profiling in tea varieties responsible for high or low cold resistance capability under controlled cold acclimation was investigated.
From the DEG comparisons between SCZ, with a high cold resistance, and YH9, with a low cold resistance, we conclude that at least two expression features associate with higher cold tolerance. First, less expression fluctuation in a short-term CS may reflect higher tolerance. This could be seen from less DEGs (879 vs. 1589) in SCZ than in YH9 after short-term (6 h) CS. Second, more DEGs could be induced by a long-term cold acclimation in the greater cold tolerant variety, which may result in a better preparation for freezing stress, and thus a higher cold tolerance. This is demonstrated by 2383 and 381 DEGs in SCZ and YH9, respectively, after 7-day long-term cold acclimation. The response of DEGs during the long-term cold acclimation, not the short-term CS, appears to be crucial for acquiring higher cold resistance. Future studies on the function of these 25 DEGs in SCZ during the long-term cold acclimation should be of highest priority, including analysis by overexpression and/or knock-out mutations. However, the transformation of tea plants is still a big challenge.
Cold acclimation is a complicated biological process that is expected to integrate multiple signals. Here, we identified DEGs involved in many pathways, partly confirming that complexity. Signal transduction pathway is known to play crucial roles in response to the stress of low temperatures [35]. Evidences have established that Ca2+ influx was a very important change in membrane during cold stress [36]. Some genes have been shown to sense changes in intracellular Ca2+ levels via phosphorylation to initiate downstream signaling processes [37]. In our study, we found related gene regulation for Ca2+ signaling. Calmodulin (CaM) gene was specifically found in SCZ and up-regulated during cold stress. Two calcineurin B-like protein (CBL) genes, three calcium-dependent protein kinase (CDPK) genes, one calmodulin-like protein (CML) genes and three CBL-interacting protein kinase (CIPK) genes were up-regulated during cold treatment in SCZ while only two CBP genes were up-regulated in YH9 (Additional file 4: Table S3). In addition, fifteen receptor like kinase genes for phosphorylation were induced during cold treatment in SCZ and six were induced in YH9. Therefore, Ca2+ signaling-related genes might contribute to higher cold tolerance in SCZ than YH9.
FLS2, encoding a type of LRR receptor-like protein kinase, which exhibited a higher expression level in only SCZ, was a DEG involved in plant-pathogen related immunity in the KEGG database. It is known that abiotic stresses induce plant immunity gene expression [38], perhaps as a way to minimize biotic infestation after abiotic tissue damage. We speculate that cold-induced plant immunity may result from a common signaling process like those for other abiotic stresses. This signaling could include components in plant hormone cross-talk. A LRR receptor-like protein kinase (GsLRPK) in Glycine soja is a positive regulator to cold stress tolerance. Overexpression of GsLRPK in Arabidopsis can increase the expression of many COR gene markers [39].
MAPK cascades are involved in plant signal transduction in response to cold [40], which could relay and amplify signals via three types of reversibly phosphorylated kinases leading to the phosphorylation of substrate proteins [41]. Here, we identified one MAPK gene was up-regulated in SCZ while down-regulated in YH9 (Additional file 4: Table S3).
We found that the enriched GO terms for DEGs included the “abscisic acid-activated signaling pathway”. ABA signaling pathway has been identified as a central regulator of abiotic stress response in plants, triggering major changes in gene expression and adaptive physiological responses [42]. Zeaxanthin epoxidase (ZEP), 9-cis-epoxycarotenoid dioxygenase (NCED) and abscisic aldehyde oxidase (AAO) are three key enzymes involving in ABA synthesis [43]. In our study, there are two ZEP genes were up-regulated during cold treatment in SCZ while down-regulated or constant in YH9. NCDE 1 gene was up-regulated but not significant in SCZ. SnRKs allowed intensifying activation of the ABA-induced signaling pathway through a double negative regulatory system (PYR/PYL-| PP2C-| SnRK2) [44]. Here, one SnRK2.8 gene was dramatically up-regulated during CS and CA in SCZ (Additional file 4: Table S3). Expression of most key genes in plant hormone signaling pathways were significantly altered in expression (Fig. 6), suggesting a huge change in hormone signaling. Plant hormone pathways are known to cross-talk under stress [45]. Hence, the CA and FA results observed agree with hormonal cross-talk manner that can increase the cold tolerance and thereby minimize cold damage.
Transcription factors (TFs) play important functions in plant development and stress tolerance. Our transcriptomic data depicted some TFs that govern the massive and highly coordinated transcriptional changes under short/long term cold stress in SCZ and YH9 (Additional file 5: Table S4). Consistent with findings in other plant exposed to cold stress, some of the major TFs of SCZ and YH9 were AP2, ERF, bHLH, bZIP, MYB, NAC, WRKY, MADS, CAMTA, B3-ARF, ZAT, C2C2-Dof, indicating the complexity of the regulatory pathways during cold acclimation [24, 46]. Transcripts encoding TFs like ERF (CSA013480), NAC (NAC86, 94), WRKY (WRKY42, 50), MYB86, bHLH (bHLH36, 48, 102, 116, 118), MADS- M-type, B3-ARF, and ZAT were significantly upregulated into a higher level in tolerant SCZ than sensitive YH9 when expose to cold stress. These TFs are reported to be linked to cold stress resistance in plants possibly via activating ABA-independent and ABA-dependent pathway in plant [47–50]. The highly up-regulated expression of TFs in SCZ could play a vital role in higher cold tolerance in SCZ.
CBF/dehydration-responsive element-binding factor (DREB1) from AP2/ERF super-family (CBF1/DREB1B, CBF2/DREB1C, and CBF3/DREB1A) in Arabidopsis plays a prominent role in cold acclimation [7, 51]. CBF expression was positively correlated with cold tolerance, and expression of downstream CORs can be activated by binding with DRE/CRT cis-acting elements [52]. Here, we found consistent results in tea plant. Expression of three CBF genes were found to be significantly induced during cold acclimation. The CBFs expression level in SCZ was significantly higher than that in YH9, the CBF pathway may play an important role in tea plants. Many COR genes were induced during the cold acclimation in both cultivars (Additional file 6: Table S5). Most COR genes have similar expression levels, which indicates that these COR genes are conservative in regulating of defense responses in tea plant. COR genes encoding LEA2 (MSTRG.46934), HSP70 (CSA030367), PRP (CSA014200, CSA008621), CIPs, PEIs, TLPs (CSA000129, CSA001426), ChiA (CSA012225) and endoglucanase (endoglucanase 6, 8, 11, 14, 17) had higher transcript abundance in SCZ than YH9. Therefore, both CBFs and these CORs may contribute to the higher cold tolerance after cold acclimation in SCZ.
ROS is a toxic compound that can cause oxidative damage to proteins, DNA, lipids and other cellular components [53, 54]. ROS-scavenging enzymes, such as SOD, POD, APX, monodehydroascorbate reductase (MDAR), glutathione peroxidase (GPX) and glutathione S-transferase (GST), were found in our transcriptome’s data, but only POD (CSA023255, CSA026001) and GST (CSA006422) were up-regulated in SCZ. This indicates that SCZ suffers less cold damage than YH9 during cold acclimation, consistent with observed higher cold tolerance in SCZ. Flavonoids has ROS-scavenging activity that protects against oxidative damage and controls ROS levels under abiotic stresses [55]. This was supported by increased flavonoids in winter sweet spinach [56] and transcription of genes for biosynthesis of flavonoids correlated with freezing tolerance after cold acclimation in in Arabidopsis [57]. In this study, the flavoid associated genes involved in flavonoid biosynthesis were detected between SCZ and YH9 (Additional file 7: Table S6). During CA treatment, flavonol synthase (FLS) was significantly up-regulated only in SCZ with high fold change, while anthocyanidin synthase (ANS) was up-regulated in YH9. Shen et al. also found that FLS expression was up-regulated under low temperature stress in SCZ [58]. Thus, both FLS and ANS could contribute to enhance cold tolerance in tea plant but with difference. The increased fatty acid desaturation in membrane lipids allows functional membrane fluidity to be maintained at low temperature [19, 59]. SAD, FAD and LTP8 are responsible for fatty acids unsaturation. In this study, SAD and LTP8 were both significantly up-regulated in SCZ during CA and FA treatment. Interestingly, the expression of FADs had different pattern in two cultivars. We speculate that the higher SAD and LTP8 expression were contribute to higher cold resistant in SCZ (Additional file 8: Table S7).
To maintain osmotic equilibrium, plants accumulate a series of compatible solutes, including soluble sugars and low molecular weight compounds such as proline and glycine betaine during cold stress response [2, 60, 61]. In our previous studies, we found SCZ exhibited a higher accumulation level of soluble sugars, particularly sucrose than YH9 during cold acclimation [26]. In this study, inductions of sucrose biosynthesis-related genes (SUS, SPS) and most of TPS were considerably higher in SCZ than YH9. The enzymes β-amylase (BMY) degrades starch to soluble sugar which leads to increased maltose, glucose, fructose and sucrose levels after further conversion. Cold stress also induces BMY transcript level or activity [62]. Here, BMY was significantly up-regulated in both cultivars, but the expression level in SCZ was higher than that in YH9. HXK is known to phosphorylate glucose and fructose, and participate in sugar signal transduction by modulating the abundances of diverse gene transcripts and integrating stress response substrates, including ABA and ethylene [63, 64]. HXK was induced in Jatropha curcas when exposed to cold stress [65]. Similarly, HXK was induced more in SCZ than YH9. To date, it is known that the proline biosynthesis is catalyzed by P5CS and P5CR in plants [66]. In this study, expression level of P5CS and P5CR were higher in SCZ than YH9 (Additional file 9: Table S8). Therefore, these genes are also involved in cold acclimation for conveying better tolerance.
Photoinhibition is a decline in photochemical activity when the available light exceeds the real receptive capacity of the photosynthesis. Exposure to low temperature has been found to cause photoinhibitory stress in chilling-sensitive plants [67]. In spinach, photoinhibition has been seen to lead to the accumulation of osmoprotectant polyamines by increasing S-adenosylmethionine decarboxylase activity [68]. In maize, light is necessary for effective cold acclimation under low temperature [69], suggesting an involvement of photoinhibition. Here, we identified many acclimation-induced DEGs enriched in photosynthesis, a novel discovery for the tea plant. The expression of light capture complex genes was decreased, suggesting a decline in photochemical activity. The expression of light-harvesting chlorophyll Lhca2 in photosystem I (Fig. 4) may be crucial for cold tolerance. Under cold treatment, the decreased expression of Lhca2 in cold-resistant SCZ contrasted with the increased expression in cold-susceptible YH9 suggesting that Lhca2 expression could be a good indicator of cold resistance. Here, we conclude that low expression of Lhca2 suggests photoinhibition and indicates a higher cold tolerance in tea plants. This is further supported by a recent finding that photoinhibition is necessary for acquiring cold resistance in maize, a chilling-sensitive plant [69]. The reduced expression of Lhca2 may lead to low activity of LHCA2 protein in photosynthesis system I, suggesting that the absorption of light is reduced and that light suppression at low temperatures is alleviated, thereby improving cold resistance.
Conclusion
This study provides a comparative transcriptomic analysis of how gene expression differs in cold acclimated leaves of two tea varieties with different tolerances to cold stress.
A responsive molecular model of cold stress in the CSS was summarized in Fig. 7. The cold signal is perceived and then mainly transduced further by Ca2+ signaling and ABA signaling resulting in the activation of downstream TFs. The activation of COR genes through involvement of TFs leads to trigger a series of physiological and cell response, and improved tolerance to cold stress. The greater transcriptional differentiation during cold acclimation in SCZ may explain its better cold tolerance compared to YH9. Genes involved in photoinhibition, hormone signaling, and plant immunity are now identified as logical targets for further investigation and for future development of improved cold-tolerance in tea cultivars.
Fig. 7.

A schematic signaling and involved genes for achieving cold tolerance in tea plant (SCZ). MAPK, mitogen-activated protein kinase; CaM, calmodulin; CBL, calcineurin B-like Ca2+ sensors; CIPK, CBL-interacting protein kinase-calcineurin B-like protein; CDPK, Ca2+ − dependent protein kinase; FLS, LRR receptor-like serine/threonine-protein kinase; ZEP, zeaxanthin epoxidase; SnRK2.8, SNF1-related protein kinase 2.8; CBF, C-repeat binding factors; ERF, ethylene response factor; NAC, NAC domain transcription factor; WRKY, WRKY transcription factor; MYB86, MYB transcription factor86; bHLH, basic helix-loop-helix protein; MADS-M-type, MADS-box domain protein; B3-ARF, B3 domain transcription factor; ARF, auxin response factor; ZAT, zinc finger protein; LEA2, late embryogenesis abundant protein 2; HSP70, heat shock protein 70; PRP, proline-rich protein; CIP, cold-induced protein; PEI, pectinesterase inhibitors; TLP, thaumatin-like protein; ChiA, chitinase; SUS, sucrose synthase; SPS, sucrose phosphate synthase; TPS, trehalose phosphate synthase; BMY, beta-amylase; HXK, hexokinase; P5CS, proline-5-carboxylate synthetase; P5CR, proline-5-carboxylate reductase; POD, peroxidase; GST, glutathione S-transferase; SAD, stearoy-l ACP desaturase; LTP8, lipid-transfer protein 8
Methods
Plant materials and treatments
Cuttings of C. sinensis var. sinensis cv. Shuchazao and Camellia sinensis var. assamica cv. Yinghong9 were obtained from the Dechang Tea Plantation in Anhui (116° 56′ 24″ E, 31° 27′ N) and the Tea Research Institute of Guangdong Academy of Agricultural Sciences (113° 22′ 48″ E, 24° 10′ 12″ N), China, respectively. One-year-old clonal plants propagated from cuttings were transferred to a growth chamber with temperature cycles of 25 °C in the day and 20 °C at night, 12 h photoperiod, and 70% relative humidity for 1 month. Subsequently, they were subjected to treatments for cold acclimation and de-acclimation. Ten one-year-old tea plants were collected from the aforementioned growth chamber and used as non-acclimation controls. The following cold acclimation treatments were applied in this study: CS, cold stimulus, was conducted by exposing SCZ and YH9 plants to 10 °C for 6 h; CA, chilling acclimation, was conducted by exposing SCZ and YH9 plants to low temperature (10 °C day, 4 °C night) for 7 days. Afterwards, FA, freezing acclimation, was conducted by exposing SCZ and YH9 plants to lower temperature (4 °C day, 0 °C night) for another 7 days. Lastly, the plants were exposed to normal temperature (25 °C day, 20 °C night) for 7 days for de-acclimation. After each treatment, the fresh second leaves immediately next to the leaf bud and third leaves downward were collected, and immediately frozen in liquid nitrogen. They were stored at − 80 °C until use. Two independent experiments were conducted for each tea cultivar, and 10 biological replicates were used in each treatment.
Electrolyte leakage assay and Fv/Fm
SCZ and YH9 were exposed to CA for 1 week, FA for another 1 week and then followed by − 5 °C freezing treatment for 12 h. The tea plants in the control group (NA) were not treated with CA and FA but were exposed to − 5 °C for 12 h. The relative electrolyte leakage rate and chlorophyll fluorescence Fv/Fm value were assayed following methods described previously [26]. The detached leaf discs of SCZ and YH9 were exposed to − 5 °C for 12 h, and Fv/Fm images of leaf discs from NA or FA were obtained with Imaging-Pam chlorophyll fluorescence imager (Walz, Germany).
RNA extraction and sequencing
The RNAs from collected tissues were extracted using methods described previously [70]. Briefly, the frozen leaves were ground into powder in liquid nitrogen. Then the powder was used for RNA extraction [70]. After preparation, RNA quality and quantity were measured using an agarose gel and the Nanodrop 2500 (Thermo Fisher Scientific, US). RNA-Seq libraries were constructed as described previously [70] and sequenced on the Illumina platform HiSeq X 10. The sequencing format was paired-end 150 bp. The reads are publicly available at the SRA database of NCBI under the project accession number SRP108833.
Transcript assembly and expression level analysis
All raw reads were cleaned to remove low quality reads as described previously [71], and combined together to build a transcript assembly using the publicly available tea draft genome [29] as the reference sequence, as described previously [71]. Newly discovered genes, that were not annotated in the draft genome, were annotated against publicly available databases using methods described previously [70]. Transcript levels were calculated in FPKM, and differentially expressed genes were defined as exhibiting at least two-fold change in transcript abundance and statistical significance levels of p < 0.05 relative to a control by using the tool DESeq2 [31] according to the methods described previously [70].
Metabolic pathway analysis of DEGs
To analyze DEG-associated functions, we searched the KEGG pathway database using the online tool KAAS (http://www.genome.jp/) [72] based on the Blast bidirectional best hit algorithm with default settings to assign a candidate ko number for each gene. We then mapped the gene to a candidate pathway using KEGG mapper. The sequence databases for Arabidopsis thaliana and Populus trichocarpa were used as the references. The functional annotation of pertinent DEGs was investigated in the KEGG protein cluster and the UniProt database (http://www.uniprot.org/uniprot/) by protein similarity using the Blastp algorithm with default settings from the corresponding database. DEG functions from two databases were then manually merged. Pathway enrichment analysis was conducted with the tool GOseq [33] according to the methods and scripts described previously [71].
RT-qPCR validation
Real-time quantitative PCR analyses were conducted with mRNA from the leaves of ten plants from three experimental replicates. The method followed the procedures described in a previous report [26]. The primers for the RT-qPCR are described in Additional file 10: Table S9.
Additional files
Table S1. Summary of RNA-seq, assembly and annotation. (DOCX 25 kb)
Table S2. Differentially expressed genes and functions in photosynthesis. (DOCX 22 kb)
Figure S1. RT-qPCR validation of selected DEG. (DOCX 452 kb)
Table S3. Differentially expressed genes related to signal transduction in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 22 kb)
Table S4. Differentially expressed transcription factors (TFs) in response to cold treatment of SCZ and YH 9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 28 kb)
Table S5. Differentially expressed cold response genes in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 21 kb)
Table S6. Differentially expressed genes related to free radical scavengers and flavonoid in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 19 kb)
Table S7. Differentially expressed genes related to fatty acid in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 13 kb)
Table S8. Differentially expressed genes related to osmoprotectans in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 15 kb)
Table S9. Primers used in the RT-qPCR. (DOCX 23 kb)
Acknowledgements
We thank Dr. Enhua Xia from Anhui Agricultural University for his advice on data analysis and Professor Zhaoliang Zhang for Providing constructive comments to this manuscript.
Abbreviations
- ABA
Abscisic acid
- APX
Ascorbate Peroxidase
- ARF
Auxin response factor
- BAK
Brassinosteroid insensitive 1-associated receptor kinase 1
- bHLH
Basic helix-loop-helix protein
- BMY
Beta-amylase
- CA
a 7-day long-term chill acclimation
- CaM
Calmodulin
- CBF
C-repeat-binding factors
- CBL
Calcineurin B-like Ca2+ sensors
- CBP
Calcium-binding protein
- CDPK
Ca2+ − dependent protein kinase
- ChiA
Chitinase
- CIP
Cold-induced protein
- CIPK
CBL-interacting protein kinase-calcineurin B-like protein
- CS
a 6 h short-term cold stimulus
- CSA
Camellia sinensis var. assamica
- CSS
Camellia sinensis var. sinensis
- DA
a 7-day long-term de-acclimation
- DEGs
Differentially expressed genes
- ERF
Ethylene response factor
- FA
a 7-day long-term freezing acclimation
- FLS2
LRR receptor-like serine/threonine-protein kinase
- Fv/Fm
Chlorophyll fluorescence
- GA
Gibberellin
- GO
Gene ontology
- GR
Glutathione reductase
- HSP
Heat shock protein
- HXK
Hexokinase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LEA2
Late embryogenesis abundant protein 2
- Lhca2
Light-harvesting chlorophyll gene 2
- LTP8
Lipid-transfer protein 8
- MAPKs
Mitogen-activated protein kinases
- MYB86
MYB transcription factor86
- NA
No-acclimation
- NAC
NAC domain transcription factor
- P5CR
Proline-5-carboxylate reductase
- P5CS
Proline-5-carboxylate synthetase
- PBS
Serine/threonine-protein kinase PBS
- PEI
Pectinesterase inhibitors
- PRP
Proline-rich protein
- REC
Relative electrical conductivity
- ROS
Reactive oxygen species
- SAD
Stearoy-l ACP desaturase
- SCZ
CSS cv. Shuchazao
- SnRK2.8
SNF1-related protein kinase 2.8
- SOD
Superoxide dismutase
- SPS
Sucrose phosphate synthase
- SUS
Sucrose synthase
- TLP
Thaumatin-like protein
- TPS
Trehalose phosphate synthase
- WRKY
WRKY transcription factor
- YH9
CSA cv. Yinghong9
- ZAT
Zinc finger protein
- ZEP
Zeaxanthin epoxidase
Authors’ contributions
YL, and XW designed the study. YL, QB, XZ and CJ conducted the experiments. XW, YL and JLB. analyzed the data. XW, YL. JLB. and CW. prepared the manuscript. All authors consent to the manuscript.
Funding
This study was financially supported by Key R&D Program of China (Project No. 2018YFD1000601), Natural Science Foundation of China (Project No. 31270729), Special Project for Central Guiding Science and Technology Innovation of Region in Anhui Province (Project No. 2016080503B024) and Changjiang Scholars and Innovative Research Team in University (Project No. IRT1101). The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The data sets supporting the results of this article are available in the NCBI’s.
Sequence Read Archive (SRA) database (accession number: SRP108833.
https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP108833&go=go).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yeyun Li, Xuewen Wang and Qiuyan Ban should be regarded as joint first authors.
Contributor Information
Yeyun Li, Email: lyy@ahau.edu.cn.
Xuewen Wang, Email: xwwang@uga.edu.
Qiuyan Ban, Email: 807071355@qq.com.
Xiangxiang Zhu, Email: zhuxiangxiang0222@163.com.
Changjun Jiang, Email: jiangcj249@163.com.
Chaoling Wei, Email: weichl@ahau.edu.cn.
Jeffrey L. Bennetzen, Email: maize@uga.edu
References
- 1.Yadav SK. Cold stress tolerance mechanisms in plants. A review. Agron Sustain Dev. 2011;30:515–527. [Google Scholar]
- 2.Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 3.Baier Margarete, Bittner Andras, Prescher Andreas, van Buer Jörn. Preparing plants for improved cold tolerance by priming. Plant, Cell & Environment. 2018;42(3):782–800. doi: 10.1111/pce.13394. [DOI] [PubMed] [Google Scholar]
- 4.Li PH, Weiser CJ, Huystee RV. The relation of cold resistance to the status of phosphorus and certain metabolites in red-osier dogwood (cornus stolonifera michx.) Plant Cell Physiol. 1966;7(3):475–484. [Google Scholar]
- 5.Thomson D, Thomson R, Thompson ET. Immunization by the oral route in respiratory infections: with special reference to influenza, colds, and their complications. Brit Med J. 1936;1(3918):258. doi: 10.1136/bmj.1.3918.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng W, Casao MC, Wang P, Sato K, Hayes PM, Finnegan EJ, et al. Direct links between the vernalization response and other key traits of cereal crops. Nat Commun. 2015;6:5882. doi: 10.1038/ncomms6882. [DOI] [PubMed] [Google Scholar]
- 7.Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. P Natl Acad Sci USA. 2004;101(42):15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maksimov EG, Mironov KS, Trofimova MS, Nechaeva NL, Todorenko DA, Klementiev KE, et al. Membrane fluidity controls redox-regulated cold stress responses in cyanobacteria. Photosynth Res. 2017;133(1–3):215–223. doi: 10.1007/s11120-017-0337-3. [DOI] [PubMed] [Google Scholar]
- 9.Ma J, Wang D, She J, Li J, Zhu JK, She YM. Endoplasmic reticulum-associated N-glycan degradation of cold-upregulated glycoproteins in response to chilling stress in Arabidopsis. New Phytol. 2016;212(1):282–296. doi: 10.1111/nph.14014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Li A, Li W. How membranes organize during seed germination: three patterns of dynamic lipid remodelling define chilling resistance and affect plastid biogenesis. Plant Cell Environ. 2015;38(7):1391–1403. doi: 10.1111/pce.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14(8):1675. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wightman F. Modern Chromatographic Methods for the Identification and Quantification of Plant Growth Regulators and Their Application to Studies of the Changes in Hormonal Substances in Winter Wheat During Acclimation to Cold Stress Conditions. Boston: Springer; 1979. pp. 327–377. [Google Scholar]
- 13.Zhong JS, Robbett M, Poire A, Preston JC. Successive evolutionary steps drove Pooideae grasses from tropical to temperate regions. New Phytol. 2018;217(2):925–938. doi: 10.1111/nph.14868. [DOI] [PubMed] [Google Scholar]
- 14.Chinnusamy V, Zhu JK, Sunkar R. Gene regulation during cold stress acclimation in plants. Methods Mol Biol. 2010;639:39–55. doi: 10.1007/978-1-60761-702-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J, Lim CJ, Shen M, Park HJ, Cha JY, Iniesto E, et al. Epigenetic switch from repressive to permissive chromatin in response to cold stress. P Natl Acad Sci USA. 2018;115(23):E5400–E5409. doi: 10.1073/pnas.1721241115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hepworth J, Antoniou-Kourounioti RL, Bloomer RH, Selga C, Berggren K, Cox D, et al. Absence of warmth permits epigenetic memory of winter in Arabidopsis. Nat Commun. 2018;9(1):639. doi: 10.1038/s41467-018-03065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, Ding Y, Yang S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018;23(7):623–637. doi: 10.1016/j.tplants.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Ding ZT, Ma QP, Wang Y. The differences between two tea varieties in their response to natural cold conditions. J Hortic Sci Biotechnol. 2016;91(5):506–513. [Google Scholar]
- 20.Li WQ, Wang RP, Li MY, Li LX, Wang CM, Welti R, et al. Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis thaliana. J Biol Chem. 2008;283(1):461–468. doi: 10.1074/jbc.M706692200. [DOI] [PubMed] [Google Scholar]
- 21.Doltchinkova V, Angelova P, Ivanova E, Djilianov D, Moyankova D, Konstantinova T, et al. Surface electric charge of thylakoid membranes from genetically modified tobacco plants under freezing stress. J Photochem Photobiol B. 2013;119(119C):22–30. doi: 10.1016/j.jphotobiol.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Hao XY, Tang H, Wang B, Yue C, Wang L, Zeng JM, et al. Integrative transcriptional and metabolic analyses provide insights into cold spell response mechanisms in young shoots of the tea plant. Tree Physiol. 2018. 10.1093/treephys/tpy038. [DOI] [PubMed]
- 23.Hao XY, Wang B, Wang L, Zeng JM, Yang YJ, Wang XC. Comprehensive transcriptome analysis reveals common and specific genes and pathways involved in cold acclimation and cold stress in tea plant leaves. Sci Hortic. 2018;240:354–368. [Google Scholar]
- 24.Wang XC, Zhao QY, Ma CL, Zhang ZH, Cao HL, Kong YM, et al. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics. 2013;14(1):415. doi: 10.1186/1471-2164-14-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vyas D, Kumar S. Tea (Camellia sinensis (L.) O. Kuntze) clone with lower period of winter dormancy exhibits lesser cellular damage in response to low temperature. Plant Physiol Bioch. 2005;43(4):383–388. doi: 10.1016/j.plaphy.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Ban QY, Wang XW, Pan C, Wang YW, Kong L, Jiang HG, et al. Comparative analysis of the response and gene regulation in cold resistant and susceptible tea plants. PLoS One. 2017;12(12):e0188514. doi: 10.1371/journal.pone.0188514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen J, Wang Y, Chen C, Ding Z, Hu J, Zheng C, et al. Metabolite profiling of tea (Camellia sinensis L.) leaves in winter. Sci Hortic. 2015;192:1–9. [Google Scholar]
- 28.Zhang Y, Zhu X, Chen X, Song C, Zou Z, Wang Y, et al. Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 2014;14(1):271. doi: 10.1186/s12870-014-0271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia EH, Zhang HB, Sheng J, Li K, Zhang QJ, Kim C, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 2017;10(6):866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zipfel C. Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol. 2008;20(1):10–16. doi: 10.1016/j.coi.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Janská A, Marík P, Zelenková S, Ovesná J. Cold stress and acclimation – what is important for metabolic adjustment? Plant Biol. 2010;12(3):395–405. doi: 10.1111/j.1438-8677.2009.00299.x. [DOI] [PubMed] [Google Scholar]
- 36.DeFalco TA, Bender KW, Snedden WA. Breaking the code: Ca2+ sensors in plant signalling. Biochem J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- 37.Reddy ASN, Ali GS, Celesnik H, Day IS. Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011;23(6):2010–2032. doi: 10.1105/tpc.111.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nejat N, Mantri N. Plant immune system. Crosstalk between responses to biotic and abiotic stresses the missing link in understanding plant Defence. Curr Issues Mol Biol. 2017;23:1–16. doi: 10.21775/cimb.023.001. [DOI] [PubMed] [Google Scholar]
- 39.Yang L, Wu KC, Gao P, Liu XJ, Li GP, Wu ZJ. GsLRPK, a novel cold-activated leucine-rich repeat receptor-like protein kinase from Glycine soja, is a positive regulator to cold stress tolerance. Plant Sci. 2014;215–216:19–28. doi: 10.1016/j.plantsci.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 40.Teige M, Scheikl E, Eulgem T, Doczi R, Ichimura K, Shinozaki K, et al. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Cristina M, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61(1):621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- 42.Danquah A, Zelicourt A, Colcombet J, Hirt H. The role of aba and mapk signaling pathways in plant abiotic stress responses. Biotechnol Adv. 2014;32(1):40–52. doi: 10.1016/j.biotechadv.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Finkelstein RR, Rock CD. Abscisic acid biosynthesis and response. Arabidopsis Book. 2013;11(11):e0166. doi: 10.1199/tab.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy A. Hormone crosstalk in plants. J Exp Bot. 2015;66(16):4853–4854. doi: 10.1093/jxb/erv339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, et al. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 2013;161(4):1783–1794. doi: 10.1104/pp.112.210773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajni P, Romit S, Pradeep S, Gopal S, Sanjay K, Ram KS. Transcriptional profiling of contrasting genotypes revealed key candidates and nucleotide variations for drought dissection in Camellia sinensis (L.) O. Kuntze. Sci Rep-UK. 2019;9:7487. doi: 10.1038/s41598-019-43925-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu ZG, Jiang W, Chen SL, Mantri N, Tao ZM, Jiang CX. Insights from the Cold Transcriptome and Metabolome of Dendrobium officinale: Global Reprogramming of Metabolic and Gene Regulation Networks during Cold Acclimation. Front Plant Sci. 2016;7:1653. 10.3389/fpls.2016.01653. [DOI] [PMC free article] [PubMed]
- 49.Wang J, Yang Y, Liu XH, Huang J, Wang Q, Gu JH, et al. Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genomics. 2014;15(1):203. doi: 10.1186/1471-2164-15-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheruiyot EK, Mumera LM, Ng'Etich WK, Hassanali A, Wachira F. Polyphenols as potential indicators for drought tolerance in tea (Camellia sinensis L.) Biosci Biotechnol Biochem. 2007;71(9):2190–2197. doi: 10.1271/bbb.70156. [DOI] [PubMed] [Google Scholar]
- 51.Novillo F, Medina J, Salinas J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. P Natl Acad Sci USA. 2007;104(52):21002–21007. doi: 10.1073/pnas.0705639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakashima K, Yamaguchi-Shinozaki K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol Plantarum. 2006;126:62–71. [Google Scholar]
- 53.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki N, Mittler R. Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Plant Physiol. 2006;126:45–51. [Google Scholar]
- 55.Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77(3):367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe M, Ayugase J. Effect of low temperature on flavonoids, oxygen radical absorbance capacity values and major components of winter sweet spinach ( Spinacia oleracea, L.) J Sci Food Agr. 2015;95(10):2095–2104. doi: 10.1002/jsfa.6925. [DOI] [PubMed] [Google Scholar]
- 57.Schulz E, Tohge T, Zuther E, Fernie AR, Hincha DK. Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ. 2015;38:1658–1672. doi: 10.1111/pce.12518. [DOI] [PubMed] [Google Scholar]
- 58.Shen J, Zhang D, Zhou L, Zhang XZ, Liao J, Duan Y, et al. Transcriptomic and Metabolomic Profiling of Camellia sinensis L. cv. 'Suchazao' Exposed to Temperature Stresses Reveals Modification in Protein Synthesis and Photosynthetic and Anthocyanin Biosynthetic Pathways. Tree Physiol. 2019. 10.1093/treephys/tpz059. [DOI] [PubMed]
- 59.Martz F, Kiviniemi S, Palva TE, Sutinen ML. Contribution of omega-3 fatty acid desaturase and 3-ketoacyl-ACP synthase II (KASII) genes in the modulation of glycerolipid fatty acid composition during cold acclimation in birch leaves. J Exp Bot. 2006;57(4):897–909. doi: 10.1093/jxb/erj075. [DOI] [PubMed] [Google Scholar]
- 60.Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. Trehalose metabolism and signaling. Annu Rev Plant Biol. 2008;59(1):417–441. doi: 10.1146/annurev.arplant.59.032607.092945. [DOI] [PubMed] [Google Scholar]
- 61.Naidu B.P., Paleg L.G., Aspinall D., Jennings A.C., Jones G.P. Amino acid and glycine betaine accumulation in cold-stressed wheat seedlings. Phytochemistry. 1991;30(2):407–409. [Google Scholar]
- 62.Kaplan F, Guy CL. Beta-amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004;135:1674–1684. doi: 10.1104/pp.104.040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smeekens S, Ma J, Hanson J, Rolland F. Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol. 2010;13(3):273–278. doi: 10.1016/j.pbi.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 64.Xiao WY, Sheen J, Jang JC. The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol. 2000;44(4):451–461. doi: 10.1023/a:1026501430422. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Xin H, Guo J, Gao Y, Liu C, Dai D. Genome-wide screening of hexokinase gene family and functional elucidation of HXK2 response to cold stress in Jatropha curcas. Mol Biol Rep. 2019;46:1649–1660. doi: 10.1007/s11033-019-04613-0. [DOI] [PubMed] [Google Scholar]
- 66.Hare PD, Cress WA, Staden JV. Proline synthesis and degradation: a model system for elucidating stress-related signal transduction. J Exp Bot. 1999;50:413–434. [Google Scholar]
- 67.Pospíšil P. Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front Plant Sci. 2016;7:1950. doi: 10.3389/fpls.2016.01950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He L, Nada K, Kasukabe Y, Tachibana S. Enhanced susceptibility of photosynthesis to low-temperature Photoinhibition due to interruption of chill-induced increase of S-Adenosylmethionine decarboxylase activity in leaves of spinach (Spinacia oleracea L.) Plant Cell Physiol. 2002;43(2):196–206. doi: 10.1093/pcp/pcf021. [DOI] [PubMed] [Google Scholar]
- 69.Szalai G, Majláth I, Pál M, Gondor OK, Rudnóy S, Oláh C, et al. Janus-faced nature of light in the cold acclimation processes of maize. Front Plant Sci. 2018;9:850. doi: 10.3389/fpls.2018.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W, Xiang F, Zhong M, Zhou L, Liu H, Li S, et al. Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis) Sci Rep. 2017;7:1693. doi: 10.1038/s41598-017-01949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y, Dong J, Bennetzen JL, Zhong M, Yang J, Zhang J, et al. Integrating transcriptome and microRNA analysis identifies genes and microRNAs for AHO-induced systemic acquired resistance in N. tabacum. Sci Rep. 2017;7(1):12504. doi: 10.1038/s41598-017-12249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS. an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue):W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of RNA-seq, assembly and annotation. (DOCX 25 kb)
Table S2. Differentially expressed genes and functions in photosynthesis. (DOCX 22 kb)
Figure S1. RT-qPCR validation of selected DEG. (DOCX 452 kb)
Table S3. Differentially expressed genes related to signal transduction in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 22 kb)
Table S4. Differentially expressed transcription factors (TFs) in response to cold treatment of SCZ and YH 9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 28 kb)
Table S5. Differentially expressed cold response genes in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 21 kb)
Table S6. Differentially expressed genes related to free radical scavengers and flavonoid in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 19 kb)
Table S7. Differentially expressed genes related to fatty acid in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 13 kb)
Table S8. Differentially expressed genes related to osmoprotectans in response to cold treatment of SCZ and YH9 leaves. Significant (P < 0.05) decrease of transcript abundance is highlighted in blue and bold, significant transcript increase is highlighted in red and bold. (XLSX 15 kb)
Table S9. Primers used in the RT-qPCR. (DOCX 23 kb)
Data Availability Statement
The data sets supporting the results of this article are available in the NCBI’s.
Sequence Read Archive (SRA) database (accession number: SRP108833.
https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP108833&go=go).