Abstract
Persistent neurogenesis is maintained throughout development and adulthood in the mouse olfactory epithelium (OE). Despite this, the identity and origin of different embryonic OE progenitors, their spatiotemporal induction and contribution to patterning during development, has yet to be delineated. Here, we show that the embryonic OE contains a novel nestin-expressing radial glia-like progenitor (RGLP) that is not found in adult OE, which is antigenically distinct from embryonic CNS radial glia. Nestin-cre-mediated lineage tracing with three different reporters reveals that only a subpopulation of nestin-expressing RGLPs activate “CNS-specific” nestin regulatory elements, and produce spatially restricted olfactory receptor neurons (ORNs) in zone 1 of the OE, and vomeronasal receptor neurons restricted to the VR1 zone. This dorsal-medial restriction of transgene-activating cells is also seen in the embryonic OE of Nestin-GFP transgenic mice, in which green fluorescent protein (GFP) is found in a subpopulation of GFP+Mash1+ neuronal progenitors, despite the fact that endogenous Nestin expression is found in RGLPs throughout the OE. Embryonic OE progenitors produce three biologically distinct colony subtypes in vitro, a subpopulation of which include nestin-expressing RGLPs during in vitro colony formation. When generated from Nestin-cre/ZEG mice, neurogenic colonies also produce GFP+Mash1+ progenitors and ORNs. We thus identify a novel neurogenic precursor, the RGLP of the OE and vomeronasal organ (VNO), and provide the first evidence for intrinsic differences in the origin and spatiotemporal potential of distinct progenitors during development of the OE and VNO.
Keywords: radial glia, neural progenitors, Nestin-cre, olfactory progenitors, neurogenesis, ensheathing cell
Introduction
To understand the mechanisms that drive nervous system development and regeneration, we need to establish when and where specific cell types originate, understand the lineage contribution of each progenitor, and how the interaction of sequentially generated cells impacts the patterning of different nervous system regions. In the adult nervous system, regeneration and adult neurogenesis is restricted to exclusive niches, one of which is the mouse olfactory epithelium (OE) (Graziadei and Graziadei, 1979; Schwob, 2002), in which newly differentiated olfactory receptor neurons (ORNs) reintegrate into existing circuitry and maintain the sense of smell in the adult organism (Farbman, 1990; Roskams et al., 1996).
ORN precursor activation in adult OE is regulated by LIF (leukemia inhibitory factor), BMP4 (bone morphogenetic protein 4), and basic FGF, in which globose basal cells (GBCs) serve as transit amplifying neuronal precursors for olfactory receptor neurons that activate Mash1 before neurogenin1 in both development and regeneration (Cau et al., 1997; Nan et al., 2001; Calof et al., 2002; Getchell et al., 2002; Bauer et al., 2003). In contrast, horizontal basal cells (HBCs) are more quiescent than GBCs, express EGFR (epidermal growth factor receptor), are epidermal growth factor (EGF)/TGFα-responsive in vitro and in vivo, similar to stem cells in many other tissues (Farbman and Buchholz, 1996; Goldstein and Schwob, 1996; Getchell et al., 2000), and a subpopulation of selected EGF-responsive HBCs form multipotent clonal colonies in vitro (Carter et al., 2004), and demonstrate limited multipotency in vivo (Leung et al., 2007). The adult OE is developmentally laminated epithelium, with progenitors at the basal lamina, and mature neurons in the apical OE (Farbman, 1992). In contrast, the embryonic OE is highly dynamic, with multilayered highly neurogenic regions located adjacent to two-cell-layered, preneurogenic regions, containing potentially different classes of progenitor. Our spatial and temporal understanding of the interrelationship between different embryonic OE neuroglial progenitors has been stalled by a paucity of identifiable genes we can use to distinguish, and assay the potential of, individual candidate progenitors, a common problem in stem cell-bearing tissues (Weissman et al., 2001).
The application of genetic fate mapping with progenitor-specific promoters driving site-specific recombinases (like Cre) now allows us to address fundamental questions of progenitor lineage contribution more definitively than traditional methods of fate mapping (like retroviral tracing) (Joyner and Zervas, 2006). Although this approach has been valuable in evaluating progenitor contributions in the adult OE (Leung et al., 2007), these approaches have yet to be used for testing potential in the embryonic olfactory system. In the developing CNS, radial glia serve as both structural scaffolding for migrating neuroblasts and as embryonic neural progenitors (Anthony et al., 2004). Multipotent radial glia are identified by shared expression of a select group of antigens (Gotz et al., 2005), and lineage tracing using Cre driven by these radial glial gene promoters and enhancers [e.g., Nestin, brain lipid binding protein (BLBP)] has revealed their spatiotemporal lineage contribution during CNS development (Anthony et al., 2004; Imayoshi et al., 2006). Here, we test for the existence of OE-based radial glia progenitors, and rely on in vivo genetic lineage tracing combined with in vitro progenitor assays to test the neurogenic potential of embryonic nestin transgene-activating OE progenitors. We reveal the existence of a distinct neural progenitor phenotype unique to the embryonic OE, and demonstrate a previously unappreciated spatial regulation of olfactory receptor neuron and vomeronasal receptor neuron genesis developmentally.
Materials and Methods
Tissue preparation.
Adult and postnatal mice were anesthetized with Xylaket: 25 mg/ml ketamine HCl (MTC Pharmaceuticals, Cambridge, Ontario, Canada), 2.5 mg/ml xylazine (Bayer, Tarrytown, NY), 15% ethanol, 0.55% NaCl (120 mg/kg ketamine and 12 mg/kg xylazine), perfused with cold PBS and 4% paraformaldehyde (PFA) in PBS and postfixed in 4% PFA at 4°C (Carter et al. 2004). Embryos were immersion-fixed in 4% PFA overnight. The day of vaginal plug was defined as embryonic day 0.5 (E0.5). Tissues were cryoprotected in sucrose, embedded in Tissue-Tek medium (OCT; Sakura Finetek, Torrance, CA), and frozen in liquid nitrogen. Twelve-micrometer sections were stored at −20°C for subsequent analysis (MacDonald et al., 2005; Carson et al., 2006).
Immunohistochemistry.
Protocols for immunohistochemistry have been described previously (Au and Roskams, 2003; MacDonald et al., 2005; Carson et al., 2006). Sections were postfixed in 4% PFA, permeabilized in 0.1% Triton X-100/PBS, and blocked with 4% normal serum before primary antibody incubation. Secondary antibodies (1:200) used were biotin, Alexa 594, or Alexa 488 conjugated (Invitrogen, Carlsbad, CA). Before blocking, Sus4 detection required a 60 s incubation of the sections with 0.12% trypsin/EDTA (Invitrogen), followed by washing in PBS (Carson et al., 2006). Colabeling with Sus4 and proliferating cell nuclear antigen (PCNA) was performed sequentially with fixation in 4% PFA after the Sus4 primary and secondary antibodies, followed by washing and reblocking before antigen retrieval (MacDonald et al., 2005) and the PCNA primary and secondary antibodies. Nestin monoclonal antibody (rat 401 clone), but not nestin polyclonal rabbit antibody (Nestin clone 20), required antigen retrieval in 0.01 m citric acid microwaved for 10 min before blocking. Cre detection required antigen retrieval before blocking/permeabilization in 10% serum/0.1% Triton X-100/PBS. Cre antibodies were incubated in blocking solution and washed in 0.1% Triton X-100/PBS. Cre signals were amplified using the Vectastain ABC kit (Vector Laboratories, Burlingame, CA) and Amplex Red ELISA kit 2, horseradish peroxidase conjugate (Invitrogen), following the manufacturer's instructions. For codetection of antigens with Cre, tissues were reblocked in 10% serum/0.1% Triton X-100/PBS before sequential primary antibody incubation. Primary antibodies used were as follows: rabbit anti-mouse nestin (Nestin clone 20; 1:500), mouse anti-rat βIII-tubulin [neuron-specific tubulin (NST); TUJ1; 1:500] (both Covance, Princeton, NJ); mouse anti-rat nestin (rat 401 clone; 1:100) (used in Fig. 6I,K; supplemental Fig. S1C,F,I,K,M, available at www.jneurosci.org as supplemental material), mouse anti-Mash1 (1:100; BD Pharmingen, San Diego, CA); mouse anti-rat RC2 (1:100; Developmental Studies Hybridoma Bank, Iowa City, IA) (developed by M. Yamamoto, University of Tsukuba, Tsukuba, Japan), rabbit anti-Cre (1:5000; EMD Biosciences, San Diego, CA); mouse anti-PCNA (clone PC10; 1:5000), mouse anti-S100β (clone SH-B1; 1:400) (all Sigma-Aldrich, St. Louis, MO); goat anti-human Doublecortin (Dcx) (C-18; 1:200; Santa Cruz Biotechnology, Santa Cruz, CA; Millipore Bioscience Research Reagents, Temecula, CA), rabbit anti-BLBP (1:2000; Millipore Bioscience Research Reagents), anti-mouse olfactory cell adhesion molecule (OCAM) (1:400; R&D Systems, Minneapolis, MN), rabbit anti-green fluorescent protein (GFP) (1:400; Abcam, Cambridge, MA), or mouse anti-GFP (1:100; Millipore Bioscience Research Reagents). Note: All GFP panels show anti-GFP immunofluorescence, with the exception of the GFP-containing panels in Figure 7, E–N, which indicate endogenous GFP fluorescence, which was confirmed using immunoperoxidase (VIP) immunohistochemistry with anti-GFP antibodies. Gift antibodies were as follows: mouse anti-rat Sus4 (1:100) from Dr. J. Schwob (Tufts University, Boston, MA), rabbit anti-BLBP (1:2000) from N. Heintz (Howard Hughes Medical Institute, The Rockefeller University, New York, NY), goat polyclonal olfactory marker protein (OMP) (1:5000) from Frank L. Margolis (University of Maryland, Baltimore, MD), and rabbit anti-nGLAST (1:100) from J. Rothstein (The Johns Hopkins University, Baltimore, MD). Nuclei were stained with 0.5 μg/ml diaminopyridine imidazole (DAPI) and sections coverslipped in Vectashield (Vector Laboratories) for fluorescent antigens or Aquapolymount for VIP. All images were visualized with an Axioskop 2 MOT microscope (Carl Zeiss, Jena, Germany) using a SPOT camera (Diagnostic Instruments, Sterling Heights, MI) with Northern Eclipse software (Empix Imaging, Mississauga, Ontario, Canada) and compiled using Adobe Photoshop 7.0.
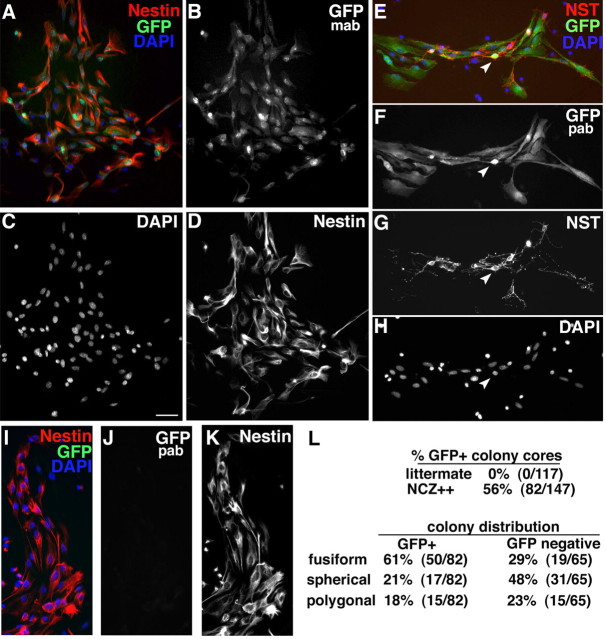
Figure 6.
E13.5 colonies from Nestin-cre/ZEG OE contain predominantly neuronal, GFP-expressing progeny. After 10 d in vitro with FGF, dissociated E13.5 OE cells from Nestin-cre/ZEG (NCZ++) mice produced primarily fusiform colonies with cells coexpressing Nestin (red) (A, D) and GFP (green; detected with monoclonal anti-GFP) (A, B). E, F, Most cells in fusiform GFP+ colony cores express GFP (green; detected with a polyclonal anti-GFP) and some coexpress βIII NST (red; arrowhead) (E, G), with cells negative for either antigen indicated by DAPI-stained (blue) nuclei (E, H). GFP-negative colony cores from NCZ++ OE (I–K) do not express GFP (I, J), but do express Nestin (I, K). L, Only NCZ++ colony cores, and not littermate negative controls, contained GFP+ cells. The majority of GFP+ colonies initiated be E13.5 progenitors were neurogenic fusiform, whereas the GFP-negative colonies contained a higher proportion of multipotent spherical colonies; n = 3–4, 117 and 147 total colonies for littermates and NCZ++, respectively. Scale bar: (in C) A–K, 50 μm.
Figure 7.
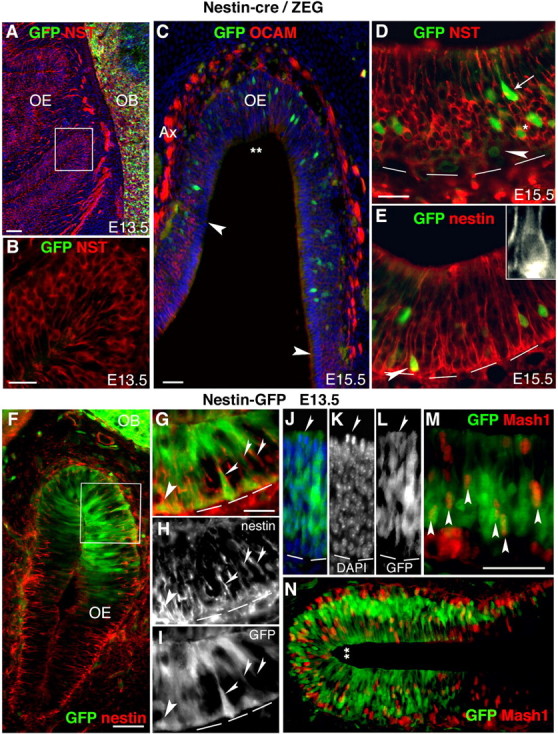
Regional restriction of GFP+ reporter cells in both embryonic Nestin-cre/ZEG mice and Nestin-GFP transgenic mice. A, In E13.5 Nestin-cre/ZEG mice, GFP (green) is readily detected in the CNS OB, but not the OE, in which NST (red) (A, B) is highly expressed in olfactory receptor neuron axons. C, First detected at E15.5 in the dorsal-medial OE and separate from emerging regions of OCAM expression (indicated by arrowheads), are the progeny of Nestin transgene-activation, GFP+ cells. D, GFP+ cells either coexpress NST (asterisk), or are NST negative, both above (arrow) and below (arrowhead) the NST+ ORNs. E, Rare GFP+ cells, close to the basal lamina, coexpress nestin (red; arrowhead; inset, nestin alone). F, E13.5 Nestin-GFP embryos highly express Nestin transgene-activated GFP that is restricted to a subpopulation of cells in the dorsal-medial OE, despite endogenous Nestin expression throughout the OE. GFP can be detected in a subpopulation of Nestin+ cells and cell processes spanning the OE (thick and thin arrowheads, respectively) (G–I), surrounding cells undergoing cytokinesis at the apical OE (arrowheads) (J–L), and in neuronal precursors expressing Mash1 (arrowheads) (M, N). A–D show anti-GFP immunofluorescence, and E–N show endogenous GFP fluorescence. Ax, Axon bundles; double asterisks indicate dorsal recess. The boxes show regions for higher magnification. The dotted lines indicate basal lamina. Scale bars: (in A, C, F, M) A, C, F, M, N, 50 μm; (in B, D, G) B, D, E, G–L, 25 μm.
Immunocytochemistry.
Immunocytochemistry was performed as described previously (Au and Roskams, 2003; Carter et al., 2004), with cells plated onto collagen:laminin (5:2 μg/cm2)-coated glass coverslips and nuclei stained with DAPI. For nuclear antigen detection, cells were fixed for 3 min in −20°C methanol and washed in PBS before blocking. The percentage of antigen-positive cells per colony was determined by counting as follows: (number of antigen-positive cells/total number of cells per colony measured by counting DAPI-stained nuclei) × 100.
Histochemistry.
LacZ histochemistry was performed on cryosections postfixed in 4% PFA, permeabilized in 0.1% Triton X-100, and washed in PBS before adding staining buffer (2 mm MgCl2, 0.01% deoxycholate, 0.02% Nonidet P-40, and 100 mm NaPO4, pH 7.3) containing 1 mg/ml X-gal, 5 mm potassium ferrocyanide, and 5 mm potassium ferricyanide. Staining proceeded at 37°C protected from light for 1–10 h. Negative controls did not demonstrate β-galactosidase staining and included CD-1 nontransgenic mice, Nestin-cre or OMP-cre transgenic mice without the ZEG transgene. Results were confirmed using antibodies to β-galactosidase; however, the detection level and reproducibility were not as good as that with histochemistry.
OE and subventricular zone cell isolation.
E13.5 OE and subventricular zone (SVZ) (for controls) were dissected from the same mice and transferred to serum-free DMEM/F12 or PBS plus 0.6% glucose, respectively (Reynolds and Weiss, 1992). Pooled OE tissue or E13.5 ganglionic emini were minced into 1 mm3 pieces, triturated 20 times with a polished Pasteur pipette, washed, filtered (40 μm), and rewashed before counting cells. Cells were counted using a hemocytometer and trypan blue exclusion, resuspended in media (0.7–3 × 105 cells/ml), and centrifuged at 100 × g for 1 min before plating, to pellet tissue aggregates.
In vitro progenitor assays.
Cells from both ganglionic emini and OE were cultured in CNS neurosphere media: Neurocult, 1× proliferation supplement (Stem Cell Technologies, Vancouver, British Columbia, Canada), 20 ng/ml growth factors (GFs) (EGF, basic FGF, or EGF plus FGF; Sigma-Aldrich), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine (Invitrogen). A single-cell suspension was determined by inspecting the cells on a hemocytometer at the time of counting and after plating, by observing the cells directly in each well. OE cells were plated at clonal density; initially into uncoated 12-well plates (Falcon; BD Biosciences Discovery Labware, Bedford, MA) to perform colony counts, and later onto collagen:laminin-coated coverslips for immunocytochemistry, in 1.5 ml of media with 0.7–2 × 105 cells/ml. SVZ cells were plated at 10–25 cells/μl, 5000–10,000 cells per well. SVZ cells derived from the same embryo as the OE cells, served as positive controls for our culture conditions and growth factor efficacy. Clonal density was first determined by mixing equal cell numbers from ubiquitous β-actin driven GFP-expressing mice (Richter et al., 2005) with GFP-nonexpressing mice to determine the density at which mostly only GFP-expressing or -nonexpressing colonies were obtained (data not shown).
Primary cultures were grown for 7–10 d in vitro (DIV) before counting nonadherent neurospheres or semiadherent colonies (OE). A neurosphere was counted if >50 μm in diameter, semiadherent colonies composed of more than eight cells were counted. Pooled OE colonies were passaged and plated at their primary cell density (or as close as possible where cell numbers were limiting) using primary culture conditions with fresh media and growth factors. Self-renewal was assessed by counting neurospheres or colonies after an additional 7–10 DIV.
Transgenic mice.
Nestin-cre mice were obtained from The Jackson Laboratory (Bar Harbor, ME) (C57BL/6) (Tronche et al., 1999) and were crossed with ZEG reporter mice obtained from C. Lobe (Sunnybrook and Women's College Health Science Centre, Toronto, Ontario, Canada) (Novak et al., 2000). Female ZEG reporters were either C57BL/6 or CD-1 strains. Nestin-cre/ZEG transgenic mice, in which Cre recombinase expression is under the control of the nestin promoter and second intron regulatory elements of the rat nestin gene (Zimmerman et al., 1994), express GFP throughout the CNS. Transgenic reporter lines Gt(ROSA)26Sortm(EYFP)Cos (The Jackson Laboratory; expressing enhanced yellow fluorescent protein from the ROSA26 locus) (Srinivas et al., 2001), produced a similar reporter expression pattern after Nestin-cre-mediated excision. An independently derived Nestin-cre line (no. 2472; FVB/N strain) crossed with a reporter, Gtrosa26tm1Sor (Soriano et al., 1999) (from R. Slack, University of Ottawa, Ottawa Health Research Institute, Ottawa, Ontario, Canada), showed strong β-galactosidase expression in the forebrain and developing CNS (Berube et al., 2005). Mice were genotyped by PCR for Cre, GFP, and/or LacZ histochemistry. Cre primers were as follows: NLS CreA, 5′-CCCGGCAAAACAGGTAGTTA-3′; NLS CreS, 5′-CATTTGGGCCAGCTAAACAT-3′ (94°C for 30 s, 55°C for 30 s, 72°C for 90 s; 454 bp product). GFP primers were as follows: XFPf, 5′-AAGTTCATCTGCACCACCG-3′; XFPr, 5′-TCCTTGAAGAAGATGGTGCG-3′ (35 cycles of 94°C for 30 s, 60°C for 60 s, 72°C for 60 s; 173 bp product; each for 35 cycles).
OMP-cre transgenic mice were generated after pronuclear injection of an 11 kb EcoR1-digested fragment, containing both 5′ and 3′ flanking regions of the OMP coding sequence (Danciger et al., 1989). The pG-ROMP plasmid, containing an 11 kb EcoRI genomic rat OMP fragment (kindly provided by Frank Margolis), was digested with AatII/AarI to remove the OMP gene, and a 1057 bp AatII/Eco31l fragment from PCR amplified NLS-Cre (plasmid kindly provided by Dr. Corrine Lobe) was subcloned into the vector. Expression and efficiency of Cre-lox recombination was verified by the specific colocalization of Cre with OMP and OMP with GFP proteins in the OE of OMP-cre/ZEG mice.
Nestin-GFP transgenic mice were obtained from Grigori Enikolopov (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) (Mignone et al., 2004) and drive expression of enhanced GFP via an identical nestin promoter (5.8 kb) and enhancer (1.8 kb second intron) to that of the Nestin-cre mice used here (Zimmerman et al., 1994; Yaworsky and Kappen, 1999). Mice were geno/phenotyped by PCR for GFP sequences and by fluorescence microscopy for the detection of GFP in the developing CNS. Schematics of Nestin-cre, Nestin-GFP, and OMP-cre constructs are shown in supplemental Figure S3 (available at www.jneurosci.org as supplemental material).
In vitro progenitor assays, Nestin-cre/ZEG mice. E13.5 embryos were geno/phenotyped by PCR for Cre, and LacZ histochemistry. All Nestin-cre/ZEG embryos (but not littermate controls) expressed readily detectable endogenous GFP in the CNS. The OE from Nestin-cre/ZEG or an equal number of littermate control embryos were pooled from each litter, in which the average number of cells/embryo was 2.6 × 105. From a total of four litters assayed, 15 of 44 embryos were Nestin-cre/ZEG. Single-cell suspensions were plated at 0.7 × 105 cells/ml into CNS neurosphere media (as above) onto collagen:laminin-coated glass coverslips in 12-well plates. Cells were processed for immunocytochemistry and counting of semiadherent colony subtypes after 10 DIV. Immunocytochemistry was performed as described above, with cells fixed in 4% PFA for 5 min and washed in PBS before blocking. Mouse monoclonal (Invitrogen) and rabbit polyclonal antibodies (Millipore Bioscience Research Reagents; both 1:100) reliably detected GFP in vitro, using GFP-expressing olfactory ensheathing cells (Richter et al., 2005) as positive controls, and littermate OE cells and secondary antibody only as negative controls.
Statistics.
Values are means ± SEM. Variance between groups was determined using ANOVA and statistical significance using Tukey's honestly significant difference post hoc test. Correlations were determined using Pearson's coefficient.
Results
The OE contains radial glia-like cells present during early development
The OE experiences three distinct phases of neurogenesis: embryonic establishment [E10 to postnatal day 0 (P0)], postnatal expansion (P1–P30), and adult maintenance (P30 to death). During each phase, the OE undergoes a dynamic reorganization of its active progenitor subcompartments both in the apical-basal and rostral-caudal planes. However, it is not known whether distinct OE zones are produced in different time windows. Understanding the nature of how different progenitors contribute to this patterning will enable us to test the regulation of these distinct phases of developmental OE neurogenesis.
During development, the majority of dividing cells are found in the most apical OE cell layer, and gradually transition to a more basal site with postnatal to adult OE maturation (Smart, 1971). This organization is highly reminiscent of the organization of embryonic CNS ventricular zone progenitors (Gotz et al., 2005), in which the apical OE would be most similar to the embryonic ventricular zone. We thus tested whether embryonic OE might contain progenitors that morphologically or antigenically resemble multipotent CNS radial glia. Nestin, an intermediate filament protein characteristic of CNS neuroepithelial stem cells (Hockfield and McKay, 1985), was detected in embryonic OE as early as E10 (Murdoch and Roskams, 2007), but was more readily detected at E13.5, in cells with a radial glia-like morphology similar to those found in embryonic olfactory bulb (OB) that were detected in every developing turbinate of the embryonic OE (Fig. 1A). Nestin detection in the embryonic CNS matches the reported radial glial expression pattern (Anthony et al., 2004), and similar patterns of nestin expression were detected in both the CNS and the OE using two independent polyclonal and monoclonal anti-nestin antibodies (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). These antibodies were also able to immunoprecipitate a single band corresponding to nestin from postnatal brain, embryonic OE, and olfactory ensheathing cells (supplemental Fig. S1, available at www.jneurosci.org as supplemental material).
Figure 1.
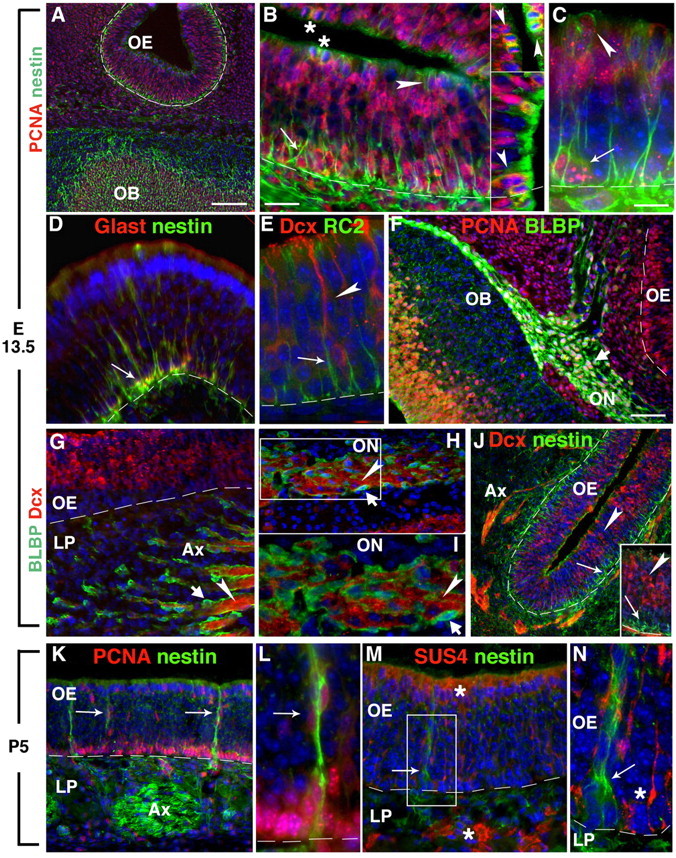
Radial glia-like progenitors in E13.5 OE express radial glia antigens. A, Nestin-expressing cells (green) are detected in the OB and OE at E13.5. B, C, Processes of nestin-expressing cells span the height of the OE and surround PCNA+ nuclei (red) from the basal (arrows) and apical OE (arrowheads). Cells undergoing cytokinesis in the apical OE (asterisk in B) are detected across the nasal cavity from each other throughout the OE (inset, arrowheads). D, Nestin is coexpressed with the glutamate transporter, GLAST (red; arrow), in OE-spanning processes, having similar morphology to RC2+ cells (green; arrow) (E), which are adjacent to cells expressing the immature neuronal antigen Dcx (red; arrowhead). F, BLBP (green; stubby arrow) is restricted to immature proliferating (PCNA+; red) OECs along the olfactory nerve (ON), and surrounding Dcx+ (arrowhead) axon bundles (G) in the underlying LP in which individual migratory Dcx+ cells (H, I) are detected in the ON (arrowheads), ensheathed by BLBP+ OECs (stubby arrows). J, Dcx+ (arrowheads) neurons do not coexpress nestin (arrows) in the OE or LP. K, L, By P5, process-bearing nestin-expressing PCNA+ (red) cells are infrequent in the OE (arrows), but nestin+ cells in the lamina propria are predominantly OECs surrounding axon bundles (Ax). M, N, Nestin (arrow) is not coexpressed with Sus4 (red; asterisk) in sustentacular cells in the OE or Bowman's gland cells in the LP. The dotted line indicates basal lamina. The boxes outline magnified area in adjacent picture. Scale bars: A, 100 μm; (in B) B, D, G, H, M, 25 μm; (in C) C, E, I, L, N, 10 μm; (in F) F, J, K, 50 μm.
At E13.5, the majority of embryonic nestin+ radial glia-like progenitors (RGLPs) coexpressed PCNA, a protein associated with the replication fork during S-phase (Fig. 1A–C) (Waseem and Lane, 1990), and were anchored at the OE basement membrane and apical surface, with nestin-rich processes spanning the OE (Fig. 1B,C). The most intense accumulation of nuclear PCNA, indicative of the peak of S-phase, was found in the vertically elongated nuclei of nestin+ RGLPs situated at the base of the OE (Fig. 1C). In contrast, nestin+ RGLPs undergoing cytokinesis segregated PCNA into their cytoplasm, and were found in clusters at the apical OE surface, most frequently located directly across opposing neurogenic epithelial surfaces (Fig. 1B). Some nestin+ RGLPs also coexpressed the glutamate transporter, GLAST (glutamate-aspartate transporter) (Fig. 1D) (Furuta et al., 1997). RC2, an antigen also detected in CNS radial glia (Misson et al., 1988), is distributed throughout a subpopulation of OE RGLPs, which form a scaffold for immature Dcx-expressing ORNs within the OE (Fig. 1E). Although Dcx is most highly enriched in migrating neuroblasts in the CNS, it is found in nonmitotic immature ORNs and their axons in the lamina propria (Fig. 1E,G,J), and neuroblast-like cells of unknown origin, encased by olfactory ensheathing cells (OECs) within the olfactory nerve (Fig. 1H,I). In contrast to CNS radial glia, BLBP, a fatty acid-binding protein associated with CNS neurogenesis (Feng et al., 1994), was not detected at any developmental stage within the OE. Instead, BLBP is highly expressed in PCNA+ cells surrounding the olfactory nerve (Fig. 1F) and in the OEC ensheathments of Dcx-positive axons in the lamina propria (Fig. 1G). Additional lineage tracing with a BLBP-cre/Rosa26 cross has revealed that reporter-positive cells are, indeed, segregated to the glial lineage of the lamina propria (Murdoch and Roskams, 2007).
At P5, the OE contained only rare isolated OE-spanning nestin+ RGLPs that coexpressed PCNA (Fig. 1K,L). These nestin-expressing cells were at no time detectable in the adult (even during neurogenesis after OE lesion) (data not shown) and at no time examined coexpressed Sus4, a marker for sustentacular cells and Bowman's glands and ducts (Fig. 1M,N). At all stages of development tested (including P5), a subpopulation of OECs surrounding axon bundles in the lamina propria (LP) expressed nestin (Fig. 1K; supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Thus, embryonic OE contains RGLPs that share some similarities with CNS radial glia, but also unique cells segregated to the glial lineage in the lamina propria, that express BLBP and nestin.
E13.5 OE forms novel semiadherent colonies containing unique progenitors in vitro
Because the E13.5 OE contains a high proportion of proliferating, nestin-expressing RGLPs in vivo compared with later developmental time windows (data not shown), we used in vitro assays of progenitor activity to test whether E13.5 OE would yield multipotent neurosphere- or colony-forming nestin-expressing cells similar to the SVZ (Mignone et al., 2004). OE-derived colonies were plated at clonal density and expanded for 10 d before counting and assay of colony composition by immunocytochemistry. The first antigenically distinguishable adherent cells expressed nestin and PCNA as early as 2.5 h in vitro, and expanded over 48 h to where most cells within developing colony cores were mitotic nestin+ cells (Fig. 2A,B). In contrast to the nonadherent neurospheres commonly obtained from embryonic SVZ, E13.5 OE formed three-dimensional semiadherent colonies that we classified into the following three distinct morphological subtypes: fusiform, polygonal, and spherical (Fig. 2C–E). Each colony subtype had a colony core (20 to >200 μm in diameter) containing cells of distinct sizes and morphologies, central to a lawn of surrounding adherent cells of multiple phenotypes. Fusiform colony cores contained loosely packed, teardrop-shaped clusters of large cells (Fig. 2C,F,G), whereas polygonal colony cores contained a higher density of compacted smaller cells, often corralled on their periphery by tightly juxtaposed, elongated bipolar cells (Fig. 2D,H,I). Spherical colonies contain the most compact cells in a raised sphere at the core of each colony, with small blast-like cells emerging on the surface of the surrounding flattened, adherent cell lawn (Fig. 2E,J).
Figure 2.
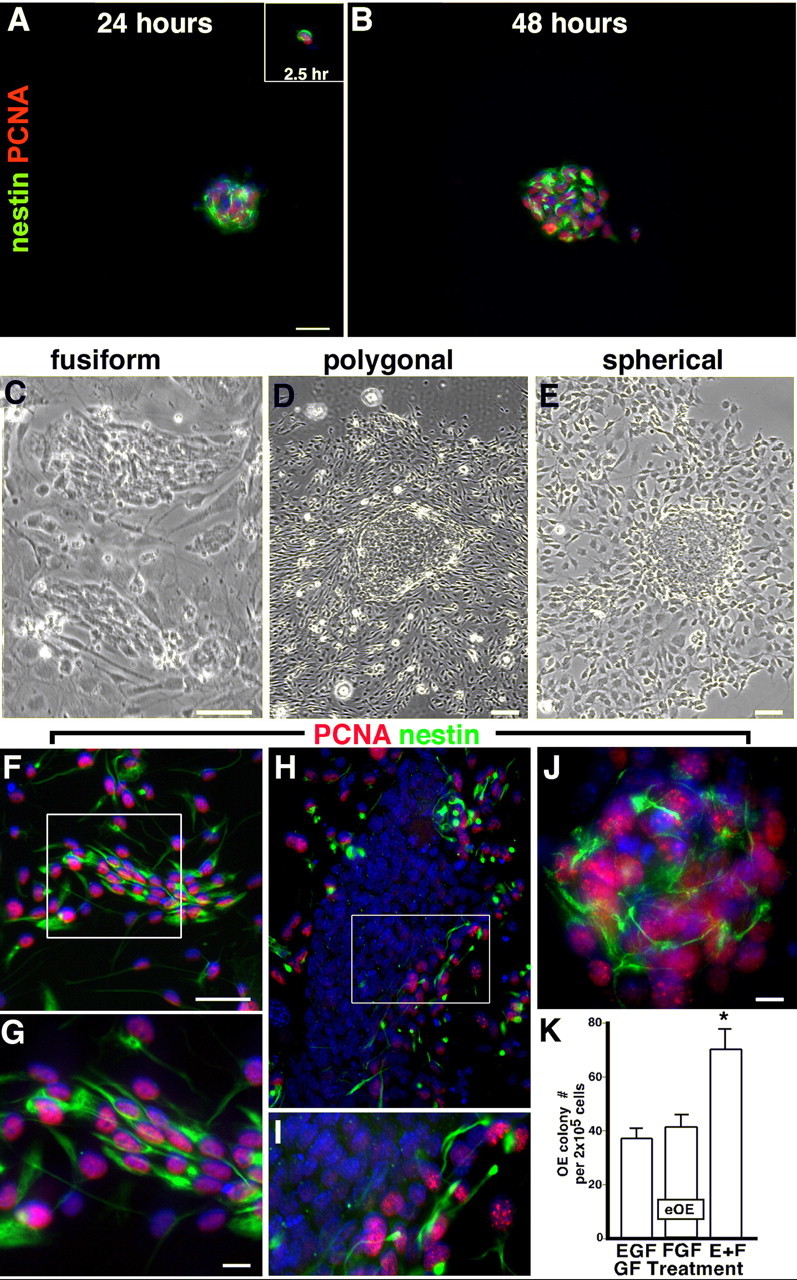
E13.5 OE forms novel semiadherent colonies in vitro containing subpopulations of dividing nestin+ progenitors. A, B, E13.5 OE cultured for 2.5 h using CNS SVZ neurosphere media with EGF, FGF, or both, demonstrated mostly (A, inset) single nestin+ cells (green) with PCNA+ nuclei (red), which divide to form larger cell clusters, not detected at 2.5 h, that retain nestin+/PCNA+ cells after 24 h (A) and 48 h (B) of culture. After 10 d in vitro, E13.5 OE produced semiadherent colonies with morphologies that are fusiform (C), polygonal (D), or spherical (E). F–J, Cores of all colony subtypes contain nestin+/PCNA+ cells in varying proportions. K, EGF plus FGF produces significantly more E13.5 OE colonies than either GF alone (*p < 0.001; n = 3, 3–6 wells per experiment). Error bars indicate SEM. The boxes show regions for higher magnification. The blue nuclear stain is DAPI. Scale bars: (in A) A, B, 100 μm; J, 80 μm; all others, 50 μm. The same magnifications are in C, F, H, and G and I. E+F, EGF plus FGF; eOE, embryonic olfactory epithelium.
By 10 d in vitro, >90% of all colonies contained bipolar, nestin-expressing cells that morphologically resemble radial glia either within, or immediately adjacent to, their colony cores (Fig. 2F–J). Fusiform colony cores contained a significantly higher percentage of nestin-expressing bipolar cells (85 ± 0.7%; p < 0.001) (Fig. 3L) compared with spherical (43 ± 2.6%) or polygonal (31 ± 3.6%) cores. In polygonal colonies, nestin-expressing cells were mostly found circling the core (Fig. 2H,I). Similarly, mitotic (PCNA+) nestin+ cells were the most abundant in fusiform, and least abundant in polygonal, colonies (Fig. 2F–J). The percentage of PCNA+ cells was significantly greater in fusiform colony cores (81 ± 0.8%) compared with either spherical (41 ± 2.8%) or polygonal (27 ± 3.2%; p < 0.001; 14–40 total colonies assayed per subtype). Nestin-expressing cells almost always coexpressed PCNA at 10 d in vitro (R2 = 0.99–0.87).
Figure 3.
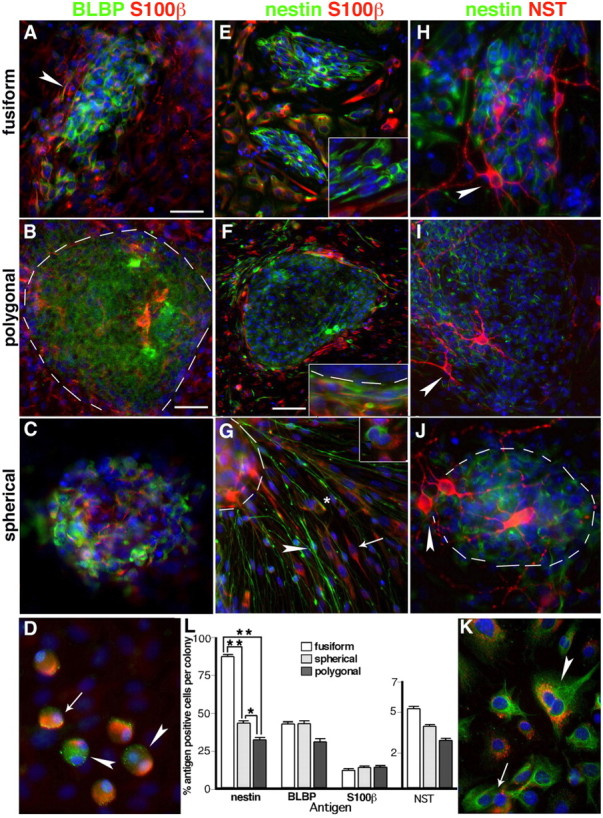
Cells in E13.5 OE colony cores and their progeny express neuronal, glial, and radial glial antigens. A–C, Fusiform, polygonal, and spherical colony cores contain cells expressing BLBP (green) and/or S100β (red). D, BLBP and S100β are asymmetrically expressed in dividing (arrowheads) and nondividing (arrow) spherical colony progeny cells. E–J, Nestin (green) is expressed in the cores of each colony subtype, and S100β (red) expressing cells (E–G) are outside colony cores and clustered at their edges (E, F, insets). G, Nestin+ cells with radial glia-like morphology (arrowhead) radiate out from a spherical colony, and are distinct from either nestin+/S100β+ (asterisk) or nestin−/S100β+ (arrow) cells with glial morphology. Dividing cells segregate nestin and S100β to individual daughter cells (G, inset). H–J, NST+ (red) nestin-negative neurons (arrowheads) are atop nestin+ cells found in all colony subtype cores. K, A dividing colony progeny cell coexpresses nestin and perinuclear NST (arrowhead), which can be distributed to individual daughter cells after division (arrow). L, The percentage of nestin, BLBP, S100β, or NST-positive cells in cores of individual colony subtypes cultured in EGF plus FGF for 10 DIV. There are significantly more nestin+ cells in fusiform colony cores compared with spherical or polygonal cores, and significantly more in spherical cores than polygonal cores (**p < 0.001, *p < 0.01; n = 3, average of 33 colonies tested per antigen). Error bars indicate SEM. The blue nuclear stain is DAPI. The dotted line indicates the edge of colony core. Scale bars: (in A) A, C–E, G, H, J, K, insets, 25 μm; (in B) B, I, 50 μm; F, 100 μm.
The frequency of E13.5 OE semiadherent colonies was only 3% of CNS neurospheres, in which 0.35% of cells from E13.5 ganglionic emini formed neurospheres, similar to previous reports (data not shown) (Hitoshi et al., 2002). Only 0.02% of E13.5 OE cells yielded colonies, with EGF and FGF together increasing colony production by 180% compared with either EGF or FGF alone (Fig. 2K). FGF specifically enhanced the production of spherical colonies (52% of all colonies in FGF alone), with the remainder of colonies equally contributed by fusiform and polygonal colonies (Table 1). FGF also enhanced the production of elongated, nestin+/S100β− radial glia-like cells from spherical colony cores, depending on density of plating (shown in Fig. 3G). Overall, colony yield was less in EGF alone, which significantly increased the production of spherical (44% of total), and polygonal colonies (40%), at the expense of fusiform colonies (only 16% of the total) (Table 1) (p < 0.001). Thus, E13.5 OE contains cells that seed highly proliferative, semiadherent colonies with three distinct morphologies and mitogen sensitivities that contain nestin+ lineage+/− cells at different frequencies. None of these colony phenotypes could be obtained, under identical conditions, from the adult OE, which instead yields slowly dividing adherent colonies (Carter et al., 2004; Murdoch and Roskams, 2007).
Table 1.
Distribution of OE E13.5 colony subtypes with FGF or EGF
| GF | Colony subtype |
||
|---|---|---|---|
| Spherical (%) | Fusiform (%) | Polygonal (%) | |
| FGF | 52 ± 6 | 25 ± 5 | 23 ± 2 |
| EGF | 44 ± 4* | 16 ± 2 | 40 ± 4* |
Values are means ± SEM. E13.5 colony subtypes were assessed after 10 d in vitro in serum-free cultures with basic FGF or EGF. In EGF, spherical and polygonal colonies are significantly greater than fusiform.
*p < 0.001; n = 5–6 independent experiments.
Cells derived from E13.5 OE colonies express neuronal, glial, and radial glial antigens
To test the gliogenic and neurogenic potential of embryonic OE-derived colonies, we next tested for the expression of combinations of developmentally regulated neuronal and glial antigens found in vivo, in different colony subtypes. BLBP was primarily found in cells in the center of fusiform colony cores, and at the edges of polygonal and spherical colony cores (Fig. 3A–C). The majority of BLBP-expressing cells at 10 d in vitro did not coexpress S100β, a calcium binding protein found in glia (Au and Roskams, 2003), which was expressed by cells immediately adjacent to BLBP-expressing cells in polygonal and spherical colonies, and at the edges of fusiform colonies (Fig. 3A–C). A subpopulation of glioblast-like cells adjacent to spherical colonies coexpressed BLBP/S100β and asymmetrically distribute BLBP and S100β in dividing cells (Fig. 3D).
Fusiform colonies primarily contain cells expressing nestin, with rare peripheral cells expressing the glial and neuronal lineage markers S100β and βIII NST (Fig. 3E,H). Lower percentages of nestin+ cells were found in polygonal and spherical colonies, compared with fusiform (Fig. 3E–J,L). A large proportion of nestin−/S100β+ cells appeared to be produced at the periphery of all colony subtypes (Fig. 3E–G). In comparison, cells radiating out from some spherical colony cores consist of nestin+ lineage-negative cells with radial glia-like morphology that are phenotypically and antigenically distinct from the underlying S100β+ cells that are larger and resemble OECs (Fig. 3G). βIII NST is first detected in nestin+ precursors, in which it can be distributed equally or asymmetrically, to daughter cells (Fig. 3K). Typically, NST+ neuronal cell bodies cluster at the edges of colony cores, with extensive processes that surround and penetrate the colony (Fig. 3H–J). Fusiform colonies contain two to three times more nestin-expressing cells and the highest percentage of NST+ cells, compared with spherical or polygonal cores (Fig. 3L). Concomitant with this, when we tested for the presence of Mash1-expressing cells, they were most frequently associated with fusiform colonies (80.8 ± 5%; three experiments, 77 total colonies counted) and spherical colonies (70.3 ± 3.8%; n = 3, 159 total colonies assayed) at a higher frequency than polygonal colonies. These data indicate the highest percentage of proliferating, nestin+ cells and neurons in neurogenic fusiform colony cores, compared with spherical (neuro/gliogenic, radial gliogenic) or polygonal (mostly gliogenic) colonies.
A subpopulation of nestin-expressing RGLPs are ORN precursors
To test whether nestin+ progenitors demonstrate neurogenic potential in vivo, we crossed Nestin-cre transgenic mice, in which Cre recombinase is under the control of the “CNS-specific” regulatory elements of the 5.8 kb rat nestin promoter and 1.8 kb second intron enhancer (Zimmerman et al., 1994), with a ZEG (LacZ/enhanced GFP) reporter line (Novak et al., 2000). ZEG mice express β-galactosidase until Cre excision removes β-galactosidase and a STOP transcription signal, allowing for GFP expression in Cre-expressing cells and their progeny. To ensure that ORNs faithfully drive (and not silence) the ZEG transgene in a temporal or zonal manner, we generated OMP-cre transgenic mice, in which Cre expression is driven by regulatory sequences controlling mature ORN-specific expression of the OMP gene (Danciger et al., 1989). When OMP-cre mice were crossed with ZEG reporters, Cre expression was only detected in the mature ORN layer of the OE at both P14 (data not shown) and adult OE (Fig. 4A–C), but the level of expression was highly variable (Fig. 4B), where Cre was only detected in a subpopulation of OMP+ ORNs at any given time (Fig. 4C). However, excision by Cre had clearly occurred, resulting in GFP expression coincident with OMP expression in mature ORNs and vomeronasal receptor neurons (VNRNs), and throughout their axon bundles (Fig. 4D,E) (data not shown).
Figure 4.
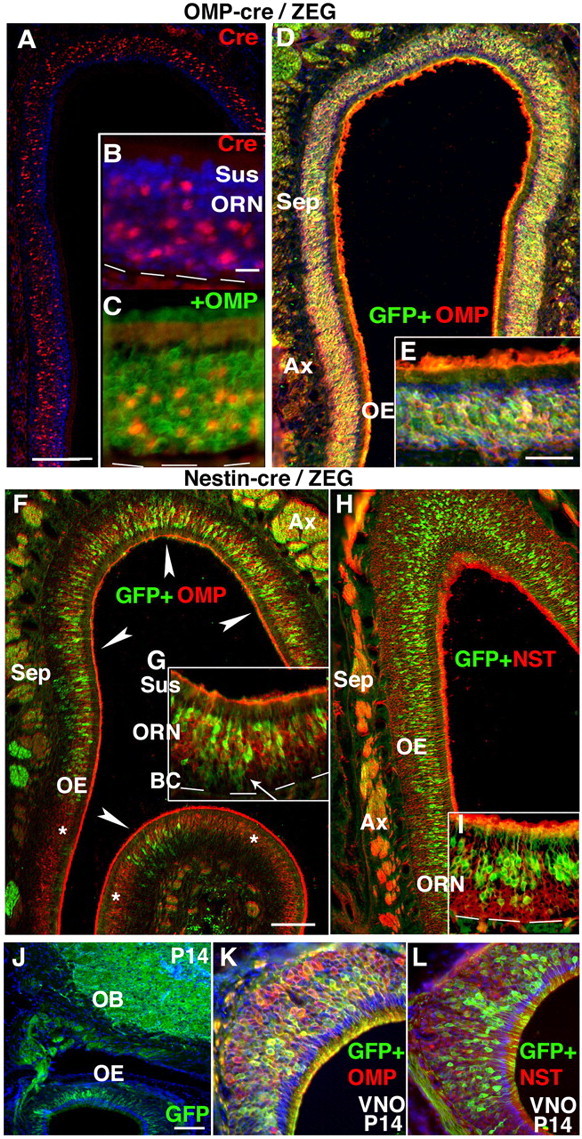
Nestin-Cre/ZEG lineage tracing reveals zonally restricted production of olfactory and vomeronasal receptor neurons. A–E, In adult OMP-cre/ZEG mice, Cre transgene expression (red) is detected in the neuronal layer throughout the OE. Cre is expressed at variable levels (B) in a subpopulation of OMP+ (green) ORNs (C). D, E, Throughout the OE, GFP reporter expression overlaps with OMP+ ORNs. F–L, Nestin-cre/ZEG mice express GFP (green) (F) in a subpopulation of ORNs in restricted zones. Most GFP+ ORNs coexpress OMP (red) (F, G), and a smaller subpopulation coexpress NST (red) (H, I). J, At P14, endogenous GFP is detected in the OB, OE, and OMP (red) (K) and NST (red) (L) in neuronal subpopulations in the VNO. The dotted line indicates the basal lamina. Asterisks indicate ORNs that do not express GFP.Sus, Sustentacular cells; Sep, septum; Ax, axon bundles; BC, basal cells. The asterisks indicate dorsal recess. Scale bars: (in A, F) A, D and F, H, respectively, 100 μm; (in B) B, C, K, L, 50 μm; (in E) E, G, I, 50 μm; J, 50 μm.
In contrast, in Nestin-cre/ZEG mouse crosses at P14 and adult, only a subpopulation of mature OMP+ ORNs expressed GFP in a spatially restricted pattern. GFP was markedly absent from many OE zones (Fig. 4F), despite widespread expression in the CNS (Fig. 4J). Serial reconstruction of adult Nestin-cre/ZEG OE revealed GFP+ ORNs are mostly restricted to the OE zone most commonly referred to as zone 1, the most dorsal-medial zone (Ressler et al., 1993) (Fig. 5B), which also contained some GFP+/OMP− cells in the adult basal progenitor and immature ORN layers (Fig. 4G). GFP was occasionally detected in a subpopulation of immature ORNs, which coexpressed NST (Fig. 4H,I). GFP expression was not detected in sustentacular cells or horizontal basal cells of the OE, or OECs or Bowman's glands of the lamina propria of Nestin-cre/ZEG mice (Fig. 4F–I) (data not shown). GFP expression was also more readily detected in a distinct subset of OMP+ and NST+ neurons of the vomeronasal organ (VNO), restricted to zones consistent with the nGi/V1R VNO subregion (Fig. 4K,L). This OE and VNO GFP expression pattern was consistent when C57BL/6 Nestin-cre mice were crossed with ZEG reporters from either C57BL/6 or CD-1 strains.
Figure 5.
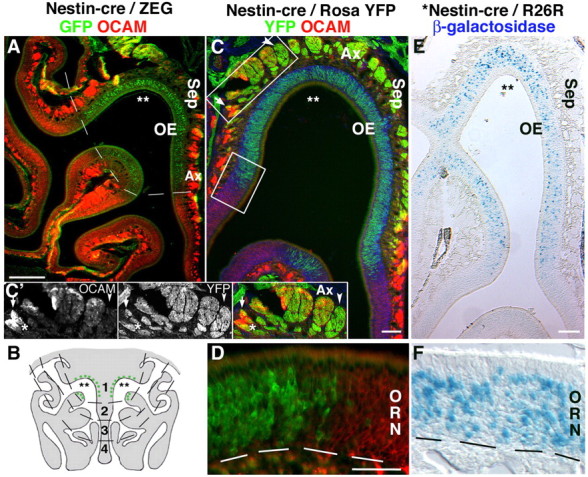
Nestin regulatory elements direct reporter expression to a subpopulation of cells in the OCAM-negative dorsal-medial zone. A, Nestin-cre/ZEG and Nestin-cre/Rosa YFP (C, D) mice express GFP/YFP (green) in the dorsal-medial OE, zone 1 (zones 1–4 indicated in B), devoid of OCAM expression (red) (A, C, D). C′, Inset, ORN axons segregate to form axon bundles (Ax) that are mostly either OCAM+ (arrows) or YFP+ (arrowheads), or OCAM+/YFP+. E, F, Identical patterns of reporter expression (β-galactosidase; blue), in a subpopulation of zone 1 neurons, are seen in an independently derived Nestin-cre line when crossed with a Rosa26R reporter mouse. Sep, Septum. The asterisks indicate dorsal recess. The dotted lines indicate zone 1 (A), zones 1–4 (B), and basal lamina (D, F). Scale bars: A, 200 μm; C, E, 50 μm; (in D) D, F, 50 μm.
To confirm the regional restriction of GFP expressing cells in the Nestin-cre/ZEG mice, we tested for coexpression with OCAM, a cell adhesion protein found throughout the OE, but excluded from zone 1 (Yoshihara et al., 1997). GFP+ ORNs in the dorsal-medial OE were devoid of OCAM expression (Fig. 5A,B), a pattern that was consistent in crosses of this Nestin-cre line with an alternative reporter line, Gt(ROSA)26Sortm(EYFP)cos (Srinivas et al., 2001), that expresses yellow fluorescent protein (YFP) from the Rosa26 locus (Fig. 5C,D). These data also clearly demonstrate a zonal segregation of axons within axon bundles of the OE, in which some axon bundles appear to derive exclusively from zone 1 (Fig. 5C), whereas others at the interface are mixed, with mesaxon groups of OCAM+/GFP− axons distinct from each other. We also crossed an independently derived Nestin-cre line in a FVB/N strain background that used identical nestin regulatory elements (Berube et al., 2005) with a Rosa26 line, and obtained reporter expression in a subpopulation of ORNs within zone 1 (Fig. 5E,F). Together, these results demonstrate that cells that can activate nestin transgene expression show a consistent and restricted pattern of ORN-specific expression in only the dorsal-medial OE that is not attributable to differences in transgene integration site, copy number, mouse line, or strain. To ensure that the ZEG transgene was not silenced in some immature olfactory receptor neurons, or in specific OE zones, β-galactosidase histochemical staining of Nestin-cre/ZEG mice detected LacZ in cells of all ORN developmental stages, in basal cells, sustentacular cells, and Bowman's glands cells, in all regions devoid of olfactory receptor neuron GFP expression (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). In addition, OMP-cre/ZEG mice showed little LacZ staining in mature ORNs, in which β-galactosidase excision had occurred, whereas sustentacular, basal, and Bowman's gland cells remained LacZ+ (supplemental Fig. S3, available at www.jneurosci.org as supplemental material).
These results indicate that, at E13.5, the OE contains regions (like zone 1) that are induced, committed, and highly neurogenic, but also contains other GFP− regions that are also highly neurogenic, and with some regions still containing multipotent progenitors that have yet to be committed. To test this, we next plated OE cells from E13.5 Nestin-cre/ZEG mice at clonal density, to test whether embryonic progenitors in vitro demonstrate a similar pattern of neurogenic potential to that observed in vivo (Fig. 6). Because of the low level of expression of endogenous GFP in some cells, we used anti-GFP immunodetection to enhance GFP detection. After 10 d in vitro, 56% of all colonies derived from Nestin-cre/ZEG mice contained GFP+ cells (Fig. 6A,B,E,F,L). All fusiform colonies were neurogenic, and all GFP+ colonies contained NST+/GFP+ cells (Fig. 6E–G). Within the GFP+ colonies, ∼90% of the cells expressed detectable levels of GFP (Fig. 6A,B,E,F), and occasional cells that appeared to be GFP− (or low) tended to be situated at the colony core. GFP+ colonies were predominantly neurogenic fusiform, containing a robust population of GFP+/nestin+ or GFP+/NST+ cells (Fig. 6A–H,L). Twenty-nine percent of GFP-negative colonies were also fusiform and contained nestin+/GFP− bipolar RGLPs (Fig. 6I–L), but the predominant GFP-negative colony phenotype (48%), presumably from progenitors situated outside zone 1, was the more multipotent spherical (Fig. 6L).
These in vitro/vivo results collectively suggest that, as early as E13.5, a subset of committed neurogenic progenitors is predestined to generate subsets of GFP+ ORNs, with other progenitors destined to seed other GFP-negative OE regions. They also indicate that the subpopulation of nestin-expressing neurogenic RGLPs that drive the second intron enhancer elements of the Nestin-cre transgene, give rise to zonally restricted ORNs and VNRNs, and are highly represented when assayed in vitro at E13.5. Despite the clear contribution of E13.5 OE progenitors to the production of GFP+ neurons in vitro, the OE of Nestin-cre/ZEG crosses at E13.5 was negative for GFP (Fig. 7A,B), with GFP+ cells first detectable at E15.5 (Fig. 7C–E). To test whether transgene-expressing precursors were present, but that GFP was not detectable because of a lag between the time taken to activate Cre and drive excision to produce GFP to a detectable level, we next used the identical nestin regulatory elements to directly drive GFP expression, and tested for coexpression and distribution of nestin+/GFP+ cells (Mignone et al., 2004). In the Nestin-GFP mice, GFP was detected throughout the E13.5 CNS, as previously shown (Fig. 7F) (data not shown), but was restricted to the dorsal-medial OE, in which it was found in a subpopulation of nestin-expressing cells, although endogenous nestin protein was evident throughout the rest of the embryonic OE (Fig. 7F–I). Because GFP is broken down inefficiently in neurons (see GENSAT) (Maskos et al., 2002; Gong et al., 2003), its retention allows us to test whether GFP+ cells that are the progeny of transgene-expressing precursors, also express markers characteristic of different ORN developmental stages. In the Nestin-GFP E13.5 OE, GFP was also found in dividing apical cells (Fig. 7J–L), a subpopulation of Mash1+ neuronal progenitors with elongated nuclei of a migratory phenotype (Fig. 7M,N), and ORNs in the dorsal-medial region. These data indicate that only a subpopulation of nestin-expressing cells activate the regulatory elements of this nestin transgene in the OE, and generate at least some GFP+ ORNs via Mash1-expressing intermediate precursors.
Discussion
The identity and spatiotemporal regulation of embryonic OE progenitors at different ages is not well understood. Here, our search for a unique embryonic OE progenitor has revealed a distinct nestin+/lineage− precursor that shares antigenic and morphologic characteristics with, but is molecularly distinct from, multipotent CNS radial glia. Nestin-cre-mediated lineage analysis demonstrates only a subset of nestin-expressing precursors of embryonic OE and VNO drive nestin transgene expression to produce ORNs and VNRNs in a zonally restricted pattern, whose neuronal restriction is recapitulated in colonies derived from E13.5 OE. These data suggest common conserved regulatory mechanisms of neurogenesis between different chemosensory neuron progenitors.
In the embryonic OE, there are distinct apical and basal subsets of progenitors that become progressively restricted to only the basal OE in the adult (Smart, 1971; Murdoch and Roskams, 2007) (Fig. 1). The enrichment of progenitors undergoing cytokinesis at the apical embryonic OE is highly reminiscent of the early embryonic cortex, in which apical OE corresponds to the ventricular zone and the basal OE to the cortical plate (Gotz et al., 2005). Embryonic OE progenitors at different mitotic stages could thus be regulated by local cues, in which mesenchymally derived neural inducers could stimulate mitosis or differentiation in basal, but not apical, progenitors (LaMantia et al., 2000), which are more likely to respond to signals within the apical OE. This highly conserved morphological organization of progenitors lead us to test whether any molecular similarities exist between the embryonic ventricular zone and OE. We found nestin was expressed by a previously unidentified population of mitotic, lineage-negative, epithelium-spanning embryonic OE cells that display many of the morphological and antigenic characteristics of CNS radial glia, including coexpression of RC2 and GLAST. We refer to these as RGLPs. A small population of RGLPs remain by P5, but are undetectable in the adult OE (Fig. 1).
Although nestin is clearly expressed by CNS progenitors, its reported expression in the OE [in the endfeet of adult rat sustentacular cells in vivo, purified lamina propria-derived OECs, basal cell lines, and OE-derived neurospheres in vitro (Doyle et al., 2001; Au and Roskams, 2003; Zhang et al., 2004)] has varied depending on animal model (rat, mouse), age, or approach used, and when considered alone, is insufficient to indicate a progenitor-like activity. In contrast to the CNS, the neurogenic radial glial protein, BLBP, is expressed only by cells in the OEC lineage that ensheathe the axons of ORNs (Fig. 1) (Murdoch and Roskams, 2007). BLBP expression could be induced by Notch 1 activation (in OEC gliogenic precursors) (Carson et al., 2006), and maintained in axon-ensheathing OECs, a hypothesis supported by lineage tracing in BLBP-cre/Rosa26 crosses (Anthony et al., 2004; Murdoch and Roskams, 2007).
Nestin is also the earliest detectable antigen in adherent cells forming colonies from E13.5 OE, in which the majority of actively cycling cells after 10 d are bipolar nestin-expressing cells similar to the RGLPs found in vivo (Fig. 2), and may represent the earliest OE progenitor identified to date. Embryonic OE yields three distinct colonies of a three-dimensional (semiadherent) phenotype. Colony heterogeneity, coupled with distinct morphological and antigen expression profiles of progeny after 10 DIV (Figs. 2, 3), suggests each colony subtype is likely formed by progenitors at different stages of induction or commitment. All colonies contain mitotic nestin+ cells, in which nestin+ cells at a distance from the colony core assume a more differentiated glial (BLBP or S100β-expressing) phenotype. Fusiform colonies contain the highest percentage of mitotic bipolar PCNA+, nestin+ cells, NST+ neurons, and Mash1+ mitotic cells (data not shown). That fusiform colony and NST+ ORN production is specifically enhanced by FGF (Table 1), a known regulator of neurogenesis in both the CNS (Vescovi et al., 1993) and OE (Calof et al., 1998), suggests that fusiform colonies contain the highest proportion of transit amplifying neurogenic precursors. Polygonal colonies preferentially respond to EGF, which specifically enhances gliogenesis developmentally (Kuhn et al., 1997; Qian et al., 2000) (Table 1), and contain a high proportion of BLBP+/S100β− expressing cells, in close proximity to expansive populations of S100β+ OEC-like cells (Au and Roskams, 2003; Carson et al., 2006). Polygonal colonies are thus more likely founded by progenitors committed to (or default toward) gliogenesis. Finally, bipotential spherical colonies expand in response to EGF and FGF together (Table 1), in which FGF specifically enhances the production of radially arrayed bipolar nestin+ RGLPs. Spherical colonies contain a high percentage of nestin+ mitotic cells, with NST+ neurons and blast-like cells of both neuronal and glial lineages loosely attached to the surface of the surrounding adherent cell layer (Figs. 2, 3). Given that spherical colonies are also mostly absent from P5 cultures, they may be formed by a population of more primitive embryonic multipotent progenitors that do not persist into the adult.
Are the nestin+ RGLPs detected in vivo, and enriched in multipotent colonies in vitro, the multipotent progenitors of the embryonic OE? If so, then Nestin-cre/ZEG lineage tracing should reveal this. Instead, Nestin-cre-expressing cells produce only ORNs regionally restricted to the OCAM-negative region classically defined as zone 1 (Ressler et al., 1993), in addition to the VR1 region of the VNO (Figs. 4, 5). This OCAM-negative Nestin transgene-expressing zone also corresponds to the dorsomedial D-zone (Oka et al., 2003; Iwema et al., 2004; Miyamichi et al., 2005; Kobayakawa et al., 2007). Although ZEG transgene silencing in other OE zones could account for this restriction, it is unlikely, because many ORNs outside zone 1 continue to express LacZ, and OMP-cre/ZEG mice demonstrate Cre-mediated excision in mature ORNs in all OE zones. Also, despite the expression of endogenous nestin in RGLPs throughout the OE (from as early as E10), the use of additional reporters and alternative Nestin-cre mice lines in a different strains consistently demonstrate nestin transgene-activating cells are restricted to the same discrete zones of the OE and VNO (Figs. 5, 7). In each of these lines, we also reveal that ORN axon targeting may have a spatiotemporal component that parallels the time course of induction, in which axons derived from GFP+ ORNs are bundled together in discrete mesaxons groups. The majority of axon bundles associated with zone 1 are composed entirely of GFP+ mesaxons, and, at the margins between zones, large axon bundles contain discrete mesaxons compartments containing axons of either GFP+ or OCAM+, but rarely both (Fig. 5).
Although GFP is easily detected throughout the CNS of E13.5 Nestin-cre/ZEG mice, the earliest time we detected OE GFP reporter expression was E15.5 (Fig. 7). This could be attributable to a lag time between transgene activation and Cre-mediated excision, followed by sufficient time for GFP accumulation (Miyoshi and Fishell, 2006). In support of this, progenitor assays (Fig. 6) indicate that the E13.5 OE of Nestin-cre/ZEG mice is highly enriched in progenitors that produce colonies containing GFP+/nestin+ progenitors and GFP+/NST+ neurons. Some neurogenic colonies from E13.5 OE were devoid of GFP+ cells and likely represent colonies derived from progenitors in the rest of the GFP-negative OE in vivo (Fig. 7). That some multipotent spherical colonies derived from E13.5 OE of Nestin-cre/ZEG mice also produce a subpopulation of GFP+ neurons (in addition to GFP-negative non-neuronal cells) suggests that spherical colonies are formed by a more hierarchical progenitor, and that even in more multipotent progenitors, this nestin transgene is committed to be driven only in a subpopulation of neuronal precursors. The parallels between in vivo and in vitro data support a model of spatiotemporal induction of different OE regions, in which nontransgene expressing colonies (Fig. 6) represent progenitors that are already induced to form other zones, or have yet to be induced.
Although it is also possible that these nestin regulatory elements are misexpressed by a subpopulation of developing ORNs, when we use the same elements to directly drive GFP, transgene-activating GFP+ cells at E13.5 are already segregated to the dorsomedial OE, some of which are mitotic, and coexpress either nestin or Mash1. This indicates that the identity of the zones that produce GFP+ ORNs/VNRNs (Figs. 4, 5) is induced before E13.5 (Fig. 7) and is distinct from the rest of the OE in terms of committed, colony-forming progenitors (Fig. 6). Although the Nestin second intron enhancer is required for CNS-specific expression, it is not sufficient for full expression in all nestin-expressing CNS or PNS progenitors (Zimmerman et al., 1994; Yaworsky and Kappen, 1999; Johansson et al., 2002; Mignone et al., 2004) and it can be differentially regulated in the CNS by class III POU domain-containing proteins (Brn 1, 2, 4) (Josephson et al., 1998). Here, the restricted activation of this nestin transgene to neuronal precursors in restricted OE/VNO zones suggests that regional subpopulations of nestin-expressing embryonic ORN/VNRN precursors may use different transcriptional mechanisms to regulate nestin expression (and expression of other zonally restricted proteins, like chemosensory receptors) in different spatiotemporal windows. One of these may be the POU domain transcription factor Brn-2, which is highly induced in a restricted manner in the dorsomedial OE from E12.5 to E14.5 (Hagino-Yamagishi et al., 1998), and may contribute to the transcriptional activation of nestin intron 2 enhancer in subpopulations of precursors in the OE and the VNO.
Here, we identify a unique chemosensory progenitor, nestin-expressing RGLPs, that may represent one of the earliest olfactory/vomeronasal progenitors identified to date. Progenitors representing different stages of neuronal or glial commitment in the embryonic OE also form self-organizing colonies in vitro in a novel assay that appears to recapitulate the spatiotemporal developmental patterning seen in vivo. A combination of transgenic mouse crosses using the same Nestin enhancer elements reveal strain-independent distinct spatial and temporal molecular variations among OE and VNO progenitor subpopulations that may share a common developmental origin, or a common mode of induction and regulation. In so doing, we also reveal that ORNs derived from reporter-expressing or OCAM-expressing zones are segregated within axon bundle subcompartments that reflect their spatiotemporal origin.
Footnotes
This work was supported by The Jack Brown Family Foundation (A.J.R.). Studentship funding was provided by the Heart and Stroke Foundation of Canada, The Canadian Stroke Network, and The Michael Smith Foundation for Health Research (B.M.). We thank the following colleagues for the reagents that made this project possible: Frank Margolis (OMP antibody and genomic OMP construct); Todd Anthony and Nat Heintz (BLBP antibody and BLBP-cre/Rosa mice); Paul Orban, Ruth Slack, and Jackie Vanderluit (Nestin-cre lines); Grigori Enikolopov (Nestin-GFP mice); Corrine Lobe (ZEG mice and NLS-Cre construct); and Jeff Rothstein (nGlast antibody). We also thank Sam Weiss and Derek van der Kooy for sharing technical information. We thank Michael Underhill, Ian Tietjen, and members of the Roskams' laboratory for critical review of this manuscript, Nicole Janzen for mouse colony maintenance, and Erin Currie for excellent technical assistance.
References
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Au E, Roskams AJ. Olfactory ensheathing cells of the lamina propria in vivo and in vitro. Glia. 2003;41:224–236. doi: 10.1002/glia.10160. [DOI] [PubMed] [Google Scholar]
- Bauer S, Rasika S, Han J, Mauduit C, Raccurt M, Morel G, Jourdan F, Benahmed M, Moyse E, Patterson PH. Leukemia inhibitory factor is a key signal for injury-induced neurogenesis in the adult mouse olfactory epithelium. J Neurosci. 2003;23:1792–1803. doi: 10.1523/JNEUROSCI.23-05-01792.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115:258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calof AL, Mumm JS, Rim PC, Shou J. The neuronal stem cell of the olfactory epithelium. J Neurobiol. 1998;36:190–205. doi: 10.1002/(sici)1097-4695(199808)36:2<190::aid-neu7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Calof AL, Bonnin A, Crocker C, Kawauchi S, Murray RC, Shou J, Wu HH. Progenitor cells of the olfactory receptor neuron lineage. Microsc Res Tech. 2002;58:176–188. doi: 10.1002/jemt.10147. [DOI] [PubMed] [Google Scholar]
- Carson C, Murdoch B, Roskams AJ. Notch 2 and Notch 1/3 segregate to neuronal and glial lineages of the developing olfactory epithelium. Dev Dyn. 2006;235:1678–1688. doi: 10.1002/dvdy.20733. [DOI] [PubMed] [Google Scholar]
- Carter LA, MacDonald JL, Roskams AJ. Olfactory horizontal basal cells demonstrate a conserved multipotent progenitor phenotype. J Neurosci. 2004;24:5670–5683. doi: 10.1523/JNEUROSCI.0330-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Danciger E, Mettling C, Vidal M, Morris R, Margolis F. Olfactory marker protein gene: its structure and olfactory neuron-specific expression in transgenic mice. Proc Natl Acad Sci USA. 1989;86:8565–8569. doi: 10.1073/pnas.86.21.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle KL, Khan M, Cunningham AM. Expression of the intermediate filament protein nestin by sustentacular cells in mature olfactory neuroepithelium. J Comp Neurol. 2001;437:186–195. doi: 10.1002/cne.1278. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Olfactory neurogenesis: genetic or environmental controls? Trends Neurosci. 1990;13:362–365. doi: 10.1016/0166-2236(90)90017-5. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Cambridge, UK: Cambridge UP; 1992. Cell biology of olfaction. [Google Scholar]
- Farbman AI, Buchholz JA. Transforming growth factor-alpha and other growth factors stimulate cell division in olfactory epithelium in vitro. J Neurobiol. 1996;30:267–280. doi: 10.1002/(SICI)1097-4695(199606)30:2<267::AID-NEU8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Furuta A, Rothstein JD, Martin LJ. Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J Neurosci. 1997;17:8363–8375. doi: 10.1523/JNEUROSCI.17-21-08363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getchell TV, Narla RK, Little S, Hyde JF, Getchell ML. Horizontal basal cell proliferation in the olfactory epithelium of transforming growth factor-alpha transgenic mice. Cell Tissue Res. 2000;299:185–192. doi: 10.1007/s004419900149. [DOI] [PubMed] [Google Scholar]
- Getchell TV, Shah DS, Partin JV, Subhedar NK, Getchell ML. Leukemia inhibitory factor mRNA expression is upregulated in macrophages and olfactory receptor neurons after target ablation. J Neurosci Res. 2002;67:246–254. doi: 10.1002/jnr.10090. [DOI] [PubMed] [Google Scholar]
- Goldstein BJ, Schwob JE. Analysis of the globose basal cell compartment in rat olfactory epithelium using GBC-1, a new monoclonal antibody against globose basal cells. J Neurosci. 1996;16:4005–4016. doi: 10.1523/JNEUROSCI.16-12-04005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB, Haubensak W, Attardo A, Denk W. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Graziadei PP, Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- Hagino-Yamagishi K, Saijoh Y, Yamazaki Y, Yazaki K, Hamada H. Transcriptional regulatory region of Brn-2 required for its expression in developing olfactory epithelial cells. Brain Res Dev Brain Res. 1998;109:77–86. doi: 10.1016/s0165-3806(98)00064-9. [DOI] [PubMed] [Google Scholar]
- Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockfield S, McKay RD. Identification of major cell classes in the developing mammalian nervous system. J Neurosci. 1985;5:3310–3328. doi: 10.1523/JNEUROSCI.05-12-03310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Ohtsuka T, Metzger D, Chambon P, Kageyama R. Temporal regulation of Cre recombinase activity in neural stem cells. Genesis. 2006;44:233–238. doi: 10.1002/dvg.20212. [DOI] [PubMed] [Google Scholar]
- Iwema CL, Fang H, Kurtz DB, Youngentob SL, Schwob JE. Odorant receptor expression patterns are restored in lesion-recovered rat olfactory epithelium. J Neurosci. 2004;24:356–369. doi: 10.1523/JNEUROSCI.1219-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson CB, Lothian C, Molin M, Okano H, Lendahl U. Nestin enhancer requirements for expression in normal and injured adult CNS. J Neurosci Res. 2002;69:784–794. doi: 10.1002/jnr.10376. [DOI] [PubMed] [Google Scholar]
- Josephson R, Muller T, Pickel J, Okabe S, Reynolds K, Turner PA, Zimmer A, McKay RD. POU transcription factors control expression of CNS stem cell-specific genes. Development. 1998;125:3087–3100. doi: 10.1242/dev.125.16.3087. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, Mori K, Sakano H. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450:503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- Kuhn HG, Winkler J, Kempermann G, Thal LJ, Gage FH. Epidermal growth factor and fibroblast growth factor-2 have different effects on neural progenitors in the adult rat brain. J Neurosci. 1997;17:5820–5829. doi: 10.1523/JNEUROSCI.17-15-05820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMantia AS, Bhasin N, Rhodes K, Heemskerk J. Mesenchymal/epithelial induction mediates olfactory pathway formation. Neuron. 2000;28:411–425. doi: 10.1016/s0896-6273(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10:720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- MacDonald JL, Gin CS, Roskams AJ. Stage-specific induction of DNA methyltransferases in olfactory receptor neuron development. Dev Biol. 2005;288:461–473. doi: 10.1016/j.ydbio.2005.09.048. [DOI] [PubMed] [Google Scholar]
- Maskos U, Kissa K, St Cloment C, Brulet P. Retrograde trans-synaptic transfer of green fluorescent protein allows the genetic mapping of neuronal circuits in transgenic mice. Proc Natl Acad Sci USA. 2002;99:10120–10125. doi: 10.1073/pnas.152266799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone JL, Kukekov V, Chiang AS, Steindler D, Enikolopov G. Neural stem and progenitor cells in nestin-GFP transgenic mice. J Comp Neurol. 2004;469:311–324. doi: 10.1002/cne.10964. [DOI] [PubMed] [Google Scholar]
- Misson JP, Edwards MA, Yamamoto M, Caviness VS., Jr Identification of radial glial cells within the developing murine central nervous system: studies based upon a new immunohistochemical marker. Brain Res Dev Brain Res. 1988;44:95–108. doi: 10.1016/0165-3806(88)90121-6. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25:3586–3592. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Fishell G. Directing neuron-specific transgene expression in the mouse CNS. Curr Opin Neurobiol. 2006;16:577–584. doi: 10.1016/j.conb.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Murdoch B, Roskams AJ. Olfactory epithelium progenitors: insights from transgenic mice and in vitro biology. J Mol Histol. 2007;38:581–599. doi: 10.1007/s10735-007-9141-2. [DOI] [PubMed] [Google Scholar]
- Nan B, Getchell ML, Partin JV, Getchell TV. Leukemia inhibitory factor, interleukin-6, and their receptors are expressed transiently in the olfactory mucosa after target ablation. J Comp Neurol. 2001;435:60–77. doi: 10.1002/cne.1193. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Oka Y, Kobayakawa K, Nishizumi H, Miyamichi K, Hirose S, Tsuboi A, Sakano H. O-MACS, a novel member of the medium-chain acyl-CoA synthetase family, specifically expressed in the olfactory epithelium in a zone-specific manner. Eur J Biochem. 2003;270:1995–2004. doi: 10.1046/j.1432-1033.2003.03571.x. [DOI] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Richter MW, Fletcher PA, Liu J, Tetzlaff W, Roskams AJ. Lamina propria and olfactory bulb ensheathing cells exhibit differential integration and migration and promote differential axon sprouting in the lesioned spinal cord. J Neurosci. 2005;25:10700–10711. doi: 10.1523/JNEUROSCI.3632-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskams AJ, Bethel MA, Hurt KJ, Ronnett GV. Sequential expression of Trks A, B, and C in the regenerating olfactory neuroepithelium. J Neurosci. 1996;16:1294–1307. doi: 10.1523/JNEUROSCI.16-04-01294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE. Neural regeneration and the peripheral olfactory system. Anat Rec. 2002;269:33–49. doi: 10.1002/ar.10047. [DOI] [PubMed] [Google Scholar]
- Smart IH. Location and orientation of mitotic figures in the developing mouse olfactory epithelium. J Anat. 1971;109:243–251. [PMC free article] [PubMed] [Google Scholar]
- Soriano P, MacGregor GR, Zambrowicz BP. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Vescovi AL, Reynolds BA, Fraser DD, Weiss S. bFGF regulates the proliferative fate of unipotent (neuronal) and bipotent (neuronal/astroglial) EGF-generated CNS progenitor cells. Neuron. 1993;11:951–966. doi: 10.1016/0896-6273(93)90124-a. [DOI] [PubMed] [Google Scholar]
- Waseem NH, Lane DP. Monoclonal antibody analysis of the proliferating cell nuclear antigen (PCNA). Structural conservation and the detection of a nucleolar form. J Cell Sci. 1990;96:121–129. doi: 10.1242/jcs.96.1.121. [DOI] [PubMed] [Google Scholar]
- Weissman IL, Anderson DJ, Gage F. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu Rev Cell Dev Biol. 2001;17:387–403. doi: 10.1146/annurev.cellbio.17.1.387. [DOI] [PubMed] [Google Scholar]
- Yaworsky PJ, Kappen C. Heterogeneity of neural progenitor cells revealed by enhancers in the nestin gene. Dev Biol. 1999;205:309–321. doi: 10.1006/dbio.1998.9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Fujita H, Hayashi H, Kagamiyama H, Mori K. OCAM: a new member of the neural cell adhesion molecule family related to zone-to-zone projection of olfactory and vomeronasal axons. J Neurosci. 1997;17:5830–5842. doi: 10.1523/JNEUROSCI.17-15-05830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Klueber KM, Guo Z, Lu C, Roisen FJ. Adult human olfactory neural progenitors cultured in defined medium. Exp Neurol. 2004;186:112–123. doi: 10.1016/j.expneurol.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Zimmerman L, Parr B, Lendahl U, Cunningham M, McKay R, Gavin B, Mann J, Vassileva G, McMahon A. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron. 1994;12:11–24. doi: 10.1016/0896-6273(94)90148-1. [DOI] [PubMed] [Google Scholar]