Abstract
The mid-hindbrain boundary (MHB) harbors an important organizing center for the adjacent brain regions. Here, we present evidence that the receptor protein tyrosine phosphatase λ (RPTPλ) is part of the complex molecular network that maintains and shapes the MHB region. RPTPλ is expressed in a tight band of cells in the caudal midbrain, anterior to the transverse ring of Wnt1 expression. Forced expression of RPTPλ across the mid-hindbrain region repressed expression of Wnt1, whereas RNA interference-mediated knock-down of RPTPλ resulted in expansion and distortion of the Wnt1 domain. When ectopically expressed in the mesencephalon, RPTPλ specifically inhibited the induction of Wnt1 expression after subsequent stimulation with Fgf8. Reduced Wnt1 expression after RPTPλ transfection correlated with a decrease in Ras- mitogen-activated protein kinase activity at the MHB. We further show that in the embryonic midbrain, RPTPλ can bind to β-catenin, a central component of the canonical Wnt signaling pathway. Overexpression of RPTPλ suppressed the activity of a β-catenin responsive promoter in the midbrain and reduced progenitor cell proliferation. Cotransfection of Wnt1 or of a stabilized form of β-catenin together with RPTPλ partially rescued the RPTPλ-mediated proliferation defect. Together, these data suggest that RPTPλ may play a dual role in the control of midbrain development: as a negative modulator of Fgf8-induced Wnt1 expression at the MHB, which may help to confine the Wnt1 domain to it characteristic tight ring at the MHB; and as an inhibitor of canonical Wnt signaling through interaction with and presumably sequestration of β-catenin.
Keywords: RPTPψ, Fgf8, Wnt1, β-catenin, midbrain, chick
Introduction
The mid-hindbrain (MHB) (isthmic) organizer, located near the constriction between the developing mesencephalic and metencephalic vesicles, acts as a secondary organizer for the control of growth and pattern formation of the adjacent midbrain and hindbrain (Nakamura et al., 1986; Martinez and Alvarado-Mallart, 1990; Martinez et al., 1991). Its activity is regulated by an interdependent network of nuclear factors and signaling proteins, which includes the transcription factors En1/En2, Pax2/Pax5, Lmx1b, Hes1/Hes3, iro1/iro7, and members of the Fgf and Wnt families of secreted proteins (Bally-Cuif et al., 1992; Rowitch and McMahon, 1995; Araki and Nakamura, 1999; Shamim et al., 1999; Adams et al., 2000; Hirata et al., 2001; Itoh et al., 2002; Matsunaga et al., 2002; O'Hara et al., 2005).
Fibroblast growth factor 8 (Fgf8), expressed in the rostral hindbrain adjacent to the MHB, is of central importance for midbrain and hindbrain development. Beads soaked in recombinant Fgf8 and implanted in prosomeres p1, p2 of the diencephalon, in the midbrain or hindbrain can mimic the inductive capacity of isthmic transplants (Crossley et al., 1996; Martinez et al., 1999; Shamim et al., 1999; Irving and Mason, 2000). Conversely, zebrafish ace mutants, which lack functional Fgf8, and mice hypomorphic for Fgf8 fail to maintain gene expression at the MHB and display massive defects in midbrain and cerebellar development (Meyers et al., 1998; Reifers et al., 1998). An important target of Fgf8 at the MHB is the secreted glycoprotein Wnt1. After isthmic organizer formation, Wnt1 expression is restricted to a narrow band of cells in the caudal mesencephalon, anterior to the Fgf8 expression domain (Bally-Cuif et al., 1995; Hollyday et al., 1995). Wnt signaling is critically involved in mid-hindbrain development, because targeted deletion or tissue-specific overexpression of Wnt1 at the MHB lead to massive perturbation of mid-hindbrain growth (Thomas and Capecchi, 1990; McMahon et al., 1992; Panhuysen et al., 2004).
Reversible protein phosphorylation on tyrosine residues is a key mechanism underlying intracellular signal transduction. Protein tyrosine phosphatases (PTPs) antagonize the activity of protein tyrosine kinases (PTKs) and thereby limit the duration and intensity of the PTK signal (Stoker and Dutta, 1998; den Hertog, 1999; Paul and Lombroso, 2003; Tonks, 2006). Receptor protein tyrosine phosphatase λ (RPTPλ; also called RPTPψ), a member of the type IIb family of receptor tyrosine phosphatases, is expressed in a spatially and temporally dynamic manner in the embryo and has been shown recently to be necessary for somitogenesis and convergent extension during gastrulation (Aerne et al., 2003; Aerne and Ish-Horowicz, 2004). We reported previously the developmental expression of RPTPλ in the CNS and have shown that its expression is particularly dynamic anterior to the isthmic constriction (Badde et al., 2005). Here, we have taken a gain-of-function and loss-of-function approach to show that RPTPλ can modulate mesencephalic development through a dual mechanism: it inhibits the induction of Wnt1 expression by Fgf8, which may help to restrict the Wnt1 domain to its characteristic tight ring at the MHB, and binds to β-catenin and appears to thereby negatively modulate canonical Wnt signaling.
Materials and Methods
Expression constructs and in ovo electroporation.
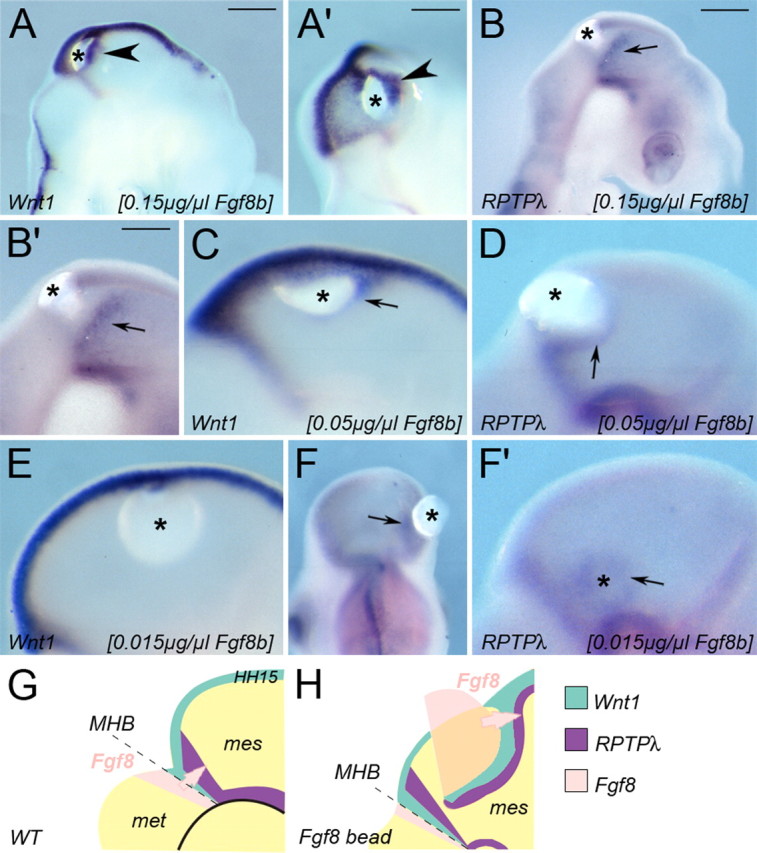
cDNAs encoding full-length mouse RPTPλ [National Center for Biotechnology Information (NCBI) accession number U55057] or RPTPλ-ΔIC (corresponding to nucleotides 352–2701 of NCBI accession number U55057) were fused to a triple HA-tag and cloned into the expression vector pMES, which includes an internal ribosomal entry site (IRES)-green fluorescent protein (GFP) cassette (Swartz et al., 2001) or into the vector pMIWIII, which lacks IRES-GFP (Suemori et al., 1990). The coding sequence of chick RPTPλ was cloned using degenerate primers based on published sequences of the mouse, rat, and human orthologs and found to be mostly identical to NCBI accession number AY147868 (sequences are available on request). The coding sequences of chick Fgf8b, mouse Wnt1, or mouse En1 were cloned into pMIWIII. For dnRasN17-pMES, a dominant-negative form of the Ras protein, which contains a serine to asparagine mutation at residue 17, was inserted into pMES (Clontech). Axin-pMES carried the full-length coding region of chick Axin, a generous gift from F. Costantini (Columbia University Medical Center, New York, NY) (Zeng et al., 1997). For caβ-catenin-pMIWIII, a stabilized form of β-catenin was cloned into pMIWIII (Funayama et al., 1995). For the rescue experiment with RPTPλ and Wnt1, the coding sequence of GFP in RPTPλ-pMES was replaced by a single ClaI site (RPTPλ-pMEC). The coding region of Wnt1 was then cloned into this ClaI site resulting in RPTPλ-Wnt1-pMEC. Unless otherwise noted, 1–2 μg/μl of each construct were injected into the neural tube of Hamburger–Hamilton stage 9–10 (HH9–HH10) chick embryos (White Leghorn) (Hamburger and Hamilton, 1992). Electroporation was performed as described, except that five pulses of 10 V for 50 ms were given to facilitate DNA uptake (Schulte et al., 1999). To visualize DNA uptake, 0.5–0.8 μg/μl of plasmids expressing enhanced GFP (GFP-pMIWIII), Discosoma red fluorescent protein (dsRed-pCMV), or a nuclear version of red fluorescent protein (nRFP-pCAGGS) were coelectroporated. To control for possible unspecific effects of the procedure, pMES (which carries IRES-GFP) or GFP-pMIWIII was introduced. For consecutive electroporation into the same region of the neural tube (see Fig. 3A), GFP with or without the full-length form of RPTPλ or the catalytically inactive allele RPTPλ-ΔIC were introduced into the mesencephalic vesicle at HH9–10. Six hours after initial transfection, a time window known to be sufficient for robust transgene expression (Momose et al., 1999), an expression plasmid carrying the coding sequence of Fgf8b together with a plasmid carrying dsRed were electroporated into the same area that had received the first electroporation 6 h earlier. Only embryos that exhibited extensive coexpression of both fluorescent markers, indicating that both transgenes were targeted to the same region of the neural tube, were chosen for additional analysis. For TOP:dgfp reporter analysis, 1 μg/μl of the TOP:dgfp reporter construct, which drives expression of destabilized green fluorescent protein off the TOPFLASH promoter/enhancer (Korinek et al., 1997; Dorsky et al., 2002), were electroporated into the mesencephalic vesicle alone or in combination with 1.5 μg/μl RPTPλ-pMIWIII or 1.5 μg/μl RPTPλ-ΔIC-pMIWIII. GFP expression was monitored and documented 24 h later.
Figure 3.
Altered Wnt1 expression after RPTPλ misexpression at the MHB. A, Schematic representation of the experimental protocol. B, Double immunohistochemical labeling using antibodies against GFP (green) and the HA-epitope (red) on a frozen section through the MHB region of an embryo electroporated with RPTPλ-HA-pMIWIII and GFP-pMIWIII. HA-specific staining is concentrated to cellular plasma membranes. C, Expression of Wnt1 after misexpression of GFP. D, E, Different example of Wnt1 expression after misexpression of RPTPλ-HA together with GFP. F, Immunohistochemical analysis of RPTPλ-ΔIC-HA expression as detected by expression of the HA-fusion epitope. Expression is concentrated to cellular plasma membranes. G, Wnt1 expression after transfection with RPTPλ-ΔIC-HA. In C–E and G, Wnt1 expression is shown in dark blue, and GFP is shown in turquoise. The arrowheads in C–E and G mark the transverse Wnt1 domain at the MHB. Scale bars: B, F, 20 μm; (in C) C, D, E, G, 100 μm. IC, Isthmic constriction.
Bead implantation.
Heparin-coated acrylic beads (Sigma-Aldrich) were soaked in 0.015–0.15 mg/ml mouse recombinant Fgf8b protein (R&D Systems) overnight at 4°C, washed in sterile PBS, and split with forceps. Appropriately sized bead fragments were implanted into the lateral wall of the mesencephalic vesicle of HH11 chick embryos. The embryos were collected 24 h after bead implantation and processed for in situ hybridization.
RNA in situ hybridization.
To ensure specificity of the probe, a partial sequence of chick RPTPλ was used for all in situ hybridization experiments (Badde et al., 2005). RNA probes for cGrg4, cSprouty2, and cSef were cloned from chick HH15–20 whole head total RNA by reverse transcriptase-PCR with gene-specific primers (primer sequences are available on request). The cDNAs used to generate in situ probes for cEn1, cFgf8, cLmx1b, cOtx2, cPax2, cPax5, or cWnt1 were gifts from C. Tabin (Harvard Medical School, Boston, MA), C. Logan (University of Calgary, Calgary, Alberta, Canada), or H. Rohrer (Max Planck Institute for Brain Research, Frankfurt, Germany). In situ hybridization on whole embryos was performed as described previously (Schulte et al., 1999). For open book preparation, the mesencephalic vesicle was opened along the dorsal midline and flat-mounted. Two-color in situ hybridization was performed as described with the exception that the first transcripts were detected using 5-bromo-4-chlor-indolyl-phosphate (BCIP) and nitroblue–tetrazolium–chloride as chromophores, which results in a dark blue/purple precipitate, whereas the second transcripts were detected with BCIP alone, which results in a bright blue/turquoise precipitate (Schulte and Cepko, 2000). Specimens were photographed with a Zeiss StemiSV11 microscope and a Canon PowerShot G5 camera or with a Leica MZ12 microscope and a Leica MPS60 camera. Brightness and contrast were adjusted in Adobe Photoshop (Adobe Systems). In Figure 1, C, D, D′, I′, and I″ were false colored in Adobe Photoshop for better visual contrast.
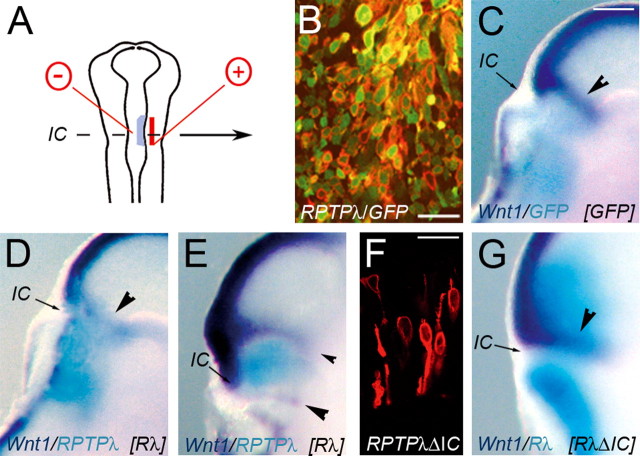
Figure 1.
Expression and regulation of RPTPλ at the MHB. A, Expression of RPTPλ in a WT chick embryo at HH15. B, Expression of Wnt1 at HH17. C, Two-color in situ hybridization showing expression of RPTPλ in dark blue and Wnt1 in turquoise at HH14. The ring of Wnt1 expression at the MHB is bordered by RPTPλ-expressing cells at its rostral side. D, D′, Open book preparation of a HH19 (E3.5) chick midbrain showing RPTPλ expression in dark blue and Wnt1 expression in turquoise. RPTPλ and Wnt1 expression occurs in tightly associated, partially overlapping domains at the MHB; anterior is to the top, and posterior is to the bottom. D′, Higher magnification of the boxed area in D. E–I″, Whole-mount in situ hybridizations on HH15–HH16 chick embryos, electroporated with different expression constructs. In all panels, detected transcripts are indicated in the bottom left corner, and misexpressed transgenes are indicated in brackets in the bottom right corner. E, Expression of RPTPλ (dark blue) was not altered after misexpression of GFP (turquoise; 1.5 μg/μl). F, Transfection of 1 μg/μl of the expression plasmid Fgf8b-pMIWIII together with GFP-pMIWIII effectively repressed RPTPλ expression at the MHB. F′, Two-color in situ hybridization on the embryo shown in F for RPTPλ (dark blue) and GFP (turquoise). G, G′, Transfection of 0.05 μg/μl of Fgf8b-pMIWIII together with GFP-pMIWIII (turquoise) induced RPTPλ expression (dark blue) in the dorsal midbrain. G′ is a higher magnification of the boxed area shown in G. H, RPTPλ expression at the MHB was lost after misexpression of the dominant-negative RasN17 together with GFP. H′, Two-color in situ hybridization on the embryo shown in H (RPTPλ, dark blue; GFP, turquoise). I, Forced expression of Fgf8b (together with GFP) in the MHB region shifted the ring of RPTPλ expression rostrally around the transfected area. The image is an overlay of the GFP fluorescence photographed immediately after harvesting of the embryo and the RPTPλ expression domain as detected subsequently by in situ hybridization (dark blue). I′, I″, Spatial relationship of RPTPλ (purple) and Wnt1 (turquoise) expression in the embryo shown in I. I″ is a higher magnification of the boxed area in I′. In all panels, upregulation of Wnt1 is indicated by arrowheads, and expression of RPTPλ (or lack thereof) is indicated by arrows. The asterisks in D mark upregulation of RPTPλ expression in the mesencephalic alar plate after HH18. C, D, D′, I′, and I″ are false colored for better visual contrast. fp, Floor plate; mes, mesencephalon; met, metencephalon. Scale bars: A, B, E–G, H–I, 500 μm; C, 250 μm.
Immunohistochemical staining.
Localization of ectopically expressed RPTPλ-HA or RPTPλ-ΔIC-HA was visualized on cryosections using a polyclonal anti-HA antibody (1:1000; Roche Diagnostics). GFP expression was confirmed using a polyclonal antibody (1:1000; Invitrogen). The mitotic index was determined with a polyclonal anti-phosphorylated Histone H3 (pH3) antibody (1:1000; Upstate Biotechnology). All antibodies were diluted in PBS containing 5% Chemiblocker (Millipore Bioscience Research Reagents) and 0.5% Triton X-100 before use. Microscope fields with a magnification of 40× were documented either with an Axioplan2 (Zeiss), a FluoView 1000 confocal microscope (Olympus), or a LSM5 Pascal confocal microscope (Zeiss). Immunohistochemical detection of diphosphorylated Erk1/2 with the monoclonal anti-dpErk antibody (clone M-8159 Sigma-Aldrich) was performed as described by Corson et al. (2003).
RNA interference.
For RNA interference (RNAi)-mediated knock-down of RPTPλ or Wnt1 expression, the psiSTRIKE-U6 Hairpin Cloning System was used (Promega). The short hairpin RNA target sequences directed against nonoverlapping regions of the cRPTPλ cytoplasmic domain were GTCAACATGACCAAAGCAA (RPTPλ_si-a) and GCTTCAAGCAGGAGTATGA (RPTPλ_si-b). The efficiency and specificity of the RNAi targeting constructs were assessed by luciferase reporter assay. Human embryonic kidney 293T (HEK293T) cells were cotransfected with the RNAi targeting constructs RPTPλ_si-a, RPTPλ_si-b, targeting constructs containing randomized sequences of RPTPλ_si-a or RPTPλ_si-b, respectively (GAACTACGTAAACCAGACA or GCATAGACGAATCGGTATG), or an unrelated targeting construct (GAAGGTACACGAATTATGT) together with a psiCHECK-2 reporter construct, which contained the intracellular portion of cRPTPλ fused to Renilla luciferase (1.5 μg of each construct/six-well plate; Promega). After 48 h, luciferase activity was measured using the Dual-Luciferase Reporter 1000 Assay System (Promega) in a GloMax96 plate Luminometer (Promega). For in vivo analysis, RPTPλ_si-a or RPTPλ_si-b was electroporated into the neural tube at a concentration of 1.5 μg/μl together with GFP-pMIWIII (0.8–0.5 μg/μl) at the developmental ages indicated. Short hairpin RNA target sequences against Wnt1 were GGTCATCTACGGCAACAAA and GCGCCTCGAGGGTCATCTA. Simultaneous transfection of both RNA targeting constructs (1.5 μg/six-well dish) repressed the activity of a psiCHECK-2 reporter construct, which contained the coding region of chick Wnt1 fused to Renilla luciferase, to 34% compared with a targeting construct carrying the randomized sequence CGAATCAGTCAGTCCAGAA (data not shown).
Immunoprecipitation.
RPTPλ-HA-pMES or RPTPλ-ΔIC-HA-pMES was electroporated into the midbrain at HH10. After 24 h, the midbrain was dissected in ice-cold PBS and lysed in 150 mm NaCl, 10 mm Na-Phosphate buffer, pH 7.2, 1% NP-40, 1% Desoxycholate, 0.1% SDS, 50 mm Na-F, 0.2 mm Na-orthovanadate, and protease inhibitors (Complete tablets; Roche Diagnostics). Precipitation of the transgenes was achieved using a monoclonal, agarose-coupled anti-HA antibody (Sigma-Aldrich). Immunoprecipitation and Western blot analysis were performed as described previously (Mühleisen et al., 2006). Primary antibodies used for Western blot analysis were rat anti-HA high-affinity antibody (1:1000; Roche Diagnostics) or polyclonal anti-β-catenin (1:1000; Upstate Biotechnology). For stripping off the first set of antibodies, the blots were incubated in 0.1 m glycine, pH 2.5, for 30 min at 37°C.
Analysis of cell proliferation and programmed cell death.
RPTPλ-HA-pMES or pMES were electroporated into the midbrain of HH9 chick embryos. After 24 h, bromodeoxyuridine (BrdU) was injected into the midbrain vesicle; labeling duration was 2 h. Detection of BrdU-positive cells was performed on 15-μm-thick cryosections using the BrdU Labeling and Detection Kit I (Roche Diagnostics) and colabeling with a GFP-specific antibody to visualize transfected cells. For analysis of programmed cell death, RPTPλ-pMIWIII together with nRFP-pCAGGS, or nRFP-pCAGGS alone, was electroporated into the midbrain vesicle. Detection of apoptotic cells was performed on cryosections using the Fluorescein In Situ Cell Death Detection Kit (Roche Diagnostics).
Results
RPTPλ is expressed in a domain tightly associated with the ring of Wnt1 expression at the MHB and is regulated by Fgf8
We reported previously that between the 19 and 40 somite stages (HH13–HH19), RPTPλ is expressed in a tight, transverse ring anterior to the isthmic constriction and to the transverse domain of Wnt1 expression (Fig. 1A,B) (Badde et al., 2005). To correlate the precise location of this ring to that of Wnt1 at the organizer, RPTPλ- and Wnt1-specific transcripts were visualized together in the same specimen by two-color in situ hybridization. Expression of the two molecules was found in tightly associated domains at HH14 (Fig. 1C) and HH19 (Fig. 1D,D′). Although both expression domains appear to slightly overlap, in all specimens analyzed, RPTPλ-specific transcripts were found rostral to the ring of Wnt1 expression that abuts the MHB. Therefore, during this developmental period, the Wnt1 domain at the MHB is bordered by a ring of RPTPλ-expressing cells at its anterior side. Based on these expression data, the RPTPλ expression domain at the MHB may lie within the future preisthmic domain, the caudal-most aspect of the mesencephalic alar plate, which is characterized by coexpression of Otx2, Pax2, and possibly Wnt1 (Hidalgo-Sánchez et al., 2005).
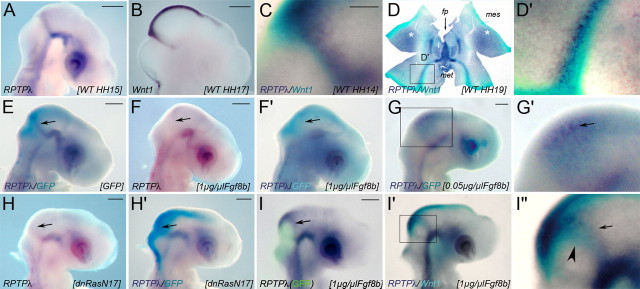
As a first step to understanding RPTPλ function at the MHB, we investigated whether its expression was regulated by the secreted protein Fgf8. Fgf8 is expressed in rhombomere 1 adjacent to the isthmic organizer and can induce organizer characteristic gene expression and cell fate changes when ectopically applied in the caudal diencephalon or mid-hindbrain region (Crossley et al., 1996; Martinez et al., 1999). Transfection of 1 μg/μl to 0.1 μg/μl of an expression plasmid carrying the splicing isoform Fgf8b into the mid-hindbrain region of chick embryos has been shown previously to induce ectopic cerebellar development, whereas transfection of 0.01 μg/μl promoted tectal growth (Sato et al., 2001). We introduced an Fgf8b expression plasmid at different concentrations into the mid-hindbrain region together with an expression plasmid carrying GFP for visualization (Fig. 1F–G′). Expression of Wnt1 and RPTPλ at the MHB were unaltered after transfection with the GFP expressing plasmid alone compared with wild-type (WT) control embryos, demonstrating that in ovo electroporation per se does not affect gene expression at the MHB (Fig. 1E) (data not shown) (n = 36 of 37 for RPTPλ; n = 12 of 12 for Wnt1). Consistent with our observation that RPTPλ is not expressed in the hindbrain region during normal development, we found that widespread electroporation across the mid-hindbrain territory of 1 μg/μl of the Fgf8b-pMIWIII expression plasmid, a concentration known to stimulate cerebellar development, effectively repressed RPTPλ expression at the MHB (n = 9 of 10) (Fig. 1F,F′). Electroporation of 0.01 μg/μl of the Fgf8b expression plasmid, a concentration that enhanced midbrain growth in a previous study (Sato et al., 2001), did not alter RPTPλ expression (n = 7 of 7) (data not shown). However, RPTPλ-specific transcripts were upregulated in the midbrain vesicle at an intermediate concentration of 0.05 μg/μl Fgf8b-pMIWIII (n = 3 of 5) (Fig. 1G,G′). Fgf8b misexpression at either concentration was unable to stimulate RPTPλ expression in the hindbrain, supporting the idea that metencephalic tissue is not permissive for RPTPλ expression (n = 18 of 18) (Fig. 1F–G′) (data not shown). Together, these results suggest that RPTPλ expression at the isthmic organizer requires a distinct Fgf8 signaling level, which is below the level required for metencephalic development but above the level that stimulates tectal growth. Fgf8 signal transduction at the MHB appears to be mediated primarily through the Ras-mitogen-activated protein (MAP) kinase pathway (Corson et al., 2003; Sato and Nakamura, 2004). To confirm the dependency of RPTPλ expression on Fgf8 signaling at the MHB, we misexpressed a dominant-negative form of Ras (dnRasN17), which was shown previously to effectively block Fgf8 signal transduction at the isthmic organizer (Sato and Nakamura, 2004). RPTPλ expression was consistently lost in cells forced to express dnRasN17, demonstrating that RPTPλ expression at the MHB indeed requires Fgf8/Ras-MAP kinase pathway activity (Fig. 1H,H′) (n = 8 of 9).
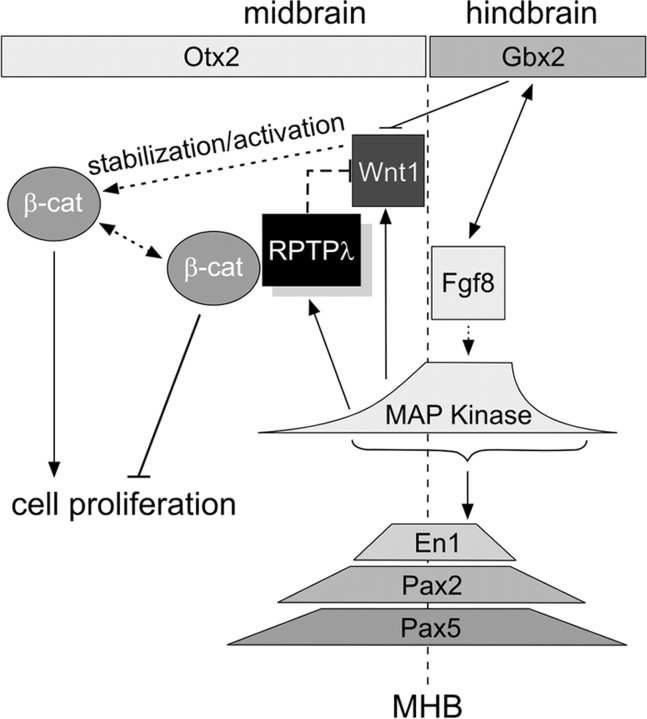
Next, we targeted the Fgf8b expression plasmid (at a concentration of 1 μg/μl) to a narrow region surrounding the isthmic organizer and analyzed the expression of RPTPλ and Wnt1 together in the same specimen 24 h later (Fig. 1I–I″). The domain of RPTPλ expression was shifted rostrally around the region targeted for Fgf8 misexpression (recognizable by the fluorescence of the coelectroporated GFP; n = 9 of 10) (Fig. 1I, arrow), consistent with the idea that RPTPλ expression is inhibited by strong Fgf8b signals but induced at a distance from the Fgf8b source, where Fgf8b protein levels are expected to be lower. When Wnt1- and RPTPλ-specific transcripts were visualized together, both expression domains were arranged in concentric circles, with the RPTPλ domain (arrow) always located anterior to the Wnt1 domain (Fig. 1I′,I″, arrowhead) (n = 6 of 6). Fgf8-induced upregulation of Wnt1 and RPTPλ thus mimics their normal spatial relationship at the MHB, where the Wnt1 domain separates the Fgf8 and RPTPλ expression domains (Badde et al., 2005) (Fig. 2G). This raises the possibility that the threshold of Fgf8 signaling differs from the induction of Wnt1 and RPTPλ expression at the MHB, and that Wnt1 expression requires higher levels of Fgf8 signaling than expression of RPTPλ. To investigate this in more detail, we narrowed down the concentrations of Fgf8 necessary for inducing Wnt1 and RPTPλ expression, respectively, by implanting fragments of beads soaked in different concentrations of Fgf8b. As reported previously, Wnt1 expression was induced around a bead fragment soaked in 0.15 mg/ml Fgf8b (Fig. 2A,A′, arrowheads) (n = 12 of 12) (Crossley et al., 1996). Induction of RPTPλ expression was also consistently observed, yet its expression domain was located at a distance to the Fgf8b soaked bead and must therefore lie distal of the ring of Wnt1 expression, which generally surrounded the Fgf8 source (Fig. 2B,B′, arrows) (n = 15 of 15). When the Fgf8b concentration was lowered to 0.05 mg/ml, some ectopic Wnt1 expression could still be observed surrounding the Fgf8 soaked bead, and the ring of RPTPλ-specific transcripts was located closer to the bead (Fig. 2C,D) (n = 5 of 8 for Wnt1; n = 7 of 7 for RPTPλ). Fgf8b at 0.015 mg/ml was not sufficient to induce Wnt1 expression (Fig. 2E) (n = 1 of 3) but could still elicit robust ectopic RPTPλ expression (Fig. 2F,F′) (n = 4 of 4). Although additional work is clearly needed to fully understand Fgf receptor signaling dynamics at the MHB, these results strongly suggest that transcriptional activation of Wnt1 and RPTPλ requires different levels of Fgf8 signaling.
Figure 2.
Fgf8-induced upregulation of Wnt1 and RPTPλ mimics their normal spatial relationship at the MHB. A′–F′, Expression of Wnt1 (A, A′, C, E) and RPTPλ (B, B′, D, F, F′) after implantation of beads soaked in different concentrations of recombinant Fgf8b. Wnt1 expression (A, A′) is ectopically upregulated in the mesencephalon closely around a partial bead soaked in 0.15 mg/ml Fgf8b, RPTPλ expression only at a distance to the bead (B, B′). A′ is a back view of the embryo shown in A. C, D, Ectopic expression surrounding beads soaked in 0.05 mg/ml Fgf8b. E, F, F′, Expression surrounding beads soaked in 0.015 mg/ml Fgf8b. In F′, the Fgf8b-releasing bead was removed for better visual clarity. G, Schematic drawing of the gene expression patterns of Fgf8, Wnt1, and RPTPλ at the MHB of a normal chick embryo (HH15). H, Schematic drawing of Wnt1 and RPTPλ expression around a Fgf8-releasing bead fragment (pink) implanted into the mesencephalic alar plate, which acts as ectopic Fgf8 source in the dorsal midbrain. The asterisks in all panels indicate the location of Fgf8b-releasing beads. Scale bar, 100 μm.
Overexpression of RPTPλ suppresses Wnt1 expression at the MHB
To examine the function of RPTPλ at the MHB, we misexpressed the murine homolog of RPTPλ in the area surrounding the isthmic constriction (Fig. 3A). By fusing the coding region of RPTPλ to a triple HA-tag, the ectopically expressed RPTPλ protein could be distinguished from its endogenous form by the use of an HA-specific antibody. As expected for a transmembrane protein, RPTPλ-HA staining was concentrated to cellular plasma membranes, suggesting that the recombinant protein was processed normally (Fig. 3B). When GFP alone was ectopically expressed in the MHB region, Wnt1 expression was unaltered (Fig. 3C) (n = 15 of 16). In contrast, after ectopic expression of RPTPλ-HA, the normal Wnt1 expression pattern was markedly disturbed (Fig. 3D,E) (n = 17 of 20). In embryos in which RPTPλ-HA was misexpressed in a broad region surrounding the isthmic constriction, Wnt1 expression was reduced or lost from the electroporated side of the embryo, suggesting that strong RPTPλ expression inhibits Wnt1 expression at the MHB (Fig. 3D, arrowhead) (n = 12 of 20). In some embryos, particularly in those where RPTPλ-HA expression was directly targeted to the organizer (and just to the region where Wnt1 would normally be expressed), a rostral displacement or split of the Wnt1 expression domain around the RPTPλ-HA expressing cells was observed (Fig. 3E, arrowheads) (n = 5 of 20). Identical results were obtained when an untagged form of the protein was misexpressed, indicating that the presence of the triple-HA tag was not critical for RPTPλ function (data not shown). To test for the possible contribution of the cytoplasmic part of RPTPλ, the domain most likely to be essential for its phosphatase activity and the interaction with components of intracellular signal transduction pathways, we constructed truncated forms of RPTPλ in which a triple HA-tag replaced the juxtamembrane and phosphatase domains (RPTPλ-ΔIC-HA). When overexpressed in the mesencephalic vesicle, RPTPλ-ΔIC-HA staining was concentrated to cellular plasma membranes, indicating correct targeting of the truncated protein (Fig. 3F). In contrast to the full-length form of RPTPλ, electroporation of RPTPλ-ΔIC-HA did not perturb Wnt1 expression (Fig. 3G) (n = 21 of 22), indicating that the cytoplasmic domain was required for RPTPλ function at the MHB.
To further characterize the potential role of RPTPλ in modulating gene expression at the MHB, we analyzed the expression of other MHB marker genes after electroporation of RPTPλ-HA. The expression domain of Fgf8 was normal 24 h after misexpression of RPTPλ (n = 6 of 6) (supplemental Fig. 1A,B, available at www.jneurosci.org as supplemental material). The altered Wnt1 expression that we observed 24 h after RPTPλ transfection can therefore not be a secondary effect of an altered Fgf8 expression profile at the MHB. In addition, forced expression of RPTPλ did not affect expression of Lmx1b (n = 16 of 17), En1 (n = 8 of 8), Pax2 (n = 9 of 11), Pax5 (n = 9 of 10), Otx2 (n = 4 of 4), Sef1 (n = 7 of 8), sprouty2 (n = 7 of 7), or Grg4 (n = 5 of 5) within 24 h after transfection (supplemental Fig. 1C–K, available at www.jneurosci.org as supplemental material) (data not shown).
RPTPλ interferes with Fgf8-mediated Wnt1 induction
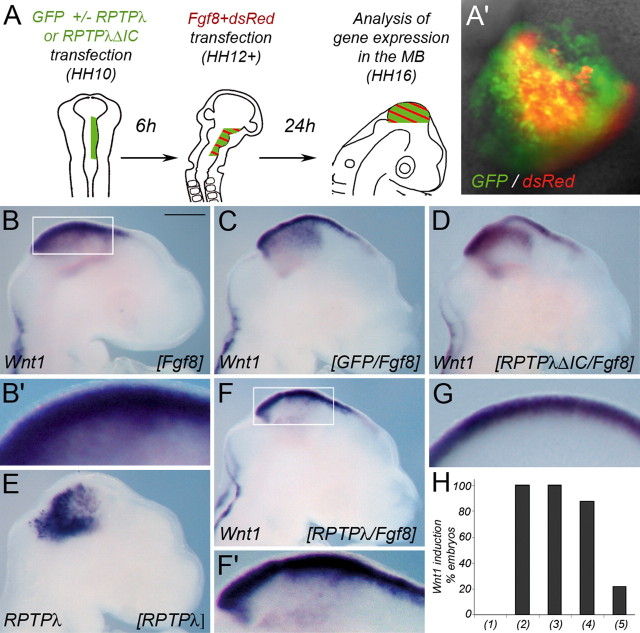
Next, we took advantage of the fact that Fgf8 can mimic the organizer's activity when ectopically expressed in adjacent brain regions and performed two consecutive rounds of in ovo electroporation within the mesencephalic alar plate (Fig. 4A,A′). Electroporation of the Fgf8b expression plasmid into an embryo without previous manipulation reproducibly led to ectopic Wnt1 expression (Fig. 4B,B′) (n = 5 of 5). Mock-transfection with GFP or dsRed did not alter Wnt1 expression (Fig. 4G). Overexpression of Fgf8b in embryos already experiencing 6 h of GFP or RPTPλ-ΔIC expression also resulted in robust upregulation of Wnt1 expression (Fig. 4C,D) (n = 5 of 5 for GFP; n = 7 of 8 for RPTPλ-ΔIC). In contrast, when full-length RPTPλ was misexpressed in the mesencephalic vesicle (Fig. 4E), only 22% of the transfected embryos exhibited robust upregulation of Wnt1 after Fgf8b electroporation (Fig. 4F,F′) (n = 4 of 18). Because overexpression of RPTPλ at the MHB had specifically perturbed the expression of Wnt1 but not that of MHB marker genes like En1, Pax2, or Pax5 (supplemental Fig. 1, available at www.jneurosci.org as supplemental material), we analyzed expression of these molecules after sequential RPTPλ/Fgf8b electroporation. In agreement with our previous results, expression of all three molecules was strongly induced by Fgf8, regardless of previous overexpression of RPTPλ (supplemental Fig. 1L–O, available at www.jneurosci.org as supplemental material) (n = 6 of 7 for En1; n = 5 of 5 for Pax2; n = 5 of 5 for Pax5). Together, these results suggest that strong RPTPλ expression specifically interferes with the ability of cells in the embryonic mesencephalon to initiate Wnt1 expression in response to Fgf8 signals. Considering that, during normal development (or in response to local Fgf8 application), RPTPλ expression always extended over the Wnt1 domain at its rostral side, our data suggest that RPTPλ may function to rostrally restrict the domain of Wnt1 expression at the MHB.
Figure 4.
Misexpression of RPTPλ interferes with transcriptional Wnt1 activation after ectopic Fgf8 expression. A, Schematic drawing of the experimental procedure. At HH10, RPTPλ plus GFP, RPTPλ-ΔIC plus GFP, or GFP alone were introduced into the right side of the midbrain vesicle. After 6 h (at HH12+), two expression plasmids carrying Fgf8 and dsRed, respectively, were electroporated into the same side of the midbrain vesicle. The embryos were allowed to develop for an additional 24 h. A′, Example of an embryo chosen for additional analysis, based on the massive coexpression of the red and green fluorescent proteins in the midbrain vesicle. B, B′, Ectopic expression of Fgf8 in the midbrain without previous manipulation leads to widespread upregulation of Wnt1. C, D, Fgf8 induced Wnt1 expression is not affected by previous introduction of GFP (C) or RPTPλ-ΔIC (D). E, Example of an embryo electroporated with RPTPλ-pMES indicating mosaic expression of the transgene. F, F′, Transcriptional upregulation of Wnt1 is mostly prevented after ectopic expression of RPTPλ 6 h before Fgf8 transgene introduction. B′, F′, Higher magnification of the boxed areas shown in B and F. G, Expression of Wnt1 in an embryo electroporated with GFP followed by misexpression of dsRed 6 h later demonstrating normal expression in the mesencephalic roof plate under these experimental conditions. H, Quantification of the results: (1), Percentage of embryos exhibiting strong ectopic expression of Wnt1 24 h after transfection with GFP or dsRed alone; (2), percentage of embryos exhibiting strong ectopic expression of Wnt1 24 h after transfection with Fgf8b (together with dsRed) without previous manipulation; (3), percentage of embryos exhibiting strong ectopic expression of Wnt1 24 h after transfection with GFP followed by Fgf8b (+dsRed); (4), percentage of embryos exhibiting strong ectopic expression of Wnt1 24 h after transfection with RPTPλ-ΔIC-HA (+GFP) followed by Fgf8b (+dsRed); (5), percentage of embryos exhibiting strong ectopic expression of Wnt1 24 h after transfection with RPTPλ-HA (+GFP) followed by Fgf8b (+dsRed). Scale bar, 500 μm.
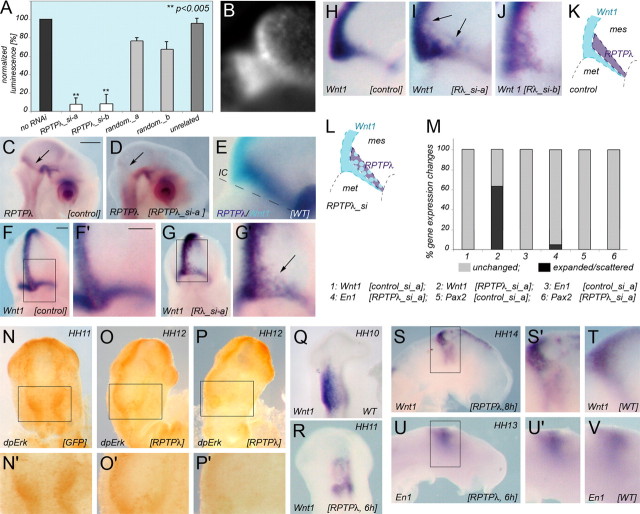
If RPTPλ expression at the MHB indeed serves to restrict Wnt1 expression, one would expect that reducing RPTPλ activity at the MHB should allow Wnt1 expression to expand anteriorly. To test this idea, two RNAi constructs targeted to nonoverlapping sequences in the intracellular domain of RPTPλ were designed. The activity and specificity of the constructs were confirmed by luciferase reporter assays compared with the effects of an RNAi construct targeted to an unrelated protein or with that of RNAi constructs containing randomized targeting sequences (Fig. 5A). Repression by unrelated or randomized sequences was negligible (Fig. 5A). Electroporation of either RPTPλ_si-a (n = 8 of 9) (Fig. 5D) or RPTPλ_si-b (n = 6 of 9) (data not shown) but not of the randomized control targeting construct random_a (n = 5 of 5) (Fig. 5C) into the MHB region at HH10 markedly reduced the endogenous expression of RPTPλ. Knock-down of RPTPλ also led to an anterior expansion of Wnt1-positive cells into the domain where RPTPλ is normally expressed (Fig. 5G–G″,I,J) (n = 9 of 14). This effect was more pronounced near the dorsal midline than in the ventral midbrain, presumably because the domain of RPTPλ expression is broader and any effect of knock-down of its expression may therefore be more profound in the dorsal midbrain (Fig. 5E,K). Wnt1 expression at the MHB, however, was not only expanded rostrally, but expression was also scattered within the rostral half of its normal domain in some embryos (Fig. 5F–J). We do not know the molecular basis for this effect at present. A possible explanation may come from the fact that RPTPλ and Wnt1 expression at the MHB appear to slightly overlap during normal development (Figs. 1C-D′, 5E). It is therefore possible that RPTPλ stabilizes the Wnt1 expression border in the small region where both proteins are normally coexpressed. In addition, because the extracellular part of RPTPλ contains multiple cell adhesion-like motives, reduced RPTPλ expression at the MHB may alter cell adhesive properties critical for the maintenance of the coherent cellular organization within its normal expression domain. It is worth pointing out that disorganization of the Wnt1 expression domain was also observed after targeted deletion of Fgf receptor 1 from the MHB territory (Trokovic et al., 2003).
Figure 5.
RNAi-mediated knock-down of RPTPλ and Ras-MAP kinase pathway activation after RPTPλ overexpression. A, Efficiency and specificity of the RNAi targeting constructs as assessed by luciferase reporter assays. Experimental details are described in Material and Methods. Renilla luciferase activity of the reporter construct psiCHECK2-RPTPλ (Promega) without cotransfection of an RNAi targeting construct served as reference and was set at 100%. Cotransfection of either RPTPλ_si-a or RPTPλ_si-b together with psiCHECK2-RPTPλ reduced Renilla luciferase activity to <10%. Cotransfection of randomized targeting sequences or of an unrelated targeting construct did not significantly affect Renilla activity. Error bars represent SD. **p < 0.005, Student's t test. B, Representative example of GFP fluorescence 24 h after transfection of RPTPλ_si-a together with GFP across the MHB. C, D, F–J, Analysis of gene expression after RNAi-mediated knock-down of RPTPλ expression. C, RPTPλ expression after transfection with the randomized RNAi targeting construct random_a. D, Expression of RPTPλ after transfection with RPTPλ_si-a. E, Two-color in situ hybridization on a wild-type embryo showing expression of RPTPλ (dark blue) and Wnt1 (turquoise). Note that the RPTPλ expression domain is broader in the dorsal than in the ventral midbrain. F, H, Expression of Wnt1 in embryos transfected with the control targeting construct random_a. G, I, J, Expression of Wnt1 after transfection with RPTPλ_si-a (G, I) or RPTPλ_si-b (J). F′ and G′ are higher magnifications of the boxed areas in F and G. K, Schematic drawing of the normal gene expression patterns of Wnt1 (turquoise) and RPTPλ (purple) at the MHB. L, Schematic drawing of Wnt1 expression after RPTPλ_si transfection. M, Quantification of the results. Gray bars indicate unchanged expression, and black bars indicate expanded and/or scattered expression of Wnt1. N–P, DpErk1/2 staining at the MHB 6 h after electroporation of GFP (N, N′) or RPTPλ (O–P′). N′, O′, and P′ are higher magnifications of the boxed areas in N, O, and P. Q–T, Wnt1 expression 6 h (R) and 8 h (S, S′) after RPTPλ transfection and in age-matched controls (Q, T). U, V, En1 expression 8 h after RPTPλ transfection (U, U′) and in an age-matched control (V). Scale bars: C, 500 μm; F, F′, 100 μm. IC, Isthmic constriction; mes, mesencephalon; met, metencephalon.
At least three signaling pathways, the Ras-MAP kinase, phosphatidyl inositol 3 kinase, and phospholipase Cγ pathway, can be activated after Fgf receptor stimulation, and of the three, the Ras-MAP kinase pathway has been demonstrated to be important for mid-hindbrain development (Corson et al., 2003; Sato and Nakamura, 2004; Dailey et al., 2005). Because signal transduction in each of these pathways requires protein phosphorylation on tyrosine residues, it is possible that RPTPλ, because of its tyrosine phosphatase activity, interrupts signal transduction through dephosphorylation of intermediate signaling components. To test whether RPTPλ impinges on Ras-MAP kinase signaling, we visualized phosphorylation of the pathway intermediate Erk1/2 with the phosphorylation-specific antibody dpErk1/2 (Corson et al., 2003). Six hours after transfection of 2 μg/μl RPTPλ-pMES but not after transfection of pMES alone, dpErk1/2 labeling intensity was slightly reduced in the right electroporated side of the electroporated embryos (Fig. 5N–P) (n = 6 of 9). Repression of Wnt1 expression by RPTPλ could also be detected 6 and 8 h after RPTPλ transfection (Fig. 5Q–T) (n = 10 of 18). In contrast to Wnt1 but in agreement with our previous results (supplemental Fig. 1, available at www.jneurosci.org as supplemental material), we did not observe any major disturbance of En1 expression 6 or 8 h after RPTPλ transfection (Fig. 5U,V) (data not shown) (n = 11 of 11). These findings indicate that the influence of RPTPλ on Wnt1 expression at the MHB may, at least in part, result from modulation of Ras-MAP kinase signaling intensity by RPTPλ.
RPTPλ interacts with β-catenin in the embryonic midbrain and inhibits the activity of a β-catenin responsive promoter
RPTPλ expression in the developing midbrain is highly dynamic during the first 4 d of chick development (Figs. 1, 6A–C) (Badde et al., 2005). Before HH12 (16 somites), RPTPλ is expressed throughout the mesencephalic vesicle (Fig. 6A) (Badde et al., 2005). Between HH12 and HH19, RPTPλ expression is lost from the midbrain alar plate but is maintained in a tight transverse domain rostral to the Wnt1 domain (Figs. 1A,C, 6B) (Badde et al., 2005). Thereafter, RPTPλ expression is induced once again in the mesencephalic vesicle and covers the entire mesencephalic alar plate from late E4 onwards, without crossing into the isthmic organizer region (Figs. 1D, 6C, arrowhead) (Badde et al., 2005). Because of this prominent “on-off” expression in the midbrain alar plate, we speculated that RPTPλ may contribute to midbrain development by mechanisms in addition to restricting Wnt1 expression at the MHB organizer.
Figure 6.
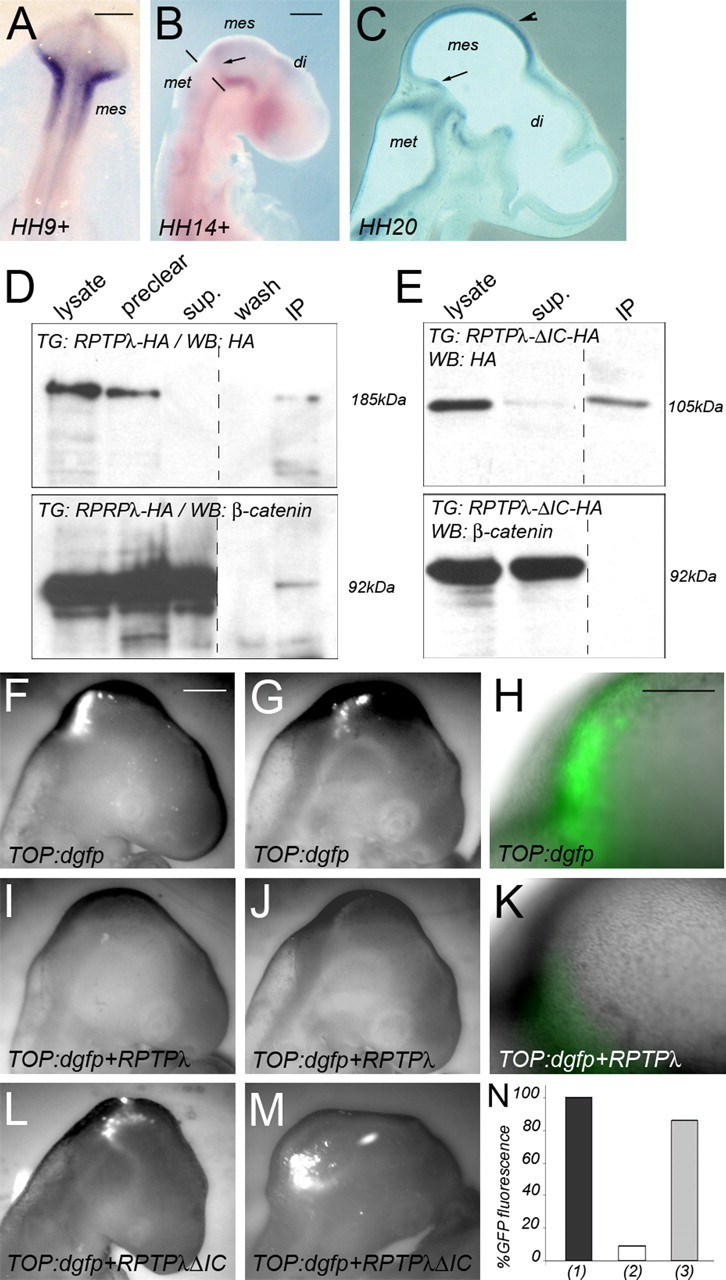
RPTPλ binds to β-catenin in the mesencephalon and inhibits the activity of a β-catenin responsive promoter in vivo. A–C, On-off-on expression of RPTPλ in the mesencephalic vesicle between HH9+ and HH20 as described by Badde et al. (2005). The dashed line in B marks the isthmic constriction, the arrow in B and C marks the location of the transverse ring of RPTPλ expression at the MHB, and the arrowhead in C marks the rostral-to-caudal upregulation of RPTPλ in the midbrain alar plate. di, Diencephalon; mes, mesencephalon; met, metencephalon. D, Immunoprecipitation using an HA-specific antibody on RPTPλ-HA transfected HH16 midbrain tissue demonstrating coprecipitation of β-catenin with RPTPλ-HA. E, As in D, except tissue transfected with RPTPλ-ΔIC-HA was used. No coprecipitation with β-catenin was observed. Top, Western Blot analysis with the HA-specific antibody detecting RPTPλ-HA (D) or RPTPλ-ΔIC-HA (E), respectively. Bottom, Detection of β-catenin on the blots shown above after stripping and reprobing with a β-catenin-specific antibody. The blots were cut to remove the marker lane. Preclear, Supernatant after precipitation with an unspecific antibody or with agarose-coupled protein-G; sup, supernatant after precipitation with the HA-specific antibody; IP, immunoprecipitate. F–M, GFP fluorescence 24 h after transfection of a TOP:dgfp reporter into the MHB region. F–H, Different examples of GFP expression in embryos 24 h after electroporation with TOP:dgfp. I–K, Different examples of GFP expression after electroporation of TOP:dgfp together with RPTPλ-pMIW. L, M, Examples of GFP expression after electroporation of TOP:dgfp together with RPTPλ-ΔIC-pMIW. F, G, I, J, L, and M are monochrome images. N, Quantification of the results: (1), percentage of embryos exhibiting GFP fluorescence 24 h after transfection with TOP:dgfp alone; (2), percentage of embryos exhibiting GFP fluorescence 24 h after transfection with TOP:dgfp together with RPTPλ-HA; (3), percentage of embryos exhibiting GFP fluorescence 24 h after transfection with TOP:dgfp together with RPTPλ-ΔIC-HA. Scale bars: A, B, 200 μm; F, 500 μm; H, 250 μm.
The juxtamembrane domain of RPTPλ exhibits a high degree of homology to the cytoplasmic domain of E-cadherin (Wang et al., 1996). Moreover, the human homolog of RPTPλ, PCP2, colocalizes with β-catenin at sites of cell-to-cell contact in human adenocarcinoma cells and coprecipitates with β-catenin in PC-12 cells overexpressing both proteins (Yan et al., 2002). The interaction of both proteins occurs independently of the phosphorylation state of β-catenin, but β-catenin can be dephosphorylated on tyrosine residues by PCP2 (Yan et al., 2002). We therefore investigated whether β-catenin and RPTPλ may physically interact in the developing mesencephalon. We overexpressed either RPTPλ-HA or RPTPλ-ΔIC-HA in the mesencephalic vesicle for 24 h and subsequently immunoprecipitated RPTPλ bound proteins using an antibody directed against the HA-epitope (Fig. 6D,E). Overexpressed RPTPλ-HA was detected as a discrete band of ∼185 kDa recognized by the HA-specific antibody (Fig. 6D, top panel, lysate). After stripping and reprobing of the blot, endogenous β-catenin expression could also be readily detected (Fig. 6D, bottom panel, lysate). Precipitation with an unspecific antibody or with agarose-coupled protein-G did not deplete RPTPλ-HA or β-catenin from the extracts (Fig. 6, preclear). After immunoprecipitation with the HA-specific antibody, the 185 kDa band was cleared from the supernatant (Fig. 6D, top panel, sup) and enriched in the immunoprecipitate (Fig. 6D, top panel, IP) (n = 5 of 5). β-Catenin was also present in the immunoprecipitates (Fig. 6D, bottom panel, IP) (n = 5 of 5). Neither β-catenin nor RPTPλ could be precipitated with an antibody not directed against HA or β-catenin (data not shown). When the truncated allele RPTPλ-ΔIC-HA, which lacks the cytoplasmic part of the protein and thus the putative interaction domain with β-catenin, was electroporated into the mesencephalic vesicle, a specific band of 105 kDa was detected (Fig. 6E, top panel, lysate). After immunoprecipitation with the HA-specific antibody, RPTPλ-ΔIC-HA could be successfully immunoprecipitated but failed to enrich β-catenin (Fig. 6E, IP) (n = 5 of 5). These data demonstrate that RPTPλ can coprecipitate β-catenin from mesencephalic extracts via its intracellular domain.
We next investigated whether the observed interaction of β-catenin with RPTPλ was biologically relevant for canonical Wnt signaling. β-Catenin plays an essential role in the structural organization of cells by linking the cytoplasmic domain of type I cadherins, such as E-cadherin, to the actin cytoskeleton and, in addition, functions as a key component of the canonical Wnt signaling pathway. Loss of E-cadherin-mediated cell adhesion increased canonical Wnt signaling, presumably because, in the absence of E-cadherin, more β-catenin is available for Wnt signal transduction (Orsulic et al., 1999; Stockinger et al., 2001). We therefore wondered whether overexpression of RPTPλ in the mesencephalon may sequester a portion of β-catenin at the plasma membrane, making it unavailable for Wnt signaling. To test this idea, we examined the activity of the β-catenin responsive reporter TOP:dgfp in the midbrain, alone or in the presence of ectopically expressed RPTPλ. TOP:dgfp contains four consensus Lef-binding sites and a minimal c-Fos promoter-driving expression of a destabilized form of GFP (dGFP) (Dorsky et al., 2002). Transgenic zebrafish that stably express dGFP under control of this promoter exhibit intense GFP expression throughout the entire midbrain alar plate, demonstrating that the canonical Wnt/β-catenin pathway is active in this region of the neural tube (Dorsky et al., 2002). When the TOP:dgfp reporter was introduced into the MHB region of HH11 chick embryos by in ovo electroporation, strong GFP expression in the midbrain alar plate could be observed 24 h later (at HH15–16), demonstrating that the midbrain alar plate is a domain of active Wnt/β-catenin signaling in the chick at this developmental age (Fig. 6F–H,N) (n = 8 of 8). Therefore, at HH15–HH16, neither RPTPλ nor Wnt1 are expressed in the mesencephalic alar plate, but the Wnt/β-catenin pathway is active. We therefore reasoned that misexpression of RPTPλ in the midbrain alar plate at HH15–H16 should allow us to analyze a possible influence of RPTPλ on canonical Wnt signal transduction that is independent of its ability to influence Wnt1 expression at the MHB. When RPTPλ was coelectroporated with TOP:dgfp, GFP fluorescence intensity was reduced severely (Fig. 6I–K,N) (n = 10 of 11). Transfection of RPTPλ-ΔIC, which lacks the intracellular domain critical for coprecipitation of β-catenin, did not suppress GFP fluorescence (Fig. 6L–N) (n = 6 of 7). These results demonstrate that RPTPλ can suppress the activation of this β-catenin responsive reporter in vivo.
The normal growth and development of the mid-hindbrain region depends on activation of the canonical Wnt signaling pathway (McMahon and Bradley, 1990; Thomas and Capecchi, 1990; Brault et al., 2001; Panhuysen et al., 2004). To investigate whether ectopic RPTPλ expression or RNAi-mediated knock-down of expression altered midbrain growth, RPTPλ expression was targeted to the right side of the mesencephalic vesicle by in ovo electroporation into HH9 embryos, and the consequence of this manipulation on midbrain growth was assessed 24 h later (Fig. 7). We chose HH9 embryos for these experiments, because this developmental time is before the downregulation of endogenous RPTPλ expression in the mesencephalon, which occurs after HH12 (Fig. 6A–C) (Badde et al., 2005). Introduction of RPTPλ-pMES into the HH9 mesencephalic vesicle therefore maintains RPTPλ expression in the midbrain beyond HH12, whereas introduction of an RNAi targeting construct leads to precocious loss of RPTPλ expression. The homeodomain Meis2 was used as a marker for the tectal anlage, because its transcripts are excluded from the roof plate, allowing us to unambiguously determine the location of the dorsal midline and estimate the size of the left and right halves of the mesencephalic vesicle. After forced expression of GFP or of the truncated allele RPTPλ-ΔIC-HA together with GFP, the right and left halves of the tectal anlage appeared to have normal size (Fig. 7A,B) (n = 5 of 5 for GFP; n = 6 of 6 for RPTPλ-ΔIC-HA). However, in embryos that had been electroporated with an expression plasmid containing RPTPλ, the size of the right electroporated tectal anlage was reduced severely when compared with the left, nonelectroporated side of the same embryo (Fig. 7C,C′) (n = 15 of 16). Conversely, the right tectal anlage was slightly enlarged in embryos transfected with the specific RNAi targeting construct RPTPλ_si-a (Fig. 7D) (n = 6 of 7).
Figure 7.
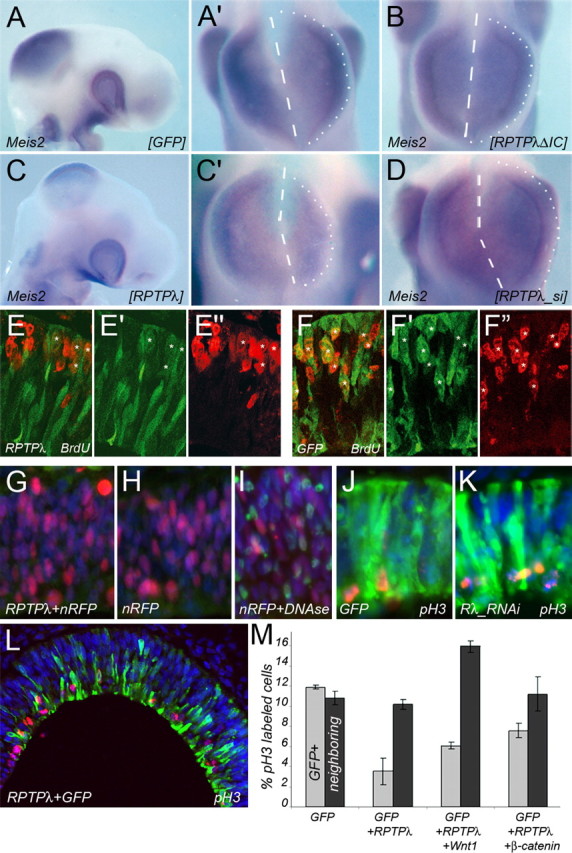
RPTPλ modulates mesencephalic growth. A–D, Whole-mount in situ hybridization with a Meis2-specific probe on HH15–HH16 embryos after electroporation with pMES (A, A′), RPTPλ-ΔIC-pMES (B), RPTPλ-pMES (C, C′), or RPTPλ_si-a (D). In A′, B, C′, and D, the embryos are shown from the top. The dashed lines define the location of the dorsal midline as observed by the lack of Meis2 expression, and the dotted lines mark the perimeter of the right side of the mesencephalic vesicle. E, F, Cross sections through HH16 midbrains showing BrdU incorporation after ectopic expression of RPTPλ together with GFP (E–E″) or GFP alone (F–F″). E, F, Overlaid channels of E′ and E″ or F′ and F″. E′, F′, GFP expression. E″, F″, BrdU incorporation. G–I, TUNEL labeling of HH16 midbrains electroporated with nRFP together with RPTPλ (G) or nRFP alone (H). I, DNase treatment before TUNEL labeling as control for the validity of the experimental procedure. J, K, Cross sections through HH16 midbrains labeled for pH3 (red) and GFP (green) and counterstained with DAPI (blue). J, Misexpression of GFP. K, Misexpression of RPTPλ_si-a together with GFP. L, Cross section through an HH16 midbrain electroporated with RPTPλ together with GFP and stained for GFP (green), pH3 (red), and DAPI (blue). Fewer transfected (GFP+) cells are labeled for pH3 than nontransfected, neighboring (GFP−) cells. For more examples, see supplemental Figure 2 (available at www.jneurosci.org as supplemental material). M, Quantification of the percentage of pH3-positive cells transfected with combinations of the gene products indicated (gray bars) and the percentage of pH3-positive cells among the nontransfected, neighboring cell population under each condition (black bars). Error bars indicate SD. Scale bar (A), 500 μm.
To determine whether this growth reduction was the result of decreased cell proliferation and/or enhanced cell death, BrdU pulse-labeling experiments were performed. We delivered RPTPλ-HA using the electroporation vector pMES, which contains an IRES-GFP cassette allowing us to visualize individual RPTPλ-HA transfected cells by their GFP fluorescence (Swartz et al., 2001). Two hours after BrdU injection, 51% (±1.85 SEM) of the cells forced to express GFP, but only 27.2% (±1.41 SEM; p < 0.002, Student's t test) of RPTPλ/GFP expressing cells had incorporated the BrdU label (Fig. 7E–F″). Similar to mock transfected or nontransfected embryos, apoptotic cell nuclei were rarely observed 24 h after misexpression of RPTPλ, indicating that RPTPλ electroporation does not compromise cell survival within this time frame (Fig. 7G–I) (n = 3). Knock-down of RPTPλ expression at HH9 slightly increased the percentage of mitotic cells, visualized by detecting pH3 from 11.7% (±1.29 SEM) after introduction of GFP alone to 14.2% (±1.45 SEM) after transfection of the RNAi targeting construct RPTPλ_si-a together with GFP (Fig. 7J,K) (supplemental Fig. 2E, available at www.jneurosci.org as supplemental material) (n = 4). In conclusion, these results suggest that RPTPλ functions as negative modulator of mesencephalic growth primarily through modulation of progenitor cell proliferation.
Because RPTPλ could precipitate (and possibly sequester) the Wnt-signaling intermediate β-catenin from mesencephalic extracts (Fig. 6D,E), we wondered whether cotransfection of β-catenin or Wnt1 together with RPTPλ would rescue the proliferation defect observed after introduction of RPTPλ alone. To test this idea, we introduced either pMES (expressing only GFP), RPTPλ-pMES (expressing RPTPλ together with GFP), RPTPλ-Wnt1-pMEC together with GFP-pMIW (which results in coexpression of RPTPλ, Wnt1, and GFP), or RPTPλ-pMES together with caβ-catenin-pMIW (which results in coexpression of RPTPλ, GFP, and a stabilized form of β-catenin) into the mesencephalic vesicle at HH9 and determined the mitotic index of midbrain progenitor cells 24 h later (Fig. 7L,M) (supplemental Fig. 2A–D, available at www.jneurosci.org as supplemental material) (n = 9 for pMES, RPTPλ-pMES; n = 3 for RPTPλ-Wnt1-pMEC, RPTPλ-pMES plus β-catenin-pMIW). Transfection of RPTPλ together with GFP significantly reduced the percentage of mitotic cells compared with GFP transfected controls. Cotransfection of Wnt1 or stabilized β-catenin together with RPTPλ and GFP partially restored the mitotic index (Fig. 7M, gray bars). Therefore, the negative influence of RPTPλ on midbrain progenitor cell proliferation could, at least in part, be alleviated by cotransfection of Wnt1 or of the Wnt1-signaling intermediate β-catenin. To determine whether RPTPλ modulates midbrain progenitor cell proliferation in a cell-autonomous or noncell-autonomous manner, we compared the percentage of pH3-positive cell nuclei in the transfected (GFP expressing) cell population under each of these experimental conditions with the percentage of pH3+ cells in nontransfected (GFP-negative) neighboring cells (Fig. 7M, black bars). The mitotic index of cells that were located in close proximity to cells transfected with RPTPλ plus GFP did not differ from that of the neighbors of GFP transfected cells, suggesting that RPTPλ acts cell autonomously on midbrain progenitor cell proliferation. We also did not observe a significant difference in the percentage of pH3+ cell nuclei in neighbors of RPTPλ/GFP/β-catenin transfected cells compared with neighbors of GFP-expressing cells. In contrast, the percentage of pH3+ nuclei in cells located near RPTPλ/Wnt1/GFP transfected cells was clearly increased. This observation is consistent with the notion that the secreted molecule Wnt1 may influence cell proliferation in a cell-nonautonomous manner.
Our finding that overexpression of RPTPλ interfered with Fgf8-mediated induction of Wnt1 expression at the MHB as well as with the activation of the Wnt/β-catenin-dependent TOP:dgfp reporter in the mesencephalon raises the question of whether the reduced TOP:dgfp reporter activity in RPTPλ transfected embryos may be a secondary consequence of their diminished Wnt1 expression or, as suggested above, may reflect a more direct inhibition of Wnt/β-catenin signaling, presumably through interaction of RPTPλ with β-catenin. To distinguish between these possibilities, we first decreased Wnt1 expression at the MHB through RNAi-mediated knock-down and monitored TOP:dgfp activity in the mesencephalic vesicle 24 h later (supplemental Fig. 3A–H′, available at www.jneurosci.org as supplemental material) (n = 15). Although electroporation of Wnt1-specific RNAi targeting constructs reduced Wnt1 expression in vivo, GFP expression of the TOP:dgfp reporter appeared normal. Second, we blocked Wnt signaling in the midbrain through forced expression of the known signaling antagonist Axin (Zeng et al., 1997). Wnt1 expression at the MHB was normal 24 h after transfection (supplemental Fig. 3I,J, available at www.jneurosci.org as supplemental material) (n = 10). Based on these observations, we suggest that RPTPλ may modulate Wnt1 expression and signal transduction in the midbrain through two separate mechanisms.
The midbrain alar plate undergoes massive expansion between HH12 and HH19 when RPTPλ expression is absent from this tissue. Cells around the MHB in embryonic day 10.5 (E10.5) to E11.5 mouse embryos are less mitotically active than cells in the midbrain and rhombomere 1 (Trokovic et al., 2005). A similar domain of reduced proliferation also exists at the chick MHB at HH15 and HH20 (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). Although the ring of RPTPλ expression seen during this developmental period does not cover this domain entirely, RPTPλ expression nevertheless appears to localize within this domain.
Discussion
Here, we present evidence based on gain-of-function and knock-down experiments that RPTPλ can counteract transcriptional activation of Wnt1 in the embryonic midbrain. In addition, we show that RPTPλ can physically interact with β-catenin in the midbrain, inhibit the activity of a β-catenin responsive promoter, and suppress mesencephalic cell proliferation. RPTPλ therefore emerges as a potential modulator of Wnt1 expression and signaling at the MHB organizer.
RPTPλ is part of the complex molecular interactions that maintain and shape the MHB region
A complex spatiotemporal pattern of gene expression has been described at the developing mid-hindbrain boundary. The MHB is bordered by a narrow, transverse ring of Wnt1 expression at its anterior side and by a ring of Fgf8 expression at its posterior side. Several proteins, including Pax2/5 and En1/2, are expressed in nested, symmetric gradients across the MHB region. Fgf8 signaling induces activation of the Ras-MAP kinase pathway, which is required for isthmic organizer activity (Sato and Nakamura, 2004). Fgf feedback inhibitors, such as Spry2, Sef, and Mkp3, interfere with Ras-MAP kinase signaling downstream of receptor tyrosine kinases at different steps within the signal transduction pathway and are thought to limit the lateral expansion of the Fgf signal at the MHB (Gross et al., 2001; Nutt et al., 2001; Fürthauer et al., 2002; Lin et al., 2002; Tsang et al., 2002; Kawakami et al., 2003; Echevarria et al., 2005). Maximal Ras-MAP kinase activation occurs within the domain of Fgf8 expression but tapers off into the mesencephalic and metencephalic vesicles (Corson et al., 2003; Sato and Nakamura, 2004). The shapes of the Pax2/5 and En1/2 expression domains reflect the gradient of Ras-MAP kinase activity at the MHB. In contrast, Wnt1 expression is confined to a tight ring of cells in the mesencephalon and is absent from the metencephalon. Metencephalic tissue is not permissive for Wnt1 expression, explaining the asymmetric nature of the Wnt1 expression domain (Kikuta et al., 2003). It is yet unclear, however, as to why Wnt1 expression is restricted to such a discrete band of cells and does not, like that of Pax2/5 or En1/2, taper off into the mesencephalic vesicle. As we show here, the receptor tyrosine phosphatase λ is expressed in a ring of cells rostral to the Wnt1 domain and inhibits Wnt1 expression when overexpressed. RPTPλ thus has the potential to restrict Wnt1 expression to its characteristic tight ring at the MHB.
RPTPλ expression itself is induced by Fgf8b in a concentration-dependent manner. When Fgf8-induced expression of Wnt1 and RPTPλ was monitored successively in the same specimen, ectopic expression of RPTPλ could always be observed anterior to the ring of Wnt1 expression. Induction of Wnt1 and RPTPλ expression by Fgf8 thus mimics their normal spatial relationship at the MHB, where the Wnt1 expression also separates the domain of RPTPλ expression from that of Fgf8. In addition, after implantation of Fgf8b releasing beads, ectopic RPTPλ expression could be observed at a Fgf8b concentration that was not sufficient to induce Wnt1 expression. We conclude from these results that different dosages of the Fgf8 signal from the MHB are required for transcriptional induction of RPTPλ and Wnt1, respectively. Examples for dosage dependency of Fgf function during embryogenesis have been described previously and include telencephalic and cerebellar development, patterning of the foregut, and skeletal development (Sato et al., 2001; Storm et al., 2003, 2006; Hajihosseini et al., 2004).
RPTPλ specifically inhibited Wnt1 expression but not that of other MHB marker genes when overexpressed in the MHB region. In addition, when cells in the midbrain were forced to express RPTPλ, they failed to upregulate Wnt1 expression after subsequent stimulation by Fgf8, yet they readily expressed En1, Pax2, or Pax5 under identical experimental conditions. RPTPλ therefore appears to specifically interfere with Fgf8-induced transcriptional activation of Wnt1 at the MHB. How can this high degree of functional specificity be achieved mechanistically? Expression of many MHB marker genes, including Otx2, Gbx2, Pax2/5, En1/2, and Wnt1, is negatively affected by overexpression of a dominant-negative form of Ras (Sato and Nakamura, 2004). Yet, we observed recently after transfection of different concentrations of a constitutive active form of Ras (caRas) that transcriptional activation of Wnt1 at the MHB could only be induced at a narrow concentration range of caRas, whereas transcriptional activation of En1 was observed over a concentration range spanning more than an order of magnitude (Vennemann et al., 2008). Induction of Wnt1 at the organizer thus appears to be more sensitive to changes in the strength of Ras-MAP kinase pathway activation than other MHB marker genes. This raises the possibility that tight, local regulation of Ras-MAP kinase activity may be a prerequisite for Wnt1 expression at the MHB and that RPTPλ may contribute to this regulation through modulation of Ras-MAP kinase signaling. Notably, in mouse embryos in which Fgf8 expression was specifically lost in the MHB territory from the five somite stage onwards, the transverse ring of Wnt1 expression in the caudal mesencephalon was also absent, whereas the expression domains of Fgf8, Otx2, Pax2, or En1 appeared normal at early stages of embryogenesis (Chi et al., 2003). Genetic loss of Fgf8 at the MHB, however, caused massive cell death, an effect we did not observe within 24 h after RPTPλ transfection. We cannot rule out that RPTPλ inhibits Wnt1 expression by interfering with still unknown factor(s) that are specifically required for transcriptional Wnt1 activation in Fgf8 responding cells. Regardless of the exact molecular nature of this inhibition, the experiments described here suggest a model in which Fgf8 induces Wnt1 and RPTPλ expression in the midbrain at different signaling thresholds and consequently at different distances to the MHB, so that the Wnt1 domain is always bordered by RPTPλ-expressing cells at its rostral side. One function of RPTPλ at the MHB may thus be to act as a Ras-MAP kinase feedback inhibitor to restrict the anterior expansion of the transverse Wnt1 domain (Fig. 8).
Figure 8.
Proposed model for the function of RPTPλ at the MHB. The MHB (dashed vertical line) is located at the border between the expression domains of Otx2 (rostral) and Gbx2 (caudal). Once the MHB is established during early somitogenesis, Fgf8, Wnt1, En2, Pax2, and Pax5 function in an interdependent, positive feedback loop, which is necessary for organizer maintenance. RPTPλ expression is induced by Fgf8 at a signaling threshold that differs from that required for Wnt1 induction, resulting in a band of RPTPλ expression anterior to the Wnt1 domain. RPTPλ counteracts Fgf8-mediated transcriptional activation of Wnt1, which constrains the Wnt1 domain to its typical tight ring anterior to the Otx2-Gbx2 boundary. Presumably, as a consequence of the high degree of homology between the juxtamembrane domain of RPTPλ with cadherins, RPTPλ can bind to and sequester β-catenin and thereby negatively influence canonical Wnt signaling.
RPTPλ binds to β-catenin and modulates canonical Wnt1 signaling at the MHB
In addition to restricting Wnt1 expression at the MHB, RPTPλ also seems to play a role in modulating Wnt1 signal transduction. In the canonical or Wnt/β-catenin pathway, Wnt binding to frizzled receptors and LRP coreceptors inhibits degradation of the cytoplasmic pool of β-catenin, allowing it to translocate to the nucleus, where it acts as coactivator for LEF/TCF transcription factors to promote target gene expression (Logan and Nusse, 2004). In addition to its role in Wnt signaling, β-catenin is a structural link between cadherins/α-catenin and the actin cytoskeleton. Cadherin bound β-catenin can be freed to participate in Wnt signaling and vice versa, a switch that appears to be regulated by phosphorylation of β-catenin on tyrosine residues (Brembeck et al., 2004; Nelson and Nusse, 2004; Schambony et al., 2004; Lilien and Balsamo, 2005). It was shown previously that β-catenin interacts with the juxtamembrane domain of the human homolog of RPTPλ and can be dephosphorylated by this protein in vitro (Yan et al., 2002). As shown here, a similar interaction of RPTPλ and β-catenin also occurs in the embryonic chick midbrain. In addition, when full-length RPTPλ was overexpressed, we found that two functional readouts of canonical Wnt signaling in the midbrain, activation of a β-catenin responsive promoter and neuroepithelial cell proliferation, were reduced. This proliferation defect could be partially reversed by cotransfection of Wnt1 or of a stabilized form of β-catenin. It is intriguing to speculate that RPTPλ may antagonize canonical Wnt signaling through binding and potentially dephosphorylation of β-catenin and ultimately serve as a negative regulator of cell proliferation in the developing midbrain (Fig. 8). Decreasing Wnt1 expression at the MHB through transfection of Wnt1-specific RNAi targeting constructs did not notably reduce β-catenin responsive reporter activity in the midbrain, nor did misexpression of Axin, a known inhibitor of the Wnt signaling pathway, in the mesencephalon disturb the transverse ring of Wnt1 expression at the organizer. These observations suggest that RPTPλ acts on Wnt1 expression and signal transduction through two independent mechanisms.
How can this dual effect be explained? Protein tyrosine phosphatases antagonize the activity of protein tyrosine kinases and influence a broad spectrum of physiological processes. For instance, RPTPλ (also termed RPTPψ) is required for Delta/Notch-dependent oscillatory gene expression in the presomitic mesoderm as well as for convergent extension during gastrulation through an as yet unknown mechanism (Aerne and Ish-Horowicz, 2004). It is therefore possible that RPTPλ impinges on multiple signaling pathways through dephosphorylation of different signaling components. In addition, the ring of RPTPλ expression at the MHB may coincide with that of another receptor tyrosine phosphatase, RPTPζ/β, a keratan sulfate modified protein, which has been suggested to modify signal transduction at the MHB through influencing the diffusion of morphogenetic signals of the Wnt and Fgf families (Canoll et al., 1993; Hamanaka et al., 1997). Although the precise role of RPTPλ in any of these processes will only be fully understood once its substrates are identified, our results suggest that RPTPλ is part of the complex molecular network that governs development of the midbrain region.
Footnotes
This work was supported by the International Max-Planck Research School Program in Structure and Function of Biological Membranes (Frankfurt, Germany) (A.B.). We thank C. Tabin (Harvard Medical School, Boston, MA), C. Logan (University of Calgary, Calgary, Alberta, Canada), C. Krull (University of Michigan, Ann Arbor, MI), H. Rohrer (Max-Planck Institute for Brain Research, Frankfurt, Germany), F. Costantini (Columbia University Medical Center, New York, NY), and R. Dorsky (University of Utah, Salt Lake City, UT) for reagents; C. Ziegler for excellent technical assistance; G. O'Sullivan for critically reading this manuscript; and A. Vennemann, E. Dohle, and A. Poplawski for experimental help.
References
- Adams KA, Maida JM, Golden JA, Riddle RD. The transcription factor Lmx1b maintains Wnt1 expression within the isthmic organizer. Development. 2000;127:1857–1867. doi: 10.1242/dev.127.9.1857. [DOI] [PubMed] [Google Scholar]
- Aerne B, Ish-Horowicz D. Receptor tyrosine phosphatase psi is required for Delta/Notch signalling and cyclic gene expression in the presomitic mesoderm. Development. 2004;131:3391–3399. doi: 10.1242/dev.01222. [DOI] [PubMed] [Google Scholar]
- Aerne B, Stoker A, Ish-Horowicz D. Chick receptor tyrosine phosphatase Psi is dynamically expressed during somitogenesis. Gene Expr Patterns. 2003;3:325–329. doi: 10.1016/s1567-133x(03)00038-3. [DOI] [PubMed] [Google Scholar]
- Araki I, Nakamura H. Engrailed defines the position of dorsal di-mesencephalic boundary by repressing diencephalic fate. Development. 1999;126:5127–5135. doi: 10.1242/dev.126.22.5127. [DOI] [PubMed] [Google Scholar]
- Badde A, Bumsted-O'Brien KM, Schulte D. Chick receptor protein tyrosine phosphatase lambda/psi (cRPTPlambda/cRPTPpsi) is dynamically expressed at the midbrain-hindbrain boundary and in the embryonic neural retina. Gene Expr Patterns. 2005;5:786–791. doi: 10.1016/j.modgep.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Alvarado-Mallart RM, Darnell DK, Wassef M. Relationship between Wnt-1 and En-2 expression domains during early development of normal and ectopic met-mesencephalon. Development. 1992;115:999–1009. doi: 10.1242/dev.115.4.999. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Cholley B, Wassef M. Involvement of Wnt-1 in the formation of the mes/metencephalic boundary. Mech Dev. 1995;53:23–34. doi: 10.1016/0925-4773(95)00421-1. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. Essential role of BCL9–2 in the switch between beta-catenin's adhesive and transcriptional functions. Genes Dev. 2004;18:2225–2230. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canoll PD, Barnea G, Levy JB, Sap J, Ehrlich M, Silvennoinen O, Schlessinger J, Musacchio JM. The expression of a novel receptor-type tyrosine phosphatase suggests a role in morphogenesis and plasticity of the nervous system. Dev Brain Res. 1993;75:293–298. doi: 10.1016/0165-3806(93)90035-9. [DOI] [PubMed] [Google Scholar]
- Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal Fgf8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martinez S, Martin GR. Midbrain development induced by FGF8 in the chick embryo. Nature. 1996;380:66–68. doi: 10.1038/380066a0. [DOI] [PubMed] [Google Scholar]
- Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- den Hertog J. Protein-tyrosine phosphatases in development. Mech Dev. 1999;85:3–14. doi: 10.1016/s0925-4773(99)00089-1. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241:229–237. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Echevarria D, Martinez S, Marques S, Lucas-Teixeira V, Belo JA. Mkp3 is a negative feedback modulator of Fgf8 signaling in the mammalian isthmic organizer. Dev Biol. 2005;277:114–128. doi: 10.1016/j.ydbio.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Funayama N, Fagotto F, McCrea P, Gumbiner BM. Embryonic axis induction by the armadillo repeat domain of beta-catenin: evidence for intracellular signaling. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürthauer M, Lin W, Ang SL, Thisse B, Thisse C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol. 2002;4:170–174. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- Gross I, Bassit B, Benezra M, Licht JD. Mammalian sprouty proteins inhibit cell growth and differentiation by preventing ras activation. J Biol Chem. 2001;276:46460–46468. doi: 10.1074/jbc.M108234200. [DOI] [PubMed] [Google Scholar]
- Hajihosseini MK, Lalioti MD, Arthaud S, Burgar HR, Brown JM, Twigg SR, Wilkie AO, Heath JK. Skeletal development is regulated by fibroblast growth factor receptor 1 signalling dynamics. Development. 2004;131:325–335. doi: 10.1242/dev.00940. [DOI] [PubMed] [Google Scholar]
- Hamanaka H, Maeda N, Noda M. Spatially and temporally regulated modification of the receptor-like protein tyrosine phsophatase ζ/β isoforms with keratan sulphate in the developing chick brain. Eur J Neurosci. 1997;9:2297–2308. doi: 10.1111/j.1460-9568.1997.tb01647.x. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. (1951) Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hidalgo-Sánchez M, Martínez-de-la-Torre M, Alvarado-Mallart RM, Puelles L. A distinct preisthmic hitogenetic domain is defined by overlap of Otx2 and Pax2 gene expression in the avian caudal midbrain. J Comp Neurol. 2005;483:17–29. doi: 10.1002/cne.20402. [DOI] [PubMed] [Google Scholar]
- Hirata H, Tomita K, Bessho Y, Kageyama R. Hes1 and Hes3 regulate maintenance of the isthmic organizer and development of the mid/hindbrain. EMBO J. 2001;20:4454–4466. doi: 10.1093/emboj/20.16.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollyday M, McMahon JA, McMahon AP. Wnt expression patterns in chick embryo nervous system. Mech Dev. 1995;52:9–25. doi: 10.1016/0925-4773(95)00385-e. [DOI] [PubMed] [Google Scholar]
- Irving C, Mason I. Signalling by FGF8 from the isthmus patterns anterior hindbrain and establishes the anterior limit of Hox gene expression. Development. 2000;127:177–186. doi: 10.1242/dev.127.1.177. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kudoh T, Dedekian M, Kim CH, Chitnis AB. A role for iro1 and iro7 in the establishment of an anteroposterior compartment of the ectoderm adjacent to the midbrain-hindbrain boundary. Development. 2002;129:2317–2327. doi: 10.1242/dev.129.10.2317. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Rodríguez-León J, Koth CM, Büscher D, Itoh T, Raya A, Ng JK, Esteban CR, Takahashi S, Henrique D, Schwarz MF, Asahara H, Izpisúa Belmonte JC. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat Cell Biol. 2003;5:513–519. doi: 10.1038/ncb989. [DOI] [PubMed] [Google Scholar]
- Kikuta H, Kanai M, Ito Y, Yamasu K. gbx2 Homeobox gene is required for the maintenance of the isthmic region in the zebrafish embryonic brain. Dev Dyn. 2003;228:433–450. doi: 10.1002/dvdy.10409. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Lilien J, Balsamo J. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of beta-catenin. Curr Opin Cell Biol. 2005;17:459–465. doi: 10.1016/j.ceb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Lin W, Fürthauer M, Thisse B, Thisse C, Jing N, Ang SL. Cloning of the mouse Sef gene and comparative analysis of its expression with Fgf8 and Spry2 during embryogenesis. Mech Dev. 2002;113:163–168. doi: 10.1016/s0925-4773(02)00018-7. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Martinez S, Alvarado-Mallart RM. Expression of the homeobox Chick-en gene in chick/quail chimeras with inverted mes-metencephalic grafts. Dev Biol. 1990;139:432–436. doi: 10.1016/0012-1606(90)90312-7. [DOI] [PubMed] [Google Scholar]
- Martinez S, Wassef M, Alvarado-Mallart RM. Induction of a mesencephalic phenotype in the 2-day-old chick prosencephalon is preceded by the early expression of the homeobox gene en. Neuron. 1991;6:971–981. doi: 10.1016/0896-6273(91)90237-t. [DOI] [PubMed] [Google Scholar]
- Martinez S, Crossley PH, Cobos I, Rubenstein JL, Martin GR. FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development. 1999;126:1189–1200. doi: 10.1242/dev.126.6.1189. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Katahira T, Nakamura H. Role of Lmx1b and Wnt1 in mesencephalon and metencephalon development. Development. 2002;129:5269–5277. doi: 10.1242/dev.129.22.5269. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Joyner AL, Bradley A, McMahon JA. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Momose T, Tonegawa A, Takeuchi J, Ogawa H, Umesono K, Yasuda K. Efficient targeting of gene expression in chick embryos by microelectroporation. Dev Growth Differ. 1999;41:335–344. doi: 10.1046/j.1440-169x.1999.413437.x. [DOI] [PubMed] [Google Scholar]
- Mühleisen TW, Agoston Z, Schulte D. Retroviral misexpression of cVax disturbs retinal ganglion cell axon fasciculation and intraretinal pathfinding in vivo and guidance of nasal ganglion cell axons in vivo. Dev Biol. 2006;297:59–73. doi: 10.1016/j.ydbio.2006.04.466. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Nakano KE, Igawa HH, Takagi S, Fujisawa H. Plasticity and rigidity of differentiation of brain vesicles studied in quail-chick chimeras. Cell Differ. 1986;19:187–193. doi: 10.1016/0045-6039(86)90095-3. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Dingwell KS, Holt CE, Amaya E. Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. 2001;15:1152–1166. doi: 10.1101/gad.191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara FP, Beck E, Barr LK, Wong LL, Kessler DS, Riddle RD. Zebrafish Lmx1b.1 and Lmx1b.2 are required for maintenance of the isthmic organizer. Development. 2005;132:3163–3173. doi: 10.1242/dev.01898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J Cell Sci. 1999;112:1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- Panhuysen M, Vogt Weisenhorn DM, Blanquet V, Brodski C, Heinzmann U, Beisker W, Wurst W. Effects of Wnt1 signaling on proliferation in the developing mid-/hindbrain region. Mol Cell Neurosci. 2004;26:101–111. doi: 10.1016/j.mcn.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Paul S, Lombroso PJ. Receptor and nonreceptor protein tyrosine phosphatases in the nervous system. Cell Mol Life Sci. 2003;60:2465–2482. doi: 10.1007/s00018-003-3123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifers F, Böhli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, McMahon AP. Pax-2 expression in the murine neural plate precedes and encompasses the expression domains of Wnt-1 and En-1. Mech Dev. 1995;52:3–8. doi: 10.1016/0925-4773(95)00380-j. [DOI] [PubMed] [Google Scholar]
- Sato T, Nakamura H. The Fgf8 signal causes cerebellar differentiation by activating the Ras-ERK signaling pathway. Development. 2004;131:4275–4285. doi: 10.1242/dev.01281. [DOI] [PubMed] [Google Scholar]
- Sato T, Araki I, Nakamura H. Inductive signal and tissue responsiveness defining the tectum and the cerebellum. Development. 2001;128:2461–2469. doi: 10.1242/dev.128.13.2461. [DOI] [PubMed] [Google Scholar]
- Schambony A, Kunz M, Gradl D. Cross-regulation of Wnt signaling and cell adhesion. Differentiation. 2004;72:307–318. doi: 10.1111/j.1432-0436.2004.07207002.x. [DOI] [PubMed] [Google Scholar]
- Schulte D, Cepko CL. Two homeobox genes define the domain of EphA3 expression in the developing chick retina. Development. 2000;127:5033–5045. doi: 10.1242/dev.127.23.5033. [DOI] [PubMed] [Google Scholar]
- Schulte D, Furukawa T, Peters MA, Kozak CA, Cepko CL. Misexpression of the Emx-related homeobox genes cVax and mVax2 ventralizes the retina and perturbs the retinotectal map. Neuron. 1999;24:541–553. doi: 10.1016/s0896-6273(00)81111-3. [DOI] [PubMed] [Google Scholar]
- Shamim H, Mahmood R, Logan C, Doherty P, Lumsden A, Mason I. Sequential roles for Fgf4, En1 and Fgf8 in specification and regionalisation of the midbrain. Development. 1999;126:945–959. doi: 10.1242/dev.126.5.945. [DOI] [PubMed] [Google Scholar]
- Stockinger A, Eger A, Wolf J, Beug H, Foisner R. E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J Cell Biol. 2001;154:1185–1196. doi: 10.1083/jcb.200104036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker A, Dutta R. Protein tyrosine phosphatases and neural development. BioEssays. 1998;20:463–472. doi: 10.1002/(SICI)1521-1878(199806)20:6<463::AID-BIES4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Storm EE, Rubenstein JL, Martin GR. Dosage of Fgf8 determines whether cell survival is positively or negatively regulated in the developing forebrain. Proc Natl Acad Sci USA. 2003;100:1757–1762. doi: 10.1073/pnas.0337736100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, Rubenstein JL. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- Suemori H, Kadodawa Y, Goto K, Araki I, Kondoh H, Nakatsuji N. A mouse embryonic stem cell line showing pluripotency of differentiation in early embryos and ubiquitous beta-galactosidase expression. Cell Differ Dev. 1990;29:181–186. doi: 10.1016/0922-3371(90)90120-l. [DOI] [PubMed] [Google Scholar]
- Swartz ME, Eberhart J, Pasquale EB, Krull CE. EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development. 2001;128:4669–4680. doi: 10.1242/dev.128.23.4669. [DOI] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990;346:847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Trokovic R, Trokovic N, Hernesniemi S, Pirvola U, Vogt Weisenhorn DM, Rossant J, McMahon AP, Wurst W, Partanen J. FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. EMBO J. 2003;22:1811–1823. doi: 10.1093/emboj/cdg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokovic R, Jukkola T, Saarimäki J, Peltopuro P, Naserke T, Weisenhorn DM, Trokovic N, Wurst W, Partanen J. Fgfr1-dependent boundary cells between developing mid- and hindbrain. Dev Biol. 2005;278:428–439. doi: 10.1016/j.ydbio.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Tsang M, Friesel R, Kudoh T, Dawid IB. Identification of Sef, a novel modulator of FGF signalling. Nat Cell Biol. 2002;4:165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- Vennemann A, Agoston Z, Schulte D. Differential and dose-dependent regulation of gene expression at the mid-hindbrain boundary by Ras-MAP kinase signaling. Brain Res. 2008;1206:33–43. doi: 10.1016/j.brainres.2008.01.100. [DOI] [PubMed] [Google Scholar]
- Wang H, Lian Z, Lerch MM, Chen Z, Xie W, Ullrich A. Characterization of PCP-2, a novel receptor protein tyrosine phosphatase of the MAM domain family. Oncogene. 1996;12:2555–2562. [PubMed] [Google Scholar]
- Yan HX, He YQ, Dong H, Zhang P, Zeng JZ, Cao HF, Wu MC, Wang HY. Physical and functional interaction between receptor-like protein tyrosine phosphatase PCP-2 and beta-catenin. Biochemistry. 2002;41:15854–15860. doi: 10.1021/bi026095u. [DOI] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL, III, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]