Abstract
γ-Secretase is an unusual membrane-embedded protease, which cleaves the transmembrane domains (TMDs) of type I membrane proteins, including amyloid-β precursor protein and Notch receptor. We have previously shown the existence of a hydrophilic pore formed by TMD6 and TMD7 of presenilin 1 (PS1), the catalytic subunit of γ-secretase, within the membrane by the substituted cysteine accessibility method. Here we analyzed the structure of TMD8, TMD9, and the C terminus of PS1, which encompass the conserved PAL motif and the hydrophobic C-terminal tip, both being critical for the catalytic activity and the formation of the γ-secretase complex. We found that the amino acid residues around the PAL motif and the extracellular/luminal portion of TMD9 are highly water accessible and located in proximity to the catalytic pore. Furthermore, the region starting from the luminal end of TMD9 toward the C terminus forms an amphipathic α-helix-like structure that extends along the interface between the membrane and the extracellular milieu. Competition analysis using γ-secretase inhibitors revealed that the TMD9 is involved in the initial binding of substrates, as well as in the subsequent catalytic process as a subsite. Our results provide mechanistic insights into the role of TMD9 in the formation of the catalytic pore and the substrate entry, crucial to the unusual mode of intramembrane proteolysis by γ-secretase.
Keywords: Alzheimer's disease, amyloid beta, intramembrane-cleaving protease, presenilin, secretase, substituted cysteine accessibility method, Aβ peptide, structure
Introduction
Intramembrane-cleaving proteases (I-CLiPs) are a family of polytopic membrane-embedded enzymes responsible for hydrolyzing substrates within the hydrophobic transmembrane domains (TMDs) (Wolfe and Kopan, 2004). Various I-CLiPs have so far been identified, including site 2 protease (S2P), signal peptide peptidase (SPP), rhomboid, and γ-secretase. γ-Secretase is an atypical aspartic protease that cleaves a set of type-1 single-span membrane proteins including amyloid precursor protein to form amyloid-β (Aβ) peptides in Alzheimer's disease (Tomita and Iwatsubo, 2006; Selkoe and Wolfe, 2007). γ-Secretase requires the formation of multimeric membrane protein complex comprised of nicastrin, Aph-1, and Pen-2 in addition to PS1 to exert the proteolytic activity (Takasugi et al., 2003), which hampered the conventional structural analysis, such as x-ray crystallography. Recently, a glimpse into the whole structure of the γ-secretase complex was obtained by single particle analysis (Lazarov et al., 2006; Ogura et al., 2006). However, because of the low resolution, these structures have not been helpful in deciphering the molecular mechanism of intramembrane proteolysis by γ-secretase. We have adopted the substituted cysteine accessibility method (SCAM) to analyze the structure of PS1. SCAM has been used to obtain structural information about various multipass membrane proteins, by forming covalent modification of sulfhydryl reagents to substituted cysteine (Cys) residues (Karlin and Akabas, 1998; Kaback et al., 2001). Using SCAM, we and others have shown that TMD6 and TMD7 of PS1 constitute the hydrophilic catalytic pore within membrane, and that the structural change of the pore correlates to the cleavage specificity (Sato et al., 2006; Tolia et al., 2006; Isoo et al., 2007). The recent x-ray crystallographic analyses of rhomboid and S2P revealed that their active sites reside in a water-filled “cavity” within the lipid bilayer (Y. Wang et al., 2006; Wu et al., 2006; Feng et al., 2007), suggesting that the hydrophilic active site located in the lipid bilayer is a structure common to I-CLiPs. Considering that the substrates for I-CLiPs also are embedded within the membrane, they should have access to the catalytic site through a “lateral gate” facing a hydrophobic environment on the analogy of the translocon system (Lemberg and Martoglio, 2004; Lemberg and Freeman, 2007; Selkoe and Wolfe, 2007). However, no such functional domain has been identified within the γ-secretase, although pharmacological and chemical biological studies indicate the presence of a substrate binding site within γ-secretase that are distinct from the catalytic site (Esler et al., 2002; Das et al., 2003; Tian et al., 2003; Kornilova et al., 2005). In this study, we further analyzed the structure and the functional role of the more distal portion of the C-terminal fragment (CTF) of PS1, which contains two hydrophobic regions as well as the PAL motif, the latter being required for the proteolytic activity (Tomita et al., 2001; Wang et al., 2004).
Materials and Methods
Polyclonal antibody G1Nr5 was raised against recombinant human PS1 N terminus protein. G1L3, which recognizes a hydrophilic loop 6, was described previously (Tomita et al., 1999). Anti-CD44ICD, anti-PS1NT (Thinakaran et al., 1998), and PS-C3 (Honda et al., 1999) were kindly provided by Drs. H. Saya (Keio University, Shinjuku, Tokyo, Japan), G. Thinakaran (University of Chicago, Chicago, IL), and A. Takashima (RIKEN, Wako, Saitama, Japan), respectively. N-[N-(3,5-difluorophenacetyl)-l-alanyl]-(S)-phenylglycine tert-butyl ester (DAPT) was synthesized as described previously (Dovey et al., 2001; Kan et al., 2003). {1S-Benzyl-4R-[1-(1S-carbamoyl-2-phenylethylcarbamoyl)-1S-3-methylbutylcarbamoyl]-2R-hydroxy-5-phenylpentyl}carbamic acid tert-butyl ester (L-685,458) (Shearman et al., 2000) and peptide 15 (pep15) (Das et al., 2003) were purchased from Bachem and Ito Life Science, respectively. All 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) reagents (Toronto Research Chemicals) were dissolved in DMSO at 200 mm before use or stored at −80°C until use. The abbreviations used for MTS cross-linkers are as follows: M2M, 1,2-ethanediyl bismethanethiosulfonate; M3M, 1,3-propanediyl bismethanethiosulfonate; M4M, 1,4-butanediyl bismethanethiosulfonate; M6M, 1,6-hexanediyl bismethanethiosulfonate; M8M, 3,6-dioxaoctane-1,8-diyl bismethanethiosulfonate; M11M, 3,6,9-trioxaundecane-1,11-diyl bismethanethiosulfonate; M14M, 3,6,9,12-tetraoxatetradecane-1,14-diyl bismethanethiosulfonate; M17M, 3,6,9,12,15-pentaoxaheptadecane-1,17-diyl bismethanethiosulfonate. cDNAs encoding APPNL, PS1, and Cys-less PS1 were described previously (Sato et al., 2006). Single- or double-Cys mutant (mt) PS1 were generated using long PCR-based protocol. Maintenance of embryonic fibroblasts derived from Psen1/Psen2 double knock-out (DKO) mouse cells (Herreman et al., 2000), retroviral infection system (Kitamura et al., 2003), and generation of stable infectants were done as previously described (Watanabe et al., 2005). Microsome preparation, immunoblot analysis, and quantitation of Aβ by two-site ELISAs were performed as previously described (Tomita et al., 1997, 1999, 2001; Hayashi et al., 2004). Biotinylation experiment using N-biotinylaminoethyl methanethiosulfonate (MTSEA-biotin) in intact cell (labeling from extracellular/luminal side) or microsome fraction (labeling from both extracellular/luminal and cytosolic sides) was performed as previously described (Sato et al., 2006; Isoo et al., 2007). The relative extents of biotinylation of PS1 fragments were calculated from the band intensities. For cross-linking experiments, resuspended microsomes incubated with MTS cross-linkers (10 mm) for 2 h at room temperature were mixed with sample buffer containing N-ethylmaleimide and then directly subjected to immunoblot analysis. For a competition assay, 2-sulfonatoethyl methanethiosulfonate (MTSES) or 2-(trimethylammonium)-ethyl methanethiosulfonate (MTSET) was preincubated with intact cells or microsomes for 15 min at room temperature and washed once before the biotinylation. γ-Secretase inhibitors were preincubated with intact cells or microsomes for 30 min at room temperature, before the biotinylation or cross-linking experiments. The inhibitors were used at concentrations that completely abolish the proteolytic activity of γ-secretase (Morohashi et al., 2006; Sato et al., 2006).
Results
SCAM analysis of hydrophobic region 9 and the PAL motif of PS1
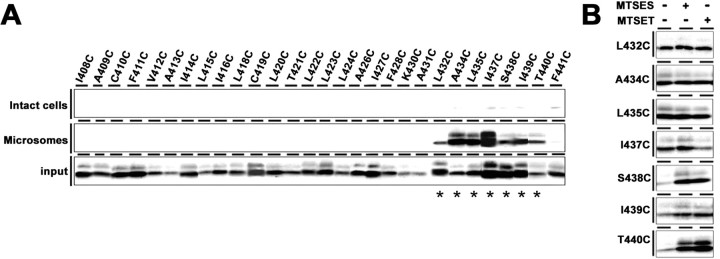
Because of the relatively weak hydrophobicity of the hydrophobic region 10, the topology and geometry of the “TMD9” as well as of the extreme C terminus of PS1 had long been controversial. Recently, however, several cell-based topological studies revealed that the hydrophobic region 9 (I408–F428) and 10 (A434–V453) of PS1 (Henricson et al., 2005) penetrate the membrane as TMD8 and TMD9, respectively, and that the extreme C terminus is exposed to the extracellular/luminal side (Laudon et al., 2005; Oh and Turner, 2005; Spasic et al., 2006). Moreover, the PAL motif that resides at the N-terminal region of the hydrophobic region 10 is highly conserved in PS and SPP proteins and is required for the enzymatic activity (Tomita et al., 2001; Wang et al., 2004; Nakaya et al., 2005; J. Wang et al., 2006), although the mechanistic role of this motif remains unknown. To analyze the water accessibility of these regions, we first generated mutant PS1 carrying a single cysteine in Cys-less PS1 (single-Cys mt PS1) at 34 consecutive amino acid residues in and around the hydrophobic region 9 and PAL motif (I408–F441) (Fig. 1). Cys substitution of some residues abolished the expression or endoproteolysis of PS1 polypeptides, or Aβ generation (i.e., G417C, L425C, K429C, P433C, and P436C) (supplemental Fig. 1A,D, available at www.jneurosci.org as supplemental material). No single-Cys mt PS1 that harbored the γ-secretase activity as a holoprotein have so far been identified. Consistent with the previous results, mutants at the PAL motif (i.e., P433C and P436C) lost γ-secretase activity, which were excluded from further structural analyses. SCAM analysis of the remaining “active” single-Cys mt in intact cells revealed that none of the mutants were biotinylated by MTSEA-biotin from the extracellular side (Fig. 2A; supplemental Fig. 2A, available at www.jneurosci.org as supplemental material). We next examined the microsome labeling of single-Cys mt PS1 and found that all single-Cys mt PS1 starting from L432C to T440C were biotinylated, whereas no other mutants were labeled. Next, we examined the effects of other membrane-impermeable MTS derivatives, i.e., the negatively charged MTSES and the bulkier, positively charged MTSET, to analyze the steric and electrostatic environment around the biotinylated residues. The labeling of I437C by MTSEA-biotin was decreased by preincubation with MTSET, suggesting that I437 faces an open hydrophilic environment (Fig. 2B). However, preincubation with the charged MTS derivatives did not decrease the biotinylation of A434C, L435C, S438C, I439C, or T440C. These data suggest that all residues within the hydrophobic region 9 are inaccessible to water and face the hydrophobic environment as membrane-embedded TMD8. In contrast, the consecutive residues around the PAL motif (L432–T440) sit in a narrow, water-accessible cleft that is exclusively accessible from the cytosolic side.
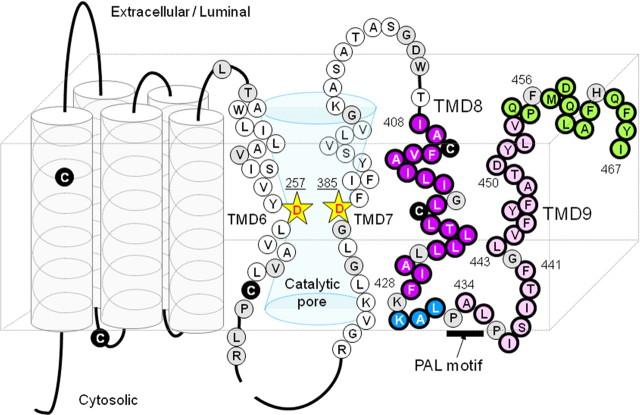
Figure 1.
Locations of the PS1 cysteine mutations used in this study. A schematic depiction of human PS1 based on the nine-TMD topology model is shown. Catalytic aspartates are shown by yellow stars. Endogenous Cys residues that were replaced with serine in Cys-less PS1 are indicated as black circles. Amino acid residues substituted with Cys are shown by a circle with a single-letter character representing the original amino acids. Single-Cys mt was systematically substituted with residues in the predicted hydrophobic region 9, hydrophilic loop, hydrophobic region 10, and the C-terminal region, which are indicated by purple, blue, pink, and green circles, respectively. Cys substitutions that resulted in a loss of γ-secretase activity are indicated as gray circles.
Figure 2.
SCAM analysis of single-Cys mt PS1 in the hydrophobic region 9 and the PAL motif. A, Biotin-labeling experiment using MTSEA-biotin in intact cells (top) and microsomes (middle). Bottom, Amount of PS1 CTF in the input fraction. Biotinylated mutants are indicated by asterisks below the panel. B, Labeling competition by MTSES and MTSET was performed using microsomes. Locations of Cys mutations are shown on the left.
SCAM analysis of hydrophobic region 10 and the extreme C terminus of PS1
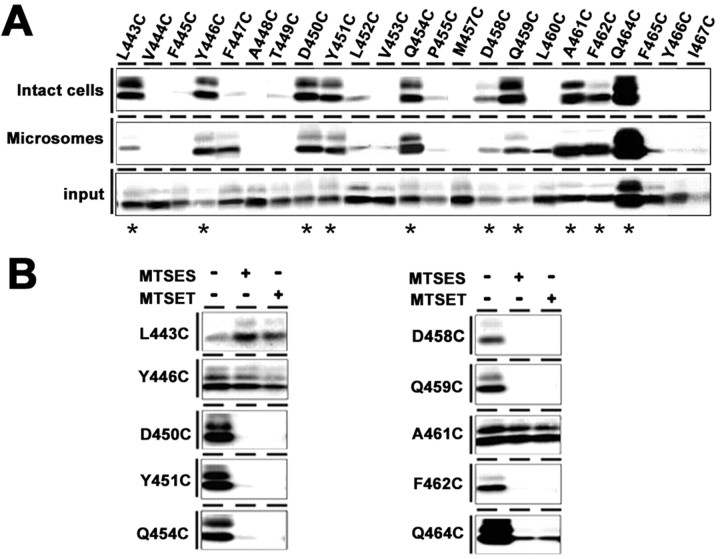
We further generated single-Cys mt PS1 at 26 consecutive amino acid residues starting from the remaining C-terminal half of the hydrophobic region 10 to the extreme C terminus of PS1 (G442–I467) (Fig. 1). Again, some mutants (i.e., G442C, F456C, and H463C) lost the protein expression or γ-secretase activity and thus were excluded from further analyses (supplemental Fig. 1B,D, available at www.jneurosci.org as supplemental material). Labeling experiments using intact cells revealed that L443C, Y446C, D450C, Y451C, Q454C, D458C, Q459C, A461C, F462C, and Q464C were reactive with MTSEA-biotin from the extracellular side (Fig. 3A; supplemental Fig. 2B, available at www.jneurosci.org as supplemental material). Moreover, the labeling of D450C, Y451C, Q454C, D458C, Q459C, F462C, and Q464C were inhibited by preincubation with MTSES or MTSET, suggesting that the region C-terminal to D450, except for A461, faces an open hydrophilic environment (Fig. 3B). In contrast, the biotinylation of L443C, Y446C, and A461C was not decreased by preincubation with the charged MTS derivatives. We next prepared microsome fractions from DKO cells expressing single-Cys mt PS1 of the hydrophobic region 10 and subjected them to labeling experiments (Fig. 3A; supplemental Fig. 2B, available at www.jneurosci.org as supplemental material). All residues that were labeled in intact cells were biotinylated, whereas the accessibilities of some residues (i.e., F447C and L460C) were different from those by intact cell labeling. Given that the PAL motif was accessible exclusively from the cytosolic side, it is most reasonable to conclude that the hydrophobic region 10 spans the membrane as TMD9 in the active γ-secretase complex, and that several residues at the C terminus of PS1 are exposed to the extracellular side.
Figure 3.
SCAM analysis of single-Cys mt PS1 around the hydrophobic region 10 and the extreme C terminus. A, Biotin-labeling experiment using MTSEA-biotin in intact cells (top) and microsomes (middle). Bottom, Amount of PS1 CTF in the input fraction. Biotinylated mutants are indicated by asterisks below the panel. B, Labeling competition by MTSES and MTSET was performed using intact cells. Locations of Cys mutations are shown on the left.
Cross-linking experiments using MTS cross-linkers
We have previously found that TMD6 and TMD7 are located in close proximity within the catalytic pore, because L250C and I387C were directly cross-linkable (Sato et al., 2006). To gain more insights into the structural characteristics of the C-terminal region in relation to the catalytic pore, we performed a systematic cross-linking experiment using MTS cross-linkers. M2M, M3M, M4M, M6M, M8M, M11M, M14M, and M17M are sulfhydryl-to-sulfhydryl cross-linking reagents with spacer arms of 5.2, 6.5, 7.8, 10.4, 13, 16.9, 20.8, and 24.7 Å long, respectively (Loo and Clarke, 2001b). First, cross-linking experiments using microsome fractions revealed that a subset of longer cross-linkers caused slow migration of N-terminal fragment (NTF) or CTF of single-Cys mt PS1 (L250C, A260C, L383C, I387C, and L435C) (supplemental Fig. 3A–E, available at www.jneurosci.org as supplemental material). No cross-linked products were observed in the following single-Cys mt: L432C, Y446C, and D450C (data not shown). Immunoblot analysis using single-Cys mt PS1 carrying M292D, which remains as a holoprotein (Steiner et al., 1999), revealed that these shifts neither represent the formation of homodimer of PS fragments nor the cross-linking of other components (i.e., nicastrin, Aph-1, and Pen-2) with PS1 (data not shown). The cross-linked products of ∼20 kDa with CTF in L383C, I387C, and L435C mutants were decreased by preincubation with L-685,458 (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). However, no antibodies against representative substrates of γ-secretase (i.e., APP, Notch, and CD44) reacted with this cross-linked product. These bands might represent the cross-linking of PS1 CTF with the endogenous unknown γ-secretase substrates or binding proteins harboring Cys. The characterization of these cross-linked proteins is underway.
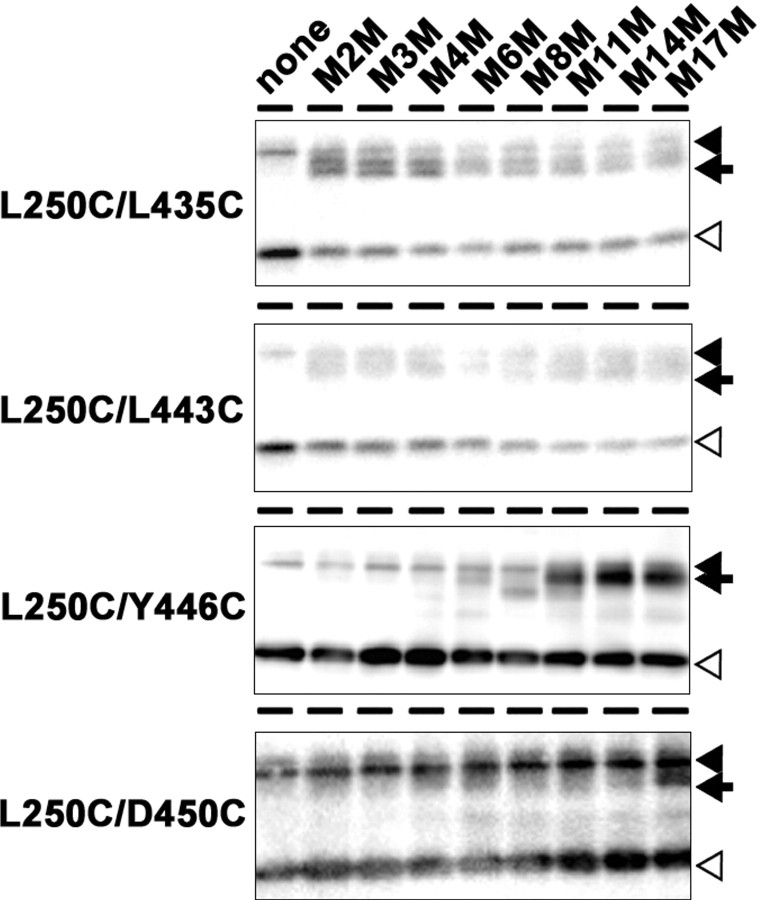
We next generated a series of double-Cys mutant PS1 harboring a pair of Cys, located in NTF and CTF, respectively, in a single molecule (e.g., L250C/Y446C) in an attempt to locate TMD9 and the extreme C terminus relative to TMD6 (i.e., L250 and A260; summarized in Table 1). We chose several residues (i.e., around the PAL motif: L432 and L435; TMD9: L443, Y446, D450, and Q454; C terminus: D458, Q459, and A461) to examine the in cis-cross-linking with Cys introduced into TMD6 (i.e., L250 or A260). All double-Cys mutants retained γ-secretase activity except for L250C/Q454C and A260C/L443C, and only the active mutants were subjected to further cross-linking analyses (supplemental Fig. 1C, available at www.jneurosci.org as supplemental material). Coincubation of L250C/L435C, L250C/L443C, L250C/Y446C, and L250C/D450C double-Cys mutants with MTS cross-linkers decreased the amount of NTF and CTF, whereas a band close in size to that of a holoprotein (∼45 kDa) emerged [Fig. 4, arrows; supplemental Fig. 5 (arrows), available at www.jneurosci.org as supplemental material]. These bands reacted with both anti-PS1 N- and C-terminal antibodies, but not with antibodies to other γ-secretase components (data not shown). The same results were obtained using two different antibodies for PS1 NTF and CTF (i.e., G1Nr5 and PS1NT for NTF, G1L3 and PS-C3 for CTF), suggesting that these polypeptides correspond to an NTF–CTF heterodimer, as previously observed (Sato et al., 2006). Notably, in L250C/L435C and L250C/L443C, NTF–CTF heterodimer was formed by M2M with a spacer arm of only 5.2 Å. Longer cross-linkers, e.g., M8M and M17M, were required for the NTF–CTF cross-linking in L250C/Y446C and L250C/D450C, respectively. In contrast, L432, D458, Q459, and A461 were not cross-linked to L250 (supplemental Fig. 6A–H, available at www.jneurosci.org as supplemental material). Collectively, these data indicate that the PAL motif and L443 in the middle of TMD9 are located in close proximity to TMD6 (i.e., ≤5.2 Å apart), and face the same hydrophilic pore.
Table 1.
Summary of the cross-linking experiments in double-Cys PS1 mt using different lengths of MTS cross-linkers
| MTS cross-linker | Distance (Å) | |
|---|---|---|
| L250C/L432C (PAL) | - | |
| L250C/L435C (PAL) | M2M | ≤5.2 |
| L250C/L443C (TMD9) | M2M | ≤5.2 |
| L250C/Y446C (TMD9) | M8M | ≤16.9 |
| L250C/D450C (TMD9) | M17M | ≤24.7 |
| L250C/Q454C (TMD9) | No activity | |
| L250C/D458C (C-term) | - | |
| L250C/Q459C (C-term) | - | |
| L250C/A461C (C-term) | - | |
| A260C/L443C (TMD9) | No activity |
“MTS cross-linkers” indicate the abbreviated name of the shortest cross-linker that enabled the production of an NTF–CTF heterodimer (e.g., M2M). ″-″ represents the absence of the heterodimer formation by any cross-linkers. ″Distance″ indicates the length of the spacer arm of the MTS cross-linkers. C-term, Most C-terminal region. “No activity” indicates PS1 mutants that lack γ-secretase activity.
Figure 4.
Cross-linking experiment using MTS cross-linkers. Cross-linking experiments of double-Cys mt in microsomes and immunoblot analysis using anti-PS1NT antibody. Locations of Cys mutations are shown on the left. PS1 full-length protein, NTF, and cross-linked product (NTF–CTF heterodimer) are shown by black arrowheads, white arrowheads, and black arrows, respectively.
Reactivity of substituted cysteines in the presence of γ-secretase inhibitors
Transition-state analog-type aspartyl protease inhibitors that target to the catalytic aspartates directly bind PS1 NTF and CTF (Li et al., 2000; Shearman et al., 2000; Esler et al., 2002). Helical peptide-based inhibitors mimicking the TMD structure bind PS fragments as well, but are unable to compete for the binding of transition-state analog inhibitors (Das et al., 2003; Kornilova et al., 2005). The dipeptidic inhibitor, DAPT, targets PS1 CTF in yet a third manner, and its binding site appears to be distinct from, but overlap with, that of transition-state analogues and helical peptides, suggesting that DAPT binds to a site involved in the substrate transfer (Kornilova et al., 2003, 2005; Micchelli et al., 2003; Morohashi et al., 2006). To examine the different binding sites for different types of inhibitors as well as the potential conformational changes that take place during their binding, we analyzed the accessibility of substituted Cys in the presence of representative γ-secretase inhibitors [i.e., transition-state analog (L-685,458), helical peptide (pep15), and dipeptidic compound (DAPT)].
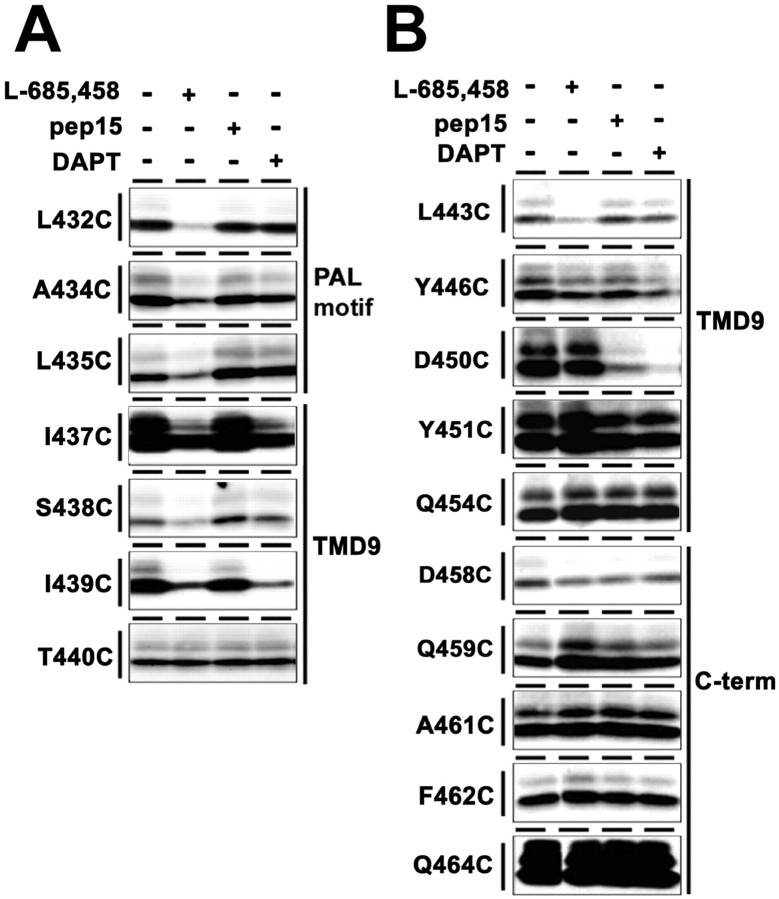
The competition assays revealed that the accessibility of a series of single-Cys mt PS1 around the PAL motif (L432C, A434C, L435C, I437C, S438C, and I439C) was significantly reduced by preincubation with L-685,458 (Fig. 5A). Moreover, the labeling of several L-685,458-sensitive residues (i.e., I437C, S438C, and I439C) was decreased by preincubation with DAPT, too. In contrast, the biotinylation of TMD9 and the extreme C terminus from the extracellular side was not affected by any of the γ-secretase inhibitors, except for L443C and D450C: the labeling of L443C was significantly and exclusively reduced by preincubation with L-685,458, whereas the biotinylation of D450C was sensitive to preincubation with pep15 and DAPT, but not with L-685,458 (Fig. 5B). It is well established that TMDs of several receptors and transporters rotate to cause structural rearrangements after treatment by agonists or antagonists (Horenstein et al., 2001; Loo and Clarke, 2001a). However, we observed no labeling of additional single-Cys mt PS1 that had originally been inaccessible to MTSEA-biotin under a condition in which the activity of γ-secretase is completely inhibited (data not shown). Therefore, it may be reasonable to speculate that the γ-secretase inhibitors tested here did not cause a rotation of TMDs. Altogether, our SCAM approach identified that the water accessibility of certain residues in PS1 is competed by various types of γ-secretase inhibitors, whose competition profiles were different from each other.
Figure 5.
Labeling competition by γ-secretase inhibitors. A, B, Biotin labeling of single-Cys mt PS1 was conducted after preincubation with L-685,458, pep15, or DAPT in microsomes (A) or intact cells (B). Locations and predicted topology of Cys mutations are shown on the left and right, respectively.
Discussion
The structure of the C-terminal region of PS1 in an active γ-secretase complex
In this study, we applied SCAM, in combination with cross-linking experiments, to the identification of amino acid residues in the C-terminal region of PS1 that are water accessible from either side of the membrane (Fig. 6A). None of the residues around the hydrophobic region 9 (I408–A431) showed water accessibility, suggesting that they are buried within the lipid bilayer as an authentic TMD (TMD8) and do not participate in the formation of the hydrophilic pore. In contrast, the consecutive residues around the PAL motif (L432–T440) were water accessible, but mostly with limited access. We speculate that this region sits in a narrow hydrophilic cleft that is only accessible from the cytosolic side, forming a v-shaped hydrophilic structure with I437 at the outermost tip. The cross-linking experiments revealed that this structure is located very close to the catalytic site within the pore. Notably, among the Cys mutants in TMD8 and TMD9, G417C and G442C lost the γ-secretase activity. Glycine is known as a “helix-breaking” residue for TMDs and gives helix flexibility in the lipid bilayer (Li and Deber, 1994; Ulmschneider and Sansom, 2001). Moreover, these glycines are almost completely conserved in PS homologues identified so far (supplemental Fig. 7, available at www.jneurosci.org as supplemental material). We speculate that TMD8 forms a bending structure at G417, which may be required for the γ-secretase activity. G442 may also form a hinge in the middle of TMD9 to provide the PAL motif and the extracellular/luminal half of TMD9 with flexibility.
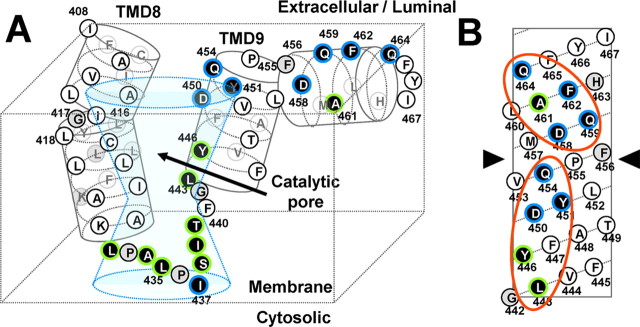
Figure 6.
Hypothetical structure around TMD8, TMD9, and the extreme C terminus of PS1 revealed in this study. A, Summary of SCAM analysis and a schematic depiction of the configuration of the TMD8 and TMD9 in relation to the catalytic pore. Cys mutants that were labeled by MTSEA-biotin are shown by a white letter in a black circle. Residues whose labeling was effectively competed by MTSES or MTSET are shown in a blue frame, and the unaffected residues are in a green frame. Residues that were not labeled by MTSEA-biotin or not analyzed are shown by black letters in white and gray circles, respectively. B, An α-helical net representation of the C terminus of human PS1 starting at the N-terminal glycine 442. Consecutive hydrophilic surfaces are shown by orange circles. An arrowhead indicates a putative helix kink.
Several residues within the extracellular/luminal side of TMD9 (G442-Q454) were highly hydrophilic: L443 and Y446 were water accessible with steric restrictions; D450, Y451, and Q454 were facing the hydrophilic environment, but with unlimited access. Importantly, all these residues were located on the same interface in an α-helical model (Fig. 6B). Moreover, the cross-linking experiments revealed that L443, Y446, and D450 were located in proximity to TMD6, suggesting that these residues face the catalytic pore. Of note, the minimum “cross-linkable” distances to L250 of these residues increased toward the C terminus. These data suggest that the luminal half of TMD9 forms a short α-helix that faces the catalytic pore, which is tilted away from the pore.
D458, Q459, A461, F462, and Q464 at the C terminus of PS1 also exhibited unlimited water access with the exception of A461. These residues were aligned on the same surface in an α-helical model, but in a surface different from that formed by the extracellular/luminal half of TMD9 (Fig. 6B). These surfaces appear to be connected by a proline that generates a helix kink (von Heijne, 1991). In contrast, the residues predicted to locate at the opposite side of the helix showed no water accessibility. This proline is highly conserved among species, suggesting the functional importance of the proline residue. After a kink at P455, the C terminus of PS1 (i.e., F456-Q464) may form a membrane-associated, short amphipathic α-helix that faces an aqueous environment. Interestingly, Cys substitution of F456, located next to P455 and also highly conserved, lost the γ-secretase activity. Phenylalanine is frequently found at the interface between the hydrophobic core and the phosphate region of the lipid (Killian and von Heijne, 2000). Furthermore, the extreme C terminus of PS1 (i.e., F465–I467) were located in a hydrophobic environment. This is in agreement with the previous finding that the relative hydrophobicity of these C-terminal residues is required for the interaction of PS1 with the TMD of nicastrin and the activity of γ-secretase, supporting the view that they are buried within the membrane (Bergman et al., 2004; Kaether et al., 2004). Thus, we speculate that the C-terminal short helix is tethered to the lipid bilayer by virtue of the hydrophobic nature of the extreme C terminus. This structure is reminiscent of a “slide helix,” an amphipathic feature that contributes to the control of the conformation and activity of Kir6.2 potassium channels (Antcliff et al., 2005; Enkvetchakul et al., 2007). Notably, sequential deletion of the C-terminal residues of PS1 gradually decreased the γ-secretase activity, and the deletion of residues 451–467, which correspond to the amphipathic region identified here, totally abolished the activity (Tomita et al., 1999; Bergman et al., 2004), implicating the auxiliary role of the short C-terminal helix in the assembly and activity of γ-secretase.
Implications for the mechanism of intramembrane proteolysis
Several lines of evidence suggest that the intramembrane proteolysis is executed by the following consecutive steps: (1) binding of the substrate to the initial docking site, (2) substrate transfer to the catalytic environment through the lateral gate, and (3) hydrolysis of the scissile bond. By a series of competition assays using pharmacologically well characterized inhibitors, we identified a set of amino acid residues within PS1 that may play critical roles in the proteolytic mechanism (Fig. 7).
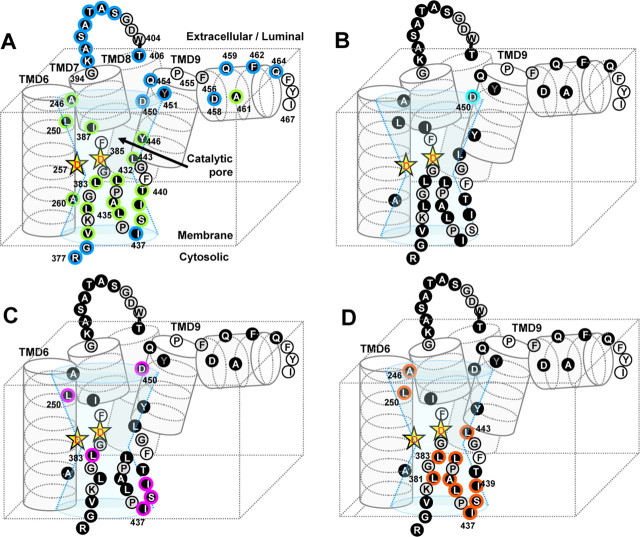
Figure 7.
The model of the catalytic pore of γ-secretase and the locations of γ-secretase inhibitor-sensitive residues in PS1. A, Schematic depiction of the structure around the catalytic pore coupled with our previous result (Sato et al., 2006) in the same manner as that in Figure 6A. Residues whose labeling was diminished by the presence of pep15 (B), DAPT (C), and L-685,458 (D) are shown in cyan, pink, and orange frames, respectively.
To examine the structural changes after substrate binding and to locate the substrate binding site, we performed a competition assay using the helical peptide-type inhibitor that is predicted to directly target the initial binding site, which may presumably reside in the interface of PS1 NTF/CTF (Das et al., 2003; Kornilova et al., 2005). The labeling of D450C by MTSEA-biotin was inhibited by preincubation with pep15 and DAPT, but not with L-685,458 (Fig. 7B). However, pep15 had no effect on the biotinylation of any other Cys mutants. Notably, the extracellular/luminal half of TMD9 harboring D450 appears to form a membrane-embedded short α-helix partitioned by the helix-breaking residues and face the hydrophilic catalytic pore. Strikingly, Cys mutants located at the hydrophobic helical interface on this region (i.e., V444C, A448C, and I453C) resulted in the augmentation of enzymatic activity (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). This whole structure is highly reminiscent of TMD5 of rhomboid that faces the catalytic cavity and is supposed to constitute the lateral gate for substrate entry (Baker et al., 2007). Of note, loosening the hydrophobic interaction between TMD2 and TMD5 of rhomboid enhanced the proteolytic activity. Together, these results indicate that D450 and the extracellular/luminal helix of TMD9 of PS1 may comprise a part of the initial docking site or the lateral gate for substrate.
DAPT competed for labeling of Cys residues located C-terminal to the PAL motif (I437C, S438C, and I439C) as well as of D450C, all of which are located in close proximity to the catalytic pore (Fig. 7C). Consistent with the previous pharmacological results, the biotinylation of all DAPT-sensitive residues was diminished by preincubation of L-685,458 or pep15. It is noteworthy that DAPT has been shown to exhibit an inhibitor profile specific to γ-secretase and directly target the PS1 CTF, despite that γ-secretase and SPP share the GxGD and the PAL motif (Morohashi et al., 2006; Fuwa et al., 2007). Intriguingly, the DAPT-sensitive residues identified in this study (i.e., I437, S438, I439, and D450) are not conserved in SPP. Thus, we hypothesize that these residues comprise a protein surface for the DAPT binding site that had been shown to reside within the PS1 CTF. Further competition analysis using dibenzazepine, a potent inhibitor that directly targets the PS1 NTF and SPP (Fuwa et al., 2007), would provide more precise pharmacostructural information about this point.
The PAL motif is reported to be critical for γ-secretase activity and the binding of the transition state analog inhibitor L-685,458 (J. Wang et al., 2006). Consistent with this notion, the labeling of consecutive residues around the PAL motif, as well as of L443 in TMD9, were inhibited by preincubation with L-685,458 (Fig. 7D). Moreover, the cross-linking study revealed that all these regions are located in close proximity around the catalytic aspartates. Importantly, mutagenesis studies revealed that the replacement of P433 or A434 with tiny amino acid residues (e.g., Ala, Gly, and Ser) was tolerated for the preservation of the γ-secretase activity, whereas substitution of P433 with bulky amino acids (e.g., Val, Leu, or Ile) abolished the activity (Nakaya et al., 2005; J. Wang et al., 2006). These results favor the notion that the requirement of the PAL motif in intramembrane proteolysis is independent of the proline-mediated reverse turn structure or the hydrophobicity. Together with our finding, we propose that the hydrophilic and smooth surface structure formed by the residues around the PAL motif directly functions as a subsite and constitutes the “catalytic plug” within the pore together with the GxGD motif (Sato et al., 2006).
Based on the results of SCAM analysis in combination with the competition assay using mechanism-based γ-secretase inhibitors, we propose the functional role of PS1 in the intramembrane proteolysis as follows (supplemental Fig. 8, available at www.jneurosci.org as supplemental material). The substrate (1) initially binds to the extracellular/luminal helix of TMD9 that comprises the lateral gate, (2) is transferred through the C-terminal side of the PAL motif, and (3) is recruited to the catalytic site composed of the extracellular/luminal half of TMD6, the GxGD motif, the PAL motif, and the N-terminal end of TMD9. However, we are not able to exclude the possibility that the binding of inhibitors caused conformational changes that made the Cys residue inaccessible to water. Nonetheless, further SCAM analysis, as well as other biochemical and structural studies, will reveal the whole picture of the γ-secretase cleavage, in which substrates are introduced through the lateral gate and subjected to cleavage within the hydrophilic catalytic pore.
Footnotes
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (C.S., T.T., T.I.), by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (T.T., T.I.), by Targeted Proteins Research Program of the Japan Science and Technology Corporation (JST) (T.T., T.I.), and by Core Research for Evolutional Science and Technology of JST (C.S., T.T., T.I.), Japan. We thank Drs. G. Thinakaran (University of Chicago, Chicago, IL), A. Takashima (RIKEN, Wako, Saitama, Japan), and H. Saya (Keio University, Shinjuku, Tokyo, Japan) for antibodies, B. De Strooper (Katholieke Universiteit Leuven, Leuven, The Netherlands) for DKO cells, T. Kitamura (The University of Tokyo, Minato, Tokyo, Japan) for retroviral infection system, and Takeda Pharmaceutical Company for Aβ ELISA. We are also grateful to our lab members for helpful discussions and technical assistance.
References
- Antcliff JF, Haider S, Proks P, Sansom MS, Ashcroft FM. Functional analysis of a structural model of the ATP-binding site of the KATP channel Kir6.2 subunit. EMBO J. 2005;24:229–239. doi: 10.1038/sj.emboj.7600487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci USA. 2007;104:8257–8262. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A, Laudon H, Winblad B, Lundkvist J, Näslund J. The extreme C terminus of presenilin 1 is essential for γ-secretase complex assembly and activity. J Biol Chem. 2004;279:45564–45572. doi: 10.1074/jbc.M407717200. [DOI] [PubMed] [Google Scholar]
- Das C, Berezovska O, Diehl TS, Genet C, Buldyrev I, Tsai JY, Hyman BT, Wolfe MS. Designed helical peptides inhibit an intramembrane protease. J Am Chem Soc. 2003;125:11794–11795. doi: 10.1021/ja037131v. [DOI] [PubMed] [Google Scholar]
- Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, et al. Functional γ-secretase inhibitors reduce β-amyloid peptide levels in brain. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- Enkvetchakul D, Jeliazkova I, Bhattacharyya J, Nichols CG. Control of inward rectifier K channel activity by lipid tethering of cytoplasmic domains. J Gen Physiol. 2007;130:329–334. doi: 10.1085/jgp.200709764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esler WP, Kimberly WT, Ostaszewski BL, Ye W, Diehl TS, Selkoe DJ, Wolfe MS. Activity-dependent isolation of the presenilin-γ-secretase complex reveals nicastrin and a γ substrate. Proc Natl Acad Sci USA. 2002;99:2720–2725. doi: 10.1073/pnas.052436599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- Fuwa H, Takahashi Y, Konno Y, Watanabe N, Miyashita H, Sasaki M, Natsugari H, Kan T, Fukuyama T, Tomita T, Iwatsubo T. Divergent synthesis of multifunctional molecular probes to elucidate the enzyme specificity of dipeptidic γ-secretase inhibitors. ACS Chem Biol. 2007;2:408–418. doi: 10.1021/cb700073y. [DOI] [PubMed] [Google Scholar]
- Hayashi I, Urano Y, Fukuda R, Isoo N, Kodama T, Hamakubo T, Tomita T, Iwatsubo T. Selective reconstitution and recovery of functional γ-secretase complex on budded baculovirus particles. J Biol Chem. 2004;279:38040–38046. doi: 10.1074/jbc.M405597200. [DOI] [PubMed] [Google Scholar]
- Henricson A, Käll L, Sonnhammer EL. A novel transmembrane topology of presenilin based on reconciling experimental and computational evidence. FEBS J. 2005;272:2727–2733. doi: 10.1111/j.1742-4658.2005.04691.x. [DOI] [PubMed] [Google Scholar]
- Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B. Total inactivation of γ-secretase activity in presenilin-deficient embryonic stem cells. Nat Cell Biol. 2000;2:461–462. doi: 10.1038/35017105. [DOI] [PubMed] [Google Scholar]
- Honda T, Yasutake K, Nihonmatsu N, Mercken M, Takahashi H, Murayama O, Murayama M, Sato K, Omori A, Tsubuki S, Saido TC, Takashima A. Dual roles of proteasome in the metabolism of presenilin 1. J Neurochem. 1999;72:255–261. doi: 10.1046/j.1471-4159.1999.0720255.x. [DOI] [PubMed] [Google Scholar]
- Horenstein J, Wagner DA, Czajkowski C, Akabas MH. Protein mobility and GABA-induced conformational changes in GABA(A) receptor pore-lining M2 segment. Nat Neurosci. 2001;4:477–485. doi: 10.1038/87425. [DOI] [PubMed] [Google Scholar]
- Isoo N, Sato C, Miyashita H, Shinohara M, Takasugi N, Morohashi Y, Tsuji S, Tomita T, Iwatsubo T. Abeta42 overproduction associated with structural changes in the catalytic pore of γ-secretase: common effects of Pen-2 N-terminal elongation and fenofibrate. J Biol Chem. 2007;282:12388–12396. doi: 10.1074/jbc.M611549200. [DOI] [PubMed] [Google Scholar]
- Kaback HR, Sahin-Tóth M, Weinglass AB. The kamikaze approach to membrane transport. Nat Rev Mol Cell Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- Kaether C, Capell A, Edbauer D, Winkler E, Novak B, Steiner H, Haass C. The presenilin C-terminus is required for ER-retention, nicastrin-binding and gamma-secretase activity. EMBO J. 2004;23:4738–4748. doi: 10.1038/sj.emboj.7600478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan T, Tominari Y, Morohashi Y, Natsugari H, Tomita T, Iwatsubo T, Fukuyama T. Solid-phase synthesis of photoaffinity probes: highly efficient incorporation of biotin-tag and cross-linking group. Chem Commun. 2003;7:2244–2245. doi: 10.1039/b306970b. [DOI] [PubMed] [Google Scholar]
- Karlin A, Akabas MH. Substituted-cysteine accessibility method. Methods Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- Kornilova AY, Das C, Wolfe MS. Differential effects of inhibitors on the γ-secretase complex. Mechanistic implications. J Biol Chem. 2003;278:16470–16473. doi: 10.1074/jbc.C300019200. [DOI] [PubMed] [Google Scholar]
- Kornilova AY, Bihel F, Das C, Wolfe MS. The initial substrate-binding site of γ-secretase is located on presenilin near the active site. Proc Natl Acad Sci USA. 2005;102:3230–3235. doi: 10.1073/pnas.0407640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudon H, Hansson EM, Melén K, Bergman A, farmery MR, Winblad B, Lendahl U, von Heijne G, Näslund J. A nine-transmembrane domain topology for presenilin I. J Biol chem. 2005;280:35352–35360. doi: 10.1074/jbc.M507217200. [DOI] [PubMed] [Google Scholar]
- Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H. Electron microscopic structure of purified, active γ-secretase reveals an aqueous intramembrane chamber and two pores. Proc Natl Acad Sci USA. 2006;103:6889–6894. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberg MK, Freeman M. Cutting proteins within lipid bilayers: rhomboid structure and mechanism. Mol Cell. 2007;28:930–940. doi: 10.1016/j.molcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Lemberg MK, Martoglio B. On the mechanism of SPP-catalysed intramembrane proteolysis; conformational control of peptide bond hydrolysis in the plane of the membrane. FEBS Lett. 2004;564:213–218. doi: 10.1016/S0014-5793(04)00192-9. [DOI] [PubMed] [Google Scholar]
- Li SC, Deber CM. A measure of helical propensity for amino acids in membrane environments. Nat Struct Biol. 1994;1:368–373. doi: 10.1038/nsb0694-368. [DOI] [PubMed] [Google Scholar]
- Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio-Mower J, Harrison T, Lellis C, Nadin A, Neduvelil JG, Register RB, Sardana MK, Shearman MS, Smith AL, Shi XP, Yin KC, Shafer JA, Gardell SJ. Photoactivated γ-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Cross-linking of human multidrug resistance P-glycoprotein by the substrate, tris-(2-maleimidoethyl)amine, is altered by ATP hydrolysis. Evidence for rotation of a transmembrane helix. J Biol Chem. 2001a;276:31800–31805. doi: 10.1074/jbc.M103498200. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Determining the dimensions of the drug-binding domain of human P-glycoprotein using thiol cross-linking compounds as molecular rulers. J Biol Chem. 2001b;276:36877–36880. doi: 10.1074/jbc.C100467200. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Esler WP, Kimberly WT, Jack C, Berezovska O, Kornilova A, Hyman BT, Perrimon N, Wolfe MS. γ-Secretase/presenilin inhibitors for Alzheimer's disease phenocopy Notch mutations in Drosophila. FASEB J. 2003;17:79–81. doi: 10.1096/fj.02-0394fje. [DOI] [PubMed] [Google Scholar]
- Morohashi Y, Kan T, Tominari Y, Fuwa H, Okamura Y, Watanabe N, Sato C, Natsugari H, Fukuyama T, Iwatsubo T, Tomita T. C-terminal fragment of presenilin is the molecular target of a dipeptidic γ-secretase-specific inhibitor DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester) J Biol Chem. 2006;281:14670–14676. doi: 10.1074/jbc.M513012200. [DOI] [PubMed] [Google Scholar]
- Nakaya Y, Yamane T, Shiraishi H, Wang HQ, Matsubara E, Sato T, Dolios G, Wang R, De Strooper B, Shoji M, Komano H, Yanagisawa K, Ihara Y, Fraser P, St George-Hyslop P, Nishimura M. Random mutagenesis of presenilin-1 identifies novel mutants exclusively generating long amyloid beta-peptides. J Biol Chem. 2005;280:19070–19077. doi: 10.1074/jbc.M501130200. [DOI] [PubMed] [Google Scholar]
- Ogura T, Mio K, Hayashi I, Miyashita H, Fukuda R, Kopan R, Kodama T, Hamakubo T, Iwastubo T, Tomita T, Sato C. Three-dimensional structure of the γ-secretase complex. Biochem Biophys Res Commun. 2006;343:525–534. doi: 10.1016/j.bbrc.2006.02.158. [DOI] [PubMed] [Google Scholar]
- Oh YS, Turner RJ. Evidence that the COOH terminus of human presenilin 1 is located in extracytoplasmic space. Am J Physiol Cell Physiol. 2005;289:C576–C581. doi: 10.1152/ajpcell.00636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Morohashi Y, Tomita T, Iwatsubo T. Structure of the catalytic pore of γ-secretase probed by the accessibility of substituted cysteines. J Neurosci. 2006;26:12081–12088. doi: 10.1523/JNEUROSCI.3614-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Shearman MS, Beher D, Clarke EE, Lewis HD, Harrison T, Hunt P, Nadin A, Smith AL, Stevenson G, Castro JL. L-685,458, an aspartyl protease transition state mimic, is a potent inhibitor of amyloid β-protein precursor γ-secretase activity. Biochemistry. 2000;39:8698–8704. doi: 10.1021/bi0005456. [DOI] [PubMed] [Google Scholar]
- Spasic D, Tolia A, Dillen K, Baert V, De Strooper B, Vrijens S, Annaert W. Presenilin-1 maintains a nine-transmembrane topology throughout the secretory pathway. J Biol Chem. 2006;281:26569–26577. doi: 10.1074/jbc.M600592200. [DOI] [PubMed] [Google Scholar]
- Steiner H, Romig H, Pesold B, Philipp U, Baader M, Citron M, Loetscher H, Jacobsen H, Haass C. Amyloidogenic function of the Alzheimer's disease-associated presenilin 1 in the absence of endoproteolysis. Biochemistry. 1999;38:14600–14605. doi: 10.1021/bi9914210. [DOI] [PubMed] [Google Scholar]
- Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T. The role of presenilin cofactors in the γ-secretase complex. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- Thinakaran G, Regard JB, Bouton CM, Harris CL, Price DL, Borchelt DR, Sisodia SS. Stable association of presenilin derivatives and absence of presenilin interactions with APP. Neurobiol Dis. 1998;4:438–453. doi: 10.1006/nbdi.1998.0171. [DOI] [PubMed] [Google Scholar]
- Tian G, Ghanekar SV, Aharony D, Shenvi AB, Jacobs RT, Liu X, Greenberg BD. The mechanism of γ-secretase: multiple inhibitor binding sites for transition state analogs and small molecule inhibitors. J Biol Chem. 2003;278:28968–28975. doi: 10.1074/jbc.M300905200. [DOI] [PubMed] [Google Scholar]
- Tolia A, Chávez-Gutiérrez L, De Strooper B. Contribution of presenilin transmembrane domains 6 and 7 to a water-containing cavity in the γ-secretase complex. J Biol Chem. 2006;281:27633–27642. doi: 10.1074/jbc.M604997200. [DOI] [PubMed] [Google Scholar]
- Tomita T, Iwatsubo T. γ-Secretase as a therapeutic target for treatment of Alzheimer's disease. Curr Pharm Des. 2006;12:661–670. doi: 10.2174/138161206775474206. [DOI] [PubMed] [Google Scholar]
- Tomita T, Maruyama K, Saido TC, Kume H, Shinozaki K, Tokuhiro S, Capell A, Walter J, Grünberg J, Haass C, Iwatsubo T, Obata K. The presenilin 2 mutation (N141I) linked to familial Alzheimer disease (Volga German families) increases the secretion of amyloid β protein ending at the 42nd (or 43rd) residue. Proc Natl Acad Sci USA. 1997;94:2025–2030. doi: 10.1073/pnas.94.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T, Takikawa R, Koyama A, Morohashi Y, Takasugi N, Saido TC, Maruyama K, Iwatsubo T. C terminus of presenilin is required for overproduction of amyloidogenic Aβ42 through stabilization and endoproteolysis of presenilin. J Neurosci. 1999;19:10627–10634. doi: 10.1523/JNEUROSCI.19-24-10627.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T, Watabiki T, Takikawa R, Morohashi Y, Takasugi N, Kopan R, De Strooper B, Iwatsubo T. The first proline of PALP motif at the C terminus of presenilins is obligatory for stabilization, complex formation, and γ-secretase activities of presenilins. J Biol Chem. 2001;276:33273–33281. doi: 10.1074/jbc.M011152200. [DOI] [PubMed] [Google Scholar]
- Ulmschneider MB, Sansom MS. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Proline kinks in transmembrane α-helices. J Mol Biol. 1991;218:499–503. doi: 10.1016/0022-2836(91)90695-3. [DOI] [PubMed] [Google Scholar]
- Wang J, Brunkan AL, Hecimovic S, Walker E, Goate A. Conserved “PAL” sequence in presenilins is essential for γ-secretase activity, but not required for formation or stabilization of γ-secretase complexes. Neurobiol Dis. 2004;15:654–666. doi: 10.1016/j.nbd.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Wang J, Beher D, Nyborg AC, Shearman MS, Golde TE, Goate A. C-terminal PAL motif of presenilin and presenilin homologues required for normal active site conformation. J Neurochem. 2006;96:218–227. doi: 10.1111/j.1471-4159.2005.03548.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Tomita T, Sato C, Kitamura T, Morohashi Y, Iwatsubo T. Pen-2 is incorporated into the γ-secretase complex through binding to transmembrane domain 4 of presenilin 1. J Biol Chem. 2005;280:41967–41975. doi: 10.1074/jbc.M509066200. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Kopan R. Intramembrane proteolysis: theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]