Abstract
NMDA receptors are glutamate-gated ion channels that play important roles in synaptic transmission and excitotoxicity. The functional NMDA receptor is thought to be a heterotetramer composed mainly of two NR1 and two NR2 subunits. Although it is generally accepted that only correctly assembled NMDA receptors can pass the ER quality control, the mechanism underlying this process is not well understood. Using truncated and chimeric NMDA receptor subunits expressed in heterologous cells and cortical neurons, we found that the third membrane domains (M3) of both NR1 and NR2B contain signals that cause the unassembled subunits to be retained in the ER. M3 of both NR1 and NR2B and, M4 of NR1, are necessary for masking ER retention signals found in M3. Thus, our data reveal a critical role of the membrane domains in the assembly of functional NMDA receptors.
Keywords: glutamate receptors, ion channel assembly, subunit interaction, ER retention, masking, intracellular trafficking
Introduction
NMDA receptors are ionotropic glutamate receptors that play critical roles in excitatory synaptic transmission and excitotoxicity (Cull-Candy et al., 2001; Lau and Zukin, 2007). These receptors are likely heterotetramers, assembled from NR1, NR2, and NR3 subunits. Coassembly of NR1 and NR2 subunits is necessary for the formation of a functional channel (Wenthold et al., 2003). A single NR1 subunit exists in eight splice forms, whereas there are four distinct NR2 subunits (NR2A–D). The subunits share the same topology: an extracellular N terminus, four putative membrane domains (M1–M4) of which the second (M2) is a re-entrant loop, and an intracellular C terminus. Several previous studies have indicated that abnormalities in the regulation of NMDA receptors may be involved in a number of psychiatric and neurological disorders (Cull-Candy et al., 2001; Lau and Zukin, 2007). These could occur at multiple steps including synthesis, subunit assembly and endoplasmic reticulum (ER) processing, trafficking and degradation of receptors.
The major NR1 splice variant (NR1-1) and the NR2 subunits are retained in the ER when expressed alone in heterologous cells and neurons, but when expressed together, they form functional receptors on the cell surface (McIlhinney et al., 1998; Okabe et al., 1999; Fukaya et al., 2003). Previous studies investigating the mechanisms underlying the ER retention of NMDA receptor subunits focused on their C termini and used chimeras of a transmembrane protein tac (interleukin-2 receptor subunit) and NR1 and NR2B C termini. These experiments have shown that an NR1-1 C-terminal (Ct) chimera is retained in the ER because of an RRR motif in its C1 cassette and that the C termini of other NR1 splice variants attached to tac are not retained because they lack the RRR motif (NR1-2 and NR1-4) or because the C2′ cassette can mask the RRR motif (NR1–3) (Standley et al., 2000; Scott et al., 2001; Xia et al., 2001). The NR2B C terminus appended to tac was retained in the ER, suggesting the presence of an ER retention signal; however, the particular motif has not yet been identified (Hawkins et al., 2004). It remains unclear how the ER retention signals in the C termini of NR1 and NR2 are masked after assembly. Previous studies have shown that the HLFY motif immediately following NR2B M4 in full-length NR2B subunit (Hawkins et al., 2004) or, at least, the presence of any two amino acid residues after NR2 M4 (Yang et al., 2007) is required for the export of NR1/NR2 complexes from the ER. This raises the possibility that this motif may be involved in masking of ER retention signals.
In this study, we focused on the identification and masking of ER retention signals in the NR1-1/NR2B receptor. We found that both NR1 and NR2B M3 possess ER retention signals, in addition to the C-terminal signals described previously. Moreover, NR1 and NR2B M3 together with NR1 M4 are necessary for negating the ER retention signals in the assembled NR1/NR2B receptor. These findings identify a critical role of membrane domains in the assembly of NMDA receptors.
Materials and Methods
Cell culture and transfection.
Human embryonic kidney 293 (HEK293) and COS7 cells were cultured in MEM medium (Invitrogen, San Diego, CA) with 10% fetal bovine serum (Sigma, St. Louis, MO). To transfect the cells, equal amounts of cDNAs for NR1 and NR2 subunits (total 1 μg) were mixed with 2 μl of Lipofectamine 2000 (Invitrogen) and were added to cells for 5 h. If any variant of NR1 and NR2B vectors were cotransfected, media was supplemented with 20 mm MgCl2 and 3 mm kynurenic acid to suppress excitotoxicity. Cells were used for experiments within 24–48 h after transfection.
The expression vectors for green fluorescent protein (GFP)-NR2B, constructed previously by inserting a GFP gene between the fifth and sixth codons after the sequence for the signal peptide, and for the yellow fluorescent protein (YFP)-NR1-1a, made previously by inserting a YFP gene between the third and fourth amino acid residues after the signal peptide, were used (Luo et al., 2002). MYC-NR2B and FLAG-NR2B expression constructs were also described previously (Hawkins et al., 1999; Papadakis et al., 2004). All mutations, including incorporation of stop codons, were generated using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). All constructs were verified by DNA sequencing.
For simplicity, the generated truncation variants of NMDA receptor subunits are referred in the text as preMx or postMx, depending on whether the stop codon was introduced before or after the referred M. The exact amino acid numbers are as follows: NR2B844stop (NR2B postM4), NR2B792stop (NR2B preM4), NR2B655stop (NR2B postM3), NR2B629stop (NR2B preM3), NR2B584stop (NR2B postM1), NR1838stop (NR1 postM4), NR1812stop (NR1 preM4), NR1655stop (NR1 postM3), NR1629stop (NR1 preM3), and NR1590stop (NR1 postM1) (GenBank accession numbers M91562 and U08261). The NR2B-M4-C-terminal peptide begins at amino acid number NR2B793; the NR2B signal peptide sequence was inserted at the beginning of its sequence.
Cortical neuronal cultures.
All experiments involving animals were performed according to the National Institutes of Health guidelines. Cortices were dissected from embryonic day 18 Sprague Dawley rats and incubated with 0.25% trypsin and 0.01% DNase. A total of 0.25 million cells per well were plated onto poly-ornithine/fibronectin-coated coverslips in Neurobasal medium containing 2% FBS and 2% B27 supplement (Invitrogen) in six-well culture plates. The culture media were changed to Neurobasal medium plus B27 after 3 d in vitro (DIV). The neurons were transfected at 6–7 DIV using Lipofectamine 2000 (Invitrogen). Two days later, neurons were processed for immunostaining.
Electrophysiology.
Whole-cell voltage-clamp recordings were performed with a patch-clamp amplifier Axopatch 200B (Molecular Devices, Foster City, CA) after capacitance and series resistance (<20 MΩ) compensation of 80%. Agonist-induced responses were low-pass filtered at 2 kHz by an eight-pole Bessel filter, digitally sampled at 5 kHz and analyzed using pClamp software version 9.2 (Molecular Devices). Patch-glass pipettes (3–5 MΩ) were filled with an intracellular solution containing (in mm) 125 gluconic acid, 15 CsCl, 5 EGTA, 10 HEPES, 3 MgCl2, 0.5 CaCl2, and 2 ATP-Mg salt, pH 7.2. The extracellular solution contained (in mm) 160 NaCl, 2.5 KCl, 10 HEPES, 10 glucose, and 0.2 CaCl2, pH 7.3. Agonist application was done using a multibarrel pipette controlled by a stepper motor (SF-77b; World Precision Instruments, Sarasota, FL). Experiments were performed at 23–26°C at a holding potential of −60 mV.
Immunofluorescence.
Intracellular labeling was performed at room temperature. Briefly, cells were fixed with 4% paraformaldehyde (PFA) for 30 min and permeabilized in 0.1% Triton X-100 (TX-100) in PBS for 5 min. Cells were then blocked with 10% goat serum in PBS for 1 h and incubated with primary (rabbit anti-calreticulin antibody; Affinity BioReagents, Golden, CO; 1:1000) and secondary (Alexa Fluor 555 goat anti-rabbit IgG; Invitrogen; 1:1000) antibodies for 1 h.
For cell surface labeling, COS7 cells were washed twice in PBS and blocked with 0.2% bovine serum albumin in PBS on ice for 15 min. This blocking solution was also used for diluting the primary (rabbit anti-GFP antibody; Millipore, Billerica, MA; 1:500) and secondary (Alexa Fluor 555 goat anti-rabbit IgG) antibodies. After incubation on ice for 30 min in the primary antibody, the cells were briefly washed in cold PBS followed by incubation on ice with the secondary antibody for another 30 min. The stained cells were then washed and fixed in 4% PFA in PBS for 20 min and mounted with ProLong Antifade mounting reagent (Invitrogen).
Images from stained COS7 cells and neurons were captured using a Nikon (Tokyo, Japan) Eclipse E1000 fluorescence microscope; a 63× oil objective was used for colocalization studies and a 40× oil objective for surface labeling experiments. The percentages of the cells exhibiting surface labeling were determined by counting of visible cells by an individual. This analysis was followed by the quantifications of intensity of the labeling using MetaMorph software (Universal Imaging, West Chester, PA). The average intensities of surface and total fluorescence were determined for regions outlined around the transfected cells. Intensity measurements are expressed in arbitrary units per unit area. Data are expressed as the mean ± SEM; each value is an average of three independent experiments performed under the same conditions including the setting of microscope parameters. The labeling on nontransfected cells was considered as background. Statistical comparisons were made using an unpaired t test or a one-way ANOVA followed by Newman–Keuls multiple comparison test.
Immunoprecipitation.
COS7 cells grown on 10 cm plates were transfected using the calcium phosphate coprecipitation method (CalPhos Mammalian Transfection kit; Clontech Mountain View, CA). Two days later, cells were washed in PBS and collected by centrifugation (1000 × g for 10 min at 4°C). The pellets were solubilized in 2.5 ml of solubilization solution (1% sodium deoxycholate in 50 mm Tris-HCl, pH 9.0) for 30 min at 37°C and were centrifuged at 45,000 × g for 30 min at 4°C. The resulting supernatants (500 μl) were incubated with 5 μg of each antibody [rabbit or mouse IgG, mouse anti-NR1 (clone 54.2), mouse anti-MYC, and rabbit anti-GFP (Millipore)] and with a 50 μl aliquot of protein A/G agarose beads (Pierce, Rockford, IL) at 4°C overnight. The beads were washed four times with 1 ml of TBST buffer (0.1% TX-100/TBS) and boiled in 2× SDS loading buffer (25 μl). Proteins were loaded onto 4–20% Tris-glycine gels (Invitrogen), transferred to polyvinylidene difluoride membranes, incubated with primary and secondary antibodies, and detected with ECL using BioMax MR x-ray films (Eastman Kodak, Rochester, NY).
Results
NR1 and NR2B subunits truncated after M4 are retained in the ER
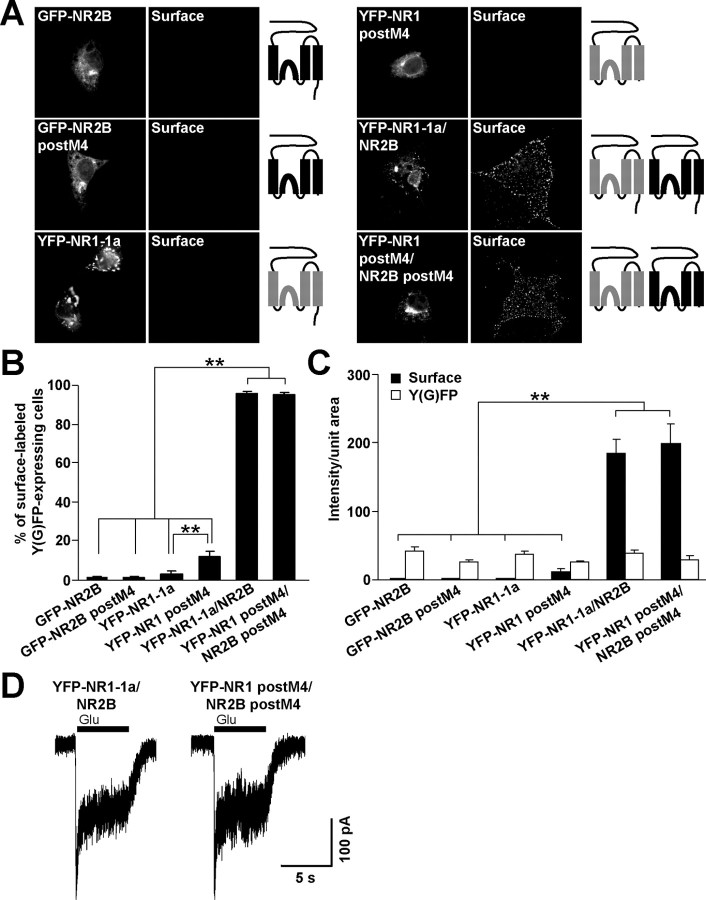
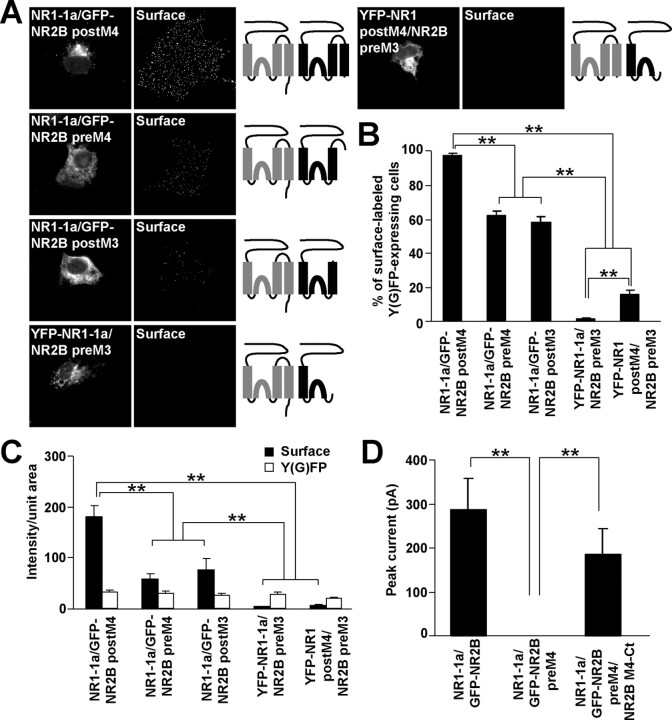
It has been shown previously that the C termini of both NR1-1 and NR2B subunits appended to tac are retained in the ER (Standley et al., 2000; Scott et al., 2001; Xia et al., 2001; Hawkins et al., 2004), indicating the presence of one or more retention sites in NR1-1 and NR2B C termini. However, NR2B subunits truncated immediately after M4 are retained intracellularly suggesting that there is at least one additional retention signal in the remaining part of the molecule (Hawkins et al., 2004). In this study, we sought to identify that site(s). Because NR2B with a complete truncation of the C terminus does not form functional channels after assembly with NR1 (Hawkins et al., 2004) (data not shown), we used an NR2B subunit truncated six amino acid residues after M4 (NR2B postM4). Similarly, NR1 subunits truncated five amino acid residues after M4 (NR1 postM4), shown previously to form functional channels (Vissel et al., 2001), were used in our experiments. We expressed full-length and truncated NMDA receptor subunits in COS7 cells, which lack native NMDA receptor subunits, and performed the surface labeling experiments with anti-GFP antibodies in nonpermeabilizing conditions. The vast majority of COS7 cells expressing either full-length GFP-NR2B or YFP-NR1-1a alone were not surface labeled, as expected; similar data were obtained for GFP-NR2B postM4 (Fig. 1A,B). Surprisingly, most COS7 cells expressing YFP-NR1 postM4 also showed no detectable surface staining, although the percentage of COS7 cells expressing YFP-NR1 postM4 on the surface was significantly higher compared with full-length YFP-NR1-1a subunits (Fig. 1B), indicating that YFP-NR1 postM4 is not completely retained in the cell. These data show that both NR2B and NR1 subunits truncated after M4 possess signals that inhibit their cell surface expression.
Figure 1.
NR2B and NR1 subunits truncated after M4 are localized intracellularly. A, COS7 cells transfected with indicated cDNAs were surface stained with rabbit anti-GFP antibodies. Schematic diagrams show the structure of NR2B (black) and NR1 (gray) subunits. B, The percentages of cells having surface staining were determined for >300 Y(G)FP-expressing cells for each combination of NMDA receptor subunits in three experiments. The bar graphs represent mean ± SEM. **p < 0.001, ANOVA. C, Data represent mean ± SEM of fluorescence intensities per unit area obtained for surface (black) or total Y(G)FP (white) expression. More than 45 Y(G)FP-expressing cells for each combination of cDNAs were analyzed in three experiments. **p < 0.001, ANOVA. D, Whole-cell patch-clamp recordings were performed from HEK293 cells cotransfected with YFP-NR1-1a/NR2B or YFP-NR1 postM4/NR2B postM4. Representative traces for both combinations are shown. Currents were elicited with 5 s applications of 1 mm glutamate and 1 mm glycine (indicated by filled bar). Quantitative analysis of the NMDA receptor mediated currents revealed that the mean peak current amplitudes were not significantly different between YFP-NR1-1a/NR2B and YFP-NR1 postM4/NR2B postM4; n = 8; p > 0.05, unpaired t test.
As expected, the coexpression of YFP-NR1-1a/NR2B and YFP-NR1 postM4/NR2B postM4 resulted in robust surface labeling in nearly all YFP-expressing cells (Fig. 1A,B). Subsequent analysis revealed that the average fluorescent intensity per unit area of the surface labeling was significantly lower for the cells transfected with single full-length or truncated NMDA receptor subunits compared with cells expressing YFP-NR1-1a/NR2B or YFP-NR1 postM4/NR2B postM4 (Fig. 1C). No significant difference was seen in the fluorescent intensities of the surface labeling between YFP-NR1-1a/NR2B- and YFP-NR1 postM4/NR2B postM4-expressing cells and among the intensities of total GFP or YFP fluorescence of the full-length and truncated GFP-NR2B or YFP-NR1 subunits (Fig. 1C). Therefore, the C termini of NR1-1 and NR2B subunits do not affect surface expression of assembled complexes in heterologous cells.
To ensure that the truncation of C termini does not affect the formation of functional ion channels, we transfected YFP-NR1-1a/NR2B and YFP-NR1 postM4/NR2B postM4 into HEK293 cells and performed whole-cell patch-clamp recording. NMDA receptor-mediated currents were evoked by 5 s application of 1 mm glutamate and 1 mm glycine at a holding potential of −60 mV. Both combinations of NMDA receptor subunits were able to form functional channels. Statistical analysis of the peak current magnitudes revealed no significant difference between YFP-NR1-1a/NR2B and YFP-NR1 postM4/NR2B postM4 receptors (Fig. 1D). This observation is consistent with the quantification of the surface labeling performed for the same combinations of NMDA receptor subunits (Fig. 1C).
Because the YFP-NR1 postM4 and GFP-NR2B postM4 did not reach the cell surface when expressed by themselves, we investigated the compartment in which the truncated NMDA receptor subunits were localized. Supplemental Figure 1 (available at www.jneurosci.org as supplemental material) shows that the distribution of YFP-NR1 postM4 and GFP-NR2B postM4, expressed in COS7 cells, closely matched the staining for the ER marker calreticulin. These data suggest that both NR1 and NR2B subunits lacking their C termini contain ER retention signals that can be masked by assembly into an NR1/NR2B complex.
NR2B and NR1 M3 possess ER retention signals
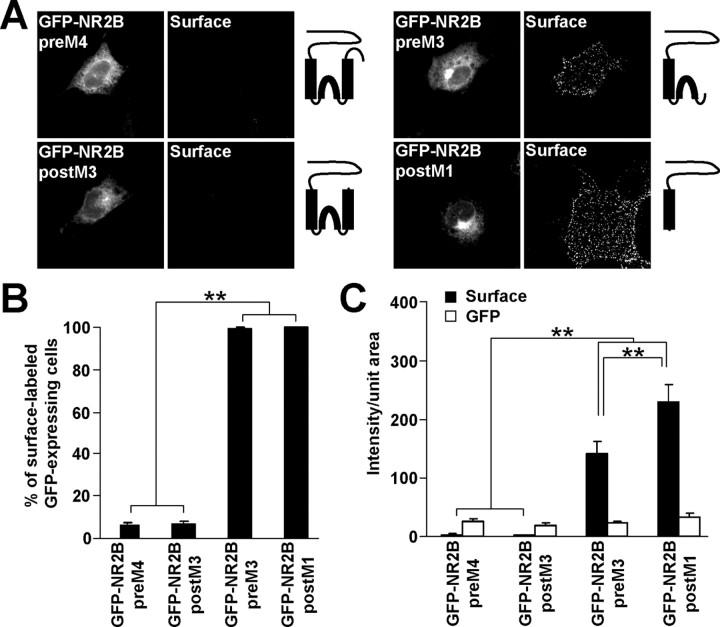
To map the ER retention signals preceding the C terminus of the NR2B subunit, truncations were made before M4 (GFP-NR2B preM4), after M3 (GFP-NR2B postM3), before M3 (GFP-NR2B preM3), and after M1 (GFP-NR2B postM1). The resulting constructs were expressed in COS7 cells, and surface receptors were labeled with anti-GFP antibodies. Immunofluorescent microscopy revealed that only a small fraction of COS7 cells expressing GFP-NR2B preM4 or GFP-NR2B postM3 exhibited detectable surface staining, in contrast to nearly all cells expressing GFP-NR2B preM3 and GFP-NR2B postM1 on the surface (Fig. 2A,B). Quantitative fluorescent measurements showed that surface labeling of GFP-NR2B preM4 and GFP-NR2B postM3 was significantly smaller than that of GFP-NR2B preM3 and GFP-NR2B postM1 (Fig. 2C). The intensities of total GFP fluorescence were not significantly different among the GFP-NR2B truncations (Fig. 2C). These results suggest that NR2B M3 contains an ER retention signal that prevents the unassembled NR2B postM4 subunit from reaching the cell surface.
Figure 2.
NR2B M3 contains an ER retention signal. A, COS7 cells transfected with GFP-NR2B truncations were stained for surface GFP. Schematic diagrams show the structure of GFP-NR2B truncation constructs. B, The percentages of cells having surface staining were determined for >300 GFP-expressing cells for each construct in three experiments. The bar graph represents mean ± SEM. C, Data represent mean ± SEM of fluorescent intensities per unit area obtained for surface (black) or total GFP (white) expression. More than 45 GFP-expressing cells for each construct were analyzed in three experiments. **p < 0.001, ANOVA.
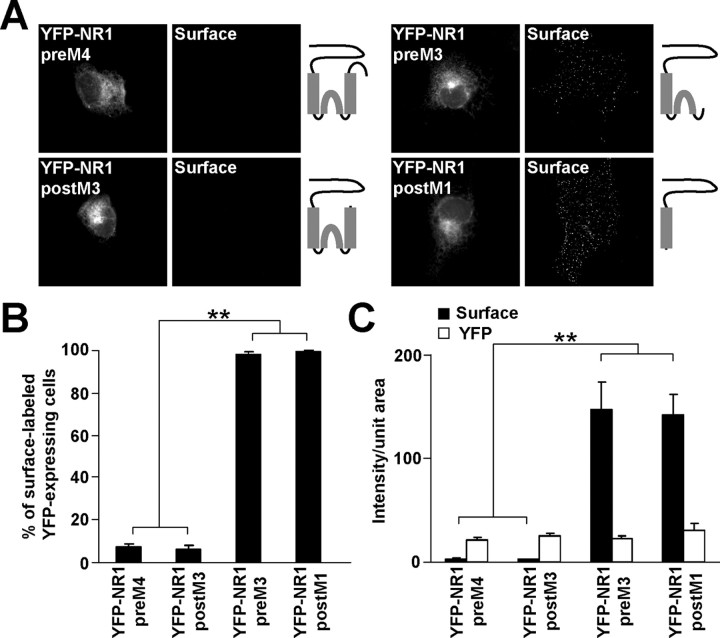
Because we showed that NR1 postM4, like NR2B postM4, contains an ER retention signal (Fig. 1, supplemental Fig. 1, available at www.jneurosci.org as supplemental material), truncations were made in YFP-NR1-1a to identify the location of the signal. The results of the surface labeling of COS7 cells expressing these NR1 truncations were similar to those obtained for NR2B truncations. YFP-NR1 preM4- and YFP-NR1 postM3-expressing cells did not exhibit surface staining, in contrast to YFP-NR1 preM3- and YFP-NR1 postM1-expressing cells, which were heavily surface labeled (Fig. 3A,B). Quantification of these results revealed a dramatic difference in the intensity of the surface labeling between YFP-NR1 preM4, YFP-NR1 postM3 and YFP-NR1 preM3, YFP-NR1 postM1 (Fig. 3C); the intensities of total YFP fluorescence were not significantly different among the YFP-NR1 truncations (Fig. 3C).
Figure 3.
NR1 M3 contains an ER retention signal. A, Live COS7 cells expressing YFP-NR1 truncations were labeled with polyclonal anti-GFP antibodies. Schematic diagrams show the structure of YFP-NR1 truncations. B, The percentages of surface labeled cells were determined for >300 YFP-positive cells for each construct in three experiments. Data represent mean ± SEM. C, Surface (black) and total (white) expression of YFP-NR1 truncations is shown. Data represent mean ± SEM of fluorescence intensities per unit area obtained from at least 45 YFP-expressing cells for each construct in three experiments. **p < 0.001, ANOVA.
Because GFP-NR2B postM3 and YFP-NR1 postM3 were not trafficked to the cell surface, we asked if they were retained in the ER. The intracellular labeling of COS7 cells showed an extensive colocalization between these constructs and the ER marker calreticulin (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Together, our data suggest that M3 of both NR1 and NR2B contain ER retention signals.
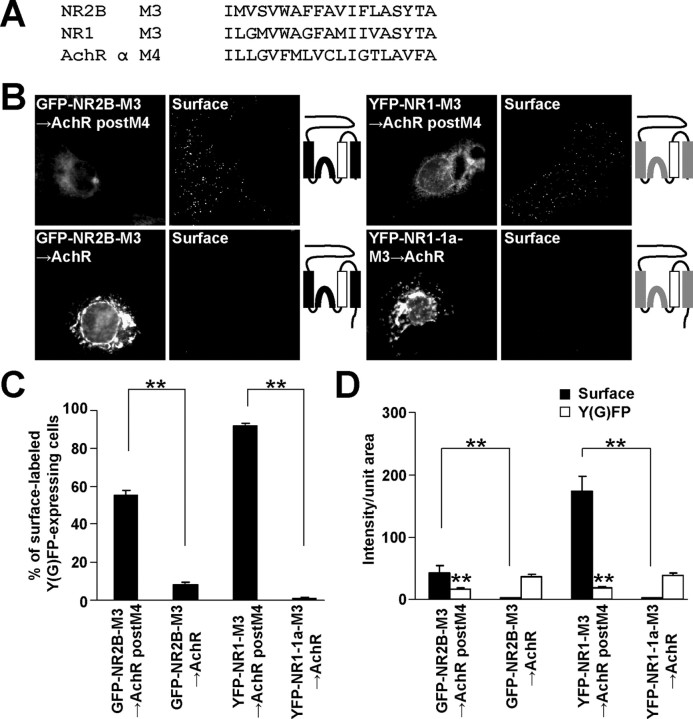
To confirm our results obtained with truncation constructs, we substituted M3 in both NR1 and NR2B subunits for the acetylcholine receptor (AchR) α M4 (Fig. 4A), which has the same orientation in the membrane as that of M3 in NMDA receptor subunits (Witzemann et al., 1990) and does not contain any ER retention signal (Wang et al., 2002). More than 50% of COS7 cells expressing GFP-NR2B-M3→AchR postM4 were surface labeled compared with ∼10% of those expressing full-length GFP-NR2B-M3→AchR (Fig. 4B,C). Fluorescent intensities of the surface labeling were significantly different between these two individually expressed NR2B chimeras (Fig. 4D). These results corroborate our observation that the full-length NR2B subunit contains two different ER retention signals: one in NR2B M3 and one in the C terminus. Even more pronounced differences were seen for the surface expressions of YFP-NR1-M3→AchR postM4 and YFP-NR1-1a-M3→AchR (Fig. 4C,D). Interestingly, the total expression levels of both GFP-NR2B-M3→AchR postM4 and YFP-NR1-M3→AchR postM4 were significantly lower when compared with their full-length variants (Fig. 4D); this may reflect that the C termini prevent the degradation of the chimeric subunits in the cells. Our observations support the presence of ER retention signals in M3 of both NR1 and NR2B.
Figure 4.
The substitution of M3 for AchR α M4 facilitates the surface expression of NR1 postM4 and NR2B postM4. A, The amino acid sequences of the NR2B M3, NR1 M3 and AchR α M4 are shown. B, COS7 cells transfected with indicated cDNAs were immunostained for surface Y(G)FP. Schematic diagrams of membrane topology of NR2B (black) and NR1 (gray) subunits with replaced M3 for AchR α M4 (white) are shown. C, Plotted data represent mean ± SEM of the percentages of surface labeled Y(G)FP-expressing cells determined for each chimera from >300 cells in three experiments. D, Quantifications of NMDA-AchR chimeric receptor surface (black) and total Y(G)FP (white) expressions are shown. The surface and total expression levels of both GFP-NR2B-M3→AchR postM4 and YFP-NR1-M3→AchR postM4 were compared with their full-length variants (in mean ± SEM of fluorescence intensities per unit area; n >45 cells in three experiments). **p < 0.001, unpaired t test.
NR2B M3 is necessary for overcoming the NR1 M3 ER retention signal
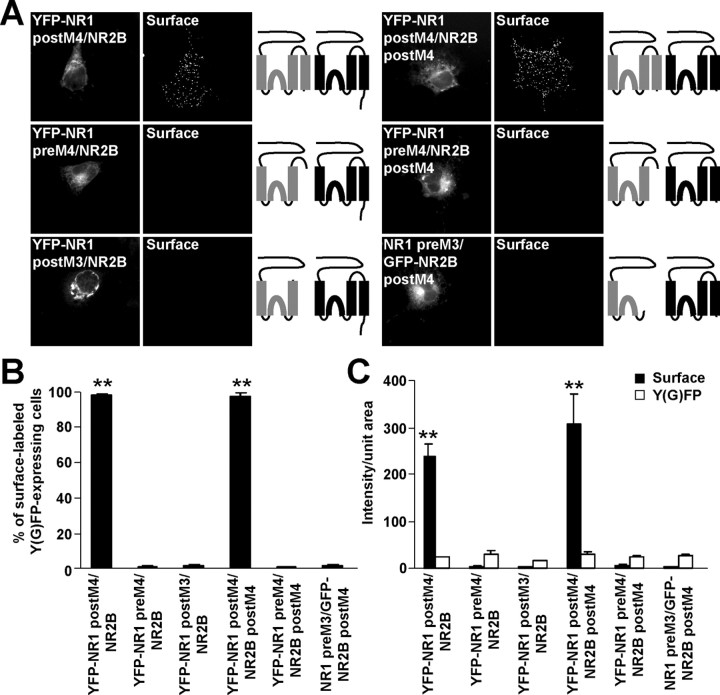
The mechanisms underlying the masking of ER retention signals after assembly of NR1 and NR2 remain mostly unexplored. Our results suggesting that M3 of both NR1 and NR2B subunits contain ER retention signals raise the interesting possibility that these signals are mutually masked during assembly. To address this, we transfected the NR2B truncation constructs together with NR1-1a or NR1 postM4 subunits into COS7 cells. When expressed alone, GFP-NR2B preM4 and GFP-NR2B postM3 truncations were retained in the ER (Fig. 2). However, when expressed with NR1-1a subunits, they were transported to the cell surface (Fig. 5A), although their surface expression was decreased when compared with that of NR1-1a/GFP-NR2B postM4 (Fig. 5B,C). The total GFP fluorescence intensities were not significantly different among cells expressing NR1-1a/GFP-NR2B postM4, NR1-1a/GFP-NR2B preM4, and NR1-1a/GFP-NR2B postM3 (Fig. 5C). These results suggest that ER retention signals from both NR1 and NR2B M3 are masked after NR1/NR2B interaction, even when NR2B M4 and an extracellular loop between M3 and M4 are not present.
Figure 5.
NR2B M3 is a necessary structural component for negating the NR1 M3 ER retention signal. A, COS7 cells transfected with indicated combinations of cDNAs were stained for surface GFP. Schematic diagrams of membrane topology of NR1 (gray) and NR2B (black) subunits are shown. B, Bar graph represents the percentages of transfected COS7 cells exhibiting the surface staining in mean ± SEM; n >300 cells for each combination of cDNAs in three experiments. C, Quantification of surface (black) and total Y(G)FP (white) expression of indicated combinations of cDNAs. Data show mean ± SEM of fluorescence intensities of >45 transfected cells measured in three experiments. D, GFP-NR2B preM4 can form a functional channel after coexpression with NR2B M4-Ct and NR1-1a. Currents evoked by the application of 1 mm glutamate and 1 mm glycine were recorded from HEK293 cells expressing indicated combinations of cDNAs. The mean peak current amplitudes ± SEM are shown; n = 6. **p < 0.001, ANOVA.
In contrast, NR2B preM3, which in the absence of NR1 subunit exhibits robust surface expression (Fig. 2), did not facilitate surface expression of YFP-NR1-1a or YFP-NR1 postM4 (Fig. 5A). The significant difference in the percentage of surface labeled cells expressing YFP-NR1-1a/NR2B preM3 and YFP-NR1 postM4/NR2B preM3 (Fig. 5B) likely reflects different properties of surface expression of individually expressed YFP-NR1-1a and YFP-NR1 postM4 (Fig. 1B). The surface and total YFP fluorescent intensities were not significantly different between cells expressing YFP-NR1-1a/NR2B preM3 and YFP-NR1 postM4/NR2B preM3 (Fig. 5C).
It has been shown previously that both NR1 and NR2A subunits truncated before M4 can form a functional channel when separate NR1 and NR2A M4-Ct peptides are coexpressed (Schorge and Colquhoun, 2003). Next, we used this approach to learn whether truncated GFP-NR2B preM4 also can form a functional channel. Similarly, we found that HEK293 cells expressing NR1-1a/GFP-NR2B preM4 exhibited no responses to 1 mm glutamate and 1 mm glycine at a membrane potential of −60 mV; however, the functional properties of truncated NR1-1a/GFP-NR2B preM4 receptors were rescued when the NR2B M4-Ct peptide was coexpressed. Interestingly, the peak current magnitudes were not significantly different between NR1-1a/NR2B and NR1-1a/GFP-NR2B preM4/NR2B M4-Ct (Fig. 5D). Our results, together with previously published data, show that the truncations of NMDA receptor subunits before M4 do not affect the ability to form functional channels when corresponding M4-Ct peptides are present. As we showed above, the NR1/GFP-NR2B preM4 complex reached the cell surface without the M4-Ct peptide, although functional channels were not formed. This suggests that NR2B M4 is an important structural component for the formation of the functional channel but is not necessary for trafficking of the receptors to the cell surface.
The lack of surface staining of YFP-NR1-1a and YFP-NR1 postM4 coexpressed with NR2B preM3 suggests that NR2B M3 is a necessary structural component for masking of the NR1 M3 ER retention signal or that the truncated NR2B preM3 subunit cannot form a molecular complex with the NR1 subunit. To address this, coimmunoprecipitation analysis was performed from COS7 cell lysates transfected with GFP-NR2B truncations and full-length NR1-1a subunits (supplemental Fig. 2A, available at www.jneurosci.org as supplemental material). These experiments showed not only that GFP-NR2B postM3 could physically associate with the NR1-1a subunit, as expected from immunofluorescence data, but also that both GFP-NR2B preM3 and GFP-NR2B postM1 interacted with NR1-1a. Therefore, these results show that NR2B M3 is involved in masking of the NR1 M3 ER retention signal.
The presence of NR1 M4 is necessary for masking the NR2B M3 ER retention signal
Our results, together with previously published data, indicate that the full-length NR2B subunit contains at least two ER retention signals, one in NR2B M3 and one in the C terminus, which must be masked before a functional NR1/NR2B receptor leaves the ER. We next investigated which structural parts of NR1 subunits are necessary for negating the NR2B ER retention signals. We coexpressed full-length NR2B subunit with the NR1 truncation constructs, YFP-NR1 postM4, YFP-NR1 preM4, and YFP-NR1 postM3, which, in the absence of NR2B, are mainly retained in the ER (Fig. 1, 3). Indeed, the coexpression of YFP-NR1 postM4 with full-length NR2B resulted in robust surface staining of NR1, whereas the coexpression of NR2B did not facilitate surface expression of YFP-NR1 preM4 or YFP-NR1 postM3 (Fig. 6A). These findings indicate that NR1 M4 is an important structural component for the masking of the NR2B ER retention signal(s). To investigate this further, we coexpressed NR2B postM4, which contains only the NR2B M3 ER retention signal, with YFP-NR1 postM4, YFP-NR1 preM4 and NR1 preM3 and performed surface staining. The results were very similar to those obtained for the full-length NR2B subunit: strong surface staining with coexpression of YFP-NR1 postM4 with NR2B postM4, but lack of surface staining for other combinations that are missing NR1 M4 (Fig. 6A). Statistical analysis showed a significant difference in both percentages of surface labeled cells and fluorescent intensities of surface expression between NMDA receptor combinations containing and lacking NR1 M4 (Fig. 6B,C). The fluorescent intensities of total protein expressions were not significantly different among the studied combinations of NMDA receptor subunits (Fig. 6C). Thus, we conclude that NR1 M4 is a necessary structural domain for overcoming the NR2B M3 ER retention signal.
Figure 6.
NR1 M4 is necessary for masking the NR2B M3 ER retention signal. A, Live cells transfected with indicated combinations of NR1 and NR2B constructs were immunostained with anti-GFP antibodies. Schematic drawings of membrane topology of NR1 (gray) and NR2B (black) subunits are shown. B, Data represent the percentages of surface labeled cells expressing indicated NMDA receptor subunits (in mean ± SEM; n >300 cells for each combination of cDNAs in three experiments). C, Each vertical bar represents the mean ± SEM of fluorescent intensities per unit area (n >45 cells for each combination of NMDA receptor subunits in three experiments). **p < 0.001, ANOVA.
We then examined the physical interaction between full-length NR2B and NR1 truncations. Coimmunoprecipitation experiments showed that NR2B subunit could physically interact with all of the NR1 truncations constructs (YFP-NR1 preM4, YFP-NR1 postM3, YFP-NR1 preM3, and YFP-NR1 postM1) (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material), although the NR1/NR2B complexes lacking NR1 M4 were not able to reach the cell surface (Fig. 6 A–C). Together, our results support a model in which the interaction of both the NR2B and NR1 M3 and NR1 M4 in the NR1/NR2B complex results in functional masking of the ER retention signals found in NR2B and NR1 M3.
Replacement of M3 in both NR2B and NR1 for AchR α M4 and of M4 in NR1 for AchR α M3 abolishes the masking of the ER retention signals in NR1 and NR2B subunits
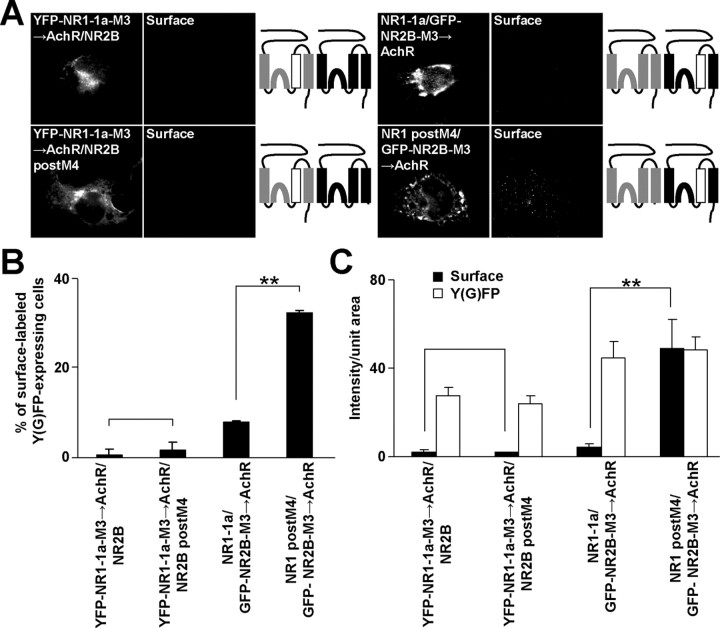
Our truncation studies show that the M3 domains of both NR1 and NR2B contain ER retention signals and that M3 of NR2B, together with M4 of NR1, are involved in masking these signals. Using truncation studies we were unable to determine whether or not M3 of NR1 was also involved in masking. To address this and to further explore the masking of ER retention signals, we studied the NMDA-AchR chimeras. We first performed coimmunoprecipitations of chimeric and native NMDA receptor subunits from COS7 cell lysates. Supplemental Figure 3 (available at www.jneurosci.org as supplemental material) shows that the replacement of M3 in both NR2B and NR1 for AchR α M4 did not affect the ability of the subunits to interact with full-length NR1-1a and NR2B subunits.
We then cotransfected full-length YFP-NR1-1a-M3→AchR with NR2B or NR2B postM4 into COS7 cells and stained them with anti-GFP antibodies in nonpermeabilizing conditions. Neither NR2B nor NR2B postM4 was able to traffic with YFP-NR1-1a-M3→AchR to the cell surface (Fig. 7 A–C). This observation shows that NR1 M3 is necessary for the masking of the NR2B M3 ER retention signal.
Figure 7.
M3 of NR1 and NR2B are necessary for the negating of the ER retention signals in M3 of NR2B and NR1. A, COS7 cells transfected with indicated combinations of NMDA receptor subunits were immunostained for surface GFP. Schematic drawings of membrane topology of NR1 (gray) and NR2B (black) subunits with replaced M3 for AchR α M4 (white) are shown. B, The percentages of surface-labeled transfected cells determined for each NR1/NR2 combination from >300 cells in three experiments are shown in mean ± SEM. C, Data represent mean ± SEM of surface (black) and total (white) fluorescence intensities per unit area of indicated combinations of subunits; n >45 cells in three experiments. **p < 0.001, unpaired t test.
Next, GFP-NR2B-M3→AchR was expressed together with NR1-1a or NR1 postM4, and GFP surface labeling was measured. The coexpression of full-length NR1-1a subunit did not promote surface expression of GFP-NR2B-M3→AchR; however, NR1 postM4 was able to significantly facilitate the surface expression of GFP-NR2B-M3→AchR in ∼30% of GFP-expressing cells (Fig. 7 A–C). This increase in surface staining of GFP-NR2B-M3→AchR is consistent with our finding that NR1 postM4 can reach the cell surface by itself to a limited extent (Fig. 1B). Together, our results strongly corroborate the idea that both NR1 and NR2B M3 play critical roles in ER retention as well as in the masking of the ER retention signals.
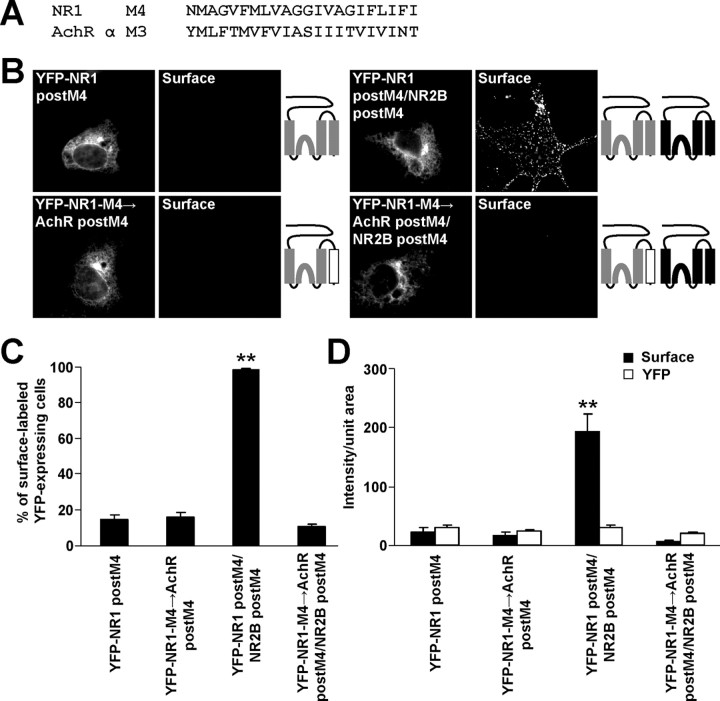
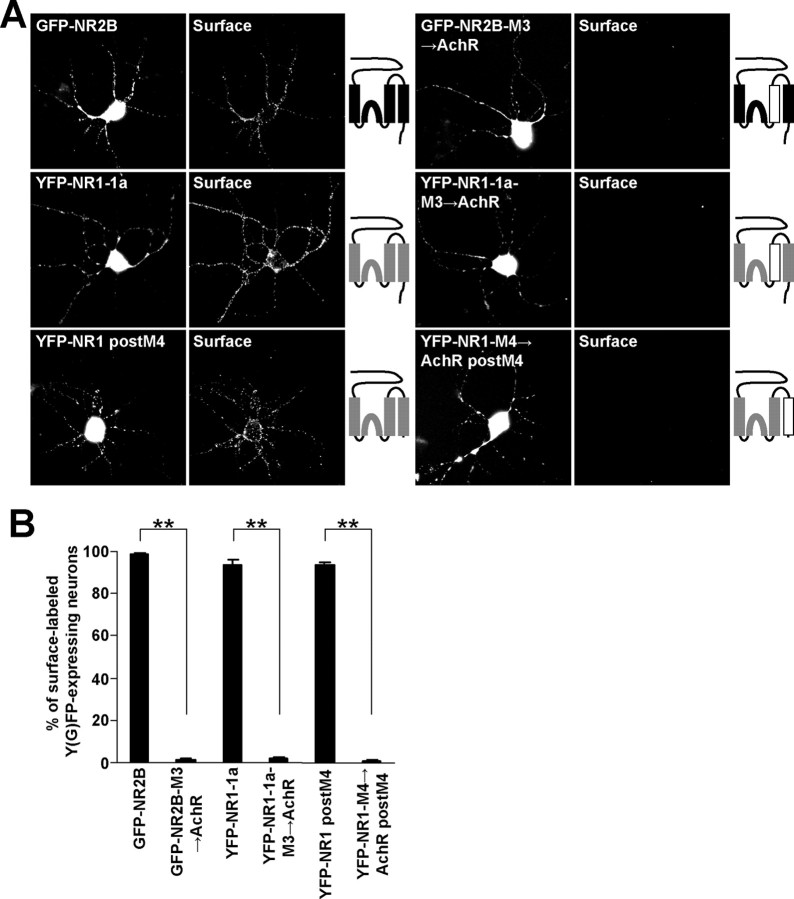
Finally, to confirm that the structure of the NR1 M4 is critical for the masking of the ER retention signals, the NR1 M4 in the NR1 postM4 construct was replaced for AchR α M3 (YFP-NR1-M4→AchR postM4) (Fig. 8A), which has the same orientation in the membrane as NR1 M4 (Witzemann et al., 1990). When expressed alone, this chimera trafficked to the surface in a small fraction of transfected cells, similar to the surface expression of NR1 postM4 (Fig. 8 B–D). However, the coexpression of NR2B postM4 with these constructs resulted in the strong surface targeting only for YFP-NR1 postM4/NR2B postM4 complexes. YFP-NR1-M4→AchR postM4/NR2B postM4 receptors exhibited very low surface expression (Fig. 8 B–D). Together with experiments performed in cortical neuronal cultures (Fig. 9A,B), our data show that M3 of both NR1 and NR2B together with M4 of NR1 are necessary for the masking of the ER retention signals found in M3 of NR1 and NR2B.
Figure 8.
The replacement of NR1 M4 for AchR α M3 inhibits surface targeting of the NMDA receptors. A, The amino acid sequences of the NR1 M4 and AchR α M3 are shown. B, COS7 cells transfected with indicated combinations of cDNAs were immunostained for surface YFP. Schematic diagrams of membrane topology of NR2B (black) and NR1 (gray) subunits with replaced M4 for AchR α M3 (white) are illustrated. C, Bar graph represents the percentages of YFP-expressing COS7 cells exhibiting the surface staining in mean ± SEM; n >300 cells in three experiments. D, Measurements of cell surface (black) and total (white) expressions of indicated NMDA receptor subunit variants. Each vertical bar represents the mean ± SEM of fluorescent intensities per unit area; n >45 cells in three experiments. **p < 0.001, ANOVA.
Figure 9.
Replacement of both NR2B and NR1 M3 for AchR α M4 and NR1 M4 for AchR α M3 inhibits surface targeting of the NMDA receptors in cortical neurons. A, Cortical neurons transfected with indicated combinations of cDNAs were immunostained for surface Y(G)FP. Schematic diagrams of membrane topology of NR2B (black) and NR1 (gray) subunits with replaced M3 or M4 for AchR α M (white) are shown. B, Bar graph represents the percentages of Y(G)FP-expressing cortical neurons exhibiting the surface staining in mean ± SEM; n >100 neurons in three experiments. **p < 0.001, unpaired t test.
Discussion
ER retention has evolved as a mechanism for preventing the expression of unassembled or improperly assembled membrane proteins from reaching the cell surface. This is particularly critical for receptors whose number and composition on the cell surface determines how the cell responds to particular stimuli. In the present study, we investigated ER retention of the NMDA receptor, a molecule that is essential for neurotransmission. We show that the M3 domains of the NMDA receptor subunits NR1 and NR2B contain ER retention signals, which, together with the previously identified C-terminal retention signals, control surface expression of the NMDA receptor. We also find that the same structures, as well as M4 of NR1, are involved in masking of these retention sites and allow the release of the functional receptor from the ER.
M3 of NR1 and NR2B regulate ER retention of unassembled subunits
Our initial finding that removal of the entire C termini still resulted in ER retention of both NR1 and NR2B subunits indicated that additional retention signals were present in the proximal areas of both subunits. Our serial truncation studies and replacement of M3 with M4 of the AchR α subunit show that M3 of both NR1 and NR2B subunits plays a role in ER retention of the unassembled subunit. Replacement of M3 with M4 of the AchR α and removal of the C termini led to surface expression of both NR1 and NR2B indicating that either the M3 or the C-terminal retention signals can alone cause ER retention of the individual subunits.
Our identification of ER retention associated with membrane domains is not unprecedented; membrane domains of the T cell antigen receptor chains (Call and Wucherpfennig, 2005), microsomal cytochrome P450 (Murakami et al., 1994), ubiquitin-conjugating enzyme UBC6 (Yang et al., 1997) and the AchR α subunit (Wang et al., 2002) have been reported to contain ER retention signals. A discrete ER retention motif has been described only in the case of the AchR α subunit [PL(Y/F)(F/Y)XXN motif in M1]. Mutagenesis of amino acids in NR2B M3 (supplemental Fig. 4, available at www.jneurosci.org as supplemental material) led to a slightly increased surface expression compared with NR2B postM4, but significantly less surface expression than that obtained with NR2B-M3→AchR postM4. These findings suggest that the structural integrity of M3 of both NR1 and NR2B is important for ER retention, and it is unlikely that a specific motif within the M3 is responsible for retention.
Masking of ER retention signals in NR1/NR2 complexes
Previous studies indicate that the functional NMDA receptor is arranged as a dimer of dimers with a NR1-NR1-NR2-NR2 orientation (Schorge and Colquhoun, 2003). Previous data suggest that NMDA receptor assembly starts with the formation of the NR1/NR2 dimers (Furukawa et al., 2005). By analogy to AMPA receptors, the first step in the assembly of the NMDA receptor might be the physical interaction between N termini of NR1 and NR2 followed by interactions among other regions (Leuschner and Hoch, 1999; Ayalon and Stern-Bach, 2001). Our coimmunoprecipitation experiments indicate that N-terminal regions of both NR2B and NR1 subunits containing M1 are sufficient for direct physical interactions with full-length NR1-1a and NR2B subunits. These findings support previous studies that reported that the N termini of NR1 and NR2 subunits are important for the oligomerization of NMDA receptors (Meddows et al., 2001).
Our results indicate that both the NR1-1 and the NR2B subunits have two distinct ER retention signals, present in M3 and in the C termini, raising questions about the roles of these individual signals as well as how they are negated with the assembly of the NMDA receptor. Interestingly, the coexpression of the NR1-1a/NR2B receptors lacking either the NR1-1 or the NR2B C terminus or both C termini did not significantly affect the levels of surface staining compared with the surface expression of full-length NR1-1a/NR2B receptors. This shows that mutual masking of the C-terminal ER retention signals, proposed for GABAB receptor and potassium channels (Zerangue et al., 1999; Margeta-Mitrovic et al., 2000, 2001), is not the mechanism involved in the masking of the NMDA C-terminal ER retention signals. This conclusion is in agreement with previous papers showing that NR1 and NR2B C termini do not interact (Hawkins et al., 2004) and that functional NR1/NR2A receptors lacking both C termini are formed in HEK293 cells (Vissel et al., 2001) and in Xenopus oocytes (Zheng et al., 1999).
Using the truncated and chimeric NMDA receptor subunits, we showed that M3 of both NR1 and NR2B is necessary, not only for retention of the unassembled subunits in the ER, but also for masking the ER retention signals in M3. Furthermore, our data show that NR1 M4 is a critical component for the masking of ER retention signals in NR1/NR2B complexes. Meddows et al. (2001) showed that the NR1 subunit truncated before M4 coassembled with NR2A and reached the cell surface, in contrast to our results using analogous constructs of NR2B. However, Meddows et al. (2001) found weak surface expression of NR1 preM4/NR2A complexes compared with that of full-length NR1/NR2A, supporting our conclusion that only a small fraction of the NR1 preM4/NR2 complexes reach the cell surface.
What is the mechanism underlying the masking of the NR1 and NR2B M3 ER retention signals in the assembled complex? Given their proximity in the membrane, it is likely that NR2B M3 interacts directly with both M3 and M4 of NR1 causing structural changes that result in release from ER retention. This model is supported by the observation that NR1 subunits truncated before M4 can form functional channels when separate NR1 M4-C-terminal peptides and NR2A subunits are present (Schorge and Colquhoun, 2003). Because it is unknown if the NR1 C terminus can interact with other regions of the NMDA receptor subunits, we suggest that NR1 M4 interacts with other membrane domains in the functional receptor and thus helps to negate the ER retention signals in NR1 and NR2B M3.
Although our results indicate that M3 and M4 (NR1 only) are involved in ER masking, we cannot rule out the possibility that the modifications that we made in these domains affects the assembly mechanism itself, for example, by destabilizing the formation of dimers or tetramers. In their study of AMPA receptor assembly, Ayalon and Stern-Bach (2001) found that although the N terminus is important for the initial assembly event, more distal regions of the molecule are also required; they proposed that the N terminus is associated with dimer:dimer formation and the association of the dimers is mediated by C-terminal determinants.
Multiple ER retention signals are present in NMDA receptor subunits
Our data indicate that ER retention signals in the C termini are capable of retaining the entire molecule when the M3 retention is removed. This was best illustrated when M3 of NR2B or NR1-1 was replaced by the AchR α M4. In both cases, surface staining was present in the absence of the C termini, but absent when the C termini were intact. The question remains, therefore, as to how the C-terminal ER retention signals are negated in the assembled NR1/NR2 complex. As shown previously, the NR1 ER retention motifs are splice variant-dependent based on expression of an RXR retention motif and a COPII (coat protein complex II) or PDZ binding domain that serves as an export motif (Standley et al., 2000; Scott et al., 2001; Xia et al., 2001; Mu et al., 2003). Therefore, most splice variants of NR1 are not ER-retained in their unassembled form. However, we show that NR1 with the entire C terminus truncated is only slightly released from the ER, whereas some splice variants are robustly expressed on the cell surface. This suggests that export signals in the C termini are able to override the ER retention of M3 in the NR1 subunit. Similarly, we suggest that during the assembly of NR1/NR2, the membrane domain interactions create a powerful export signal that overrides the C-terminal ER retention because it is unlikely that a direct interaction among C termini and M3 is involved. Why does a single subunit require two distinct retention signals that apparently do not directly interact? One possibility is that the different ER retention signals dominate at different levels of assembly. For example, one site may be blocked after assembly of the initial NR1/NR2 dimer, and the second site may be blocked when the tetramer is properly assembled and folded. Interestingly, however, elimination of the C termini did not significantly affect surface expression of functional NMDA receptors. An alternative possibility is that the two retention sites may play a role in targeting the subunits to specific microcompartments within the ER. It is plausible that each ER retention signal could bind a specific factor responsible for the retention and that various extracellular and intracellular stimuli can regulate the interactions with these factors resulting in the tightly regulated assembly of the receptors depending on the actual need for replenishment of the surface receptors. Finally, the two ER retention signals may simply represent redundancy to ensure that this critical molecule is properly assembled and folded before it is released from the ER.
Footnotes
This work was supported by the National Institute on Deafness and Other Communication Disorders Intramural Program. We thank L. Ge and Y.-X. Wang for excellent technical assistance and R. A. Al-Hallaq, R. S. Petralia, and C. C. Swanwick for critical reading of this manuscript.
References
- Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–113. doi: 10.1016/s0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Call ME, Wucherpfennig KW. The T cell receptor: critical role of the membrane environment in receptor assembly and function. Annu Rev Immunol. 2005;23:101–125. doi: 10.1146/annurev.immunol.23.021704.115625. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Fukaya M, Kato A, Lovett C, Tonegawa S, Watanabe M. Retention of NMDA receptor NR2 subunits in the lumen of endoplasmic reticulum in targeted NR1 knockout mice. Proc Natl Acad Sci USA. 2003;100:4855–4860. doi: 10.1073/pnas.0830996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Hawkins LM, Chazot PL, Stephenson FA. Biochemical evidence for the co-association of three N-methyl-d-aspartate (NMDA) R2 subunits in recombinant NMDA receptors. J Biol Chem. 1999;274:27211–27218. doi: 10.1074/jbc.274.38.27211. [DOI] [PubMed] [Google Scholar]
- Hawkins LM, Prybylowski K, Chang K, Moussan C, Stephenson FA, Wenthold RJ. Export from the endoplasmic reticulum of assembled N-methyl-d-aspartic acid receptors is controlled by a motif in the c terminus of the NR2 subunit. J Biol Chem. 2004;279:28903–28910. doi: 10.1074/jbc.M402599200. [DOI] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8:413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- Leuschner WD, Hoch W. Subtype-specific assembly of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their n-terminal domains. J Biol Chem. 1999;274:16907–16916. doi: 10.1074/jbc.274.24.16907. [DOI] [PubMed] [Google Scholar]
- Luo JH, Fu ZY, Losi G, Kim BG, Prybylowski K, Vissel B, Vicini S. Functional expression of distinct NMDA channel subunits tagged with green fluorescent protein in hippocampal neurons in culture. Neuropharmacology. 2002;42:306–318. doi: 10.1016/s0028-3908(01)00188-5. [DOI] [PubMed] [Google Scholar]
- Margeta-Mitrovic M, Jan YN, Jan LY. A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron. 2000;27:97–106. doi: 10.1016/s0896-6273(00)00012-x. [DOI] [PubMed] [Google Scholar]
- Margeta-Mitrovic M, Jan YN, Jan LY. Ligand-induced signal transduction within heterodimeric GABA(B) receptor. Proc Natl Acad Sci USA. 2001;98:14643–14648. doi: 10.1073/pnas.251554798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlhinney RA, Le Bourdelles B, Molnar E, Tricaud N, Streit P, Whiting PJ. Assembly intracellular targeting and cell surface expression of the human N-methyl-d-aspartate receptor subunits NR1a and NR2A in transfected cells. Neuropharmacology. 1998;37:1355–1367. doi: 10.1016/s0028-3908(98)00121-x. [DOI] [PubMed] [Google Scholar]
- Meddows E, Le Bourdelles B, Grimwood S, Wafford K, Sandhu S, Whiting P, McIlhinney RA. Identification of molecular determinants that are important in the assembly of N-methyl-d-aspartate receptors. J Biol Chem. 2001;276:18795–18803. doi: 10.1074/jbc.M101382200. [DOI] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD. Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron. 2003;40:581–594. doi: 10.1016/s0896-6273(03)00676-7. [DOI] [PubMed] [Google Scholar]
- Murakami K, Mihara K, Omura T. The transmembrane region of microsomal cytochrome P450 identified as the endoplasmic reticulum retention signal. J Biochem (Tokyo) 1994;116:164–175. doi: 10.1093/oxfordjournals.jbchem.a124489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Miwa A, Okado H. Alternative splicing of the C-terminal domain regulates cell surface expression of the NMDA receptor NR1 subunit. J Neurosci. 1999;19:7781–7792. doi: 10.1523/JNEUROSCI.19-18-07781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis M, Hawkins LM, Stephenson FA. Appropriate NR1-NR1 disulfide-linked homodimer formation is requisite for efficient expression of functional, cell surface N-methyl-d-aspartate NR1/NR2 receptors. J Biol Chem. 2004;279:14703–14712. doi: 10.1074/jbc.M313446200. [DOI] [PubMed] [Google Scholar]
- Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci. 2003;23:1151–1158. doi: 10.1523/JNEUROSCI.23-04-01151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standley S, Roche KW, McCallum J, Sans N, Wenthold RJ. PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron. 2000;28:887–898. doi: 10.1016/s0896-6273(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Vissel B, Krupp JJ, Heinemann SF, Westbrook GL. A use-dependent tyrosine dephosphorylation of NMDA receptors is independent of ion flux. Nat Neurosci. 2001;4:587–596. doi: 10.1038/88404. [DOI] [PubMed] [Google Scholar]
- Wang JM, Zhang L, Yao Y, Viroonchatapan N, Rothe E, Wang ZZ. A transmembrane motif governs the surface trafficking of nicotinic acetylcholine receptors. Nat Neurosci. 2002;5:963–970. doi: 10.1038/nn918. [DOI] [PubMed] [Google Scholar]
- Wenthold RJ, Prybylowski K, Standley S, Sans N, Petralia RS. Trafficking of NMDA receptors. Annu Rev Pharmacol Toxicol. 2003;43:335–358. doi: 10.1146/annurev.pharmtox.43.100901.135803. [DOI] [PubMed] [Google Scholar]
- Witzemann V, Stein E, Barg B, Konno T, Koenen M, Kues W, Criado M, Hofmann M, Sakmann B. Primary structure and functional expression of the alpha-, beta-, gamma-, delta- and epsilon-subunits of the acetylcholine receptor from rat muscle. Eur J Biochem. 1990;194:437–448. doi: 10.1111/j.1432-1033.1990.tb15637.x. [DOI] [PubMed] [Google Scholar]
- Xia H, Hornby ZD, Malenka RC. An ER retention signal explains differences in surface expression of NMDA and AMPA receptor subunits. Neuropharmacology. 2001;41:714–723. doi: 10.1016/s0028-3908(01)00103-4. [DOI] [PubMed] [Google Scholar]
- Yang M, Ellenberg J, Bonifacino JS, Weissman AM. The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J Biol Chem. 1997;272:1970–1975. doi: 10.1074/jbc.272.3.1970. [DOI] [PubMed] [Google Scholar]
- Yang W, Zheng C, Song Q, Yang X, Qiu S, Liu C, Chen Z, Duan S, Luo J. A three amino acid tail following the TM4 region of the N-methyl-d-aspartate receptor (NR) 2 subunits is sufficient to overcome endoplasmic reticulum retention of NR1-1a subunit. J Biol Chem. 2007;282:9269–9278. doi: 10.1074/jbc.M700050200. [DOI] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- Zheng X, Zhang L, Wang AP, Bennett MV, Zukin RS. Protein kinase C potentiation of N-methyl-d-aspartate receptor activity is not mediated by phosphorylation of N-methyl-d-aspartate receptor subunits. Proc Natl Acad Sci USA. 1999;96:15262–15267. doi: 10.1073/pnas.96.26.15262. [DOI] [PMC free article] [PubMed] [Google Scholar]