Abstract
The 5-HT1A receptor (5-HT1AR) is the most extensively characterized serotonin (5-HT) receptor mainly because of its involvement in the mode of action of antidepressants. The 5-HT1AR is confined to the somatodendritic domain of central neurons, where it mediates serotonin-evoked hyperpolarization. Our previous studies underlined the role of the short 5-HT1AR C-terminal domain in receptor targeting to dendrites. We used this 17 aa region as bait in a yeast two-hybrid screen, and identified, for the first time, an intracellular protein interacting with the 5-HT1AR. This protein is homologous to the yeast Yif1p, previously implicated in vesicular trafficking between the endoplasmic reticulum (ER) and the Golgi apparatus, but not yet characterized in mammals. We confirmed 5-HT1AR–Yif1B interaction by glutathione S-transferase pull-down experiments using rat brain extracts and transfected cell lines. Yif1B is highly expressed in the brain, and specifically in raphe 5-HT1AR-expressing neurons. Colocalization of Yif1B and 5-HT1AR was observed in small vesicles involved in transient intracellular trafficking. Last, inhibition of endogenous expression of Yif1B in primary neuron cultures by small interfering RNA specifically prevented the addressing of 5-HT1AR to distal portions of the dendrites, without affecting other receptors, such as sst2A, P2X2, and 5-HT3A receptors. Together, our results provide strong evidence that Yif1B is a member of the ER/Golgi trafficking machinery, which plays a key role in specific targeting of 5-HT1AR to the neuronal dendrites. This finding opens up new pathways for the study of 5-HT1AR regulation by partner proteins and for the development of novel antidepressant drugs.
Keywords: Yif1B, 5-HT1A receptor, dendrite, trafficking, GPCR, intermediate compartment
Introduction
Serotonin [5-hydroxytryptamine (5-HT)] is a neuromodulator acting through a large family of receptors for the regulation of a wide range of physiological functions in the CNS as well as in the periphery (Barnes and Sharp, 1999). Among these receptors, the 5-HT1A receptor (5-HT1AR) is a Gi/Go-protein-coupled receptor (GPCR) that modulates adenylyl cyclase and MAP (mitogen-activated protein) kinase activities (Kushwaha et al., 2006; Turner et al., 2007), and acts as autoreceptor and heteroreceptor in brain (Lanfumey and Hamon, 2004). It is localized at the plasma membrane of soma and dendrites (Sotelo et al., 1990; Kia et al., 1996; Riad et al., 2000), where it controls the excitability pattern (Haj-Dahmane et al., 1991). In addition, the 5-HT1AR is critically involved in the mode of action of antidepressant drugs, because its delayed desensitization is responsible, at least in part, for the delayed therapeutic efficacy in chronic treatment (Le Poul et al., 1995).
Because the 5-HT1AR localization in dendritic membrane is closely related to its inhibiting effect on raphe neuron firing, it was important to investigate the mechanisms responsible for this localization. In contrast to other classes of receptors, little data are available on the intracellular trafficking and sorting of the serotonin receptors. Numerous studies on GPCRs have revealed that interaction of their carboxyl C-terminal domain with PDZ domain-containing proteins plays a critical role in the regulation of their desensitization/resensitization and trafficking (for review, see Bockaert et al., 2004). However, the 5-HT1AR C terminus is a very short domain without any PDZ recognition motif. In previous studies, we have demonstrated that the C terminus of the 5-HT1AR is implicated in its basolateral localization in epithelial cells (Darmon et al., 1998) and its dendritic targeting in neurons (Jolimay et al., 2000). Furthermore, we showed that a dileucine motif in the short cytosolic C-terminal domain is necessary for 5-HT1AR export from the endoplasmic reticulum (ER) to the plasma membrane (Carrel et al., 2006). However, the precise molecular mechanism by which the C terminus regulates receptor export to the dendrites remained to be clarified. Thus, we searched for proteins that would interact with 5-HT1AR C terminus and contribute to 5-HT1AR trafficking in neurons. Using the yeast two-hybrid screening procedure, we identified Yif1B, a homolog of the yeast Yif1p, as the first known protein interacting with the 5-HT1AR C terminus. We confirmed this interaction by glutathione S-transferase (GST) pull-down experiments performed on Yif1B-transfected cell lines and brain extracts. Coexpression of 5-HT1AR and Yif1B in the rat brain and their subcellular colocalization in transfected LLC-PK1 cells strongly support this partnership. Small interfering RNA (siRNA) inhibition of endogenous Yif1B expression in cultured neurons from rat embryos clearly demonstrated its decisive role in 5-HT1AR dendritic targeting.
Materials and Methods
Antibodies.
The following primary antibodies were used: rabbit anti-green fluorescent protein (GFP) antibody (Millipore Bioscience Research Reagents; 1:1000), mouse monoclonal anti-α-tubulin antibody (GE Healthcare; 1:5000), rabbit anti-actin antibody (Sigma; 1:2000) sheep anti-tryptophan hydroxylase (TPH) antibody (Ehret et al., 1991) (1:1000), mouse monoclonal anti-Flag M2 antibody (Sigma; 1:3000), rabbit anti-calregulin antibody (Santa Cruz Biotechnology; 1:100), mouse monoclonal anti-CTR433 antibody (Jasmin et al., 1989) (median Golgi; 1:200), rabbit anti-giantin antibody (CRP; 1:2000), and affinity-purified anti-5-HT1A rabbit antibody (1:500) (el Mestikawy et al., 1990). The secondary antibodies used were CY3-conjugated donkey anti-rabbit IgG (1:1600 dilution; Jackson ImmunoResearch), Alexa Fluor 488-, 568-, and 546-conjugated antibodies from Invitrogen (1:1600), and HRP-conjugated anti-rabbit and anti-mouse antibodies (Sigma). Yif1B antiserum was produced by immunization of rabbits with the N-terminal cytoplasmic 12TPRLRKWPSKRRV24 peptide (see supplemental Fig. S1, available at www.jneurosci.org as supplemental material) coupled to ovalbumin (NeoMPS). The antibody was affinity purified as described previously (Doucet et al., 1998).
Animals.
Adult male Sprague Dawley rats (250–300 g body weight) were housed in agreement with the institutional guidelines for use of animals and their care, in compliance with national and international laws and policies (Council directives no. 87-848, October 19, 1987, Ministère de l'Agriculture et de la Forêt, Service Vétérinaire de la Santé et de la Protection Animale, permission 75-116 to M.H. and 75-974 to M.D.)
Plasmid constructs.
The C-terminal cytoplasmic domain (amino acids 406–422) of the rat 5-HT1AR (Albert et al., 1990) was PCR-amplified and cloned in a yeast two-hybrid vector (Hybrigenics) and in pGEX-6p1 (GE Healthcare), for GST-tagged fusion protein production. The rat 5-HT1AR sequence (Albert et al., 1990) without the stop codon was inserted between the HindIII and AgeI sites of the pEGFP-N1 plasmid (Clontech) generating the 5-HT1A-enhanced GFP (eGFP) construct. The rat 5-HT1AΔ407 plasmid was constructed by PCR inserting a stop codon at amino acid 407 and resulted in a truncated 5-HT1AR (Jolimay et al., 2000). The plasmid encoding the C terminus (CT1A) fused to cyan fluorescent protein (CFP) was obtained by cloning the last 18 aa of the rat 5-HT1A receptor between the SacII and the BamHI sites of the peCFP-C1 vector (Clontech). The cDNA of Yif1B was synthesized with reverse transcriptase from rat brain RNA, amplified with the forward primer 5′-AAGCATGCACGCGACAGGTTTG-3′ (nucleotides 38–59 of NM_198734) and reverse primer 5′-CAGTTCACCGTACAAGGTGGAA-3′ (nucleotides 960–981), and cloned into pCB6 vector (Brewer and Roth, 1991) downstream of the CMV promoter, or in frame downstream of the flag epitope of the pFlag-CMV-6a vector (Sigma). The 425 nt probe for Northern blot was amplified with forward primer 5′-AAGCATGCACGCGACAGGTTTG-3′ (nucleotides 38–59) and reverse primer 5′-TGTCCTGTTGGTACTGGACCTC-3′ (nucleotides 441–462), and cloned into TA cloning vector (Invitrogen). The Yif1B siRNAs were double-stranded stealth RNAs from Invitrogen: siRNA(Yif1B-1), 5′-CCAGCCAUGGCUUUCAUAACCUACA-3′ (nucleotides 509–533 of Yif1B); the control siRNA(contr-1) was 5′-CCAGUACUUCGUACUCCAAUCGACA-3′; siRNA(Yif1B-2), 5′-CGGUACUCAUGUACUGGCUCACCUU-3′ (nucleotides 937–961 of Yif1B), and the control siRNA(contr-2) was 5′-CGGACUCAUGCGGUCACUCCAUCUU-3′. All plasmid constructs were checked by sequencing the entire insert. The plasmids encoding sst2A-eGFP (Lelouvier et al., 2008), P2X2-eGFPR (Boué-Grabot et al., 2003), and enhanced yellow fluorescent protein (eYFP)-5-HT3A (Grailhe et al., 2004) have already been used and described.
Yeast two-hybrid screening.
A rat hippocampus random-primed cDNA library transformed into the Y187 yeast strain and containing 10,000,000 independent fragments was used for mating. High mating efficiency was obtained by using a specific mating method (United States patents no. 6187535, 6531284, and 6913886). The screen was first performed on a small scale to adapt the selective pressure to the intrinsic property of the bait. No autoactivation of the bait was observed. Then, the full-scale screen was performed in conditions ensuring a minimum of 50,000,000 interactions tested, to cover five times the primary complexity of the yeast-transformed cDNA library (Rain et al., 2001). Fifty-nine million interactions were actually tested with the C-terminal cytoplasmic domain (amino acids 406–422) of rat 5-HT1AR. After selection on medium lacking leucine, tryptophan, and histidine, 210 positive clones were picked, and the corresponding prey fragments were amplified by PCR and sequenced at their 5′ and 3′ junctions. Sequences were then filtered and contiged as described previously (Formstecher et al., 2005) and compared with the latest release of the GenBank database using BLASTN (Altschul et al., 1997). A Predicted Biological Score was attributed to assess the reliability of each interaction, as described previously (Wojcik et al., 2002). Briefly, the Predicted Biological Score relies on two different levels of analysis: first, a local score takes into account the redundancy and independence of prey fragments, as well as the distributions of reading frames and stop codons in overlapping fragments; second, a global score takes into account the interactions found in all the screens performed at Hybrigenics using the same library. In addition, potential false-positives were flagged by a specific “E” Predicted Biological Score. This was done by discriminating prey proteins containing “highly connected” domains, previously found several times in screens performed on libraries derived from the same organism. The Predicted Biological Scores have been shown to positively correlate with the biological significance of interactions (Wojcik et al., 2002).
Northern blot experiments.
The mRNA Northern blot NBA (Normalized By Amount of RNA; BioChain Institute) was hybridized using [α-32P]dCTP (GE Healthcare) labeled probe made following the Rediprime II Random Prime Labeling System (GE Healthcare). After exposure to a MP Hyperfilm (GE Healthcare) for 24 h, the blot was scanned with a digital camera.
Western blot experiments.
LLC-PK1 cells expressing Yif1B were grown on 90 mm dishes, and tissues were dissected from adult male rats and then homogenized by sonication in sample buffer (Laemmli, 1970). The protein content in supernatants was measured using the BCA protein assay kit for reducing agents (Pierce). Proteins (1 μg) were analyzed by SDS-PAGE followed by blotting to polyvinylidene difluoride (PVDF) membrane, and incubation with Yif1B affinity-purified antiserum (1:500), and reprobed with a rabbit actin antibody (Sigma; 1:2000). For normalization with α-tubulin, the proteins were blotted to nitrocellulose membrane, which was then incubated with anti-α-tubulin monoclonal antibody (1:1000).
Cell culture and transfections.
LLC-PK1 cells and COS-7 cells were grown in DMEM GlutaMax I (Invitrogen) supplemented with 4.5 g/L and 1 g/L glucose, respectively, 10% fetal bovine serum, 10 U/ml penicillin G, and 10 g/ml streptomycin (Jolimay et al., 2000; Carrel et al., 2006). LLC-PK1 and COS-7 cells (50% confluent) were transfected using Lipofectin reagent (Invitrogen) and FuGENE (Roche), respectively, according to the manufacturers' protocols. For GST pull-down experiments, LLC-PK1 cells were transfected by electroporation using Gene Pulser Xcell electroporation system (Bio-Rad; 135 V, 1800 μF in 200 μl of DMEM containing 5 × 106 cells and 10 μg of plasmid DNA). Hippocampal neuron cultures from 18-d-old rat embryos were performed according to the procedure described previously (Boué-Grabot et al., 2004a,b). Hippocampal neurons were transfected after 7–8 d in vitro (DIV) with Lipofectamine 2000 (Invitrogen) (Boué-Grabot et al., 2004a,b). For cotransfection experiments in the presence of siRNA, the plasmids (2 μg) and the siRNA (2 μl of a 20 mm solution) were thoroughly mixed before the addition of Lipofectamine 2000 (1.25 μl/coverslip). All cells were used 48 h after transfection.
GST pull-down assay.
GST-tagged fusion proteins were produced in the BL21DE3 strain of Escherichia coli (Stratagene). After induction at 28°C with 0.5 mm isopropyl-β-d-thiogalactopyranoside, the bacterial pellet was sonicated in PBS, 0.1% Triton X-100, and protease inhibitors (Sigma) and centrifuged at 14,000 × g for 15 min. Supernatants were incubated overnight with glutathione-Sepharose beads (GE Healthcare) at 4°C, then washed in PBS. Extracts from LLC-PK1 cells expressing Yif1B or from hippocampus and cerebellum dissected from Sprague Dawley rats were homogenized by sonication in buffer A (20 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm EDTA, 0.5% NP-40, protease inhibitors, 100 mg of tissue/ml of buffer). After centrifugation at 15,000 × g for 10 min, 100–200 μg of proteins from cell lysates or 1 mg of proteins from tissue homogenates were incubated with glutathione-Sepharose beads coupled to the different GST fusion proteins in buffer A containing 2.5 mg/ml bovine serum albumin, overnight at 4°C. After washing three times, the proteins retained on the beads were eluted in sample buffer (Laemmli, 1970) and loaded on SDS-PAGE gel, followed by blotting to PVDF membrane and incubation with Yif1B affinity-purified antiserum (1:500).
Immunofluorescence.
For brain section immunofluorescence, rats were deeply anesthetized with pentobarbital (60 mg/kg, i.p.) and perfused with 4% paraformaldehyde (PFA) in PBS. Brains were postfixed in the same fixative for 24 h, and sections (30 μm) were obtained using a vibratome. For immunofluorescence on transfected cells or neurons, coverslips with attached cells were washed three times with PBS+ (PBS containing 0.1 mm CaCl2 and 0.1 mm MgCl2) at 37°C, and fixed for 15 min at 37°C with PBS+ containing 4% paraformaldehyde and 4% sucrose. For brain slices and coverslips, immunofluorescence experiments were performed as described previously (Boué-Grabot et al., 2004a,b). Immunofluorescence images were generated using a Leica TCS-400 laser scanning confocal microscope (100× oil-immersion lens) or a TCS SP2 AOBS laser scanning confocal microscope (63× oil-immersion lens). The percentage of colocalization was calculated on confocal images acquired with a pixel size of 100 nm (objective 63×, zoom 2.5), without any saturation of the two labels, and was calculated with the “colocalization threshold” program of ImageJ (Costes et al., 2004). The total dendritic tree length was measured with the “length measurement” of the Lucia 4.7 software (Nikon).
Quantification of dendrite fluorescence.
Contrast and brightness of confocal images were chosen to ensure that all relevant pixels were within linear range and were maintained identical for all measurements. For double-labeling experiments, pictures were generated using Adobe Photoshop 7.0. Fluorescence profiles along dendrites were generated using the Lucia 4.71 software (Nikon). For the comparison of 5-HT1A-eGFPR distribution, all neurons showing intact morphology along their longest dendrite, with unambiguous visual identification of the axon, were analyzed (one dendrite per neuron). The variability of distribution in individual neurons was eliminated by using the cumulated fluorescence profiles obtained for 20 neurons in each group.
Results
Identification of Yif1B as a 5-HT1AR C terminus-interacting protein using the yeast two-hybrid assay
To identify proteins interacting specifically with 5-HT1AR, we performed a yeast two-hybrid screening of a rat hippocampus cDNA library using the 5-HT1AR cytoplasmic C terminus (residues 406–422) as bait. Five clones corresponded to amino acids 22–311 of a protein recently named Yif1B (accession number: NP_942029), and were scored B as a Predicted Biological Score (Wojcik et al., 2002). This protein, not yet characterized in mammals, appeared as the ortholog of the Saccharomyces cerevisiae protein Yif1p, a 35.5 kDa transmembrane protein that was first described as a Yip1p-interacting factor (Matern et al., 2000). Yip1p is localized in the Golgi membrane and ER-derived anterograde transport COPII vesicles (Otte et al., 2001). Yif1p was shown to interact with Yip1p and with transport Rab GTPases, and to play a critical role in ER–Golgi trafficking (Matern et al., 2000).
Rattus norvegicus Yif1B is a 311 aa protein. Transmembrane prediction with TMHMM2.0 (Krogh et al., 2001) showed that it contains five transmembrane segments clustered in the C-terminal moiety, a long hydrophilic N-terminal domain within the cytoplasm, and a very short C terminus turned to the ER lumen (Fig. 1 A). Bioinformatics analysis revealed that Yif1B protein is well conserved across species, with the following identity scores to Rattus norvegicus: Saccharomyces cerevisiae 28%, Xenopus laevis 76%, Mus musculus 97%, and Homo sapiens 89% (Fig. 1 B). Yif1p is present as only one gene in yeast and Caenorhabditis elegans and corresponds to two closely related genes in mammals. In rat, Yif1A and Yif1B display 50% identity in amino acids clustered after the first 60 aa (as shown in the alignment in supplemental Fig. S1, available at www.jneurosci.org as supplemental material). These two related genes belong also to a family already described as a five-passed transmembrane protein family sharing homology in the transmembrane domains (Shakoori et al., 2003)
Figure 1.
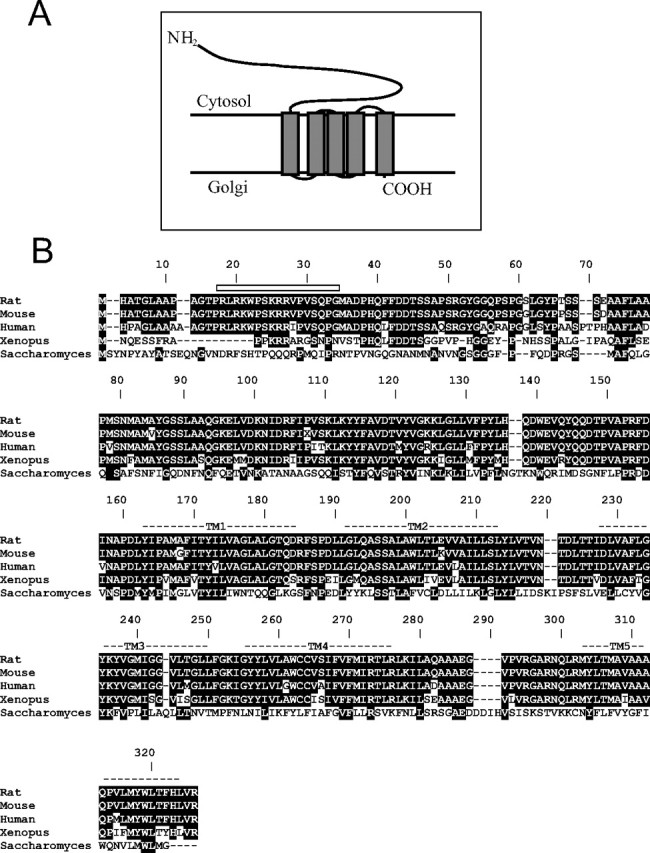
Interspecies comparison of Yif1B sequence. A, Schematic predicted topology of the Yif1B protein in Golgi membrane with five transmembrane domains and a large cytoplasmic N terminus. B, The amino acid sequence of Yif1B from rat (Yip1-interacting factor homolog B, Rattus norvegicus, accession numbers: NP_942029 and XP_214879) is aligned with those of homologous proteins in mouse (Yip1-interacting factor homolog B, Mus musculus, accession number: NP_084163.1), human (YIF1B protein, Homo sapiens, accession number: AAH91477.2), Xenopus (LOC443676 protein, Xenopus laevis, accession number: AAH73660.1), and yeast (Yif1p, Saccharomyces cerevisiae, accession number: NP_014136.1). Identical amino acid residues between rat Yif1B and other Yif1Bs or the Yif1p ancestor are in white in black box. Putative transmembrane domains are marked TM1–TM5. The rat sequence (TPRLRKWPSKRRV) represents the peptide used for the production of specific polyclonal anti-Yif1B antibody in rabbit.
High expression of Yif1B in brain tissue
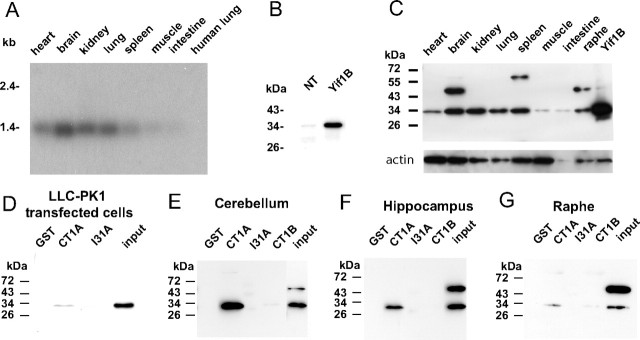
The expression of Yif1B in various tissues was analyzed by Northern blot. The hybridization revealed a single band of ∼1.4 kb corresponding to the size expected from the length of the sequenced cDNA (1193 bp) plus the polyA-tail (∼200 residues). As shown in Figure 2 A, Yif1B mRNA was expressed in a wide range of tissues, but was particularly abundant in brain. In addition, we detected high levels of Yif1B mRNA in rat heart, kidney, and lung and lower levels in spleen, muscle, and intestine. The negative data with human lung mRNA (Fig. 2 A) confirmed the specificity of our probe under the used hybridization conditions.
Figure 2.
Yif1B mRNA and protein expression in rat tissues and direct interaction of Yif1B from rat brain homogenates with the 5-HT1AR C-tail. A, Yif1B mRNA expression in rat tissues was analyzed by Northern blot. Each lane represents 3 μg of mRNA from each rat tissue; the human lung is used as a control for the hybridization specificity. The 32P-labeled Yif1B probe consisted of 425 bp in the 5′end of the rat cDNA. B, C, Yif1B protein expression in LLC-PK1 cells untransfected or transfected with Yif1B (B) and in rat tissues (C) was analyzed by Western blot using anti-Yif1B affinity-purified polyclonal antibody (1:1000). Each lane represents 1 μg of total protein, and the same Western blot was also revealed with actin antibody at the bottom. D–G, GST pull-down experiments on Yif1B-transfected LLC-PK1 cells (D) and brain tissue homogenates: cerebellum (E), hippocampus (F), or raphe (G). Extracts were incubated with beads coupled to GST alone or to GST fused with CT1A, I31A, or CT1B. Interaction was analyzed by Western blot with anti-Yif1B affinity-purified polyclonal antibody (1:1000). “Input” represents 2.5 μg of proteins (tissue extract prepared as in C). Results are representative of at least three independent experiments.
The pattern of expression of Yif1B protein was well correlated to that of its mRNA in all tissues. Western blots were performed on 1 μg of proteins from various tissues using the polyclonal antibody obtained after immunization of a rabbit with an N-terminal peptide from the protein (Fig. 1 A). The antiserum, tested on LLC-PK1 cells transfected with rat Yif1B, recognized an ∼34 kDa band (Fig. 2 B) corresponding to the size expected from the open reading frame of Yif1B. The faint labeling corresponding to the same size was also observed in untransfected cells that most probably reflected cross-reaction of rabbit antibodies with porcine Yif1B protein endogenously expressed in LLC-PK1 cell line. Yif1B was particularly abundant in whole brain extract, kidney, and spleen, present at lower levels in heart and lung (Fig. 2 C). Loading control experiment performed using actin antibody shows that the level of Yif1B is low in the muscle in comparison with that of actin. Yif1B was also well detected in raphe extracts. The ∼45 kDa band observed in brain and raphe (Fig. 2 C) corresponded most probably to a nonspecific cross-reaction of the antibody with brain tissues on Western blot.
Direct interaction of Yif1B with the 5-HT1AR C terminus is revealed by GST pull-down experiments
Direct interaction between 5-HT1AR and Yif1B was assessed by GST pull-down experiments on extracts of cells transfected with Yif1B or rat brain tissue homogenates as a source of native Yif1B. We used a fusion protein made of the 5-HT1AR cytoplasmic C terminus coupled to GST (GST-CT1A) and for controls, a fusion protein made of GST coupled to the third intracellular loop of 5-HT1AR (I31A) and a fusion protein made of GST coupled to the cytoplasmic C terminus of the 5-HT1B receptor, which did not interact with Yif1B in yeast two-hybrid screening. GST-CT1A, but not GST, GST-I31A, or GST-CT1B, was able to pull down Yif1B from the three regions tested (Fig. 2 E–G) (hippocampus, raphe, and cerebellum), and from LLC-PK1 cells transfected with Yif1B cDNA (Fig. 2 D). The nonspecific band at ∼45 kDa observed on Western blots of hippocampus or cerebellum extracts is not detected when the protein was retained by the GST-CT1A beads (Fig. 2 E,F), attesting thereby to the specificity of such interaction.
Together, the yeast two-hybrid and the GST pull-down data demonstrate that the C terminus of 5-HT1AR and Yif1B are capable of direct physical interaction.
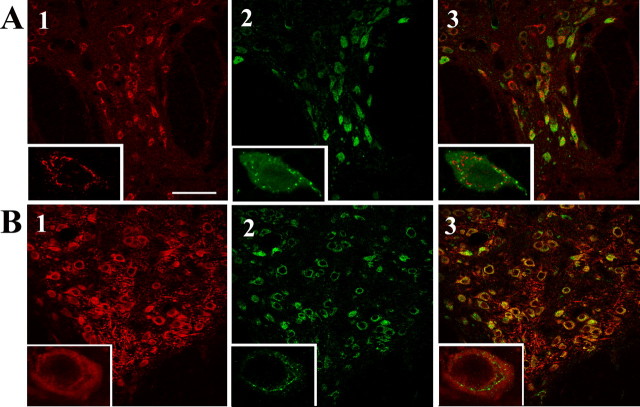
Yif1B is expressed in serotoninergic neurons of the rat brain
In the next series of experiments, we checked for the presence of Yif1B in brain areas known to express 5-HT1AR, such as the anterior raphe (Sotelo et al., 1990). Direct immunolabeling of Yif1B and 5-HT1AR could not be performed, because their respective antibodies were both raised in rabbits. This was overcome by labeling serotonin neurons of the anterior raphe either with sheep antibodies directed against TPH (the first enzyme for serotonin synthesis) or rat antibodies directed against serotonin. This was combined with Yif1B immunofluorescence labeling using crude rabbit antibodies. Microphotographs in Figure 3 show that Yif1B is well expressed in serotoninergic neurons labeled either with anti-5-HT (Fig. 3 A) or with anti-TPH antibodies (Fig. 3 B). Superimposed immunofluorescence images clearly showed that in these neurons, Yif1B is present in intracellular vesicles different from those labeled by the 5-HT antibody (Fig. 3 A). The specificity of the Yif1B labeling on slices was attested by control experiments with the serum saturated with the immunization peptide or with the 45 kDa band seen on Western blot (data not shown).
Figure 3.
Yif1B expression in serotoninergic neurons in the dorsal raphe nucleus. A, B, Rat dorsal raphe nucleus sections immunolabeled with anti-TPH antibody (A; red) or with anti-serotonin antibody (B; red) and polyclonal crude anti-Yif1B antiserum (1:5000; in green). Overlay shows the superposition of the two labels. Insets correspond to enlargement of a double-labeled cell. Scale bar, 75 μm.
Yif1B colocalizes with 5-HT1AR in vesicles of the intermediate compartment
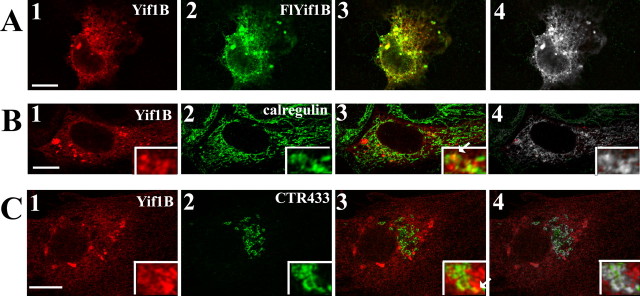
The nature of the intracellular compartment where Yif1B is localized was investigated in COS-7 and LLC-PK1 transfected cell lines using specific markers of the ER (calregulin) and of the median Golgi apparatus (CTR433). In these cell lines, Yif1B antibody revealed a punctate immunolabeling pattern. The specificity of this peculiar pattern was confirmed by using a Flag epitope-tagged Yif1B cDNA (Fig. 4 A1,A2). The colocalized pixels are shown in white (Fig. 4 A4). Subsequent double-immunofluorescence experiments showed that the punctate labeling observed for Yif1B partially colocalized with calregulin (Fig. 4 B), a marker of the ER, and with CTR433 (Fig. 4 C), a marker of the Golgi apparatus. The percentage of colocalization (Fig. 4 B4,C4) was quantified as the ratio of the colocalized pixels to those labeled by Yif1B and reached 47.44% (±2.23, SEM; n = 26) for the ER, and 24.45% (±1.063, SEM; n = 52) for the Golgi apparatus. Therefore, Yif1B appeared to be present in vesicles partially overlapping the ER and the Golgi apparatus and partially excluded from these compartments (Yif1B+/ER−/Golgi−) as expected for vesicles belonging to the intermediate compartment (IC). Such localization is in agreement with the function of Yif1p in yeast (Matern et al., 2000).
Figure 4.
Yif1B subcellular localization. A–C, Transfection of COS-7 cells (A) with Flag-Yif1B or Yif1B and of LLCPK1 cells with Flag Yif1B cDNA (B, C). Yif1B labeling (anti-Yif1B affinity-purified polyclonal antibody; 1:1000) is in red (A1, B1, C1). Flag, calregulin, and CTR433 labeling are shown in green (A2, B2, C2), and colocalized pixels are shown in white on the superposed labeling (A4, B4, C4). Some punctate colocalizations are shown by arrows in insets. Scale bars, 10 μm.
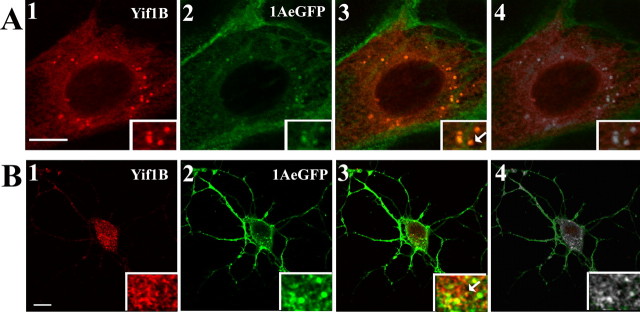
Colocalization between the 5-HT1AR and Yif1B was investigated in LLC-PK1 cells stably expressing the 5-HT1AR coupled to GFP (5-HT1A-eGFP), and transiently transfected with Yif1B. As shown in Figure 5 A, the 5-HT1A-eGFP was mostly localized at the plasma membrane, as previously shown (Carrel et al., 2006). Interestingly, 5-HT1A-eGFP was also found to colocalize with Yif1B in vesicle-like puncta (Fig. 5 A). The quantification of the colocalization (Fig. 5 A4) showed that only 12.95% of the 5-HT1A-labeled pixels were colocalized with Yif1B (±1.86, SEM; n = 68). The level of colocalization was highly heterogeneous, probably depending on the amount of intracellular receptor in the trafficking pathway, which may vary from cell to cell, and with the level of expression. In Yif1B-overexpressing cells, the 5-HT1AR was highly colocalized with Yif1B (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). In hippocampal neurons also, Yif1B colocalized with the 5-HT1AR, mostly in the soma but also in small dots in the dendrites (Fig. 5 B). These results provide strong evidence that Yif1B partially colocalizes with the 5-HT1AR in intracellular vesicles, where Yif1B can play a role in trafficking. The partial colocalization probably reflects a transient interaction occurring during the intracellular trafficking of the receptor.
Figure 5.
Yif1B colocalization with 5-HT1AR. A, Transfection of 5-HT1A-eGFP-stable LLC-PK1 cells with Yif1B. Yif1B immunofluorescence is shown in red (anti-Yif1B affinity-purified polyclonal antibody; 1:1000; A1), and eGFP autofluorescence is shown in green (A2). B, Primary cultures of rat hippocampal neurons (DIV 7) were cotransfected with a plasmid encoding the 5-HT1A-eGFPR and Yif1B. Superposition of labels shown in 1 and 2 is visible in 3. In 4, the colocalized pixels are shown in white. Arrows show Yif1B and 5-HT1AR colocalization in insets. Scale bars, 10 μm.
Inhibition of Yif1B expression prevents the distal dendritic targeting of 5-HT1AR
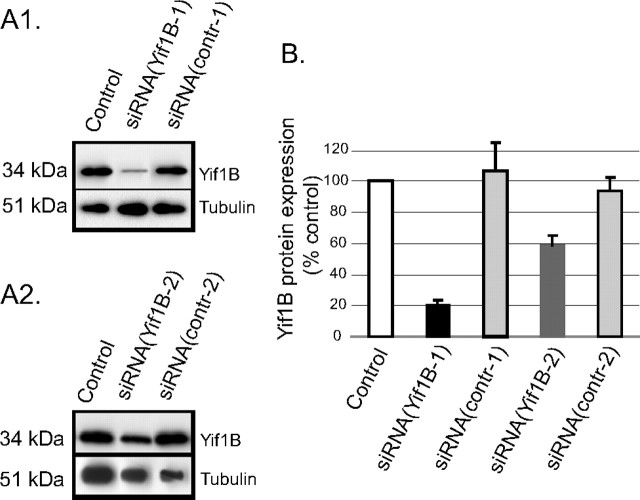
To delineate the implication of Yif1B in 5-HT1AR trafficking, we inhibited the endogenous expression of Yif1B and analyzed the effects on 5-HT1AR localization. We first selected a small double-stranded siRNA sequence that was effective in inhibiting rat Yif1B expression in LLC-PK1 stably transfected cells (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). Then we inhibited Yif1B endogenous expression in primary cultures of hippocampal neurons in which the recombinant 5-HT1AR remained mainly confined to the somatodendritic compartment (Jolimay et al., 2000). Inhibition of endogenous Yif1B expression reached ∼80% 48 h after transfection with a double-stranded siRNA as assessed by Western blot (Fig. 6 A1) and relative quantification with respect to α-tubulin (Fig. 6 B). As controls, we used neurons transfected with eYFP as transfection itself could, on its own, modify protein expression level. The siRNA (Yif1B-1 or contr-1) was also cotransfected with eYFP. The chosen siRNA sequence was therefore effective to inhibit the endogenous Yif1B expression in neurons 48 h after transfection. Another siRNA [siRNA(Yif1B-2)] was also tested and proved efficient in inhibiting the Yif1B endogenous expression (Fig. 6 A2,B).
Figure 6.
Downregulation of the protein Yif1B by siRNA in primary cultures of rat hippocampal neurons. A1, A2, Yif1B was detected by Western blots (anti-Yif1B affinity-purified polyclonal antibody; 1:1000) of protein extracts, 48 h after transfection of neurons at DIV 7; α-tubulin was detected by Western blots on the same sample (mouse antibody; 1:1000) to normalize the amount of extract. A1, siRNA(Yif1B-1); A2, siRNA(Yif1B-2). All neurons were cotransfected with a control plasmid (encoding eYFP). “Control” corresponds to eYFP alone, “siRNA(Yif1B-1/2)” to eYFP plus siRNA(Yif1B-1/2), and “siRNA(contr-1/2)” to eYFP plus siRNA(contr-1/2). B, Quantification of Yif1B protein expression (normalized with reference to α-tubulin expression) in transfected neurons (n = 5, in 3 independent experiments). Bars represent mean, and error bars represent SEM.
The siRNA(Yif1B-1) was used to inhibit Yif1B expression in neurons cotransfected with 5-HT1A-eGFPR. In line with previous studies (Carrel et al., 2006), neurons transfected with 5-HT1A-eGFPR alone or with siRNA(contr-1) displayed immunofluorescence in dendrites, where it overlapped α-tubulin labeling (Fig. 7 A). The 5-HT1A-eGFPR labeling of neurons transfected with both 5-HT1A-eGFPR and Yif1B(siRNA-1) was confined to the proximal portion of the dendrites. To ensure that this effect was not caused by changes in dendrite structure, we measured the labeling of α-tubulin and 5-HT1A-eGFPR in transfected neurons along the longest dendrite for each neuron as exemplified in Figure 7 B. The curves depicted in Figure 7 C represent cumulated fluorescence intensity for 5-HT1A-eGFPR and tubulin along dendrites in transfected neurons versus the distance from the soma. Immunofluorescence analysis showed that 5-HT1A-eGFPR and α-tubulin had a similar distribution in neurons transfected with 5-HT1A-eGFPR alone or with siRNA(contr-1). Indeed, the total dendritic length of neurons is the same in the three different conditions as shown in supplemental Figure S4 (available at www.jneurosci.org as supplemental material). In addition, inhibition of Yif1B did not change the pattern of expression of Golgi markers as shown in supplemental Figure S5 (available at www.jneurosci.org as supplemental material). In the case of cotransfection with siRNA (Yif1B-1), the pattern of α-tubulin labeling was not significantly different from that of control neurons, whereas that of 5-HT1A-eGFPR extended along dendrites on a much shorter distance than α-tubulin. The clear-cut difference observed between the corresponding curves demonstrates that Yif1B interferes with the 5-HT1AR transport to the distal part of the dendrites without affecting tubulin distribution.
Figure 7.
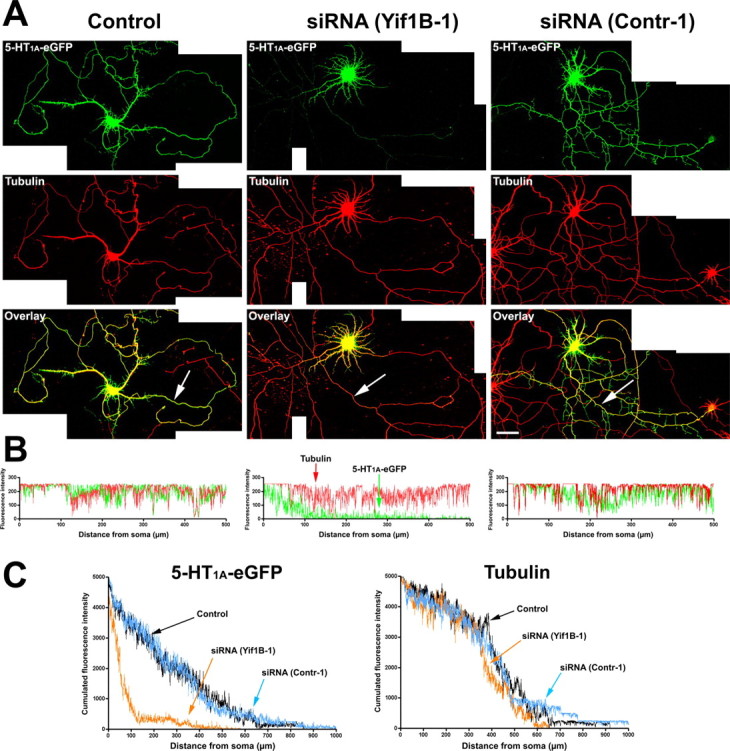
Altered 5-HT1A receptor distribution in the distal part of dendrites after Yif1B RNA interference. A, Primary cultures of rat hippocampal neurons (7 DIV) were transfected with a plasmid encoding the 5-HT1A-eGFPR (Control, left), cotransfected with the 5-HT1A-eGFPR plus the siRNA(Yif1B-1) (middle), or cotransfected with the 5-HT1A-eGFPR plus the siRNA(contr-1) (right). Immunofluorescence was performed with anti-GFP antibody to enhance the GFP signal (green, top) or anti-α-tubulin antibody (red, middle). Bottom, Overlay. Note the drastic reduction of 5-HT1A-eGFPR fluorescence in the distal part of dendrites in siRNA(Yif1B-1)- but not in siRNA(Contr-1)-transfected neurons. B, 5-HT1A-eGFPR (green) and tubulin (red) fluorescence profiles along the longest dendrites (arrows) of the corresponding neurons. C, Cumulated fluorescence profiles for each group (60 neurons analyzed). siRNA(Yif1B-1) reduces 5-HT1A-eGFPR fluorescence in the distal part of dendrites without affecting their average length, as shown by the tubulin fluorescence distribution. Scale bar, 50 μm.
Similar experiments were performed on neurons cotransfected with other receptors, i.e., the somatostatin sst2A receptor, the purinergic P2X2, or the serotonin 5-HT3A, each receptor belonging to a different class of receptors: seven-transmembrane GPCR, two-transmembrane nucleotide receptor, or four-transmembrane channel receptor, respectively. No effect of the siRNA(Yif1B-1) was observed on the dendritic localization of sst2A, P2X2 receptors, or 5-HT3A localization (Fig. 8 Ab–Ad), in comparison with the effect observed on the 5-HT1AR (Fig. 8 Aa). For each receptor, we represented the cumulated fluorescence intensity of the receptor along dendrites in transfected neurons versus the distance from the soma (Fig. 8, right). Except for the 5-HT1AR, no effect of Yif1B inhibition could be visualized. This confirms the specific effect of Yif1B in targeting 5-HT1AR to neuronal dendrites.
Figure 8.
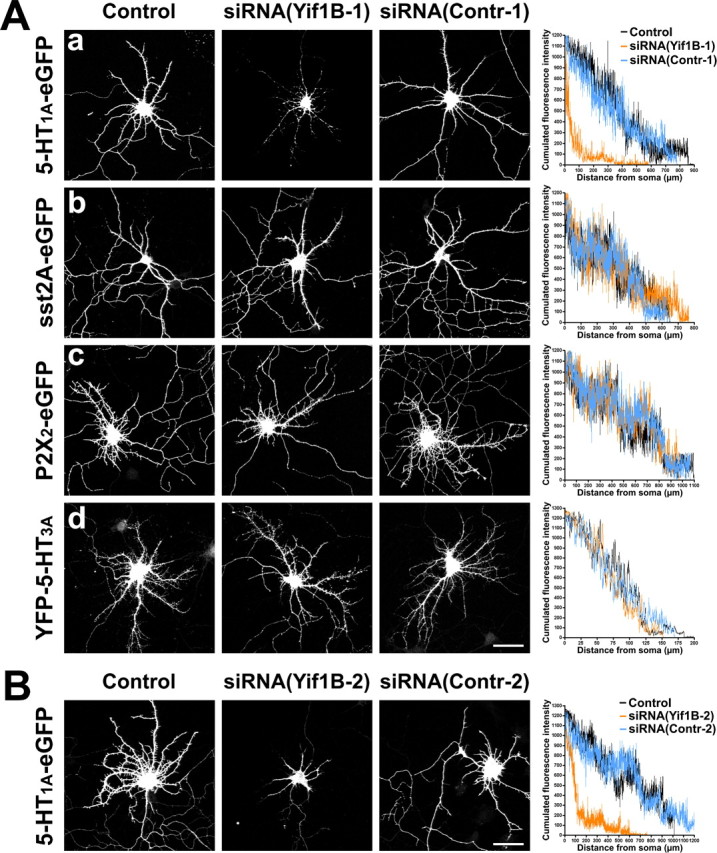
Yif1B RNA interference effect is specific for the 5-HT1A receptor. A, No effect of Yif1B RNA interference on sst2A, P2X2, and 5-HT3A receptor distribution in the dendritic tree. Primary cultures of rat hippocampal neurons (7 DIV) were transfected with plasmids encoding the following receptors: 5-HT1A-eGFP (a), sst2A-eGFP (b), P2X2-eGFP (c), and YFP-5-HT3A (d), and either transfected alone (left), cotransfected with siRNA(Yif1B-1) (middle), or cotransfected with siRNA(Contr-1) (middle right). Immunofluorescence was performed with anti-GFP antibodies to enhance the GFP signal. Cumulated fluorescence profiles along the longest dendrite for each group (15 neurons analyzed) are shown on the right. Note the drastic reduction of fluorescence induced by siRNA(Yif1B-1) in the distal part of dendrites in 5-HT1A-eGFP-transfected neurons but not in sst2A-eGFP-, P2X2-eGFP-, or YFP-5-HT3A-transfected neurons. The higher variability in fluorescence intensities compared with Figure 7 is attributable to the lower number of neurons analyzed. Scale bar, 50 μm. B, Inhibition of 5-HT1A-eGFP receptor distribution in the dendritic tree by another siRNA specific to Yif1B (nucleotides 937–961). Neurons were transfected with a plasmid encoding 5-HT1A-eGFP (left), cotransfected with the siRNA(Yif1B-2) (middle), or cotransfected with the control siRNA, siRNA(Contr-2) (middle right). As above, cumulated fluorescence profiles along the longest dendrite for each group (15 neurons analyzed) are shown on the right. Scale bar, 50 μm.
As a control for the specificity of siRNA, inhibition was made by transfecting a second siRNA(Yif1B-2) in neurons together with 5-HT1A-eGFPR. Endogenous expression of Yif1B was also efficiently inhibited by ∼40%, as shown by Western blot experiments (Fig. 6 A2). The effect of this second siRNA on 5-HT1A-eGFPR targeting to dendrites was the same as that observed for the first siRNA, by comparing Figure 8 Aa and Figure 8 B. The inhibition of 5-HT1A-eGFPR dendritic targeting was similar with the two siRNAs, suggesting that the effect was specific to the Yif1B sequence and did not result from an off-target effect of a particular siRNA sequence, as has already been described in neurons (Alvarez et al., 2006).
5-HT1A receptor distribution in the dendritic tree is dependent on the C-terminal segment
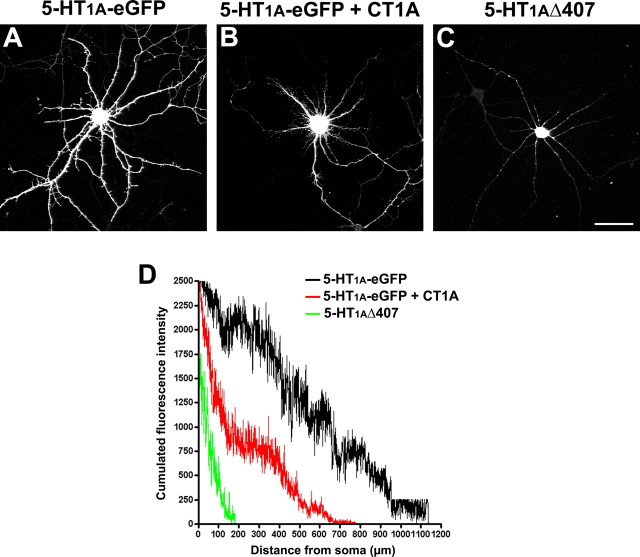
We analyzed the localization of a 5-HT1AR without its C terminus (5-HT1AΔ407) (Jolimay et al., 2000) and showed that it is localized in the soma, which confirms that the C terminus plays a role on the dendritic 5-HT1A-eGFPR localization, probably via the interaction between Yif1B and the C terminus of the 5-HT1AR. In Figure 9 is plotted the cumulated fluorescence intensity of the 5-HT1A-eGFPR and 5-HT1AΔ407 receptor along dendrites in transfected neurons versus the distance from the soma; comparison of the two plots shows clearly that the truncated 5-HT1AR is restricted to the cell body. We also performed a competition experiment by overexpressing the C terminus of the 5-HT1AR together with the full-length 5-HT1AR and analyzed the localization of the 5-HT1A-eGFPR. In Figure 9, the cumulated fluorescence intensity of the 5-HT1A-eGFPR cotransfected with CT1A-CFP reveals that its dendritic localization is impaired in comparison with the 5-HT1A-eGFPR alone. This experiment suggests that the overexpression of the 5-HT1AR C terminus prevented the interaction between the full-length receptor and Yif1B, and confined the 5-HT1AR in a proximal dendritic compartment. This result confirms that the role of Yif1B in the 5-HT1AR dendritic targeting is mediated by a direct interaction between the C terminus of 5-HT1AR and Yif1B.
Figure 9.
5-HT1A receptor distribution in the dendritic tree is dependent on the C-terminal segment. Primary cultures of rat hippocampal neurons (7 DIV) were transfected with the following plasmids: 5-HT1A-eGFP (A), 5-HT1A-eGFP plus CT1A (B), or 5-HT1AΔ407 (C). Immunofluorescence was performed with anti-GFP antibodies to enhance the GFP signal (A, B) or with anti-5-HT1A antibodies (C). D, Cumulated fluorescence profiles along the longest dendrite for each group (30 neurons analyzed). Note that cotransfection with an excess CT1A reduces the 5-HT1A-eGFP fluorescence in distal dendrites (B), whereas the complete absence of the C-terminal segment virtually abolishes the presence of the mutant receptor in dendrites (C). Scale bar, 50 μm.
Discussion
In the present study, we identified for the first time a protein (Yif1B), which interacts with the 5-HT1AR and plays a key role in its targeting to neuronal dendrites. Direct interaction between Yif1B and the 5-HT1AR C terminus was demonstrated by yeast two-hybrid screening and GST pull-down assays. Such interaction probably takes place in vesicles located in the IC (between the endoplasmic reticulum and the Golgi apparatus), where Yif1B is confined. Yif1B is expressed in serotoninergic neurons of the raphe area, and inhibition of its expression in cultured neurons prevents the transport of 5-HT1AR to the distal part of the dendrites, but of no other receptors that we have tested (somatostatin sst2A, purinergic P2X2, and serotonin 5-HT3A). This result suggests that a ubiquitous protein plays, in addition to its ubiquitous role, a specific function in a specialized cell, such as the dendritic targeting of a specific receptor in a neuron.
Yif1B is the ortholog of a yeast protein involved in ER-to-Golgi trafficking
Yif1B and Yif1A are the two mammalian orthologs of the Saccharomyces cerevisiae protein Yif1p. As reflected by its name (Yip1p-interacting factor 1), Yif1p was first described as a partner of another yeast protein named Yip1p (Matern et al., 2000). Via their C-terminal moiety, these two proteins form a tight complex in Golgi membranes, and both interact with the transport of GTPases Ypt1p and Ypt31p (Yang et al., 1998). Furthermore, deletion experiments, which resulted in yeast death and alteration in conditional mutants, highlighted the role of Yif1p in ER-to-Golgi protein trafficking and secretion. Indeed, Yif1p is localized in COPII vesicles that are responsible for the anterograde transport from ER to Golgi (Otte et al., 2001). It has also been shown that Yif1p interacts weakly with Yipt1, the yeast ortholog of rab1 (Calero et al., 2002). More recently, the Yip1p–Yif1p complex has been more precisely implicated in the fusion process between ER-derived vesicles and the Golgi apparatus (Barrowman et al., 2003).
In comparison with the data available in yeast, the functions of Yif1p mammalian orthologs are poorly documented. Few studies showed that Yif1A (also called Yif1 as the first described Yif1p ortholog) is an early secretory pathway protein cycling between ER and Golgi, with a Golgi localization depending on its interaction with Yip1A (Yip1p ortholog) (Breuza et al., 2004; Jin et al., 2005). On the other hand, the only study mentioning a protein corresponding to Yif1B showed that it belongs, together with Yip1p and Yif1p, to a family of ER and Golgi transmembrane proteins. Indeed, Yif1A and Yif1B on one side and Yip1p, Yip4p, and Yip5p on the other side have been considered as members of the same protein family (FinGER) sharing a common structure with an N-terminal hydrophilic region, followed by five conserved transmembrane regions (Shakoori et al., 2003).
This is the first study demonstrating a role for Yif1B in mammals. Northern or Western blot analysis of Yif1B expression in several tissues showed that, although ubiquitous, Yif1B is notably expressed at higher levels in the brain, where it may play a specific role. Subcellular distribution patterns of recombinant Yif1B in different mammalian cell lines substantiate its involvement in the ER-to-Golgi trafficking, in mammals as well.
Yif1B interacts with the short C terminus of the 5-HT1A receptor
Our data clearly show that Yif1B interacts with the short C terminus of the 5-HT1A receptor in brain extracts and in transfected mammalian cell lines. Yif1B represents the first partner protein identified for the 5-HT1A receptor.
No protein interacting with the C terminus of class 1 serotonin receptors has been identified before. These receptors contain a short C terminus with no known binding consensus sequences, in contrast to class 2 (5-HT2A and 5-HT2C) serotonin receptors, which interact with several proteins through a PDZ ligand in their long C terminus domain. These receptors interact with a network including scaffolding proteins containing one or several PDZ domains, signaling proteins, and proteins of the cytoskeleton (Bécamel et al., 2002, 2004) involved in the regulation of receptor function (Gavarini et al., 2006).
Characterization of Yif1B as a routing protein interacting with the 5-HT1A receptor C terminus can therefore be considered as a first step toward the molecular identification of 5-HT1A receptor regulatory pathways. In addition, it was recently shown that genetic manipulation (downregulation in knock-out mice or overexpression in transgenic mice) of a protein interacting with the 5-HT1B serotonin receptor can modify the response to antidepressant agents (Svenningsson et al., 2006). Thus, it will be of interest to assess whether inhibition of Yif1B could influence 5-HT1A receptor desensitization that follows administration of antidepressants as mentioned before.
Yif1B is implicated in 5-HT1A trafficking in dendrites
Our results show that Yif1B plays a major role in the targeting of 5-HT1A receptors to the neuronal dendrites. An 80% decrease of endogenous Yif1B protein content in cultured neurons by siRNA caused a marked impairment of 5-HT1AR transport to the distal portion of dendrites. Under these conditions, 5-HT1AR was restricted to the proximal dendrites, with no alteration in the general structure of the dendrites as estimated from tubulin labeling. A control experiment performed with a second siRNA sequence, siRNA(Yif1B-2), confirmed the specificity of siRNA inhibition on the 5-HT1AR localization and excluded any off target resulting from siRNA inhibition (Alvarez et al., 2006). We used two controls to further assess the direct implication of the 5-HT1AR C terminus in the interaction with Yif1B. The first one consisted of a truncated 5-HT1AR, which remained confined in the soma of neurons, because it lacks the 5-HT1AR C terminus. Another control performed with competitive overexpression of the 5-HT1AR CT1A confirmed that a direct interaction between the 5-HT1AR CT1A and Yif1B was necessary for the proper dendritic targeting of 5-HT1AR.
This is the first report of a protein implicated in the specific transport of a serotonin receptor along the neuronal dendritic tree. Indeed, among the GPCR-interacting proteins that have been identified so far, several of them play a role in membrane localization or selective retention at presynaptic or postsynaptic sites. This is particularly the case for PICK1 (Xia et al., 1999), GRIP (Wyszynski et al., 1999), SHANK (Lim et al., 1999) and Homer (Brakeman et al., 1997; Ango et al., 2002), known to interact with glutamate receptors. On the other hand, some neurotransmitter receptors were shown to interact with intracellular trafficking proteins. For example, dopamine D1 receptor has been shown to interact with Drip78, an ER protein, and with COP-1, a coatomer protein (Bermak et al., 2002). Overexpression or ER sequestration of Drip78 results in ER retention of the D1 receptor. Yif1B is the first protein reported to be involved in intracellular trafficking of a GPCR and to mediate its targeting to distal dendrites. Also, this role seems to be receptor-specific, because inhibition of Yif1B expression did not alter the distribution of sst2 somatostatin, P2X2 purinergic, or 5-HT3A serotonin receptors.
A further question raised by this study is how Yif1B controls dendritic targeting of 5-HT1AR. Yif1B is a transmembrane protein localized in vesicles of the IC, which can participate in the ER–Golgi trafficking as its yeast ortholog. Although widely distributed, Yif1B may assume specific functions in specialized cells such as neurons, in addition to a more general function common to all cell types. This specialized function could depend on Yif1B level of expression and its interaction with surrounding proteins. Therefore, specific receptor targeting to neuronal dendrites could imply the interaction of Yif1B with dendrite-specific IC. In this context, another IC protein, Rab1, has also been implicated in the intracellular trafficking of some GPCRs to the plasma membrane. Experiments inhibiting rab1 expression led to the characterization of rab1-dependent and -independent pathways from the ER to the plasma membrane depending on the GPCR studied (Wu et al., 2003; Filipeanu et al., 2006). Similarly, concerning another family of receptors, the regulation of a nicotinic acetylcholine receptor subtype trafficking has been recently described in Caenorhabditis elegans, revealing that its traffic depended on UNC-50, an endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-associated protein (Eimer et al., 2007). Furthermore, UNC-50 displays an organization and size similar to Yif1B, i.e., five transmembrane segments clustered in the C-terminal moiety, a long hydrophilic N-terminal domain within the cytoplasm, and the C terminus turned to the ER lumen. But no sequence homology could be found between Yif1B and UNC-50, confirming that they belong to different protein families exhibiting similarities: they are both ubiquitous ERGIC proteins that are also involved in the specific trafficking of one type of receptor, in a differentiated cell type.
Although these small proteins regulating the intracellular traffic are ubiquitous, they may play specific roles according to the cell type or the receptors that transit from the ER to the plasma membrane (Sivars et al., 2003). The existence in mammals of several proteins (Yif1A and Yif1B, or Yip1p, Yip4p, and Yip5p), as opposed to only one in yeast, may reflect the diversity of intracellular vesicular trafficking in differentiated cells (Shakoori et al., 2003). Such an idea is supported by differences in IC protein composition observed between soma and neurites of differentiated PC12 cells. Indeed, Rab1, P58, and COPI IC proteins mainly colocalize in the ER exit sites (ERESs) at the soma, whereas only Rab1 is present in dynamic vesicular-tubular structures at the neurites, suggesting the existence of neurite-specific ERESs (Sannerud et al., 2006).
In neurons, a secretory pathway has been identified in neuronal dendrites, as far as in dendritic spines (Pierce et al., 2000, 2001). Furthermore, the existence of dendritic ERESs may enable endogenous proteins to exit from the ER at different levels in the dendritic tree (Aridor et al., 2004). In addition, proteins of the secretory pathway, such as Sar1, affect dendritic growth both in vivo in Drosophila mutants, and in vitro, in the early differentiation of primary neuronal cultures (Ye et al., 2007). Yif1B would similarly participate in the dendritic differentiation, including the distal dendritic targeting of the receptor, but without affecting the dendritic neuronal morphology, because we have shown that inhibition of Yif1B does not change the length of the dendritic tree, nor the morphology of the Golgi apparatus.
Yif1B could belong to the pool of chaperone proteins involved in the exit from the ER (through IC vesicles) of proteins finally targeted to the dendritic cell membrane, such as 5-HT1AR. Therefore, the identification of Yif1B as a 5-HT1AR-specific dendritic targeting protein makes an initial ground for the discovery of other members of the intracellular machinery involved in the dendritic targeting of the 5-HT1AR, and more generally of other GPCRs with a short C terminus.
Footnotes
This work was supported by grants from Inserm, Université Pierre et Marie Curie, and European Community NewMood (LSHM-CT-2003-503474) Program. D.C. was a recipient of fellowships from the French Ministère de la Recherche and the Fondation pour la Recherche Médicale during performance of this work. We are grateful to Dr. Michel Maitre (Inserm U575, Strasbourg, France) and Dr. Michel Bornens (Unité Mixte de Recherche 144 Centre National de la Recherche Scientifique/Institut Curie, Paris, France) for their kind gifts of anti-tryptophan hydroxylase antibodies and anti CTR-433 antibody, respectively. We are indebted to Dr. Nacira Tabti for critical reading of this manuscript.
References
- Albert PR, Zhou QY, Van Tol HH, Bunzow JR, Civelli O. Cloning, functional expression, and mRNA tissue distribution of the rat 5-hydroxytryptamine1A receptor gene. J Biol Chem. 1990;265:5825–5832. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez VA, Ridenour DA, Sabatini BL. Retraction of synapses and dendritic spines induced by off-target effects of RNA interference. J Neurosci. 2006;26:7820–7825. doi: 10.1523/JNEUROSCI.1957-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ango F, Robbe D, Tu JC, Xiao B, Worley PF, Pin JP, Bockaert J, Fagni L. Homer-dependent cell surface expression of metabotropic glutamate receptor type 5 in neurons. Mol Cell Neurosci. 2002;20:323–329. doi: 10.1006/mcne.2002.1100. [DOI] [PubMed] [Google Scholar]
- Aridor M, Guzik AK, Bielli A, Fish KN. Endoplasmic reticulum export site formation and function in dendrites. J Neurosci. 2004;24:3770–3776. doi: 10.1523/JNEUROSCI.4775-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes NM, Sharp T. A review of central 5-HT receptors and their function. Neuropharmacology. 1999;38:1083–1152. doi: 10.1016/s0028-3908(99)00010-6. [DOI] [PubMed] [Google Scholar]
- Barrowman J, Wang W, Zhang Y, Ferro-Novick S. The Yip1p.Yif1p complex is required for the fusion competence of endoplasmic reticulum-derived vesicles. J Biol Chem. 2003;278:19878–19884. doi: 10.1074/jbc.M302406200. [DOI] [PubMed] [Google Scholar]
- Bécamel C, Alonso G, Galéotti N, Demey E, Jouin P, Ullmer C, Dumuis A, Bockaert J, Marin P. Synaptic multiprotein complexes associated with 5-HT(2C) receptors: a proteomic approach. EMBO J. 2002;21:2332–2342. doi: 10.1093/emboj/21.10.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécamel C, Gavarini S, Chanrion B, Alonso G, Galéotti N, Dumuis A, Bockaert J, Marin P. The serotonin 5-HT2A and 5-HT2C receptors interact with specific sets of PDZ proteins. J Biol Chem. 2004;279:20257–20266. doi: 10.1074/jbc.M312106200. [DOI] [PubMed] [Google Scholar]
- Bermak JC, Li M, Bullock C, Weingarten P, Zhou QY. Interaction of gamma-COP with a transport motif in the D1 receptor C-terminus. Eur J Cell Biol. 2002;81:77–85. doi: 10.1078/0171-9335-00222. [DOI] [PubMed] [Google Scholar]
- Bockaert J, Fagni L, Dumuis A, Marin P. GPCR interacting proteins (GIP) Pharmacol Ther. 2004;103:203–221. doi: 10.1016/j.pharmthera.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Boué-Grabot E, Barajas-López C, Chakfe Y, Blais D, Bélanger D, Emerit MB, Séguéla P. Intracellular cross talk and physical interaction between two classes of neurotransmitter-gated channels. J Neurosci. 2003;23:1246–1253. doi: 10.1523/JNEUROSCI.23-04-01246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boué-Grabot E, Emerit MB, Toulmé E, Séguéla P, Garret M. Cross-talk and co-trafficking between rho1/GABA receptors and ATP-gated channels. J Biol Chem. 2004a;279:6967–6975. doi: 10.1074/jbc.M307772200. [DOI] [PubMed] [Google Scholar]
- Boué-Grabot E, Toulmé E, Emerit MB, Garret M. Subunit-specific coupling between gamma-aminobutyric acid type A and P2X2 receptor channels. J Biol Chem. 2004b;279:52517–52525. doi: 10.1074/jbc.M410223200. [DOI] [PubMed] [Google Scholar]
- Brakeman PR, Lanahan AA, O'Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- Breuza L, Halbeisen R, Jenö P, Otte S, Barlowe C, Hong W, Hauri HP. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J Biol Chem. 2004;279:47242–47253. doi: 10.1074/jbc.M406644200. [DOI] [PubMed] [Google Scholar]
- Brewer CB, Roth MG. A single amino acid change in the cytoplasmic domain alters the polarized delivery of influenza virus hemagglutinin. J Cell Biol. 1991;114:413–421. doi: 10.1083/jcb.114.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero M, Winand NJ, Collins RN. Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors. FEBS Lett. 2002;515:89–98. doi: 10.1016/s0014-5793(02)02442-0. [DOI] [PubMed] [Google Scholar]
- Carrel D, Hamon M, Darmon M. Role of the C-terminal di-leucine motif of 5-HT1A and 5-HT1B serotonin receptors in plasma membrane targeting. J Cell Sci. 2006;119:4276–4284. doi: 10.1242/jcs.03189. [DOI] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon M, Langlois X, Suffisseau L, Fattaccini CM, Hamon M. Differential membrane targeting and pharmacological characterization of chimeras of rat serotonin 5-HT1A and 5-HT1B receptors expressed in epithelial LLC-PK1 cells. J Neurochem. 1998;71:2294–2303. doi: 10.1046/j.1471-4159.1998.71062294.x. [DOI] [PubMed] [Google Scholar]
- Doucet E, Hamon M, Emerit MB. Immunolabelling of the rat intestinal tract with antibodies specific to the long form of the 5-hydroxytryptamine3 receptor. Neuroscience. 1998;87:691–707. doi: 10.1016/s0306-4522(98)00184-5. [DOI] [PubMed] [Google Scholar]
- Ehret M, Pevet P, Maitre M. Tryptophan hydroxylase synthesis is induced by 3′,5′-cyclic adenosine monophosphate during circadian rhythm in the rat pineal gland. J Neurochem. 1991;57:1516–1521. doi: 10.1111/j.1471-4159.1991.tb06346.x. [DOI] [PubMed] [Google Scholar]
- Eimer S, Gottschalk A, Hengartner M, Horvitz HR, Richmond J, Schafer WR, Bessereau JL. Regulation of nicotinic receptor trafficking by the transmembrane Golgi protein UNC-50. EMBO J. 2007;26:4313–4323. doi: 10.1038/sj.emboj.7601858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Mestikawy S, Riad M, Laporte AM, Vergé D, Daval G, Gozlan H, Hamon M. Production of specific anti-rat 5-HT1A receptor antibodies in rabbits injected with a synthetic peptide. Neurosci Lett. 1990;118:189–192. doi: 10.1016/0304-3940(90)90623-h. [DOI] [PubMed] [Google Scholar]
- Filipeanu CM, Zhou F, Fugetta EK, Wu G. Differential regulation of the cell-surface targeting and function of beta- and alpha1-adrenergic receptors by Rab1 GTPase in cardiac myocytes. Mol Pharmacol. 2006;69:1571–1578. doi: 10.1124/mol.105.019984. [DOI] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V, et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavarini S, Bécamel C, Altier C, Lory P, Poncet J, Wijnholds J, Bockaert J, Marin P. Opposite effects of PSD-95 and MPP3 PDZ proteins on serotonin 5-hydroxytryptamine2C receptor desensitization and membrane stability. Mol Biol Cell. 2006;17:4619–4631. doi: 10.1091/mbc.E06-03-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grailhe R, de Carvalho LP, Paas Y, Le Poupon C, Soudant M, Bregestovski P, Changeux JP, Corringer PJ. Distinct subcellular targeting of fluorescent nicotinic alpha 3 beta 4 and serotoninergic 5-HT3A receptors in hippocampal neurons. Eur J Neurosci. 2004;19:855–862. doi: 10.1111/j.1460-9568.2004.03153.x. [DOI] [PubMed] [Google Scholar]
- Haj-Dahmane S, Hamon M, Lanfumey L. K+ channel and 5-hydroxytryptamine1A autoreceptor interactions in the rat dorsal raphe nucleus: an in vitro electrophysiological study. Neuroscience. 1991;41:495–505. doi: 10.1016/0306-4522(91)90344-n. [DOI] [PubMed] [Google Scholar]
- Jasmin BJ, Cartaud J, Bornens M, Changeux JP. Golgi apparatus in chick skeletal muscle: changes in its distribution during end plate development and after denervation. Proc Natl Acad Sci U S A. 1989;86:7218–7222. doi: 10.1073/pnas.86.18.7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zhang Y, Zhu H, Ahmed K, Fu C, Yao X. Human Yip1A specifies the localization of Yif1 to the Golgi apparatus. Biochem Biophys Res Commun. 2005;334:16–22. doi: 10.1016/j.bbrc.2005.06.051. [DOI] [PubMed] [Google Scholar]
- Jolimay N, Franck L, Langlois X, Hamon M, Darmon M. Dominant role of the cytosolic C-terminal domain of the rat 5-HT1B receptor in axonal-apical targeting. J Neurosci. 2000;20:9111–9118. doi: 10.1523/JNEUROSCI.20-24-09111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kia HK, Miquel MC, Brisorgueil MJ, Daval G, Riad M, El Mestikawy S, Hamon M, Vergé D. Immunocytochemical localization of serotonin1A receptors in the rat central nervous system. J Comp Neurol. 1996;365:289–305. doi: 10.1002/(SICI)1096-9861(19960205)365:2<289::AID-CNE7>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kushwaha N, Harwood SC, Wilson AM, Berger M, Tecott LH, Roth BL, Albert PR. Molecular determinants in the second intracellular loop of the 5-hydroxytryptamine-1A receptor for G-protein coupling. Mol Pharmacol. 2006;69:1518–1526. doi: 10.1124/mol.105.019844. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lanfumey L, Hamon M. 5-HT1 receptors. Curr Drug Targets CNS Neurol Disord. 2004;3:1–10. doi: 10.2174/1568007043482570. [DOI] [PubMed] [Google Scholar]
- Lelouvier B, Tamagno G, Kaindl AM, Roland A, Lelievre V, Le Verche V, Loudes C, Gressens P, Faivre-Baumann A, Lenkei Z, Dournaud P. Dynamics of somatostatin type 2A receptor cargoes in living hippocampal neurons. J Neurosci. 2008;28:4336–4349. doi: 10.1523/JNEUROSCI.4379-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poul E, Laaris N, Doucet E, Laporte AM, Hamon M, Lanfumey L. Early desensitization of somato-dendritic 5-HT1A autoreceptors in rats treated with fluoxetine or paroxetine. Naunyn Schmiedebergs Arch Pharmacol. 1995;352:141–148. doi: 10.1007/BF00176767. [DOI] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- Matern H, Yang X, Andrulis E, Sternglanz R, Trepte HH, Gallwitz D. A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J. 2000;19:4485–4492. doi: 10.1093/emboj/19.17.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte S, Belden WJ, Heidtman M, Liu J, Jensen ON, Barlowe C. Erv41p and Erv46p: new components of COPII vesicles involved in transport between the ER and Golgi complex. J Cell Biol. 2001;152:503–518. doi: 10.1083/jcb.152.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JP, van Leyen K, McCarthy JB. Translocation machinery for synthesis of integral membrane and secretory proteins in dendritic spines. Nat Neurosci. 2000;3:311–313. doi: 10.1038/73868. [DOI] [PubMed] [Google Scholar]
- Pierce JP, Mayer T, McCarthy JB. Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr Biol. 2001;11:351–355. doi: 10.1016/s0960-9822(01)00077-x. [DOI] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schächter V, Chemama Y, Labigne A, Legrain P. The protein-protein interaction map of Helicobacter pylori . Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Riad M, Garcia S, Watkins KC, Jodoin N, Doucet E, Langlois X, el Mestikawy S, Hamon M, Descarries L. Somatodendritic localization of 5-HT1A and preterminal axonal localization of 5-HT1B serotonin receptors in adult rat brain. J Comp Neurol. 2000;417:181–194. [PubMed] [Google Scholar]
- Sannerud R, Marie M, Nizak C, Dale HA, Pernet-Gallay K, Perez F, Goud B, Saraste J. Rab1 defines a novel pathway connecting the pre-Golgi intermediate compartment with the cell periphery. Mol Biol Cell. 2006;17:1514–1526. doi: 10.1091/mbc.E05-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoori A, Fujii G, Yoshimura S, Kitamura M, Nakayama K, Ito T, Ohno H, Nakamura N. Identification of a five-pass transmembrane protein family localizing in the Golgi apparatus and the ER. Biochem Biophys Res Commun. 2003;312:850–857. doi: 10.1016/j.bbrc.2003.10.197. [DOI] [PubMed] [Google Scholar]
- Sivars U, Aivazian D, Pfeffer SR. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature. 2003;425:856–859. doi: 10.1038/nature02057. [DOI] [PubMed] [Google Scholar]
- Sotelo C, Cholley B, El Mestikawy S, Gozlan H, Hamon M. Direct immunohistochemical evidence of the existence of 5-HT1A autoreceptors on serotoninergic neurons in the midbrain raphe nuclei. Eur J Neurosci. 1990;2:1144–1154. doi: 10.1111/j.1460-9568.1990.tb00026.x. [DOI] [PubMed] [Google Scholar]
- Svenningsson P, Chergui K, Rachleff I, Flajolet M, Zhang X, El Yacoubi M, Vaugeois JM, Nomikos GG, Greengard P. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- Turner JH, Garnovskaya MN, Raymond JR. Serotonin 5-HT1A receptor stimulates c-Jun N-terminal kinase and induces apoptosis in Chinese hamster ovary fibroblasts. Biochim Biophys Acta. 2007;1773:391–399. doi: 10.1016/j.bbamcr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Wojcik J, Boneca IG, Legrain P. Prediction, assessment and validation of protein interaction maps in bacteria. J Mol Biol. 2002;323:763–770. doi: 10.1016/s0022-2836(02)01009-4. [DOI] [PubMed] [Google Scholar]
- Wu G, Zhao G, He Y. Distinct pathways for the trafficking of angiotensin II and adrenergic receptors from the endoplasmic reticulum to the cell surface: Rab1-independent transport of a G protein-coupled receptor. J Biol Chem. 2003;278:47062–47069. doi: 10.1074/jbc.M305707200. [DOI] [PubMed] [Google Scholar]
- Wyszynski M, Valtschanoff JG, Naisbitt S, Dunah AW, Kim E, Standaert DG, Weinberg R, Sheng M. Association of AMPA receptors with a subset of glutamate receptor-interacting protein in vivo . J Neurosci. 1999;19:6528–6537. doi: 10.1523/JNEUROSCI.19-15-06528.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Yang X, Matern HT, Gallwitz D. Specific binding to a novel and essential Golgi membrane protein (Yip1p) functionally links the transport GTPases Ypt1p and Ypt31p. EMBO J. 1998;17:4954–4963. doi: 10.1093/emboj/17.17.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]