Abstract
Neurons acquire their molecular, neurochemical, and connectional features during development as a result of complex regulatory mechanisms. Here, we show that a ubiquitous, multifunctional protein cofactor, Chip, plays a critical role in a set of neurons in Drosophila that control the well described posteclosion behavior. Newly eclosed flies normally expand their wings and display tanning and hardening of their cuticle. Using multiple approaches to interfere with Chip function, we find that these processes do not occur without normal activity of this protein. Furthermore, we identified the nature of the deficit to be an absence of Bursicon in the hemolymph of newly eclosed flies, whereas the responsivity to Bursicon in these flies remains normal. Chip interacts with transcription factors of the LIM-HD (LIM-homeodomain) family, and we identified one member, dIslet, as a potential partner of Chip in this process. Our findings provide the first evidence of transcriptional mechanisms involved in the development of the neuronal circuit that regulates posteclosion behavior in Drosophila.
Keywords: LIM, wing expansion, Bursicon, behavior, neuroendocrine, neurosecretion
Introduction
Transcriptional mechanisms that regulate neuronal development are the focus of study in many systems. A family of LIM-homeodomain (LIM-HD) transcription factors and their cofactors have been implicated in various aspects of neuronal development in both vertebrates and invertebrates. In the vertebrate spinal cord, LIM-HD protein Islet1 acts in concert with Lhx3/4 or Lhx1 to regulate multiple aspects of motoneuron specification and pathfinding (Shirasaki and Pfaff, 2002; Thaler et al., 2004). In the vertebrate forebrain, Lhx2 functions as a “cortical selector” gene (Mangale et al., 2008). It is also required for the specification of part of the amygdala (Remedios et al., 2004) and for multiple aspects of olfactory system development, including the regulation of survival, receptor expression, and axon guidance (Hirota and Mombaerts, 2004; Kolterud et al., 2004; Saha et al., 2007). Striking parallels are found in Drosophila: Apterous (Ap), an orthologue of Lhx2, acts as a selector gene for dorsal identity in the developing wing disc (Blair et al., 1994) and is required for axonal pathfinding, neurotransmitter identity, and receptor expression in the nervous system (Lundgren et al., 1995; Benveniste et al., 1998; Baumgardt et al., 2007). dIslet regulates neurotransmitter identity and pathfinding of specific classes of embryonic neurons and acts in concert with Lim3 to confer target specificity in motor neurons (Thor and Thomas, 1997; Thor et al., 1999).
None of these functions is possible without a ubiquitous and versatile cofactor, Chip. Each LIM-HD molecule binds to Chip, which dimerizes, forming a functional tetramer (Gill, 2003). In the wing disc, the LIM only gene (dLmo/Beadex) is capable of binding Chip with a higher affinity than Ap and thus acts as a dominant-negative protein, preventing the formation of Ap:Chip tetramers (see Fig. 1) (Milán et al., 1998; Weihe et al., 2001). Chip-null mutations result in lethality before third-instar larval stage, whereas embryos derived from Chip mutant germ line clones fail to hatch (Morcillo et al., 1997). We predicted that overexpressing dLmo in neurons would sequester Chip from its partners and enable us to identity novel functions of Chip in neurons. Pan-neuronal expression of dLmo using ELAV (embryonic lethal and abnormal vision) GAL4 resulted in lethality of the progeny at 25°C. When maintained at 18°C, a few flies survived, which failed to expand their wings after eclosion. This directed our interest to the well described posteclosion behavior of Drosophila, in which newly eclosed flies expand their wings and the cuticle becomes tanned and hard. The crustacean cardioactive peptide (CCAP)-expressing neurons of the ventral ganglion are known to regulate this behavior by secreting the neurohormone Bursicon (Honegger et al., 2002; Dewey et al., 2004; Luan et al., 2006a). Using multiple approaches to interfere with Chip function, we uncover a novel function of Chip in these neurons in the regulation of posteclosion behavior.
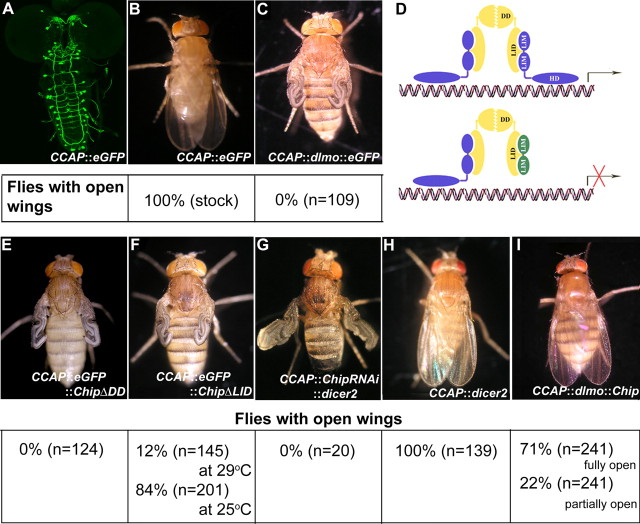
Figure 1.
dLmo overexpression in CCAP-expressing neurons produces a wing expansion phenotype. A, CCAP::eGFP marks a set of neurons in the brain and ventral ganglia, seen in a third-instar larval preparation. B, Flies with this genotype are able to expand their wings normally after eclosion. C, Overexpressing dLmo using this driver generates flies that are unable to expand their wings after eclosion, and this phenotype is fully penetrant. D, As illustrated in the schematic, dLmo (green) is expected to bind cofactor Chip (yellow) and prevent formation of the active LIM-HD:Chip tetramer. E, F, Consistent with this model, flies overexpressing truncated constructs of Chip, such as ChipΔDD (E) or ChipΔLID (F) also fail to open their wings. G, H, A ChipRNAi line, together with Dicer 2 overexpression, also produces this phenotype (G), whereas Dicer 2 alone has no effect (H). I, Chip is able to rescue the dLmo overexpression phenotype, providing conclusive evidence of the role of Chip in this process.
Materials and Methods
Fly stocks and culture conditions.
Fly stocks included CCAP Gal4 (16) on II (F. Rob Jackson, Tufts University School of Medicine, Boston, MA), UAS-Chip FL, UAS Chip-ΔDD, UAS Chip-ΔLID (D. van Meyel, McGill University, Montreal, Quebec, Canada), UAS-lim3, UAS-islet (J. I. Pueyo, University of Sussex, Brighton, UK), EP1306 (U. Heberlein, University of California, San Francisco, San Francisco, CA), UAS-dimmed (P. Taghert, Washington University School of Medicine, St. Louis, MO), UAS 2x eGFP (H. Keshishian, Yale University, New Haven, CT), and Gal80ts lines (Bloomington Stock Center). All fly stocks were reared on standard cornmeal medium containing yeast at 25°C unless otherwise stated. UAS-Chip RNA interference (RNAi) stock (GD 30454) and UAS-dicer2 (stock number 60009) were obtained from Vienna Drosophila RNAi Centre (VDRC), Austria (Dietzl et al., 2007).
Staging of pupae.
For developmental studies, white prepupae [0 h after puparium formation (APF)] were collected and allowed to develop on moist filter paper at 25°C unless specified. This stage lasts for an hour; hence the error in staging is ±30 min. Ages of pupae grown at other temperatures were normalized with respect to growth at 25°C. Wild-type pupae took ∼100 h to eclose when grown at 25°C in our laboratory.
Gal80ts experiment.
The cross was grown at 18°C to antagonize Gal4 action; transferring cultures to the restrictive temperature of Gal80ts (29°C) resulted in Gal4 being able to regulate genes downstream of UAS. Heat shock was given to the pupae (after the staging) at different time points. Each time point denotes the normalized age of pupae at 25°C when they were shifted to higher temperature (29°C).
Immunohistochemistry: antibodies.
Rabbit anti-green fluorescent protein (GFP) (1:10,000; Invitrogen; catalog #A11122) and mouse anti-Islet (1:20; DSHB 40.3A4). Secondary antibodies from Invitrogen, Alexa 488 goat anti-rabbit (A11008), and Alexa 555 goat anti-mouse (A-21424) were used were used at 1:400 dilution. Immunohistochemistry was performed as described previously (Jhaveri et al., 2000) and imaged using an Olympus FV1000 confocal microscope. Serial optical frontal sections of the brain were acquired at intervals of 1 μm and saved as stacks. Two-dimensional projections were generated by stacking appropriate sections for each channel using Olympus Fluoview 1000 (version 1.4a) viewer software (confocal). Image processing, including pseudocolouring and labeling, were done using Adobe Photoshop 7.0 and WCIF ImageJ (1.37c) software.
burs (bursicon α) and pburs (bursicon β) probes. Genes were amplified from fly genomic DNA using primers containing a T7 binding sequence. Primers were as follows: CG13419 (burs), forward, 5′-CGCGGATCCATGCTGCGCCACCTGCTC-3′; reverse, 5′-AATACGACTCACTATAGGGCTATTGCAGAGCAATGCG-3′; CG15284 (pburs), forward, 5′-CGCGGATCCATGCATGTCCAGGAACTG-3′; reverse, 5′-TAATACGACTCACTATAGGGTTAACGTGTGAAATCGCC-3′. In vitro transcription was performed using the PCR product as template per the manufacturer's instruction (Roche).
In situ hybridization.
The tissue was dissected in cold PBS, fixed in 4% paraformaldehyde (PFA) for 2 h at room temperature, and washed with PBS containing 0.3% Triton X-100 (0.3% PTX) with frequent changes for 1 h. The samples were prehybridized at 65°C for 1 h in hybridization buffer (50% formamide, 5× SSC, 1% SDS, 0.5 mg/ml salmon sperm DNA, and 5× Denhardt's solution) and then hybridized with digoxygenin-labeled RNA probe in hybridization buffer at 65°C for 60 h. After hybridization, the samples were washed with graded dilutions of hybridization buffer in 0.3% PTX at 65°C (75, 50, and 25%). The final wash was with 0.3% PTX at room temperature, followed by incubation with anti-digoxigenin Fab fragments conjugated to alkaline phosphatase (Roche) diluted at 1:5000 in 0.3% PTX, overnight at 4°C. Samples were washed with several changes of 0.3% PTX for 1 h and further washed overnight. The color reaction was performed using NBT (nitroblue tetrazolium) and BCIP (5-bromo-4-chloro-3-indolyl phosphate) (Roche) per the manufacturer's instructions. After color development, samples were transferred through serial grades of glycerol in water and mounted in 70% of glycerol for imaging using an Axiocam attached to a Zeiss Axioplan microscope.
Drosophila neck ligation and hemolymph transfer assay for Bursicon release.
Newly eclosed adults were collected at 5 min intervals. Flies were immobilized on an ice-chilled Petri plate and ligated at the neck with a thin silk thread, and the head was cut away. They were maintained at room temperature in a humid chamber for 60 min (±10 min). Ligated flies that tanned or expanded their wings during this interval were discarded. For collection of hemolymph, a small hole was made in the thorax and/or abdomen of a fly at 30 min (±10 min) after eclosion with a glass capillary needle and the thorax was compressed using forceps. A drop of hemolymph was collected in another glass capillary needle and was injected into the host neck-ligated fly by connecting the capillary to a glass syringe. The volume of solution injected was ∼65–144 nl. Male flies were used as the host because it is easy to observe the cuticular darkening and tanning at abdominal tergites. Injected flies were kept in a moist Petri dish, and cuticular darkening and tanning were examined at 1 h after injection.
For injection of hemolymph/8-bromo-cAMP into intact flies, newly eclosed adults of the mutants (males and females) were collected and immobilized as above. They were injected with PBS or 0.1 m 8-bromo-cAMP or hemolymph from wild-type flies, maintained at room temperature in a humid chamber for 60 min, and examined for wing expansion.
Results
Functional Chip protein is required in CCAP neurons for posteclosion wing expansion behavior
We used the EP1306 line (Milán et al., 1998; Zeng et al., 1998; Tsai et al., 2004) to drive ectopic expression of dLmo using CCAP-Gal4. This gave rise to flies that failed to expand their wings after eclosion (Fig. 1). To verify whether the wing expansion defect seen with dLmo overexpression was indeed attributable to impairment of Chip function, we used several different approaches. Truncated forms of Chip can act in a dominant-negative manner when expressed in cells requiring Chip function (Milán and Cohen, 1999; van Meyel et al., 1999). Consistent with this, two such truncated versions independently recapitulated the dLmo overexpression phenotype. Expression of either Chip-ΔDD, a protein that lacks the dimerization domain (DD), or Chip-ΔLID, which lacks the LIM interaction domain (LID), produced flies that did not open their wings after eclosion (Fig. 1). Chip-ΔDD gave a fully penetrant phenotype at 25°C, whereas Chip-ΔLID gave a stronger phenotype at 29°C than at 25°C (Fig. 1). It is well known that the level of expression from a UAS promoter is dependent on its insertion site in the genome. The ChipΔLID line may require the increased Gal4 levels produced at 29°C, to drive sufficient levels of the transgene to cause a phenotype. However, it is also possible that, compared with Chip-ΔDD, higher levels of Chip-ΔLID are, in fact, required to produce the phenotype. Chip-ΔLID contains the dimerization domain and is therefore able to self-dimerize, leaving fewer molecules free to bind the endogenous Chip. In contrast, Chip-ΔDD lacks the dimerization domain and can be expected to act more efficiently by binding with the partner(s) of Chip.
To confirm the role of Chip in the control of posteclosion wing expansion, a Chip RNAi line was obtained from VDRC, Austria (GD 30454). When expressed together with Dicer 2, flies from this line also fail to open their wings after eclosion, whereas Dicer 2 alone did not cause a phenotype. Finally, we found that the wing expansion defect caused by expression of dLmo can be rescued by coexpressing Chip protein in CCAP neurons (Fig. 1). These results indicate that Chip-mediated functions are required for normal posteclosion behavior.
Time window of Chip requirement
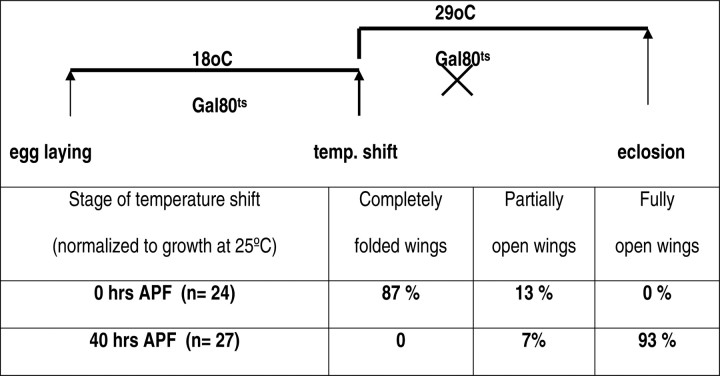
Chip is known to be ubiquitously expressed at multiple stages of Drosophila development (Morcillo et al., 1997). To identify the stage at which Chip function is required, we used the Gal80ts system to control the timing of dLmo overexpression. At the permissive temperature (18°C), the Gal80 protein binds to Gal4, preventing its action in turning on genes downstream of UAS. At restrictive temperatures (29°C), Gal80 is inactivated, leaving Gal4 available to act on UAS-dLmo. As expected, misexpression of dLmo in the CCAP neurons for the entire pupal period (shift up at 0 h APF) led to a fully penetrant wing expansion defect. Gal4-induced misexpression of dLmo after 40 h (shift up at 40 APF), however, does not affect posteclosion behavior (Table 1).
Table 1.
Chip function is required during pupal stages for normal posteclosion wing expansion
Flies maintained at 18° C express Gal80ts, which interferes with Gal4 function. Consequently, dLmo overexpression does not occur. Raising the temperature to 29° C destabilizes Gal80ts and permits the overexpression of dLmo. Such a shift in temperature performed at the beginning of pupal formation (0 h APF) severely affects the posteclosion wing expansion. However, raising the temperature at a stage corresponding to 40 h APF does not affect wing expansion and produces a normal phenotype. Genotype: EP1306/+ or EP1306/Y; CCAP-Gal4/+; tubGal80ts.
These experiments indicate that Chip function is likely to be required during the pupal stages to regulate posteclosion behavior. These data do not allow us to accurately pinpoint the time of requirement of Chip function in CCAP neurons because of the lag time for inactivation of Gal80 and restoration of Gal4 function. In addition to this, the time subsequently required for dLmo induction would cause a delay in the time at which Chip function is compromised. In a different system, the estimate for the lag time of Gal4 function on temperature shift up is a few hours (McGuire et al., 2003). With these caveats, we tentatively conclude that Chip function is necessary in the CCAP neurons in the earlier one-half of pupal life for generation of wing expansion behavior that occurs on eclosion.
Islet rescues the dLmo-induced wing expansion defect
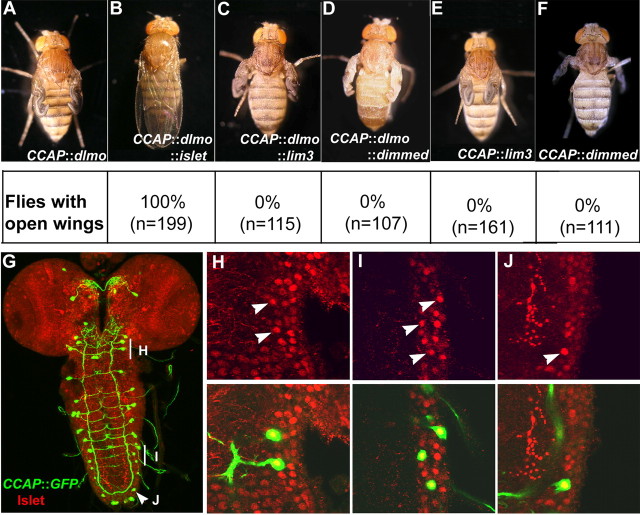
To identify potential partners of Chip in regulating posteclosion behavior, we coexpressed three known interactors of Chip (van Meyel et al., 1999; Allan et al., 2005) in CCAP neurons together with dLmo. We hypothesized that the correct partner would be able to overcome the effect of dLmo overexpression. Among the candidates we tested, only dIslet was able to rescue wing expansion, whereas dLim3 (a LIM-HD protein) and Dimmed [a bHLH (basic helix-loop-helix) protein] were not able to do so (Fig. 2). Furthermore, dLim3 or Dimmed expressed alone (i.e., without dLmo) produced a defect in wing expansion when expressed in CCAP neurons (0% open wings; n > 100). This is consistent with the expectation that any protein capable of binding Chip can sequester it away from its correct partner(s) and produce phenotypes similar to those seen with dLmo. In contrast, dIslet expressed alone resulted in normal wing opening (100% open wings; n = 134). Because dIslet is known to bind Chip (van Meyel et al., 1999), this suggests that Islet may function as the partner of Chip in CCAP neurons in regulating posteclosion behavior.
Figure 2.
dIslet, but not other partners of Chip, can rescue the wing expansion phenotype. A–D, The wing expansion phenotype in dLmo-overexpressing flies (A) is rescued by expression of dIslet (B) but not dLim3 (C) or Dimmed (D). E, F, dLim3 (E) or Dimmed (F) expressed alone produce files with closed wings as expected. G–J, Distribution of CCAP-expressing cells (green) and Islet (red) in the third-instar larval brain (G), with regions shown at high magnification marked (H–J). Single confocal sections reveal that Islet expression overlaps with CCAP-positive neurons in the subesophageal ganglia (H) and abdominal segments (I, J).
Additional support for such a function comes from the expression of dIslet in third-instar larval brains. dIslet is widely expressed in the nervous system. In particular, neurons in the subesophageal, thoracic, as well as abdominal ganglia show colocalization of GFP (indicating CCAP-Gal4 expression) with dIslet immunoreactivity (n = 5 animals) (Fig. 2). The subesophageal and thoracic neurons do not express Bursicon but are implicated in regulating its release from the neurons in the abdominal ganglia, which are known to be Bursicon-expressing (Luo et al., 2005). Null mutants of islet are embryonic lethal, preventing a direct examination of whether wing expansion can occur in the absence of Islet (Thor and Thomas, 1997). Nonetheless, our findings raise the hypothesis that Islet may interact with Chip to regulate Bursicon production or release, which would then affect posteclosion behavior.
burs (burs α)- and pburs (burs β)-expressing neurons are unaffected by dLmo overexpression
The neurohormone Bursicon is expressed in a subset of CCAP neurons and is responsible for initiating wing expansion and tanning and hardening of the cuticle after eclosion (Honegger et al., 2002; Dewey et al., 2004; Luan et al., 2006a). Bursicon is active as a heterodimer (Luo et al., 2005; Mendive et al., 2005), and both subunits are coexpressed in paired neurons in four abdominal segments (A1–A4) of third-instar larval/early pupal CNS. The burs α subunit is expressed in a larger subset of CCAP neurons (seven pairs) (Luo et al., 2005). We analyzed the expression of burs or pburs transcripts in dLmo-overexpressing flies by in situ hybridization. Although this technique does not offer a quantitative measure of the transcripts, the numbers and positions of burs- or pburs-expressing neurons were normal at this stage of development (Fig. 3). Thus, it seems unlikely that Chip acts directly in controlling Bursicon production.
Figure 3.
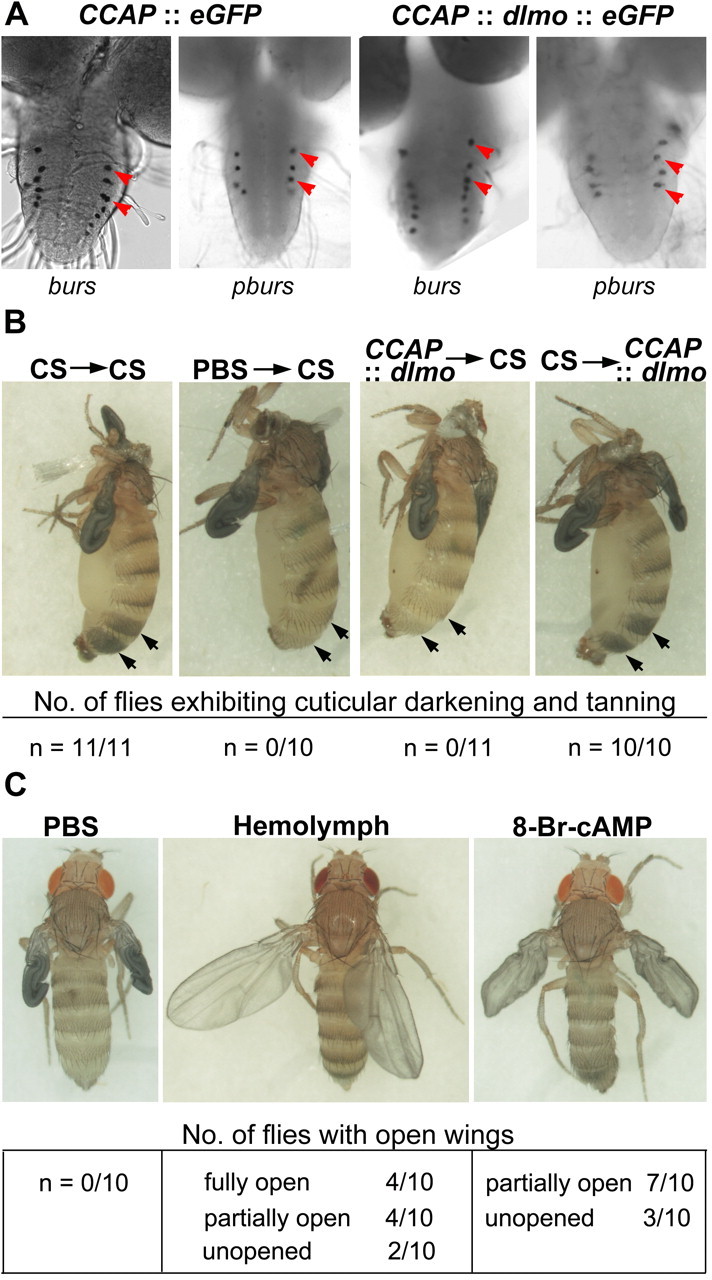
dLmo overexpression interferes with the release, but not the production or the responsiveness to Bursicon in the posteclosion hemolymph. A, The burs and pburs expression appears normal in dLmo-overexpressing third-instar larvae. B, Neck ligation of wild-type flies prevents tanning of the cuticle after eclosion. Injecting posteclosion hemolymph from wild-type flies, but not PBS, into such neck-ligated preparations, rescues this tanning phenotype (arrows). Injection of posteclosion hemolymph from dLmo-overexpressing flies fails to rescue the tanning of neck-ligated wild-type flies, indicating a deficiency of Bursicon in the hemolymph. However, injection of wild-type hemolymph into neck-ligated dLmo-overexpressing flies produces normal tanning, indicating that the response to Bursicon is normal in these flies. C, Intact dLmo-overexpressing flies do not expand their wings when injected with PBS after eclosion. However, wild-type hemolymph is able to rescue the wing expansion defect. This effect is also mimicked by injection of 8-Br-cAMP, implicating both a signal from the head and a Bursicon-cAMP-mediated pathway in regulating posteclosion wing expansion.
dLmo overexpression results in an absence of Bursicon in the hemolymph at the time of eclosion
We used a well established assay to test for the presence of Bursicon in hemolymph. Neck ligation of flies immediately after eclosion prevents wing expansion and tanning and hardening of the cuticle. Of these posteclosion behaviors, the cuticle tanning and hardening deficit in neck-ligated flies can be rescued by transfer of hemolymph from a newly eclosed, intact wild-type fly (Cottrell, 1962; Fraenkel and Hsiao, 1965; Kimura and Truman, 1990). This assay is very sensitive to the presence of Bursicon in the hemolymph, because a 30-fold dilution of the hemolymph can also produce tanning (Fraenkel and Hsiao, 1963).
Whereas hemolymph from control Canton S (CS) rescued cuticle tanning when transferred into neck ligated CS flies, hemolymph from dLmo-overexpressing flies failed to do so. In the reverse experiment, using neck-ligated dLmo-overexpressing flies, we found that tanning of the cuticle was rescued by hemolymph from control CS flies (Fig. 3). Both phenomena were highly robust, in that all the flies tested showed consistent results. Together, these data indicate that hemolymph from dLmo-overexpressing flies has inadequate levels of Bursicon but that these files are able to respond to the Bursicon in CS hemolymph. The lack of Bursicon activity in the hemolymph could be attributable to underproduction of one of the subunits. Also, although we could detect burs and pburs transcripts in the appropriate neurons at the third-instar larva stage, we cannot rule out a subsequent loss of expression in CCAP neurons during the pupal stages. However, the production of Bursicon could be normal, as our in situ experiments suggest, but there could be a defect in release, resulting in inadequate levels of the neurohormone in the hemolymph at the time of eclosion.
Rescue of wing expansion in neck-ligated flies appears to be more complicated, because it is thought to be regulated by an additional inhibitory signal from the head, which appears to act in a highly restricted time window, from 40 min before eclosion to 10 min after eclosion. This inhibition apparently persists in neck-ligated flies, suggesting that the presence of an intact head is required so that the inhibitory control is removed and Bursicon can mediate wing expansion (Baker et al., 1999; Baker and Truman, 2002).
Therefore, we attempted to rescue wing expansion in dLmo-overexpressing intact flies (i.e., without neck ligation). We found that, in CS, hemolymph was indeed capable of rescuing wing expansion in these flies. Eight of 10 flies showed complete or partial rescue. That wing expansion in the remaining flies could not be rescued suggests a higher threshold for Bursicon for this function than for tanning. Moreover, injection of 8-bromo-cAMP partially mimicked this effect (Fig. 3), consistent with previous reports of Bursicon action via the cAMP pathway downstream of its receptor DLGR2 (Luo et al., 2005; Mendive et al., 2005).
In summary, the dLmo-overexpressing flies appear similar to flies that have inadequate levels of secreted Bursicon, but other aspects such as the response to normal hemolymph or 8-bromo-cAMP are normal.
Discussion
In this study, we used a selective overexpression strategy to identify a novel function of Chip in a set of neurons that control a stereotyped behavioral program. Chip is a widely expressed multidomain cofactor molecule that interacts with many transcription factors. It can function both as a transcriptional coactivator and a bridging factor between proteins that bind to distal enhancers and the core transcription machinery (Matthews and Visvader, 2003). Identifying specific functions of such a protein is confounded by the superposition of a multitude of effects. The mutations in Chip cause early lethality precluding the examination of later functions. Because the molecule exists as a part of multiple complexes, even simultaneously within the same cell (Ramain et al., 2000), altering the level of one class of interactors can potentially disrupt several functions. We have elucidated a highly specific role of Chip in a particular class of neurons in Drosophila and implicated a known LIM-HD partner of Chip, Islet, in this function.
We propose that the defect is attributable to a failure in the release rather than in the production or the responsiveness to the neurohormone Bursicon. How might Bursicon release be controlled as a result of Chip function in development? The hemolymph transfer experiments provide a unique insight into this puzzle. The literature describes a model wherein posteclosion wing expansion requires a combination of a neural signal from the brain as well as Bursicon release. Our results extend our understanding of how this interplay of activity and secreted factors is set up in development. It appears that, several days before eclosion, Chip is able to regulate an as-yet-unidentified event in the CCAP neurons, such that the hemolymph contains adequate levels of Bursicon after eclosion. The CCAP-expressing neurons are divided into at least two interacting subpopulations, only one of which secretes Bursicon. The other subpopulation does not secrete Bursicon but is implicated in regulating its release (Luan et al., 2006a,b). Chip may therefore mediate the formation of proper connectivity among CCAP neurons, which eventually ensures timely Bursicon release several days later. Supporting this scenario, Chip has been reported to regulate axon pathfinding and proper innervation of targets in other systems (van Meyel et al., 2000; Komiyama and Luo, 2007). Our data are suggestive of Chip requirement in the early period of puparium formation, which fits well with a report that CCAP neurons undergo extensive remodeling in this period of metamorphosis (Zhao et al., 2008). The importance of this connectivity is underscored by the identification of several other genes in a gain-of-function screen, which displayed a simultaneous disruption of both posteclosion wing expansion and the pattern of CCAP neuron innervation (Zhao et al., 2008). Therefore, our findings motivate an examination of Chip function in regulating the connectivity of CCAP neurons, a role that directly links this key aspect of neuronal development with the control of posteclosion behavior in Drosophila.
Footnotes
This work was supported by Wellcome Trust Senior Fellowship 056684/Z/99/Z and a Swarnajayanti Fellowship (Department of Science and Technology, Government of India) (S.T.), and a Kanwal Rekhi Career Development Award (Tata Institute of Fundamental Research Endowment Fund) (P.H.). We thank M. Panicker for support and discussions, and anonymous reviewers for very helpful criticisms.
References
- Allan DW, Park D, St Pierre SE, Taghert PH, Thor S. Regulators acting in combinatorial codes also act independently in single differentiating neurons. Neuron. 2005;45:689–700. doi: 10.1016/j.neuron.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Baker JD, Truman JW. Mutations in the Drosophila glycoprotein hormone receptor, rickets, eliminate neuropeptide-induced tanning and selectively block a stereotyped behavioral program. J Exp Biol. 2002;205:2555–2565. doi: 10.1242/jeb.205.17.2555. [DOI] [PubMed] [Google Scholar]
- Baker JD, McNabb SL, Truman JW. The hormonal coordination of behavior and physiology at adult ecdysis in Drosophila melanogaster. J Exp Biol. 1999;202:3037–3048. doi: 10.1242/jeb.202.21.3037. [DOI] [PubMed] [Google Scholar]
- Baumgardt M, Miguel-Aliaga I, Karlsson D, Ekman H, Thor S. Specification of neuronal identities by feedforward combinatorial coding. PLoS Biol. 2007;5:e37. doi: 10.1371/journal.pbio.0050037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste RJ, Thor S, Thomas JB, Taghert PH. Cell type-specific regulation of the Drosophila FMRF-NH2 neuropeptide gene by Apterous, a LIM homeodomain transcription factor. Development. 1998;125:4757–4765. doi: 10.1242/dev.125.23.4757. [DOI] [PubMed] [Google Scholar]
- Blair SS, Brower DL, Thomas JB, Zavortink M. The role of apterous in the control of dorsoventral compartmentalization and PS integrin gene expression in the developing wing of Drosophila. Development. 1994;120:1805–1815. doi: 10.1242/dev.120.7.1805. [DOI] [PubMed] [Google Scholar]
- Cottrell CB. The imaginal ecdysis of blowflies. The control of cuticular hardening and darkening. J Exp Biol. 1962;39:395–411. [Google Scholar]
- Dewey EM, McNabb SL, Ewer J, Kuo GR, Takanishi CL, Truman JW, Honegger HW. Identification of the gene encoding bursicon, an insect neuropeptide responsible for cuticle sclerotization and wing spreading. Curr Biol. 2004;14:1208–1213. doi: 10.1016/j.cub.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Fraenkel G, Hsiao C. Hormonal and nervous control of tanning in the fly. Science. 1962;138:27–29. doi: 10.1126/science.138.3536.27. [DOI] [PubMed] [Google Scholar]
- Fraenkel G, Hsiao C. Tanning in the adult fly: a new function of neurosecretion in the brain. Science. 1963;141:1057–1058. doi: 10.1126/science.141.3585.1057. [DOI] [PubMed] [Google Scholar]
- Gill GN. Decoding the LIM development code. Trans Am Clin Climatol Assoc. 2003;114:179–189. [PMC free article] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci U S A. 2004;101:8751–8755. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger HW, Market D, Pierce LA, Dewey EM, Kostron B, Wilson M, Choi D, Klukas KA, Mesce KA. Cellular localization of bursicon using antisera against partial peptide sequences of this insect cuticle-sclerotizing neurohormone. J Comp Neurol. 2002;452:163–177. doi: 10.1002/cne.10357. [DOI] [PubMed] [Google Scholar]
- Jhaveri D, Sen A, Rodrigues V. Mechanisms underlying olfactory neuronal connectivity in Drosophila—the atonal lineage organizes the periphery while sensory neurons and glia pattern the olfactory lobe. Dev Biol. 2000;226:73–87. doi: 10.1006/dbio.2000.9855. [DOI] [PubMed] [Google Scholar]
- Kimura KI, Truman JW. Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J Neurosci. 1990;10:403–411. doi: 10.1523/JNEUROSCI.10-02-00403.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolterud A, Alenius M, Carlsson L, Bohm S. The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development. 2004;131:5319–5326. doi: 10.1242/dev.01416. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Luo L. Intrinsic control of precise dendritic targeting by an ensemble of transcription factors. Curr Biol. 2007;17:278–285. doi: 10.1016/j.cub.2006.11.067. [DOI] [PubMed] [Google Scholar]
- Luan H, Lemon WC, Peabody NC, Pohl JB, Zelensky PK, Wang D, Nitabach MN, Holmes TC, White BH. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J Neurosci. 2006a;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006b;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren SE, Callahan CA, Thor S, Thomas JB. Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development. 1995;121:1769–1773. doi: 10.1242/dev.121.6.1769. [DOI] [PubMed] [Google Scholar]
- Luo CW, Dewey EM, Sudo S, Ewer J, Hsu SY, Honegger HW, Hsueh AJ. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein receptor LGR2. Proc Natl Acad Sci U S A. 2005;102:2820–2825. doi: 10.1073/pnas.0409916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, Li Y, Flanagan LA, Tole S, Monuki ES. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Mendive FM, Van Loy T, Claeysen S, Poels J, Williamson M, Hauser F, Grimmelikhuijzen CJ, Vassart G, Vanden Broeck J. Drosophila molting neurohormone Bursicon is a heterodimer and the natural agonist of the orphan receptor DLGR2. FEBS Lett. 2005;579:2171–2176. doi: 10.1016/j.febslet.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Milán M, Cohen SM. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLmo. Mol Cell. 1999;4:267–273. doi: 10.1016/s1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- Milán M, Diaz-Benjumea FJ, Cohen SM. Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev. 1998;12:2912–2920. doi: 10.1101/gad.12.18.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Baylies MK, Dorsett D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramain P, Khechumian R, Khechumian K, Arbogast N, Ackermann C, Heitzler P. Interactions between chip and the achaete/scute-daughterless heterodimers are required for pannier-driven proneural patterning. Mol Cell. 2000;6:781–790. doi: 10.1016/s1097-2765(05)00079-1. [DOI] [PubMed] [Google Scholar]
- Remedios R, Subramanian L, Tole S. LIM genes parcellate the embryonic amygdala and regulate its development. J Neurosci. 2004;24:6986–6990. doi: 10.1523/JNEUROSCI.0001-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B, Hari P, Huilgol D, Tole S. Dual role for LIM-homeodomain gene Lhx2 in the formation of the lateral olfactory tract. J Neurosci. 2007;27:2290–2297. doi: 10.1523/JNEUROSCI.5571-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Koo SJ, Kania A, Lettieri K, Andrews S, Cox C, Jessell TM, Pfaff SL. A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron. 2004;41:337–350. doi: 10.1016/s0896-6273(04)00011-x. [DOI] [PubMed] [Google Scholar]
- Thor S, Thomas JB. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 1997;18:397–409. doi: 10.1016/s0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Tsai LT, Bainton RJ, Blau J, Heberlein U. Lmo mutants reveal a novel role for circadian pacemaker neurons in cocaine-induced behaviors. PLoS Biol. 2004;2:e408. doi: 10.1371/journal.pbio.0020408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meyel DJ, O'Keefe DD, Jurata LW, Thor S, Gill GN, Thomas JB. Chip and Apterous physically interact to form a functional complex during Drosophila development. Mol Cell. 1999;4:259–265. doi: 10.1016/s1097-2765(00)80373-1. [DOI] [PubMed] [Google Scholar]
- van Meyel DJ, O'Keefe DD, Thor S, Jurata LW, Gill GN, Thomas JB. Chip is an essential cofactor for apterous in the regulation of axon guidance in Drosophila. Development. 2000;127:1823–1831. doi: 10.1242/dev.127.9.1823. [DOI] [PubMed] [Google Scholar]
- Weihe U, Milán M, Cohen SM. Regulation of Apterous activity in Drosophila wing development. Development. 2001;128:4615–4622. doi: 10.1242/dev.128.22.4615. [DOI] [PubMed] [Google Scholar]
- Zeng C, Justice NJ, Abdelilah S, Chan YM, Jan LY, Jan YN. The Drosophila LIM-only gene, dLMO, is mutated in Beadex alleles and might represent an evolutionarily conserved function in appendage development. Proc Natl Acad Sci U S A. 1998;95:10637–10642. doi: 10.1073/pnas.95.18.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Gu T, Rice HC, McAdams KL, Roark KM, Lawson K, Gauthier SA, Reagan KL, Hewes RS. A Drosophila gain-of-function screen for candidate genes involved in steroid-dependent neuroendocrine cell remodeling. Genetics. 2008;178:883–901. doi: 10.1534/genetics.107.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]