Abstract
The gene encoding DM20 emerged in cartilaginous fish, descending from a bilaterian ancestor of the M6 proteolipid gene family. Its proteolipid protein (PLP) isoform appeared in amphibians, contains an additional 35 amino acids, and, in the mammalian CNS, is the dominant myelin protein in which it confers an essential neuroprotective function. During development, the DM20 isoform is prominent in a number of tissues, and plp/DM20 transcripts are detected in multiple progenitor populations, including those that continue to express plp/DM20 as they differentiate into myelinating oligodendrocytes. The locus also encodes isoforms with extended leader sequences that accumulate in the cell bodies of several types of neurons. Here, to locate and characterize regulatory sequences controlling the complex plp/DM20 transcription program, putative regulatory sequences, suggested by interspecies conservation, were ligated individually to a minimally promoted eGFPlacZ reporter gene. These constructs were inserted in single copy at a common site adjacent to the hypoxanthine-guanine phosphoribosyltransferase locus in embryonic stem cells and their in vivo expression programs were compared in transgenic mice. Most expressed developmental and cell-specific subprograms accommodated within the known expression phenotype of the endogenous plp/DM20 locus, thus defining multiple components of the combinatorial mechanism controlling its normal temporal and cell-specific program. Along with previously characterized nervous system enhancers, those described here should help expose the content and configuration of elements that are operational in multiple glial and neuronal lineages. The transgenic lines derived here also provide effective markers for multiple stages of glial and neuronal lineage progression.
Keywords: plp/DM20, enhancers, neural crest, myelin, oligodendrocyte, Schwann cell
Introduction
Myelin is elaborated in the CNS and PNS by oligodendrocytes and Schwann cells, respectively. Proteolipid protein (PLP), a tetraspan integral membrane protein, is upregulated as myelin elaboration commences to become the dominant protein in mature CNS myelin of mammals (Verity and Campagnoni, 1988; Kanfer et al., 1989; LeVine et al., 1990; Yoshida and Colman, 1996). PLP contributes to both myelin compaction and interlamellar spacing and also confers a neuroprotective role; impaired axonal transport and progressive axonal degeneration are observed in myelinated fibers in which PLP/DM20 is either lacking or experimentally substituted with P0 (Yin et al., 2006; H. B. Werner et al., 2007)
The plp/DM20 locus also encodes the more ancient DM20 isoform that lacks a 35 amino acid cytoplasmic loop. DM20 transcripts are detected in the blastocyst (Skoff et al., 2004), seminiferous tubules, myocardial cells, thymus, and spleen (Campagnoni et al., 1992; Pribyl et al., 1996). In the mouse nervous system, DM20 is expressed in neural progenitors as early as embryonic day 9.5 (E9.5) and in oligodendrocyte progenitors by E14.5 (Timsit et al., 1992, 1995; Yu et al., 1994; Fanarraga et al., 1996; Dickinson et al., 1997; Spassky et al., 1998; Gudz et al., 2006). DM20 also accumulates in multiple glial cell types, including olfactory bulb ensheathing cells (OECs), spinal and autonomic ganglia satellite cells, Schwann cells, and enteric glia (Griffiths et al., 1989, 1995; Puckett et al., 1987; Dickinson et al., 1997). Additionally, a short intron 1 exon can be spliced into both the plp and DM20 mRNAs generating soma-restricted (sr) proteins that accumulate in multiple neuronal populations (Bongarzone et al., 1999; Jacobs et al., 2004).
plp/DM20 coding mutations along with deletions or duplications that affect PLP accumulation cause a spectrum of diseases, including Pelizaeus–Merzbacher disease (PMD) and spastic paraplegia type 2 (SPG2) (Willard and Riordan, 1985; Saugier-Veber et al., 1994; Sistermans et al., 1998; Woodward et al., 1998; Mimault et al., 1999). Consequently, plp/DM20 regulation has been investigated extensively, and long sequence domains capable of directing expression to oligodendrocytes, Schwann cells, and neuronal precursors have been identified (Nadon et al., 1994; Readhead et al., 1994; Fuss et al., 2000; Li et al., 2002; Mallon et al., 2002; Delaunay et al., 2008). However, the location and function of plp/DM20 enhancers remain essentially unknown. Here we interrogated the in vivo regulatory potential of multiple plp/DM20 sequences containing evolutionarily conserved non-protein-coding modules as well as the anti-silencer enhancer (ASE) sequence shown previously to have in vitro regulatory activity (Wight et al., 1997; Meng et al., 2005).
Constructs bearing such putative regulatory sequences were ligated to a common reporter gene and docked in single copy, and common orientation 5′ of hypoxanthine-guanine phosphoribosyltransferase (HPRT) (Bronson et al., 1996) and transgenic lines were derived. All transgenes revealed unique expression programs initiating at different stages of development and in different populations of glia and/or neurons. This investigation demonstrates that control of plp/DM20 transcription is realized through a complex combinatorial mechanism in which multiple enhancers, including some with partially redundant targeting activities, serve different lineages and developmental stages.
Materials and Methods
Interspecies sequence comparisons.
A University of California, Santa Cruz (UCSC) browser (available at http://genome.ucsc.edu/) reveals multiple regions of high conservation around PLP (Fig. 1). Design of the PLP constructs used in this study was based on the Mouse May 2004 (mm5) assembly (Mouse Genome Sequencing Consortium, 2002). The genomic location of each construct is shown in supplemental Table 1 (available at www.jneurosci.org as supplemental material) and is based on the updated Mouse July 2007 assembly.
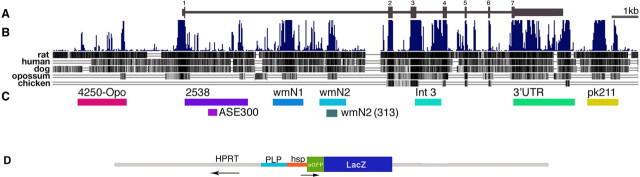
Figure 1.
A, Organization of the plp/DM20 locus; exons are numbered. B, Vertical lines indicate relative sequence conservation across multiple vertebrate species as provided by the UCSC browser. Alignment against the mouse sequence is shown. Sequence conservation of mouse versus rat, human, dog, opossum, and chicken are shown in separate tracks below. C, The plp/DM20 conserved non-protein-coding sequences that were introduced into reporter constructs, along with their designation, are underlined. D, These were cloned in their normal 5′–3′ orientation in front of a minimal hsp promoter driving an eGFPlacZ reporter, and the resulting constructs were inserted upstream and in opposite orientation to HPRT.
Isolation of mouse plp/DM20 genomic DNA.
Mouse bacterial artificial chromosome (BAC) clones RP23-344F8 and RP23-290F7, containing the PLP locus, were obtained from the BACPAC Resource (Oakland Research Institute, Oakland, CA). BAC DNA was prepared from 1L bacterial cultures in Luria broth containing12.5 mg/ml chloramphenicol. DNA was isolated using a midiprep kit following the instructions of the manufacturer (Qiagen).
Coordinates of plp/DM20 sequences introduced into reporter constructs along with PCR primers are presented in supplemental Table 1 (available at www.jneurosci.org as supplemental material). Sequences were amplified using the BD Advantage Polymerase mix (BD Biosciences) in the following touchdown program: initial denaturation for 3 min at 96°C, 44 cycles of amplification with denaturation at 96°C for 45 s, annealing temperatures beginning at 70°C, decreasing by 0.3°C per cycle, for 45 s, extension at 72°C for periods ranging from 45 s to 2.5 min for the longest sequence, and final extension at 72° for 7 min.
Generation of reporter constructs.
PCR products were digested and subcloned into a multiple cloning site upstream of the heat shock protein (hsp) promoter in an hspeGFPLacZ Entry vector, pHEL, modified from Denarier et al. (2005). BD InFusion cloning was used according to the instructions of the manufacturer. Restriction sites for insert digestion were incorporated into the primers listed in supplemental Table 1 (available at www.jneurosci.org as supplemental material). These reporter constructs were cloned using the LR clonase reaction kit (Invitrogen) into a “Gateway” Destination vector bearing HPRT homology arms for recombination in embryonic stem (ES) cells at the HPRT locus. The final destination vectors were amplified, sequenced across the insert, and linearized by Age1 or SalI. The ES cells, described by Farhadi et al. (2003), bear a deletion of the promoter and exon 1 of HPRT, and recombination with the homology arms in the destination vector restores the gene and allows them to survive under hypozanthine, aminopterin, thymidine (HAT) selection (Bronson et al., 1996).
Sequencing.
Positive clones were sequenced at the McGill University and Genome Quebec Innovation Center (Montreal, Quebec, Canada) using the forward primer 5′-CGCTTGTCTCTGGATGGAAC-3′ located in the hsp promoter and the reverse primer 5′-AGCCTGGGCAACAGAGAAATATC-3′ located in the HPRT homology arm. Sequences were analyzed using MacVector 7.2.
Histochemistry, fluorescence microscopy, and immunocytochemistry.
β-Galactosidase histochemistry was performed as described previously (Farhadi et al., 2003). For fluorescence microscopy, fixation was performed with 4% paraformaldehyde for 1 h at 4°C, and tissues were observed as whole mounts. For immunocytochemistry, mature mice were perfused transcardially with 4% paraformaldehyde, and recovered tissues were immersion fixed overnight at 4°C. E14.5 fetuses were recovered and similarly immersion fixed. After cryoprotection in 30% sucrose for 48 h, tissues were embedded in OCT compound and frozen in precooled isopentane. Cryostat sections, 14 μm thick, were postfixed in ice-cold acetone for 20 min, rinsed in PBS, and incubated overnight at 37°C in 0.8 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining solution. The slides then were rinsed briefly in PBS, blocked with 0.3% H2O2 followed by 10% BSA in PBS for 20 min, and incubated overnight with mouse anti-NeuN (neuron-specific nuclear protein; 1:100; Millipore Bioscience Research Reagents), rabbit anti-NG2 (1:100; PharMingen), rat anti-PDGF receptor α (PDGFRα) (1:50; Millipore), or rabbit anti-GFAP (1:1000; DakoCytomation). Slides were rinsed in PBS, incubated for 1 h in biotinylated anti-rabbit, mouse, or rat Ig (1:200; GE Healthcare), and developed using DAB (0.5 mg/ml) (Sigma). As a negative control, primary antibodies were omitted. Digital images were captured using a Magnafire CCD camera (Optronics) or a Carl Zeiss AxioCam color camera and an Axiovert 100 microscope.
Sciatic nerve regeneration.
To induce peripheral nerve regeneration, adult mice were anesthetized, and, using watchmakers forceps, the right sciatic nerve was crushed (two times for 10 s with a 10 s pause between crushes) at the level of the sciatic notch. Seven weeks after crush, mice were killed by an anesthetic overdose, and their sciatic nerves were recovered for histochemical analysis.
Transgenic production/breeding programs.
Constructs were transfected into BPES11 or BPES5 (4250-opo) cells, and HAT-resistant clones were recovered. To derive chimeras, these were injected into C57BL/6 blastocysts and transplanted into CBA/B6F1 pseudopregnant females (Farhadi et al., 2003). Transgenic lines were established by breeding male chimeras with C57BL/6 females and maintained through subsequent backcrosses to C57BL/6 mice. Samples analyzed for expression phenotypes were obtained primarily from heterozygous females, in which ∼50% of the cells have inactivated the X chromosome bearing the reporter construct (H. C. Friedman and A. C. Peterson, unpublished data).
Results
Our search for plp/DM20 enhancers focused on six evolutionarily conserved non-protein-coding sequences (Fig. 1) (supplemental Table 1, available at www.jneurosci.org as supplemental material) and the previously characterized intron 1 ASE. These were ligated to a common hspeGFPlacZ reporter gene (Fig. 1), inserted in single copy 5′ of the HPRT locus, and subsequently evaluated for expression programs in transgenic lines. From the common HPRT insertion site, all reporter constructs expressed in the nervous system in apparently unique cell-specific programs. Three expressed in myelinating glia (Fig. 2A), and three expressed in various neuronal populations (Fig. 2B) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). The two ASE-containing constructs expressed in multiple neuronal populations (Fig. 2C) and in undefined spinal cord cells (supplemental Fig. 2, available at www.jneurosci.org as supplemental material).
Figure 2.
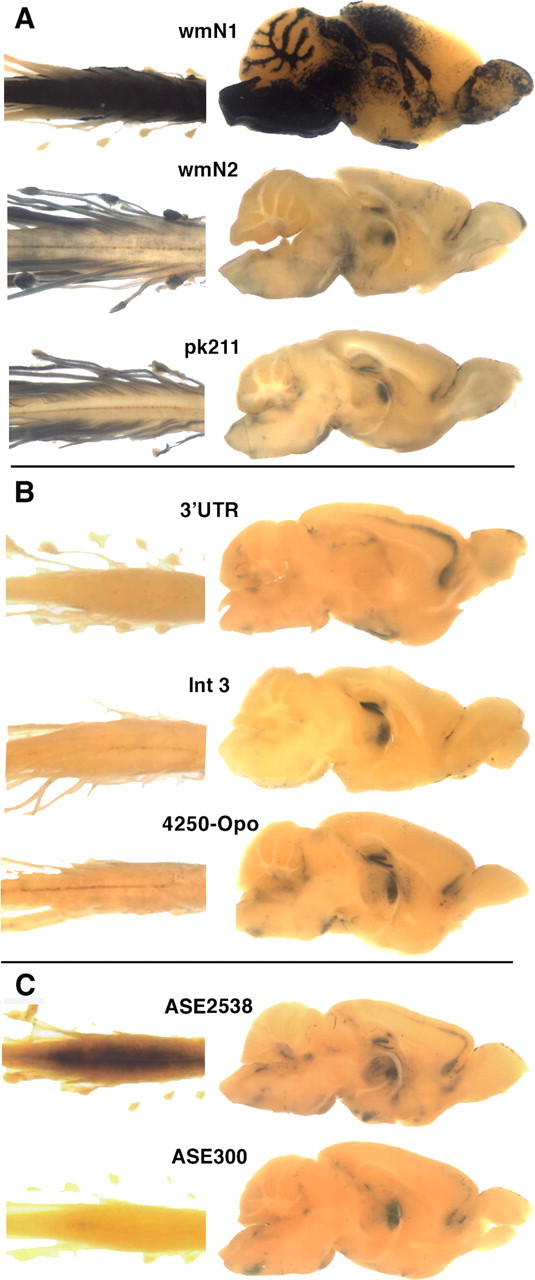
Histochemical detection of β-galactosidase activity in whole mounts of spinal cord/spinal root and dorsal root ganglia (left) and in midsagittal sections of brain (right). All samples were obtained from postweaning mice. Construct designations are above samples. A, Transgenic lines demonstrating expression in myelinating cells. B, Transgenic lines demonstrating expression in neuronal populations. C, Transgenic lines bearing constructs containing the ASE sequence.
The 1171 bp wmN1 sequence, located in intron 1, contains 584 bp of conserved sequence. In postweaning mice [examined up to postnatal day 120 (P120)], white matter tracts of the brain and spinal cord and the superficial layer of the olfactory bulb labeled intensely, indicating transgene expression in oligodendrocytes and OECs. To determine when the wmN1 enhancer activates during oligodendrocyte lineage progression, E15.5, E19, P0, P3, and P7 mice were examined. Apart from the emerging olfactory bulb, no labeling was observed in the developing CNS at E15.5. However, by E19 and at P0, a few labeled cells were present in the cervical spinal cord, and weak and diffuse labeling was present in most regions of future spinal cord white matter. By P3, the future spinal cord white matter extending from the brainstem (Fig. 3) to the lumbar level (data not shown) contained intensely labeled cells with the morphology of myelinating oligodendrocytes (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). In the brain, labeled cells emerged in the brainstem at P7, with remaining white matter tracks closely following. This overall developmental program coincides precisely with initiation of myelination in the mouse CNS, leading to the conclusion that activation of the wmN1 enhancer occurs at a late stage of oligodendrocyte maturation when multiple myelin genes upregulate (Foran and Peterson, 1992; Solly et al., 1996). Beyond the oligodendrocyte lineage, intense labeling was present in the superficial layer of the olfactory bulb at all ages examined, consistent with expression in OECs. Also, at P0, unknown cells within the deep cerebellar and neighboring tegmental nuclei labeled. Weak labeling also was encountered in fetal chondrocytes and diffusely distributed cells in DRGs. In contrast to the wmN2 and pk21l constructs (see below), wmN1 did not direct expression in peripheral nerves (Fig. 2A). In postnatal mice, heart and large blood vessels also labeled (blood vessel labeling shown in multiple figures), but, because this pattern of expression has been observed with multiple HPRT docked constructs, it likely represents a regulatory program contributed by enhancers associated with the HPRT docking site.
Figure 3.
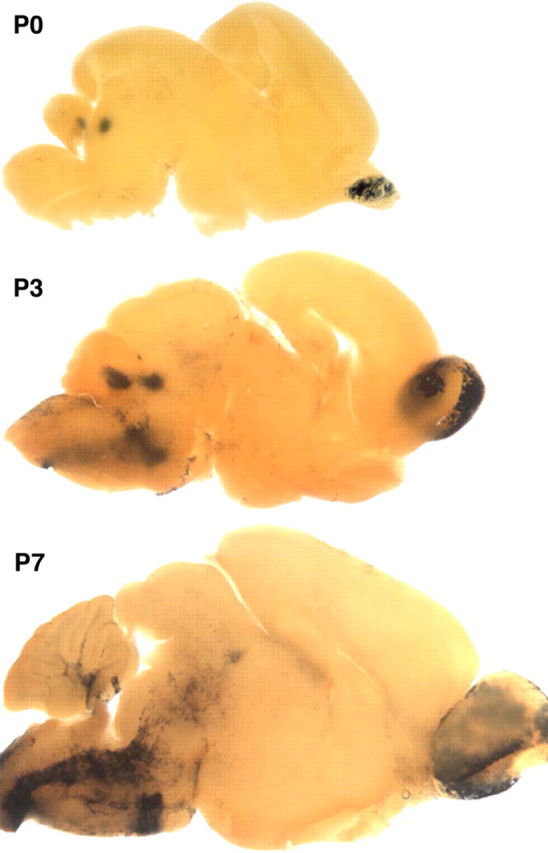
The wmN1 construct is expressed in myelinating oligodendrocytes and not in oligodendrocyte precursor cells. At P0, expression is limited to the superficial layer of the olfactory bulb and the deep cerebellar and tegmental nuclei. Future white matter does not contain expressing cells. Consistent with the known program of myelination, at P3, expressing cells are visible in brainstem, and, at P7, more rostral regions and future deep cerebellar white matter are labeled.
The 1034 bp wmN2 sequence, also derived from intron 1, contains a highly conserved 269 bp sequence. In mature mice, wmN2 was found to drive high-level expression in both peripheral nerves and DRGs (Fig. 2A). Consistent with expression in myelin-forming Schwann cells, teased sciatic nerve fibers revealed a strong fluorescent signal in the cytoplasmic compartments of myelin sheaths (data not shown). Also, in mice <2 months of age, after long substrate incubation times, white matter tracks in brain and spinal cord labeled weakly. Such CNS labeling was most obvious with P14 samples, barely detectable at P30, and extinguished by P60, suggesting that the wmN2 sequence supports low-level expression in the oligodendrocyte lineage limited to the period when CNS myelin is elaborated. In contrast, the glomerular layer of the olfactory bulb and DRGs labeled intensely at all postnatal ages examined (up to P120). Labeled cells also were encountered in mature mice in the rostral cortex, the hippocampus, diencephalon, and thalamus.
Because wmN2 demonstrated robust activity in mature Schwann cells, it was of interest to characterize its role during earlier development. Transgene-bearing fetuses were examined at E8.5, E9.5 E12.5, E14.5, and E16.5 (Fig. 4). At E8.5, only rare labeled cells were detected, and these were within the mesencephalic folds of the developing CNS (data not shown). However, by E9.5, intense label was obvious in the prosencephalic neural crest, trigeminal placode, epibranchial placodes, otic placode, and in neural crest cells migrating to form DRGs. Less intense labeling was detected in sympathetic ganglia. Thus, wmN2 drives strong expression in migrating neural crest and in the earliest neural crest-derived structures. By E12.5, robust labeling was restricted to DRGs and peripheral nerves, although weak labeling was detectable in the ventral part of the cervical neural tube. By E14.5, wmN2 activity had expanded along the length of the neural tube, giving rise to intensely labeled cells primarily limited to the ventral domain. Beyond the developing nervous system, multiple tissues, including the thymus, heart, aorta, gut, tongue, lower jaw, Meckel's cartilage, and the submandibular gland, express DM20 (Nadon et al., 1997). With the apparent exception of the thymus, these developing organs also labeled in fetuses at E14.5 and older.
Figure 4.
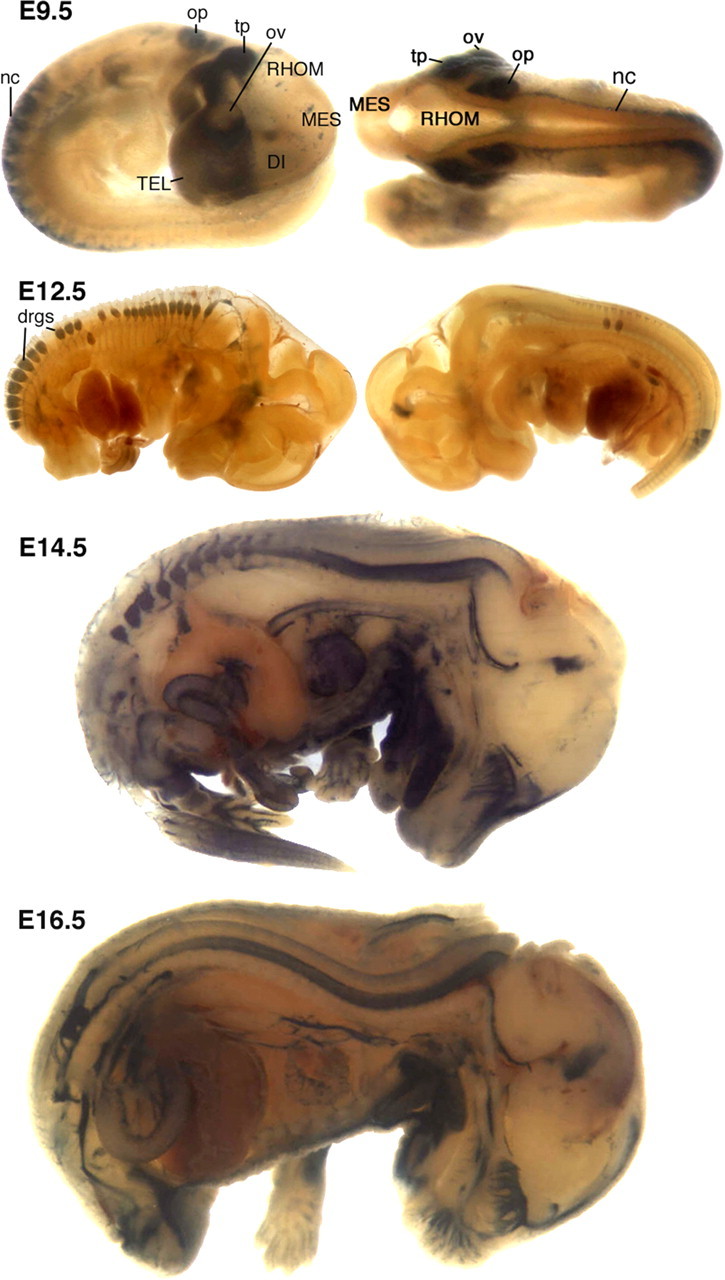
Expression of wmN2 in fetal mice. At E9.5, intense label was obvious in the prosencephalic neural crest, trigeminal placode (tp), epibranchial placodes, otic placode (op), and in neural crest (nc) cells migrating to form DRGs. Less-intense labeling was detected in sympathetic ganglia. At E12.5, DRG (drgs) labeled, as did spinal roots and the proximal segments of peripherally projecting mixed nerves. In the CNS, trace labeling was observed in the ventral neural tube and in the presumed ependymal layer of the mesencephalic vesicle. At E14.5, DRGs, spinal roots, peripheral nerves, and the surface of the olfactory bulb and cribiform plate are labeled. In addition, the tongue, part of the lower jaw, heart, dorsal aorta, and gut labeled. At E16.5, a similar expression pattern is observed, with the addition of labeled cells in the apparent midline of the dorsal neural tube and a reduction of label in heart. DI, Diencephalon; MES, mesencephalon; ov, optic vesicle; RHOM, rhombencephalon; TEL, telencephalon.
To reveal the distribution of cells expressing the wmN2-regulated construct within the neural tube at higher resolution, cross sections were examined. Labeled cells were primarily constrained within the ventrolateral neural tube, being most dense at the midline (Fig. 5A). Because oligodendrocyte progenitors are emerging within the E14.5 neural tube (Pringle and Richardson, 1993) and because such progenitors express DM20 (Timsit et al., 1995), we considered it probable that such transgene-expressing cells were oligodendrocyte progenitors. To confirm this possibility, cross sections were processed to reveal both β-galactosidase and the well established oligodendrocyte progenitor marker PDGFRα. Numerous transgene-expressing cells colabeled, confirming that oligodendrocyte progenitors are among some of the cells expressing the wmN2 transgene (Fig. 5B). We also show that transgene-expressing cells in the DRGs of such mice are not neurons but rather are typical DRG satellite cells with flat profiles in intimate contact with the perikarya of DRG neurons (Fig. 5C).
Figure 5.
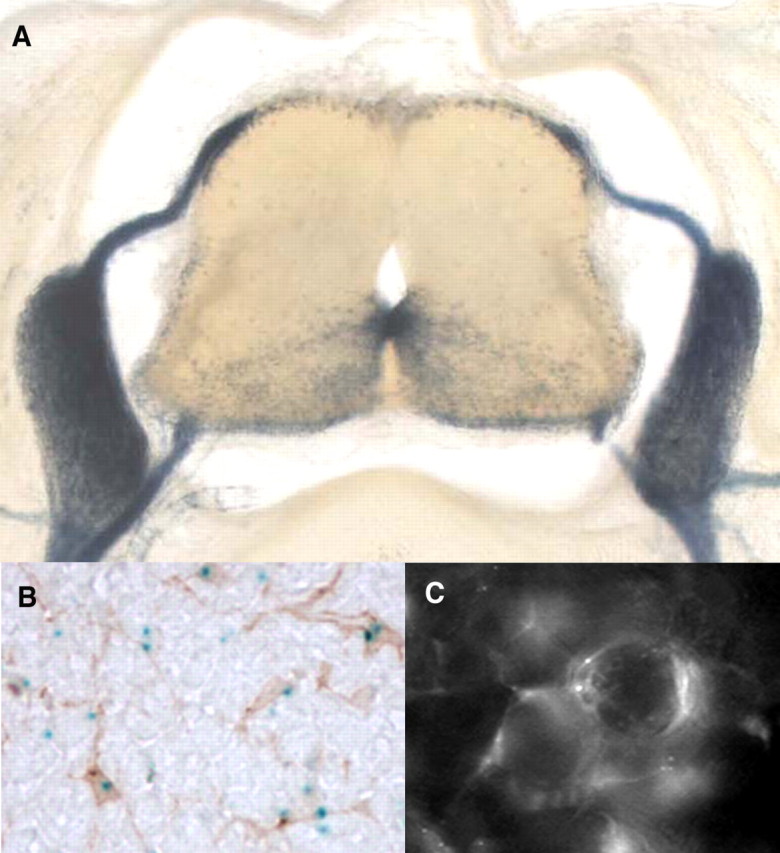
Characterization of cell populations expressing the wmN2 construct. A, Cross section, E14.5 embryo, demonstrating intense β-galactosidase histochemical labeling in spinal roots, DRGs, peripheral nerves, and the ventral portion of the neural tube. B, Neural tube cross section colabeled for β-galactosidase (histochemistry) and PDGFRα (immunohistochemistry) contains multiple colabeled cells, confirming that some transgene-expressing cells are within the oligodendrocyte lineage. C, Whole-mount preparation of a dorsal root ganglia from a 4-month-old wmN2 male chimera examined by fluorescence microscopy for eGFPlacZ reporter gene expression. Fluorescing cells, closely apposed to the large cell bodies of DRG neurons, are characteristic of DRG satellite cells.
The plp/DM20 sequences evaluated here for regulatory capacity were designed to include conserved non-protein-coding modules. However, in these initial constructs, regions of conservation were flanked by relatively long, nonconserved sequences. To determine whether the module of sequence conservation within the wmN2 sequence alone can confer regulatory function, its flanking sequences were removed. At E9.5, a reporter construct bearing the 269 bp conserved module within a 313 bp wmN2 subsequence expressed like the longer construct in neural crest and neural crest derivatives (supplemental Fig. 4, available at www.jneurosci.org as supplemental material).
A second enhancer with strong Schwann cell activity was mapped to 1209 bp of 3′ flanking sequence. Designated pk211, it contains 605 bp of conservation and drove expression in Schwann cells and DRG satellite cells in postnatal mice (P8 and P60 examined) (Fig. 2A). Like wmN2, spinal roots, DRGs, and mixed nerves labeled as early as E12.5, and expression was detected in the ventral neural tube at E14.5 and E16.5 but at low levels (data not shown). Additionally, in postweaning mice, spinal cord gray matter labeled weakly, and cells in the cerebellar nucleus, dentate gyrus, and a hippocampus-related nucleus labeled intensely (Fig. 2A).
To determine whether the Schwann cell enhancer activity emanating from wmN2 and pk211 extended to Schwann cells remyelinating regenerated peripheral nerves, reporter gene expression was examined in regenerated sciatic nerves 7 weeks after crush injury. In both lines, proximal and regenerated distal nerve segments labeled robustly with equivalent staining intensity, demonstrating that both enhancers are active in remyelinated nerves (data not shown).
Of the five remaining sequences characterized for function, the 3′ untranslated region (3′ UTR), Int3, and 4250-opo sequences were selected, as above, on the basis of sequence conservation. None drove expression in glia, but all three constructs were expressed in recognizable neuronal populations (Fig. 2B). The 3′ UTR sequence encompasses the entire 1623 bp 3′ UTR and extends 683 bp into conserved flanking sequences. In postnatal mice at P8 and P90, intense labeling was observed in a single layer of cortical pyramidal cells, the dentate gyrus, thalamic nuclei, hypothalamus, amygdala, and the cerebellar nucleus and granular cells. The 1053 bp Int3 sequence contains 579 bp of conservation located in intron 3. In mature mice, prominent labeling was restricted to the dentate gyrus and pyramidal layer of the hippocampus, whereas thalamic nuclei labeled weakly. The 4250-opo sequence extends 1938 bp from −4170 to −2232 from the transcription start site and contains 310 bp of conservation. Mice at P9, P14, P18, P60, P115, and P120 exhibited strong labeling throughout the spinal cord gray matter and in a manner similar to the 3′ UTR construct, in a single layer of cortical pyramidal cells. Also labeled were hippocampus, striatum, and numerous ventral neuronal nuclei.
In contrast to the above constructs selected on the basis of interspecies sequence conservation, the ASE2538 and ASE300 sequences were designed to include the well characterized 100 bp intron 1 ASE (Wight et al., 1997). ASE is not prominently conserved but is part of a SINE repeating element. Although it has been shown to confer enhancer activity in the context of reporter constructs transfected into oligodendroglial cell lines (Meng et al., 2005), constructs containing the ASE2538 and ASE300 sequences did not express in the oligodendrocytes of the transgenic mice derived here. Rather, both ASE2538 and ASE300 conferred postnatal expression to a subpopulation of spinal cord cells concentrated in the lumbar enlargement near the central canal in ventrolateral gray matter with mice bearing the longer sequence staining more intensely. In addition, both expressed in presumed neuronal populations in frontal cortex, hippocampus, deep cerebellum, and brainstem (supplemental Fig. 2, available at www.jneurosci.org as supplemental material).
Discussion
In this investigation, we located and partially characterized the function of several plp/DM20-associated enhancers. In five cases, they conferred expression subprograms accommodated within the known developmental and cell-specific accumulation programs of PLP, DM20, srPLP, and srDM20. Expression was conferred to myelinating oligodendrocytes and OECs by wmN1 and to Schwann cells and their progenitors, satellite, and OECs by wmN2 and pk211. wmN2 also conferred expression to the neural crest and its derivatives, oligodendrocyte progenitors, and oligodendrocytes actively elaborating myelin sheaths. Variously restricted neuronal subpopulation activity was conferred by the 3′ UTR, 4250-opo, int3, and ASE2583/ASE300. The ASE2583/ASE300 sequences also conferred expression in cells of unknown lineage in the lumbar spinal cord.
In common with most functional investigations based on reporter constructs, the plp/DM20 sequences investigated here were isolated from possible negative and/or positive influences emerging from the integrated plp/DM20 regulatory machinery. In such isolation, they may confer regulatory activities that could be constrained or enhanced in the context of the endogenous locus. Although the transgenesis strategy used here docks the reporter gene in single copy at a common insertion site and thus overcomes the variability associated with randomly inserted transgenes, the insertion site is adjacent to the ubiquitously expressed HPRT locus. We have observed expression in heart, large blood vessels, and several neuronal populations, including the hippocampus, with numerous HPRT docked constructs, including an hspeGFPlacZ construct to which no additional regulatory sequences were added (Denarier et al., 2005). Notably, when this same hspeGFPlacZ construct carries with it robust enhancers such as wmN1, wmN2, and pk211, these HPRT-directed expression programs are attenuated or extinguished (supplemental Fig. 5, available at www.jneurosci.org as supplemental material). However, neuronal populations reported to accumulate srPLP and srDM20 also expressed the hspeGFPlacZ construct alone, leaving the significance of plp/DM20 regulated reporter gene expression in such cells open to interpretation. Nonetheless, the constructs investigated here each expressed in unique constellations of cell populations, suggesting that significant regulatory activity was conferred through the majority, if not all, of the plp/DM20 sequences evaluated in this investigation.
In support of the above conclusion, the expression programs reported for several randomly integrated plp/DM20 transgenes fit well with the present findings. Specifically, the strong oligodendrocyte and Schwann cell enhancers identified within intron 1, wmN1 and wmN2, respectively, are contained within randomly inserted transgenes that appropriately reflect their regulatory activity (Spassky et al., 1998; Fuss et al., 2000). The proximal promoter sequence extending to −2.5 kb has no independent targeting ability (Li et al., 2002), but addition of the wmN1 and wmN2 containing intron 1 sequence leads to strong transgene expression in both oligodendrocytes and Schwann cells (Mallon et al., 2002). A Cre-containing transgene regulated by plp/DM20 sequences extending 5′ to −3.74 kb and the same intron 1 sequence demonstrated a similar expression program (Delaunay et al., 2008).
Sequence conservation is a common theme of regulatory system evolution (Pennacchio et al., 2006), and all conserved plp/DM20 modules examined here appear to confer regulatory activity. For the wmN2 sequence, we show that the complex expression program observed at midfetal development (Mallon et al., 2002) is conferred by activity emerging from the conserved 269 bp module alone. Nonetheless, significant regulatory sequences may remain unrecognized. Notably, regulatory elements with capacity to bind a common transcription factor are not always conserved at the sequence level (Odom et al., 2007; ENCODE Project Consortium et al., 2007), and, thus, regions without obvious conservation may confer additional or redundant plp/DM20 expression phenotypes. Also, several prominently conserved modules located in more distal plp/DM20 flanking regions remain to be investigated.
Previous investigations of plp/DM20 transcriptional regulation identified the 100 bp, intron 1 ASE. ASE contains several AP-1 like binding sites (Dobretsova et al., 2004), differentially binds nuclear proteins from diverse cell types, and functions in reporter constructs with both the plp/DM20 proximal promoter, and, in a position-dependent manner, with a thymidine kinase (TK) promoter when transfected into a plp/DM20-expressing oligodendrocyte cell line (Meng et al., 2005). In contrast, neither the ASE2538 nor ASE300 constructs expressed in oligodendrocytes, Schwann cells, satellite cells, or OECs in transgenic mice. Rather, both expressed in unknown cells in the spinal cord of postnatal mice and in a few, likely neuronal, populations in the brain. This discrepancy may be related to the different promoter used in the present constructs or some other incompatibility with the reporter gene or the HPRT docking site. Alternatively, ASE may confer its in vivo regulatory activity elsewhere than in the major glial lineages.
Our observations support the view that plp/DM20 is regulated by a complex combinatorial mechanism involving multiple enhancers and constellations of transcription factors. Moreover, the factor repertoire appears to change as development proceeds, e.g., wmN2 demonstrates activity in oligodendrocyte progenitors and myelin elaborating oligodendrocytes but downregulates in mature oligodendrocytes. Similar stage-specific activities were observed for multiple derivatives of the SCE1 enhancer of the myelin basic protein locus (Denarier et al., 2005). This may accommodate the disparity in regulatory output detected using in vivo and in vitro preparations. Specifically, a −2.4 kb to exon 2 plp/DM20 sequence conferred oligodendrocyte expression in multiple transgenic lines. This was subdivided, incorporated into TK-promoted luciferase reporters, and transfected into NIH/3T3 fibroblasts and the plp/DM20-expressing oligodendrocyte cell line N20.1 (Wight et al., 1997). Although one such subsequence, corresponding to the ASE2538 construct, markedly enhanced expression, it did so in both cell types. Most importantly, the regions of conservation within the wmN1 and wmN2 sequences were included intact among these transfected constructs, but they did not upregulate expression. This striking difference between the in vivo and in vitro output of these enhancer-containing constructs suggests, first, that the N20.1 cell line drives transcription of plp/DM20 using a unique factor repertoire, and, second, that exposing additional features of the plp/DM20 regulatory mechanism may require experimental access, either in vivo or in vitro, to differentiating oligodendrocytes.
An additional developmental change in plp/DM20 regulation is observed in the ratio of PLP and DM20 isoform accumulation. Oligodendrocyte precursors preferentially accumulate DM20, but, as myelin elaboration initiates, PLP becomes prominent. In vitro, this isoform switch appears to be mediated by splicing enhancers (Wang et al., 2007). The choice of splice and transcription initiation sites also is associated with alternate promoter usage (Roberts et al., 1998; Vouyiouklis et al., 2000; Kornblihtt, 2005, 2006). plp/DM20 has two transcription initiation sites spaced 30 bp apart (Milner et al., 1985; Baumgartner et al., 1999), and these are used selectively in different tissues, over the course of development and in response to injury (Kamholz et al., 1992; Scherer et al., 1992). wmN1, a particularly strong enhancer in mature oligodendrocytes, activates at the initiation of CNS myelin formation, coincident with the switch in preferred isoform accumulation. Recruiting wmN1 at this maturation stage would accommodate the coincident increase in PLP accumulation. The additional coincidence of wmN1 activation with the transition in preferred splice site also could signal a direct relationship between these regulatory activities.
So far, high-mobility group (Sox8, Sox10), basic helix–loop–helix (Olig1, Olig2), and homeodomain (Nkx6.2, Nkx2.2) factors have been linked to myelin gene enhancer activity (Awatramani et al., 1997; Lu et al., 2000; Zhou et al., 2001; Stolt et al., 2004; Denarier et al., 2005; Ghislain and Charnay, 2006; Liu et al., 2007). As enhancers from the coordinately regulated myelin gene family emerge, characterization of their element content and configuration should reveal shared components and common features of the code controlling myelin gene transcription (Farhadi et al., 2003; Denarier et al., 2005; Dionne, 2006). Also, because wmN2 is active in neural crest, its derivatives, and in oligodendrocyte and Schwann cell progenitors, its further investigation may reveal transcription factors that participate in the transition to committed glial progenitors (Deneen et al., 2006). To that objective, several conserved enhancers from the Sox10 gene were recently located and shown to confer a developmental program sharing features with wmN2 (T. Werner et al., 2007). Thus, both Sox10 and plp/DM20 genes may engage common transcription factors including Sox10 itself.
Our findings with reporter constructs demonstrate that some plp/DM20 enhancers confer partially redundant regulatory functions potentially accounting for the low frequency at which mutations in transcription factor binding sites have been reported in patients with outright PMD or SPG2. Although multiple noncoding mutations have been associated with disease, the majority of these lead to disruptions in splicing (Hobson et al., 2000). Nonetheless, in PMD or SPG2 cases in which partial plp/DM20 duplications or deletions are encountered, knowledge of the enhancers located in this investigation may contribute to diagnosis and prognostic accuracy.
This investigation demonstrates that interspecies sequence conservation, combined with a controlled strategy of in vivo functional analysis, can be an effective means of locating and characterizing regulatory sequence. The transgenic mice derived for this investigation express a fusion gene reporter supporting sensitive histochemical and fluorescent detection systems, thus offering flexible markers applicable to multiple glial and neuronal lineages, in some cases extending to progenitor cells. Finally, because construct insertion is controlled, the regulatory programs described here should be transferable to any coding sequence regulated by the same enhancer/promoter combinations.
Footnotes
This work was supported by the Genome Quebec and Genome Canada “GRID” program and by an Inserm/FRSQ travel grant (C.G.-Z.). We gratefully acknowledge France Bourdeau, Tess Fernandez, Irene Tretjakoff, and Priscila Valera for excellent technical support and Salvador Martinez for helpful comments on this manuscript.
References
- Awatramani R, Scherer S, Grinspan J, Collarini E, Skoff R, O'Hagan D, Garbern J, Kamholz J. Evidence that the homeodomain protein Gtx is involved in the regulation of oligodendrocyte myelination. J Neurosci. 1997;17:6657–6668. doi: 10.1523/JNEUROSCI.17-17-06657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner BG, Deppe A, Rettenberger G, Leeb T, Hameister H, Brenig B. Molecular analysis of the porcine proteolipid protein (PLP) gene. Mamm Genome. 1999;10:895–899. doi: 10.1007/s003359901110. [DOI] [PubMed] [Google Scholar]
- Bongarzone ER, Campagnoni CW, Kampf K, Jacobs EC, Handley VW, Schonmann V, Campagnoni AT. Identification of a new exon in the myelin proteolipid protein gene encoding novel protein isoforms that are restricted to the somata of oligodendrocytes and neurons. J Neurosci. 1999;19:8349–8357. doi: 10.1523/JNEUROSCI.19-19-08349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci U S A. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnoni CW, Garbay B, Micevych P, Pribyl T, Kampf K, Handley VW, Campagnoni AT. DM20 mRNA splice product of the myelin proteolipid protein gene is expressed in the murine heart. J Neurosci Res. 1992;33:148–155. doi: 10.1002/jnr.490330119. [DOI] [PubMed] [Google Scholar]
- Delaunay D, Heydon K, Cumano A, Schwab MH, Thomas JL, Suter U, Nave KA, Zalc B, Spassky N. Early neuronal and glial fate restriction of embryonic neural stem cells. J Neurosci. 2008;28:2551–2562. doi: 10.1523/JNEUROSCI.5497-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denarier E, Forghani R, Farhadi HF, Dib S, Dionne N, Friedman HC, Lepage P, Hudson TJ, Drouin R, Peterson A. Functional organization of a Schwann cell enhancer. J Neurosci. 2005;25:11210–11217. doi: 10.1523/JNEUROSCI.2596-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Dickinson PJ, Griffiths IR, Barrie JM, Kyriakides E, Pollock GF, Barnett SC. Expression of the dm-20 isoform of the plp gene in olfactory nerve ensheathing cells: evidence from developmental studies. J Neurocytol. 1997;26:181–189. doi: 10.1023/a:1018584013739. [DOI] [PubMed] [Google Scholar]
- Dionne N. McGill University; 2006. Structure and function of Module 3, a conserved enhancer of the myelin basic protein gene. PhD thesis. [Google Scholar]
- Dobretsova A, Kokorina NA, Wight PA. Potentiation of myelin proteolipid protein (Plp) gene expression is mediated through AP-1-like binding sites. J Neurochem. 2004;90:1500–1510. doi: 10.1111/j.1471-4159.2004.02683.x. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanarraga ML, Dickinson PJ, Sommer I, Montague P, Kyriakides E, Griffiths IR. Evidence that some oligodendrocyte progenitors in the developing optic pathway express the plp gene. Glia. 1996;18:282–292. doi: 10.1002/(sici)1098-1136(199612)18:4<282::aid-glia3>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Farhadi HF, Lepage P, Forghani R, Friedman HC, Orfali W, Jasmin L, Miller W, Hudson TJ, Peterson AC. A combinatorial network of evolutionarily conserved myelin basic protein regulatory sequences confers distinct glial-specific phenotypes. J Neurosci. 2003;23:10214–10223. doi: 10.1523/JNEUROSCI.23-32-10214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foran DR, Peterson AC. Myelin acquisition in the central nervous system of the mouse revealed by an MBP-Lac Z transgene. J Neurosci. 1992;12:4890–4897. doi: 10.1523/JNEUROSCI.12-12-04890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss B, Mallon B, Phan T, Ohlemeyer C, Kirchhoff F, Nishiyama A, Macklin WB. Purification and analysis of in vivo-differentiated oligodendrocytes expressing the green fluorescent protein. Dev Biol. 2000;218:259–274. doi: 10.1006/dbio.1999.9574. [DOI] [PubMed] [Google Scholar]
- Ghislain J, Charnay P. Control of myelination in Schwann cells: a Krox20 cis-regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 2006;7:52–58. doi: 10.1038/sj.embor.7400573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths IR, Mitchell LS, McPhilemy K, Morrison S, Kyriakides E, Barrie JA. Expression of myelin protein genes in Schwann cells. J Neurocytol. 1989;18:345–352. doi: 10.1007/BF01190837. [DOI] [PubMed] [Google Scholar]
- Griffiths IR, Dickinson P, Montague P. Expression of the proteolipid protein gene in glial cells of the post-natal peripheral nervous system of rodents. Neuropathol Appl Neurobiol. 1995;21:97–110. doi: 10.1111/j.1365-2990.1995.tb01035.x. [DOI] [PubMed] [Google Scholar]
- Gudz T, Komuro H, Macklin WB. Glutamate stimulates oligodendrocyte progenitor migration mediated via an αv integrin/myelin proteolipid protein complex. J Neurosci. 2006;26:2458–2466. doi: 10.1523/JNEUROSCI.4054-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson GM, Davis AP, Stowell NC, Kolodny EH, Sistermans EA, de Coo IF, Funanage VL, Marks HG. Mutations in noncoding regions of the proteolipid protein gene in Pelizaeus-Merzbacher disease. Neurology. 2000;55:1089–1096. doi: 10.1212/wnl.55.8.1089. [DOI] [PubMed] [Google Scholar]
- Jacobs EC, Bongarzone ER, Campagnoni CW, Campagnoni AT. Embryonic expression of the soma-restricted products of the myelin proteolipid gene in motor neurons and muscle. Neurochem Res. 2004;29:997–1002. doi: 10.1023/b:nere.0000021244.38279.c4. [DOI] [PubMed] [Google Scholar]
- Kamholz J, Sessa M, Scherer S, Vogelbacker H, Mokuno K, Baron P, Wrabetz L, Shy M, Pleasure D. Structure and expression of proteolipid protein in the peripheral nervous system. J Neurosci Res. 1992;31:231–244. doi: 10.1002/jnr.490310204. [DOI] [PubMed] [Google Scholar]
- Kanfer J, Parenty M, Goujet-Zalc C, Monge M, Bernier L, Campagnoni AT, Dautigny A, Zalc B. Developmental expression of myelin proteolipid, basic protein, and 2′,3′-cyclic nucleotide 3′-phosphodiesterase transcripts in different rat brain regions. J Mol Neurosci. 1989;1:39–46. doi: 10.1007/BF02896855. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- LeVine SM, Wong D, Macklin WB. Developmental expression of proteolipid protein and DM20 mRNAs and proteins in the rat brain. Dev Neurosci. 1990;12:235–250. doi: 10.1159/000111853. [DOI] [PubMed] [Google Scholar]
- Li S, Moore CL, Dobretsova A, Wight PA. Myelin proteolipid protein (Plp) intron 1 DNA is required to temporally regulate Plp gene expression in the brain. J Neurochem. 2002;83:193–201. doi: 10.1046/j.1471-4159.2002.01142.x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, Qiu M. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302:683–693. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Mallon BS, Shick HE, Kidd GJ, Macklin WB. Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J Neurosci. 2002;22:876–885. doi: 10.1523/JNEUROSCI.22-03-00876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Zolova O, Kokorina NA, Dobretsova A, Wight PA. Characterization of an intronic enhancer that regulates myelin proteolipid protein (Plp) gene expression in oligodendrocytes. J Neurosci Res. 2005;82:346–356. doi: 10.1002/jnr.20640. [DOI] [PubMed] [Google Scholar]
- Milner RJ, Lai C, Nave KA, Lenoir D, Ogata J, Sutcliffe JG. Nucleotide sequences of two mRNAs for rat brain myelin proteolipid protein. Cell. 1985;42:931–939. doi: 10.1016/0092-8674(85)90289-2. [DOI] [PubMed] [Google Scholar]
- Mimault C, Giraud G, Courtois V, Cailloux F, Boire JY, Dastugue B, Boespflug-Tanguy O. Proteolipoprotein gene analysis in 82 patients with sporadic Pelizaeus-Merzbacher disease: duplications, the major cause of the disease, originate more frequently in male germ cells, but point mutations do not. The Clinical European Network on Brain Dysmyelinating Disease. Am J Hum Genet. 1999;65:360–369. doi: 10.1086/302483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Nadon NL, Arnheiter H, Hudson LD. A combination of PLP and DM20 transgenes promotes partial myelination in the jimpy mouse. J Neurochem. 1994;63:822–833. doi: 10.1046/j.1471-4159.1994.63030822.x. [DOI] [PubMed] [Google Scholar]
- Nadon NL, Miller S, Draeger K, Salvaggio M. Myelin proteolipid DM20: evidence for function independent of myelination. Int J Dev Neurosci. 1997;15:285–293. doi: 10.1016/s0736-5748(97)00009-9. [DOI] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pribyl TM, Campagnoni CW, Kampf K, Kashima T, Handley VW, McMahon J, Campagnoni AT. Expression of the myelin proteolipid protein gene in the human fetal thymus. J Neuroimmunol. 1996;67:125–130. doi: 10.1016/0165-5728(96)00058-6. [DOI] [PubMed] [Google Scholar]
- Pringle NP, Richardson WD. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development. 1993;117:525–533. doi: 10.1242/dev.117.2.525. [DOI] [PubMed] [Google Scholar]
- Puckett C, Hudson L, Ono K, Friedrich V, Benecke J, Dubois-Dalcq M, Lazzarini RA. Myelin-specific proteolipid protein is expressed in myelinating Schwann cells but is not incorporated into myelin sheaths. J Neurosci Res. 1987;18:511–518. doi: 10.1002/jnr.490180402. [DOI] [PubMed] [Google Scholar]
- Readhead C, Schneider A, Griffiths I, Nave KA. Premature arrest of myelin formation in transgenic mice with increased proteolipid protein gene dosage. Neuron. 1994;12:583–595. doi: 10.1016/0896-6273(94)90214-3. [DOI] [PubMed] [Google Scholar]
- Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugier-Veber P, Munnich A, Bonneau D, Rozet JM, Le Merrer M, Gil R, Boespflug-Tanguy O. X-linked spastic paraplegia and Pelizaeus-Merzbacher disease are allelic disorders at the proteolipid protein locus. Nat Genet. 1994;6:257–262. doi: 10.1038/ng0394-257. [DOI] [PubMed] [Google Scholar]
- Scherer SS, Vogelbacker HH, Kamholz J. Axons modulate the expression of proteolipid protein in the CNS. J Neurosci Res. 1992;32:138–148. doi: 10.1002/jnr.490320203. [DOI] [PubMed] [Google Scholar]
- Sistermans EA, de Coo RF, De Wijs IJ, Van Oost BA. Duplication of the proteolipid protein gene is the major cause of Pelizaeus-Merzbacher disease. Neurology. 1998;50:1749–1754. doi: 10.1212/wnl.50.6.1749. [DOI] [PubMed] [Google Scholar]
- Skoff RP, Bessert DA, Cerghet M, Franklin MJ, Rout UK, Nave KA, Carlock L, Ghandour MS, Armant DR. The myelin proteolipid protein gene modulates apoptosis in neural and non-neural tissues. Cell Death Differ. 2004;11:1247–1257. doi: 10.1038/sj.cdd.4401498. [DOI] [PubMed] [Google Scholar]
- Solly SK, Thomas JL, Monge M, Demerens C, Lubetzki C, Gardinier MV, Matthieu JM, Zalc B. Myelin/oligodendrocyte glycoprotein (MOG) expression is associated with myelin deposition. Glia. 1996;18:39–48. doi: 10.1002/(SICI)1098-1136(199609)18:1<39::AID-GLIA4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Spassky N, Goujet-Zalc C, Parmantier E, Olivier C, Martinez S, Ivanova A, Ikenaka K, Macklin W, Cerruti I, Zalc B, Thomas JL. Multiple restricted origin of oligodendrocytes. J Neurosci. 1998;18:8331–8343. doi: 10.1523/JNEUROSCI.18-20-08331.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- Timsit S, Martinez S, Allinquant B, Peyron F, Puelles L, Zalc B. Oligodendrocytes originate in a restricted zone of the embryonic ventral neural tube defined by DM-20 mRNA expression. J Neurosci. 1995;15:1012–1024. doi: 10.1523/JNEUROSCI.15-02-01012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit SG, Bally-Cuif L, Colman DR, Zalc B. DM-20 mRNA is expressed during the embryonic development of the nervous system of the mouse. J Neurochem. 1992;58:1172–1175. doi: 10.1111/j.1471-4159.1992.tb09378.x. [DOI] [PubMed] [Google Scholar]
- Verity AN, Campagnoni AT. Regional expression of myelin protein genes in the developing mouse brain: in situ hybridization studies. J Neurosci Res. 1988;21:238–2348. doi: 10.1002/jnr.490210216. [DOI] [PubMed] [Google Scholar]
- Vouyiouklis DA, Barrie JA, Griffiths IR, Thomson CE. A proteolipid protein-specific pre-mRNA (Ppm-1) contains intron 3 and is up-regulated during myelination in the CNS. J Neurochem. 2000;74:940–948. doi: 10.1046/j.1471-4159.2000.0740940.x. [DOI] [PubMed] [Google Scholar]
- Wang E, Dimova N, Cambi F. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 2007;35:4164–4178. doi: 10.1093/nar/gkm387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, Orfaniotou F, Dhaunchak A, Brinkmann BG, Möbius W, Guarente L, Casaccia-Bonnefil P, Jahn O, Nave KA. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci. 2007;27:7717–7730. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Hammer A, Wahlbuhl M, Bösl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight PA, Dobretsova A, Macklin WB. Regulation of murine myelin proteolipid protein gene expression. J Neurosci Res. 1997;50:917–927. doi: 10.1002/(SICI)1097-4547(19971215)50:6<917::AID-JNR2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Willard HF, Riordan JR. Assignment of the gene for myelin proteolipid protein to the X chromosome: implications for X-linked myelin disorders. Science. 1985;230:940–942. doi: 10.1126/science.3840606. [DOI] [PubMed] [Google Scholar]
- Woodward K, Kendall E, Vetrie D, Malcolm S. Pelizaeus-Merzbacher disease: identification of Xq22 proteolipid-protein duplications and characterization of breakpoints by interphase FISH. Am J Hum Genet. 1998;63:207–217. doi: 10.1086/301933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Baek RC, Kirschner DA, Peterson A, Fujii Y, Nave KA, Macklin WB, Trapp BD. Evolution of a neuroprotective function of central nervous system myelin. J Cell Biol. 2006;172:469–478. doi: 10.1083/jcb.200509174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Colman DR. Parallel evolution and coexpression of the proteolipid proteins and protein zero in vertebrate myelin. Neuron. 1996;16:1115–1126. doi: 10.1016/s0896-6273(00)80138-5. [DOI] [PubMed] [Google Scholar]
- Yu WP, Collarini EJ, Pringle NP, Richardson WD. Embryonic expression of myelin genes: evidence for a focal source of oligodendrocyte precursors in the ventricular zone of the neural tube. Neuron. 1994;12:1353–1362. doi: 10.1016/0896-6273(94)90450-2. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]