Abstract
NMDA receptors (NMDARs) are important for neuronal development and circuit formation. The NMDAR subunits NR2A and NR2B are biophysically distinct and differentially expressed during development but their individual contribution to structural plasticity is unknown. Here we test whether NR2A and NR2B subunits have specific functions in the morphological development of tectal neurons in living Xenopus tadpoles. We use exogenous subunit expression and endogenous subunit knockdown to shift synaptic NMDAR composition toward NR2A or NR2B, as shown electrophysiologically. We analyzed the dendritic arbor structure and found evidence for both overlapping and distinct functions of NR2A and NR2B in dendritic development. Control neurons develop regions of high local branch density in their dendritic arbor, which may be important for processing topographically organized inputs. Exogenous expression of either NR2A or NR2B decreases local branch clusters, indicating a requirement for both subunits in dendritic arbor development. Knockdown of endogenous NR2A reduces local branch clusters, whereas knockdown of NR2B has no effect on branch clustering. Analysis of the underlying branch dynamics shows that exogenous NR2B-expressing neurons are more dynamic than control or exogenous NR2A-expressing neurons, demonstrating subunit-specific regulation of branch dynamics. Visual experience-dependent increases in dendritic arbor growth rate seen in control neurons are blocked in both exogenous NR2A- and NR2B-expressing neurons. These experiments indicate that NR2A and NR2B have subunit-specific properties in dendritic arbor development, but also overlapping functions, indicating a requirement for both subunits in neuronal development.
Keywords: structural plasticity, branch dynamics, in vivo imaging, visual system, activity dependent, glutamatergic transmission
Introduction
Synaptic activity mediated by NMDA receptors (NMDARs) plays key roles in brain development including refining topographic projections, organizing functional circuits and regulating dendritic growth (Li et al., 1994; Cline, 2001; Ruthazer and Cline, 2004). The effects of NMDAR-mediated synaptic activity on dendritic arbor growth have been intensely investigated, motivated by an interest in understanding the mechanisms by which NMDAR-activity affects developmental plasticity. Synaptic NMDAR-mediated responses promote dendritic arbor growth (Dailey and Smith, 1996; Sin et al., 2002; Niell et al., 2004; Lee et al., 2005), whereas blocking NMDARs reduces dendritic growth (Rajan and Cline, 1998; Rajan et al., 1999). Furthermore, genetic disruption of individual NMDAR subunits impairs map formation (Li et al., 1994; Kutsuwada et al., 1996; Iwasato et al., 1997, 2000; Lee et al., 2005), possibly by affecting dendrites of target neurons. Although these studies establish a role for NMDARs in dendrite development, and suggest specific roles for particular subunits (Kutsuwada et al., 1996), the role of NMDAR types, defined by their subunit composition, is unclear.
NMDARs consist of two obligate NR1 subunits and two NR2A-D or NR3A-B subunits, whose expression is spatially and developmentally regulated (Cull-Candy and Leszkiewicz, 2004). NR2A and NR2B have different biophysical properties, resulting in fast decay times for NR2A- and threefold to fourfold slower decay times for NR2B-containing receptors (Monyer et al., 1994; Vicini et al., 1998), and consequently in different Ca2+ influx (Sobczyk et al., 2005). NR2A and NR2B also differ in their interactions with different postsynaptic density proteins (Sans et al., 2000; Krapivinsky et al., 2003; Barria and Malinow, 2005; Kim et al., 2005; Al-Hallaq et al., 2007). During development, NMDAR decay kinetics shorten because NR2B is expressed in immature neurons, whereas NR2A-containing receptors become the electrophysiologically predominant subunit at later stages of development (Carmignoto and Vicini, 1992; Hestrin, 1992; Monyer et al., 1994; Cline et al., 1996; Shi et al., 1997; Kew et al., 1998; Roberts and Ramoa, 1999).
Although NMDAR subunit composition is hypothesized to confer distinct roles in nervous system development (Cull-Candy and Leszkiewicz, 2004), the functional significance of the developmental change in NR2A and NR2B expression remains unclear. The proposed role in regulating critical period plasticity (Carmignoto and Vicini, 1992; Flint et al., 1997; Roberts and Ramoa, 1999) was not supported by the normal barrel cortex critical period observed in NR2A knock-out mice (Lu et al., 2001). Evidence for opposing roles of NR2A- and NR2B-containing receptors in regulating synaptic plasticity (Liu et al., 2004; Massey et al., 2004) was complicated by drug specificity (Neyton and Paoletti, 2006) and conflicts with other findings, including data showing overlapping roles of NR2A and NR2B in synaptic plasticity (Tang et al., 1999; Lu et al., 2001; Kohr et al., 2003; Barria and Malinow, 2005; Berberich et al., 2005; Toyoda et al., 2005; Weitlauf et al., 2005; Morishita et al., 2007).
We tested whether subunit composition of synaptic NMDARs affects dendritic arbor development in vivo by controlling expression of NR2A and NR2B in single optic tectal neurons in the visual circuit of intact albino Xenopus tadpoles and visualized their development and experience-dependent plasticity by two-photon time-lapse imaging. We find that although some features of dendritic arbor development are more strongly affected by NR2A or NR2B, others indicate overlapping roles for the two subunits in neuronal development.
Materials and Methods
Expression vectors and morpholinos.
The Xenopus laevis cDNA sequences of NR2A and NR2B were cloned from a cDNA library screen (GenBank accession numbers EU275164 and EU275165). Based on the complete sequences of X. laevis NR2A and NR2B cDNA, the amino acid sequence and all major functional regions and residues are conserved between Xenopus and rat NR2 subunits (R. Ewald and H. Cline, unpublished observation). X. laevis NR2A and rat NR2B cDNA (a gift from Dr. Robert Malinow, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) were subcloned in the bidirectional double-promoter expression vector BiCs2GFP (a generous gift from Dr. Dave Turner, University of Michigan, Ann Arbor, MI). BiCs2GFP contains two cytomegalovirus promoters on opposite strands. One promoter drives the expression of enhanced green fluorescent protein (EGFP) and the other drives one of the NR2 subunits or nothing for control cells. Protein expression from BiCs2GFPNR2A and BiCs2GFPNR2B was verified by Western blot. The NR2A and NR2B morpholino (MO) sequences and their respective five-mispair controls (Gene-Tools, Philomath, OR) were based on the cloned X. laevis NR2A and NR2B cDNA sequences and tagged with carboxyfluorescein or lissamine.
The NR2A and NR2B morpholino sequences are as follows: NR2A-MO, AAACAAACATCCCCATCTTGTCCAG; MisNR2A-MO, AAAgAAAgATCCCgATCTTcTCgAG; NR2B-MO, GCCTCATCTTCAACTCTTACACTCC; MisNR2B-MO, GCgTCATgTTCAAgTCTTAaACTaC. The plasmid pBαact-Gal4VP16-UAS-EGFP (Koster and Fraser, 2001) (a generous gift from Dr. Scott Fraser, California Institute of Technology, Pasadena, CA) was used to visualize the morphology of MO-transfected cells.
Electrophysiology.
Cells in stage 47 tadpole brains were transfected with the expression vectors (2–4 μg/μl) or the carboxyfluorescein-labeled MOs (500 μm) by whole-brain electroporation (stimulation settings, five pulses, 1.4 ms duration, 50V) (Haas et al., 2002). After 3 d, the brains were prepared for electrophysiological recordings as described by Wu et al. (1996). Animals were anesthetized in 0.02% 3-aminobenzoic acid (MS-222; Sigma, St. Louis, MO). The brain was dissected into HEPES-buffered extracellular saline. For recordings of evoked NMDA receptor mediated synaptic currents, the composition of the saline was (in mm) 115 NaCl, 2 KCl, 3 CaCl2, 3 MgCl2, 5 HEPES, 10 glucose, 0.01 glycine, 0.1 picrotoxin, and 20 2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo[f]quinoxaline-2,3-dione (NBQX), pH 7.2 with NaOH, and osmolarity 255 mOsm. For recordings of AMPA mEPSC, the composition of the saline was the same except we used 4 KCl and included 1 μm tetrodotoxin (TTX). Ifenprodil (9 μm) was added as stated. The brains were stabilized in a submerged perfusion chamber and maintained at room temperature. Cells were visualized with a Nikon (Tokyo, Japan) E600FN fluorescence microscope and a 60× water-immersion lens. Recordings were limited to GFP-expressing or carboxyfluorescein-labeled cells except for recordings from control AMPA mEPSCs that were recorded from untransfected cells. Recordings were taken from cells located in the middle region of the optic tectum, avoiding the extreme caudal and rostral boundaries where developmental differences in expression of NR2A and NR2B subunits affect synaptic NMDA responses (Cline et al., 1996). No difference was seen in recordings from cells electroporated with control MOs for NR2A and NR2B, therefore, results were pooled. Whole-cell voltage-clamp recordings were made with glass micropipettes (8–10 MΩ) filled with intracellular saline [(in mm) for evoked recordings: 90 Cs-methane sulfonate, 5 MgCl2, 20 TEA, 10 EGTA, 20 HEPES, 2 ATP, 0.3 GTP, pH 7.2 with CsOH, osmolarity 255 mOsm; for AMPA mEPSC recordings: same except 80 Cs-methane sulfonate, osmolarity 250 mOsm]. Input resistances averaged ∼1.5 GΩ, and series resistances ∼50 MΩ (Wu et al., 1996; Aizenman et al., 2002). For the evoked NMDA responses, a bipolar stimulating electrode (FHC, Bowdoinham, ME) was placed in the optic chiasm to evoke retinotectal synaptic responses. The NMDA responses were recorded at +55 mV with 6 s between stimuli. The AMPA mEPSCs were recorded at a holding potential of −600 mV. Signals were measured with an Axopatch 2-D amplifier, and digitized with a Digidata 1200 A-D board (all equipment was from Molecular Devices, Palo Alto, CA). Traces were recorded with P-Clamp8 software, and digitized at 10kHz. NBQX was obtained from Tocris (Ellisville, MO), picrotoxin and ifenprodil from Sigma, and TTX from Alomone Labs (Jerusalem, Israel).
Data analysis: electrophysiology.
The data were analyzed with Axograph software (Molecular Devices). To obtain the decay time constant τ, a single exponential was fitted to the digitized data (Monyer et al., 1992) within a window from the peak to 50% of the peak. All AMPA mEPSC events were analyzed that were detected within the first 4 min of recording or the first 100 events, whichever came first. For the cumulative frequency plots the first 20 events of each recorded cell were used.
Image acquisition.
Stage 46–47 X. laevis tadpoles were anesthetized in 0.02% MS-222 (Sigma) and single cells were electroporated (Haas et al., 2001; Bestman et al., 2006) via a glass electrode (tip diameter ≤1 μm) filled with the BiCs2 expression vectors (1–1.5 μg/μl), or a mixture of MOs (500 μm) and the Gal4VP16-UAS-EGFP plasmid (1–1.5 μg/μl). Electroporation settings for DNA constructs alone were 1 s train, 1 ms pulse, 200 Hz at 1–2 μA, negative voltage; and for Mos plus DNA were (twice) 1 s train, 1 ms pulse, 200 Hz at 1–2 μA, first train at negative, second train at positive voltage. The tadpoles were imaged the next day, and then imaged again according to their time-lapse imaging protocol, with a custom-built two-photon laser-scanning microscope that was modified from an Olympus Fluoview confocal scan box mounted on an Olympus (Center Valley, PA) BX50WI microscope. The light source was a Tsunami femtosecond-pulsed Ti:sapphire laser pumped by a 10 W solid-state Millenia X laser (both from Newport Spectra, Physics, Mountain View, CA) at 905–910 nm. Images were acquired with the Olympus Fluoview acquisition software and a 40× water-immersion lens, and images were collected to capture the entire extent of the dendritic arbor at 1.5 μm steps in the z direction.
Imaging protocols.
Before and after electroporation and in between the daily imaging sessions the tadpoles were kept on a 12 h dark/light cycle in Steinberg's rearing solution. In between all imaging points the tadpoles were allowed to swim freely, and in case of the branch dynamics experiment they were left at ambient light during the imaging session. Before imaging the animals were anesthetized in 0.02% MS-222 (Sigma) and placed into a Sylgard imaging chamber with a glass coverslip on top. For the daily imaging experiments, the tadpoles were imaged once daily over 3 d, 1 d after electroporation. For the branch dynamics experiment, the tadpoles were imaged every 2 h over 8 h the day after electroporation. For the visual stimulation experiments we used the setup and imaging protocol described by Sin et al. (2002). The light stimulus intensity was 0.031mW/cm2 as measured with an IL1400BL photometer (International Light Technologies, Peabody, MA). The tadpoles were imaged the day after electroporation (t0), placed in a dark box for 4 h and imaged again (t4). Then they were put into a chamber with four rows of light-emitting diodes flashing in sequence to simulate a continuous motion-stimulus for 4 h before a final image was taken (t8).
Image analysis.
The three dimensional dendritic arbor of the cells was manually reconstructed from the raw two-photon z-stack series using the drawing program ObjectImage (http://simon.bio.uva.nl/object-image.html). Custom macros developed by Dr. Edward Ruthazer (McGill University, Montreal, Canada; http://clinelab.cshl.edu/methods.html) were used to determine total dendritic branch length and branch tip number, and for the three-dimensional (3D) Sholl analysis (1 μm radius interval, averaged over 5 μm bins). For the pixel density analysis, a square of 63 × 63 pixels was centered on a single pixel within the 512 × 512 pixel image window of the drawn object, and the total number of pixels in the square was counted to determine the density value of the pixel in the center. The box was moved across the image window to determine a density value for every pixel. The individual slices were then convolved to yield density values for the 3D stack. For interbranch tip distance and interbranch point distance analyses, the x, y, z coordinates of the branch tips and branch points were determined and the distances were calculated with the formula d = √[(x − xn)2 + (y − yn)2 + (z − zn)2], where x, y, z are the coordinates of one branch tip/point, and xn, yn, zn are the coordinates of all other branch tips/points. To avoid oversampling branch points, distances smaller than 0.5 μm (the maximum x–y resolution) were not counted. The normalized change in branch tip number (BTN) in the knockdown experiment was calculated as the difference between the branch tip numbers on day 3 and day 1, and was normalized to the initial branch tip number on day 1.
For the branch dynamics experiment, branch identities were assigned to follow individual branch behavior across time points. Branch dynamics were analyzed with custom macros (Dr. Edward Ruthazer, McGill University, Montreal, Canada). We analyzed branch behavior according to dynamic events that occurred in relation to a reference time point. In this analysis, addition events appear at one time point and persist through the next, retraction events are branches that were present before the time point and are lost at the reference time point, maintained branches are present before, during, and after the time point, and transient events are branches that are only present at the reference time point. The fraction of these events of all branches at every single time point was calculated, and then averaged across all cells. To determine the persistence of branches present at t0, we calculated the fraction of branches that were present at t0 and were still present at any other time point. We calculated the fraction of transient branches at any single time point by determining the fraction of transient branches (as categorized over the entire 8 h imaging period) of all branches present at t2, t4, and t6. The number of appearances of transient branches counted how often transient branches appeared during the experiment as a fraction of all transient branches.
For the visual stimulation experiment, the growth rate in the dark and light was calculated as total dendritic branch length [TDBL(t4–t0)] and TDBL(t8–t4), respectively. Branch identities were assigned so behavior of individual branches could be followed in response to the dark or the visual stimulation. To calculate total branch length addition, the lengths of newly added branches and the extension of existing branches were added. To calculate total branch length retraction, the lengths of lost and the shortening of existing branches were summed. Branch extensions and retractions were calculated by subtracting branch lengths of branches present at least at two time points and that changed more than ±1.5 μm.
Results
Exogenous expression and knockdown of NR2A and NR2B shifts synaptic NMDAR responses in tectal neurons
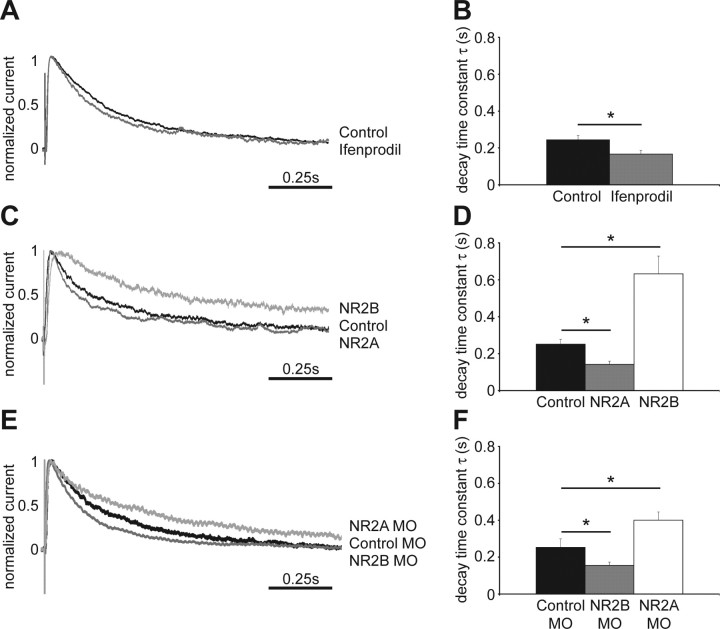
The X. laevis cDNA sequence information for NR2A and NR2B demonstrate a high degree of conservation of NMDAR subunits across species, and electrophysiological recordings and biochemical data provide strong evidence for the expression of both NR2A and NR2B subunits in tectal neurons of stage 47 X. laevis tadpoles (Cline et al., 1996; Aizenman and Cline, 2007) (Ewald and Cline, unpublished observation). We recorded evoked whole-cell responses from stage 47 tectal cells using an open brain preparation, where retinal axons in the optic chiasm are stimulated (Wu et al., 1996). The decay time constant τ in control neurons was 0.244 ± 0.023 s (Fig. 1A,B), consistent with previous studies in Xenopus tectal neurons (Cline et al., 1996). To test the relative contribution of NR2B-containing NMDARs to synaptic NMDAR currents, we applied the NR2B-specific antagonist ifenprodil at 9 μm. At this concentration, ifenprodil effectively blocks NR1/NR2B-only-containing NMDARs, but not NR1/NR2A-only-containing receptors. However, inclusion of the NR2A subunit in the receptor to generate NR1/NR2A/NR2B-containing heteromers significantly reduces effectiveness of the ifenprodil block (Williams, 2001). We found a small but significant acceleration of the decay time constant τ (0.167 ± 0.02 s) (Fig. 1A,B), suggesting that the majority of synaptic NMDAR contain the NR2A subunit and are composed of either NR1/NR2A/NR2B or NR1/NR2A receptors.
Figure 1.
Exogenous expression or knockdown of NMDAR subunits shifts the kinetics of NMDAR-mediated EPSCs. A, Normalized averaged traces of evoked NMDAR-mediated EPSCs recorded from control tectal neurons and cells exposed to ifenprodil. Retinal ganglion cell axons were stimulated at the optic chiasm and responses from tectal cells were recorded by whole-cell patch clamp. B, Quantification of the decay time constant τ (control, 0.244 ± 0.023 s; Ifenprodil, 0.167 ± 0.02 s; *p < 0.03, n = 9–14 cells in each group). C, Normalized averaged traces of evoked NMDAR-mediated EPSCs from control and exogenous NR2A- and NR2B-expressing tectal neurons. D, Quantification of the decay time constant τ (control, 0.251 ± 0.027; NR2A, 0.142 ± 0.018; *p < 0.02; NR2B, 0.632 ± 0.097, *p < 0.01; n = 5–9 cells in each group). E, Normalized averaged traces of evoked NMDAR-mediated EPSCs from cells expressing control-, NR2A- and NR2B-MOs. F, Quantification of the decay time constant τ (control MO, 0.253 ± 0.046; NR2B MO, 0.155 ± 0.019, *p < 0.05; NR2A MO, 0.4 ± 0.045, *p < 0.05; n = 11–25 cells in each group). Error bars are SEM and all statistical tests are Mann–Whitney U tests.
To shed light on the respective contribution of NR2A- and NR2B-containing receptors to the morphological development of tectal cells, we shifted the synaptic NMDAR composition within these neurons to become more NR2A- or NR2B-like with exogenous expression of recombinant vectors or morpholino antisense oligonucleotide-mediated knockdown of the endogenous subunits. Previous experiments suggest that the endogenous levels of the other NMDAR subunits remain unaltered despite such manipulations (Morikawa et al., 1998; Tang et al., 1999; Philpot et al., 2007). To assess the effect of NR2 subunit manipulation on synaptic NMDAR composition, we recorded evoked retinotectal synaptic currents from transfected cells, identified by EGFP coexpression or by the carboxyfluorescein label of the MOs.
Sample traces of responses of isolated NMDAR currents show that exogenous NR2B-expressing cells have significantly longer decay times than NR2A-expressing cells or control neurons, expressing only EGFP (Fig. 1C). The decay time constant τ is significantly longer in recombinant NR2B-expressing neurons than for control cells (Fig. 1D). In contrast, NR2A-expressing cells have a much shorter decay time compared with NR2B transfected neurons (Fig. 1C) and their τ is significantly faster than that of control cells (Fig. 1D). The responses of exogenous NR2A- and NR2B-expressing cells are characteristic of each receptor subunit, as seen for example in recordings from thalamocortical slices from wild-type and NR2A −/− mice (Lu et al., 2001). These results indicate that retinotectal synaptic responses in control cells are mediated by NMDARs composed of both NR2A and NR2B subunits, and that it is possible to shift their relative ratio in the synapse to show more NR2A- or NR2B-like features by exogenous expression of the receptor subunits. The recordings also indicate that both recombinant NR2A and NR2B associate with endogenous NR1 to form functional NMDARs that are incorporated into synapses.
We further tested whether we could shift the endogenous NMDAR composition by targeted knockdown of NR2A and NR2B subunits with MOs. Compared with neurons that were transfected with 5-mispair control MOs, knockdown of NR2A results in evoked NMDAR synaptic responses with longer decay times (Fig. 1E,F) typical of NR2B-containing receptors. Similarly, knockdown of the NR2B subunit results in evoked synaptic NMDA currents with significantly shorter decay time constants than control cells (Fig. 1E,F). These results show that it is also possible to shift the synaptic NMDAR subunit composition toward NR2A- or NR2B-like electrophysiological phenotypes by MO-mediated knockdown.
Exogenous NR2A and NR2B expression regulates the local dendritic arbor architecture
To investigate whether the different NMDAR subtypes play distinct functional roles in dendritic arbor development we used a daily imaging protocol that captures gross morphological changes of the dendritic tree in vivo (Wu and Cline, 1998). Single neurons were transfected by electroporation (Haas et al., 2001; Bestman et al., 2006) with recombinant NMDAR subunits or MOs, and coexpressed EGFP as a morphological marker. Cells were imaged once daily for three consecutive days starting the day after electroporation (Figs. 2A, 3A).
Figure 2.
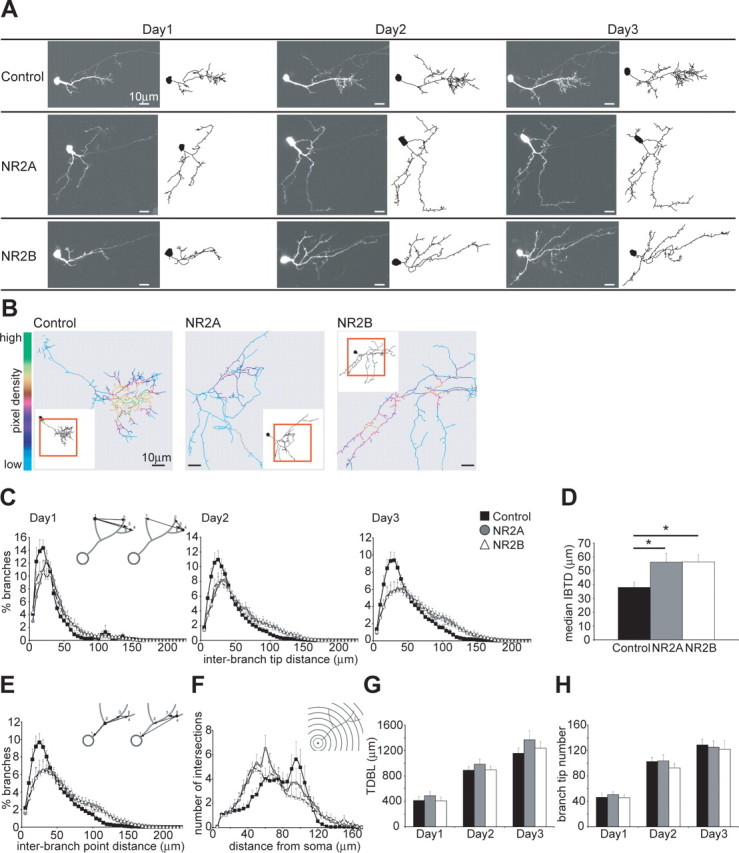
Exogenous expression of NR2A and NR2B decreases local dendritic branch clusters. A, Images and drawings of representative control and exogenous NR2A- and NR2B-expressing cells imaged once daily over 3 d. Axons present in the images are not shown in the drawings. B, Pixel density analysis shows areas of high pixel density with yellow and green colors, and areas of low pixel density with turquoise and blue colors, and highlights the differences in dendritic arbor architecture between control and NR2-expressing neurons. The inset shows the entire reconstructed cell and a red box surrounds the area seen in the pixel density analysis. C, Analysis of the IBTDs (see inset diagram) as neurons develop over 3 d. The distances are measured for every branch tip from one branch tip to every other branch tip on the dendritic tree. D, The median IBTD of each cell was calculated and the average per group is shown. NR2A- and NR2B-expressing cells have longer IBTDs than control cells (control, 37.94 ± 3.80 μm; NR2A, 56.30 ± 6.32 μm, *p < 0.03; NR2B, 56.45 ± 5.23 μm, *p < 0.02). E, Analysis of the interbranch point distance measured from one branch point to every other branch point (see diagram inset) confirms the different branch arrangement. F, Three-dimensional Sholl analysis (see diagram inset) characterizes arbor structure and complexity. G, H, Quantification of the average TDBL (G) and of the total BTN (H). The general growth parameters are similar between the groups on all 3 d: TDBL on days 1, 2, and 3, respectively (in μm), control, 412.97 ± 62.88, 889.40 ± 55.3, and 1156.08 ± 81.26; NR2A, 488.96 ± 62.87, 984.43 ± 80.11, and 1369.29 ± 139.58; NR2B, 406.57 ± 61.58, 896.54 ± 61.03, and 1236.25 ± 103.37; BTN on days 1, 2, and 3, respectively, control, 46.3 ± 6.83, 102.6 ± 6.54, and 128.8 ± 9.07; NR2A, 50.6 ± 4.8, 103.8 ± 9.38, and 125.2 ± 9.8; NR2B, 45.8 ± 4.35, 92.6 ± 6.59, and 122 ± 13.74; n = 10 cells per group. Error bars are SEM and all statistical tests are Mann–Whitney U tests.
Figure 3.
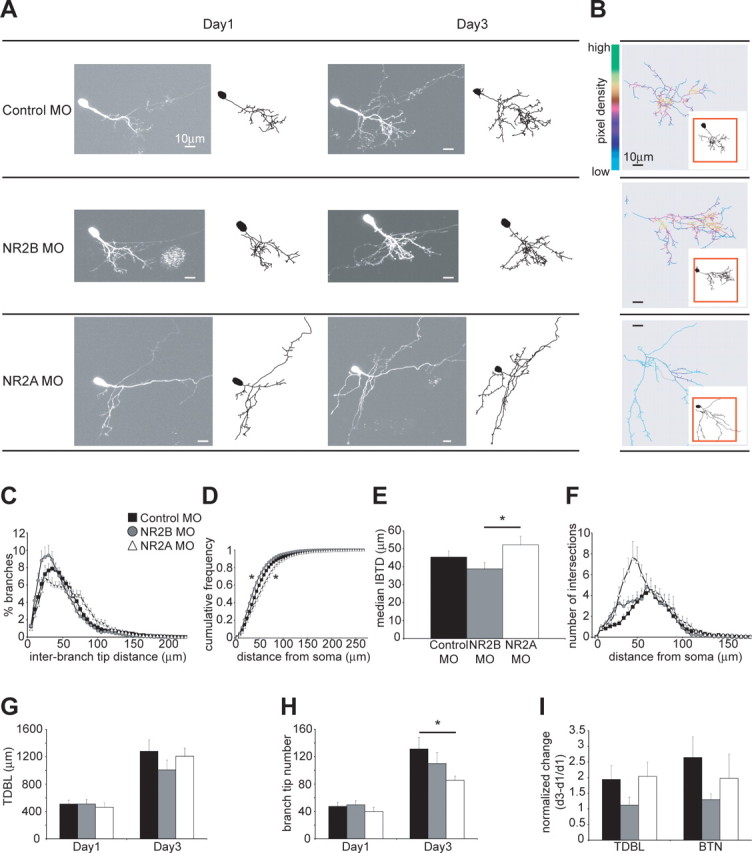
Knockdown of NR2A and NR2B leads to distinct rearrangements of the dendritic arbor architecture. A, Images and drawings of representative neurons imaged on day 1 and day 3 after single-cell electroporation of control, NR2A and NR2B MOs. Axons present in the images are not shown in the drawings. B, Pixel density analysis of the dendritic arbor highlights areas of high and low branch density. C, IBTD analysis shows that cells expressing NR2B MO have shorter IBTDs, whereas neurons expressing NR2A MO have longer IBTDs. Control cells have IBTDs that are in between the two groups. D, Cumulative frequency plot of the IBTDs (control MO vs NR2A MO, *p < 0.01; control MO vs NR2B MO, *p < 0.05; NR2A MO vs NR2B MO, *p < 0.0001). E, Quantification of the averaged median IBTD for each group (control MO, 45.40 ± 3.31 μm; NR2A MO, 52.19 ± 4.77 μm; NR2B MO, 38.78 ± 3.62 μm; *p = 0.05). F, Three-dimensional Sholl analysis of dendritic arbor structure and complexity. G, TDBL is similar at day 1 and day 3 for all three groups [days 1 and 3, respectively (in μm): control MO, 509.77 ± 55.1, 1280.69 ± 166.77; NR2B MO, 511.06 ± 63.15, 1007.61 ± 144.48; NR2A MO, 462.49 ± 63.29, 1208.19 ± 118.39; n = 9–14 cells per group]. H, BTNs are smaller for cells expressing NR2A MO compared with controls on day 3 [days 1 and 3, respectively: control MO, 47.29 ± 5.71, 131.21 ± 16.97; NR2B MO, 49.6 ± 5.94, 109.9 ± 16.1; NR2A MO, 39.89 ± 6.21, 85.33 ± 6.13; *p < 0.05). I, The change in TDBL and BTN over 3 d normalized to the starting size at day 1 shows no significant difference in growth rate or rate of branch tip additions [TDBL and BTN, respectively: control, 1.94 ± 0.44, 2.64 ± 0.66; NR2B MO, 1.21 ± 0.25, 1.3 ± 0.19; NR2A MO, 2.04 ± 0.46, 1.98 ± 0.77]. Error bars are SEM. The statistical tests are Mann–Whitney U tests, and the Kolmogorov–Smirnov test for the cumulative frequency plots.
Neurons that express exogenous NR2A or NR2B look morphologically different from control cells. Dendritic arbors of control neurons form local clusters of branches, whereas the branches of recombinant NR2A- and NR2B-expressing cells appear more evenly distributed along the arbor, as illustrated by the example cells (Fig. 2A). Pixel-density analysis highlights the areas of local branch clusters. Yellow and green colors indicate areas of high pixel density and thus branch clustering, whereas turquoise and blue colors show areas of low pixel density and therefore little clustering. Figure 2B illustrates a representative control cell showing areas of high branch density, whereas exogenous NR2A- and NR2B-expressing cells lack these local branch clusters and are clearly more sparsely branched.
We quantified local branch clusters in dendritic arbors by analyzing interbranch tip distances (IBTD), where the distances from one branch tip to every other branch tip of the arbor are calculated. If a branch terminates further away from other branches, it will have longer interbranch tip distances and if it is in a cluster of branches it will have shorter interbranch tip distances (Fig. 2C, diagram inset). Control cells have shorter interbranch tip distances compared with exogenous NR2A- and NR2B-expressing cells and the phenotypes become more pronounced over the imaging period (Fig. 2C). This indicates that control cells have dendritic regions that become more densely arborized over a 3 d period, resulting in more local clusters of branches. Both recombinant NR2A- and NR2B-expressing cells have longer interbranch tip distances than control cells, indicating that their branches are more spaced out and less clustered (Fig. 2C). The median interbranch tip distance was calculated for every cell and the average for each group further shows that exogenous NR2A- and NR2B-expressing cells have significantly larger interbranch tip distances than control cells (Fig. 2D). Because branch tips are organized in the three-dimensional space around the dendritic shaft, there is the possibility that the arrangement of branch points could be similar between groups despite the branch tips being organized differently. We determined the distance of every branch point to every other branch point (Fig. 2E, diagram inset) and find that, consistent with the interbranch tip distance analysis, recombinant NR2A- and NR2B-expressing cells have fewer short interbranch point distances and many long interbranch point distances, whereas control cells have many short and fewer long inter branch point distances (Fig. 2E). Both analyses therefore show that exogenous NR2A- and NR2B-expressing cells develop a dendritic arbor of lower branching complexity, compared with control cells that develop local branch clusters.
To further analyze the distribution of branches across the complete dendritic arbor we performed a three-dimensional Sholl analysis, where a set of concentric spheres is drawn around the cell soma and the number of branch intersections at each sphere is counted, revealing arbor complexity (Fig. 2F, diagram inset). Over 3 d, recombinant NR2A- and NR2B-expressing cells develop a different dendritic structure than control neurons. They start branching closer to the cell body and tend to extend branches further into the neuropil than control cells (Fig. 2F). Control cells in contrast are characterized by a gradual increase in branch density with increasing distances from the cell body and a sharp drop in branches at approximately 100 μm from the soma (Fig. 2F) indicating a perimeter of maximal branching and extension into the tectal neuropil.
These data demonstrate that shifts in the synaptic NR2A and NR2B ratio by exogenous expression lead to similar structural changes in dendritic arbors in vivo. To determine whether these structural changes are caused by changes in the growth behavior of tectal cells we assessed their general growth parameters. Total dendritic branch length and branch-tip number are good indicators of the growth behavior of a neuron (Wu et al., 1999). Over 3 d the cells of the three groups grew comparably and there was no difference in their total dendritic arbor size (Fig. 2G) or branch tip number (Fig. 2H). Notably, the starting size of the total dendritic arbor and branch tip number was similar for all groups (Fig. 2G,H, day 1). Therefore, recombinant NR2A- and NR2B-expressing cells cannot be distinguished from control cells according to these general growth parameters, indicating that exogenous expression of NR2A and NR2B subunits neither stunts nor promotes dendritic arbor growth, but rather increases the maximal extent of the dendrites and modifies the branching pattern within the arbor.
These data show that exogenous expression of either NR2A or NR2B controls the establishment of local branch clusters within the dendritic arbor without affecting the growth of the neuron. This suggests that signaling through NR2A- and NR2B-containing receptors impacts the mechanisms controlling sites to form and maintain dendritic branches, which results in a clustering of branches in control neurons. Exogenous expression of either NR2A or NR2B results in similar phenotypes, where branches are not clustered, despite the different underlying synaptic electrophysiology. Therefore, we conclude that the process that results in branch clustering depends on finely tuned NMDAR signaling achieved by the mixture of NR2A-, NR2B-, and NR2A/NR2B-containing NMDARs in the neuron.
Knockdown of endogenous NR2A and NR2B affects dendritic branch clustering
Analysis of dendritic arbor development in cells where endogenous levels of NR2A and NR2B were knocked down with MOs reveals additional insights into the roles of the two subunits, which differentiate between NR2A- and NR2B-containing receptors. As illustrated by the example cells and the pixel-density analysis (Fig. 3A,B), control neurons transfected with five-mispair MOs and cells transfected with NR2B MOs appear to have more clustered branches than cells electroporated with NR2A MOs. Indeed, interbranch tip distance analysis shows that cells where NR2B is knocked down have more short and fewer long interbranch tip distances, indicating a clustered branch phenotype (Figs. 3C,D). Neurons with knocked down levels of NR2A, in contrast, have fewer short and more long interbranch tip distances, which is indicative of a dendritic arbor where branches are more spaced apart (Fig. 3C,D). Cells expressing control MOs have a phenotype that is in between the two experimental groups (Fig. 3C) and significantly different from each one, as shown by the cumulative frequency plot (Fig. 3D). The median interbranch tip distances (Fig. 3E) further demonstrate the difference in phenotype between cells with knocked down levels of NR2A or NR2B. Three-dimensional Sholl analysis shows that neurons expressing NR2A MOs start branching closer to the cell body than NR2B MO and control MO cells (Fig. 3F). These data support the conclusion that NR2A- and NR2B-containing NMDARs play a role in the structural plasticity of dendritic arbors in vivo.
As seen with exogenous expression of the NR2 subunits, general growth parameters of cells electroporated with MOs are similar between the groups (Fig. 3G–I). The neurons have comparable starting sizes of their dendritic arbor and show similar growth rates so they remain close in size at day 3 (Fig. 3G,I). This is also true for the number of branch tips of control and NR2B MO-expressing cells (Fig. 3H,I). The absolute number of branch tips at day 3 is significantly smaller for NR2A MO electroporated neurons than controls (Fig. 3H). However, the rate of change in branch tip number is not different (Fig. 3I).
Together, these data demonstrate a more differentiated picture of how NR2A- and NR2B-containing NMDARs impact dendritic structural development. Knockdown of endogenous NR2A protein levels decreases local branch clusters, whereas neurons with knocked down levels of NR2B, but endogenous levels of NR2A develop regions of clustered branches. This suggests that signaling through NR2A-containing receptors in particular is critical for the establishment and maintenance of higher order branch structure in dendrites.
Exogenous NR2B expression decreases dendritic branch stability
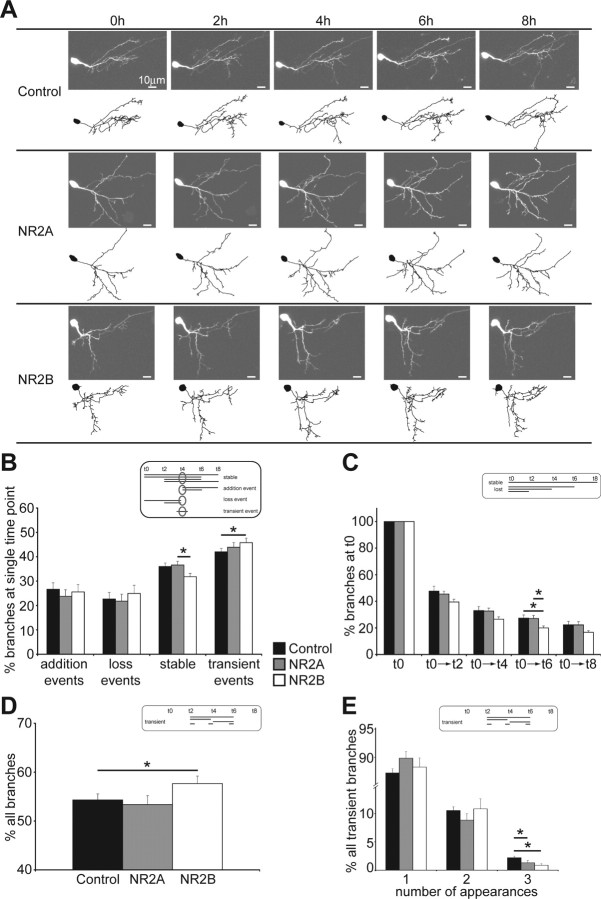
Small local changes in neuronal branch dynamics can add up to have a big global impact on the structure of a neuron (Ruthazer et al., 2003; Javaherian and Cline, 2005; Haas et al., 2006). Therefore, we tested whether changes in branch dynamics can account for the observed rearrangement of the dendritic arbor in cells where the NMDAR subunit composition is shifted. We imaged single neurons every 2 h over 8 h, 1 d after electroporation (Fig. 4A) and examined the dynamic behavior of their branches. Growth rates between time points were comparable and not different between groups. We analyzed the dynamic events at every time point and categorized branches into stable, added, lost and transient branches (Fig. 4B, diagram inset with time point t4 as an example). This analysis revealed that at any individual time point exogenous NR2B-expressing cells have significantly fewer stable branches than exogenous NR2A-expressing cells (Fig. 4B), and they show a trend to have fewer stable branches than controls (p = 0.06). In addition, cells expressing recombinant NR2B have more transient events than control neurons (Fig. 4B). These data show that branches of exogenous NR2B-expressing cells are not as stable as branches of control or exogenous NR2A-expressing cells.
Figure 4.
Exogenous NR2B-expressing cells are more dynamic than control or exogenous NR2A-expressing cells. A, Images and drawings of representative cells imaged every 2 h over 8 h, 1 d after single-cell electroporation. Axons present in the images are not shown in the drawings. B, Quantification of branch dynamics in relation to a reference time point (see diagram inset with time point t4 as an example) categorizes dynamic events in the context of the time points immediately before and after the reference time point [addition, loss, stable, transient, respectively (in percent): control, 26.67 ± 2.64, 22.67 ± 2.71, 36.02 ± 1.47, 42.01 ± 1.37; NR2A, 23.78 ± 2.7, 21.8 ± 2.85, 36.6 ± 1.56, 43.88 ± 1.89; NR2B, 25.56 ± 3.03, 24.93 ± 3.37, 31.82 ± 1.31, 45.77 ± 1.9; *p < 0.05]. C, Analysis of the persistence of branches present at t0 [see diagram inset; t0→t2, →t4, →t6, and →t8, respectively (in percent): control, 47.65 ± 3.65, 32.99 ± 3.11, 27.32 ± 2.41, 22.31 ± 2.55; NR2A, 45.44 ± 2.16, 32.7 ± 2.21, 27.06 ± 2.3, 22.21 ± 2.44; NR2B, 39.52 ± 1.99, 26.55 ± 1.76, 19.98 ± 1.52, 16.7 ± 1.16; *p < 0.05]. D, Analysis of the fraction of all transient branches at any single time point (see diagram inset; control, 54.56 ± 1.67%; NR2A, 53.63 ± 2.87%; NR2B, 57.55 ± 2.0%; *p < 0.05). E, Analysis of the life times of transient branches according to their number of appearances during the imaging protocol [number of appearances 1, 2, 3 (in percent): control, 87.25 ± 0.74, 10.55 ± 0.64, 2.2 ± 0.28; NR2A, 89.85 ± 1.19, 8.85 ± 1.13, 1.3 ± 0.4; NR2B, 88.28 ± 1.61, 10.85 ± 1.76, 0.87 ± 0.31; *p < 0.05]. Error bars are SEM and all statistical tests are Mann–Whitney U tests.
To examine the stability of branches further, we analyzed the persistence of branches that were present at the first time point t0, and were still present at later time points (Fig. 4C, diagram inset). Exogenous NR2B-expressing cells lose their branches at a more rapid rate than control or exogenous NR2A-expressing neurons (Fig. 4C). This suggests that branches of recombinant NR2B-expressing cells have shorter life times and is consistent with the observation, above, that the branches in NR2B-expressing cells are not as stable as controls. We further examined for each time point the fraction of transient branches that are added and lost during the 8 h experiment and whose life times could therefore be captured (Fig. 4D, diagram inset). We found that exogenous NR2B-expressing cells have slightly, but significantly, more of these transient branches than recombinant NR2A-expressing or control cells (Fig. 4D). Because this transient category consists of a heterogeneous population of transient branches with different life times (Fig. 4E, diagram inset) we analyzed their duration by counting their number of appearances during the imaging protocol. We found that, although ∼90% of all transient branches in all three experimental groups appear only at one time point, both exogenous NR2A- and NR2B-expressing cells have fewer transient branches with long life times (Fig. 4E). This in turn suggests that branches of recombinant NR2A- and especially NR2B-expressing cells are maintained over different time spans than branches of control cells.
Together, the branch dynamics data show that exogenous NR2B-expressing cells are more dynamic than control and exogenous NR2A-expressing cells because they lose their branches at a faster rate, have more transient and fewer maintained branches at single time points and have fewer transient branches with longer life times. In contrast, the dynamics of recombinant NR2A-expressing cells resemble control cells, which is perhaps not surprising given that synaptic NMDAR in control cells seem to be predominated by NR2A (Fig. 1).
Exogenous NR2B expression blocks experience-dependent structural plasticity
Despite the differences in branch dynamics, recombinant NR2A- and NR2B-expressing cells acquire similar structural phenotypes over time. NMDAR-mediated synaptic activity is important for dendritic growth (Rajan and Cline, 1998; Sin et al., 2002) and appears to be important for branch location within the arbor (Figs. 2, 3), which can potentially impact a neuron's function within the neuropil. Therefore, we examined the effect of exogenous expression of NR2A or NR2B on activity-dependent dendritic structural plasticity. We used an experimental protocol in which tadpoles reared in a 12 h ambient light/dark cycle are placed in the dark for 4 h and then are subjected to a 4 h period of visual stimulation (Sin et al., 2002). In response to the visual stimulation, the dendritic arbor growth rate increases compared with 4 h of light deprivation (Sin et al., 2002) (Fig. 5A–C, control). The tadpoles were freely swimming during the visual stimulation, so there is no consistent directionality to the stimulus. We assessed the total branch length that was added, which consists of new branch additions and existing branch extensions, as well as the total branch length that was retracted, which consists of lost branches and the shortening of existing branches, during 4 h periods in the dark and the light (Fig. 5D). Control cells add significantly more branch length during the light period, and this is mainly because of the contribution of newly added branches (Fig. 5D). Exogenous NR2B-expressing cells do not show increased growth rates in the light compared with the dark (Fig. 5B,C) and the total branch length added and retracted was similar in both conditions (Fig. 5D). This is consistent with our results showing that exogenous NR2B expression decreases branch stability (Fig. 4). The population of exogenous NR2A-expressing cells shows a heterogeneous response to visual stimulation with 8 of 15 neurons increasing and 7 of 15 decreasing their growth rate with visual stimulation (Fig. 5B), canceling out any net effect in the group of cells imaged (Fig. 5C,D). Neither initial total dendritic branch length, nor initial branch tip number correlated with the growth behavior of the recombinant NR2A-expressing cells. The reason for the heterogeneous responses to visual stimulation therefore remains unclear. These results demonstrate that exogenous NR2B expression prevents cells from exhibiting normal experience-dependent structural plasticity, suggesting that recombinant NR2B expression restricts dendritic arbor growth and branch clustering in response to sensory input.
Figure 5.
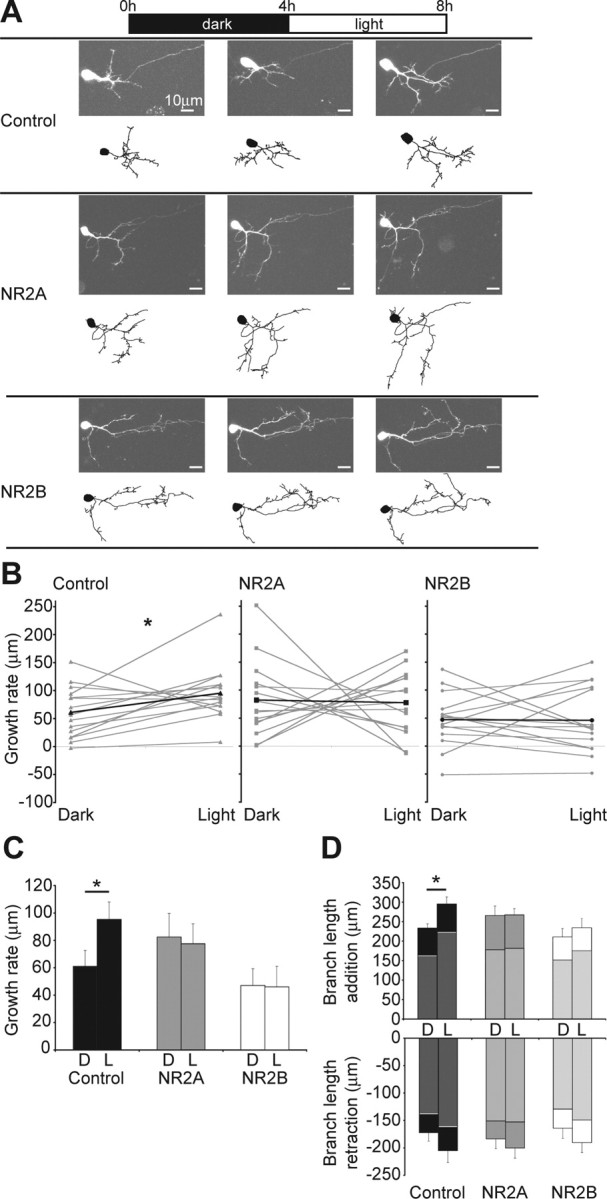
Exogenous expression of NR2B blocks an activity-dependent increase in growth rate. A, Images and drawings of representative neurons imaged 1 d after single-cell electroporation. The neurons were imaged before and after a 4 h period in the dark and then after a 4 h period of visual stimulation. Axons present in the images are not shown in the drawings. B, Growth rates in the dark and light of all analyzed cells are shown in gray with the average highlighted in black. C, Average growth rates for the groups of cells imaged in the dark and in the light [dark and light, respectively (in μm): control, 60.99 ± 11.67, 95.27 ± 12.73, *p < 0.05; NR2A, 82.35 ± 17.36, 77.53 ± 14.58; NR2B, 46.98 ± 12.27, 45.92 ± 15.18; n = 15 cells per group]. D, Total branch length additions and retractions in the dark and in the light. The average branch length added by new branches or retracted through the loss of old branches is highlighted in gray. The remaining difference to the total represents the branch length that was added or retracted through extension or shortening of existing branches [dark and light, respectively (in μm): total branch length addition, control, 162.72 ± 10.55, 223.17 ± 21.06; NR2A, 177.59 ± 15.40, 181.56 ± 15.42; NR2B, 151.4 ± 18.61, 174.95 ± 20.63, *p < 0.02; total branch length retraction, control, −172.31 ± 15.4, −204.97 ± 21.92; NR2A, −183.67 ± 17.82, −200.23 ± 18.61; NR2B, −164.04 ± 18.65, −190.47 ± 18.19]. D, Dark, L, light. Error bars are SEM. The Wilcoxon signed-rank test was used to compare growth rates and the Mann–Whitney U test to compare branch length additions and retractions.
Shifting NMDAR subunit composition weakens synaptic strength
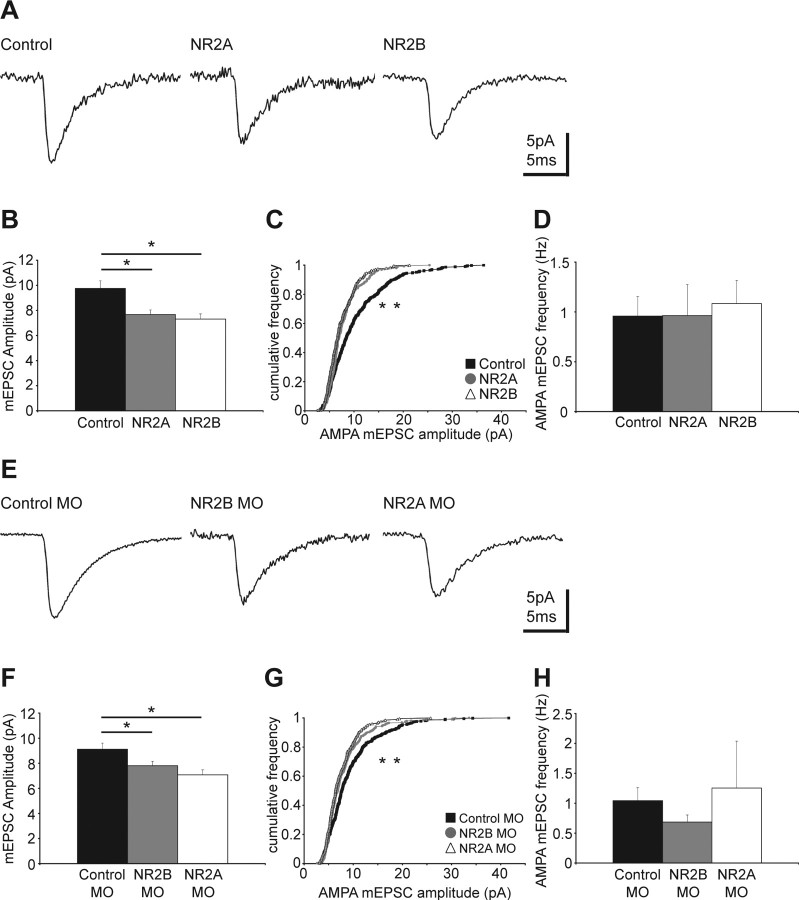
We investigated whether exogenous expression or targeted knockdown of NR2A and NR2B subunits impacts glutamatergic synaptic transmission. AMPA receptor (AMPAR) trafficking plays a key role in regulating synaptic strength and is at least partially dependent on NMDAR activity (Malinow and Malenka, 2002). We therefore recorded AMPAR-mediated miniature EPSCs (mEPSCs) and found that exogenous expression of either NR2A or NR2B significantly reduces AMPA mEPSC amplitudes by ∼21–25%, respectively, compared with control cells (Fig. 6A–C). We did not observe a significant change in the average AMPA mEPSC frequency (Fig. 6D). Similarly, knockdown of the endogenous NR2B and NR2A subunits significantly decreases AMPA mEPSC amplitude by ∼14–22%, respectively, compared with cells electroporated with control MO (Fig. 6E–G). Again the average frequency of the events was not significantly different between groups (Fig. 6H). The reduction in AMPA mEPSC amplitude without a change in frequency indicates that the number of synaptic AMPARs is reduced whereas the number of synapses is unchanged. Therefore, glutamatergic synaptic strength is weakened in neurons that have a shifted NMDAR composition.
Figure 6.
Exogenous expression and knockdown of NR2A and NR2B decrease AMPAR-mediated synaptic transmission. A, Representative traces of AMPAR-mediated mEPSCs in cells expressing exogenous NR2A or NR2B. B, The average AMPA mEPSC amplitudes of exogenous NR2A- and NR2B-expressing neurons are smaller compared with controls (control, 9.75 ± 0.61 pA; NR2A, 7.67 ± 0.37 pA, *p < 0.02; NR2B 7.31 ± 0.41 pA, *p < 0.01; n = 12–20 per group). C, Cumulative frequency plot of the AMPA mEPSC amplitudes shows the distribution toward smaller amplitudes in exogenous NR2-expressing cells (*p < 0.001 for both NR2A and NR2B vs control). D, The average frequency of AMPA mEPSC events is similar between control and exogenous NR2A- and NR2B-expressing cells (control, 0.96 ± 0.2 Hz; NR2A, 0.96 ± 0.31 Hz; NR2B, 1.08 ± 0.23 Hz). E, Representative traces of AMPA mEPSCs in cells expressing control-, NR2B- and NR2A-morpholinos. F, AMPA mEPSC amplitudes are reduced in NR2B MO and NR2A MO expressing cells (control MO, 9.12 ± 0.49 pA; NR2B MO, 7.80 ± 0.35 pA, *p < 0.05; NR2A MO, 7.08 ± 0.39 pA, *p < 0.01; n = 10–20 per group). G, Cumulative frequency plot of the AMPA mEPSC amplitudes shows the distribution toward smaller values for neurons expressing NR2B MO (*p < 0.001) and NR2A MO (*p < 0.001). H, The average frequency of AMPA mEPSC events is not significantly different between cells expressing control-, NR2A-, or NR2B-MO (control MO, 1.04 ± 0.22 Hz; NR2B MO, 0.68 ± 0.12 Hz; NR2A MO, 1.25 ± 0.79 Hz). Error bars are SEM. The Mann–Whitney U test was used to compare the amplitudes and frequencies, and the Kolmogorov–Smirnov test to compare the cumulative frequency plots.
Discussion
Shifting NMDAR subunit composition and its effect on structural plasticity
We manipulated NMDAR subunit composition directly by exogenous expression or knockdown of NR2A or NR2B subunits. Electrophysiological recordings demonstrated that evoked NMDAR-mediated synaptic currents were shifted to be more NR2A- or more NR2B-like. In control neurons, when synaptic NMDAR composition is regulated by endogenous mechanisms, dendritic arbors develop areas of high local branch density. In contrast, exogenous expression of either NR2A or NR2B decreases the formation of local branch clusters, despite differences in synaptic NMDAR-mediated currents. These observations suggest that a shift in NMDAR composition by exogenous expression abrogates the ability of the cell to establish local branch clusters that is normally achieved by signaling through endogenous receptors composed of both NR2A and NR2B. Knockdown of NR2A leaves synaptic responses dominated by NR2B; structurally these dendrites do not develop areas of high branch density. In contrast, knockdown of NR2B results in endogenous NR2A becoming the main component of synaptic NMDARs. This is the only manipulation that results in dendritic arbors with areas of high branch density comparable with controls, and is consistent with the observation that synaptic NMDAR are predominantly mediated by NR2A-containing receptors. It is surprising that the morphological effect of NR2B knockdown does not match the effect seen with NR2A expression. However, the morphological phenotype is likely a read-out of multiple intracellular signaling events impacting neuronal development over the 3-d period of the experiment. This suggests that the mechanisms guiding the development of the dendritic arbor are more intricate than direct signaling from NMDARs to a uniform downstream signaling pathway. Shifting the NMDAR composition by either exogenous expression or MO-mediated knockdown could have different effects on cellular processes like synapse development, receptor trafficking, postsynaptic density assembly and signaling within the postsynaptic density that ultimately result in different structural phenotypes.
Functional implications of structural changes in dendritic arbor architecture
The impact of shifting NMDAR subunit composition on the formation of local branch clusters has important functional implications. In the retinotectal projection, axons from neighboring retinal ganglion cells terminate in adjacent and partially overlapping regions of the tectal neuropil, producing a well described topographic map (Sakaguchi and Murphey, 1985). Previous work showed that synaptic inputs stabilize tectal cell dendritic branches (Niell et al., 2004). Furthermore, areas with more convergent coactive inputs have enhanced dendritic branching, for example, in the optic tectum and in the columnar organization of the barrel and visual cortex (Harris and Woolsey, 1981; Katz and Constantine-Paton, 1988; Katz et al., 1989; Kossel et al., 1995). Indeed, experiments examining axonal arbor refinement have shown that correlated activity promotes branch stabilization, whereas uncorrelated activity promotes branch elimination, and both dynamic processes are NMDAR-dependent (Ruthazer et al., 2003). Here, we show that neurons expressing exogenous NR2 lack local areas of clustered dendritic branching. As a result, these cells are less likely to form synapses with convergent coactive axons, and more likely to form synapses with axons from more disparate parts of the retina. Furthermore, modeling studies suggest that the location of coactive synapses and NMDAR-mediated synaptic responses play a significant role in determining the response of a neuron to synaptic inputs (Mel, 1993; Niell, 2005). This suggests that these cells are not receiving topographically organized inputs, consistent with their inability to show experience-dependent structural plasticity. Therefore the development of local branch clusters in neuronal dendritic arbors likely reflects the recognition of and responses to patterns of synaptic inputs.
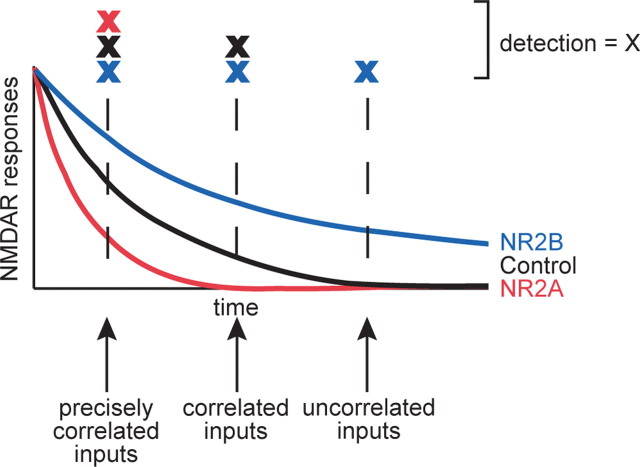
NMDARs as coincidence detectors
The NMDAR functions as a coincidence detector in which the decay time of the synaptic current is thought to determine the time window over which inputs would be recognized as coincident. Receptors with NR1/NR2A or NR1/NR2B subunit composition decrease or increase this integration window, respectively. The long decay time of NR1/NR2B receptors allows detection of relatively uncorrelated inputs, whereas the short decay time of NR1/NR2A receptors would detect only precisely coincident inputs (Fig. 7). Control cells with NR1/NR2A/NR2B subunit composition detect a wider range of correlated inputs (Fig. 7), which may be important for the development of clusters of dendritic branches. Our data suggest that heterotrimeric receptors promote the development of dendritic arbors with local branch clusters.
Figure 7.
NMDARs as coincidence detectors. Model of the influence of the NR2A and NR2B subunits on the NMDAR as a coincidence detector. NR2A confers a short, NR2B a long integration window for detection of coincident inputs, whereas control cells with signaling through both receptors have an intermediate window. We suggest that the ability of control cells to detect a wider range of correlated inputs that carry useful information for the neuron is critical for the establishment of normal dendritic arbor structure. The detection of inputs is marked by X.
NMDAR subunit composition and experience-dependent structural plasticity
Neurons that express exogenous NR2A and NR2B fail to show a consistent increase in dendritic growth rate with visual stimulation, suggesting a deficiency in the proper integration of synaptic information. This is consistent with the idea that exogenous expression of NR2A and NR2B impairs the neuron's ability to distinguish signal from noise. Visual stimulation produces highly correlated synaptic activity. NR2A-expressing neurons may only respond to precisely correlated inputs, therefore extensions into areas of high afferent coactivity would be expected to promote increased growth rates, whereas branching into areas of low coactivity would lead to branch elimination, consistent with the heterogeneous growth rates seen with visual stimulation in these cells. NR2B-expressing neurons may not distinguish changes in synaptic coactivity with visual stimulation compared with dark, resulting in a similar growth response under both conditions. Consequently, these data support a model where NR2A and NR2B together fine tune NMDAR responses.
NMDAR subunit composition and branch dynamics
Examination of dynamic branch behavior shows that NR2A- and NR2B-containing NMDARs have preferred functional roles in the structural development of a neuron. Analysis of branch additions and losses indicates that exogenous NR2B-expressing neurons have more transient branches with shorter life times than control and recombinant NR2A-expressing cells. Normally, NR2B-containing receptors predominate early in development (Carmignoto and Vicini, 1992; Hestrin, 1992; Monyer et al., 1994; Cline et al., 1996; Roberts and Ramoa, 1999), perhaps enabling immature neurons to detect temporally imprecise inputs. The branch dynamics of exogenous NR2B-expressing neurons are reminiscent of the rapid and dynamic growth characteristic of immature neurons (Wu et al., 1999), where exploratory branching predominates over branch stabilization. As maturing neurons add NR2A-containing receptors, NMDAR decay kinetics tighten, and like exogenous NR2A-expressing neurons, mature cells show less dynamic behavior (Wu et al., 1999), suggesting a preferred role for NR2A in branch stabilization. Support of this idea is also provided by our NR2A knockdown experiments, suggesting that NR2A promotes branch clustering.
Downstream signaling candidates of NMDARs
Shifting the NMDAR composition by either exogenous expression or knockdown weakens AMPAR-mediated synaptic transmission, indicating that signaling through both subunits is important for the establishment of AMPAR-mediated glutamatergic transmission because neither has a subunit-specific effect on AMPAR-mediated transmission. This result also suggests that NMDARs may be important for at least two distinct events: the establishment of AMPAR-mediated transmission and the organization of dendritic branch clusters. Previous work demonstrated that AMPAR-mediated synaptic transmission is required for the stabilization of newly added dendritic branches (Haas et al., 2006), and therefore normal arbor development. It is unlikely that the effect of NMDARs on arbor development that we report here is an indirect effect of impaired AMPAR transmission because neurons with NR2B knocked-down acquire a dendritic arbor with normal local branch clusters despite reduced AMPAR-mediated transmission.
Several downstream targets of NMDAR-signaling that might affect structural plasticity have been identified, including CaMKII and Rho GTPases. Expression of constitutively active CaMKII in tectal neurons decreases dendritic growth rate and stabilizes the dendritic arbor (Wu and Cline, 1998), whereas inhibiting CaMKII increases dendritic growth rate (Wu and Cline, 1998; Zou and Cline, 1999). NMDAR subunit composition does not appear to affect overall dendritic growth rate, suggesting that CaMKII signaling is not a predominant player in the structural phenotype we observe. The Rho family of small GTPases are also downstream targets of NMDAR-signaling that control cytoskeletal rearrangements and dendritic arbor development in optic tectal neurons (Li et al., 2000, 2002; Sin et al., 2002). Previous studies demonstrated that NMDAR-mediated synaptic activity enhances dendritic arbor growth by decreasing RhoA activity and increasing Rac and Cdc42 activity (Li et al., 2000, 2002; Sin et al., 2002). Although the structural modifications we report here must include cytoskeletal rearrangements, there is no clear correlation between the structural effects of changing NMDAR subunit composition and effects of Rho GTPase activity. Despite reports of preferential interactions of several postsynaptic density proteins with either NR2A or NR2B (Sans et al., 2000; Kim et al., 2005; Al-Hallaq et al., 2007), subunit-specific signaling pathways that link individual NMDAR subunits to structural plasticity effectors have not yet been characterized.
In conclusion, our results support aspects of two models that have been proposed for the functional roles of NR2A and NR2B. One model explaining experience-dependent synaptic plasticity focuses on the dynamic regulation of the ratio of NR2A and NR2B (Bear, 2003). This is consistent with our evidence that signaling through both NR2A and NR2B is required for normal dendritic arbor development and experience-dependent structural plasticity, supporting the concept that the relative ratio of NR2A and NR2B is important for fine-tuning NMDAR responses. The second model assigns distinct functions to NR2A and NR2B underlying synaptic plasticity (Liu et al., 2004; Massey et al., 2004; Barria and Malinow, 2005; Kim et al., 2005), and we find subunit-specific differences, for example, in the regulation of branch dynamics. Our study suggests that features of both models regulate dendritic arbor development. Furthermore, our results emphasize the interdependence of receptors and signaling molecules such as NR2A and NR2B and stress the importance of analyzing single molecules within the complex context of their intact biological environment.
Footnotes
This work was supported by the National Institutes of Health and a Watson School of Biological Sciences Engelhorn Fellowship. We thank past and present members of the Cline laboratory for valuable discussions.
References
- Aizenman CD, Cline HT. Enhanced visual activity in vivo forms nascent synapses in the developing retinotectal projection. J Neurophysiol. 2007;97:2949–2957. doi: 10.1152/jn.00452.2006. [DOI] [PubMed] [Google Scholar]
- Aizenman CD, Munoz-Elias G, Cline HT. Visually driven modulation of glutamatergic synaptic transmission is mediated by the regulation of intracellular polyamines. Neuron. 2002;34:623–634. doi: 10.1016/s0896-6273(02)00674-8. [DOI] [PubMed] [Google Scholar]
- Al-Hallaq RA, Conrads TP, Veenstra TD, Wenthold RJ. NMDA di-heteromeric receptor populations and associated proteins in rat hippocampus. J Neurosci. 2007;27:8334–8343. doi: 10.1523/JNEUROSCI.2155-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. doi: 10.1016/j.neuron.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Bear MF. Bidirectional synaptic plasticity: from theory to reality. Philos Trans R Soc Lond B Biol Sci. 2003;358:649–655. doi: 10.1098/rstb.2002.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich S, Punnakkal P, Jensen V, Pawlak V, Seeburg PH, Hvalby O, Kohr G. Lack of NMDA receptor subtype selectivity for hippocampal long-term potentiation. J Neurosci. 2005;25:6907–6910. doi: 10.1523/JNEUROSCI.1905-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman JE, Ewald RC, Chiu SL, Cline HT. In vivo single-cell electroporation for transfer of DNA and macromolecules. Nat Protoc. 2006;1:1267–1272. doi: 10.1038/nprot.2006.186. [DOI] [PubMed] [Google Scholar]
- Carmignoto G, Vicini S. Activity-dependent decrease in NMDA receptor responses during development of the visual cortex. Science. 1992;258:1007–1011. doi: 10.1126/science.1279803. [DOI] [PubMed] [Google Scholar]
- Cline HT. Dendritic arbor development and synaptogenesis. Curr Opin Neurobiol. 2001;11:118–126. doi: 10.1016/s0959-4388(00)00182-3. [DOI] [PubMed] [Google Scholar]
- Cline HT, Wu GY, Malinow R. In vivo development of neuronal structure and function. Cold Spring Harb Symp Quant Biol. 1996;61:95–104. [PubMed] [Google Scholar]
- Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE. 2004;2004:re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- Dailey ME, Smith SJ. The dynamics of dendritic structure in developing hippocampal slices. J Neurosci. 1996;16:2983–2994. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint AC, Maisch US, Weishaupt JH, Kriegstein AR, Monyer H. NR2A subunit expression shortens NMDA receptor synaptic currents in developing neocortex. J Neurosci. 1997;17:2469–2476. doi: 10.1523/JNEUROSCI.17-07-02469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas K, Sin WC, Javaherian A, Li Z, Cline HT. Single-cell electroporation for gene transfer in vivo. Neuron. 2001;29:583–591. doi: 10.1016/s0896-6273(01)00235-5. [DOI] [PubMed] [Google Scholar]
- Haas K, Jensen K, Sin WC, Foa L, Cline HT. Targeted electroporation in Xenopus tadpoles in vivo–from single cells to the entire brain. Differentiation. 2002;70:148–154. doi: 10.1046/j.1432-0436.2002.700404.x. [DOI] [PubMed] [Google Scholar]
- Haas K, Li J, Cline HT. AMPA receptors regulate experience-dependent dendritic arbor growth in vivo. Proc Natl Acad Sci USA. 2006;103:12127–12131. doi: 10.1073/pnas.0602670103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RM, Woolsey TA. Dendritic plasticity in mouse barrel cortex following postnatal vibrissa follicle damage. J Comp Neurol. 1981;196:357–376. doi: 10.1002/cne.901960302. [DOI] [PubMed] [Google Scholar]
- Hestrin S. Developmental regulation of NMDA receptor-mediated synaptic currents at a central synapse. Nature. 1992;357:686–689. doi: 10.1038/357686a0. [DOI] [PubMed] [Google Scholar]
- Iwasato T, Erzurumlu RS, Huerta PT, Chen DF, Sasaoka T, Ulupinar E, Tonegawa S. NMDA receptor-dependent refinement of somatotopic maps. Neuron. 1997;19:1201–1210. doi: 10.1016/s0896-6273(00)80412-2. [DOI] [PubMed] [Google Scholar]
- Iwasato T, Datwani A, Wolf AM, Nishiyama H, Taguchi Y, Tonegawa S, Knopfel T, Erzurumlu RS, Itohara S. Cortex-restricted disruption of NMDAR1 impairs neuronal patterns in the barrel cortex. Nature. 2000;406:726–731. doi: 10.1038/35021059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaherian A, Cline HT. Coordinated motor neuron axon growth and neuromuscular synaptogenesis are promoted by CPG15 in vivo. Neuron. 2005;45:505–512. doi: 10.1016/j.neuron.2004.12.051. [DOI] [PubMed] [Google Scholar]
- Katz LC, Constantine-Paton M. Relationships between segregated afferents and postsynaptic neurones in the optic tectum of three-eyed frogs. J Neurosci. 1988;8:3160–3180. doi: 10.1523/JNEUROSCI.08-09-03160.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, Gilbert CD, Wiesel TN. Local circuits and ocular dominance columns in monkey striate cortex. J Neurosci. 1989;9:1389–1399. doi: 10.1523/JNEUROSCI.09-04-01389.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew JN, Richards JG, Mutel V, Kemp JA. Developmental changes in NMDA receptor glycine affinity and ifenprodil sensitivity reveal three distinct populations of NMDA receptors in individual rat cortical neurons. J Neurosci. 1998;18:1935–1943. doi: 10.1523/JNEUROSCI.18-06-01935.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Dunah AW, Wang YT, Sheng M. Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron. 2005;46:745–760. doi: 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Kohr G, Jensen V, Koester HJ, Mihaljevic AL, Utvik JK, Kvello A, Ottersen OP, Seeburg PH, Sprengel R, Hvalby O. Intracellular domains of NMDA receptor subtypes are determinants for long-term potentiation induction. J Neurosci. 2003;23:10791–10799. doi: 10.1523/JNEUROSCI.23-34-10791.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossel A, Lowel S, Bolz J. Relationships between dendritic fields and functional architecture in striate cortex of normal and visually deprived cats. J Neurosci. 1995;15:3913–3926. doi: 10.1523/JNEUROSCI.15-05-03913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Direct imaging of in vivo neuronal migration in the developing cerebellum. Curr Biol. 2001;11:1858–1863. doi: 10.1016/s0960-9822(01)00585-1. [DOI] [PubMed] [Google Scholar]
- Krapivinsky G, Krapivinsky L, Manasian Y, Ivanov A, Tyzio R, Pellegrino C, Ben-Ari Y, Clapham DE, Medina I. The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron. 2003;40:775–784. doi: 10.1016/s0896-6273(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Kutsuwada T, Sakimura K, Manabe T, Takayama C, Katakura N, Kushiya E, Natsume R, Watanabe M, Inoue Y, Yagi T, Aizawa S, Arakawa M, Takahashi T, Nakamura Y, Mori H, Mishina M. Impairment of suckling response, trigeminal neuronal pattern formation, and hippocampal LTD in NMDA receptor epsilon 2 subunit mutant mice. Neuron. 1996;16:333–344. doi: 10.1016/s0896-6273(00)80051-3. [DOI] [PubMed] [Google Scholar]
- Lee LJ, Lo FS, Erzurumlu RS. NMDA receptor-dependent regulation of axonal and dendritic branching. J Neurosci. 2005;25:2304–2311. doi: 10.1523/JNEUROSCI.4902-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Erzurumlu RS, Chen C, Jhaveri S, Tonegawa S. Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell. 1994;76:427–437. doi: 10.1016/0092-8674(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Li Z, Van Aelst L, Cline HT. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci. 2000;3:217–225. doi: 10.1038/72920. [DOI] [PubMed] [Google Scholar]
- Li Z, Aizenman CD, Cline HT. Regulation of rho GTPases by crosstalk and neuronal activity in vivo. Neuron. 2002;33:741–750. doi: 10.1016/s0896-6273(02)00621-9. [DOI] [PubMed] [Google Scholar]
- Liu L, Wong TP, Pozza MF, Lingenhoehl K, Wang Y, Sheng M, Auberson YP, Wang YT. Role of NMDA receptor subtypes in governing the direction of hippocampal synaptic plasticity. Science. 2004;304:1021–1024. doi: 10.1126/science.1096615. [DOI] [PubMed] [Google Scholar]
- Lu HC, Gonzalez E, Crair MC. Barrel cortex critical period plasticity is independent of changes in NMDA receptor subunit composition. Neuron. 2001;32:619–634. doi: 10.1016/s0896-6273(01)00501-3. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- Massey PV, Johnson BE, Moult PR, Auberson YP, Brown MW, Molnar E, Collingridge GL, Bashir ZI. Differential roles of NR2A and NR2B-containing NMDA receptors in cortical long-term potentiation and long-term depression. J Neurosci. 2004;24:7821–7828. doi: 10.1523/JNEUROSCI.1697-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mel BW. Synaptic integration in an excitable dendritic tree. J Neurophysiol. 1993;70:1086–1101. doi: 10.1152/jn.1993.70.3.1086. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Morikawa E, Mori H, Kiyama Y, Mishina M, Asano T, Kirino T. Attenuation of focal ischemic brain injury in mice deficient in the epsilon1 (NR2A) subunit of NMDA receptor. J Neurosci. 1998;18:9727–9732. doi: 10.1523/JNEUROSCI.18-23-09727.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita W, Lu W, Smith GB, Nicoll RA, Bear MF, Malenka RC. Activation of NR2B-containing NMDA receptors is not required for NMDA receptor-dependent long-term depression. Neuropharmacology. 2007;52:71–76. doi: 10.1016/j.neuropharm.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Neyton J, Paoletti P. Relating NMDA receptor function to receptor subunit composition: limitations of the pharmacological approach. J Neurosci. 2006;26:1331–1333. doi: 10.1523/JNEUROSCI.5242-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niell CM. Theoretical analysis of a synaptotropic dendrite growth mechanism. J Theor Biol. 2005 doi: 10.1016/j.jtbi.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Niell CM, Meyer MP, Smith SJ. In vivo imaging of synapse formation on a growing dendritic arbor. Nat Neurosci. 2004;7:254–260. doi: 10.1038/nn1191. [DOI] [PubMed] [Google Scholar]
- Philpot BD, Cho KK, Bear MF. Obligatory Role of NR2A for Metaplasticity in Visual Cortex. Neuron. 2007;53:495–502. doi: 10.1016/j.neuron.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan I, Cline HT. Glutamate receptor activity is required for normal development of tectal cell dendrites in vivo. J Neurosci. 1998;18:7836–7846. doi: 10.1523/JNEUROSCI.18-19-07836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan I, Witte S, Cline HT. NMDA receptor activity stabilizes presynaptic retinotectal axons and postsynaptic optic tectal cell dendrites in vivo. J Neurobiol. 1999;38:357–368. doi: 10.1002/(sici)1097-4695(19990215)38:3<357::aid-neu5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Roberts EB, Ramoa AS. Enhanced NR2A subunit expression and decreased NMDA receptor decay time at the onset of ocular dominance plasticity in the ferret. J Neurophysiol. 1999;81:2587–2591. doi: 10.1152/jn.1999.81.5.2587. [DOI] [PubMed] [Google Scholar]
- Ruthazer ES, Cline HT. Insights into activity-dependent map formation from the retinotectal system: a middle-of-the-brain perspective. J Neurobiol. 2004;59:134–146. doi: 10.1002/neu.10344. [DOI] [PubMed] [Google Scholar]
- Ruthazer ES, Akerman CJ, Cline HT. Control of axon branch dynamics by correlated activity in vivo. Science. 2003;301:66–70. doi: 10.1126/science.1082545. [DOI] [PubMed] [Google Scholar]
- Sakaguchi DS, Murphey RK. Map formation in the developing Xenopus retinotectal system: an examination of ganglion cell terminal arborizations. J Neurosci. 1985;5:3228–3245. doi: 10.1523/JNEUROSCI.05-12-03228.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos J, II, Hell JW, Wenthold RJ. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–1271. doi: 10.1523/JNEUROSCI.20-03-01260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Aamodt SM, Constantine-Paton M. Temporal correlations between functional and molecular changes in NMDA receptors and GABA neurotransmission in the superior colliculus. J Neurosci. 1997;17:6264–6276. doi: 10.1523/JNEUROSCI.17-16-06264.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419:475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- Sobczyk A, Scheuss V, Svoboda K. NMDA receptor subunit-dependent [Ca2+] signaling in individual hippocampal dendritic spines. J Neurosci. 2005;25:6037–6046. doi: 10.1523/JNEUROSCI.1221-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YP, Shimizu E, Dube GR, Rampon C, Kerchner GA, Zhuo M, Liu G, Tsien JZ. Genetic enhancement of learning and memory in mice. Nature. 1999;401:63–69. doi: 10.1038/43432. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Zhao MG, Zhuo M. Roles of NMDA receptor NR2A and NR2B subtypes for long-term depression in the anterior cingulate cortex. Eur J Neurosci. 2005;22:485–494. doi: 10.1111/j.1460-9568.2005.04236.x. [DOI] [PubMed] [Google Scholar]
- Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR. Functional and pharmacological differences between recombinant N-methyl-d-aspartate receptors. J Neurophysiol. 1998;79:555–566. doi: 10.1152/jn.1998.79.2.555. [DOI] [PubMed] [Google Scholar]
- Weitlauf C, Honse Y, Auberson YP, Mishina M, Lovinger DM, Winder DG. Activation of NR2A-containing NMDA receptors is not obligatory for NMDA receptor-dependent long-term potentiation. J Neurosci. 2005;25:8386–8390. doi: 10.1523/JNEUROSCI.2388-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. Ifenprodil, a novel NMDA receptor antagonist: site and mechanism of action. Curr Drug Targets. 2001;2:285–298. doi: 10.2174/1389450013348489. [DOI] [PubMed] [Google Scholar]
- Wu G, Malinow R, Cline HT. Maturation of a central glutamatergic synapse. Science. 1996;274:972–976. doi: 10.1126/science.274.5289.972. [DOI] [PubMed] [Google Scholar]
- Wu GY, Cline HT. Stabilization of dendritic arbor structure in vivo by CaMKII. Science. 1998;279:222–226. doi: 10.1126/science.279.5348.222. [DOI] [PubMed] [Google Scholar]
- Wu GY, Zou DJ, Rajan I, Cline H. Dendritic dynamics in vivo change during neuronal maturation. J Neurosci. 1999;19:4472–4483. doi: 10.1523/JNEUROSCI.19-11-04472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou DJ, Cline HT. Postsynaptic calcium/calmodulin-dependent protein kinase II is required to limit elaboration of presynaptic and postsynaptic neuronal arbors. J Neurosci. 1999;19:8909–8918. doi: 10.1523/JNEUROSCI.19-20-08909.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]