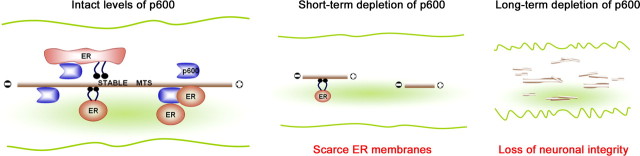
Abstract
There is an increasing body of literature pointing to cytoskeletal proteins as spatial organizers and interactors of organelles. In this study, we identified protein 600 (p600) as a novel microtubule-associated protein (MAP) developmentally regulated in neurons. p600 exhibits the unique feature to interact with the endoplasmic reticulum (ER). Silencing of p600 by RNA interference (RNAi) destabilizes neuronal processes in young primary neurons undergoing neurite extension and containing scarce staining of the ER marker Bip. Furthermore, in utero electroporation of p600 RNAi alters neuronal migration, a process that depends on synergistic actions of microtubule dynamics and ER functions. p600-depleted migrating neurons display thin, crooked, and “zigzag” leading process with very few ER membranes. Thus, p600 constitutes the only known MAP to associate with the ER in neurons, and this interaction may impact on multiple cellular processes ranging from neuronal development to neuronal maturation and plasticity.
Keywords: cytoarchitecture, cytoskeleton, development, differentiation, migration, neurite
Introduction
The rearrangement of the cytoskeleton regulates membrane dynamics and signaling, and synergistically, they orchestrate changes in cell shape, motility, and survival (Lee et al., 2001; Ross and Walsh, 2001; Dent and Gertler, 2003; Ayala et al., 2007). Enriched in neurons, microtubules (MTs) serve as the main tracks for the intracellular transport of vesicles, organelles, and, therefore, play an essential role in neurite extension and neuronal migration. The stability and dynamics of the MT network depends on a set of proteins known as MT-associated proteins (MAPs). Alterations in MAPs trigger neurodevelopmental and neurodegenerative disorders (Lee et al., 2001; Ross and Walsh, 2001; Ayala et al., 2007). Despite our understanding of how MAPs promote cell integrity as building blocks, little is known about their interactions with organelles.
The endoplasmic reticulum (ER) is a well characterized reservoir of Ca2+ in axonal and dendritic compartments of neurons that regulates neuronal development and maturation (Rizzuto, 2001; Verkhratsky, 2004). Transported on MTs, ER membranes can also localize to sites of cellular expansion such as the growth cone and leading process to mediate signaling and promote outgrowth (Terasaki et al., 1986, 1994; Waterman-Storer and Salmon, 1998). Thus far, among tens of MAPs, only one MAP, 63 kDa cytoskeleton-linking membrane protein (CLIMP-63), has been shown to interact with the ER (Klopfenstein et al., 1998, 2001; Vedrenne et al., 2005). This protein is as yet uncharacterized in the CNS.
Protein 600 (p600) encodes for a polypeptide of ≈5184 aa (600 kDa) that interacts with viral oncoproteins and the tumor suppressor retinoblastoma (DeMasi et al., 2005; Huh et al., 2005; Nakatani et al., 2005). By virtue of these associations, p600 is important for anoikis and anchorage-independent neoplastic growth (DeMasi et al., 2005; Huh et al., 2005; Nakatani et al., 2005). However, the basic functions of p600 remain unknown. Interestingly, p600 exhibits homology to BIG in Arabidopsis thaliana, a molecule that regulates intracellular transport (Gil et al., 2001), and to CALO/Pushover in Drosophila melanogaster, a calmodulin (CaM)-binding protein (Xu et al., 1998). Here, we discovered that p600 is a novel MAP expressed in developing and adult CNS neurons. We found that it acts in a unique manner to link the MT cytoskeleton with the ER. We propose that, via these interactions, p600 regulates neuronal differentiation and migration.
Materials and Methods
Western blots
The mice were killed by intraperitoneal injection of chloral hydrate. Total protein extracts of spinal cord and brain were obtained by homogenization in SDS-urea β-mercaptoethanol (0.5% SDS, 8 m urea in phosphate buffer, pH 7.4), or radioimmunoprecipitation assay (RIPA) lysis buffer (10 mm Tris-HCl, pH 7.5; 150 mm NaCl; 1 mm EDTA, pH 8.0; 1% Triton X-100) with a mixture of protease inhibitors (PMSF, leupeptin, pepstatin, apoprotinin) and 1 mm sodium orthovanadate. The protein concentration in tissues and cells was estimated by the Bradford procedure (Bio-Rad Laboratories, Hercules, CA). Proteins (20–50 μg) were fractionated on 6 or 8% SDS-PAGE and blotted on a nitrocellulose or polyvinylidene difluoride membrane for Western blot analysis. Membranes were incubated with antibodies against p600 (F4; F7), focal adhesion kinase (FAK) (C-20; Santa Cruz Biotechnology, Santa Cruz, CA), dynein heavy chain (C-19; S18; Santa Cruz Biotechnology), actin (C4; MAB1501; Millipore Bioscience Research Reagents, Temecula, CA), synaptophysin (MAB329; Millipore Bioscience Research Reagents), p150 (C1; BD Biosciences Transduction Laboratories, Lexington, KY), neurofilament (NF) (RT-97; Millipore Bioscience Research Reagents), α-tubulin (B512; Sigma-Aldrich, St. Louis, MO), Bip (Sigma-Aldrich), Tau1 (Boehringer Ingelheim, Ingelheim, Germany), calcium/CaM-dependent protein kinase IIα (CaMKIIα) (Millipore Bioscience Research Reagents), FLAG (M2; Sigma-Aldrich), tubulin (B512; Sigma-Aldrich), and acetylated tubulin (ab24610; Abcam, Cambridge, MA). Quantifications were corrected with levels of actin, α-tubulin, or FAK, and performed with the Labscan program (Image Master, 2D software, version 3.10; GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Biochemistry
Immunoprecipitations.
Immunoprecipitations were performed according to Nguyen et al. (2004) and standard protocols.
Microtubule preparation.
Brain was homogenized at a ratio of 1 ml of PME buffer (0.1 m PIPES, pH 6.9; 2 nm EGTA; 1 nm MgSO4; 1 mm DTT; 0.5 mm GTP) per gram of brain. The homogenate was then centrifuged at 40,000 × g at 4°C for 45 min. The supernatant containing cold soluble tubulin dimers and MAPs is then incubated at 37°C for 30–45 min to induce the assembly of dimers into MTs. The assembled MTs were then collected by centrifugation at 40,000 × g at 25°C. The resulting pellet of MTs was resuspended in one-fifth the volume of PME. The suspension was then incubated in an ice/water slurry for 30 min to allow the cold-labile MTs (but not insoluble aggregates) to disassemble. The solution was cleared by centrifugation at 40,000 × g at 4°C for 30 min. This supernatant (S) contained once-cycled MTs (i.e., MTs that have been assembled and disassembled once in vitro), whereas the pellet (P1) contained debris and insoluble aggregates. The fraction S is then subjected to two other cycles starting at incubation at 37°C for 30–45 min to induce the assembly of dimers into MTs (see above). The last pellet (P2) recovered after three cycles contained tubulin and MAPs.
Neurofilament preparation.
A neurofilament-enriched fraction was prepared from the spinal cord of C57BL/6 and NF-L null mice by the procedure by Nguyen et al. (2004). A complete spinal cord or one-half of the spinal cord was homogenized in 400 μl of buffer A (0.1 m NaCl; 1 mm EDTA, pH 8.0; 10 mm sodium phosphate, pH 6.5), containing 0.85 m sucrose, using a Dounce homogenizer. The volume was then adjusted to 2 ml with buffer A. The homogenate was centrifuged in a Sorvall with a TLA-55 rotor at 27,000 × g for 15 min, and the floating pad of myelinated axons was collected, re-homogenized in 1.5 ml of buffer B (0.1 m NaCl; 1 mm EDTA, pH 8.0; 10 mm sodium phosphate, pH 6.5; 1% Triton X-100). The final homogenate was then added to 500 μl of buffer B before centrifugation at 27,000 g for 1 h. The pellet enriched in neurofilaments was then resuspended in 50 μl of 1× Laemmli buffer before loading on gels.
Crude synaptosomes preparation.
Mouse brain was homogenized in 7 ml of 10 mm HEPES, 0.32 m sucrose at 1000 × g at 4°C for 10 min. Separated from the pellet (P1), the supernatant (S1) was then centrifuged again for 15 min at 12,000 × g. The resulting pellet (P2) corresponds to the crude synaptosome preparation, whereas the supernatant (S2) corresponds to the cleared soluble fraction.
Membrane preparation.
Spinal cord, brain, or cells were homogenized in 10 vol of 0.25 m sucrose dissolved in ice-cold 10 mm Tris-HCl buffer, pH 7.4, to which a mixture of protease inhibitors was added before use. The homogenate was centrifuged for 15 min at 4°C to remove cell debris. The supernatant was then centrifuged at 105,000 × g for 60 min at 4°C, and the new supernatant was saved. This fraction was referred to as “soluble cytosol.” The remaining pellet was resuspended in the above sucrose-Tris buffer and was referred to as “membrane fraction.”
Crude ER preparation.
Briefly, crude ER microsomes were isolated using a series of centrifugations and homogenization/resuspension in isotonic/hypotonic extraction buffers and calcium chloride solution (for additional details, see protocols in isolation kit of ER0100; Sigma-Aldrich). One of the intermediate fractions, called the postmitochondrial fraction, is the source of ER microsomes. The postmitochondrial fraction was then centrifuged at 100,000 × g for 60 min at 4°C. The pellet that contains the ER fraction was then washed and dissolved 8 m urea and 0.5% SDS for Western blot analysis.
Cell culture, transfection, immunofluorescence
CAD cells were cultured and transfected with the seven truncated fragments of p600 with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to Nguyen et al. (2004). These fragments named A to G were cloned in the pFLAG-CMV2 vector. In serum-free medium, CAD cells stop proliferating and exhibit morphological characteristics and molecular signatures of primary neurons such as expression of class III β-tubulin and enzymatically active tyrosine hydroxylase (Nguyen et al., 2004). Cortical neurons were isolated and cultured according to Xie et al. (2003). Neurons were transfected with Lipofectamine 2000 (Invitrogen) or electroporated with the nucleotransfection device (Amaxa Biosystems, Gaithersburg, MD). After 2–3 d in culture, cells were lysed in RIPA or 0.5% SDS, 8 m urea buffer for Western blot or fixed in 4% paraformaldehyde for immunoassaying. Staining of cells and mouse brain tissues were performed according to Nguyen et al. (2004). Antibodies used are p600 (F4; F7), Tuj1 (Covance Research Products, Princeton, NJ), NF (RT-97; N-52; NR-4; MAB1983; Roche, Basel, Switzerland; Boehringer Ingelheim; Millipore Bioscience Research Reagents), Bip (Sigma-Aldrich), inositol trisphosphate receptor (IP3R) (Santa Cruz Biotechnology), green fluorescent protein (GFP) (B2; Santa Cruz Biotechnology; Invitrogen; A-6455), Cy3-tubulin (Sigma-Aldrich), tubulin (B512; Sigma-Aldrich), and FLAG (M2; Sigma-Aldrich). Confocal microscopy photos were taken with a Nikon (Tokyo, Japan) Eclipse 2000 and Carl Zeiss (Jena, Germany) LS510.
MT stabilization and immunogold labeling
Primary cortical neurons were cultured on coverslips and fixed with prewarmed 4% paraformaldehyde/0.025% glutaraldehyde in Brinkley buffer 1980 (BRB80) (80 mm PIPES, pH 6.8; 1 mm MgCl2; 1 mm EGTA) plus 5 μg/ml Taxol for 10–20 min at 37°C. Samples were then rinsed three times with BRB80 and twice with BRB80 (+0.1% Triton X-100, 2% BSA, 0.1% azide) before permeabilization with 0.1% Triton X-100 in BRB80 plus 5 μg/ml Taxol. Samples were blocked in PBS/1% BSA plus 0.1% Triton X-100 for 30 min at room temperature (RT). Primary antibody was incubated overnight at 4°C followed by two washes with PBS/1% BSA plus 0.1% Triton X-100 and three washes with BRB80 over 30 min. The gold labeling was performed with protein A-gold (5 nm) diluted 1:70 in PBS/1% BSA plus 0.1% Triton X-100 for 2 h at RT. All antibodies and protein A-gold are diluted in 1% BSA. Antibody dilutions were centrifuged at 14,000 rpm for 1 min before labeling to avoid aggregation. After labeling, samples were washed before fixation with 2.5% glutaraldehyde/0.1 m cacodylate buffer, pH 7.4, for 30 min at RT. Coverslips were then embedded in Epon before analysis under electron microscope.
Microtubule polymerization assay
Lyophilized tubulin from Cytoskeleton (Denver, CO) was reconstituted in G-PEM buffer at 4°C, as described by the manufacturer, and centrifuged to remove tubulin aggregates. To perform the in vitro tubulin assembly assay, 1 mg/ml tubulin (100 μg) was added to 30 μg of each purified fragment of p600 in G-PEM buffer. Tubulin assembly was monitored once room temperature was reached. The turbidity measurement of tubulin polymerization was monitored in an Ultrospec 3100 Pro spectrophotometer at 340 nm.
Generation, characterization of RNA interference vectors and in utero electroporation
RNA interference (RNAi) sequences were selected based on the criteria of Ambion (Austin, TX). Complementary hairpin sequences were commercially synthesized and cloned into pSilencer 2.0 under promoter U6 (Ambion). Sequences for the mouse p600 RNAi are base pairs GCAGTACGAGCCGTTCTAC and AATGATGAGCAGTCATCTA. A random sequence without homology to any known mRNA was used for control RNAi. All RNAi constructs were tested in CAD cells, primary neuronal cultures by both Western blots and immunofluorescence staining. In utero electroporation was performed according to Sanada and Tsai (2005). The enhanced GFP (EGFP)-expressing plasmid (final concentration, 2 μg/μl; pCAG-IRES-GFP) was coinjected with the RNAi construct (final concentration, 5 μg/μl) at a concentration ratio of 2:5. For analysis of ER distribution in migrating neurons, the DsRed2-expressing plasmid (2 μg/μl; pCAG-DsRed2), the ER-targeted EGFP-expressing plasmid (pEF/Myc/ER/GFP; Invitrogen), and the RNAi construct were coinjected at a concentration ratio of 2:5:5. After the injection of the plasmids into the lateral ventricle of the embryos, electric pulses (four 50 ms square pulses of 38 V with 950 ms intervals) were applied to the embryos once or twice. For immunofluorescence, embryonic day 17 (E17) brains were fixed with 4% paraformaldehyde/PBS overnight at 4°C. The brain sections (60 μm in thickness) were prepared with a vibratome. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Images were obtained with a Carl Zeiss LSM510 confocal microscope.
Results
p600 is expressed in developing and adult CNS neurons
To assess the spatiotemporal expression pattern of p600 in the developing and mature CNS, we used two purified rabbit polyclonal antibodies raised against p600 (F4 and F7). F4 encompasses residues 3755–4160, whereas F7 was directed against residues 2656–3017. Both antibodies show similar efficiency and patterns of staining, although F7 is more potent for immunoprecipitation and Western blots; accordingly, they were used as such in the present study. The specificity of the antibodies for staining and Western blots was determined previously by peptide competition experiments (DeMasi et al., 2005; Huh et al., 2005; Nakatani et al., 2005) and, in this study, is confirmed by both Western blots and immunofluorescence staining of tissue depleted of p600 by RNA interference (see below). As demonstrated by Western blots, p600 is expressed in the mouse brain from E15 to postnatal day 1 (P1) (Fig. 1A). Expression increases progressively and up to twofold during brain development, suggesting that p600 is important for neuronal maturation. Confocal microscopy of E15 mouse brain sections double-stained with the early neuronal marker class III β-tubulin (Tuj1) indicate that p600 is enriched in early-born neurons of the intermediate zone and cortical plate (CP) (Fig. 1C). The staining pattern is consistent with the pattern exhibited by migrating neurons of the developing cortex (Moody et al., 1989; Lee et al., 1990). In the adult mouse brain, p600 is expressed in all CNS neurons, including neurons of the cortex and hippocampus (Fig. 1B,D). In situ hybridization further confirms the broad expression of p600 in adult CNS neurons with the highest levels in the pyramidal layer of the hippocampus and granular layer of the dentate gyrus (data not shown). Together, these results suggest that p600 is present in both developing neurons and mature CNS neurons.
Figure 1.
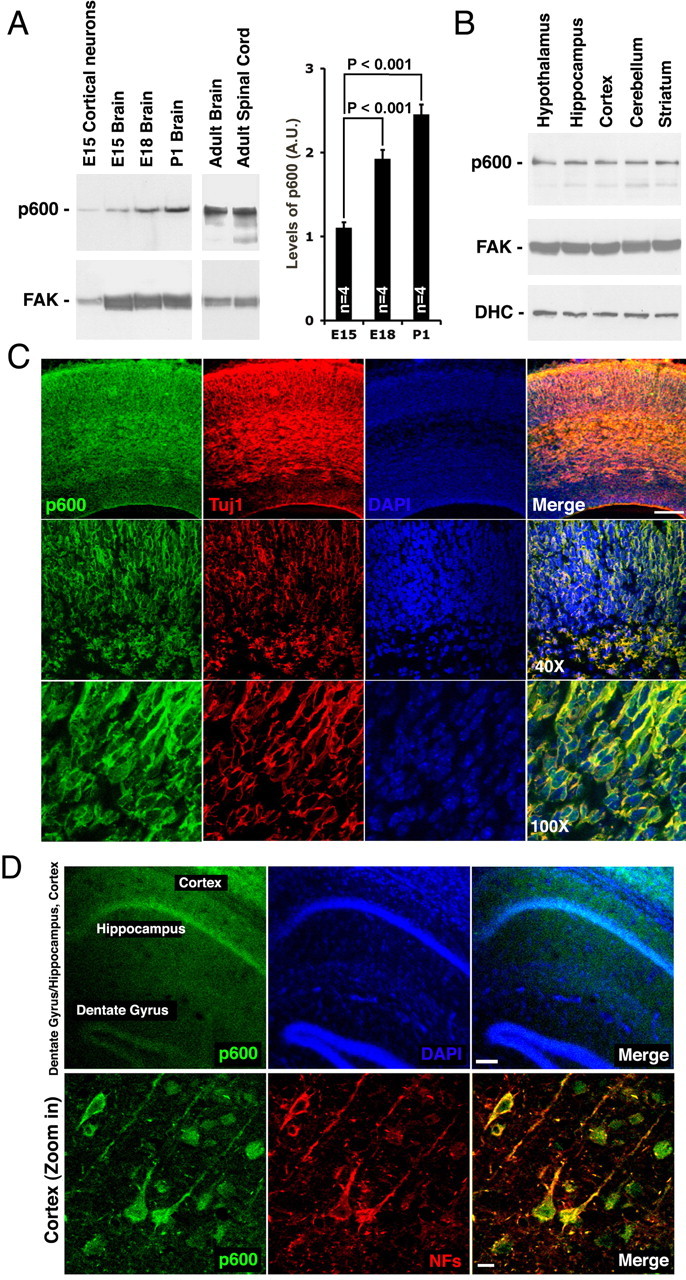
Spatiotemporal expression pattern of p600 in the mouse CNS. A, Expression of p600 protein in primary cortical neurons isolated from mouse at E15, in E15, E18, P1, adult mouse brain and adult mouse spinal cord (12 weeks of age). The n value indicates the number of brains. p < 0.001, T < t, two-tailed. Error bars indicate SD. B, Expression of p600 in specific brain regions of adult mouse. Dynein heavy chain (DHC) and FAK are used as controls for loading in A and B. C, p600 is expressed in early-born neurons of the E15 mouse neocortex as detected by costaining with the early neuronal marker class III β-tubulin (Tuj1). The staining pattern suggests that these p600-positive neurons are migrating neurons. D, p600 is expressed in neurons of the adult mouse brain as revealed by costaining with the mature neuronal marker anti-NFs. All pictures were taken with a confocal microscope. Scale bars: C, 200 μm; D, 50 and 12 μm.
p600 is a novel MAP
To define the subcellular localization of p600 and get insight into its functions, we performed several biochemical fractionations. Crude synaptosome preparations from adult brain revealed the predominance of p600 in the soluble fraction (S2; ∼37%) and lower levels in synaptic compartments (P2; ∼20%), as controlled with the synaptic protein synaptophysin (Fig. 2A). Using membrane preparations, we found an appreciable pool of p600 copurifying with membranes [pellet (P); ∼47%] (Fig. 2B). This association might be explained by the high content of hydrophobic residues (45%).
Figure 2.
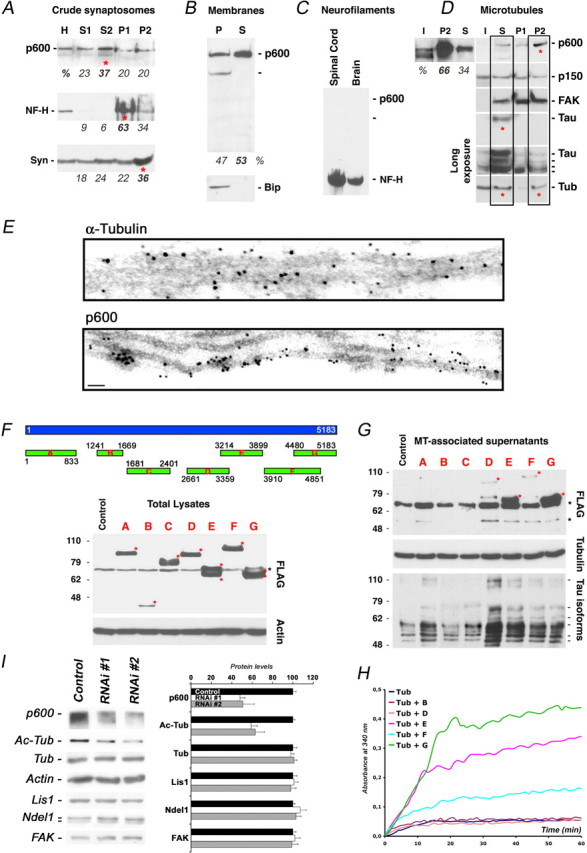
p600 is a novel MAP. A, Amounts of p600 in the cytosolic fraction (S2), pellet 1 (P1), or synaptic compartment (P2). H, Homogenate; S1, preclear lysate. The amount of each protein in each specific fraction is estimated in percentage. B, p600 is a soluble protein and cofractionates with membranes. The ER marker Bip is used as control. C, p600 is not found in preparations of NFs from adult spinal cord and brain. D, p600 is found in the active MT fraction (S) and is enriched in the fraction P2 containing tubulin and MAPs. I, Input; P1, fraction containing insoluble aggregates and debris with no tubulin. E, Immunogold labeling of MTs from cytosol-depleted primary mouse cortical neurons stabilized with Taxol using α-tubulin or p600 antibodies. Scale bar, 50 nm. F, Western blots depicting the expression of the seven FLAG-tagged fragments of p600 (designed A to G) in the neuroblastoma CAD cell line as detected with FLAG antibody (red stars). G, MT preparations from CAD cells transfected with each of one of the seven fragments of p600. Fragments E and G were found in significant amounts in MT preparations (red stars). Tubulin and actin, Loading controls; Tau, positive control; black stars refer to nonspecific bands. H, Bacterially expressed and purified fragments E, F, and G, but not B or D, enhance MT polymerization in vitro as detected by absorbance at 340 nm. Four to six independent assays were performed for each fragment. A representative assay is depicted for each fragment. I, p600 RNAi nos. 1 and 2 efficiently knockdown p600 protein. Knockdown of p600 in CAD cells by RNAi no. 1 or 2 reduces the levels of acetylated tubulin (Ac-Tub), but not tubulin or other cytoskeletal proteins. Five samples for each condition were processed for quantifications. Error bars indicate SDs.
To examine a potential association with the cytoskeleton, we performed preparations of MTs and NFs, the main intermediate filaments in CNS neurons. Interestingly, p600 did not copurify with NFs but was found in MT preparations (Fig. 2C,D). Indeed, p600 was present in the soluble fraction containing active tubulin (S; ∼34%) (i.e., the fraction containing MTs that have been assembled and disassembled three times in vitro (see Materials and Methods) and, importantly, was enriched in the P2 fraction (∼66%) containing MAPs and stable MTs (Fig. 2D). Furthermore, p600 colocalized with MTs in a punctate/vesicular tubular pattern throughout the cytosol of neuroblastoma CAD cells and primary neurons and their neuronal processes (axon and dendrites) (Fig. 3A,B). Furthermore, using immunogold electron microscopy, we found that p600 antibodies coupled with gold particles adorned Taxol-stabilized MT structures in primary hippocampal neurons depleted of cytosol (Fig. 2E). These results indicated that p600 associates with MTs.
Figure 3.
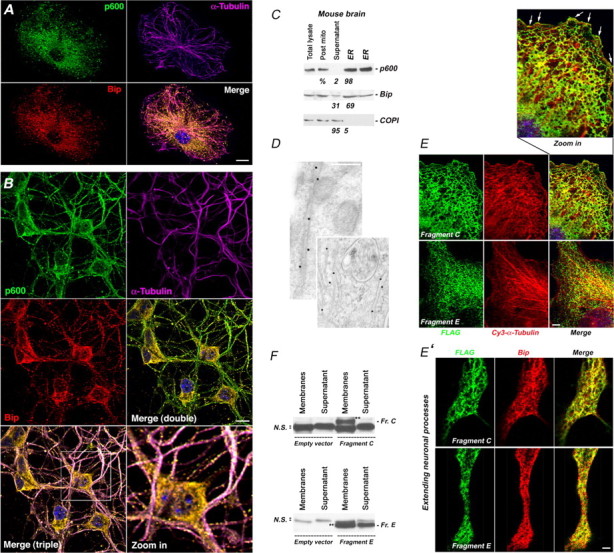
p600 associates with the ER. A, B, Colocalization of p600 with MTs and the ER marker Bip in nondifferentiated CAD cells (A) and primary mouse cortical neurons (B) in a vesicular/punctate pattern. C, Crude microsome ER preparation from adult mouse brains. Post-mito, Postmitochondrial fraction containing ER microsomes before purification; supernatant, fraction discarded before collection of microsomes. The amount of each protein in each fraction is expressed in percentage. D, Immunogold labeling of ER tubules with p600 antibodies. E, Polygonal reticular pattern of peripheral ER tubules exhibited by the fragments C and E in nondifferentiated CAD cells. These ER tubules are juxtaposing MTs up to the cell edge (white arrows). E′, Colocalization of FLAG-tagged fragment C or E with the ER marker Bip in differentiated CAD cells. F, Fragment C (Fr. C) (double black stars) is enriched in the membrane fraction, whereas fragment E (Fr. E) (double black stars) is found in both membrane and nonmembrane fractions. N.S. refers to nonspecific bands. Cells transfected with an empty vector were used as control. All pictures were taken with a confocal microscope. Scale bars: A, E′, 25 μm; B, E, 10 μm; D, 200 nm.
To determine the MT-binding domains of p600, we generated seven FLAG-tagged truncated fragments of p600 (termed fragment A to G). Because p600 does not contain any known domains with the exception of few putative SH2/SH3/PTB domains scattered in the protein (data not shown), these fragments were selected to cover most of the p600 protein. They were then expressed in CAD cells, and MT preparations were isolated (Fig. 2F). The small MAP Tau was used as a positive control for MT preparations. Only fragments E and G, containing residues 3214–3899 and 4480–5183, respectively, cofractionated in significant amounts with MTs (Fig. 2G, red stars), indicating a minimum of two MT-binding domains in the C terminus of p600. The fragments D and F containing residues 2661–3359 and 3910–4851, respectively, were recovered in small amounts in these preparations (Fig. 2G, red stars), possibly because their residues partially overlap with those in fragments E and G.
One physiological role of MAPs is to aid in the polymerization of tubulin to form stable microtubules. Therefore, a microtubule polymerization assay was performed in the presence of the fragments E, G, D, F, and B (negative control). Over time, tubulin alone, or with the fragment B or D, did not efficiently polymerize into microtubules (Fig. 2H). When fragments E, F, or G were added to tubulin, efficient microtubule polymerization was observed (Fig. 2H). Consistent with their stronger associations with MTs, fragments E and G were the most potent to induce polymerization, whereas fragment F exhibited lower activity. These results indicate that p600 can facilitate MT polymerization through MT-binding domains located in the C terminus of the protein.
To test the physiological role of p600 in cells, we generated two specific RNAi sequences (RNAi no. 1 and RNAi no. 2). Both p600 RNAis efficiently reduced the levels of p600 by 50–60%, 24 h after transfection, whereas a control RNAi sequence without homology to any mRNA did not alter levels of p600 (Fig. 2I). To test the role of p600 on MT stabilization, p600 RNAi nos. 1 and 2 vectors were transfected in CAD cells to assess the levels of acetylated tubulin, a marker for stable MTs. Levels of acetylated tubulin, but not tubulin or other cytoskeletal proteins (Lis1, Ndel1, FAK), decreased in these cells without affecting survival 48 h after transfection (Fig. 2I). Together, these results provide solid evidence of a role for p600 in stabilizing MTs in cells. Collectively, our data indicate that p600 is a novel developmentally regulated MAP.
p600 associates with the endoplasmic reticulum
Because a significant fraction of p600 copurified with membranes (Fig. 2B) and p600 stabilized MTs in a punctate pattern reminiscent of vesicular/tubular membranes (Fig. 3), we sought to determine the types of membranes with which p600 associates. We costained CAD cells and E15 primary mouse cortical neurons with antibodies that recognize p600 and diverse membranous organelles. Although p600 did not colocalize with mitochondria, Golgi, and endosome markers (data not shown), it extensively colocalized with Bip, an ER marker that decorates MTs in both mitotic cells and primary cultured neurons (Fig. 3A,B). p600 was strongly detected in ER microsomes purified from adult mouse brain (Fig. 3C). Finally, using immunogold electron microscopy, we found that p600 decorates smooth ER tubules in neuronal processes (Fig. 3D).
To define the domain(s) that target(s) p600 to the ER, we assessed the staining pattern of seven FLAG-tagged p600 fragments expressed in CAD cells by confocal microscopy. Whereas fragments A, B, D, F, and G showed diffuse staining patterns (data not shown), the fragments C and E, exhibited a tubular appearance (Fig. 3E), indistinguishable from the pattern shown by ER-associated proteins (Munro and Pelham, 1987; Du et al., 2004). These fragments formed a network of polygonal reticulum reminiscent of peripheral ER in interphase cells (Dreier and Rapoport, 2000; Shibata et al., 2006). In agreement with the fact that ER tubules are transported on and extend along MTs (Waterman-Storer and Salmon, 1998), higher magnification pictures revealed the intimate juxtaposition between the p600 fragments-expressing network and the MT cytoskeleton up to the cell edge (Fig. 3E). Costaining of fragments C and E with ER marker Bip ultimately confirms their localization to the ER (Fig. 3E′). Interestingly, fragment C, which exhibits the most distinctive ER-tubule pattern, was exclusively found in membranes (Fig. 3F). Together, this set of data indicates that p600 associates with the ER in addition to binding to MTs.
p600 is required for neurite extension
Conveyed on MTs, ER membranes can extend into areas of cellular expansion including in the growth cones of neurons, the leading process, neurites, and filipodia of adherent cells in which they can initiate signal cascades to promote extension (Dailey and Bridgman, 1989; Kocsis et al., 1994; Waterman-Storer and Salmon, 1998; Rizzuto, 2001). p600 colocalized with ER-specific proteins bearing KDEL antigens and IP3 receptors at the tip of filipodia (data not shown). p600 is also found at the growth cone (Fig. 4A), a structure modulated by dynamic MTs and ER functions that guide neurite extension. Because p600 associates with MTs and ER and its expression correlates with neuronal differentiation, we investigated the role of p600 in neurite outgrowth. In cultured primary neurons, p600 RNAi also efficiently reduced the levels of p600 and acetylated tubulin by ∼40%, 24 h after electroporation, without affecting levels of other cytoskeletal proteins (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). At the cellular level, we found that, after cotransfection of p600 RNAi no. 1 or p600 RNAi no. 2 and GFP vector (5:1 ratio) to mark the p600 RNAi-expressing hippocampal neurons [2 d in vitro (DIV2)], lack of p600 altered the stability of neuronal processes without causing total collapse, as evidenced by the GFP staining (Fig. 4B). Remarkably, in these p600 RNAi-treated neurons, the levels of the ER marker Bip in neuronal processes was reduced, whereas control RNAi-treated neurons or adjacent nontransfected neurons displayed normal Bip staining in neuronal processes. As determined with confocal microscopy, 57 ± 12% of 224 randomly selected neurons transfected with GFP/p600 RNAi did not exhibit Bip staining in more than three neuronal processes in contrast to only 9 ± 11% of 216 GFP/control RNAi transfected neurons. The silencing of p600 by RNAi nos. 1 and 2 for a longer time (48 h) totally collapsed neuronal processes (Fig. 4C; supplemental Fig. 1, available at www.jneurosci.org as supplemental material). This phenotype was confirmed on a large number of young primary neurons (DIV2) electroporated with GFP/p600 RNAi for 48 h. Sixty-eight percent ± 9% of neurons electroporated with GFP/p600 RNAi (n = 1834) did not exhibit neuronal processes in contrast to 22 ± 13% of GFP/control RNAi electroporated neurons (n = 1795). Together, these results indicate that p600, which is expressed in neuronal processes and growth cone, is important for neurite extension and stability.
Figure 4.
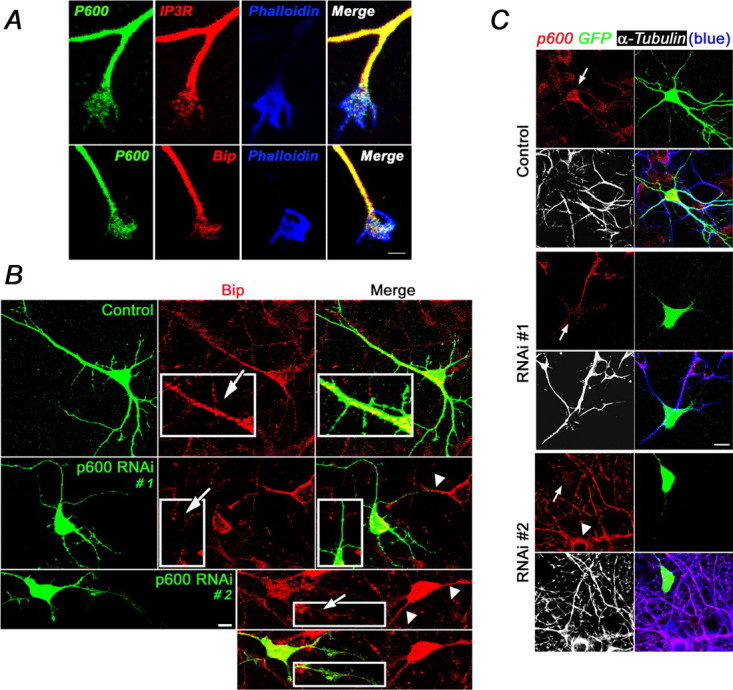
p600 is required for neurite extension. A, Colocalization of p600 with ER markers IP3R and Bip in phalloidin-positive growth cones. B, Altered neuronal processes associated with reduced staining of Bip in primary neurons cotransfected with GFP/p600 RNAi no. 1 or RNAi no. 2, 24 h after transfection when compared with adjacent nontransfected neurons (white arrowhead) or neurons cotransfected with GFP/control RNAi. C, Total collapse of neuronal processes in individual primary neurons cotransfected with GFP/p600 RNAi no. 1 or RNAi no. 2 (48 h) as evidenced by the lack of GFP and tubulin staining (black/white and blue in the fourth panel). All pictures were taken with a confocal microscope. Scale bars: A, 5 μm; B, C, 10 μm.
p600 is important for the integrity of migrating neurons and cortical neuronal positioning
MT dynamics and MT-based ER transport/signaling are also essential for stability and extension of the leading process of migrating neurons during cortical positioning in the developing brain (Ross and Walsh, 2001; Ayala et al., 2007; Ghashghaei et al., 2007; Rakic, 2007). Because p600 modulates MT dynamics, interacts with ER, and is expressed in early-born neurons containing ER membranes (Fig. 1; supplemental Fig. 2, available at www.jneurosci.org as supplemental material), it may play an important role in neuronal cortical positioning. To test this hypothesis, we generated p600 null mice. The mice die between E10.5 and E14.5 (Y. Nakatani and M. D. Nguyen, unpublished observations) with severe brain phenotype (data not shown), thereby precluding a clear demonstration of a role for p600 in neuronal positioning. This phenotype is reminiscent of the early lethality of mice lacking the lissencephaly protein, Lis1, and its binding partner, Ndel1, two MT-associated proteins involved in neuronal migration (Hirotsune et al., 1998; Sasaki et al., 2005).
To circumvent this problem, we performed in utero electroporation. The specific RNAi sequences nos. 1 and 2 for p600 or control RNAi vector were coexpressed with GFP vector (5:2 ratio) in E14 mouse neocortex by in utero electroporation, and the brains were analyzed at E17 when many neurons have reached their destination in the cortical plate. Because it is difficult to distinguish radial glial cells from neurons in the ventricular and subventricular zones, only cells that have migrated out of these zones were quantified. Neurons were counted blindly by two researchers in three distinct regions of the cortical plate (superficial, middle, and lower) and in the intermediate zone (Fig. 5A,B), according to reference (Xie and Tsai, 2004). As depicted in Figure 5A, neurons coelectroporated with control RNAi and GFP migrate normally through the cortical layers to reach their destination. Those neurons showed a robust and healthy leading process that guides them during this journey (Fig. 5C). In contrast, neurons with p600 RNAi no. 1 or 2 fail to migrate properly and accumulate in the intermediate zone, ventricular and subventricular zone, with only a very few reaching the superficial cortical plate (Fig. 5A). In the p600 RNAi nos. 1 and 2 electroporated neocortices, 69.0 ± 3.2% (n = 6; 538 cells) and 61.2 ± 2.7% (n = 6; 1009 cells) of GFP-positive cells were found in the intermediate zone, respectively, whereas 13.2 ± 2.2% (n = 5; 1175 cells) of control RNAi GFP-positive cells were found in the same region (Fig. 5A,B). Moreover, the proportions of GFP-positive neurons found in lower, middle, and superficial layers of the cortical plate from p600 RNAi no. 1 or 2 electroporated neocortices (RNAi no. 1, 14.4 ± 2.4, 10.4 ± 0.5, 6.2 ± 1.8%; RNAi no. 2, 18.0 ± 2.6, 9.9 ± 0.8, 10.9 ± 0.7%, respectively) were reduced compared with control neocortices (lower, 15 ± 3.5%; middle, 26 ± 2.3%; superficial, 45.8 ± 7.5%) (Fig. 5B). These results underscore defects in migration in p600-silenced brains. Interestingly, single-cell analysis indicated that migrating neurons depleted of p600 very often exhibited a crooked, thin, and “zigzag” leading process (Fig. 5C), a structure modulated by dynamic MTs and ER functions that directs cell migration. Of the ∼65% of cells found in the intermediate zone of the p600 RNAi electroporated neocortices (n = 4), ∼70% showed this deficiency, whereas in the superficial cortical plate from the same neocortices, ∼9% of the cells presented this feature.
Figure 5.
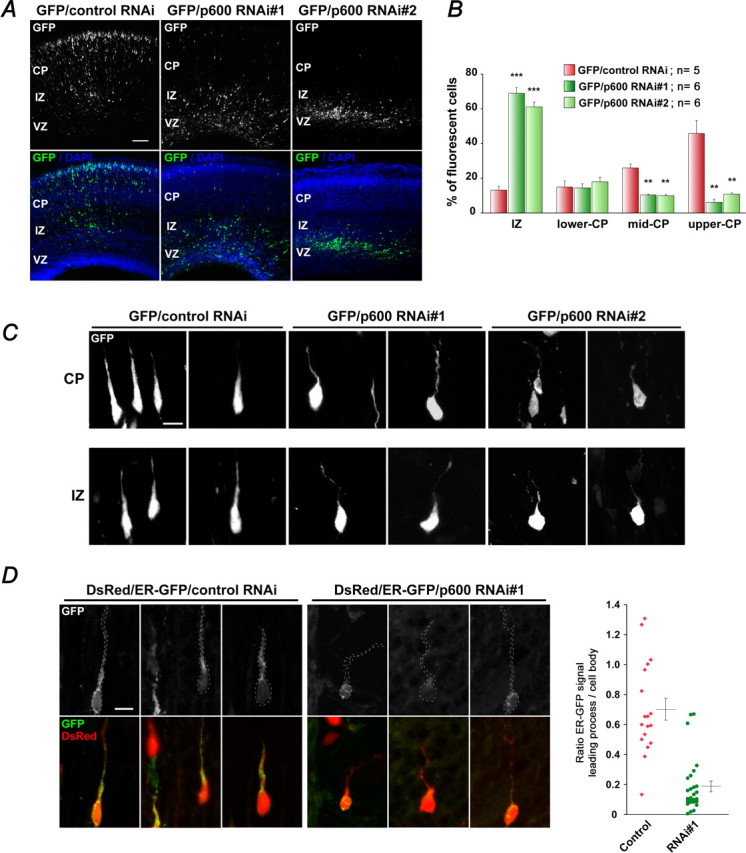
In utero electroporation of p600 RNAi in the mouse neocortex alters neuronal migration. A, E17 migratory profile of neurons coelectroporated at E14 with GFP and control RNAi or p600 RNAi no. 1 or p600 RNAi no. 2. Nuclei were stained with DAPI (blue). Representative images are shown. B, Quantification of number of migrating neurons plotted as the mean ± SEM in the intermediate zone (IZ) and lower, middle, and upper CP performed on five, six, and six embryonic brains coelectroporated with GFP/control RNAi, GFP/p600 RNAi no. 1, and GFP/p600 RNAi no. 2, respectively. **p < 0.01, T < t, two-tailed; ***p < 0.001, T < t, two-tailed. n, Number of embryonic brains analyzed. C, Healthy morphology of migrating neurons coelectroporated with GFP and control RNAi characterized by robust leading process versus the thin, crooked, zigzag leading process of migrating neurons coelectroporated with GFP and p600 RNAi no. 1. Representative images are shown. D, ER-targeting GFP signal is enriched at the perinuclear region and in the leading process of control RNAi neurons. The signal in p600-silenced neurons is predominant at the perinuclear region and, if any, in the proximal site (base) of the leading processes but mostly absent along the leading process. Quantifications of the ratio intensity of fluorescent ER-GFP signal in the leading process/cell soma indicates significant alteration in ER distribution in p600-silenced neurons. Each dot corresponds to the ratio for one single neuron. n = 18 for control RNAi-treated neurons; n = 27 for p600 RNAi-treated neurons. DsRed2 transfected with RNAi and ER-GFP delimits the morphology of neurons (outlines) having incorporated the RNAi. All pictures were taken with a confocal microscope. Scale bars: A, 100 μm; C, D, 10 μm.
To further examine the relationship between MTs and ER organization in p600-silenced migrating neurons, we coexpressed control RNAi or p600 RNAi no. 1 vector with DsRed2 (to mark the cells having incorporated the RNAi and to define the morphology of these cells) and the fluorescent ER-targeted GFP marker (see Materials and Methods) in E14 mouse neocortex by in utero electroporation. Distribution of ER membranes was analyzed at E17. In control migrating neurons, ER is distributed at perinuclear regions and along the leading process, up to the tip (Fig. 5D). In contrast, p600-depleted neurons with thin, crooked, and zigzag leading process accumulate predominantly ER membranes at the perinuclear region and, if any, in the proximal site (base) of the leading processes (Fig. 5D). Almost no ER-GFP signal was found along the leading process (Fig. 5D). Because MTs in the leading process convey ER membranes up to the tip, these results underscore alteration in the MTs–ER interface in p600-silenced migrating neurons. Thus, the failure in positioning of p600-deficient neurons is likely caused by a destabilization of MTs in the leading process and reduced localization of ER membranes within the process.
Discussion
A novel MAP associated with the ER
Cytoskeletal proteins have been extensively studied for their structural roles in neuronal development but their association with organelles is poorly understood (Rizzuto, 2001; Ross and Walsh, 2001; Ayala et al., 2007). In migrating fibroblasts, CLASPS (CLIP-associating proteins) constitute a family of crucial proteins that bind to MT plus ends, localize to the Golgi apparatus, and organize MTs at the leading edge (Galjart, 2005). Furthermore, the Golgi-associated Hook family member Hook3 also binds to MTs to locate Golgi membranes (Walenta et al., 2001). For a decade, among tens of MAPs, CLIMP-63 was the only known MAP that associates with the ER (Klopfenstein et al., 1998, 2001; Vedrenne et al., 2005). CLIMP-63 (63 kDa), which is 10 times smaller than p600, is as yet uncharacterized in the CNS and has never been studied in vivo.
Because of its humongous size, the cDNA of p600 (∼15,500 bp) cannot be cloned, and consequently, overexpression studies for nocodazole/cold resistance, MT-bundling activity, and ER fission assays are not yet possible. Nevertheless, using other established biochemical and cellular methods, we demonstrated that p600 fulfills the requirements of a MAP and exhibits the unique feature of binding to MTs and ER membranes via overlapping regions (Figs. 2, 3). Whereas MT-binding activity is found in fragments E and G, ER-directed domains reside in fragments C and E. These results indicate that p600 contains domains in fragment E that may connect MTs to ER. They also suggest a model in which p600 stabilizes MTs for ER extension (see below) (Fig. 6). This view would be consistent with the idea that MTs transport ER membranes and spatially localize them in neurons. Because MTs contribute to ER dynamics but are not essential for the maintenance of the existing network (Terasaki et al., 1986), it is possible that p600 organizes ER tubule formation and networking independently of its MT-associated activity. Of note, p600 is not a plus-binding MT protein because it does not colocalize with endogenous EB1 or ectopic EB1-GFP at the cell edge (data not shown). These data indicate that p600 is not involved in the transport of ER membranes through the TAC (“tip attachment complex”) in which ER is attached at the growing plus ends of MTs. They rather suggest that p600 is implicated in ER transport through the sliding mechanism on MTs via molecular motors and the “microtubule movement mechanism” in which ER membranes are anchored to moving MTs [for model, see Fig. 6; for the complete definition of the mechanisms, see Waterman-Storer and Salmon (1998)]. Because p600 is found at close proximity of EB1, it may help stabilizing newly grown plus-end MTs. Finally, the ER is also a prominent source of intracellular Ca2+, a potent mediator that rearranges MTs after membrane depolarization. The select colocalization of p600 with ER membranes and MTs up to the cell edge (Fig. 3) raises the intriguing possibility that p600-associated MT functions are regulated on membrane depolarization in a Ca2+-dependent manner. A fine dissection of the domains contained in fragments C, E, and G of p600, which contain the ER-directed and MT-binding domains, and a better understanding of the biochemistry and topology of p600 would help to refine the roles of p600 in these aspects.
Figure 6.
Model depicting the interaction between p600, MTs, and ER. p600 associates with stable MTs and binds to ER. p600 may couple MT stability and dynamics to ER transport, tubule extension, and morphogenesis. A short-term loss of p600 destabilizes MT, reduces anterograde ER transport, and causes retraction of existing ER in neuronal processes, including the leading process. Over time, the disruption of the MTs–ER interface can alter the integrity of these processes, thereby impeding neurite outgrowth and neuronal migration during development. In this model, ER membranes are transported via “sliding mechanism” (with molecular motors) and “MT movement mechanism” (ER membranes attached to MTs).
By virtue of its association with ER membranes, p600 also has direct access to nuclear membrane and might couple transcription to ER signaling. Indeed, p600 was initially found in the nucleus and isolated as a retinoblastoma-binding protein (DeMasi et al., 2005; Huh et al., 2005; Nakatani et al., 2005). Finally, p600, recently named ZUBR1 (zinc finger, UBR1 type 1), may be critical for protein degradation because it has a zinc finger/UBR box, and its newly discovered murine homolog, UBR4, participates in the N-end rule pathway (Tasaki et al., 2005). This hypothesis would be consistent with the association of p600 with the ER, a cellular compartment essential for protein folding and degradation.
p600 is important for neurite outgrowth and neuronal migration
p600 expression increases during neuronal development (Fig. 1). MT dynamics coupled to ER trafficking are essential for neuronal differentiation and migration. Consistent with the association of p600 with both MTs and ER, we found that depletion of p600 in primary neurons by two RNAi sequences reduced progressively the stability of extending neurites and, ultimately, culminated in collapse of neurites (Fig. 4; supplemental Fig. 1, available at www.jneurosci.org as supplemental material). It remains unclear whether the lack of Bip staining is the result of lack of ER generation/fission, absence of transport toward the periphery, or simply a step preceding and contributing to the collapse. Interestingly, the defect in neurite outgrowth observed of p600-depleted primary neurons associated with scarce Bip staining is reminiscent of the alteration of the leading process of p600-silenced migrating neurons (Figs. 4, 5). These latter neurons exhibit crooked, thin, and zigzag leading process with fewer ER members. We attributed the defect in neurite outgrowth and neuronal migration to the disruption of the MT–ER interface maintained by p600 (see model in Fig. 6). This alteration would reduce the anterograde transport of ER membranes and cause their retraction within the neuronal processes. Because p600 also associates with the Ca2+ transducer molecule calmodulin (Nakatani et al., 2005) and the ER, a prominent reservoir of Ca2+, failure of p600-deficient cells to migrate may partly be attributable to the lack of integration of Ca2+ signaling. By transducing Ca2+, p600 may also interact with DCAMKL1 (doublecortin calcium/calmodulin-dependent protein kinase-like kinase 1), a MAP with a CaMKII domain important for neuronal migration (Deuel et al., 2006; Koizumi et al., 2006). Simultaneously, p600 may also regulate actin dynamics at the extremity of the leading process. A role for p600 in neurite outgrowth and neuronal migration is reminiscent of the defects in axonal elongation and neuronal migration in mice with disrupted Tau and MAP1b genes (Takei et al., 2000). Our data would also be consistent with the finding that mutations in BIG2, a protein involved in membrane transport and trafficking, alter neuronal migration in a human neurodevelopmental disorder (Sheen et al., 2004).
In addition to a direct stabilizing effect on MTs, p600 may also indirectly stabilize MTs via FAK, a molecule that regulates focal adhesion sites and is inactivated in fibroblasts depleted of p600 (Nakatani et al., 2005). Indeed, silencing p600 suppresses formation of ruffled membranes in mitotic cells, which are critical in initiating the formation of focal contacts at the leading edge of migrating cells (Rinnerthaler et al., 1988; Ezratty et al., 2005). It remains unclear how p600 coordinates membranes ruffling, establishes focal adhesion contacts, and signals in that context, but it is reasonable to propose that p600, via polymerization and stabilization of MTs, sustains the disassembly of focal adhesion sites at the leading edge of migrating neurons, thereby favoring cell motility and neuronal migration. In addition, p600 may control ER tubule extension and membrane morphogenesis during neuronal migration.
In summary, we identified a first MAP to associate with the ER in CNS neurons that impacts on neuronal development. p600 may have strong implications in physiological cellular processes that heavily rely on ER-dependent Ca2+ transduction and cytoskeletal dynamics such as neuronal plasticity. Additional studies on p600 will uncover its importance in other neuronal processes.
Footnotes
This work was supported by an Operating Grant from the Canadian Institutes of Health Research (CIHR) (M.D.N.), the Human Frontier Science Program Organization (HFSPO) (Y.N.), and a Grant-in-Aid for Young Scientist from the Japanese Ministry of Education, Culture, Sports, Science and Technology (K.S.). M.D.N. holds a Career Development Award from HFSPO, a New Investigator Award from the CIHR, a scholarship from the Alberta Heritage Foundation for Medical Research, and the Brenda Strafford Foundation Chair in Alzheimer research. We are grateful to Drs. M. Colico, J. De Masi, K. W. Huh, H. Kurooka, H. Konishi, S. Tsukita for reagents; Dr. L. H. Tsai for partly hosting this study; and Drs. G. Zamponi, S. McFarlane, and N. Syed for critical reading of this manuscript.
References
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Dailey ME, Bridgman PC. Dynamics of the endoplasmic reticulum and other membranous organelles in growth cones of cultured neurons. J Neurosci. 1989;9:1897–1909. doi: 10.1523/JNEUROSCI.09-06-01897.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMasi J, Huh KW, Nakatani Y, Munger K, Howley PM. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc Natl Acad Sci USA. 2005;102:11486–11491. doi: 10.1073/pnas.0505322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Gertler FB. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- Deuel TA, Liu JS, Corbo JC, Yoo SY, Rorke-Adams LB, Walsh CA. Genetic interactions between doublecortin and doublecortin-like kinase in neuronal migration and axon outgrowth. Neuron. 2006;49:41–53. doi: 10.1016/j.neuron.2005.10.038. [DOI] [PubMed] [Google Scholar]
- Dreier L, Rapoport TA. In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J Cell Biol. 2000;148:883–898. doi: 10.1083/jcb.148.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Ferro-Novick S, Novick P. Dynamics and inheritance of the endoplasmic reticulum. J Cell Sci. 2004;117:2871–2878. doi: 10.1242/jcs.01286. [DOI] [PubMed] [Google Scholar]
- Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- Galjart N. CLIPs and CLASPs and cellular dynamics. Nat Rev Mol Cell Biol. 2005;6:487–498. doi: 10.1038/nrm1664. [DOI] [PubMed] [Google Scholar]
- Ghashghaei HT, Lai C, Anton ES. Neuronal migration in the adult brain: are we there yet? Nat Rev Neurosci. 2007;8:141–151. doi: 10.1038/nrn2074. [DOI] [PubMed] [Google Scholar]
- Gil P, Dewey E, Friml J, Zhao Y, Snowden KC, Putterill J, Palme K, Estelle M, Chory J. BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 2001;15:1985–1997. doi: 10.1101/gad.905201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- Huh KW, DeMasi J, Ogawa H, Nakatani Y, Howley PM, Munger K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci USA. 2005;102:11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DR, Kappeler F, Hauri HP. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DR, Klumperman J, Lustig A, Kammerer RA, Oorschot V, Hauri HP. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal alpha-helical segment. J Cell Biol. 2001;153:1287–1300. doi: 10.1083/jcb.153.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocsis JD, Rand MN, Lankford KL, Waxman SG. Intracellular calcium mobilization and neurite outgrowth in mammalian neurons. J Neurobiol. 1994;25:252–264. doi: 10.1002/neu.480250306. [DOI] [PubMed] [Google Scholar]
- Koizumi H, Tanaka T, Gleeson JG. Doublecortin-like kinase functions with doublecortin to mediate fiber tract decussation and neuronal migration. Neuron. 2006;49:55–66. doi: 10.1016/j.neuron.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter A. The expression and posttranslational modification of a neuron-specific beta-tubulin isotype during chick embryogenesis. Cell Motil Cytoskeleton. 1990;17:118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Moody SA, Quigg MS, Frankfurter A. Development of the peripheral trigeminal system in the chick revealed by an isotype-specific anti-beta-tubulin monoclonal antibody. J Comp Neurol. 1989;279:567–580. doi: 10.1002/cne.902790406. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Konishi H, Vassilev A, Kurooka H, Ishiguro K, Sawada J, Ikura T, Korsmeyer SJ, Qin J, Herlitz AM. p600, a unique protein required for membrane morphogenesis and cell survival. Proc Natl Acad Sci USA. 2005;102:15093–15098. doi: 10.1073/pnas.0507458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MD, Shu T, Sanada K, Lariviere RC, Tseng HC, Park SK, Julien JP, Tsai LH. A NUDEL-dependent mechanism of neurofilament assembly regulates the integrity of CNS neurons. Nat Cell Biol. 2004;6:595–608. doi: 10.1038/ncb1139. [DOI] [PubMed] [Google Scholar]
- Rakic P. The radial edifice of cortical architecture: from neuronal silhouettes to genetic engineering. Brain Res Rev. 2007;55:204–219. doi: 10.1016/j.brainresrev.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnerthaler G, Geiger B, Small JV. Contact formation during fibroblast locomotion: involvement of membrane ruffles and microtubules. J Cell Biol. 1988;106:747–760. doi: 10.1083/jcb.106.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R. Intracellular Ca2+ pools in neuronal signalling. Curr Opin Neurobiol. 2001;11:306–311. doi: 10.1016/s0959-4388(00)00212-9. [DOI] [PubMed] [Google Scholar]
- Ross ME, Walsh CA. Human brain malformations and their lessons for neuronal migration. Annu Rev Neurosci. 2001;24:1041–1070. doi: 10.1146/annurev.neuro.24.1.1041. [DOI] [PubMed] [Google Scholar]
- Sanada K, Tsai LH. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, Muramatsu M, Miyagawa S, Hiraiwa N, Yoshiki A, Wynshaw-Boris A, Hirotsune S. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen VL, Ganesh VS, Topcu M, Sebire G, Bodell A, Hill RS, Grant PE, Shugart YY, Imitola J, Khoury SJ, Guerrini R, Walsh CA. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat Genet. 2004;36:69–76. doi: 10.1038/ng1276. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Voeltz GK, Rapoport TA. Rough sheets and smooth tubules. Cell. 2006;126:435–439. doi: 10.1016/j.cell.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Takei Y, Teng J, Harada A, Hirokawa N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J Cell Biol. 2000;150:989–1000. doi: 10.1083/jcb.150.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M, Chen LB, Fujiwara K. Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol. 1986;103:1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M, Slater NT, Fein A, Schmidek A, Reese TS. Continuous network of endoplasmic reticulum in cerebellar Purkinje neurons. Proc Natl Acad Sci USA. 1994;91:7510–7514. doi: 10.1073/pnas.91.16.7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedrenne C, Klopfenstein DR, Hauri HP. Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell. 2005;16:1928–1937. doi: 10.1091/mbc.E04-07-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky A. Endoplasmic reticulum calcium signaling in nerve cells. Biol Res. 2004;37:693–699. doi: 10.4067/s0716-97602004000400027. [DOI] [PubMed] [Google Scholar]
- Walenta JH, Didier AJ, Liu X, Kramer H. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J Cell Biol. 2001;152:923–934. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol. 1998;8:798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- Xie Z, Tsai LH. Cdk5 phosphorylation of FAK regulates centrosome-associated miocrotubules and neuronal migration. Cell Cycle. 2004;3:108–110. [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- Xu XZ, Wes PD, Chen H, Li HS, Yu M, Morgan S, Liu Y, Montell C. Retinal targets for calmodulin include proteins implicated in synaptic transmission. J Biol Chem. 1998;273:31297–31307. doi: 10.1074/jbc.273.47.31297. [DOI] [PubMed] [Google Scholar]