Abstract
Mrg class G-protein-coupled receptors (GPCRs) are expressed exclusively in sensory neurons in the trigeminal and dorsal root ganglia. Pharmacological activation of Mrg proteins is capable of modulating sensory neuron activities and elicits nociceptive effects. In this study, we illustrate a control mechanism that allows the Runx1 runt domain transcription factor to generate compartmentalized expression of these sensory GPCRs. Expression of MrgA, MrgB, and MrgC subclasses is confined to an “A/B/C” neuronal compartment that expresses Runx1 transiently (or does not express Runx1), whereas MrgD expression is restricted to a “D” compartment with persistent Runx1 expression. Runx1 is initially required for the expression of all Mrg genes. However, during late development Runx1 becomes a repressor for MrgA/B/C genes. As a result, MrgA/B/C expression persists only in the Runx1− “A/B/C” compartment. In Δ446 mice, in which Runx1 lacks the C-terminal repression domain, expression of MrgA/B/C genes is dramatically expanded into the Runx1+ “D” compartment. MrgD expression, however, is resistant to Runx1-mediated repression in the “D” compartment. Therefore, the creation of Runx1+ and Runx1− compartments, in conjunction with different responses of Mrg genes to Runx1-mediated repression, results in the compartmentalized expression of MrgA/B/C versus MrgD genes. Within the MrgA/B/C compartment, MrgB4-expressing neurons innervate exclusively the hairy skin. Here we found that Smad4, a downstream component of bone morphological protein-mediated signaling, is required selectively for the expression of MrgB4. Our study suggests a new line of evidence that specification of sensory subtypes is established progressively during perinatal and postnatal development.
Keywords: Runx1, nociceptors, Mrg class G-protein-coupled receptors, nociceptive ion channels and receptors, cell type specification, dorsal root ganglia
Introduction
The mouse genome encodes 50 Mrg (also named Mrgpr/SNSR) class G-protein-coupled receptors (GPCRs), 12 of which are expressed exclusively in the somatic sensory neurons located in the trigeminal ganglia and dorsal root ganglia (DRG) (Dong et al., 2001; Lembo et al., 2002; Choi and Lahn, 2003; Zylka et al., 2003; Zhang et al., 2005; Burstein et al., 2006). These Mrg genes are divided into four subclasses: A (MrgA1-A8), B (MrgB4 and B5), C (MrgC11), and D (MrgD) (Zylka et al., 2003; Zhang et al., 2005). In adult mice, these Mrg subclasses exhibit compartmentalized expression, as indicated by the nonoverlapping expression of MrgD with other Mrg genes (Dong et al., 2001; Zylka et al., 2003, 2005) (see below). Most interestingly, MrgD+ and MrgB4+ neurons innervate distinct peripheral targets, skin epidermis and the hairy skin, respectively (Zylka et al., 2005; Liu et al., 2007). Pharmacological activation of Mrg proteins is able to modulate neuronal activities and evoke painful responses (Grazzini et al., 2004; Cai et al., 2007; Crozier et al., 2007). However, how compartmentalized expression of Mrg genes is established during development is poorly understood.
A series of recent studies has shown that the runt domain transcription factor Runx1 plays a pivotal role in controlling the development of sensory neurons, particularly those involved with nociception (Theriault et al., 2005; Chen et al., 2006; Kramer et al., 2006; Marmigere et al., 2006; Zhong et al., 2006; Woolf and Ma, 2007; Yoshikawa et al., 2007). Runx1 is initially expressed in most nociceptors (Levanon et al., 2002; Chen et al., 2006). Subsequently, Runx1 expression is extinguished in most peptidergic nociceptors (Chen et al., 2006), and persists primarily in nonpeptidergic nociceptors (Chen et al., 2006; Kramer et al., 2006). Genetic studies demonstrate that Runx1 is required for the expression of nearly two-dozen ion channels and receptors, including the whole family of Mrg genes (Chen et al., 2006) (see Fig. 1).
Figure 1.
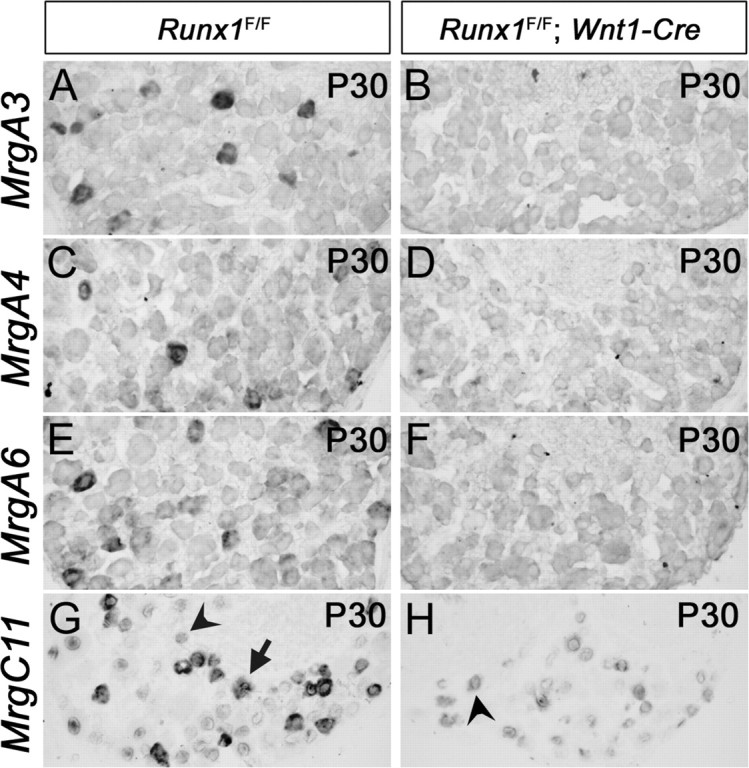
Loss of Mrg gene expression in Runx1F/F; Wnt1-Cre mice [mentioned as Runx1 −/−(Wnt1) mice in text]. In situ hybridizations with indicated Mrg probes were performed on sections through P30 T12 thoracic DRG.
In this study, we will illustrate a control mechanism that allows Runx1 to establish compartmentalized expression for Mrg class sensory GPCRs. Accordingly, expression of MrgA, MrgB, and MrgC subclasses is confined to an “A/B/C” neuronal compartment that expresses Runx1 transiently (or does not express Runx1), whereas MrgD expression is restricted to a “D” compartment with persistent Runx1 expression.
Materials and Methods
Animals.
The generation of Runx1 conditional mutant mice, Wnt1-Cre transgenic mice, Δ446 mice, Smad4 conditional null mice, MrgD-GFP mice and SNS-Cre transgenic mice has been described previously (Jiang et al., 2000; Yang et al., 2002; Agarwal et al., 2004; Nishimura et al., 2004; Growney et al., 2005; Zylka et al., 2005). The morning that vaginal plugs were observed was considered as E0.5. PCR-based genotyping for conditional null mice has been described previously (Chen et al., 2006). The following primers were used for the MrgD-GFP mutant allele, 5′-ATG GTG AGC AAG GGC GAG GAG-3′ and 5′-TCG CGC TTC TCG TTG GGG TCT TTG-3′; for the MrgD-GFP wild-type allele, 5′-ATG AAC TCC ACT CTT GAC AG-3′ and 5′-CAC TGG TGT TTG TTG GGA TG-3′; for the Δ446 mutant allele, 5′-TCG CTT TCA AGG TGG TGG CA-3′ and 5′-TCC GGA GCC GTT GAG AGT C-3′; and for the wild-type Runx1 allele, 5′-TGT CTC TGC ATC GCA GGA CT-3′ and 5′-TGT GCG TTC CAA GTC AGT TGT-3′.
In situ hybridization and immunostaining.
The in situ hybridization (ISH) procedure and the probes used in this study have been described previously (Chen et al., 2006). For ISH combined with anti-GFP fluorescent immunostaining, GFP was detected before the ISH procedure. Frozen sections were dried at room temperature for 10 min, postfixed in 4% paraformaldehyde for 15 min, washed 3 times with PBS for 5 min each, incubated with anti-GFP primary antibody (1:500 in PBT; Invitrogen, Carlsbad, CA) for 30 min, washed three times with PBS for 5 min each, and incubated with Alexa-488-conjugated secondary antibody (1:200 in PBT; Invitrogen) for 30 min. Note that all solutions were prepared under RNase-free conditions. To avoid the masking of fluorescent signal by the subsequent ISH dye signal, all sections were photographed by fluorescence microscopy, followed by regular ISH procedure using an alkaline phosphatase (AP)-conjugated antibody (1:2000; Roche, Indianapolis, IN) and 5-bromo-4-chloro-indolyl-phosphate/nitroblue–tetrazolium (BCIP/NBT) substrates for development. The bright-field images of opaque in situ signals were inverted into pseudo-fluorescent images, and then merged with GFP fluorescent images.
ISH combined with anti-Runx1 (from Dr. Thomas Jessell, Columbia University, New York, NY) or IB4 fluorescent staining has been described previously (Chen et al., 2006), with the following modification. Fluorescent signals of Runx1 and IB4 were photographed first, followed by color development of ISH signals with BCIP/NBT substrates. The bright-field images of ISH signals were inverted and then merged with fluorescent images. As noted above, this sequential photographing avoids the masking of low-level fluorescent signals by nonfluorescent ISH signals, leading to a more sensitive detection of the coexpression of Runx1 or IB4 with genes of interest. For example, in this study, we were able to detect high or medium levels of Runx1 protein in virtually all MrgD+ cells, whereas the previous procedure (in which ISH signals developed first, followed by Runx1 immunostaining) failed to show medium levels of Runx1 expression in a fraction of MrgD+ neurons (Chen et al., 2006).
For double color ISH, two probes were labeled with fluorescein- or digoxigenin-UTP. The probes were detected with peroxidase (POD)-conjugated anti-digoxigenin antibody (1:400; Roche) and AP-conjugated anti-fluorescein antibody (1:1000; Roche). The fluorescent signal development for POD consists of three sequential amplification steps: (1) TSA Biotin System (1:100; PerkinElmer, Wellesley, MA), (2) Vectastain ABC Kit (Vector Laboratories, Burlingame, CA), and (3) TSA Fluorescein System (1:50; PerkinElmer). The fluorescent signals were photographed, followed by development for AP with BCIP/NBT substrates. The bright-field images of nontransparent purple signals were inverted, and then merged with fluorescent images. Because the signal of the second probe was developed with nonfluorescent BCIP/NBT substrates, this modified double color ISH procedure is more sensitive than the procedure that involves fluorescent substrates for both probes (Dong et al., 2001; Zylka et al., 2003). For example, the colocalization of MrgA3 and MrgB4 shown in this study was not detected in the previous study (Zylka et al., 2003).
Cell counting.
To compare total DRG neurons, T12 thoracic and L4/L5 lumbar DRG were dissected from three pairs of mutant and control mice, fixed, embedded, sectioned at 12 μm thickness. One of six adjacent sets of sections was hybridized with the pan-neuronal probe SCG10, and the number of SCG10+ neurons was counted. Only cells containing nuclei were counted. To determine the percentages of neurons expressing molecular markers, six adjacent sets of sections were prepared from each T12 DRG and probed separately with six different probes, one of which was the pan-neuronal marker SCG10 so that percentages of DRG neurons expressing a gene can be calculated. Four or more independent T12 DRG or L4/L5 lumbar DRG were used for each counting. The difference between wild-type and mutant samples was subjected to a Student's t test, with p < 0.05 considered significant.
Results
Loss of Mrg gene expression in conditional Runx1 null mutants
We recently reported that Runx1 is necessary for the expression of two Mrg subclasses, class B (MrgB4 and MrgB5) and class D (MrgD) (Chen et al., 2006). To determine whether Runx1 controls the expression of class A (MrgA1-A8) and class C (MrgC11) genes, we analyzed Runx1 conditional knock-out mice by crossing floxed Runx1 mice (Runx1F) with Wnt1-Cre transgenic mice (Jiang et al., 2000; Growney et al., 2005). We have previously reported that in Runx1F/F;Wnt1-Cre mice [referred here to as Runx1−/−(Wnt1)], Runx1 is removed in sensory precursors, therefore representing a complete null mutation in nociceptors (Chen et al., 2006). We found that expression of MrgA1-A8 was eliminated in Runx1−/−(Wnt1) mice at every stage examined, from embryonic day 16.5 (E16.5) to postnatal day 30 (P30) (Fig. 1A–F) (data not shown). High levels of MrgC11 expression were also eliminated in adult Runx1−/−(Wnt1) mice (Fig. 1H vs G, arrow), but low levels of expression were independent of Runx1 (Fig. 1H vs G, arrowheads). We have previously shown that neuronal survival is not affected in Runx1−/−(Wnt1) mice (Chen et al., 2006). Therefore, the loss of Mrg gene expression is unlikely to be caused by a loss of neuronal cells. Altogether, these data imply a requirement of Runx1 for the expression of all Mrg class GPCR genes.
Compartmental expression of Mrg genes in adult DRG
To understand how Mrg gene expression is regulated, we examined in detail the cellular compartments that express Mrg subclasses by using double color in situ hybridizations. As reported previously, adult MrgD+ neurons did not coexpress MrgA3 (Fig. 2A) or MrgB4 (Fig. 2B) (Dong et al., 2001; Zylka et al., 2005). Here we found that the expression of two class A members, MrgA3 and MrgA4, overlapped extensively with each other (Fig. 2C, arrows). Expression of MrgB4 was confined to neurons that exhibited a low level of MrgA3 (Fig. 2D, arrows); neurons with elevated MrgA3 expression did not coexpress MrgB4 (Fig. 2D, arrowheads). It was reported that in adult DRG, elevated MrgC11 expression is excluded from MrgD+ neurons (also see Fig. 3C), but overlaps with MrgA3+ neurons (Zylka et al., 2005). Consistent with the restriction of MrgB4 expression to a subset of MrgA3+ neurons, MrgB4 was expressed in a subset of MrgC11+ neurons (Fig. 2E, arrows). Previous studies failed to detect a coexpression of MrgB4 with MrgA3 or MrgC11 (Zylka et al., 2005); the discrepancy may be attributable to a difference in sensitivity between using nonfluorescent substrates and fluorescent substrates (see Materials and Methods)
Figure 2.
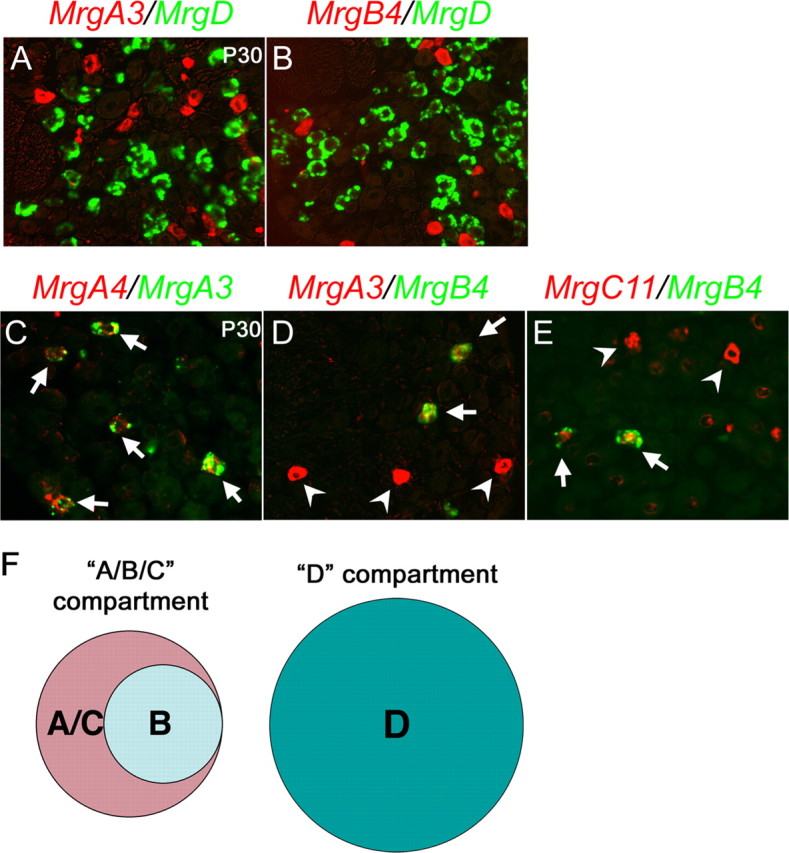
Compartmental expression of Mrg genes. A–E, Double color in situ hybridizations with indicated probes on sections through P30 T12 thoracic DRG of wild-type mice. F, Schematics of two Mrg compartments. The “A/B/C” compartment expresses MrgA3/A4 (“A”), MrgB4 (“B”), and MrgC11 (“C”) in a partially overlapping manner, and the “D” compartment expresses MrgD (“D”).
Figure 3.
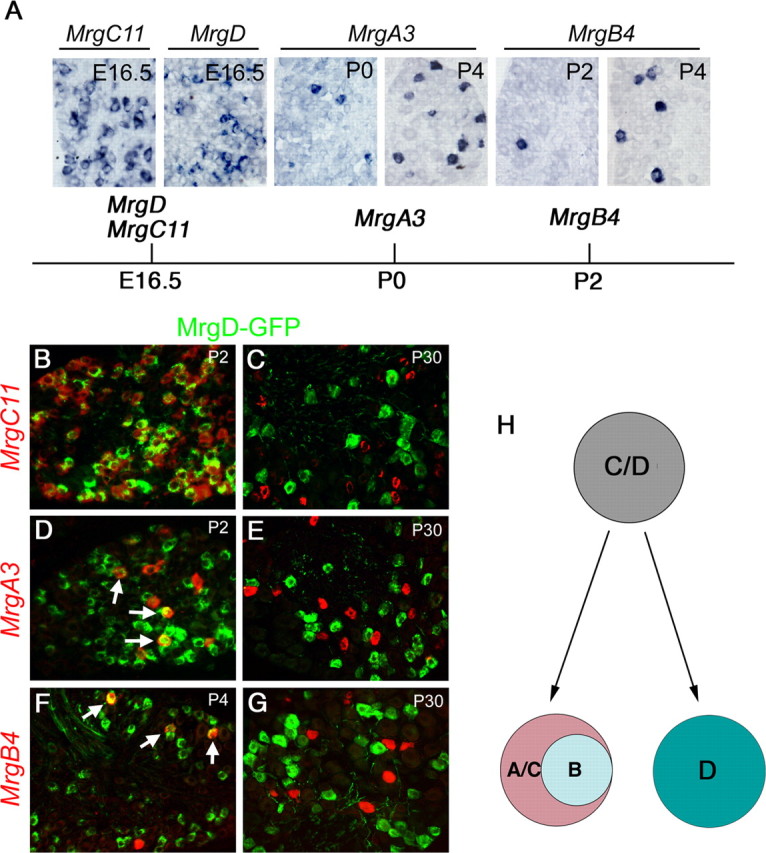
Progressive segregation of Mrg compartments. A, In situ hybridizations on sections through T12 thoracic wild-type DRG at indicated stages and the schematics for the onset of expression of Mrg genes. B–G, Double staining of GFP protein (green) and Mrg mRNA (red) on sections through thoracic DRG at various stages of MrgDGFP/+ heterozygous mice. H, Schematics for progressive compartmental segregation of Mrg gene expression.
In summary, neurons expressing Mrg genes in adult DRG can be divided into two compartments: the “D” compartment that expresses MrgD and the “A/B/C” compartment that shows a partially overlapping expression of MrgA, MrgB, and MrgC genes (Fig. 2F).
Progressive segregation of Mrg expression compartments
We next asked when the segregation of the “A/B/C” and “D” neuronal compartments emerges, by examining their expression at early developmental stages. We found that robust expression of MrgC11 and MrgD was already detected at E16.5, whereas expression of MrgA3 and MrgB4 was initiated at P0 and P2, respectively (Fig. 3A). To further examine the relationship among Mrg+ neurons, we performed a transient fate-mapping experiment by using the MrgDGFP knock-in mice, in which the MrgD coding region is replaced by the gene for green fluorescent protein (GFP) (Zylka et al., 2005). In adult MrgDGFP/+ heterozygous mice, double staining detected a complete overlap between GFP and MrgD mRNA, suggesting that GFP expression can be used to faithfully mark MrgD+ neurons (Zylka et al., 2005) (data not shown).
In P2 MrgDGFP/+ heterozygous mice, ∼96.3% (734 of 762) of MrgD+ neurons, marked by GFP expression, coexpressed MrgC11 (Fig. 3B). We also found that at P2 and P4, GFP protein was detected in 79.4% (131 of 165) and 82.3% (51 of 62) of MrgA3+ and MrgB4+ neurons, respectively (Fig. 3D,F, arrows). Because adult MrgA3+ and MrgB4+ neurons coexpress MrgC11 (Fig. 2), we concluded that most Mrg+ neurons at early developmental stages coexpress MrgD and MrgC11. By P30, GFP protein, however, can no longer be detected in MrgA3+, MrgB4+ or MrgC11+ neurons in MrgDGFP/+ heterozygous mice (Fig. 3C,E,G). These data suggest that the “D” compartment retains MrgD expression and extinguishes MrgC11 expression, and the “A/B/C” compartment retains MrgC11 expression, extinguishes MrgD expression, and acquires the expression of MrgA and MrgB genes (summarized in Fig. 3H).
Dynamic Runx1 expression marks two Mrg expression compartments
To gain insights into how Runx1 regulates Mrg genes, we examined Runx1 expression in Mrg+ neurons at multiple developmental stages. Expression of MrgD was confined predominantly to Runx1+ neurons, from P7 to P30 (Fig. 4A,B). Unexpectedly, despite the loss of expression of MrgA3 and MrgB4 in Runx1−/−(Wnt1) mice, Runx1 protein was not detected in MrgA3+ or MrgB4+ neurons at neonatal and adult stages, including P2 and P4 when the expression of these two genes is actively being established (Fig. 4C–F). It was reported previously that Runx1 is expressed in most, if not all, nociceptors at E12.5-E14.5, but is extinguished in ∼50% of nociceptors during perinatal and postnatal development (Chen et al., 2006). The simplest interpretation is that Runx1 is expressed transiently in immature nociceptors that are fated to become MrgA3+ or MrgB4+ neurons.
Figure 4.
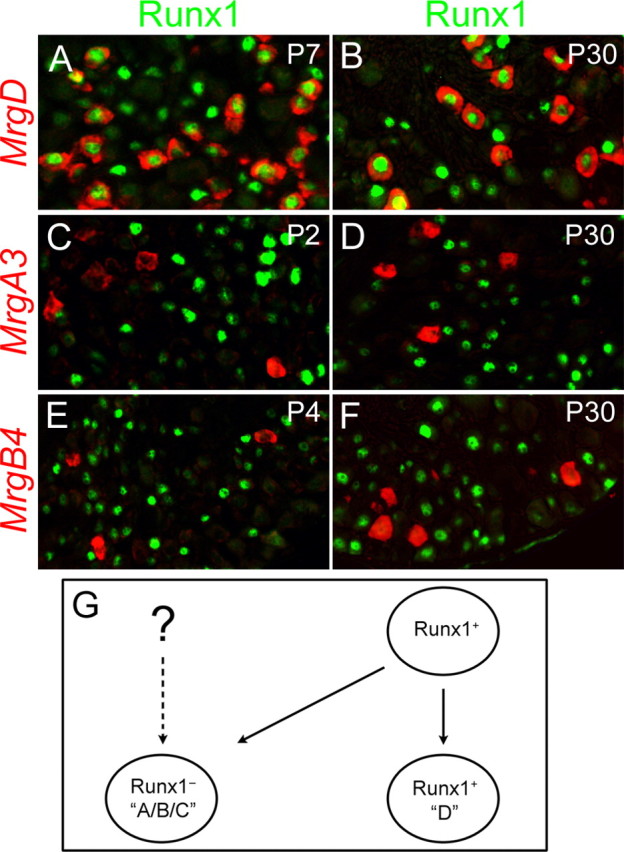
Dynamic Runx1 expression marks distinct Mrg compartments. A–F, Double staining of Runx1 protein (green) and Mrg mRNAs (red) on sections through thoracic wild-type DRG at various stages. G, Schematics for the origin of Mrg compartments. Runx1 expression in immature MrgA/B/C+ neurons is inferred from the fact that Runx1 is expressed in most, if not all, TrkA+ neurons at early embryonic stages (Chen et al., 2006), and TrkA signaling is required for the expression of most Mrg genes (Luo et al., 2007). However, it remains a possibility that Runx1 is never expressed in some MrgA/B/C+ neurons (the dashed line).
In summary, among Mrg+ neurons, persistent Runx1 expression marks the “D” neuronal compartment that expresses MrgD, whereas Runx1 is probably expressed transiently in the “A/B/C” compartment that expresses MrgA, MrgB, and MrgC subclasses (Fig. 4G). However, it is possible that Runx1 might never be expressed in some MrgA3+ or MrgB4+ neurons, thereby non-autonomously controlling the expression of these sensory GPCRs (Fig. 4G). Previous studies show that Mrg+ neurons express Ret, but not TrkA (Dong et al., 2001; Zylka et al., 2003). The lack of Runx1 expression in MrgA/B/C+ neurons implies that Runx1-negative cells include both TrkA+ neurons (Chen et al., 2006) and Ret+ neurons.
Δ446/Δ446 mice
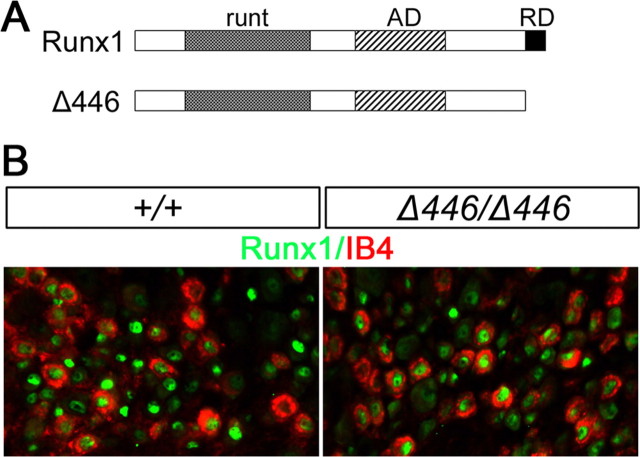
Runx1 protein contains an N-terminal DNA-binding runt domain, a middle transcriptional activation domain, and a C-terminal peptide VWRPY that is capable of binding the Groucho class transcriptional repressor complex (Fig. 5A). Runx1 therefore can act as either a transcriptional repressor or activator (Durst and Hiebert, 2004). To determine whether Runx1 repressor activity contributes to nociceptor phenotype specification, including compartmentalized expression of Mrg genes, we analyzed Δ446 mice (Nishimura et al., 2004). Δ446 encodes a truncated Runx1 protein that lacks the C-terminal repression motif, the VWRPY peptide, but retains the activation domain (Fig. 5A) (Nishimura et al., 2004). In the Δ446 mutant allele, the DNA sequence that encodes the truncated Δ446 protein was inserted into the Runx1 locus. As a result, the expression of Δ446 is under the control of the endogenous Runx1 promoter. Homozygous Δ446/Δ446 mutant mice survive to adulthood (Nishimura et al., 2004). Total neuron numbers in T12 thoracic DRG, detected by the expression of the pan-neuronal marker SCG10 (Stein et al., 1988), were not affected by this Runx1 mutation, with 1244 ± 53 per set of T12 DRG sections in Δ446/Δ446 mice versus 1276 ± 35 in wild-type mice (p = 0.27) (see Materials and Methods). The number of DRG neurons that were labeled by the isolectin B4 (IB4) of Griffonia simplicifolia (Silverman and Kruger, 1988) were also not changed, with 645 ± 15 per set of T12 DRG sections in Δ446/Δ446 mice versus 643 ± 16 in wild-type mice (p = 0.45).
Figure 5.
Sensory neuron development in Δ446/Δ446 mice. A, Schematics for full-length Runx1 protein and the truncated Δ446 protein. The wild-type Runx1 protein (“Runx1”) contains the DNA-binding motif “runt,” the activation domain “AD,” and the C-terminal repression domain “RD.” Δ446 is a truncated Runx1 protein that lacks the RD. B, Double staining of IB4 (red) and full-length or truncated Runx1 protein (green) on sections through T12 thoracic DRG of wild-type and Δ446/Δ446 mice.
Expression of the truncated Δ446 protein can still be detected by the anti-Runx1 antibody (Fig. 5B). Double staining of Δ446 and IB4 in T12 thoracic DRG revealed that ∼64.0 ± 3.6% of IB4+ neurons expressed Δ446 in Δ446/Δ446 mice, which was not different from 65.4 ± 2.3% of IB4+ neurons that expressed the full-length Runx1 protein in wild-type mice (p = 0.18). The percentages of Δ446+ or Runx1+ neurons that were labeled by IB4 were also comparable, with 50.3 ± 6.9% in Δ446/Δ446 mice versus 46.4 ± 2.7% in wild-type mice (p = 0.15). These data suggest that deletion of the C-terminal repression domain affects neither neuronal survival nor Runx1 expression.
Runx1 is required to suppress TrkA and the precursor gene encoding the α-CGRP protein in prospective nonpeptidergic neurons (marked by the staining of IB4), as indicated by the derepression of these two genes in Runx1−/−(Wnt1) mice (Chen et al., 2006; Yoshikawa et al., 2007). Expression of TrkA and α-CGRP, however, was not expanded in IB4+ neurons in Δ446/Δ446 mice (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). These data suggest that the C-terminal repression domain is not involved with Runx1-mediated suppression of TrkA and α-CGRP.
Expression of a set of ion channels and receptors, which is eliminated in conditional Runx1 null mice (Chen et al., 2006), was also not affected in Δ446/Δ446 mice, including TRP channels (TRPA1, TRPM8, and TRPC3), the ATP-gated channel P2X3, the sodium channel (SNS2/Nav1.9), and the GDNF receptor Ret (supplemental Figs. 1, 2, available at www.jneurosci.org as supplemental material), implying that Runx1-mediated activation of these nociceptive molecules does not operate through the C-terminal repression domain.
Expansion of MrgA/B/C gene expression in Δ446/Δ446 mice
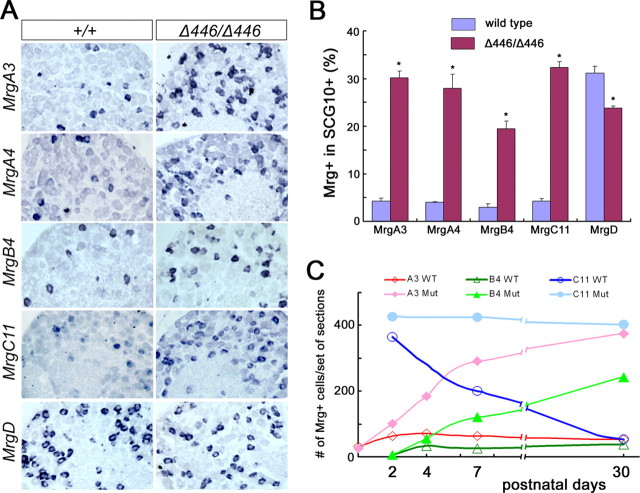
We next examined the expression of Mrg genes in Δ446/Δ446 mice. We found that the expression of class A, B and C Mrg genes was dramatically expanded in adult thoracic DRG of Δ446/Δ446 homozygous mice, including MrgA2-A7, MrgB4, MrgB5, and MrgC11 (Fig. 6A) (data not shown). The percentage of MrgA3+ neurons in P30 T12 thoracic DRG increased from 4.2 ± 0.6 in wild-type mice to 30.1 ± 1.5 in Δ446/Δ446 mice, a 7.2-fold increase (p < 0.0006). Similarly, the percentage of MrgA4+ neurons increased from 4.0 ± 0.1–28.0 ± 3.0, a 7.0-fold increase (p < 0.003), the percentage of MrgB4+ neurons increased from 2.9 ± 0.8–19.4 ± 1.7, a 6.7-fold increase (p < 0.004), and the percentage of MrgC11+ neurons increased from 4.2 ± 0.6–32.3 ± 1.2 (p < 0.0002) (Fig. 6B). An expansion of MrgA3 and MrgB4 expression was also observed in lumbar DRG (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). In contrast, we did not observe increased expression of MrgD, the most abundantly expressed family member. In fact, the percentage of MrgD+ neurons was slightly but significantly reduced, from 31.1 ± 1.4 in wild-type mice to 23.9 ± 0.4 in Δ446/Δ446 mice (p < 0.005) (Fig. 6A,B). These data suggest that, during removal of the Runx1 repressor domain, there is a selective expansion of neurons that express MrgA/B/C genes.
Figure 6.
Expansion of Mrg genes in Δ446/Δ446 mice. A, In situ hybridizations with indicated Mrg probes on sections through T12 thoracic DRG of P30 wild-type and Δ446/Δ446 mice. B, Percentages of Mrg+ neurons in T12 thoracic DRG of P30 wild-type and Δ446/Δ446 mice (*p < 0.005). C, MrgA3+, MrgB4+, and MrgC11+ neuron numbers in T12 thoracic DRG of wild-type and Δ446/Δ446 mice at multiple developmental stages. Six adjacent sets of sections were prepared from each DRG, and numbers of positive neurons in one of the six sets were presented.
We next determined when the derepression of MrgA3, MrgB4 and MrgC11 expression was established, by examining their expression at multiple developmental stages. In wild-type mice, expression of MrgA3 and MrgB4 started at P0 and P2, respectively, and reached peak levels at approximately P4 (Fig. 6C). In Δ446/Δ446 mice, onset of MrgA3 and MrgB4 expression was also established at P0 and P2, respectively (Fig. 6C). Expansion of MrgA3 expression occurred from P2 to beyond P7, and from P4 to beyond P7 for MrgB4 (Fig. 6C), implying a progressive activation of these GPCR genes. The lack of precocious expression in Δ446/Δ446 mice also suggests that the late onset of MrgA3 and MrgB4 expression in wild-type mice is not attributable to Runx1-mediated repression at embryonic stages. Instead, the activator activity for these GPCR genes is established progressively during neonatal development.
In wild-type mice, MrgC11 expression is extinguished from MrgD+ neurons from P2 to adulthood (Fig. 3). In Δ446/Δ446 mice, the numbers of MrgC11+ neurons in T12 thoracic DRG did not exhibit gross reduction at stages from P2 to P7 to P30 (Fig. 6C), implying that Runx1 repression domain is required for postnatal extinguishment of MrgC11 expression (see also below).
Expansion of MrgA/B/C expression in Δ446/Δ446 mice is confined to the IB4+;MrgD+ neuronal compartment
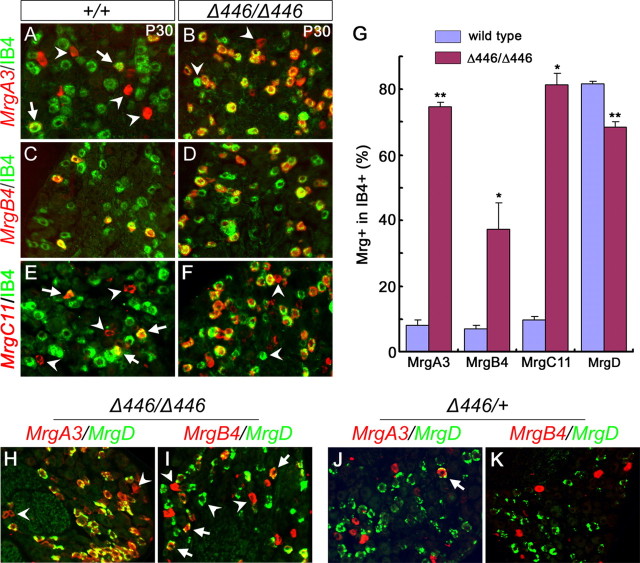
We next asked which population of neurons shows derepression of these GPCR genes in Δ446/Δ446 mice. In wild-type thoracic DRG, MrgA3 and MrgC11 were expressed in both IB4+ and IB4− neurons (Fig. 7A,E, arrows vs arrowheads), whereas MrgB4 (Fig. 7C) and MrgD (Dong et al., 2001) are expressed exclusively in IB4+ neurons. In Δ446/Δ446 homozygous mice, double staining of IB4 and Mrg mRNAs showed that expansion of MrgA3, MrgB4 and MrgC11 expression was confined to IB4+ neurons (Fig. 7B,D,F). The percentage of IB4+ neurons expressing MrgA3 increased from 8.2 ± 1.4 in wild-type mice to 74.7 ± 1.2 in Δ446/Δ446 mice (p < 0.0001), that of MrgB4+ neurons increased from 6.9 ± 1.2–37.4 ± 8.0 in Δ446/Δ446 mice (p < 0.02), and that of MrgC11+ neurons increased from 9.7 ± 1.2–81.4 ± 3.5 (p < 0.02) (Fig. 7G). However, the numbers of MrgA3+, MrgA4+, or MrgC11+ neurons in the IB4− compartment were not significantly changed (data not shown). Expansion of MrgA3 and MrgB4 expression in lumbar DRG was also confined to IB4+ neurons (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). The percentage of MrgD+ neurons in T12 thoracic IB4+ neuronal population, however, is slightly but significantly reduced, from 81.6 ± 1.0 in P30 wild-type mice to 68.5 ± 1.7 in Δ446/Δ446 mice (p < 0.005) (Fig. 7G).
Figure 7.
Expansion of MrgA3, MrgB4, and MrgC11 expression in IB4+;MrgD+ neurons in Δ446/Δ446 mice. A–F, Double staining of IB4 (A–F, green) and MrgA3 mRNA (A, B, red), MrgB4 mRNA (C, D, red) or MrgC11 mRNA (E, F, red) on sections through thoracic DRG of P30 wild-type and Δ446/Δ446 mice. G, The percentage of IB4+ neurons that express MrgA3, MrgB4, MrgC11, or MrgD in T12 thoracic DRG of P30 wild-type and Δ446/Δ446 homozygous mice (*p < 0.02; **p < 0.005). H–K, Double color in situ hybridization on sections through T12 thoracic DRG of P30 Δ446/Δ446 homozygous mice (H, I) and P30 Δ446/+ heterozygous mice (J, K).
In wild-type DRG, the majority of IB4+ neurons express MrgD, and these MrgD+ neurons normally do not coexpress MrgA3, MrgB4 or MrgC11 (Fig. 2) (Zylka et al., 2003). In adult Δ446/Δ446 mice, nearly all MrgD+ neurons in thoracic DRG coexpressed MrgA3 (Fig. 7H) and MrgA4 (data not shown). Similarly, a portion of MrgD+ neurons coexpressed MrgB4 (Fig. 7I). In addition, because 81.4% and 68.5% of IB4+ neurons express MrgC11 and MrgD, respectively, we concluded that at least 49.9% [81.4-(100–68.5) = 49.9] of IB4+ neurons coexpress MrgC11 and MrgD in adult Δ446/Δ446 mice. These results suggest that Runx1 actively suppresses the expression of MrgA/B/C genes in MrgD+ neurons. However, we noted that a small number of MrgA3+ or MrgB4+ cells lacked MrgD expression in Δ446/Δ446 mice (Fig. 7H,I, arrowheads). The simplest interpretation is that in Δ446/Δ446 mice, MrgD expression is not expanded into the prospective “A/B/C” compartment that normally lack persistent Runx1 expression.
In summary, removal of the C-terminal Runx1 repression domain leads to a unidirectional expansion of MrgA/B/C genes into the “D” neuronal compartment.
Discussion
Mechanism of compartmentalized expression of Mrg class sensory GPCRs
Our studies suggest a model that leads to compartmentalized expression of Mrg genes. This model contains two key components. First, Runx1 is initially expressed in most embryonic nociceptors at E12.5-E14.5, but is extinguished in ∼50% of cells during perinatal/postnatal development (Chen et al., 2006), leading to the creation of Runx1+ and Runx1− neuronal compartments. Second, different Mrg family members exhibit different responses to Runx1-mediated repression. Runx1 is initially required for the expression of all Mrg genes, but at an undefined developmental stage Runx1 becomes a repressor of MrgA/B/C. Accordingly, expression of MrgA/B/C is confined to the Runx1− “A/B/C” compartment, and is actively suppressed in the Runx1+ “D” compartment. During removal of the Runx1 repressor domain in Δ446/Δ446 mice, the truncated Runx1 protein is converted into an activator, leading to a dramatic expansion of MrgA and MrgB expression in the “D” compartment. The truncated Δ446 protein is required for the expansion because a null mutation in Runx1−/−(Wnt1) mice leads to a loss, rather than an expansion, of MrgA and MrgB expression. MrgC11 is initially expressed in the “D” compartment. Subsequent extinguishment of MrgC11 expression in this compartment relies on Runx1-mediated repression. Expression of MrgD, however, is essentially insensitive to Runx1-mediated repression, thereby allowing sustained MrgD expression in the Runx1+ “D” compartment. In summary, the selective extinguishment of Runx1 is critical for the establishment of the “A/B/C” compartment (by avoiding Runx1-mediated suppression of MrgA/B/C genes at late developmental stages), whereas persistent Runx1 expression allows a singular MrgD expression in the “D” compartment (by actively suppressing MrgA/B/C expression and possibly maintaining MrgD expression).
A few outstanding issues remain to be solved. First, Runx1 protein is not detected in MrgA3+ or MrgB4+ neurons at neonatal stages, raising the question about whether Runx1 indirectly or even non-autonomously controls the expression of these GPCR genes. Second, the signal that is responsible for Runx1 extinguishment in the “A/B/C” compartment is unknown. Third, it should be noted that in rat DRG, MrgA and MrgD gene expression overlaps extensively (Zylka et al., 2003), mimicking the expression pattern present in Δ446/Δ446 mice. The mechanism for species-specific expression pattern is unclear. One attractive possibility is that the rat MrgA promoter loses the capacity to bind to the Runx1 repressor complex, leading to a concurrent Runx1-mediated activation of MrgA and MrgD genes.
Runx1 uses distinct pathways to suppress gene expression in nociceptors
Runx1 is required to suppress several peptidergic neuron markers in IB4+ nonpeptidergic nociceptors, including TrkA and CGRP (Chen et al., 2006; Yoshikawa et al., 2007). Here we found that Runx1 also actively suppresses MrgA/B/C genes in these neurons. Interestingly, Runx1 uses distinct mechanisms to suppress these two categories of genes. Runx1-mediated suppression of MrgA/B/C, but not TrkA/CGRP, is dependent on the C-terminal repression domain that is known to interact with the Groucho-repressor complex (Durst and Hiebert, 2004; Nishimura et al., 2004). Removal of this repression domain in Δ446/Δ446 mice leads to a derepression of MrgA/B/C (Figs. 6, 7), without causing a concurrent derepression of TrkA/CGRP in IB4+ neurons (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). In the immune system, Runx1 is able to use multiple repression domains to suppress T cell receptor expression (Durst and Hiebert, 2004; Telfer et al., 2004). Runx1 could in principle use a different repression domain to suppress TrkA/CGRP. Alternatively, Runx1 may activate a downstream pathway that indirectly suppresses peptidergic neuron markers. A support for the latter scenario is the finding that Ret-mediated signaling is required for postnatal suppression of TrkA in IB4+ neurons (Luo et al., 2007). Accordingly, the loss of Ret expression after conditional Runx1 knock-out may explain the derepression of TrkA and CGRP in Runx1−/−(Wnt1) mice (Chen et al., 2006; Yoshikawa et al., 2007), whereas a normal expression of Ret in Δ446/Δ446 mice (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) may explain the lack of TrkA derepression. Regardless, Runx1 operates through distinct pathways to suppress peptidergic differentiation and MrgA/B/C expression.
Progressive specification of sensory neuron subtypes
Our studies, combined with a series of recent studies, suggest that specification of sensory subtypes is progressively established during perinatal and postnatal development. Most embryonic nociceptors (and thermoceptors) initially express both Runx1 and TrkA (Chen et al., 2006). To our knowledge, there are no known molecular markers that are able to divide TrkA+ neurons into distinct subgroups at E12.5. It is from E14.5 to postnatal stages that nociceptors are progressively segregated into Ret+, TrkA+ and Ret+;TrkA+ subclasses, with persistent Runx1 expression is confined to a portion of Ret+ neurons (Bennett et al., 1996, 1998; Molliver et al., 1997; Chen et al., 2006). Nociceptors that express distinct profiles of TRP class thermal receptors also emerge during perinatal/postnatal development (Hjerling-Leffler et al., 2007). Here we further found that Mrg+ sensory neurons initially coexpress MrgD and MrgC11 at embryonic stages. During postnatal development, future MrgC11+ neurons extinguish MrgD and activate MrgAand/or MrgB genes. Conversely, future MrgD+ neurons switch off MrgC11, leading to a mutually exclusive expression of MrgA/B/C and MrgD.
Intriguingly, MrgD+ afferents innervate skin epidermis (Zylka et al., 2005), whereas MrgB4+ afferents innervate exclusively the hairy skin (Liu et al., 2007). Such topographically distinct innervation raises the hypothesis that segregation of MrgD+ and MrgB4+ nociceptors might partly depend on specific target-derived signals. Indeed, Ret signaling is required for the expression of MrgB4 (and MrgA3), but not MrgD (Luo et al., 2007). Hair follicles also release other signaling molecules, such as bone morphological proteins or BMPs (Blanpain and Fuchs, 2006). Indeed, a conditional knock-out of Smad4, encoding a key component of BMP-mediated signaling (Yang et al., 2002), is required selectively for the expression of MrgB4 (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). Future studies will be directed to investigate how target-derived signals interface with intrinsic factors, such as Runx1, in generating sensory cell diversity.
Footnotes
The work was supported by National Institutes of Health (NIH)–National Institute of Neurological Disorders and Stroke Training Grant 5T32NS007473-09 (F.-C.Y.), NIH–National Institute of Dental and Craniofacial Research Grant 1R01DE018025 (Q.M.), and NIH–National Institute of Neurological Disorders and Stroke Grant 5P01NS047572 (Q.M.). Q.M. is a Claudia Adams Barr Scholar. We thank Drs. Gary Gilliland and Nancy Speck for the Runx1 conditional knock-out mice, Chuxia Deng for conditional Smad4 null mice, David Rowitch for Wnt1-Cre mice, Tom Jessell for the Runx1 antibody, and Keith Ligon for critical comments on this manuscript.
References
- Agarwal N, Offermanns S, Kuner R. Conditional gene deletion in primary nociceptive neurons of trigeminal ganglia and dorsal root ganglia. Genesis. 2004;38:122–129. doi: 10.1002/gene.20010. [DOI] [PubMed] [Google Scholar]
- Bennett DL, Averill S, Clary DO, Priestley JV, McMahon SB. Postnatal changes in the expression of the trkA high-affinity NGF receptor in primary sensory neurons. Eur J Neurosci. 1996;8:2204–2208. doi: 10.1111/j.1460-9568.1996.tb00742.x. [DOI] [PubMed] [Google Scholar]
- Bennett DL, Michael GJ, Ramachandran N, Munson JB, Averill S, Yan Q, McMahon SB, Priestley JV. A distinct subgroup of small DRG cells express GDNF receptor components and GDNF is protective for these neurons after nerve injury. J Neurosci. 1998;18:3059–3072. doi: 10.1523/JNEUROSCI.18-08-03059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein ES, Ott TR, Feddock M, Ma JN, Fuhs S, Wong S, Schiffer HH, Brann MR, Nash NR. Characterization of the Mas-related gene family: structural and functional conservation of human and rhesus MrgX receptors. Br J Pharmacol. 2006;147:73–82. doi: 10.1038/sj.bjp.0706448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Jiang J, Chen T, Hong Y. Sensory neuron-specific receptor agonist BAM8–22 inhibits the development and expression of tolerance to morphine in rats. Behav Brain Res. 2007;178:154–159. doi: 10.1016/j.bbr.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Chen CL, Broom DC, Liu Y, de Nooij JC, Li Z, Cen C, Samad OA, Jessell TM, Woolf CJ, Ma Q. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Choi SS, Lahn BT. Adaptive evolution of MRG, a neuron-specific gene family implicated in nociception. Genome Res. 2003;13:2252–2259. doi: 10.1101/gr.1431603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozier RA, Ajit SK, Kaftan EJ, Pausch MH. MrgD activation inhibits KCNQ/M-currents and contributes to enhanced neuronal excitability. J Neurosci. 2007;27:4492–4496. doi: 10.1523/JNEUROSCI.4932-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106:619–632. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- Grazzini E, Puma C, Roy MO, Yu XH, O'Donnell D, Schmidt R, Dautrey S, Ducharme J, Perkins M, Panetta R, Laird JM, Ahmad S, Lembo PM. Sensory neuron-specific receptor activation elicits central and peripheral nociceptive effects in rats. Proc Natl Acad Sci USA. 2004;101:7175–7180. doi: 10.1073/pnas.0307185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerling-Leffler J, Alqatari M, Ernfors P, Koltzenburg M. Emergence of functional sensory subtypes as defined by transient receptor potential channel expression. J Neurosci. 2007;27:2435–2443. doi: 10.1523/JNEUROSCI.5614-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, Arber S. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–393. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Lembo PM, Grazzini E, Groblewski T, O'Donnell D, Roy MO, Zhang J, Hoffert C, Cao J, Schmidt R, Pelletier M, Labarre M, Gosselin M, Fortin Y, Banville D, Shen SH, Strom P, Payza K, Dray A, Walker P, Ahmad S. Proenkephalin A gene products activate a new family of sensory neuron-specific GPCRs. Nat Neurosci. 2002;5:201–209. doi: 10.1038/nn815. [DOI] [PubMed] [Google Scholar]
- Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Vrontou S, Rice FL, Zylka MJ, Dong X, Anderson DJ. Molecular genetic visualization of a rare subset of unmyelinated sensory neurons that may detect gentle touch. Nat Neurosci. 2007;10:946–948. doi: 10.1038/nn1937. [DOI] [PubMed] [Google Scholar]
- Luo W, Wickramasinghe SR, Savitt JM, Griffin JW, Dawson TM, Ginty DD. A Hierarchical NGF signaling cascade controls Ret-dependent and Ret-independent events during development of nonpeptidergic DRG neurons. Neuron. 2007;54:739–754. doi: 10.1016/j.neuron.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Marmigere F, Montelius A, Wegner M, Groner Y, Reichardt LF, Ernfors P. The Runx1/AML1 transcription factor selectively regulates development and survival of TrkA nociceptive sensory neurons. Nat Neurosci. 2006;9:180–187. doi: 10.1038/nn1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliver DC, Wright DE, Leitner ML, Parsadanian AS, Doster K, Wen D, Yan Q, Snider WD. IB4-binding DRG neurons switch from NGF to GDNF dependence in early postnatal life. Neuron. 1997;19:849–861. doi: 10.1016/s0896-6273(00)80966-6. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Fukushima-Nakase Y, Fujita Y, Nakao M, Toda S, Kitamura N, Abe T, Okuda T. VWRPY motif-dependent and -independent roles of AML1/Runx1 transcription factor in murine hematopoietic development. Blood. 2004;103:562–570. doi: 10.1182/blood-2003-06-2109. [DOI] [PubMed] [Google Scholar]
- Silverman JD, Kruger L. Lectin and neuropeptide labeling of separate populations of dorsal root ganglion neurons and associated “nociceptor” thin axons in rat testis and cornea whole-mount preparations. Somatosens Res. 1988;5:219–246. doi: 10.3109/07367228809144630. [DOI] [PubMed] [Google Scholar]
- Stein R, Orit S, Anderson DJ. The induction of a neural-specific gene, SCG10, by nerve growth factor in PC12 cells is transcriptional, protein synthesis dependent, and glucocorticoid inhibitable. Dev Biol. 1988;127:316–325. doi: 10.1016/0012-1606(88)90318-1. [DOI] [PubMed] [Google Scholar]
- Telfer JC, Hedblom EE, Anderson MK, Laurent MN, Rothenberg EV. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J Immunol. 2004;172:4359–4370. doi: 10.4049/jimmunol.172.7.4359. [DOI] [PubMed] [Google Scholar]
- Theriault FM, Nuthall HN, Dong Z, Lo R, Barnabe-Heider F, Miller FD, Stifani S. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J Neurosci. 2005;25:2050–2061. doi: 10.1523/JNEUROSCI.5108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf CJ, Ma Q. Nociceptors-noxious stimulus detectors. Neuron. 2007;55:353–364. doi: 10.1016/j.neuron.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002;32:80–81. doi: 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Senzaki K, Yokomizo T, Takahashi S, Ozaki S, Shiga T. Runx1 selectively regulates cell fate specification and axonal projections of dorsal root ganglion neurons. Dev Biol. 2007;303:663–674. doi: 10.1016/j.ydbio.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Zhang L, Taylor N, Xie Y, Ford R, Johnson J, Paulsen JE, Bates B. Cloning and expression of MRG receptors in macaque, mouse, and human. Brain Res Mol Brain Res. 2005;133:187–197. doi: 10.1016/j.molbrainres.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Zhong J, Pevny L, Snider WD. “Runx”ing towards sensory differentiation. Neuron. 2006;49:325–327. doi: 10.1016/j.neuron.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Dong X, Southwell AL, Anderson DJ. Atypical expansion in mice of the sensory neuron-specific Mrg G protein-coupled receptor family. Proc Natl Acad Sci USA. 2003;100:10043–10048. doi: 10.1073/pnas.1732949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45:17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]