Abstract
The epithelium of the mouse vomeronasal organ (VNO) consists of apical and basal layers of neuronal cell bodies. Vomeronasal sensory neurons (VSNs) with cell bodies in the basal layer express the G-protein subunit Gαo and members of the V2R superfamily of vomeronasal receptor genes and project their axons to the posterior accessory bulb (AOB). V2R+ VSNs also express particular patterns of a family of nine nonclassical class I major histocompatibility Mhc genes, the H2-Mv genes. The function of H2-Mv molecules remains unknown. H2-Mv molecules have been reported to be associated with V2R molecules and have been proposed to participate in pheromone detection. Here, we find that a substantial fraction of V2R+ VSNs does not express these nine H2-Mv genes. The cell bodies of H2-Mv+ and H2-Mv− VSNs reside in the lower and upper sublayers of the basal layer, respectively. This spatial segregation is maintained at the level of the AOB: H2-Mv+ and H2-Mv− VSNs project their axons to the posterior and anterior subdomains of the posterior AOB, respectively. By generating a C-terminal green fluorescent protein fusion protein with M10.2 in gene-targeted mice, we observe subcellular localization of M10.2 not only in dendrites but also in axons of VSNs. Our results reveal a tripartite organization of the VNO and AOB, question the generality of the requirement of these nine H2-Mv molecules for V2R surface expression, and suggest that H2-Mvs can function in both dendrites and axons.
Keywords: accessory olfactory bulb, axonal projection, Mhc, sensory neuron, V2R, vomeronasal organ
Introduction
The vomeronasal organ (VNO) is a chemosensory organ in higher vertebrates and resides at the base of the nasal septum. It is thought to be specialized in the detection of pheromones that mediate behaviors such as mating and aggression (Keverne, 1999; Dulac and Torello, 2003; Halpern and Martinez-Marcos, 2003), but the VNO has neither a specificity nor the exclusivity for pheromone detection: “common” odorants can also be detected by the VNO (Sam et al., 2001; Trinh and Storm, 2003), and, conversely, molecules with pheromonal effects can also be detected by the main olfactory epithelium (Brennan and Zufall, 2006; Zufall and Leinders-Zufall, 2007). Vomeronasal sensory neurons (VSNs) express genes of two superfamilies that encode polypeptides with a putative seven-transmembrane-domain structure: vomeronasal receptor genes V1R (Dulac and Axel, 1995; Rodriguez et al., 2002) and V2R (Herrada and Dulac, 1997; Matsunami and Buck, 1997; Ryba and Tirindelli, 1997). The cell bodies of V1R+ and V2R+ VSNs reside in nonoverlapping layers of the VNO epithelium: apical and basal, respectively. The spatial restriction of V1R and V2R gene expression correlates with differential expression of two G-protein subunits, Gαi2 and Gαo (Berghard and Buck, 1996; Jia and Halpern, 1996). This duality is maintained at the level of the accessory olfactory bulb (AOB): axons of VSNs from the apical and basal layers synapse in, respectively, the anterior (aAOB) and posterior (pAOB) halves of the AOB.
V1R+ VSNs respond to volatile compounds that have pheromonal activity in phenomena such as puberty delay, estrus induction, and intermale aggression (Leinders-Zufall et al., 2000; Boschat et al., 2002; Del Punta et al., 2002a; Stowers and Marton, 2005). V2R+ VSNs respond to nonvolatile ligands such as major histocompatibility Mhc class I peptides, which function as individuality signals underlying mate recognition (Leinders-Zufall et al., 2004), ESP1, a male-specific 7 kDa peptide secreted from the extraorbital lacrimal gland (Kimoto et al., 2005), and MUPs, major urinary proteins (Chamero et al., 2007).
Others (Loconto et al., 2003) and we (Ishii et al., 2003) have shown that V2R+ VSNs express another multigene family, termed H2-Mv, representing nonclassical class I genes of the Mhc. Each of the nine H2-Mv genes is expressed in a subset of VSNs; individual VSNs can coexpress multiple H2-Mv genes. These H2-Mvs are coexpressed in a combinatorial manner with the V2R superfamily. It has been proposed that H2-Mvs function as escort molecules in the transport of V2Rs to the cell surface and are restricted to dendrites (Loconto et al., 2003). However, H2-Mv genes are not present in the opossum genome, which nonetheless contains 70–120 V2R genes with an intact open reading frame (Shi and Zhang, 2007; Young and Trask, 2007).
Here, we reveal a tripartite compartmentalization of the mouse VNO and AOB, with three neuronal populations: V1R+ VSNs, V2R+ H2-Mv− VSNs, and V2R+ H2-Mv+ VSNs. A substantial fraction of V2R receptors is unlikely to be dependent on these H2-Mv molecules for surface expression because the genes are not coexpressed.
Materials and Methods
In situ hybridization.
Male mice were used for all experiments. In situ hybridization (ISH) was performed as described (Ishii et al., 2004). RNA probes were prepared for H2-Mv, V2r1b, V2ra, V2rb, V2rc, and V2rf1–3 (Ishii et al., 2003), V1rb1 (Rodriguez et al., 2002), V2rf4 (nucleotide 21286287–21286501 from GenBank accession number NT_039500.7), V2ro (nucleotide 430–947 from GenBank accession number XM_001472549.1), pan-specific Mhc class I (α3 domain of H2-Q1, nucleotide 655–841 from GenBank accession number NM_010390.2), and tauVenus (bovine tau, nucleotide 102–621 from GenBank accession number NM_174106.2; Venus, nucleotide 1543–2262 from GenBank accession number DQ289580.1). The probe V2rf1–3 is identical to the probe V2rf in the study by Ishii et al. (2003). The mixed H2-Mv probe was prepared by mixing all nine H2-Mv probes after each probe was tested by ISH. To enhance the probe specificity of M10.2 and M9, cRNAs without antigen from other M10s and M1 were added to M10.2–digoxigenin (DIG) and M9–DIG probes in the hybridization mixture, respectively. In the ISH conditions used, sequence homology of <80% does not result in cross-hybridization. The bovine tau probe does not cross-react with murine tau. Probe specificity was determined by analysis of related genes in the mouse genome sequence and, in some cases, confirmed by multicolor ISH. There are two nomenclatures for H2-Mv genes (Ishii et al., 2003; Loconto et al., 2003).
Generation of M10.2–internal ribosomal entry site (IRES)–tauVenus, M10.2::green fluorescent protein (GFP), and V2rf2–IRES–barley lectin (BL) strains.
A bacterial artificial chromosome containing M10.2 was isolated from RCPI-22, a 129SvEvTAC library (Children's Hospital Oakland Research Institute, Oakland, CA). A 8.5 kb SmaI fragment containing exons 3–6 of M10.2 was subcloned in pBluescript. A PacI site was generated by recombinant PCR immediately after the stop codon of M10.2. The IRES–tauVenus–loxP-+ACE-cre/neo-loxP (ACNF) cassette, in which GFP of the IRES–tauGFP–ACNF (Bozza et al., 2002) was replaced with Venus (Nagai et al., 2002), was inserted in this PacI site to assemble the M10.2–IRES–tauVenus–ACNF targeting vector. For the M10.2::GFP–ACNF targeting vector, a PacI site was generated by recombinant PCR immediately before the stop codon of M10.2. The GFP–ACNF cassette, in which loxP-neo-loxP (LNL) of GFP–LNL (Feinstein et al., 2004) was replaced with ACNF, was inserted in the PacI site, generating the M10.2::GFP–ACNF targeting vector.
For the V2rf2 targeting vector, an 8.5 kb EcoRV–PvuII fragment containing the transmembrane domain exon and 3′ nontranslated region of V2rf2 was subcloned, and a PacI site was generated one nucleotide after the stop codon of V2rf2. The IRES–BL–ACNF cassette, in which tauGFP of the IRES–tauGFP–ACNF was replaced with barley lectin (Horowitz et al., 1999), was inserted in the PacI site, generating the V2rf2–IRES–BL–ACNF targeting vector. The vectors were linearized and electroporated into E14 embryonic stem (ES) cells as described previously (Mombaerts et al., 1996). G418-resistant clones were screened by Southern blot hybridization with probes that are 5′ external to the targeting vector of both M10.2 (nucleotides 22488475–22487976 from GenBank accession number NT_039649.6, EcoRI digestion of genomic DNA) and V2rf2 (nucleotides 5141453–5142292 from GenBank accession number NW_000028.1, PacI and KpnI digestions of genomic DNA). Homologous recombinant ES cells were injected into C57BL/6 blastocysts, and chimeras were bred with wild-type C57BL/6 mice. The M10.2–tauVenus, M10.2::GFP, and V2rf2–BL strains are in a mixed (129 × C57BL/6) background. The strains will be publicly available from The Jackson Laboratory (Bar Harbor, ME), as follows: M10.2–tauVenus with strain name B6;129P2–H2–M10.2tm1Mom/MomJ or stock number 6725; M10.2::GFP with strain name B6;129P2–H2–M10.2tm2Mom/MomJ or stock number 7878; and V2rf2–BL with strain name B6;129P2–Vmn2r81tm1Mom/MomJ or stock number 6727.
Immunohistochemistry.
All mice used for analysis are male. Wild-type mice are C57BL/6J, and mutant mice are in a mixed (129 × C57BL/6) background. Dissection and sectioning were performed as described (Ishii et al., 2004) except that postfixation with 4% paraformaldehyde/PBS was performed for 2 h for both VNO and AOB samples, and the decalcification step was omitted for AOB samples. Sections were fixed in 4% paraformaldehyde/PBS for 15 min and blocked in 5% horse serum and 0.1% Triton X-100/PBS for 60 min, followed by incubation with primary antibodies at 4°C overnight. After washing in 0.1% Triton X-100/PBS, sections were incubated with secondary antibodies at room temperature for 2 h and counterstained with TOTO-3 (Invitrogen, Carlsbad, CA). Primary antibodies were the rabbit anti-V2R2 antibody (Martini et al., 2001), chicken anti-GFP antibody (Abcam, Cambridge, MA) used as an anti-Venus antibody, goat anti-wheat germ agglutinin antibody (Vector Laboratories, West Grove, PA) used as an anti-barley lectin antibody, and rabbit anti-Gαo antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies were Alexa 555 donkey anti-rabbit IgG, Alexa 546 donkey anti-goat IgG (Invitrogen), FITC donkey anti-chicken IgG, and cyanine 5 donkey anti-rabbit IgG (Jackson ImmunoResearch). Sections were analyzed with a Zeiss (Oberkochen, Germany) LSM510 confocal microscope.
Gene nomenclature.
The genes we call V2rf1, V2rf2, V2rf3, and V2rf4 are identical to mV2R56, mV2R58, mV2R57, and mV2R55 in the study by Yang et al. (2005) and to Vmn2r82, Vmn2r81, Vmn2r80, and Vmn2r83 in the study by Young and Trask (2007). Moreover, V2rf2 is identical to EC1–V2R (Loconto et al., 2003). V2r1b (Del Punta et al., 2002b) is identical to mV2R19 (Yang et al., 2005) and to Vmn2r26 (Young and Trask, 2007). For H2-Mv genes, we continue to use the nomenclature used by Ishii et al. (2003), as follows: M10.1 (M10.2 in the study by Loconto et al., 2003), M10.2 (M10.1), M10.3 (M10.3), M10.4 (M10.5), M10.5 (M10.7), M10.6 (M10.8), M1 (M1), M9 (M9), and M11 (M7.2).
Results
Maturation of H2-Mv expression during postnatal development
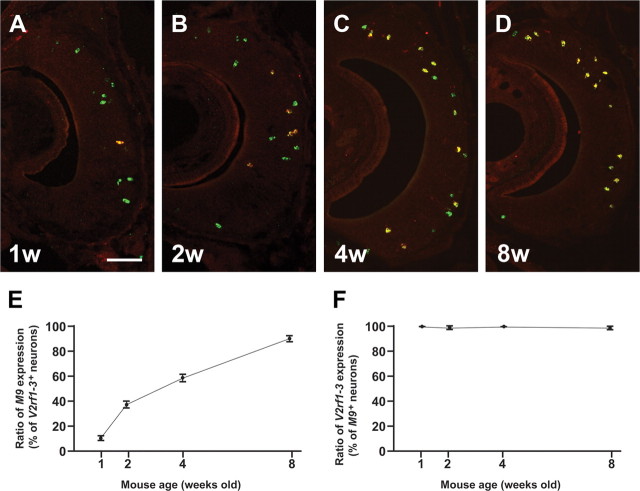
The onset of H2-Mv expression follows that of V2Rs during postnatal development (Ishii et al., 2003). To define the temporal expression pattern in more detail, we examined M9 and V2rf1–3 mRNA coexpression in mice of ages 1, 2, 4, and 8 weeks by ISH (Fig. 1A–D). The M9 probe is specific to M9 of the H2-Mv family, and the V2rf1–3 probe detects three V2R genes: V2rf1, V2rf2, and V2rf3. We find that, in mice that are 1 week of age, ∼10% of V2rf1–3+ cells coexpress M9. This fraction increases to ∼37% at 2 weeks and to ∼59% at 4 weeks, and reaches ∼90% at 8 weeks (Fig. 1E). The converse is not the case: close to 100% of M9+ cells are labeled with the V2rf1–3 probe at all stages (Fig. 1F). Thus, M9 expression appears to undergo a process of maturation at the population level of VSNs. To exclude this confounding factor, mice are at least 8 weeks old in the analyses below, unless mentioned otherwise.
Figure 1.
Onset of M9 expression follows expression the V2rf1–3 subfamily. A–D, Two-color ISH with the M9 probe (red) and the V2rf1–3 subfamily probe (green) on coronal sections of the VNO of wild-type mice at 1 week (1w, A), 2 weeks (2w, B), 4 weeks (4w, C), and 8 weeks (8w, D). VSNs that are labeled with both probes appear yellow. E, Ratio of M9 expression among V2rf1–3+ neurons increases with age: 10 ± 2.6% at 1 week, 37 ± 5.3% at 2 weeks, 59 ± 6.5% at 4 weeks, and 90 ± 4.4% at 8 weeks. F, Ratio of V2rf1–3 expression among M9+ neurons remains consistent at all ages: 100% at 1 week, 98.7 ± 1.5 at 2 weeks, 100% at 4 weeks, and 99.3% ± 0.58 at 8 weeks. Average ± SD for three mice are shown in E and F. Scale bar, 100 μm.
Distinct layer localization of V2R+ VSNs that are H2-Mv+ and H2-Mv−
Some VSNs are not labeled with a mixed ISH probe for the nine H2-Mv genes (Fig. 2A,B): for instance, cells revealed by an ISH probe for GFP as expressing V2r1b in V2r1b–IRES–tauGFP mice (Del Punta et al., 2002b) and cells revealed by a specific ISH probe for V2rf4. The position of the cell bodies of H2-Mv+ and H2-Mv− VSNs within the VNO epithelium is distinct, as revealed by using probe V2r1b (detecting nine related V2R genes) for H2-Mv− VSNs (Fig. 2C) and probe V2rf1–3 (detecting V2rf1, V2rf2, and V2rf3 but not V2rf4) for H2-Mv+ VSNs (Fig. 2D), together with Gαi2 and mixed H2-Mv probes. We find that ∼90% of VSNs that are labeled with the V2r1b probe are negative for the H2-Mv mixed probe in mice of 8–10 weeks. In contrast, >98% of VSNs labeled with the V2rf1–3 probe are positive for the mixed H2-Mv probe. V2r1b+ cell bodies reside between Gαi2+ cell bodies and H2-Mv+ cell bodies (Fig. 2C), and V2rf1–3+ cell bodies are located most basally among cell bodies labeled with the mixed H2-Mv probe (Fig. 2D). We analyzed the distribution of V1R+ cell bodies, V2R+ H2-Mv− cell bodies, and V2R+ H2-Mv+ cell bodies along the apical–basal dimension of the VNO epithelium with probes for V1rb1, V2r1b, and V2rf1–3, as representative genes, respectively (Fig. 2E,F). V2r1b+ cell bodies are located more apically compared with V2rf1–3+ cell bodies, and, in turn, V1rb1+ cell bodies are localized more apically (Fig. 2F).
Figure 2.
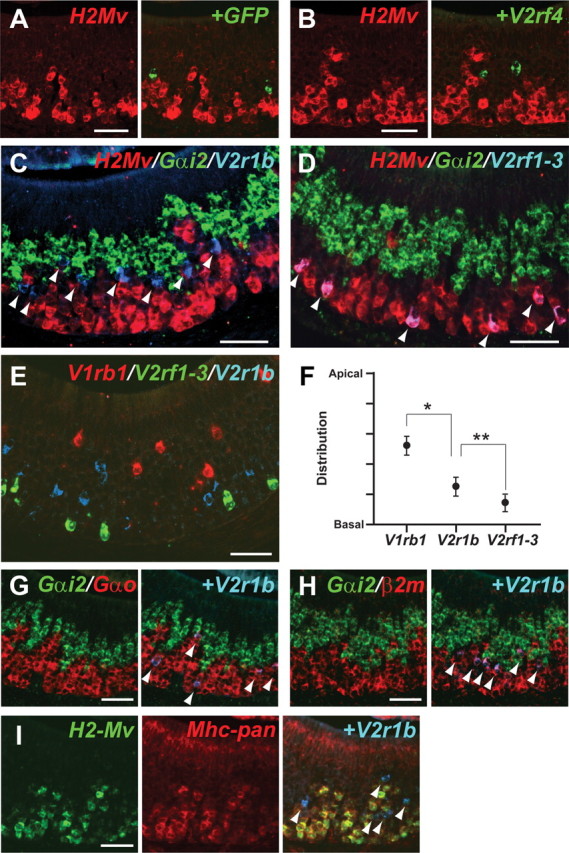
Two- and three-color ISH reveal three layers of VSNs. A, Two-color ISH with the mixed H2-Mv probe (red) and the GFP probe (green) on a VNO section of a V2r1b–IRES–tauGFP mouse shows nonoverlapping expression. B, Two-color ISH with the mixed H2-Mv probe (red) and the V2rf4-specific probe (green) shows nonoverlapping expression. C, D, Three-color ISH with the mixed H2-Mv mixed probe (red), the Gαi2 probe (green), and the V2r1b probe (blue) (C) or the V2rf1–3 probe (blue) (D). Overlay of red and blue results in purple. E, Three-color ISH with the V1rb1 probe (red), the V2rf1–3 probe (green), and the V2r1b probe (blue). F, The apical–basal positions of V1rb1-, V2rf1–3-, and V2r1b-labeled cell bodies in E were measured, with 0 for basal and 1 for apical. They are distributed in distinct layers: V1rb1, 0.52 ± 0.12 (n = 89); V2r1b, 0.26 ± 0.13 (n = 196); V2rf1–3, 0.15 ± 0.11 (n = 159). *p < 0.01, **p < 0.01. G, H, Three-color ISH with the Gαi2 probe (green), the V2r1b probe (blue), and the Gαo probe (red) (G) or the β2m probe (red). H shows overlap of V2r1b signals with both Gαo and β2-microglobulin. I, Three-color ISH with the mixed H2-Mv mixed probe (green), the Mhc-pan probe from H2-Q1 (red), and the V2r1b probe. V2r1b+ neurons are also Mhc-pan−. Arrowheads point to V2r1b+ cell bodies in C and G–I and to V2rf1–3+ cell bodies in D. All samples were from 8- to 10-week-old male mice. Wild-type mice were used in B–I. Scale bars, 50 μm.
We then examined V2r1b+ cells, which are not labeled with the mixed H2-Mv probe, further with ISH probes for Gαo and β2-microglobulin (β2m) and with a pan-Mhc class I probe (Mhc-pan) (Fig. 2G–I). β2m is associated in a protein complex with most Mhc class I molecules. The pan-specific Mhc class I probe corresponds to a sequence in the α3 region of H2-Q1 that has >92% homology with most known class Ia genes and class Ib genes (H2-Q and H2-T) and 77–89% homology to H2-M, which includes the nine members of the H2-Mv family and three other class I Mhc genes. We find that V2r1b+ cells are unmistakably positive for the Gαo probe, such that there is no obvious gap between the Gαi2 and Gαo layers (Fig. 2G). Although most V2r1b+ cells are negative for the H2-Mv mixed probe, they are labeled with the β2m probe (Fig. 2H). V2r1b+ cells are negative for the Mhc-pan probe (Fig. 2I). Incidentally, the Mhc-pan probe labels H2-Mv+ cells, most likely by cross-hybridization with the expressed H2-Mvs, and also labels sustentacular cells, which probably express other Mhc class I genes. Thus, it appears that no known Mhc class I gene, classical or nonclassical, is expressed in V2r1b+ cells and in VSNs with cell bodies in the upper part of the basal layer.
M10.2 is expressed in most H2-Mv+ neurons
An ISH probe for M10.2 detects the largest number of VSNs among all H2-Mv members (Fig. 3D,E). To visualize M10.2+ VSNs and their axonal projections to the AOB, we generated a mouse strain with a targeted mutation in the M10.2 locus that results in cotranslation of bicistronic messages encoding M10.2 along with a fusion protein between tau and Venus (Nagai et al., 2002), a variant of yellow fluorescent protein (Fig. 3A). The M10.2–IRES–tauVenus mutation is abbreviated as M10.2–Venus. In coronal sections through the VNO of a M10.2–Venus mouse, large numbers of cell bodies in the basal layer of the epithelium express strongly Venus by immunohistochemistry (IHC) (Fig. 3B), consistent with the ISH results (Fig. 3D).
Figure 3.
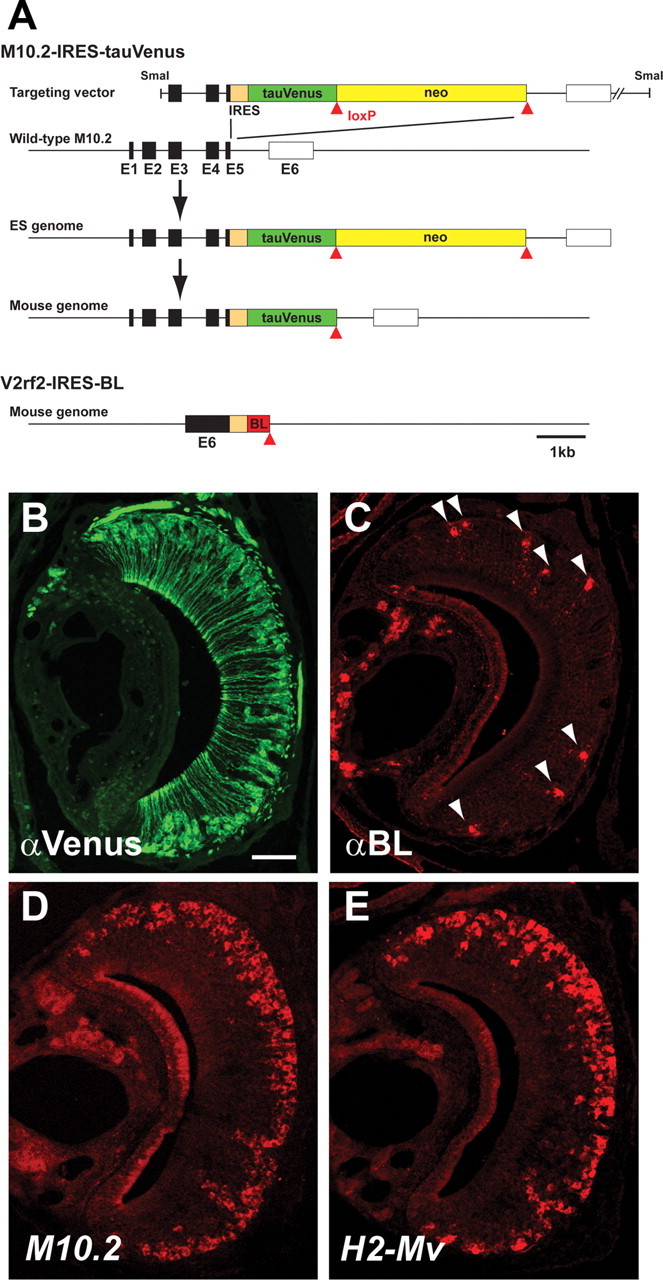
Targeted mutagenesis of the M10.2 and V2rf2 loci. A, The IRES–tauVenus–ACNF cassette was inserted immediately after the stop codon of M10.2 by homologous recombination in ES cells. The ACNF cassette, a self-excising neo gene, was removed during transmission through the male germ line, leaving a single loxP site (red triangle) behind in the locus. The IRES–BL was inserted after the stop codon of V2rf2 in the mouse genome according to a similar genetic design. Filled and open boxes represent coding and noncoding exons, respectively. E, Exon. B, IHC for Venus expression in a coronal section of the VNO from an M10.2–Venus mouse. C, IHC for BL in a coronal section of the VNO of a V2rf2-mouse. Arrowheads point to labeled cell bodies of VSNs. D, ISH with the M10.2 probe in a wild-type mouse. E, ISH with the mixed H2-Mv probe in a wild-type mouse. Scale bar, 100 μm.
To characterize the expression pattern of M10.2–Venus, we performed ISH with M10.2 and bovine tau probes on VNO sections from heterozygous M10.2–Venus mice (Fig. 4A). We observe that the ISH signals overlap completely. This result, together with a similar expression pattern of M10.2 in wild-type mice (Fig. 3D), indicates that the expression pattern of M10.2 is not altered by the targeted IRES–tauVenus insertion. The overlap further indicates that M10.2 expression is bi-allelic, in contrast to mono-allelic expression of V2r1b (Del Punta et al., 2002b). More than 95% of the VSNs that are labeled with the H2-Mv mixed probe are also positive for the tauVenus probe (Fig. 4B). IHC with an anti-Venus antibody and an anti-V2R2 antibody (Martini et al., 2001), which presumably labels most or all basal VSNs, shows that nearly all H2-Mv+ neurons are situated in the lower part of the basal layer (Fig. 4C), confirming and extending the ISH observations of Figure 2.
Figure 4.
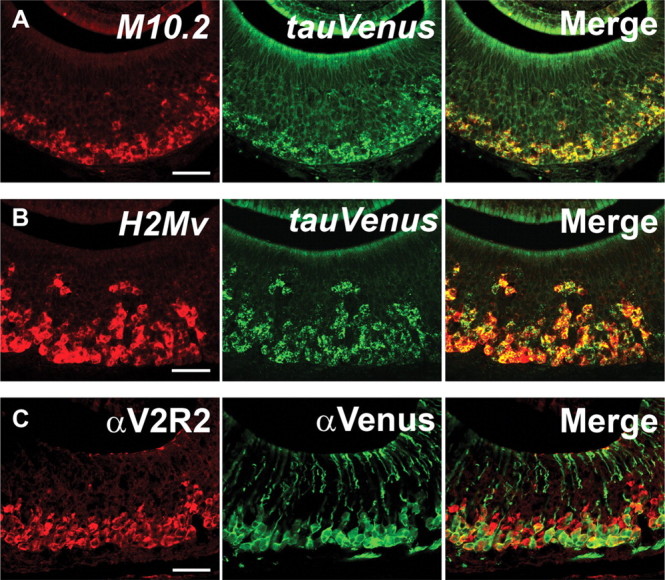
M10.2–Venus expression in the VNO. A, Two-color ISH of an M10.2–Venus heterozygous mouse. Unaltered expression pattern of M10.2 by the targeted insertion of IRES–tauVenus was confirmed by two-color ISH with the M10.2 probe (red) and the tauVenus probe (green) on a coronal section of the VNO. All cell bodies are both red and green, thus yellow in overlay, indicating bi-allelic expression of M10.2. B, Two-color ISH in an M10.2–Venus homozygous mouse. More than 95% of cell bodies that are labeled with the mixed H2-Mv probe (red) are also labeled with the tauVenus probe (green) by two-color ISH. C, Two-color IHC in an M10.2–Venus homozygous mouse with anti-V2R2 and anti-Venus antibodies. The cell bodies of M10.2+ neurons reside in the lower sublayer of the basal layer. All mice are 10 weeks old. Scale bars, 50 μm.
M10.2+ neurons project to the most posterior region within the posterior AOB
The M10.2–Venus mice provides the opportunity, which was thus far not available, to visualize specifically the axonal projections of H2-Mv+ VSNs. In whole-mount specimens, bright Venus fluorescence is observed in the pAOB, whereas faint fluorescence can also be observed in the most anterior part of the aAOB (Fig. 5A). Sagittal sections of AOB of a M10.2–Venus mice were stained by IHC with anti-Venus antibody and with anti-Gαo antibody, which labels the entire pAOB. We find that the posterior two-thirds of the pAOB are innervated by M10.2–Venus axons (Fig. 5B). The Venus+ subdomain is separated from the Venus− subdomain along the anterior–posterior axis, although the border is not as sharp as for Gαo (Fig. 5B,C). Together with the observation that M10.2+ neurons represent most of H2-Mv+ neurons, we conclude that the pAOB consists of an H2-Mv− subdomain in the anterior third and a H2-Mv+ subdomain in the posterior two-thirds. A relatively faint Venus IHC signal was observed in the area around the most anterior part of the aAOB (Fig. 5D), as is also observed in whole-mount view (Fig. 5A). The origin of this signal is not known.
Figure 5.
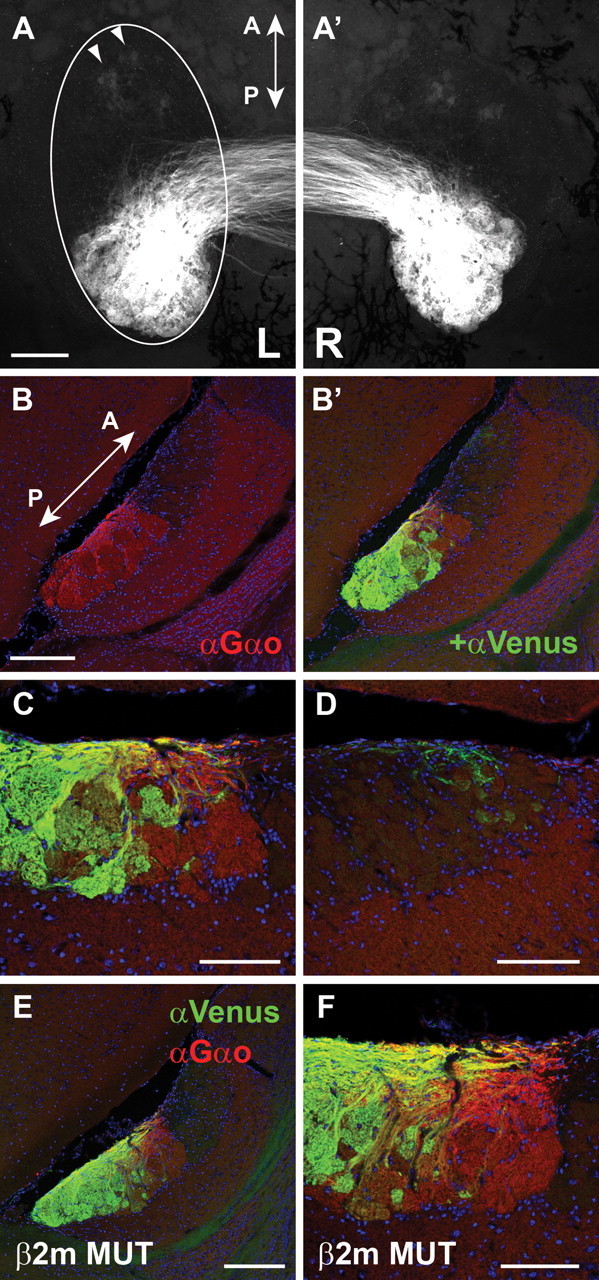
Axonal projections of M10.2–Venus+ VSNs to the AOB. (A, A′) Whole-mount dorsal view of the AOB shows major axonal projections to the pAOB with minor projections to the aAOB (arrowheads), in an M10.2–Venus homozygous mouse. Left and right AOBs are shown in A and A′, respectively. B, B′, IHC in an M10.2–Venus homozygous mouse. A sagittal section of the AOB was stained with anti-Gαo (red) and anti-Venus (green) antibodies. The Venus signal localizes the most posterior two-thirds of the pAOB, and weak signals are also observed in the most anterior region of the aAOB. C, Higher-magnification view of the middle area of the AOB in B. D, Higher-magnification view of the aAOB in B. Weak signals of diffusely projecting axons are observed. E, F, IHC with anti-Gαo and anti-Venus antibodies on sagittal sections of the AOB from a β2-microglobulin mutant mouse in an M10.2–Venus heterozygous background. No obvious difference is observed compared the higher-magnification views of the β2-microglobulin mutant mouse in F with the wild-type mouse in C. Sections were counterstained with TOTO-3 to visualize nuclei in B–F. Mice are 10–12 weeks old. A, Anterior; P, posterior. Scale bars: A, B, E, 200 μm; C, D, F, 100 μm.
No obvious role for β2-microglobulin in axonal projections
β2-microglobulin is essential for expression of most Mhc class I molecules on the cell surface (Koller et al., 1990; Zijlstra et al., 1990; Neefjes and Momburg, 1993). It has been proposed that H2-Mv and β2-microglobulin molecules form a VSN-specific, multi-molecular complex and that they participate in transport of V2Rs to the cell surface (Loconto et al., 2003). β2-microglobulin mutant mice reportedly exhibit a defect of V2R expression on dendritic terminals (Loconto et al., 2003), as evaluated with the anti-VN4/V2R antiserum (Martini et al., 2001). However, we were unable to confirm this defect (our unpublished observations). In any case, our observation that some of the V2R+ VSNs are H2-Mv− raises the possibility that transport of some V2Rs to the cell surface is independent of H2-Mv. A possible role of H2-Mvs in the formation of segregation of axonal projections between H2-Mv+ and H2-Mv− neurons in the AOB was tested in β2-microglobulin mutant mice, crossed with M10.2–Venus mice. We do not observe an obvious difference between wild-type (Fig. 5B,C) and mutant (Fig. 5E,F) mice.
Thus, it is possible that not all V2Rs require β2-microglobulin and H2-Mvs for surface expression.
Axonal projections of VSNs expressing a particular V2R gene
We previously reported a mouse strain with a targeted mutation V2r1b–IRES–tauGFP (abbreviated as V2r1b–GFP mice) (Del Punta et al., 2002b). With a similar genetic design, we generated a novel mouse strain with a targeted mutation in the V2rf2 gene such that BL is cotranslated with V2rf2 from bicistronic messages (Fig. 3A). The V2rf2–IRES–barley lectin strain is henceforth abbreviated as V2rf2–BL. Barley lectin is an axonal marker and has also been used for trans-synaptic labeling (Horowitz et al., 1999; Yoshihara et al., 1999; Zou et al., 2001). In coronal sections of the VNO, scattered VSNs are revealed by immunohistochemistry with an anti-BL antibody (Fig. 3C). In AOB sections, BL+ axons coproject to the same glomeruli as GFP+ axons of V2rf2–IRES–tauGFP mice (data not shown).
We determined the location of glomeruli for V2r1b and V2rf2 in the AOB of V2r1b–GFP and V2rf2–BL mice, respectively. V2r1b glomeruli are located anterior to V2rf2 glomeruli. V2rf2 glomeruli reside within the anterior region of the M10.2+ subdomain (Fig. 6A), and V2r1b glomeruli reside within the anterior region of the pAOB (Fig. 6B,C), which corresponds to the M10.2− subdomain. Together, VSNs expressing a particular V2R project their axons to a domain of the AOB along the anterior–posterior axis (Fig. 6D), with a relative order that corresponds to the layers in the VNO epithelium along the apical–basal axis.
Figure 6.
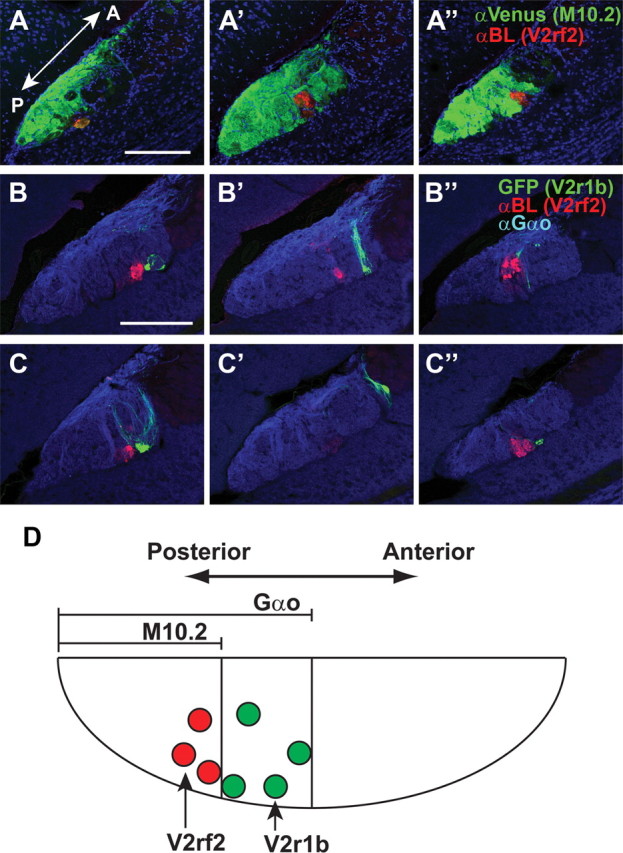
Axonal projections of V2R1b+ and V2rf2+ VSNs to the AOB. A, A′, A″, IHC in a mouse doubly heterozygous for M10.2–tauVenus and V2rf2-BL. Three distinct sagittal sections from the AOB were stained with anti-Venus (green) and anti-BL (red) antibodies. B, B′, B″, C, C′, C″, IHC in mice doubly heterozygous for V2r1b–GFP and V2rf2–BL. Three distinct sagittal sections from the AOB of two mice (B, C) were stained with anti-BL (red) and anti-Gαo (blue) antibodies. D, Summary of the subdomains of the AOB. Sections were counterstained with TOTO-3 in A–D. All mice are 10 weeks old. A, Anterior; P, posterior. Scale bars, 200 μm.
M10.2 protein localization in the VNO and AOB
IHC with an antibody directed against M10.7 (Loconto et al., 2003; M10.5 in our nomenclature) has revealed the presence of M10.5 at the VNO dendrite tip but not at synaptic terminals in the AOB, suggesting a role of H2-Mv in dendrites but not in axons. To examine further the H2-Mv subcellular localization, we generated a novel mouse strain in which GFP is fused to the C-terminal end of M10.2 (Fig. 7A). This mouse is abbreviated henceforth as the M10.2::GFP. In the VNO of 5-week-old mice, M10.2::GFP signal by IHC is abundantly present at microvilli in the VNO (Fig. 7B,C). Interestingly, M10.2::GFP is also clearly detected in the glomeruli of the AOB (Fig. 7D,E). IHC in the AOB of M10.2::GFP is very similar to that of M10.2–Venus, indicating that the expression of M10.2 is not altered by the targeted insertion of GFP.
Figure 7.
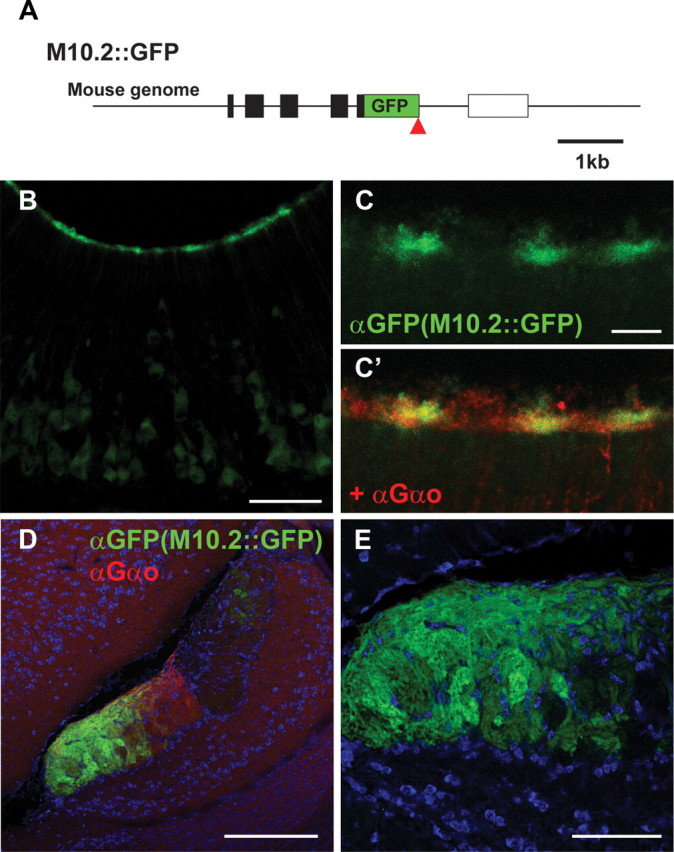
M10.2 subcellular localization. A, Generation of M10.2::GFP fusion by gene targeting. GFP was inserted by homologous recombination after the M10.2 coding sequence, such that GFP is fused to C terminal of M10.2. Symbols are same as those in Figure 3A. B, C, IHC with anti-GFP antibody on coronal VNO sections of an M10.2::GFP mouse. Anti-Gαo antibody was also used to visualize microvilli (C′). M10.2::GFP localizes to microvilli. D, IHC with anti-GFP and anti-Gαo antibodies on sagittal sections of the AOB of an M10.2::GFP mouse. E, A close-up view of the GFP signal in the AOB in D. All mice are 5 weeks old. Scale bars: B, 50 μm; C, 10 μm; D, 200 μm; E, 100 μm.
Thus, M10.2 protein localizes both in dendrites and axons of VSNs.
Discussion
Our developmental analysis of M9 and V2rf1–3 coexpression reveals that M9 expression does not reach a stable level until adult stage, when the ratio is nearly 1:1. Postnatal onset of other H2-Mvs and coexpression patterns of other combinations of H2-Mv and V2r (Ishii et al., 2003; Loconto et al., 2003) suggest that expression of H2-Mv genes undergoes a “maturation” until it is stable in adulthood. This maturation may imply that H2-Mvs function in adult mice rather than in developing mice.
VSNs expressing V2Rs, such as V2r1b and V2rf4, are negative for the mixed H2-Mv probe, although they express β2-microglobulin, a polypeptide that is typically but not always associated with class I Mhc molecules. We offer five interpretations for these observations. First, these VSNs indeed do not express any class I Mhc gene. Second, the expression level of some H2-Mvs is below the sensitivity of ISH. Third, class I Mhc genes other than the nine H2-Mvs are expressed in these VSNs. However, even the Mhc-pan probe derived from H2-Q1 does not reveal expression of class I Mhc genes in V2r1b+ neurons. Fourth, expression of class I Mhc genes is induced in certain conditions. Fifth, β2-microglobulin itself functions as an escort protein for surface expression of V2Rs in VSNs that do not express any of the nine H2-Mvs.
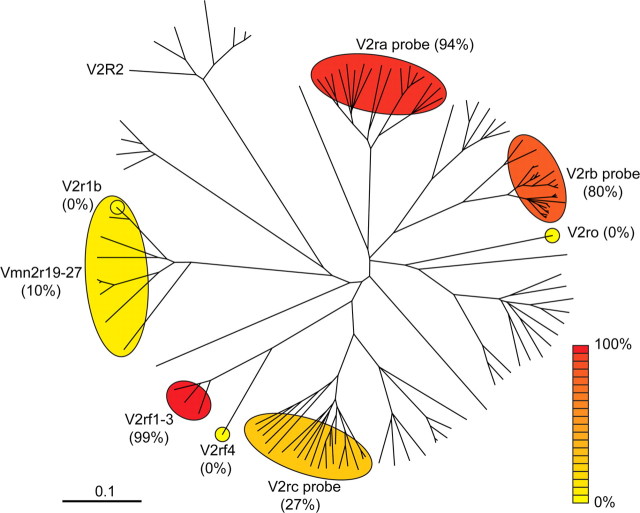
We constructed an unrooted tree of amino acid sequences from the most recent analysis of the mouse V2R repertoire (Young and Trask, 2007) and is shown in Figure 8. We overlaid the information that is available about ratios of coexpression with H2-Mv genes. Interestingly, V2rf4 is not coexpressed with H2-Mv genes, whereas the closely related genes V2rf1, V2rf2, and V2rf3 are nearly always coexpressed with H2-Mv genes. Such striking differences can perhaps be developed as an experimental model to understand the mechanisms of coexpression.
Figure 8.
An unrooted tree representing mouse V2R genes with ratios of H2-Mv coexpression. Amino acid sequences reported by Young and Trask (2007) were aligned using ClustalX, with 1000 bootstraps. V2R genes that are detected with a given probe are shown within a circle. The ratios of V2R+ cells that are colabeled with a mixed H2-Mv probe by ISH are represented in various colors, from low (yellow) to high (red). Probes should detect the following V2R genes: V2ra (Vmn2r8–17, 84–89, 121), V2rb (Vmn2r28–52), V2rc (Vmn2r91–110), V2rf1–3 (Vmn2r80–82), V2r1b (Vmn2r19–27), V2rf4 (specific for Vmn2r83), and V2ro (specific for Vmn2r65).
H2-Mv− VSNs are localized in the upper sublayer of the basal layer, thus in the middle layer of the epithelium of the VNO. Our studies thus reveal a tripartite organization of layers in the epithelium of the mouse VNO, defined by gene expression patterns: V1R+/Gαi2+, V2R+/Gαo+/H2-Mv−, and V2R+/Gαo+/H2-Mv+ layers. In rat, expression of V2Rs is also regionalized into two to three layers along the basal–apical axis (Herrada and Dulac, 1997).
Because M10.2 is expressed in the majority of H2-Mv+ neurons, the M10.2–Venus mice are a useful, and thus far unique, strain to visualize H2-Mv+ neurons and their axons. We find that M10.2+ VSNs project their axons to the most posterior two-third of the pAOB, revealing a novel compartmentalization of the pAOB. Consistent with the division of the pAOB into an anterior H2-Mv− subdomain and a posterior H2-Mv+ subdomain, glomeruli for V2r1b+ VSNs, which are H2-Mv−, are distributed within the anterior region of the pAOB, and glomeruli for V2rf2+ VSNs, which are H2-Mv+, reside within the anterior part of the M10.2+ domain, thus in the middle third of the pAOB. Together, our observations indicate that the pAOB exhibits also a tripartite organization, with subdomains arranged along the anterior–posterior axis. The biological significance of this compartmentalization is not clear, but it could subserve aspects of the processing of chemosensory information.
A correlation between positions of cell bodies and axonal projection sites is also known for the main olfactory system: odorant receptor (OR) expression zones in main olfactory epithelium correlate roughly with the dorsal–ventral positions of glomeruli in the main olfactory bulb (Alenius and Bohm, 1997; Yoshihara and Mori, 1997; Yoshihara et al., 1997; Vassalli et al., 2002; Nakatani et al., 2003; Miyamichi et al., 2005). However, there is a fundamental difference in the axis of organization: the compartments of OR gene expression occupy a part of the surface area of epithelium (“zones”), but the three compartments of V1R/V2R gene expression occupy the entire surface area of the epithelium of the VNO and are organized in a dimension that is perpendicular to the surface (“layers”).
There is some evidence that H2-Mv proteins interact with V2Rs and that H2-Mvs promote cell surface localization of V2Rs in a heterologous cell expression system and in mouse (Loconto et al., 2003). In contrast, in our hands, V2R IHC signals along dendritic processes are also present in β2-microglobulin mutant mice and indistinguishable from wild-type mice (our unpublished observations). Insofar as H2-Mvs are dependent on β2-microglobulin for surface expression, our observations suggest that H2-Mvs do not function always as essential chaperone molecules for V2R molecules. Perhaps H2-Mvs may be involved in sorting or recycling of V2R molecules in endosomes. Immunoelectron microscopy will be necessary to examine definitively the subcellular localization of V2R proteins. Additional arguments against a generic chaperone function of H2-Mvs are that some V2Rs are not coexpressed with H2-Mvs (this study) and that H2-Mv genes are absent from the opposum genome, which has nonetheless many intact V2R genes (Shi and Zhang, 2007). The H2-Mv-defined subdomains in the AOB do not depend on β2-microglobulin expression, as indicated by a normal projection of M10.2–Venus+ axons to the AOB in β2m-deficient mice. Although H2-Mv can form a complex with β2m (Olson et al., 2005), H2-Mv function may not require β2m, because some class I Mhc molecules such as MIC and ZAG can function without β2m (Rodgers and Cook, 2005). Finally, the behavioral defects observed in β2m-deficient mice are difficult to interpret in terms of defective H2-Mv/V2R function, given the ubiquitous expression of β2-microglobulin in nearly every cell of a mouse.
Our observations of M10.2::GFP protein in both the dendrites and axons are consistent with dual roles of H2-Mv molecules. Human and mouse class I and class II MHC molecules have been tagged with GFP, by transfection in cultured cells or by targeted mutagenesis, and no deleterious effects of the tagging on subcellular location or biological function have been observed (Wubbolts et al., 1996; Grommé et al., 1999; Boes et al., 2002; Zwart et al., 2005). Because H2-Mv genes are expressed in a variegated manner in V2R+ VSNs and some V2R+ VSNs do not express H2-Mv genes, H2-Mvs may have supportive or modulatory functions for particular but not all types of V2R molecules or have functions that are independent of V2Rs. Combinatorial coexpression of H2-Mvs and V2Rs (Ishii et al., 2003; Loconto et al., 2003) and association of H2-Mv with V2R molecules (Loconto et al., 2003) suggest that H2-Mvs may function, in some cases, in a V2R complex in VSN microvilli and may be involved in the reception of pheromone ligand as a receptor or a modulatory factor or in signaling directly or indirectly. The “maturation” of H2-Mv expression in adulthood could alter the recognition ability of sensory cues by modifying V2R functions or by adding chemosensory receptor functions of H2-Mvs. Mhc peptide ligands can function as chemosignals in VSNs (Leinders-Zufall et al., 2004; Boehm and Zufall, 2006). V2R+ VSNs respond to Mhc peptides in a manner similar to peptide recognition by class I Mhc molecules, and these peptides function as a mate recognition cue in the context of the pregnancy-block effect. Therefore, it is tempting to speculate that Mhc peptide ligands may bind to H2-Mvs, although the only H2-Mv tested, M10.4 in our nomenclature, does not appear to bind classical Mhc peptide ligands (Olson et al., 2005).
The best known function of class I Mhc molecules is to present peptide antigen from self and non-self origin to T-cell antigen receptors (Neefjes and Momburg, 1993). Because M10.2::GFP is expressed in dendrites and axons, essentially throughout the VSN, it cannot be excluded that the main function of H2-Mvs is in immune defense. However, over the past decade, class I Mhc molecules have been implicated in the refinement of synapse formation and plasticity in the visual system and the hippocampus (Corriveau et al., 1998; Huh et al., 2000; Boulanger and Shatz, 2004) and in the maintenance of synapses of motoneurons (Oliveira et al., 2004). Mhc molecules are localized postsynaptically in dendrites of hippocampal neurons and appear to be involved in homeostatic regulation of synaptic function and morphology in response to neural activity (Goddard et al., 2007). A candidate Mhc class I receptor is expressed in subsets of neurons throughout the brain (Syken et al., 2006). The presence of M10.2 protein at axon terminals suggests that H2-Mvs may also have synaptic functions and modify synaptic connectivity or plasticity at an adult stage, perhaps resulting in dynamic functional changes of a subpopulation of V2R+ H2-Mv+ VSNs. Because V2R proteins are not detected in the AOB by the anti-VN4 antibody (Martini et al., 2001), the function of H2-Mvs at axon terminals may be independent of V2R molecules. H2-Mv protein operating at presynaptic sites, in contrast to class I Mhc molecules functioning at postsynaptic sites in the classical models for neuronal Mhc expression, may reveal novel functions of class I Mhc molecules in neural connectivity.
Definitive answers about the various proposed functions of H2-Mv molecules await the generation of mice that lack all nine H2-Mv genes.
Footnotes
This work was supported by a postdoctoral fellowship from the Human Frontier Science Program and R03 grant support from National Institutes of Health/National Institute on Deafness and Other Communication Disorders (NIH/NIDCD) (T.I.). P.M. acknowledges the generous grant support from NIH/NIDCD. We thank Akiko Ishii for expert technical assistance, Roberto Tirindelli for anti-VN4 and anti-V2R2 antibodies, and Duancheng Wen and Wei Tang for blastocyst injections of ES cells.
References
- Alenius M, Bohm S. Identification of a novel neural cell adhesion molecule-related gene with a potential role in selective axonal projection. J Biol Chem. 1997;272:26083–26086. doi: 10.1074/jbc.272.42.26083. [DOI] [PubMed] [Google Scholar]
- Berghard A, Buck LB. Sensory transduction in vomeronasal neurons: evidence for Gαo, Gαi2, and adenylyl cyclase II as major components of a pheromone signaling cascade. J Neurosci. 1996;16:909–918. doi: 10.1523/JNEUROSCI.16-03-00909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T, Zufall F. MHC peptides and the sensory evaluation of genotype. Trends Neurosci. 2006;29:100–107. doi: 10.1016/j.tins.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Boes M, Cerny J, Massol R, Op den Brouw M, Kirchhausen T, Chen J, Ploegh HL. T-cell engagement of dendritic cells rapidly rearranges MHC class II transport. Nature. 2002;418:983–988. doi: 10.1038/nature01004. [DOI] [PubMed] [Google Scholar]
- Boschat C, Pelofi C, Randin O, Roppolo D, Luscher C, Broillet MC, Rodriguez I. Pheromone detection mediated by a V1r vomeronasal receptor. Nat Neurosci. 2002;5:1261–1262. doi: 10.1038/nn978. [DOI] [PubMed] [Google Scholar]
- Boulanger LM, Shatz CJ. Immune signalling in neural development, synaptic plasticity and disease. Nat Rev Neurosci. 2004;5:521–531. doi: 10.1038/nrn1428. [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan PA, Zufall F. Pheromonal communication in vertebrates. Nature. 2006;444:308–315. doi: 10.1038/nature05404. [DOI] [PubMed] [Google Scholar]
- Chamero P, Marton TB, Logan DW, Flanagan K, Cruz JR, Saghatelian A, Cravatt BF, Stowers L. Identification of protein pheromones that promote aggressive behaviour. Nature. 2007;450:899–902. doi: 10.1038/nature05997. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–520. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysocki CJ, Ogawa S, Zufall F, Mombaerts P. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature. 2002a;419:70–74. doi: 10.1038/nature00955. [DOI] [PubMed] [Google Scholar]
- Del Punta K, Puche A, Adams NC, Rodriguez I, Mombaerts P. A divergent pattern of sensory axonal projections is rendered convergent by second-order neurons in the accessory olfactory bulb. Neuron. 2002b;35:1057–1066. doi: 10.1016/s0896-6273(02)00904-2. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat Rev Neurosci. 2003;4:551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proc Natl Acad Sci USA. 2007;104:6828–6833. doi: 10.1073/pnas.0702023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grommé M, Uytdehaag FGCM, Janssen H, Calafat J, van Binnendijk RS, Kenter MJH, Tulp A, Verwoerd D, Neefjes J. Recycling MHC class I molecules and endosomal peptide loading. Proc Natl Acad Sci USA. 1999;96:10326–10331. doi: 10.1073/pnas.96.18.10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern M, Martinez-Marcos A. Structure and function of the vomeronasal system: an update. Prog Neurobiol. 2003;70:245–318. doi: 10.1016/s0301-0082(03)00103-5. [DOI] [PubMed] [Google Scholar]
- Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Horowitz LF, Montmayeur JP, Echelard Y, Buck LB. A genetic approach to trace neural circuits. Proc Natl Acad Sci USA. 1999;96:3194–3199. doi: 10.1073/pnas.96.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Hirota J, Mombaerts P. Combinatorial coexpression of neural and immune multigene families in mouse vomeronasal sensory neurons. Curr Biol. 2003;13:394–400. doi: 10.1016/s0960-9822(03)00092-7. [DOI] [PubMed] [Google Scholar]
- Ishii T, Omura M, Mombaerts P. Protocols for two- and three-color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J Neurocytol. 2004;33:657–669. doi: 10.1007/s11068-005-3334-y. [DOI] [PubMed] [Google Scholar]
- Jia C, Halpern M. Subclasses of vomeronasal receptor neurons: differential expression of G proteins (Giα2 and Goα) and segregated projections to the accessory olfactory bulb. Brain Res. 1996;719:117–128. doi: 10.1016/0006-8993(96)00110-2. [DOI] [PubMed] [Google Scholar]
- Keverne EB. The vomeronasal organ. Science. 1999;286:716–720. doi: 10.1126/science.286.5440.716. [DOI] [PubMed] [Google Scholar]
- Kimoto H, Haga S, Sato K, Touhara K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature. 2005;437:898–901. doi: 10.1038/nature04033. [DOI] [PubMed] [Google Scholar]
- Koller BH, Marrack P, Kappler JW, Smithies O. Normal development of mice deficient in β2M, MHC class I proteins, and CD8+ T cells. Science. 1990;248:1227–1230. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- Leinders-Zufall T, Lane AP, Puche AC, Ma W, Novotny MV, Shipley MT, Zufall F. Ultrasensitive pheromone detection by mammalian vomeronasal neurons. Nature. 2000;405:792–796. doi: 10.1038/35015572. [DOI] [PubMed] [Google Scholar]
- Leinders-Zufall T, Brennan P, Widmayer P, S PC, Maul-Pavicic A, Jager M, Li XH, Breer H, Zufall F, Boehm T. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- Loconto J, Papes F, Chang E, Stowers L, Jones EP, Takada T, Kumanovics A, Fischer Lindahl K, Dulac C. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell. 2003;112:607–618. doi: 10.1016/s0092-8674(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Martini S, Silvotti L, Shirazi A, Ryba NJ, Tirindelli R. Co-expression of putative pheromone receptors in the sensory neurons of the vomeronasal organ. J Neurosci. 2001;21:843–848. doi: 10.1523/JNEUROSCI.21-03-00843.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25:3586–3592. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nakatani H, Serizawa S, Nakajima M, Imai T, Sakano H. Developmental elimination of ectopic projection sites for the transgenic OR gene that has lost zone specificity in the olfactory epithelium. Eur J Neurosci. 2003;18:2425–2432. doi: 10.1046/j.1460-9568.2003.02998.x. [DOI] [PubMed] [Google Scholar]
- Neefjes JJ, Momburg F. Cell biology of antigen presentation. Curr Opin Immunol. 1993;5:27–34. doi: 10.1016/0952-7915(93)90077-6. [DOI] [PubMed] [Google Scholar]
- Oliveira AL, Thams S, Lidman O, Piehl F, Hokfelt T, Karre K, Linda H, Cullheim S. A role for MHC class I molecules in synaptic plasticity and regeneration of neurons after axotomy. Proc Natl Acad Sci USA. 2004;101:17843–17848. doi: 10.1073/pnas.0408154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson R, Huey-Tubman KE, Dulac C, Bjorkman PJ. Structure of a pheromone receptor-associated MHC molecule with an open and empty groove. PLoS Biol. 2005;3:e257. doi: 10.1371/journal.pbio.0030257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Del Punta K, Rothman A, Ishii T, Mombaerts P. Multiple new and isolated families within the mouse superfamily of V1r vomeronasal receptors. Nat Neurosci. 2002;5:134–140. doi: 10.1038/nn795. [DOI] [PubMed] [Google Scholar]
- Ryba NJ, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Sam M, Vora S, Malnic B, Ma W, Novotny MV, Buck LB. Odorants may arouse instinctive behaviours. Nature. 2001;412:142. doi: 10.1038/35084137. [DOI] [PubMed] [Google Scholar]
- Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L, Marton TF. What is a pheromone? Mammalian pheromones reconsidered. Neuron. 2005;46:699–702. doi: 10.1016/j.neuron.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Syken J, Grandpre T, Kanold PO, Shatz CJ. PirB restricts ocular-dominance plasticity in visual cortex. Science. 2006;313:1795–1800. doi: 10.1126/science.1128232. [DOI] [PubMed] [Google Scholar]
- Trinh K, Storm DR. Vomeronasal organ detects odorants in absence of signaling through main olfactory epithelium. Nat Neurosci. 2003;6:519–525. doi: 10.1038/nn1039. [DOI] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Wubbolts R, Fernandez-Borja M, Oomen L, Verwoerd D, Janssen H, Calafat, Tulp A, Dusseljee S, Neefjes J. Direct vesicular transport of MHC class II molecules from lysosomal structures to the cell surface. J Cell Biol. 1996;135:611–622. doi: 10.1083/jcb.135.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Shi P, Zhang YP, Zhang J. Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics. 2005;86:306–315. doi: 10.1016/j.ygeno.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Mori K. Basic principles and molecular mechanisms of olfactory axon pathfinding. Cell Tissue Res. 1997;290:457–463. doi: 10.1007/s004410050953. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Fujita H, Hayashi H, Kagamiyama H, Mori K. OCAM: A new member of the neural cell adhesion molecule family related to zone-to-zone projection of olfactory and vomeronasal axons. J Neurosci. 1997;17:5830–5842. doi: 10.1523/JNEUROSCI.17-15-05830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara Y, Mizuno T, Nakahira M, Kawasaki M, Watanabe Y, Kagamiyama H, Jishage K, Ueda O, Suzuki H, Tabuchi K, Sawamoto K, Okano H, Noda T, Mori K. A genetic approach to visualization of multisynaptic neural pathways using plant lectin transgene. Neuron. 1999;22:33–41. doi: 10.1016/s0896-6273(00)80676-5. [DOI] [PubMed] [Google Scholar]
- Young JM, Trask B. V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet. 2007;23:212–215. doi: 10.1016/j.tig.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. β2-Microglobulin deficient mice lack CD4–8+ cytolytic T cells. Nature. 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- Zou Z, Horowitz LF, Montmayeur JP, Snapper S, Buck LB. Genetic tracing reveals a stereotyped sensory map in the olfactory cortex. Nature. 2001;414:173–179. doi: 10.1038/35102506. [DOI] [PubMed] [Google Scholar]
- Zufall F, Leinders-Zufall T. Mammalian pheromone sensing. Curr Opin Neurobiol. 2007;17:483–489. doi: 10.1016/j.conb.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Zwart W, Griekspoor A, Kuijl C, Marsman M, van Rheenen J, Janssen H, Calafat J, van Ham M, Janssen L, van Lith M, Jalink K, Neefjes J. Spatial segregation of HLA-DM/HLA-DR interactions within MIIC and phagosome-induced immune escape. Immunity. 2005;22:221–233. doi: 10.1016/j.immuni.2005.01.006. [DOI] [PubMed] [Google Scholar]