Abstract
Gene expression changes are a hallmark of the neuropathology of Huntington's disease (HD), but the exact molecular mechanisms of this effect remain uncertain. Here, we report that in vitro models of disease comprised of primary striatal neurons expressing N-terminal fragments of mutant huntingtin (via lentiviral gene delivery) faithfully reproduce the gene expression changes seen in human HD. Neither viral infection nor unrelated (enhanced green fluorescent protein) transgene expression had a major effect on resultant RNA profiles. Expression of a wild-type fragment of huntingtin [htt171-18Q] also caused only a small number of RNA changes. The disease-related signal in htt171-82Q versus htt171-18Q comparisons was far greater, resulting in the differential detection of 20% of all mRNA probe sets. Transcriptomic effects of mutated htt171 are time- and polyglutamine-length dependent and occur in parallel with other manifestations of polyglutamine toxicity over 4–8 weeks. Specific RNA changes in htt171-82Q-expressing striatal cells accurately recapitulated those observed in human HD caudate and included decreases in PENK (proenkephalin), RGS4 (regulator of G-protein signaling 4), dopamine D1 receptor (DRD1), DRD2, CNR1 (cannabinoid CB1 receptor), and DARPP-32 (dopamine- and cAMP-regulated phosphoprotein-32; also known as PPP1R1B) mRNAs. HD-related transcriptomic changes were also observed in primary neurons expressing a longer fragment of mutant huntingtin (htt853-82Q). The gene expression changes observed in cultured striatal neurons are not secondary to abnormalities of neuronal firing or glutamatergic, dopaminergic, or brain-derived neurotrophic factor signaling, thereby demonstrating that HD-induced dysregulation of the striatal transcriptome might be attributed to intrinsic effects of mutant huntingtin.
Keywords: polyglutamine disease, Huntington's disease, neurodegenerative disease, gene expression profiling, striatum, transcription
Introduction
Huntington's disease (HD) is a member of a family of nine inherited polyglutamine neurodegenerative disorders associated with dramatic changes in motor, cognitive and/or psychiatric performance, progression to severe systemic wasting, and eventually, death (Bates, 2002). Although the genetic causes for these diseases are known, the multipartite cellular mechanisms have thus far made them intractable to fully understand or treat (Luthi-Carter, 2007).
HD pathology includes a dramatic atrophy of the caudate with extensive degeneration of its GABAergic medium spiny output neurons. Widespread HD-related atrophy of the cerebral cortex has also been described through both postmortem neuropathology and morphometric imaging studies. Cortical pathology and electrophysiologic studies in animal models further indicate dysfunction of corticostriatal synapses (DiFiglia et al., 1997; Cepeda et al., 2003; Cummings et al., 2006). Although not as prominent a feature in HD as in Parkinson's disease, abnormalities of the dopaminergic substantia nigra pars compacta have also been observed (Yohrling et al., 2003).
One mechanism hypothesized to play a major role in HD and other polyglutamine diseases is the dysregulation of transcription (Luthi-Carter and Cha, 2003). A consistent pattern of RNA-level changes in human HD caudate and HD mouse striatum from an early disease stage indicates a primary and universal effect of mutant huntingtin on gene expression. The mechanism underlying transcriptomic effects of mutant huntingtin has been debated, however, with evidence for changes in the activities of soluble DNA-binding transcription factors (Dunah et al., 2002; Zuccato et al., 2003), abnormalities of chromatin biochemistry and organization (Steffan et al., 2001; Helmlinger et al., 2006; Ryu et al., 2006; Stack et al., 2007), and aggregate-driven nuclear transcription factor sequestration (Boutell et al., 1999; Kazantsev et al., 1999).
In parallel with nuclear effects of mutant huntingtin, many other disease-related cellular consequences have been demonstrated. These include the dysregulation of glutamatergic and dopaminergic signaling (Zeron et al., 2002; Cepeda et al., 2003; Cummings et al., 2006; Johnson et al., 2006; Tang et al., 2007), the dysfunction of calcium homeostasis (Panov et al., 2002; Brustovetsky et al., 2005; Tang et al., 2005), metabolic abnormalities (Weydt et al., 2006; Lee et al., 2007; Valenza et al., 2007; van der Burg et al., 2008), and decreases in brain-derived neurotrophic factor (Zuccato et al., 2003; Gauthier et al., 2004). These events can also lead to downstream changes in gene expression (Lee et al., 2007; Strand et al., 2007). Importantly, the manifestation of disease arises from a combination of nuclear, non-nuclear, and transsynaptic huntingtin effects (Landles and Bates, 2004; Luthi-Carter, 2007), and the interrelationships of these effects need to be understood in greater detail to access better therapeutic strategies (Handley et al., 2006).
To distinguish neuronal effects of mutant huntingtin that may be independent of brain circuitry, we investigated gene expression abnormalities arising in vitro after expression of 171 or 853 aa fragments of mutant huntingtin in primary striatal neurons via lentiviral vectors. Here, we describe the use of these models to uncover a polyglutamine-length-dependent pathway leading to transcriptomic dysfunction in the medium spiny GABAergic neurons that are most prominently affected in HD.
Materials and Methods
Lentiviral vectors.
Lentiviral vectors encoding the first 171 aa of human huntingtin containing either 18, 44, 66, or 82 CAG repeats under control of the PGK (phosphoglycerate kinase) promoter [SIN-PGK-htt171-18Q/44Q/66Q/82Q-WPRE], the first 853 aa of human huntingtin under control of a TRE-regulated promoter (SIN-TRE-htt853-18Q/82Q-WPRE), the enhanced green fluorescent protein (EGFP) (SIN-PGK-GFP-WPRE), or the tTA transactivator (SIN-PGK-tTA-WPRE) were produced in human embryonic kidney 293T (HEK293T) cells with a four-plasmid system as described previously (Zala et al., 2005). (Sequencing of the transgenes used for this study revealed the deletion of one repeat, accounting for the difference from the previously reported htt19Q.) The viruses were resuspended in PBS with 1% of BSA and matched for particle content to 1500 ng of p24 antigen per milliliter as measured by ELISA (RETROtek; Gentaur).
Primary cultures and RNA extraction.
Dissociated neuronal cultures were prepared from ganglionic eminences of embryonic day 16 rat embryos, yielding a majority population of neuronal nuclear antigen (NeuN)-positive neurons and some residual astroglial cells (Zala et al., 2005). Ninety-five percent of NeuN-positive cells also coexpressed dopamine- and cAMP-regulated phosphoprotein-32 (DARPP-32; also known as PPP1R1B) (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). On day 1 in vitro, cells were infected with lentiviral expression vectors at a titer of 1 ng of p24/10,000 cells and harvested for RNA after 2, 4, 6, or 8 weeks. The time points used for the microarray measurements (PGK-htt171-82Q at 6 weeks or TRE-htt853-82Q at 4 weeks) correspond to a stage in which huntingtin-positive protein inclusions are present, but no significant cell death is observed by criteria of terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) positivity, caspase-3 activation, or diminished expression of pan-neuronal genes (Zala et al., 2005) (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). Death of htt82Q-expressing cells is, however, observed at time points of 8 weeks and greater. RNA samples from independent culture samples were prepared with the RNeasy Mini System (Qiagen) and comprised biological replicates for quantitative real-time PCR (QPCR) and microarray studies.
Microarray sample processing.
Biotinylated cRNAs were prepared from 1 μg of total RNA using the CodeLink Expression Assay Reagent Kit (GE Healthcare) (htt171 series) or Affymetrix One-Cycle Target Labeling Kit (htt853 model). cRNA (11 μg) was hybridized to Rat Expression 230A or 230 2.0 Genechips (for htt171 or htt853 experiments, respectively) and processed, stained, and scanned according to the manufacturer's recommendations. The quality of input RNAs and cRNAs was verified with the Bioanalyzer 2100 (Agilent Technologies) before use in each step of the procedure.
Microarray analysis.
Microarray quality control was performed using the software package affyPLM from Bioconductor (www.bioconductor.org). Microarrays with a median normalized unscaled SE ≥1.05 were excluded. After quantification of gene expression by robust multiarray analysis (Irizarry et al., 2003) using the affy package, spike probe sets were removed, and differential gene expression was determined using the limma package. For the htt853-82Q versus htt853-18Q, differential expression was determined by direct two-group comparison (n = 6 of 6 arrays). For the htt171 experimental series, differential gene expression between each of the three treatment groups (EGFP, n = 3 arrays; htt171-18Q, n = 4 arrays; htt171-82Q, n = 3 arrays) and the uninfected group (n = 3 arrays) was determined with one-way ANOVA using the limma package. We used the uninfected sample group as baseline and fitted three coefficients representing the difference between each of the three treatment groups and the uninfected sample group (treatment-contrasts parametrization). Differential expression between 82Q and 18Q was determined by calculating the corresponding contrast from the fitted coefficients. p values were corrected for multiple testing using a false discovery rate (FDR) approach (Benjamini and Hochberg, 1995). Gene rankings reflected the order of ascending p values. Affymetrix annotations (version 24) were used for probe-set-to-gene assignments; unannotated probe sets were discarded. Where multiple probe sets were assigned to the same gene, the probe set with the smallest p value was included. The comparison of differential expression in each HD model to gene expression changes detected in human HD caudate was performed using the annotationTools package (Kuhn et al., 2008) as described in the study by Kuhn et al. (2007), with human microarray data from the study by Hodges et al. (2006).
Quantitative real-time PCR.
Custom Taqman Low-Density Arrays were obtained from Applied Biosystems. Reverse-transcription reactions were performed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Replicate QPCR assays were performed with cDNA equivalent to 1 ng of total RNA per reaction in Taqman Universal PCR Master Mix on microfluidics cards in a 7900HT Real-Time PCR System (all from Applied Biosystems). Data are presented for the following inventoried assays: ADORA2a (Rn00583935_m1), proenkephalin 1 (PENK1) (Rn00567566_m1), regulator of G-protein signaling 4 (RGS4) (Rn00568067_m1), neurogranin (NRGN) (Rn00480741_m1), Purkinje-cell protein 4 (PCP4) (Rn00564515_m1), visinin-like 1 (VSNL1) (Rn00562148_m1), and nuclear transcription factor Y gamma subunit (NFYC) (Rn00563487_m1). Expression values were calculated as described in the study by Runne et al. (2007), using β-actin (assay ID Rn00667869_m1) as the normalization control. Similar results were obtained after normalization to 18S ribosomal RNA (assay ID 4342379-18srRNA).
Multielectrode recordings.
Primary neurons were cultured on MEA60 four-well multielectrode arrays (Ayanda Biosystems), and spontaneous electrical activity was recorded using the MEA60 data acquisition system (Multi Channel Systems).
BDNF assays.
BDNF was measured by ELISA (Emax ImmunoAssay System; Promega; sensitivity, 15.6 pg/ml). No BDNF was detected in uninfected striatal cell lysates (∼100 μg of total protein in triplicate) or culture medium (100 μl of 20× concentrated medium, in triplicate). For cellular lysates, parallel samples of cultured primary cortical neuron or HEK293 cells were included as positive and negative controls, respectively. Striatal cells had assay values indistinguishable from negative control samples, whereas BDNF was detected at 215 pg/ml in cortical neuron lysates (in triplicate), corresponding to 320 pg of BDNF per milligram of total cellular protein. Similar results were obtained in independent batches of neuronal cultures.
Results
Transcriptomic effects of lentiviral expression are polyglutamine dependent
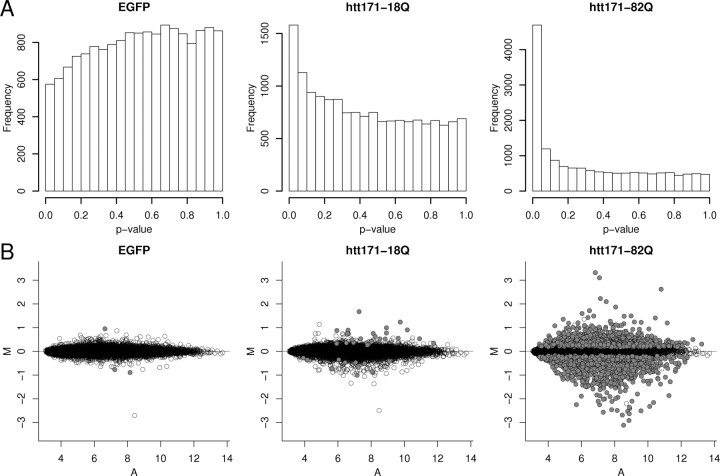
We first investigated the potential effects of our lentiviral expression vector or transprotein per se on neuronal RNA profiles. Uninfected neurons were compared with EGFP-, htt171-18Q-, and htt171-82Q-expressing cells after 6 weeks in vitro using microarray analysis. Global profiles of each lentiviral-infected group was compared with uninfected cells to examine the differences in overall signal characteristics. Uncorrected p value distributions and signal/change ratio plots for these comparisons are shown in Figure 1. GFP expression had very little effect on endogenous RNA levels, as evidenced by only 3 (of 10,222) probe sets meeting a cutoff criterion of FDR p < 0.05 (supplemental Table 1, available at www.jneurosci.org as supplemental material), an even distribution of p values, and little deviation from zero in the signal/change ratio plot. Expression of an N-terminal huntingtin fragment with 18Q had a slightly larger effect, as assessed by 32 (of 10,222) probe sets meeting the criterion of FDR p < 0.05 (supplemental Table 1, available at www.jneurosci.org as supplemental material), an increased distribution of p values near zero, and more deviation from zero in the signal/change ratio plot. An N-terminal huntingtin fragment with 82Q had a much greater effect on endogenous RNA expression than the other two conditions. This was demonstrated by 2151 (of 10,222; 21%) of probe sets meeting the criterion of FDR p < 0.05, an obvious peak of small p values, and large deviations from zero in the signal/change ratio plots. These criteria all indicated that the major gene expression effect detected in the neurons was attributable to the expansion of the polyQ tract in huntingtin.
Figure 1.
Comparison of overall differential expression signal in EGFP-, htt171-18Q-, and htt171-82Q-expressing cells. A, The relative distributions of uncorrected p values in the three conditions. B, The signal (A) and differential expression (M) distributions, with probe sets meeting FDR p < 0.05 indicated by filled circles.
Polyglutamine-length dependence of htt171 effects and confirmation of disease-related effects in a second model system
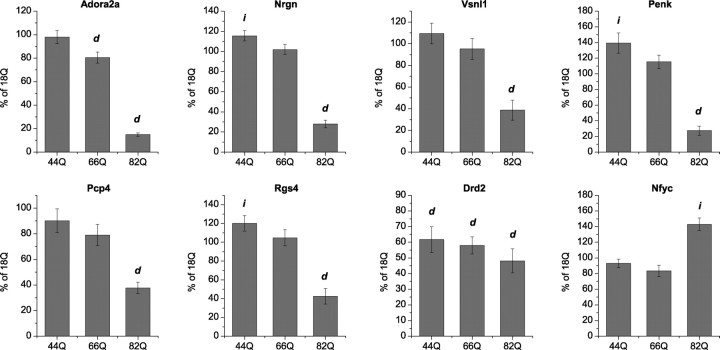
Disease-specific changes in gene expression were extracted from the comparison of htt171-82Q- versus htt171-18Q-expressing cells. These comprised 2009 probe sets at a cutoff of FDR p < 0.05 (of 10,222; 20%) (Table 1; supplemental Table 2, available at www.jneurosci.org as supplemental material), representing 1242 decreases and 767 increases in RNA expression. To assess the strict polyglutamine-length dependence of the effects of mutant htt171 fragments, we performed quantitative real-time PCR assays for selected RNAs (ADORA2A, PENK, RGS4, NRGN, PCP4, VSNL1, and NFYC) after 6 weeks of expression of htt171 transproteins with 18Q, 44Q, 66Q, or 82Q repeats (Fig. 2). Htt171-82Q had the greatest disease-related effect on RNA levels (significantly different from 18Q for all), whereas htt171-66Q significantly diminished the expression of only ADORA2a and DRD2. Htt171-44Q showed a decrease in only the DRD2 RNA, but resulted in levels of RGS4 and PENK that were slightly higher than in htt171-18Q-expressing cells. In total, these results show the expected pattern of polyglutamine-length-dependent effects on gene expression.
Table 1.
mRNA changes concordant in striatal neuron HD models and human HD caudate
| N171 log2 fold change | N853 log2 fold change | Human log2 fold change | Gene symbol | Gene name |
|---|---|---|---|---|
| −2.24 | −0.33 | −0.78 | GUCY1A3 | Guanylate cyclase 1, soluble, α 3 |
| −1.84 | −0.24 | −0.85 | PDE2A | Phosphodiesterase 2A, cGMP-stimulated |
| −1.88 | −1.56 | RGS4 | Regulator of G-protein signaling 4 | |
| −2.11 | −0.42 | −1.28 | NGEF | Neuronal guanine nucleotide exchange factor |
| −2.19 | −0.34 | −1.38 | PENK | Proenkephalin |
| −1.95 | −1.22 | PCP4 | Purkinje cell protein 4 | |
| −2.37 | −0.42 | −1.60 | GPR88 | G-protein-coupled receptor 88 |
| −1.48 | −1.34 | RASD2 | RASD family, member 2 | |
| −1.24 | −0.55 | RGS2 | Regulator of G-protein signaling 2, 24 kDa | |
| −0.90 | −0.58 | ATP2B2 | ATPase, Ca2+ transporting, plasma membrane 2 | |
| −2.55 | −0.94 | −0.40 | NPY | Neuropeptide Y |
| −1.25 | −1.52 | LPL | Lipoprotein lipase | |
| −1.13 | −1.23 | TPBG | Trophoblast glycoprotein | |
| −1.16 | −0.33 | SLC24A3 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | |
| −1.69 | −0.85 | NPTX1 | Neuronal pentraxin I | |
| −0.88 | −1.00 | PLCB1 | Phospholipase C, β 1 (phosphoinositide-specific) | |
| 1.26 | 0.96 | HSPB1 | Heat shock 27 kDa protein 1 | |
| 2.91 | 0.89 | CP | Ceruloplasmin (ferroxidase) | |
| −1.54 | −0.67 | ARPP-21 | CAMP-regulated phosphoprotein, 21 kDa | |
| −1.13 | −1.51 | PLK2 | Polo-like kinase 2 | |
| −1.23 | −0.65 | VSNL1 | Visinin-like 1 | |
| −1.38 | −0.75 | PPP1R1B | Protein phosphatase 1, regulatory (inhibitor) subunit 1B (DARPP-32) | |
| −1.48 | −0.62 | HRH3 | Histamine receptor H3 | |
| −0.82 | −1.11 | PRKCB1 | Protein kinase C, β 1 | |
| −1.06 | −0.31 | −0.67 | GAD1 | Glutamate decarboxylase 1 (brain, 67 kDa) |
| −0.81 | −0.55 | OLFM1 | Olfactomedin 1 | |
| −1.03 | −1.49 | DRD1 | Dopamine receptor D1 | |
| −1.02 | −0.69 | VMP | Vesicular membrane protein p24 | |
| −0.84 | −0.79 | ATP2A2 | ATPase, Ca2+ transporting, cardiac muscle, slow-twitch 2 | |
| −0.69 | −1.09 | SYT1 | Synaptotagmin I | |
| −0.54 | −1.23 | CPLX1 | Complexin 1 | |
| −1.33 | −0.40 | −1.31 | SYNPR | Synaptoporin |
| −0.70 | −1.06 | GUCY1B3 | Guanylate cyclase 1, soluble, β 3 | |
| −1.46 | −1.30 | NRGN | Neurogranin (protein kinase C substrate, RC3) | |
| −0.55 | −0.76 | TESC | Tescalcin | |
| −0.53 | −0.74 | ACTN2 | α Actinin-2 | |
| −0.69 | −0.80 | GNG7 | Guanine nucleotide binding protein (G protein), γ 7 | |
| −1.33 | −1.12 | KCNA1 | Potassium voltage-gated channel, shaker-related subfamily, member 1 | |
| −0.63 | −0.29 | −0.39 | DUSP14 | Dual-specificity phosphatase 14 |
| −1.41 | −1.42 | PCP4L1 | Purkinje-cell protein-4-like 1 | |
| 0.46 | 0.39 | ACADVL | Acyl-coenzyme A dehydrogenase, very long chain | |
| 0.76 | 0.43 | GSN | Gelsolin (amyloidosis, Finnish type) | |
| −0.80 | −0.49 | −0.41 | SNCA | Synuclein, α (non-A4-component of amyloid precursor) |
| −0.56 | −0.70 | NELL2 | NEL-like 2 | |
| −0.72 | −1.71 | CNR1 | Cannabinoid receptor 1 (brain) | |
| −1.33 | −0.28 | −0.33 | NNAT | Neuronatin |
| −1.10 | −0.85 | CAMKV | CaM kinase-like vesicle-associated | |
| −0.65 | −0.89 | CKMT1 | Creatine kinase, mitochondrial 1A/B | |
| −0.87 | −1.08 | ITPR1 | Inositol 1,4,5-triphosphate receptor, type 1 | |
| 0.62 | 0.84 | HSPA1B | Heat shock 70 kDa protein 1B | |
| −0.41 | −1.23 | ADORA2A | Adenosine A2a receptor |
Selected genes differentially expressed in primary neuron HD models and human HD meeting criteria of FDR p < 0.05 in htt171-82Q (compared to htt171-18Q), FDR p < 0.05 in human HD grades 0–2 [compared with human controls; microarray data from Hodges et al. (2006)], and top 500 genes in htt853-82Q (compared with htt853-18Q; blank fields indicate that no concordant regulation was observed).
Figure 2.
Polyglutamine-dependent regulation of selected striatal RNAs in primary neurons by htt171. d, Significantly decreased expression; i, significantly increased expression compared with htt171-18Q (p < 0.05 by Student's two-tailed t test; assays in triplicate from independent RNA samples: 18Q, 82Q, n = 5; 44Q, 66Q, n = 4). Bar plots represent percentage of htt171-18Q expression normalized to β-actin; error bars represent SEM.
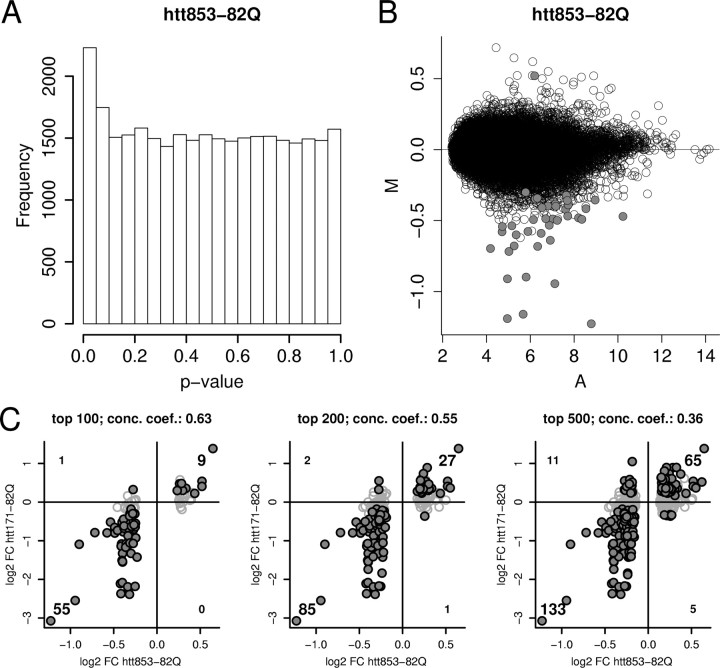
We further validated our in vitro system for assessing polyglutamine-regulated changes in gene expression by examining the effects of a mutant huntingtin fragment of a different length. Expression of the first 853 aa of huntingtin under the control of a TRE-regulated promoter (htt853-82Q vs htt853-18Q) also resulted in significant changes in gene expression (Fig. 3A,B). Although the 4 week time point in the htt853 model showed a more modest overall effect than the 6 week time point in the htt171-82Q model, the differential expression results exhibited a strong correlation with the htt171-82Q data (concordance coefficient, 0.63 for top 100 genes) [Table 1; Fig. 3C; supplemental Table 3, available at www.jneurosci.org as supplemental material (see also below)]. These results show that different lentiviral vectors expressing different fragments of mutant huntingtin show similar polyglutamine-related effects.
Figure 3.
Comparison of overall differential expression signal in htt853-82Q- versus htt853-18Q-expressing cells (n = 6 arrays for each condition) and concordance between htt853-82Q and htt171-82Q model data. A, The distribution of uncorrected p values (compare with Fig. 1). B, The signal (A) and differential expression (M) distributions, with probe sets meeting FDR p < 0.05 (40 probe sets of 31,042 total) indicated by filled circles. Lower stringency cutoffs show 2230 probe sets meet the criterion of nominal p < 0.05, and 473 probe sets meet the criterion of FDR p < 0.33. The top 100, 200, and 500 unique probe sets show a significant transcriptomic relationship to the htt171-82Q model (as shown in C). C, Plots top 100, 200, or 500 differentially regulated genes from the htt853-82Q versus htt853-18Q expression analysis for RNAs detected by both Rat 230A and Rat 230 2.0 arrays. Genes meeting criteria of FDR p < 0.05 in htt171-82Q versus htt171-18Q comparison are indicated by filled black circles. The high fractions of genes showing concordant regulations (top right quadrant for upregulated genes and bottom left for downregulated genes) versus discordantly regulated genes (in top left and bottom right quadrants) represents the strong association between models. Concordance coefficient (conc. coeff.): 0.63 for top 100 genes; 0.55 for top 200 genes; 0.36 for top 500 genes.
RNA changes show largest effects on neuronal signaling, neurite outgrowth, protein degradation, and RNA splicing pathways
To assess potential biological effects of these transcriptomic changes, we asked whether the differentially expressed RNAs were significantly enriched in defined Gene Ontology (GO) categories. In agreement with previous observations in HD mice and HD patient brains, top GO hits among genes downregulated in htt171-82Q samples could be linked to neuronal signaling, including cell communication, synaptic transmission, and monovalent and divalent cation transport (supplemental Table 4, available at www.jneurosci.org as supplemental material). There was also significant overrepresentation of GO terms representing gene sets involved in neuron differentiation and neuron and neurite development, and morphogenesis in the set of downregulated genes. Genes upregulated in htt171-82Q samples showed a high representation of GO categories related to protein degradation, including modification-dependent/ubiquitin-dependent protein catabolic processes, suggesting cellular responses to mutant protein accumulation (Yamamoto et al., 2006). Another interesting set of GO categories populated by upregulated genes was categories associated with RNA splicing/RNA processing. This is particularly intriguing given that mutant and wild-type huntingtin have been reported to have differential affinities for WW-domain proteins involved in RNA splicing (Passani et al., 2000).
In vitro gene expression effects of huntingtin fragments reproduce those found in human HD caudate
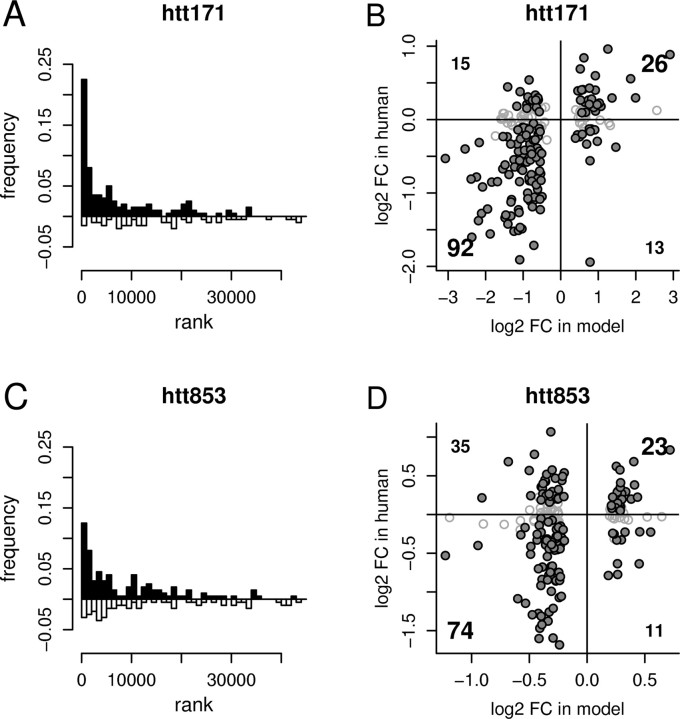
To examine the extent to which transcriptomic changes in our in vitro models of HD accurately recapitulate the transcriptomic changes of the human HD brain, we compared our results to microarray data from low pathologic grade human HD caudate (Hodges et al., 2006). We assessed the interaction of these two data sets using a concordance coefficient method that we developed previously, which tallies the fraction of same-direction gene expression changes (e.g., increased or decreased in model and human HD) and penalizes for opposite-direction changes (e.g., increased in model and decreased in human, or vice versa). This analysis showed that our in vitro models are very similar to HD brain [for htt171-82Q: concordance coefficient, 0.45; p < 0.0026 (Fig. 4A,B); for htt853-82Q: concordance coefficient, 0.255; p = 0.0142 (Fig. 4C,D)], with the 6 week htt171-82Q in vitro model showing a particularly strong HD profile, as evidenced by its transcriptomic concordance coefficient being equivalent to the highest-scoring transgenic mouse models of HD (Kuhn et al., 2007). Also, like the HD mice, a higher concordance in downregulated than in upregulated genes was observed in htt171-82Q-expressing cells (0.53 vs 0.24, respectively). Specific gene expression changes concordant with those of human HD brain are presented in Table 1 and supplemental Table 2, available at www.jneurosci.org as supplemental material.
Figure 4.
Concordance of HD models with human HD caudate. A, C, The distributions of the top 200 differentially expressed RNAs in htt171-82Q- versus htt171-18Q-expressing cells or htt853-82Q- versus htt853-18Q-expressing cells relative to the differential expression of their orthologs in human HD (see Materials and Methods). Same-sign regulations (increased or decreased in both mouse and human HD) are shown above the x-axis in black; opposite-sign regulations (increased in mouse and decreased in human, or vice versa) are shown below the x-axis in white. B, D, Scatterplots of differential expression in the htt171-82Q model or htt853-82Q model versus human HD. Large numbers in the top right and bottom left quadrants summarize same-sign (concordant) regulations (increased or decreased in both mouse and human HD); small numbers summarize opposite-sign regulations (increased in mouse and decreased in human, or vice versa) in the top left and bottom right quadrants. Filled gray circles indicate RNAs whose human orthologs show statistically significant regulation by the criterion of FDR p < 0.05; open circles (outlined in gray) indicate RNAs for which the orthologous expression measure in human is considered nonsignificant (FDR p ≥ 0.05).
Transcriptomic changes are progressive
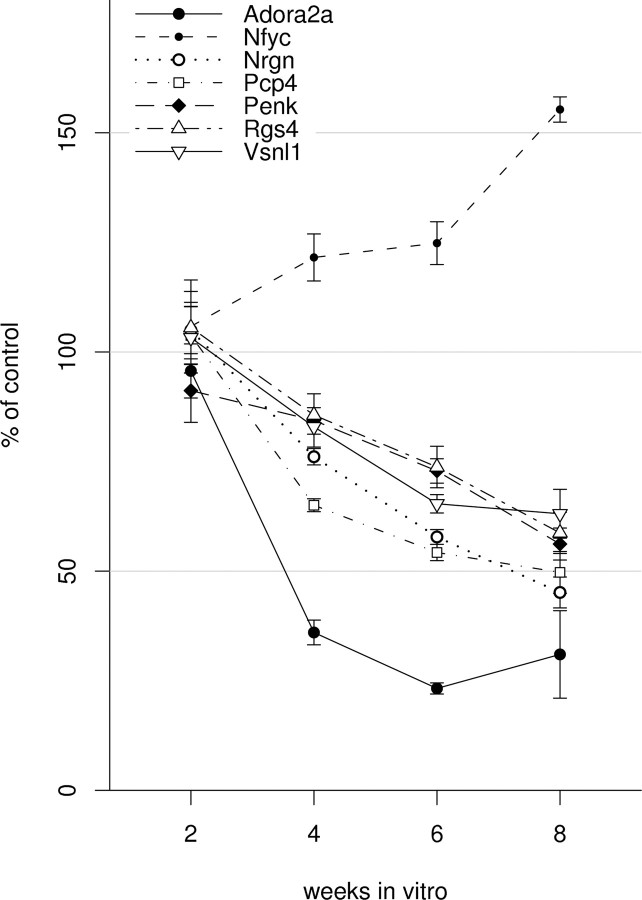
A distinct evolution of transcriptomic effects parallels the deterioration of neuronal function in HD models, and magnitudes of changes in mRNA levels increase with pathological grade in human HD brain. To evaluate the time dependence of htt171-82Q effects on gene expression in our in vitro HD model, we conducted quantitative real-time PCR assays at 2 week intervals for RNAs that are consistently dysregulated in mouse and/or human HD brain (Hodges et al., 2006; Kuhn et al., 2007). Mutant polyQ-associated gene expression changes became significant at 4 weeks and increased in magnitude through 8 weeks, ranging from a decrease to 23% of control (ADORA2A) to an increase to 155% of control (NFYC) [expressed as htt171-82Q- vs htt171-18Q-expressing cells (Fig. 5)]. The steep onset of gene expression changes just after the appearance of inclusions and shallow but continued progression to cell death is reminiscent of the timeline of molecular pathologies observed in HD mice (Luthi-Carter et al., 2002).
Figure 5.
Time course of expression changes in the htt171-82Q in vitro model of HD as measured by QPCR (n = 5 per condition) (see Materials and Methods). Progressive changes are observed between 2 and 8 weeks in vitro.
Gene expression effects are not attributable to abnormal neuronal firing
Changes in neuronal circuitry have been observed in mouse and cell models of HD. Although we had isolated ganglionic eminence-derived cells from other neurons in the brain, we wanted to rule out any transsynaptic effects that might nonetheless be present as the driving force for gene expression changes. To confirm that gene expression changes did not result from the aberrant spontaneous firing of neurons in the culture, we assessed their electrical behavior using multielectrode arrays. Indeed, our DARPP32-positive cells behaved as expected for an inhibitory GABAergic neuron culture and did not show any spontaneous firing (0 of 256 channels on four multielectrode arrays). In contrast, we observed spontaneous firing in one of every six channels on multielectrode array cultures of dissociated primary hippocampal neurons (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material). Therefore, we conclude that the changes in gene expression observed in htt171-82Q-expressing cells are not attributable to changes in neuronal firing behavior.
Gene expression effects are not attributable to a decrease in BDNF production
Because BDNF has been heavily implicated in the etiology of HD, and BDNF signaling can also influence gene expression, we asked whether changes in BDNF protein levels might underlie gene expression changes in our in vitro model of HD. Although it is clear that most striatally delivered BDNF is synthesized in cortical neurons, there has been some debate over whether striatal cells also produce this neurotrophic factor. In addition, it is possible that the cell culture conditions used in our experiment might result in nonphysiologic BDNF expression. Therefore, we assayed BDNF by ELISA in uninfected primary neurons and in their culture medium, reflecting BDNF accumulation over ∼1 week. No BDNF was detected in striatal cells [consistent with previous data (Zala et al., 2005)], whereas a BDNF level of 320 pg/mg protein was detected in primary cortical neurons. Together with the above data, these results show that gene expression changes in our striatal neuron model of HD are not a result of large transsynaptic events or BDNF signaling. These findings support the view that some aspects of HD-related striatal dysfunction are caused by the effects of mutant huntingtin within medium spiny striatal neurons.
Discussion
Dating from the establishment of animal models of HD using excitotoxic lesions, the issue of cortical and striatal interactions in Huntington's disease has received much attention and has been heavily debated. This study provides clear evidence that intracellular polyQ-driven events that closely resemble HD can occur within striatal neurons in the absence of brain circuitry. These results suggest that intrinsic effects of mutant huntingtin in striatal neurons may be sufficient to cause some aspects of striatal dysfunction.
The balance with which intrinsic and extrinsic events determine striatal cell fates in vivo will nonetheless require further examination. The findings of the present study by no means rule out possible neurotrophic or excitotoxic roles of the HD cortex in vivo. Neither can we completely rule out that the culture conditions used here reproduce a (presumably non-BDNF-driven) neurotrophic signaling activity that dysfunctions in striatal neurons.
Recent data on the possible involvement of BDNF in HD are very persuasive. Previous reports indicate that BDNF transcription is inhibited through a decreased cytoplasmic targeting of the neural-restrictive silencing factor (Zuccato et al., 2003). In parallel, dysregulated expression of TrkB (tyrosine-related kinase B)/Ntrk2 and other BDNF receptors is observed in human HD brain (Zuccato et al., 2008), and a decrease of Ntrk2 mRNA is also observed in our in vitro lentiviral model studied here (albeit without any detectable BDNF protein). An HD-related decrease in axonal transport of BDNF has also been proposed to result from mutant huntingtin's inhibition of forward vesicular movement along microtubules (Gauthier et al., 2004). In addition, striata of BDNF null mutant mice have a molecular phenotype that is strikingly similar to human HD (Strand et al., 2007). Moreover, BDNF is neuroprotective in chemical and genetic models of HD, including the very model we study in these experiments. Although the sum of the data implicating BDNF is very impressive, it is nonetheless likely that extracortical mechanisms also govern striatal neurodegeneration in HD.
Recent studies using regionally restrictive promoters to drive mutant huntingtin expression have suggested that pathological cell–cell interactions contribute to cortical pathogenesis in an HD mouse model (Gu et al., 2005) and also to a major interdependence of cortical and striatal cells in HD pathogenesis (Gu et al., 2007). Moreover, previous studies by our group also show that HD-like pathology and typical motor deficits can be seen in the striata of primates expressing mutant huntingtin in the striatum only (Palfi et al., 2007). Therefore, it is also quite plausible that the range of effects of mutant huntingtin comprises cellularly independent, parallel, and combinatorial toxicities that may all be involved in the pathogenesis of HD. In fact, one could postulate that the strong reproducibility of striatal degeneration despite a wide range of clinical HD phenotypes arises from functionally overlapping pathways that ensure the nearly universal demise of this cell population.
The present findings focus on the transcriptomic level changes in polyQ-affected cells. Although there is considerable evidence to suggest that these changes are primary targets of mutant huntingtin through its direct interactions with transcription and chromatin remodeling factors, it is possible that some of these changes are secondary events, or possibly even reflect nonpathogenic compensatory changes. We cannot discriminate between these possibilities at the present time, but nonetheless, we believe that these transcriptomic indicators can serve as robust readouts for the specific molecular and cellular mechanisms that regulate polyQ-dependent effects and responses. This perspective is well-supported in animal model data, which show that gene expression changes generally correlate well with behavioral measures in animals with early-to-moderate disease signs and that the RNA changes worsen with disease progression (Luthi-Carter et al., 2002; Hodges et al., 2007; Kuhn et al., 2007). Future testing of the effects of individual gene manipulations will eventually allow us to establish the link between transcriptomic changes and particular cellular effects.
Qualitatively, the profiles of gene expression changes in our primary striatal neuron models show a strong similarity to human HD but are somewhat different from those reported in cell lines. One factor may be that our ganglionic eminence cultures have a different basal pattern of gene expression that changes the transcriptomic background on which the polyglutamine-dependent changes arise. Another important difference may be the chronic nature of the polyQ exposure, which accumulates toxicities over weeks instead of days in our models. Indeed, cell line gene expression profiles of polyQ toxicity typically show signatures of stress-related and extramitochondrial metabolic genes (Sipione et al., 2002; Bae et al., 2005; Apostol et al., 2006; Yamamoto et al., 2006; Lee et al., 2007) more prominently than changes in neurotransmitter signaling pathways. Our view is that different cellular models represent different balances of polyQ effects, with the cell lines showing a higher contribution from protein aggregation and cellular stress-related effects, and the mice and primary neurons reflecting perturbations of cellular communication and neuronal connectivity. Because both sets of mechanisms are likely contributors to the disease process, the appropriate choice of model system can be made depending on which aspect(s) of disease one intends to study.
With respect to disease-related events that lead to dysregulation of the transcriptome, both our in vitro lentiviral models of HD show good correlation to the disease-related effects detected in human HD brain. This finding is consistent with our studies of HD mice, which also found that mutant N-terminal huntingtin fragments had qualitatively indistinguishable effects from full-length mutant huntingtin (Kuhn et al., 2007), except for a slower evolution of transcriptomic changes. The strong correlation between the gene expression patterns in the N171-82Q in vitro model and human caudate suggests that this model may be an excellent system for testing the mechanisms underlying transcriptome-level effects of mutant huntingtin in HD.
Footnotes
This work was supported by the Swiss National Science Foundation and the Ecole Polytechnique Fédérale de Lausanne. We are grateful for equipment and technical support from Ayanda Biosystems and the Lausanne DNA Array Facility. We thank Dr. Beena Pillai for critical reading of this manuscript. Microarray data reported in this manuscript have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus database, series entries GSE1918 and GSE12481.
References
- Apostol BL, Illes K, Pallos J, Bodai L, Wu J, Strand A, Schweitzer ES, Olson JM, Kazantsev A, Marsh JL, Thompson LM. Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum Mol Genet. 2006;15:273–285. doi: 10.1093/hmg/ddi443. [DOI] [PubMed] [Google Scholar]
- Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y, Hayward SD, Moran TH, Montell C, Ross CA, Snyder SH, Sawa A. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Bates G. Huntington's disease. Ed 3. Oxford: Oxford UP; 2002. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Boutell JM, Thomas P, Neal JW, Weston VJ, Duce J, Harper PS, Jones AL. Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum Mol Genet. 1999;8:1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- Brustovetsky N, LaFrance R, Purl KJ, Brustovetsky T, Keene CD, Low WC, Dubinsky JM. Age-dependent changes in the calcium sensitivity of striatal mitochondria in mouse models of Huntington's disease. J Neurochem. 2005;93:1361–1370. doi: 10.1111/j.1471-4159.2005.03036.x. [DOI] [PubMed] [Google Scholar]
- Cepeda C, Hurst RS, Calvert CR, Hernández-Echeagaray E, Nguyen OK, Jocoy E, Christian LJ, Ariano MA, Levine MS. Transient and progressive electrophysiological alterations in the corticostriatal pathway in a mouse model of Huntington's disease. J Neurosci. 2003;23:961–969. doi: 10.1523/JNEUROSCI.23-03-00961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings DM, Milnerwood AJ, Dallérac GM, Waights V, Brown JY, Vatsavayai SC, Hirst MC, Murphy KP. Aberrant cortical synaptic plasticity and dopaminergic dysfunction in a mouse model of Huntington's disease. Hum Mol Genet. 2006;15:2856–2868. doi: 10.1093/hmg/ddl224. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pagès M, Dompierre JP, Rangone H, Cordelières FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, Saudou F. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Gu X, Li C, Wei W, Lo V, Gong S, Li SH, Iwasato T, Itohara S, Li XJ, Mody I, Heintz N, Yang XW. Pathological cell-cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;46:433–444. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Gu X, André VM, Cepeda C, Li SH, Li XJ, Levine MS, Yang XW. Pathological cell-cell interactions are necessary for striatal pathogenesis in a conditional mouse model of Huntington's disease. Mol Neurodegener. 2007;2:8. doi: 10.1186/1750-1326-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley OJ, Naji JJ, Dunnett SB, Rosser AE. Pharmaceutical, cellular and genetic therapies for Huntington's disease. Clin Sci (Lond) 2006;110:73–88. doi: 10.1042/CS20050148. [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Tora L, Devys D. Transcriptional alterations and chromatin remodeling in polyglutamine diseases. Trends Genet. 2006;22:562–570. doi: 10.1016/j.tig.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL, et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Hodges A, Hughes G, Brooks S, Elliston L, Holmans P, Dunnett SB, Jones L. Brain gene expression correlates with changes in behavior in the R6/1 mouse model of Huntington's disease. Genes Brain Behav. 2007;7:288–299. doi: 10.1111/j.1601-183X.2007.00350.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Rajan V, Miller CE, Wightman RM. Dopamine release is severely compromised in the R6/2 mouse model of Huntington's disease. J Neurochem. 2006;97:737–746. doi: 10.1111/j.1471-4159.2006.03762.x. [DOI] [PubMed] [Google Scholar]
- Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Goldstein DR, Hodges A, Strand AD, Sengstag T, Kooperberg C, Becanovic K, Pouladi MA, Sathasivam K, Cha JH, Hannan AJ, Hayden MR, Leavitt BR, Dunnett SB, Ferrante RJ, Albin R, Shelbourne P, Delorenzi M, Augood SJ, Faull RL, et al. Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Luthi-Carter R, Delorenzi M. Cross-species and cross-platform gene expression studies with the Bioconductor-compliant R package ‘annotationTools’. BMC Bioinformatics. 2008;9:26. doi: 10.1186/1471-2105-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landles C, Bates GP. Huntingtin and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series. EMBO Rep. 2004;5:958–963. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Walters RH, Murphy RM. Reconsidering the mechanism of polyglutamine peptide aggregation. Biochemistry. 2007;46:12810–12820. doi: 10.1021/bi700806c. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R. Huntington's and other polyglutamine diseases: many effects of single gene mutations. Drug Discov Today Dis Mech. 2007;4:111–119. [Google Scholar]
- Luthi-Carter R, Cha J-HJ. Mechanisms of transcriptional dysregulation in Huntington's disease. Clin Neurosci Res. 2003;3:165–177. [Google Scholar]
- Luthi-Carter R, Hanson SA, Strand AD, Bergstrom DA, Chun W, Peters NL, Woods AM, Chan EY, Kooperberg C, Krainc D, Young AB, Tapscott SJ, Olson JM. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum Mol Genet. 2002;11:1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- Palfi S, Brouillet E, Jarraya B, Bloch J, Jan C, Shin M, Condé F, Li XJ, Aebischer P, Hantraye P, Déglon N. Expression of mutated huntingtin fragment in the putamen is sufficient to produce abnormal movement in non-human primates. Mol Ther. 2007;15:1444–1451. doi: 10.1038/sj.mt.6300185. [DOI] [PubMed] [Google Scholar]
- Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- Passani LA, Bedford MT, Faber PW, McGinnis KM, Sharp AH, Gusella JF, Vonsattel JP, MacDonald ME. Huntingtin's WW domain partners in Huntington's disease post-mortem brain fulfill genetic criteria for direct involvement in Huntington's disease pathogenesis. Hum Mol Genet. 2000;9:2175–2182. doi: 10.1093/hmg/9.14.2175. [DOI] [PubMed] [Google Scholar]
- Runne H, Kuhn A, Wild EJ, Pratyaksha W, Kristiansen M, Isaacs JD, Régulier E, Delorenzi M, Tabrizi SJ, Luthi-Carter R. Analysis of potential transcriptomic biomarkers for Huntington's disease in peripheral blood. Proc Natl Acad Sci U S A. 2007;104:14424–14429. doi: 10.1073/pnas.0703652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Lee J, Hagerty SW, Soh BY, McAlpin SE, Cormier KA, Smith KM, Ferrante RJ. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc Natl Acad Sci U S A. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipione S, Rigamonti D, Valenza M, Zuccato C, Conti L, Pritchard J, Kooperberg C, Olson JM, Cattaneo E. Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses. Hum Mol Genet. 2002;11:1953–1965. doi: 10.1093/hmg/11.17.1953. [DOI] [PubMed] [Google Scholar]
- Stack EC, Del Signore SJ, Luthi-Carter R, Soh BY, Goldstein DR, Matson S, Goodrich S, Markey AL, Cormier K, Hagerty SW, Smith K, Ryu H, Ferrante RJ. Modulation of nucleosome dynamics in Huntington's disease. Hum Mol Genet. 2007;16:1164–1175. doi: 10.1093/hmg/ddm064. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Strand AD, Baquet ZC, Aragaki AK, Holmans P, Yang L, Cleren C, Beal MF, Jones L, Kooperberg C, Olson JM, Jones KR. Expression profiling of Huntington's disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J Neurosci. 2007;27:11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TS, Slow E, Lupu V, Stavrovskaya IG, Sugimori M, Llinás R, Kristal BS, Hayden MR, Bezprozvanny I. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington's disease. Proc Natl Acad Sci U S A. 2005;102:2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TS, Chen X, Liu J, Bezprozvanny I. Dopaminergic signaling and striatal neurodegeneration in Huntington's disease. J Neurosci. 2007;27:7899–7910. doi: 10.1523/JNEUROSCI.1396-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenza M, Leoni V, Tarditi A, Mariotti C, Björkhem I, Di Donato S, Cattaneo E. Progressive dysfunction of the cholesterol biosynthesis pathway in the R6/2 mouse model of Huntington's disease. Neurobiol Dis. 2007;28:133–142. doi: 10.1016/j.nbd.2007.07.004. [DOI] [PubMed] [Google Scholar]
- van der Burg JM, Bacos K, Wood NI, Lindqvist A, Wierup N, Woodman B, Wamsteeker JI, Smith R, Deierborg T, Kuhar MJ, Bates GP, Mulder H, Erlanson-Albertsson C, Morton AJ, Brundin P, Petersén A, Björkqvist M. Increased metabolism in the R6/2 mouse model of Huntington's disease. Neurobiol Dis. 2008;29:41–51. doi: 10.1016/j.nbd.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD, Cui L, Beyer RP, Easley CN, Smith AC, Krainc D, Luquet S, Sweet IR, Schwartz MW, La Spada AR. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohrling GJ, Farrell LA, Hollenberg AN, Cha JH. Mutant huntingtin increases nuclear corepressor function and enhances ligand-dependent nuclear hormone receptor activation. Mol Cell Neurosci. 2003;23:28–38. doi: 10.1016/s1044-7431(03)00032-0. [DOI] [PubMed] [Google Scholar]
- Zala D, Benchoua A, Brouillet E, Perrin V, Gaillard MC, Zurn AD, Aebischer P, Déglon N. Progressive and selective striatal degeneration in primary neuronal cultures using lentiviral vector coding for a mutant huntingtin fragment. Neurobiol Dis. 2005;20:785–798. doi: 10.1016/j.nbd.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Zeron MM, Hansson O, Chen N, Wellington CL, Leavitt BR, Brundin P, Hayden MR, Raymond LA. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Marullo M, Conforti P, MacDonald ME, Tartari M, Cattaneo E. Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington's disease. Brain Pathol. 2008;18:225–238. doi: 10.1111/j.1750-3639.2007.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]