Abstract
PSD-95 is an abundant postsynaptic density (PSD) protein involved in the formation and regulation of excitatory synapses and dendritic spines, but the underlying mechanisms are not comprehensively understood. Here we report a novel PSD-95-interacting protein Preso that regulates spine morphogenesis. Preso is mainly expressed in the brain and contains WW (domain with two conserved Trp residues), PDZ (PSD-95/Dlg/ZO-1), FERM (4.1, ezrin, radixin, and moesin), and C-terminal PDZ-binding domains. These domains associate with actin filaments, the Rac1/Cdc42 guanine nucleotide exchange factor βPix, phosphatidylinositol-4,5-bisphosphate, and the postsynaptic scaffolding protein PSD-95, respectively. Preso overexpression increases the density of dendritic spines in a manner requiring WW, PDZ, FERM, and PDZ-binding domains. Conversely, knockdown or dominant-negative inhibition of Preso decreases spine density, excitatory synaptic transmission, and the spine level of filamentous actin. These results suggest that Preso positively regulates spine density through its interaction with the synaptic plasma membrane, actin filaments, PSD-95, and the βPix-based Rac1 signaling pathway.
Keywords: PSD-95, WW, PDZ, FERM, actin, spine
Introduction
PSD-95 is an abundant postsynaptic scaffolding protein implicated in the regulation of the formation, maturation, maintenance, and plasticity of excitatory synapses and dendritic spines (Scannevin and Huganir, 2000; Sheng and Sala, 2001; Kim and Sheng, 2004; Funke et al., 2005; Craig et al., 2006; Tada and Sheng, 2006; Sheng and Hoogenraad, 2007). PSD-95 is thought to regulate the polymerization of filamentous actin (F-actin), the main cytoskeleton in dendritic spines (Rao and Craig, 2000; Oertner and Matus, 2005; Tada and Sheng, 2006), and spine morphogenesis through various mechanisms. For instance, PSD-95 directly interacts with actin-regulatory proteins such as Kalirin-7 [a guanine nucleotide exchange factor (GEF) for Rac1], SPAR [a GTPase-activating protein (GAP) for Rap], and IRSp53 (a Rac1 effector) (Pak et al., 2001; Penzes et al., 2001; Soltau et al., 2004; Choi et al., 2005). PSD-95 indirectly interacts though the postsynaptic scaffold Shank with actin-regulatory proteins such as βPix (a GEF for Rac1 and Cdc42) and cortactin (an activator of the Arp2/3 actin nucleating complex) (Hering and Sheng, 2003; Park et al., 2003; Zhang et al., 2005). However, our current understanding of the molecular mechanisms underlying PSD-95-dependent spine regulation may not be comprehensive.
The band 4.1 superfamily of proteins is characterized by the FERM (4.1, ezrin, radixin, and moesin) domain (also known as N-ERMAD), a ∼300-amino-acid-long module that mediates protein anchoring to the plasma membrane (Bretscher et al., 2000, 2002; Ramesh, 2004). Members of the band 4.1 superfamily include ERM proteins (ezrin, radixin, and moesin), merlin, protein 4.1, talin, focal adhesion kinase, myosin VII, and protein phosphatases. ERM proteins function as membrane-cytoskeletal linkers contributing to the assembly and maintenance of specialized plasma membrane domains. ERM proteins also regulate signaling pathways and membrane transport (Bretscher et al., 2000, 2002; Ramesh, 2004).
Neuronal functions of FERM domain proteins have been studied previously. ERM proteins regulate axonal development (Dickson et al., 2002; Mintz et al., 2003) and growth cone morphology and motility (Paglini et al., 1998). 4.1N, a brain-enriched protein 4.1 (Walensky et al., 1999), interacts with PIKE, a nuclear GTPase, and is involved in PIKE-dependent activation of phosphatidylinositol 3-kinase (Ye et al., 2000). At neuronal synapses, 4.1N interacts with and regulates surface expression of AMPA and dopamine receptors (Shen et al., 2000; Binda et al., 2002; Hayashi et al., 2005). 4.1N also binds to CASK (calcium/calmodulin-dependent serine kinase), a PDZ (PSD-95/Dlg/ZO-1) scaffold that interacts with and promotes actin nucleation on the synaptic adhesion molecule neurexin (Cohen et al., 1998; Biederer and Sudhof, 2001; Biederer et al., 2002). These results indicate that synaptic FERM domain proteins anchor synaptic receptors and adhesion molecules to actin filaments. However, little is known about the molecular mechanisms that regulate the integrated interactions between actin filaments, the synaptic plasma membrane and embedded membrane proteins, and synaptic scaffolding and signaling proteins.
We here report a novel and brain-specific FERM and PDZ domain protein Preso (PSD-95-interacting regulator of spine morphogenesis) that directly interacts with PSD-95. Results from overexpression, knockdown, and dominant-negative inhibition suggest that Preso regulates dendritic spine morphogenesis.
Materials and Methods
Yeast two-hybrid.
Yeast two-hybrid screens (human brain cDNA library) and assays were performed as described previously (Lee et al., 2006). For bait constructs, the Preso C terminus [amino acids 1316–1322; wild-type (WT) and point mutants] and the N-terminal region containing the PDZ domain (amino acids 32–437) of Preso were subcloned into pBHA (British Biotechnology). Other PDZ domains in pGAD10 have been described previously (Choi et al., 2005).
Expression and short hairpin RNA constructs.
For Flag-tagged Preso expression constructs, the following regions of human Preso (KIAA0316) were subcloned into p3xFLAG-CMV7.1 (Sigma): full length (amino acids 1–1322), ΔC (1–1318), ΔWW (68–1322), ΔWW+C (68–1318), ΔWW+PDZ (186–1322), ΔWW+PDZ+C (186–1318), WW (1–71), WW+PDZ (1–168), WW+PDZ+FERM (1–437), and ΔFERM (deletion of 203–442). The Preso short hairpin RNA (shRNA) knockdown construct (sh-Preso) was generated by annealing oligonucleotides containing nucleotides 1811–1829 of rat Preso (GenBank accession number XM_228906.4; 5′-CCTTGTGTCCCAAAGAGCA-3′) and subcloning it into pSUPER.gfp/neo (Oligoengine). Deletion variants of βPix have been described previously (Park et al., 2003).
Antibodies.
Preso polyclonal antibodies were generated using glutathione S-transferase (GST) fusion proteins of Preso (human) as immunogen: amino acids 4–580 for 1495 (guinea pig) antibodies and amino acids 618–1320 for 1496 (guinea pig) and 1488 (rabbit) antibodies. Specific antibodies were affinity purified using polyvinylidene difluoride membranes (GE Healthcare). Antibodies for PSD-95, PSD-93, SAP97, SAP102, enhanced green fluorescent protein (EGFP), and βPix have been described previously (Park et al., 2003; Choi et al., 2005). Other antibodies were purchased: PSD-95 from Affinity BioReagents; and synaptophysin, synapsin I, β-tubulin, MAP2, and Flag from Sigma.
PIP2 binding assay.
Phosphatidylinositol-4,5-bisphosphate (PIP2) binding of the Preso FERM domain was examined as described previously (Barret et al., 2000). Briefly, large multilamellar liposomes were prepared from phosphatidylcholine (PC) and PIP2 in buffer comprising 20 mm HEPES, pH 7.4, and 0.2 mm EGTA. Purified GST–FERM (Preso amino acids 173–437) preincubated in HEPES buffer was incubated with liposomes (final concentrations of 25 μg/ml proteins and 0.5 mg/ml lipid). Mixtures were subsequently centrifuged for 30 min at 100,000 × g, 4°C, followed by immunoblot analysis. To further determine the specificity of PIP2 binding, neomycin (Sigma), a high-affinity PIP2 ligand, was preincubated with liposomes before mixing with proteins.
F-actin cosedimentation assay.
F-actin binding assay was performed as described by the manufacturer (Cytoskeleton). In brief, protein preparations (GST–WW and α-actinin) were incubated with freshly polymerized actin (F-actin) for 30 min at room temperature. After incubation, reaction solution was subject to ultracentrifugation (150,000 × g) to pellet F-actin and proteins bound to F-actin. The pellet and supernatant were analyzed by Coomasie blue staining.
Pull-down and immunoprecipitation.
For GST pull-down, the last seven residues of Preso were subcloned into pGEX4T-1. For in vitro coimmunoprecipitation, transfected HEK293T cells were extracted in PBS containing 1% Triton X-100 and pulled down by Flag-agarose (Sigma). For in vivo coimmunoprecipitation, deoxycholate extracts of the crude synaptosomal fraction of adult (6 weeks) rat brain were immunoprecipitated with Preso (1488, 10 μg/ml), or rabbit IgG (control), antibodies, and immunoblotted with the following antibodies; Preso (1496, 1 μg/ml), PSD-95 (1:1000), PSD-93 (1:1000), SAP97 (1:1000), and SAP102 (1:1000).
Neuron culture, transfection, and immunohistochemistry.
Cultured hippocampal neurons were prepared from embryonic day 18 rat brain. Dissociated neurons on poly-l-lysine-coated (1 mg/ml) coverslips were placed in Neurobasal medium supplemented with B27 (Invitrogen), 0.5 mm l-glutamine, 12.5 μm glutamate, and penicillin–streptomycin (Invitrogen) for 3 h and grown in fresh medium without glutamate. Neurons were transfected by calcium phosphate precipitation (mammalian transfection kit; Invitrogen). For immunostaining, neurons were permeabilized by PBS containing 0.2% Triton X-100 and incubated with primary antibodies against Preso (1496; 2 μg/ml), PSD-95 (1:500), MAP2 (1:500), EGFP (1:500), synapsin I (1:500), and Flag (1 μg/ml), followed by cyanine 3-, cyanine 5-, or FITC-conjugated secondary antibodies (Jackson ImmunoResearch). Rat brain sections (50 μm) permeabilized by 50% ethanol were immunostained with Preso (1496; 5 μg/ml) antibodies. F-actin in COS-7 cells was visualized using rhodamine–phalloidin (1:500; Invitrogen).
Image acquisition and quantification.
Fluorescent images were acquired using a confocal microscope (LSM510). The same parameter settings were used for all scans. Each experiment was repeated two to three times, and images were blindly analyzed using MetaMorph (Universal Imaging). Dendritic spines were defined as protrusions 0.5–3 μm length, with or without a head. Spine density was measured by counting the number of spines on 12–24 neurons (∼200 μm total dendritic length per neuron). For spine dimensions, ∼700–1300 spines were measured by manually drawing a vertical line from the base of the neck to the furthest point on the spine head for length and by drawing a maximal line perpendicular to the length for width. The density and dimensions of spines from single neurons were averaged to obtain a population mean and SEM. Statistical significance was determined by Student's t test or ANOVA Tukey's test. To measure the spine localization of endogenous Preso, its immunofluorescence intensity in a PSD-95-positive spine was compared with that of an adjacent dendritic trunk. For exogenous Preso cotransfected with PSD-95, Preso signals (R) at spines and adjacent dendritic trunks were normalized to those of PSD-95–EGFP (G) to obtain the spine localization index of Preso ([R/G]spine/[R/G]dendrite). The spine to dendrite ratios of Preso localization were obtained from ∼30 measurements per neuron in 9–12 neurons. For colocalization of Preso variants with F-actin in COS cells, area of Preso overlapping with F-actin were analyzed using the colocalization module of MetaMorph.
Electrophysiology.
Cultured hippocampal neurons were transfected with Preso overexpression or knockdown constructs [days in vitro (DIV) 10–17]. EGFP-expressing neurons were whole-cell voltage clamped at −60 mV using an Axopatch 200B amplifier (Molecular Devices). Miniature EPSCs (mEPSCs) were analyzed using the Mini Analysis Program (Igor).
Results
Identification of Preso as a novel PSD-95-interacting protein
Yeast two-hybrid screens with the PDZ domains of PSD-95 and a human brain cDNA library yielded a novel protein designated Preso (GenBank accession numbers KIAA0316 and NP055543; 1322 amino acids). Preso contains protein interaction domains, including WW (domain with two conserved Trp residues), PDZ, and FERM domains in the N-terminal half and a C-terminal PDZ domain-binding motif, which interacts with the PDZ domains of PSD-95 (Fig. 1A). Homologs of Preso were found in Caenorhabditis elegans and Drosophila, which exhibited higher amino acid sequence identity in WW, PDZ, and FERM domains (C. elegans, GenBank accession number T33637, 691 amino acids, 27% identity; Drosophila, GenBank accession number AAF55127, 1363 amino acids, 21% identity) (Fig. 1A).
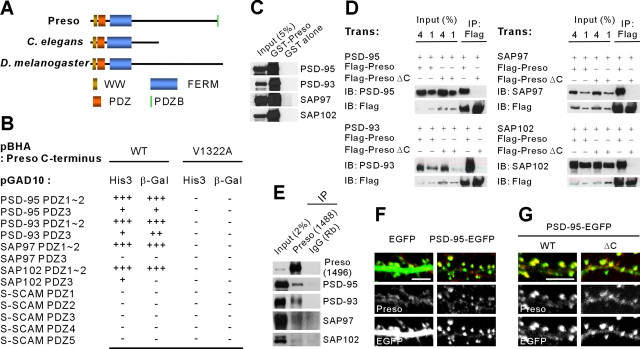
Figure 1.
Interaction between PSD-95 and Preso. A, Domain structure of Preso (human) and its C. elegans and Drosophila homologs. PDZB, PDZ domain-binding motif. B, Preso interacts with the PDZ domains of PSD-95 family proteins in the yeast two-hybrid assay. The Preso C terminus, WT (last 7 residues), and a point mutant lacking the PDZ interaction (V1322A), in pBHA (bait vector) was tested for binding to PDZ domains in pGAD10 (prey vector). PDZ domains from S-SCAM (synaptic scaffolding molecule) were used as negative controls. HIS3 activity: +++, >60%; ++, 30–60%; +, 10–30%; −, no significant growth. β-Galactosidase activity: +++, <45 min; ++, 45–90 min; +, 90–240 min; −, no significant activity. C, GST fusion proteins of Preso (amino acids 1316–1322) pull-down PSD-95 family proteins expressed in HEK293T cells. D, Preso forms a complex with PSD-95 family proteins in HEK293T cells. Cell lysates double transfected (Trans) with Flag–Preso and PSD-95 family proteins were immunoprecipitated (IP) with Flag antibodies and immunoblotted (IB) with the indicated antibodies. ΔC, A mutant that lacks the last four residues and thus PSD-95 binding. E, Preso forms a complex with PSD-95 and PSD-95 relatives in brain. Detergent lysates of the crude synaptosomal fraction of adult rat brain were immunoprecipitated with the Preso antibodies (1488) or rabbit IgG (Rb; control) and immunoblotted with the indicated antibodies. F, PSD-95 overexpression in cultured hippocampal neurons promotes spine localization of endogenous Preso. Cultured neurons transfected with PSD-95–EGFP or EGFP alone (control; DIV 15–16) were stained for EGFP and Preso. Spine localization of Preso was determined by comparing the immunofluorescence intensity of Preso at a dendritic spine versus an adjacent dendritic trunk. Scale bar, 5 μm. G, PSD-95 promotes spine localization of Preso through its PDZ interaction with Preso. Cultured neurons were double transfected with PSD-95–EGFP and Flag–Preso (WT or ΔC; DIV 15–16) and stained for EGFP (PSD-95) and Flag (Preso). PSD-95 was cotransfected with Preso to ensure that the amount of endogenous PSD-95 was not a limiting factor for spine localization of exogenously expressed Preso. Spine localization of Preso was determined by comparing the immunofluorescence intensity of Preso, normalized to that of PSD-95–EGFP (see Materials and Methods), in a PSD-95-positive dendritic spine with that in adjacent dendritic trunk. Scale bar, 5 μm.
In yeast two-hybrid assays, Preso interacted with all known PSD-95 family proteins (PSD-95/SAP90, PSD-93/chapsyn-110, SAP97, and SAP102) in vitro (Fig. 1B). The Preso C terminus bound with high affinity to the first two PDZ domains. Preso binding to PSD-95 family proteins was further confirmed in pull-down and in vitro coimmunoprecipitation assays (Fig. 1C,D). In rat brain, Preso formed a complex with all known PSD-95 family proteins (Fig. 1E), consistent with the in vitro results. Details on the Preso antibodies used for coimmunoprecipitation experiments are described below (Fig. 2D).
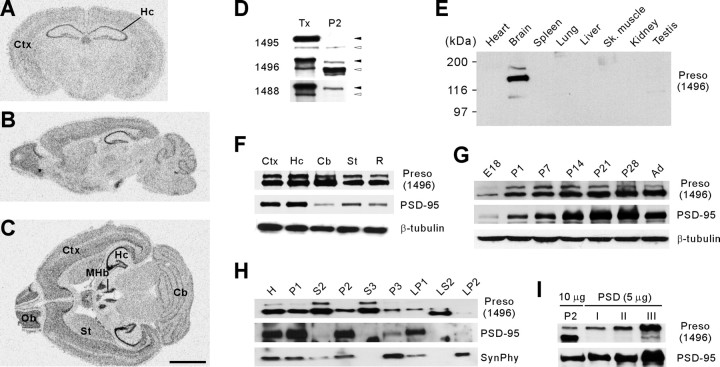
Figure 2.
Expression patterns of Preso mRNAs and proteins. A–C, Distribution patterns of Preso mRNAs in the brain. Coronal (A), sagittal (B), and horizontal (C) sections of adult mouse brain (6 weeks) were hybridized with a Preso riboprobe. Ctx, Cortex; Hc, hippocampus; MHb, medial habenular nucleus; St, striatum; Ob, olfactory bulb; Cb, cerebellum. Scale bar, 6 mm. D, Three different Preso antibodies (1495, 1496, and 1488) differentially recognize two Preso bands (upper and lower, indicated by filled and open arrowheads, respectively). Tx, Preso proteins expressed in transfected HEK293T cells. P2, Crude synaptosomal fraction from adult rat brain. E, Brain-specific expression of Preso proteins, revealed by immunoblot analysis of adult rat tissue homogenates. Sk, Skeletal. F, Widespread distribution of Preso proteins in adult rat brain regions. R, Other regions of the brain. PSD-95 and β-tubulin were used as controls. G, Gradual increase in Preso expression during postnatal rat brain development. E, Embryonic day; P, postnatal day; Ad, adult (6 weeks). H, Distribution of Preso in subcellular fractions of rat brain. H, Homogenates; S2, supernatant after P2 precipitation; S3, cytosol; P3, light membranes; LP1, synaptosomal membranes; LS2, synaptosomal cytosol; LP2, synaptic vesicle-enriched fraction. Synaptophysin (SynPhy) was used as control. I, Enrichment of Preso in PSD fractions. Postsynaptic density fractions extracted with Triton X-100 once (PSD I), twice (PSD II), and Triton X-100 and a strong detergent Sarcosyl (PSD III) were immunoblotted with Preso and PSD-95 antibodies. Note that the upper, but not the lower, Preso band is selectively enriched.
We next tested whether PSD-95 interaction promotes synaptic localization of Preso, which is partly synaptic (see below). In cultured hippocampal neurons, overexpression of PSD-95 markedly increased spine localization of endogenous Preso, as determined by the relative distribution of Preso at a spine versus a nearby dendritic trunk (EGFP alone, 1.12 ± 0.06, n = 9; PSD-95–EGFP, 1.91 ± 0.08, n = 10; ***p < 0.001 compared with EGFP alone, Student's t test) (Fig. 1F). In addition, in neurons double transfected with Preso and PSD-95, WT Preso showed a greater spine localization than a mutant Preso that lacks PSD-95-binding C terminus (Preso ΔC) (WT Preso, 0.96 ± 0.06, n = 11; Preso ΔC, 0.59 ± 0.03, n = 9; ***p < 0.001, Student's t test) (Fig. 1G). These results suggest that PSD-95 promotes synaptic localization of Preso through their PDZ interaction.
PDZ-interacting peptides are often phosphorylated at the −2 position, and this phosphorylation inhibits their PDZ interactions (Hung and Sheng, 2002). The Preso C- terminal sequence is KIKETTV (supplemental Fig. S1A, available at www.jneurosci.org as supplemental material). A phosphomimetic mutation at the −2 position of the Preso C terminus (KIKEDTV; T1320D) abolished the coprecipitation of Preso with PSD-95 in HEK293T cells, similar to results with the Preso ΔC mutant (supplemental Fig. S1B, available at www.jneurosci.org as supplemental material). In addition, significantly lower levels of the Preso T1320D mutant are localized to dendritic spines compared with WT Preso (supplemental Fig. S1C,D, available at www.jneurosci.org as supplemental material). These results suggest that the interaction of Preso with PSD-95 may be regulated by phosphorylation.
Expression patterns of Preso mRNAs and proteins
Because Preso is a novel protein, we examined expression patterns of Preso mRNAs and proteins. Preso mRNAs were widely expressed in various mouse brain regions, including cortex, hippocampus, cerebellum, olfactory bulb, and medial habenular nucleus (Fig. 2A–C). For the analysis of protein expression, we generated polyclonal Preso antibodies (1495, 1496, and 1488) using immunogens containing two independent regions of Preso. These antibodies revealed two major Preso bands in the brain (∼170 and 140 kDa, respectively), which were differentially recognized by the antibodies (Fig. 2D). The upper band had a molecular weight similar to Preso proteins expressed in heterologous cells. Immunoblot analysis revealed that Preso proteins are predominantly expressed in the brain, widespread in various brain regions, and gradually increased during postnatal rat brain development (Fig. 2E–G). Preso proteins were detected in various subcellular fractions of rat brain, including crude synaptosomes (P2), cytosol (S3), and light membranes (P3) (Fig. 2H). The upper, but not the lower, Preso band was enriched in postsynaptic density (PSD) fractions including the PSD III fraction (Fig. 2I), indicative of a very tight association with the PSD.
When immunofluorescence staining of Preso was performed in cultured hippocampal neurons, Preso was detected in MAP2 (dendritic marker)-positive dendrites, with some signals in MAP2-negative axons (supplemental Fig. S2A, available at www.jneurosci.org as supplemental material). In dendrites, Preso signals were detected in discrete clusters, which were localized at PSD-95-positive synaptic sites (Preso-positive PSD-95, 72%) as well as PSD-95-negative extrasynaptic sites (supplemental Fig. S2B, available at www.jneurosci.org as supplemental material), consistent with its widespread biochemical distribution in rat brain fractions. In brain slices, Preso signals were observed in various rat brain regions, including hippocampus, cortex, and cerebellum (supplemental Fig. S3, available at www.jneurosci.org as supplemental material), similar to the distribution patterns Preso mRNAs. Preso signals were detected in dendrites of principal neurons, including hippocampal CA1 and CA3 pyramidal cells and cerebellar Purkinje cells.
Preso associates with PIP2 and actin filaments
Because many FERM domains bind PIP2 (Niggli et al., 1995; Hirao et al., 1996), we tested whether the FERM domain of Preso associates with PIP2. GST fusion proteins fused to the FERM domain of Preso cosedimented with PIP2-containing, but not control, liposomes (Fig. 3A). Preincubation of PIP2-containing liposomes with neomycin, a high-affinity PIP2 ligand, inhibited the cosedimentation with the Preso FERM domain, suggesting the Preso FERM domain specifically associates with PIP2.
Figure 3.
Preso associates with PIP2 and actin filaments. A, Association of the Preso FERM domain with PIP2. GST fusion proteins containing the FERM domain of Preso (GST-Preso-FERM) cosedimented with liposomes containing PC plus PIP2 but not with those with PC alone (control). Neomycin, a high-affinity PIP2 ligand, was added to confirm the specificity of PIP2 binding to the Preso FERM domain. B, Schematic diagram of Preso deletion variants. C–G, The WW domain of Preso is sufficient and required for F-actin association. COS-7 cells transfected with Flag–Preso deletion variants were stained for Flag (for Preso) and F-actin (rhodamine–phalloidin). Cells were visualized within ∼12 h of transfection to minimize overexpression of the proteins. Scale bar, 10 μm. H, Quantification of the colocalization of Preso variants with F-actin from the results in C–G. Area of Preso overlapping with F-actin was compared with the total area of Preso to obtain the ratio of F-actin-positive Preso/total Preso. Mean ± SEM (n = 3 cells, respectively; ***p < 0.001, ANOVA Tukey's test). I, Preso does not coprecipitate with F-actin. GST fusion proteins containing the WW domain of Preso (GST-WW; amino acids 1–71) or α-actinin (a positive control) were mixed with F-actin, precipitated by ultracentrifugation, and Coomasie blue stained. P, Pellet; S, supernatant.
A number of FERM domain-containing proteins often associate with actin filaments in addition to PIP2 (Bretscher et al., 2002; Ramesh, 2004), suggesting that Preso might interact with F-actin. During expression in COS-7 cells, Preso colocalized with F-actin-rich structures, such as lamellipodia and stress fibers (Fig. 3B,C). Deletion of the WW domain of Preso (ΔWW) eliminated the actin colocalization of Preso, whereas the WW domain (WW) alone was sufficient for actin colocalization (Fig. 3B,D–G). Quantification analysis showed significantly reduced levels of F-actin colocalization with Preso ΔWW compared with other Preso variants with intact WW domains (Fig. 3H). However, Preso did not coprecipitate with F-actin in a sedimentation assay, whereas α-actinin, a positive control, was brought down by F-actin (Fig. 3I). The results suggest that Preso indirectly associates with actin filaments through the WW domain.
Preso overexpression leads to an increase in spine density
Because Preso associates with actin filaments, a main cytoskeleton in dendritic spines (Rao and Craig, 2000; Oertner and Matus, 2005; Tada and Sheng, 2006), we examined the effects of Preso overexpression on dendritic spines. Preso overexpression in cultured hippocampal neurons significantly increased the linear density of dendritic spines, as defined by dendritic protrusions 0.5–3.0 μm long (with or without the head) (Fig. 4A,B). However, the length and width of dendritic spines were not changed (Fig. 4A,C,D). This suggests that Preso positively regulates dendritic spine density but not morphology.
Figure 4.
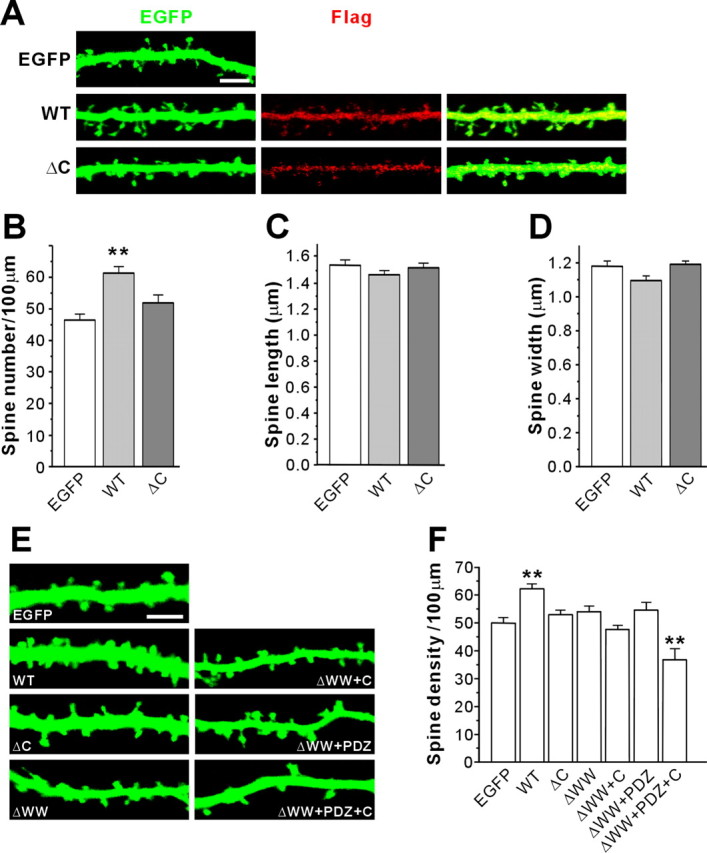
Dendritic spine density is regulated by Preso overexpression and dominant-negative inhibition. A, Effects of Preso overexpression on dendritic spines. Cultured hippocampal neurons were transfected with Flag–Preso (WT or ΔC) plus EGFP or EGFP alone (control) (DIV 9–16) and visualized by immunofluorescence staining for Flag and EGFP. Scale bar, 5 μm. B–D, Preso overexpression increases spine density (B; number of spines in 100 μm dendrites) but not length (C) or width (D) in a manner requiring the PSD-95-binding C terminus. Mean ± SEM (n = 12 neurons for EGFP, 15 for WT, and 11 for ΔC, **p < 0.01, ANOVA Tukey's test). E, Effects of Preso deletion variants on spine density. Neurons double transfected with Preso deletion variants and EGFP (DIV 9–16) were stained for EGFP and Preso (data not shown). Scale bar, 5 μm. F, Quantification of the results from E. Deletion of the WW domain reverses Preso-dependent spine promotion, similar to the C-terminal deletion. In addition, Preso ΔWW+PDZ+C reduces spine density in a dominant-negative manner. Mean ± SEM (n = 20 for EGFP, 18 for WT, 24 for ΔC, 13 for ΔWW, 12 for ΔWW+C, 12 for ΔWW+PDZ, and 16 for ΔWW+PDZ+C; **p < 0.01, ANOVA Tukey's test).
In contrast, overexpression of Preso mutants (Preso ΔC and Preso ΔWW), which lack the C-terminal PDZ-binding motif and the WW domain, respectively, did not show spine-promoting effects (Fig. 4A–F). This suggests that Preso promotes spine density in a manner requiring its binding to PSD-95 as well as actin filaments. Moreover, a bigger deletion in Preso (Preso ΔWW+PDZ+C) caused a significant reduction in spine density in a dominant-negative manner (Fig. 4E,F), further supporting the importance of Preso in spine regulation.
Preso knockdown causes a decrease in spine density
We next observed the effects of Preso knockdown on dendritic spines, using an shRNA construct that reduces endogenous Preso expression (sh-Preso) by ∼80% in cultured neurons (supplemental Fig. S4A,B, available at www.jneurosci.org as supplemental material). Preso knockdown in cultured hippocampal neurons significantly reduced dendritic spine density (Fig. 5A,B). However, Preso knockdown had no effects on spine length and width (Fig. 5A,C,D). In addition, Preso knockdown reduced the number of excitatory synapses, as defined by synapsin I (a presynaptic marker)-positive PSD-95 clusters (Fig. 5A,E). In contrast, cotransfection of neurons with sh-Preso and an sh-Preso-resistant Preso expression construct, which restores Preso expression levels to 126% (n = 10 cells) of original levels, fully reversed the Preso knockdown-induced reductions in spine density and excitatory synapse number (Fig. 5A–E). These results suggest that Preso is required for the maintenance of dendritic spines.
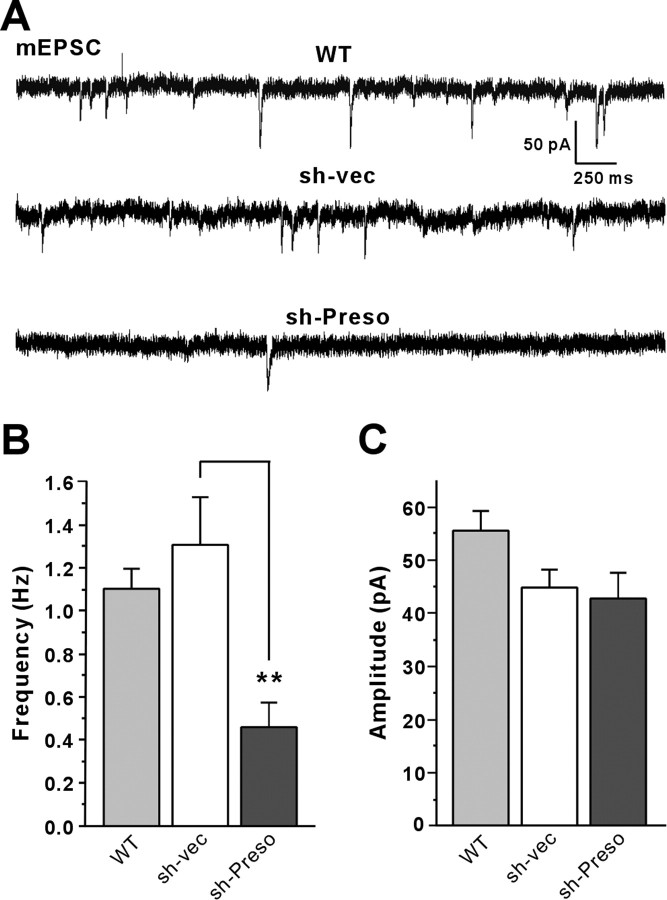
Figure 5.

Preso knockdown decreases the number of dendritic spines and excitatory synapses. A, Effects of Preso shRNA knockdown on dendritic spines and excitatory synapses. Cultured neurons were transfected with a Preso knockdown construct (sh-Preso), empty pSuper vector (sh-vec), or sh-Preso plus rescue Preso expression construct (Rescue; human; DIV 9–16) and stained for EGFP (for shRNA transfection), PSD-95 (an excitatory postsynaptic marker), and synapsin I (a presynaptic marker). Scale bar, 5 μm. B–D, Preso knockdown reduces spine number (B) but not length (C) or width (D). Preso knockdown also reduces the number of excitatory synapses (E), as defined by synapsin I-positive PSD-95 clusters. Note that the human Preso expression construct, which differs from rat Preso by 2 nt in the 19 bp shRNA target region, fully rescues the negative effects of Preso knockdown on spine density and excitatory synapse number. Mean ± SEM (n = 22 for sh-vec, 22 for sh-Preso, and 30 for rescue; ***p < 0.001, ANOVA Tukey's test).
Preso knockdown suppresses excitatory synaptic transmission
To determine whether Preso-dependent regulation of spine density and excitatory synapses accompanies a reduction in excitatory synaptic transmission, we tested whether Preso overexpression or knockdown affects excitatory synaptic transmission, as measured by mEPSCs. Overexpression of Preso in cultured hippocampal neurons did not affect the frequency or amplitude of mEPSCs compared with control neurons expressing EGFP alone (sh-vec) (Fig. 6A–C), contrary to the increase in spine density by Preso overexpression (Fig. 4). This suggests that Preso overexpression is not sufficient for the functional maturation of induced spines. Importantly, however, Preso knockdown led to a significant decrease in the frequency, but not amplitude, of mEPSCs (Fig. 6A–C), consistent with the Preso knockdown-induced decreases in spine density and excitatory synapse number (Fig. 5). These results suggest that Preso is required for the maintenance of excitatory synaptic transmission.
Figure 6.
Preso knockdown decreases excitatory synaptic transmission. A, Effects of Preso overexpression and knockdown on mEPSCs. Cultured neurons were transfected with WT Preso expression construct plus sh-vec, sh-vec, or sh-Preso (DIV 10–17), followed by mEPSC measurements. WT Preso was cotransfected with sh-vec to visualize transfected live neurons, using EGFP proteins expressed from sh-vec and to compare the effects of Preso overexpression with those of sh-vec. B, C, Quantification of the results from A. Note that Preso overexpression does not affect mEPSCs, whereas Preso knockdown decreases the frequency, but not amplitude, of mEPSCs. Mean ± SEM (n = 14 for WT, n = 16 for sh-vec, and 11 for sh-Preso; **p < 0.01, ANOVA Tukey's test).
Preso interacts with βPix and requires the PDZ and FERM domains for spine regulation
To understand the functions of the PDZ domain of Preso, we attempted another yeast two-hybrid screen using the N-terminal region of Preso containing the PDZ domain as bait. We isolated βPix, a GEF for Rac1 and Cdc42 small GTPases, as a novel binding partner of the Preso PDZ domain. Preso formed a complex with βPix in HEK293T cells (Fig. 7A,B). Deletion of the last four residues in βPix (ΔETNL) slightly reduced Preso association of βPix (Fig. 7A,B). Another βPix mutant that lacks the leucine zipper (LZ) domain showed a greater reduction in Preso association. Deletion of the region containing LZ and the last four residues completely abolished Preso association, suggesting that both the LZ domain and the last four residues of βPix are involved in the interaction with Preso. Molecular modeling predicted that the Glu (E) residue at the −3 position in the βPix C terminus forms a strong salt bridge with R102 in the Preso PDZ domain (Fig. 7C). Indeed, a point mutation (R102A) in the PDZ domain of Preso abolished its βPix interaction (Fig. 7D). In brain, Preso antibodies brought down βPix proteins, and, conversely, βPix coprecipitated with Preso (Fig. 7E), suggesting that Preso and βPix form a complex in vivo.
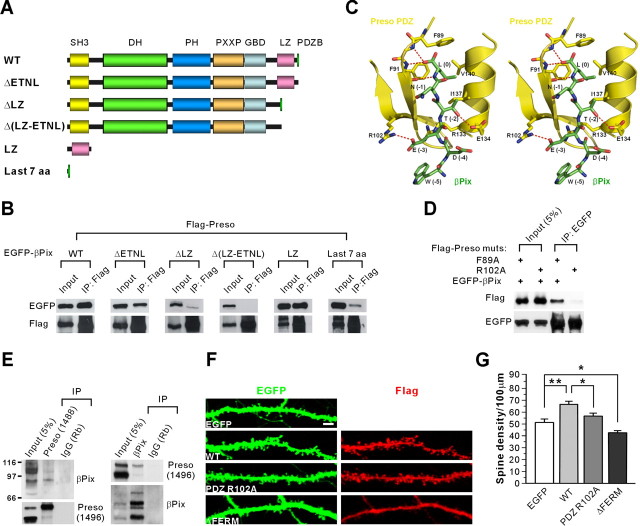
Figure 7.
Preso interacts with βPix and increases spine density in a manner requiring the PDZ and FERM domains. A, Diagram of deletion variants of EGFP–βPix used in the coimmunoprecipitation with Flag–Preso. SH3, Src homology 3; DH, Dbl homology; PH, pleckstrin homology; PXXP, proline-rich region; GBD, GIT-binding domain; aa, amino acids. B, βPix requires the LZ and C-terminal domains for the interaction with Preso. Lysates of HEK293T cells double transfected with Flag–Preso plus EGFP–βPix (WT and mutants) were immunoprecipitated (IP) with Flag–agarose and immunoblotted with Flag and EGFP antibodies. C, Molecular modeling and stereo view of the interaction between the βPix C terminus, which ends with WDETNL (green), and the Preso PDZ domain (yellow). The model was built using the structure of the PDZ domain of human RGS-3 (Protein Data Bank identification number 2F5Y), which has a high amino acid sequence identity to the Preso PDZ domain. Note that residues at 0, −2, and −3 positions are important for the PDZ interaction. Especially, the Glu (E) residue at the −3 position is highly likely to form a salt bridge with R102 in the Preso PDZ domain. D, R102 residue in the Preso PDZ domain is critical for the interaction with the βPix C terminus. Lysates of HEK293T cells double transfected with Flag–Preso (point mutants; F89A or R102A) plus EGFP-βPix were immunoprecipitated with EGFP antibodies and immunoblotted with EGFP and Flag antibodies. E, Preso forms a complex with βPix in the brain. Detergent lysates of the crude synaptosomal fraction of adult (6 weeks) rat brain were immunoprecipitated with Preso antibodies (1488), βPix antibodies (1254), or rabbit IgG (Rb; control) and immunoblotted with βPix (1257) and Preso (1496) antibodies. F, Preso requires the PDZ and FERM domains for the enhancement of spine density. Neurons double transfected with Preso mutants (PDZ R102A or ΔFERM that lacks the FERM domain) plus EGFP (DIV 9–16) were stained for EGFP and Preso. Scale bar, 5 μm. G, Quantification of the results from F. PDZ mutant (PDZ R102A) did not increase significantly. Mean ± SEM (n = 17 for EGFP, 15 for WT, 20 for PDZ R102A, and 14 for ΔFERM; *p < 0.05, **p < 0.01, ANOVA Tukey's test).
We next determined whether the PDZ domain of Preso is required for Preso-induced enhancement of spine density. Overexpression of a Preso variant with a point mutation in the PDZ domain (PDZ R102A), which lacks βPix interaction, did not induce a significant increase in spine density in cultured neurons, contrary to WT Preso (Fig. 7F,G). In addition, the spine density in neurons expressing PDZ R102A was significantly smaller than that in neurons expressing WT Preso (Fig. 7F,G). In parallel experiments, a mutant Preso that lacks the PIP2-binding FERM domain (ΔFERM) induced a reduction in spine density in a dominant-negative manner (Fig. 7F,G). These results suggest that Preso positively regulates dendritic spine density in a manner requiring the PDZ and FERM domains.
Knockdown or inhibition of Preso reduces spine content of F-actin
Our data indicate that Preso associates with F-actin (Fig. 3) and directly interacts with βPix (Fig. 7), a positive regulator of the actin cytoskeleton in dendritic spines (Zhang et al., 2003). We thus investigated whether reduced expression or inhibition of Preso affects F-actin levels in dendritic spines. Interestingly, Preso knockdown significantly reduced the spine level of F-actin, whereas that of PSD-95 was unaffected (Fig. 8A,B). In addition, overexpression of Preso ΔWW+PDZ+C, which reduces spine density in a dominant-negative manner (Fig. 4E,F), significantly reduced the spine level of F-actin but not PSD-95 (Fig. 8A,B). These results indicate that Preso is important for the maintenance of F-actin stability in dendritic spines.
Figure 8.
Knockdown or inhibition of Preso reduces F-actin levels in dendritic spines. A, Preso shRNA knockdown or overexpression of a dominant-negative Preso (ΔWW+PDZ+C) in neurons reduces spine levels of F-actin but not that of PSD-95. Cultured neurons were transfected with a Preso knockdown construct (sh-Preso), an empty pSuper vector (sh-vec), or a Preso mutant construct (sh-vec+ΔWW+PDZ+C; DIV 9–16) and stained for EGFP, F-actin (by rhodamine–phalloidin), and PSD-95. Spine levels represent average fluorescence intensities of F-actin or PSD-95 signals in a spine. Scale bar, 5 μm. B, Quantification of the results from A. Mean ± SEM (n = 29 for sh-vec, 21 for sh-Preso, and 27 for ΔWW+PDZ+C; ***p < 0.001, ANOVA Tukey's test).
Discussion
The present study identifies Preso as a novel binding partner of PSD-95 that contains WW, PDZ, FERM, and C-terminal PDZ-binding domains. Functionally, Preso positively regulates dendritic spine density in a manner requiring the four domains of Preso. How might these domains contribute to Preso-dependent spine regulation? First, the WW domain of Preso may contribute to spine regulation by associating with actin filaments, a principal cytoskeletal component of dendritic spines (Rao and Craig, 2000; Oertner and Matus, 2005; Tada and Sheng, 2006). This is consistent with the reported F-actin association of various spine regulatory proteins, including SPAR, cortactin, drebrin, profilin, neurabin, spinophilin, abp1, α-actinin, and myosins II/VI (Pak et al., 2001; Ackermann and Matus, 2003; Hering and Sheng, 2003; Takahashi et al., 2003; Nakagawa et al., 2004; Qualmann et al., 2004; Zito et al., 2004; Osterweil et al., 2005; Terry-Lorenzo et al., 2005; Ryu et al., 2006).
Second, the PDZ domain of Preso interacts with βPix, a GEF for Rac1 and Cdc42, which are well known for its roles in the regulation of spine morphogenesis (Yuste and Bonhoeffer, 2004; Ethell and Pasquale, 2005; Govek et al., 2005; Kennedy et al., 2005; Newey et al., 2005; Segal, 2005; Calabrese et al., 2006; Tada and Sheng, 2006; Schubert and Dotti, 2007). In addition, a signaling complex containing CaMKI (calcium/calmodulin-dependent kinase I), GIT1, Pix, Shank, Rac, and PAK (p21-activated kinase) has been strongly implicated in the regulation of F-actin polymerization and dendritic spines (Park et al., 2003; Penzes et al., 2003; Zhang et al., 2003; Hayashi et al., 2004; Zhang et al., 2005; Saneyoshi et al., 2008).
Third, the FERM domain of Preso associates with PIP2, a phospholipid concentrated in the plasma membrane, and regulates various cellular processes, including endocytosis, exocytosis, and plasma membrane-cytoskeleton interaction (Di Paolo and De Camilli, 2006). Therefore, the FERM domain of Preso may mediate the anchoring of Preso to the synaptic plasma membrane. In addition, the PIP2 binding of Preso may promote Preso-associated actin filaments to be linked to the synaptic plasma membrane. This is reminiscent of the involvement of ERM proteins in linking the plasma membrane with actin filaments (Bretscher et al., 2000, 2002; Ramesh, 2004). Last, the C terminus of Preso interacts with PSD-95, and this interaction promotes synaptic localization of Preso. Consistently, a subset of Preso proteins is tightly associated with the PSD. This suggests that PSD-95 may recruit Preso to the PSD to support its spine regulatory effects.
How might these molecular mechanisms be integrated for Preso-dependent spine regulation? A straightforward possibility is that Preso, which is capable of interacting with both PIP2 and actin filaments, might play a role in linking the synaptic plasma membrane with actin filaments, as mentioned above. In addition, Preso, which can associate with both actin filaments and PSD-95, may link actin filaments to the PSD. The PSD is likely to be in close contact with the growing ends of actin filaments and play a key role in regulating actin dynamics (Rao and Craig, 2000). Consistently, knockdown and dominant-negative inhibition of Preso reduced spine content of F-actin but not PSD-95 (Fig. 8). This is reminiscent of SPAR, a spine-promoting Rap GAP that associates with both PSD-95 and F-actin (Pak et al., 2001). A similar interaction has been observed in other FERM domain proteins, i.e., protein 4.1 links actin filaments to the PDZ-containing scaffold CASK/LIN-2 (Cohen et al., 1998; Biederer and Sudhof, 2001), and ERM proteins couple actin filaments to the tandem PDZ protein EBP50 (Reczek et al., 1997). Finally, because Preso associates with βPix, Preso might bring βPix-associated F-actin polymerizing activity to the proximity of growing actin filaments. These possibilities collectively suggest that Preso might function as a point in which the synaptic plasma membrane, actin filaments, PSD-95, and the βPix-based Rac signaling complex are integrated for spine regulation.
It should be noted that a significant fraction of Preso proteins are present in the cytosolic fraction (Fig. 2H), despite the fact that Preso associates with synaptically concentrated actin filaments and PSD-95, although βPix has been shown to be present in both crude synaptosomal as well as extrasynaptosomal rat brain fractions (Park et al., 2003). This suggests that the subcellular localization of extrasynaptic Preso might be regulated. In support of this possibility, a mutant Preso mimicking the C-terminal phosphorylation shows reduced PSD-95 binding and spine localization (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). It has been shown that PIP2 binding to the FERM domain recruits dormant ERM proteins from the cytosol to the plasma membrane and activates them by unmasking the F-actin binding site for ERM-dependent cross-linking of actin filaments with the plasma membrane (Bretscher et al., 2000, 2002; Ramesh, 2004). Therefore, the subcellular localization of Preso proteins in neurons might be regulated by levels of local PIP2 and protein phosphorylation in an activity-dependent manner.
In conclusion, our study identifies a novel PSD-95-interacting FERM and PDZ domain protein Preso. Preso regulates dendritic spines in a manner requiring its four distinct domains involved in the association with actin filaments, βPIX, PIP2, and PSD-95. Future studies will focus on genetic confirmation of Preso functions and exploration of detailed molecular mechanisms underlying Preso-dependent spine regulation.
Footnotes
This work was supported by the National Creative Research Initiative Program of the Korean Ministry of Science and Technology (E.K.) and the 21C Frontier Research and Development Program in Neuroscience of the Korean Ministry of Science and Technology (Grant M103KV010023-07K2201-02510) (H.K.). We thank the Kazusa DNA Research Institute for the KIAA0316 cDNA clone.
References
- AckermannM, Matus A (2003) Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci 6:1194–1200. [DOI] [PubMed] [Google Scholar]
- BarretC, Roy C, Montcourrier P, Mangeat P, Niggli V (2000) Mutagenesis of the phosphatidylinositol 4,5-bisphosphate (PIP(2)) binding site in the NH(2)-terminal domain of ezrin correlates with its altered cellular distribution. J Cell Biol 151:1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BiedererT, Sudhof TC (2001) CASK and protein 4.1 support F-actin nucleation on neurexins. J Biol Chem 276:47869–47876. [DOI] [PubMed] [Google Scholar]
- BiedererT, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC (2002) SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297:1525–1531. [DOI] [PubMed] [Google Scholar]
- BindaAV, Kabbani N, Lin R, Levenson R (2002) D2 and D3 dopamine receptor cell surface localization mediated by interaction with protein 4.1N. Mol Pharmacol 62:507–513. [DOI] [PubMed] [Google Scholar]
- BretscherA, Chambers D, Nguyen R, Reczek D (2000) ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annu Rev Cell Dev Biol 16:113–143. [DOI] [PubMed] [Google Scholar]
- BretscherA, Edwards K, Fehon RG (2002) ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol 3:586–599. [DOI] [PubMed] [Google Scholar]
- CalabreseB, Wilson MS, Halpain S (2006) Development and regulation of dendritic spine synapses. Physiology (Bethesda) 21:38–47. [DOI] [PubMed] [Google Scholar]
- ChoiJ, Ko J, Racz B, Burette A, Lee JR, Kim S, Na M, Lee HW, Kim K, Weinberg RJ, Kim E (2005) Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. J Neurosci 25:869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CohenAR, Woods DF, Marfatia SM, Walther Z, Chishti AH, Anderson JM (1998) Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J Cell Biol 142:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CraigAM, Graf ER, Linhoff MW (2006) How to build a central synapse: clues from cell culture. Trends Neurosci 29:8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di PaoloG, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443:651–657. [DOI] [PubMed] [Google Scholar]
- DicksonTC, Mintz CD, Benson DL, Salton SR (2002) Functional binding interaction identified between the axonal CAM L1 and members of the ERM family. J Cell Biol 157:1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EthellIM, Pasquale EB (2005) Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol 75:161–205. [DOI] [PubMed] [Google Scholar]
- FunkeL, Dakoji S, Bredt DS (2005) Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem 74:219–245. [DOI] [PubMed] [Google Scholar]
- GovekEE, Newey SE, Van Aelst L (2005) The role of the Rho GTPases in neuronal development. Genes Dev 19:1–49. [DOI] [PubMed] [Google Scholar]
- HayashiML, Choi SY, Rao BS, Jung HY, Lee HK, Zhang D, Chattarji S, Kirkwood A, Tonegawa S (2004) Altered cortical synaptic morphology and impaired memory consolidation in forebrain- specific dominant-negative PAK transgenic mice. Neuron 42:773–787. [DOI] [PubMed] [Google Scholar]
- HayashiT, Rumbaugh G, Huganir RL (2005) Differential regulation of AMPA receptor subunit trafficking by palmitoylation of two distinct sites. Neuron 47:709–723. [DOI] [PubMed] [Google Scholar]
- HeringH, Sheng M (2003) Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci 23:11759–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HiraoM, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S, Tsukita S (1996) Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol 135:37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HungAY, Sheng M (2002) PDZ domains: structural modules for protein complex assembly. J Biol Chem 277:5699–5702. [DOI] [PubMed] [Google Scholar]
- KennedyMB, Beale HC, Carlisle HJ, Washburn LR (2005) Integration of biochemical signalling in spines. Nat Rev Neurosci 6:423–434. [DOI] [PubMed] [Google Scholar]
- KimE, Sheng M (2004) PDZ domain proteins of synapses. Nat Rev Neurosci 5:771–781. [DOI] [PubMed] [Google Scholar]
- LeeHW, Ko J, Kim E (2006) Analysis of PDZ domain interactions using yeast two-hybrid and coimmunoprecipitation assays. Methods Mol Biol 332:233–244. [DOI] [PubMed] [Google Scholar]
- MintzCD, Dickson TC, Gripp ML, Salton SR, Benson DL (2003) ERMs colocalize transiently with L1 during neocortical axon outgrowth. J Comp Neurol 464:438–448. [DOI] [PubMed] [Google Scholar]
- NakagawaT, Engler JA, Sheng M (2004) The dynamic turnover and functional roles of alpha-actinin in dendritic spines. Neuropharmacology 47:734–745. [DOI] [PubMed] [Google Scholar]
- NeweySE, Velamoor V, Govek EE, Van Aelst L (2005) Rho GTPases, dendritic structure, and mental retardation. J Neurobiol 64:58–74. [DOI] [PubMed] [Google Scholar]
- NiggliV, Andréoli C, Roy C, Mangeat P (1995) Identification of a phosphatidylinositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett 376:172–176. [DOI] [PubMed] [Google Scholar]
- OertnerTG, Matus A (2005) Calcium regulation of actin dynamics in dendritic spines. Cell Calcium 37:477–482. [DOI] [PubMed] [Google Scholar]
- OsterweilE, Wells DG, Mooseker MS (2005) A role for myosin VI in postsynaptic structure and glutamate receptor endocytosis. J Cell Biol 168:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PagliniG, Kunda P, Quiroga S, Kosik K, Cáceres A (1998) Suppression of radixin and moesin alters growth cone morphology, motility, and process formation in primary cultured neurons. J Cell Biol 143:443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PakDT, Yang S, Rudolph-Correia S, Kim E, Sheng M (2001) Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron 31:289–303. [DOI] [PubMed] [Google Scholar]
- ParkE, Na M, Choi J, Kim S, Lee JR, Yoon J, Park D, Sheng M, Kim E (2003) The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J Biol Chem 278:19220–19229. [DOI] [PubMed] [Google Scholar]
- PenzesP, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA (2001) The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29:229–242. [DOI] [PubMed] [Google Scholar]
- PenzesP, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL (2003) Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF Kalirin. Neuron 37:263–274. [DOI] [PubMed] [Google Scholar]
- QualmannB, Boeckers TM, Jeromin M, Gundelfinger ED, Kessels MM (2004) Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. J Neurosci 24:2481–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RameshV (2004) Merlin and the ERM proteins in Schwann cells, neurons and growth cones. Nat Rev Neurosci 5:462–470. [DOI] [PubMed] [Google Scholar]
- RaoA, Craig AM (2000) Signaling between the actin cytoskeleton and the postsynaptic density of dendritic spines. Hippocampus 10:527–541. [DOI] [PubMed] [Google Scholar]
- ReczekD, Berryman M, Bretscher A (1997) Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the Ezrin-Radixin-Moesin family. J Cell Biol 139:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RyuJ, Liu L, Wong TP, Wu DC, Burette A, Weinberg R, Wang YT, Sheng M (2006) A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron 49:175–182. [DOI] [PubMed] [Google Scholar]
- SaneyoshiT, Wayman G, Fortin D, Davare M, Hoshi N, Nozaki N, Natsume T, Soderling TR (2008) Activity-dependent synaptogenesis: regulation by a CaM-kinase kinase/CaM-kinase I/betaPIX signaling complex. Neuron 57:94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ScannevinRH, Huganir RL (2000) Postsynaptic organization and regulation of excitatory synapses. Nat Rev Neurosci 1:133–141. [DOI] [PubMed] [Google Scholar]
- SchubertV, Dotti CG (2007) Transmitting on actin: synaptic control of dendritic architecture. J Cell Sci 120:205–212. [DOI] [PubMed] [Google Scholar]
- SegalM (2005) Dendritic spines and long-term plasticity. Nat Rev Neurosci 6:277–284. [DOI] [PubMed] [Google Scholar]
- ShenL, Liang F, Walensky LD, Huganir RL (2000) Regulation of AMPA receptor GluR1 subunit surface expression by a 4.1N-linked actin cytoskeletal association. J Neurosci 20:7932–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ShengM, Hoogenraad CC (2007) The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem 76:823–847. [DOI] [PubMed] [Google Scholar]
- ShengM, Sala C (2001) PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci 24:1–29. [DOI] [PubMed] [Google Scholar]
- SoltauM, Berhörster K, Kindler S, Buck F, Richter D, Kreienkamp HJ (2004) Insulin receptor substrate of 53 kDa links postsynaptic shank to PSD-95. J Neurochem 90:659–665. [DOI] [PubMed] [Google Scholar]
- TadaT, Sheng M (2006) Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol 16:95–101. [DOI] [PubMed] [Google Scholar]
- TakahashiH, Sekino Y, Tanaka S, Mizui T, Kishi S, Shirao T (2003) Drebrin-dependent actin clustering in dendritic filopodia governs synaptic targeting of postsynaptic density-95 and dendritic spine morphogenesis. J Neurosci 23:6586–6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry-LorenzoRT, Roadcap DW, Otsuka T, Blanpied TA, Zamorano PL, Garner CC, Shenolikar S, Ehlers MD (2005) Neurabin/protein phosphatase-1 complex regulates dendritic spine morphogenesis and maturation. Mol Biol Cell 16:2349–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WalenskyLD, Blackshaw S, Liao D, Watkins CC, Weier HU, Parra M, Huganir RL, Conboy JG, Mohandas N, Snyder SH (1999) A novel neuron-enriched homolog of the erythrocyte membrane cytoskeletal protein 4.1. J Neurosci 19:6457–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YeK, Hurt KJ, Wu FY, Fang M, Luo HR, Hong JJ, Blackshaw S, Ferris CD, Snyder SH (2000) Pike. A nuclear gtpase that enhances PI3kinase activity and is regulated by protein 4.1N. Cell 103:919–930. [DOI] [PubMed] [Google Scholar]
- YusteR, Bonhoeffer T (2004) Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci 5:24–34. [DOI] [PubMed] [Google Scholar]
- ZhangH, Webb DJ, Asmussen H, Horwitz AF (2003) Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol 161:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZhangH, Webb DJ, Asmussen H, Niu S, Horwitz AF (2005) A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci 25:3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZitoK, Knott G, Shepherd GM, Shenolikar S, Svoboda K (2004) Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton. Neuron 44:321–334. [DOI] [PubMed] [Google Scholar]