Abstract
Disrupted-in-Schizophrenia-1 (DISC1), identified by positional cloning of a balanced translocation (1;11) with the breakpoint in intron 8 of a large Scottish pedigree, is associated with a range of neuropsychiatric disorders including schizophrenia. To model this mutation in mice, we have generated Disc1tr transgenic mice expressing 2 copies of truncated Disc1 encoding the first 8 exons using a bacterial artificial chromosome (BAC). With this partial simulation of the human situation, we have discovered a range of phenotypes including a series of novel features not previously reported. Disc1tr transgenic mice display enlarged lateral ventricles, reduced cerebral cortex, partial agenesis of the corpus callosum, and thinning of layers II/III with reduced neural proliferation at midneurogenesis. Parvalbumin GABAergic neurons are reduced in the hippocampus and medial prefrontal cortex, and displaced in the dorsolateral frontal cortex. In culture, transgenic neurons grow fewer and shorter neurites. Behaviorally, transgenic mice exhibit increased immobility and reduced vocalization in depression-related tests, and impairment in conditioning of latent inhibition. These abnormalities in Disc1tr transgenic mice are consistent with findings in severe schizophrenia.
Keywords: bacterial artificial chromosome, behavior, Disc1, neuroanatomy, schizophrenia, transgenic mice
Introduction
Disrupted-in-Schizophrenia-1 (DISC1) is truncated from intron 8 by a balanced translocation (1;11) in a large Scottish family (Millar et al., 2000), which cosegregates with major mental illness including schizophrenia, depression and bipolar disorders (St Clair et al., 1990; Blackwood et al., 2001). Independent linkage/association studies now implicate DISC1 in schizophrenia, severe affective disorders and autistic spectrum disorders of diverse populations (Devon et al., 2001; Ekelund et al., 2001; Cannon et al., 2005; Hennah et al., 2005, 2008; Sachs et al., 2005; Thomson et al., 2005; Zhang et al., 2005; Chen et al., 2007; Kilpinen et al., 2008). However, robust evidence for functional variants is still lacking, and genetic heterogeneity is likely.
DISC1 is a coiled-coil protein forming complexes with proteins including PDE4, NDEL1, LIS1, and 14-3-3ε, and is involved in nucleus-centrosome association, neuronal proliferation, differentiation, and migration (Ozeki et al., 2003; Brandon et al., 2004; Millar et al., 2005). The C terminus of DISC1 binds NDEL1. DISC1 mutant truncated after exon 8 fails to bind NDEL1, inhibits neurite outgrowth in vitro (Ozeki et al., 2003), and impairs cortical development in vivo (Kamiya et al., 2005). The N terminus of DISC1 binds all PDE4 isoforms (Murdoch et al., 2007), and PDE4B is independently implicated in schizophrenia and mood disorders (Millar et al., 2005). Mice mutated in the Pde4 binding region of Disc1 are defective in prepulse inhibition and latent inhibition (Clapcote et al., 2007).
How DISC1 truncation results in psychiatric illness is unclear. No truncated DISC1 protein is detected in lymphoblasts from the Scottish family (Millar et al., 2005), and no brains from the translocation carriers have become available for examination. It seems likely that a single copy of the normal DISC1 is insufficient for proper brain development and function. However, if the truncated DISC1 protein is produced from cDNA deletion constructs (Ozeki et al., 2003; Hikida et al., 2007; Pletnikov et al., 2008), it may act in a dominant-negative manner (Kamiya et al., 2005). It is also clear that disruption of DISC1 binding partners alters brain development. For instance, deletion or mutation of PAFAH1B1 encoding LIS1 results in lissencephaly in humans (Reiner et al., 1993; Vallee and Tsai, 2006). In mice, Pafah1b1−/− embryos die shortly after implantation, and Pafah1b1+/− mice display cortical and hippocampal disorganization due to delayed neuronal migration (Assadi et al., 2003). Ywhae encodes 14-3-3ε that binds/stabilizes phosphorylated Ndel1, and Ywhae−/− mutants die at birth with defects similar to Pafah1b1+/− mice (Toyo-oka et al., 2003). Deletion of Lis1 binding partners (NdeI and Ndel1) either dramatically reduces cerebral cortex (Feng and Walsh, 2004) or is embryonic lethal with neuronal migration defects (Sasaki et al., 2005).
To explore the role of Disc1 in brain development, we have generated Disc1tr transgenic mice with a ∼148 kb artificial chromosome (BAC) comprising Disc1 exons 1–8. This mimics to a high degree the genetics in the Scottish family. The transgenic mice display a wide variety of schizophrenia-related phenotypes.
Materials and Methods
BAC clone and validation.
The RPCI-23 BAC library was constructed by cloning EcoRI genomic fragments of C57BL/6J mice into the pBACe3.6 vector (http://bacpac.chori.org). The RP23–236F19 clone was kindly supplied by Dr. de Jong (Children's Hospital Oakland Research Institute, Oakland, CA), with end sequences available (AZ705991 and AZ705988). The clone was verified by pulsed-field gel electrophoresis (PFGE) and PCR with primers from MWG-Biotech. The T7 end was defined by a 950 bp product with primers BACT7For (5′-CGCAAGATGTGGCGTGTTACGG-3′) and TsnaxRev (5′-GGCTGCTCACAACCTACACACG-3′), and the Sp6 end by a 1099 bp band with Disc1In9For (5′-AAGGTAGAACCAGGTGGCTTCC-3′) and BACSp6Rev (5′-CGTGATAGCCGTTGTATTCAGC-3′). The BAC insert was further validated by PCR with primers for the Disc1 promoter (245 bp with PromFor 5′-TATCAACTTCAGChCGCATCCGC-3′ and PromRev 5′-TCATAACCTCGCCTCTGG-3′), exon 2 (626 bp with E2For 5′-GACAATCTGAGAGGCTGACTGG-3′ and E2Rev 5′-GTTGCTCAGTAGGTAGTCCTGC-3′), and intron 5 (545 bp with In5For 5′-AGAGTCTTGTGGTTGGATGGCG-3′ and In5Rev 5′-TGAATACAGCACCAGGCTCTGC-3′).
Modification of RP23–236F19.
We performed homologous recombination as described (Yang et al., 1997), for in-frame fusion of EGFP cDNA to the end of Disc1 exon 8. A homologous recombination cassette was constructed in the pSV1 vector to comprise (1) 1026 bp Disc1 intron7-exon8 region with primers In7BamFor (5′-aaaaggatccTTGACTAACTACTGTTGCCAGG-3′) and E8NcoIRev (5′-bcagtccatggATAGGGCCAGCATCTTGG-3′), (2) 745 bp NcoI-EcoRI fragment of EGFP cDNA (gift from Dr. Ian Chambers, University of Edinburgh, Edinburgh, UK), (3) 148 bp BglII-SalI PolyA from pSG5, (4) 1785 bp Disc1 intron 8 (XhoI-EcoRV) with primers In8EcoFor (5′-atgtgaattcTAGTGGTGTCAGGCAGGTGTGG-3′) and In8RVRev (5′-taatgatatcGATGAGGAATACCACAGACGCC-3′), and (5) blunted BamHI fragment of the RecA. The cassette was transformed into the BAC clone. The first round homologous recombination was screened for a 1076 bp PCR product with primers In7HRFor (5′-ACAGGTGATGTGTGTGGA-GTCC-3′), and EGFPRev (5′-ATGCCGTTCTTCTGCTTGTCGG-3′). The second round homologous recombination was confirmed by the appearance of the same 1076 bp PCR product, together with a 2443 bp PCR fragment with primers EGFPFor (5′-TCCTGCTGGAGTTCGTG-ACC-3′) and In8HRRev (5′-GTCACACAGGAATAAGCCACGG-3′).
PFGE.
Both the original and EGFP-modified BAC DNA were digested with ClaI, MluI, NotI, NruI, PvuI, SalI, and XhoI, and run on 1% agarose in 0.5× TBE at 14°C, 150 V, 10-10s for 18 h and 5-5s for 6 h, with Midrange I PFG marker (N3551S, Biolabs) and Kb ladder (N3232L, Biolabs), to verify the predicted restriction patterns. The modified DNA was diagnosed definitively by the appearance of a 4.5 kb XhoI band instead of 4.9 kb in the un-modified BAC DNA. The 148,730 bp NruI fragment, with 1335 bp vector sequence at the T7 site and 219 bp vector sequence at the Sp6 site, was purified from PFGE for pronuclear injection.
Generation of Disc1tr transgenic mice.
All experimental procedures were conducted in accordance with the United Kingdom Animals (Scientific Procedures) Act of 1986 and were approved by the Ethical Review Committee, University of Aberdeen, and the UK Home Office (London). The purified NruI fragment at ∼5 ng/μl was injected into fertilized eggs superovulated from F1 mice of CBA/CaCrl and C57BL/6JCrl (Charles River UK) mice. Transgenic founders were identified by a 319 bp EGFP product with EGFPFor (5′-ACCATCTTCTTCAAGGACGACG-3′) and EGFPRev (5′-TGCTCAGGTAGTGGTTGTCG-3′), and by a 591 bp fragment with primers 5′-ATAATAAGCGGATGAATGGC-3′ and 5′-CTGCTCACAACCTACACACG-3′. The copy number was determined by semiquantitative PCR for 17, 21, 25 and 30 cycles, on the ratio of a 517 bp band from the endogenous Disc1 (In13For, 5′-CTACAACACAGAGCCTTGCTGC-3′ and E14Rev, 5′-AGCAGTAGCAGCGGCATTGG-3′), with a 706 bp fragment from the transgene (E8For, 5′-TTGCTGGAAGCCAAGATGCTGG-3′ and EGFPRTR2, 5′-TCACGAACTCCAGCAGGACC-3′). Experiments were performed on M19 transgenic mice and wild-type (WT) littermates from heterozygote × WT littermate breeding, on the genetic background of 50% CBA/CaCrl and 50% C57BL/6JCrl, unless specified otherwise.
RT-PCR.
The mRNA was extracted from E17.5 embryonic and adult brains using RNAzol B (Biogenesis). Reverse transcription (RT) was performed with 1 μg of total RNA using Omniscript kits (QIAGEN) at 37°C for 1 h. RT-PCR was performed for 30 cycles with primers for the transgene (696 bp, 5′-TGTGACCTGATGGCACTGGTGG-3′ and 5′-GTTGCCGTCCTCCTTGAAGTCG-3′), and for endogenous Disc1 (363 bp, 5′-TTGCTGGAAGCCAAGATGCTGG-3′ and 5′-CTTCACGCCTATGGCTTCGC-3′), and for the housekeeping gene Hprt (352 bp, 5′-CCTGCTGGATTACATTAAAGCACTG-3′ and 5′-GTCAAGGGCATATCCAACAACAAAC-3′). To further define relative expression levels, RT-PCR was performed in triple duplicates at 12, 15, 18, 21, 24, 27, and 30 cycles, in the presence of a common forward primer (E8For: 5′-TTGCTGGAAGCCAAGATGCTGG-3′) derived from exon 8 of Disc1, a reverse primer (E11Rev: 5′-TCCTCGTCCTCTGCGTGTG-3′) from exon 11 for the endogenous Disc1 mRNA, and a reverse primer (EGFPRTR2, 5′-TCACGAACTCCAGCAGGACC-3′) for Disc1tr-EGFP only. The intensity of the PCR products was quantified by ImageJ.
In situ hybridization was performed to compare the spatial and temporal expression of endogenous Disc1 and the transgene
To detect endogenous gene expression, a 394 bp RT-PCR product comprising exons 12–14 of the mouse Disc1 was amplified and cloned into XbaI-XhoI sites of pBluescript SK− vector with primers Disc1E12XbaFor (5′-ctagtctagaTGCGAAGCCATAGGCGTGAAG-3′) and Disc1E14XhoRev (5′-tatccgctcgagCATCCTGTAGACATCTCCTGAG-3′). The plasmid DNA was linearized with XbaI or XhoI, and DIG-labeled antisense or sense probe was transcribed with T3 or T7 RNA polymerase respectively (Roche). The probe for the transgene expression was reversely transcribed from the entire EGFP coding sequence. The hybridization was performed as described (Nishida et al., 2002).
Morphometric and histological analyses.
Adult mice were humanely killed with a lethal dose of sodium pentobarbitone, and brains were dissected, postfixed in cold paraformaldehyde (4%) for 24 h and imaged under a Zeiss stereomicroscope with AxioVision Rel. 4.5. After cryoprotection with 30% sucrose in PBS overnight, brains were sectioned coronally on a microtone at 40 μm and kept in 30% glycerol at 4°C before use. Newborn brains were freshly dissected, snap-frozen in OCT, processed in 12 μm serial coronal sections on a cryostat (CM1850; Leica Microsystems) and mounted on Polysine slides (VWR). One set of sections from each brain was stained with cresyl violet and imaged using an Axiovert 40CFL microscope for anatomical examination. Images were morphometrically quantified with AxioVision Rel. 4.5 software (Zeiss). Data were analyzed by one-way ANOVA and presented as mean ± SEM. p < 0.05 was considered to be statistically significant.
Neuronal culture and neurite outgrowth.
Primary neuronal culture was conducted as described previously (Lang et al., 2006). Briefly, 24-well plates were precoated with poly-l-ornithine (Sigma) for 1 h followed with fibronectin (Invitrogen) for 2 h. The newborns were genotyped by PCR. Their cortices were individually dissected, trypsinized, and mechanically dissociated into single cell suspension. Cells were seeded in the precoated 24-well plates (2 × 105/well), and cultured in Neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen) plus 2 mm glutamine (Sigma) at 37°C with 5% CO2. Twelve hours after plating, tissue debris was removed and the medium was renewed.
Cell images were taken randomly around the center of each well 26 h after culture, using an Axiovert 40CFL microscope with a 20× objective lens. Cells were quantified with AxioVision Rel. 4.5 software for the number of neurites on individual cells and grouped into one, two, three, or more neurites. For the length of neurites, rings with radius at 20, 40, 60, and 80 μm, respectively, were applied to each cell, and cells were categorized accordingly. Data were analyzed by one-way ANOVA and presented as mean ± SEM. *p < 0.05, **p < 0.01.
Birthdating of BrdU labeling.
Time-mated pregnant females were injected with a pulse of BrdU (i.p., 50 mg/kg body weight) at E15.5. Newborn brains were processed as described above. Sections were treated with 0.4% pepsin (Sigma-Aldrich) in PBS for 30 min at 37°C, denatured with 2N HCl for 30 min at 37°C, and neutralized with 0.1 m sodium borate, pH 8.5, for 10 min at room temperature. BrdU incorporation assay (BD Biosciences) was executed according to the manufacturer's instructions.
Immunohistochemistry.
Sections were processed immunohistochemically as described previously (Lang et al., 2006). The primary antibodies included mouse anti-BrdU (1:200; BD Biosciences) and rabbit anti-parvalbumin (1:1500; Swant). For immunofluorescent staining, the secondary antibodies were Texas red-conjugated donkey anti-mouse IgG (1:1000; Invitrogen). For the bright-field staining, the secondary antibody was biotinylated goat anti-rabbit antibody (1:400; Sigma-Aldrich), and sections were developed with Extra-kit (Sigma). Images were taken under an Axiovert 40CFL microscope with a 5× objective lens. BrdU- or parvalbumin-positive cells were quantified with AxioVision Rel. 4.5 software and analyzed by one-way ANOVA.
Latent inhibition.
The latent inhibition procedures were conducted in accordance with the local Animal Care Committee and the EC regulations for animal use in research (86/609/EEC). Eleven WT littermate males and 14 Disc1tr transgenic males at 9–10 months old were housed individually under standard conditions (20−21°C, 60–65% relative humidity), with ad libitum access to water and food. They were divided into 4 experimental groups of preexposed WT (pe-WT, n = 5), non-preexposed WT (npe-WT, n = 6), preexposed transgenic (pe-Tg, n = 7), and non-preexposed transgenic (npe-Tg, n = 7). The tests took place during the light phase of a 12 h light:12 h dark cycle in two square boxes A and B. Both boxes were equipped for tone delivery (10 s at 70 dB, 1500 Hz) and were enclosed in a sound-attenuating cubicle, whereas only Box A was designed to deliver 2 s of 0.2 mA electric shocks through the metallic grid floor. Both were equipped with infrared beams (at 1 cm above floor) to detect horizontal movements.
Before electric shocks, the preexposed mice were acclimatized for 2 min to Box A and received 20 × 10 s tone with 20 s intervals on day 1, and 15 × 10 s tone with 20 s intervals on day 2, whereas non-preexposed ones were placed in Box A for the same durations each day with no tone. Then, all mice were given 5 sets of repeated conditioning (10 s tone + 2 s electric shock + 20 s interval). The session was terminated after a further 40 s interval. On day 3, animals were tested for retention in Box B, with 2 min habituation, followed by 2 min continuous tone and 2 min posttone habituation. The horizontal locomotor activity was monitored by the numbers of infrared beam breaks. Data were analyzed statistically using one-way or two-way ANOVA for either repeated or not repeated measures followed by a post hoc test when required. Differences with a p < 0.05 value were considered as significant.
Modified Porsolt swim test and tail suspension test.
Porsolt swim test (PST) was conducted by placing individual mice (∼3 month) in a 5 L glass beaker with 3 L fresh tap water (22°C) for 6 min. A mouse is regarded as immobile when floating motionless or making only adjustments necessary to keep its head above the water. The modified tail suspension test (TST) was performed for 6 min, by securing the mouse tail to the edge of a shelf ∼20 cm above a large cage with deep bedding. Mice were considered immobile when they hung passively and completely motionless. Both tests were videotaped and examined by two independent observers. The immobility in the first 2 min, the last 4 min, or the whole 6 min trial was assessed. During the TST, mouse vocalizations also were recorded with a bat detector, and analyzed by BatSound Standard-Sound Analysis version 3.31, for the amplitude, frequency and nature of calls. The number of squeaks was counted in each period. Data were analyzed by one-way ANOVA and presented as mean ± SEM. p < 0.05 was considered to be statistically significant.
Results
Truncated Disc1tr-EGFP transgene and expression
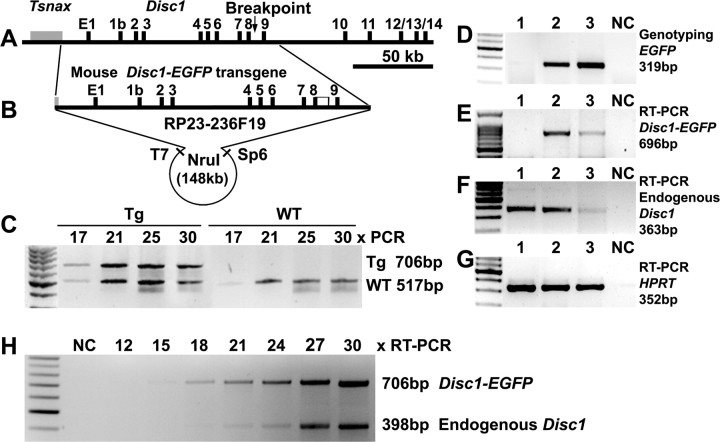
It is known that genomic DNA constructs reproduce gene expression more faithfully than cDNA-based constructs, and that mouse transgenes express more efficiently than human ones in mice. To genetically model the DISC1 truncation (Fig. 1A), we characterized a mouse BAC RP23–236F19 containing Disc1 exons 1–9 with its entire upstream sequences (Fig. 1B). To facilitate the identification of the transgene, we fused an EGFP cDNA to the end of exon 8 followed by a SV40 polyA signal. The modified BAC DNA was microinjected into fertilized mouse eggs, and 3 Disc1tr transgenic founders (M19, M20, and M22) were generated that contained the EGFP fragment. Most studies were performed on the M19 heterozygotes and WT littermate controls unless specified otherwise, as the M22 offspring did not express the transgene mRNA, and the M20 female founder failed to transmit the transgene with reduced litter sizes (6.3 ± 0.7 mice/litter, n = 6) compared with other breeding pairs (8.7 ± 0.5 mice/litter, n = 29 litters, p < 0.01). The M19 transgenic heterozygotes contained two copies of the truncated Disc1 on the background of two copies of full-length Disc1 (Fig. 1C), closely mimicked the genetic ratio (1:1) in the Scottish family.
Figure 1.
The truncated mouse Disc1 transgene and expression. A, Genomic organization of the mouse Disc1 locus, with an arrow corresponding to the breakpoint in the Scottish family. B, A BAC clone RP23–236F19 containing ∼148 kb mouse Disc1 genomic DNA, starting from the 3′UTR of Tsnax, and ending with 16.7 kb of Disc1 intron 9. The BAC was fused to an EGFP at the end of exon 8 followed by a PolyA. The NruI fragment (148,730 bp) was purified for microinjection. C, M19 heterozygous transgenics contained 2 copies of the transgene, as similar intensity of PCR products were obtained from the truncated and endogenous Disc1 genes at 17–30 cycles. D, Two E17.5 embryos (lanes 1–2) and a transgenic mother (lane 3) were genotyped by PCR with EGFP primers. E–G, RT-PCR with primers for the transgene (E), the endogenous Disc1 (F), and a housekeeping HPRT (G), showing comparable levels of the truncated (E) and full-length (F) Disc1 mRNA. H, Relative expression of endogenous Disc1 and Disc1tr-EGFP demonstrated by RT-PCR at 12, 15, 18, 21, 24, 27, and 30 cycles. The average ratio of the RT-PCR products 706 bp:398 bp at 18–30 cycles was 1.09 ± 0.19, showing comparable abundance of endogenous and truncated Disc1 mRNA (Fig. 1H). NC, Negative control.
The transgene expression was assessed by RT-PCR and in situ hybridization. RT-PCR suggested that M19 transgenic mice produced comparable levels of endogenous Disc1 and Disc1tr-EGFP transcripts in E17.5 (Fig. 1E–G, lane 2) or adult (Fig. 1E–G, lane 3) brains. To further determine relative expression levels, RT-PCR was performed in triple duplicates at 12, 15, 18, 21, 24, 27, and 30 cycles, in the presence of three primers: a common forward primer (E8For) with an endogenous Disc1-specific reverse primer (E11Rev), and an EGFP-specific reverse primer (EGFPRTR2), which amplified a 706 bp band from Disc1tr-EGFP transcripts and a 398 bp fragment from endogenous Disc1. The molecular ratio of the RT-PCR products 706 bp:398 bp at 18–30 cycles was 1.09 ± 0.19, showing a similar abundance of Disc1tr-EGFP and endogenous Disc1 mRNA (Fig. 1H).
To characterize spatial and temporal expression patterns of the transgene, we performed in situ hybridization on E17.5 and 2-month-old brain sections with antisense Disc1 and EGFP probes. The DIG-labeled Disc1 probe was reversely transcribed from exons 12–14 of the Disc1 which was not present in the transgene, whereas the EGFP probe was derived from the entire EGFP coding sequence. In E17.5 developing brain, both the Disc1 and EGFP hybridization signals were localized predominantly in the cerebral cortex and hippocampus (Fig. 2A–F). In 2-month-old brain, the full-length and truncated Disc1 mRNA were found in the cerebellum (data not shown), hippocampus, and cerebral cortex, including cingulate and piriform cortex (Fig. 2G–L). In the hippocampus, both the Disc1 and EGFP probes detected expression in the pyramidal layer of CA1–CA3, and granule layer of the dentate gyrus (Fig. 2M–P). These data suggest that the 148 kb BAC expressed Disc1tr-EGFP largely in the same anatomical sites as endogenous Disc1.
Figure 2.
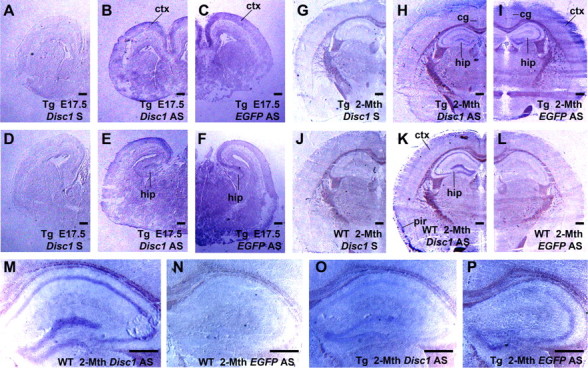
Comparable temporal and spatial patterns of expression of truncated and full-length Disc1. In situ hybridization was performed using DIG-labeled sense (S) and antisense (AS) RNA probes on E17.5 (A–F) and 2-month-old brain sections (G–P). Whereas the Disc1 AS probe hybridized to the endogenous full-length Disc1 transcripts, EGFP AS probe detected the Disc1tr-EGFP messages only. Both the endogenous and transgenic transcripts (purple) were detected at the regions of cerebral cortex (ctx), including cingulate (cg) and piriform (pir) cortex, CA1, CA2, CA3, and dentate gyrus of the hippocampus (hip). Scale bars, 500 μm.
Dilated lateral ventricles and reduced cerebral cortex in Disc1tr transgenic mice
Schizophrenic symptoms usually begin in late adolescence or early adulthood, and neuroanatomic changes in lateral ventricles and cerebral cortex are seen in schizophrenic patients. We analyzed neuroanatomy in sexually mature, 2-month-old transgenic mice. Mouse brains were processed histologically from 11 Disc1tr transgenics (6 male and 5 female) and 15 WT littermates (8 male and 7 female). Sections were imaged at the plane where the anterior commissure crossed the midline (Fig. 3A,B). Sizes of the cerebral cortex and corpus callosum were quantified. The transgenic lateral ventricles were found to be dilated by ∼44% (p < 0.05). In addition, we have detected a subtle (∼4%) but significant (p < 0.05) reduction in the thickness of the dorsolateral frontal cortex (Fig. 3C,D) of transgenic mice. Remarkably, this reduction largely resulted from the thinning of cortical layers II/III, which was reduced by ∼17% (Fig. 3C,D). Severe but consistent neuropathologies were observed in the untransmittable transgenic founder M20 (see supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Compared with a littermate control, the lateral ventricles were enlarged 2.3- to 3.0-fold (see supplemental Fig. 1A,B,D,E, available at www.jneurosci.org as supplemental material). The frontal cortex was reduced by 16% in thickness. The layers II/III and V, where pyramidal neurons resided, were particularly affected (see supplemental Fig. 1G,H, available at www.jneurosci.org as supplemental material). These phenotypes are consistent with mild cytoarchitectural abnormalities reported in schizophrenia (Harrison, 1999; Lewis and Levitt, 2002).
Figure 3.
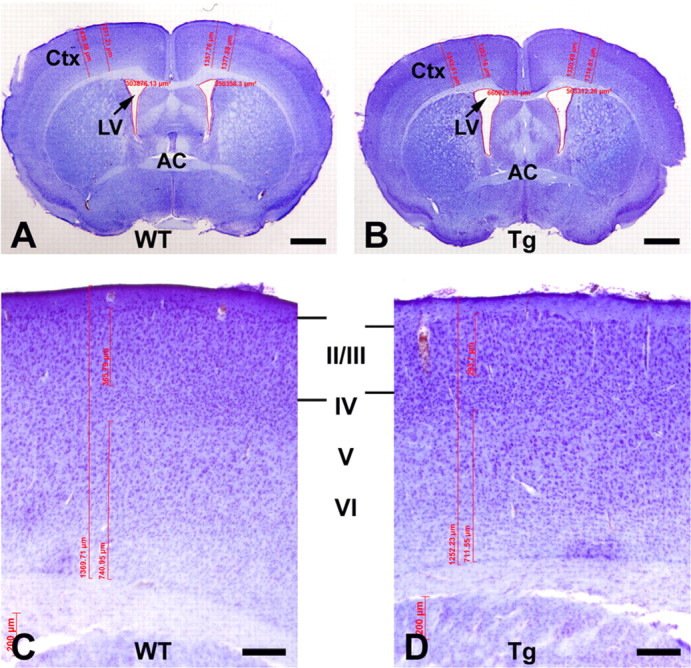
Enlarged lateral ventricles (LV) and reduced cerebral cortex (Ctx) in Disc1tr transgenic (Tg) mice. WT (A) and Tg (B) brain sections were Nissl stained, imaged at the level where the anterior commissure (AC) crossed the midline, and quantified with AxioVision Rel. 4.5. The LV was significantly (p < 0.05) enlarged in Tg brains (0.819 ± 0.079 mm2, n = 11) compared with WT littermates (0.568 ± 0.043 mm2, n = 15). C, D, Magnified view of the cerebral cortex from WT and Tg representatives showing changes in layers II–III. Statistical analyses detected moderate but significant (p < 0.05) reduction in Tg cerebral cortex (1353.6 ± 19.9 μm, n = 11) compared with WT littermates (1409.2 ± 10.0 μm, n = 15). Cortical layers II/III was thinned (p < 0.01) from 347.2 ± 4.5 μm in WT to 302.7 ± 3.4 μm in Tg mice. Scale bars: A, B, 1 mm; C, D, 200 μm.
Reduced neuronal proliferation in the developing transgenic brain
In the mouse, cortical neurogenesis starts from E10.5 and is largely completed by E17.5. Cells in the ventricular zone of the dorsolateral telencephalon undergo a maximum of 11 cell divisions, and neurons at different layers are generated in a cell cycle number-dependent manner (Estivill-Torrus et al., 2002). To explore the cellular mechanisms causing thinned layers II/III in Disc1tr transgenic mice, we performed birth-dating experiments during midneurogenesis.
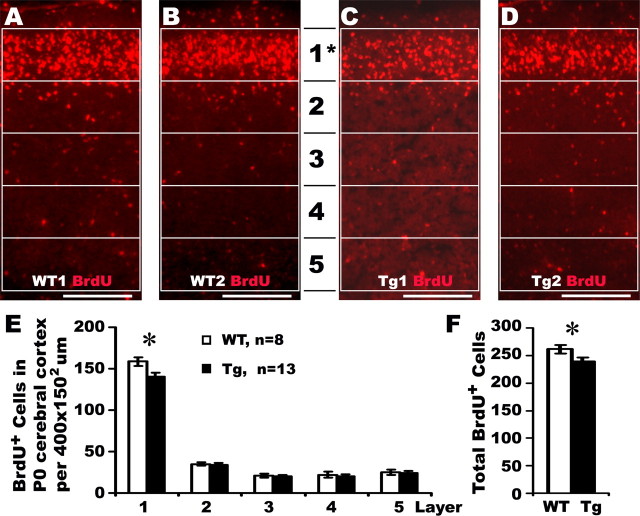
A pulse of BrdU was injected into pregnant females at E15.5, when layer II/III neurons were formed. Four independent litters of newborn brains were processed with anti-BrdU antibody. BrdU-positive cells were quantified from equally divided areas (400 μm wide ×150 μm high) of newborn cortex of 13 transgenic heterozygotes and 8 WT littermates at the lateral ventricle level (Fig. 4A–D). Transgenic newborns showed a modest but significant reduction of BrdU-labeled cells in the outermost layer (arbitrary layer 1) of the cortex, corresponding to layers II–III in adult brain, whereas BrdU-positive cells in other layers (2–5) were not significantly different. The total number of BrdU-positive cells was also significantly reduced in transgenic newborns (Fig. 4E) (239.2 ± 6.8, n = 13, p < 0.05) compared with WT littermates (261.5 ± 7.50, n = 8). These data indicate that truncated Disc1 causes reduced neuronal proliferation at midneurogenesis, which contributes to subtle alterations in the cytoarchitecture of the cerebral cortex in the Disc1tr transgenic mice.
Figure 4.
Reduced neurogenesis in Disc1tr transgenic (Tg) embryos. A–D, A pulse of BrdU was injected into four E15.5 pregnant females and newborn brains were processed with an anti-BrdU antibody. For each brain, four images were taken from the cerebral cortex at the left and right sides of two consecutive sections with the largest lateral ventricles. Images were arbitrarily divided into 5 layers as shown, and BrdU-positive cells were quantified from each area (400 μm wide × 150 μm high). A and B were from two WT littermates, C and D from two Tg newborns. E, Statistical analyses revealed significant reduction of BrdU-positive cells in the arbitrarily assigned layer 1 of Tg mice (140.4 ± 4.9, n = 13), compared with their WT littermates (158.6 ± 5.3, n = 8). F, The total number of BrdU-incorporated cells was also significantly reduced in Tg mice compared with that in WT littermates. Scale bars, 200 μm. *p < 0.05.
Partial agenesis of corpus callosum in Disc1tr transgenic adults
The corpus callosum consists of nerve fibers projecting from cortical neurons to communicate between the two hemispheres. To evaluate the consequence of lamination changes in Disc1tr transgenic cortex, we have compared the thickness of the corpus callosum between 11 transgenic (6 male and 5 female) and 15 WT littermates (8 male and 7 female). At the rostral brain where the anterior commissure crosses the midline (Fig. 5A,B), transgenic corpus callosum (184.2 ± 6.1 μm, n = 11) was significantly thinner (∼20%) than that in WT littermates (228.3 ± 5.3 μm, n = 15, p < 0.01). In the caudal brain at the level of the subcommissural organ (SCO), all 15 WT littermates displayed a thick layer of the corpus callosum crossing the midline (Fig. 5C). However, in the majority (9/11) of transgenic brains, the corpus callosum failed to cross the midline above the SCO (Fig. 5D). These data demonstrate clearly a partial agenesis of the corpus callosum both rostrally and caudally in Disc1tr transgenic brains.
Figure 5.

Partial agenesis of the corpus callosum (CC) in 2-month-old transgenic brains. Coronal sections of WT (A, C) and Tg (B, D) brains were Nissl-stained. A, B, Images represented average thickness of rostral CC in WT (A) and Tg (B) brains where the AC crossed the midline, showing significant reduction of the CC in 11 Tg brains compared with 15 WT. At the SCO level (arrowed in C and D), a thick layer of the CC crossed the midline in all 15 WT mice (C), whereas Tg CC failed to cross the midline in 9 of 11 cases (D). Scale bars: A, B, 200 μm; C, D, 1 mm.
Effect of truncated Disc1 on neurite outgrowth in vitro
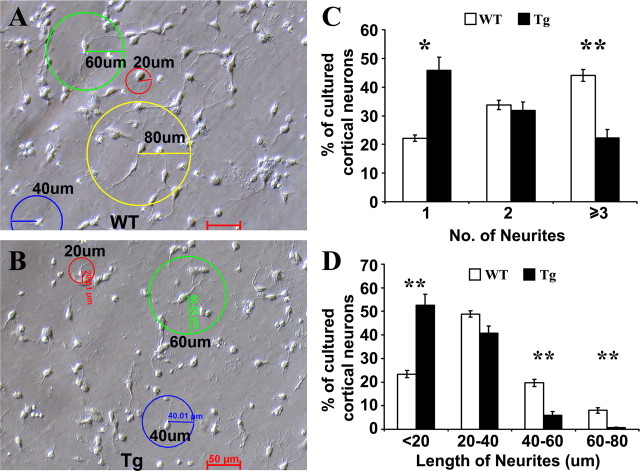
Disc1 and its complex members modulate neurite outgrowth (Ozeki et al., 2003; Pletnikov et al., 2008). To further examine the origin of cortical and callosal abnormalities of Disc1tr transgenic mice, we cultured primary cortical neurons from two litters of newborn mice. We examined 845 neurons from 4 transgenic and 1817 neurons from 8 WT littermates after 26 h of culture (Fig. 6). In WT littermate cultures, 44.1% of neurons were found to have 3 or more neurites, and this was reduced to 22.3% in transgenic cultures (Fig. 6C). Meanwhile, the proportion of cells with a single neurite increased from 22.2% to 45.8%. We also measured the length of neurites in 845 transgenic and 1817 WT littermate neurons, and categorized them into 4 groups according to their length (Fig. 6D). Neurons with >40 μm neurites were reduced from 20.5% in WT littermates to 6.9% in transgenic cultures. On the other hand, cells with <20 μm of neurites have risen by 23.6% in transgenic mice. These data suggest that reduced neurite outgrowth may contribute to the neuropathology observed in the Disc1tr transgenic mice.
Figure 6.
Fewer and shorter neurites in cultured transgenic (Tg) neurons. WT (A) and Tg (B) newborn cortex were dissociated and neurons were cultured for 26 h in vitro. Images (6–8 fields/mouse) were randomly taken under a 20× objective lens. Neurons (1817 WT and 845 Tg) were quantified for the number (C) and length of neurites with 20 (red), 40 (blue) 60 (green), and 80 μm (yellow) rings (D), respectively. The data were presented as mean ± SEM. *p < 0.05, **p < 0.01. Scale bars, 50 μm.
Parvalbumin cells in Disc1tr transgenic prefrontal cortex
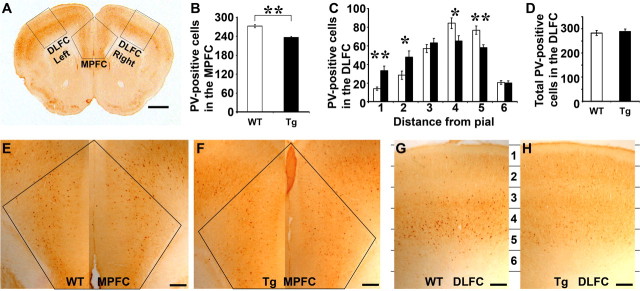
Appropriate neuronal synchronization is crucial for brain function, and a subgroup of GABA inhibitory interneurons, parvalbumin-expressing cells, is reduced in schizophrenic cortex (Lewis et al., 2005). Reduced parvalbumin immunoreactivity also was reported in transgenic mice expressing a dominant-negative DISC1 cDNA under the control of a CaMKII promoter (Hikida et al., 2007). We investigated parvalbumin expression in 2-month-old mice (Fig. 7). Consistent with the DN-DISC1 mice, we found a significant reduction (13%, p < 0.01) in the number of parvalbumin cells in the medial prefrontal cortex (MPFC) (Fig. 7B,E,F) of Disc1tr transgenic mice (236.4 ± 2.9, n = 11) compared with WT littermates (272.2 ± 4.7, n = 14).
Figure 7.
Parvalbumin (PV) interneurons in the prefrontal cortex of WT and Tg brains. A, A brain section stained with anti-PV, showing areas of MPFC and DLFC where magnified images (E–H) were taken for counting PV cells. B, Statistical analyses of PV cells in the MPFC of WT and Tg mice as illustrated in E (WT) and F (Tg). C, PV-positive cells in 6 arbitrarily assigned layers (1386 μm wide × 267 μm high) of the DLFC as illustrated in G and H. D, Total number of PV cells in the DLFC showing no difference between WT and Tg mice. Scale bars: A, 1 mm; E–H, 200 μm.
Additionally, we noticed that there were differences in the patterns of parvalbumin staining at the dorsolateral frontal cortex (DLFC) (Fig. 7G,H). Parvalbumin-positive cells were then quantified in arbitrarily divided 6 layers (∼267 μm height each). In WT littermates, most parvalbumin cells were clustered in the inner layers. However, transgenic mice had significantly reduced parvalbumin cells in inner layers (4–5), with a significant increase of parvalbumin cells in outer layers (Fig. 7C). The truncated Disc1 did not seem to affect specification, but lamination, of parvalbumin cells, as the total number of parvalbumin cells in the DLFC was not significantly altered between transgenic (288.3 ± 9.9, n = 11, 6 male and 5 female) and WT (281.5 ± 9.6, n = 14, 8 male and 6 female) mice (Fig. 7D).
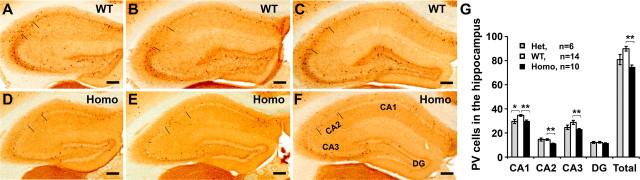
Reduced parvalbumin neurons in Disc1tr hippocampus
Neuropathological studies have demonstrated deficits of GABAergic interneurons in the hippocampus of postmortem schizophrenic patients. Glutamate decarboxylase 67 (GAD67) encoding an enzyme synthesizing GABA is strikingly downregulated in the hippocampus of schizophrenia and bipolar patients (Benes et al., 2007). Independently, a profound deficit in the relative density of parvalbumin-immunoreactive neurons was found in all subfields of schizophrenic hippocampus (Zhang and Reynolds, 2002). To examine the GABAergic signaling in Disc1tr transgenic mice, we have compared parvalbumin neurons in the hippocampus of 14 WT littermates and 6 heterozygous and 10 homozygous transgenic mice. A significant reduction was observed in the CA1 region of the heterozygous mice (p < 0.05), with trends of reductions also in the CA3 and total numbers of parvalbumin neurons (Fig. 8G). The reduction of parvalbumin cells was significant statistically in all subfields of the homozygous hippocampus with the exception of the dentate gyrus (Fig. 8). Similarly, the untransmittable transgenic founder M20 displayed dramatic reductions of parvalbumin neurons in the CA1, CA2, and CA3 regions compared with a control mouse (see supplemental Fig. 2, available at www.jneurosci.org as supplemental material).
Figure 8.
PV interneurons are reduced in the hippocampus of Disc1tr transgenic mice. A–F, Brain sections from 14 WT (A–C), 6 heterozygous (data not shown), and 10 homozygous (D–F) transgenic mice were stained with anti-PV. PV-positive cells at the CA1, CA2, CA3, and dentate gyrus (DG) were quantified from 6 comparable images of each mouse brain as shown. G, Statistical analyses of the mean PV cells in each area of the hippocampus, showing significant reduction of the PV interneurons in the CA1 of the heterozygotes, and in the CA1–CA2–CA3 of the homozygotes. Scale bars: 200 μm. *p < 0.05, **p < 0.01.
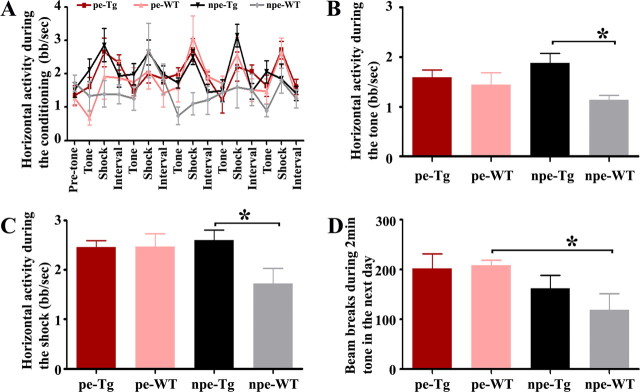
Defects in conditioning of latent inhibition
Schizophrenic patients often have defects in prepulse inhibition (Braff et al., 2001) and latent inhibition (Rascle et al., 2001). Although ENU Disc1 mutants have profound deficits in latent inhibition and prepulse inhibition (Clapcote et al., 2007), transgenic mice with ectopic promoters do not show robust changes in prepulse inhibition (Hikida et al., 2007; Pletnikov et al., 2008).
We evaluated Disc1tr transgenic mice using a latent inhibition test of fear learning (Fig. 9). When animals were habituated to the test boxes with or without tone delivery before the test, there was no difference on horizontal locomotor activity (data not shown). During the conditioning phase in the second day, factorial ANOVA on the horizontal activity revealed a significant genotype/exposure interaction (Fig. 9A). A reliable difference was detected between the non-preexposed WT littermate and transgenic groups during the tone periods, suggesting that only the WT littermates acquired predictability of the tone (Fig. 9B). Similarly, the non-preexposed WT littermates had significantly reduced activity during the shock (p < 0.05), whereas the non-preexposed transgenic mice remained highly active (Fig. 9C).
Figure 9.
Disc1tr transgenic mice are defective in conditioning of latent inhibition. A, The horizontal activity (mean ± SEM) in numbers of beam breaks per second (bb/s) during the different phases of conditioning. B, Pooled activity (bb/s) during the 5 × 10 s tone period. C, Pooled activity (bb/s) during the 5 × 2 s shock period. D, Total number of beam breaks (mean ± SEM) during the 120 s retention test on the following day. Note that only the npe-WT group showed considerable “freezing” during the tone (B), shock (C), or retention (D) period. *p < 0.05. npe, Non-preexposed; pe, preexposed to tone.
On the following day, animals were transferred to a different box and given 2 min tone to examine the retention (Fig. 9D). Again, only the non-preexposed WT littermates significantly reduced their activity during the 2 min tone, whereas independent of the preexposure, transgenic mice were not significantly different from each other. These data demonstrate that latent inhibition is established in WT littermates, but no conditioning is attained by the Disc1tr transgenic mice.
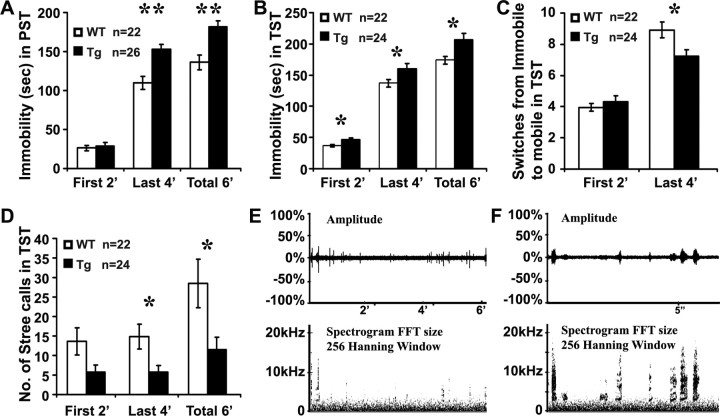
Increased immobility in depression tests
Schizophrenia is often associated with depressive disorders. In the Scottish schizophrenic family, ∼35% of the carriers develop schizoaffective, bipolar or major depressive disorders (Blackwood et al., 2001). TST and PST are common behavioral tests for depression-related behavior in animals, and a longer immobility in either of the tests is viewed as increased depressiveness. We first videotaped 26 Disc1tr transgenic mice (13 male and 13 female) and 22 WT littermates (12 male and 10 female) in a 6 min PST. Significantly increased immobility was seen in the last 4 min and the total 6 min of the PST in the transgenic group (Fig. 10A).
Figure 10.
Increased immobility (A–C) and reduced vocalization (D–F) of Disc1tr transgenic mice in depression tests. Disc1tr transgenic (Tg) mice and WT littermates were tested individually in 6 min PST (A) or TST (B–F). Transgenic mice showed increased immobility in PST (A) and TST (B), with a reduced number of switches from immobile to mobile status in the last 4 min of the TST (C). D, Disc1tr transgenic mice made significantly fewer stress calls. E, An example of vocalizations (squeaks) recorded by a bat detector showing amplitude and frequency (kHz) of calls during the 6 min TST. F, Magnified view of the calls. *p < 0.05, **p < 0.01.
This was further verified by the TST on an independent cohort of 24 transgenic (13 male and 11 female) and 22 WT littermates (12 male and 10 female) (Fig. 10B). Transgenic mice again were significantly less active throughout the 6 min TST. In addition, a reduced number of switches from immobile to mobile phases were observed in the transgenic group in the last 4 min of the TST (Fig. 10C). Disc1tr transgenic mice did not exhibit reduced locomotor activity in the latent inhibition test (Fig. 9) or open field test (data not shown). The increased immobility in PST and TST is unlikely due to a general hypoactivity of the Disc1tr transgenic mice.
Reduced stress calls
Schizophrenia is associated with social and communication deficits. Mice can produce a variety of social vocalizations, such as mating calls at ultrasonic frequencies beyond human hearing (30–110 kHz) (Holy and Guo, 2005), and postpartum/distress calls (0–30 kHz) audible to humans (Whitney, 1970; Whitney and Nyby, 1983). Under stressful conditions such as TST, mice squeak.
During the TST, variation in individual mouse vocalizations was evident, and the number of squeaks was counted (Fig. 10D). WT littermates squeaked frequently (28.5 ± 6.2, n = 22) during the 6 min TST, and WT males (39.7 ± 9.6, n = 12) called more than twice as often as WT females (15.0 ± 5.1, n = 10, p < 0.05). Disc1tr transgenic females tended to make fewer calls (10.2 ± 4.0, n = 11) compared with WT females. However, transgenic males squeaked 3 times less frequently (12.8 ± 4.7, n = 13, p < 0.05) than WT littermate males. The reduced calls were particularly prominent in the last 4 min of the test; WT males made on average 22.3 ± 4.4 calls and transgenic males only 5.2 ± 2.1 calls (p < 0.01).
To examine frequency of the calls during the TST, we recorded vocalizations with a bat detector (Fig. 10E). No ultrasound vocalizations were detected during the tail suspension, and all calls were within the audible range (0–20 kHz) (Fig. 10E,F), similar to postpartum/distress calls (Whitney, 1970; Whitney and Nyby, 1983). These data indicate strongly that Disc1tr transgenic mice have substantially reduced vocal communication under stress conditions.
Discussion
We have generated Disc1tr transgenic mice expressing 2 copies of a Disc1tr-EGFP fusion gene in an ∼148 kb native mouse Disc1 genomic environment, which drive transgene expression at the endogenous Disc1 expression sites in the cerebellum, cerebral cortex and hippocampus. Disc1tr transgenic mice display an array of schizophrenia-related abnormalities (Table 1). Similar to phenotypes described in most other mouse models, we observe enlarged lateral ventricles and reduced cerebral cortex. Consistent with the findings in ENU mutants (Clapcote et al., 2007), Disc1tr transgenic mice show deficits in conditioning of latent inhibition, and longer immobility in depression-related tests. They also resemble aspects of DN-DISC1 mice, and have reduced parvalbumin neurons in the medial prefrontal cortex (Hikida et al., 2007) and reduced neurite outgrowth in culture (Pletnikov et al., 2008). What is more, Disc1tr transgenic mice exhibit a remarkable series of novel phenotypes not previously reported. The novel features include the thinning of the cortical layers II/III, selective decrease of neural proliferation in the developing cortex, partial agenesis of the corpus callosum, reduced parvalbumin neurons in the hippocampus and displaced parvalbumin cells at the frontal cortex, as well as reduced vocalizations under stress conditions.
Table 1.
Phenotypes of DISC1/Disc1 mouse models
| Phenotype |
Koike et al. (2006) |
Clapcote et al. (2007) |
Hikida et al. (2007) |
Pletnikov et al. (2008) |
This study |
|
|---|---|---|---|---|---|---|
| 129 mice | 31L | 100P | DN-DISC1 | Inducible DISC1 | Disc1tr | |
| Brain volume | NS | ↓6% | ↓13% | NS | NS | ↓ in male |
| Lateral ventricles | ? | ? | ? | ↑ | ↑ | ↑ |
| Cerebral cortex | NS | ↓ | ↓ | NS | NS | ↓ |
| Neural proliferation | ? | ? | ? | ? | ? | ↓ |
| Neurite outgrowth | ? | ? | ? | ? | ↓ | ↓ |
| Corpus callosum | ? | ? | ? | ? | ? | ↓ |
| Parvalbumin in MPF cortex | ? | ? | ? | ↓ | ? | ↓ |
| Parvalbumin in hippocampus | ? | ? | ? | ? | ? | ↓ |
| Open field: horizontal activity | NS | NS | ↑↑ | ↑ | ↑ in male | NS |
| Open field: vertical activity | NS | NS | ↑ | ↑ | ? | ? |
| Anxiety | NS | NS | NS | NS | NS | ? |
| Aggression | ? | ? | ? | ? | ↑ in male | ? |
| Sociability | ? | ↓ | NS | NS | ↓ in male | ? |
| Spatial learning and memory | ? | NS | NS | NS | ↓ in female | ? |
| Working memory | ↓ | ↓ | ↓↓ | ? | ? | ? |
| Prepulse inhibition (PPI) | NS | ↓ | ↓↓ | ↓ | NS | ? |
| Latent inhibition (LI) | ? | ↓ | ↓↓ | ? | ? | ↓ |
| Immobility in forced swim test | ? | ↑ | NS | ↑ | ? | ↑ |
| Immobility in tail suspension test | ? | ? | ? | ? | ? | ↑ |
| Stress calls | ? | ? | ? | ? | ? | ↓ in male |
| Rolipram in PPI | ? | NS | +++ | ? | ? | ? |
| Bupropion in PPI | ? | +++ | NS | ? | ? | ? |
| Clozapine in LI | ? | NS + | ? | ? | ? | |
| Clozapine on horizontal activity | ? | +++ | ++ | ? | ? | ? |
| Bupropion in FST | ? | ++ | ? | ? | ? | ? |
?, Not known; NS, no statistical difference; ↓, reduced; ↑, increased. +, ++, +++, positive effect with p < 0.05, 0.01, and 0.001, respectively.
Some of the symptoms of schizophrenia such as hallucinations and delusions cannot be assessed easily in animals. For other abnormalities such as cognitive dysfunction and affective symptoms, some proxy measures have been developed to test animals. For example, prepulse inhibition and latent inhibition are commonly used to detect attention-related deficits, whereas PST and TST can indicate a depression-related phenotype. Recently reported DISC1 transgenics or mutants exhibit some abnormalities consistent with a schizophrenia phenotype (Table 1). For example, strain 31L has a predominant mood disorder-like phenotype with reduced Pde4b activity, whereas the 100P strain shows profound deficits in prepulse inhibition and latent inhibition (Clapcote et al., 2007). However, neither 129 mice with spontaneous Disc1 truncation (Koike et al., 2006, Ishizuka et al., 2007) nor DN-DISC1 mice using an ectopic (Hikida et al., 2007) or inducible promoter (Pletnikov et al., 2008) display significant changes in prepulse inhibition. We have demonstrated that Disc1tr transgenic mice are defective in both of these symptom-related tests. In latent inhibition, the non-preexposed Disc1tr transgenic mice fail to “freeze” during the tone, shock, or postshock tone periods. In PST and TST, they have longer immobility, with reduced switches from immobile to mobile status. Remarkably, they make fewer stress calls during the TST. The significance of this last observation is unknown. It is interesting to speculate that it may be a novel indicator of the presence of communication deficits and/or other negative symptoms that resemble those found in schizophrenia.
The changes we have seen in Disc1tr transgenic brains are consistent with neuropathology in schizophrenia. Examinations on first-episode and unmedicated schizophrenic patients consistently show selective regional deficits in brain volume and ventricular enlargement (Harrison, 1999; Honea et al., 2005; Ross et al., 2006); the latter also appears to be a common feature of Disc1 mutants and transgenic mice reported so far. Morphometric analyses of Disc1tr transgenic brains reveal a significant dilation of the lateral ventricles. Although the scale of reduction is not as dramatic as in ENU mutants (Clapcote et al., 2007), quantitative analyses of brain surface areas show a significant reduction in Disc1tr transgenic mice (see Fig. 3 and supplemental Fig. 1, available at www.jneurosci.org as supplemental material). In contrast to DN-DISC1 mice, we do not observe compensational changes (Hikida et al., 2007). Instead, the neuropathology appears to be dosage-related, and most dramatic phenotypes are observed in the M20 female founder (supplemental Figs. 2, 3, available at www.jneurosci.org as supplemental material) and in some of the M19 homozygotes (Fig. 8 and supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
One of the most prominent cellular features of schizophrenia that has emerged from postmortem studies is a consistent reduction of parvalbumin neurons (Lewis et al., 2005). Parvalbumin cells belong to a subgroup of GABAergic inhibitory interneurons, which are vital for neuronal synchronization. Along with the GABA-synthesizing enzyme GAD67, parvalbumin expression is consistently reduced in schizophrenic brains. We detected significant reduction of parvalbumin neurons in the medial prefrontal cortex, similar to the findings reported in DN-DISC1 transgenic mice (Hikida et al., 2007). Additionally, parvalbumin neurons also were reduced in the hippocampus of Disc1tr transgenic mice. These appear to be anatomically specific, as the total number of parvalbumin cells at the reticular nucleus of the thalamus or the dorsolateral frontal cortex remained unchanged. However, in the frontal cortex, the distribution of parvalbumin-positive cells is altered. In contrast to a more concentrated localization in the inner half of WT littermates, they are more evenly spread throughout the layers in the transgenic brains. The causes and functional consequences of these abnormalities are yet to be determined. However, dorsolateral frontal cortex-dependent cognitive functions are compromised in schizophrenia (Miller and Cohen, 2001).
Disc1 is critical for cortical neurogenesis, and the reduction of Disc1tr brain volume is attributed mainly to the differences in the cerebral cortex. This is paralleled by a reduction in the thickness of cortex in both transgenic males and females. Surprisingly, no significant changes have been detected in the cortex of DN-DISC1 mice (Hikida et al., 2007; Pletnikov et al., 2008), and it is not clear whether this is related to the ectopic promoters used. The reduced cerebral cortex we observe largely results from the thinning of layers II/III. The differences are statistically significant on morphometric analyses of WT and Disc1tr transgenic brains. These are precisely the layers altered in schizophrenia (Harrison, 1999).
These cortical layers contain pyramidal neurons, which are involved in interhemispheric communication. Indeed a partial agenesis of the corpus callosum is seen in Disc1tr transgenic mice. In the rostral brain, the corpus callosum is thinned, and in the caudal brain, it stops crossing the midline before the appearance of the SCO. It is worth noting that some mouse strains (I/LnJ, 129/J, and BALB/c) have agenesis of the corpus callosum (Livy and Wahlsten, 1991). However, the background strains (C57BL/6J and CBA/Ca) of the Disc1tr transgenic mice reported in this study are known to have an intact corpus callosum. Consistent with our findings, a complete agenesis of the corpus callosum is rare in schizophrenia (Motomura et al., 2002; Chinnasamy et al., 2006; Paul et al., 2007). However, mild alterations in interhemispheric callosal connections may be relatively common in schizophrenia and in autism (Innocenti et al., 2003; Miyata et al., 2007). Partial agenesis of the corpus callosum may result from decreased numbers of cortical neurons and/or reduced neurite outgrowth. This is supported by our observation that Disc1tr transgenic neurons have fewer and shorter neurites in primary culture, which also echoes earlier studies that truncated DISC1 inhibits neurite outgrowth (Ozeki et al., 2003; Pletnikov et al., 2008).
We have presented evidence that truncated Disc1 selectively reduces proliferation at the outermost cortex during midneurogenesis. This corresponds to the peak of Disc1 expression in embryos (Schurov et al., 2004) and the reduced layers II/III we observe in transgenic adults. A more dramatic reduction of cortical neurogenesis is reported in NdeI null mutants, with reduced proliferation and retarded migration (Feng and Walsh, 2004). Interestingly, the newly identified Disc1 binding partner DBZ (or Su48 or Zfp365) is also a coiled-coil protein (Hattori et al., 2007). It associates with centrosomes and is involved in proliferation (Wang et al., 2006). Ectopic expression of Su48 causes abnormal mitosis, whereas injection of an anti-Su48 antibody leads to mitotic failure. Su48 associates with NdeI (Hirohashi et al., 2006), and the latter is shown to be vital for cortical development (Feng and Walsh, 2004). Furthermore, NdeI interacts with Lis1, and Lis1 regulates mitosis in cultured mammalian cells (Faulkner et al., 2000). Our data add to accumulating evidence that Disc1 complexes play critical roles in the cortical genesis. Alterations in the Disc1 gene lead to failure of normal neuronal proliferation, reduced neurite outgrowth, and decreased/displaced parvalbumin neurons. Consequently, these lead to thinned layers II/III, reduced cortical size, enlarged ventricles and behavioral changes, all of which are consistent with schizophrenia-like phenotypes.
Footnotes
This work was supported by an award (Ref. NS_AU_044) from the Translational Medicine Research Collaboration—a consortium made up of the Universities of Aberdeen, Dundee, Edinburgh, and Glasgow, the four associated National Health Service (NHS) Health Boards (Grampian, Tayside, Lothian, and Greater Glasgow & Clyde), Scottish Enterprise, and Wyeth Pharmaceuticals. We also acknowledge University of Aberdeen for funding the early phase of the project, BDF Newlife for the cryostat, and NHS Grampian for the imaging facility. We thank Professor Pieter de Jong (Children's Hospital Oakland Research Institute, Oakland, CA) for the RP23-236F19 clone and Dr. Ian Chambers (University of Edinburgh, UK) for the EGFP construct. We thank Professors Steve Logan and Michael Greaves for support, and Dr, Gillian Auld for coordinating the project.
References
- Assadi AH, Zhang G, Beffert U, McNeil RS, Renfro AL, Niu S, Quattrocchi CC, Antalffy BA, Sheldon M, Armstrong DD, Wynshaw-Boris A, Herz J, D'Arcangelo G, Clark GD. Interaction of reelin signaling and Lis1 in brain development. Nat Genet. 2003;35:270–276. doi: 10.1038/ng1257. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff DL, Geyer MA, Swerdlow NR. Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology. 2001;156:234–258. doi: 10.1007/s002130100810. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran-Jimeniz B, Camargo LM, Oliver KR, Beher D, Shearman MS, Whiting PJ. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Hennah W, van Erp TG, Thompson PM, Lonnqvist J, Huttunen M, Gasperoni T, Tuulio-Henriksson A, Pirkola T, Toga AW, Kaprio J, Mazziotta J, Peltonen L. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- Chen QY, Chen Q, Feng GY, Lindpaintner K, Wang LJ, Chen ZX, Gao ZS, Tang JS, Huang G, He L. Case-control association study of Disrupted-in-Schizophrenia-1 (DISC1) gene and schizophrenia in the Chinese population. J Psychiatr Res. 2007;41:428–434. doi: 10.1016/j.jpsychires.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Chinnasamy D, Rudd R, Velakoulis D. A case of schizophrenia with complete agenesis of the corpus callosum. Australas Psychiatry. 2006;14:327–330. doi: 10.1080/j.1440-1665.2006.02299.x. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, Lerch JP, Trimble K, Uchiyama M, Sakuraba Y, Kaneda H, Shiroishi T, Houslay MD, Henkelman RM, Sled JG, Gondo Y, Porteous DJ, Roder JC. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Devon RS, Anderson S, Teague PW, Burgess P, Kipari TM, Semple CA, Millar JK, Muir WJ, Murray V, Pelosi AJ, Blackwood DH, Porteous DJ. Identification of polymorphisms within Disrupted in Schizophrenia 1 and Disrupted in Schizophrenia 2, and an investigation of their association with schizophrenia and bipolar affective disorder. Psychiatr Genet. 2001;11:71–78. doi: 10.1097/00041444-200106000-00003. [DOI] [PubMed] [Google Scholar]
- Ekelund J, Hovatta I, Parker A, Paunio T, Varilo T, Martin R, Suhonen J, Ellonen P, Chan G, Sinsheimer JS, Sobel E, Juvonen H, Arajärvi R, Partonen T, Suvisaari J, Lönnqvist J, Meyer J, Peltonen L. Chromosome 1 loci in Finnish schizophrenia families. Hum Mol Genet. 2001;10:1611–1617. doi: 10.1093/hmg/10.15.1611. [DOI] [PubMed] [Google Scholar]
- Estivill-Torrus G, Pearson H, van Heyningen V, Price DJ, Rashbass P. Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development. 2002;129:455–466. doi: 10.1242/dev.129.2.455. [DOI] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O'Connell CB, Wang Y, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Harrison PJ. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain. 1999;122:593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- Hattori T, Baba K, Matsuzaki S, Honda A, Miyoshi K, Inoue K, Taniguchi M, Hashimoto H, Shintani N, Baba A, Shimizu S, Yukioka F, Kumamoto N, Yamaguchi A, Tohyama M, Katayama T. A novel DISC1-interacting partner DISC1-Binding Zinc-finger protein: implication in the modulation of DISC1-dependent neurite outgrowth. Mol Psychiatry. 2007;12:398–407. doi: 10.1038/sj.mp.4001945. [DOI] [PubMed] [Google Scholar]
- Hennah W, Tuulio-Henriksson A, Paunio T, Ekelund J, Varilo T, Partonen T, Cannon TD, Lönnqvist J, Peltonen L. A haplotype within the DISC1 gene is associated with visual memory functions in families with a high density of schizophrenia. Mol Psychiatry. 2005;10:1097–1103. doi: 10.1038/sj.mp.4001731. [DOI] [PubMed] [Google Scholar]
- Hennah W, Thomson P, McQuillin A, Bass N, Loukola A, Anjorin A, Blackwood D, Curtis D, Deary IJ, Harris SE, Isometsä ET, Lawrence J, Lönnqvist J, Muir W, Palotie A, Partonen T, Paunio T, Pylkkö E, Robinson M, Soronen P, et al. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.22. in press. [DOI] [PubMed] [Google Scholar]
- Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, Kong S, Wu D, Xue R, Andradé M, Tankou S, Mori S, Gallagher M, Ishizuka K, Pletnikov M, Kida S, Sawa A. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirohashi Y, Wang Q, Liu Q, Li B, Du X, Zhang H, Furuuchi K, Masuda K, Sato N, Greene MI. Centrosomal proteins Nde1 and Su48 form a complex regulated by phosphorylation. Oncogene. 2006;25:6048–6055. doi: 10.1038/sj.onc.1209637. [DOI] [PubMed] [Google Scholar]
- Holy TE, Guo Z. Ultrasonic songs of male mice. PLoS Biol. 2005;3:2177–2186. doi: 10.1371/journal.pbio.0030386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. Am J Psychiatry. 2005;162:2233–2245. doi: 10.1176/appi.ajp.162.12.2233. [DOI] [PubMed] [Google Scholar]
- Innocenti GM, Ansermet F, Parnas J. Schizophrenia, neurodevelopment and corpus callosum. Mol Psychiatry. 2003;8:261–274. doi: 10.1038/sj.mp.4001205. [DOI] [PubMed] [Google Scholar]
- Ishizuka K, Chen J, Taya S, Li W, Millar JK, Xu Y, Clapcote SJ, Hookway C, Morita M, Kamiya A, Tomoda T, Lipska BK, Roder JC, Pletnikov M, Porteous D, Silva AJ, Cannon TD, Kaibuchi K, Brandon NJ, Weinberger DR, et al. Evidence that many of the DISC1 isoforms in C57BL/6J mice are also expressed in 129S6/SvEv mice. Mol Psychiatry. 2007;12:897–899. doi: 10.1038/sj.mp.4002024. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, Nieminen-von Wendt T, von Wendt L, Paunio T, Peltonen L. Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 2008;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- Koike H, Arguello PA, Kvajo M, Karayiorgou M, Gogos JA. Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc Natl Acad Sci U S A. 2006;103:3693–3697. doi: 10.1073/pnas.0511189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B, Song B, Davidson W, MacKenzie A, Smith N, McCaig CD, Harmar AJ, Shen S. Expression of the human PAC1 receptor leads to dose-dependent hydrocephalus-related abnormalities in mice. J Clin Invest. 2006;116:1924–1934. doi: 10.1172/JCI27597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Livy DJ, Wahlsten D. Tests of genetic allelism between four inbred mouse strains with absent corpus callosum. J Hered. 1991;82:459–464. doi: 10.1093/oxfordjournals.jhered.a111128. [DOI] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Annu Rev Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, Thomson PA, Hill EV, Brandon NJ, Rain JC, Camargo LM, Whiting PJ, Houslay MD, Blackwood DH, Muir WJ, Porteous DJ. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Miyata J, Hirao K, Namiki C, Fukuyama H, Okada T, Miki Y, Hayashi T, Murai T. Interfrontal commissural abnormality in schizophrenia: tractography-assisted callosal parcellation. Schizophr Res. 2007;97:236–241. doi: 10.1016/j.schres.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Motomura NH, Satani S, Inaba M. Monozygotic twin cases of the agenesis of the corpus callosum with schizophrenic disorder. Psychiatry Clin Neurosci. 2002;56:199–202. doi: 10.1046/j.1440-1819.2002.00944.x. [DOI] [PubMed] [Google Scholar]
- Murdoch H, Mackie S, Collins DM, Hill EV, Bolger GB, Klussmann E, Porteous DJ, Millar JK, Houslay MD. Isoform-selective susceptibility of DISC1/phosphodiesterase-4 complexes to dissociation by elevated intracellular cAMP levels. J Neurosci. 2007;27:9513–9524. doi: 10.1523/JNEUROSCI.1493-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Flanagan JG, Nakamoto M. Domain-specific olivocerebellar projection regulated by the EphA-ephrin-A interaction. Development. 2002;129:5647–5658. doi: 10.1242/dev.00162. [DOI] [PubMed] [Google Scholar]
- Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH, Ross CA, Sawa A. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul LK, Brown WS, Adolphs R, Tyszka JM, Richards LJ, Mukherjee P, Sherr EH. Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity. Nat Rev Neurosci. 2007;8:287–299. doi: 10.1038/nrn2107. [DOI] [PubMed] [Google Scholar]
- Pletnikov MV, Ayhan Y, Nikolskaia O, Xu Y, Ovanesov MV, Huang H, Mori S, Moran TH, Ross CA. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008;13:173–186. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- Rascle C, Mazas O, Vaiva G, Tournant M, Raybois O, Goudemand M, Thomas P. Clinical features of latent inhibition in schizophrenia. Schizophr Res. 2001;51:149–161. doi: 10.1016/s0920-9964(00)00162-6. [DOI] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein b-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neurobiology of schizophrenia. Neuron. 2006;52:139–153. doi: 10.1016/j.neuron.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Sachs NA, Sawa A, Holmes SE, Ross CA, DeLisi LE, Margolis RL. A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol Psychiatry. 2005;10:758–764. doi: 10.1038/sj.mp.4001667. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, Muramatsu M, Miyagawa S, Hiraiwa N, Yoshiki A, Wynshaw-Boris A, Hirotsune S. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurov IL, Handford EJ, Brandon NJ, Whiting PJ. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicated its role in neurodevelopment. Mol Psychiatry. 2004;9:1100–1110. doi: 10.1038/sj.mp.4001574. [DOI] [PubMed] [Google Scholar]
- St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Wray NR, Millar JK, Evans KL, Hellard SL, Condie A, Muir WJ, Blackwood DH, Porteous DJ. Association between the TRAX/DISC locus and both bipolar disorder and schizophrenia in the Scottish population. Mol Psychiatry. 2005;10:657–668. doi: 10.1038/sj.mp.4001669. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, Hirotsune S, Wynshaw-Boris A. 14-3-3 epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- Vallee RB, Tsai JW. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 2006;20:1384–1393. doi: 10.1101/gad.1417206. [DOI] [PubMed] [Google Scholar]
- Wang Q, Du X, Meinkoth J, Hirohashi Y, Zhang H, Liu Q, Richter M, Greene MI. Characterization of Su48, a centrosome protein essential for cell division. Proc Natl Acad Sci U S A. 2006;103:6512–6517. doi: 10.1073/pnas.0601682103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney G. Ontogeny of sonic vocalizations of laboratory mice. Behav Genet. 1970;1:269–273. doi: 10.1007/BF01074658. [DOI] [PubMed] [Google Scholar]
- Whitney G, Nyby J. Sound communication among adults. In: Willott JF, editor. The auditory psychobiology of the mouse. Springfield, IL: C. C. Thomas; 1983. pp. 98–129. [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in Tg mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Zhang X, Tochigi M, Ohashi J, Maeda K, Kato T, Okazaki Y, Kato N, Tokunaga K, Sawa A, Sasaki T. Association study of the DISC1/TRAX locus with schizophrenia in a Japanese population. Schizophr Res. 2005;79:175–180. doi: 10.1016/j.schres.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Zhang ZJ, Reynolds GP. A selective decrease in the relative density of parvalbumin-immunoreactive neurons in the hippocampus in schizophrenia. Schizophr Res. 2002;55:1–10. doi: 10.1016/s0920-9964(01)00188-8. [DOI] [PubMed] [Google Scholar]