Abstract
Immediate-early genes (IEGs) are tightly coupled to cellular activity and play a critical role in regulating synaptic plasticity. While encoding spatial experience, hippocampal principal cells express IEGs in a behaviorally dependent and cell-specific manner. This expression can be detected through the use of cellular compartment analysis of temporal activity by fluorescence in situ hybridization to generate estimates of cellular activity that match direct neuronal recording under comparable conditions. During rest, IEG expression continues to occur in a small number of cells, and the role of this basal expression is unknown. Imaging IEGs expressed during exploration and adjacent rest periods reveals that “constitutive” IEG expression during rest is not random. Rather, consistent with proposed memory consolidation mechanisms, it recapitulates a subset of the pattern generated by recent experience.
Keywords: Arc, Arg3.1, Homer 1a, reactivation, consolidation, memory
Introduction
During spatial exploration, hippocampal principal cells display robust high-frequency activity that is thought to encode an episode's context (O'Keefe and Nadel, 1978). This patterned activity triggers the transcription of immediate-early genes (IEGs) such as Arc, which is coupled to neuronal activity during attentive behavior (Guzowski et al., 1999) and essential for enduring synaptic plasticity and memory consolidation (Guzowski et al., 2000; Plath et al., 2006). The tight association between neuronal activity and Arc expression results from Arc's critical role in regulating AMPA receptor trafficking (Chowdhury et al., 2006) and homeostatic scaling of synapses containing AMPA receptors (Sheperd et al., 2006), processes that may be essential for information storage. Moreover, Arc and Homer 1a (H1a) are coregulated by physiological activity in individual neurons (Vazdarjanova et al., 2002), suggesting that coordinated IEG cascades are essential for modifying neuronal networks mediating memory.
Lasting memory, however, also requires continual consolidation of encoded representations into more permanent states. This may involve coordinated reactivation of memory traces while the hippocampus is “off-line” (i.e., not processing external stimuli), a phenomenon repeatedly observed during resting states after spatial processing (Wilson and McNaughton, 1994; Kudrimoti et al., 1999; Hoffman and McNaughton, 2002) and associated with upregulation of IEGs. This activity may contribute to “constitutive” IEG expression that occurs when animals remain under controlled conditions in their home cage (Guzowski et al., 1999; Vazdarjanova et al., 2002; Ramirez-Amaya et al., 2005). Investigating this possibility, however, critically requires discriminating IEG expression during resting states and adjacent spatial processing episodes in the same animal.
Previous data indicate that Arc and H1a provide this discrimination. Although both IEGS are transcribed immediately after neuronal activity, intranuclear Arc appears within 5 min, whereas H1a is detected only after 30 min, because of large introns atypical of IEGs (Vazdarjanova et al., 2002). Cytoplasmic accumulation of these IEGs is similarly staggered: cytoplasmic Arc appears 30 min after activity, whereas cytoplasmic H1a requires 60 min (Bottai et al., 2002). Thus, monitoring Arc and H1a transcription permits the detection of three episodes of activity. This novel tool permits testing the hypothesis that off-line reactivation of network activity contributes to basal IEG expression.
Materials and Methods
Subjects and behavioral procedures.
The consistency of IEG transcription was assessed during three discrete episodes in 18 Fischer 344 rats aged 12–13 months. The procedure for the spatial exploration trials is similar to those described previously (Guzowski et al., 1999; Vazdarjanova et al., 2002). Briefly, rats were housed with ad libitum access to food and water with a 12 h reversed light cycle. At least 2 h after the commencement of the dark cycle, rats were shuttled to a dimly lit room containing several distinct landmarks, and placed within a 61 × 61 cm box divided into nine equal grids. Rats were gently picked up and randomly placed within a different grid every 15 s until every grid was visited to ensure all aspects of the environment were sampled equally. Rats were then shuttled back to their colony room cages, where they remained undisturbed until the exploration was repeated in the same environment. The current study differs from previous work in the addition of a third rest episode (Fig. 1a). In one group [rest-explore-explore (R-E-E)], the rest episode occurred before two exploration episodes, whereas in a second group [explore-rest-explore (E-R-E)], the rest episode occurred between the two explorations. A control group [rest-rest-rest (R-R-R)] remained undisturbed in their home cages (n = 6/group).
Figure 1.
A timeline shows gene expression patterns indicative of cellular activity (top) and each group's behavior (bottom) during each episode. Cells were labeled with digoxigenin-conjugated Homer 1a (H1a, red), fluorescein-conjugated Arc (green), and counterstained with TO-PRO-3 iodide (blue). Cells active 60 min before the animals were killed exhibit cytoplasmic H1a (left), when E-R-E animals explored a novel environment and R-E-E animals rested. Thirty minutes before being killed, R-E-E animals explored and E-R-E animals rested. Cells active during this time exhibit cytoplasmic Arc and intranuclear H1a (middle). Five minutes before the animals were killed, active cells exhibit intranuclear Arc. Both R-E-E and E-R-E animals explored at this time (right). A third group (R-R-R) remained undisturbed in their home cages.
Tissue processing and microscopy.
Immediately after the second exploration, rats were decapitated under isofluorane anesthesia. Brains were removed and quick-frozen in a beaker of isopentane bathed in dry ice/ethanol within 180 s to maintain mRNA integrity. Hemisections from 8–10 rats were mounted with Tissue-Tek OCT compound such that all experimental conditions were represented on each slide. Coronal sections (20 μm) were cut through the right dorsal hippocampus (−3.2 to −3.8 mm from bregma) (Paxinos and Watson, 1986), thaw-mounted on Superfrost Plus slides (VWR, West Chester, PA), dried, and stored at −70°C.
Fluorescence in situ hybridization was performed as described previously (Guzowski et al., 1999; Vadarjanova et al., 2002). Briefly, riboprobes for Arc and H1a were synthesized using a commercially available transcription kit (Maxiscript; Ambion, Austin, TX). Arc riboprobes were conjugated to fluorescein-labeled UTP, whereas H1a was conjugated to digoxigenin-labeled UTP (Roche Molecular Biochemicals, Nutley, NJ). Riboprobes were purified on a G-50 spin column (Roche Diagnostics, Indianapolis, IN) and verified by gel electrophoresis. Slides were thawed, fixed in 4% paraformaldehyde, treated with 0.5% acetic anhydride/1.5% triethanolamine, incubated in methanol and acetone (1:1), equilibrated in SSC, and treated with 100 μl of prehybridization buffer (Sigma, St. Louis, MO) for 30 min at room temperature. Each slide was then treated with ∼100 ng of each riboprobe [diluted in hybridization buffer (GE Healthcare Bio-Sciences, Piscataway, NJ), denatured at 90°C, and chilled on ice] and incubated in a humid chamber overnight at 56°C. After a graduated series of posthybridization washes in SSC, slides were bathed in RNase A (10 mg/ml) at 37°C and endogenous peroxidases were quenched in 2% H2O2, blocked with tyramide signal amplification blocking buffer (PerkinElmer, Boston, MA) containing 5% normal sheep serum. Slides were then incubated with anti-digoxigenin-HRP antibody (Roche Molecular Biochemicals) at room temperature for 2 h, washed with Tris-buffered saline containing 0.05% Tween 20, and the HRP-antibody conjugate was detected using CY3 (cyanine-3) signal amplification kit (PerkinElmer). Endogenous peroxidases were quenched again and slides were incubated with anti-fluorescein-HRP antibody (Roche Molecular Biochemicals) at room temperature for 2 h and detected using a fluorescein-tyramide signal amplification kit (PerkinElmer). After nuclear counterstaining with TO-PRO-3 iodide (Invitrogen, Eugene, OR), coverslips were applied with Vectashield anti-fade media (Vector Laboratories, Burlingame, CA) and sealed with nail polish.
Images were collected using a Zeiss (Oberkochen, Germany) 510 Meta laser scanning confocal microscope at 40× under oil-immersion. For consistency, photomultiplier tube assignments, pinhole size, and contrast values remained constant for each slide. Images of CA1 were acquired by taking z-stacks (optical thickness, 1.1 μm; interval, 0.7 μm) throughout the thickness of 2–3 slides per animal.
Image analysis.
Given the histological activity record that Arc and H1a provide (Fig. 1b), the overlap between ensembles participating in two bouts of spatial exploration was quantified using cellular compartment analysis of temporal activity by fluorescence in situ hybridization (catFISH), as described previously (Guzowski et al., 1999; Vazdarjanova et al., 2002). In R-E-E animals, cells responsive to both explorations express nuclear H1a and both cytoplasmic and nuclear Arc, whereas in E-R-E animals they express cytoplasmic H1a and nuclear Arc. For consistency, R-R-R animals were paired with either E-R-E or R-E-E animals mounted on the same slide, and overlap was assessed using identical IEG expression patterns. The overlap in R-R-R animals did not change based on which episodes were used to assess repeated activation (data not shown).
Reactivation was quantified by the proportion of cells transcribing IEGs during both exploration periods and rest (i.e., containing nuclear and cytoplasmic H1a, as well as nuclear and cytoplasmic Arc). An estimate of random overlap was also generated for each animal as the product of the proportion of cells active during both explorations and the proportion active during rest. For instance, if 40% of cells in an individual animal transcribed IEGs during two explorations in the same environment, and 10% of cells transcribed IEGs during rest, the expected random overlap between these two active populations would be 4% (0.4 × 0.1 = 0.04). The estimated overlap based on random chance and the overlap observed in each animal was then compared using a paired t test.
Results
Spatial exploration induces IEGs in comparable numbers of CA1 principal cells in all groups
Exploration induced a comparable (p = 0.97) increase in transcription in both E-R-E (episode 1, 41.9 ± 3.6%; episode 3, 42.2 ± 3.1%; all data presented as mean ± SEM) and R-E-E (episode 2, 37.8 ± 1.9; episode 3, 39.3 ± 2.8) animals (p < 0.001 vs R-R-R). In addition, ∼80–90% of cells activated by the first exploration were reactivated by the second exploration (R-E-E, 31.6 ± 1.1%; E-R-E, 33.2 ± 1.4%) (Fig. 2a). These findings are consistent with previous data (Guzowski et al., 1999) and indicate that three-episode catFISH can effectively detect the transcriptional response induced by spatial exploration with comparable sensitivity during any combination of three behavioral episodes.
Figure 2.
A, The proportion of CA1 principal cells transcribing IEGs across episodes. During exploration (hatched bars), IEG transcription is significantly higher in both E-R-E (white) and R-E-E (light gray) animals than controls (R-R-R; dark gray), whereas transcription during rest is comparable with controls. Most cells active during the first exploration are active in the second (behavioral overlap). B, The proportion of cells active in rest and both explorations (solid), and the overlap expected by random chance (cross-hatched). Significantly more cells were active during all episodes in E-R-E than R-R-R animals, and in R-R-R and E-R-E animals this overlap is significantly greater than expected by random chance, suggesting transcription recapitulates previous experience both in the exploration and home cage environments (mean ± SEM; **p < 0.001 vs R-R-R; ††p < 0.001 between behavioral groups; ‡p < 0.05 vs chance overlap).
The proportion of cells transcribing IEGs during rest is not altered by behavioral history
Caged control animals showed low levels of IEG expression in 10–15% of neurons during all three episodes (episode 1, 14.4 ± 3.3%; episode 2, 8.9 ± 2.2%; episode 3, 13.8 ± 3.8%), comparable with previous reports (Guzowski et al., 1999; Vazdarjanova et al., 2002). Similar transcription levels occurred during rest in both E-R-E (12.8 ± 2.9; p = 0.82 vs R-R-R) and R-E-E (12.50 ± 1.56%; p = 0.09 vs R-R-R) animals. Thus, if memory-trace reactivation influences IEG transcription during rest, it does so by means other than increasing the number of cells transcribing IEGs.
IEG expression during rest is nonrandom
Although constitutive transcription remains low in R-R-R animals, it is not random: the same cells repeatedly transcribe IEGs (t(5) = 2.25; p < 0.05). Moreover, if reactivation contributes to constitutive expression without increasing the number of cells engaged in transcription, IEG expression during subsequent rest episodes should predominantly occur in cells engaged during the previous exploration, and this effect should be absent if rest occurs before exploration. As predicted by this hypothesis (Fig. 2b), IEG expression during rest largely occurred in cells that responded to exploration in E-R-E animals (t(5) = 6.25; p < 0.001), whereas the probability of a cell transcribing IEGs during both rest and exploration in R-E-E animals was equivalent to random chance (t(5) = 1.65; p = 0.09).
Discussion
Constitutive expression (i.e., IEG expression that occurs when animals remain at rest and are not actively engaged in processing the external environment) is often treated as a reflection of spontaneous synaptic activity occurring during rest. However, the current data suggest that IEG expression during rest is a small but reliable signal that can match the pattern of transcription generated in response to previous environments (Fig. 3). Moreover, the absence of this effect in R-E-E animals strongly suggests that the IEG expression patterns observed here reflect bona fide reactivation, rather than a sustained transcriptional response. The pattern of IEG transcription in the home cage is environmentally specific (i.e., occurring in response to the cues associated with the home cage), because the same cells repeatedly transcribe IEGs, provided animals remain in this environment. Given this specificity, if the overlap in IEG expression between rest and a preceding exploration were the result of a sustained response to the memory of cues after those cues are removed, one would expect to see the same overlap occurring in animals moving from the home cage to an exploration environment (i.e., R-E-E animals). The overlap between the neuronal population active during rest and those neurons active during exploration in R-E-E animals, however, is random. Thus, the reactivation of activity patterns generated by previous experience is consistent with the IEG transcription patterns observed here, suggesting that IEGs contribute not only to encoding contextual episodes, but also to “off-line” consolidation.
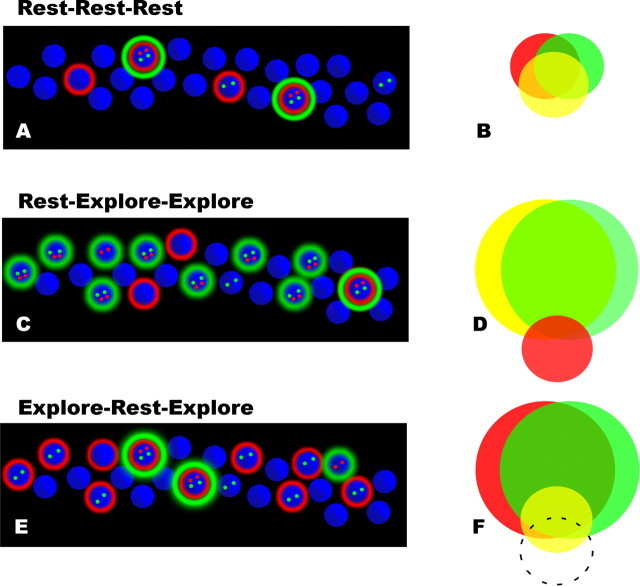
Figure 3.
A schematic diagram illustrates the main observations made in the experiment. At left, schematic illustrations show cells (blue) displaying typical patterns of Homer 1a (H1a; red) and Arc (green) expression in the three groups examined in this experiment. At right, Venn diagrams depict the degree of overlap in the active cell populations during episodes 1 (red), 2 (yellow), and 3 (green). A, B, In rest-rest-rest animals (A), a small number of neurons (∼10%) actively transcribe IEGs during any episode, and the degree of overlap in the activities during each episode is far greater than expected by random chance (B). C, In R-E-E animals, exploration induces a robust increase in cells expressing intranuclear H1a (indicating activity 30 min before the animal was killed), and both cytoplasmic and nuclear Arc (indicating activity at 30 and 5 min before the animal was killed, respectively), corresponding to the time of the two explorations. Relatively little cytoplasmic H1a, corresponding to activity during the initial rest period, can be observed. D, In addition, ∼40% of CA1 principle cells are active during the first exploration (yellow), and ∼90% of cells active during the first exploration are again active after returning to the same environment (green). Moreover, the degree of overlap between the population of cells that are active during rest (red) and those that are active during exploration is no greater than would be expected by random chance. E, F, Exploration induces a similar increase in IEG transcription in E-R-E animals (E), appearing as a large number of cells expressing cytoplasmic H1a (indicating activity 60 min before the animal was killed), and intranuclear Arc (indicating activity 5 min before the animal was killed) with only ∼10% of cells transcribing IEGs during the intervening rest period. In these animals (F), ∼40% of CA1 principle cells are active during the first exploration (red), and ∼90% of these cells are also active during the second exploration (green), similar to R-E-E animals. However, the majority of cells that are active during rest in E-R-E animals are the same cells activated by exploration (yellow). This degree of overlap is significantly greater than would be expected by random chance, and is a significant shift relative to the degree of overlap observed in R-E-E animals (dashed circle).
This molecular index of reactivation also provides the boundary conditions that constrain the number of cells simultaneously engaged in reactivation in CA1 networks. During a single rest episode, ∼10% of CA1 pyramidal cells transcribe IEGs, which reflects ∼25% of the initial cellular response to exploration. Thus, IEG reactivation does not increase the number of cells transcribing IEGs during rest. Rather, although the absolute number of cells engaging in constitutive expression remains relatively constant, the activity is shifted to those cells that were active during a preceding exploration episode. There is, however, a small trend suggesting that IEG expression is marginally greater during rest episodes that follow exploration, consistent with previous experiments [Guzowski et al. (1999), their Fig. 4, and Guzowski et al. (2006), their Fig. 1a]. These trends suggest that transcription related to the recapitulation of the exploration environment does, in fact, occur in addition to recapitulation related to the resting environment. However, because the activity “normally” observed during rest is a small fraction of that induced by exploration, it is likely that activity specific to the rest environment is more difficult to detect.
Although imaging methods cannot capture fine temporal structure within ensemble activity, they demonstrate that during rest, IEG transcription is induced in the same cells that were active during a previous exploration session. Because the high-frequency activity inherent in place cell discharge is sufficient to induce IEG transcription in the same proportion of cells that express place fields during exploration (Guzowski et al., 1999; Vazdarjanova et al., 2002), it is reasonable to conclude that the transcription observed during rest is the product of the recapitulation of this activity. IEG reactivation thus provides a histological record of which cells were active both during exploration and a subsequent rest episode.
In this way, IEG reactivation is reminiscent of rate-based reactivation measures (Pavlides and Winson, 1989; Battaglia et al., 2005). Although persistently increased firing is not sufficient for activity pattern reactivation, these measures produce comparable results. Both rate- and correlation-based reactivation measures exhibit similar decay times, are most robust during sharp waves, and covary across sessions (Battaglia et al., 2005) and, thus, have been argued to capture similar physiological phenomena. Moreover, although the dissociation between rate and pattern reactivation places limits on the use of IEG expression as an indicator of electrical activity, the fact that Arc and H1a are mechanistically linked to forms of plasticity considered essential for information storage indicates that IEG expression can identify subpopulations of neurons that are engaged in the molecular process of plasticity. Thus, IEG methods can extract information from the same process by providing a binary “on/off” indication of which cells are simultaneously active during a single rest episode across a large number of distributed neuronal populations. This distributed “snapshot” of activity is in many ways complementary to the long-term continuous activity observable through single-unit recording.
Regardless of the potential distinctions in the phenomena captured through electrophysiological vs molecular indices of reactivation, the critical conclusion to be made from the current data are that IEG expression during rest is not “noise” generated by spontaneous activity. Rather, IEG transcription patterns during rest can match those elicited by previous environments, indicating that constitutive IEG expression can reflect an information processing signal that changes in predictable ways depending on an animal's behavioral history.
Footnotes
This work was supported by National Institute on Aging Grant AG009219, the McKnight Brain Research Foundation, the state of Arizona and Arizona Department of Health Services, and Natural Sciences and Engineering Research Council of Canada Grant PDF20154427. We thank P. Lipa, M. Tatsuno, and K. Gothard for helpful discussion of these results.
References
- Battaglia FP, Sutherland GR, Cowen SL, McNaughton BL, Harris KD. Firing rate modulation: a simple statistical view of memory trace reactivation. Neural Netw. 2005;18:1280–1291. doi: 10.1016/j.neunet.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Bottai D, Guzowski JF, Schwarz MK, Kang SH, Xiao B, Lanahan A, Worley PF, Seeburg PH. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate early gene expression. J Neurosci. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman KL, McNaughton BL. Coordinated reactivation of distributed memory traces in primate neocortex. Science. 2002;297:2070–2073. doi: 10.1126/science.1073538. [DOI] [PubMed] [Google Scholar]
- Kudrimoti HS, Barnes CA, McNaughton BL. Reactivation of hippocampal cell assemblies: effects of behavioral state, experience, and EEG dynamics. J Neurosci. 1999;19:4090–4101. doi: 10.1523/JNEUROSCI.19-10-04090.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe J, Nadel L. The hippocampus as a cognitive map. Oxford: Clarendon; 1978. [Google Scholar]
- Pavlides C, Winson J. Influences of hippocampal place cell firing in the awake state on the activity of these cells during subsequent sleep episodes. J Neurosci. 1989;9:2907–2918. doi: 10.1523/JNEUROSCI.09-08-02907.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Ramirez-Amaya V, Marrone DF, Gage FH, Worley PF, Barnes CA. Integration of new neurons into functional neural networks. J Neurosci. 2005;26:12237–12241. doi: 10.1523/JNEUROSCI.2195-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF. Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. J Neurosci. 2002;22:10067–10071. doi: 10.1523/JNEUROSCI.22-23-10067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL. Reactivation of hippocampal ensemble memories during sleep. Science. 1994;265:676–679. doi: 10.1126/science.8036517. [DOI] [PubMed] [Google Scholar]