Abstract
Herpesviruses are thought to have evolved in very close association with their hosts. This is notably the case for cytomegaloviruses (CMVs; genus Cytomegalovirus) infecting primates, which exhibit a strong signal of co-divergence with their hosts. Some herpesviruses are however known to have crossed species barriers. Based on a limited sampling of CMV diversity in the hominine (African great ape and human) lineage, we hypothesized that chimpanzees and gorillas might have mutually exchanged CMVs in the past. Here, we performed a comprehensive molecular screening of all 9 African great ape species/subspecies, using 675 fecal samples collected from wild animals. We identified CMVs in eight species/subspecies, notably generating the first CMV sequences from bonobos. We used this extended dataset to test competing hypotheses with various degrees of co-divergence/number of host switches while simultaneously estimating the dates of these events in a Bayesian framework. The model best supported by the data involved the transmission of a gorilla CMV to the panine (chimpanzee and bonobo) lineage and the transmission of a panine CMV to the gorilla lineage prior to the divergence of chimpanzees and bonobos, more than 800,000 years ago. Panine CMVs then co-diverged with their hosts. These results add to a growing body of evidence suggesting that viruses with a double-stranded DNA genome (including other herpesviruses, adenoviruses, and papillomaviruses) often jumped between hominine lineages over the last few million years.
Keywords: cytomegalovirus, hominine, host switch, codivergence, dsDNA virus
1. Introduction
Herpesviruses (family Herpesviridae) are a family of large enveloped double-stranded DNA (dsDNA) viruses that infect many vertebrates, including humans and nonhuman primates (NHPs; McGeoch et al. 2008). The broad distribution of herpesviruses combined with infection being generally asymptomatic has been considered as indicative of long-term co-evolution with their mammalian host (McGeoch et al. 2008). This hypothesis is supported by the recent identification of an endogenous herpesvirus element in the NHP tarsier genome, with insertion estimated to have occurred > 56 million years (My) ago (Aswad and Katzourakis 2014). Congruent topologies and similar relative branch lengths in phylogenetic trees of herpesviruses and their mammalian hosts further suggest that co-divergence largely shaped the evolution of these viruses: their diversification has largely been driven by their host diversification (McGeoch, Rixon, and Davison 2006).
Although herpesvirus evolution appears to be closely tied to that of their host, cross-species transmission still appears possible. Herpesviridae are divided into three distinct subfamilies: Alpha-, Beta-, and Gammaherpesvirinae. In addition to the genetic structure, sequence and cell type tropism differences defining these subfamilies, they also appear to differ in their capacity for cross-species transmission. Transmission is frequently documented for members of the Alphaherpesvirinae and the Gammaherpesvirinae subfamilies (Huff and Barry 2003; Schrenzel et al. 2003; Oya et al. 2004; Russell, Stewart, and Haig 2009). Herpesvirus B (Cercopithecine alphaherpesvirus 1), an alphaherpesvirus closely related to human herpes simplex virus 1 (HSV-1), naturally infects macaques (Macaca mulatta), but is easily transmitted to humans where it often causes fatal encephalomyelitis (Huff and Barry 2003; Oya et al. 2004). Conversely, HSV-1 has been shown to result in life-threatening infections in white-faced saki monkeys (Pithecia pithecia; Schrenzel et al. 2003). Similarly, the bovine and ovine malignant catarrhal fever viruses (Alcelaphine gammaherpesvirus 1 and Ovine gammaherpesvirus 2), as well as endemic gammaherpesviruses viruses of wildebeest and sheep, respectively, frequently infect bison, cattle, deer, pigs, and water buffalo where they can cause fatal lymphoproliferative disease (Russell, Stewart, and Haig 2009).
Evidence for cross-species transmission of betaherpesviruses, including the most studied subfamily members, cytomegaloviruses (CMVs; genus Cytomegalovirus), is much more rare (Murthy et al. 2013). Although in vitro experiments have shown that CMVs from rodents and NHPs can infect human cell lines (Lafemina and Hayward 1988; Michaels et al. 1997; Lilja and Shenk 2008) and that human CMV can infect primary fibroblasts of chimpanzees (Perot, Walker, and Spaete 1992), in vivo transmission (both from experimental as well as natural infection) of CMVs between closely related host species has never been observed even for closely interacting predator–prey NHP species in the wild. Together, this information supports a difference of CMVs compared with members of other herpesvirus subfamilies, with CMVs being strongly restricted to their natural host in nature (Murthy et al. 2013; Burwitz et al. 2016; Anoh et al. 2018; James et al. 2018).
Consistent with this model of restricted CMV cross-species transmission, phylogenetic analysis of CMV sequences has identified a strong signal for co-speciation of CMVs with their Old and New World primate hosts (Leendertz et al. 2009; Anoh et al. 2018; James et al. 2018). At such evolutionary timescales CMV diversification is most often explained by host diversification. However, a striking exception was observed for CMVs infecting hominines (African great apes and humans; Leendertz et al. 2009). In this case, CMV sequences from Western chimpanzees (Pan troglodytes verus [P.t.v.]) and Western lowland gorillas (Gorilla gorilla gorilla [G.g.g.]) clustered into two different clades (CG1 and CG2), both of which contained chimpanzee and gorilla CMVs. Based on in-depth phylogenetic analyses, CG1 and CG2 appeared to be the co-speciational clades for gorilla and chimpanzee CMVs, respectively (Leendertz et al. 2009). Rare ancestral transmission events between hosts belonging to the chimpanzee and gorilla lineages were proposed to account for the presence of viruses belonging to the CG1 or CG2 clades in the non-co-speciational primate (chimpanzee and gorilla, respectively).
This unexpected yet statistically well-supported model (hereafter referred to as the ‘transmission model’) was based on a small dataset comprising CMVs from twenty-five Western chimpanzees (P.t.v.) and seven Western lowland gorillas (G.g.g.), representing two of the nine subspecies/species of African great apes (Leendertz et al. 2009). This study provides the first large-scale extensive analysis of CMVs in all 9 taxa of African non-human great ape subspecies/species using 675 fecal samples collected in the wild at 20 sites in 11 sub-Saharan African countries (Fig. 1). This analysis identifies new CMV variants belonging to previously characterized as well as potentially novel CMV species. When combined with human CMV sequence information, this study provides a complete picture of the evolution of CMVs in hominines.
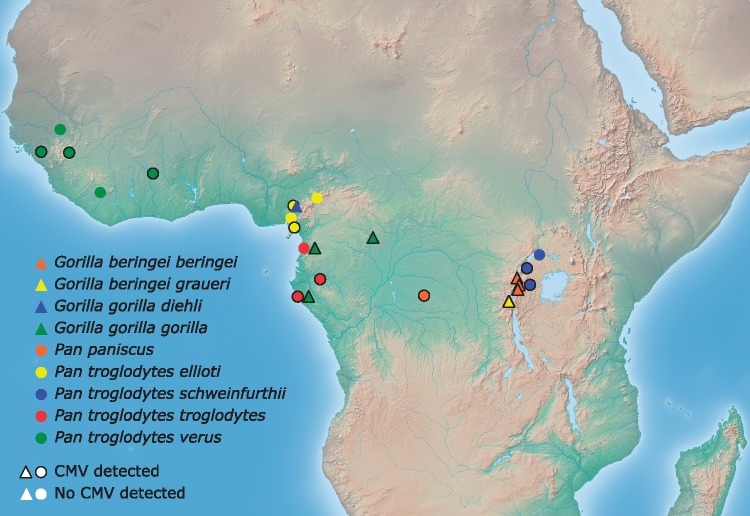
Figure 1.
African great ape sampling site locations. The twenty sites represented on this map are (ordered from North to South within African great ape subspecies): G.b.b., Virunga National Park (DRC); Bwindi Impenetrable National Park* (Uganda), Volcanoes National Park (Rwanda); G.b.g., Kahuzi Biega National Park (DRC); G.g.d., Takamanda National Park (Cameroon); G.g.g., Dzanga-Sangha Special Reserve (Central African Republic), Campo Ma’an National Park* (Cameroon), Loango National Park* (Gabon); P.p., Salonga National Park (DRC); P.t.e., Gashaka Gumti National Park (Nigeria), Mbe Mountains Community Forest (Nigeria), Korup National Park (Cameroon), Mount Cameroon National Park (Cameroon); P.t.s., Budongo Forest (Uganda), Kibale National Park (Uganda), Bwindi Impenetrable National Park* (Uganda); P.t.t., Campo Ma’an National Park* (Cameroon), Lope National Park (Gabon), Loango National Park* (Gabon); P.t.v., Kayan (Senegal), Sangaredi (Guinea), Sobeya (Guinea), Comoe-GEPRENAF (Côte d’Ivoire), East Nimba (Liberia). DRC: Democratic Republic of the Congo; * samples were obtained from two African great apes species at this site.
2. Results and discussion
We analyzed a total of 675 fecal samples, which represented (i) all chimpanzee subspecies: Pan troglodytes ellioti (P.t.e.), Pan troglodytes schweinfurthii (P.t.s.), Pan troglodytes troglodytes (P.t.t.), and P.t.v., (ii) bonobos (Pan paniscus [P.p.]), and (iii) all gorilla subspecies: Gorilla beringei beringei (G.b.b.), Gorilla beringei graueri (G.b.g.), Gorilla gorilla diehli (G.g.d.), and G.g.g., using a generic nested PCR that targets CMV DNA (PCR1; Table 1; Ehlers et al. 2007; Prepens et al. 2007). We sequenced all products of expected size.
Table 1.
PCR primers used in this study.
| PCR (target) | Round | Primer number | Sequence | Tm (°C) | Product length (bp) |
|---|---|---|---|---|---|
| 1 (UL55 CDSa of betaherpesviruses) | I | 2743-sb | CGCAAATCGCAGA(N/I)KC(N/I)TGGTG | 46 | 250 |
| 2746-asc | TGGTTGCCCAACAG(N/I)ATYTCRTT | ||||
| II | 2744-s | TTCAAGGAACTCAGYAARAT(N/I)AAYCC | 46 | 240 | |
| 2745-as | CGTTGTCCTC(N/I)CC(N/I)ARYTG(N/I)CC | ||||
| 2 (UL56 CDS of betaherpesviruses) | I | 3903-s | CCTGTCGCACAATGTGGACATG | 46 | 250 |
| 3903-as | CAGCTGTTTTCCGAA(N/I)GTTTCRTTAT | ||||
| II | 3904-s | TGGCCTACGCYTGYGAYAACG | 46 | 179 | |
| 3904-as | GCGAACGTGC(N/I)TCCACATCTCC | ||||
| 3 (UL55 CDS of primate CMVs) | I | 7393-s | CTGGTGGTCTTCTGGCAGG | 60 | 421 |
| 7393-as | GCACCTTGACRCTGGTCT | ||||
| II | 7393-s | CTGGTGGTCTTCTGGCAGG | 60 | 418 | |
| 7394-as | CCTTGACGCTGGTCTGGTT | ||||
| 4 (UL55 to UL56 CDS of primate CMVs) | I | 7470-s | CGTGTACCCCAGYGAGTG | 55 | 2400 |
| 7470-as | GGCGATGGGYTTGTYGTA | ||||
| II | 7471-s | CAGCGAGTGGATGGTGGT | 55 | 2400 | |
| 7471-as | ATGGGCTTGTYGTARATGGC | ||||
| 5 (UL55 CDS of bonobo CMVs) | I | 7484-s | CAAGCCCACCAAGGARGAC | 55 | 1300 |
| 7484-as | CAGCACGTCGCCCATGAA | ||||
| II | 7485-s | TCATGGTGGTCTACAARCGC | 55 | 1250 | |
| 7485-as | ATGGGCTTGTRGTAGATGGC | ||||
| 6 (UL55 CDS of bonobo CMVs) | I | 7470-s | CGTGTACCCCAGYGAGTG | 55 | 1326 |
| 7499-as | TGTGGGTGTTGGTGTAGTCG | ||||
| II | 7471-s | CAGCGAGTGGATGGTGGT | 55 | 1272 | |
| 7500-as | TCTCGTAGCTGTCCTCGTGA | ||||
| 7 (cytB CDS of vertebrates) | I | 258-s | CCATCCAACATCTCAGCATGATGAAA | 50 | 359 |
| 258-as | GCCCCTCAGAATGATATTTGTCCTCA |
Coding sequence.
Sense.
Antisense.
We identified sixteen CMV-positive chimpanzee samples (overall detection rate: 4.5%) in all four subspecies (P.t.e.: 6.3%; P.t.s.: 2.7%; P.t.t.: 9.6%; P.t.v.: 3.0%), nineteen positive bonobo samples (57.6%), and forty-five positive gorilla samples (overall detection rate: 16.0%) in three of the four subspecies (G.g.g.: 9.4%; G.b.b.: 22.7%; G.b.g.: 22.8%), with CMV only not being detected in Cross River gorillas (G.g.d.; Table 2). Since the detection rate of HCMV in stool samples of humans is much lower than seroprevalence (Anoh et al. 2018), it is likely that the seroprevalence of CMVs in African great apes is much higher than the detection rates in stool samples reported here. The detection rates varied considerably in gorillas, with western gorilla subspecies showing lower values (G.g.d.: 0%; G.g.g.: 9.4%) than eastern subspecies (G.b.b.: 22.7%; G.b.g.: 22.8%); such variation may be due to the relative small sample sizes in our study or reflect biological processes, for example, contrasted local demographic histories for the different gorilla populations. Together with the previously reported findings (Leendertz et al. 2009), our results indicate that CMVs circulate in wild populations of all African great ape subspecies, reaching variable but overall high prevalence.
Table 2.
Great ape CMV screening results.
| Species | Country | Site | Tested | CMV1 positive | CMV2 positive | CMV1 or CMV2 positive | Percentage CMV1/2 positive (95% CI) |
|---|---|---|---|---|---|---|---|
| Genus Pan | 390 | 18 | 17 | 35 | 9.0 (6.0–11.6) | ||
| Pan paniscus (P.p.) | 33 | 10 | 9 | 19 | 58 (40.5–64.7) | ||
| Democratic Republic of the Congo | Salonga National Park | 33 | 10 | 9 | 19 | ||
| Pan troglodytes ellioti (P.t.e.) | 63 | 2 | 2 | 4 | 6.3 (0.3–12.4) | ||
| Nigeria | Mbe Mountains Community Forest | 17 | 2 | 0 | 2 | ||
| Gashaka Gumti National Park | 12 | 0 | 0 | 0 | |||
| Cameroon | Mount Cameroon National Park | 17 | 0 | 2 | 2 | ||
| Korup National Park | 17 | 0 | 0 | 0 | |||
| Pan troglodytes schweinfurthii (P.t.s.) | 75 | 1 | 1 | 2 | 2.7 (0–6.3) | ||
| Uganda | Bwindi Impenetrable National Park | 40 | 0 | 1 | 1 | ||
| Budongo Forest | 25 | 0 | 0 | 0 | |||
| Kibale National Park | 10 | 1 | 0 | 1 | |||
| Pan troglodytes troglodytes (P.t.t.) | 52 | 4 | 1 | 5 | 9.6 (1.5–17.7) | ||
| Cameroon | Campo Ma'an National Park | 25 | 2 | 1 | 3 | ||
| Gabon | Loango National Park | 23 | 1 | 0 | 1 | ||
| Lope National Park | 4 | 1 | 0 | 1 | |||
| Pan troglodytes verus (P.t.v.) | 167 | 1 | 4 | 5 | 3.0 (0.4–5.6) | ||
| Cote d'Ivoire | Comoe-GEPRENAF | 31 | 0 | 1 | 1 | ||
| Guinea | Sobeya | 38 | 1 | 2 | 3 | ||
| Sangaredi | 35 | 0 | 1 | 1 | |||
| Liberia | East Nimba | 28 | 0 | 0 | 0 | ||
| Senegal | Kayan | 34 | 0 | 0 | 0 | ||
| Genus Gorilla | 281 | 24 | 21 | 45 | 16.0 (11.4–20.0) | ||
| Gorilla beringei beringei (G.b.b.) | 97 | 16 | 6 | 22 | 22.7 (14.3–31.0) | ||
| Democratic Republic of the Congo | Virunga National Park | 31 | 3 | 3 | 6 | ||
| Rwanda | Volcanoes National Park | 18 | 2 | 0 | 2 | ||
| Uganda | Bwindi Impenetrable National Park | 48 | 11 | 3 | 14 | ||
| Gorilla beringei graueri (G.b.g.) | 79 | 6 | 12 | 18 | 22.8 (14.8–33.9) | ||
| Democratic Republic of the Congo | Kahuzi Biega National Park | 79 | 6 | 12 | 18 | ||
| Gorilla gorilla diehli (G.g.d.) | 56 | 0 | 0 | 0 | 0 (0–8.0) | ||
| Cameroon | Takamanda National Park | 56 | 0 | 0 | 0 | ||
| Gorilla gorilla gorilla (G.g.g.) | 53 | 2 | 3 | 5 | 9.4 (1.7–19.2) | ||
| Gabon | Loango National Park | 29 | 1 | 3 | 4 | ||
| Cameroon | Campo Ma'an National Park | 4 | 0 | 0 | 0 | ||
| Central African Republic | Dzanga-Sangha Special Reserve | 20 | 1 | 0 | 1 |
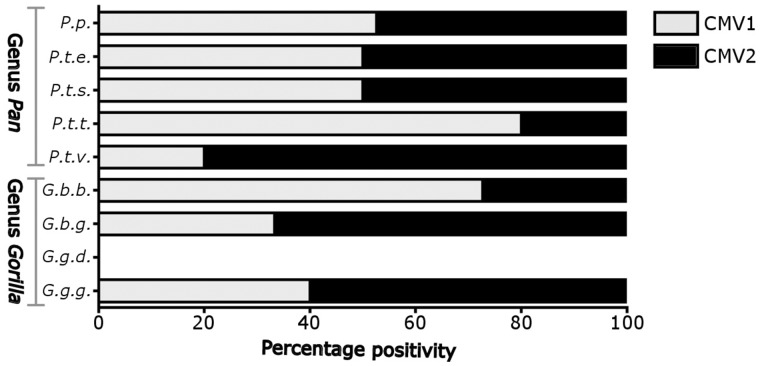
Our earlier study identified two distinct types of African great ape CMVs, CMV1 and CMV2 belonging to the above mentioned clades CG1 and CG2, respectively (Leendertz et al. 2009). We compared the CMV sequences identified in the present study with the published great ape CMV1 and CMV2 sequences. All sequences (n = 80) could be attributed to either CMV1 (n = 42) or CMV2 (n = 38) (Table 2). The eight African great ape species/subspecies which were positive for CMV also appeared to be infected with both CMV1 and CMV2 (Table 1; Fig. 2). For chimpanzees, bonobos, and gorillas, CMV1 and CMV2 detection rates did not markedly differ, reaching 50, 52.6, and 53.3 per cent, respectively for CMV1; and 50, 47.3, and 46.7 per cent for CMV2 (Fig. 2). Therefore, patterns of detections rates (and presumably prevalence) of CMV1 and CMV2 do not reflect the assumed origin of the viruses within one ape species followed by transmission to another as proposed by the transmission model (Leendertz et al. 2009). Our results contrast with previous observations for human adenovirus species B (HAdV-B), which was originally transmitted from gorillas to chimpanzees and is still present at a much higher prevalence in its original gorilla host (55 vs. 11%; Hoppe et al. 2015).
Figure 2.
CMV1 and CMV2 in stools of great ape subspecies. Fecal samples (n = 675) from animals belonging to nine great ape subspecies, P.p. (n = 33), P.t.e. (n = 63), P.t.s. (n = 75), P.t.t. (n = 52), P.t.v. (n = 167), G.b.b. (n = 97), G.b.g. (n = 79), G.g.d. (n = 56), and G.g.g. (n = 53), were analyzed with generic nested PCR1 for the presence of CMVs. The positive samples were sequenced to determine the presence of CMV1 and CMV2. The figure represents percentage positivity of CMV1 (gray) and CMV2 (black). P.p., Pan paniscus; P.t.e., Pan troglodytes ellioti; P.t.s., Pan troglodytes schweinfurthii; P.t.t., Pan troglodytes troglodytes; P.t.v., Pan troglodytes verus; G.b.b., Gorilla beringei beringei; G.b.g., Gorilla beringei graueri; G.g.d., Gorilla gorilla diehli; and G.g.g., Gorilla gorilla gorilla.
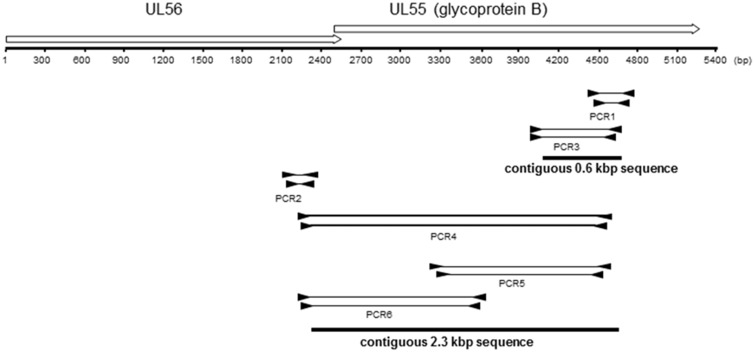
Though CMV1 and CMV2 were clearly distinguishable from one another, the short sequences (0.2 kb) generated from the initial PCR analysis did not exhibit enough genetic variation for in-depth phylogenetic analyses. A preliminary analysis in a maximum likelihood (ML) framework indeed revealed very low support for a vast majority of branches. Therefore, we attempted to generate longer sequences from the fecal samples. Although contiguous regions (contigs) of <0.6 kb were generated with PCR3 in combination with PCR1, PCR4 in combination with PCR1 and PCR2 was unable to amplify longer products (Fig. 3), which was likely due to low copy number and/or limited DNA quality. Given the impossibility to generate a sequence dataset suitable to investigate CMV host subspecies-level evolution and phylogeography, we created a Microreact project based on the abovementioned ML tree to allow us and others to formulate testable hypotheses, should longer sequences be generated. This project can be consulted at: https://microreact.org/project/0qicEkhgV.
Figure 3.
Map of targeted open reading frames (ORFs) and diagram of PCR strategy. Nested primers (black triangles) were used to amplify parts of the UL55 or UL56 ORFs. The amplified fragments are represented by thin solid lines between the primer binding sites. Fragments were sequenced and assembled to final contiguous sequences of 0.6 and 2.3 kbp. At the top of the figure, the genomic locus spanning ORFs UL55 and UL56 is depicted with open arrows. The arrowhead indicates the direction of transcription. The start of the ruler corresponds with the first base of the ORF UL56.
Bonobo CMVs were detected for the first time in this study. To obtain additional sequence information for CMVs from this African great ape species, three blood samples were obtained from captive bonobos. Using PCR1, we identified bonobo CMV1 and CMV2 sequences in two of the three blood samples that were indistinguishable from the respective CMV1 and CMV2 sequences of the wild bonobos. Using PCR5 and PCR6, we were able to amplify a CMV1 and a CMV2 sequence of ∼2.3 kb from these samples, comprising the UL55/UL56 gene loci (Fig. 3). We also tried to obtain larger genomic fragments using hybridization capture. Although this method has already been used to generate CMV genomes (Lassalle et al. 2016) and has already been implemented to generate alphaherpesvirus genomes in our laboratory (Burrel et al. 2017) it did not allow us to collect more information from these samples.
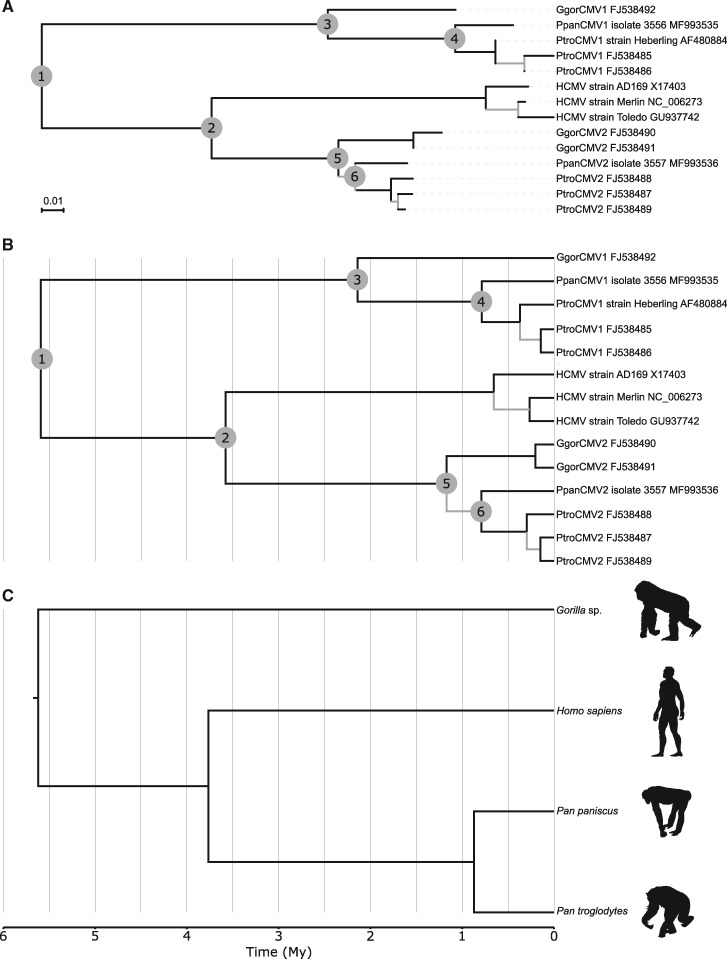
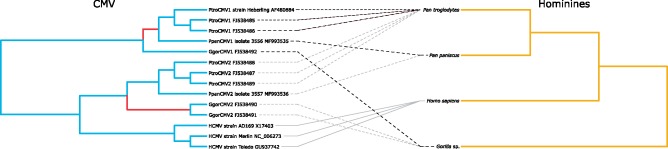
Bonobo CMVs were used to further refine our understanding of CMV evolution within hominines. We first performed phylogenetic analyses in a ML framework, using an alignment comprising the new bonobo CMV sequences and a selection of available hominine CMV sequences (Fig. 4A). The ML tree revealed that bonobo CMV1 and CMV2 were closely related sister taxa of chimpanzee CMV1 and CMV2, respectively. Although this placement did not definitely exclude the transmission model, it was also compatible with an alternative model, wherein the CMV1 and CMV2 lineages independently co-diverged with their African great ape hosts (hereafter ‘co-divergence model’). The potential co-divergence patterns are best illustrated with a tanglegram (Fig. 5).
Figure 4.
Host and CMV phylogenetic trees. (A) ML CMV tree. The scale is in aa substitution per site. (B) Bayesian CMV timetree. (C) Host timetree derived from the divergence dates published by Prado-Martinez et al. (2013) based on genomic analyses. For (A) and (B): branches supported by SH-like aLRT < 0.90 or posterior probability < 0.95 are gray; numbered nodes are discussed in the text. (B) and (C) have been drawn to the same scale that is, node depths are immediately comparable.
Figure 5.
Tanglegram of host and CMV phylogenetic trees. Associations corresponding to the parallel co-divergence in CMV1 and CMV2 according to the co-divergence model are represented with black- and gray-dashed lines, respectively. Branches likely to harbor a transmission event according to the transmission model appear in red in the CMV phylogenetic tree.
Depending on the hypothesis considered (transmission or co-divergence model) different nodes in the phylogenetic tree will correspond to the same host-driven divergence event. For example, the transmission model assumes that Node 1 corresponds to the unique hypothetical CMV that infected the ancestor of all hominines; in contrast the co-divergence model assumes that the ancestor of all hominines was already infected by two hypothetical CMVs represented by Nodes 3 and 5 (Fig. 4A and B). Divergent assumptions on node ages translate into specific predictions regarding node height ratios. We determined these ratios from posterior sets of trees generated by Bayesian Markov chain Monte Carlo (BMCMC) analyses under various uncalibrated clock models (strict, lognormal relaxed, and exponential relaxed). For all models we also estimated marginal likelihoods (Table 3). Using Bayes factor (BF) comparison, strict and lognormal relaxed clock models were nearly indistinguishable and appeared as performing better, although not decisively better according to our criteria (2 ln BF > 10), than the exponential relaxed clock model. Irrespective of the model, node height ratios lent support to the transmission model (Table 4). Median estimates of the ratio Nodes 1/2 fell very close to the ratio derived from host divergence events and the latter was always comprised within the 95% highest posterior density (HPD) intervals of the former. On the contrary, median estimates and 95% HPD intervals of the ratios Nodes 3/4 and Nodes 5/6 appeared as incompatible with the predictions of the co-divergence model. These analyses therefore suggested that the transmission model is a plausible explanation to the observed pattern of CMV genetic diversity in hominines; conversely, the co-divergence model did not seem to adequately describe the evolution of CMVs in this lineage.
Table 3.
Log marginal likelihood values for models with uncalibrated molecular clocks.
| Model | lnL | 2 ln BFa |
|---|---|---|
| Strict clock | −2,132.9 | – |
| Lognormal relaxed clock (uncorrelated) | −2,133.0 | 0.2 |
| Exponential relaxed clock (uncorrelated) | −2,136.2 | 6.6 |
BF calculations all correspond to comparisons to the best model using the same sampling approach (strict clock). 2 ln BF > 0 indicates a better performance of the strict clock model; 2 ln BF > 10 indicates decisive support. The values presented here were all obtained using stepping stone sampling; values obtained with path sampling were very similar.
Table 4.
Height ratios in uncalibrated molecular clock analyses.
| Molecular clock model |
Model and host divergence ratio of referencea | ||||||
|---|---|---|---|---|---|---|---|
| Strict clock |
Lognormal relaxed clock (uncorrelated) |
Exponential relaxed clock (uncorrelated) |
|||||
| Ratiob | Median | 95% HPDc | Median | 95% HPDc | Median | 95% HPDc | |
| Nodes 1/2d | 1.64 | 1.29–2.13 | 1.64 | 1.16–2.35 | 1.39 | 1.00–2.94 | Transmission 1.51 |
| Nodes3/4d | 2.72 | 1.84–4.34 | 2.66 | 1.70–4.66 | 2.11 | 1.26–5.89 | Co-divergence CMV1 6.43 |
| Nodes 5/6d | 2.43 | 1.00–2.27 | 1.42 | 1.00–2.44 | 1.38 | 1.00–3.97 | Co-divergence CMV2 6.43 |
This column gives the expected ratio according to the relevant model of diversification. Ratios determined from the molecular clock analyses should be close to the expected ratio of the model(s) of host/CMV evolution compatible with the data; the data support the transmission model.
Ratios were determined from the indicated node heights in posterior sets of trees generated by uncalibrated molecular clock analyses (height unit: aa substitutions per site).
95% highest posterior density.
According to the transmission model Nodes 1 and 2, respectively correspond to the last common ancestors of all hominines and of the panine and human lineages; according to the co-divergence model Nodes 3 and 5 and 4 and 6, respectively correspond to the last common ancestors of all hominines and of the panine lineage.
To further explore the ability of the two models to account for the observed pattern, we formally compared them, taking advantage of their divergent assumptions on node ages to run BMCMC analyses under clock models with different multiple calibrations, for which marginal likelihoods were also estimated (Table 5). BF comparisons identified the transmission model as the best explanation and it was significantly better than two of the three competing co-divergence models, including the model with simultaneous co-divergence of both CMV1 and CMV2.
Table 5.
Log marginal likelihood values for models with different calibration schemes.
| Model | ln L | 2 ln BFa |
|---|---|---|
| Transmission | −2124.3 | – |
| Co-divergence CMV1 and CMV2 | −2132.7 | 16.8 |
| Co-divergence CMV1 | −2126.2 | 3.8 |
| Co-divergence CMV2 | −2131.9 | 15.2 |
BF calculations all correspond to comparisons to the best model (transmission model). 2 ln BF > 0 indicates a better performance of the transmission model; 2 ln BF > 10 indicates decisive support. The values presented were all obtained using stepping stone sampling; values obtained with path sampling were very similar. All models were run using a lognormal relaxed clock, which we previously identified as one of the two best-performing clock models.
Considering both uncalibrated and calibrated molecular clock analyses, the addition of bonobo CMVs therefore clearly confirmed the transmission model. This model requires that there is (or was) opportunity for virus transmission between the panine and gorilla lineages. Currently, chimpanzees and gorillas live in sympatry in rainforests from Central Africa. The diets of chimpanzees and gorillas overlap significantly and this sometimes results in groups of both species foraging the same fruit trees on the same day (Walsh et al. 2007). Exploiting the same resources provides a plausible route for viral transmission, whether oral-fecal or via contaminated food items. For example, fruit wedges have recently been shown to be contaminated with the genetic material of NHP-infecting viruses, including herpesviruses (Smiley Evans et al. 2016), thereby suggesting cross-species CMV transmission is possible in natural settings.
Molecular clock analyses allowed us to date the bidirectional CMV transmission events (Fig. 4B and 4C). Transmission of CMV1 from gorilla to panine (chimpanzee/bonobos) hosts may have occurred as early as 2.19 My ago (Node 3; 95% HPD: 1.32–3.15 My), while CMV2 transmission from panine hosts to gorillas could have happened 1.20 My ago (Node 5; 95% HPD: 0.68–1.77 My). Interestingly, both events unambiguously predated the divergence of bonobos and chimpanzees (0.87 My ago), and the divergence of bonobo and chimpanzee CMV1 and CMV2 were almost perfectly synchronous with the divergence of their host (Node 4: 0.82 My [0.40–1.26 My]; Node 6: 0.82 My [0.42–1.27 My]), indicating co-divergence of these CMVs with their hosts. In summary, our analyses show a unique and complex evolution of CMVs within their hominine hosts that is closely linked to diversification events of their respective hosts but is also marked by two ancient transmission events between the gorilla and panine lineages.
Until recently, Plasmodium falciparum, HIV-1 and SIVgor were the clearest examples of cross-hominine transmission events, respectively between gorillas and humans, chimpanzees and humans and chimpanzees and gorillas (reviewed in Sharp and Hahn 2011; Loy et al. 2017). These transmission events shared the characteristic of being relatively recent: HIV-1 emergences happened during the 20th century, SIVgor during the last few centuries and P. falciparum in the last 10,000 years (Wertheim and Worobey 2009; Sharp and Hahn 2011; Loy et al. 2017). In the last few years, the notion that specialized hominine-infecting parasites (in an ecological sense) may find their origins in much more ancient transmission events gained much momentum. This is particularly striking when considering viruses with a dsDNA genome: (i) papillomavirus Types 16 and 58 have recently been suggested to originate in archaic humans (>30,000 years ago; Pimenoff, de Oliveira, and Bravo 2016; Chen et al. 2017); (ii) human herpes virus simplex 2 (HSV-2) is thought to have been transmitted from panine to archaic human ancestors 1.6 My ago (Wertheim et al. 2014); and (iii) the gorilla-borne HAdV-B was transmitted from gorillas to humans at least twice (>300,000 years ago) and from gorillas to panines as early as 2.9 My ago (Hoppe et al. 2015). In hominines, the diversity of several dozens of lineages of dsDNA viruses whose evolution is thought to involve a combination of co-speciation and infrequent host switches (including other adenoviruses, herpesviruses, papillomaviruses, and polyomaviruses) still remains to be characterized. Accumulating information about cross-hominine transmission events such as those confirmed in this study will allow us to investigate the temporal dynamics of co-speciation and host switch rates during the last few million years, a period during which the different hominine lineages have interacted in very complex ways.
3. Materials and methods
3.1 Sample collection, DNA isolation, and PCR methods
In total 675 stool samples were collected at 20 sites in 11 sub-Saharan African countries from 9 great ape subspecies (Fig. 1), P.p., n = 33; P.t.e., n = 63; P.t.s., n = 75; P.t.t., n = 52; P.t.v., n = 167; G.b.b., n = 97; G.b.g., n = 79; G.g.d., n = 56; and G.g.g., n = 53. Sampling authorization was obtained from responsible local authorities. Except for G.b.b. and G. b. graueri, fecal samples were collected opportunistically from non-habituated communities; we did not try to determine the number of individuals that were sampled. DNA was isolated using the Stool DNA Kit (Roboklon, Berlin, Germany). Additionally, blood samples were collected from three captive bonobos from the Wilhelma Zoological garden in Stuttgart, Germany, and DNA isolated with the Qiagen blood and tissue kit (Qiagen, Hilden, Germany).
For PCR, the nested primer sets were based on conserved sequence regions of betaherpesviruses (PCR1 and 2) or solely, on primate CMVs (PCR3 and 4) and bonobo CMVs (PCR5 and 6), and are listed in Table 1. For generic amplification of CMV glycoprotein B (UL55 - gB) sequence (0.2 kb; PCR1) and UL56 sequence (0.14 kb; PCR2), PCR was carried out as previously described (Murthy et al. 2013). PCR 3 was used to obtain extended CMV UL55 sequences from bonobos, chimpanzees and gorillas, and was performed using the same cycler settings as PCR 1 and 2 with an exception of annealing temperature. PCR 4 was used for amplification of 2.3 kb sequences (extending from UL55 to UL56) of bonobos, chimpanzees and gorillas, and PCR5 for amplification of 1.3 kb UL55 sequences of bonobo CMVs. Both were performed with the TaKaRa-Ex PCR system (TaKaRa Bio) according to the manufacturer’s instructions. PCR 6 amplified 1.2 kb of bonobo CMVs and was performed with the AmpliTaq Gold PCR system (Applied Biosystems, Warrington, UK). PCR (PCR 7; Table 1). To confirm host species, the cytochrome b sequence was amplified using AmpliTaq Gold PCR system (Applied Biosystems) with PCR7 primers (Table 1). Sequencing reactions were performed with the Big Dye terminator cycle sequencing kit (Applied Biosystems) and products analyzed on a 377 automated DNA sequencer (Applied Biosystems).
3.2 Bioinformatic and phylogenetic analysis
Short sequences determined during the screening phase were only used to confirm that CMVs, or CMV1 or CMV2, had been detected. This was done using BLAST (Altschul et al. 1990) for CMV sequences and by aligning sequences and running a ML analysis with PhyML with smart model selection (Guindon et al. 2005, 2010; Lefort, Longueville, and Gascuel 2017) using the SPR tree search and assessing branch robustness with Shimodaira-Hasegawa-like approximate likelihood ratio test (SH-like aLRT; Anisimova et al. 2011). Although many branches in the resulting tree were poorly supported, it provided a unique opportunity to co-plot information on host species/subspecies and geographical origin, which we did using Microreact (Argimon et al. 2016). The project is available at: https://microreact.org/project/0qicEkhgV.
The longer bonobo CMV1 and CMV2 amino acid (aa) sequences were aligned with a set of twelve references hominine CMV aa sequences using Muscle (Edgar 2004) as implemented in SeaView v4 (Gouy, Guindon, and Gascuel 2010). We identified conserved blocks in the alignment using Gblocks (Talavera and Castresana 2007) as implemented in SeaView. This alignment was back-translated to the original nucleotide alignment and examined for evidence of recombination using RDP4 with default settings and requiring that at least two methods agree to validate a recombination event (Martin et al. 2015). We identified unambiguous recombination events, leading us to reduce the alignment to the largest block not comprising any breakpoint likely to affect our analyses. This block covered a total of 933 nucleotide positions (311 aa positions), all located in the coding sequence of the UL55 gene. At these positions no recombination was detectable, except between very closely related CMV strains infecting the same host species (HCMV and ProCMV1). HCMV is known to recombine frequently. UL55 however exhibits the fourth highest linkage disequilibrium score in the HCMV genome (Lassalle et al. 2016). Lassalle et al. (2016) suggest that recombination methods similar to those employed here can lead to false positive detection events for genes which like UL55 show high diversity and rate variation across their sequences. Therefore, it seems plausible that a number of the recombination events that we detected be artifacts, all the more so since the recombinant sequences themselves were not generated by this study (also raising the untestable question of in vitro recombination). Our decision to focus all following analyses on this relatively recombination-free block of aa sequences was a conservative one.
We performed model selection using ProtTest v2.4 (Darriba et al. 2011); model likelihoods were compared using the Bayesian information criterion and the selected model was JTT+G. We then ran phylogenetic analyses in ML and Bayesian frameworks. We reconstructed a ML tree using PhyML v3 (Guindon et al. 2010) using the BEST tree search and assessing branch robustness with SH-like aLRT. This ML tree, a host tree and their tip associations were used to generate a tanglegram with TreeMap v3b (Jackson and Charleston 2004).
We also ran BMCMC analyses using BEAST v1.8.2 (Drummond et al. 2012). In a first set of analyses, we tested a strict clock, a lognormal relaxed clock and an exponential relaxed clock, always modeling the tree shape using a birth–death model (multiple independent runs were performed for all models). We checked run convergence and appropriate sampling behavior using Tracer v1.7 (Rambaut et al. 2018). To be able to compare model performance we also estimated their marginal likelihoods using path and stepping stone sampling. BF comparisons were considered to convincingly support a model when 2 ln BF > 10. Posterior sets of trees (PST) were used to calculate node height ratios relevant to the transmission and co-divergence models. All heights were extracted from PST using TreeStat v1.8.2.
We then ran an additional set of BMCMC analyses, this time using four calibrated models which differed only with respect to their calibration points (all models used a lognormal relaxed clock; see Table 3). To be able to compare these different models at least two calibration points per model had to be defined, imposing a constraint on some relative branch lengths. All calibration points can be seen in Fig. 4; the respective dates are all derived from a large African great ape genomic study (Prado-Martinez et al. 2013). The first model was defined to fit the transmission model: the age of Node 1 was calibrated to correspond to the time to the most recent common ancestor (tMRCA) of all hominines using a normal distribution of mean 5.6 My and SD 0.5 My; the age of Node 2 was calibrated to fit the time to the MRCA of humans and panines using a normal distribution of mean 3.7 My and SD 0.35 My. The second model was defined to fit a scenario of complete co-divergence within the CMV1 and CMV2 lineages: the age of Nodes 3 and 5 was set to fit the tMRCA of all hominines using a normal distribution of mean 5.6 My and standard deviation (SD) 0.5 My while the age of Nodes 4 and 6 was calibrated to correspond to the divergence of all panines using a normal distribution of mean 0.87 My and SD 0.08 My. The third and fourth model used the same calibrations as the second model but only applied it to one of the CMV lineages that is, CMV1 or CMV2. Marginal likelihoods of the models were also estimated using path and stepping stone sampling. Run validation and model comparison were performed as mentioned earlier. PST from multiple runs were combined using LogCombiner v1.8.2 and summarized onto the maximum clade credibility tree identified with TreeAnnotator v1.8.2. Branch robustness was assessed using their posterior probability in PST.
Two exemplary XML files corresponding to one of the uncalibrated analyses performed under a lognormal relaxed clock and one of the calibrated analyses performed under a lognormal relaxed clock are available as Supplementary Material.
3.3 Provisional nomenclature, abbreviations, and nucleotide sequence accession numbers for the novel herpesviruses
The viruses from which the novel sequences originated were named after the host species name and the herpesvirus genus to which the virus was tentatively assigned, for example, Panpaniscuscytomegalovirus, PpanCMV. The genotypic variants of PpanCMV that were related more closely to CCMV than to HCMV (CG1) were named PpanCMV1, while those closely related to HCMV (CG2) were named PpanCMV2. The previously published variants of gorilla CMV (GgorCMV1 and 2), chimpanzee CMV (PtroCMV1 and 2), and orangutan CMV (PpygCMV1) were named accordingly (Leendertz et al. 2009). All novel viruses and previously reported viruses, whose UL55 sequences were used for phylogenetic comparison, are listed with their abbreviations and GenBank accession numbers in Table 6.
Table 6.
CMV sequences used in phylogenetic analysis, accession numbers and abbreviations.
| Virus name | Host species/subspecies | GenBank accession number | Abbreviation used in phylogenetic tree |
|---|---|---|---|
| Human CMV (Human herpesvirus 5) | |||
| Strain Merlin | Homo sapiens | NC_006273 | HCMV strain Merlin NC_006273 |
| Strain Toledo | Homo sapiens | GU937742 | HCMV strain Toledo AC146905 |
| Strain AD169 | Homo sapiens | X17403 | HCMV strain AD169 X17403 |
| Great ape CMVs | |||
| PpanCMV1 | Pan paniscus | MF993535 | PpanCMV1 isolate 3556 MF993535 |
| PpanCMV 2 | Pan paniscus | MF993536 | PpanCMV2 isolate 3557 MF993536 |
| Pan troglodytes cytomegalovirus 1.1 | Pan troglodytes verus | FJ538485 | PtroCMV1 FJ538485 |
| Pan troglodytes cytomegalovirus 1.2 | Pan troglodytes verus | FJ538486 | PtroCMV1 FJ538486 |
| Panine betaherpesvirus 2 | Pan troglodytes verus | AF480884 | PtroCMV1 strain Heberling AF480884 |
| Pan troglodytes cytomegalovirus 2.1 | Pan troglodytes verus | FJ538487 | PtroCMV2 FJ538487 |
| Pan troglodytes cytomegalovirus 2.2 | Pan troglodytes verus | FJ538488 | PtroCMV2 FJ538488 |
| Pan troglodytes cytomegalovirus 2.3 | Pan troglodytes verus | FJ538489 | PtroCMV2 FJ538489 |
| Gorilla gorilla cytomegalovirus 1.1 | Gorilla gorilla gorilla | FJ538492 | GgorCMV1 FJ538492 |
| Gorilla gorilla cytomegalovirus 2.1 | Gorilla gorilla gorilla | FJ538490 | GgorCMV2 FJ538490 |
| Gorilla gorilla cytomegalovirus 2.2 | Gorilla gorilla gorilla | FJ538491 | GgorCMV2 FJ538491 |
Supplementary Material
Acknowledgements
We are very grateful to the national authorities of Cameroon, the Central African Republic, Côte d’Ivoire, the Democratic Republic of Congo, Gabon, Guinea, Liberia, Nigeria, Rwanda, Senegal, and Uganda for granting authorizations to and supporting the great ape research programs which allowed for the collection of the fecal samples used in this study. We also thank F. Aubert, C. Boesch, K. Corogenes, L. D’Auvergne, T. Desarmeaux, O. Diotoh, A. Dunn, G. Hohmann, I. Imong, K. J. Jeffery, D. Kujirakwinja, N. Maldonado, G. Maretti, V. Mihindou, G. Mitamba, R. Nishuli, S. Regnaut, A. Tickle, and K. Zuberbuehler for their help with sample collection. Samples from Bwindi Impenetrable National Park, Campo Ma’an National Park, Loango National Park, Gashaka Gumti National Park, Mbe Mountains Community Forest, Korup National Park, Mount Cameroon National Park, Budongo Forest, Kibale National Park, Kayan, Sangaredi, Sobeya, GEPRENAF, and East Nimba were specifically collected as part of the Pan African Program: The Cultured Chimpanzee (PanAf). For the PanAf program, Sample collection was completed with the generous support of the following government agencies: Ministere de la Recherche Scientifique et de l’Innovation & Ministere des Forets et de la Faune (Cameroon), Ministere des Eaux et Forets (Cote d’Ivoire), Institut Congolais pour la Conservation de la Nature & Ministere de la Recherche Scientifique (DRC), Agence Nationale des Parcs Nationaux & Centre National de la Recherche Scientifique (Gabon), Ministere de l'Agriculture de l'Elevage et des Eaux et Forets (Guinea), Forestry Development Authority (Liberia), National Park Service & Nigeria Conservation Society of Mbe Mountains (Nigeria), Direction des Eaux, Forêts et Chasses (Senegal), Uganda National Council for Science and Technology, Uganda Wildlife Authority & Makerere University Biological Field Station (Uganda). Funding for the PanAf was generously provided by the Max Planck Society Innovation Fund, and Heinz L. Krekeler Foundation.
Conflict of interest: None declared.
References
- Altschul S. F. et al. (1990) ‘Basic Local Alignment Search Tool’, Journal of Molecular Biology, 215: 403–10. [DOI] [PubMed] [Google Scholar]
- Anisimova M. et al. (2011) ‘Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-Based Approximation Schemes’, Systematic Biology, 60: 685–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anoh A. E. et al. (2018) ‘Cytomegaloviruses in a Community of Wild Nonhuman Primates in Taï National Park, Côte D’Ivoire’, Viruses, 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argimon S. et al. (2016) ‘Microreact: Visualizing and Sharing Data for Genomic Epidemiology and Phylogeography’, Microbial Genomics, 2: e000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad A., Katzourakis A. (2014) ‘The First Endogenous Herpesvirus, Identified in the Tarsier Genome, and Novel Sequences from Primate Rhadinoviruses and Lymphocryptoviruses’, PLoS Genetics, 10: e1004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrel S. et al. (2017) ‘Ancient Recombination Events between Human Herpes Simplex Viruses’, Molecular Biology and Evolution, 34: 1713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwitz B. J. et al. (2016) ‘Cross-Species Rhesus Cytomegalovirus Infection of Cynomolgus Macaques’, PLoS Pathogens, 12: e1006014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. et al. (2017) ‘Ancient Evolution and Dispersion of Human Papillomavirus 58 Variants’, Journal of Virology, 91: e01285-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D. et al. (2011) ‘ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution’, Bioinformatics (Oxford, England), 27: 1164–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J. et al. (2012) ‘Bayesian Phylogenetics with BEAUti and the BEAST 1.7’, Molecular Biology and Evolution, 29: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput’, Nucleic Acids Research, 32: 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B. et al. (2007) ‘Identification of Novel Rodent Herpesviruses, Including the First Gammaherpesvirus of Mus musculus’, Journal of Virology, 81: 8091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010) ‘SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building’, Molecular Biology and Evolution, 27: 221–4. [DOI] [PubMed] [Google Scholar]
- Guindon S. et al. (2005) ‘PHYML Online–A Web Server for Fast Maximum Likelihood-Based Phylogenetic Inference’, Nucleic Acids Research, 33: W557–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Hoppe E. et al. (2015) ‘Multiple Cross-Species Transmission Events of Human Adenoviruses (HAdV) during Hominine Evolution’, Molecular Biology and Evolution, 32: 2072–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff J. L., Barry P. A. (2003) ‘B-Virus (Cercopithecine Herpesvirus 1) Infection in Humans and Macaques: Potential for Zoonotic Disease’, Emerging Infectious Diseases, 9: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Charleston M. A. (2004) ‘A Cophylogenetic Perspective of RNA-Virus Evolution’, Molecular Biology and Evolution, 21: 45–57. [DOI] [PubMed] [Google Scholar]
- James S. et al. (2018) ‘DNA Polymerase Sequences of New World Monkey Cytomegaloviruses: Another Molecular Marker with Which to Infer Platyrrhini Systematics’, Journal of Virology, 92: e00980-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafemina R. L., Hayward G. S. (1988) ‘Differences in Cell-Type-Specific Blocks to Immediate Early Gene Expression and DNA Replication of Human, Simian and Murine Cytomegalovirus’, Journal of General Virology, 69: 355–74. [DOI] [PubMed] [Google Scholar]
- Lassalle F. et al. (2016) ‘Islands of Linkage in an Ocean of Pervasive Recombination Reveals Two-Speed Evolution of Human Cytomegalovirus Genomes’, Virus Evolution, 2: vew017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leendertz F. H. et al. (2009) ‘Novel Cytomegaloviruses in Free-Ranging and Captive Great Apes: Phylogenetic Evidence for Bidirectional Horizontal Transmission’, Journal of General Virology, 90: 2386–94. [DOI] [PubMed] [Google Scholar]
- Lefort V., Longueville J. E., Gascuel O. (2017) ‘SMS: Smart Model Selection in PhyML’, Molecular Biology and Evolution, 34: 2422–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja A. E., Shenk T. (2008) ‘Efficient Replication of Rhesus Cytomegalovirus Variants in Multiple Rhesus and Human Cell Types’, Proceedings of the National Academy of Sciences of the United States of America, 105: 19950–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy D. E. et al. (2017) ‘Out of Africa: Origins and Evolution of the Human Malaria Parasites Plasmodium falciparum and Plasmodium vivax’, International Journal for Parasitology, 47: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P. et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, Virus Evolution, 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Rixon F. J., Davison A. J. (2006) ‘Topics in Herpesvirus Genomics and Evolution’, Virus Research, 117: 90–104. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J. et al. (2008), ‘Molecular Evolution of the Herpesvirales’, in Origin and Evolution of Viruses, 2nd edn Amsterdam: Elsevier, pp. 447–75. [Google Scholar]
- Michaels M. G. et al. (1997) ‘Distinguishing Baboon Cytomegalovirus from Human Cytomegalovirus: Importance for Xenotransplantation’, The Journal of Infectious Diseases, 176: 1476–83. [DOI] [PubMed] [Google Scholar]
- Murthy S. et al. (2013) ‘Absence of Frequent Herpesvirus Transmission in a Nonhuman Primate Predator-Prey System in the Wild’, Journal of Virology, 87: 10651–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oya C. et al. (2004) ‘Specific Detection and Identification of Herpes B Virus by a PCR-Microplate Hybridization Assay’, Journal of Clinical Microbiology, 42: 1869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perot K., Walker C. M., Spaete R. R. (1992) ‘Primary Chimpanzee Skin Fibroblast Cells Are Fully Permissive for Human Cytomegalovirus Replication’, Journal of General Virology, 73: 3281–4. [DOI] [PubMed] [Google Scholar]
- Pimenoff V. N., de Oliveira C. M., Bravo I. G. (2016) ‘Transmission between Archaic and Modern Human Ancestors during the Evolution of the Oncogenic Human Papillomavirus’, Molecular Biology and Evolution, 34: 16–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Martinez J. et al. (2013) ‘Great Ape Genetic Diversity and Population History’, Nature, 499: 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prepens S. et al. (2007) ‘Discovery of Herpesviruses in Multi-Infected Primates Using Locked Nucleic Acids (LNA) and a Bigenic PCR Approach’, Virology Journal, 4: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. et al. (2018) ‘Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7’, Systematic Biology, 67: 901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell G. C., Stewart J. P., Haig D. M. (2009) ‘Malignant Catarrhal Fever: A Review’, Veterinary Journal (London, England: 1997), 179: 324–35. [DOI] [PubMed] [Google Scholar]
- Schrenzel M. D. et al. (2003) ‘Naturally Occurring Fatal Herpes Simplex Virus 1 Infection in a Family of White-Faced Saki Monkeys (Pithecia pithecia pithecia)’, Journal of Medical Primatology, 32: 7–14. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Hahn B. H. (2011) ‘Origins of HIV and the AIDS Pandemic’, Cold Spring Harbor Perspectives in Medicine, 1: a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley Evans T. et al. (2016) ‘Detection of Viruses Using Discarded Plants from Wild Mountain Gorillas and Golden Monkeys’, American Journal of Primatology, 78: 1222–34. [DOI] [PubMed] [Google Scholar]
- Talavera G., Castresana J. (2007) ‘Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks From Protein Sequence Alignments’, Systematic Biology, 56: 564–77. [DOI] [PubMed] [Google Scholar]
- Walsh P. D. et al. (2007) ‘Potential for Ebola Transmission Between Gorilla and Chimpanzee Social Groups’, TheAmerican Naturalist, 169: 684–9. [DOI] [PubMed] [Google Scholar]
- Wertheim J. O., Worobey M. (2009) ‘Dating the Age of the SIV Lineages That Gave Rise to HIV-1 and HIV-2’, PLoS Computational Biology, 5: e1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J. O. et al. (2014) ‘Evolutionary Origins of Human Herpes Simplex Viruses 1 and 2’, Molecular Biology and Evolution, 31: 2356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.