Abstract
The neuronal intermediate filament protein peripherin is a component of ubiquitinated inclusions and of axonal spheroids in amyotrophic lateral sclerosis (ALS). Overexpression of peripherin causes motor neuron degeneration in transgenic mice and variations within the peripherin gene have been identified in ALS cases. We have shown previously the abnormal expression of a neurotoxic peripherin splice variant in transgenic mice expressing mutant superoxide dismutase-1. These findings indicated that abnormalities of peripherin splicing may occur in ALS. In the current study, peripherin splice variants were identified by reverse transcription-PCR of human neuronal RNA and comparisons in expression made between control and ALS spinal cord using Western blot analysis and immunocytochemistry. Using this approach we have identified a novel peripherin transcript retaining introns 3 and 4 that results in a 28 kDa splice isoform, designated Per 28. Using an antibody specific to Per 28, we show that this isoform is expressed at low stoichiometric levels from the peripherin gene, however causes peripherin aggregation when its expression is upregulated. Importantly we show an upregulation of Per 28 expression in ALS compared with controls, at both the mRNA and protein levels, and that Per 28 is associated with disease pathology, specifically round inclusions. These findings are the first to establish that peripherin splicing abnormalities occur in ALS, generating aggregation-prone splice isoforms.
Keywords: peripherin, splice variant, isoform, ALS, intron retention, intermediate filament
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disease affecting motor neurons of the motor cortex, brainstem, and spinal cord. ∼10–15% of cases are inherited [familial (fALS)] with the remainder occurring sporadically [sporadic (sALS)]. Mutations in the gene encoding superoxide dismutase-1 (SOD1) are causative of ∼20% of fALS cases, corresponding to 1–2% of all ALS cases (Rosen et al., 1993). Sporadic and familial ALS are clinically and pathologically indistinguishable, sharing a number of intraneuronal inclusion bodies that are characteristic of the disease (for review, see Hays, 2006). These include ubiquitinated inclusions that comprise skein-like inclusions, round inclusions and Lewy body-like inclusions (Ince et al., 1998a; Hays, 2006). Hyaline conglomerate inclusions are also ubiquitinated, but appear to be more closely linked with fALS cases carrying mutations in SOD1 (Ince et al., 1998b; Hays et al., 2006). Peripherin, a neuronal intermediate filament protein, is associated with ubiquitinated inclusions, specifically round inclusions and Lewy body-like inclusions, hyaline conglomerate inclusions, as well as with axonal spheroids, large swellings that occur in proximal axons of diseased motor neurons (Corbo and Hays, 1992; Migheli et al., 1993; He and Hays, 2004; Xiao et al., 2006). Together with the association of peripherin with intraneuronal inclusions, there is also a generalized increased in peripherin immunoreactivity in both the perikarya and axons of motor neurons affected by the disease (Robertson et al., 2002; Robertson et al., 2003; Xiao et al., 2006). Overexpression of peripherin induces motor neuron degeneration in transgenic mice and variations within the peripherin gene have been identified in a few ALS cases. This combined evidence supports a role for peripherin in the pathogenesis of ALS.
We have shown previously the deregulated expression of peripherin splice variants in transgenic mouse models of ALS (Robertson et al., 2003). In particular, we showed the abnormal expression of a neurotoxic splice variant of peripherin, Per 61, in motor neurons of mutant SOD1 transgenic mice (Robertson et al., 2003). This splice variant was generated by the retention of intron 4 and its upregulated expression induced peripherin aggregate formation and motor neuronal death (Landon et al., 1989, 2000; Robertson et al., 2003). These findings indicated that abnormalities of peripherin splicing may be relevant to the disease mechanisms in ALS.
In this regard, we set out to find the human equivalent of Per 61. However, in mouse, intron 4 is 96 bp in length, whereas in human it is 91 bp. As such, the complete retention of intron 4 in human would lead to a frameshift and generation of a C-terminally truncated protein of 32 kDa. Therefore the splicing event that generates Per 61 in mouse cannot occur in human. Nevertheless, the existence of a human peripherin expressed sequence tag (EST) sequence retaining part of intron 4 indicated that read through into this intron could occur. In our attempt to amplify transcripts retaining intron 4, we inadvertently identified a transcript retaining introns 3 and 4. This transcript encodes a C-terminally truncated protein of 28 kDa derived from translation of the 5′ end of peripherin up to a stop codon 30 bp into intron 3. This peripherin splice variant, designated Per 28, is the first to be identified in human and is completely distinct from Per 61 in mouse. We have shown that there is upregulated expression of Per 28 at both the mRNA and protein levels in ALS, and that this may be associated with inclusion body formation. Collectively our findings establish that splicing abnormalities of peripherin occur in ALS generating an aggregation-prone splice variant, Per 28.
Materials and Methods
RNA and semiquantitative RT-PCR.
Human RNA samples for reverse transcription (RT)-PCR were obtained commercially either from Clontech (Palo Alto, CA) or Ambion (Austin, TX) and treated with DNase before RT-PCR to remove any genomic DNA contamination of the RNA sample. Control RT-PCR experiments with omission of the reverse transcriptase were performed in parallel with all experiments to further ensure results obtained were not caused by genomic contamination. Premium total RNA from human dorsal root ganglia was from Clontech (catalog number CR2496). Control total RNA samples from human lumbar spinal cord was pooled from 49 male/female Caucasians age 15 to 66 (catalog number 64113-1; Clontech), and from a normal 65-year-old male Caucasian (catalog number B6840; Ambion). Total RNA from ALS human lumbar spinal cord (catalog number B6162; Ambion) was from two 70-year-old male Caucasians. The cDNAs were synthesized from 1 μg of total RNA with oligo-(dT)20 using the SuperScript III First-Strand Synthesis System for RT-PCR from Invitrogen (Burlington, Ontario, Canada) following the manufacturer's protocol. The ratio of mRNA transcripts was estimated using semiquantitative RT-PCR normalized using primers specific for β-actin (Ambion) and choline acetyltransferase (ChAT) (for primer sequences, see Table 1). For amplifying the normal peripherin gene transcript PRPH, primers were located in exons 3 and 6 (Table 1). For PCR, 1 μl of template cDNA solution was placed in 25 μl of reaction solution containing 1x PCR buffer (20 mm Tris-HCl, pH 8.4, 50 mm KCl), 200 μm dNTP, 2 mm MgCl2,5% DMSO, 2 μm each primer, and 2 U Platinum TaqDNA Polymerase (Invitrogen). The amplification conditions for the peripherin splice variant consisted of initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 30 s in a GeneAmp PCR system 9700 (Applied Biosystems, Streetsville, Ontario, Canada). Cycle numbers for the RT-PCR amplifications of β-actin, ChAT, and PRPH were 25, 35, and 30 cycles, respectively.
Table 1.
Primer sequences
| Name | Sequence | Location | Length |
|---|---|---|---|
| F1 | 5′-GTGGACGATGCCACTCTGTC-3′ | Exon 3 | |
| R1 | 5′-GGTCCGCGTACTGAGAAGTG-3 | Intron 4/ Exon 5 junction | 701 and 339 bp |
| F2 | 5′-TCGACTTCTCCATGGCCGAG-3′ | Exon 1 | |
| R2 | 5′-CGAGTCCAGAGGCCAGGTTTC-3 | Intron 3 | 683 bp |
| PRPH-Fwd | 5′-GTGGACGATGCCACTCTGTC-3′ | Exon 3 | |
| PRPH-Rvs | 5′-CTGGTACTCCCTCAGGTGC-3′ | Exon 6 | 534 bp |
| ChAT-Fwd | 5′-CTCCAATTGGCCTGCTGACGTC | Exon 7 | |
| ChAT-Rvs | 5′-GGACTTGTCGTACCAGCGATTG-3′ | Exon 8 | 233 bp |
Fwd, Forward; Rvs, reverse.
Cloning of full-length Per 3,4 cDNA using 5′ and 3′ RACE.
According to the sequence of intron 4 of peripherin obtained from the University of California at Santa Cruz (UCSC) Genome Browser (accession number, NM_006262), we designed 5′ and 3′ gene-specific primers (GSPs): an antisense primer (GSP1) for 5′-rapid amplification of cDNA ends (RACE) PCR (5′-GAC AGG TCC GCG TAC TGA GAA GTG G-3′) and a sense primer (GSP2) for 3′-RACE PCR (5′-GTC CAA GGT GCA AGA GCC GGG AGG-3′). 5′-RACE-ready cDNA was synthesized by combining 1 μg of total RNA with BD SMART II A Oligonucleotide (BD Biosciences, Mississauga, Ontario, Canada) and 5′-RACE coding sequence (CDS) primer; 3′-RACE-ready cDNA was synthesized by combining 1 μg of total RNA with 3′-CDS primer. CDS primers were supplied by the manufacturer. The detailed procedure is exactly as described in the BD SMARTTM RACE cDNA Amplification Kit user manual (BD Biosciences; catalog number 634914). Fifty microliters of RACE PCR solution consisted of 2.5 μl of 5′-RACE-ready cDNA or 3′-RACE-ready cDNA, 34.5 μl of PCR-grade water, 5 μl of 10x Advantage 2 PCR buffer, 1 μl of 10 mm dNTP, 1 μl of 50x BD Advantage 2 Polymerase Mix, 5 μl of 10x Universal Primer Mix, and 1 μl of 10 μm GSP1 or GSP2. Thermal cycling was followed using the touchdown PCR program: five cycles at 94°C for 30 s, 72°C for 3 min; five cycles at 94°C for 30 s, 70°C for 30 s, 72°C for 3 min; and 30 cycles at 94°C for 30 s, 68°C for 30 s, 72°C for 3 min. The 5′-RACE and 3′-RACE PCR products were analyzed by electrophoresis on 1.2% (w/v) agarose/ethidium bromide gels and purified using the MiniElute Gel Extraction kit (catalog number 28604; Qiagen, Mississauga, Ontario, Canada). The isolated fragments were cloned directly into a topoisomerase (TOPO) cloning vector: pCR2.1-TOPO (catalog number K4510-20; Invitrogen). The inserts were sequenced with M13 forward and reverse primers on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). The full-length cDNAs were generated by blunt-end ligation of the 5′ fragment and 3′ fragment, then subcloned into the BamH1/EcoRI sites of pcDNA3.1 (catalog number V795-20; Invitrogen) and subjected to sequence analysis for final verification.
Cloning of full-length human peripherin gene PRPH.
A probe specific to the human peripherin gene was used to identify bacterial artificial chromosome (BAC) clone number 977B10 from the RPCI-11 Human BAC Clones available from the Canadian Institutes of Health Research Genome Resource Facility. The BAC DNA was extracted from a bacterial culture inoculate using alkaline lysis and the full-length human peripherin gene was obtained from 977B10 BAC DNA by simultaneous digestion with EcoRI and EcoRV. A band corresponding to ∼5000 bp was purified using the MiniElute Gel Extraction Kit (Qiagen), subcloned into the EcorR1/EcoRV site of pcDNA3.1(−), and subjected to sequence analysis for verification.
Transient transfection.
A human adrenal carcinoma cell line, SW13 vimentin (−) [SW13vim(−)], was transfected using Lipofectamine 2000 (Invitrogen) following manufacturer instructions. Ectopic expression of peripherin was detected using peripherin polyclonal antibody (AB1530; Millipore, Temecula, CA).
Preparation of siRNA targeting Per 3,4.
The selected small interfering RNA (siRNA) sequences for Per 3,4 was from nucleotides 129–137 for RNAi-1 and 262–280 for RNAi-2, relative to the start of intron 3. The sequences were designed using an siRNA design program (Dharmacon, Lafayette, CO) and a BLAST search revealed no substantial homology of the chosen sequences to other genes. The oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA) and diluted to 1 μg/μl. The forward and reverse primers were annealed and cloned into linearized pSupressorNeo vector according to the manufacturer's protocol (Imgenex, San Diego, CA).
Immunocytochemistry of cultured cells.
SW13vim(−) cells grown on glass coverslips were fixed in methanol for 5 min at −20°C and rehydrated in PBS. Immunocytochemistry was performed using an antibody recognizing peripherin (AB1530) and used at 1:1000 diluted in PBS. Antibody distribution was visualized by epifluorescence microscopy after incubation with secondary antibodies, with Alexa Fluor 594 diluted 1:350 in PBS (Invitrogen).
Immunoblotting.
Cells were harvested in 62.5 mm Tris, pH 6.8, containing 2% SDS and 10% glycerol, and assayed for total protein using the bicinchoninic acid assay (Sigma, St. Louis, MO). Loadings of 10–100 μg of protein were analyzed on 10% (w/v) SDS-polyacrylamide gels and then blotted to polyvinyldiflouride membrane. For immunoblotting, membranes were incubated with antibodies recognizing peripherin (MAB1527 or AB1530) diluted 1:5,000 in blocking solution (3% skim milk powder in TBS-Tween). To detect the Per 28 splice variant, a rabbit polyclonal antiserum was raised to a synthetic peptide corresponding to the sequence derived from translation of the first 30 bp of intron 3 (VSGPGIRGGF) and used at a dilution of 1:3000. Antibody binding was revealed with the ECL detection system (PerkinElmer, Waltham, MA).
Primary motor neuron cultures.
The same protocol was used as described previously for our studies on Per 61 (Robertson et al., 2003). In brief, dissociated spinal cord cultures were prepared as described previously (Durham et al., 1997; Robertson et al., 2001, 2003). The Per 3,4 cDNA in pcDNA 3.1 at 100 ng/ul was microinjected into motor neuron nuclei along with the fluorescent marker dextran-FITC (15 mg/ml; Invitrogen). Viability was assessed daily by counting the number of motor neurons containing the marker. The number of viable motor neurons counted on each day was normalized to the number present on day 1 after microinjection. Experiments were performed in triplicate. For immunocytochemistry, cultures were fixed in 4% paraformaldehyde in PBS for 15 min, and then in blocking solution [as for the SW13vim(−) cells] for 30 min. Double labeling was performed using the Per 28 antibody (diluted 1:1000) and monoclonal peripherin antibody (MAB1527, diluted 1:1000) followed by secondary antibody detection, Alexa Fluor 594 or 488, respectively.
Human spinal cord tissue.
Triton X-100 (TX-100) extractions were prepared from ∼100 mg of lumbar spinal cord tissue taken from four sporadic ALS cases and four controls. The control cases comprised one with no indication of neurological disease and three with neurological disease (two multisystems atrophy and one corticobasal degeneration). Briefly, samples were homogenized in 1 ml high salt buffer (HSB; 50 mm Tris, pH7.5, 750 mm NaCl, 5 mm EDTA) containing protease inhibitor mixture (Sigma) and centrifuged at high speed in an Eppendorf (Mississauga, Ontario, Canada) microcentrifuge at 4°C for 10 min. The resulting pellet was rehomogenized in HSB containing 1% TX-100 and centrifuged as before. This extraction was repeated twice. The final pellet was then homogenized in HSB containing 1M sucrose and centrifuged as before to remove myelin. Equal protein amounts from the different samples were resolved by 10% SDS-PAGE and probed by immunoblotting with Per 28-specific antisera or polyclonal peripherin antibody (AB1530). For immunocytochemistry, paraffin embedded sections (6 μm) of lumbar spinal cord were rehydrated through a series of washes in graded ethanol and finally in water. Sections were pretreated with 10 mm sodium citrate, pH 6.0, for epitope retrieval and incubated with Per 28 splice variant-specific antisera diluted 1:1000 in DakoCytomation (Mississauga, Ontario, Canada) Antibody Diluent overnight at 4°C. Antibody labeling was revealed using the DakoCytomation EnvisonTM System according to the manufacturer's instructions using 3,3′-diaminobenzidine as chromagen. Labeled sections were visualized using a Leica (Richmond Hill, Ontario, Canada) DM 6000 microscope and digital images obtained with a MicroPublisher 3.3 RTV color camera and Openlab imaging software (Improvision, Lexington, MA).
Results
Identification of a peripherin transcript retaining introns 3 and 4
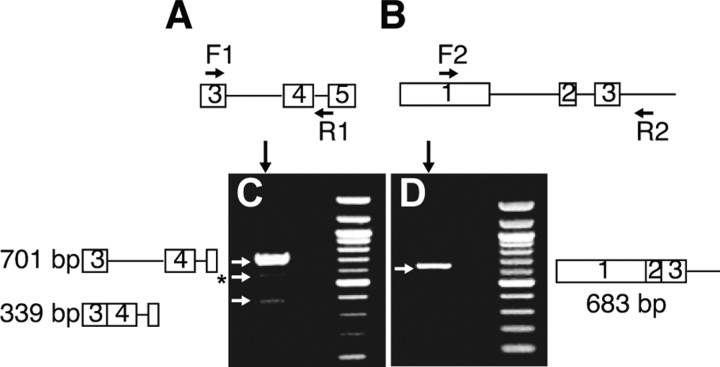
We identified two EST sequences corresponding to splice variants of human peripherin that retain part of intron 4 or intron 3 (BE786797 and BI832203, respectively) using the UCSC Genome Browser. Primers for RT-PCR were designed according to the retained sequence of intron 4 because we had found that peripherin splicing events involving retention of intron 4 occurred in mutant SOD1 transgenic mice (Robertson et al., 2003). Premium total RNA extracted from human dorsal root ganglia (Clontech) was chosen as the source for RT-PCR to identify peripherin splice variants because peripherin is highly expressed in this tissue (Wong and Oblinger, 1990). Because our aim was to identify splicing events involving intron retention, a number of steps were taken to avoid possible confounding results caused by genomic contamination. First, all of the RNA samples were treated with DNase before RT-PCR. Second, control experiments were performed in which reverse transcriptase was omitted showing that there was no PCR amplification of genomic DNA. Third, the first strand synthesis was performed using oligo-(dT)20 to amplify mRNA only. Fourth, parallel RT-PCR experiments were performed using off-target primers located in exon 1 and intron 3 (Fig. 1B). The product generated from these reactions had introns 1 and 2 spliced out (Fig. 1D, arrow), confirming that our samples were not contaminated with genomic DNA. Using a forward primer located in exon 3 of peripherin and a reverse primer in intron 4 (Fig. 1A, Table 1), one major product of 701 bp was generated by RT-PCR, which was shown by sequencing to correspond to a transcript retaining introns 3 and 4 (Fig. 1C). Two minor products were also detected, one of 339 bp corresponded to a transcript retaining intron 4 only, the other (Fig. 1C, asterisk) could not be identified. These transcripts were in low abundance as only the constitutively spliced peripherin isoform (in which all introns are spliced out) was detected when RT-PCR was performed using primers located in the 5′ and 3′ untranslated regions (data not shown). Therefore 5′ and 3′ RACE using gene-specific primers in intron 4 (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) was used to obtain the full-length transcript retaining introns 3 and 4, which was called Per 3,4 cDNA.
Figure 1.
Identification of a peripherin transcript retaining introns 3 and 4. A, Schematic showing locations of forward primer F1 in exon 3 and reverse primer R1 in intron 4/exon 5 used for RT-PCR amplification of transcripts retaining intron 4. B, Off-target primer locations F1 in exon 1 and R2 in intron 3 used to control for genomic contamination. C, A major RT-PCR product of 701 bp was amplified from human dorsal root ganglion RNA using the primer pair indicated in A corresponding to a transcript retaining introns 3 and 4; a minor transcript of 339 bp encompassing intron 4 was also identified, as well as product of 550 bp, which could not be identified. D, The primer pair in B amplified a product of 683 bp corresponding to a transcript in which introns 1 and 2 were spliced out, providing additional confirmation that the samples were not contaminated with genomic DNA. All RT-PCR products were confirmed by sequencing.
Expression of Per 3,4 in transfected cells
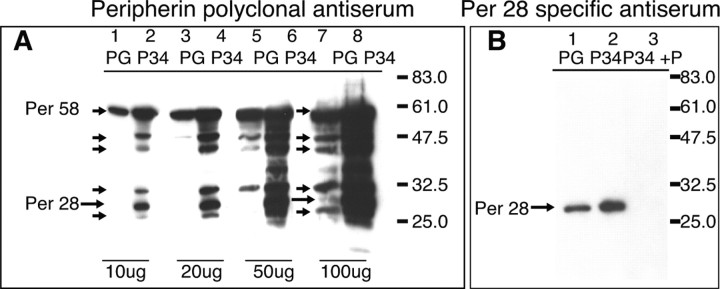
Expression of the Per 3,4 cDNA was compared with the normal full-length human peripherin gene in transfected SW13vim(−) cells, a human cell line lacking an endogenous intermediate filament network (Sarria et al., 1990). The constitutive expression product from the peripherin gene, in which all intronic sequences are spliced out, has a molecular weight of ∼58 kDa (Per 58) according to SDS-PAGE (Fig. 2A). For Per 3,4, the retention of intron 3 introduces a premature stop codon 30 bp downstream from the start of the intron, predicting a peripherin species of 28 kDa with a unique 10 amino acid sequence at the C-terminus. Immunoblots of cell lysates from transfected cells probed with commercially available peripherin antibody (AB1530; Millipore) showed the expected expression of Per 58 from the peripherin gene (Fig. 2A, lane 1). In lysates from cells expressing the Per 3,4 cDNA, a species of ∼28 kDa was apparent, which corresponded to the molecular weight predicted from the cDNA sequence and was designated Per 28 (Fig. 2A, lane 2, large arrow). The identity of this species was confirmed using a specific antibody raised to the unique 10 amino acid sequence derived from translation of the initial 30 bp of intron 3 (Fig. 2B). There was also expression of Per 58 from the Per 3,4 cDNA, as well as expression of four other minor species of ∼48, ∼45, ∼32, and ∼25 kDa (Fig. 2A, lane 2, small arrows). Expression of Per 58 from the Per 3,4 cDNA indicates that the Per 3,4 transcript undergoes splicing to remove introns 3 and 4 in transfected SW13vim(−) cells. The processing events that generated the four additional peripherin species are unknown, but unlikely to involve post-translational modifications because they are absent in similar loadings of lysates from cells expressing the peripherin gene (Fig. 2A, compare lanes 1, 2).
Figure 2.
Expression of Per 3,4 cDNA in transfected cells. A, Immunoblot of increasing loadings (10, 20, 50, and 100 μg) of cell lysates derived from SW13vim(−) cells transfected with the peripherin gene (PG; lanes 1, 3, 5, and 7) or the Per 3,4 cDNA (P34; lanes 2, 4, 6, and 8), probed with peripherin polyclonal antiserum, AB1530. Samples were harvested after 48 h expression. The 28 kDa species predicted from the Per 3,4 cDNA sequence is indicated by the large arrow in lane 2 (Per 28). This species was undetectable in similar loadings of PG transfected cells (lane 1). Note also the additional peripherin species of ∼48, 45, 32.5, and 25 kDa, indicated by small arrows (lane 2). All of these species were apparent in the Per 3,4 transfected cell lysates at 10 μg loadings. The only species apparent in 10 μg loadings of the peripherin gene transfected cell lysates was of 58 kDa, corresponding to the constitutively spliced peripherin species, Per 58 (lane 1). This was also expressed from the Per 3,4 cDNA, indicating that Per 3,4 is itself alternatively spliced in SW13vim(−) cells. However, as protein loadings were increased, the additional peripherin species observed in lane 2 also became apparent in the peripherin gene transfected cell lysates (indicated by arrows in lane 7). B, Validation of Per 28 kDa species as product derived from transcript retaining intron 3. Total protein lysates from SW13vim(−) cells expressing PG or P34 probed by immunoblotting with the Per 28-specific antibody revealed a single band of 28 kDa (indicated by arrow). This band could be competed out when the peptide used to generate Per 28 antibody was included in the antibody incubations, confirming the specificity of the antibody for Per 28 (lane 3, P34 +P).
As protein loadings were increased, we found that all of the species expressed from Per 3,4 cDNA were also expressed at low levels from the peripherin gene (Fig. 2A, lane 7, arrows). This included Per 28, verified using the specific antisera (Fig. 2B, lane 1). However, Per 28 was expressed at a relatively higher level from the Per 3,4 cDNA than from the wild-type peripherin gene. These findings indicated that the additional peripherin species are in fact normal expression products from the peripherin gene, but are expressed at lower levels and at different relative stoichiometric levels compared with expression from the Per 3,4 cDNA.
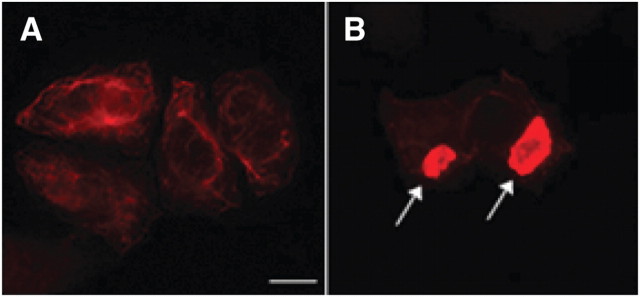
Changes in peripherin isoform expression causes aggregation
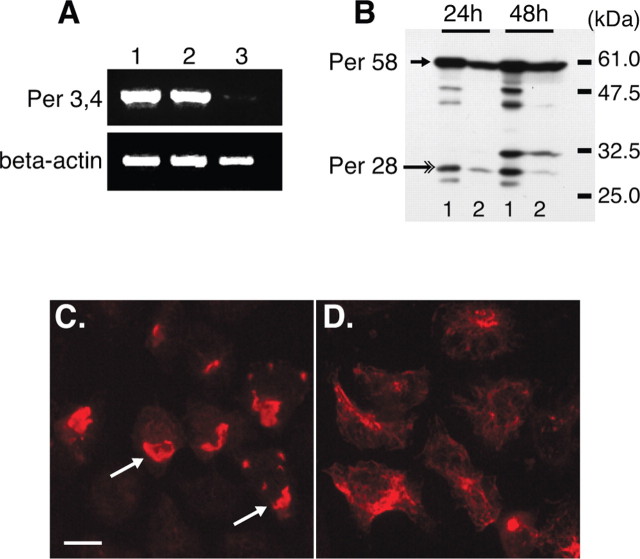
In SW13vim(−) cells expressing the human peripherin gene peripherin displayed the normal filamentous networks that we described previously for the mouse gene (Fig. 3A) (Beaulieu et al., 1999b). In contrast, expression of the Per 3,4 cDNA led to the generation of large aggregates (Fig. 3B, arrows). As the same peripherin species are expressed from the peripherin gene as from the Per 3,4 cDNA, this indicated that a change in the relative ratios of peripherin isoform expression induces peripherin aggregate formation. To verify this, we designed an interfering RNA targeting intron 3, which when coexpressed with the Per 3,4 cDNA would lead to a downregulation of Per 28. Of two siRNAs designed, siRNA2, but not siRNA1, downregulated expression of transcripts retaining intron 3 (Fig. 4A, compare lanes 2, 3) and correspondingly reduced expression of Per 28 while maintaining expression of Per 58 (Fig. 4B, compare lanes 1, 2, Per 28 indicated with large arrow). This change in the relative expression levels of peripherin isoforms caused Per 3,4 to form filaments instead of aggregates (Fig. 4, compare C, D), indicating that alterations in the relative expression levels of peripherin isoforms determines whether peripherin forms filaments or aggregates.
Figure 3.
A, B, Immunofluorescence labeling of SW13vim(−) cells transfected with the normal peripherin gene (A) compared with the Per 3,4 cDNA (B), labeled with polyclonal peripherin antibody (AB1530). Note the normal filamentous distribution of peripherin in A and the large amorphous aggregates (indicated by arrows) in B. Scale bar, 10 μm.
Figure 4.
Downregulation of Per 28 using RNAi prevents aggregate formation. A, RT-PCR of RNA isolated from SW13vim(−) cells transfected with Per 3,4 (lane 1); Per 3,4 and RNAi-1 (lane 2); and Per 3,4 and RNAi-2 (lane 3). Samples were normalized to β-actin. Note the downregulation of Per 3,4 message in cells cotransfected with RNAi-2 (lane 3). B, Immunoblot of total cell lysates of cells transfected with Per 3,4 (lanes 1) or cotransfected with Per 3,4 and RNAi-2 (lane 2), probed with AB1530. Note the reduction in expression of Per 28 at 24 and 48 h after expression (lane 2, large arrow) and the maintained expression of the normal peripherin product (lanes 1, 2, small arrow). C, D, Immunofluorescence labeling of SW13vim(−) cells expressing Per 3,4 (C) or Per 3,4 and RNAi-2 (D), labeled with AB1530. Note the peripherin aggregates in C and the predominance of filamentous networks in D. Scale bar, 30 μm.
Expression of Per 3,4 forms inclusion bodies in primary motor neurons
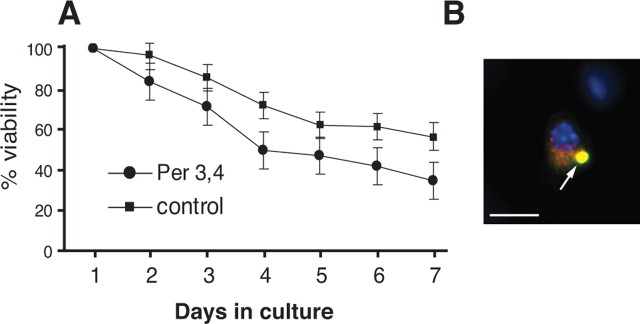
We had shown previously that expression of the mouse splice variant Per 61 formed punctate aggregates and was neurotoxic to motor neurons in culture (Robertson et al., 2003). We therefore tested the effects of Per 3,4 expression on motor neuron viability using intranuclear microinjection of the Per 3,4 plasmid or plasmid alone, as described previously (Fig. 5A) (Robertson et al., 2003). Expression of Per 3,4 in motor neurons led to the formation of singular inclusion bodies in the perikarya, very different from the punctate aggregates we previously observed for Per 61 (Fig. 5B) (Robertson et al., 2003), but more reminiscent of peripherin pathology observed in ALS (Xiao et al., 2006). The viability assay indicated that although Per 3,4 was associated with some neurotoxicity, the effect was not as dramatic as that observed previously for Per 61 (Fig. 5A) (Robertson et al., 2003).
Figure 5.
Expression of Per 3,4 transcript in primary motor neurons. A, Motor neuron viability curve showing that expression of Per 3,4 induced significantly increased neurotoxicity compared with control cultures in which empty vector was injected alone. B, Example of a microinjected motor neuron expressing Per 3,4 double labeled with Per 28 antibody in red and monoclonal peripherin antibody in green, with the nuclei labeled with DAPI. The inclusion body is indicated by an arrow. Scale bar, 20 μm.
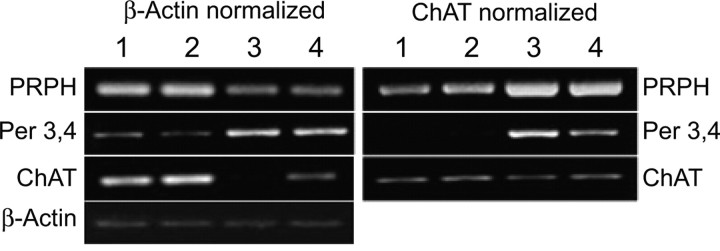
Per 3,4 transcript expression is upregulated in ALS
Because upregulated expression of Per 28 induced peripherin to aggregate and form structures resembling peripherin-immunoreactive inclusion bodies in ALS, we investigated whether there was an upregulation of Per 3,4 transcript expression in ALS spinal cord tissue compared with controls using semiquantitative RT-PCR. Because the cellular profile is different in disease spinal cord tissue compared with normal tissue (loss of motor neurons and proliferation of non-neuronal inflammatory cells, i.e., microglia and astrocytes in disease tissue), two sets of primers were used for normalization: β-actin to normalize for total mRNA, and ChAT to normalize for total neuronal mRNA. Findings showed a dramatic upregulation of the Per 3,4 transcript in ALS spinal cord compared with controls whether β-actin or ChAT was used for normalization (Fig. 6, compare lanes 1, 2 with 3, 4). The normal peripherin transcript (PRPH) was also upregulated when samples were normalized to ChAT, consistent with our previous findings showing an upregulation of peripherin expression in ALS (Robertson et al., 2003).
Figure 6.
Upregulated expression of Per 3,4 mRNA in ALS versus control tissue. RT-PCR of total RNA isolated from spinal cords of control (lanes 1, 2) and ALS (lanes 3, 4) cases. Samples were normalized to β-actin or to ChAT (to normalize for neuronal content). Note the reduction in ChAT RT-PCR product in ALS samples (lanes 3, 4) compared with controls (lanes 1, 2) in the reactions normalized to β-actin, consistent with loss of motor neurons in ALS. The normal peripherin message was amplified using the primers shown in Table 1 (PRPH). PRPH appeared decreased in ALS samples (lanes 3, 4) compared with controls (lanes 1, 2) when RT-PCRs were normalized to β-actin, but increased when normalized to ChAT. An increase in Per 3,4 expression was detected in ALS samples compared with controls whether the samples were normalized to β-actin or to ChAT.
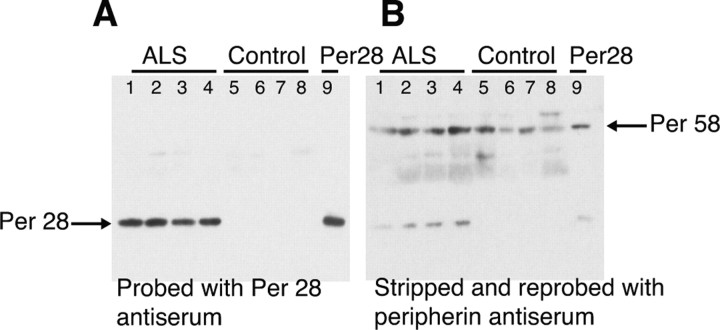
Per 28 expression is upregulated in ALS
We next tested whether the increase in expression of the Per 3,4 transcript in ALS tissue led to a corresponding increased expression of Per 28. Equivalent amounts of TX-100 preparations from lumbar spinal cord of four ALS cases and four control cases were probed by immunoblotting using the Per 28-specific antisera (Fig. 7A). The control samples were comprised of one non-neurological disease case (lane 8) and three with neurological diseases other than ALS (two multisystem atrophy, lanes 5 and 6, and one corticobasal degeneration, lane 7). The details of the cases used for the study are shown in supplemental Table 1 (available at www.jneurosci.org as supplemental material). The results in Figure 7A show that there is a pronounced increase in expression of Per 28 in all four of the ALS cases with expression below detectable limits in the control cases. Reprobing of the blots with commercially available peripherin antibody (AB1530) showed the increase in Per 58 expression in the ALS cases relative to controls that we have reported previously (Fig. 7B) (Robertson et al., 2003). However, this generalized increase in peripherin expression was not sufficient to account for the substantive increase in signal detected for Per 28 in the ALS samples.
Figure 7.
Upregulated expression of Per 28 in ALS versus control tissue. A, Triton X-100 (30 μg) preparations from lumbar spinal cord of four independent ALS and control cases were probed by immunoblotting with the Per 28 kDa splice variant-specific antiserum. Per 28 was detected in the ALS samples but not in the controls. B, The immunoblot in A was stripped and reprobed with AB1530, the commercially available antiserum, revealing the constitutively spliced Per 58 kDa peripherin species in all samples.
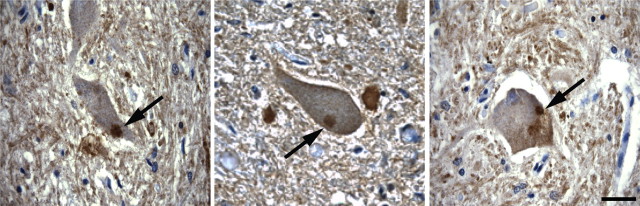
Association of Per 28 with peripherin pathology in ALS
We have shown that there is an upregulation of the Per 3,4 splicing intermediate in ALS spinal cord compared with controls, and a corresponding increased expression of Per 28, the major expression product from the Per 3,4 transcript. Because we have shown that upregulated expression of Per 3,4 induces peripherin aggregate formation, we investigated whether Per 28 was associated with peripherin pathology in ALS. Immunocytochemical labeling of ALS spinal cord tissue with the Per 28 antibody showed distinct labeling of peripherin aggregates resembling round inclusions in the cytoplasm of motor neurons (Fig. 8). There was no labeling of control spinal cord. These findings indicated that not only was there increased expression of Per 28 in ALS, but that it was also associated with disease pathology.
Figure 8.
Immunocytochemical labeling of ALS lumbar spinal cord with Per 28-specific antiserum. A–C, Three examples of motor neurons containing Per 28 immunoreactive inclusion bodies (arrows). Scale bar, 20 μm.
Discussion
Peripherin is a type III intermediate filament protein associated with the major disease pathologies in ALS including ubiquitinated round inclusions and Lewy body-like inclusions (He and Hays, 2004; Xiao et al., 2006). Overexpression of peripherin in transgenic mice induces motor neuron degeneration that is accompanied by the presence of intraneuronal aggregates of peripherin (Beaulieu et al., 1999a). We have shown previously the expression of a neurotoxic splice variant of peripherin in motor neurons of transgenic mice expressing mutant SOD1 (Robertson et al., 2003). Although overexpression or ablation of peripherin in these mice did not affect disease course (Lariviere et al., 2003), it is now known that motor neuron degeneration caused by mutant SOD1 in transgenic mice is multifactorial, involving both neuronal and non-neuronal components (Clement et al., 2003) and discussed in (Xiao et al., 2006). Furthermore, mutations in SOD1 are causative of only 1–2% of all fALS, whereas peripherin abnormalities are found broadly in both fALS and sALS (Corbo and Hays, 1992; Migheli et al., 1993; He and Hays, 2004). The neurotoxic splice variant we described previously in mouse, Per 61, was generated through the in-frame retention of intron 4, which is 96 bp in length and introduces a 32 amino acid insertion within a highly conserved domain of the peripherin protein (Landon et al., 1989, 2000; Robertson et al., 2003). Although a Per 61 antibody labeled diseased motor neurons in ALS and immunoprecipitated a higher molecular weight peripherin species, the transcript underlying the generation of this species remains to be identified (Robertson et al., 2003). Indeed the splicing event that generates Per 61 in mouse cannot occur in human, as intron 4 in human is 91 bp in length and its complete retention would introduce a premature stop codon and the generation of a truncated peripherin species of ∼32 kDa. Nevertheless, evidence from EST sequences indicated that read through into intron 4 could occur in human. As such, it was through our attempts to identify human peripherin transcripts retaining intron 4 that we identified the Per 3,4 transcript, which retains both introns 3 and 4. Because this transcript was derived from intron retention, we were particularly cautious of spurious results obtained from possible genomic contamination. To confirm that this transcript was not caused by genomic contamination all of our RNA samples were pretreated with DNase before RT-PCR, controls omitting reverse transcriptase showed no amplification of genomic DNA, oligo-(dT)20 was used for first strand synthesis and the use of off-target primers showed the proper splicing of introns 1 and 2. This transcript was in low relative abundance compared with the constitutively spliced transcript, encoding Per 58, and could only be amplified using gene-specific primers within either introns 3 or 4. Intron retention is the least frequent of alternative splicing events in mammals, and as such, is the least studied (Clark and Thanaraj, 2002; Kan et al., 2002). The retention of intron 3 leads to the introduction of a premature stop codon 30 bp downstream from the start of the intron, generating a truncated protein of ∼28 kDa with a unique 10 amino acid sequence at the C-terminus, designated Per 28. Using a specific antisera generated to the unique 10 amino acid sequence, we have established the existence of Per 28 as an alternatively spliced isoform expressed from the Per 3,4-cDNA and at low levels from the wild-type human peripherin gene. Additional minor peripherin species were also identified of ∼25, ∼32, ∼45, and ∼48 kDa expressed from the Per 3,4-cDNA and at substoichiometric levels from the peripherin gene. We have positively identified the 45 kDa species as being derived from the use of an downstream, in-frame alternative initiation codon within the peripherin transcript (McLean et al., 2008) and we are currently investigating the processing events leading to the generation of the other species. Remarkably, the deregulated expression of peripherin isoforms from the Per 3,4-cDNA induced peripherin to form aggregates instead of filaments, raising the possibility that deregulated peripherin splice isoform expression may contribute to peripherin aggregation and inclusion body formation in ALS. This premise was supported by experiments in which the peripherin splicing profile of Per 3,4 was reverted using RNAi to downregulate the expression of Per 28 relative to Per 58, consequently inducing peripherin to form filaments instead of aggregates. We have provided direct evidence that there is a similar deregulation of peripherin splicing in ALS by showing an upregulation of the Per 3,4 mRNA relative to the constitutively spliced peripherin transcript in ALS lumbar spinal cord compared with controls, and that this corresponded with upregulated expression of Per 28. We also showed the association of Per 28 with round inclusion bodies in affected motor neurons, providing additional support for the relevance of abnormalities in peripherin splicing contributing to the pathogenic mechanism(s) underlying ALS by generating aggregation-prone isoforms.
To test whether the inclusions formed by Per 28 were neurotoxic, we expressed the Per 3,4 cDNA in primary motor neurons by intranuclear microinjection. Although Per 28 expression formed inclusions in the perikarya of motor neurons, the presence of these inclusions was only associated with mild neurotoxicity. This is distinct from our earlier findings on the mouse splice variant Per 61, which was extremely neurotoxic when expressed in motor neurons (Robertson et al., 2003). However, Per 28 is very different from Per 61. Per 28 encodes the N-terminal region of peripherin up to the distal end of coil 1b of the α helical rod domain and has a unique amino sequence, VSGPGIRGGF, at the C-terminus. In contrast, Per 61 incorporates a 32 amino acid insertion within coil 2 of the rod domain, but otherwise has the same sequence as Per 58 (Landon et al., 1989, 2000). Expression of Per 28 in transfected SW13 cells or primary motor neurons was associated with the formation of singular inclusion bodies, whereas Per 61 formed punctate aggregates, dispersed throughout the cytoplasm (Robertson et al., 2003). Therefore based on structural considerations alone, it would be anticipated that the biological effects of Per 61 compared with Per 28 would be entirely different.
Here, we have identified a novel human peripherin splice variant, Per 28, that is encoded by a transcript retaining introns 3 and 4. Upregulated expression of Per 28 induces formation of peripherin inclusions, both in transfected SW13vim(−) cells and in primary motor neurons. Importantly we have shown using a splice variant-specific antibody that Per 28 expression is upregulated in ALS spinal cord, and that Per 28 is associated with disease pathology, specifically round inclusions. These findings are the first to demonstrate unequivocally that abnormalities of peripherin splicing occur in ALS and that these splice variants may be intrinsically involved in the generation of disease pathology.
It is interesting to speculate how peripherin splicing abnormalities could occur in ALS. In this regard, the nuclear DNA/RNA binding protein TAR DNA binding protein-43 (TDP-43) has been identified previously as a component of ubiquitinated inclusions in ALS (Arai et al., 2006; Neumann et al., 2006), and we have shown that peripherin and TDP-43 are colocalized to the same inclusions (Sanelli et al., 2007). One of the known functions of TDP-43 is to act as a splicing factor (Buratti and Baralle, 2001), and it is therefore tempting to speculate that the deregulated splicing we have observed for peripherin may be causally linked to abnormalities of TDP-43. This will be the subject of future investigations.
Footnotes
J. Robertson was supported by grants from the Canadian Institutes of Health Research (CIHR), The ALS Society of Canada, The UK Motor Neurone Disease Association, The ALS Association (ALSA), and The Muscular Dystrophy Association (MDA), and holds a Canada Research Chair in ALS. S.X. was supported by a CIHR Postdoctoral Training Fellowship. J. Ravits was supported by National Institutes of Health Grant R21.NS051738, the Benaroya Foundation, and the Juniper Foundation. M.J.S. was supported by ALS Canada, the Scottish Rite Charitable Foundation, ALSA, and the MDA.
References
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Nguyen MD, Julien JP. Late onset of motor neurons in mice overexpressing wild-type peripherin. J Cell Biol. 1999a;147:531–544. doi: 10.1083/jcb.147.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Robertson J, Julien JP. Interactions between peripherin and neurofilaments in cultured cells: disruption of peripherin assembly by the NF-M and NF-H subunits. Biochem Cell Biol. 1999b;77:41–45. [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Clark F, Thanaraj TA. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum Mol Genet. 2002;11:451–464. doi: 10.1093/hmg/11.4.451. [DOI] [PubMed] [Google Scholar]
- Clement AM, Nguyen MD, Roberts EA, Garcia ML, Boillee S, Rule M, McMahon AP, Doucette W, Siwek D, Ferrante RJ, Brown RH, Jr, Julien JP, Goldstein LS, Cleveland DW. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- Corbo M, Hays AP. Peripherin and neurofilament protein coexist in spinal spheroids of motor neuron disease. J Neuropathol Exp Neurol. 1992;51:531–537. doi: 10.1097/00005072-199209000-00008. [DOI] [PubMed] [Google Scholar]
- Durham HD, Roy J, Dong L, Figlewicz DA. Aggregation of mutant Cu/Zn superoxide dismutase proteins in a culture model of ALS. J Neuropathol Exp Neurol. 1997;56:523–530. doi: 10.1097/00005072-199705000-00008. [DOI] [PubMed] [Google Scholar]
- Hays AP. Pathology of amyotrophic lateral sclerosis. In: Mitsumoto H, Przedborski S, Gordon PH, editors. Amyotrophic lateral sclerosis. New York: Taylor and Francis Group; 2006. pp. 43–80. [Google Scholar]
- Hays AP, Naini A, He CZ, Mitsumoto H, Rowland LP. Sporadic amyotrophic lateral sclerosis and breast cancer: hyaline conglomerate inclusions lead to identification of SOD1 mutation. J Neurol Sci. 2006;242:67–69. doi: 10.1016/j.jns.2005.11.016. [DOI] [PubMed] [Google Scholar]
- He CZ, Hays AP. Expression of peripherin in ubiquinated inclusions of amyotrophic lateral sclerosis. J Neurol Sci. 2004;217:47–54. doi: 10.1016/j.jns.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Ince PG, Lowe J, Shaw PJ. Amyotrophic lateral sclerosis: current issues in classification, pathogenesis and molecular pathology. Neuropathol Appl Neurobiol. 1998a;24:104–117. doi: 10.1046/j.1365-2990.1998.00108.x. [DOI] [PubMed] [Google Scholar]
- Ince PG, Tomkins J, Slade JY, Thatcher NM, Shaw PJ. Amyotrophic lateral sclerosis associated with genetic abnormalities in the gene encoding Cu/Zn superoxide dismutase: molecular pathology of five new cases, and comparison with previous reports and 73 sporadic cases of ALS. J Neuropathol Exp Neurol. 1998b;57:895–904. doi: 10.1097/00005072-199810000-00002. [DOI] [PubMed] [Google Scholar]
- Kan Z, States D, Gish W. Selecting for functional alternative splices in ESTs. Genome Res. 2002;12:1837–1845. doi: 10.1101/gr.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landon F, Lemonnier M, Benarous R, Huc C, Fiszman M, Gros F, Portier MM. Multiple mRNAs encode peripherin, a neuronal intermediate filament protein. EMBO J. 1989;8:1719–1726. doi: 10.1002/j.1460-2075.1989.tb03564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landon F, Wolff A, de Nechaud B. Mouse peripherin isoforms. Biol Cell. 2000;92:397–407. doi: 10.1016/s0248-4900(00)01099-6. [DOI] [PubMed] [Google Scholar]
- Lariviere RC, Beaulieu JM, Nguyen MD, Julien JP. Peripherin is not a contributing factor to motor neuron disease in a mouse model of amyotrophic lateral sclerosis caused by mutant superoxide dismutase. Neurobiol Dis. 2003;13:158–166. doi: 10.1016/s0969-9961(03)00036-6. [DOI] [PubMed] [Google Scholar]
- McLean J, Xiao S, Miyazaki K, Robertson J. A novel peripherin isoform generated by alternative translation is required for normal filament network formation. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2007.05198.x. in press. [DOI] [PubMed] [Google Scholar]
- Migheli A, Pezzulo T, Attanasio A, Schiffer D. Peripherin immunoreactive structures in amyotrophic lateral sclerosis. Lab Invest. 1993;68:185–191. [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Robertson J, Beaulieu JM, Doroudchi MM, Durham HD, Julien JP, Mushynski WE. Apoptotic death of neurons exhibiting peripherin aggregates is mediated by the proinflammatory cytokine tumor necrosis factor-alpha. J Cell Biol. 2001;155:217–226. doi: 10.1083/jcb.200107058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J, Kriz J, Nguyen MD, Julien JP. Pathways to motor neuron degeneration in transgenic mouse models. Biochimie. 2002;84:1151–1160. doi: 10.1016/s0300-9084(02)00025-1. [DOI] [PubMed] [Google Scholar]
- Robertson J, Doroudchi MM, Nguyen MD, Durham HD, Strong MJ, Shaw G, Julien JP, Mushynski WE. A neurotoxic peripherin splice variant in a mouse model of ALS. J Cell Biol. 2003;160:939–949. doi: 10.1083/jcb.200205027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van Den Bergh R, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Sanelli T, Xiao S, Horne P, Bilbao J, Zinman LH, Robertson J. Evidence that TDP-43 is not the major ubiquitinated target in the pathological inclusions of ALS. J Neuropathol Exp Neurol. 2007;66:1147–1153. doi: 10.1097/nen.0b013e31815c5edd. [DOI] [PubMed] [Google Scholar]
- Sarria AJ, Nordeen SK, Evans RM. Regulated expression of vimentin cDNA in cells in the presence and absence of a preexisting vimentin filament network. J Cell Biol. 1990;111:553–565. doi: 10.1083/jcb.111.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Oblinger MM. Differential regulation of peripherin and neurofilament gene expression in regenerating rat DRG neurons. J Neurosci Res. 1990;27:332–341. doi: 10.1002/jnr.490270312. [DOI] [PubMed] [Google Scholar]
- Xiao S, McLean J, Robertson J. Neuronal intermediate filaments and ALS: a new look at an old question. Biochim Biophys Acta. 2006;1762:1001–1012. doi: 10.1016/j.bbadis.2006.09.003. [DOI] [PubMed] [Google Scholar]