SUMMARY
Self-renewal and pluripotency of the embryonic stem cell (ESC) state are established and maintained by multiple regulatory networks that comprise transcription factors and epigenetic regulators. While much has been learned regarding transcription factors, the function of epigenetic regulators in these networks is less well defined. We conducted a CRISPR-Cas9 mediated loss-of-function genetic screen that identified two new epigenetic regulators, TAF5L and TAF6L, components/co-activators of the GNAT-HAT complexes for the mouse ESC (mESC) state. Detailed molecular studies demonstrate that TAF5L/TAF6L transcriptionally activate c-Myc and Oct4, and their corresponding MYC and CORE regulatory networks. Besides, TAF5L/TAF6L predominantly regulate their target genes through H3K9ac deposition and c-MYC recruitment that eventually activate the MYC regulatory network for self-renewal of mESCs. Thus, our findings uncover a novel role of TAF5L/TAF6L in directing the MYC regulatory network that orchestrates gene expression programs to control self-renewal for the maintenance of mESC state.
INTRODUCTION
Embryonic stem cells (ESCs) isolated from inner cell mass of the blastocyst during embryonic development (Thomson, 1998) have two unique properties: self-renewal– the ability to proliferate in the same state indefinitely, and pluripotency– the capacity to differentiate into all lineages of the organism (Zwaka and Thomson, 2005). Induced pluripotent stem cells (iPSCs) are molecularly and functionally equivalent to ESCs; however, iPSCs can be derived from diverse cell types through ectopic expression of specific factors (Takahashi and Yamanaka, 2006). These unique properties of ESCs and iPSCs make them attractive possible routes toward future cellular therapies (Robinton and Daley, 2012).
Self-renewal and pluripotency of the “ESC state” are established and maintained by the concerted action of signaling pathways that respond to external stimuli and intrinsically activate critical “transcription regulatory networks” (Young, 2011). Each of the transcription regulatory networks comprises transcription factors (TFs), co-factors and chromatin/epigenetic regulators, and they form interconnected regulatory networks that control chromatin architecture and gene expression programs (Orkin and Hochedlinger, 2011; Young, 2011). Although much has been learned regarding specific transcription factors, the epigenetic components that establish and maintain the ESC state are less well defined.
Comprehensive studies using protein–DNA interaction data and expression of target genes have revealed three key functionally distinct regulatory modules/networks in mouse ESCs (mESCs) that maintain the ESC state: CORE (active), MYC (active) and Polycomb/PRC (repressive) (Chen et al., 2008; Kim et al., 2008; 2010). The CORE module is composed of “core” ESC TFs– OCT4, SOX2 and NANOG (OSN)– that cooperate with other ESC TFs, co-activators and chromatin regulators to activate ESC-specific genes and repress lineage-specific genes (Kim et al., 2010; Young, 2011), whereas the PRC module is composed of both PRC1 and PRC2 components and represses mainly lineage-specific genes to maintain the ESC state (Boyer et al., 2006; Laugesen and Helin, 2014). The MYC regulatory module/network is c-MYC centered and includes partner co-activators and chromatin modifiers. The MYC module primarily controls self-renewal of mESCs through cell cycle, metabolism, ribosome biogenesis and protein synthesis–associated genes, but also regulates pluripotency-related genes to prevent differentiation to maintain the ESC state (Fagnocchi and Zippo, 2017; Fagnocchi et al., 2016; Scognamiglio et al., 2016; Smith et al., 2010).
c-MYC has been implicated in controlling chromatin architecture and epigenetic landscapes of ESCs in multiple ways. First, TIP60 (KAT5)-p400, a MYST (Moz/Morf, Ybf2, Sas3, Sas2 and Tip60) family member of the histone acetyltransferase (HAT) complex, interacts with c-MYC and functionally belongs to the MYC module (Kim et al., 2010). Notably, TIP60-p400 plays a crucial role in maintaining the ESC state through Nanog (Fazzio et al., 2008a). Likewise, GCN5/KAT2A (a catalytic component of SAGA and TFTC HAT complexes of the GNAT (GCN5 N-acetyltransferase) family) belongs to the MYC module in mESCs (Kim et al., 2010). These findings suggest that MYC acts with distinct HAT complexes through diverse mechanisms because different HAT complexes act on different substrates for gene regulation. For instance, TIP60-HAT acetylates H4K16, whereas GNAT-HAT preferentially acetylates H3K9 (Lee and Workman, 2007). c-MYC and the MYC module are both positively correlated with active H3ac, H4ac and H3K4me3 marks and are negatively correlated with repressive H3K27me3 (Kim et al., 2008; 2010). Although the connection between c-MYC and HAT complexes has been established, the precise molecular mechanism by which c-MYC and HAT cooperatively regulate gene expression and maintain the ESC state is not well understood. Second, c-MYC targets several pluripotency factors, especially core ESC TF- SOX2, and numerous chromatin-associated genes (Kim et al., 2010) for gene regulation to maintain the ESC state. Third, c-MYC also mediates gene repression of developmental genes through PRC2/H3K27me3 for mESC identity (Fagnocchi et al., 2016; Varlakhanova et al., 2011). Nonetheless, the repressive role of c-MYC is less well characterized compared to its pervasive role in gene activation.
Previously, short hairpin RNA (shRNA)-based knockdown screening approaches identified epigenetic factors necessary for maintenance of the mESC state (Bilodeau et al., 2009; Cooper and Brockdorff, 2013; Das et al., 2014; Fazzio et al., 2008b; Hu et al., 2009; Kagey et al., 2010). However, this approach remains technically challenging due to incomplete gene knockdown and the off-target activity of shRNAs, often making interpretation of the phenotypic changes difficult (Jackson and Linsley, 2010). Recently, clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9–mediated gene-specific knockouts (KOs) have been used in multiple biological systems to circumvent partial knockdown and off-target effects of shRNAs (Hsu et al., 2014). Moreover, CRISPR-Cas9–mediated functional genetic screens successfully identified novel genes that are required for various biological processes (Shalem et al., 2015; 2014; Wang et al., 2014; Zhou et al., 2014). However, to date, CRISPR-Cas9 mediated functional genetic screens have not described to identify new epigenetic factors that control the mESC state.
Here, we performed a CRISPR-Cas9 mediated loss-of-function genetic screen, which identified two novel epigenetic regulators for the mESC state with high confidence: TAF5L and TAF6L, components of GNAT-HAT complexes. Through integrated molecular and genomics approaches– we have implicated the role of TAF5L/TAF6L in transcriptional activation of c-Myc and Oct4; and their respective MYC and CORE regulatory modules/networks. Furthermore, we revealed a detailed mechanism that shows TAF5L/TAF6L primarily regulate their target genes expression through H3K9ac deposition and c-MYC recruitment, which ultimately trigger the MYC regulatory network for self-renewal of mESCs. Thus, TAF5L/TAF6L establish a link between HAT complexes and c-MYC/MYC regulatory network to fine-tune gene expression programs for the maintenance of mESC state.
RESULTS
CRISPR-Cas9 mediated loss-of-function genetic screen identifies potential candidate epigenetic genes for the mESC state
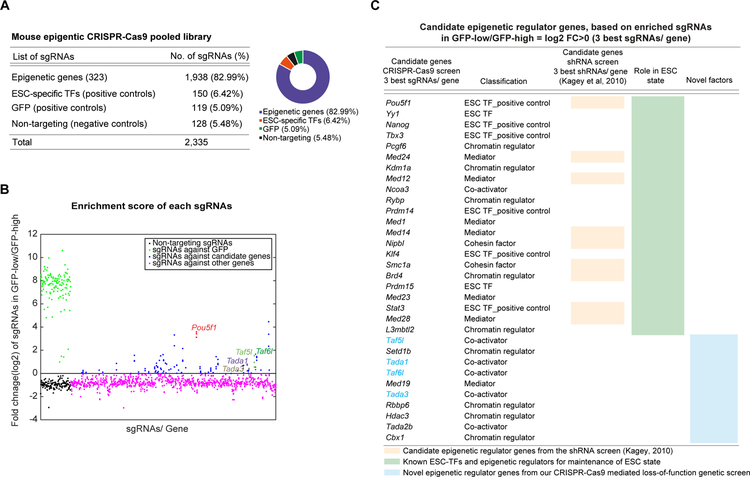
The pooled sgRNAs used for the CRISPR-Cas9 screen targeted 323 genes of different classes of epigenetic regulators and ESC-specific TFs (Figure S1A) (Table S1). We used six single guide RNAs (sgRNAs) per gene, targeting coding sequences from prior published work (Shalem et al., 2014) (Table S1). The “epigenetic CRISPR-Cas9 pooled library” contained total 2335 sgRNAs; including 1938 sgRNAs targeting all 323 epigenetic and ESC TF genes (Table S1, Figure 1A), 128 non-targeting sgRNAs as negative controls, 119 sgRNAs targeting GFP (of the Oct4-GFP reporter) and 150 sgRNAs targeting coding sequence of known mESC-specific TFs as positive controls (Table S1, Figure 1A). The pooled library was transduced into an Oct4-GFP reporter (OCT4 is the master regulator of ESCs) mESC line, which constitutively expressed Cas9 (Figures S1A–S1C). The Oct4-GFP reporter was used as a “readout” for the screen to measure the change in GFP levels upon perturbation of any of candidate epigenetic regulator genes by their corresponding sgRNAs. Lentiviral transduction of the pooled library was performed at low multiplicity (MOI) to ensure that selected cells contained roughly one sgRNA per cell (Figure S1A). After drug selection, the GFP-low and the GFP-high cells were sorted, genomic DNA was isolated, and next-generation sequencing (NGS) was employed to enumerate the sgRNAs in each cell populations (Figures S1A, S1D). The entire screen was performed in triplicate.
Figure 1. CRISPR-Cas9 mediated loss-of-function genetic screen identifies potential candidate epigenetic genes for the mESC state.
(A) Mouse epigenetic CRISPR-Cas9 pooled library distribution.
(B) Dot plot analysis shows enrichment scores of sgRNAs by comparing their frequency in the GFP-low cells over the GFP-high cells. Enrichment scores of the three-best individual sgRNAs per gene are represented. sgRNAs are ranked based on their corresponding target gene names, alphabetically from left to right on the X-axis. sgRNAs targeting GFP (positive controls, in green), non-targeting sgRNAs (negative controls, in black), and sgRNAs targeting all candidate genes (in blue) are shown. The sgRNAs targeting novel candidate epigenetic genes (Taf5l, Taf6l, Tada1 and Tada3) are labelled. The sgRNAs targeting Pou5f1/Oct4 (master regulator of mESCs, used as positive control; in red) provide high enrichment scores.
(C) List of candidate epigenetic regulator and TF genes ranked by enrichment scores. Candidate genes that have been previously identified through either shRNA screens (orange column) or individual functional studies (green column) related to mESC state are shown. Novel candidate epigenetic genes are also presented (blue column); among them Taf5l, Taf6l, Tada1 and Tada3 were selected for further validation.
Related to Figure S1.
We calculated an “enrichment score” of each sgRNA by comparing its frequency in GFP-low versus GFP-high cells (Figure 1B). Enrichment scores were built based on the three-best individual sgRNAs per gene (Table S2). The highest and lowest enrichment scores were obtained from GFP-targeting sgRNAs (positive controls) and non-targeting sgRNAs (negative controls), respectively, indicating the screen was technically successful (Figure 1B). Based on the enrichment scores, we identified several known and novel distinct epigenetic regulator genes as candidates (24 out of 323 epigenetic genes in the pooled library) (Figures 1B, 1C); including epigenetic regulator genes (Pcgf6, Kdm1a, Rybp, L3mbtl2, Smc1a, Nipbl, Brd4), ESC TF genes (Oct4, Nanog, Tbx3, Prdm14, Yy1, Prdm14, Klf4), and mediator genes (Med24, Med12, Med1, Med14, Med23) (Figure 1C), which have been previously linked to the mESC state (Whyte et al., 2012).
TAF5L and TAF6L are novel epigenetic genes for the mESC state
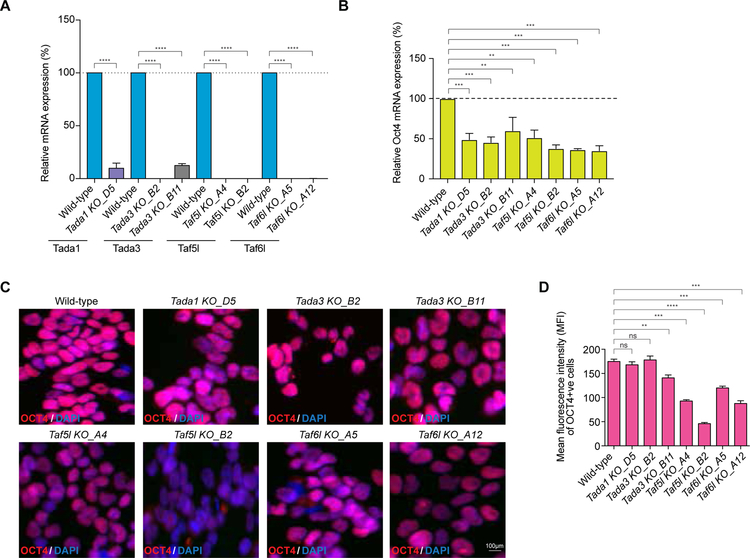
As novel candidate epigenetic genes, we selected Tada1, Tada3, Taf5l and Taf6l for further validation (Figures 1B, 1C). These factors belong to the GNAT family of HAT complexes (Lee and Workman, 2007). Each of the factors was highly expressed in undifferentiated mESCs, except Tada1, which was moderately expressed. However, the expression levels of Tada3, Taf5l and Taf6l were significantly reduced upon differentiation (24 and 48 hrs after differentiation) (Figure S1E), suggesting their possible roles in controlling the mESC state. Validation experiments were performed in two settings to target Tada1, Tada3, Taf5l and Taf6l genes in wild-type mESCs. First, we used two “individual” sgRNAs (with highest enrichment scores from the screen) that target coding sequences of selected genes and create “indels” (insertions or deletions of bases) (Figure S2A). Second, we used “paired” sgRNAs that target exon/s at the N-terminal end of each of the selected candidate genes (Table S3) to create “homozygous deletion” or KO clones (Figures S2D–S2G). These KO clones were confirmed by genotyping PCR, Sanger sequencing and quantitative RT-PCR (RT-qPCR) (Figures 2A, S2D–S2G).
Figure 2. Validation confirms TAF5L and TAF6L are the new epigenetic genes for the mESC state.
(A) RT-qPCR data showing mRNA expression levels of Tada1, Tada3, Taf5l and Taf6l in their corresponding KOs compared to wild-type. mRNA levels were normalized to GAPDH. Paired sgRNAs were used to target exon/s at the N-terminal of Tada1, Tada3, Taf5l and Taf6l candidate epigenetic genes to create homozygous/biallelic deletion or KO clones.
(B) Endogenous Oct4 mRNA expression levels in the Tada1, Tada3, Taf5l and Taf6l KOs.
(C) Immunofluorescence staining of OCT4 in the Tada1, Tada3, Taf5l and Taf6l KOs. Scale bar is 100µm.
(D) Quantification of mean fluorescence intensity (MFI) of OCT4 in the Tada1, Tada3, Taf5l and Taf6l KOs.
Data are represented as mean ± SEM (n = 3); p-values were calculated using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; and ns (non-significant).
Related to Figure S2.
The individual sgRNAs (that create indels) for all the target genes (Tada1, Tada3, Taf5l and Taf6l) showed significant reduction of the endogenous levels of Oct4 with different degrees, except Tada3 (Figures S2A–S2C). Also, individual sgRNAs targeting Taf5l and Taf6l showed substantial reduction of GFP levels of the Oct4-GFP reporter (Figure S2H), consistent with the primary outcome of the screen. Moreover, Taf5l and Taf6l KO clones consistently displayed a significant reduction of the endogenous Oct4 both at mRNA and protein levels (Figures 2B–2D, S2I). SSEA1, a mouse pluripotency marker was also reduced in Taf5l and Taf6l KO mESCs (Figures S2J). Thus, TAF5L and TAF6L were chosen as novel epigenetic regulators for further mechanistic studies.
TAF5L and TAF6L maintain gene expression programs of the mESC state
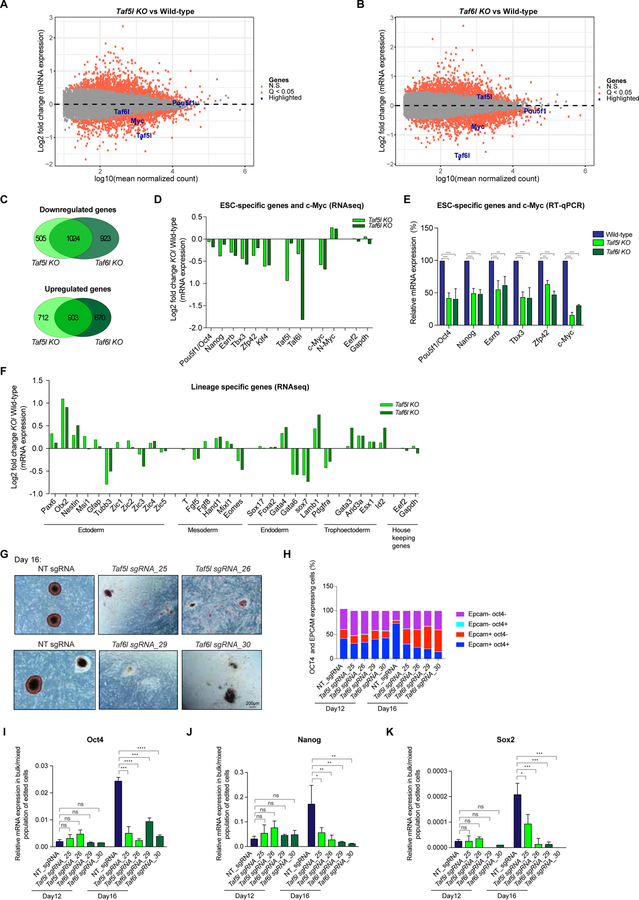
Next, we asked the role of TAF5L and TAF6L in gene expression programs in mESCs. Transcriptomic profiling (RNAseq) performed in the absence of Taf5l and Taf6l revealed differentially expressed genes (P-value (Q-value) <0.05) in Taf5l and Taf6l KOs compared to wild-type mESCs (Figures 3A, 3B, S3A) (Table S5), with a substantial overlap among upregulated, as well as downregulated genes from Taf5l KO and Taf6l KO (Figure 3C). Expression of mESC-specific genes, including Oct4/Pou5f1, Nanog, Esrrb, Tbx3, Zfp42 and Klf4, as well as c-Myc, was downregulated (Figures 3D). Furthermore, quantitative RT-PCR (RT-qPCR) data confirmed a significant reduction of several mESC-specific genes, including Oct4, and dramatic reduction of c-Myc in Taf5l and Taf6l KO ESCs (Figures 3E, S3B–S3C). The marked reduction of c-MYC was also monitored at the protein level in Taf5l and Taf6l KOs (Figures S3D). We also validated our findings in two independent ESC lines (J1 and CJ7) (Figures 3E, S3B, S3C). Ectoderm, mesoderm and endoderm lineage-specific genes were both up and downregulated without an obvious pattern; trophoectoderm (TE) lineage-specific genes were consistently upregulated in Taf5l and Taf6l KOs (Figure 3F), implying that loss of Taf5l and Taf6l biases mESCs toward the TE lineage. Importantly, mRNA expression of Taf5l and Taf6l itself was markedly downregulated in their corresponding KOs, as expected (Figures 3A, 3B, 3D). We observed that exogenous expression of TAF6L fully rescued Oct4 and c-Myc expression in the Taf6l KO, whereas exogenous expression of TAF5L partially rescued Oct4 and c-Myc expression in the Taf5l KO (Figures S3E–S3H), suggesting that TAF5L and TAF6L regulate Oct4 and c-Myc gene expression to different extents.
Figure 3. TAF5L and TAF6L are required for gene expression programs of mESC state; and for somatic cell reprogramming/iPSCs generation.
(A, B) Scatter plots showing differentially expressed genes from Taf5l KO (A) and Taf6l KO (B), compared to wild-type mESCs. Orange dots are significantly up and downregulated genes in Taf5l and Taf6l KOs with a q-value <0.05. Grey dots are unaltered genes in KOs compared to wild-type. Genes of interest are labelled in the scatter plot.
(C) Venn diagrams represent overlapped up or downregulated genes between Taf5l KO and Taf6l KO.
(D) mRNA expression levels of mouse ESC-specific genes, c-Myc and N-Myc in Taf5l and Taf6l KOs compared to wild-type (from RNAseq). Eef2 and Gapdh used as internal controls.
(E) mRNA expression levels of mouse ESC-specific genes and c-Myc in Taf5l and Taf6l KOs compared to wild-type (from RT-qPCR). mRNA levels were normalized to GAPDH.
(F) mRNA expression levels of lineage-specific genes in Taf5l and Taf6l KOs compared to wild-type (from RNAseq).
(G) AP staining of transgene-independent iPSC colonies following reprogramming (at day 16). Scale bar is 200µm.
(H) FACS analysis showing quantification of relative percentages of OCT4 and EPCAM expressing cells after reprogramming at day 12 and day 16.
(I-K) mRNA expression levels of ESC Core TFs– endogenous Oct4, Nanog, Sox2 in bulk/mixed populations of edited cells after reprogramming at day 12 and day 16, upon perturbation of Taf5l and Taf6l using sgRNAs. Non-targeting sgRNA used as control.
Data are represented as mean ± SEM (n = 3); p-values were calculated using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; and ns (non-significant).
Related to Figure S3.
To interrogate the possible functions of TAF5L and TAF6L in mESCs, gene ontology (GO) analysis was performed on differentially expressed genes from Taf5l and Taf6l KOs. Taf5l KO- and Taf6l KO-upregulated genes were considerably enriched for several biological processes, including cell-cell adhesion, cytoskeleton organization and cell migration related to differentiation of mESCs; in contrast Taf5l KO- and Taf6l KO-downregulated genes were significantly enriched with cell cycle, transcription, DNA replication, ribosome biogenesis/translation, nucleosome assembly and chromatin-silencing biological functions related to self-renewal of mESCs (Figures S3I–S3L). Taken together, these data demonstrate that TAF5L and TAF6L are required to maintain the gene expression programs of the mESC state.
TAF5L and TAF6L are required for efficient somatic cell reprogramming/iPSCs generation
Enforced expression of ESC-specific TFs OCT4, SOX2, KLF4 and c-MYC (OSKM) reprograms somatic cells to iPSCs that are molecularly and functionally equivalent to ESCs (Takahashi and Yamanaka, 2006). Epigenetic regulators serve as critical controllers of the reprogramming process and iPSC generation (Papp and Plath, 2013). Since we observed downregulation of mESC-specific genes (Oct4, Klf4) and c-Myc in mESCs lacking Taf5l and Taf6l (Figures 3D, 3E, S3C, S3D), we sought to determine whether TAF5L and TAF6L influence somatic cell reprogramming/iPSC generation. We used doxycycline-inducible OKSM driven reprogrammable mouse embryonic fibroblasts (MEFs) that carry a Oct4-GFP reporter (Stadtfeld et al., 2009). The reprogrammable MEFs were transduced with Taf5l and Taf6l targeting sgRNAs, and non-targeting (control) sgRNAs; and measured the efficiency of generation of stable transgene-independent iPSC colonies. Indel frequencies of targeted Taf5l and Taf6l alleles were quantified by amplicon sequencing before and after the reprogramming. More than 80% of Taf5l and Taf6l alleles were edited (Figure S3S). The high indel frequencies of Taf5l and Taf6l alleles correlate with higher fractions of edited cells. Interestingly, Taf5l and Taf6l targeted reprogrammable MEFs generated similar percentages of OCT4+EPCAM+ expressing cells as non-targeted (control) cells in the “presence of doxycycline” at day 12 (when reprogrammed cells are ready to acquire full pluripotency but not yet established) (Figure 3H). However, shortly after “doxycycline withdrawal” at day16 (when reprogrammed cells already acquire full pluripotency in a transgene-independent manner and become iPSCs), Taf5l and Taf6l bulk edited, reprogrammed cells displayed reduced fractions of iPSCs and increased fractions of differentiated cells (Figures 3G, S3M, S3N), along with decreased percentages of OCT4+ EPCAM+ and OCT4+ SSEA1+ expressing cells (Figures 3H, S3O). Furthermore, RNA expression analysis from Taf5l and Taf6l bulk edited, reprogrammed cells demonstrated no significant changes of ESC-specific genes (endogenous Oct4, Nanog, Sox2) and TE-specific genes (Cdx2, Arid3a, Esx1) at day 12 (Figures 3I–3K, S3P–S3R). Nonetheless, at day 16, Taf5l and Taf6l bulk edited, reprogrammed cells exhibited reduced levels of ESC-specific genes (endogenous Oct4, Nanog, Sox2) and elevated levels of TE-specific genes (Cdx2, Arid3a, Esx1) (Figures 3I–3K, S3P–S3R), similar to gene expression changes in Taf5l and Taf6l KO mESCs (Figures 3D–3F). Taken together, these data suggest that both TAF5L and TAF6L are critically required for somatic cell reprogramming/iPSC generation induced by OSKM, precisely at the final stages of acquisition and/or maintenance of the iPSCs.
Genome-wide distribution of TAF5L and TAF6L binding sites in mESCs
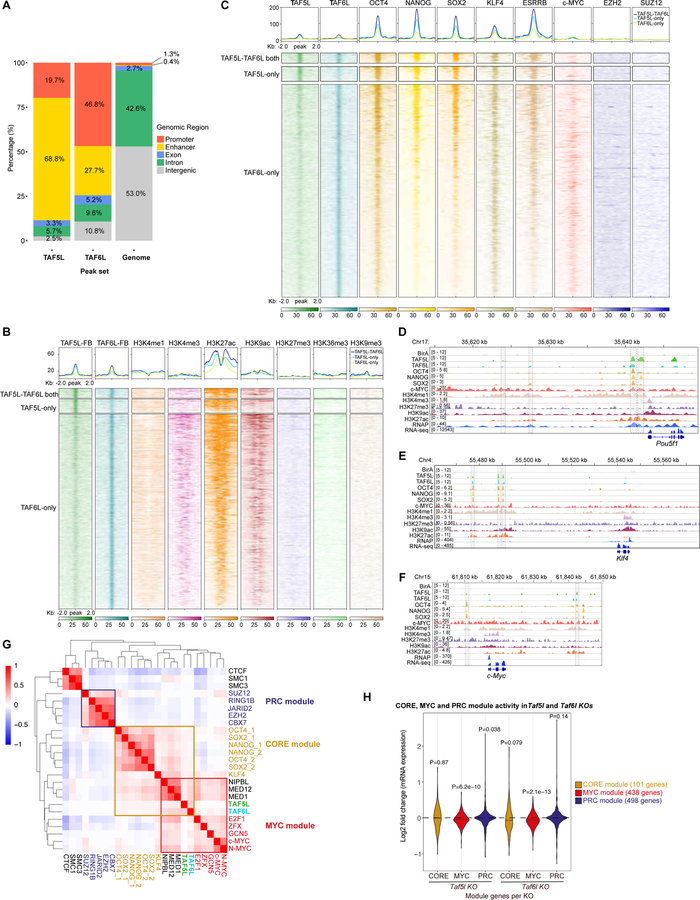
To understand the mechanisms by which TAF5L and TAF6L regulate gene expression and the mESC state, we determined their genome-wide occupancy using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq). As we could not identify any antibodies that specifically recognize endogenous TAF5L and TAF6L, we generated flag-biotin (FB)-tagged TAF5L and TAF6L mESC lines (Figure S4A) and performed in vivo biotinylation-mediated ChIP-seq (Bio-ChiP-seq) of TAF5L and TAF6L (Das et al., 2014; Kim et al., 2009). The majority of TAF5L and TAF6L binding sites were distributed at the promoter and enhancer regions in mESCs (Figure 4A). Of note, mass spectroscopy analyses of streptavidin pull-down of biotinylated TAF5L-FB and TAF6L-FB identified their associated proteins (Figures S4B, S4C), including components of GNAT-HAT complexes (Lee and Workman, 2007). Hence, proteomics supports that FB-tagged versions of TAF5L and TAF6L are functional and suitable for Bio-ChIP-seq.
Figure 4. TAF5L and TAF6L belong to the MYC and CORE regulatory modules but mainly regulate the MYC module activity.
(A) A bar chart shows the genome-wide binding distribution of TAF5L-FB and TAF6L-FB.
(B) A heat map displaying co-occupancy between different active and repressive histone marks with TAF5L-TAF6L both, TAF5L-only and TAF6L-only binding regions.
(C) A heat map representing co-occupied regions of ESC-TFs (OCT4, NANOG, SOX2, KLF4, ESRRB); components of the PRC2 complex (EZH2, SUZ12); and c-MYC binding sites with TAF5L-TAF6L both, TAF5L-only and TAF6L-only binding regions.
(D-F) Genomic tracks of ChIP intensities of several factors, including TAF5L and TAF6L, and histone marks binding at Oct4/pou5f1, Klf4 and c-Myc gene loci. RNAseq tracks represent expression of these genes.
(G) A target correlation map of binding loci shows the degree of co-occupancy between selected TFs and chromatin/epigenetic regulators; three clusters or modules are presented: CORE, MYC and PRC. The colour scale depicts the Pearson correlation coefficient, and the clustering tree is derived from hierarchical clustering. Red colour indicates more frequent co-occupancy between factors.
(H) A violin plot representing changes in CORE, MYC and PRC module gene expression in Taf5l and Taf6l KOs compared to wild-type.
Related to Figure S4.
To determine the probable functions of TAF5L and TAF6L through their direct binding to the target genes, we performed GO analysis of the TAF5L and TAF6L binding target genes. TAF5L binding target genes were enriched for positive regulation of transcription and stem cells maintenance biological functions; while TAF6L binding target genes were highly enriched for positive regulation of transcription, cell cycle, cell division and stem cell maintenance biological functions (Figures S4D, S4E). Furthermore, metagene analysis showed that both TAF5L and TAF6L binding target genes were highly and moderately expressed in mESCs (Figures S4F, S4G) implying that TAF5L and TAF6L most likely positively regulate their target genes for the maintenance of mESC state.
Both TAF5L and TAF6L belong to the MYC and CORE regulatory modules but predominantly regulate the MYC module activity
To interrogate the significance of TAF5L and TAF6L binding, we correlated their occupancy with different active (H3K4me1, H3K4me3, H3K9ac, H3K27ac and H3K36me3) and repressive (H3K27me3 and H3K9me3) histone marks (Zhou et al., 2010), as well as with ESC-TFs (OCT4, NANOG, SOX2, KLF4, ESRRB), components of the PRC2 complex (EZH2, SUZ12) and c-MYC binding sites. TAF5L and TAF6L binding regions overlapped with H3K4me1, H3K9ac, H3K27ac and H3K4me3 active histone marks occupancy (Figure 4B). In addition, TAF5L and TAF6L binding sites overlapped with active TFs such as OCT4, NANOG, SOX2, KLF4, ESRRB and c-MYC occupied sites, but not with repressive EZH2 and SUZ12 binding sites (Figure 4C). Similarly, co-occupancy of TAF5L, TAF6L, OCT4, NANOG, SOX2, c-MYC, H3K4me1, H3K9ac, H3K27ac and H3K4me3 was observed at a few ESC-specific genes (Oct4/Pou5f1, Nanog, Klf4, Esrrb); as well as at c-Myc, Taf5l and Taf6l gene loci (Figures 4D–4F, S4H–S4K). Importantly, binding of TAF5L and TAF6L at these mESC-specific genes and c-Myc loci (Figures 4D–4F, S4H–S4L), and their reduced gene expression in Taf5l and Taf6l KOs (Figures 3D, 3E, S3C), collectively indicate that TAF5L and TAF6L positively and directly regulate ESC-specific genes and c-Myc.
To obtain a global view of TAF5L and TAF6L binding in the mESC genome and shared their occupancy with ESC-TFs and other epigenetic regulators, we combined previously published genome-wide binding datasets of several ESC factors (Das et al., 2014; Kim et al., 2010) together with the TAF5L and TAF6L binding dataset. Hierarchical clustering and a target correlation heat map showed the degree of co-occupancy between factors, and revealed three distinct ESC regulatory modules– CORE, MYC and PRC, as defined previously (Kim et al., 2010). TAF5L and TAF6L binding sites were clustered mainly with the MYC module. However, binding sites of TAF5L and TAF6L were also correlated with the CORE module through NIPBL/MED1/MED12 binding (Figure 4G).
Because TAF5L and TAF6L appear to be new members of the MYC and CORE modules, we monitored “module activity” by examining gene expression changes of the MYC and CORE module genes (i.e., MYC and CORE targeted/bound genes) in Taf5l and Taf6l KOs. MYC module activity was significantly reduced in Taf5l and Taf6l KOs, whereas CORE module activity was modestly reduced only in the Taf6l KO (Figure 4H). Taken together, these data indicate that TAF5L and TAF6L belong to the MYC and CORE regulatory modules, but they predominantly regulate the MYC module activity.
TAF5L/TAF6L regulate target gene expression through H3K9ac deposition and predominant recruitment of c-MYC over OCT4
Since both TAF5L and TAF6L belong to the MYC and CORE regulatory modules, we asked whether and how TAF5L and TAF6L cooperate with c-MYC (of MYC module) and OCT4 (of CORE module) for transcriptional gene regulation. To address this, genome-wide binding data of c-MYC and OCT4 were generated from wild-type, Taf5l KO and Taf6l KO mESCs. The global occupancy of c-MYC was significantly reduced in Taf5l and Taf6l KOs compared to wild-type mESCs, whereas global occupancy of OCT4 was unchanged in these KOs (Figure S5A). The substantial loss of global binding of c-MYC was correlated with dramatic reduction of c-Myc levels in Taf5l and Taf6l KOs; however, global binding of OCT4 was not compromised by a modest reduction of Oct4 levels in Taf5l and Taf6l KOs (Figures 2B, 3D, 3E, S3C, S3D, S5A). Besides, a significant decrease of c-MYC binding was observed at TAF5L bound genes in the Taf5l KO, and at TAF6L bound genes in the Taf6l KO (Figures 5A, 5B). In contrast, OCT4 binding was considerably reduced only at TAF6L bound genes in the Taf6l KO compared to wild-type (Figures 5C, 5D).
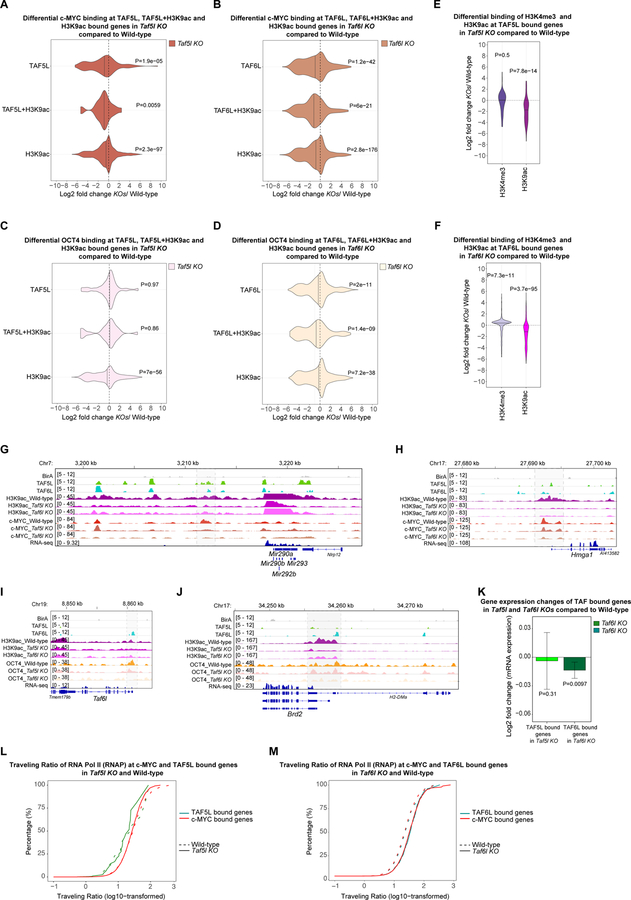
Figure 5. Predominantly TAF5L/TAF6L modulate H3K9ac deposition and c-MYC recruitment at the TAF5L/TAF6L target genes to activate their gene expression through RNA Pol II pause release.
(A) Differential binding of c-MYC at the TAF5L, TAF5L+H3K9ac and H3K9ac bound genes in Taf5l KO compared to wild-type.
(B) Differential binding of c-MYC at the TAF6L, TAF6L+H3K9ac and H3K9ac bound genes in Taf6l KO compared to wild-type.
(C) Differential binding of OCT4 at the TAF5L, TAF5L+H3K9ac and H3K9ac bound genes in Taf5l KO compared to wild-type.
(D) Differential binding of OCT4 at the TAF6L, TAF6L+H3K9ac and H3K9ac bound genes in Taf6l KO compared to wild-type.
(E) Differential binding of H3K9ac and H3K4me3 at the TAF5L bound genes in Taf5l KO compared to wild-type.
(F) Differential binding of H3K9ac and H3K4me3 at the TAF6L bound genes in Taf6l KO compared to wild-type.
(G, H) Genomic tracks of H3K9ac and c-MYC binding at miR 290–295 cluster and Hmga1 gene loci from wild-type, Taf5l and Taf6l KOs. Genomic tracks of BirA (control), TAF5L and TAF6L binding are presented at the same gene loci. Highlighted regions show changes of H3K9ac and c-MYC binding.
(I, J) Genomic tracks of H3K9ac and OCT4 binding at Taf6l and Brd2 gene loci from wild-type, Taf5l and Taf6l KOs. Genomic tracks of BirA (control), TAF5L and TAF6L binding are presented at the same gene loci. Highlighted regions show changes of H3K9ac and OCT4 binding.
(K) Gene expression changes of TAF5L and TAF6L bound genes in Taf5l and Taf6l KOs, respectively.
(L) The traveling ratio (TR) of RNA Pol II (RNAP) at c-MYC and TAF5L bound genes in Taf5l KO and wild-type.
(M) The traveling ratio (TR) of RNA Pol II (RNAP) at c-MYC and TAF6L bound genes in Taf6l KO and wild-type.
Related to Figure S5.
Next, we measured changes of H3K9ac and H3K4me3 marks in Taf5l and Taf6l KOs compared to wild-type, as TAF5L & TAF6L are components of the GNAT-HAT complexes that are related to H3K9ac modification and gene activation (Lee and Workman, 2007), and c-MYC occupancy is correlated with the H3K4me3 signature (Kim 2008; 2010). Our analysis exhibited a substantial reduction of H3K9ac globally (Figure S5B), as well as at TAF5L and TAF6L bound genes in the absence of Taf5l and Taf6l (Figures 5E, 5F). Nonetheless, binding of H3K4me3 was slightly increased globally, but its binding was unaltered at TAF5L and TAF6L bound genes in Taf5l and Taf6l KOs (Figures S5B, 5E, 5F).
Based on these data, we hypothesized that TAF5L/TAF6L deposit H3K9ac and recruit c-MYC and/or OCT4 at target sites. Further analysis revealed a significant reduction of c-MYC occupancy at the common bound gene loci of TAF5L and H3K9ac (TAF5L+H3K9ac), TAF6L and H3K9ac (TAF6L+H3K9ac) in Taf5l KO and Taf6l KO, respectively (Figures 5A, 5B). Likewise, c-MYC occupancy was also reduced at H3K9ac bound genes both in Taf5l and Taf6l KOs (Figures 5A, 5B). Moreover, reduction of H3K9ac and c-MYC occupancy at the TAF5L and TAF6L binding sites of specific genes loci (ESC-specific miR 290–295 cluster and Hmga1) in Taf5l and Taf6l KOs was also observed (Figures 5G, 5H). On the other hand, OCT4 occupancy was mainly reduced at common bound gene loci of TAF6L and H3K9ac (TAF6L+H3K9ac) in Taf6l KO; as well as at H3K9ac bound genes both in Taf5l and Taf6l KOs (Figures 5C, 5D). These events were also observed at specific gene loci, such as Taf6l, and Brd2 (Figure 5I, 5J).
Lastly, we checked gene expression changes of TAF5L and TAF6L bound genes in the Taf5l KO and Taf6l KO, respectively. TAF6L bound genes were significantly downregulated in Taf6l KO, whereas expression of TAF5L bound genes was unchanged in Taf5l KO (Figure 5K). Taken together, our data suggest that TAF5L/TAF6L regulate their target genes expression through H3K9ac deposition and predominate recruitment of c-MYC rather than OCT4, which might occur through indirect recruitment of c-MYC by TAF5L/TAF6L. Of note, we did not detect physical association of TAF5L/TAF6L and c-MYC (Figures S4B, S4C). However, it has been shown that TAF5L/TAF6L physically interacts with c-MYC through other co-factors of the GNAT-HAT complexes in mESCs (Kim et al., 2010). Our data also indicate that TAF6L plays a more critical role compared to TAF5L in controlling gene expression. These inferences are consistent with the respective module activities, where both TAF5L and TAF6L impact the MYC module activity, yet only TAF6L regulates OCT4/CORE module activity (Figure 4H).
TAF5L/TAF6L function with c-MYC to activate target genes expression through RNA Pol II pause release
TAF5L and TAF6L regulate target gene expression principally through c-MYC (Figure 5). c-MYC plays a crucial role in RNA Pol II pause release at target genes for active transcription in mESCs (Rahl et al., 2010). We speculated that TAF5L and TAF6L might function with c-MYC to activate target genes expression through RNA Pol II pause release. To gain insight into this mechanism, we generated binding data for RNA Pol II (RNAP) from Taf5l KO, Taf6l KO and wild-type. Next, we calculated the traveling ratio (TR) of RNAP that evaluates the ratio of RNAP density/occupancy at promoter regions over transcribed/gene body regions (Rahl et al., 2010). We observed a substantial increase in the TR of RNAP at the TAF6L and c-MYC target genes in Taf6l KO compared to wild-type, whereas no such difference in the TR of RNAP was observed at the TAF5L and c-MYC target genes in Taf5l KO (Figures 5L, 5M). Similarly, the TR of RNA Pol II-Ser 2p (transcription elongation) and RNA Pol II-Ser 5p (transcription initiation) was also significantly increased at the TAF6L and c-MYC bound genes in Taf6l KO (Figures S5D, S5F). A small, but significant increase of the TR of RNA Pol II-Ser 2p (but not RNA Pol II-Ser 5p) was observed at the TAF5L and c-MYC bound genes in Taf5l KO (Figures S5C, S5E). Overall, these data suggest that primarily TAF6L functions with c-MYC for RNAP pause release at the promoters of TAF6L and c-MYC target genes for activation.
TAF5L/TAF6L activates MYC regulatory network to maintain self-renewal of mESCs
Several lines of evidence demonstrate that TAF5L and TAF6L not only transcriptionally regulate c-Myc expression (Figures 3E, 4F), but also collaborate with c-MYC to control expression of their target genes (Figures 5). As the connection between TAF5L/TAF6L and c-MYC is robust, we hypothesized that TAF5L/TAF6L might function with c-MYC in the same pathway to control the mESC state. c-MYC maintains the ESC state— preferentially self-renewal of ESCs— by regulating cell cycle, cell proliferation, ribosome biogenesis and metabolism (Chappell and Dalton, 2013; Fagnocchi and Zippo, 2017). Therefore, we asked whether TAF5L and TAF6L are involved in controlling these biological processes.
First, cell cycle profiling showed lengthening of the G0/G1 phase, shortening of the S-phase (related to DNA synthesis) and an unaltered G2/M phase in Taf5l and Taf6l KOs (Figure 6A, S6A). Additionally, expression of the cell cycle gene set was downregulated in Taf5l and Taf6l KOs (Figure 6B), particularly gene sets and individual genes that are related to G1/S-phase, S-phase and DNA replication (Figures S6B–S6F). Moreover, the cell cycle gene set, including G1/S-phase, S-phase and DNA replication gene sets, revealed a reduction of H3K9ac and c-MYC binding at their gene loci in the absence of Taf5l and Taf6l, both globally and at individual gene loci (Figures 6C–6G, S6G, S6H). These data suggest that TAF5L and TAF6L regulate the cell cycle, specifically G1/S-phase, S-phase and DNA replication– similar to the function of c-MYC in controlling cell cycle (Cartwright, 2005; Scognamiglio et al., 2016; Singh and Dalton, 2009).
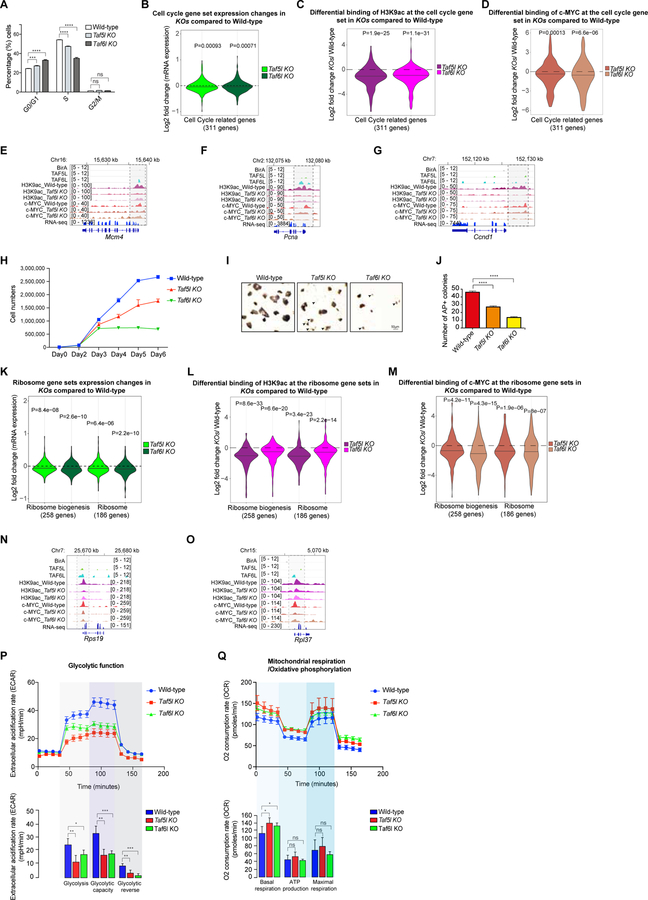
Figure 6. TAF5L/TAF6L maintain self-renewal of mESCs through MYC regulatory module/network.
(A) Quantitative analysis of different cell cycle phases from wild-type, Taf5l and Taf6l KOs.
(B) Cell cycle gene set expression changes in Taf5l and Taf6l KOs compared to wild-type.
(C, D) Differential binding of H3K9ac (C) c-MYC (D) at the cell cycle gene set in Taf5l and Taf6l KOs compared to wild-type.
(E-G) Genomic tracks of H3K9ac and c-MYC binding at Mcm4, Pcna and Ccnd1 gene loci from wild-type, Taf5l and Taf6l KOs. Genomic tracks of BirA, TAF5L and TAF6L binding are presented at the same gene loci. RNAseq tracks show the expression of these genes in mESCs. Highlighted regions display changes of H3K9ac and c-MYC occupancy.
(H) Cell proliferation assay from wild-type, Taf5l and Taf6l KOs (day0 to day6).
(I) Alkaline phosphatase (AP)+ colonies from wild-type, Taf5l and Taf6l KO mESCs. Arrowheads indicate differentiating cells. Scale bar: 50µm.
(J) Quantification of numbers of AP+ colonies (12-well scale).
(K) Expression changes of two ribosome gene sets in Taf5l and Taf6l KOs compared to wild-type.
(L, M) Differential binding of H3K9ac (L) and c-MYC (M) at the ribosomes gene sets in Taf5l and Taf6l KOs compared to wild-type.
(N-O) Genomic tracks of H3K9ac and c-MYC binding at ribosome (Rps19 and Rpl37) gene loci from wild-type, Taf5l and Taf6l KOs. Genomic tracks of BirA, TAF5L and TAF6L binding are presented at the same loci. RNAseq tracks show the expression of these genes in mESCs. Highlighted regions show changes of H3K9ac and c-MYC occupancy.
(P) Glycolytic function measurement using extracellular acidification rate (ECAR) from wild-type, Taf5l and Taf6l KOs.
(Q) Oxidative phosphorylation activity presented based on oxygen consumption rate from wild-type, Taf5l and Taf6l KOs.
Data are represented as mean ± SEM (n = 3); p-values were calculated using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; and ns (non-significant).
Second, we observed a reduction in cell proliferation in Taf5l and Taf6l KOs (Figure 6H), consistent with a cell cycle defect. Alkaline phosphatase (AP) staining revealed smaller and fewer dome-shaped AP+ colonies, with a fraction of differentiated cells in Taf5l and Taf6l KO mESCs (Figures 6I, 6J). Moreover, we observed relatively fewer AP+ colonies and more differentiated cells in Taf6l KO compared to Taf5l KO (Figures 6I, 6J). These findings suggest that TAF5L and TAF6L regulate cell proliferation of mESCs in a manner comparable to c-MYC (Eilers and Eisenman, 2008; Singh and Dalton, 2009).
Third, we observed reduced gene expression of ribosome gene sets (Figure 6K) and MYC module (MYC targeted) ribosome genes (Figure S6I) in Taf5l and Taf6l deleted mESCs. Ribosome gene sets showed a reduction of H3K9ac and c-MYC binding at their gene loci in the absence of Taf5l and Taf6l, both globally and at individual gene loci (Figures 6L–6O). These data suggest that TAF5L and TAF6L regulate ribosome biogenesis through c-MYC, similar to the function of c-MYC in ribosome biogenesis (Kim et al., 2008; van Riggelen et al., 2010).
Fourth, we assessed two major metabolic pathways, glycolysis and oxidative phosphorylation (OxPhos). Glycolytic function was significantly reduced in the absence of Taf5l and Taf6l (Figure 6P), and correlated with reduced expression of glucose transporter genes (important for glucose uptake) in Taf5l and Taf6l KOs (Figure S6J). A high rate of glucose uptake and glycolysis is required for rapid proliferation of mESCs, similar to cancer cells (Vander Heiden et al., 2009). In contrast, mitochondrial respiration/OxPhos function was unaffected, except for an upregulation of basal respiration level in Taf5l and Taf6l KOs (Figure 6Q), which may be linked to the low level of ongoing differentiation (Figure 6I) (Shyh-Chang et al., 2013). These functional data suggest that TAF5L and TAF6L play a pivotal role in glycolysis metabolism. Recent evidence also underlines the importance of c-MYC in regulating the glycolytic metabolism of both human and mouse ESCs (Cao et al., 2015; Gu et al., 2016).
Lastly, we examined whether loss of c-Myc function phenocopies loss of Taf5l and Taf6l function in mESCs. Gene expression analysis demonstrated a partial but significant overlap between differentially expressed genes in c-Myc, Taf5l and Taf6l KOs (Figure S7A–S7E), which implies that TAF5L/TAF6L work together with c-MYC and regulate gene expression. However, a significant fraction of non-overlapping differentially expressed genes was observed in c-Myc, Taf5l and Taf6l KOs (Figure S7E), which indicates that TAF5L/TAF6L and c-MYC work independently as well.
Taken together, our data demonstrate that TAF5L/TAF6L activates the MYC regulatory network through multidimensional control of cell cycle, cell proliferation, ribosome biogenesis and metabolism to maintain self-renewal of mESCs.
DISCUSSION
TAF5L and TAF6L mediated gene regulation
Here, we identified TAF5L and TAF6L as novel epigenetic regulators for the mESC state (Figures 1, 2). TAF5L and TAF6L are components/co-activators of different STAGA- (or SAGA), PCAF- and TFTC-HAT complexes that belong to the GNAT family of HAT complexes (Carrozza et al., 2003; Lee and Workman, 2007). GCN5/KAT2A is the catalytic component of STAGA- and TFTC-HAT complexes, whereas PCAF/KAT2B is the catalytic component of the PCAF-HAT complex (Lee and Workman, 2007). HAT complexes exhibit different functions depending on their precise composition. Specific subunits of HAT complexes may select particular histone residues for acetylation; and/or guide complexes to the genome to recruit other TFs, co-activators and general transcription machinery for gene activation (Lee and Workman, 2007). We demonstrate that both TAF5L and TAF6L are part of the STAGA-, PCAF- and TFTC-HAT complexes (Figure S4B–S4C), and belong to the MYC and CORE regulatory modules, but predominantly regulate the MYC module activity (Figures 4G–4H). Previous studies reveal a functional relationship between GCN5/KAT2A of HAT complexes and c-MYC (Hirsch et al., 2015; Kim et al., 2010). However, molecular mechanisms underlying this relationship are ill-defined. Our findings close this gap in understanding. We show that c-Myc is a direct downstream target of TAF5L/TAF6L, and TAF5L/TAF6L transcriptionally activate c-Myc gene expression (Figures 3, 4). In addition, mechanistic studies illustrate how TAF5L and TAF6L modulate H3K9ac deposition and recruit c-MYC at the TAF5L and TAF6L target genes to activate the MYC gene regulatory network/module (Figures 5, 6). Likewise, TAF5L/TAF6L transcriptionally activate Oct4 (Figures 3, 4); and particularly, TAF6L controls H3K9ac deposition and OCT4 recruitment at the TAF6L bound genes to trigger the CORE regulatory network (Figure 5, 6I). These events are consistent with the observation that both TAF5L and TAF6L influence MYC module activity, whereas only TAF6L affects OCT4/CORE module activity (Figure 4H). Overall, our data indicate that TAF5L/TAF6L regulate both c-Myc and the MYC module, as well as Oct4 and the CORE module; but predominately regulate c-Myc and the MYC network/module for self-renewal of mESCs (Figures 3, 4, 5, 6).
Our systematic analyses show that TAF6L plays a more critical role than TAF5L in gene regulation based on the followings: 1) TAF6L controls the MYC module activity to a greater degree than TAF5L (Figure 4H); 3) H3K9ac deposition and c-MYC binding/recruitment are more dependent on TAF6L than TAF5L at their bound genes for activation (Figures 5A, 5B, 5E, 5F, S5A, S5B); 4) primarily TAF6L functions with c-MYC for RNAP pause release at the promoters of TAF6L and c-MYC target genes for activation (Figures 5L, 5M, S5C–S5F); 5) only TAF6L controls OCT4 (of CORE module) recruitment at the TAF6L bound genes, and regulates the CORE module activity (Figures 4H, 5C, 5D, 5I, 5J); 6) the correlation between gene expression and binding data of TAF5L and TAF6L demonstrates that only TAF6L bound genes are significantly downregulated in the absence of Taf6l (Figure 5K). However, we cannot exclude a positive role for TAF5L in controlling some critical ESC-specific genes, such as Oct4, Nanog, Klf4, Esrrb and c-Myc (Figures 3, 4, 5). Indeed, metagene and GO analyses show that TAF5L bound genes are high/moderately expressed in the mESCs, and they are related to positive regulation of transcription and stem cells maintenance functions, same as TAF6L (Figures S4D–S4G). Moreover, TAF5L also regulates the cell cycle, DNA replication and ribosomal genes, similar to TAF6L (Figures 6, S6); suggesting that TAF5L works with TAF6L to regulate important gene sets and individual genes that control the mESC state.
TAF5L and TAF6L predominantly regulates self-renewal of mESCs
Our data provide evidence that TAF5L/TAF6L work together with c-MYC to activate the MYC regulatory network that regulates cell cycle, DNA replication, ribosome biogenesis and metabolism (particularly glycolysis) to maintain mainly cellular proliferation and self-renewal of mESCs (Figures 4, 5, 6, 6H–6I) (Fagnocchi and Zippo, 2017; Shyh-Chang et al., 2013; Singh and Dalton, 2009; van Riggelen et al., 2010). However, low-level differentiation of mESCs was also detected in the absence of Taf5l and Taf6l (Figure 6I)– caused by upregulation of TE-specific genes and/or downregulation of ESC-specific genes (e.g., Pou5f1/Oct4, Nanog, Klf4, Tbx3) in Taf5l and Taf6l KOs (Figures 3D–3F), possibly through ESC-specific genes as they are the direct targets of TAF5L/TAF6L and part of the CORE regulatory network (Figures 4, 5). Overall, we demonstrate a predominant self-renewal defect, and a low-level of differentiation in the absence of Taf5l and Taf6l (Figures 6H–6J), which is associated with TAF5L/TAF6L directed predominant regulation of the c-MYC/MYC network over the OCT4/CORE network in the mouse ESCs (Figures 3, 4, 5, 6).
Several studies demonstrate that MYC regulates cell cycle, DNA replication, ribosome biogenesis and metabolism for stem cell proliferation and self-renewal of ESCs (Chen et al., 2008; Eilers and Eisenman, 2008; Kim et al., 2008). Furthermore, MYC controls proliferation and self-renewal, but not pluripotency/differentiation in the ground-state of mESCs using 2i+LIF condition (Scognamiglio et al., 2016). ESCs and cancer cells/cancer stem cells are linked to self-renewal and proliferation, based on overlapping ESC/CORE and MYC module activities (Kim et al., 2010; Wong et al., 2008). While the majority of the studies emphasize the central role of MYC in proliferation and self-renewal, others demonstrate the role of MYC in differentiation/pluripotency of ESCs as well (Smith et al., 2010; Varlakhanova et al., 2011; 2010).
The self-renewal defect observed in c-Myc and N-Myc double knockout (DKO) ESCs, but not in single KO ESCs maintained in serum+LIF or 2i+LIF conditions (Scognamiglio et al., 2016; Varlakhanova et al., 2010), suggests overlapping functions of c-MYC and N-MYC. We demonstrate a self-renewal defect in Taf5l and Ta6l single KO mESCs (Figures 6H–6J), which is similar to the self-renewal defect in c-Myc and N-Myc DKO mESCs, but not that seen in c-Myc single KO mESCs (Scognamiglio et al., 2016; Varlakhanova et al., 2010). These observations imply that TAF5l/TAF6L regulate self-renewal through both c-MYC and N-MYC (of the MYC regulatory network) and they have a compensatory function in mESCs.
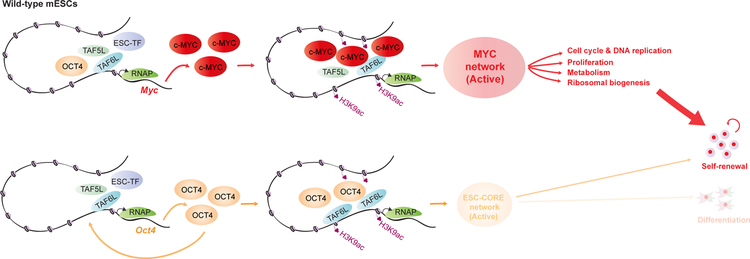
In conclusion, we demonstrate that TAF5L/TAF6L transcriptionally activate c-Myc and Oct4, and their respective MYC and CORE regulatory modules. Furthermore, we reveal a mechanism by which TAF5L/TAF6L predominantly activates the MYC regulatory module/network that fine-tunes gene expression programs to control self-renewal for the maintenance of mESC state (Figure 7).
Figure 7. A model represents the detailed function of TAF5L and TAF6L in mESCs.
The proposed model describes TAF5L and TAF6L transcriptionally activate c-Myc gene expression. Also, TAF5L/TAF6L regulate their target genes through H3K9ac deposition and c-MYC recruitment that eventually activate the MYC regulatory network for self-renewal of mESCs. Similarly, TAF5L and TAF6L transcriptionally activate Oct4 gene expression. Particularly, TAF6L modulate H3K9ac deposition and Oct4 recruitment at their target sites to activate ESC/CORE regulatory network that controls self-renewal and a low-level of pluripotency/differentiation of mESCs. Our findings suggest that TAF5L/TAF6L predominantly activates c-Myc and the MYC regulatory network over Oct4 and the CORE regulatory network to control mainly self-renewal for the maintenance of mESC state. TAF5L/TAF6L mediated gene activation of c-Myc and Oct4, and their corresponding activated MYC and CORE networks are shown. MYC network display in larger bold font (bright red), compared to CORE network (light orange)– represents predominant MYC network activity over CORE network. The thick arrowhead (red) depicts the activated MYC network that primarily directs the self-renewal of mESCs. Simultaneously, the thin arrowhead (light orange) illustrates an activated CORE network that controls a low-level of self-renewal and differentiation of mESCs.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact:
Stuart Orkin (orkin@bloodgroup.tch.harvard.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse embryonic stem cells (mESCs)
Mouse ESCs (mESCs) were cultured in mouse ESC media that contains DMEM (Dulbecco’s modified Eagle’s medium) (Thermo Fisher Scientific) supplemented with 15% fetal calf serum (FCS) (Omega Scientific), 0.1mM b-mercaptoethanol (Sigma-Aldrich), 2mM L-glutamine (Thermo Fisher Scientific), 0.1mM nonessential amino acid (Thermo Fisher Scientific), 1% of nucleoside mix (Merck Millipore), 1000 U/ml recombinant leukemia inhibitory factor (LIF/ESGRO) (Merck Millipore), and 50U/ml Penicillin/Streptomycin (Thermo Fisher Scientific). mESCs were cultured at 37ºC, 5% CO2.
Mouse embryonic fibroblasts (MEFs)
Reprogrammable mouse embryonic fibroblasts (MEFs) cultures were prepared from rtTA3-OKSM embryos (embryonic day-E13.5) that were heterozygous for the Oct4-GFP reporter, OKSM cassette and rtTA3 (Stadtfeld et al., 2009). These Doxycycline-inducible-OKSM driven reprogrammable MEFs harbouring Oct4-GFP reporter– were cultured with DMEM supplemented with 10% fetal bovine serum (FBS) (Omega Scientific) and 2% penicillin-streptomycin at 37ºC, 5% CO2. For reprogramming into iPSCs, MEFs were cultured at 37ºC, 5% CO2 in mESC media supplemented with 2µg/ml doxycycline (dox) (Sigma-Aldrich) and 50ug/ml ascorbic acid (Sigma-Aldrich).
Induced pluripotent stem cells (iPSCs)
iPSCs were cultured on irradiated MEFs (Thermo Fisher Scientific) at 37ºC, 5% CO2 in mESC media.
Human embryonic kidney cells (HEK293T cells)
HEK293T cells were cultured with DMEM supplemented with 10% fetal bovine serum (FBS) (Omega Scientific) and 2% penicillin-streptomycin (Thermo Fisher Scientific). These cells were cultured at 37ºC, 5% CO2.
METHOD DETAILS
Mouse epigenetic CRISPR-Cas9 pooled library design
A list of epigenetic genes was generated through literature mining, which cover different classes of epigenetic regulators and ESC-specific TFs (Supplementary Table 1). Positive controls were selected as genes with known roles in embryonic stem cell state (25 genes). All possible sgRNAs targeting GFP (of Oct4-GFP reporter) were also included as positive controls. Previously published non-targeting sgRNAs were included as negative controls (Canver et al., 2017). sgRNAs (6 sgRNAs/ gene) targeting coding sequences of all the selected genes were retrieved from the mouse GeCKOv2.0 library (Sanjana et al., 2014). In total, the epigenetic CRISPR-Cas9 pooled library consisted of 2,335 sgRNAs, including 1,938 sgRNAs targeting the 323 identified epigenetic and ESC TF genes, 150 sgRNAs targeting the 25 genes with known roles in embryonic stem cell state, 119 sgRNAs targeting GFP and 128 non-targeting sgRNAs.
Mouse epigenetic CRISPR-Cas9 pooled library construction
All the sgRNA oligonucleotides of the library were synthesized as previously described (Shalem et al., 2014) using a B3 synthesizer (CustomArray, Inc.), pooled together, PCR amplified and cloned into Esp3I-digested plentiGuide-Puro (Addgene plasmid ID: 52963) lentiviral vector, using a Gibson assembly master mix (New England Biolabs). Gibson assembly products were transformed into electrocompetent cells (E. cloni, Lucigen) and plated on 245mm x 245mm square LB-agar plates to obtain sufficient number of bacterial colonies at a ∼50× library coverage. Bacterial colonies were collected from the plates, genomic DNA was isolated and plasmid libraries were prepared for high-throughput sequencing to confirm the representation of each sgRNAs in the pooled library.
Lentiviral production
HEK293T cells were seeded onto 15cm dishes ~24hrs prior to transfection. Cells were transfected at 80% confluence in 16ml of media with 8.75μg of VSVG, 16.25μg of psPAX2, and 25μg of the CRISPR-Cas9 pooled lentiviral plasmids, using 150μg of linear polyethylenimine (PEI) (Sigma-Aldrich). Media was changed with fresh media 16–24hrs of post-transfection. Lentiviral supernatant was collected at 48 and 72hrs post-transfection and subsequently concentrated by ultracentrifugation (24000 rpm, 4ºC, 2hrs) (Beckman Coulter SW32).
CRISPR-Cas9 mediated screen in mESCs
Oct4-GFP reporter mESCs with stably expressed Cas9 were transduced at low multiplicity of infection (MOI) to avoid more than one lentiviral integration per cell. Test transductions were performed to estimate the amount of virus required to ensure 30% transduction rate. 10μg/ml blasticidin (InvivoGen) and 1μg/ml puromycin (Sigma-Aldrich) were added 24hrs after transduction to select for lentiviral library integrants (puromycin resistant) in cells with Cas9 (blasticidin resistant). Cells were selected for next 5days, before FACS sorting. The GFP-low and GFP-high cells were sorted, genomic DNA was isolated, and libraries were prepared for deep sequencing to enumerate the existence of sgRNAs in these cell populations. The screening was performed in biological triplicates.
Generation of mESC KO deletion clones
Paired sgRNAs were designed to delete critical coding exons of the selected candidate genes. These sgRNAs were cloned into lentiguide-Puro (Addgene ID: 52963) plasmid, using Golden Gate Cloning approach as mentioned previously. Wild-type (J1) mESCs were transduced with Cas9-blast virus (generated from pLentiCas9-Blast, Addgene ID: 52962) and selected with 10μg/ml blasticidin (InvivoGen) to generate the J1 mESC cell line with stable expression of Cas9 (mESC+Cas9). 50,000 of mESC+Cas9 cells were transfected with 500ng of each sgRNAs (5 and 3’ sgRNAs) using Lipofectamine 2000 (Thermo Fisher Scientific), and cells were selected with 1μg/ml puromycin (Sigma-Aldrich) for 3–4 days. Next, puromycin-resistant cells were re-plated on a 15cm plate as single cells to grow individual clones. The individual clones were picked, expanded and genotyped PCR was performed to detect the homozygous/biallelic deletion or knockout (KO) mESC clones.
Somatic cell reprogramming/iPSCs generation
Reprogramming was performed by seeding Doxycycline-inducible-OKSM driven reprogrammable MEFs harbouring Oct4-GFP reporter, onto gelatin-coated 6-well plates (Stadtfeld et al., 2009). These cells were transduced with lentiviral particles of sgRNAs carrying Cas9 and mCherry. 24hrs post-transduction, mCherry+ MEFs were sorted, ~50,000 mCherry+ MEFs were plated on irradiated MEFs at 6-well format, and cultured in mESC media with 2µg/ml doxycycline (Sigma-Aldrich) and 50ug/ml ascorbic acid (Sigma-Aldrich). At day12, doxycycline was withdrawn, and cells were cultured for an additional 4 days with mESC media to obtain transgene-independent iPSCs. However, we obtained different percentages of mixed populations of iPSCs and differentiated cells after the reprogramming upon perturbation of Taf5l and Taf6l using their targeting sgRNAs. Non-targeting sgRNA was used as a control. The bulk/mixed populations of iPSCs and differentiated cells after reprogramming were used for RT-PCR analysis, FACS and Alkaline Phosphatase (AP) staining. Indel frequencies of Taf5l and Taf6l genes were assessed by targeted amplicon sequencing before (from mCherrey+ reprogrammable MEFs transduced with viral particles of Taf5l and Taf6l targeting sgRNAs that carrying mCherry and Cas9) and after (mixed populations of iPSCs and differentiated cells) reprogramming. Indel frequencies of Taf5l and Taf6l alleles were quantified using the web-based tool– CRISPRESSO2 (http://crispresso.pinellolab.partners.org/).
Flow cytometry
Cells were dissociated using trypsin, washed with 1XPBS, followed by sorting. i) During the CRISPR-Cas9 screening, mESCs were sorted based on GFP-low and GFP-high; ii) percentages (%) of GFP (high and low) cells were quantified from Oct4-GFP reporter mESCs upon depletion of Taf5l and Taf6l using their targeting sgRNAs; iii) SSEA1+ve cells were measured from wild-type, Taf5l and Taf6l KOs; iv) During reprogramming, reprogrammable MEFs were also sorted based on mCherry.
Intracellular FACS
Cells were dissociated using trypsin; then fixed and permeabilized using eBioscience Fixation & Permeabilization Buffer Set (88–8824, eBioscience). Permeabilized Cells were incubated with PE-Conjugated-OCT3/4 Monoclonal Antibody (1:100; 12–5841-80: eBioscience™) and its Isotype control (1:100; 12–4321-80:eBioscience™) for overnight at 4°C. Next day, cells were washed 3 times and resuspended in 200μl of permeabilization wash buffer, and analysed through FACS Calibur.
Immunofluorescence microscopy
mESCs were fixed with 4% paraformaldehyde. The primary antibody against OCT4 (N-19: sc-8628, Santa Cruz) was used, and the secondary antibody was obtained from Jackson ImmunoResearch (705–585-003). Cells were imaged using a Nikon Eclipse fluorescence microscope. Mean fluorescence intensity (MFI) of OCT4+ cells were quantified using Image J.
Alkaline Phosphatase (AP) staining
AP staining of ESCs and iPSCs was performed according to the manufacture’s protocol (Vector Red Substrate Kit, SK-5100, Vector Lab).
Cell Cycle analysis
Cell cycle analysis was performed by BrdU staining using the APC BrdU Flow Kit (BD). Cells were incubated with BrdU for 15 minutes and analysed using a BD Accuri C6 Plus flow cytometer.
Cell proliferation assay
~10,000 mESCs were plated on each well of the 6-well plates and cultured in mouse ESC (mESC) media up to 6 days. Cells were dissociated using trypsin, washed with 1XPBS, stained with trypan blue, and the live cells were counted using Countess™ II Automated Cell Counter (thermos Fisher Scientific) at Day 2, 3, 4, 5 and 6.
Measurement of metabolic functions in mESCs
A Seahorse Bioscience XFe24 Extracellular Flux Analyzer (Agilent) was used to measure– the oxygen consumption rate (OCR), related to OxPhos; and extracellular acidification rate (ECAR), related to glycolytic function from mESCs in XFe24 FluxPak Mini cell culture microplates as described previously (Sun et al., 2015). mESCs were seeded onto gelatin-coated XFe24 microplates at a density of 1 × 105 per well in 500µl of mESC media, and then incubated at 37°C with 5% CO2. After cell attachment (~5 hours later) in mESC media, the media was replaced with 500ul of Seahorse assay media. Measurements of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were performed after equilibration of cells at 37°C in an anaerobic incubator for 1 hour. OCR was measured in cells that were treated with oligomycin (1μM), FCCP (1μM) and rotenone (1μM) (Agilent Seahorse XF Cell Mito Stress Test Kit (103015–100, Agilent); ECAR was measured in cells that were treated with glucose (10mM), oligomycin (1μM) and 2-DG (50mM) (Agilent Seahorse XF Glycolysis Stress Test Kit (103020–100, Agilent) according to the manufacturer’s instructions. OCR is reported in the unit of picomoles per minute and ECAR is reported in milli-pH units (mpH) per minute. OCR and ECAR were normalized to protein content (determined via Bradford assay) in each well. Basal respiration, ATP production, maximum respiratory capacity (related to OxPhos); and glycolysis, glycolytic capacity and glycolytic reserve (related to glycolytic function) were calculated as per the manufacturer’s instructions.
Generation of Flag-Biotin (FB) tagged mESC lines
Mouse Taf5l and Taf6l ORFs were synthesized (IDT), and cloned into BamHI-digested pEF1a-Flagbio(FB)-puro vector, using Gibson Assembly Master Mix (NEB). Positive clones were analysed by Sanger sequencing. 10µg of Taf5l-FB and Taf6l-FB constructs were electroporated into 5×106 J1 wild-type mESCs, which constitutively express BirA ligase (neomycin). The electroporated cells were plated on a 15cm dish, with mESC media. After ~24hrs, media was replaced with fresh mESC media containing 1μg/ml of puromycin (Sigma-Aldrich) and 1μg/ml of neomycin (Sigma-Aldrich), and cells were selected for 4–5days. Individual ESC colonies were picked, expanded, and tested by western blot using streptavidin-HRP (GE healthcare) (dilution 1:2000 in 5% BSA) to detect the clones that expressed TAF5L-FB and TAF6L-FB separately.
Pull down of Flag-Biotin tagged TAF5L and TAF6L, and identification of their interacting partners through mass spectrometry
One-step affinity purification with streptavidin-beads was performed as previously reported (Kim et al., 2009). In brief, nuclear extracts were obtained from 300millions cells using NE-PER reagents (Thermo Fisher Scientific) and quantified using DC Protein Assay (Biorad). 5mg of nuclear extract from mESCs either expressing only BirA (control) or BirA plus biotin-tagged protein of interest (for example, TAF5L-FB/TAF6L-FB) were diluted in 10 ml of IP150 or IP350 buffer (20mM Tris-HCl (pH 7.5), 0.3% (vol/vol) NP-40, 1mM EDTA, 10% (vol/vol) glycerol (vol/vol), 1mM DTT, 0.2mM PMSF and protease inhibitor cocktail (Sigma 8340, 1: 1000), and 150 mM or 350 mM NaCl). 100ul Dybabeads MyOne Streptavidin T1 beads (Thermo Fisher Scientific) were added and incubated with rotation overnight at 4ºC. Beads were washed 4 times with 10ml of IP buffer and eluted in 250ul NP40 Lysis buffer (50mM Tris-HCl, 150mM NaCl, 1% NP-40, and 5mM EDTA) supplemented with 1% SDS. Samples were concentrated using TCA, boiled in Laemmli buffer for 5 min, and resolved on a 10% SDS-polyacrylamide gel. Whole lane LC-MS/MS sequencing and peptide identification were performed at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School.
Western Blot
Protein extract was mixed with Laemmli buffer, boiled for 5 minutes, and resolved on a 4–12% gradient Bis-Tris gel (Bio-Rad). Proteins are on the gel transferred to the PVDF membrane, and specific antibodies were used to detect proteins of interest.
RNA isolation and RT-qPCR
RNA was isolated from cells using RNeasy Mini Kit (Qiagen), and cDNA was prepared using iScript cDNA Synthesis Kit (Bio-Rad). RT-qPCR was performed using iQ SYBR Green Supermix (Bio-Rad) on Bio-Rad iCycler RT-PCR detection system.
RNA-seq
Total RNA was isolated, and ribosomal RNAs were depleted using ribosomal RNA depletion kit (rRNA Depletion Kit, NEB). Ribosomal depleted RNA was used to make the RNAseq libraries using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB). All the libraries were checked through a Bio-analyzer for quality control purpose. Single End (SE) 100bp reads were generated using a HiSeq-2500 sequencer (Illumina). RNAseq were performed in triplicates.
ChIP-seq
Chromatin Immunoprecipitation (ChIP) was performed as described previously (Das et al., 2014; Kim et al., 2009). For bioChIP reactions, streptavidin beads (Dynabeads MyOne Streptavidin T1- Invitrogen) were used for the precipitation of chromatin, and 2% SDS was applied for first washing step. All other steps were same as conventional ChIP protocol. BirA expressing J1 mESCs were used as a control. Conventional ChIP reactions were performed as described previously (Das et al., 2014; 2015). Input genomic DNA was used for the reference sample. Briefly, cells were tryipsinized from 15cm dishes, washed twice with 1XPBS, and crosslinked with 37% formaldehyde solution (Calbiochem) to a final concentration of 1% for 8 min at room temperature with gentle shaking. The reaction was quenched by adding 2.5M glycine to a final concentration of 0.125M. Cells were washed twice with 1XPBS, and cell pellet was resuspended in SDS-ChIP buffer (20 mM Tris-HCl pH 8.0, 150mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100 and protease inhibitor), and chromatin was sonicated to around 200–500 bp. Sonicated chromatin was incubated with 5~10µg of antibody overnight at 4°C. After overnight incubation, protein A/G Dynabeads magnetic beads (Thermo Fisher Scientific), were added to the ChIP reactions and incubated for 2–3 hours at 4°C to immunoprecipitate chromatin. Subsequently, beads were washed twice with 1 ml of low salt wash buffer (50mM HEPES pH 7.5, 150mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate), once with 1ml of high salt wash buffer (50mM HEPES pH 7.5, 500mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate), once with 1ml of LiCl wash buffer (10mM Tris-HCl pH 8.0, 1mM EDTA, 0.5% sodium deoxycholate, 0.5% NP-40, 250 mM LiCl), and twice with 1ml of TE buffer (10mM Tris-HCl pH 8.0, 1mM EDTA, pH 8.0). The chromatin was eluted twice in SDS elution buffer (1% SDS, 10mM EDTA, 50mM Tris-HCl, pH 8.0) at 72ºC, each time with 150µl of SDS elution buffer. Eluted chromatin was reverse-crosslinked at 65°C overnight. The next day, an equal volume of TE was added (300µl). ChIP DNA was treated with 1µl of RNaseA (10mg/ml) for 1hr, and with 3µl of proteinase K (20mg/ml) for 3hrs at 37ºC, and purified using phenol-chloroform extraction, followed by QIAquick PCR purification spin columns (Qiagen). Finally, ChIP-DNA was eluted from the column with 40µl of water. For several factors, we used multiple ChIPs. At the end, all eluted ChIP-DNA samples were pooled and precipitated to enrich the ChIP-DNA material to make the libraries for high-throughput sequencing. Input ChIP samples were reserved before adding the antibodies, and these input samples were processed from reverse cross-linking step until the end, same as other ChIP samples.
2–10 ng of purified ChIP DNA was used to prepare sequencing libraries, using NEBNext ChIP-seq Library Prep Master Mix Set for Illumina (NEB) and NEBNext Ultra DNA Library Prep Kit for Illumina (NEB) according to the manufacturer’s instructions. All libraries were checked through a Bio-analyser for quality control purpose. ChIP sequencing (50bp SE reads) was performed using Hiseq-2500 (Illumina).
QUANTIFICATION AND STATISTICAL ANALYSIS
CRISPR-Cas9 screen data analysis
sgRNA sequences were enumerated from the GFP-low and GFP-high populations. An enrichment score for each sgRNA was calculated as the log2 transformation of the median number of occurrences of a given sgRNA in the GFP-low population divided by the median number of occurrences of the same sgRNA in the GFP-high population. The enrichment score for each gene was calculated as the median of the 3 most enriched sgRNA targeting each gene across three biological replicates.
RNA-seq analysis
Reads were mapped to the mm9 mouse genome assembly and the GENCODE version M1 mouse transcriptome (Mudge and Harrow, 2015) using the STAR RNA-seq read alignment program (v2.5.3a) (Dobin et al., 2013) with default parameters. Bigwig tracks were generated using STAR to generate RPM-normalized wiggle files. Subsequently the wigToBigWig tool (v4) from the UCSC genome browser Kent tools (Kent et al., 2010) was used to convert the wiggle files to BigWig format. Gene expression quantification, both raw counts and TPM (Transcripts Per Million) values, were obtained using Salmon (v0.8.2) (Patro et al., 2017). Differential gene expression analysis was done using the DESeq2 (v1.14.1) (Love et al., 2014) R statistical programming language (v3.3.2) software package from the Bioconductor project (v3.4) (Huber et al., 2015) using the GENCODE version M1 mouse transcriptome. DESeq2 was also used to calculate the normalized counts and the FPKM (Fragments Per Kilobase per Million mapped reads) expression values. MA plots were created employing the ggplot2 R package (v2.2.1) that used log2 fold changes and mean normalized counts calculated by the DESeq2 R package.
GO (Gene Ontology)
Gene Ontology (Ashburner et al., 2000) analysis was performed using the NIH DAVID website (v6.8) (Dennis et al., 2003); default settings and the whole genome were used as background. Genes were identified using their Ensembl Gene IDs for the DAVID analysis, and their associated gene names were obtained from the GENCODE vM1 mouse transcriptome annotation and added to the resulting GO term enrichment tables.
ChIP-seq analysis
The 50bp single end (SE) reads were mapped to the mm9 mouse genome assembly using Bowtie 2 (v2.3.2) (Langmead and Salzberg, 2012). BigWig tracks were generated using the ‘bamCoverage’ program from the DeepTools software suite (v2.5.3) (Ramírez et al., 2016), with reads extended to 300bp and tracks normalized using the RPKM (Reads Per Kilobase per Million mapped reads) algorithm. Peak calling was done with the MACS2 program (v2.1.1.20160309) (Zhang et al., 2008). For most ChIP-seq datasets default MACS2 settings were used. For the TAF5L-FB and TAF6L-FB datasets, MACS2 was run with the “nomodel” option. Artifact peaks were filtered out based on the mouse ENCODE project (Mouse ENCODE Consortium et al., 2012), They were removed using “bedtools intersect” from the BEDTools software suite (v2.26.0) (Quinlan and Hall, 2010). Mapping of peaks to the nearest gene was done using “bedtools closest”. Genomic distribution of peaks was performed using “bedtools intersect” using annotated genomic regions taken from the UCSC mm9 known gene transcriptome as implemented in the Bioconductor “TxDb.Mmusculus.UCSC.mm9.knownGene” package, as well as mouse enhancers taken from the literature (Whyte et al., 2013). As a single peak can overlap to the multiple genomic regions, the following hierarchy was used to prevent counting the same peak multiple times.
Heat-map
ChIP-seq datasets were generated using the “computeMatrix” and “plotHeatmap” programs of the DeepTools software suite (v.2.5.3) (Ramírez et al., 2016) for ChIP-seq heatmaps and profile plots of histone marks and transcription factors binding. Regions from −2kb to +2kb around the centre of the peaks were used, split into 50bp bins (default for “computeMatrix”), with each bin receiving the mean of the BigWig signal scores across it.
Hierarchical clustering
Hierarchical clustering of genome-wide binding profiles for different transcription factors was accomplished as follows: First, 500bp tiling windows were created along the entire mm9 genome using “bedtools makewindows” from the BEDTools software suite (v2.26.0) (Quinlan and Hall, 2010). These genomic windows were overlapped with the peak BED files for all the transcription factors using “bedtools annotate”, resulting in a peak overlap matrix. This matrix was loaded into the R statistical program (v3.3.2) and converted into a binary matrix, thus counting each window-peak overlap only once even if the window overlaps multiple peaks from the same transcription factor ChIP-seq dataset. Empty rows, corresponding to universally unbound genomic windows, were discarded. This binary matrix was subsequently used to calculate a Pearson correlation matrix between transcription factors. Finally, the correlation matrix was hierarchically clustered and plotted using the pheatmap (v1.0.8) R package, using default settings for the hierarchical clustering (Euclidean distance metric and complete linkage clustering).
Module activity
Differential expression of CORE, MYC and PRC module genes were plotted based on the log2 fold changes (log2FC) between Taf5l/Taf6l KO and wild-type normalized count-based gene expression, as calculated by the DESeq2 R package (v1.14.1) (Love et al., 2014). Log2FC for module genes were plotted as violin plots using the ggplot2 (v2.2.1) R package, and the statistical significance of differential expression was calculated using the Wilcoxon rank sum test with continuity correction, as implemented in the R statistical program (v3.3.2).
Violin Plots/ differential binding
Differential binding analyses were performed using the DiffBind (v2.2.12) (Ross-Innes et al., 2012) R package from the Bioconductor project (v3.4) (Huber et al., 2015). For each KO versus wild-type (WT) differential binding comparison, peak sets were merged between KO and WT and the differential binding signal was calculated from the BAM file reads of the merged peak sets. Violin plots of the KO versus WT log2 fold change of normalized read densities, as calculated by DiffBind, were generated using the ggplot2 (v2.2.1) R package. Statistical significance of overall differential binding was calculated using the Wilcoxon rank sum test with continuity correction as implemented in the R statistical program (v3.3.2).
Calculation of traveling ration of RNAP, RNAP-Ser 5p and RNAP-Ser 2p
Traveling Ratio calculations were performed according to the Rahl et al. 2010 (Rahl et al., 2010). The GENCODE version M1 mouse transcriptome was used (Mudge and Harrow, 2015), and the transcripts were filtered to remove very short transcripts (<1kb genomic length). In accordance with Rahl et al. 2010, promoters were defined as the region from 30bp upstream of the Transcription Start Site (TSS-30bp) to 300bp downstream of it (TSS+300bp). Gene bodies were defined as TSS+300bp to TES (Transcription End Site). Promoter and gene body RPKMs were calculated using “bedtools intersect” (v2.26.0) to count overlapping RNAP/RNAP-Ser 5p/RNAP-Ser 2p reads with transcript promoters & gene bodies, followed by processing with Linux “awk” command (GNU Awk 4.0.2) to calculate the RPKMs. Traveling Ratios (TRs) were calculated per transcript from promoter & gene body RPKMs using the Linux “awk” command. Transcript-level TRs were subsequently converted into gene-level TRs by selecting the highest-expressed transcript per gene. This was done using R (v3.3.2), by selecting the promoter with highest RPKM per gene, followed by the gene body with highest RPKM for this promoter, to obtain one canonical transcript per gene. For comparability across the WT and both Taf5l & Taf6l KOs, the canonical transcripts were determined in the WT samples, with the same transcript set being used for both WT and KOs.
Metagene profile plots of RNAP2 binding along the gene body were created using the “computeMatrix” and “plotProfile” tools of the DeepTools software suite (v2.5.3) (Ramírez et al., 2016), whereby the gene body length was scaled to 10kb with 2kb flanking regions, and mean BigWig scores were calculated per bin.
Statistical analysis
All statistical analyses for the RNA-seq and ChIP-seq data were performed using the R statistical program (v3.3.2).
Supplementary Material
Figure S1. CRISPR-Cas9 mediated loss-of-function genetic screen identifies potential candidate epigenetic genes for the mESC state
(A) Schematic diagram represents an outline of the CRISPR-Cas9 screen.
(B) Representation of the epigenetic CRISPR-Cas9 pooled library. The pooled library comprised of 2,335 sgRNAs was synthesized and cloned into a lentiviral vector, and deep-sequenced to ensure representation of the sgRNAs. Each dot represents an sgRNA and its corresponding read count in the pooled library. Y axis: sgRNAs; X axis: number of reads per sgRNAs.
(C) Oct4-GFP reporter mESC line (Yeom et al., 1996 and Szabo et al., 2002) that constitutively expresses Cas9. Cas9 expression was confirmed by Western blot.
(D) FACS analysis represents percentage (%) of GFP-ve (GFP-low) cells from Oct4-GFP reporter mESCs either with no lentiviral infection (control) or with lentiviral infection of epigenetic CRISPR-Cas9 pooled library at low multiplicity (MOI).
(E) mRNA expression levels of selected novel candidate epigenetic genes (Taf5l, Taf6l, Tada1 and Tada3), ESC specific genes (Pou5f1/Oct4, Nanog, Sox2, Esrrb, Prdm14), and HAT complex genes (Kat2a and Kat2b) during differentiation– from undifferentiated ESCs to differentiated state (0 to 24 to 96 hrs).
Data are represented as mean ± SEM (n = 3); p-values were calculated using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; and ns (non-significant).
Related to Figure 1.
Figure S2. Validation confirms TAF5L and TAF6L are the new epigenetic genes for the mESC state
(A) mRNA expression levels of Tada1, Tada3, Taf5l and Taf6l after delivery of individual sgRNAs that target coding sequences of these genes and create indels. Non-targeting sgRNA (NT sgRNA) used as control. mRNA levels were normalized to GAPDH. The two best sgRNAs (for each gene) from the screen were used.
(B) Endogenous Oct4 mRNA expression levels in the Tada1, Tada3, Taf5l and Taf6l edited cells.
(C) Immunofluorescence staining of OCT4 in the Tada1, Tada3, Taf5l and Taf6l edited cells. Scale bar is 100µm.
(D-G) Schematic diagram illustrating the strategy to generate homozygous/biallelic deletion or KO clones. Paired sgRNAs (5’ and 3’ sgRNAs) flanking critical exons of Taf5l (D), Taf6l (E), Tada1 (F) and Tada3 (G) were introduced to create homozygous deletions. Two PCRs (deletion PCR and internal PCR) were used for each targeted gene to identify biallelic/homozygous deletions. Sanger sequencing was used to confirm the biallelic deletions. The KO clones show the same deletion breakpoints in both alleles.
(H) Percentages of GFP-high and GFP-low cells of Oct4-GFP reporter mESCs upon delivery of non-targeting (NT) or Taf5l and Taf6l targeting sgRNAs.
(I-J) Percentages of OCT4-positive (I) and SSEA1-positive (J) cells in wild-type, Taf5l KO and Taf6l KO mESCs.
Data are represented as mean ± SEM (n = 3); p-values were calculated using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; and ns (non-significant).
Related to Figure 2.
Figure S3. TAF5L and TAF6L are required for gene expression programs of mESC state; and for somatic cell reprogramming/iPSCs generation
(A) PCA plot of RNAseq data from three biological replicates of wild-type, Taf5l KO and Taf6l KO.
(B) mRNA expression levels of Taf5l and Taf6l in wild-type (CJ7) and their corresponding KOs.
(C) mRNA expression levels of Oct4 and c-Myc from wild-type (CJ7), Taf5l KO and Taf6l KO (from RT-qPCR).
(D) Protein levels of c-MYC in wild-type (J1) and KOs, using Western blot.
(E, F) Oct4 mRNA expression levels upon exogenous expression of TAF5L and TAF6L in their corresponding KOs and wild-type.
(G, H) c-Myc mRNA expression levels upon exogenous expression of TAF5L and TAF6L in their corresponding KOs and wild-type.
(I-L) Gene ontology (GO) term analysis (biological process) of up and downregulated genes from Taf5l and Taf6l KOs.
(M) Morphology and GFP fluorescence of transgene-independent iPSC colonies (at day 16 after reprogramming) that were generated upon perturbation of Taf5l and Taf6l using sgRNAs. Non-targeting sgRNA used as a control. Scale bar is 1000 µm.
(N) Percentages of pluripotent iPSCs, partially differentiated and differentiated cells after reprogramming (at day 16) upon perturbation of Taf5l and Taf6l using sgRNAs. Non-targeting sgRNA used as a control.
(O) FACS analysis of OCT4+ and SSEA1+ iPSCs at day 16.
(P-R) mRNA expression levels of Cdx2, Arid3a and Esx1 in bulk/mixed populations of edited cells after reprogramming at day 12 and day 16, upon perturbation of Taf5l and Taf6l using sgRNAs. Non-targeting sgRNA used as control.
(S) Percentages of indels (insertions and deletions) of the targeted regions of Taf5l and Taf6l genes were quantified using targeted amplicon sequencing. Indel frequencies of Taf5l and Taf6l genes were measured before (from mCherry+ reprogrammable MEFs, as MEFs were transduced with viral particles of sgRNAs carrying mCherry and Cas9) and after (bulk/mixed populations of edited cells) reprogramming at day 16. Percentages indicate the edited reads over the total number of reads.
Data are represented as mean ± SEM (n = 3); p-values were calculated using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; and ns (non-significant).
Related to Figure 3.
Figure S4. TAF5L and TAF6L belong to the MYC and CORE regulatory modules but mainly regulate the MYC module activity
(A) Generation of flag-biotin (FB)-tagged TAF5L and TAF6L mESC lines. The biotinylated forms of TAF5L and TAF6L were detected using streptavidin.
(B, C) Interacting partners of TAF5L and TAF6L are displayed. These interacting partners were identified through pull down of the biotinylated version of TAF5L and TAF6L using streptavidin beads, followed by mass spectrometry.
(D, E) Gene ontology (GO) term analysis (biological processes) of TAF5L and TAF6L binding target genes.
(F, G) Distribution of TAF5L and TAF6L binding target genes around the five different metagenes– ranked by their expression levels in mESCs.
(H-K) Genomic tracks of ChIP intensities of several factors, including TAF5L and TAF6L, and histone marks binding at Esrrb, Nanog, Taf5l and Taf6l gene loci. RNAseq tracks represent expression of these genes.
(L) ChIP-qPCR analyses of TAF5L and TAF6L binding at Oct4, c-Myc and Actin gene loci. Relative enrichment is shown as % of input (all the factors multiplied with 106).
Related to Figure 4.
Figure S5. Predominantly TAF5L/TAF6L modulate H3K9ac deposition and c-MYC recruitment at the TAF5L/TAF6L target genes to activate their gene expression through RNA Pol II pause release
(A) Global differential binding of OCT4 and c-MYC in Taf5l and Taf6l KOs compared to wild-type.
(B) Global differential binding of H3K9ac and H3K4me3 in Taf5l and Taf6l KOs compared to wild-type.
(C) The traveling ratio (TR) of RNA Pol II-Ser 2p at c-MYC and TAF5L bound genes in Taf5l KO and wild-type.
(D) The traveling ratio (TR) of RNA Pol II-Ser 2p at c-MYC and TAF6L bound genes in Taf6l KO and wild-type.
(E) The traveling ratio (TR) of RNA Pol II-Ser 5p at c-MYC and TAF5L bound genes in Taf5l KO and wild-type.
(F) The traveling ratio (TR) of RNA Pol II-Ser 5p at c-MYC and TAF6L bound genes in Taf6l KO and wild-type.
Related to Figure 5.
Figure S6. TAF5L/TAF6L maintain self-renewal of mESCs through MYC regulatory module/network
(A) FACS plots of cell cycle profiling from wild-type, Taf5l KO and Taf6l KO.
(B) Gene expression changes of different phases of the cell cycle and DNA replication gene sets in Taf5l and Taf6l KOs compared to wild-type.
(C-F) Gene expression changes of several individual genes related to different phases of the cell cycle and DNA replication in Taf5l and Taf6l KOs compared to wild-type.
(G, H) Differential binding of H3K9ac (G) and c-MYC (H) at the cell cycle phase and DNA replication gene sets in Taf5l and Taf6l KOs compared to wild-type.
(I) Gene expression changes of ribosome genes of MYC module in Taf5l and Taf6l KOs compared to wild-type.
(J) mRNA expression levels of glucose transporter genes in Taf5l and Taf6l KOs compared to wild-type.
Related to Figure 6.
Figure S7. TAF5L/TAF6L and c-MYC work together, as well as independently for gene regulation
(A) Strategy to generate homozygous deletion or KO clone of c-Myc in mESCs. Paired sgRNAs (5’ and 3’ sgRNAs) were used to target exon 2 of c-Myc that create homozygous deletion/KO of c-Myc. Two PCRs (deletion PCR and internal PCR) were used to identify biallelic/homozygous deletion.
(B) Sanger sequencing showing a biallelic 26bp deletion at the c-Myc coding sequence.
(C) The protein sequence of wild-type c-MYC, and the predicted protein sequence upon 26bp deletion of c-Myc gene (c-Myc KO– clone C5) are shown. The 26bp deletion created a frameshift mutation that resulted in a premature stop codon.
(D) c-MYC protein levels detected by Western blot in wild-type, c-Myc KO, Taf5l KO and Taf6l KO mESCs.
(E) Venn diagram illustrates overlapping and non-overlapping differentially expressed genes in c-Myc, Taf5l and Taf6l KOs.
Related to Figure 6.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Streptavidin-HRP | GE Healthcare | Cat#RPN1231; |
| Oct-3/4 (N-19) antibody | Santa Cruz | Cat#sc-8628; RRID: AB_653551 |
| c-Myc antibody | Cell Signaling Technology | Cat#9402; RRID: AB_2151827 |
| H3K4me3 antibody | Abcam | Cat#ab1012; RRID: AB_442796 |
| H3K9ac antibody | Abcam | Cat#ab4441; RRID: AB_2118292 |
| RNA Pol II CTD, phos. Serine 5 [3E8] antibody | Chromotek | Cat#3e8–5; RRID: AB_2631404 |
| RNA Pol II CTD, phos. Serine 2 [3E10] antibody | Chromotek | Cat#3e10–5; RRID: AB_2631403 |
| RNA Pol II CTD, unphosphorylated CTD [1C7] antibody | Chromotek | Cat#1c7–5; RRID: AB_2631402 |
| Bacterial and Virus Strains | ||
| E.cloni 10G ELITE electrocompentent cells | Lucigen | Cat#60052–4 |
| Critical Commercial Assays | ||
| QIAGEN MinElute PCR purification kit | QIAGEN | Cat#28006 |
| APC BrdU Flow Kit | BD | Cat#552598 |
| Vector Red Substrate Kit | Vector Lab | Cat#SK-5100 |
| Agilent Seahorse XF Cell Mito Stress Test Kit | Agilent | Cat#103015–100 |
| iQ SYBR Green Supermix | Bio-Rad | Cat#1708882 |
| iScript cDNA Synthesis Kit | Bio-Rad | Cat#1708891 |
| RNeasy Mini Kit | QIAGEN | Cat#74106 |
| rRNA Depletion Kit | NEB | Cat#E6310L |
| NEBNext Ultra Directional RNA Library Prep Kit | NEB | Cat#E7420L |
| NEBNext ChIP-seq Library Prep Master Mix Set for Illumina | NEB | Cat#6240L |
| NEBNext Ultra DNA Library Prep Kit for Illumina | NEB | Cat#E7370L |
| Dynabeads MyOne Streptavidin T1 beads | Thermo Fisher Scientific | Cat#65601 |
| Dynabeads Protein A | Thermo Fisher Scientific | Cat#10002D |
| Dynabeads Protein G | Thermo Fisher Scientific | Cat#10004D |
| NE-PER reagents | Thermo Fisher Scientific | Cat#78833 |
| Deposited Data | ||
| RNA-seq and ChIP-seq data | This paper | GEO: GSE113335 |
| ChIP-seq data | Das et al., 2014 | GEO: GSE43231 |
| Mendeley dataset | This paper | https://doi.org/10.17632/gkp97zr5zy.1 |
| Experimental Models: Cell Lines | ||
| J1 mouse embryonic stem cells | ATCC | Cat#SCRC-1010, RRID:CVCL_6412 |
| Taf5l, Taf6l, c-Myc KO mESCs | This paper | N/A |
| HEK293T | ATCC | Cat#CRL-3216; RRID: CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mouse embryonic fibroblast isolated from ROSA26-rtTA, Collagen-OKSM, Oct4-GFP mouse embryos | Stadtfeld et al., 2010 | JAX: 011001; 008214; 006965 |
| Recombinant DNA | ||
| Lentiguide-Puro | Sanjana et al., 2014 | Addgene Plasmid # 52963 |
| LentiCRISPR-mCherry V6 | Unpublished | Addgene Plasmid #99154 |
| LentiCas9-Blast | Sanjana et al., 2014 | Addgene Plasmid #52962 |
| Software and Algorithms | ||
| BEDTools (2.26.0) | Quinlan and Hall, 2010 | http://bedtools.readthedocs.io/en/latest/ |
| Bowtie 2 (2.3.2) | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MACS2 (2.1.1.20160309) | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| DeepTools (2.5.3) | Ramírez et al., 2016 | https://deeptools.readthedocs.io/en/develop/ |
| STAR (2.5.3a) | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Salmon (0.8.2) | Patro et al., 2017 | https://combine-lab.github.io/salmon/ |
| wigToBigWig, Kent tools (v4) | Kent et al., 2010 | http://hgdownload.cse.ucsc.edu/admin/exe/ |
| DESeq2 R package (1.14.1; Bioconductor 3.4) | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| DiffBind R package (2.2.12; Bioconductor 3.4) | Ross-Innes et al., 2012 | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| R statistical program (3.3.2) | N/A | https://www.R-project.org/ |
| ggplot2 R package (2.2.1) | N/A | http://ggplot2.tidyverse.org |
| pheatmap R package (1.0.8) | N/A | https://cran.r-project.org/web/packages/pheatmap/index.html |
| NIH DAVID website (6.8) | Dennis et al., 2003 | https://david.ncifcrf.gov/ |
| Mendeley dataset | This paper | https://doi.org/10.17632/gkp97zr5zy.1 |
ACKNOWLEDGMENTS
We thank the Dana Farber Cancer Institute FACS core facility and Monash University FACS core facility. We thank Xiaofeng Wang for Illumina HiSeq2500 high-throughput Sequencing at Harvard Medical School, as well as the Genewiz and Novogene high-throughput sequencing facility, China. We are also thankful to the Monash Biomedical Imaging facility. This work was supported by funding from NIH Grant HLBI U01HL100001 to S.H.O. P.P.D. is supported by National Health and Medical Research Council (NHMRC) of Australia (GNT1159461). J.K. is supported by a NIH grant R01GM112722; M.C.C. is supported by funding from a National Institute of Diabetes and Digestive and Kidney Diseases award (F30-DK103359); L.P. is supported by a National Human Genome Research Institute (NHGRI) Career Development Award (R00HG008399); E.A. is supported by the NIH Director’s New Innovator Award (DP2DA043813); and J.M.P. was supported by a Sylvia and Charles Senior Medical Viertel Fellowship. S.H.O is an Investigator of the Howard Hughes Medical Institute (HHMI).
Footnotes
The authors declare no conflicts of interest.
DATA AND SOFTWARE AVAILABILITY
The accession number for all the NGS data is- GSE113335
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, and Young RA (2009). SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23, 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. [DOI] [PubMed] [Google Scholar]
- Canver MC, Lessard S, Pinello L, Wu Y, Ilboudo Y, Stern EN, Needleman AJ, Galactéros F, Brugnara C, Kutlar A, et al. (2017). Variant-aware saturating mutagenesis using multiple Cas9 nucleases identifies regulatory elements at trait-associated loci. Nature Publishing Group 49, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Guo WT, Tian S, He X, Wang XW, Liu X, Gu KL, Ma X, Huang D, Hu L, et al. (2015). miR-290/371-Mbd2-Myc circuit regulates glycolytic metabolism to promote pluripotency. The EMBO Journal 34, 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Utley RT, Workman JL, and Côté J (2003). The diverse functions of histone acetyltransferase complexes. Trends in Genetics 19, 321–329. [DOI] [PubMed] [Google Scholar]
- Cartwright P (2005). LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132, 885–896. [DOI] [PubMed] [Google Scholar]
- Chappell J, and Dalton S (2013). Roles for MYC in the Establishment and Maintenance of Pluripotency. Cold Spring Harbor Perspectives in Medicine 3, a014381–a014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. (2008). Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell 133, 1106–1117. [DOI] [PubMed] [Google Scholar]
- Cooper S, and Brockdorff N (2013). Genome-wide shRNA screening to identify factors mediating Gata6 repression in mouse embryonic stem cells. Development 140, 4110–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Hendrix DA, Apostolou E, Buchner AH, Canver MC, Beyaz S, Ljuboja D, Kuintzle R, Kim W, Karnik R, et al. (2015). PRC2 Is Required to Maintain Expression of the Maternal Gtl2-Rian-Mirg Locus by Preventing De Novo DNA Methylation in Mouse Embryonic Stem Cells. CellReports 1–57. [DOI] [PMC free article] [PubMed]
- Das PP, Shao Z, Beyaz S, Apostolou E, Pinello L, De Los Angeles A, O’Brien K, Atsma JM, Fujiwara Y, Nguyen M, et al. (2014). Distinct and Combinatorial Functions of Jmjd2b/Kdm4b and Jmjd2c/Kdm4c in Mouse Embryonic Stem Cell Identity. Molecular Cell 53, 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, and Lempicki RA (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4, P3. [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, and Eisenman RN (2008). Myc’s broad reach. Genes Dev 22, 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagnocchi L, and Zippo A (2017). Multiple Roles of MYC in Integrating Regulatory Networks of Pluripotent Stem Cells. Front. Cell Dev. Biol 5, 981–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagnocchi L, Cherubini A, Hatsuda H, Fasciani A, Mazzoleni S, Poli V, Berno V, Rossi RL, Reinbold R, Endele M, et al. (2016). A Myc-driven self-reinforcing regulatory network maintains mouse embryonic stem cell identity. Nat Commun 7, 11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, and Panning B (2008a). An RNAi Screen of Chromatin Proteins Identifies Tip60-p400 as a Regulator of Embryonic Stem Cell Identity. Cell 134, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, and Panning B (2008b). An RNAi Screen of Chromatin Proteins Identifies Tip60-p400 as a Regulator of Embryonic Stem Cell Identity. Cell 134, 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Gaeta X, Sahakyan A, Chan AB, Hong CS, Kim R, Braas D, Plath K, Lowry WE, and Christofk HR (2016). Glycolytic Metabolism Plays a Functional Role in Regulating Human Pluripotent Stem Cell State. Cell Stem Cell 19, 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch CL, Coban Akdemir Z, Wang L, Jayakumaran G, Trcka D, Weiss A, Hernandez JJ, Pan Q, Han H, Xu X, et al. (2015). Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev 29, 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, and Zhang F (2014). Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, and Elledge SJ (2009). A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, and Linsley PS (2010). Recognizing and avoiding siRNA off- target effects for target identification and therapeutic application 1–11. [DOI] [PubMed]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, and Karolchik D (2010). BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cantor AB, Orkin SH, and Wang J (2009). Use of in vivo biotinylation to study protein–protein and protein–DNA interactions in mouse embryonic stem cells. Nat Protoc 4, 506–517. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, and Orkin SH (2008). An Extended Transcriptional Network for Pluripotency of Embryonic Stem Cells. Cell 132, 1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, and Orkin SH (2010). A Myc Network Accounts for Similarities between Embryonic Stem and Cancer Cell Transcription Programs. Cell 143, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, and Helin K (2014). Chromatin Repressive Complexes in Stem Cells, Development, and Cancer. Stem Cell 14, 735–751. [DOI] [PubMed] [Google Scholar]
- Lee KK, and Workman JL (2007). Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol 8, 284–295. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse ENCODE Consortium, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. (2012). An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge JM, and Harrow J (2015). Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome 26, 366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, and Hochedlinger K (2011). Chromatin Connections to Pluripotency and Cellular Reprogramming. Cell 145, 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, and Plath K (2013). Epigenetics of Reprogramming to Induced Pluripotency. Cell 152, 1324–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, and Young RA (2010). c-Myc Regulates Transcriptional Pause Release. Cell 141, 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton DA, and Daley GQ (2012). The promise of induced pluripotent stem cells in research and therapy. Nature 481, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scognamiglio R, Cabezas-Wallscheid N, Thier MC, Altamura S, Reyes A, Prendergast ÁM, Baumgärtner D, Carnevalli LS, Atzberger A, Haas S, et al. (2016). Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell 164, 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, and Zhang F (2015). High-throughput functional genomics using CRISPR–Cas9. Nature Publishing Group 16, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, and Cantley LC (2013). Stem cell metabolism in tissue development and aging. Development 140, 2535–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, and Dalton S (2009). The Cell Cycle and Myc Intersect with Mechanisms that Regulate Pluripotency and Reprogramming. Stem Cell 5, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Singh AM, and Dalton S (2010). Myc Represses Primitive Endoderm Differentiation in Pluripotent Stem Cells. Stem Cell 7, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, and Hochedlinger K (2009). A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nature Methods 7, 53–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YBY, Qu X, Howard V, Dai L, Jiang X, Ren Y, Fu P, Puelles VG, Nikolic-Paterson DJ, Caruana G, et al. (2015). Smad3 deficiency protects mice from obesity-induced podocyte injury that precedes insulin resistance. Kidney Int 88, 286–298. [DOI] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Thomson JA (1998). Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, and Felsher DW (2010). MYC as a regulator of ribosome biogenesis and protein synthesis 1–9. [DOI] [PubMed]
- Vander Heiden MG, Cantley LC, and Thompson CB (2009). Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, and Knoepfler PS (2010). myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation 80, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova N, Cotterman R, Bradnam K, Korf I, and Knoepfler PS (2011). Myc and Miz-1 have coordinate genomic functions including targeting Hox genes in human embryonic stem cells. Epigenetics Chromatin 4, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, and Young RA (2012). Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, and Chang HY (2008). Module Map of Stem Cell Genes Guides Creation of Epithelial Cancer Stem Cells. Cell Stem Cell 2, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA (2011). Control of the Embryonic Stem Cell State. Cell 144, 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, and Bernstein BE (2010). Charting histone modifications and the functional organization of mammalian genomes. Nature Publishing Group 12, 7–18. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, and Wei W (2014). High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491. [DOI] [PubMed] [Google Scholar]
- Zwaka TP, and Thomson JA (2005). Differentiation of human embryonic stem cells occurs through symmetric cell division. Stem Cells 23, 146–149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. CRISPR-Cas9 mediated loss-of-function genetic screen identifies potential candidate epigenetic genes for the mESC state
(A) Schematic diagram represents an outline of the CRISPR-Cas9 screen.
(B) Representation of the epigenetic CRISPR-Cas9 pooled library. The pooled library comprised of 2,335 sgRNAs was synthesized and cloned into a lentiviral vector, and deep-sequenced to ensure representation of the sgRNAs. Each dot represents an sgRNA and its corresponding read count in the pooled library. Y axis: sgRNAs; X axis: number of reads per sgRNAs.
(C) Oct4-GFP reporter mESC line (Yeom et al., 1996 and Szabo et al., 2002) that constitutively expresses Cas9. Cas9 expression was confirmed by Western blot.
(D) FACS analysis represents percentage (%) of GFP-ve (GFP-low) cells from Oct4-GFP reporter mESCs either with no lentiviral infection (control) or with lentiviral infection of epigenetic CRISPR-Cas9 pooled library at low multiplicity (MOI).
(E) mRNA expression levels of selected novel candidate epigenetic genes (Taf5l, Taf6l, Tada1 and Tada3), ESC specific genes (Pou5f1/Oct4, Nanog, Sox2, Esrrb, Prdm14), and HAT complex genes (Kat2a and Kat2b) during differentiation– from undifferentiated ESCs to differentiated state (0 to 24 to 96 hrs).
Data are represented as mean ± SEM (n = 3); p-values were calculated using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; and ns (non-significant).
Related to Figure 1.
Figure S2. Validation confirms TAF5L and TAF6L are the new epigenetic genes for the mESC state
(A) mRNA expression levels of Tada1, Tada3, Taf5l and Taf6l after delivery of individual sgRNAs that target coding sequences of these genes and create indels. Non-targeting sgRNA (NT sgRNA) used as control. mRNA levels were normalized to GAPDH. The two best sgRNAs (for each gene) from the screen were used.
(B) Endogenous Oct4 mRNA expression levels in the Tada1, Tada3, Taf5l and Taf6l edited cells.
(C) Immunofluorescence staining of OCT4 in the Tada1, Tada3, Taf5l and Taf6l edited cells. Scale bar is 100µm.
(D-G) Schematic diagram illustrating the strategy to generate homozygous/biallelic deletion or KO clones. Paired sgRNAs (5’ and 3’ sgRNAs) flanking critical exons of Taf5l (D), Taf6l (E), Tada1 (F) and Tada3 (G) were introduced to create homozygous deletions. Two PCRs (deletion PCR and internal PCR) were used for each targeted gene to identify biallelic/homozygous deletions. Sanger sequencing was used to confirm the biallelic deletions. The KO clones show the same deletion breakpoints in both alleles.
(H) Percentages of GFP-high and GFP-low cells of Oct4-GFP reporter mESCs upon delivery of non-targeting (NT) or Taf5l and Taf6l targeting sgRNAs.
(I-J) Percentages of OCT4-positive (I) and SSEA1-positive (J) cells in wild-type, Taf5l KO and Taf6l KO mESCs.
Data are represented as mean ± SEM (n = 3); p-values were calculated using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; and ns (non-significant).
Related to Figure 2.
Figure S3. TAF5L and TAF6L are required for gene expression programs of mESC state; and for somatic cell reprogramming/iPSCs generation
(A) PCA plot of RNAseq data from three biological replicates of wild-type, Taf5l KO and Taf6l KO.
(B) mRNA expression levels of Taf5l and Taf6l in wild-type (CJ7) and their corresponding KOs.
(C) mRNA expression levels of Oct4 and c-Myc from wild-type (CJ7), Taf5l KO and Taf6l KO (from RT-qPCR).
(D) Protein levels of c-MYC in wild-type (J1) and KOs, using Western blot.
(E, F) Oct4 mRNA expression levels upon exogenous expression of TAF5L and TAF6L in their corresponding KOs and wild-type.
(G, H) c-Myc mRNA expression levels upon exogenous expression of TAF5L and TAF6L in their corresponding KOs and wild-type.
(I-L) Gene ontology (GO) term analysis (biological process) of up and downregulated genes from Taf5l and Taf6l KOs.
(M) Morphology and GFP fluorescence of transgene-independent iPSC colonies (at day 16 after reprogramming) that were generated upon perturbation of Taf5l and Taf6l using sgRNAs. Non-targeting sgRNA used as a control. Scale bar is 1000 µm.
(N) Percentages of pluripotent iPSCs, partially differentiated and differentiated cells after reprogramming (at day 16) upon perturbation of Taf5l and Taf6l using sgRNAs. Non-targeting sgRNA used as a control.
(O) FACS analysis of OCT4+ and SSEA1+ iPSCs at day 16.
(P-R) mRNA expression levels of Cdx2, Arid3a and Esx1 in bulk/mixed populations of edited cells after reprogramming at day 12 and day 16, upon perturbation of Taf5l and Taf6l using sgRNAs. Non-targeting sgRNA used as control.
(S) Percentages of indels (insertions and deletions) of the targeted regions of Taf5l and Taf6l genes were quantified using targeted amplicon sequencing. Indel frequencies of Taf5l and Taf6l genes were measured before (from mCherry+ reprogrammable MEFs, as MEFs were transduced with viral particles of sgRNAs carrying mCherry and Cas9) and after (bulk/mixed populations of edited cells) reprogramming at day 16. Percentages indicate the edited reads over the total number of reads.
Data are represented as mean ± SEM (n = 3); p-values were calculated using ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; and ns (non-significant).
Related to Figure 3.
Figure S4. TAF5L and TAF6L belong to the MYC and CORE regulatory modules but mainly regulate the MYC module activity
(A) Generation of flag-biotin (FB)-tagged TAF5L and TAF6L mESC lines. The biotinylated forms of TAF5L and TAF6L were detected using streptavidin.
(B, C) Interacting partners of TAF5L and TAF6L are displayed. These interacting partners were identified through pull down of the biotinylated version of TAF5L and TAF6L using streptavidin beads, followed by mass spectrometry.
(D, E) Gene ontology (GO) term analysis (biological processes) of TAF5L and TAF6L binding target genes.
(F, G) Distribution of TAF5L and TAF6L binding target genes around the five different metagenes– ranked by their expression levels in mESCs.
(H-K) Genomic tracks of ChIP intensities of several factors, including TAF5L and TAF6L, and histone marks binding at Esrrb, Nanog, Taf5l and Taf6l gene loci. RNAseq tracks represent expression of these genes.
(L) ChIP-qPCR analyses of TAF5L and TAF6L binding at Oct4, c-Myc and Actin gene loci. Relative enrichment is shown as % of input (all the factors multiplied with 106).
Related to Figure 4.
Figure S5. Predominantly TAF5L/TAF6L modulate H3K9ac deposition and c-MYC recruitment at the TAF5L/TAF6L target genes to activate their gene expression through RNA Pol II pause release
(A) Global differential binding of OCT4 and c-MYC in Taf5l and Taf6l KOs compared to wild-type.
(B) Global differential binding of H3K9ac and H3K4me3 in Taf5l and Taf6l KOs compared to wild-type.
(C) The traveling ratio (TR) of RNA Pol II-Ser 2p at c-MYC and TAF5L bound genes in Taf5l KO and wild-type.
(D) The traveling ratio (TR) of RNA Pol II-Ser 2p at c-MYC and TAF6L bound genes in Taf6l KO and wild-type.
(E) The traveling ratio (TR) of RNA Pol II-Ser 5p at c-MYC and TAF5L bound genes in Taf5l KO and wild-type.
(F) The traveling ratio (TR) of RNA Pol II-Ser 5p at c-MYC and TAF6L bound genes in Taf6l KO and wild-type.
Related to Figure 5.
Figure S6. TAF5L/TAF6L maintain self-renewal of mESCs through MYC regulatory module/network
(A) FACS plots of cell cycle profiling from wild-type, Taf5l KO and Taf6l KO.
(B) Gene expression changes of different phases of the cell cycle and DNA replication gene sets in Taf5l and Taf6l KOs compared to wild-type.
(C-F) Gene expression changes of several individual genes related to different phases of the cell cycle and DNA replication in Taf5l and Taf6l KOs compared to wild-type.
(G, H) Differential binding of H3K9ac (G) and c-MYC (H) at the cell cycle phase and DNA replication gene sets in Taf5l and Taf6l KOs compared to wild-type.
(I) Gene expression changes of ribosome genes of MYC module in Taf5l and Taf6l KOs compared to wild-type.
(J) mRNA expression levels of glucose transporter genes in Taf5l and Taf6l KOs compared to wild-type.
Related to Figure 6.
Figure S7. TAF5L/TAF6L and c-MYC work together, as well as independently for gene regulation
(A) Strategy to generate homozygous deletion or KO clone of c-Myc in mESCs. Paired sgRNAs (5’ and 3’ sgRNAs) were used to target exon 2 of c-Myc that create homozygous deletion/KO of c-Myc. Two PCRs (deletion PCR and internal PCR) were used to identify biallelic/homozygous deletion.
(B) Sanger sequencing showing a biallelic 26bp deletion at the c-Myc coding sequence.
(C) The protein sequence of wild-type c-MYC, and the predicted protein sequence upon 26bp deletion of c-Myc gene (c-Myc KO– clone C5) are shown. The 26bp deletion created a frameshift mutation that resulted in a premature stop codon.
(D) c-MYC protein levels detected by Western blot in wild-type, c-Myc KO, Taf5l KO and Taf6l KO mESCs.
(E) Venn diagram illustrates overlapping and non-overlapping differentially expressed genes in c-Myc, Taf5l and Taf6l KOs.
Related to Figure 6.