Abstract
NALCN is a conserved cation channel, which conducts a permanent sodium leak current and regulates resting membrane potential and neuronal excitability. It is part of a large ion channel complex, the “NALCN channelosome”, consisting of multiple proteins including UNC80 and UNC79. The predominant neuronal expression pattern and its function suggest an important role in neuronal function and disease. So far, biallelic NALCN and UNC80 variants have been described in a small number of individuals leading to infantile hypotonia, psychomotor retardation, and characteristic facies 1 (IHPRF1, OMIM 615419) and 2 (IHPRF2, OMIM 616801), respectively. Heterozygous de novo NALCN missense variants in the S5/S6 pore-forming segments lead to congenital contractures of the limbs and face, hypotonia, and developmental delay (CLIFAHDD, OMIM 616266) with some clinical overlap. In this study, we present detailed clinical information of 16 novel individuals with biallelic NALCN variants, 1 individual with a heterozygous de novo NALCN missense variant and an interesting clinical phenotype without contractures, and 12 individuals with biallelic UNC80 variants. We report for the first time a missense NALCN variant located in the predicted S6 pore-forming unit inherited in an autosomal-recessive manner leading to mild IHPRF1. We show evidence of clinical variability, especially among IHPRF1-affected individuals, and discuss differences between the IHPRF1-and IHPRF2 phenotypes. In summary, we provide a comprehensive overview of IHPRF1 and IHPRF2 phenotypes based on the largest cohort of individuals reported so far and provide additional insights into the clinical phenotypes of these neurodevelopmental diseases to help improve counseling of affected families.
Introduction
The “NALCN channelosome” is a large ion channel complex consisting of multiple proteins, including NALCN, UNC80, UNC79, and G protein-coupled receptors (GPCRs), which are predominantly expressed in neurons (Cochet-Bissuel et al. 2014). NALCN forms the channel pore of the complex and conducts a permanent sodium leak. UNC80 and UNC79 contribute to the stabilization and neuronal localization of the “channelosome” and the different subunits regulate each other’s expression in C. elegans and D. melanogaster, but the conservation of this regulation has not yet been demonstrated in mammals. In humans, the “NALCN channelosome” regulates resting membrane potential and neuronal excitability and it has been implicated in different neurological diseases.
The gene encoding NALCN is located on chromosome 13q32.3–33.1 and monoallelic (de novo) and biallelic (recessive) variants lead to different clinical phenotypes with some overlapping features. Heterozygous NALCN variants lead to congenital contractures of the limbs and face, muscular hypotonia, and global developmental delay (CLIFAHDD, OMIM 616266) (Chong et al. 2015), while biallelic NALCN variants lead to infantile hypotonia, psychomotor retardation, and characteristic facies 1 (IHPRF1, OMIM 615419) (Al-Sayed et al. 2013; Koroglu et al. 2013; Gal et al. 2016; Angius et al. 2018; Bourque et al. 2018; Campbell et al. 2018; Takenouchi et al. 2018). The gene encoding UNC80 is located on chromosome 2q34 and biallelic UNC80 variants are associated with infantile hypotonia, psychomotor retardation, and characteristic facies 2 (IHPRF2, OMIM 616801) (Perez et al. 2016; Shamseldin et al. 2016; Stray-Pedersen et al. 2016; Valkanas et al. 2016; Boerkoel and du Souich 2017; He et al. 2018; Obeid et al. 2018). To our knowledge, variants in the UNC79 gene, which is located on chromosome 14q32.12, have not yet been associated with any disease.
Here, we present the so far largest cohort of novel individuals with NALCN and UNC80 variants, including 16 individuals with biallelic NALCN variants, 1 individual with a de novo NALCN variant displaying an interesting clinical phenotype and 12 individuals with biallelic UNC80 variants, of which scarce clinical information as well as the variant information of one individual was previously published (Cherot et al. 2018). The goal of this study was to further delineate and specify the clinical spectra of IHPRF1 and IHPRF2, to broaden the spectrum of biallelic NALCN and UNC80 variants, and to increase the understanding of these diseases to improve genetic counseling.
Methods
Patients
Clinical information of the index individuals was provided by the parents and the evaluating clinicians. Written informed consent was obtained from the parents of the individuals for participation in study. The study was performed according to the Declaration of Helsinki protocols and was approved by local institutional review boards [ethical votum 08–3663 and 5360/13 for the Technical University Munich; HPA-01-R-015 (project no:943/24.04.2017) for the Research ethics committee at Prince Sultan Medical Military City; EA2/107/14 for the Charité-Universitatsmedizin Berlin].
Chromosomal analysis
Microarray analyses for molecular karyotyping were normal in individuals 1, 4, 6, 7, 8, 9, and 23 [Individual 1: Agilent Human Genome CGH Microarray 244A (Agilent, Santa Clara, CA, USA), individual 4: CytoChip Oligo 4 × 180 k (Agilent, Santa Clara, CA, USA), individuals 6, 7, 8, and 23: Illumina OmniExpress-24 (Illumina, San Diego, CA, USA), and individual 9: Agilent Human Genome CGH Microarray 4 × 180K (hg18) (Agilent, Santa Clara, CA, USA)]. For individuals 10 and 19, chromosomal microarray analysis was performed using both copy number and single-nucleotide polymorphism (SNP) probes on a whole genome array (Affymetrix CytoScan HD platform; Affymetrix, Santa Clara, CA, USA). No array analysis was performed for individuals 2, 3, 5, 11, 12, 13, 14, 15, 16, 17, 18, 20, 21, 22, 24, 25, 26, 27, 28, and 29.
High-throughput sequencing
DNA from peripheral blood lymphocytes or from dry blood spots in filter cards (CentoCard®) was obtained from the index individual and parents and extracted by the standard extraction procedures. For individual 1, WES was performed as previously described (Bramswig et al. 2015). For individuals 2 (WES-Solo), 10 (WES-trio), 11 (WES-Solo-Illum), 13 (WES-Solo), 18 (WES-Solo), 19–20 (WES with affected sib), and 21 (WES-Solo), exome capture was carried out with the Nextera Rapid Capture Exome Kit (Illumina, Inc., San Diego, CA). Sequencing used either NextSeq500 or HiSeq4000 sequencers (both Illumina), and average coverage was targeted to 100x. The bioinformatics pipeline eventually resulting in called variants was performed as described previously (Trujillano et al. 2017). The corresponding list of variants was evaluated with an inhouse developed tool (CentoFit) for prioritization of variants. Confirmation of WES-derived variants as well as determination of genotypes in additional family members (individuals 3, 12, and 22) was based on Sanger sequencing. Individual 5 was included in a next generation sequencing panel study comprising 3089 genes known to be causative for one or more Mendelian diseases. The coding parts of 3089 genes (http://doro.charite.de/MENDEL_public/HPOv2_Gen_Panel.pdf) targeted were enriched (Sure Select XT, Agilent) followed by sequencing with Illumina’s HiSeq system (single-end method). Sequence reads were mapped to the haploid human reference genome release (GRCh37) using BWA MEM. Single-nucleotide variants (SNVs) and short insertions and deletions (indels) were called with the FreeBayes toolkit version 0.9.20 and resulted in a high-quality exome variant set (Li and Durbin 2009; DePristo et al. 2011; Li 2013). Variant annotation was performed with Jannovar (Jäger et al. 2014). We removed sequence variants that occurred with a frequency above 1% in large population studies (Project Consortium et al. 2012; Lek et al. 2016). The remaining sequence variants were prioritized based on predicted pathogenicity and clinical relevance using the computational method PhenIX (Phenotypic Interpretation of eXomes) (Zemojtel et al. 2014). We entered the following human phenotype ontology (Robinson et al. 2008) terms: global developmental delay (HP:0001236), mild short stature (HP:0003502), muscular hypotonia (HP:0001252), and recurrent infections (HP:0002719). In individual 4, the brother of individual 5, the variants were confirmed by Sanger Sequencing. For individuals 6, 7, and 8, WES was performed with Roche Medexome library preparations (Roche Diagnostic, Basel, Switzerland) and Illumina NextSeq 2 × 150 bp sequencing kits (Illumina, San Diego, CA, USA) according to manufacturers’ specifications. For individual 9, WES was performed using TruSeq Exome (Illumina, San Diego, CA, USA). Raw data generated by the NextSeq 500 sequencer were demultiplexed and converted into fasq files using bcl2fastq 2.18. Reads quality was assessed using fastqc 0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Obtained sequences were mapped to the human reference assembly genome hg19 using the Maximum Exact Matches algorithm in Burrows–Wheeler Aligner 0.7.15 (BWA) (Li and Durbin 2009). On average, 98% of target sequences were successfully covered with a depth > 20X. Alignment quality was evaluated using qualimap 2.2.1 (Okonechnikov et al. 2016). The Genome Analysis Software Kit 3.7 (GATK) pipeline was applied following the germline version of GATK best-practice recommendations to performed variant calling (Van der Auwera et al. 2013). Variant annotation, mining, and manual review were performed using VarAFT (http://varaft.eu). For individual 23, sequencing was performed with Illumina TruSight One library preparation and Illumina NextSeq 2 × 150 bp sequencing kit according to manufacturers’ specifications. For individuals 14, 15, 16, 17, 24, 25, 26, 27, 28, and 29, WES was performed in at least one affected family member as reported previously (Dixon-Salazar et al. 2012). In these individuals, we focused on homozygous, rare (< 0.1% allele frequency in our exome database), and potentially deleterious variants (genomic evolutionary rate profile score > 4 or phastCons > 0.9).
RNA isolation and reverse transcription
The fibroblast cell line of individual 9 was grown and maintained in DMEM media under the standard tissue culture conditions. Total RNA was extracted from the patient’s cells using the illustraRNAspin Mini (GE Healthcare, Buckinghamshire, UK). Complementary DNAs (cDNA) were synthesized using the SuperScript II Reverse Transcriptase (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instruction. To determine the consequence of the variant for the NALCN transcript, we performed PCR experiments on cDNAs using primers located in exons 24/25 (forward, 5′-GC ACCTACTTTGCAGATT G-3′) and in exon 26 (reverse, 5′-GCCTCAAAATGTACC TGCTG-3′).
Results
In “Results”, we have included case reports for the entire cohort including 16 individuals with biallelic NALCN variants, 1 individual (individual 8) with a heterozygous NALCN variant presenting with an interesting phenotype, and 12 individuals with biallelic UNC80 variants. Some case reports are concise, while others are more comprehensive to give deeper insight into the clinical phenotype of these individuals. The entire cohort was assembled through word of mouth. Extensive clinical information of all 29 individuals is provided in Supplemental Tables 1 (individuals with NALCN variants) and 2 (individuals with UNC80 variants).
Clinical description of individuals with NALCN variants
Individual 1 is the child of healthy, non-consanguineous parents with an uneventful family history. All her siblings (sister, paternal half-sister, and maternal half-brother) are healthy. She was born at 39+3 weeks of gestation with a weight of 2760 g (– 1.39 SD), a length of 49 cm (– 0.95 SD), and an OFC of 34 cm (– 0.46 SD). She showed muscular hypotonia and hyperbilirubinemia, for which she received phototherapy. She sat at the age of 24 months. At the age of 4.5 years, she displayed atonic seizures with gaze palsy and epileptic EEG activity and was treated with levetiracetam. She presented with constipation and sleeping problems. Her first assessment was at the age of 5 6/12 years when her height was 98 cm (– 2.92 SD), her weight was 12 kg (– 3.62 SD, BMI: 12.49 kg/m2), and her OFC was 51 cm (– 0.7 SD). She presented with severe intellectual disability (ID), and she had not developed any speech. Facial dysmorphisms included a long face, a high forehead, fine hair, a small mouth, and thin upper vermillion (Fig. 1a, b). Subsequently, she developed uncontrollable seizures despite antiepileptic treatment with lamotrigine and valproate. However, the introduction of ketogenic diet at the age of 6 7/12 years led to a significant improvement of seizure control and weight gain. She became more attentive and progressed in her development. Her last assessment was at the age of 11 7/12 years; her height was about 120 cm (– 4.26 SD), her weight 21 kg (– 3.5 SD; BMI: 14.58 kg/m2), and her OFC 51 cm (– 1.57 SD). Seizure control was further improved with ketogenic diet, so that valproate was slowly reduced; she only received lamotrigine and did not show any more seizures. She still showed sleeping problems, sleep studies revealed Cheyne–Stokes-like breathing and central apnea without hypoxemia. She was able to walk for a few steps with support and orthotics and she did not speak. She displayed recurrent hand flapping. Array analysis was normal. Whole exome sequencing (WES) revealed compound heterozygous variants in the NALCN gene [NM_052867.2; variant 1, maternal: chr13:g.101103207G > A [GRCh38/hg38], c.3022C > T, p.(Arg1008Ter); variant 2, paternal: chr13:g.101104901delG [GRCh38/hg38], c.2629delC, p.(Gln877Asnfs*16)].
Fig. 1.

Individuals with biallelic NALCN variants. a, b Individual 1 at the age of 11 7/12 years presenting with long face, high forehead, strabismus, fine hair and large ears. c, d Individual 3 at the age of 3 10/12 years displaying hypotonic facies, high forehead, strabismus, sparse hair, and large ears. e and f individual 4 at the age of 3 6/12 years (e) and 4 6/12 years (f) showing fine hair and facial hypotonia (open-mouth appearance). g Individual 5 at the age of 3 years also displaying fine hair and facial hypotonia (open-mouth appearance). h, i Individual 14 at the age of 4 years with high forehead, deep-set eyes, and thin upper lip vermillion. j Individual 15 at the age of 2 9/12 years presenting with long face, fine hair, strabismus, and thin upper lip vermillion. k, l Individual 17 at the age of 3 years showing facial hypotonia, triangular face with high forehead, deep-set eyes, and large ears
Individual 2 is a 7-year-old boy born to consanguineous parents originating from Lebanon. He has an elder healthy sister and a younger, similarly affected sister (individual 3). Family history also mentions two affected cousins. The index presented from birth with mild hypotonia. At the time of last examination (7 years) he presents severe global developmental delay and muscular hypotonia. His metabolic work up was normal. His younger sister (individual 3) has a similar presentation with hypotonia, which was noticed from birth (Fig. 1c, d). Her last examination at 3 10/12 years showed severe global developmental delay and no speech. WES analysis was performed in individual 2 resulting in the identification of a homozygous likely pathogenic variant in the NALCN gene [NM_052867.2; chr13:g.101103173dupA [GRCh38/hg38], c.3056dupT, p.(Leu1019Phefs*30)]. Sanger sequencing confirmed this finding in individual 2 and his affected sister (individual 3).
Individual 4 is the son of healthy, unrelated parents of German origin. Individual 5 is his younger brother. The father of the children has a divergent strabismus; the mother has a heterozygous Factor V Leiden variant. Individual 4 was born by secondary caesarean section due to a pathological cardiotocography (CTG) at 40+1 weeks of gestation with a weight of 3595 g (+ 0.35 SD), a length of 53 cm (+0.57 SD) and an OFC of 37 cm (+ 1.85 SD). After birth he had hyperbilirubinemia, which required no further treatment. At the age of 3 months he showed motor developmental delay. He was able to sit at 8 months and started walking at 2 1/12 years. His speech development was mildly delayed. He was able to speak two-word sentences at the age of 2 3/12 years. Starting from the age of 5 months he showed poor weight gain. Tests for celiac disease were negative. His weight stabilized at the age of 2 years under a high caloric diet. He developed a chronic cough, which improved later under treatment with inhalative cortisone, but no immunodeficiency was found. Because his blood titers were low he received additional vaccination. He had problems falling asleep, which improved later under treatment with melatonin. Monitoring in a sleep laboratory showed periodic breathing without saturation loss, sleep disturbance due to apnea-associated arousals and periodic breathing during sleep was observed. The respiratory pattern was reminiscent of Cheyne-Stokes respiration: flattening breathing, apnea, and deep breathing. He showed recurring increases of thyroid-stimulating hormone (TSH). Array CGH, as well as CFTR gene and NKX2–1 gene analyses, were normal. Sleep EEG, somatosensory evoked potential (SSEP), nerve conduction studies, and cranial MRT did not show any pathological results. His first genetic assessment was at the age of 2 4/12 years. He presented with global development delay, muscular hypotonia, and a gluteal heamangioma. He had facial muscular hypotonia and an open-mouth appearance (Fig. 1e, f). He showed fusion of two teeth (International Standards Organization Designation System: 72 and 73). At the age 4 6/12 years, his height was 109.5 cm (+ 0.28 SD), his weight 19 kg [+ 0.35 SD; BMI: 15.85 kg/m2 (+ 0.23 SD)], and his OFC 52 cm (+ 0.47 SD). His motor and speech development had further improved, but was still delayed regarding gross and fine motor skills and expressive language. His muscular hypotonia appeared to be more pronounced in his lower extremities. It seemed that he had an increased muscle tone before executing a movement. He had problems to concentrate on tasks for a longer period of time.
Individual 5 was born by secondary caesarean section due to a pathological CTG (recurrent early and late decelerations) at 35+1 weeks of gestation with a weight of 2200 g (– 2.29 SD), a length of 47 cm (– 1.89 SD), and an OFC of 34 cm (– 0.90 SD). After birth, he developed a mild respiratory distress syndrome (IRDS), which was treated by endotracheal administration of artificial surfactant and breathing support with nasal continuous positive airway pressure (CPAP) for 2.5 days. Hyperbilirubinemia was treated with phototherapy. Recurrent respiratory infections were observed and pneumonia occurred at the age of one month and at the age of 7 months, which required hospitalization. His TSH was intermittently increased. His first genetic assessment was at the age of 10 months. He presented with muscular hypotonia, including facial hypotonia and open-mouth appearance, strabismus convergens, a gluteal hemangioma, and one truncal café au lait spot. His motor development was delayed. At this time, he was able to lift his head in ventral position. At the age of 1 2/12 years, he had his third pneumonia and he continued to have multiple respiratory infections. Therefore, he received prophylactic Palivizumab and Cotrimoxazol treatment, as well as inhalative and nasal cortisone. Because his blood titers were low, he received additional vaccination. No immunodeficiency was found, and echocardiography, cranial ultrasound, EEG, and bronchoscopy were normal. Chromosome analysis was normal and genetic test for Prader–Willi syndrome showed normal methylation of the SNRPN locus. PCR analysis for Fragile X syndrome did not show a CGG expansion in FMR1. Normal enzyme activity of acid alpha-glucosidase excluded Morbus Pompe. Transferrin electrophoresis gave normal results. He had frequent vomiting during the night, which improved under omeprazole. Monitoring in a sleep laboratory showed periodic breathing without saturation loss, but sleep disturbance due to apnea-associated arousals. At the age of 2 years, he developed a non-spastic muscular talipes equinus, which was treated with an ankle–foot orthosis, and he then achieved to walk at the age of 2 10/12 years. At the age of 2 2/12 years, his OFC was greater than 2 SD, the cranial MRT showed no pathological results. At the age of 3 years his height was 103.5 cm (+ 1.79 SD), his weight 15.4 kg [+ 0.27 SD; BMI: 14.38 kg/m2 (– 1.43 SD)], and his OFC 54 cm (+ 2.61 SD) (Fig. 1g). His speech development was delayed and he had slurred speech. He had general muscle hypotonia, especially truncal hypotonia, and it seemed that he had an increased muscle tone before executing a movement. His weight gain was stable under a high caloric diet including supplement food. Sequencing (HiSeq, Illumina) of 3089 disease related genes in individual 5 identified compound heterozygosity for two novel variants in NALCN [NM_052867.2; variant 1, maternal: chr13:g.101068744G > T [GRCh38/ hg38], c.4281C > A, p.(Phe1427Leu); variant 2, paternal: chr13:g.101074512A> G [GRCh38/hg38], c.4103 + 2T > C]. Sanger sequencing in individual 4 confirmed these variants.
Individual 6 is a 7-year-old female born to healthy related Tunisian parents. She was born full term with birth weight 2800 g (–1.81 SD), length 50 cm (–1 SD), and OFC 33 cm (–1.69 SD). She had poor sucking and feeding difficulties requiring tube feeding from the age of 7 years on. Breathing difficulties during the first months of life were explained by a turbinate hypertrophy. She had epilepsy (absences) from the age of 7 months treated with valproate. At the age of 7 years, individual 6 is a bedridden girl with severe intellectual disability (she sat at 18 months, stood with support at 7 years, and never walked or spoke). Physical examination revealed truncal hypotonia, dystonia of limbs and mouth, significant growth failure (height and weight – 3 SD), and microcephaly (OFC – 3 SD). She did not have any facial dysmorphic features, but a strabismus. WES revealed a homozygous variant in the NALCN gene [NM_052867.2; chr13: g.101083738G > A [GRCh38/hg38], c.3556C > T, p.(Gln1186Ter)].
Individual 7 is an 18.5-year-old female born to healthy unrelated Tunisian parents. One sibling died at the age of 6 years with the same clinical features as individual 7. Individual 7 was born full term after a pregnancy characterized by poor fetal active movements with a birth weight 2700 g (– 1.83 SD), length 50 cm (– 0.77 SD), and OFC 35 cm (0.08 SD). She had neonatal hypotonia and poor sucking. Feeding difficulties required a gastrostomy at age 4 years. She had severe epilepsy from the age of 5 months (generalized tonic-clonic seizures) and choreic movements of upper limbs noticed from the age 3 years. Epilepsy is currently controlled by valproate, topiramate, and clobazam. Individual 7 never walked or spoke. At the age of 18.5 years, she was a bedridden female with severe intellectual disability and hand stereotypies. Physical examination revealed spastic hypertonia, mild dysmorphic features (long face, large ears, strabismus, small teeth, long fingers, a unique palmar fold, and equinovarus deformation of feet), significant failure to thrive (weight – 3 SD, height – 4.5 SD), and microcephaly (OFC – 3 SD). WES identified a homozygous variant in NALCN [NM_052867.2; chr13:g.101107719dupT [GRCh38/hg38], c.2435dupA, p.(Glu813Glyfs*23)].
Individual 8 is a 27-year-old male born to healthy unrelated Moroccan parents. He was born full term with small parameters (unknown). He had early hypotonia, feeding difficulties, and severe gastro-esophageal reflux. He had global developmental delay: he spoke first words and walked at age of 4 years. He experienced an acute motor regression at 17 years characterized by progressive gait impairment. Brain MRI revealed cerebellar atrophy. He did not show any contractures. At the age of 27 years, his weight was 41 kg (– 2.5 SD), his height 161 cm (– 2 SD), and his OFC 55 cm (– 0.5 SD). Dysmorphic features were limited to strabismus and long fingers. With WES, we identified a de novo variant in the NALCN gene [NM_052867.2; chr13:g.101068031T > A [GRCh38/hg38], c.4333A > T, p.(Ile1445Leu)]. A heterozygous deletion in the second allele is unlikely since, (1) there was no deletion in NALCN detected in chromosomal array analysis and (2) in addition to this variant, 5 heterozygous variants in NALCN were called, all common in GnomAD (c.4416A > C, c.3955–32_3955–31dupCT, c.3570T > C, c.1764 + 21dupT, c.1593C > T) and no homozygous variant. In addition, we identified a de novo NPAS4 variant (OMIM 608554, Supplemental Table 1), and its significance remains unknown.
Individual 9 is the second child of first cousin parents. Her older brother as well as the rest of family members are healthy. The pregnancy was uneventful except for a decrease of fetal movements. The individual was born at 39 weeks of gestation with a weight of 3400 g (0.14 SD), length of 48 cm (– 1.41 SD), and an OFC of 36 cm (1.08 SD). At birth, she showed muscular hypotonia and required a brief oxygen therapy for respiratory distress. She also received phototherapy for hyperbilirubinemia in the neonatal period. She then developed global hypotonia and muscle wasting. By the age of 6 years, she was not able to sit without support. She presented with severe ID and had not developed any speech. However, the eye contact as well as communication by sounds, gestures, and smiling was present. She had stereotypical movements like hand rubbing and bringing hands to the mouth. Bruxism as well as episodes of hyperventilation was also noted. Facial dysmorphisms included midface hypoplasia, everted lower lip, convergent strabism, and high forehead. Her other medical problems included asthma, constipation, and growth retardation. She did not show any seizures. At the last assessment at age of 6 10/12 years, her weight was 14.7 kg (– 2.5 SD, BMI 11.8 kg/m2), height was 111 cm (– 1.4 SD), and her OFC was 51 cm (– 0.2 SD). CGH-array analysis was normal. Analysis of MECP2 and MEF2C genes did not reveal any pathogenic variant. A homozygous splice site variant in the NALCN gene was identified by WES [NM_052867.2, chr13 :101104289_101104292delACTT [GRCh38/hg38], c.2889 + 3_6delAAGT (IVS25 + 3_6)]. Deleterious effect of this variant was confirmed by RT-PCR on skin biopsy. In the patient, a slightly shorter product than expected was amplified (observed fragment of 169 bp for an expected fragment of 193 bp in a control cDNA). Sanger sequencing of this abnormal product revealed a deletion of 24 bp in the cDNA at the end of exon 25, putatively leading to the deletion of 8 amino acids from the NALCN protein [p.(Val956_Leu-963del)]. This deletion would affect the S3 transmembrane domain of repeat III. Another homozygous variant in the POLR2D (NM_004805.3) gene was identified in individual 9 [c.200A> G; p.(Tyr67Cys); rs142734461]. Several in silico tools predict this particular variant as potentially deleterious, but no disease has been associated with POLR2D. Parents were heterozygous for this variant. This variant was not found in her healthy brother.
Individual 10 is a 5-year-old boy born to consanguineous parents from Saudi Arabia, with a negative family history. He was born at 40 weeks of gestation with a birth weight of 2700 g (–2.1 SD). He presented with axial hypotonia and neurodevelopmental delay and later developed generalized tonic-clonic seizures. At the age of 5 years he was nonverbal and could sit with the support of his hands. His measurements showed a low BMI and microcephaly: height 102 cm (–1.72 SD), weight 11.2 kg (BMI: 10.77 kg/m2) and OFC 47.5 cm (–2.73 SD). Chromosomal microarray was negative, with several regions showing absence of heterozygosity, raising the suspicion of an autosomal-recessive disease. Trio-based WES was performed for individual 10 and his parents, with the identification of this homozygous variant in NALCN [NM_052867.2; chr13:g.101073631G > A [GRCh38/hg38], c.4150C > T, p.(Arg1384Ter)].
Individual 11 is a female 4-year-old individual, born to consanguineous parents from Saudi Arabia. Antenatal scan showed oligohydramnios. She was born by emergency Caesarean section due to fetal distress. Her birth weight was 1400 g (– 3.89 SD). She was intubated and ventilated immediately after birth because of respiratory distress syndrome. In the neonatal period she had hypotonia and feeding difficulty that resulted in severe failure to thrive. She presented with global neurodevelopmental delay. She cannot sit without support and she has no speech. She developed microcephaly and scoliosis. She has dysmorphic features including high forehead, long eyelashes, down-slanting palpebral fissures, and prominent chin. Neuromuscular assessment showed hypotonia, dystonia, microcephaly, scoliosis and lax joints. She had no seizures; however, her EEG was diffusely slow which was compatible with encephalopathy. Her younger brother is similarly affected (individual 12). He also has failure to thrive, hypotonia, long face with a high forehead and protruding chin, long eyelashes and down-slanting palpebral fissures. WES analysis detected this homozygous variant in the NALCN gene [NM_052867.2; chr13:g.101378624C > T [GRCh38/hg38], c.321G > A, p.(Trp107Ter)] in individual 11, which was also confirmed in her brother (individual 12) by Sanger sequencing.
Individual 13 is the first child of consanguineous parents from Egypt, with a negative family history. She was born at 39 weeks of gestation with a birth weight of 1950 g (–3.33 SD). At the age of 3 years, she presented with severe neurodevelopmental delay, abnormal movements, dysmorphic facial features including a triangular face with a high forehead, large ears, fine hair, and convergent strabismus. She was not able to speak; she could sit. Metabolic workup and karyotype were both normal. By WES analysis we identified the homozygous NALCN variant [NM_052867.2; chr13:g.101104426delT [GRCh38/hg38], c.2758delA, p.(Ile920Leufs*7)].
Individual 14 and 15 were siblings, the only offspring of healthy consanguineous Egyptian parents. They presented at the ages of 4 years and 2 9/12 years with severe developmental delay, failure to thrive, dysmorphic facial features (individual 14, Fig. 1h, i; individual 15, Fig. 1j), hypotonia with elicited reflexes, autistic features and abnormal movements. Epilepsy was manifested around the age of 1 year in both of them. Genetic investigations including karyotyping, the metabolic workup was normal. Mild defective myelination was detected in their MRI. WES revealed the homozygous NALCN variant [NM_052867.2; chr13:g.101104616delC [GRCh38/hg38], c.2671delG, p.(Val891Serfs*2)]. Individual 14 passed away near the age of 5.
Individual 16 was a 3-year-old male born to consanguineous parents (first cousins) from Egypt. There was a positive family history with a similarly affected deceased cousin; however, no genetic investigations were performed in this relative. He was born at term with a birth weight of 3500 g (–0.28 SD), birth length of 50 cm (–1.09 SD) and OFC of 34.2 cm (–0.8 SD). He presented at the age of 3 years with severe psychomotor retardation, he was not able to speak or sit. In addition, he showed failure to thrive, epilepsy, autistic behavior and dysmorphic facial features. WES revealed the homozygous NALCN variant [NM_052867.2; chr13:g.101104616delC [GRCh38/hg38], c.2671delG, p.(Val891Serfs*2)]. He passed away at the age of 5 6/12 years with chest infection.
Individual 17 was a 3-year-old female from Egypt, descended from healthy consanguineous parents. No similarly affected members in the family were recorded. She was born at 38 weeks of gestation with a birth weight of 3500 g (+ 0.76 SD), birth length of 49 cm (– 0.5 SD) and OFC of 34 cm (– 1 SD). She was referred to the genetics clinic because of severe psychomotor retardation; failure to gain weight and recurrent infections started at 6 months of age. On examination, she showed severe ID, autistic features, irritability, muscular hypotonia and she had dysmorphic facial features (Fig. 1k, l). All metabolic workup and karyotyping were normal; brain MRI showed mild cortical atrophic changes and thin corpus callosum. WES revealed homozygous NALCN variant [NM_052867.2; chr13:g.101376807delC [GRCh38/hg38], c.537delG, p.(Trp179Ter)].
Clinical description of individuals with UNC80 variants
Individual 18 is a 1-year-old boy born to consanguineous parents from Syria. He has three older, unaffected siblings. He was born after an uneventful pregnancy with a birth weight 3500 g (– 0.64 SD). At birth, he showed neonatal hypotonia and he presented with jaundice, which resolved within 72 h. The parents noticed a developmental delay at the age of about 4 months. He was brought to genetic consultation at the age of 10 months. He presented with severe global neurodevelopmental delay, hypotonia, and failure to thrive. His OFC was 44.5 cm (–1.07 SD). Dysmorphic facial features included triangular face, high forehead, large ears, convergent strabismus, and arched upper lip. Delayed tooth eruption and left single palmar crease were noticed. Brain MRI and EEG were unremarkable. Diagnostic WES analysis revealed a homozygous, likely pathogenic variant in the UNC80 gene [NM_032504.1; chr2:g.209972258C > T [GRCh38/hg38], c.8116C > T, p.(Arg2706Ter)].
Individual 19 is a 3-year-old boy born to consanguineous parents from Saudi Arabia. He had two older siblings who died undiagnosed with spasticity and severe developmental delay. He was born at 39 weeks of gestation, his birth weight was 2230 g (– 2.9 SD), length 45 cm (– 3 SD), and OFC was 33 cm (– 1.77 SD). He showed neonatal hypotonia, bilateral undescended testes, and metatarsal adduction of the right foot. His hypotonia progressed to severe muscular hypotonia at the age of 5 months; he did not achieve any developmental milestone, had global neurodevelopmental delay, tonic-clonic seizures at the age of 4 months, and failure to thrive. He developed spastic paraparesis. Brain MRI showed diffusion restriction in both parietooccipital cortex (Supplemental Table 2). He had many hospital admissions due to pneumonia and died at age of 10 months with cardiopulmonary arrest at home. His younger sister (individual 20) is a 1-year-old girl who was born at term, her birth weight was 2240 g (– 3.13 SD), length 45 cm (– 3.26 SD). She presented at the age of 4 months with global neurodevelopmental delay, muscular hypotonia, and failure to thrive. Microcephaly with brachycephaly and dysmorphic facial features was noted. At the age of 9 months, she showed severe failure to thrive with a weight of 3.9 kg (BMI 8.95 kg/m2), height 66 cm (– 1.72 SD), and OFC 41.6 cm (– 2.1 SD). She had global developmental delay and showed muscle wasting. Array analysis in individual 19 was normal, except for excess homozygosity. WES was performed in both affected siblings and asymptomatic parents. The UNC80 variant [NM_032504.1; chr2:g.209777479C > T [GRCh38/hg38], c.520C > T, p.(Arg174Ter)] was found to co-segregate with the phenotype as it was present in the homozygous state in both affected siblings, and the unaffected parents were both heterozygotes.
The same homozygous UNC80 variant [NM_032504.1; chr2:g.209777479C > T [GRCh38/hg38], c.520C > T, p.(Arg174Ter)] was found in two other affected relatives. The female individual 21 was born at full term, birth weight was 2100 g (– 2.97 SD), length was 44 cm (– 3.23 SD), and OFC 30.5 cm (– 3.15 SD). Her parents were first cousins from Saudi Arabia. Individual 21 presented prenatally with intrauterine growth retardation (IUGR). Later, she presented with severe failure to thrive, global developmental delay, and hypotonia. She has mild hydronephrosis. Her affected sister (individual 22) was born at 36 weeks of gestation; prenatally, IUGR had been noted. Her birth weight was 2400 g (– 0.79 SD), length 46 cm (– 0.92 SD), and OFC 31 cm (– 1.53 SD). She later presented with microcephaly and joint contractures (elbow, wrist, and hip). Cardiac echo showed a small ASDII at the age of 5 months. She showed severe failure to thrive, global developmental delay, and recurrent aspiration pneumonia, with difficult feeding requiring nasogastric tube feeding.
Individual 23 (Chérot et al. 2018) was the second child of healthy parents. He was born full term with normal parameters [birth weight 3080 g (– 1.46 SD), birth length 48 cm (– 2.2 SD), birth OFC 35 cm (– 0.69 SD)]. On his second day of life, he had mild respiratory distress, mild hypotonia, and jaundice. Individual 23 showed signs of infection, but the cause was not identified. He showed clinical improvement after antibiotic therapy. At the age of 1 month, he had to be hospitalized because of poor weight gain due to poor sucking and little appetite. He did not require tube feeding. At the age of 4 years, he was still thin (weight 13 kg, – 2 SD) and ate small and enriched meals. Axial hypotonia and involuntary choreic movements of upper limbs have been noticed since the first months of life. Individual 23 has been able to catch objects from the age of 15 months, but at 3 years of age, he was unable to make stacks or play simple puzzles. He also had severe language delay, since he did not speak a word at the age of 3 years. He had mild dysmorphism (enophthalmia, long face with a high forehead, and pectus excavatum) and marked pink skin under his fingernails. WES revealed compound heterozygous variants in the UNC80 gene [NM_032504.1; variant 1, paternal: chr2:g.209825974delT [GRCh38/ hg38], c.2399delT, p.(Leu800Trpfs*19); variant 2, maternal: chr2:g.209888128G > T [GRCh38/hg38], c.520C > T, p.(Arg174Ter)]. The variant details with scarce clinical information were previously reported (Chérot et al. 2018).
Individuals 24 and 25 were the two only male siblings from healthy, consanguineous Egyptian parents. They were presented at the ages of 7 years and 1 3/12 years, respectively. A maternal history of recurrent miscarriages was recorded. Both individuals presented with severe psychomotor retardation, dysmorphic facial features (individual 25, Fig. 2a, b), and autistic behavior. Excessive abnormal limb movement was prominent since early life. At first presentation, both siblings could not use their hands or respond to their names. The last examination of individual 24 was at the age of 10 years when he presented with the following measurements: height 114 cm (– 3.7 SD), weight 17 kg (BMI: 12.31 kg/m2), and OFC 49.5 cm (– 2.1 SD). The last examination of individual 25 was at the age of 3 8/12 years when he presented with the following measurements: height 92 cm (– 2 SD), weight 12 kg (BMI: 14.81 kg/m2), and OFC 47 cm (– 2.8 SD). WES revealed the following homozygous variant in the UNC80 gene [NM_032504.1; chr2:209817940_209817941delAC [GRCh38/hg38], c.1681_1682delAC, p.(Thr561Argfs*33)]. Individual 25 passed away at the age of 4 years.
Fig. 2.
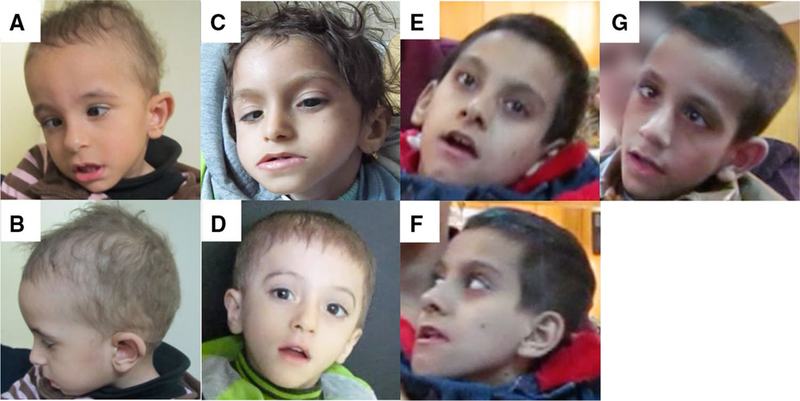
Individuals with biallelic UNC80 variants. a, b Individual 25 at the age of 3 6/12 years showing triangular face, high forehead, strabismus, fine hair, thin upper lip, and everted lower lip. c Individual 26 at the age of 1 9/12 years displaying triangular face with high forehead, fine hair, and strabismus. d Individual 27 at the age of 4 years presenting with triangular face, fine hair, and thin upper lip. e, f Individual 28 at the age of 8 years with long face, deep-set eyes, and facial hypotonia. g Individual 29 at the age of 7 years with long face, deep-set eyes, strabismus, and open-mouth appearance
Individual 26 was the first child in the family; a female individual born to consanguineous Egyptian parents (first cousins). They had a positive family history of similarly affected paternal cousin, who passed away at the age of 3 years. Individual 16 presented at the age of 8 months with severe psychomotor retardation and failure to thrive. Axial hypotonia was noted at the age of 2 months. Limbs had mixed hypotonia and rigidity with brisk reflexes, abnormal movement, and agitation. She did not respond to her name or recognize her parents. Clenched fist was noted with inability to catch objects. Her last examination was at the age of 1 9/12 years with the following measurements: height 68 cm (– 4.8 SD), weight 4.6 kg (BMI: 9.95 kg/ m2), and OFC 37 cm (– 7.2 SD); she displayed dysmorphic facial features (Fig. 2c). Brain MRI showed a thin corpus callosum. WES revealed this homozygous variant in the UNC80 gene [NM_032504.1; chr2:g.209929933C > T [GRCh38/hg38], c.5671C > T, p.(Arg1891Ter)]. Individual 26 suffered from recurrent infections and passed away at the age of 2 7/12 years.
Individual 27 was a 1 10/12-year-old boy born to healthy first cousins from Egypt. Severe motor and cognitive impairment, failure to thrive, dysmorphic facial features (Fig. 2d), and excessive abnormal limb movement simulating chore-oathetosis (Supplemental Video 1) were noted. He also showed axial hypotonia, mixed limb hypotonia, and rigidity with preserved reflexes. Recurrent infections were recorded. Brain MRI showed mild fronto-parietal cortical atrophy; extensive metabolic work up was normal. His last examination was at the age of 4 years when he presented with the following measurements: height 87 cm (– 3.2 SD), weight 9.1 kg (BMI: 12.02 kg/m2), and OFC 46 cm (– 3.7 SD). WES revealed the following homozygous variant in UNC80 [NM_032504.1; chr2:g.209970959T > G [GRCh38/hg38], c.8058 + 2T > G, p.?].
Individuals 28 and 29 were female and male siblings born to healthy, consanguineous parents (first cousins) from Upper Egypt. There was a positive family history with a similar male sibling passing away at the age of 1 6/12 years with chest infection. Both individuals presented with global developmental delay, failure to thrive, severe intellectual disability, dysmorphic facial features, axial hypotonia, and abnormal and involuntary limb movements. Excessive crying and irritability was noted during early life. They were both nonverbal; they did not experience seizures. MRI brain revealed mild cortical changes and thin corpus callosum. Their last examination took place at the age of 8 years (individual 28, Fig. 2e, f) and 9 years (individual 29, Fig. 2g) (see Supplemental Table 2 for measurements). WES revealed the following variant in UNC80 [NM_032504.1; chr2:g.209786065G> A [GRCh38/hg38], c.601–1G > A, p.?]. Both siblings passed away early at the ages of 10 years (individual 28) and 9 6/12 years (individual 29).
A schematic representation of the NALCN ion channel including all reported biallelic variants is displayed in Fig. 3a, and an overview of our UNC80 variants is shown in Fig. 3b. A reader-friendly overview of the important clinical features including the present and previously published individuals with biallelic NALCN and UNC80 variants is provided in Table 1.
Fig. 3.

Schematic representation of the NALCN ion channel (a) and UNC80 (b) including all variants of the reported individuals (NALCN, UniProt ID Q8IZF0). Light blue circles indicate compound heterozygous variants; dark blue circles indicate homozygous variants. The green circle shows the predicted location of the NALCN missense variant inherited in an autosomal-recessive manner in individuals 4 and 5, their paternal variant c.4103 + 2T > C is not displayed. The red circle demonstrates the predicted location of the denovo heterozygous NALCN missense variant identified in individual 8. All variants are listed according to NM_052867.2 (NALCN) and NM_032504.1 (UNC80)
Table 1.
Clinical features of individuals with biallelic NALCN and UNC80 variants
| Individuals 1–7, 9–17 (NALCN) |
Previously published individuals with biallelic NALCN variants* |
Total (NALCN) | Individu- als 18–29 (UNC80) |
Previously published individuals with biallelic UNC80 vari- ants** |
Total (UNC80) | |
|---|---|---|---|---|---|---|
| Neurodevelopment | ||||||
| Neonatal hypotonia | 16/16 | 13/15 | 29/31 | 12/12 | 19/21 | 31/33 |
| Feeding difficulties | 15/16 | 17/17 | 32/33 | 11/12 | 5/8 | 16/20 |
| Failure to thrive | 16/16 | 8/14 | 24/30 | 12/12 | 18/20 | 30/32 |
| ID, severe ID | 16/16, 14/16 | 19/19, 15/19 | 35/35, 29/35 | 11/11, 11/11 | 22/22, 22/22 | 33/33, 33/33 |
| Speech impairment | 16/16 | 19/19 | 35/35 | 11/11 | 22/22 | 33/33 |
| Motor delay | 16/16 | 19/19 | 35/35 | 11/11 | 16/16 | 27/27 |
| Hypotonia | 16/16 | 19/19 | 35/35 | 10/12 | 22/22 | 32/34 |
| Hypertonia (e.g., extremities) | 2/16 | 2/2 | 4/18 | 3/12 | n.a | 3/12 |
| Extrapyramidal/ abnormal movements | 12/16 | n.a | 12/16 | 8/12 | (Dystonic posture of limbs) 9/9 | 17/19 |
| Seizures | 7/16 | 13/19 | 20/35 | 1/12 | 12/22 | 13/34 |
| Early death | 2/16 | 2/19 | 4/35 | 5/12 | n.a | 5/12 |
| Measurements | ||||||
| Normal birth weight | 10/16 | 8/10 | 18/26 | 9/12 | 12/13 | 21/25 |
| Short stature (height ≤ – 2 SD) | 11/15 | 2/4 | 13/19 | 6/9 | 12/15 | 18/24 |
| Low weight (BMI ≤ 15 kg/m2) | 12/15 | 7/8 | 19/23 | 8/8 | (weight < 2 SD) 11/13 | 19/21 |
| Microcephaly (OFC ≤ – 2 SD) | 12/16 | 2/5 | 14/21 | 7/10 | 13/22 | 20/32 |
| Other features | ||||||
| Breathing abnormalities | 5/16 | 6/7 | 11/23 | 0/12 | n.a | 0/12 |
| Chronic constipation | 14/16 | 17/17 | 31/33 | 10/12 | 6/9 | 16/21 |
| Sleeping difficulties | 10/16 | 4/5 | 14/21 | 10/12 | 5/6 | 15/18 |
| Increased tendency to infections | 9/16 | n.a | 9/16 | 8/12 | n.a | 8/12 |
| Strabismus | 12/16 | 8/8 | 20/24 | 11/12 | 13/13 | 24/25 |
| Hand flapping | 7/16 | n.a | 7/16 | 3/12 | 3/3 | 6/15 |
| Contractures | (Camptodactyly) 2/16 | (Extremities) 2/2 | 4/18 | (Joints) 1/12 | (Joints) 10/10 | 11/24 |
Individual 8 with the de novo heterozygous NALCN variant was excluded. There were overlapping facial features, but no recognizable facial phenotype. Facial features are listed in Supplemental Tables 1 and 2 (n.a not available, ID intellectual disability, BMI body mass index, OFC occipital frontal circumference).
Including Al Sayed et al., Koroglu et al., Gal et al., Takenouchi et al., Angius et al., Campbell et al., and Bourque et al.
Including Perez et al., Shamseldin et al., Stray-Pedersen et al., Valkanas et al., Boerkoel et al., He at al., and Obeid et al.
Discussion
In this study, we present the clinical phenotypes of 16 individuals with biallelic NALCN variants, 1 individual with a de novo NALCN variant, and 12 individuals with biallelic UNC80 variants. We report the largest cohort of individuals with biallelic NALCN variants, compare our cohort with clinical data of all previously published IHPRF1-affected individuals providing the first comprehensive overview of the clinical phenotype associated with biallelic NALCN variants. In addition, we greatly enlarge the number of published individuals with biallelic UNC80 variants. This allowed us to greatly refine their clinical phenotypes and determine differences in the clinical phenotypes of IHPRF1-and IHPRF2-affected individuals. This lead us to propose the new term ‘autosomal-recessive NALCN channelopathies’ summarizing all the previously mentioned, but also including the novel clinical features observed in our cohort of IHPRF-affected individuals. To illustrate our findings, we have included a thorough discussion of all previously published clinical features and our novel, formerly unpublished clinical details below.
Comparison of the clinical phenotypes of IHPRF1-and IHPRF2-affected individuals
All individuals (16/16) with biallelic NALCN variants presented with neonatal hypotonia, failure to thrive, ID, and muscular hypotonia (Table 1, Supplemental Table 1). More specifically, most individuals displayed severe ID (14/16); 14/16 individuals were nonverbal, and 13/16 individuals did not walk (Supplemental Table 1). More than half of the individuals had chronic constipation (14/16), extrapyramidal/abnormal movements (12/16), strabismus (12/16), sleeping difficulties (10/16), increased tendency to infections (9/16), and some individuals presented with seizures (7/16). Immunological profiling was performed in most individuals with increased tendency to infections; these immunological profiles were all normal. There was no evidence for splenic dysfunction. The infections mostly included upper respiratory infections and (aspiration) pneumonia. We suspect the increased tendency to infections to arise from their failure to thrive. Analysis of the measurements showed that most individuals were born with normal birth weight (10/16), postnatally most individuals showed short stature (11/15), low BMI (12/15), and microcephaly (12/16). Some IHPRF1-affected individuals showed breathing abnormalities (5/16), and 2/16 individuals died early at the ages of 4 8/12 years and 5 6/12 years, 1 of them in the course of a chest infection. Contractures were only identified in 2/16 individuals, including mild camptodactyly in individual 2 and clenched hands and camptodactyly in individual 16. Some facial features were shared among the individuals (Fig. 1, Supplemental Table 1), but there was no recognizable facial phenotype. Interestingly, individual 1 strongly benefitted from a ketogenic diet. After introduction of the ketogenic diet, she showed better weight gain, progress in development, and seizure control. Her antiepileptic medication was reduced from a dual therapy with lamotrigine and valproate to only lamotrigine and she has remained without obvious seizures. It remains to be determined whether other individuals with biallelic NALCN variants might also benefit from the ketogenic diet, but it might represent a helpful treatment option.
In summary, the overall distribution of clinical features is similar to previously published individuals with biallelic NALCN variants, although we did not find any mention of extrapyramidal/abnormal movements or increased tendency to infections in the previous publications (Table 1). Hence, we could refine the clinical phenotype of individuals with biallelic NALCN variants, now termed autosomal-recessive NALCN channelopathy, and we provide the first comprehensive overview of the clinical features of all published IHPRFl-affected individuals (Table 1).
Individuals with biallelic UNC80 variants showed a less variable clinical phenotype (Table 1, Supplemental Table 2). All individuals (12/12) presented with neonatal hypotonia, failure to thrive, and severe ID; 7/7 individuals were nonverbal; and 7/7 individuals did not walk (Supplemental Table 2). Most IHPRF2-affected individuals displayed feeding difficulties (11/12), hypotonia (10/12), extrapyramidal/abnormal movements (8/12) (Supplemental Movie 1), chronic constipation (10/12), sleeping difficulties (10/12), increased tendency to infections (8/12), and strabismus (11/12). Most individuals were born with normal weight (9/12), postnatally they mostly presented with short stature (6/9) and microcephaly (7/10); all individuals had a low BMI (8/8). In contrast to individuals with IHPRF1, individuals with IHPRF2 did not show any breathing abnormalities (0/12), which we will discuss in more detail below. Only 1 individual (individual 18) presented with seizures (1/12) and only 1 individual (individual 21) showed contractures (1/12; elbow, wrist, and hip). These findings are in contrast to previously published individuals with biallelic UNC80 variants [12/22 individuals with seizures, 10/10 individuals with joint contractures; Table 1]. One reason might be that many individuals in our cohort were still young at the age of last examination (3 individuals < 1 year, 2 individuals n.a.) and 5/12 individuals passed away early (range 10 months to 10 years), e.g., after recurrent infections/pneumonia and cardiopulmonary arrest.
Taken together, the comparison of our IHPRF1-and IHPRF2 cohorts with each other and with previously published individuals showed that the clinical presentation of IHPRF1 seems to be more variable than of IHPRF2, e.g., with regard to the severity of ID. Early death was not previously discussed in these patients, but in our cohort, 12.5% of the IHPRF1-and 41.7% of the IHPRF2-affected individuals died early, which is a very important piece of information for counseling of affected families.
Breathing abnormalities in IHPRF1-affected individuals
Breathing abnormalities were only documented in IHRPF1-affected, but not in IHPRF2-affected individuals. Work in different animal models has shown that NALCN has many physiological roles (Cochet-Bissuel et al. 2014). More specifically, Nalcn-deficient mice (null mice) showed an abnormal breathing pattern and died within 24 h after birth (Lu et al. 2007). These mice showed only sporadic respiration with apnea for a few seconds followed by deep breathing. This pattern was described as reminiscent of Cheyne-Stokes respiration in humans (Lu et al. 2007). Interestingly, 5/16 individuals with IHPRF1 in our cohort also showed breathing abnormalities; individual 1 displayed Cheyne-Stokes-like respiration with central apneas without hypoxemia; and individuals 4 and 5 were diagnosed with periodic breathing with central apneas without hypoxemia. Individual 5 needs to be monitored during the night. Abnormal breathing patterns were also observed in newborns with heterozygous NALCN variants (Chong et al. 2015), recurrent episodes of apneic spells in 2/3 individuals with homozygous splice site NALCN variants (Gal et al. 2016), and central apneas during sleep in 3/3 individuals reported by Campbell and colleagues (Campbell et al. 2018). Periodic breathing in awake and sleep states with three-to-four rapid breaths followed by central apneas was also documented in 1 recently published individual with a homozygous nonsense mutation in NALCN (Bourque et al. 2018). Based on these data, we recommend sleep studies for all individuals with monoallelic and biallelic NALCN variants.
Differential diagnoses
Breathing abnormalities were documented in 5/16 individuals with biallelic NALCN variants. In general, breathing abnormalities are a very rare clinical sign and they are found in only a small number of diseases/syndromes. For example, breathing abnormalities are observed in Pitt-Hopkins syndrome (OMIM 610954), in central hypoventilation syndrome (OMIM 209880), and types of Joubert syndrome. All the aforementioned syndromes have distinct clinical features, so that breathing abnormalities in combination with muscular hypotonia, failure to thrive, and developmental delay/ID or the occurrence of contractures are strongly suggestive of a ‘NALCN-channelopathy’ and should lead to analysis of the NALCN gene. In addition, considerable clinical overlap was noted between IHPRF1-and IHPRF2-affected individuals and individuals with biallelic PYCR2 mutations causing hypomyelinating leukodystrophy 10 (HLD10, OMIM 616420) (Zaki et al. 2016). Individuals with HLD10 also present with failure to thrive, muscular hypotonia, hyperkinetic movements, and severe developmental delay/ID and many affected individuals passed away within the first decade of life. Differing clinical features are postnatal, progressive microcephaly, which has not been documented to this degree in individuals with biallelic NALCN or UNC80 mutations, and cerebral atrophy. Taken together, HLD10 is an important differential diagnosis.
Spectrum of variants and clinical implications
Most biallelic NALCN variants were homozygous frameshift (7/16 individuals) or nonsense variants (5/16 individuals) (Supplemental Table 1). However, we also identified compound heterozygous variants, e.g., two truncating NALCN variants in individual 1 and a paternal putative splice site variant and a maternal missense variant in individuals 4 and 5. Interestingly, the maternally derived missense variant was located in the predicted S6 pore-forming segment of domain IV of NALCN. Typically, monoallelic missense variants in or close to the S5/S6 pore-forming unit of NALCN lead to CLIFAHDD (Chong et al. 2015). Three IHPRF1-affected individuals with homozygous (Al-Sayed et al. 2013) or compound heterozygous (Campbell et al. 2018) missense NALCN variants have been previously described, but these variants were located in an S3 segment, extracellular, or cytoplasmic domain. Interestingly, individuals 4 and 5 show a milder phenotype than the remaining 14 individuals with biallelic NALCN variants in our cohort. While they shared many of the IHPRF1-typical clinical features, such as neonatal hypotonia, failure to thrive, and hypotonia, they did not present with severe but with mild ID; they started walking at the age of 2 1/12 and 2 10/12 years and showed (mildly) delayed speech impairment. Unlike individuals with CLIFAHDD, individuals 4 and 5 did not display any contractures. To the best of our knowledge, this is the first missense variant in a predicted S6 segment, which is inherited in an autosomal-recessive manner leading to a mild form of IHPRF1.
Individual 8 presented with a de novo NALCN missense variant, no other possibly pathogenic variants were identified in WES analysis. A heterozygous deletion was very unlikely, since the array analysis showed no deletion in NALCN and 6 other heterozygous benign variants were carried and no homozygous variant. In addition to IHPRF1-typical clinical features, such as neonatal hypotonia, feeding difficulties, failure to thrive, and severe ID, he showed cerebellar atrophy and cerebellar syndrome. The NALCN missense variant [NM_052867.2; variant 1, maternal: chr13:g.101068744G > T [GRCh38/hg38], c.4281C > A, p.(Phe1427Leu)] is located in the predicted pore-forming S6 transmembrane segment of domain IV, a typical location for missense variants leading to CLIFAHDD syndrome (Chong et al. 2015). Interestingly, the initial publication describing CLIFAHDD syndrome reported on 1 individual (Family D in Chong et al. 2015) with a variant affecting the neighboring amino acid (p.Ile1446Met) (Chong et al. 2015). However, a comparison of the clinical features of these 2 individuals revealed marked differences. While individual 8 displayed cerebellar atrophy and did not show any contractures, the previously published individual (Family D in Chong et al. 2015) presented with multiple contractures, but cerebellar atrophy was not listed in the MRI findings. Other individuals with NALCN missense variants in the aforementioned pore-forming transmembrane segments displaying cerebellar hypoplasia/atrophy and/or ataxia have been published (Aoyagi et al. 2015; Chong et al. 2015; Karakaya et al. 2016; Wang et al. 2016; Vivero et al. 2017), but all of them also showed contractures, with the exception of 1 individual, who showed a decreased range of motion in all large joints (patient #1 published by Vivero et al. 2017) (Vivero et al. 2017). In addition, we identified a de novo NPAS4 variant in individual 8. This gene encodes for a transcription factor, which has been shown to play a role in the development and redistribution of inhibitory synapses in rat and mouse hippocampal cultures (Lin et al. 2008; Bloodgood et al. 2013). Npas4 knockout mice appear to be hyperactive and prone to seizures (Lin et al. 2008) and have learning and memory deficits (Ramamoorthi et al. 2011). Variants in the NPAS4 gene have not been associated with any disease, so that the significance of this finding remains unknown. To our knowledge, individual 8 is only the second reported individual with a missense variant in the predicted S5/S6 pore-forming transmembrane segment without contractures. Our finding supports the notion that the variability of the clinical phenotype caused by heterozygous NALCN missense variants in the predicted pore-forming segments is even higher than previously expected.
From the three members of the NALCN-UNC80-UNC79 complex, disease-causing variants have been found only in NALCN and UNC80, which lead us to search the CentoMD database (Centogene’s proprietary database), aiming to identify undiagnosed patients with homozygous high impact variants in UNC79. However, none were found. Data from ExAc and Gnomad databases clearly show that UNC79 is intolerant for missense and loss of function variants (missense z=4.02, LoF pLi = 1). The Z score indicates the deviation of observed variant counts from the expected number. Positive Z scores indicate increased constraint (intolerance to variation). The pLI is similarly calculated, and genes with pLi close to 1 are considered very intolerant to LoF. Lu et al. (2010) stated that Unc79−/− mice have disrupted breathing rhythms, fail to nurse, and die within the first days of life. Taken together with data from the Unc79 knockout mouse, one can speculate that biallelic LoF variants in UNC79 might be incompatible with life. For UNC80, with biallelic LoF variants leading to a severe neurological phenotype, as we demonstrated here, a pLi score = 0.14 is reported (missense, z =0.71).
On the contrary, the NALCN gene seems also intolerant to missense variation. Others and we are reporting on mono-and biallelic missense changes in NALCN and its associated neurological phenotype. The gene has a very high Z score of 5.48 (ExAc database). The precise mechanism leading to NALCN channelopathies still needs to be established.
In summary, we report on the so far largest IHPRF1-and IHPRF2 cohorts including 16 and 12 individuals, respectively. We provide the first comprehensive overview of the clinical phenotype of IHPRF1-affected individuals allowing for a refined analysis of the IHPRF1-associated clinical phenotype and propose the term autosomal-recessive NALCN channelopathies for the entirety of all observed features. We present the first 2 individuals in which a NALCN missense variant located in a predicted S6 pore-forming segment is inherited in an autosomal-recessive pattern and leads to a comparably mild clinical phenotype; and we report on 1 individual with a heterozygous de novo NALCN variant in a predicted pore-forming S6 segment without any sign of contractures supporting the notion that CLIFAHDD is even more variable than previously expected.
Supplementary Material
Acknowledgements
We are grateful to the families for participating in this study and giving their consent for publication. We thank Sabine Kaya for excellent technical assistance. NE is participant in the Clinician Scientist Program, funded by the Berlin Institute of Health (BIH) and the Charité. LS received a Medical Student Research Grant from the BIH. This work was in part supported by the German Ministry of Research and Education [grant numbers 01GS08164 (HE), 01GS08167 (DW), 01GS08163 (TMS), German Mental Retardation Network] as part of the National Genome Research Network, the Broad Institute (U54HG003067 to E. Lander and UM1HG008900 to D. MacArthur), the Yale Center for Mendelian Disorders (U54HG006504 to R. Lifton and M. Gunel), NIH grants R01NS048453, R01NS052455, the Simons Foundation for Autism Research Initiative and Howard Hughes Medical Institute (to JGG). We would like to thank Prince Abdullah Ben Khalid Celiac Disease Research Chair, Vice Deanship of Research Chairs, King Saud University, Riyadh, Kingdom of Saudi Arabia for their support.
Footnotes
Compliance with ethical standards
Conflict of interest Arndt Rolfs is founder and shareholder of CENTOGENE AG, a commercial company offering genetic testing service. Aida Bertoli-Avella and Peter Bauer are employees of CENTOGENE AG.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00439-018-1929-5) contains supplementary material, which is available to authorized users.
References
- Al-Sayed MD, Al-Zaidan H, Albakheet A et al. (2013) Mutations in NALCN cause an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay. Am J Hum Genet 93:721–726. 10.1016/j.ajhg.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angius A, Cossu S, Uva P et al. (2018) Novel NALCN biallelic truncating mutations in siblings with IHPRF1 syndrome. Clin Genet. 10.1111/cge.13162 [DOI] [PubMed] [Google Scholar]
- Aoyagi K, Rossignol E, Hamdan FF et al. (2015) A gain-of-function mutation in NALCN in a child with intellectual disability, ataxia, and arthrogryposis. Hum Mutat 36:753–757. 10.1002/humu.22797 [DOI] [PubMed] [Google Scholar]
- Bloodgood BL, Sharma N, Browne HA et al. (2013) The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature 503:121–125. 10.1038/nature12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel C, du Souich C (2017) UNC80 Deficiency. University of Washington, Seattle: [PubMed] [Google Scholar]
- Bourque DK, Dyment DA, MacLusky I et al. (2018) Periodic breathing in patients with NALCN mutations. J Hum Genet. 10.1038/s10038-018-0484-1 [DOI] [PubMed] [Google Scholar]
- Bramswig NC, Ludecke HJ, Alanay Y et al. (2015) Exome sequencing unravels unexpected differential diagnoses in individuals with the tentative diagnosis of Coffin-Siris and Nicolaides-Baraitser syndromes. Hum Genet 134:553–568. 10.1007/s00439-015-1535-8 [DOI] [PubMed] [Google Scholar]
- Campbell J, Fitzpatrick DR, Azam T et al. (2018) NALCN dysfunction as a cause of disordered respiratory rhythm with central apnea. Pediatrics 141:S485–S490. 10.1542/peds.2017-0026 [DOI] [PubMed] [Google Scholar]
- Cherot E, Keren B, Dubourg C et al. (2018) Using medical exome sequencing to identify the causes of neurodevelopmental disorders: Experience of 2 clinical units and 216 patients. Clin Genet 93:567–576. 10.1111/cge.13102 [DOI] [PubMed] [Google Scholar]
- Chong JX, McMillin MJ, Shively KM et al. (2015) De novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia, and developmental delay. Am J Hum Genet 96:462–473. 10.1016/j.ajhg.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochet-Bissuel M, Lory P, Monteil A (2014) The sodium leak channel, NALCN, in health and disease. Front Cell Neurosci 8:1–17. 10.3389/fncel.2014.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M, Banks E, Poplin RE et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. 10.1038/ng.806.A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon-Salazar TJ, Silhavy JL, Udpa N et al. (2012) Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med 4:138ra78–138ra78. 10.1126/scitranslmed.3003544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal M, Magen D, Zahran Y et al. (2016) A novel homozygous splice site mutation in NALCN identified in siblings with cachexia, strabismus, severe intellectual disability, epilepsy and abnormal respiratory rhythm. Eur J Med Genet 59:204–209. 10.1016/j.ejmg.2016.02.007 [DOI] [PubMed] [Google Scholar]
- He Y, Ji X, Yan H et al. (2018) Biallelic UNC80 mutations caused infantile hypotonia with psychomotor retardation and characteristic facies 2 in two Chinese patients with variable phenotypes. Gene 660:13–17. 10.1016/j.gene.2018.03.063 [DOI] [PubMed] [Google Scholar]
- Jäger M, Wang K, Bauer S et al. (2014) Jannovar: a Java library for exome annotation. Hum Mutat 35:548–555. 10.1002/humu.22531 [DOI] [PubMed] [Google Scholar]
- Karakaya M, Heller R, Kunde V et al. (2016) Novel mutations in the nonselective sodium leak channel (NALCN) lead to distal arthrogryposis with increased muscle tone. Neuropediatrics 47:273–277. 10.1055/s-0036-1584084 [DOI] [PubMed] [Google Scholar]
- Köroglu f, Seven M, Tolun A (2013) Recessive truncating NALCN mutation in infantile neuroaxonal dystrophy with facial dysmorphism. J Med Genet 50:515–520. 10.1136/jmedgenet-2013-101634 [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v1 [q-bio.GN] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL et al. (2008) Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455:1198–1204. 10.1038/nature07319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Su Y, Das S et al. (2007) The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell 129:371–383. 10.1016/j.cell.2007.02.041 [DOI] [PubMed] [Google Scholar]
- Lu B, Zhang Q, Wang H et al. (2010) Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron 68:488–499. 10.1016/j.neuron.2010.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeid T, Hamzeh AR, Saif F et al. (2018) Identification of a novel homozygous UNC80 variant in a child with infantile hypotonia with psychomotor retardation and characteristic facies-2 (IHPRF2). Metab Brain Dis. 10.1007/s11011-018-0200-z [DOI] [PubMed] [Google Scholar]
- Okonechnikov K, Conesa A, García-Alcalde F (2016) Qualimap 2: advanced multi-sample quality control for high-through put sequencing data. Bioinformatics 32:292–294. 10.1093/bioinformatics/btv566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Y, Kadir R, Volodarsky M et al. (2016) UNC80 mutation causes a syndrome of hypotonia, severe intellectual disability, dyskinesia and dysmorphism, similar to that caused by mutations in its interacting cation channel NALCN. J Med Genet 53:397–402. 10.1136/jmedgenet-2015-103352 [DOI] [PubMed] [Google Scholar]
- Project Consortium G, Consortium Participants are arranged by project role G, by institution alphabetically then et al. (2012) An integrated map of genetic variation from 1092 human genomes. Nature. 10.1038/nature11632 [DOI]
- Ramamoorthi K, Fropf R, Belfort GM et al. (2011) Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science 334(80):1669–1675. 10.1126/science.1208049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Köhler S, Bauer S et al. (2008) The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 83:610–615. 10.1016/j.ajhg.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin HE, Faqeih E, Alasmari A et al. (2016) Mutations in UNC80, encoding part of the UNC79-UNC80-NALCN channel complex, cause autosomal-recessive severe infantile encephalopathy. Am J Hum Genet 98:210–215. 10.1016/j.ajhg.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray-Pedersen A, Cobben JM, Prescott TE et al. (2016) Biallelic mutations in UNC80 cause persistent hypotonia, encephalopathy, growth retardation, and severe intellectual disability. Am J Hum Genet 98:202–209. 10.1016/j.ajhg.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenouchi T, Inaba M, Uehara T et al. (2018) Biallelic mutations in NALCN: expanding the genotypic and phenotypic spectra of IHPRF1. Am J Med Genet Part A 176:431–437. 10.1002/ajmg.a.38543 [DOI] [PubMed] [Google Scholar]
- Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K et al. (2017) Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet 25:176–182. 10.1038/ejhg.2016.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkanas E, Schaffer K, Dunham C et al. (2016) Phenotypic evolution of UNC80 loss of function. Am J Med Genet Part A 170:3106–3114. 10.1002/ajmg.a.37929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C et al. (2013) From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinform 43:1–33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivero M, Cho MT, Begtrup A et al. (2017) Additional de novo missense genetic variants in NALCN associated with CLIFAHDD syndrome. Clin Genet 91:929–931. 10.1111/cge.12899 [DOI] [PubMed] [Google Scholar]
- Wang Y, Koh K, Ichinose Y et al. (2016) A de novo mutation in the NALCN gene in an adult patient with cerebellar ataxia associated with intellectual disability and arthrogryposis. Clin Genet 90:556–557. 10.1111/cge.12851 [DOI] [PubMed] [Google Scholar]
- Zaki MS, Bhat G, Sultan T et al. (2016) PYCR2 Mutations cause a lethal syndrome of microcephaly and failure to thrive. Ann Neurol 80:59–70. 10.1002/ana.24678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Kohler S, Mackenroth L et al. (2014) Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci Transl Med 6:252ra123–252ra123. 10.1126/scitranslmed.3009262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


