Abstract
Glial cells exist throughout the nervous system, and play essential roles in various aspects of neural development and function. Distinct types of glia may govern diverse glial functions. To determine the roles of glia requires systematic characterization of glia diversity and development. In the adult Drosophila central brain, we identify five different types of glia based on its location, morphology, marker expression, and development. Perineurial and subperineurial glia reside in two separate single-cell layers on the brain surface, cortex glia form a glial mesh in the brain cortex where neuronal cell bodies reside, while ensheathing and astrocyte-like glia enwrap and infiltrate into neuropils, respectively. Clonal analysis reveals that distinct glial types derive from different precursors, and that most adult perineurial, ensheathing, and astrocyte-like glia are produced after embryogenesis. Notably, perineurial glial cells are made locally on the brain surface without the involvement of gcm (glial cell missing). In contrast, the widespread ensheathing and astrocyte-like glia derive from specific brain regions in a gcm-dependent manner. This study documents glia diversity in the adult fly brain and demonstrates involvement of different developmental programs in the derivation of distinct types of glia. It lays an essential foundation for studying glia development and function in the Drosophila brain.
Keywords: glial cells, glial subtype, cell lineage, adult brain, MARCM, Drosophila
Introduction
Proper development and function of neural circuitry require glial cells. Although glial cells were originally believed to have just supporting roles in nourishing and insulating neurons, recent studies have revealed that glia contribute to virtually all aspects of nervous system development and function [for review, see Lemke (2001), Villegas et al. (2003), and Parker and Auld (2006)]. However, our understanding of glial diversity and origin is far behind their neuronal counterparts.
Drosophila, with its accessibility to genetic, molecular, and behavior analysis, has emerged as a powerful model system for studying brain development and function and its underlying molecular mechanisms. In contrast to vertebrates, in which neurons are outnumbered by glial cells, Drosophila have fewer neurons and exhibits a lower glia–neuron ratio with even fewer glial cells (Ito et al., 1995; Pfrieger and Barres, 1995).
The larval ventral ganglion constitutes a relatively simple model system for studying glial development and function up to individual cells. Based on the position of glial cell bodies and morphology, glial cells in the Drosophila ventral ganglion are classified into three types; surface-, cortex-, and neuropil-associated glial cells (Ito et al., 1995). They are derived from specific neuroglioblasts and glioblasts (Schmidt et al., 1997; Schmid et al., 1999). The glial cell missing (gcm) gene governs the derivation of all embryonic glial cells except for one specific type of glia, midline glia (Hosoya et al., 1995; Jones et al., 1995; Vincent et al., 1996). Loss of gcm causes deficits in glial production while ectopic GCM transform neurons to glia. Thus, gcm is necessary and sufficient for driving glial development during embryogenesis.
Embryonic glia also play essential roles in regulating neuronal viability and morphogenesis. Neuronal loss and connectivity defects are observed following ablation of glial cells (Booth et al., 2000; Hidalgo and Booth, 2000), although survival and functional differentiation of glial cells also require signals from neurons (Hidalgo et al., 2001). In addition, glial cells located at the midline, midline glia, secrete and present molecules to guide the growth cone of axons (Kidd et al., 1999).
Defects in glial function affect various adult behaviors of Drosophila, including circadian rhythm, courtship behavior, and longevity (Ewer et al., 1992; Buchanan and Benzer, 1993; Kretzschmar et al., 1997; Suh and Jackson, 2007; Grosjean et al., 2008). Disruption of glial function in mature brains also induces neural degeneration (Xiong and Montell, 1995; Kretzschmar et al., 1997). In addition, glia play an essential role in the clearance of degenerating axons in both pupal and adult brains (Awasaki and Ito, 2004; Watts et al., 2004; Awasaki et al., 2006; Hoopfer et al., 2006; MacDonald et al., 2006). Further elucidation of these glial functions and their underlying mechanisms requires better characterization of glial cells in the adult brain and their derivation through postembryonic development of the Drosophila brain. In this study, we show five different types of glial cells locate in the adult fly brain with specific patterns and are produced by distinct developmental programs.
Materials and Methods
Fly strains.
To label specific subtypes of glia, we screened in total ∼3500 GAL4 enhancer trap strains made by NP consortium (NP lines) (Hayashi et al., 2002): NP6293 (perineurial glia), NP2276 (subperineurial glia), NP577 and NP2222 (cortex glia), NP3233 and NP1243 (astrocyte-like glia), and NP6520 (ensheathing glia). Note that although these lines label specific subtypes of glia preferentially, they are not pure lines exclusively labeling only specific subtype of glia; every line also labels a subset of neurons. NP1243 labels ensheathing glia and cortex glia weakly and NP6520 also labels cortex glia weakly. Two different repo-GAL4 lines were used as pan-glial lines localized on X and third chromosome, respectively (Sepp et al., 2001; Lai and Lee, 2006). gcm ΔP1 (Jones et al., 1995) and Df(2L)132 (Kammerer and Giangrande, 2001) were used for gcm mutant and deletion chromosome uncovering gcm and gcm2, respectively. To make a repo-GAL80 line, GAL80 from tubP-GAL80 (Lee and Luo, 1999) was subcloned into pCaSpeR4 vector, which carries the repo-promoter (Lai and Lee, 2006), and transgenic flies were generated with this construct. (1) hs-FLP; UAS-FRT-rCD2-FRT-mCD8::GFP was used for FLP-out labeling. (2) hs-FLP; FRTG13, tubP-GAL80, repo-GAL80/CyO, (3) repo-GAL4; G13, UAS-mCD8::GFP/CyO, and (4) repo-GAL4; G13, UAS-nlsGFP/CyO were used for systematic MARCM analysis of glia. (5) gcm-GAL4, FRTG13, UAS-nlsGFP/CyO was used for MARCM analysis of gcm-expressing cells. (6) repo-GAL4; FRT40A, tubP-GAL80/CyO; UAS-nlsGFP/TM3 Sb, (7) hs-FLP; FRT40A, (8) hs-FLP; FRT40A, gcm ΔP1 /CyO, and (9) hs-FLP; FRT40A, Df(2L)132/CyO were used for examining gcm mutant phenotypes in glia. (10) repo-LexA::GAD; FRTG13/CyO; UAS-mCD8::RFP, LexA-operatot-rCD2::GFP/TM3 Sb and (11) hs-FLP; FRTG13, tubP-GAL80, repo-GAL80/CyO; NP6520/TM3 Sb were used for dual-expression control MARCM analysis.
Immunohistochemistry.
Brains were dissected, fixed, and processed as described previously (Lee and Luo, 1999; Ito et al., 2003). Antibodies used in this study include rat anti-Elav (DSHB; Developmental Studies Hybridoma Bank), 1:200; mouse anti-Repo (DSHB), 1:100; rabbit anti-Repo (Halter et al., 1995) (gift from G. M. Technau, University of Mainz, Mainz, Germany), 1:600; mouse monoclonal nc82 (anti-Bruchpilot, DSHB), 1:20; rat anti-mCD8 (Caltag), 1:100; rabbit anti-GFP (Invitrogen), 1:1000; Alexa-488-, Alexa-568-, or Alexa-647-conjugated anti-mouse, -rabbit, and -rat antibody (Invitrogen), 1:1000.
FLP-out and MARCM analysis.
Details of FLP-out, standard MARCM, and dual-expression control MARCM labeling were shown in Wong et al. (2002), Lee and Luo (1999), and Lai and Lee (2006), respectively. To label cells with the FLP-out system, pupae possessing hs-FLP, FLP-out cassette, and appropriate GAL4 were incubated at 32°C for 20 min and labeled cells were examined in adult brain. To perform MARCM analysis, newly hatched larvae were collected for a 4 h interval and cultured at the density of 60–80 larvae per vial at 18°C. Mitotic recombination was induced by heat shock at 38°C for 15 min at certain developmental periods and animals were raised at 18°C after heat shock. One, three, and five days after larval hatching larvae at 18°C were used as mid-first-, mid-second-, and early third-instar larvae, respectively. Pupae of 1 and 3 d after puparium formation at 18°C were used as early pupae and midpupae, respectively.
Microscopic observation.
Confocal serial scanning images were obtained at 1.0 or 1.5 μm intervals using LSM510 laser microscope (Carl Zeiss). To reconstruct the stack of images, serial sections were projected using LSM image Browser V3.1 software (Carl Zeiss). Resulting images were processed with Photoshop CS software (Adobe Systems).
Results
Classification of Repo-positive glial cells in the adult Drosophila central brain
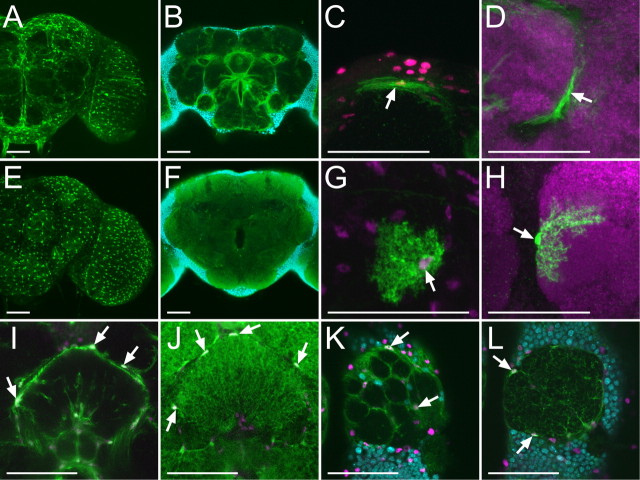
In the Drosophila embryonic CNS, glial cells are classified into three categories based on the position of their cell bodies, cell morphology, and characteristic labeling pattern of the GAL4 enhancer trap lines: surface-, cortex-, and neuropil-associated glia (Ito et al., 1995). Except for midline glial cells, which are finally eliminated by apoptosis during the pupal stage, all other glial cells in the CNS express a homeodomain protein, Repo (Xiong et al., 1994; Halter et al., 1995; Awad and Truman, 1997). To analyze glial cells in the adult central brain, we therefore focused on the Repo-positive glial cells. Nuclei of Repo-positive glial cells were observed on the brain surface, in the cortex, and around and within neuropils (Fig. 1 A,D–F). In addition, when glial membrane were revealed with membrane-tagged GFP (mCD8::GFP) driven by repo-GAL4, surface, cortex, and neuropils of the adult brain were labeled (Fig. 1 B,C). These observations suggest that glial cells in the adult central brain could also be classified into three categories like those of the larval CNS.
Figure 1.
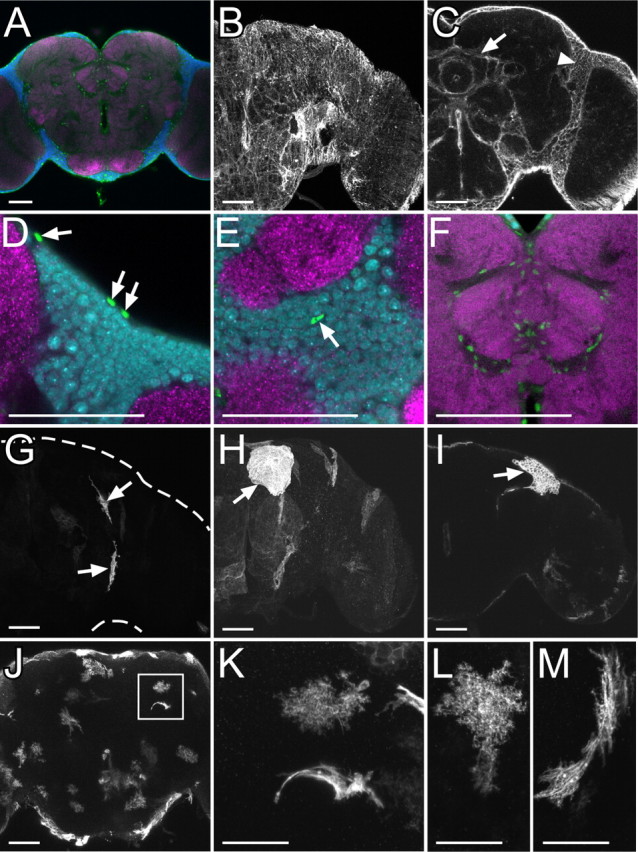
Repo-positive glial cells in the adult central brain. A, Glial nuclei labeled by anti-Repo antibody (green), neuronal nuclei labeled by anti-Elav (blue), and neuropil labeled by nc82 (magenta). B, C, Glial membrane labeled with mCD8::GFP driven by repo-GAL4 on brain surface (B) and inner brain (C). Arrow and arrowhead show glial membrane forming borders among substructures of neuropil and brain cortex, respectively. D–F, Glial nuclei (green) on brain surface (D), cortex (E), and neuropil (F). Glial nuclei (arrows in D and E), neuronal nuclei, and neuropil were labeled as shown (A). G–I, Single cells of small surface glia (G), large surface glia (H), and cortex glia (I) labeled by FLP-out combined with repo-GAL4. J–M, Single cells of neuropil glia labeled by FLP-out combined with repo-GAL4. A dendritic and fibrous lamellar morphology of neuropil glia was labeled around neuropil (J). Two different types of neuropil glia arranged nearby (square in J) are shown at a high magnification (K). L, M, High-magnification images of a dendritic (L) and fibrous lamellar morphology (M) of neuropil glia. Scale bars: A–J, 50 μm; K–M, 25 μm.
To examine their morphologies, we labeled small subsets of glial cells in the adult brain using a UAS-FLP-out system combined with repo-GAL4. On the brain surface, two morphologically different types of glial cells were identified (Fig. 1 G,H). One was a small and slender oblong cell and the other was a large sheet-like cell. These two types of glial cells were present throughout the entire brain surface. In the cortex region, cells with mesh-like morphology were evident (Fig. 1 I). Each mesh-like cell occupies a distinct part of brain cortex. Although cells labeled in the brain cortex possessed a common mesh-like structure, the size of each cell varied. In the neuropil region, two types of glial cells were revealed (Fig. 1 J–M). One type displayed dendritic morphology with tiny spines (Fig. 1 L), while the other exhibited fibrous lamellar morphology (Fig. 1 M). Both types of glia were found in most, if not all, neuropils.
Based on their location and characteristic morphology, these glial cells are classified as surface, cortex, and neuropil glial cells of the adult brain, respectively. Further, we identified two morphologically distinct types of surface and neuropil glia and one type of cortex glia in the adult central brain.
Organization of each type of glial cells in the adult central brain
In the embryonic CNS, enhancer-trap lines that label specific types of glial cells have been identified (Ito et al., 1995; Beckervordersandforth et al., 2008). Several genes selectively expressed in specific classes of embryonic glia are known (Beckervordersandforth et al., 2008). We reasoned that a given type of adult glia should also share a specific gene expression pattern. To target adult glia for better characterization of class-specific organization, development, and function, we searched for GAL4 enhancer-trap lines that label each type of glial cells in the adult fly brain.
We identified two GAL4 lines that preferentially label the two different types of surface glial cells (Fig. 2 A,D). Although both lines express GAL4 in the cells throughout the entire brain surface, the distribution of the cell nuclei is quite different; one labels cell nuclei densely on the brain surface (Fig. 2 A) and the other labels cell nuclei sparsely (Fig. 2 D). When single cells within each group of GAL4-expressing cells were labeled with the UAS-FLP-out system, a small and slender oblong cell and a large sheet-like cell were labeled, respectively (Fig. 2 B,E). Their nuclei are all positive for Repo expression (Fig. 2 C,F, arrows). These results indicate that the small oblong and the large sheet-like glial cells correspond to the two types of surface glia that have been independently revealed through the above single-cell analysis of repo-GAL4-positive adult glia. Notably, these distinct types of surface glia form separate glial layers on the adult brain surface.
Figure 2.
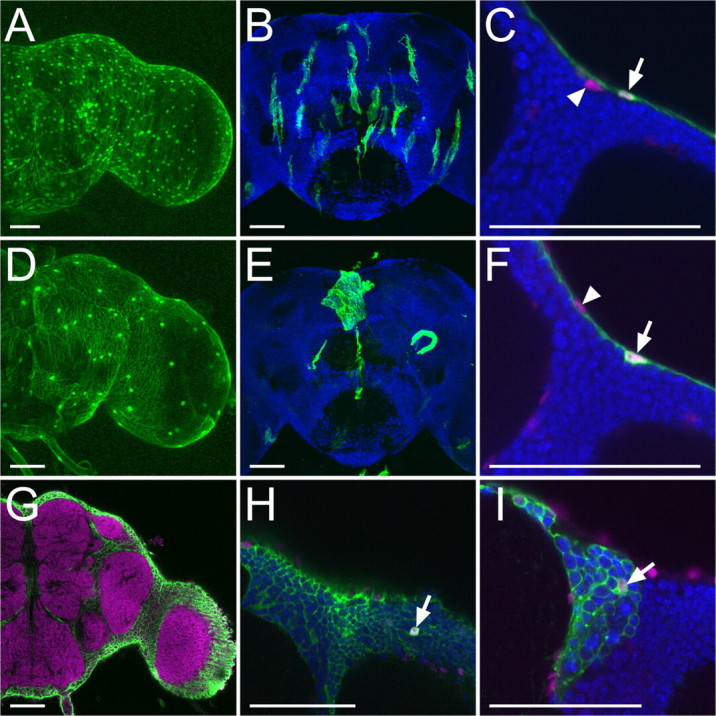
Surface and cortex glia of adult brain. A–F, Perineurial glia (A–C, green) and subperineurial glia (D–F, green) labeled with UAS-GFP (A, C, D, F) and UAS-FRT-mCD8::GFP (FLP-out) (B, E), driven by NP6293 (A–C) and NP2276 (D–F), respectively. Glial nuclei and neuronal nuclei were labeled with anti-Repo (magenta) and anti-Elav (blue), respectively. Note that GAL4-negative, Repo-positive surface nuclei (arrowheads, C and F) were labeled under and over the layer of perineurial glia and subperineurial glia, respectively (arrows in C and F). G–I, Cortex glial cells (green) labeled with UAS-GFP driven by NP2222 (G, H) and with FLP-out combined with NP2222 (I). Neuropils were labeled with nc82 (magenta in G). Glial nuclei (magenta) and neuronal nuclei (blue) were labeled with anti-Repo and anti-Elav, respectively (H, I). Arrows show nuclei of cortex glia. Scale bars, 50 μm.
To determine the spatial relationship of these two separate glial layers, we located GAL4-negative, Repo-positive surface nuclei in brains with only one surface glial layer selectively labeled using these subtype-specific glial GAL4 drivers (Fig. 2 C,F, arrowheads). When small oblong surface glial cells were marked, Repo-positive, GFP-negative surface nuclei consistently reside underneath the layer of GFP-positive small glial cells (Fig. 2 C). Alternatively, the arrangement of GFP-negative, Repo-positive glial nuclei situated above the large GFP-labeled glial layer resembled the arrangement of small oblong glial nuclei (Fig. 2 F). These results indicate that the small oblong glia form the apical glial layer and the large sheet-like glia constitute the basal glial layer on the adult brain surface. Thus, they correspond to the perineurial and subperineurial glial cells of the adult brain, respectively.
We found GAL4 lines that label glia throughout the entire brain cortex (Fig. 2 G). Cells with mesh-like morphology that look indistinguishable from one another were labeled following FLP-out-activation of a UAS-reporter gene in single cells with these GAL4 lines (Fig. 2 I, analogous to Fig. 1 I). Despite recovery of multiple independent cortex glial GAL4 enhancer-trap lines, we failed to identify lines that label cells in only subregions of the brain cortex. Thus, the glial mesh of the brain cortex is probably composed of a single type of glial cells, which elaborate locally and together constitute the entire cortex mesh. When neuronal cells were marked with anti-Elav antibody, neuronal cell bodies were arranged in the holes formed by the mesh-like glial processes (Fig. 2 H,I), suggesting that the cortex glia enwrap individual neuronal cell bodies in the adult brain and one cortex glial cell can enwrap many neuronal cell bodies.
We also identified several GAL4 enhancer-trap lines that selectively mark glial cells associating with neuropils (Fig. 3). Among them, two patterns of labeling are discernible. Whereas cell bodies of both types are consistently located on the interface between neuropil and cortex regions or among neuropil subregions (Fig. 3 A,E), their membranous processes elaborate differentially. One preferentially outlines the neuropils or their subcompartments (Fig. 3 A,B), while the other apparently fills the interior of the neuropils (Fig. 3 F). Both types of glia are found in all neuropil areas, including the fan-shaped body and antennal lobe (Fig. 3 I–L). In addition, the glial type that ensheaths neuropils surrounds neural bundles like the great commissure and antennocerebral tracts (data not shown).
Figure 3.
Two subtypes of neuropil glia of adult central brain. Ensheathing (A–D, I, K) and astrocyte-like (E–H, J, L) glia labeled with NP1243 and NP6520, respectively. Entire and single cells of neuropil glial subtype were labeled with UAS-GFP (A, E, and I–L), UAS-mCD8::GFP (B, F), and UAS-FLP-out (C, D, G, H), respectively. Arrows show nuclei of each subtype of glial cells labeled with anti-Repo (magenta in C, G, and I–L). Neuronal nuclei (blue) were labeled with anti-Elav (B, F, K, L) and neuropil (magenta) was labeled with nc82 (D, H). Scale bars, 50 μm.
When single cells of each type were labeled using the UAS-FLP-out system, we obtained cells with either fibrous lamellar or dendritic morphology (Fig. 3 C,D,G,H). Nuclei of both types of cells were also labeled with anti-Repo antibody (Fig. 3 C,G). These indicate that the fibrous lamellar and dendritic glial cells are identical to the two types of neuropil glia labeled by repo-GAL4 we described earlier (compare Figs. 1 L,M and 3 C,G). Whereas the fibrous type of glia extends processes along the outer surface of neuropil, the dendritic type elaborates inside the neuropil (Fig. 3 D,H). Judging from their morphologies, the fibrous glia possibly provides insulation among neighboring neural compartments and the dendritic ones potentially associate with synapses to modulate neuronal connections similar to vertebrate astrocytes. Thus, we name them ensheathing glia and astrocyte-like glia, respectively.
From our screening of GAL4 enhancer-trap lines, we have identified several lines that preferentially label each type of glial cells in the adult brain. The distribution pattern of each GAL4 line suggests that each type of glial cells has a different function within the adult brain. Since the expression of enhancer trap lines reflects endogenous gene regulation, the identification of these subtype-specific glial enhancer-trap lines suggests that each type of glial cell possesses unique sets of gene expression and molecular features.
Clonal analysis of adult glial cells to determine their developmental origins
Holometabolous insects undergo metamorphosis, and most of their adult tissues, including the brain, are generated after larval hatching. Clonal analysis by MARCM, a positive-labeling genetic mosaic technique, has greatly facilitated the analysis of postembryonic neurogenesis. Briefly, using a subtype-specific GAL4 driver, one can target a specific subset of brain cells for labeling by inducing mitotic recombination in their precursors. Analysis of the patterns of clones induced at different developmental times then allows one to describe the lineage relationship and further determine when a particular precursor is proliferating.
To understand the postembryonic development of adult glia, we targeted Repo-positive adult brain cells for systematic MARCM analysis using repo-GAL4. We induced mitotic recombination with brief heat-shock pulses at various time points through postembryonic development and examined the glial cells labeled in the adult brain. Based on the morphology and position of labeled glial cells, we determined what types of glial cells were labeled. Although two different types of surface glia, perineurial and subperineurial glia, are readily distinguishable by size and morphology, two types of neuropil glia, ensheathing and astrocyte-like glia, are more difficult to distinguish by morphology alone when both are labeled closely and densely. Thus, in this set of experiments, we classified labeled glia into four types: perineurial, subperineurial, cortex, and neuropil glia. We determined their lineage relationship of subtypes of neuropil glia with more sophisticated MARCM techniques (see later section).
Perineurial and neuropil glial cells were frequently labeled in adult brains after brief induction of mitotic recombination during larval and pupal stages (Fig. 4 A–L). In contrast, the subperineurial and cortex glial cells were rarely labeled following induction at various larval or pupal stages (data not shown). These results suggest that the perineurial and neuropil glial cells are extensively generated during postembryonic development, whereas the majority of subperineurial and cortex glial cells are possibly made before larval hatching.
Figure 4.
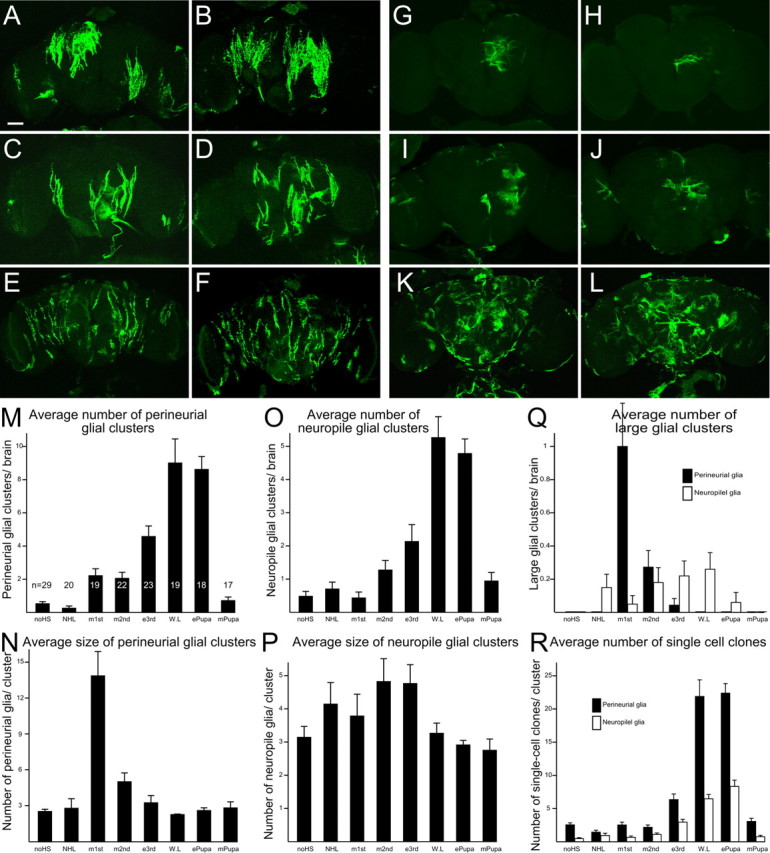
Systematic MARCM analysis of glial proliferation during postembryonic development. A–L, Perineurial (A–F) and neuropil (G–L) glial cells labeled by heat-shock-induced mitotic recombination in mid-first-instar larvae (A, B), newly hatched larvae (G, H), early third-instar larvae (C, D, I, J) and early pupae (E, F, K, L). Scale bar, 50 μm. M–R, Quantification of glial cells that were labeled by MARCM system with heat-shock-induced mitotic recombination at different developmental stages (x-axis). Average number (M, O) and size (N, P) of perineurial glia (M, N) and neuropil glia (O, P) clusters that were induced at different developmental stage (y-axis). Q, Average number of large perineurial glia (>10 cells) and neuropil glia (>8) clusters per brain. R, Average number of single cell clones of perineurial and neuropil glia. Same samples were used for analysis in M–R. Numbers of examined brains are shown in M. Genotypes: hs-FLP/repo-GAL4; FRTG13, UAS-mCD8::GFP/FRTG13, tubP-GAL80, repo-GAL80 (A–L) and hs-FLP/repo-GAL4; FRTG13, UAS-nlsGFP/FRTG13, tubP-GAL80, repo-GAL80 (M–R).
Interestingly, perineurial and neuropil glial cells were labeled separately with characteristic patterns. First, with mild heat shock at the first-instar larval stage, one could readily obtain mosaic brains that exclusively carried clones consisting of only perineurial or neuropil glial cells (Fig. 4 A,H). This suggests distinct subsets of larval precursors in the production of perineurial or neuropil glia (Fig. 4 A,H). Second, labeled glial cells were arranged differentially depending on the induction timing of the mitotic recombination (Fig. 4 A–L). Whereas large clusters of perineurial glial cells were labeled following induction of mitotic recombination in the first-instar larvae, smaller clusters were obtained by analogous induction at later larval stages (Fig. 4, compare A,B and C,D). As for the neuropil glia, locally arranged clusters were also labeled following mild heat shock before the wandering larval stage (Fig. 4 G–J). Labeled cells tended to form cell clusters, suggesting that cells of the same clonal origin do not migrate randomly. Thus, many of the glial clusters are potentially derived from a single progenitor. The clustering phenomenon was no longer evident with mitotic recombination induced at the wandering larval or early pupal stages. In these mosaic brains, although many labeled perineurial and neuropil glia remained, they were widely distributed and mostly present singly or in very small clusters (Fig. 4 E,F,K,L). Finally, labeled cells were rarely obtained when mitotic recombination was induced after midpupal development (data not shown). Given the independent origins for the different types of adult glia, quantitative analyses of the stage-specific clone inductions were conducted separately for the perineurial glia and the neuropil glia.
Postembryonic development of perineurial glial cells
Postmitotic cells might increase exponentially through multiple rounds of symmetric divisions or derive sequentially from self-renewing progenitors via asymmetric divisions. The analysis of cluster size distribution should help distinguish between these two possibilities. For example, one would expect production of single-cell or two-cell clones through the entire process from asymmetric divisions or only near the end of gliogenesis from symmetric division depending on when the terminal mitoses for mature glial cells occurred. However, we found it challenging to count glial cell numbers when they were labeled with a membrane marker, because, unlike neurons, glial cells lack cell-body domains and closely contact with one another.
To better quantify the sizes of glial clusters, we repeated the stage-specific induction of MARCM clones and resorted to nuclear-GFP, driven by repo-GAL4, for highlighting glial nuclei while marking the whole cells. We identified all the clusters and determined the size of each cluster in the adult brains undergoing somatic recombination at specific developmental time points (Fig. 4 M,N,R). We defined a single cluster as a group of cells among which physical contact can extend from one to another, and counted the number of nuclei within one cluster as its cluster size. If cells proliferate exponentially, the cluster size should decrease exponentially as clones are induced at later time points while the total cluster number should increase drastically as the pool of dividing glia expanded with time. On the other hand, if cells proliferate by asymmetric division of self-renewing progenitors, the cluster size should decrease gradually as clones are induced at later time points while the total cluster number should not change because the number of self-renewing progenitors remains constant. Consistent with the predictions made based on an exponential model of cell proliferation, we obtained a few large clusters of glia following clone induction in early larval stages versus many small clusters of glia in the brains that received heat shock at late larval stages (Fig. 4 M,N). Furthermore, we rarely obtained single-cell clones of glia except when mitotic recombination was induced in wandering larvae or early pupae before the end of gliogenesis around the midpupal stage when the frequency of clones reduced to a background level (Fig. 4 R). These results suggest that perineurial glial cells are made through exponential cell proliferation, and that most perineurial glial precursors enter the final round of proliferation around the wandering larval or early pupal stage.
Given that most, if not all, of the adult perineurial glial cells are born after larval hatching and that this postembryonic process of gliogenesis continues until the midpupal stage, perineurial glia probably only exist as precursors in the developing Drosophila nervous system. To locate such perineurial glial precursors, we examined the lineages of proliferating perineurial glia in wandering larvae. Since mitotic recombination at the mid-first-instar larval stage could label large perineurial glial clusters in the adult brain (Fig. 4 N), we induced mitotic recombination at this stage and examined the labeled cells in the wandering larvae. Cell clusters covering the surface of the larval brain were observed by this labeling (Fig. 5 A,B). Whereas the perineurial glia have slender and oblong morphology extending along the dorsoventral axis in the adult brain (Figs. 1 G, 2 B, 5 C), these larval cells extend filopodial processes radially (Fig. 5 B, arrows), indicating that they are the morphologically immature perineurial glia of the adult. These results suggest that perineurial glia proliferate and mature on the brain surface locally and expand as the brain surface enlarges during development.
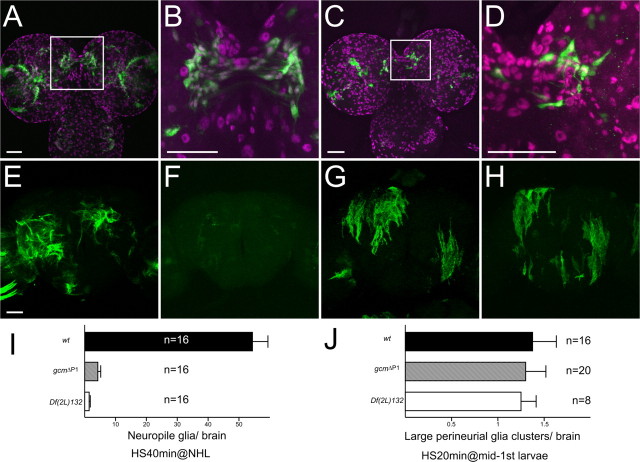
Figure 5.
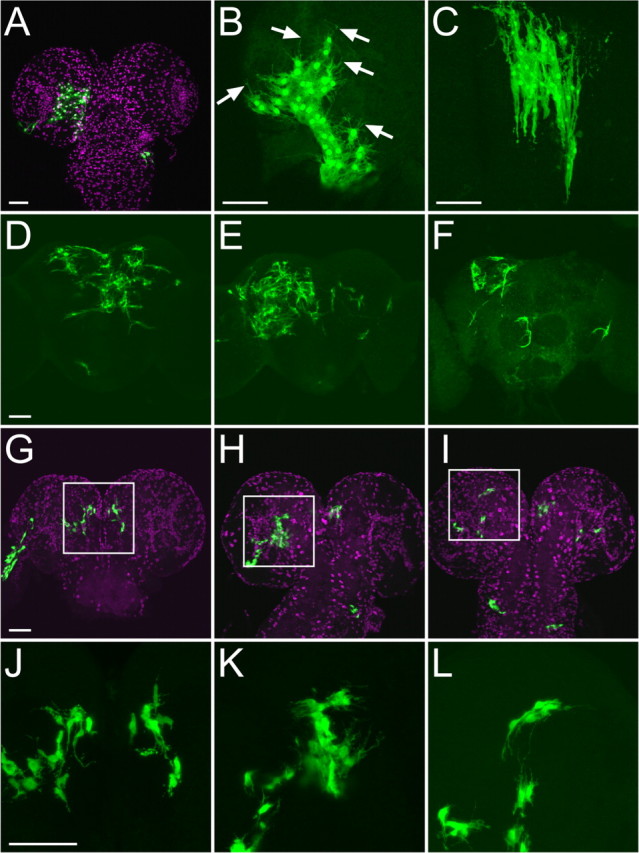
Precursors of perineurial and neuropil glia. Perineurial glia cluster in a wandering larva (A, B) and adult (C), which were labeled by MARCM system with heat-shock-induced mitotic recombination in mid-first-instar larvae. Arrows show filopodial processes extending from larval perineurial glia. Multiple neuropil glia clusters in adults (D–F) and wandering larvae (G–L), which were labeled by MARCM with long heat-shock (40 min) induced mitotic recombination in NHL. Note that labeled cells were localized in posterior-medial (G, J), lateral (H, K), and dorsal (I, L) interface between brain cortex and neuropil. J–L, High-magnification images of G–I, respectively. Glial nuclei were labeled with anti-Repo (magenta, A and G–I). Scale bars, 50 μm.
Postembryonic development of neuropil glial cells
We also analyzed postembryonic proliferation of neuropil glia with MARCM (Fig. 4 O–R). Although more neuropil glial clusters were obtained with mitotic recombination induced at later developmental stages (Fig. 4 O), there was no significant change in the average size of clusters generated at different times (Fig. 4 P). Large clusters (>8 cells) could be induced at comparable frequencies through larval development except mid-first larval stage (Fig. 4 Q). In contrast, the frequency in obtaining clusters with >10 perineurial glial cells reduced drastically after early larval stages (Fig. 4 Q). In addition, the average cell number per cluster of neuropil glia was smaller than that of the perineurial glial cells (Fig. 4, compare N, P). These results suggest that the proliferation pattern of neuropil glia is different from that of perineurial glia. However, as in perineurial glia, we rarely obtained single-cell clones of neuropil glia until late larval or early pupal stages (Fig. 4 R). This result argues against a model where neuropil glia are generated by asymmetric cell division. Instead, neuropil glia are also derived through multiple rounds of symmetric cell division, but each progenitor undergoes fewer divisions than that of perineurial glia, and the timing of proliferation appears irregular, although the final mitoses to derive postmitotic neuropil glia remain rather synchronized around puparium formation.
We further examined precursors of neuropil glia in the wandering larvae. Following a 40 min heat shock in NHL (newly hatched larvae), we consistently obtained clusters of neuropil glia in every mosaic brain (Fig. 5 D–F). When such mosaic brains were examined at the wandering larval stage, the labeled Repo-positive cells on the interface between neuropil and cortex were consistently clustered in at least three specific regions, the posteromedial, lateral, and dorsal interface (Fig. 5 G–L). In addition, whereas mature neuropil glia acquire a fibrous lamellar or dendritic morphology, these cells are round-shaped with filopodial processes (Fig. 5 J–L). Mature neuropil glia should reside not only in the neuropil–cortex interface but also among the neuropils that lie apart from the brain cortex (Figs. 1 F, 3 B,F). Furthermore, the analogously induced neuropil glial clones are more broadly distributed when examined at the adult stage (Fig. 5, compare D–F and G–I). These results collectively suggest that precursors of neuropil glia proliferate in the specific regions of the larval brain and migrate to maturate upon metamorphosis.
The function of gcm in postembryonic gliogenesis
During embryogenesis, glial cell missing (gcm) plays an essential role in glial development. Is gcm also essential for postembryonic gliogenesis of perineurial and neuropil glia? To address this question, we first examined gcm expression in the developing brain with gcm-GAL4. Three clusters of gcm-positive cells were found in the interface between neuropil and cortex of the larval brain. Their morphology and location are similar to the clusters of neuropil glia precursors (compare Figs. 5 G–L and 6 A,B). gcm expression could be detected beginning in early third instar, and it becomes prominent in the wandering larval stage (data not shown). The cells expressing gcm spread along the cortex–neuropil interface during early pupal stage and the expression disappears after the midpupal stage (data not shown). In addition, cells labeled with gcm-GAL4 in the interface are also recognized with anti-Repo antibody (Fig. 6 B,D). Furthermore, analogous patterns of neuropil glial clones were obtained in the mosaic larval brain when MARCM experiments were performed with either repo-GAL4 or gcm-GAL4 (Fig. 6 C,D, compared with Fig. 5 G,J). These results suggest that precursors of neuropil glia express gcm transiently. In contrast, we could not detect any cells labeled with gcm-GAL4 on the brain surface during larval and pupal stage. In addition to the transient expression in the neuropil glial precursors, gcm-GAL4 persistently labels two clusters of neurons bilaterally in the central brain (data not shown).
Figure 6.
Function of gcm on postembryonic development of perineurial and neuropil glia. A, B, Cells labeled with GFP driven by gcm-GAL4 in a wandering larva. C, D, Cells labeled by MARCM system with gcm-GAL4. Mitotic recombination was induced in NHL. Glial nuclei were labeled with anti-Repo antibody (magenta, A–D). High-magnification images of A and C are shown in B and D, respectively. Note that gcm-positive cells were labeled with anti-Repo antibody (B, D). E–H, Wild-type (E, G) and gcm ΔP1 (F, H) MARCM clones of neuropil glia (E, F) induced at NHL and perineurial glia (G, H) induced at mid-first-instar larvae, respectively. I, J, Quantification of neuropil glia (I) and perineurial glia (J) that were labeled by MARCM system. Average number of neuropil glia per brain (I) and average number of large perineurial glia cluster (>10) per brain were examined in wt, gcm ΔP1, and Df(2L)132 clones. Scale bars, 50 μm. Genotypes: gcm-GAL4/UAS-GFP (A, B); hs-FLP/+; gcm-GAL4, FRTG13, UAS-nlsGFP/FRTG13, tubP-GAL80, repo-GAL80 (C, D); hs-FLP, UAS-mCD8::GFP/repo-GAL4; FRT40A, tubP-GAL80/FRT40A or FRT40A, gcm ΔP1 (E–H); and hs-FLP/repo-GAL4; FRT40A, tubP-GAL80/FRT40A or FRT40A, gcm ΔP1 or FRT40A, Df(2L)132; UAS-nlsGFP/+ (I, J).
Next we determined the effect of gcm mutations on postembryonic development of adult glial cells using the MARCM system with repo-GAL4. Somatic recombination was induced with 40 min heat shock in NHL and with a 20 min heat shock in mid-first-instar larvae to generate clones of neuropil and perineurial glia, respectively. We obtained results that are consistent with the notion, as implied from the expression pattern, that gcm is required for proper development of neuropil glia but appears fully dispensable in perineurial glia. First, gcm ΔP1 mutant neuropil glia were barely detectable even after a 40 min heat shock that reproducibly yielded ∼50 homozygous wild-type neuropil glial cells per control mosaic brain (Fig. 6 E,F,I). In contrast, there was no significant difference in the cluster number or size of perineurial glia between control and gcm ΔP1 clones (Fig. 6 G,H,J). Second, since gcm and gcm2 possess partially redundant function in the glial and neuronal development of the optic lobe (Chotard et al., 2005), it is possible that gcm2 might compensate for the loss of gcm in the support of perineurial glial development. To examine this possibility, we determined how deletion of both gcm and gcm2 in the Df(2L)132 deficiency (Kammerer and Giangrande, 2001) might affect the clonal development of specific adult glia. Again, normal-looking mutant perineurial glial clones could be readily obtained while mutant neuropil glia were largely absent (Fig. 6 I,J). The effects of gcm mutations on the adult glia, together with expression pattern of gcm, suggest that gcm is essential for the postembryonic development of the neuropil glia but not for the perineurial glia.
The cell lineage of two different neuropil glia
Perineurial and neuropil glia are apparently derived from different precursors, as evidenced by multiple independent lines of evidence, including their differential dependence on gcm. How, then, do the two subtypes of neuropil glia differentiate? We found it challenging to determine subtype identity of neuropil glia among cells that constitute a large cluster of neuropil glia. Although some large clusters of neuropil glia appear homogeneous consisting of either ensheathing glia or astrocyte-like glia (Fig. 7 A–D), it remained unclear whether the ensheathing and astrocyte-like glia consistently arise from separate lineages.
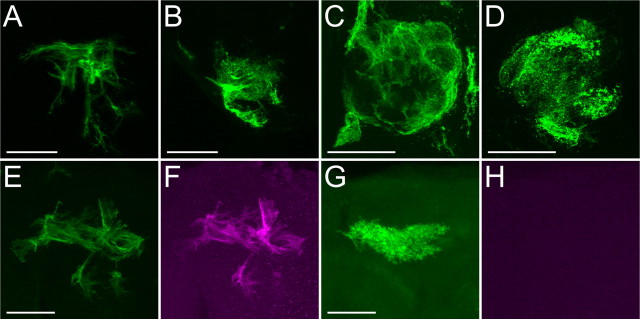
Figure 7.
Neuropil glial clones. Ensheathing glial clones (A, C, E, F) and astrocyte-like glial clones (B, D, G) labeled by standard MARCM (A–D) and dual-expression control MARCM (E–H). Note that C and D show glial clones located in the antennal lobe. Whereas the ensheathing glial clone was labeled by both rCD2::GFP driven by repo-LexA::GAD (green, E) and mCD8::RFP driven by NP6520 (magenta, F), the astrocyte-like glial cone was labeled by only rCD2::GFP driven by repo-LexA::GAD (green, G), but not by mCD8::RFP driven by NP6520 (magenta, H). Scale bars, 50 μm. Genotypes: hs-FLP/repo-GAL4; FRTG13, UAS-mCD8::GFP/FRTG13, tubP-GAL80, repo-GAL80 (A–D) and hs-FLP/repo-LexA::GAD; FRTG13/FRTG13, tubP-GAL80, repo-GAL80; NP6520/UAS-mCD8::RFP, LexA-operator-rCD2::RFP (E–H).
To address this question, we repeated clonal analysis with dual-expression-control MARCM that permits differential labeling of distinct cells in a given MARCM clone owing to independent controls over distinct reporter genes by GAL4 versus LexA::GAD, two GAL80-suppressible binary transcriptional activators (Lai and Lee, 2006). We used repo-LexA::GAD plus the ensheathing glial GAL4 driver, NP6520, in dual-expression-control MARCM, to label GAL80-minus ensheathing glia with both lexAop-rCD2::GFP and UAS-mCD8::RFP while marking the astrocyte-like glia of the same genotype with lexAop-rCD2::GFP only. If a neuropil glial cluster consisted of only ensheathing or astrocyte-like glia, one would expect expression of mCD8::RFP in all or none of the cells in the cluster. In contrast, if ensheathing and astrocyte-like glia derived from common progenitors, one would detect mCD8::RFP in a subset of rCD2::GFP-marked cells in a single cluster. Fifteen clusters of neuropil glia were obtained following induction of somatic recombination at the newly hatched larval stage. Among them, 12 clusters were found to purely consist of either ensheathing glia or astrocyte-like glia, as evidenced by the expression of mCD8::RFP in all or none of the cells in a given cluster (Fig. 7 E–H). In the other three clusters that contained both mCD8::RFP-positive and -negative cells, no matter whether ensheathing or astrocyte-like glia predominated, the minority was overwhelmed and composed of only one or two cells per cluster despite the cluster size being roughly >8 cells. Since a low level of mitotic recombination could occur even without any heat-shock induction (Fig. 4 R), it is possible that the presence of one or two heterotypic cells in the otherwise homogeneous population may occur as a consequence of multiple events of mitotic recombination. Given that ensheathing and astrocyte-like glia rarely coexist in a single cluster, the two subtypes of neuropil glia differ not only in morphology and marker expression but also with different origins of development.
Discussion
Our analysis of Repo-expressing cells reveals the presence of three classes and five types of glia in the adult Drosophila central brain. Each glial type exhibits a characteristic location and morphology. Multiple glial functions can be inferred from their different cellular organizations in the brain. The different functions of specific glial types are further supported by their differential marker expression. In addition, different glial types derive independently from distinct populations of precursors. They proliferate in different temporal and spatial patterns, and the involvement of gcm varies from one to another glial type. Identification of glial diversity lays an essential foundation for unraveling the diverse glial functions in the brain, and determining how diverse glial types are derived is integral to the understanding of how a mature brain is made.
Two layers of distinct glial cells surround the entire adult Drosophila brain. The inner one, possibly composed of a fixed number of large sheet-like subperineurial glial cells since larval hatching, constitutes the fly blood–brain barrier in both larval and adult brains (Bainton et al., 2005; Schwabe et al., 2005; Stork et al., 2008). In contrast, the outer layer consists of numerous small oblong perineurial glial cells that proliferate through postembryonic development of the brain and may remain immature until formation of the adult brain. Given its more external location and apparent adherence to the subperineurial layer, the perineurial glia possibly helps make up the adult blood–brain barrier. However, these two types of surface glia likely play nonoverlapping, complementary roles in the regulation of the permeability to the CNS, as implied from their different enhancer trap expression patterns.
The other three types of glia form independent networks inside the brain. Cortex glia enwrap individual neuronal cell bodies in the brain cortex. Each cortex glial cell send mesh-like processes to surround multiple neuronal cell bodies locally. Cortex glia may provide metabolic support for neurons and have the potential for modulating neuronal cell body functions in a spatially controlled manner. Further inside the brain lie two types of neuropil glia. The ensheathing glia outlines individual neuropils and their subcompartments, while the astrocyte-like glia extend processes into the neuropils where synapses form.
The glial boundary established by the ensheathing glia may provide the necessary insulation to prevent lateral communication between adjacent neuropils, and provide septa when a neuropil is composed of multiple independent subcompartments. In contrast, astrocyte-like glial cells potentially associate with synapses. Several electron microscopic studies have shown that glial processes are located around synapses of insect neuropils (Carlson and Saint Marie, 1990). In an analogous manner, mammalian astrocytes target processes to synapses. They express excitatory amino-acid transporters (EAAT-1 and -2), and can recover excess extracellular glutamate or aspartate to keep the local environment suitable for the next synaptic transmission (Kanai, 1997; Takahashi et al., 1997). In addition, mammalian astrocytes may release gliotransmitters in response to synaptic activity to modulate synaptic functions (Halassa et al., 2007). Whereas in the vertebrate nervous system synapses are formed not only on the dendrites and axon terminals but also on the surface of the neural cell bodies, in the invertebrate brain no synapses are formed on the neuronal cell bodies (Ito and Awasaki, 2008). Accordingly, the cortex glia and neuropil glia are associated exclusively to the neural cell bodies and synaptic neuropils, respectively. Although Drosophila glia also expresses EAAT-1 (dEAAT-1) that is concentrated in neuropils (Rival et al., 2004), it remains to be determined which function of the vertebrate astrocytes is achieved by which type of insect glial cells.
In the adult brain or ventral nerve cord of other insects, two types of glial cells associated with neuropil have been identified; type II and type III glia or interface and neuropilar glia in Musca (Sohal et al., 1972; Strausfeld, 1976), interface and neuropil glia in Acheta (Meyer et al., 1987) and neuropil cover type and neuropil glia or simple and complex neuropil glia in Manduca (Cantera, 1993; Oland et al., 1999). One type is localized cortex/neuropil interface or borders among neuropils and ensheaths neuropil, and another type extends glial processes inside the neuropils, like the ensheathing and astrocyte-like glia of adult Drosophila brain, respectively. It is, therefore, these two subtypes of neuropil glia would commonly exist in insects.
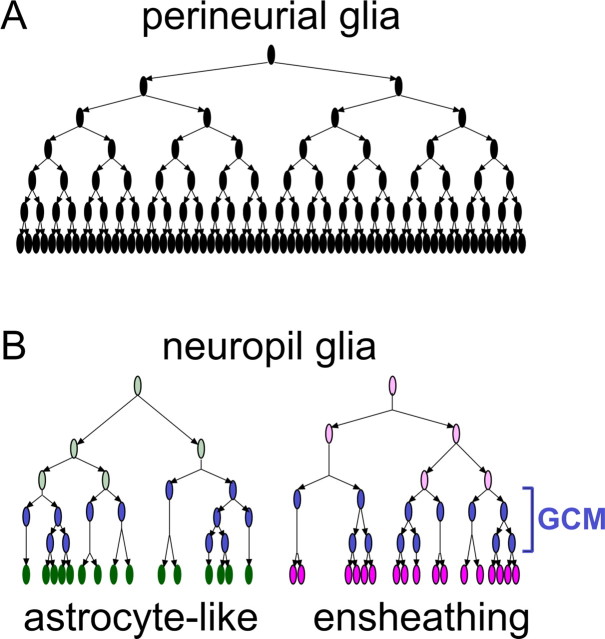
These distinct types of adult fly brain glia arise independently from each other. The proliferation of subperineurial glia is probably restricted to embryogenesis, and any postembryonic increase in the cortex glial cell number should be much less significant than the expansion of the other three types of adult glia, which primarily occurs during larval development. Furthermore, distinct populations of precursors give rise to perineurial, ensheathing, or astrocyte-like glia (Fig. 8). Notably, perineurial glia precursors distribute around the brain surface and make clones that expand locally as the brain grows in size through development. In contrast, ensheathing and astrocyte-like glial cells are derived from specific proliferation centers and do not migrate throughout the brain until metamorphosis. Most somatic clones of neuropil glia exclusively consist of ensheathing or astrocyte-like glial cells. However, the identity of the neuropil clones could not be unambiguously determined until late pupal stages, making it unclear whether the progenitors of the ensheathing and astrocyte-like glia are mixed in those proliferation centers. Perineurial glial precursors make clones of similar sizes at a given developmental stage, suggesting that they retain the same proliferation potential through development. In contrast, the clones of neuropil glia derived at the same stage vary in size, raising the possibility that neuropil glial precursors undergo stochastic proliferation, possibly, in response to some limited local cue(s). Supply of Vein, a neuregulin-like trophic factor, from neurons has been shown to promote local proliferation of the embryonic longitudinal glia (Griffiths and Hidalgo, 2004). It remains to be determined whether similar mechanisms govern the postembryonic expansion of the adult neuropil glia. Finally, all the postembryonic glial precursors apparently divide symmetrically to produce two daughters following each division; intriguingly, the final round of mitosis consistently takes place around pupal formation. This suggests a stage-specific signal may trigger the final glia-producing mitoses.
Figure 8.
Schematic model of postembryonic glial proliferation. A, B, Perineurial glia (A) and two subtypes of neuropil glia (B).
The involvement of different developmental programs in the derivation of different types of adult glia is further evidenced in the differential temporal requirement for gcm. GCM promotes all embryonic gliogenesis except for midline glia (Hosoya et al., 1995; Jones et al., 1995; Vincent et al., 1996). GCM suppresses neural differentiation through expression of Tramtrack and confers glial identity via induction of Repo and other glial genes (Jones, 2001). Transient expression of gcm also occurs in the postembryonic development of both medulla and thoracic glial lineages, and inhibition of GCM function by expression of a dominant-negative form of GCM suppresses differentiation of their glial development (Colonques et al., 2007; Soustelle and Giangrande, 2007). In addition, we show that precursors of neuropil glia express gcm transiently and the postembryonic induction of the gcm mutant clone effectively arrest the development of neuropil glia. These observations indicate that GCM plays analogous roles in promoting glial fate at different developmental stages. It is possible that the precursors of neuropil glia do not acquire full glial identity until gcm is expressed at an intermediate stage of development. In contrast, gcm is dispensable for postembryonic development of perineurial glia. Unlike midline glia in which Repo is never expressed, perineurial glial cells are strongly positive for Repo, a direct target for GCM, making it likely that GCM is involved in the development of perineurial glia. It appears more plausible that like neuropil glia transient expression of gcm also occurs in the perineurial glial precursors but at an earlier stage, and that they have already acquired glial cell fate before larval hatching.
While most glial types appear to be conserved in flies, the microglia type of glia is not obvious in the adult fly brain. It is possible that we failed to identify all major types of glia because of the restriction of our analysis to only Repo-positive cells. Nevertheless, evidence is accumulating that neuropil glia may share microglia-type functions to engulf degenerating axons in the process of programmed axon pruning or following injury of neurites (Awasaki and Ito, 2004) (M. R. Freeman, personal communication). Additional glial diversity has been suggested in vertebrates. Many subtypes of glial cells in the Drosophila embryonic ventral ganglion has also been revealed through detailed analysis of lineage as well as gene expression (Schmidt et al., 1997; Schmid et al., 1999; Beckervordersandforth et al., 2008). The identification of five major types of adult fly brain glia and the analysis of their developmental origins pave a way for further use of the Drosophila as a model system for study of glial diversity, development, and function.
Footnotes
This work was supported by Naito Foundation to T.A., a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to K.I., and the United States National Institutes of Health to T.L. We are grateful to M. R. Freeman for communicating results before publication; M. R. Freeman and Bloomington Stock Center for fly strains; G. G. Technau and Developmental Studies Hybridoma Bank for antibodies; H.-H. Yu for sharing UAS-mCD8::RFP strains before publication; the members of Ito's laboratory for screening of fly strains; members of Lee's laboratory for fruitful discussion; and B. Leung for critical reading of this manuscript.
References
- Awad TA, Truman JW. Postembryonic development of the midline glia in the CNS of Drosophila: proliferation, programmed cell death, and endocrine regulation. Dev Biol. 1997;187:283–297. doi: 10.1006/dbio.1997.8587. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14:668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LT, Schwabe T, DeSalvo M, Gaul U, Heberlein U. moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila . Cell. 2005;123:145–156. doi: 10.1016/j.cell.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth RM, Rickert C, Altenhein B, Technau GM. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mech Dev. 2008;125:542–557. doi: 10.1016/j.mod.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Booth GE, Kinrade EF, Hidalgo A. Glia maintain follower neuron survival during Drosophila CNS development. Development. 2000;127:237–244. doi: 10.1242/dev.127.2.237. [DOI] [PubMed] [Google Scholar]
- Buchanan RL, Benzer S. Defective glia in the Drosophila brain degeneration mutant drop-dead. Neuron. 1993;10:839–850. doi: 10.1016/0896-6273(93)90200-b. [DOI] [PubMed] [Google Scholar]
- Cantera R. Glial cells in adult and developing prothoracic ganglion of the hawk moth Manduca sexta . Cell Tissue Res. 1993;272:93–108. [Google Scholar]
- Carlson SD, Saint Marie RL. Structure and function of insect glia. Annu Rev Entomol. 1990;35:597–621. [Google Scholar]
- Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila . Neuron. 2005;48:237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Colonques J, Ceron J, Tejedor FJ. Segregation of postembryonic neuronal and glial lineages inferred from a mosaic analysis of the Drosophila larval brain. Mech Dev. 2007;124:327–340. doi: 10.1016/j.mod.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells' influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RL, Hidalgo A. Prospero maintains the mitotic potential of glial precursors enabling them to respond to neurons. EMBO J. 2004;23:2440–2450. doi: 10.1038/sj.emboj.7600258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean Y, Grillet M, Augustin H, Ferveur JF, Featherstone DE. A glial amino-acid transporter controls synapse strength and courtship in Drosophila . Nat Neurosci. 2008;11:54–61. doi: 10.1038/nn2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Fellin T, Haydon PG. The tripartite synapse: roles for gliotransmission in health and disease. Trends Mol Med. 2007;13:54–63. doi: 10.1016/j.molmed.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, Technau GM. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster . Development. 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, Ueda R, Uemura T, Yoshihara M, Goto S. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Booth GE. Glia dictate pioneer axon trajectories in the Drosophila embryonic CNS. Development. 2000;127:393–402. doi: 10.1242/dev.127.2.393. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Kinrade EF, Georgiou M. The Drosophila neuregulin vein maintains glial survival during axon guidance in the CNS. Dev Cell. 2001;1:679–690. doi: 10.1016/s1534-5807(01)00074-0. [DOI] [PubMed] [Google Scholar]
- Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O'Leary DD, Luo L. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y. glial cells missing: a binary switch between neuronal and glial determination in Drosophila . Cell. 1995;82:1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- Ito K, Awasaki T. Clonal unit architecture of the adult fly brain. In: Technau GM, editor. Brain development in Drosophila melanogaster. Austin: Landes Bioscience; 2008. pp. 137–158. [DOI] [PubMed] [Google Scholar]
- Ito K, Urban J, Technau G. Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Dev Genes Evol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- Ito K, Okada R, Tanaka NK, Awasaki T. Cautionary observations on preparing and interpreting brain images using molecular biology-based staining techniques. Microsc Res Tech. 2003;62:170–186. doi: 10.1002/jemt.10369. [DOI] [PubMed] [Google Scholar]
- Jones BW. Glial cell development in the Drosophila embryo. Bioessays. 2001;23:877–887. doi: 10.1002/bies.1129. [DOI] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- Kammerer M, Giangrande A. Glide2, a second glial promoting factor in Drosophila melanogaster . EMBO J. 2001;20:4664–4673. doi: 10.1093/emboj/20.17.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y. Family of neutral and acidic amino acid transporters: molecular biology, physiology and medical implications. Curr Opin Cell Biol. 1997;9:565–572. doi: 10.1016/s0955-0674(97)80035-x. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila . Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Kretzschmar D, Hasan G, Sharma S, Heisenberg M, Benzer S. The Swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila . J Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila . Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lemke G. Glial control of neuronal development. Annu Rev Neurosci. 2001;24:87–105. doi: 10.1146/annurev.neuro.24.1.87. [DOI] [PubMed] [Google Scholar]
- MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Meyer MR, Reddy GR, Edwards JS. Immunological probes reveal spatial and developmental diversity in insect neuroglia. J Neurosci. 1987;7:512–521. doi: 10.1523/JNEUROSCI.07-02-00512.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oland LA, Marrero HG, Burger I. Glial cells in the developing and adult olfactory lobe of the moth Manduca sexta . Cell Tissue Res. 1999;297:527–545. doi: 10.1007/s004410051379. [DOI] [PubMed] [Google Scholar]
- Parker RJ, Auld VJ. Roles of glia in the Drosophila nervous system. Semin Cell Dev Biol. 2006;17:66–77. doi: 10.1016/j.semcdb.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Pfrieger FW, Barres BA. What the fly's glia tell the fly's brain. Cell. 1995;83:671–674. doi: 10.1016/0092-8674(95)90178-7. [DOI] [PubMed] [Google Scholar]
- Rival T, Soustelle L, Strambi C, Besson MT, Iché M, Birman S. Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr Biol. 2004;14:599–605. doi: 10.1016/j.cub.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in Drosophila . Cell. 2005;123:133–144. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Schulte J, Auld VJ. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev Biol. 2001;238:47–63. doi: 10.1006/dbio.2001.0411. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Sharma SP, Couch EF. Fine structure of the neural sheath, glia and neurons in the brain of the housefly, Musca domestica . Z Zellforsch Mikrosk Anat. 1972;135:449–459. doi: 10.1007/BF00583429. [DOI] [PubMed] [Google Scholar]
- Soustelle L, Giangrande A. Novel gcm-dependent lineages in the postembryonic nervous system of Drosophila melanogaster . Dev Dyn. 2007;236:2101–2108. doi: 10.1002/dvdy.21232. [DOI] [PubMed] [Google Scholar]
- Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klämbt C. Organization and function of the blood–brain barrier in Drosophila . J Neurosci. 2008;28:587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld NJ. Insect brain. New York: Springer; 1976. [Google Scholar]
- Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Billups B, Rossi D, Sarantis M, Hamann M, Attwell D. The role of glutamate transporters in glutamate homeostasis in the brain. J Exp Biol. 1997;200:401–409. doi: 10.1242/jeb.200.2.401. [DOI] [PubMed] [Google Scholar]
- Villegas SN, Poletta FA, Carri NG. GLIA: A reassessment based on novel data on the developing and mature central nervous system. Cell Biol Int. 2003;27:599–609. doi: 10.1016/s1065-6995(03)00119-7. [DOI] [PubMed] [Google Scholar]
- Vincent S, Vonesch JL, Giangrande A. Glide directs glial fate commitment and cell fate switch between neurones and glia. Development. 1996;122:131–139. doi: 10.1242/dev.122.1.131. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004;14:678–684. doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Montell C. Defective glia induce neuronal apoptosis in the repo visual system of Drosophila . Neuron. 1995;14:581–590. doi: 10.1016/0896-6273(95)90314-3. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]