Abstract
Age-related hearing loss (ARHL) is the most prevalent sensory deficit in the elderly. This progressive hearing impairment leads to social isolation and is also associated with comorbidities, such as frailty, falls, and late-onset depression. Moreover, there is a growing evidence linking it with cognitive decline and increased risk of dementia. Given the large social and welfare burden that results from ARHL, and because ARHL is potentially a modifiable risk factor for dementia, there is an urgent need for therapeutic interventions to ameliorate age-related auditory decline. However, a prerequisite for design of therapies is knowledge of the underlying molecular mechanisms. Currently, our understanding of ARHL is very limited. Here, we review recent findings from research into ARHL from both human and animal studies and discuss future prospects for advances in our understanding of genetic susceptibility, pathology, and potential therapeutic approaches in ARHL.
Age-related hearing loss (ARHL), also known as presbycusis, is a complex disorder that results from the cumulative effects of aging on the auditory system. It is defined as a progressive, bilateral, symmetrical age-related sensorineural hearing loss, which is most pronounced at the higher frequencies. ARHL is the most prevalent chronic sensory deficit experienced by older adults, with approximately half of adults in their seventh decade showing hearing loss that is severe enough to affect communication (Agrawal et al. 2008), and is the third-most-common health condition affecting older adults after heart disease and arthritis (Collins 1997). Although not life-threatening, this condition is associated with significant psychological and medical morbidity, including social isolation, frailty, depression, and cognitive decline (Lin et al. 2011a, 2013; Kamil et al. 2016; Rutherford et al. 2018). In addition to the health burden, there is also a significant economic cost associated with ARHL. Stucky et al. (2010) estimated that in the year 2002 alone the combined direct medical and lost productivity costs attributable to hearing loss in adults aged ≥65 years in the United States was ∼$9.5 billion. Taking into account the expected increases in life expectancy, it is estimated that the annual cost will increase to ∼$60 billion by 2030 (Stucky et al. 2010). As such, the health, societal, and economic costs of ARHL are vast and ever-increasing. However, compared with congenital and early-onset hearing loss, our understanding of the biochemical processes and molecular biology underlying this condition are limited. Historically, this may reflect an underappreciation of the condition owing to the now outmoded assumption that ARHL is an unavoidable effect of aging, but it is also because of the practical considerations and difficulties associated with studying this chronic condition in older human patients.
ARHL PATHOLOGY AND EPIDEMIOLOGY
Historical studies of postmortem human temporal bones suggest that ARHL involves a number of auditory structures: degeneration of the mechanotransducing cochlear inner and outer hair cells (sensory presbycusis); reduced function within the stria vascularis (SV) (strial presbycusis, also known as metabolic presbycusis); and degeneration of the auditory nerve (neural presbycusis) (Schuknecht 1955; Schuknecht and Gacek 1993; Ohlemiller 2004). However, in reality, it is likely most people with presbycusis will show a “mixed” pathology. In addition to peripheral lesions it is also likely there will be changes occurring in central auditory pathways that contribute to the development and progression of ARHL. In this article, we will predominantly focus on what is known about the effects of aging in the peripheral auditory system. Although degenerative changes within the cochlea arise as a consequence of cellular aging, they also reflect the cumulative effects of additional extrinsic factors throughout the life of the individual. As such, ARHL is considered a multifactorial disorder with underlying risk factors that can be divided into several categories: biological age, gender, ethnicity, environment (e.g., noise exposure, ototoxic medications), lifestyle (e.g., smoking, drinking, diet), health comorbidities (e.g., hypertension, diabetes), and genetic predisposition (Yamasoba et al. 2013). Information pertaining to the epidemiology of ARHL has mostly arisen from large cohort studies, in which audiometric testing has been undertaken as part of the evaluation (Brant and Fozard 1990; Gates et al. 1990; Cruickshanks et al. 2003; Agrawal et al. 2008; Gopinath et al. 2009). However, although it is often difficult to directly compare data across these studies, owing to differences in cohort demographics and definitions of hearing loss, it is generally accepted that prevalence doubles with every decade of life from the second through to the seventh decade. Males are more likely to experience hearing loss than females of the same age, and African-Americans show a decreased risk compared with White or Hispanic-Americans (Lin et al. 2011b). Although these ethnic differences in hearing sensitivity are not fully understood, it is thought they might relate to differential lifetime exposure to noise or other environmental risk factors, or differences in melanin levels. Indeed, a study undertaken in albino mice reported that absence of strial melanin coincided with age-associated loss of marginal cells and endocochlear potential (EP) decline when compared with coisogenic pigmented mice (Ohlemiller et al. 2009).
Although noise exposure and ototoxic medications are able to independently cause hearing loss in an individual of any age, they are also extrinsic factors that can exacerbate progression of ARHL (Yang et al. 2015). In modern society, it can be difficult to avoid exposure to environmental noise, whether this is in the workplace, for example, factory workers and armed forces, or with the dramatic increase in the use of personal music devices. Moreover, if you are prescribed ototoxic antibiotics, or require cisplatin-based anticancer therapy, these risks are unavoidable. Conversely, lifestyle is an ARHL risk factor that is amenable to change. In a recent cross-sectional analysis, using 164,770 adults aged between 40 and 69 years of age from the U.K. Biobank Resource, it was found that smoking and passive smoking were associated with increased odds of hearing loss, odds ratio (OR) 1.15 (95% confidence interval [CI], 1.09–1.21), and 1.28 (95% CI, 1.21–1.35), respectively (Dawes et al. 2014). The investigators of this study suggest the higher OR for passive smoking compared with smoking can be explained by smokers being compared with nonsmokers, which will include passive smokers and therefore may underestimate the effects of smoking. However, moderate alcohol consumption was associated with reduced odds of hearing loss, OR 0.61 (95% CI, 0.57–0.65) (Dawes et al. 2014), which is in alignment with previous studies reporting an association between moderate alcohol intake with better hearing (Popelka et al. 2000; Fransen et al. 2008; Gopinath et al. 2010). In a European multicenter study using 4083 subjects between 53 and 67 years, in addition to smoking, they also found that high body mass index (BMI) correlated with hearing loss (Fransen et al. 2008). However, in a similar-sized study undertaken in Korea, low BMI was found to be associated with hearing loss in adults (Lee et al. 2015). Another larger study investigating the weighted prevalence and associated factors of hearing impairment in ∼16,000 Korean adults found that individuals with cardiovascular risk factors (e.g., hypertension, diabetes, smoking, increased serum cholesterol) are at risk of developing hearing impairment (Hong et al. 2015). Moreover, the Australian Blue Mountains Hearing Study found type 2 diabetes was associated with prevalent hearing loss in older adults, OR 1.55 (95% CI, 1.11–2.17) after adjusting for multiple risk factors (Mitchell et al. 2009). A meta-analysis of the effect of either type 1 or type 2 diabetes on hearing found an increased prevalence of hearing loss in diabetics compared with nondiabetics, OR 2.15 (95% CI, 1.72–2.68) and this relationship was not related to the age of participants (n = 20,194) (Horikawa et al. 2013). Although detecting associations between hearing loss and particular lifestyle choices or medical comorbidities can be revealed using large population cohorts, it can be difficult to disentangle the direct contribution of individual risk factors from indirect effects on general health, and therefore establish a causal link. However, taken together, these data suggest that a healthy lifestyle, which includes moderate alcohol intake, may afford some protection against ARHL.
Last, heritability studies among twins and longitudinal studies of family cohorts have shown that genetic predisposition forms a large and important risk factor for ARHL. Various studies report heritability indices of between 0.35 and 0.55 (Karlsson et al. 1997; Gates et al. 1999; Christensen et al. 2001). Hence, given the difficulties of accessing human cochlear tissue for biochemical studies, and as with congenital deafness, it is a genetic approach that is the most likely to provide insights into the molecular mechanisms that result in age-related decline in hearing. Here, we summarize and discuss the progress in our understanding of ARHL genetics based on recent studies undertaken in humans and animals and highlight future perspectives.
INSIGHTS INTO ARHL PATHOLOGY FROM RECENT RESEARCH: HUMAN GENETICS
As a common sensory disorder and a complex disease with a mixture of genetic and environmental components ARHL is resistant to genetic analysis by traditional linkage analysis in families. In a disorder with high heterogeneity, small-to-moderate genetic risk variants, and a significant environmental impact a familial genetic approach is of limited use. Since the first successful genome-wide association study (GWAS) conducted in age-related macular degeneration was published in 2005 (Klein et al. 2005), much hope was raised that this cohort-based approach rather than family-based analysis could reveal the genetic susceptibilities underlying common complex diseases including ARHL. Several GWAS into adult hearing status or ARHL have now been published (summarized in Table 1) (Friedman et al. 2009; Van Laer et al. 2010; Girotto et al. 2011; Nolan et al. 2013; Wolber et al. 2014; Fransen et al. 2015; Vuckovic et al. 2015; Hoffmann et al. 2016), and although many candidates have been linked to ARHL a lack of genome-wide findings that are significant and a poor replication of findings across these studies has limited their impact. One interpretation of these studies is that the heritability of ARHL has been overestimated or, alternatively, that there are thousands of rare variants in the population of small effect on ARHL risk, which would be impossible to detect by GWAS (Fransen et al. 2015).
Table 1.
Summary of genome-wide association studies into age-related hearing loss (ARHL)
| Study reference | Candidate genes highlighted by investigatorsa | Study design | Sample size | Ethnicity | Hearing phenotype | Age (yr) | Functional validation | Independent cohort replicationb |
|---|---|---|---|---|---|---|---|---|
| Friedman et al. 2009 | GRM7 (metabotropic glutamate receptor 7); CDH13 (cadherin 13) | Ca:Co | 846 Ca | European | Z-score based on PTA | 53–67 | Grm7 expression in HC and SGN | GRM7: several other studies; CDH13: one study |
| 846 Co | ||||||||
| Van Laer et al. 2010 | IQGAP2 (IQ motif-containing GTPase-activating-like protein); GRM7 (glutamate receptor 7) | QTA | 347 | Finnish Saami | Principal component analysis based on PTA | 50–75 | None | GRM7 (see above) |
| Girotto et al. 2011 | DCLK1 (doublecortin-like kinase 1); PTPRD (protein tyrosine phosphatase receptor-type delta); GRM8 (metabotropic glutamate receptor 8); CMIP (c-Maf inducing protein) | QTA | 3417 | European | Principal component analysis based on PTA | >18 | Expression studies in a subsequent study | GRM8 in Fransen et al. (2015) |
| Nolan et al. 2013 | ESRRG (estrogen-related receptor γ) | QTA | 3900 | British | Logistic regression analysis of hearing thresholds at 1 and 4 kHz | 44–45 | Esrrg in HC, SC; Esrrg−/− mice have a mild hearing loss | Variable replication within study |
| Fransen et al. 2015 | ACVR1B (activin receptor type-1B); CCBE1 (collagen and calcium-binding epidermal growth factor (EGF) domains 1) | QTA | 2161 | Belgian | Principal component analysis based on PTA | 53–67 | None | None to date |
| Wolber et al. 2014 | SIK3 (salt-inducible kinase 3) | Meta | 4939 | European Silk Road |
Principal component analysis based on PTA | 18–98 | Expression of Sik3 in stria, HC and SGN | None to date |
| Vuckovic et al. 2015 | PCDH20 (protocadherin 20); SLC28A3 (solute carrier family 28 member 3) | Meta | 2636 | Italian Silk Road |
Logistic regression analysis based on PTA | 18–89 | PCDH20 and SLC28A3 expression in HC and cochlea | Nominal replication within study |
| Hoffmann et al. 2016 | ISG20 (interferon-stimulated gene 20 kDa protein); TRIOBP (TRIO and F-actin binding protein); ILDR1 (immunoglobulin-like domain-containing receptor 1); EYA4 (EYA transcriptional coactivator and phosphatase 4) | Ca:Co | 6527 Ca | White-American (non-Hispanic) | Diagnosis of ARHL-related phenotype in electronic health records | 18–100 | SHIELD database supports expression in the inner ear | Within study replication |
| 45,882 Co |
PTA, pure tone audiogram; HC, hair cell; SC, supporting cell; SGN, spiral ganglion neuron; Ca:Co, case control; QTA, quantitative trait analysis; Meta, meta-analysis.
aGenome-wide significance indicated in bold, defined here as p < 5 × 10−8.
bReplication of association at gene level, not necessarily at SNP level.
There are, however, explanations for why common ARHL risk variants in the population may not have been detected in these studies. The power of GWAS to detect genetic risk factors relies primarily on two factors (Manolio et al. 2009; Eichler et al. 2010; Ku et al. 2010). First, it is necessary to have a good phenotyping measure, which is able to clearly delineate between patients and controls; ideally, the measure would also stratify patients into subphenotypes that are a result of the same underlying pathology. Second, GWAS require very large cohorts involving sample sizes in the thousands and tens of thousands (Spencer et al. 2009; Gibson 2010; Hong and Park 2012). For hearing as a phenotype this is problematic; a pure tone audiogram is the gold standard measure but this requires a trained audiologist, a quiet environment, and a significant amount of time. Extrapolate that out to a sample size of 10,000, and then it becomes extremely expensive to collect these data. Although it may be possible to collect large numbers of ARHL patients from clinics, the recruitment of age-matched controls with “good hearing” is more difficult. A compromise therefore often has to be made between the quality of the hearing data collected and the sample size. This explains why many of the large, population-based, publicly funded genetic cohorts have either not included hearing in the phenotypes collected, or have only collected self-reported questionnaire-based data. This has meant that hearing GWAS cohort collections have had to be initiated from individual research groups, which inevitably has limited the number and scale of the studies undertaken.
Different approaches have been taken to defining a hearing phenotype in the GWAS performed to date. Some studies have performed a quantitative population-based analysis, others used a case-control design. Various phenotype measures used include defined frequency thresholds, speech-in-noise tests, and public health records for evidence of hearing aid prescription to define the phenotype (see Table 1). The cohorts also differ greatly in the age range studied. Given these varied parameters, it is perhaps unsurprising that there is a lack of replication between individual GWAS. Compared with the samples sizes of recent successful GWAS in other disorders (see ebi.ac.uk/gwas), the studies also have reduced power because of the smaller samples caused by the difficulties in collecting large datasets as discussed above.
Despite these issues, the GWAS performed have identified a set of candidates for further analysis, providing a first step toward knowledge of ARHL pathogenesis. The strongest of these candidates is GRM7, the gene-encoding glutamate metabotropic receptor 7, a finding that has been replicated in several independent cohorts although not in all populations tested (Friedman et al. 2009; Van Laer et al. 2010; Newman et al. 2012; Luo et al. 2013). It is a G-coupled receptor activated by l-glutamate and its activation is associated with reduced release of the neurotransmitter. Because GRM7 is expressed in hair cells and spiral ganglion neurons, Friedman et al. (2009) suggested that a genetic variant in GRM7 might alter ARHL susceptibility caused by increased glutamate release at the synaptic connection between inner hair cells and auditory neurons, which could accumulate to toxic levels. A decline in the number and quality of synaptic connections between the sensory hair cells and auditory neurons is also thought to underlie “hidden hearing loss,” an effect in which individuals with normal hearing thresholds still struggle to hear when in a noisy environment (Schaette and McAlpine 2011; Liberman 2015; Viana et al. 2015; Liberman and Kujawa 2017). Based on evidence from mice, it has been suggested that this synaptopathy is a primary pathology in noise-induced hearing loss (NIHL) and ARHL (Viana et al. 2015; Liberman and Kujawa 2017). The OR for the GRM7 variant most strongly linked to ARHL was 2.56, although the overall haplotype effect was estimated at 12.01 (Friedman et al. 2009).
Single-nucleotide polymorphisms (SNPs) at two other genetic loci were recently reported to be associated at genome-wide significant levels in a study using electronic health records from 6527 patients with a diagnosis of an ARHL-related phenotype (Hoffmann et al. 2016). One variant is a missense SNP in TRIOBP, a gene-encoding TRIO and F-actin binding protein, an actin cytoskeleton regulator. Nonsense and frameshift mutations in this gene underlie autosomal recessive deafness-28 (DFNB28, MIM#609823), which present as a severe to profound deafness with a prelingual onset (Riazuddin et al. 2006; Shahin et al. 2006). In addition, characterization of Triobp isoform-specific knockout mice identify a role for TRIOBP in the formation of cochlear hair cell stereocilia rootlets, which are required to provide mechanical rigidity to the stereocilia bundle (Kitajiri et al. 2010). The second variant is intergenic, between the ISG20 and ACAN genes encoding proteins involved in interferon signaling and an extracellular matrix component, respectively. In both cases, the SNPs identified had only a very modest effect on risk of ARHL with an OR below 1.2 despite the genome-wide significance (Hoffmann et al. 2016).
Given the lack of genome-wide significance in many of the studies performed to date, researchers have sought to find the true associations among the background of false associations below this statistical threshold by a variety of strategies. These have included pathway analysis to identify enrichment of genes from pathogenic pathways, prioritizing hits in known deafness genes, investigation of gene function in mutant mouse models, and meta-analysis of data across cohorts. These methods have helped strengthen evidence for ESRRG, encoding estrogen-related receptor γ (Nolan et al. 2013) (mouse model), PCDH20 encoding protocadherin 20 (Vuckovic et al. 2015), SLC28A3 encoding the nucleoside transporter, solute carrier family 28 member 3 (Vuckovic et al. 2015), and SIK3 encoding salt-inducible kinase 3 (Wolber et al. 2014) (all meta-analysis) as having a role in adult hearing loss. However, given the known heterogeneity involved in congenital deafness (Bowl et al. 2017), the expectation is that variants in hundreds of genes contribute to ARHL. Therefore, there is still a great need for much larger genetic studies using GWAS or whole-exome or whole-genome sequencing with greater power to detect the relatively subtle effects.
INSIGHTS INTO ARHL PATHOLOGY FROM RECENT RESEARCH: ANIMAL MODELS
Given the difficulties described above of studying presbycusis in human cohorts, researchers have taken to using animal models to help determine the pathogenesis and genetics associated with ARHL. Although no one model organism faithfully displays all aspects of human presbycusis, insights relating to molecular and cellular determinants of cochlear aging have been obtained from studies undertaken in several species including chinchilla, gerbil, rat, and mouse. For example, studies in aged Mongolian gerbils have shown that from 36 months of age they show a mild (15–35 dB) threshold shift, which is most pronounced at the higher frequencies (Mills et al. 1990). Moreover, an identified decline in EP with little sensory hair cell and neuron loss, led to them being classified as a model for a strial-based ARHL (Tarnowski et al. 1991; Schmiedt et al. 2002). The decline in EP was shown to correlate with strial capillary loss and changes to the structure of the marginal cells (Gratton and Schulte 1995; Gratton et al. 1997; Spicer and Schulte 2005). In particular, it is thought that oxidative damage to mitochondria within the strial marginal cells causes reduced ATP production, which in turn reduces Na+,K+ ATPase activity, leading to a reduced EP and elevated auditory thresholds (Spicer and Schulte 2005).
ARHL studies in the rat have used the inbred Fischer 344 (F344) albino strain, of which there are two substrains (DuCrl and NHsd). The age-related hearing function and pathology showed by these two substrains are mostly similar, but there are some differences (Syka 2010). F344 rats of both substrains develop a fast, progressive high-frequency hearing loss beginning at 1 year of age, which extends to the lower frequencies by 18 months of age (Popelar et al. 2003). In addition, these rats show a gradual decline in DPOAE amplitudes beginning in young animals, such that by 12 to 18 months of age DPOAEs are no longer present (Popelar et al. 2006). However, old F344 rats have good preservation of inner and outer hair cells (except at the very apical and very basal regions of the cochlea) (Popelar et al. 2006; Bielefeld et al. 2008). They do show markedly reduced labeling for collagen fibers in the lateral wall (SV and spiral ligament [SL]) and fewer type IV fibrocytes in the SL when compared with younger F344 (Buckiova et al. 2006, 2007). Similar to the Mongolian gerbils, aged F344 rats have a less vascularized SV and show degenerative changes within the marginal cell layer of the SV. A more recent study of outbred Wistar albino rats show they also develop a hearing loss similar in progression and magnitude to F344 (Alvarado et al. 2014).
Similar to gerbils and rats, certain mouse strains also show age-related cochlear pathologies that correspond to a “strial-type” of human presbycusis suggested by Schuknecht (Schuknecht 1955; Schuknecht and Gacek 1993; Ohlemiller 2004).
MOUSE AS A MODEL FOR ARHL
The mouse has become a robust and reliable mammalian model for aging research (Vanhooren and Libert 2013). The reasons for this include the ability to strictly control both intrinsic and extrinsic factors, for example, genetic background, diet, environment, and health status. In addition, mice have a short life span, meaning the effects of age become apparent within a contracted time period.
Some of the earliest research using mice to investigate the genetics of ARHL involved the use of inbred strains. Certain strains have long been reported as having “good” hearing into old age (e.g., CAST, CBA/CaJ, CBA/J, C3H/HeH), whereas others show a progressive decline in auditory function (e.g., BALB, C57BL/6, DBA/2J). Subsequent genetic mapping studies have defined ∼20 loci that influence ARHL in laboratory strains of mice, and for four of these the underlying genetic lesions have been identified, designated as ahl alleles, that is, Cdh23753A (ahl, cadherin-23), Csrs29358506-A (ahl4, citrate synthase), Gipc3343A (ahl5, GIPC PDZ domain-containing family member), and Fscn2R109H (ahl8, fascin actin-bundling protein 2, retinal) (Noben-Trauth et al. 2003; Shin et al. 2010; Charizopoulou et al. 2011; Johnson et al. 2012; Ohlemiller et al. 2016).
The commonly used C57BL/6 strains develop progressive high-frequency hearing loss caused by a hypomorphic mutation within the cadherin 23 (Cdh23) gene, which encodes a component of the stereocilial tip-link required for gating of the mechanoelectrical transducer channel (Noben-Trauth et al. 2003; Siemens et al. 2004; Kazmierczak et al. 2007). C57BL/6J mice show cochlear pathological changes that match the sensory, neural, and strial prebycusis subtypes proposed by Schuknecht. Consistent with the hearing loss, which is evident from as early as 3 months of age, these mice progressively lose sensory hair cells (inner and outer) in an age-dependent manner proceeding from cochlear base to apex. They also show loss of basal spiral ganglion cells, loss of SL fibrocytes and thinning of the SV, although no progressive EP decline is observed at >24 months of age (Hequembourg and Liberman 2001; Ohlemiller 2006, 2009). However, although this strain has been labeled as a model of ARHL, currently there is no convincing evidence associating variants in human CDH23 with ARHL (Yang et al. 2015).
Used in forward genetic screens, the mouse has been an invaluable model organism for the discovery of genes required for hearing function, and mutations leading to congenital and early-onset hearing loss (Hrabe de Angelis et al. 2000; Nolan et al. 2000; Clark et al. 2004; Bowl et al. 2017). Similar hypothesis-generating approaches would likely yield important insights to genes involved with ARHL, but such programs require large numbers of mice to be bred, aged, and phenotyped, and are therefore prohibitively expensive for an individual research group to undertake. Recently, at the MRC Harwell Institute, a large-scale phenotype-driven screen for genes associated with age-related disease was undertaken in mice. Using the chemical mutagen ENU, pedigrees of mutagenized mice were generated and subject to recurrent screens for phenotypes as they aged (Potter et al. 2016). Importantly, the phenotyping pipeline included clickbox and auditory brainstem response (ABR) tests to assess hearing function. From ∼150 pedigrees tested, 27 were found to show late-onset phenotypes, including several that displayed late-onset progressive hearing loss. Of these, three were reported to be caused by ENU-induced mutations within novel hearing loss genes (Slc4a10, Wars2, and Zfyve26, encoding solute carrier family 4 member 10, mitochondrial tryptophanyl transfer RNA [tRNA] synthetase 2 and zinc finger FYVE-type containing 26, respectively). This shows phenotype-driven screens have the capacity to identify models of progressive and/or late-onset hearing loss, and the potential to elaborate on the genetics of this condition.
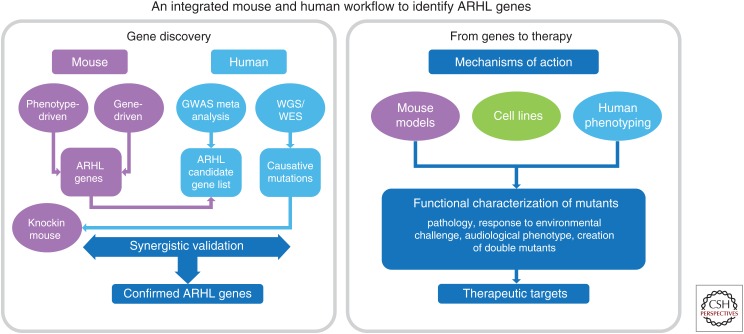
The mouse is the predominant model organism for studying mammalian auditory function (Bowl and Dawson 2015). In particular, the genetic concordance for mutations causing congenital and early-onset hearing loss is very high, in that genetic lesions identified in patients when modeled in the mouse cause hearing loss, and “hearing” genes discovered in the mouse are often found to harbor mutations in human deafness patients. At present, the concordance for ARHL genes is not so high. In a recent review, Ohlemiller et al. (2016) list ∼50 mouse and 20 human candidate genes that have been proposed to influence ARHL and/or NIHL. Interestingly, only around one quarter of the proposed human ARHL genes overlap with proposed mouse ARHL genes. This could mean the remaining candidate human genes are not required for age-related auditory function in the mouse or, more likely, the mouse models for these genes may not have been fully characterized into old age. To fully elaborate on the requirement of these genes, mice would not only need to be sufficiently aged, but they may also need to be challenged (e.g., with noise or secondary genetic alleles) to induce an auditory phenotype. Additionally, the genetic variants that increase risk of ARHL, unlike many of those that cause congenital deafness, may need to be studied in “knockin” models of the human mutation, rather than knockout models. These concerns are particularly relevant for the validation of candidate genes arising from GWAS, the vast majority of which have not yet been followed up in mouse models (Table 1). A more integrated approach is required to both validate ARHL GWAS hits in animal models and also provide insights into the mechanisms underlying ARHL (see Fig. 1).
Figure 1.
An integrated mouse and human workflow to identify age-related hearing loss (ARHL) genes. GWAS, genome-wide association studies; WES, whole-exome sequencing; WGS, whole-genome sequencing.
As additional mouse and human ARHL susceptibility alleles are identified, it will be interesting to observe whether they are “mild” alleles of genes reported for congenital/early-onset, hearing loss, or whether they are novel genes not previously linked with hearing loss. In particular, one might expect that ARHL susceptibility genes are likely to encode structural and homeostatic proteins normally required for maintaining auditory cellular structure and function into old age, and that hypomorphic alleles of these genes will provide less resilience to environmental or lifestyle ARHL risk factors. Indeed, genes linked to metabolism of reactive oxygen species, antioxidant systems, and mitochondrial function have been reported as ARHL susceptibility genes (McFadden et al. 1999; Ohlemiller et al. 2000; Van Eyken et al. 2007; Someya et al. 2008). These genes are not inner-ear specific, but instead have widespread expression and function in many cells throughout the body. Identifying why the ear is so susceptible to loss of these genes will be important for the design of therapeutic interventions.
FUTURE PERSPECTIVES
Rapid technical developments in genetics are providing researchers with more advanced tools applicable to common disease research. Whole-genome sequencing or whole-exome sequencing capture all variants present so, unlike GWAS, these approaches will be able to identify whether there are many rare variants in genes that confer ARHL susceptibility. However, even with these new tools, there is still a need of large cohorts to have sufficient power to detect ARHL genes. The U.K. Biobank study is a population-based study involving more than 500,000 people between 40 and 69 years of age, which recently released genotype data imputed to 90 million variants (Sudlow et al. 2015). Self-reported hearing data is available from all participants and a speech-in-noise test was completed in a subset of 188,000 participants (Dawes et al. 2014). Completion of this U.K. Biobank GWAS analysis in such a large cohort should determine whether there are common variants within the population that play a role in ARHL. Certainly, failure to detect any such associations in a study with such great power would indicate that either there are many thousands of very rare variants responsible, or that the heritability of ARHL has been greatly overestimated.
To date, the mouse has been underutilized as a model organism for the study of human presbycusis. However, this likely reflects the paucity of strong candidate genes arising from human association studies together with the cost of aging cohorts of mice to detect late-onset effects rather than the mouse not being a good model organism. As more genes/alleles are identified through GWAS and familial sequencing studies, future endeavors will certainly involve the generation and longitudinal phenotyping of mouse mutants carrying these exact genetic lesions. Recent advances in genome-editing technologies such as CRISPR/Cas9 that now allow the generation of mice carrying specific point mutations without leaving a “footprint” is a rapid, highly efficient, and relatively inexpensive approach to take. Validation of human ARHL-causing mutations in the mouse, or other organisms, will provide a genetic diagnosis, but equally importantly these models will also provide the opportunity to study disease progression and ascertain the pathological changes occurring within the aging cochlea, something that is not possible in humans. This understanding is a prerequisite for the design of therapeutic interventions to ameliorate age-related auditory decline. In addition, validated models can also be used for preclinical studies to test the efficacy of new therapeutic approaches. However, before embarking on the generation of a genome-edited knockin, it is important to consider the effect of the candidate human mutation in the context of the human sequence. Moreover, careful consideration should also be given to which inbred or outbred mouse strain is used to model a human genetic lesion, as a given knockin mutation on one particular genetic background may or may not show penetrance compared with when tested on a different background. Indeed, it may be that the allele should be trialed on several backgrounds.
ARHL AND DEMENTIA
One of the most significant developments in our knowledge of ARHL etiology over the last decade has been the emergence of hearing loss as a risk factor for developing dementia. A large number of studies have found a link between ARHL and cognitive decline, dementia, and Alzheimer’s disease with the hearing loss predating and predicting subsequent clinical diagnosis of dementia (Lin et al. 2013; Gurgel et al. 2014; Wei et al. 2017; Jayakody et al. 2018). There are several possible explanations for this association between the two morbidities and the resulting implications for our understanding of the mechanisms underlying the two diseases. The most widely proposed model suggests that the lack of auditory input and subsequent social isolation accelerates loss of cognitive function. Alternatively, it could be that in some individuals with ARHL, the hearing loss itself is an early manifestation of a preclinical cognitive decline; it is known that patients with dementia show pathological signs of decline for decades before a clinical diagnosis of dementia. Third, there may be common pathological pathways involved in causing both disorders. Plausible pathways that have been linked to both pathologies include oxidative damage, inflammation, vascular function, mitochondrial dysfunction, glutamate excitotoxicity, and RNA granule dysregulation (Towers et al. 2011; Wong and Ryan 2015; Ganguly et al. 2017; Maziuk et al. 2017; Du et al. 2018; Jayakody et al. 2018; Ridge and Kauwe 2018). It is also possible that more than one of these models is relevant and that, as well as common causative factors underlying hearing loss and cognitive decline with age, the resulting lack of auditory input accelerates the psychological consequences and hence the clinical impact of the pathology. Pertinent to this question, some studies have found that treatment of hearing loss with hearing aids is able to prevent or lessen the speed of cognitive decline (Dawes et al. 2015; Mamo et al. 2017; Maharani et al. 2018), although further studies are required to establish this as a preventative strategy. A positive side effect of recent publicity about the link between midlife hearing loss and the risk of dementia is that it will raise the profile of research into ARHL with research funding bodies (Livingston et al. 2017).
THERAPEUTIC APPROACHES TO TREATING OR PREVENTING ARHL
Given there is currently an insufficient understanding of the mechanisms involved in ARHL, it is unsurprising that very few clinical trials to prevent or treat ARHL have taken place. In the absence of clear biological targets for drug development, pharmaceutical companies are reluctant to invest in expensive translational research despite the size of the potential market for an ARHL therapy. In addition, the design of clinical trials to assess efficacy of therapies for ARHL is problematic. The identification of a drug that could prevent or ameliorate further progression of a mild-to-moderate hearing loss is currently a more realistic prospect than a drug or therapy, which can improve an existing hearing loss. However, to detect a significant effect on hearing loss clinical trials may need to follow participants for months or years making them long and consequently expensive to undertake. Despite these difficulties, a number of clinical and preclinical trials with pharmacological therapies for adult hearing loss are underway. The results of these early trials, even if unsuccessful at identifying an efficacious therapy, will be important in informing the design of future ARHL clinical trials. The first reported clinical trial designed to treat ARHL with a pharmacological agent was conducted by Pfizer and completed in 2013 using PF-04958242, a positive allosteric modulator of the AMPA receptor, an ionotropic glutamate receptor (Bednar et al. 2015). The study was undertaken with 44 participants with ARHL over the age of 50, and no improvement in hearing thresholds was detected 1 and 5 hours after a single dose. In the CLARITY-1 study sponsored by Autifony Therapeutics and completed in 2016, the effect of a modulator of voltage-gated potassium channels, AUT00063, on the hearing of people over the age of 50 who had been diagnosed with ARHL was assessed (see clinicaltrials.gov/ct2/show/NCT02345031). After a 4-week treatment period, no improvement in hearing as assessed by a speech-in-noise test was identified (n = 37) compared with the placebo group (n = 39) at the end of the study. A much larger study involving 550 participants is going to be completed in 2021, assessing the effect of the acetylcholinesterase inhibitor, Huperzine A on hearing function and cognitive decline over a 3-year period (see clinicaltrials.gov/ct2/show/NCT03101722). A number of other clinical trials are underway to investigate the effect of various therapies on mild-to-moderate hearing loss, including a Novartis Pharmaceuticals phase I and II study using a gene therapy approach to deliver the pro–hair cell transcription factor ATOH1 to participants with severe-to-profound hearing loss, in an attempt to improve hearing by promoting regeneration of hair cells from supporting cells (see clinicaltrials.gov/ct2/show/NCT02132130). There is some evidence this approach has been able to restore hearing in animal models of hearing loss (Izumikawa et al. 2005). Furthermore, studies with modulators of metabotropic glutamate receptor 7 in animal models of NIHL conducted by Pragma Therapeutics have shown proof-of-concept and are progressing to preclinical trials. Although these and other studies are not specifically recruiting ARHL patients, the results are likely to have implications for all types of acquired hearing loss.
CONCLUDING REMARKS
Compared with the major advances in the understanding of the genes and pathological mechanisms involved in congenital deafness our knowledge of ARHL is still limited. There are many challenges that remain in this field, but it should also be acknowledged that some advances have been made, including the identification of some strong candidate genes implicated in ARHL susceptibility. Progress is also signified by the first clinical trials for progressive adult hearing loss being undertaken. Although whole-genome sequencing is becoming increasingly affordable and will be used in ARHL, a lack of adequate research funding to undertake studies with sufficient power is probably still the major barrier to further progress in ARHL.
Footnotes
Editors: Guy P. Richardson and Christine Petit
Additional Perspectives on Function and Dysfunction of the Cochlea available at www.perspectivesinmedicine.org
REFERENCES
- Agrawal Y, Platz EA, Niparko JK. 2008. Prevalence of hearing loss and differences by demographic characteristics among US adults: Data from the National Health and Nutrition Examination Survey, 1999–2004. Arch Intern Med 168: 1522–1530. [DOI] [PubMed] [Google Scholar]
- Alvarado JC, Fuentes-Santamaria V, Gabaldon-Ull MC, Blanco JL, Juiz JM. 2014. Wistar rats: A forgotten model of age-related hearing loss. Front Aging Neurosci 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar MM, DeMartinis N, Banerjee A, Bowditch S, Gaudreault F, Zumpano L, Lin FR. 2015. The safety and efficacy of PF-04958242 in age-related sensorineural hearing loss: A randomized clinical trial. JAMA Otolaryngol Head Neck Surg 141: 607–613. [DOI] [PubMed] [Google Scholar]
- Bielefeld EC, Coling D, Chen GD, Li M, Tanaka C, Hu BH, Henderson D. 2008. Age-related hearing loss in the Fischer 344/NHsd rat substrain. Hear Res 241: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowl MR, Dawson SJ. 2015. The mouse as a model for age-related hearing loss—A mini-review. Gerontology 61: 149–157. [DOI] [PubMed] [Google Scholar]
- Bowl MR, Simon MM, Ingham NJ, Greenaway S, Santos L, Cater H, Taylor S, Mason J, Kurbatova N, Pearson S, et al. 2017. A large scale hearing loss screen reveals an extensive unexplored genetic landscape for auditory dysfunction. Nat Commun 8: 886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant LJ, Fozard JL. 1990. Age changes in pure-tone hearing thresholds in a longitudinal study of normal human aging. J Acoust Soc Am 88: 813–820. [DOI] [PubMed] [Google Scholar]
- Buckiova D, Popelar J, Syka J. 2006. Collagen changes in the cochlea of aged Fischer 344 rats. Exp Gerontol 41: 296–302. [DOI] [PubMed] [Google Scholar]
- Buckiova D, Popelar J, Syka J. 2007. Aging cochleas in the F344 rat: Morphological and functional changes. Exp Gerontol 42: 629–638. [DOI] [PubMed] [Google Scholar]
- Charizopoulou N, Lelli A, Schraders M, Ray K, Hildebrand MS, Ramesh A, Srisailapathy CR, Oostrik J, Admiraal RJ, Neely HR, et al. 2011. Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat Commun 2: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Frederiksen H, Hoffman HJ. 2001. Genetic and environmental influences on self-reported reduced hearing in the old and oldest old. J Am Geriatr Soc 49: 1512–1517. [DOI] [PubMed] [Google Scholar]
- Clark AT, Goldowitz D, Takahashi JS, Vitaterna MH, Siepka SM, Peters LL, Frankel WN, Carlson GA, Rossant J, Nadeau JH, et al. 2004. Implementing large-scale ENU mutagenesis screens in North America. Genetica 122: 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JG. 1997. Prevalence of selected chronic conditions: United States, 1990–1992. Vital Health Stat 10: 1–89. [PubMed] [Google Scholar]
- Cruickshanks KJ, Tweed TS, Wiley TL, Klein BE, Klein R, Chappell R, Nondahl DM, Dalton DS. 2003. The 5-year incidence and progression of hearing loss: The epidemiology of hearing loss study. Arch Otolaryngol Head Neck Surg 129: 1041–1046. [DOI] [PubMed] [Google Scholar]
- Dawes P, Cruickshanks KJ, Moore DR, Edmondson-Jones M, McCormack A, Fortnum H, Munro KJ. 2014. Cigarette smoking, passive smoking, alcohol consumption, and hearing loss. J Assoc Res Otolaryngol 15: 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes P, Emsley R, Cruickshanks KJ, Moore DR, Fortnum H, Edmondson-Jones M, McCormack A, Munro KJ. 2015. Hearing loss and cognition: The role of hearing AIDS, social isolation and depression. PLoS ONE 10: e0119616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Wang X, Geng M. 2018. Alzheimer’s disease hypothesis and related therapies. Transl Neurodegener 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. 2010. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11: 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen E, Topsakal V, Hendrickx JJ, Van Laer L, Huyghe JR, Van Eyken E, Lemkens N, Hannula S, Maki-Torkko E, Jensen M, et al. 2008. Occupational noise, smoking, and a high body mass index are risk factors for age-related hearing impairment and moderate alcohol consumption is protective: A European population-based multicenter study. J Assoc Res Otolaryngol 9: 9264–9276; discussion 261–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen E, Bonneux S, Corneveaux JJ, Schrauwen I, Di Berardino F, White CH, Ohmen JD, Van de Heyning P, Ambrosetti U, Huentelman MJ, et al. 2015. Genome-wide association analysis demonstrates the highly polygenic character of age-related hearing impairment. Eur J Hum Genet 23: 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RA, Van Laer L, Huentelman MJ, Sheth SS, Van Eyken E, Corneveaux JJ, Tembe WD, Halperin RF, Thorburn AQ, Thys S, et al. 2009. GRM7 variants confer susceptibility to age-related hearing impairment. Hum Mol Genet 18: 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly G, Chakrabarti S, Chatterjee U, Saso L. 2017. Proteinopathy, oxidative stress and mitochondrial dysfunction: Cross talk in Alzheimer’s disease and Parkinson’s disease. Drug Des Devel Ther 11: 797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates GA, Cooper JC Jr, Kannel WB, Miller NJ. 1990. Hearing in the elderly: The Framingham cohort, 1983–1985. Part I: Basic audiometric test results. Ear Hear 11: 247–256. [PubMed] [Google Scholar]
- Gates GA, Couropmitree NN, Myers RH. 1999. Genetic associations in age-related hearing thresholds. Arch Otolaryngol Head Neck Surg 125: 654–659. [DOI] [PubMed] [Google Scholar]
- Gibson G. 2010. Hints of hidden heritability in GWAS. Nat Genet 42: 558–560. [DOI] [PubMed] [Google Scholar]
- Girotto G, Pirastu N, Sorice R, Biino G, Campbell H, d’Adamo AP, Hastie ND, Nutile T, Polasek O, Portas L, et al. 2011. Hearing function and thresholds: A genome-wide association study in European isolated populations identifies new loci and pathways. J Med Genet 48: 369–374. [DOI] [PubMed] [Google Scholar]
- Gopinath B, Rochtchina E, Wang JJ, Schneider J, Leeder SR, Mitchell P. 2009. Prevalence of age-related hearing loss in older adults: Blue Mountains Study. Arch Intern Med 169: 415–416. [DOI] [PubMed] [Google Scholar]
- Gopinath B, Flood VM, McMahon CM, Burlutsky G, Smith W, Mitchell P. 2010. The effects of smoking and alcohol consumption on age-related hearing loss: The Blue Mountains Hearing Study. Ear Hear 31: 277–282. [DOI] [PubMed] [Google Scholar]
- Gratton MA, Schulte BA. 1995. Alterations in microvasculature are associated with atrophy of the stria vascularis in quiet-aged gerbils. Hear Res 82: 44–52. [DOI] [PubMed] [Google Scholar]
- Gratton MA, Smyth BJ, Lam CF, Boettcher FA, Schmiedt RA. 1997. Decline in the endocochlear potential corresponds to decreased Na,K-ATPase activity in the lateral wall of quiet-aged gerbils. Hear Res 108: 9–16. [DOI] [PubMed] [Google Scholar]
- Gurgel RK, Ward PD, Schwartz S, Norton MC, Foster NL, Tschanz JT. 2014. Relationship of hearing loss and dementia: A prospective, population-based study. Otol Neurotol 35: 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hequembourg S, Liberman MC. 2001. Spiral ligament pathology: A major aspect of age-related cochlear degeneration in C57BL/6 mice. J Assoc Res Otolaryngol 2: 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, Keats BJ, Yoshikawa N, Schaefer C, Risch N, Lustig LR. 2016. A large genome-wide association study of age-related hearing impairment using electronic health records. PLoS Genet 12: e1006371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EP, Park JW. 2012. Sample size and statistical power calculation in genetic association studies. Genomics Inform 10: 117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Jeon JH, Ku CR, Noh JH, Yoo HJ, Kim DJ. 2015. The prevalence and factors associated with hearing impairment in the Korean adults: The 2010–2012 Korea National Health and Nutrition Examination Survey (observational study). Medicine (Baltimore) 94: e611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa C, Kodama S, Tanaka S, Fujihara K, Hirasawa R, Yachi Y, Shimano H, Yamada N, Saito K, Sone H. 2013. Diabetes and risk of hearing impairment in adults: A meta-analysis. J Clin Endocrinol Metab 98: 51–58. [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, et al. 2000. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet 25: 444–447. [DOI] [PubMed] [Google Scholar]
- Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, Brough DE, Raphael Y. 2005. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med 11: 271–276. [DOI] [PubMed] [Google Scholar]
- Jayakody DMP, Friedland PL, Martins RN, Sohrabi HR. 2018. Impact of aging on the auditory system and related cognitive functions: A narrative review. Front Neurosci 12: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Gagnon LH, Longo-Guess C, Kane KL. 2012. Association of a citrate synthase missense mutation with age-related hearing loss in A/J mice. Neurobiol Aging 33: 1720–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamil RJ, Betz J, Powers BB, Pratt S, Kritchevsky S, Ayonayon HN, Harris TB, Helzner E, Deal JA, Martin K, et al. 2016. Association of hearing impairment with incident frailty and falls in older adults. J Aging Health 28: 644–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson KK, Harris JR, Svartengren M. 1997. Description and primary results from an audiometric study of male twins. Ear Hear 18: 114–120. [DOI] [PubMed] [Google Scholar]
- Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Muller U, Kachar B. 2007. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449: 87–91. [DOI] [PubMed] [Google Scholar]
- Kitajiri S, Sakamoto T, Belyantseva IA, Goodyear RJ, Stepanyan R, Fujiwara I, Bird JE, Riazuddin S, Riazuddin S, Ahmed ZM, et al. 2010. Actin-bundling protein TRIOBP forms resilient rootlets of hair cell stereocilia essential for hearing. Cell 141: 786–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. 2005. Complement factor H polymorphism in age-related macular degeneration. Science 308: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku CS, Loy EY, Pawitan Y, Chia KS. 2010. The pursuit of genome-wide association studies: Where are we now? J Hum Genet 55: 195–206. [DOI] [PubMed] [Google Scholar]
- Lee JS, Choi HG, Jang JH, Sim S, Hong SK, Lee HJ, Park B, Kim HJ. 2015. Analysis of predisposing factors for hearing loss in adults. J Korean Med Sci 30: 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC. 2015. Hidden hearing loss. Sci Am 313: 48–53. [DOI] [PubMed] [Google Scholar]
- Liberman MC, Kujawa SG. 2017. Cochlear synaptopathy in acquired sensorineural hearing loss: Manifestations and mechanisms. Hear Res 349: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FR, Metter EJ, O’Brien RJ, Resnick SM, Zonderman AB, Ferrucci L. 2011a. Hearing loss and incident dementia. Arch Neurol 68: 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FR, Niparko JK, Ferrucci L. 2011b. Hearing loss prevalence in the United States. Arch Int Med 171: 1851–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FR, Yaffe K, Xia J, Xue QL, Harris TB, Purchase-Helzner E, Satterfield S, Ayonayon HN, Ferrucci L, Simonsick EM, et al. 2013. Hearing loss and cognitive decline in older adults. JAMA Intern Med 173: 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston G, Sommerlad A, Orgeta V, Costafreda SG, Huntley J, Ames D, Ballard C, Banerjee S, Burns A, Cohen-Mansfield J, et al. 2017. Dementia prevention, intervention, and care. Lancet 390: 2673–2734. [DOI] [PubMed] [Google Scholar]
- Luo H, Yang T, Jin X, Pang X, Li J, Chai Y, Li L, Zhang Y, Zhang L, Zhang Z, et al. 2013. Association of GRM7 variants with different phenotype patterns of age-related hearing impairment in an elderly male Han Chinese population. PLoS ONE 8: e77153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharani A, Dawes P, Nazroo J, Tampubolon G, Pendleton N; SENSE-Cog WP1 Group. 2018. Longitudinal relationship between hearing aid use and cognitive function in older americans. J Am Geriatr Soc 10.1111/jgs.15363. [DOI] [PubMed] [Google Scholar]
- Mamo SK, Nirmalasari O, Nieman CL, McNabney MK, Simpson A, Oh ES, Lin FR. 2017. Hearing care intervention for persons with dementia: A pilot study. Am J Geriatr Psychiatry 25: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziuk B, Ballance HI, Wolozin B. 2017. Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front Mol Neurosci 10: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden SL, Ding D, Reaume AG, Flood DG, Salvi RJ. 1999. Age-related cochlear hair cell loss is enhanced in mice lacking copper/zinc superoxide dismutase. Neurobiol Aging 20: 1–8. [DOI] [PubMed] [Google Scholar]
- Mills JH, Schmiedt RA, Kulish LF. 1990. Age-related changes in auditory potentials of Mongolian gerbil. Hear Res 46: 201–210. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Gopinath B, McMahon CM, Rochtchina E, Wang JJ, Boyages SC, Leeder SR. 2009. Relationship of type 2 diabetes to the prevalence, incidence and progression of age-related hearing loss. Diabet Med 26: 483–488. [DOI] [PubMed] [Google Scholar]
- Newman DL, Fisher LM, Ohmen J, Parody R, Fong CT, Frisina ST, Mapes F, Eddins DA, Robert Frisina D, Frisina RD, et al. 2012. GRM7 variants associated with age-related hearing loss based on auditory perception. Hear Res 294: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K, Zheng QY, Johnson KR. 2003. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat Genet 35: 21–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, et al. 2000. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet 25: 440–443. [DOI] [PubMed] [Google Scholar]
- Nolan LS, Maier H, Hermans-Borgmeyer I, Girotto G, Ecob R, Pirastu N, Cadge BA, Hubner C, Gasparini P, Strachan DP, et al. 2013. Estrogen-related receptor γ and hearing function: Evidence of a role in humans and mice. Neurobiol Aging 34: 2077.e2071–2077.e2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK. 2004. Age-related hearing loss: The status of Schuknecht’s typology. Curr Opin Otolaryngol Head Neck Surg 12: 439–443. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK. 2006. Contributions of mouse models to understanding of age- and noise-related hearing loss. Brain Res 1091: 89–102. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK. 2009. Mechanisms and genes in human strial presbycusis from animal models. Brain Res 1277: 70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, McFadden SL, Ding DL, Lear PM, Ho YS. 2000. Targeted mutation of the gene for cellular glutathione peroxidase (Gpx1) increases noise-induced hearing loss in mice. J Assoc Res Otolaryngol 1: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK, Rice ME, Lett JM, Gagnon PM. 2009. Absence of strial melanin coincides with age-associated marginal cell loss and endocochlear potential decline. Hear Res 249: 1–14. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK, Jones SM, Johnson KR. 2016. Application of mouse models to research in hearing and balance. J Assoc Res Otolaryngol 17: 493–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popelar J, Groh D, Mazelova J, Syka J. 2003. Cochlear function in young and adult Fischer 344 rats. Hear Res 186: 75–84. [DOI] [PubMed] [Google Scholar]
- Popelar J, Groh D, Pelanova J, Canlon B, Syka J. 2006. Age-related changes in cochlear and brainstem auditory functions in Fischer 344 rats. Neurobiol Aging 27: 490–500. [DOI] [PubMed] [Google Scholar]
- Popelka MM, Cruickshanks KJ, Wiley TL, Tweed TS, Klein BE, Klein R, Nondahl DM. 2000. Moderate alcohol consumption and hearing loss: A protective effect. J Am Geriatr Soc 48: 1273–1278. [DOI] [PubMed] [Google Scholar]
- Potter PK, Bowl MR, Jeyarajan P, Wisby L, Blease A, Goldsworthy ME, Simon MM, Greenaway S, Michel V, Barnard A, et al. 2016. Novel gene function revealed by mouse mutagenesis screens for models of age-related disease. Nat Commun 7: 12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Khan SN, Ahmed ZM, Ghosh M, Caution K, Nazli S, Kabra M, Zafar AU, Chen K, Naz S, et al. 2006. Mutations in TRIOBP, which encodes a putative cytoskeletal-organizing protein, are associated with nonsyndromic recessive deafness. Am J Hum Genet 78: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge PG, Kauwe JSK. 2018. Mitochondria and Alzheimer’s disease: The role of mitochondrial genetic variation. Curr Genet Med Rep 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford BR, Brewster K, Golub JS, Kim AH, Roose SP. 2018. Sensation and psychiatry: Linking age-related hearing loss to late-life depression and cognitive decline. Am J Psychiatry 175: 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaette R, McAlpine D. 2011. Tinnitus with a normal audiogram: Physiological evidence for hidden hearing loss and computational model. J Neurosci 31: 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedt RA, Lang H, Okamura HO, Schulte BA. 2002. Effects of furosemide applied chronically to the round window: A model of metabolic presbyacusis. J Neurosci 22: 9643–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuknecht HF. 1955. Presbycusis. Laryngoscope 65: 402–419.14392966 [Google Scholar]
- Schuknecht HF, Gacek MR. 1993. Cochlear pathology in presbycusis. Ann Otol Rhinol Laryngol 102: 1–16. [DOI] [PubMed] [Google Scholar]
- Shahin H, Walsh T, Sobe T, Abu Sa’ed J, Abu Rayan A, Lynch ED, Lee MK, Avraham KB, King MC, Kanaan M. 2006. Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am J Hum Genet 78: 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JB, Longo-Guess CM, Gagnon LH, Saylor KW, Dumont RA, Spinelli KJ, Pagana JM, Wilmarth PA, David LL, Gillespie PG, et al. 2010. The R109H variant of fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of DBA/2J mice. J Neurosci 30: 9683–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens J, Lillo C, Dumont RA, Reynolds A, Williams DS, Gillespie PG, Muller U. 2004. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature 428: 950–955. [DOI] [PubMed] [Google Scholar]
- Someya S, Yamasoba T, Kujoth GC, Pugh TD, Weindruch R, Tanokura M, Prolla TA. 2008. The role of mtDNA mutations in the pathogenesis of age-related hearing loss in mice carrying a mutator DNA polymerase gamma. Neurobiol Aging 29: 1080–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CC, Su Z, Donnelly P, Marchini J. 2009. Designing genome-wide association studies: Sample size, power, imputation, and the choice of genotyping chip. PLoS Genet 5: e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer SS, Schulte BA. 2005. Pathologic changes of presbycusis begin in secondary processes and spread to primary processes of strial marginal cells. Hear Res 205: 225–240. [DOI] [PubMed] [Google Scholar]
- Stucky SR, Wolf KE, Kuo T. 2010. The economic effect of age-related hearing loss: National, state, and local estimates, 2002 and 2030. J Am Geriatr Soc 58: 618–619. [DOI] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. 2015. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12: e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syka J. 2010. The Fischer 344 rat as a model of presbycusis. Hear Res 264: 70–78. [DOI] [PubMed] [Google Scholar]
- Tarnowski BI, Schmiedt RA, Hellstrom LI, Lee FS, Adams JC. 1991. Age-related changes in cochleas of mongolian gerbils. Hear Res 54: 123–134. [DOI] [PubMed] [Google Scholar]
- Towers ER, Kelly JJ, Sud R, Gale JE, Dawson SJ. 2011. Caprin-1 is a target of the deafness gene Pou4f3 and is recruited to stress granules in cochlear hair cells in response to ototoxic damage. J Cell Sci 124: 1145–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eyken E, Van Camp G, Fransen E, Topsakal V, Hendrickx JJ, Demeester K, Van de Heyning P, Maki-Torkko E, Hannula S, Sorri M, et al. 2007. Contribution of the N-acetyltransferase 2 polymorphism NAT2*6A to age-related hearing impairment. J Med Genet 44: 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhooren V, Libert C. 2013. The mouse as a model organism in aging research: Usefulness, pitfalls and possibilities. Ageing Res Rev 12: 8–21. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Huyghe JR, Hannula S, Van Eyken E, Stephan DA, Maki-Torkko E, Aikio P, Fransen E, Lysholm-Bernacchi A, Sorri M, et al. 2010. A genome-wide association study for age-related hearing impairment in the Saami. Eur J Hum Genet 18: 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana LM, O’Malley JT, Burgess BJ, Jones DD, Oliveira CA, Santos F, Merchant SN, Liberman LD, Liberman MC. 2015. Cochlear neuropathy in human presbycusis: Confocal analysis of hidden hearing loss in post-mortem tissue. Hear Res 327: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuckovic D, Dawson S, Scheffer DI, Rantanen T, Morgan A, Di Stazio M, Vozzi D, Nutile T, Concas MP, Biino G, et al. 2015. Genome-wide association analysis on normal hearing function identifies PCDH20 and SLC28A3 as candidates for hearing function and loss. Hum Mol Genet 24: 5655–5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Hu Y, Zhang L, Hao Q, Yang R, Lu H, Zhang X, Chandrasekar EK. 2017. Hearing impairment, mild cognitive impairment, and dementia: A meta-analysis of cohort studies. Dement Geriatr Cogn Dis Extra 7: 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolber LE, Girotto G, Buniello A, Vuckovic D, Pirastu N, Lorente-Canovas B, Rudan I, Hayward C, Polasek O, Ciullo M, et al. 2014. Salt-inducible kinase 3, SIK3, is a new gene associated with hearing. Hum Mol Genet 23: 6407–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Ryan AF. 2015. Mechanisms of sensorineural cell damage, death and survival in the cochlea. Front Aging Neurosci 7: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba T, Lin FR, Someya S, Kashio A, Sakamoto T, Kondo K. 2013. Current concepts in age-related hearing loss: Epidemiology and mechanistic pathways. Hear Res 303: 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Schrepfer T, Schacht J. 2015. Age-related hearing impairment and the triad of acquired hearing loss. Front Cell Neurosci 9: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]