Abstract
Autism spectrum disorder (ASD) is a common disorder that causes substantial distress. Heritability studies consistently show a strong genetic contribution, raising the hope that identifying ASD-associated genetic variants will offer insights into neurobiology and ultimately therapeutics. Next-generation sequencing (NGS) enabled the identification of disruptive variants throughout protein-coding regions of the genome. Alongside large cohorts and novel statistical methods, these NGS methods revolutionized ASD gene discovery. NGS methods have also contributed substantially to functional genetic data, such as gene expression, used to understand the neurobiological consequences of disrupting these ASD-associated genes. These functional data are also critical for annotating the noncoding genome as whole-genome sequencing (WGS) begins to provide initial insights outside of protein-coding regions. NGS methods still have a major role to play, as do similarly transformative advances in stem cell and gene-editing methods, in translating genetic discoveries into a first generation of ASD therapeutics.
Autism spectrum disorder (ASD) is defined by the American Psychiatric Association’s Diagnostic and Statistical Manual, Fifth Edition (DSM5) as the co-occurrence of social impairment and restricted or repetitive behaviors or interests. These symptoms are frequently comorbid with other neuropsychiatric symptoms, including intellectual disability, developmental delay, seizures, and attention-deficit disorder. Diagnosis is based on clinical opinion, informed by standardized diagnostic instruments, including the Autism Diagnostic Interview (ADIR) and Autism Diagnostic Observation Schedule (ADOS), alongside measures of cognitive function, adaptive function, and behavior. Full evaluation requires multiple hours of assessment by a team of psychologists and physicians. At present, there is no effective biomarker such as a blood test or imaging technique.
Estimates of ASD prevalence have consistently increased since the first description in 1943. The latest estimate from the Center for Disease Control and Prevention (CDC) figures is one in 59 children (Baio et al. 2018); this is approaching to the prevalence of 2.6% (one in 38) estimated by applying diagnostic measures consistently across a large population (Kim et al. 2011). Changes in diagnostic definition and increased awareness explain much of the increase in prevalence and complicate efforts to determine whether there is also a true increase in symptomatology in the population. The prevalence is consistently about threefold higher in males than females, with a greater sex bias in “high-functioning” individuals with no comorbid cognitive impairment (Halladay et al. 2015). Despite the high prevalence, there is strong selective pressure against ASD symptoms, as shown by the reduced fecundity (Power et al. 2013).
Twin studies and sibling studies have consistently shown that ASD is one of the most highly heritable complex disorders in humans. Most studies estimate the narrow sense heritability (h2) to be between 70% and 90% (Tick et al. 2016; Sandin et al. 2017). A recent family study based on insurance claim data from about a third of the population of the United States found ASD to be the most highly heritable of the 149 disorders considered (h2 = 92%) (Wang et al. 2017). These high heritability estimates suggest that genetic factors have an important role in the causation of ASD.
There is an urgent need for therapeutic interventions capable of substantially improving symptoms that cause distress to the millions of individuals with ASD and their families. Efforts to identify such therapies are limited by the lack of understanding of the neurobiological mechanisms underlying ASD. Identifying the specific genetic variants that contribute to ASD, and understanding how these genetic variants lead to symptoms, has the potential to lead to therapies. Next-generation sequencing (NGS) has had a transformative role in accelerating this progress in ASD.
EARLY PROGRESS IN ASD GENETICS
Karyotype analysis identifies large-scale changes to chromosomes (e.g., ≥5 Mb). Special culture conditions can also identify fragile X syndrome, although methods based on polymerase chain reaction (PCR) or methylation are now used for detection. Comparison of karyotypes across 2173 autism cases prior to 2010 identified fragile X in 1.5% of ASD cases, Down syndrome (trisomy 21) in 0.9%, and large deletions/duplications (now called copy number variants [CNVs]), translocations, or inversions (copy neutral variants) in 3.4% of cases (Table 1). Of note, maternal duplications at the 15q11.2–13 locus were identified in several families (Cook et al. 1997). The vast majority of these genetic variants were de novo mutations, observed in DNA from the child’s blood, but not in DNA from either parents’ blood. Affected children often had syndromic features such as cardiovascular abnormalities or dysmorphic features and were described a “syndromic autism,” distinct from “idiopathic autism” without such features. Autism symptoms were also observed in several distinctive syndromes without chromosomal abnormalities, including tuberous sclerosis and neurofibromatosis. The extent to which ASD is a feature of each syndrome varies considerably, from ∼50% of cases with fragile X (FMR1) and tuberous sclerosis (TSC1/TSC2) to fewer than 5% of cases with neurofibromatosis (NF1) (Williams and Hersh 1998; de Vries et al. 2007; Garcia-Nonell et al. 2008).
Table 1.
Karyotype analysis of ASD
| Study | ASD samples | Karyotyped ASD samples | Copy number variant | Copy number neutral | Trisomy 21 | Fragile X |
|---|---|---|---|---|---|---|
| Gillberg and Wahlström (1985) | 66 | 66 | 1 | 0 | 1 | 8 |
| Ritvo et al. (1990) | 233 | Not stated | 2 | 1 | 5 | 2 |
| Li et al. (1993) | 104 | 104 | 0 | 3 | 2 | 8 |
| Weidmer-Mikhail et al. (1998) | 92 | 92 | 3 | 0 | 0 | 0 |
| Konstantareas and Homatidis (1999) | 127 | 28 | 7 | 2 | 0 | 0 |
| Lauritsen et al. (1999) | 437 | 145 | 1 | 3 | 0 | 0 |
| Wassink et al. (2001) | 898 | 278 | 15 | 1 | 2 | 6 |
| Kielinen et al. (2004) | 187 | 187 | 4 | 0 | 7 | 4 |
| Reddy (2005) | 433 | 421 | 9 | 5 | 0 | 3 |
| Shen et al. (2010) | 933 | 852 | 8 | 9 | 2 | 2 |
| Total | 3510 | 2173 | 50 | 24 | 19 | 33 |
| % ASD samples | 1.40% | 0.70% | 0.50% | 0.90% | ||
| % Karyotyped ASD samples | 2.30% | 1.10% | 0.90% | 1.50% | ||
ASD, Autism spectrum disorder.
Within a syndrome, the presence of distinctive features allowed affected and unaffected individuals to be distinguished within families. By comparing how the pattern of genetic variants (e.g., short tandem repeats) segregates with the syndrome in the family, linkage analysis can identify the genetic locus, which can then be resolved to a nearby gene harboring the causative variant (e.g., FMR1 in fragile X) (Verkerk et al. 1991). In addition to identifying the causative locus behind many syndromic forms of ASD, linkage analysis identified variants in the genes NLGN3 and NLGN4X on chromosome X in large families with X-linked inheritance patterns for ASD (Jamain et al. 2003; Laumonnier et al. 2004). However, apart from NLGN3 and NLGN4X, in the era before NGS, linkage analysis failed to identify other genetic loci with robust evidence for ASD association.
Sequencing of the human genome (Lander et al. 2001; Venter et al. 2001), alongside improvements in methods for detecting variants in PCR-amplified DNA, allowed single genes to be screened for genetic variation in hundreds of individuals. In disorders for which the underlying biology was partially understood, for example, infantile epilepsy (Ogiwara et al. 2009), these approaches identified associated genes. However, in ASD, these so-called “candidate gene” approaches did not result in the reliable detection of ASD-associated genes. In retrospect, these experiments often analyzed the wrong genes (Johnson et al. 2017), lacked the sample sizes to detect the rarest variants with the strongest evidence for association, and suffered from publication bias and winner’s curse (Xiao and Boehnke 2009).
EARLY PROGRESS IN ASD GENOMICS
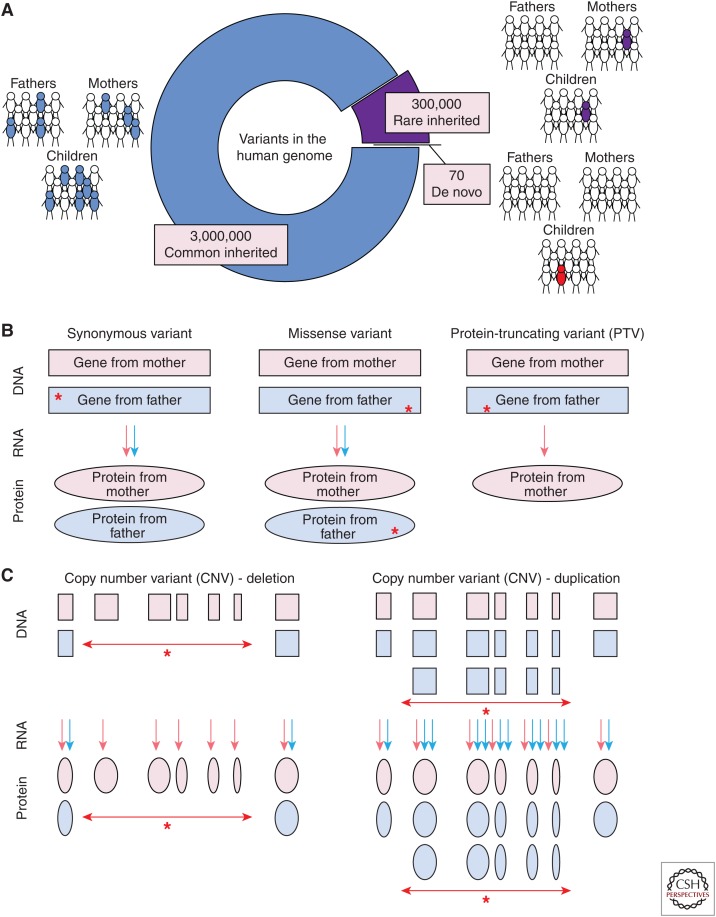
The availability of single-nucleotide polymorphism (SNP) genotyping microarrays, which could assess tens of thousands of SNPs simultaneously in a single individual, enabled two novel experimental designs: (1) genome-wide association studies (GWAS), in which thousands of common variant SNPs were assessed for enrichment in cases versus controls (Risch and Merikangas 1996); and (2) detection of large-scale deletions or duplications in DNA, called CNVs (Fig. 1) (Iafrate et al. 2004; Sebat et al. 2004; Feuk et al. 2006). Microarrays improved the accuracy and throughput of CNV detection and expanded the CNV resolution by at least 1000-fold, from the ≥5 Mb achieved by karyotype analysis to ≥50 kb.
Figure 1.
Types of genetic variants. (A) The majority of genetic variation in the human genome is common (population frequency ≥1%, blue); these variants are transmitted from parents following Mendelian inheritance patterns. A smaller proportion is rare (≤1%, purple) and also transmitted from parents, whereas only ∼70 variants are de novo (red), observed only in the child, but not in either parent. (B) The impact of single-nucleotide variants (SNVs) and small (≤50 bp) insertion/deletions (indels) depends on their location in the genome. In the 1.5% of the genome that encodes proteins (i.e., the exome) these variants can either be synonymous, with no change to the resulting protein, missense, in which a single amino acid is changed in the protein with variable functional impact, or protein-truncating variants (PTVs), which result in nonsense-mediated decay and no protein. Variants and their consequences (red stars) are shown on the father’s allele, but can also arise on the maternal allele. (C) Copy number variants (CNVs) are large (≥50 bp to millions of nucleotides) deletions, resulting in no protein, or duplications, potentially resulting in excess protein.
A landmark experiment in 2007 identified a significant excess of de novo CNVs in 195 ASD cases versus 196 controls (Sebat et al. 2007). Of note, this increased CNV burden was especially prominent in simplex families, in which a single individual is affected by ASD, compared with multiplex families, with a first-degree relative also affected by ASD. Critically, this excess of de novo CNVs in ASD was replicated in several larger, independent cohorts (Pinto et al. 2010; Gilman et al. 2011; Sanders et al. 2011), including the Simons Simplex Collection (SSC) of ∼2500 simplex families (Fischbach and Lord 2010). The SSC performed in-person assessment on cases to ensure cases met “gold standard” diagnostic criteria and also assessed the parents and siblings to exclude ASD. DNA from blood was collected from the child with ASD (the proband), both parents, and an unaffected sibling who could act as a control.
Several studies highlighted CNVs at specific genomic loci in multiple independent ASD cases, including the 16p11.2 locus (Kumar et al. 2008; Marshall et al. 2008; Weiss et al. 2008), raising the question of how many CNVs are required to show ASD association at a specific genetic locus in a genome-wide analysis. This was resolved by developing a statistical model that integrated the observed de novo variants in cases with the expected mutation rate from the unaffected sibling controls to assess the false discovery rate (FDR) (Sanders et al. 2011). Along with the observation that the 16p11.2 locus met this genome-wide threshold for association, this analysis showed that duplications at 7q11.23 were associated with ASD. Of note, deletions at 7q11.23 are the cause of Williams–Beuren syndrome, a neurodevelopmental disorder that includes hypersociability as a feature (Pober 2010), suggesting that the degree of expression of a gene or genes at this locus correlates with an individual’s sociability.
ASD GENETICS PRIOR TO NGS
The high heritability of ASD had sparked considerable interest in the genetics of ASD and the identification and replication of de novo CNVs as a major contributor to ASD offered a path to identifying the ASD-associated genetic loci with the highest effect sizes. As groundbreaking as this approach was, experimental follow-up in model systems was complicated by the size of the variants and the multiple genes involved. The CNV analysis also mapped out a route to leverage NGS methods, suggesting an experimental design (i.e., comparing gene-disrupting de novo variants in cases and controls, providing an ideal cohort, and laying out the statistical approaches to evaluate the results).
EXOME SEQUENCING IN ASD
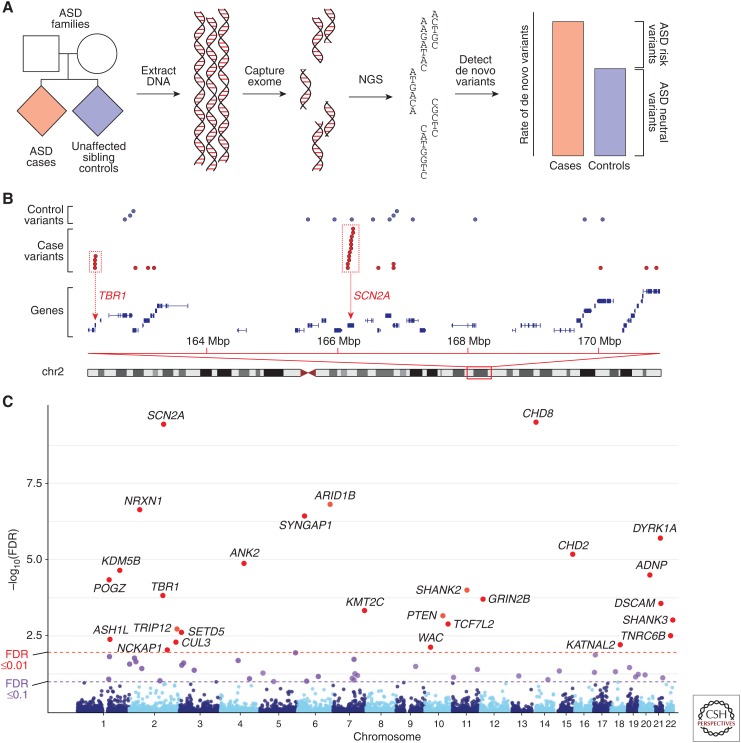
Exome capture uses thousands of DNA probes to bind and extract DNA in the 1.5% of the genome that encodes proteins, called the exome. Pairing exome capture with NGS resulted in “whole-exome sequencing” (WES), which reduced the per-sample sequencing costs sufficiently to enable multiple individuals to be analyzed simultaneously (Fig. 2). Several early studies highlighted the potential of WES to identify risk-mediating variants in individual families or small cohorts with developmental delay or ASD (Aldahmesh et al. 2011; O’Roak et al. 2011; Sirmaci et al. 2011).
Figure 2.
Exome sequencing in autism spectrum disorder (ASD). (A) Blood-derived DNA is collected from individuals with ASD (cases), both their parents, and, ideally, an unaffected sibling control. The 1.5% of the genome that encodes proteins is captured and undergoes next-generation sequencing (NGS). De novo variants are identified by comparing the child’s sequence to the parents’ and the rate of de novo variants between cases and controls is assessed. If other factors, such as parental age and sequencing quality metrics, can be excluded, then the excess of de novo variants represents risk-mediating variants identified in the cases only. (B) Distribution of de novo variants in cases (red) and controls (blue) in a region of chromosome 2. Most de novo variants are distributed across multiple genes; however, some de novo variants in cases are clustered in specific genes, such as TBR1 and SCN2A. (C) Statistical analysis of the distribution of variants across protein-coding genes in 4687 ASD cases (Sanders et al. 2015) using the “transmitted and de novo association” (TADA) test (He et al. 2013) generates a per-gene false discovery rate (FDR). The Manhattan plot shows 27 named genes at a threshold of FDR ≤0.01 (red points) and a further 36 genes at a threshold of FDR ≤0.1 (purple points). NLGN3 on chromosome X is not shown and KDM5B was excluded because of an excess of mutations in siblings.
DE NOVO PTVs
Four papers, published early in 2012, provided the first robust evidence that de novo protein-truncating variants ([PTVs], also called loss-of-function or likely gene disrupting [LGD]) (see Fig. 1), were associated with ASD (Iossifov et al. 2012; Neale et al. 2012; O’Roak et al. 2012a; Sanders et al. 2012). PTVs include single-nucleotide variants (SNVs) that lead to premature stop codons (also called nonsense variants), SNVs in the highly conserved canonical splice sites, two nucleotides on either side of an exon, and insertion/deletions (indels) that disrupt the reading frame (also called frameshift indels). All three of these types of variants are predicted to cause nonsense-mediated decay, resulting in the transcribed messenger RNA (mRNA) being degraded and no protein being produced for that allele (Fig. 1).
By comparing the rate of heterozygous de novo PTVs in cases versus controls or expected mutation rates, these four studies showed a consistent excess of de novo PTVs in ASD cases, thus showing that some of these de novo PTVs were contributing risk for ASD (Fig. 2). This observation has since been replicated in additional large independent ASD cohorts (De Rubeis et al. 2014; Dong et al. 2014; Iossifov et al. 2014), with de novo PTVs observed in 15.8% of ASD cases compared with 8.5% of controls, suggesting that such variants contribute to ASD in ∼7% of ASD cases (Sanders et al. 2015).
DE NOVO MISSENSE VARIANTS
Because missense variants can also disrupt gene function (Fig. 1), it follows that an excess of de novo variants should be expected too. Although the original four exome studies (Iossifov et al. 2012; Neale et al. 2012; O’Roak et al. 2012a; Sanders et al. 2012) showed such a trend, it required larger sample sizes to reliably show a significant excess of de novo missense variants, independent of de novo PTVs (De Rubeis et al. 2014; Iossifov et al. 2014). As expected, ASD association is strongest for missense variants predicted to disrupt protein function.
Missense variants can also alter rather than disrupt gene function, potentially identifying a different set of genes from PTVs. At present, bioinformatic approaches cannot reliably distinguish such gain-of-function variants from loss-of-function variants; however, clustering of variants at specific nucleotides or functional domains is likely to enrich for gain-of-function variants (Geisheker et al. 2017), although similar patterns can also be observed for loss-of-function variants (Ben-Shalom et al. 2017).
PATTERNS OF MUTATIONS
Exome sequencing provided some of the first large-scale analyses of germline de novo mutation patterns in humans. The O’Roak study showed that the majority of de novo variants were on the father’s allele and that mutation rate increased with paternal age (O’Roak et al. 2012a), a result predicted in 1947 (Haldane, 1947), because of the ongoing replication in the paternal germline compared with the maternal germline. This result has been replicated several times (Kong et al. 2012; Rahbari et al. 2016; Jónsson et al. 2017), also identifying a weaker correlation between maternal age and germline de novo mutation rate (Jónsson et al. 2017). It is important to correct for parental age in analyses of de novo mutation burden, especially because phenomena such as stoppage (ceasing to have children after having a child with medical problems) or age of diagnosis can lead to differences in parental age between cases and controls (Werling et al. 2018).
An alternative explanation for observing an excess of de novo mutations in ASD cases could be genomic instability/fragility, resulting in an overall increase mutation rate (similar to that observed in many cancers). Three important data points limit this as a possibility. First, the excess of mutations observed for CNVs, PTVs, and missense variants is not observed for synonymous variants (Fig. 1) (Iossifov et al. 2012, 2014; Sanders et al. 2012) or noncoding variants (Werling et al. 2018). Second, the mutation rate observed in ASD families is similar to that observed in non-ASD cohorts (Jónsson et al. 2017). Finally, the location of these mutations differs from hot spots of mutability identified in whole-genome analyses (Michaelson et al. 2012).
The exome data also replication earlier reports of differing mutation rates per nucleotide, especially for C→T mutations owing to the sensitivity of methylated cytosine (5-methylcytosine) be deaminated into tyrosine (Lynch 2010). Combining these mutational patterns with gene length allowed the expected mutation rate to be determined for each gene (Sanders et al. 2012). Considering the nucleotide, either side of the mutated nucleotide improved the accuracy of this model (Samocha et al. 2014) and further refinements were derived from the 60,000 WES samples in the Exome Aggregation Consortium (ExAC) (see exac.broadinstitute.org) (Lek et al. 2016).
GENE DISCOVERY
Along with identifying an overall excess of de novo PTVs in the 2012 exome studies, three genes (SCN2A, CHD8, and KATNAL2) had two de novo PTVs, a result that is unlikely by chance given the very low rate of such variants (one in 20 individuals) and the ∼18,000 protein coding genes in the genome and not seen in controls (O’Roak et al. 2012a; Sanders et al. 2012). Using permutation analysis, based on the rate of de novo PTVs in siblings and the per-gene mutability, the Sanders study showed that the two de novo PTVs observed in SCN2A were genome-wide significant in a cohort of 225 families (Sanders et al. 2012). Critically, this permutation analysis was able to estimate the expected rate of gene discovery as sample size increased, estimating >30 genes would be associated with ASD in a cohort of 3000 families (Sanders et al. 2015).
This statistical approach to gene discovery was refined by the creation of the “transmitted and de novo association” (TADA) test in 2013 (He et al. 2013). Using a Bayesian approach, it compares the observed and expected numbers of variants based on the per-gene mutation rate and incorporates evidence across multiple variant classes, for example, de novo PTVs, de novo missense, and rare inherited PTVs. The resulting genome-wide FDR for each gene enables the researcher to select a level of confidence appropriate to the analysis. An alternative frequentist approach generates a p-value for each gene from de novo PTVs alone (Samocha et al. 2014). As expected, these two approaches are highly correlated, with a TADA FDR threshold of ≤0.0001 being approximately equivalent to a genome-wide significant p-value of ≤1 × 10−6. Similar approaches have also been used in other disorders with a contribution from de novo variants on WES, including developmental delay (McRae et al. 2017), congenital heart disease (Jin et al. 2017), and infantile epileptic encephalopathy (Heyne et al. 2018).
As predicted, larger cohorts of ASD families yielded substantial increases in the number of ASD-associated genes. Analysis of ∼2500 SSC families identified 27 genes with at least two de novo PTVs (Iossifov et al. 2014), whereas the analysis of 3871 ASD cases (of which 2270 were families) identified 33 genes with a TADA FDR ≤0.1 (De Rubeis et al. 2014). Combining these two cohorts and integrating small de novo deletion CNVs from 4687 ASD cases into the TADA model, identified 65 ASD-associated genes (Fig. 2) with a TADA FDR ≤0.1 (Sanders et al. 2015). These studies also assessed the total number of genes that could contribute to ASD, with estimates in the range of 500–1000 genes (He et al. 2013; De Rubeis et al. 2014; Iossifov et al. 2014).
Alongside these WES cases, targeted analyses that assayed DNA with NGS from a few hundred genes only, were able to identify further variants in many of the genes identified by WES (O’Roak et al. 2012b; Stessman et al. 2017). Although these analyses bolster the evidence for many of the ASD-associated genes, the lack of statistical methods to integrate WES and targeted sequencing analysis have limited the evaluation of which genes meet strict genome-wide statistical criteria.
INHERITED HETEROZYGOUS VARIANTS IN ASD
If a de novo PTV contributes to ASD, it follows that an inherited PTV in the same gene also contributes to ASD. Finding an inherited heterozygous contribution to ASD is complicated by the ability to distinguish variants that mediate ASD risk from neutral variants that do not. Because of natural selection and the reduced fecundity in ASD (Power et al. 2013), we would expect the variants with the highest effect sizes to be the rarest. Assuming they need to occur at a similar frequency to a de novo PTV, a reference cohort of at least 50 million individuals would be required (Sanders et al. 2017), 1000-fold more than ExAC at present (Lek et al. 2016). With available tools, a slight excess in rare inherited PTVs can be detected; however, the association is too weak to have a meaningful impact on gene discovery (Sanders et al. 2015).
HOMOZYGOUS VARIANTS IN ASD
Homozygous variants at locations not observed in ExAC occur at a similar population frequency to de novo variants and therefore have the potential to mediate equivalent risk for ASD (Sanders et al. 2017). While simplex ASD families would be expected to have an increased contribution from de novo variants, consanguineous ASD families should have an excess of risk-mediating homozygous variants, although analysis of families with developmental delay shows that de novo variants are still a major risk factor even in consanguineous families (McRae et al. 2017).
Three families affected by ASD, seizures, and intellectual disability were identified with homozygous variants in the gene branched chain ketoacid dehydrogenase kinase (BCKDK), two of these were PTVs, while the third was a missense variant with loss-of-function effects (Novarino et al. 2012). Analysis of 933 ASD cases from simplex and multiplex families showed a twofold excess of homozygous (the same variant on both alleles) and compound heterozygous (different variants on each allele in the same gene) PTVs in cases compared with controls (Lim et al. 2013). Despite the frequency and relative risk of these variants being similar to those observed for de novo variants, they did not show the same degree of clustering within genes (Fig. 2); therefore, no gene met criteria for genome-wide association. Concurrent analysis of 277 consanguineous families identified two families with homozygous PTVs in the PAH gene, resulting in undiagnosed phenylketonuria, and homozygous missense variants in five genes associated with known Mendelian disorders (Yu et al. 2013).
Taken together, homozygous and compound heterozygous gene-disrupting variants are associated with ASD and probably contribute to at least 3% of ASD cases. However, these biallelic variants show less evidence of clustering in specific genes, limiting gene discovery with clear statistical thresholds at these sample sizes.
SOMATIC MUTATIONS IN ASD
De novo mutations can also arise after conception, resulting in a somatic mutation detected in only a subset of cells. Such mutations can be identified in blood samples by the proportion of sequencing reads containing the variant allele being below 50%. Approximately 10% of the de novo mutations identified in ASD cases have this signature, suggesting a postzygotic origin (Dou et al. 2017; Krupp et al. 2017; Lim et al. 2017). Mutations that arise later in development can be undetectable in the blood but have a profound impact in the brain. Analysis of 55 ASD brains identified several somatic mutations, including a de novo missense variant in the gene SETD2 in the cortex, but not cerebellum (D’Gama et al. 2015). Somatic mutations have the potential to identify regions of the brain that contribute to ASD and novel genes that might be lethal or lead to alternative diagnoses if present in the germline.
FUNCTIONAL INSIGHTS FROM DATA
The success of gene discovery in ASD offers a foundation from which to understand the neurobiology of ASD. Analysis of the genes identified (Fig. 2) point toward two major gene groups: (1) gene expression regulators (e.g., CHD8), and (2) neuronal communication (e.g., SCN2A) (De Rubeis et al. 2014; Sanders et al. 2015). The relationship between these two groups remains unclear, although the simplest explanation would be for the gene expression regulators to control the expression of the genes involved in neuronal communication (Sanders 2015).
Fragile X was one of the first syndromes associated with ASD. It is caused by a triplet expansion that silences the gene FMR1 on chromosome X preventing the protein fragile X mental retardation protein (FMRP) from being made. FMRP interacts with RNAs expressed by other genes, usually reducing the quantity of the protein they encode. Cross-linking immunoprecipitation of FMRP in the mouse brain, followed by NGS of the precipitated FMRP-bound RNA, identified mRNA gene targets (Darnell et al. 2011). These FMRP gene targets are highly enriched for ASD genes (Darnell et al. 2011; Iossifov et al. 2012, 2014; Sanders et al. 2015), suggesting a potential mechanism by which fragile X leads to ASD. Attempts to modify this deficit have so far not improved symptoms in humans, although this may be because treatment at earlier ages is required (Berry-Kravis et al. 2018).
The chromatin modifier CHD8 was one of the first genes identified by exome sequencing (O’Roak et al. 2012a) and remains one of the genes with the strongest evidence of ASD association (Fig. 2). Chromatin immunoprecipitation and NGS (ChIP-Seq) can be used to identify the regulatory targets of chromatin modifiers and transcription factors, such as CHD8. ChIP-Seq analyses of CHD8 in neural progenitor cells (Sugathan et al. 2014) and human neural stem cells, human midfetal brain at 16–19 postconception weeks, and mouse cortex at 17.5 postconception days (Cotney et al. 2015) showed that CHD8 targets are enriched for other ASD-associated genes, particularly those in the gene expression regulator group (Sanders 2015). Reducing the expression of CHD8 in neural progenitor cells (Sugathan et al. 2014) and human neural stem cells (Cotney et al. 2015) resulted in mild dysregulation of multiple genes, although most of these were not direct targets of CHD8 on ChIP-Seq. Of note, down-regulated genes that were not CHD8 targets were also enriched for ASD-associated genes, mostly in the neuronal communication group (Sanders 2015), and many are targets of RE-1 silencing transcription factor (REST), which may act as an intermediate (Katayama et al. 2016).
LARGE-SCALE NGS DATASETS
Along with functional analysis of individual genes, ASD research has leveraged large-scale NGS datasets of hundreds or thousands of samples. Analysis of gene expression in postmortem brain provides an alternative approach to identifying genes implicated in ASD, although it is hard to distinguish causal factors from consequences. Across several such analyses, two main groups of genes are consistently dysregulated: (1) genes down-regulated in ASD that are enriched for neuronal markers, they are also enriched for ASD-associated genes and common variants; and (2) genes up-regulated in ASD that are enriched for markers of microglia and astrocytes, but not enriched for variants associated with ASD (Luo et al. 2012; Ellis et al. 2016; Parikshak et al. 2016; Gandal et al. 2018). Improvements in the quality of NGS data have allowed assessment of splicing patterns as well as gene expression, suggesting reduced splicing diversity in the ASD brain (Parikshak et al. 2016).
Gene expression data from unaffected individuals and wild-type animal models have also provided insights into ASD. Assessing ASD-associated genes in the context of gene co-expression patterns in the human (Kang et al. 2011) implicated the prefrontal cortex during midfetal development (Willsey et al. 2013). Comparison to cell-type-specific marker genes suggested this signal was driven by excitatory glutamatergic neurons (Willsey et al. 2013). Similar results have been identified by other groups, along with a potential role for striatal medium spiny neurons (Parikshak et al. 2013; Xu et al. 2014; Chang et al. 2015).
In addition to improving the mutational model used for gene discovery, WES data from >60,000 individuals in ExAC has helped identify which genes are most sensitive to heterozygous disruption by PTVs in humans (Lek et al. 2016). Comparing the observed number of PTVs to the expected number from the mutation model led to a “probability of loss-of-function intolerance” (pLI) score, which is high in genes that are most likely to cause a disorder in the presence of a PTV. Comparing the de novo variants identified in ASD cases to controls showed that genes with a pLI score >0.9 explained the majority of the association signal in ASD (Kosmicki et al. 2017). This observation has the potential to improve gene discovery, since these high pLI genes have a much stronger association with ASD.
WHOLE-GENOME SEQUENCING
As the cost of NGS has continued to decrease, short-read whole-genome sequencing (WGS) is beginning to replace WES as a method to identify genetic factors associated with ASD. WGS data has several advantages over previous genomic methods (Sanders et al. 2017). Without the need to perform DNA capture, WGS offers more even coverage across the genome, allowing a more thorough analysis of protein-coding regions, resulting in the discovery of 10%–20% more coding variants than WES (Turner et al. 2016; Yuen et al. 2017; Werling et al. 2018). Although microarray and GWAS allow the detection of common noncoding variants associated with ASD, WGS can also identify rare and de novo noncoding variants. The consistent coverage also allows the detection of small CNVs and, by detecting break points, translocations, and inversions (Brandler et al. 2016; Collins et al. 2017; Werling et al. 2018).
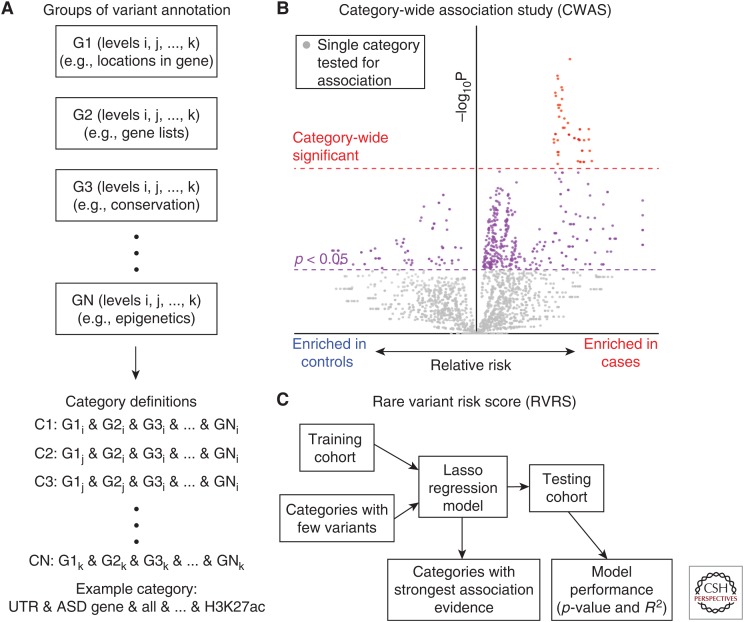
Identifying a noncoding contribution to ASD, which may provide further insight into when and where ASD pathology occurs in the brain (Sanders et al. 2017), is complicated by a limited understanding of how information is encoded in noncoding regulatory sequences. Although there was a clear hypothesis that de novo PTVs, and to a lesser extent de novo missense variants, in protein-coding genes would contribute to ASD, it is not clear which noncoding elements or noncoding annotations best predict functional impact. To overcome this limitation, the category-wide association study (CWAS) was developed, mirroring the statistical rigor of a GWAS (Werling et al. 2018). Under the CWAS framework, thousands of categories are defined by combinations of groups of annotations (Fig. 3); for example, a category might consider variants defined by H3K27ac epigenetic markers in the untranslated regions (UTRs) of ASD-associated genes. Each category is tested for enrichment of de novo variants in ASD cases versus controls and the results are displayed as a volcano plot (Fig. 3). For a category to be associated with ASD, it must reach a p-value threshold defined by the number of effective tests across all the categories (Werling et al. 2018). Applying this CWAS approach to 519 ASD cases and 519 unaffected sibling controls from the SSC without prior disruptive variants identified by WES or microarray, no noncoding annotation category met the threshold of significance (Werling et al. 2018). This result suggests that with current annotations no noncoding category has the equivalent frequency and relative risk of de novo PTVs, although a category equivalent to de novo missense variants remains possible. A similarly negative result was observed considering inherited heterozygous and homozygous variants and considering CNVs across all modes of inheritance.
Figure 3.
Category-wide association study framework. (A) Variants identified by whole-genome sequencing (WGS) are annotated against several annotation groups, for example, location relative to genes (e.g., untranslated regions [UTRs], promoters, introns), gene lists (e.g., autism spectrum disorder [ASD]-associated genes, synaptic genes, fragile X mental retardation protein [FMRP] targets), conservation (e.g., PhyloP, PhastCons), and epigenetic markers (e.g., H3K27ac, DNase, transcription factor binding sites). Annotation categories are defined by looking at the combination of one annotation selected from each annotation group, for example, de novo mutations marked by H3K27ac in the UTRs of ASD-associated genes. The total number of categories will be equivalent to the product of the number of types of annotation in each of the annotation groups. (B) Each of the annotation categories is assessed for evidence of association with ASD by comparing the number of de novo mutations in cases versus controls (or vs. null expectation based on a mutation model [Lek et al. 2016]). Association can be tested using a binomial exact test, linear regression, or permutation analysis. The impact of paternal age should be corrected. The results are plotted on a volcano plot, with relative risk in cases versus controls on the x-axis, –log10(P) on the y-axis, and each point representing an annotation category from A. An annotation category is considered significant if it exceeds the multiple comparison threshold (red), defined by looking for the number of effective tests based on simulated data (Werling et al. 2018). Conceptually, this is similar to a Manhattan plot for genome-wide association studies (GWAS). (C) Although individual categories may not reach the significance threshold, groups of categories may carry signal (i.e., more points to the right than the left of the midline in B). To identify an underlying association across multiple categories, a rare variant risk score based on a Lasso regression model is generated from a training cohort. The score is created from categories with few variants, based on the logic that highly refined categories will carry the strongest signals. The performance of the model is tested on an independent testing cohort to assess if it is a significant predictor (p-value) and the relative accuracy of the prediction (R2). If the model is significant, the categories in the model can be assessed to evaluate which annotation terms account for the prediction. Conceptually, this is similar to a polygenic risk score in a GWAS.
An alternative approach to identifying ASD-associated noncoding variants is to predict the category of noncoding variants that is most likely to mediate risk from available knowledge of the genome (Turner et al. 2016, 2017; Yuen et al. 2016; Brandler et al. 2018). This approach may suffer from the same issues as candidate gene studies, specifically a lack of biological knowledge to make accurate predictions, small samples sizes, and publication bias. Of note, the category of noncoding risk selection differs in each study, suggesting a lack of consensus on where the strongest noncoding association would be expected. What is clear is that larger cohorts of WGS data will be needed to define the noncoding contribution of de novo variants in ASD; a similar conclusion was reached in an analysis of de novo variants in conserved enhancers in developmental delay (Short et al. 2018).
CONCLUDING REMARKS
Next-generation sequencing has had a revolutionary impact in identifying high-impact genetic loci that contribute to ASD, especially through the clustering of de novo PTVs in specific genes (Fig. 2). The genes identified perform one of two broad roles, regulation of the expression of other genes or neuronal communication. This progress in gene discovery, alongside NGS-derived functional data, is already beginning to provide insights into the neurobiology of ASD, implicating midfetal development, excitatory cortical neurons, and medium spiny neurons in the striatum. Despite this progress, a coherent model of ASD pathogenesis and effective therapies remain elusive, prompting the need for further research to follow up on the leads NGS methods have been critical in finding.
ACKNOWLEDGMENTS
The research described would not be possible without the contribution of families affected by autism spectrum disorder. This work was supported by grants from the Simons Foundation for Autism Research Initiative (SFARI 402281 to S.J.S.) and the National Institutes of Health (NIH) (R01MH110928 to S.J.S.). My thanks to Kathryn Roeder and Bernie Devlin for discussions related to the figures in this manuscript.
Footnotes
Editors: W. Richard McCombie, Elaine R. Mardis, James A. Knowles, and John D. McPherson
Additional Perspectives on Next-Generation Sequencing in Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Aldahmesh MA, Mohamed JY, Alkuraya HS, Verma IC, Puri RD, Alaiya AA, Rizzo WB, Alkuraya FS. 2011. Recessive mutations in ELOVL4 cause ichthyosis, intellectual disability, and spastic quadriplegia. Am J Hum Genet 89: 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, Kurzius-Spencer M, Zahorodny W, Robinson Rosenberg C, White T, et al. 2018. Prevalence of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 sites, United States, 2014. MMWR Surveill Summ 67: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shalom R, Keeshen CM, Berrios KN, An JY, Sanders SJ, Bender KJ. 2017. Opposing effects on NaV1.2 function underlie differences between SCN2A variants observed in individuals with autism spectrum disorder or infantile seizures. Biol Psychiatry 82: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis EM, Lindemann L, Jonch AE, Apostol G, Bear MF, Carpenter RL, Crawley JN, Curie A, Des Portes V, Hossain F, et al. 2018. Drug development for neurodevelopmental disorders: Lessons learned from fragile X syndrome. Nat Rev Drug Discov 17: 280–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandler WMM, Antaki D, Gujral M, Noor A, Rosanio G, Chapman TRR, Barrera DJJ, Lin GNN, Malhotra D, Watts ACC, et al. 2016. Frequency and complexity of de novo structural mutation in autism. Am J Hum Genet 98: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandler WM, Antaki D, Gujral M, Kleiber ML, Whitney J, Maile MS, Hong O, Chapman TR, Tan S, Tandon P, et al. 2018. Paternally inherited cis-regulatory structural variants are associated with autism. Science 360: 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. 2015. Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci 18: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Redin CE, Hanscom C, Antolik C, Stone MR, Glessner JT, Mason T, Pregno G, Dorrani N, et al. 2017. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 18: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, Lord C, Courchesne E. 1997. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet 60: 928–934. [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Muhle RA, Sanders SJ, Liu L, Willsey AJ, Niu W, Liu W, Klei L, Lei J, Yin J, et al. 2015. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun 6: 6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg APAAP, Poultney CSCCS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. 2014. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries PJ, Hunt A, Bolton PF. 2007. The psychopathologies of children and adolescents with tuberous sclerosis complex (TSC): A postal survey of UK families. Eur Child Adolesc Psychiatry 16: 16–24. [DOI] [PubMed] [Google Scholar]
- D’Gama AM, Pochareddy S, Li M, Jamuar SS, Reiff RE, Lam ATTN, Sestan N, Walsh CA. 2015. Targeted DNA sequencing from autism spectrum disorder brains implicates multiple genetic mechanisms. Neuron 88: 910–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Walker MF, Carriero NJ, DiCola M, Willsey AJ, Ye AY, Waqar Z, Gonzalez LE, Overton JD, Frahm S, et al. 2014. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep 9: 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Yang X, Li Z, Wang S, Zhang Z, Ye AY, Yan L, Yang C, Wu Q, Li J, et al. 2017. Postzygotic single-nucleotide mosaicisms contribute to the etiology of autism spectrum disorder and autistic traits and the origin of mutations. Hum Mutat 38: 1002–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SE, Panitch R, West AB, Arking DE. 2016. Transcriptome analysis of cortical tissue reveals shared sets of downregulated genes in autism and schizophrenia. Transl Psychiatry 6: e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. 2006. Structural variation in the human genome. Nat Rev Genet 7: 85–97. [DOI] [PubMed] [Google Scholar]
- Fischbach GD, Lord C. 2010. The Simons Simplex Collection: A resource for identification of autism genetic risk factors. Neuron 68: 192–195. [DOI] [PubMed] [Google Scholar]
- Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM, et al. 2018. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359: 693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Nonell C, Ratera ER, Harris S, Hessl D, Ono MY, Tartaglia N, Marvin E, Tassone F, Hagerman RJ. 2008. Secondary medical diagnosis in fragile X syndrome with and without autism spectrum disorder. Am J Med Genet A 146A: 1911–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisheker MR, Heymann G, Wang T, Coe BP, Turner TN, Stessman HAF, Hoekzema K, Kvarnung M, Shaw M, Friend K, et al. 2017. Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat Neurosci 20: 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillberg C, Wahlström J. 1985. Chromosome abnormalities in infantile autism and other childhood psychoses: A population study of 66 cases. Dev Med Child Neurol 27: 293–304. [DOI] [PubMed] [Google Scholar]
- Gilman SRR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. 2011. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 70: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. 1947. The mutation rate of the gene for haemophilia, and its segregation ratios in males and females. Ann Eugen 13: 262–271. [DOI] [PubMed] [Google Scholar]
- Halladay AK, Bishop S, Constantino JN, Daniels AM, Koenig K, Palmer K, Messinger D, Pelphrey K, Sanders SJ, Singer AT, et al. 2015. Sex and gender differences in autism spectrum disorder: Summarizing evidence gaps and identifying emerging areas of priority. Mol Autism 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, Schellenberg GD, Gibbs RA, Daly MJ, Buxbaum JD, et al. 2013. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet 9: e1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyne HO, Singh T, Stamberger H, Abou Jamra R, Caglayan H, Craiu D, De Jonghe P, Guerrini R, Helbig KL, Koeleman BPC, et al. 2018. De novo variants in neurodevelopmental disorders with epilepsy. Nat Genet 50: 1048–1053. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. 2004. Detection of large-scale variation in the human genome. Nat Genet 36: 949–951. [DOI] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. 2012. De novo gene disruptions in children on the autistic spectrum. Neuron 74: 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. 2014. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, et al. 2003. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet 34: 27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, Depalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al. 2017. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet 49: 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Border R, Melroy-Greif WE, de Leeuw CA, Ehringer MA, Keller MC. 2017. No evidence that schizophrenia candidate genes are more associated with schizophrenia than noncandidate genes. Biol Psychiatry 82: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, Hardarson MT, Hjorleifsson KE, Eggertsson HP, Gudjonsson SA, et al. 2017. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549: 519–522. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature 478: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, Suyama M, Takumi T, Miyakawa T, Nakayama KI. 2016. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature 537: 675–679. [DOI] [PubMed] [Google Scholar]
- Kielinen M, Rantala H, Timonen E, Linna SL, Moilanen I. 2004. Associated medical disorders and disabilities in children with autistic disorder: A population-based study. Autism 8: 49–60. [DOI] [PubMed] [Google Scholar]
- Kim YS, Leventhal BL, Koh YJ, Fombonne E, Laska E, Lim EC, Cheon KA, Kim SJ, Kim YK, Lee H, et al. 2011. Prevalence of autism spectrum disorders in a total population sample. Am J Psychiatry 168: 904–912. [DOI] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir AA, Jonasdottir AA, et al. 2012. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantareas MM, Homatidis S. 1999. Chromosomal abnormalities in a series of children with autistic disorder. J Autism Dev Disord 29: 275–285. [DOI] [PubMed] [Google Scholar]
- Kosmicki JA, Samocha KE, Howrigan DP, Sanders SJ, Slowikowski K, Lek M, Karczewski KJ, Cutler DJ, Devlin B, Roeder K, et al. 2017. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet 49: 504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp DR, Barnard RA, Duffourd Y, Evans SA, Mulqueen RM, Bernier R, Riviere JB, Fombonne E, O’Roak BJ. 2017. Exonic mosaic mutations contribute risk for autism spectrum disorder. Am J Hum Genet 101: 369–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Dobyns WB, et al. 2008. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet 17: 628–638. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MPP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. 2004. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet 74: 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritsen M, Mors O, Mortensen PB, Ewald H. 1999. Infantile autism and associated autosomal chromosome abnormalities: A register-based study and a literature survey. J Child Psychol Psychiatry 40: 335–345. [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SY, Chen YC, Lai TJ, Hsu CY, Wang YC. 1993. Molecular and cytogenetic analyses of autism in Taiwan. Hum Genet 92: 441–445. [DOI] [PubMed] [Google Scholar]
- Lim ETT, Raychaudhuri S, Sanders SJJ, Stevens C, Sabo A, MacArthur DGG, Neale BMM, Kirby A, Ruderfer DMM, Fromer M, et al. 2013. Rare complete knockouts in humans: Population distribution and significant role in autism spectrum disorders. Neuron 77: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ET, Uddin M, De Rubeis S, Chan Y, Kamumbu AS, Zhang X, D’Gama AM, Kim SN, Hill RS, Goldberg AP, et al. 2017. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat Neurosci 20: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Sanders SJ, Tian Y, Voineagu I, Huang N, Chu SH, Klei L, Cai C, Ou J, Lowe JK, et al. 2012. Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. Am J Hum Genet 91: 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2010. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci 107: 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. 2008. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 82: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae JF, Clayton S, Fitzgerald TW, Kaplanis J, Prigmore E, Rajan D, Sifrim A, Aitken S, Akawi N, Alvi M, et al. 2017. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542: 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A, et al. 2012. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CFF, Stevens C, Wang LSS, Makarov V, et al. 2012. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485: 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, El-Fishawy P, Kayserili H, Meguid NA, Scott EM, Schroth J, Silhavy JL, Kara M, Khalil RO, Ben-Omran T, et al. 2012. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science 338: 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara I, Ito K, Sawaishi Y, Osaka H, Mazaki E, Inoue I, Montal M, Hashikawa T, Shike T, Fujiwara T, et al. 2009. De novo mutations of voltage-gated sodium channel αII gene SCN2A in intractable epilepsies. Neurology 73: 1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. 2011. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet 43: 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. 2012a. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. 2012b. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. 2013. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155: 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta LDLT, Huang J, et al. 2016. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540: 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR. 2010. Williams–Beuren syndrome. N Engl J Med 362: 239–252. [DOI] [PubMed] [Google Scholar]
- Power RA, Kyaga S, Uher R, MacCabe JH, Långström N, Landen M, McGuffin P, Lewis CM, Lichtenstein P, Svensson AC. 2013. Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse vs their unaffected siblings. JAMA Psychiatry 70: 22–30. [DOI] [PubMed] [Google Scholar]
- Rahbari R, Wuster A, Lindsay SJ, Hardwick RJ, Alexandrov LB, Al Turki S, Dominiczak A, Morris A, Porteous D, Smith B, et al. 2016. Timing, rates and spectra of human germline mutation. Nat Genet 48: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KS. 2005. Cytogenetic abnormalities and fragile-X syndrome in autism spectrum disorder. BMC Med Genet 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. 1996. The future of genetic studies of complex human diseases. Science 273: 1516–1517. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Mason-Brothers A, Freeman BJ, Pingree C, Jenson WR, McMahon WM, Petersen PB, Jorde LB, Mo A, Ritvo A. 1990. The UCLA–University of Utah epidemiologic survey of autism: The etiologic role of rare diseases. Am J Psychiatry 147: 1614–1621. [DOI] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, et al. 2014. A framework for the interpretation of de novo mutation in human disease. Nat Genet 46: 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ. 2015. First glimpses of the neurobiology of autism spectrum disorder. Curr Opin Genet Dev 33: 80–92. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. 2011. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70: 863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. 2012. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Devlin B, Roeder K, State MW, Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, et al. 2015. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87: 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Neale BM, Huang H, Werling DM, An JYY, Dong S, Abecasis G, Arguello PA, Blangero J, Boehnke M, et al. 2017. Whole genome sequencing in psychiatric disorders: The WGSPD consortium. Nat Neurosci 20: 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A. 2017. The heritability of autism spectrum disorder. JAMA 318: 1182–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. 2007. Strong association of de novo copy number mutations with autism. Science 316: 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Dies KA, Holm IA, Bridgemohan C, Sobeih MM, Caronna EB, Miller KJ, Frazier JA, Silverstein I, Picker J, et al. 2010. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics 125: e727–e735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short PJ, McRae JF, Gallone G, Sifrim A, Won H, Geschwind DH, Wright CF, Firth HV, FitzPatrick DR, Barrett JC, et al. 2018. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature 555: 611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirmaci A, Spiliopoulos M, Brancati F, Powell E, Duman D, Abrams A, Bademci G, Agolini E, Guo S, Konuk B, et al. 2011. Mutations in ANKRD11 cause KBG syndrome, characterized by intellectual disability, skeletal malformations, and macrodontia. Am J Hum Genet 89: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HAF, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, Kvarnung M, Gerdts J, Trinh S, Cosemans N, et al. 2017. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet 49: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugathan A, Biagioli M, Golzio C, Erdin S, Blumenthal I, Manavalan P, Ragavendran A, Brand H, Lucente D, Miles J, et al. 2014. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci 111: E4468–E4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tick B, Bolton P, Happe F, Rutter M, Rijsdijk FF, Happé F, Rutter M, Rijsdijk FF. 2016. Heritability of autism spectrum disorders: A meta-analysis of twin studies. J Child Psychol Psychiatry 57: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TNN, Hormozdiari F, Duyzend MHH, McClymont SAA, Hook PWW, Iossifov I, Raja A, Baker C, Hoekzema K, Stessman HAA, et al. 2016. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am J Hum Genet 98: 58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TN, Coe BP, Dickel DE, Hoekzema K, Nelson BJ, Zody MC, Kronenberg ZN, Hormozdiari F, Raja A, Pennacchio LA, et al. 2017. Genomic patterns of de novo mutation in simplex autism. Cell 171: 710–722.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP. 1991. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65: 905–914. [DOI] [PubMed] [Google Scholar]
- Wang K, Gaitsch H, Poon H, Cox NJ, Rzhetsky A. 2017. Classification of common human diseases derived from shared genetic and environmental determinants. Nat Genet 49: 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassink TH, Piven J, Patil SR. 2001. Chromosomal abnormalities in a clinic sample of individuals with autistic disorder. Psychiatr Genet 11: 57–63. [DOI] [PubMed] [Google Scholar]
- Weidmer-Mikhail E, Sheldon S, Ghaziuddin M. 1998. Chromosomes in autism and related pervasive developmental disorders: A cytogenetic study. J Intellect Disabil Res 42: 8–12. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. 2008. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358: 667–675. [DOI] [PubMed] [Google Scholar]
- Werling DM, Brand H, An JY, Stone MR, Zhu L, Glessner JT, Collins RL, Dong S, Layer RM, Markenscoff-Papadimitriou E, et al. 2018. An analytical framework for whole genome sequence association studies and its implications for autism spectrum disorder. Nat Genet 50: 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PG, Hersh JH. 1998. Brief report: The association of neurofibromatosis type 1 and autism. J Autism Dev Disord 28: 567–571. [DOI] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. 2013. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Boehnke M. 2009. Quantifying and correcting for the winner’s curse in genetic association studies. Genet Epidemiol 33: 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. 2014. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci 34: 1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TWW, Chahrour MHH, Coulter MEE, Jiralerspong S, Okamura-Ikeda K, Ataman B, Schmitz-Abe K, Harmin DAA, Adli M, Malik ANN, et al. 2013. Using whole-exome sequencing to identify inherited causes of autism. Neuron 77: 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Merico D, Cao H, Pellecchia G, Alipanahi B, Thiruvahindrapuram B, Tong X, Sun Y, Cao D, Zhang T, et al. 2016. Genome-wide characteristics of de novo mutations in autism. NPJ Genom Med 1: 160271–1602710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RKC, Merico D, Bookman M, L Howe J, Thiruvahindrapuram B, Patel RV, Whitney J, Deflaux N, Bingham J, Wang Z, et al. 2017. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 20: 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]