SUMMARY
RNA molecules perform diverse functions in mammalian cells, including transferring genetic information from DNA to protein and playing diverse regulatory roles through interactions with other cellular components. Here, we discuss how clustered regularly interspaced short palindromic repeat (CRISPR)-based technologies for directed perturbations of DNA and RNA are revealing new insights into RNA regulation. First, we review the fundamentals of CRISPR–Cas enzymes and functional genomics tools that leverage these systems. Second, we explore how these new perturbation technologies are transforming the study of regulation of and by RNA, focusing on the functions of DNA regulatory elements and long noncoding RNAs (lncRNAs). Third, we highlight an emerging class of RNA-targeting CRISPR–Cas enzymes that have the potential to catalyze studies of RNA biology by providing tools to directly perturb or measure RNA modifications and functions. Together, these tools enable systematic studies of RNA function and regulation in mammalian cells.
1. INTRODUCTION
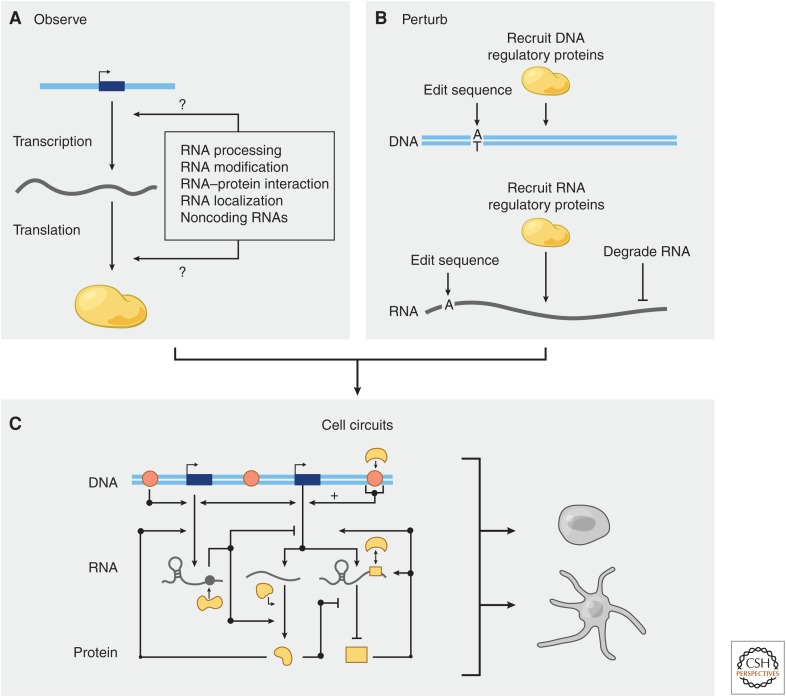
The transfer of genetic information from DNA to protein by RNA molecules is controlled by a number of regulatory processes (Fig. 1A). These include DNA sequence elements and DNA-binding proteins that control RNA production; RNA sequence elements and RNA-binding proteins that control RNA processing, degradation, localization, and translation; and small and long noncoding RNAs (lncRNAs) that guide and integrate the functions of protein effectors.
Figure 1.
Systematic perturbations to understand RNA regulation. (A) The transfer of genetic information from DNA to RNA to protein is controlled by many molecular processes that regulate RNA. New sequencing technologies have allowed us to map some of these processes at genome-wide scale, but functional characterization has lagged. (B) Clustered regularly interspaced short palindromic repeat (CRISPR) perturbations at the level of DNA or RNA will help to test the functions of RNA regulatory mechanisms. (C) Combining technologies for observation and perturbation will allow us to build models of cell circuits and understand how different components (DNA and RNA regulatory elements, DNA- and RNA-binding proteins, and posttranscriptional modifications) are integrated to control cellular functions.
In the past decade, many advances in our understanding of RNA biology were facilitated by genome-scale observational studies, including the application of sequencing-based technologies to map the locations of RNA transcription, modifications, and RNA-binding proteins (Li et al. 2016; Hentze et al. 2018). The pace at which these observations were made far outstripped the pace at which they could be functionally characterized because there were no comparably powerful technologies for RNA perturbation. As a result, the precise functions and mechanisms of most of these observed sequences, interactions, and modifications remain unknown.
The recent development of DNA and RNA perturbation tools based on clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) enzymes has opened new opportunities for efficiently manipulating specific molecular features to test their functions (Fig. 1B) (Doudna and Charpentier 2014; Hsu et al. 2014). Just as the recombinant DNA revolution in the 1970s unleashed a wave of new biotechnologies and insights into disease biology by giving us the ability to clone and manipulate human genes in plasmids, CRISPR-based tools have ushered in a new era of studies to understand human genes and gene regulation via editing of DNA and RNA directly in mammalian cells. Here, we explore how this emerging suite of CRISPR tools will enable us to dissect the regulation of and by RNA and determine how these regulatory circuits influence cellular functions (Fig. 1C).
2. A CRISPR–CAS PLATFORM FOR GENOME EDITING AND FUNCTIONAL GENOMICS
Components from CRISPR–Cas systems can be engineered and optimized to enable programmable targeting of specific DNA or RNA sequences. Typically, a Cas protein effector (e.g., Cas9, Cas12a, or Cas13) is combined with a customized guide RNA to establish a ribonucleoprotein complex capable of targeting DNA or RNA in a cell type of interest (Gasiunas et al. 2012; Jinek et al. 2012; Cong et al. 2013; Mali et al. 2013b; Zetsche et al. 2015; Abudayyeh et al. 2016; Smargon et al. 2017). In contrast to earlier DNA-editing enzymes (Kim and Kim 2014)—such as meganucleases, zinc finger nucleases, and transcription activator–like effector nucleases—CRISPR–Cas systems are targeted by Watson–Crick base-pairing between the guide RNA and target DNA or RNA, allowing rapid and flexible reprogramming by adjusting the sequence of the guide RNA. By modifying the Cas protein or RNA guide, the system can be used to recruit other molecules to act on the target sequence in different ways. In combination with appropriate cellular or molecular assays, this flexibility enables systematic forward and reverse genetic studies in mammalian cells.
In this section, we review key aspects of CRISPR biology and ongoing technology development to expand this platform, including (1) exploring the natural diversity of CRISPR systems to identify a flexible suite of targeting enzymes, (2) using CRISPR–Cas proteins to edit DNA sequence in precise ways or perturb specific regulatory processes, and (3) combining these tools with pooled assays to connect genetic perturbations with specific cellular or molecular phenotypes.
2.1. Properties of CRISPR–Cas Systems
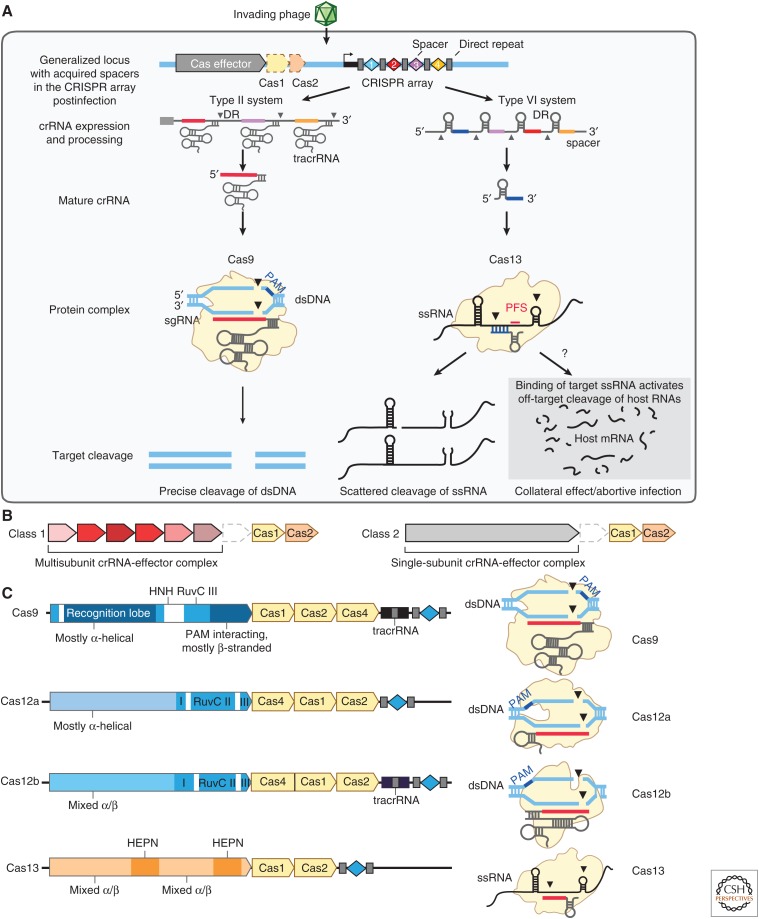
CRISPR and CRISPR-associated (CRISPR–Cas) proteins in bacteria and archaea constitute adaptive immune systems that capture fragments of genetic material from invading phages or mobile genetic elements and use these fragments to generate CRISPR RNAs (crRNAs) that guide the cleavage of matching viral sequences in future infections (Marraffini 2015). CRISPR–Cas systems display diverse sequence characteristics and architectural organization, providing a range of features that have utility in genome-editing applications (Fig. 2A). Here, we highlight the properties of a few of the most studied systems (Fig. 2B).
Figure 2.
Structure and function of CRISPR systems. (A) Depiction of the three steps of CRISPR immunity (adaptation, expression, and interference) by DNA-targeting (e.g., Cas9-containing type II) and RNA-targeting (e.g., Cas13-containing type VI) systems. Cas9 interference is mediated by RNA-guided cleavage of double-stranded DNA (dsDNA) targets. Cas13 interference results in two modes of activation in which RNA-guided targeting of single-stranded RNAs (ssRNAs) results in targeted cleavage of the transcripts as well as collateral cleavage of noncomplementary RNAs that are in proximity to the activated Cas13 complex. The collateral cleavage of Cas13 systems is thought to be part of a programmed cell death pathway to further augment immunity against pathogen infection. (B) CRISPR systems can be divided into two classes: Class 1 systems consist of multiprotein interference complexes (types I, III, and IV systems), whereas Class 2 systems contain a single-protein interference effector (types II, V, and VI systems). (C) Genetic loci for four Class 2 CRISPR systems (left), including sequences coding for effector proteins annotated with key protein domains, accessory Cas proteins involved in adaptation colored in beige, trans-activating CRISPR RNAs (tracrRNAs) (if required), and CRISPR array. The protein complexes (right) show the nucleic acid target (dsDNA or ssRNA), locations of the protospacer adjacent motif (PAM) sequence and cleavage sites, and approximate structure of the guide RNA.
2.1.1. Cas9
The most characterized single-protein Cas effector (Class 2) is Cas9. Cas9 is a dual-guide RNA-dependent endonuclease that contains two nuclease domains, RuvC and HNH, that coordinate to cleave both strands of a double-stranded DNA (dsDNA) target complementary to the crRNA spacer sequence (Doudna and Charpentier 2014; Hsu et al. 2014). Cas9 can be engineered to facilitate genome editing in eukaryotic cells (Cong et al. 2013; Mali et al. 2013b). The dual-guide RNAs, crRNA and its trans-activating crRNA (tracrRNA) (Deltcheva et al. 2011), can be covalently linked to form a single-guide RNA (sgRNA) to simplify the RNA components (Jinek et al. 2012). Targeting is dependent on a protospacer adjacent motif (PAM) sequence (e.g., Cas9 from Streptococcus pyogenes requires a 5′-NGG-3′ PAM at the 3′ end of the target sequence), restricting editing to genomic sites that contain the appropriate PAM sequence.
2.1.2. Cas12
CRISPR–Cas12a systems (originally called Cpf1) also target DNA using a single-protein endonuclease and can be engineered to facilitate genome editing in eukaryotic cells (Zetsche et al. 2015). Cas12a cleaves dsDNA using two domains, the RuvC and nuclease domains, although the RuvC domain only contains the catalytic residues necessary to cleave both strands and generate a dsDNA break (Zetsche et al. 2015; Yamano et al. 2016; Swarts et al. 2017). In contrast to Cas9, Cas12a does not require a tracrRNA (Zetsche et al. 2015) and possesses its own RNase domain capable of cleaving a pre-crRNA array into individual mature crRNAs for convenient delivery of multiple guide RNAs (Fonfara et al. 2016; Zetsche et al. 2017). Cas12a-mediated dsDNA cleavage results in 5′ staggered ends (Zetsche et al. 2015), and the PAM sequence and location differ from those used by Cas9, with most Cas12a orthologs requiring a 5′-TTTV-3′ PAM at the 5′ end of the target site and cleaving distal to the PAM (Zetsche et al. 2015).
2.1.3. Cas13
The recent discovery of single-effector CRISPR systems that target RNA (Shmakov et al. 2015, 2017) has further expanded our conception of the diversity of these bacterial enzymes. Members of the Cas13 family contain two domains with homology to higher eukaryotes and prokaryotes nucleotide-binding (HEPN) RNase domains (Anantharaman et al. 2013). Cas13a, like DNA-targeting CRISPR systems, is guided to target RNA sequences by a complementary crRNA, and hybridization between the target RNA and its complementary crRNA leads to cleavage of the target RNA at multiple sites within single-stranded regions (Abudayyeh et al. 2016). Continued protein discovery has since uncovered many Cas13 subfamilies (Cas13a, Cas13b, Cas13c, and Cas13d) (Shmakov et al. 2015, 2017; Abudayyeh et al. 2016; East-Seletsky et al. 2016, 2017; Smargon et al. 2017; Konermann et al. 2018; Yan et al. 2018). In bacteria, both Cas13a and Cas13b have a sequence constraint known as the protospacer flanking site (PFS) (Abudayyeh et al. 2016; Smargon et al. 2017). In bacteria, Cas13a from Leptotrichia shahii has a 3′H (not G) requirement (Abudayyeh et al. 2016); in mammalian cells it appears that the PFS does not affect targeting (Abudayyeh et al. 2017; Cox et al. 2017). An additional unique feature of these enzymes observed in vitro is that, upon target RNA binding, the activated Cas13 complex is capable of cleaving both the targeted transcript and other nearby noncomplementary RNAs via a “collateral” activity (Abudayyeh et al. 2016), which has been harnessed for nucleic acid detection applications (East-Seletsky et al. 2016, 2017; Gootenberg et al. 2017, 2018).
These examples illustrate the value of continued exploration of the diversity of CRISPR systems for the development of molecular tools. Many CRISPR systems remain unexplored and may harbor novel combinations of sequence flexibility, endonuclease capabilities, or other unexpected functions to facilitate genome-engineering applications.
2.2. CRISPR-Mediated Recruitment of Proteins and RNAs to Specific Genomic Sites
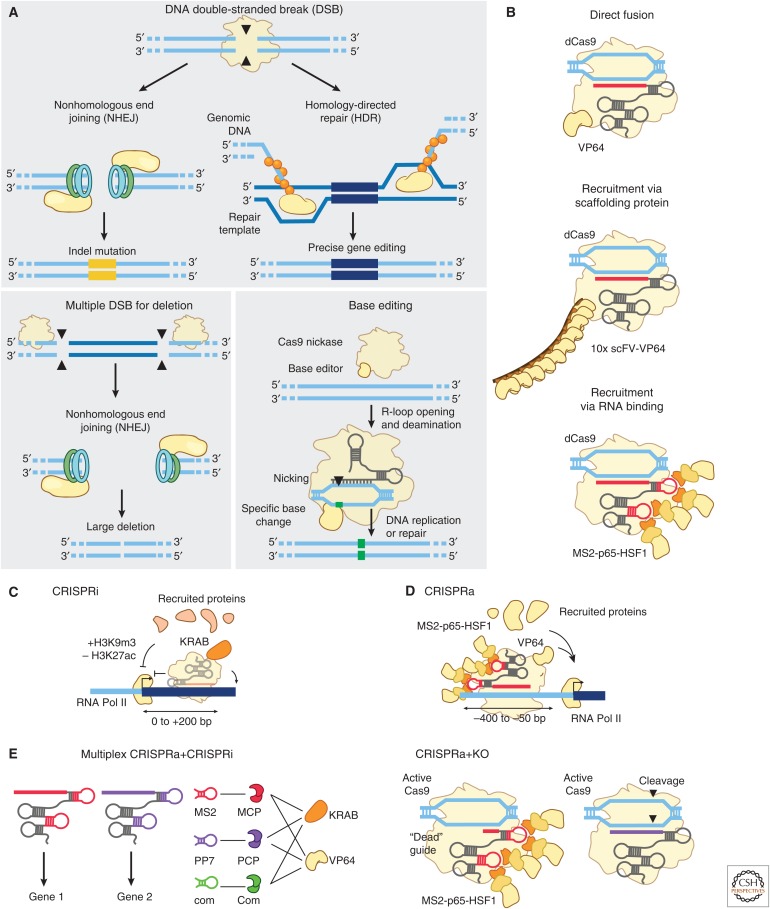
CRISPR–Cas tools have transformed studies of molecular biology and genetics by enabling direct, easily reprogrammable genome editing in mammalian cells via endogenous repair of targeted double strand breaks (Doudna and Charpentier 2014; Hsu et al. 2014; Komor et al. 2017). Edits can include creation of small indels (using one guide RNA), precise gene editing (with one guide RNA and a repair template), or larger deletions (using two guide RNAs) (Fig. 3A). Beyond cleaving genomic sequences, CRISPR effectors can be used to direct protein or RNA cargo to specific locations in the genome. In such applications, functional proteins or RNAs are adjoined either to catalytically dead Cas9 (dCas9) (Gilbert et al. 2013; Mali et al. 2013a; Qi et al. 2013), in which the active sites of the nuclease domains have been mutated, or to the guide RNA via a number of different recruitment strategies (Fig. 3B). Emerging tools in this category include a variety of base editors, which precisely convert one nucleotide to another, and “epigenome editors,” which recruit one or more proteins to alter gene regulatory processes.
Figure 3.
DNA-targeting CRISPR tools enable diverse types of perturbations. (A) CRISPR tools allow for multiple methods of DNA editing. Double-stranded breaks (DSBs) engage DNA-damage responses that can lead to either indels and gene knockout through nonhomologous end joining (NHEJ) or specific gene replacement through homology-directed repair (HDR). (Adapted from Hsu et al. 2014.) Two targeted cuts can also lead to removal of large stretches of DNA. Catalytically inactive or nicking mutants of Cas9 can be used to recruit nucleoside deaminases or other base-editing enzymes for precise, single-nucleotide edits. As an example, this figure shows a C:G to T:A editor comprised of Cas9 nickase fused to cytidine deaminase. (B) Cas proteins can recruit other domains or effector proteins through multiple designs, including direct fusion to the Cas protein, indirect recruitment via a scaffolding or conditional dimerization domain fused to the Cas protein, and/or recruitment using a guide RNA modified with a sequence motif recognized by an exogenous RNA-binding protein. (C) In CRISPR interference (CRISPRi), recruitment of repressive chromatin regulators, such as Krüppel-associated box (KRAB), result in assembly of repressive complexes and silencing of active regulatory elements including promoters and enhancers. (D) In CRISPR activation (CRISPRa), recruitment of activating chromatin regulators to promoters or other cis-acting elements can promote gene expression. Recruitment of multiple different transactivators, such as combinations of VP64, p65, and HSF1, can lead to increased gene expression. (E) These tools can be multiplexed for simultaneous and differential perturbation of genes. Activation and inhibition can be targeted to different genes by guide RNAs containing orthogonal RNA binding domains and associated regulatory proteins, or Cas9 cleavage activity can be tuned on a per-guide basis by adjusting the length of the spacer on the guide RNA.
2.2.1. Base Editing to Modify Specific Nucleotides
CRISPR-mediated base editing is performed by fusing certain enzymes—such as APOBEC, AID, or ADAT engineered to target DNA—to a Cas9 protein in which one or both catalytic domains have been genetically inactivated (Fig. 3A) (Komor et al. 2016; Nishida et al. 2016; Gaudelli et al. 2017). These fusions currently enable specific and programmable conversions of C:G to T:A base pairs or vice versa, enabling relatively efficient modification of just over half of all known pathogenic single-nucleotide variants annotated in ClinVar (Gaudelli et al. 2017), although the requirement for a PAM sequence nearby reduces this number. An advantage of these approaches is that they can edit DNA without introducing double-stranded DNA breaks, thereby avoiding certain error-prone repair mechanisms. For example, the APOBEC C-to-T base editor first enzymatically converts a target cytosine to uracil, and then creates a single-strand DNA break to engage the mismatch repair (MMR) machinery to repair the opposing guanine to adenine (Komor et al. 2016).
2.2.2. CRISPR Interference (CRISPRi) for Gene Repression
By itself, dCas9 delivery to gene promoters can physically block RNA polymerase or transcription factors from accessing DNA, leading to modest (20%–40%) repression of endogenous gene expression (Gilbert et al. 2013). Repressive activity can be dramatically improved by fusing dCas9 to repressive chromatin regulators (Gilbert et al. 2013). For example, fusion to the KRAB domain leads to deposition of repressive histone modifications (including H3K9me3) and loss of activating modifications (including H3K27ac) at gene promoters, leading to 50%–90% repression of gene expression when delivered in a critical window 0–200 bp downstream from a transcription start site (Fig. 3C) (Gilbert et al. 2013, 2014). Other repressive domains—including SID, LSD1, and DNMT3A proteins or protein domains (Konermann et al. 2013; Kearns et al. 2015; Bintu et al. 2016; Liu et al. 2016; Vojta et al. 2016)—have been explored and may have different kinetic properties for fine-tuning gene silencing.
2.2.3. CRISPR Activation (CRISPRa)
CRISPR–Cas9 tools for activation use Cas9 to recruit multiple transcriptional activators to achieve robust induction of gene expression at promoters (Bikard et al. 2013; Cheng et al. 2013; Farzadfard et al. 2013; Gilbert et al. 2013, 2014; Maeder et al. 2013; Mali et al. 2013a; Perez-Pinera et al. 2013; Chavez et al. 2015; Konermann et al. 2015). Konermann and colleagues developed an approach called “synergistic activation mediator” (SAM), in which MS2 stem loops are fused to modified Cas9 guide RNAs to recruit multiple activating proteins to the same site (Fig. 3D) (Konermann et al. 2015). Recruiting multiple activating proteins (VP64, HSF1, and p65) led to synergistic effects on gene expression compared with recruiting each protein individually (Konermann et al. 2015). In an alternative approach (“SunTag”), Tanenbaum and colleagues fused dCas9 to a repetitive GCN4 peptide-domain scaffold that is recognized by a GCN4-targeting nanobody fused to VP64, enabling simultaneous recruitment of up to 10 copies of VP64 (Tanenbaum et al. 2014). Each of these approaches appears to vary in efficiency across different genes in a way that is not yet entirely predictable, and the precise location of targeting (ideally, 50–400 bp upstream of the transcription start site) has an important effect on the efficiency of activation (Gilbert et al. 2014; Konermann et al. 2015).
Building on these basic systems for CRISPR–Cas9 recruitment, new tools have now been developed to manipulate the localization of lncRNA molecules (CRISP-Disp) (Shechner et al. 2015), modify chromosome conformation (by linking multiple Cas9 molecules to dimerization domains) (Morgan et al. 2017), and visualize specific DNA loci in living cells (via Cas9 fusions to green fluorescent protein [GFP]) (Chen et al. 2013; Ma et al. 2016; Gu et al. 2018; Maass et al. 2018). Simultaneous activation and inhibition can be achieved at different targets in the same cell by, for example, using guide RNAs containing different RNA aptamers to recruit different protein effectors (Zalatan et al. 2015) or using variable guide spacer lengths to either enable or suppress catalytic Cas9 activity at different targets (Fig. 3E) (Dahlman et al. 2015; Kiani et al. 2015). Although the repertoire of CRISPR perturbations is expanding, much work remains to understand the specific regulatory processes perturbed by each approach and to determine how these tools operate in different genomic contexts.
2.3. CRISPR-Based Forward Genetics with Pooled Cellular Assays
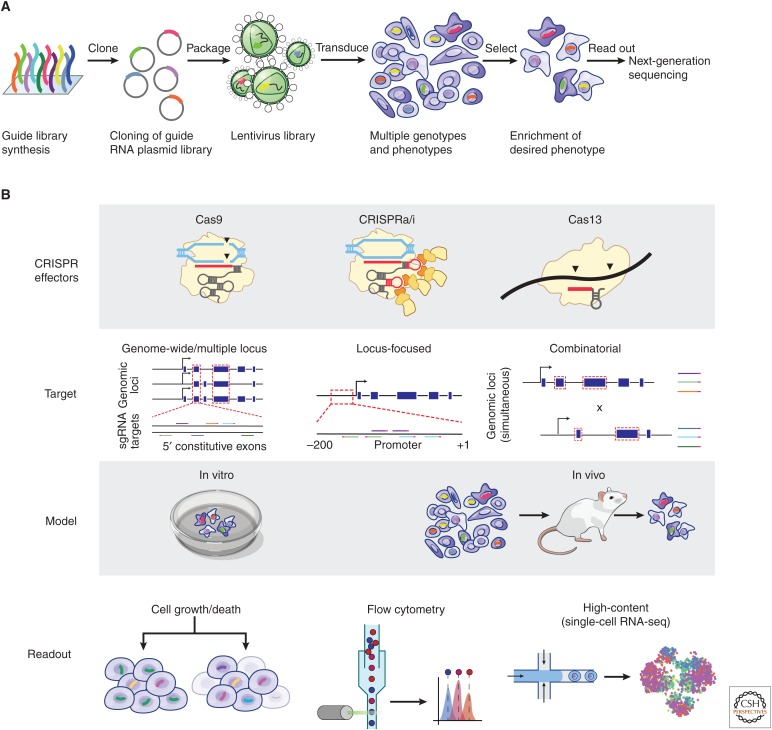
Paired with various CRISPR effectors, pooled cellular assays are a key part of the CRISPR functional genomics toolkit, enabling experiments that assay the effects of up to millions of perturbations simultaneously. Although pooled experiments were previously applied in combination with RNA interference (RNAi) (Brummelkamp et al. 2006; Ngo et al. 2006; Iorns et al. 2007), the diversity and flexibility of CRISPR systems have led to a new period of rapid innovation in the development of pooled assays for forward genetics (Shalem et al. 2014; Wang et al. 2014). In such assays, libraries of CRISPR sgRNAs are typically delivered by viral transduction such that individual cells in a population receive only one sgRNA (Fig. 4A). Sorting, selecting, or phenotyping single cells then allows associating specific genotypes (specified by a sgRNA perturbation) with phenotypes at high throughput (Fig. 4B) (Joung et al. 2017b).
Figure 4.
Forward genetics with pooled screening and CRISPR tools. (A) General workflow for pooled screening with CRISPR-based perturbations. A library of single-guide RNAs (sgRNAs) is synthesized, packaged into a viral vector, and delivered at a ratio of ∼1 sgRNA per cell. This pool of cells, each with a different genotype, is selected or sorted according to a phenotype of interest. Sequencing of the sgRNA in each cell is used to map the genotypes to the resulting phenotypes. (B) A pooled screen can be constructed from multiple different perturbations, distributions of targeting, model contexts, and phenotyping methods. Screens are amenable to many of the CRISPR-based tools, such as knockout, CRISPRa/i, base editing, or RNA-targeting tools. The targeting of guides in the screen can be distributed across the genome or transcriptome to test many different genes or focused at a specific locus to dissect regulatory elements. Multiple perturbations can also be delivered simultaneously to test epistatic mechanisms. Screens can be performed in vitro or in animal models for in vivo phenotypes. Phenotyping approaches can include enrichment or depletion caused by cell death or changes in proliferation, fluorescence-based sorting to measure gene expression or protein localization, or high-content methods such as single-cell RNA sequencing.
2.3.1. Cell Sorting and Guide Counting
Simple pooled assays involve selecting cells based on a single phenotype and counting the distribution of sgRNAs before and after the selection process using high-throughput DNA sequencing. For example, cell proliferation can serve as a selection parameter to identify genes that are essential for cell growth in culture, in vivo, in the presence of drugs, or in response to a viral infection (Shalem et al. 2014; Wang et al. 2014), and can be combined with genome-wide gene knockout or activation screens in both positive and negative selection screens (Doench et al. 2014; Gilbert et al. 2014; Koike-Yusa et al. 2014; Zhou et al. 2014; Wang et al. 2015). Cells can also be selected based on fluorescent sorting readouts or other properties, enabling forward genetic screens that examine the expression, localization or stability of specific genes (Canver et al. 2015; Parnas et al. 2015; Fulco et al. 2016; Sanjana et al. 2016; Wu et al. 2016; Stewart et al. 2017; Doench 2018).
2.3.2. High-Content Phenotyping
Recent approaches leverage new single-cell technologies to simultaneously associate CRISPR sgRNAs with multiple phenotypes. In Perturb-seq, single-cell RNA sequencing is used to assay the transcriptional state of the cell as well as determine which sgRNA is expressed in that cell, allowing studies to perturb multiple genes and characterize their effects on cells in an unbiased way that does not require designing specific assays to read out specific phenotypes (Adamson et al. 2016; Dixit et al. 2016; Jaitin et al. 2016; Datlinger et al. 2017). Similar approaches may be developed to combine CRISPR screens with other high-content readouts such as cellular morphology profiling (Caicedo et al. 2016) or single-cell measurements of protein abundance (Peterson et al. 2017; Stoeckius et al. 2017).
The development of appropriate pooled assays can therefore leverage the power of CRISPR-based forward genetics to systematically connect genes, noncoding elements, and RNAs with specific phenotypes.
Together, these technologies—including targetable CRISPR systems, methods to modify DNA sequence or localize regulatory proteins, and pooled screening assays for forward genetics—form a flexible platform for perturbation-based studies in mammalian cells. In the next section, we explore how these approaches can be applied to systematically explore regulation of and by RNA.
3. DISSECTING GENOME AND RNA REGULATION WITH CRISPR–CAS TOOLS
To illustrate how CRISPR-based approaches can illuminate mechanisms of gene regulation, we examine recent studies of the functions of two classes of regulatory elements in mammalian genomes: (1) noncoding DNA regulatory elements and (2) lncRNAs. These examples show how systematic CRISPR perturbations can establish causal relationships between regulatory elements and effects on gene expression and identify the molecular features that specify their context-specific functions in different cell types and at different genomic loci.
3.1. Functions of DNA Elements and Proteins That Regulate Transcription
Mammalian genomes are estimated to encode millions of distal noncoding DNA elements that quantitatively control the expression of specific genes in specific cell types (Roadmap Epigenomics Consortium 2015). Major consortia efforts have generated genome-wide maps of histone modifications, transcription factor binding, and DNA accessibility to identify and annotate noncoding elements that appear to have the characteristics of enhancers, promoters, and topological boundary elements (Fig. 5A) (ENCODE Project Consortium 2012; Roadmap Epigenomics Consortium 2015). However, the ability of these annotations to predict the functional role of any given element in regulating gene expression is unclear. For many years, perturbational studies of these elements were limited to knockouts of individual regulatory elements, providing only a glimpse of the complex networks responsible for regulating gene expression in different cell types. The programmable nature of CRISPR tools has opened new avenues to study DNA regulatory elements in an unbiased manner, including methods to perturb three facets of these elements: DNA sequence, recruitment of DNA-binding proteins, and 3D contacts with target genes.
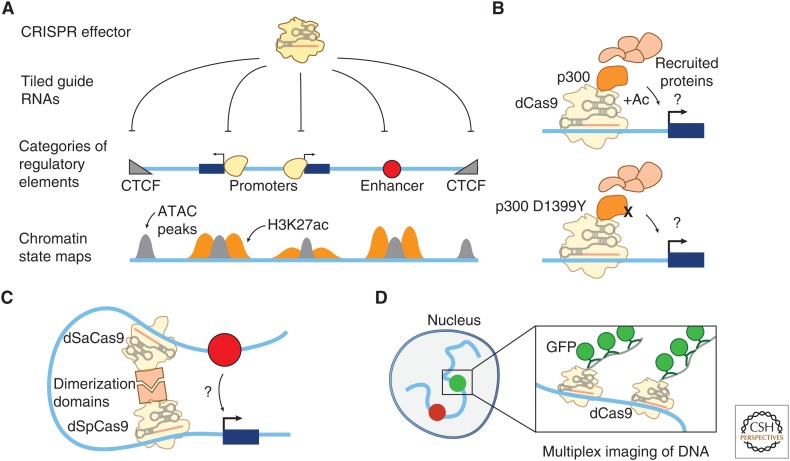
Figure 5.
CRISPR tools to study transcriptional regulation by noncoding DNA elements. (A) High-throughput CRISPR screens of noncoding regulatory elements enable testing the functions of various categories of regulatory elements. In such screens, a CRISPR perturbation method (including deletions caused by one or two single-guide RNAs [sgRNAs], CRISPRi, or CRISPRa) are combined with forward genetic screening approaches to enable high-throughput screens targeting hundreds of thousands of sequences in parallel. Different categories of elements are marked by different chromatin features: CTCF sites are marked by chromatin accessibility (as measured in ATAC-seq [gray peaks] or DNase-seq experiments) and CTCF binding. Gene promoters and enhancers are marked by chromatin accessibility and H3K27ac occupancy (orange peaks). (B) Recruitment strategies can be used to study the functions of chromatin regulatory proteins that read or write chromatin modifications. For example, studies of a protein domain of the transcriptional activator p300 (an acetyltransferase that also interacts with many other regulatory proteins) have shown that its catalytic activity is required for gene activation at several loci. (C) CRISPR can be used to engineer 3D chromatin contacts, for example, by fusing dimerization domains to dead SpCas9 and SaCas9 and recruiting each to nearby sites. (D) dCas9 can be used for programmable and multiplexed imaging of DNA loci in living cells (e.g., by using multiple sgRNAs to target dCas9 and many copies of GFP to a DNA locus).
CRISPR-based technologies are now enabling forward genetics screening approaches to define which noncoding sequences in the genome functionally regulate gene expression (Fig. 5A). An important technical consideration is that noncoding elements appear to consist of short functional sequences, such as transcription factor binding sites, interspersed with nonfunctional ones (Bulger and Groudine 2011; Spitz and Furlong 2012), making them relatively robust to indels in some locations. As such, two categories of recent approaches explore the effect of perturbations at two levels of resolution: sequence motifs (1–10 bp) and full regulatory elements (100–1000 bp).
In the first category (1–10 bp resolution), researchers have designed pooled experiments in which each cell in the pool expresses catalytically active Cas9 as well as an sgRNA, with different sgRNAs in the population tiling densely across noncoding regions around a gene of interest. Cas9 cuts at the target sequences, typically resulting in short deletions or insertions (indels). When coupled to phenotypic assays for the expression of that gene (for example, fusing the gene to GFP and sorting cells based on fluorescence), these experiments can identify short sequences (1–10 bp) important for transcriptional control. In one of the first such studies, Canver and colleagues designed 1338 sgRNAs tiled across a 12-kb enhancer known to regulate BCL11A and identified multiple sgRNAs that disrupted conserved transcription factor motifs in one of these elements (Canver et al. 2015). These approaches have now been scaled up to include thousands of sgRNAs in a single experiment (Diao et al. 2016; Korkmaz et al. 2016; Rajagopal et al. 2016; Sanjana et al. 2016).
In the second category (100–1000 bp resolution), larger-scale perturbations can be achieved by (1) using epigenome editors such as KRAB-dCas9, which has been shown to silence regulatory elements within a range of ∼100 bp (Gilbert et al. 2014; Thakore et al. 2015; Fulco et al. 2016; Klann et al. 2017; Simeonov et al. 2017) or (2) engineering larger deletions by codelivering two sgRNAs per cell (Zhu et al. 2016; Diao et al. 2017; Gasperini et al. 2017). For example, Fulco and colleagues recently used CRISPRi (KRAB-dCas9) to tile 98,000 sgRNAs across 1.2 Mb and identified nine and two elements that quantitatively tuned MYC and GATA1 expression, respectively (Fulco et al. 2016). Diao and colleagues used paired sgRNAs to tile ∼2-kb deletions across a 2-Mb region and reported 45 distal elements that regulate POU5F1 expression in human embryonic stem cells (Diao et al. 2017). Preliminary comparisons of these studies suggest that KRAB-dCas9 and 2-sgRNA deletions enable robust detection of cis-regulatory elements at a resolution of 100–1000 bp, whereas single-sgRNA deletion approaches enable finer resolution studies of regulatory sequences but are less robust in theory because of the small number of sgRNAs (often zero or one) interrogating each functional motif (Sanjana et al. 2016; Canver et al. 2017). Further work remains to understand the precise mechanisms of each perturbation method and to determine whether the screening approaches are adequately powered to detect repressive or activating elements with small effect sizes.
Technical considerations notwithstanding, CRISPR tiling studies are beginning to provide a sufficient number of noncoding perturbations to explore which elements act as enhancers, defined here as activating noncoding elements located >1 kb from any promoter region. (Notably, CRISPR tiling studies have also identified important roles for promoters in regulating other nearby genes, see Sec. 2.2.) Most of the activating elements identified in CRISPR-based screens, with a few exceptions, are marked by a chromatin signature previously thought to identify functional enhancers, including DNase accessibility and acetylation at histone 3 lysine 27 (H3K27ac) (Fulco et al. 2016; Rajagopal et al. 2016; Sanjana et al. 2016; Diao et al. 2017). However, only a small proportion of the DNase accessible elements detectably affect gene expression, suggesting that accessibility alone is not sufficient to identify a functional enhancer. Interestingly, the elements that do affect gene expression tend to have stronger quantitative accessibility and H3K27ac signals (Fulco et al. 2016), suggesting that the quantitative signals in these assays may be predictive of functional enhancers. Analysis of CRISPR tiling screens has also shown that enhancer elements frequently contact their target genes in 3D, as measured by chromosome conformation capture (Fulco et al. 2016; Sanjana et al. 2016), consistent with a model in which enhancers must physically contact promoters to activate transcription (Bulger and Groudine 2011; Spitz and Furlong 2012). Interestingly, in one study, a simple model incorporating a measure of enhancer activity (estimated from quantitative accessibility and H3K27ac signals) and a measure of contact frequency (estimated from Hi-C) was able to reasonably predict the quantitative effects of 93 noncoding elements on the expression of one gene (MYC) (Fulco et al. 2016). A later study integrated these measures in a combined H3K27ac ChIP-seq and Hi-C assay (HiChIP) and validated that this measurement accurately predicted the functions of several enhancers (Mumbach et al. 2017). Although further work will be required to test these conclusions across many genes, CRISPR perturbation-based studies have begun to reveal the regulatory architecture of enhancer-gene networks and determine how measurements of chromatin state and contact frequency predict the functional effects of noncoding elements on gene expression.
Additional CRISPR tools provide new ways to independently manipulate and test how chromatin state and 3D chromosome contacts influence enhancer function. For example, enhancer activity is thought to be determined by specific sequence motifs that recruit transcription factors and co-activators, including histone-modifying enzymes. Cas enzymes can be fused to these chromatin regulatory proteins to directly test how they affect gene expression in different genomic contexts (Fig. 5B). For example, Hilton and colleagues fused the catalytic domain of the acetyltransferase p300 to dCas9 and, by comparing the wild-type p300 domain to a mutated domain, showed that the acetyltransferase activity of p300 is required for gene activation at several loci (Hilton et al. 2015). In contrast, CRISPR-mediated recruitment of catalytically dead EZH2 (the enzymatic component of polycomb repressive complex 2) to the promoter of an active gene was unable to deposit H3K27me3 (a histone modification associated with gene silencing) but nonetheless was able to efficiently repress gene expression (O’Geen et al. 2017), indicating that dCas9-EZH2 fusions can modulate gene repression independent of the enzymatic activity of EZH2 and that H3K27me3 is not required for silencing at this locus. These CRISPR-based studies, by recruiting chromatin regulators to specific sites in the genome, circumvent the limitations of using knockdown or overexpression approaches to study the functions of chromatin regulators, which can lead to difficulties in separating direct and indirect effects on gene expression. Further studies, such as recruitment of individual or combinations of protein effectors at many locations in the genome, will be required to determine which complexes enable enhancers to activate gene transcription and how genomic context influences these regulatory functions.
CRISPR recruitment strategies can also be adapted to modulate and study 3D contact frequency between distal regulatory elements and gene promoters. The genomic determinants of contact frequency are thought to include properties of the chromatin fiber, loop-extrusion complexes, and protein–protein interactions (e.g., between distal CTCF-bound sites or interacting factors bound to enhancers and promoters [Dekker and Mirny 2016]). Recent studies have developed methods to directly manipulate these contacts (Fig. 5C), for example by using orthogonal Cas9 and Cas12 systems, each fused to a heterologous dimerization domain, to bring together two target DNA sequences (Morgan et al. 2017). In several examples, studies using CRISPR or other tools to tether two DNA sequences have shown that increasing the contact frequency between an enhancer and a target promoter leads to an increase in gene expression (Deng et al. 2012; Bartman et al. 2016; Morgan et al. 2017). Multiple CRISPR-based studies of individual CTCF sites have revealed that these sites do indeed control 3D contacts and gene expression, perhaps by tuning the contacts between genes and their enhancers (de Wit et al. 2015; Guo et al. 2015; Sanborn et al. 2015; Flavahan et al. 2016; Hnisz et al. 2016). Recent studies using CRISPR-GFP recruitment methods to image the locations and movement of DNA loci in living cells (Fig. 5D) have also provided new insights into mechanisms that control 3D chromatin contacts (Gu et al. 2018; Maass et al. 2018). Imaging of enhancers and promoters in the same 3D domain showed that enhancer activation correlates with an increased rate of chromatin diffusion in the same 3D domain in a transcription-dependent fashion, likely affecting the rate of enhancer–promoter contact formation (Gu et al. 2018). Imaging loci on different chromosomes revealed that gene loci on different chromosomes can stably associate and have stereotyped positions in the nucleus, perhaps influencing their regulation or function (Maass et al. 2018).
Together, the ability to target and manipulate specific DNA loci at scale is unlocking new insights into the functions of DNA regulatory elements, including how histone modifications, chromatin regulatory proteins, and 3D chromosome conformation influence gene expression. Because these properties of enhancers can vary dramatically across cell types, further exploration will be required to learn how these rules change in different cellular contexts. These studies to modify different aspects of transcriptional regulation also provide an important guide for understanding posttranscriptional regulation: similar CRISPR-based approaches using RNA-targeting tools could soon be applied to study the functions of RNA regulatory elements and the proteins, sequence and structural motifs, and RNA modification sites that mediate their functions.
3.2. Functions of Long Noncoding RNAs and Their Transcription
Long noncoding RNAs (lncRNAs) are a heterogeneous class of molecules, operationally defined as polyadenylated RNAs of >200 nt that do not encode peptides. Before the genomic era, biochemical and genetic studies identified a handful of lncRNAs with important molecular functions, including Xist, which orchestrates X-chromosome inactivation by spreading across the X chromosome in cis and recruiting multiple repressive chromatin regulatory complexes to silence gene transcription (Plath et al. 2002). RNA sequencing studies found that nearly the whole genome is transcribed to some extent in one cell type or another, and that these noncoding transcripts included thousands of polyadenylated and spliced lncRNAs (Kapranov et al. 2007; Djebali et al. 2012; ENCODE Project Consortium 2012). Although there are some notable exceptions, such as Xist, the function (if any) of most lncRNAs has remained elusive. CRISPR-based tools have now begun to address two key questions: Which genomic loci encoding lncRNAs have functional roles in the cell, and how do these noncoding loci accomplish these functions?
New tools for direct perturbation of lncRNA expression and sequence have provided an opportunity to systematically search for functional lncRNA loci via forward genetic screens. Like noncoding DNA elements, perturbations to lncRNAs involve different technical considerations than protein-coding genes. The relationship between RNA sequence and function is poorly understood, but is apparently flexible and can consist of functional sequences (such as sites that RNA-binding proteins recognize) interspersed with nonfunctional ones (Zappulla and Cech 2004; Guttman and Rinn 2012; Rinn and Chang 2012). High-throughput screening approaches to disrupt lncRNA function must either perturb broad regions of genomic sequence, for example to remove the promoter or some critical RNA domain of a lncRNA, or densely tile smaller mutations through each element to identify and perturb critical sequences.
Using these approaches, several groups have recently performed forward genetic screens by activating or inhibiting lncRNA promoters and assessing the effects on cellular functions (Zhu et al. 2016; Joung et al. 2017a; Liu et al. 2017). Liu and colleagues performed loss-of-function screens using CRISPRi (KRAB-dCas9) to target 16,401 lncRNA promoters and characterized their effects on cellular proliferation across nine cell lines (Liu et al. 2017). They found 499 loci where inhibition of the lncRNA promoter led to a significant effect on cellular proliferation in at least one cell line, 89% of which showed cell line–specific effects. Joung and colleagues designed gain-of-function screens using CRISPRa (SAM) to target 10,504 lncRNA promoters and identified 11 noncoding loci whose activation led to resistance to the melanoma drug vemurafinib (Joung et al. 2017a). Most of these effects appeared to be due to activation of nearby genes in cis. From these and other studies (Guttman et al. 2011; Rinn and Chang 2012; Luo et al. 2016), it is becoming clear that many noncoding genomic loci that produce lncRNAs play important roles in controlling gene expression and cellular functions, and that these roles are typically specific to certain cell types or contexts.
Importantly, however, perturbations to “lncRNA promoters” do not themselves establish that the “lncRNA transcripts” are functional. Rather, promoter perturbations might identify loci that affect a phenotype through a variety of mechanisms related to the lncRNA locus (Fig. 6), which can be broadly categorized as (1) nonlocal functions of a lncRNA, in which the RNA transcript acts at locations in the cell other than the genomic locus of the lncRNA; (2) local, sequence-specific functions of a lncRNA transcript to regulate the expression of nearby gene(s) on the same chromosome (in cis); (3) local functions to regulate nearby gene(s) via mechanisms of lncRNA transcription or splicing that do not require any specific sequences in the lncRNA transcript itself; and (4) local functions to regulate nearby gene(s) via the lncRNA promoter acting as an enhancer.
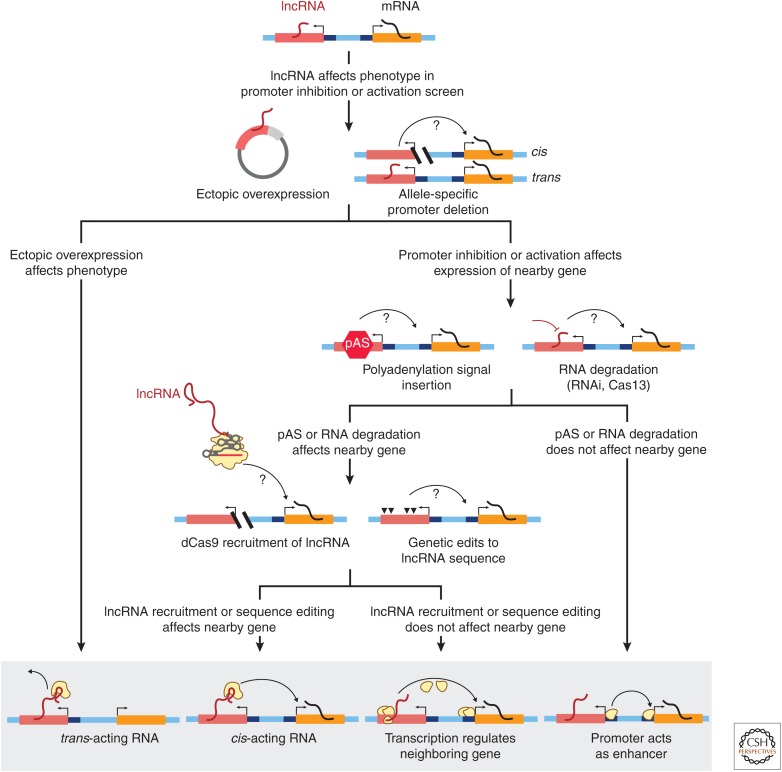
Figure 6.
CRISPR tools to dissect the functions of genomic loci that produce long noncoding RNAs (lncRNAs). Flowchart (top) shows experimental approaches to determine how lncRNA loci regulate gene expression (bottom) via one or more of the following mechanisms: (1) producing a trans-acting lncRNA that acts nonlocally (away from its genomic locus); (2) producing a cis-acting lncRNA that acts locally at its genomic locus via a sequence-dependent function of the RNA molecule; (3) affecting a nearby gene via a sequence-independent function of lncRNA transcription or splicing; and (4) regulating a neighboring gene via proteins that bind its promoter or other overlapping cis-regulatory element. A series of targeted experiments can be used to distinguish between these functions. If ectopic overexpression of the lncRNA recapitulates or rescues the screen phenotype, this supports a trans-acting mechanism. Conversely, demonstrating that allele-specific deletion affects the expression of a nearby gene in an allele-specific manner is the best demonstration that a lncRNA locus acts to regulate gene expression in cis. To distinguish between multiple potential cis-acting mechanisms, a next step involves either inserting a polyadenylation signal (pAS) just downstream from the lncRNA promoter or using RNA interference (RNAi) or Cas13 to directly degrade the RNA. If the mature RNA and lncRNA transcription are successfully eliminated but do not affect the phenotype, then DNA-mediated effects of promoters are the most likely mechanism. If these perturbations do affect the phenotype, then dCas9 recruitment of the lncRNA in combination with genetic edits to the lncRNA sequence can be used to distinguish between sequence-specific effects of the lncRNA molecule or sequence-independent effects of lncRNA transcription.
Although many lncRNAs have been hypothesized to act via the first two (RNA-mediated) mechanisms, CRISPR-based studies have surprisingly revealed that the third and fourth categories of functions—that is, effects on gene expression that are not mediated by the RNA transcript itself—occur in many loci that produce lncRNAs as well as in many loci that encode mRNAs. In a recent study, Engreitz and colleagues knocked out the promoters of 12 uncharacterized lncRNAs and six mRNAs and found, using allele-specific RNA sequencing, that promoter deletion led to an effect on the expression of a neighboring gene at five of these 12 lncRNA loci and four of the six mRNA loci (Engreitz et al. 2016). To distinguish between possible cis-regulatory mechanisms (Fig. 6), Engreitz and colleagues first inserted polyadenylation signals a few hundred bp downstream of the transcription start site, thereby leaving the DNA sequence of the promoter intact but eliminating downstream transcription and the mature lncRNA transcript (Fig. 6). In five out of six cases, polyadenylation insertion near the promoter did not affect the expression of a gene neighbor, indicating that the regulatory effect was likely caused by the lncRNA promoter acting as an enhancer. For one lncRNA locus called Blustr (bivalent locus is up-regulated by the splicing and transcription of an RNA), insertion of polyadenylation sites did in fact reduce the expression of a neighboring gene (Sfmbt2), indicating that transcription of Blustr was required for Sfmbt2 activation. However, deleting individual exons or introns of the Blustr transcript with CRISPR–Cas9 did not affect Sfmbt2 activation, suggesting that there were no specific sequences in the spliced lncRNA transcript that were individually necessary. Indeed, for none of the six loci studied did the RNA transcripts themselves appear to contribute to cis regulation of gene expression in a sequence-dependent fashion. Instead, activation depended on the lncRNA promoter acting as an enhancer or on the act of lncRNA transcription.
Together with an increasing number of other examples using similar genetic approaches—including Airn (Latos et al. 2012), Hand2 (Anderson et al. 2016), and others (Shearwin et al. 2005; Yin et al. 2015; Paralkar et al. 2016; Joung et al. 2017a)—these systematic CRISPR-based dissections of lncRNA loci point to a pervasive role for promoters and the process of transcription in affecting gene expression in a surrounding region, re-orienting how we can view the vast number of noncoding transcripts produced by mammalian genomes. Indeed, the cell-type specificity seen in lncRNA promoter perturbation screens may result at least in part from the cell type–specific functions of these promoters acting as enhancers for neighboring genes. A principle emerging from these studies is that gene loci—including those encoding lncRNAs and mRNAs—can have multiple independent functions, including one or more of these local effects on nearby genes as well as trans-acting functions of the RNA transcripts (Fig. 6).
Given these observations, an important future direction for studies of the noncoding genome will be to expand the CRISPR forward genetics toolkit to perturb specific categories of regulatory mechanisms. For example, functional RNAs might be directly identified and distinguished from promoter-mediated mechanisms via knockdown with RNA-targeting CRISPR effectors, or by tailored screens to alter RNA sequence, RNA modifications, RNA localization, or RNA-binding proteins. trans-acting lncRNAs may be directly identified via lncRNA overexpression (via expression at an ectopic locus) in forward genetic screens, or first via CRISPRa/i screens followed by overexpression of specific candidates. cis-acting lncRNAs may be directly identified (and distinguished from sequence-independent functions of transcription) via CRISPR-mediated recruitment of RNAs to specific genomic sites and dense saturation mutagenesis of lncRNA sequences in situ. Technological advances in CRISPR systems, including RNA-targeting CRISPR systems (see next section), promise to enable such precise studies to dissect the functions of lncRNAs and noncoding transcription in mammalian genomes.
4. EMERGING CRISPR TOOLS TO DIRECTLY MANIPULATE RNA
The recent introduction of RNA-targeting CRISPR tools have opened a new realm of potential applications for studying RNA regulation via direct modulation or modification of RNA. In particular, Cas13 systems enable a new toolbox of easy-to-use technologies for perturbing RNA function, overcoming limitations of previous methods such as exogenous tagging of RNA with MS2 loops (Park et al. 2010) or engineering of Pumilio RNA-binding proteins (Abil and Zhao 2015). In this section, we discuss CRISPR-based RNA-targeting technologies and how they can be applied for studying RNA biology.
4.1. Methods to Target CRISPR–Cas Proteins to Specific RNA Sequences
Several CRISPR-based approaches have been developed to enable RNA-targeted degradation or programmable protein recruitment.
Cas9: Although Cas9 is typically regarded as a DNA-targeting enzyme, certain natural Cas9 variants or engineered modifications have been shown to have the potential to also interact with and cleave RNA. Initial in vitro characterization of the Cas9 from Streptococcus pyogenes (SpCas9) showed the possibility of RNA as a binding substrate (Gasiunas et al. 2012), and later methods refined the activity in vitro via codelivery of PAM duplex-forming small oligos, termed PAMmers (O’Connell et al. 2014). This engineered adaptation of SpCas9 for RNA binding was shown to function in vivo for RNA imaging or cleavage of repeat-containing RNA (Nelles et al. 2016; Batra et al. 2017). Certain Cas9 orthologs also possess endogenous RNase activity and can cleave RNA without a PAMmer in Escherichia coli (Strutt et al. 2018) or in vitro (Rousseau et al. 2018). RNA targeting with Cas9 in mammalian cells appears to require the introduction of the exogenous synthetic modified oligo (PAMmer) (Nelles et al. 2016), rendering Cas9 challenging to use for pooled screening approaches that require genetically encodable constructs.
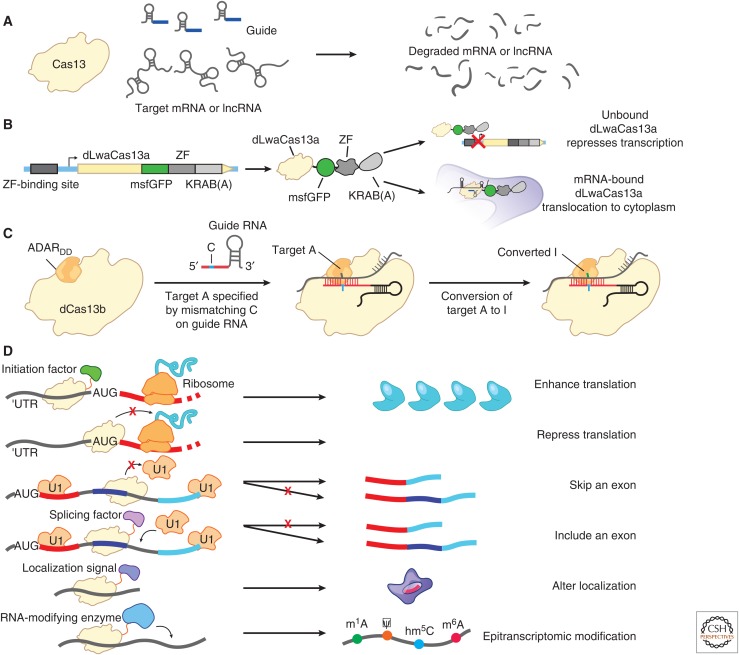
Cas13: The single-effector RNA-targeting CRISPR system Type VI and the associated endoribonuclease Cas13 (Shmakov et al. 2015; Abudayyeh et al. 2016; Smargon et al. 2017) have provided an improved tool for RNA knockdown and RNA-targeted protein recruitment (Abudayyeh et al. 2017; Cox et al. 2017). The Cas13 family has diverse features, including different crRNA structures and different levels of activity (Shmakov et al. 2015, 2017; Abudayyeh et al. 2016; East-Seletsky et al. 2016; Smargon et al. 2017; Konermann et al. 2018; Yan et al. 2018). Cas13a, the first characterized member, has been shown to cleave RNA and reduce gene expression in bacterial, plant, and mammalian cells (Fig. 7A) (Abudayyeh et al. 2016, 2017). Catalytically dead Cas13—generated by mutating the endonuclease domains—provides a flexible platform for recruitment of proteins to target RNAs in mammalian cells, including for imaging of RNA through fusion to GFP (Fig. 7B) (Abudayyeh et al. 2017), RNA editing through fusion to ADAR2 (Fig. 7C) (Cox et al. 2017), and alteration of isoforms by blocking splice sites (Konermann et al. 2018).
Figure 7.
RNA targeting applications of Cas13. (A) RNA knockdown with Cas13. (B) Image tracking using dCas13-eGFP to fluorescently visualize transcripts in cells. Modifying the dCas13-EGFP fusion with a zinc finger–Krüppel associated box (KRAB) fusion can improve visualization and lower fluorescence background signal by repressing the expression of unbound dCas13-EGFP complexes via negative feedback. (C) RNA editing of adenines to inosines by a dCas13-ADAR2 fusion. Precise editing is accomplished by creating an A:C base pair bubble by encoding a mismatch in the guide RNA. (D) Further applications of Cas13 for studying RNA regulation. Translation could be activated by recruiting translation initiation factors or repressed by blocking start codons on transcripts. Splicing has been altered by blocking splice sites on transcripts or by recruiting splicing factors. Transcript localization could be modulated by fusing dCas13 to different localization sequences. Lastly, posttranscriptional RNA modifications could be introduced by recruiting RNA-modifying enzymes, such as pseudouridine synthase, to specific bases on RNA transcripts.
4.2. Opportunities for Systematic Studies of RNA Biology
These new RNA-targeted tools open new opportunities to study many aspects of RNA biology. Here, we highlight key studies that have developed proof-of-concept tools using Cas13.
4.2.1. RNA Knockdown
In light of the challenges in distinguishing the effects of DNA- and RNA-mediated mechanisms (see Sec. 2.2), technologies for RNA knockdown with CRISPR enzymes have the potential to enable more specific studies of RNA-mediated functions because they avoid manipulating DNA sequence. To this end, we recently showed that the Cas13 platform can knock down RNAs with efficiency similar to RNA interference (Fig. 7A) (Abudayyeh et al. 2017; Cox et al. 2017). At the same time, Cas13 knockdown of a reporter transcript (luciferase) had minimal effects on the rest of the transcriptome, whereas RNAi knockdown of the same transcript led to significant effects on ∼900 other genes (Abudayyeh et al. 2017; Cox et al. 2017; Konermann et al. 2018). Thus, RNA knockdown screens with Cas13 have the potential to reduce the off-target effects that have confounded RNAi screens, and, in contrast to approaches that rely on endogenous RNAi proteins, the Cas13 enzymes can be further optimized and engineered for additional functionality or specificity. For example, adding a nuclear localization tag to Cas13a led to successful knockdown of nuclear-localized lncRNAs, while RNAi was ineffective (Abudayyeh et al. 2017). We also found that a second Cas13 enzyme, Cas13b from Prevotella sp. P5-125, was far more effective at RNA knockdown (>90% for each site targeted) than our original studies using LwaCas13a (Cox et al. 2017). Further optimizations of the system—including adapting other Cas13 proteins and guide RNAs for viral delivery, learning design rules for efficient guides, and exploring or optimizing Cas13 proteins for high stability and specificity—should soon enable RNA-targeted forward genetic screens to directly study RNA function.
4.2.2. Posttranscriptional RNA Modifications
Posttranscriptional modifications of RNA nucleotides—including N6-methyladenosine (m6A), pseudouridine, and inosine—appear to be a critical layer of posttranscriptional regulation (Schwartz 2016; Helm and Motorin 2017; Hsu et al. 2017; Lewis et al. 2017). RNA mapping technologies have revealed that these modifications can affect tens of thousands of sites in both coding and noncoding RNAs (Li et al. 2016; Schwartz 2016; Helm and Motorin 2017; Hsu et al. 2017; Lewis et al. 2017), but the functions of most of these modifications are poorly understood. We developed a Cas13 approach to perform adenine-to-inosine (A-to-I) editing, a natural modification that alters the sequence of RNA. In this approach, named RNA editing for programmable A to I replacement (REPAIR), we fused catalytically dead Cas13 to the ADAR2 enzyme to enable targetable A-to-I editing activity (Fig. 7C) (Cox et al. 2017). By extending the guide RNA and including a cytidine mismatch in the guide across from the targeted adenosine, a more optimal double-stranded RNA substrate with a bubble was created, maximizing ADAR2 enzymatic activity. This approach achieved up to 90% A-to-I editing of targeted sites on reporter transcripts and up to 40% on endogenous transcripts. Off-target editing observed with the initial system (>18,000 edited off-target sites) was reduced through a rational mutagenesis strategy, which produced a more specific version of Cas13 that retained on-target editing activity but reduced off-target activity (20 edited off-target sites) (Cox et al. 2017). Because of the high specificity, high efficiency, lack of sequence restriction, and precise modification site, REPAIR provides a useful tool for studying the effects of sequence variants and inosine modifications, and for developing therapeutics. Similar approaches with alternative RNA editing or modification enzymes, such as RNA methyltransferases, cytidine deaminases, and pseudouridine synthases, may enable precise introduction of RNA modifications and subsequent studies to connect these modifications with their cellular and molecular functions (Fig. 7D).
4.2.3. RNA Splicing, Localization, and More
RNA-targeted CRISPR tools are still in their infancy, and—just as an explosion of dCas9 fusion tools have begun to reveal biological insights into transcriptional regulation—dCas13 fusions will likely have a major impact in illuminating posttranscriptional regulation. Indeed, studies using previous RNA-targeting technologies have highlighted additional possibilities for manipulating RNA regulation with new CRISPR tools. For example, fusion of PUF proteins to the arginine- and serine-rich domain of SRSF1 or the glycine-rich domain of HNRNPA1 enabled the creation of programmable activators and repressors of splicing, respectively (Cheong and Hall 2006). RNA recruitment systems using MS2 and PP7 (or other systems that involve tagging an endogenous RNA with the recognition sequence for an exogenous RNA-binding protein) have been used to test the sufficiency of specific RNA–protein interactions for RNA function (Chen and Varani 2013; Bos et al. 2016) and to track transcripts via fluorescence (Park et al. 2010). Early studies have shown the potential for Cas13 tools to modulate the expression of specific RNA isoforms by binding and blocking splice sites (Fig. 7D) (Konermann et al. 2018). The ability to combine each of these tools with transcriptome-wide screening will enable systematic perturbation studies of many aspects of RNA biology, including translation, splicing, RNA localization, and posttranscriptional modifications (Fig. 7D).
5. CONCLUDING REMARKS
CRISPR systems have remarkable diversity and flexibility, providing many opportunities for creative innovation to expand the CRISPR toolkit. Further exploration of natural CRISPR systems, as well as other natural enzymes and systems with novel functionalities, will help to uncover new tools for manipulating RNA and DNA. Directed evolution and rational engineering of CRISPR enzymes will expand their generalizability and increase specificity. Functionalizing CRISPR proteins with additional DNA- or RNA-modifying proteins will enable precisely perturbing a diverse array of regulatory processes in human cells.
Building on recent studies of DNA regulatory elements and lncRNAs, application of CRISPR tools has the potential to reveal new principles of molecular biology. In the past decade, many advances in our understanding of RNA biology were facilitated by genome-scale “observational” studies, including the application of new sequencing-based technologies to map the locations of RNA transcription, modifications, and RNA-binding proteins. In the next decade, we anticipate that an equally transformative contribution will come from genome-scale “perturbation” studies, including systematic interrogation of each of these layers of RNA regulation. These studies will establish general principles for regulatory molecules as well as illuminate how specific genomic or molecular contexts modify their functions. Integrating combinatorial perturbations and multiplexed molecular measurements will reveal molecular circuits that control cellular behavior. In tandem with the development of appropriate phenotypic assays and delivery methods, CRISPR systems will connect these circuits with cellular phenotypes relevant to human diseases.
CRISPR studies of RNA regulation will not only reveal the functions of key molecular processes but also will help us to directly manipulate them in humans. Preclinical development for CRISPR-based therapeutics is already underway, including approaches for genome editing ex vivo (where editing is performed outside the body and edited cells are transplanted into the patient) and in vivo (where CRISPR tools are delivered to edit cells directly in the tissue of interest). Many challenges remain, including optimizing the efficiency and specificity of CRISPR editing, developing new methods for efficient and precise delivery to target tissues and cell types, and minimizing unintended consequences of these manipulations on immune responses or target cell functions. Nonetheless, overcoming these challenges promises to enable a flexible class of therapeutics that leverage a programmable suite of CRISPR tools to manipulate DNA or RNA.
Although therapeutic development efforts have largely focused on modification of protein-coding genes, approaches for targeting DNA and RNA regulatory mechanisms may prove advantageous for certain indications. For example, therapeutic targeting of DNA enhancers, which can have cell type–specific activities, may enable more precise modulation of gene function in cell types relevant to disease (Canver et al. 2015). RNA-targeted therapeutic approaches may enable specific regulation of entities that are difficult to manipulate at the level of DNA, such as degradation of toxic repeat expansion RNAs (Batra et al. 2017) or selective activation of specific RNA isoforms (Palacino et al. 2015; Konermann et al. 2018).
Although much work remains to characterize and apply CRISPR tools, such studies may ultimately yield new insights into the regulatory machinery of the cell and new strategies for treating human disease.
ACKNOWLEDGMENTS
We thank Eric Lander and Rhiannon Macrae for comments on the manuscript and Sigrid Knemeyer for assistance with figures.
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
References
- Abil Z, Zhao H. 2015. Engineering reprogrammable RNA-binding proteins for study and manipulation of the transcriptome. Mol Biosyst 11: 2658–2665. [DOI] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. 2016. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353: aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. 2017. RNA targeting with CRISPR–Cas13. Nature 550: 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. 2016. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167: 1867–1882.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L. 2013. Comprehensive analysis of the HEPN superfamily: Identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. 2016. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman CR, Hsu SC, Hsiung CC-S, Raj A, Blobel GA. 2016. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol Cell 62: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R, Nelles DA, Pirie E, Blue SM, Marina RJ, Wang H, Chaim IA, Thomas JD, Zhang N, Nguyen V, et al. 2017. Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell 170: 899–912.e810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. 2013. Programmable repression and activation of bacterial gene expression using an engineered CRISPR–Cas system. Nucleic Acids Res 41: 7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Yong J, Antebi YE, McCue K, Kazuki Y, Uno N, Oshimura M, Elowitz MB. 2016. Dynamics of epigenetic regulation at the single-cell level. Science 351: 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos TJ, Nussbacher JK, Aigner S, Yeo GW. 2016. Tethered function assays as tools to elucidate the molecular roles of RNA-binding proteins. Adv Exp Med Biol 907: 61–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. 2006. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol 2: 202–206. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo JC, Singh S, Carpenter AE. 2016. Applications in image-based profiling of perturbations. Curr Opin Biotechnol 39: 134–142. [DOI] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. 2015. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527: 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Bauer DE, Orkin SH. 2017. Functional interrogation of non-coding DNA through CRISPR genome editing. Methods 121–122: 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer EPR, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. 2015. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12: 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Varani G. 2013. Engineering RNA-binding proteins for biology. FEBS J 280: 3734–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CG, Hall TM. 2006. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci 103: 13635–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. 2017. RNA editing with CRISPR–Cas13. Science 358: 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman JE, Abudayyeh OO, Joung J, Gootenberg JS, Zhang F, Konermann S. 2015. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol 33: 1159–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C. 2017. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods 14: 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, Teunissen H, Splinter E, Wijchers PJ, Krijger PHL, de Laat W. 2015. CTCF binding polarity determines chromatin looping. Mol Cell 60: 676–684. [DOI] [PubMed] [Google Scholar]
- Dekker J, Mirny L. 2016. The 3D genome as moderator of chromosomal communication. Cell 164: 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149: 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Li B, Meng Z, Jung I, Lee AY, Dixon J, Maliskova L, Guan K-L, Shen Y, Ren B. 2016. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res 26: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Fang R, Li B, Meng Z, Yu J, Qiu Y, Lin KC, Huang H, Liu T, Marina RJ, et al. 2017. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods 14: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. 2016. Perturb-seq: Dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167: 1853–1866.e1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG. 2018. Am I ready for CRISPR? A user’s guide to genetic screens. Nat Rev Genet 19: 67–80. [DOI] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. 2014. Rational design of highly active sgRNAs for CRISPR–Cas9-mediated gene inactivation. Nat Biotechnol 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR–Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, Doudna JA. 2016. Two distinct RNase activities of CRISPR–C2c2 enable guide-RNA processing and RNA detection. Nature 538: 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, Doudna JA. 2017. RNA Targeting by functionally orthogonal type VI-A CRISPR–Cas enzymes. Mol Cell 66: 373–383.e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. 2016. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 171: 305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Perli SD, Lu TK. 2013. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol 2: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E. 2016. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532: 517–521. [DOI] [PubMed] [Google Scholar]
- Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM. 2016. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354: 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Findlay GM, McKenna A, Milbank JH, Lee C, Zhang MD, Cusanovich DA, Shendure J. 2017. CRISPR/Cas9-mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am J Hum Genet 101: 192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. 2017. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551: 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, et al. 2017. Nucleic acid detection with CRISPR–Cas13a/C2c2. Science 356: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. 2018. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360: 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Swigut T, Spencley A, Bauer MR, Chung M, Meyer T, Wysocka J. 2018. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 7: eaao3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. 2012. Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Motorin Y. 2017. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat Rev Genet 18: 275–291. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Castello A, Schwarzl T, Preiss T. 2018. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 19: 327–341. [DOI] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. 2015. Epigenome editing by a CRISPR–Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature Biotechnol 33: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton A-L, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351: 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR–Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PJ, Shi H, He C. 2017. Epitranscriptomic influences on development and disease. Genome Biol 18: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorns E, Lord CJ, Turner N, Ashworth A. 2007. Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov 6: 556–568. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I. 2016. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell 167: 1883–1896.e1815. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, et al. 2017a. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548: 343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F. 2017b. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat Protoc 12: 828–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488. [DOI] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. 2015. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 12: 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S, Chavez A, Tuttle M, Hall RN, Chari R, Ter-Ovanesyan D, Qian J, Pruitt BW, Beal J, Vora S, et al. 2015. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods 12: 1051–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. 2014. A guide to genome engineering with programmable nucleases. Nat Rev Genet 15: 321–334. [DOI] [PubMed] [Google Scholar]
- Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, Crawford GE, Reddy TE, Gersbach CA. 2017. CRISPR–Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nature Biotechnol 7: 46545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. 2014. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32: 267–273. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533: 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR. 2017. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 169: 559. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. 2013. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. 2015. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. 2018. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, Zwart W, Elkon R, Agami R. 2016. Functional genetic screens for enhancer elements in the human genome using CRISPR–Cas9. Nat Biotechnol 34: 192–198. [DOI] [PubMed] [Google Scholar]
- Latos PA, Pauler FM, Koerner MV, Şenergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al. 2012. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338: 1469–1472. [DOI] [PubMed] [Google Scholar]
- Lewis CJ, Pan T, Kalsotra A. 2017. RNA modifications and structures cooperate to guide RNA–protein interactions. Nat Rev Mol Cell Biol 18: 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Yi C. 2016. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat Methods 14: 23–31. [DOI] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. 2016. Editing DNA methylation in the mammalian genome. Cell 167: 233–247.e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. 2017. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355: aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, et al. 2016. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18: 637–652. [DOI] [PubMed] [Google Scholar]
- Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, Pederson T. 2016. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 34: 528–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass PG, Barutcu AR, Weiner CL, Rinn JL. 2018. Inter-chromosomal contact properties in live-cell imaging and in Hi-C. Mol Cell 69: 1039–1045.e1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. 2013. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10: 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. 2013a. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013b. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. 2015. CRISPR–Cas immunity in prokaryotes. Nature 526: 55–61. [DOI] [PubMed] [Google Scholar]
- Morgan SL, Mariano NC, Bermudez A, Arruda NL, Wu F, Luo Y, Shankar G, Jia L, Chen H, Hu JF, et al. 2017. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun 8: 15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Satpathy AT, Boyle EA, Dai C, Gowen BG, Cho SW, Nguyen ML, Rubin AJ, Granja JM, Kazane KR, et al. 2017. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet 49: 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. 2016. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165: 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, et al. 2006. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441: 106–110. [DOI] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, et al. 2016. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353: aaf8729. [DOI] [PubMed] [Google Scholar]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. 2014. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516: 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H, Ren C, Nicolet CM, Perez AA, Halmai J, Le VM, Mackay JP, Farnham PJ, Segal DJ. 2017. dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res 45: 9901–9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang X, Van Hoosear M, Shin Y, Chin DN, Keller CG, et al. 2015. SMN2 splice modulators enhance U1–pre-mRNA association and rescue SMA mice. Nat Chem Biol 11: 511–517. [DOI] [PubMed] [Google Scholar]
- Paralkar VR, Taborda CC, Huang P, Yao Y, Kossenkov AV, Prasad R, Luan J, Davies JOJ, Hughes JR, Hardison RC, et al. 2016. Unlinking an lncRNA from its associated cis element. Mol Cell 62: 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Buxbaum AR, Singer RH. 2010. Single mRNA tracking in live cells. Methods Enzymol 472: 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. 2015. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162: 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. 2013. RNA-guided gene activation by CRISPR–Cas9-based transcription factors. Nat Methods 10: 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S, Klappenbach JA. 2017. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol 35: 936–939. [DOI] [PubMed] [Google Scholar]
- Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. 2002. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet 36: 233–278. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal N, Srinivasan S, Kooshesh K, Guo Y, Edwards MD, Banerjee B, Syed T, Emons BJ, Gifford DK, Sherwood RI. 2016. High-throughput mapping of regulatory DNA. Nat Biotechnol 34: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau BA, Hou Z, Gramelspacher MJ, Zhang Y. 2018. Programmable RNA cleavage and recognition by a natural CRISPR–Cas9 system from Neisseria meningitidis. Mol Cell 69: 906–914.e904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, Cheng C, Regev A, Zhang F. 2016. High-resolution interrogation of functional elements in the noncoding genome. Science 353: 1545–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. 2016. Cracking the epitranscriptome. RNA 22: 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. 2005. Transcriptional interference—A crash course. Trends in Genetics 21: 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. 2015. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods 12: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. 2015. Discovery and functional characterization of diverse Class 2 CRISPR–Cas systems. Mol Cell 60: 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, et al. 2017. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol 15: 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonov DR, Gowen BG, Boontanrart M, Roth TL, Gagnon JD, Mumbach MR, Satpathy AT, Lee Y, Bray NL, Chan AY, et al. 2017. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 549: 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, Cox DB, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P, Shmakov S, Makarova KS, et al. 2017. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell 65: 618–630.e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: From enhancer binding to developmental control. Nat Rev Genet 13: 613–626. [DOI] [PubMed] [Google Scholar]
- Stewart SE, Menzies SA, Popa SJ, Savinykh N, Petrunkina Harrison A, Lehner PJ, Moreau K. 2017. A genome-wide CRISPR screen reconciles the role of N-linked glycosylation in galectin-3 transport to the cell surface. J Cell Sci 130: 3234–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P. 2017. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14: 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt SC, Torrez RM, Kaya E, Negrete OA, Doudna JA. 2018. RNA-dependent RNA targeting by CRISPR–Cas9. eLife 10.7554/eLife.32724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, van der Oost J, Jinek M. 2017. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Cell 66: 221–233.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. 2015. Highly specific epigenome editing by CRISPR–Cas9 repressors for silencing of distal regulatory elements. Nat Methods 12: 1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, Klasic M, Zoldos V. 2016. Repurposing the CRISPR–Cas9 system for targeted DNA methylation. Nucleic Acids Res 44: 5615–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR–Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou L, Wang X, Lu J, Zhang R, Liang X, Wang L, Deng W, Zeng YX, Huang H, et al. 2016. A genome-scale CRISPR–Cas9 screening method for protein stability reveals novel regulators of Cdc25A. Cell Discov 2: 16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, Koonin EV, et al. 2016. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165: 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA. 2018. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell 70: 327–339.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z, Zhao Y, Shen B, Huang X, Zhu H, et al. 2015. Opposing roles for the lncRNA Haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16: 504–516. [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. 2015. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR. 2004. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc Natl Acad Sci 101: 10024–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, et al. 2017. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat Biotechnol 35: 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. 2014. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509: 487–491. [DOI] [PubMed] [Google Scholar]
- Zhu S, Li W, Liu J, Chen C-H, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, et al. 2016. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR–Cas9 library. Nature Biotechnol 34: 1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]