Abstract
During vertebrate embryogenesis, the cranial neural crest (CNC) forms at the neural plate border and subsequently migrates and differentiates into many types of cells. The transcription factor Snai2, which is induced by canonical Wnt signaling to be expressed in the early CNC, is pivotal for CNC induction and migration in Xenopus. However, snai2 expression is silenced during CNC migration, and its roles at later developmental stages remain unclear. We generated a transgenic X. tropicalis line that expresses enhanced green fluorescent protein (eGFP) driven by the snai2 promoter/enhancer, and observed eGFP expression not only in the pre-migratory and migrating CNC, but also the differentiating CNC. This transgenic line can be used directly to detect deficiencies in CNC development at various stages, including subtle perturbation of CNC differentiation. In situ hybridization and immunohistochemistry confirm that Snai2 is re-expressed in the differentiating CNC. Using a separate transgenic Wnt reporter line, we show that canonical Wnt signaling is also active in the differentiating CNC. Blocking Wnt signaling shortly after CNC migration causes reduced snai2 expression and impaired differentiation of CNC-derived head cartilage structures. These results suggest that Wnt signaling is required for snai2 re-expression and CNC differentiation.
Subject terms: Cell lineage, Morphogen signalling, Transgenic organisms
Introduction
The cranial neural crest (CNC) cells are a transient group of multipotent stem cells that exists during early vertebrate embryogenesis. CNC development can be divided into three major stages: the induction, migration and post-migratory differentiation of the CNC. During gastrulation, the CNC is induced at the posterior neural plate border (NPB) between the neuroectoderm and the epidermis. The CNC cells continue to proliferate and undergo epithelial-mesenchymal transition (EMT), and subsequently emigrate from the closing neural tube in several streams that target distinct areas. Once the migrating CNC cells arrive at their destinations, they begin to differentiate into multiple types of cells that contribute to various tissues. Derivatives of CNC include nearly all the craniofacial structures, such as skeleton, connective tissues, muscles and the peripheral nervous system1–3.
The abilities to differentiate into multiple cell types and contribute to many tissues make CNC an intriguing subject of research in developmental biology. Recent studies suggest that several new types of cells derive from the CNC4,5, but the identification of all CNC derivatives remains a daunting task and requires new lineage-tracing tools. Moreover, CNC development is a dynamic and complex process that is tightly controlled spatially and temporally. Perturbation of CNC cells during any developmental stages may result in defects known as neurocristopathies, which are among the most common birth defects in humans6. Some of these defects are subtle, and are therefore difficult to detect with the techniques currently available. Hence the development of model systems that can be used for tracing CNC development will have a tremendous impact on the studies of CNC biology and the etiology of neurocristophathies. To this end, a number of transgenic mouse lines have been generated to facilitate tracing of the CNC lineage7. However, due to technical difficulties and possibly higher levels of functional redundancy in mice, non-mammalian vertebrates are often the preferred models for studying CNC development8. Among the non-mammalian models, zebrafish and Xenopus are particularly suitable for live imaging of tissue morphogenesis, owing to their external embryonic development, transparent epithelium and large brood size. Transgenic zebrafish lines expressing eGFP or Cre recombinase driven by the sox10 promoter have been widely used as lineage tracing tools to label neural crest derivatives9,10. More recently, a transgenic snai1b:GFP line was developed to visualize the EMT of CNC cells and screen for EMT inhibitors11. Historically, the detection of CNC in Xenopus was almost solely dependent on in situ hybridization for CNC markers such as snai2, sox9 and twist, whereas Alcian blue staining was commonly used for visualization of the head cartilage structures that derive from the CNC. These procedures are time-consuming and labor-intensive, and require fixation that prevents further manipulations of the embryos. While this manuscript was being prepared, two separate groups published the generation of pax3:GFP and sox10:GFP, the first two X. laevis transgenic lines that can be used for live imaging of CNC induction and migration, respectively12,13. However, currently no similar tool is available for X. tropicalis, a diploid species that is highly suitable for genetic studies, or for imaging CNC differentiation in any frog species.
Snai2 (a.k.a. Slug) is a zinc-finger transcription factor that is expressed in early CNC precursors and is required for the induction/specification of CNC in Xenopus14,15. A major signaling pathway that activates snai2 expression during CNC induction is the canonical Wnt (hereinafter referred to as “Wnt”) pathway, as forced activation of Wnt signaling causes ectopic expression of snai2 and other CNC markers, and blocking Wnt signaling inhibits snai2 expression and CNC induction16. Importantly, the snai2 enhancer contains an evolutionarily conserved LEF/TCF-binding site and can respond to Wnt signaling, suggesting that snai2 is a direct Wnt target gene17. After CNC induction, snai2 continues to be expressed in the pre-migratory and early migrating CNC, and plays a critical role in EMT and migration of the CNC15,18. However, recent quantitative RT-PCR and RNA-seq data show that snai2 expression drastically decreases in Xenopus embryos after the CNC cells begin to migrate19–21. Therefore, studies published to date have been focused on the roles of Snai2 in CNC induction and migration, and little is known about the expression or function of this important transcription factor at later stages of embryonic development.
Because of its specific expression and pivotal function during both CNC induction and migration, snai2 is one of the most commonly used CNC markers in frogs and other vertebrates such as chicks. The cis-regulatory elements of X. tropicalis snai2 gene have been well characterized, and a ~3.9 kb region of the promoter/enhancer sequence has been shown to contain the LEF/TCF-binding site and be able to drive CNC-specific GFP expression when transiently expressed17. Using the I-SceI meganuclease-mediated transgenic method22, we generated a transgenic line that expresses eGFP driven by this ~3.9 kb snai2 promoter/enhancer. Expression of eGFP in the snai2:eGFP transgenic embryos not only faithfully reflects the expression of endogenous snai2 in the pre-migratory and early migrating CNC, but also unveils a previously unknown expression of snai2 in the post-migratory CNC. In the snai2:eGFP transgenic tadpoles, eGFP labels multiple differentiating CNC derivatives, and subtle perturbation of CNC differentiation, such as those caused by partial knockdown of the disintegrin metalloproteinase ADAM13, can be readily detected using the snai2:eGFP transgenic embryos. We further show that Wnt signaling, which is regulated by ADAM13, is similarly activated in the differentiating CNC. Finally, blocking Wnt signaling shortly after the completion of CNC migration leads to reduction in snai2 expression and under-differentiation of CNC-derived head cartilage structures, suggesting that Wnt is required for post-migratory CNC differentiation, probably by regulating snai2 expression.
Results
Generation of the snai2:eGFP transgenic X. tropicalis line
We cloned a ~3.9 kb X. tropicalis genomic DNA upstream of the snai2 transcription start site, including the promoter and the 5′-enhancer, as described by Vallin et al.17. This snai2 promoter/enhancer sequence was inserted into a transgenic vector (Fig. S1A), and stable transgenic founders were generated as described by Ogino et al.22. At stage ~22, some of these founder embryos showed distinct fluorescence in the migrating CNC with minimal ectopic expression (Fig. S1B). This is consistent with snai2 expression in the migrating CNC, and suggests that the reporter construct had been integrated into the genome in these embryos22,23. When a potential transgenic founder was crossed with wild-type frogs, it produced heterozygous progeny (F1) that showed distinct fluorescence patterns (see below), indicating that the transgene insertion was inherited through germline transmission. We further inbred the F1 transgenic frogs to produce F2 progeny. About 71% (161/226) of all F2 embryos were eGFP-positive, and ~25% (41/161) of eGFP-positive embryos displayed stronger fluorescence than the others. The embryos with higher eGFP expression were singled out and raised to sexual maturity, and further crossing with wild-type frogs yielded 100% eGFP-positive embryos, suggesting that these frogs with higher eGFP expression were homozygotes. These results point to a single integration of the transgene, which was confirmed by whole-genome sequencing (see below). All heterozygous and homozygous snai2:eGFP transgenic frogs were healthy and fertile, and displayed normal craniofacial morphology (data not shown). In situ hybridization for snai2 and sox9 in the pre-migratory CNC (Fig. S2A,B,D), as well as snai2 and twist in the migrating CNC (Fig. S2C,E,F), also showed normal patterns, indicating that the transgene insertion did not affect CNC development. To better understand the potential impact of the transgene insertion at the molecular level, we carried out whole-genome shotgun sequencing on heterozygous snai2:eGFP transgenic embryos, and mapped the transgene insertion to a single non-coding region on Chromosome 1 (Wang and Wei, manuscript in preparation).
eGFP is expressed in the CNC lineage including differentiating CNC in snai2:eGFP transgenic embryos
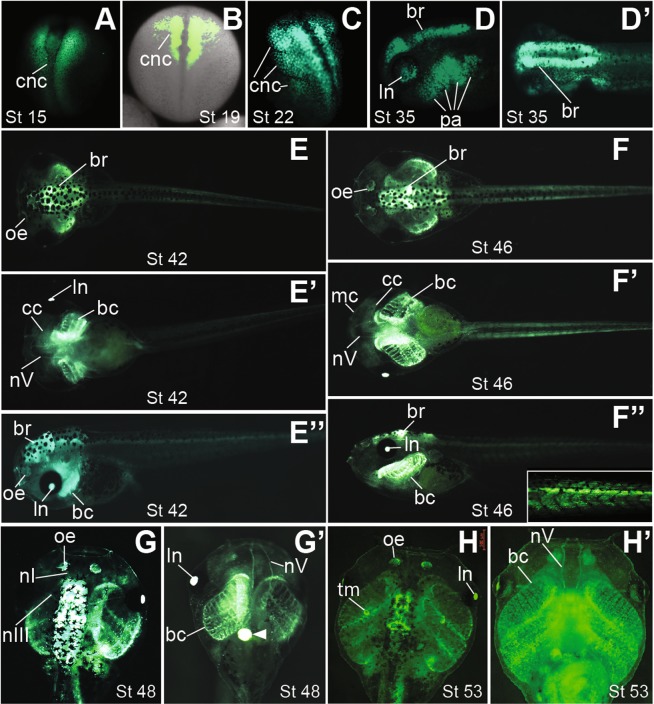
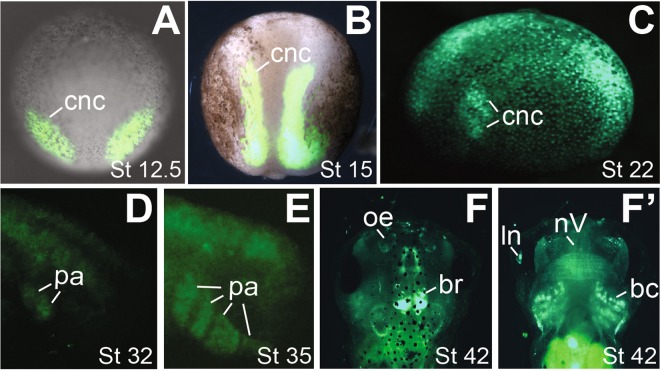
The snai2:eGFP transgenic embryos displayed highly specific fluorescence patterns. Green fluorescence was observed in the pre-migratory CNC cells as early as stage ~15 (Fig. 1A), and in the migrating CNC streams at stage 19–22 (Fig. 1B,C). Although recent reports show that snai2 expression is downregulated during CNC migration in Xenopus embryos19–21, fluorescence signals were clearly detectable in the post-migratory CNC as well as the developing lens and brain at tailbud stages in the snai2:eGFP transgenic embryos (Fig. 1D,D’). These patterns continued into swimming tadpole stages, when eGFP labeled multiple structures that are known to have CNC contributions (Fig. 1E–H’). In particular, the morphological changes were highlighted by green fluorescence in the snai2:eGFP embryos as cells in the pharyngeal arches developed from condensing mesenchyme (Fig. 1D) to partially differentiated cartilage, including Meckel’s, ceratohyal, and branchial cartilage (Fig. 1E’,F’,G’), and eventually to highly complex cartilaginous structures (Fig. 1H’). Starting from stage ~42, fluorescence was also detected in several cranial nerves, including the trigeminal and oculomotor nerves, as well as the olfactory nerves that are known to have a CNC contribution in other vertebrate species (Fig. 1E’,F’,G,G’,H’)4. Recent lineage tracing studies suggest that CNC may also contribute to some of the neurons in the olfactory epithelium in zebrafish and mice24,25. At stage ~42, fluorescence became detectable in the emerging olfactory epithelium of the snai2:eGFP tadpoles (Fig. 1E,E”), and the signal increased throughout early tadpole stages and persisted to later stages (Fig. 1F,G,H). We also observed strong fluorescence in the developing thymus, which is known to be populated by CNC cells26, at stage ~53 (Fig. 1H). Although snai2 was not shown previously to be expressed in the post-migratory CNC, the eGFP signals likely reflect the true expression of snai2 instead of simply being the remnant of early expression prior to silencing during CNC migration, as these eGFP signals remained in the differentiating CNC lineage for more than 2 weeks, whereas the half-life of eGFP is ~26 hr27. Additionally, fluorescence was found in the lens and somites of the transgenic tadpoles (Fig. 1E”,F”,G’,H), consistent with published results showing that snai2 is expressed in these tissues in various vertebrate species, ranging from fish to mice28,29.
Figure 1.
The snai2:eGFP transgenic embryos show eGFP expression in the CNC lineage at various stages. Heterozygous snai2:eGFP embryos were imaged at the indicated stages. (A–C) Dorsal view of neurula-stage embryos showing eGFP expression in pre-migratory (A), early migrating (B) and extensively migrating (C) CNC, with anterior at the top. Green fluorescence and bright-field images are merged in (B) to show the relative positions of migrating CNC streams in the whole embryo. (D,D’) Side (D) and dorsal (D’) views (with anterior to the left) of the head of a stage ~35 tadpole, with eGFP expression in the developing brain (br), lens (ln), and CNC cells forming condensing mesenchyme in the pharyngeal arches (pa). (E-E”) Dorsal (E), ventral (E’) and side (E”) views (with anterior to the left) of a stage ~42 tadpole. eGFP expression is detectable in the developing olfactory epithelium (oe), trigeminal nerves (nV), and CNC cells in the pharyngeal arches that begin to differentiate into branchial cartilage (bc) and ceratohyal cartilage (cc). (F-F”) Dorsal (F), ventral (F’) and side (F”) views (with anterior to the left) of a stage ~46 tadpole. eGFP is seen in the more differentiated trigeminal nerves and head cartilage structures, including Meckel’s cartilage (mc). Inset in (F”) is a higher-magnification image showing eGFP expression in tail somites. (G,G’) Dorsal (G) and ventral (G’) views (with anterior at the top) of a stage ~48 tadpole, with eGFP visible in both the olfactory (nI) and oculomotor (nIII) nerves. White arrowhead in (G’) indicates strong autofluorescence in the liver, which was also seen in wild-type tadpoles. (H,H’) Dorsal (H) and ventral (H’) views (with anterior at the top) of a stage ~53 tadpole. eGFP labels the thymus (tm) and highly differentiated head cartilage structures. Red scale bar in H = 500 μm.
eGFP patterns in the snai2:eGFP transgenic embryos faithfully reflect the endogenous expression of Snai2 mRNA and protein
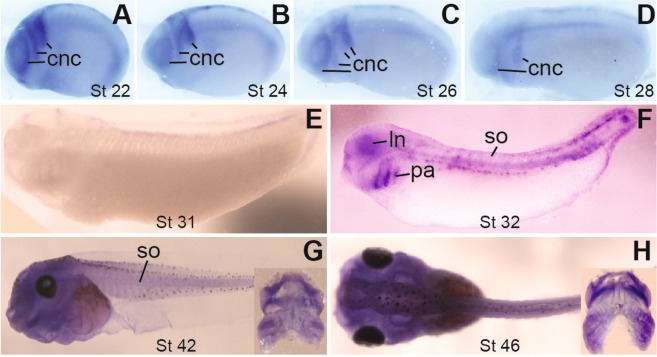
Because the stability of eGFP prevents the observation of subtle dynamics of snai2 expression in the migrating and post-migratory CNC, we carried out in situ hybridization to detect endogenous snai2 transcripts at various developmental stages. At stage ~12, snai2 mRNA was mainly expressed in the midline; there was also weak expression in the future CNC territory (Fig. S3A). About half an hour later (stage ~12.5), when the embryos approached the end of gastrulation, strong snai2 expression was detected in the newly formed CNC (Fig. S3B). At early neurula stages, snai2 continued to be expressed in the pre-migratory CNC, but the midline expression diminished (Fig. S3C). By stage ~19, CNC cells had emigrated from the closing neural tube, as shown by the in situ hybridization for snai2 (Fig. S3D–F). Therefore, the patterns of eGFP in the snai2:eGFP transgenic embryos faithfully reflect the expression of snai2 in both pre-migratory and early migrating CNC. The expression of snai2 persisted in the early migrating CNC cells, which formed distinct streams as they migrated out of the completely closed neural tube (Fig. 2A), but the intensity started to decrease thereafter and was minimal at stage ~31, several stages after CNC cells ceased migration (Fig. 2B–E). This is in line with previous reports that snai2 expression is downregulated in late migrating CNC cells19–21. However, snai2 expression started to increase again at stage ~32 and was clearly detectable in the condensing mesenchyme within the pharyngeal arches as well as the developing lens (Fig. 2F). At swimming tadpole stages, snai2 transcripts were found in the head and brain as well as the somites (Fig. 2G,H). These expression patterns are similar to those of eGFP in situ hybridization detected in snai2:eGFP tadpoles (Fig. S4A,B). Unfortunately, visualization of detailed cartilage structures was difficult, probably because the in situ probes and/or alkaline phosphatase substrate were trapped in the cavities that had formed in the head at these stages. We therefore dissected the head cartilage, and were able to detect snai2 staining in the fine cartilaginous structures (insets in Fig. 2G,H), which was highly similar to the eGFP patterns in the snai2:eGFP transgenic tadpoles (Fig. 1E’,F’). Thus, snai2 is re-expressed in the differentiating CNC cells after migration.
Figure 2.
The transcripts of snai2 are downregulated during CNC migration but upregulated again as CNC differentiates. In situ hybridization was performed for snai2 with wild-type X. tropicalis embryos at the indicated stages. The expression of snai2 in the CNC decreases from stage ~22 to ~31 (A–E), but elevates again in the CNC cells that form condensing mesenchyme in the pharyngeal arches (pa; F) and persists in the differentiating head cartilage structures (G,H). All embryos are shown with anterior to the left. (A–G) side view; (H) dorsal view. Insets in (G,H) are ventral view of head cartilage dissected from tadpoles after in situ hybridization, with anterior at the top. ln, lens; so, somites.
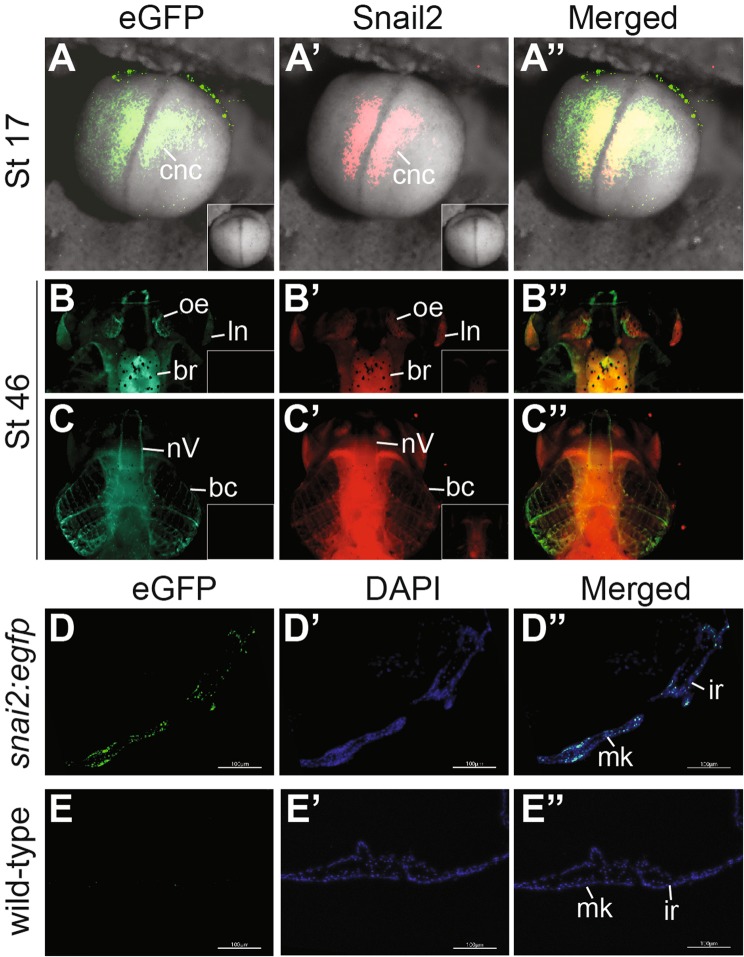
We further performed double-immunohistochemistry to determine if the spatiotemporal patterns of eGFP in the snai2:eGFP embryos reflect those of the endogenous Snai2 protein. At stage ~17, eGFP and Snai2 displayed significant co-localization in the closing neural tube, where some CNC cells just started to emigrate (Fig. 3A-A”). At stage ~46, eGFP clearly co-localized with Snai2 in the differentiating head cartilage structures, trigeminal nerves, olfactory epithelium, lens and brain (Fig. 3B–C”). It should be noted, though, that there were regions where eGFP but not Snai2 was detected, which may reflect the difference in stability between these two proteins. Images of head sections further reveal eGFP expression in various cartilage structures, including Mechel’s and infrarostral cartilage, in snai2:eGFP but not wild-type tadpoles at stage ~46 (Fig. 3D–E”). Together with the in situ hybridization data, these results indicate that Snai2 mRNA and protein are expressed in the post-migratory CNC, and that the snai2:eGFP transgenic line is suitable for tracing the CNC lineage at various developmental stages.
Figure 3.
Localization of eGFP protein in snai2:eGFP embryos. (A–C”) Co-localization of eGFP with endogenous Snai2 protein in snai2:eGFP embryos. Immunohistochemistry was carried out for eGFP (green) and Snai2 (red) simultaneously at the indicated stages in snai2:eGFP embryos and tadpoles. (A-A”) eGFP and Snai2 are co-localized in the CNC at the onset of migration. A control embryo processed with secondary antibodies only but not either primary antibody did not display any signal (insets in A and A’). Embryos are shown in dorsal view with anterior at the top. (B–C”) Dorsal (B-B”) and ventral (C-C”) views of the head of a stage ~46 tadpole showing co-localization of eGFP and Snai2 in the branchial cartilage (bc), brain (br), lens (ln), trigeminal nerve (nV), and olfactory epithelium (oe), with anterior at the top. (D–E”) Transverse sections of anterior head cartilage. Immunohistochemistry for eGFP (green) and DAPI labeling for nuclei (blue) were carried out for stage ~46 snai2:eGFP (D-D”) and wild-type (E-E”) tadpoles, and images were taken with a Zeiss Axiozoom.V16 epifluorescence microscope. Sections are shown with anterior at the bottom (tilted toward the right in D-D”). Expression of eGFP is detectable in Meckel’s (mk) and infrarostral (ir) cartilage in snai2:eGFP but not wild-type tadpoles. Scale bar = 100 μm.
CNC defects at various stages are readily detectable in live snai2:eGFP transgenic embryos
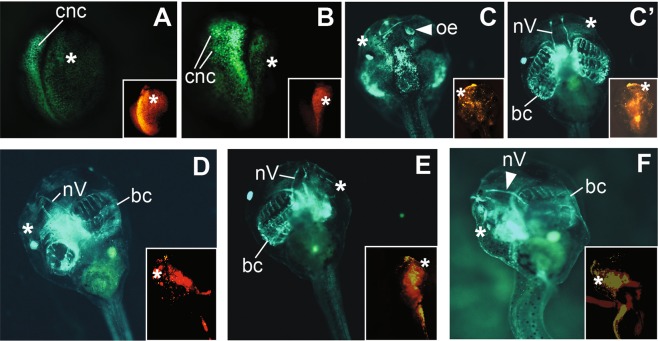
We next tested if the snai2:eGFP line can be used for real-time detection of CNC defects. To do this we carried out antisense morpholino (MO)-mediated knockdown of the disintegrin metalloproteinase ADAM13, a protease that is known to be required for normal CNC induction and migration30–32. An ADAM13 MO (MO 13-3), which has been well characterized in previous studies30,32,33, was injected into one blastomere of 8-cell stage snai2:eGFP embryos to target the dorsal-animal region; a red-fluorescence dye was co-injected as lineage tracer. As expected, many embryos that were injected with ADAM13 MO (the “morphants”) show reduced eGFP expression on the injected side prior to CNC migration (Fig. 4A), suggesting that CNC induction was inhibited by ADAM13 knockdown. At stage ~22, CNC migration was also blocked in most embryos, as shown by the lack of eGFP labeled cells that emigrate from the neural tube in some ADAM13 morphants (Fig. 4B). To investigate if ADAM13 also affects post-migratory CNC development, we selected ADAM13 morphants with no apparent defects in CNC induction or migration, and cultured them to stage ~46. At this stage, various defects in CNC derivatives, such as reduction of the head cartilage structures and/or cranial nerves, were observed in some ADAM13 morphants (Fig. 4C–F). Interestingly, some of these morphants displayed hypoplasia or impaired differentiation of specific tissues that may have CNC contribution. For example, in a morphant with intact head cartilage structures and cranial nerves, we found that the olfactory epithelium was almost completely missing on the injected side (Fig. 4C,C’), suggesting that post-migratory CNC development, but not earlier CNC induction or migration, might be affected. Hence the snai2:eGFP transgenic embryos can be used for live imaging of both early and late defects in CNC development.
Figure 4.
Phenotypes of ADAM13 knockdown displayed by snai2:eGFP embryos. Eight-cell stage heterozygous snai2:eGFP embryos were injected with 1.5 ng MO 13-3 to target ADAM13 in one dorsal-animal blastomere, and cultured to the indicated stages; a red fluorescent dye was co-injected as a lineage tracer. The injected side is denoted with a white asterisk, and structures that are present on the uninjected side but absent on the injected side are denoted with white arrowheads. Insets show red fluorescence images of the same embryos. (A,B) Dorsal view (with anterior at the top) of stage ~18 (A) and ~22 (B) embryos displaying reduced CNC domain on the injected side, as determined by eGFP expression. In (B) CNC migration is normal on the uninjected side but inhibited on the injected side. (C–F) Injected embryos that did not show apparent defects in CNC induction or migration were selected and cultured to stage ~46. (C,C’) are dorsal and ventral views (with anterior at the top), respectively, of the same tadpole. The olfactory epithelium (oe) is not detectable (C) but branchial cartilage (bc) and trigeminal nerve (nV) appear normal (C’) on the injected side of this embryo. (D,E) Embryos with under-differentiated head cartilage structures on the injected side, as compared with the uninjected side. (F) An embryo with severely defective trigeminal nerve and head cartilage structures on the injected side. (D–F) are ventral views with anterior at the top.
Wnt signaling is active in the post-migratory CNC
During CNC induction, ADAM13 functions by regulating Wnt signaling and snai2 expression30,32. Snai2 is thought to be a direct Wnt target gene at this early stage of CNC development, because its 5′-enhancer contains a LEF/TCF-binding site that can respond to Wnt signaling1,17. This LEF/TCF-binding site was part of the 3.9 kb promoter/enhancer sequence that we used to generate the snai2:eGFP transgenic line, raising the possibility that the re-expression of snai2 in the post-migratory CNC, as reflected by eGFP patterns in the snai2:eGFP embryos, is also induced by Wnt signaling. The effects of ADAM13 knockdown on post-migratory CNC development (Fig. 4C–F) further suggest that Wnt may have an important role in this later developmental process. To investigate the activity of Wnt signaling in the CNC at various developmental stages, especially during post-migratory CNC differentiation, we used a transgenic X. tropicalis Wnt reporter line that expresses destabilized eGFP driven by an artificial enhancer containing 7 LEF/TCF-binding sites. This destabilized eGFP molecule has a short half-life (~2 hr) and can precisely reflect the dynamic on-and-off patterns of endogenous Wnt activity34,35. As shown previously, there was a strong Wnt signal at the posterior NPB during CNC specification (stage ~12.5; Fig. 5A), which is critical for inducing snai2 expression and the specification of CNC lineage32,35. This Wnt signal remained in the pre-migratory CNC (Fig. 5B), and was evident in the migrating CNC streams (Fig. 5C). Interestingly, increasing Wnt signal was detected in the condensing mesenchyme in the pharyngeal arches from stage ~32 to ~35, when the CNC cells start to differentiate (Fig. 5D,E). At stage ~42, Wnt was active in the differentiating head cartilage structures, cranial nerves, olfactory epithelium, lens and brain (Fig. 5F,F’), consistent with previously reported activities and/or functions of Wnt signaling in these tissues in mice36–39. These patterns are also strikingly similar to those of eGFP in the snai2:eGFP embryos as well as endogenous Snai2 mRNA and protein (Figs 1, 2 and 3), suggesting a possible role for Wnt signaling in inducing snai2 expression, not only in the pre-migratory and migrating CNC but also in the differentiating CNC, in X. tropicalis embryos.
Figure 5.
Wnt signaling activity in the CNC lineage. Heterozygous transgenic Wnt reporter embryos were imaged at the indicated stages. Expression of eGFP is detectable in the pre-migratory (A,B), migrating (C) and differentiating (D–F’) CNC. (A,B) dorsal view with anterior at the top; green fluorescence and bright-field images are merged to show the relative positions of CNC in the whole embryo. (C–E) side view with anterior to the left (C) or right (D,E). (F,F’) dorsal and ventral views (with anterior at the top), respectively, of the same tadpole. br, brain; ln, lens; bc, branchial cartilage; nV, trigeminal nerve; oe, olfactory epithelium; pa, pharyngeal arches.
Wnt is required for head cartilage differentiation as well as snai2 and sox9 expression in the post-migratory CNC
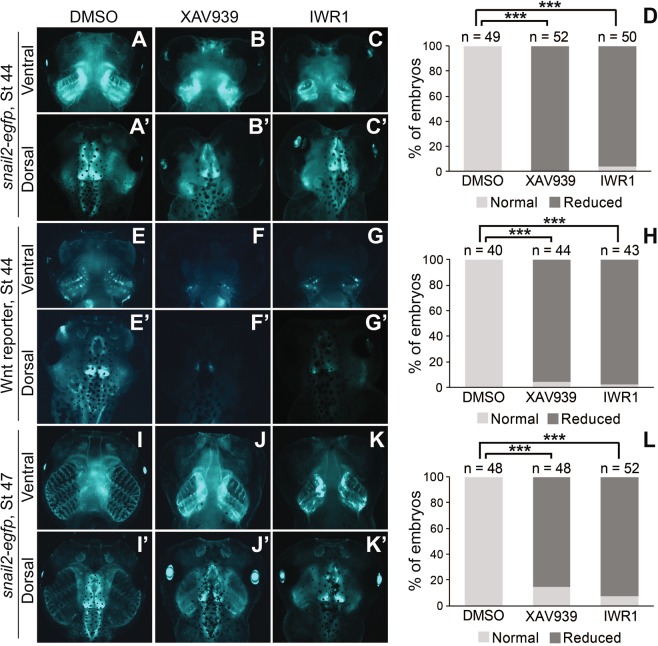
We show above that some ADAM13 morphants displayed defects in the CNC lineage after CNC migration (Fig. 4C–F). Because ADAM13 is required for Wnt signal activation during CNC induction30,32, these results suggest that Wnt may also be important for CNC differentiation. To further test this hypothesis, we treated transgenic embryos with small-molecule Wnt inhibitors starting at stage ~28, shortly after CNC completed migration. The treatment lasted until stage ~35, before CNC cells in the pharyngeal arches start to differentiate into cartilaginous structures (see Fig. 1D). Two Wnt inhibitors, which were identified in two independent screens, were used in these experiments. XAV939 stabilizes Axin, a major component of the β-catenin destruction complex, by inhibiting the tankyrases that stimulate Axin degradation40. The other compound, IWR1-endo, also elevates the protein levels of Axin, but the underlying mechanism remains unclear41. When snai2:eGFP embryos were treated with high dosage of XAV939 or IWR1-endo after CNC migration, the branchial cartilage was clearly under-differentiated at stage ~44, as compared with embryos that were treated with vehicle control (Fig. 6A–D). In contrast, eGFP expression in the brain appeared to be normal (Fig. 6A’–C’). Similarly, defects in head cartilage structures were observed in Wnt reporter embryos treated with either Wnt inhibitor (Fig. 6E–H). Notably, global expression of the destabilized eGFP was greatly downregulated in these Wnt reporter embryos (Fig. 6E–G’), confirming that endogenous Wnt signaling was inhibited. Both the snai2:eGFP and Wnt reporter tadpoles treated with Wnt inhibitors developed edema and died shortly after stage ~44. To rule out the possibility that the effects on head cartilage differentiation were secondary to the edema, we carried out similar treatment of the snai2:eGFP embryos with lower dosage of Wnt inhibitors. These tadpoles did not have edema and were alive and swimming at stage ~47. However, the head cartilage structures were still underdeveloped as compared with those of the control embryos (Fig. 6I–L). No apparent reduction in total eGFP intensity was detected in the head cartilage of the snai2:eGFP tadpoles at either stage ~44 or ~47 (Fig. 6A–C, I–K), suggesting that cell proliferation and death were not affected. The unaffected eGFP levels seen in the snai2:eGFP tadpoles upon Wnt inhibitor treatment is likely due to the stability of the eGFP protein, as endogenous snai2 was found to be downregulated (see below). These results, together with the late CNC phenotypes displayed by the ADAM13 morphants (Fig. 4C–F), indicate that Wnt signaling plays critical roles in post-migratory CNC differentiation.
Figure 6.
Wnt signaling is required for the differentiation of CNC into head cartilage structures. Snai2:eGFP or Wnt reporter embryos were treated with XAV939 (B,B’, F,F’, 20 μM; J,J’, 5 μM), IWR1-endo (C,C’, G,G’, 40 μM; K,K’, 10 μM) or DMSO (vehicle control) from stage ~28 to ~35. Embryos were washed and cultured again to the indicated stages, and images were taken with a Zeiss Axiozoom.V16 epifluorescence microscope. A representative embryo from each treatment group is shown on the left, with upper and lower panels displaying ventral and dorsal views (with anterior at the top), respectively, of the same embryos, and statistics is shown in the graphs on the right. ***P < 0.001.
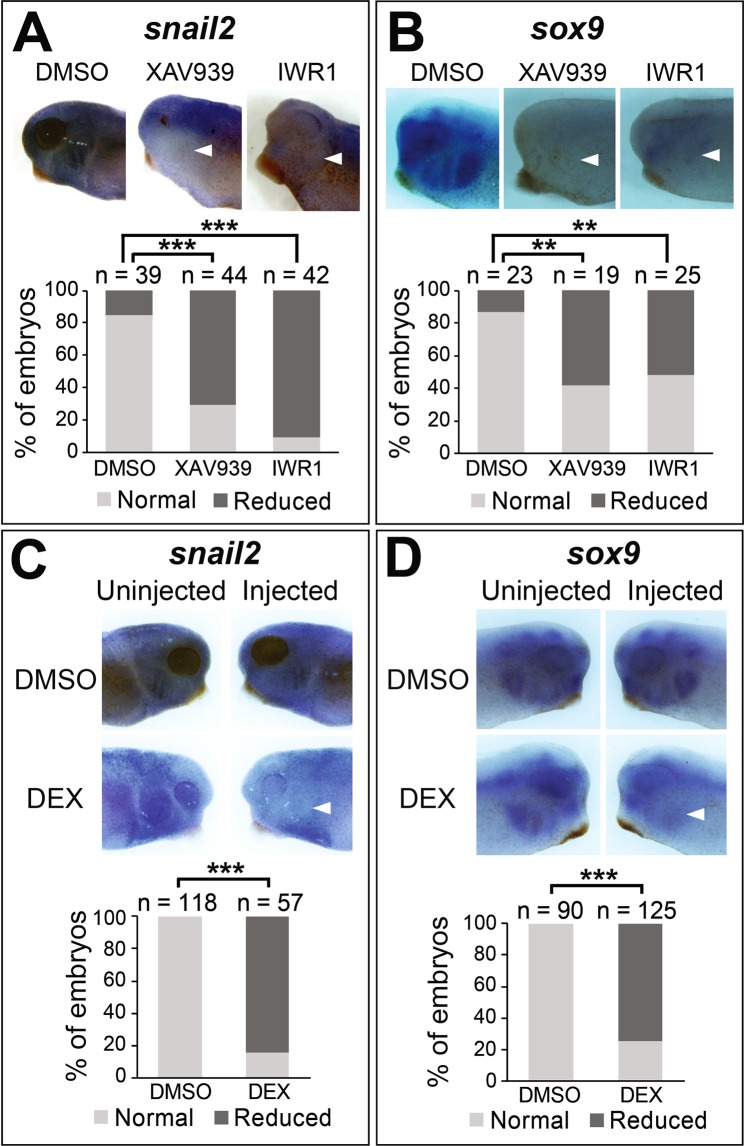
Finally, we examined directly if Wnt is responsible for inducing snai2 expression in the differentiating CNC. As shown in Fig. 7A, incubation of wild-type embryos in low-dosage XAV939 or IWR1-endo starting at stage ~28 resulted in decreased snai2 expression at stage ~35 in pharyngeal arches, suggesting that Wnt is required for snai2 expression in the post-migratory CNC that are about to differentiate into head cartilage structures. Previous studies have shown that knockdown of Snai2 causes loss of sox9 transcripts during Xenopus CNC induction14,42. Because Sox9 is a skeletogenic CNC marker and a master regulator of chondrogenesis from the CNC lineage at later stages43,44, we assessed the effects of Wnt inhibition after CNC migration on the expression of sox9. Similar to snai2, inhibition of Wnt signaling after CNC migration also reduced the expression of sox9 at stage ~35 (Fig. 7B), providing a possible mechanism for the inhibition of head cartilage differentiation as shown in Fig. 6. To validate these results, we used a hormone-inducible fusion protein, which consists of the high mobility group box of mouse LEF1, the repression domain of Drosophila Engrailed, and the hormone-binding domain of the human glucocorticoid receptor (EnR-LefΔN-GR755A)45. Upon induction with dexamethasome (DEX), this fusion protein inhibits Wnt target gene expression in Xenopus embryos45. We injected the EnR-LefΔN-GR755A mRNA into one blastomere of 2-cell stage embryos, and treated the embryos with DEX or vehicle control from stage ~28 to ~35. Similar to the treatment with Wnt inhibitors (Fig. 7A,B), induction of EnR-LefΔN-GR755A expression by DEX after CNC migration led to reduced expression of snai2 and sox9 in pharyngeal arches on the injected side (Fig. 7C,D). Taken together, these data indicate that Wnt signaling is indispensable for sox9 expression and head cartilage differentiation, possibly through inducing snai2 expression.
Figure 7.
Inhibition of Wnt signaling in the post-migratory CNC reduces snai2 and sox9 expression. A,B. Wild-type embryos were treated with XAV939 (5 μM), IWR1-endo (10 μM) or DMSO from stage ~28 to ~35, and processed for in situ hybridization for snai2 (A) or sox9 (B). (C,D) One blastomere of 2-cell stage embryos was injected with 50 pg of mRNA encoding EnR-LefΔN-GR755A. Embryos were treated with DEX or DMSO (as control) from stage ~28 to ~35, and processed for in situ hybridization for snai2 (C) or sox9 (D). A representative tadpole from each treatment group is shown in side view in the upper panels (in C,D both the uninjected and injected sides of the same embryos are displayed side by side for comparison), and statistics is shown in the graphs. White arrowheads point to reduced staining in the condensing mesenchyme in the pharyngeal arches. **P < 0.01; ***P < 0.001.
Discussion
The CNC can differentiate into many types of cells during early embryonic development1–3. Transgenic reporter animals provide powerful lineage-tracing tools for identifying CNC derivatives and understanding the mechanisms that control CNC differentiation, which are critical for the studies of CNC biology as well as the prevention and treatment of neurocristopathies. Although X. laevis and X. tropicalis have long been used to study CNC induction and migration8, little is known about CNC differentiation in these species. Instead, most of our knowledge on CNC differentiation was obtained from previous transgenic studies using other models such as mice and zebrafish7–9,36. Most recently, the first two X. laevis transgenic CNC reporter lines were generated. However, these lines are suitable for imaging CNC induction and migration, respectively, but not differentiation12,13. Here we report the first X. tropicalis transgenic CNC reporter line, which can be used not only for tracing CNC induction and early migration, but also for high-resolution live imaging of CNC differentiation. The ability of eGFP to label CNC derivatives is due to the expression of snai2 in the differentiating CNC, which has not been described before.
Snai2 is a transcription factor that is expressed in several mesodermal and ectodermal tissues, such as the CNC, lens, and somites, in all vertebrates that have been examined28,29. However, the timing of snai2 expression in the CNC varies in different species. Transcripts of snai2 are detectable in the pre-migratory CNC in frogs, reptiles and chicks, but not in fish or mice28,29,46. Snai2 in the pre-migratory CNC is required for the emigration of CNC cells from the neural tube in frogs and chicks, likely due to its ability to induce EMT18,46. At later stages, snai2 transcripts were found in the migrating CNC in essentially all vertebrate species that have been examined, including Xenopus, chicks and mice15,29,46. It has also been shown that snai2 expression diminishes toward the end of CNC migration in both Xenopus and chick embryos19–21,46. This is in line with the hypothesis that CNC cells undergo mesenchymal-to-epithelial transition, which is the reciprocal of EMT, to stop migration, allowing them to colonize various tissues in the embryo1. Currently, there is no published information on the expression of snai2 after CNC migration in any species. However, studies with mice suggest that Snai2 may function in CNC development at later stages. While whole-embryo double knockout of snai2 and its close paralog snail1 has no effect on CNC induction or early migration47, neural crest-specific loss of snail1 on the snai2-null background leads to multiple craniofacial defects that are reminiscent of conditional neural crest mutants of several other important genes48. In contrast, neither conditional knockout of snail1 in the neural crest nor global knockout of snai2 alone causes these defects48. These results imply a redundant role of Snail1 and Snai2 in late CNC development, possibly in CNC differentiation. Therefore, it is important to determine if the Snail family transcription factors are expressed and functional in the post-migratory CNC. Here we report that snai2 is re-expressed in the differentiating CNC in X. tropicalis embryos. This is supported by the fluorescence patterns displayed by the snai2:eGFP transgenic tadpoles, as well as the in situ hybridization and immunohistochemistry data. Our results are consistent with a possible role of snai2 during CNC differentiation, as implicated by the mouse study48. It remains to be examined if snai2 is similarly expressed in the differentiating CNC in mice and other vertebrates, and if this gene indeed functions in CNC differentiation.
The Wnt signaling pathway is a major inducer of Snai2 in various vertebrate species, likely through direct activation of snai2 transcription17,49. In addition, Wnt-induced GSK3β inhibition can lead to stabilization of the Snai2 protein50, which is capable of binding to its own enhancer and further stimulating snai2 expression51,52. In Xenopus embryos, Wnt signaling induces the formation of the NPB. After NPB formation, a second wave of Wnt signal activates the expression of Snai2, which is required for CNC specification within the NPB53. Both the Wnt signal and Snai2 mRNA/protein are clearly detectable throughout the pre-migratory CNC (Figs 3A’, 5A,B, S3B–D). The accumulating Snai2 in the pre-migratory CNC likely prepares the CNC cells for EMT/migration, as knockdown of Snai2 inhibits CNC migration18. It has also been shown that in pre-migratory Xenopus CNC explants, β-catenin is mainly detected in the nucleus; in contrast, in migrating CNC explants, β-catenin is redistributed to the plasma membrane, indicating a reduction of Wnt signaling in CNC cells that have emigrated from the neural tube54. Because Wnt induces snai2 expression, these observations provide a possible mechanism for the downregulation of snai2 transcripts during CNC migration. Our data further suggest that the re-expression of snai2 during CNC differentiation is also driven by Wnt signaling. A comparison between the fluorescence patterns of snai2:eGFP and Wnt reporter transgenic tadpoles shows striking similarity during CNC differentiation, and blocking Wnt signaling after CNC migration inhibits snai2 expression and head cartilage differentiation (Figs 1D–E”, 5E–F’, 6, 7). Thus, the Wnt-Snai2 axis may function reiteratively during CNC specification, emigration and differentiation.
Wnt signaling is known to be important for neural crest differentiation, but the exact roles of Wnt in this developmental process are controversial and may vary from species to species. An earlier report shows that Wnt promotes CNC differentiation into pigment cells at the expense of neurons and glia in zebrafish55. In contrast, β-catenin instructs mouse neural crest cells to adopt a sensory neuronal fate at the cost of essentially all other neural crest derivatives, presumably through mediating Wnt signaling56,57. Thus, the effects of Wnt signaling on cell fate determination during neural crest differentiation seem to be species-dependent. With regard to craniofacial morphogenesis, Wnt is crucial to the selection between chondrocytic and osteoblastic fates in the mammalian CNC. Specifically, Wnt promotes bone formation and simultaneously suppresses chondrogenesis in mice36. However, Wnt has also been shown to be required for chondrogenic differentiation in cultured mouse cells in a Sox9-dependent manner58. In zebrafish, blocking Wnt signaling after CNC migration inhibits ventral cartilage differentiation59. Although head cartilage is often used as a phenotypic readout for disrupted CNC induction or migration in Xenopus, little is known about how CNC cells differentiate into cartilaginous structures in frogs. This is probably because genes and pathways that are important for CNC differentiation, such as Wnt and sox9, often play critical roles in earlier induction and/or migration as well, and tools for temporally controlled gene inactivation in Xenopus are lacking. In the current study, we show that inhibition of Wnt signaling in X. tropicalis embryos after CNC migration leads to reduced sox9 expression and under-differentiated head cartilage structures (Figs 6,7). To our knowledge, this is the first evidence of Wnt function in Xenopus CNC differentiation. Future studies are needed to understand how Wnt affects sox9 expression and head cartilage differentiation in Xenopus.
The snai2:eGFP transgenic line is a useful tool for live imaging of CNC development in Xenopus. Normal and dysregulated CNC induction, migration and differentiation can be visualized directly in the transgenic embryos (Figs 1, 4), making them highly suitable for high-throughput screens to identify genetic and environmental factors that interfere with CNC development at any stage. The labeling of multiple CNC derivatives, such as cells in the head cartilage, cranial nerves, and thymus, by eGFP in the snai2:eGFP transgenic tadpoles (Fig. 1E–H’), further raises the possibility of using this transgenic line to identify new types of cells that derive from the CNC. For example, we detected strong eGFP expression in the brain, especially in the hindbrain (Figs 1E,F, 3B), which is consistent with the expression of Snai2 mRNA and protein (Figs 2H, 3B’) as well as eGFP expression in the Wnt reporter tadpoles (Fig. 5F). Notably, a similar GFP expression in the brain was detected in the sox10:GFP transgenic tadpoles13, suggesting that these cells may derive from the CNC. In addition, there is published evidence supporting a CNC origin for the gonadotropin releasing hormone-positive and microvillous neurons in the early zebrafish olfactory epithelium, but a most recent study suggests that all the sensory neurons in the zebrafish olfactory epithelium derive from the preplacodal ectoderm instead24,60,61. Interestingly, we observed clear eGFP expression in the olfactory epithelium of snai2:eGFP tadpoles starting at stage ~42, when the microvillous neurons just emerge62 (Fig. 1E). Whether there is a CNC contribution to the Xenopus olfactory epithelium warrants further investigation, and additional transgenic tools may be needed for this type of studies. To better trace the dynamic development of CNC cells, we are in the process of generating a new snai2:mEos3.2 line, in which the CNC lineage is labeled with the mEOS3.2 photoswitching fluorescent protein. This second-generation snai2 reporter line will allow the labeling of pre-migratory or migrating CNC cells and tracing their fates at later stages. Together, these transgenic reporter lines should have a profound impact on the studies of CNC development.
Methods
Plasmids and antibodies
Genomic DNA was prepared from X. tropicalis embryos as described63. A 3.9 kb fragment of the snai2 promoter/enhancer, as reported by Vallin et al.17, was cloned from the genomic DNA using nested PCR. To generate the transgenic construct, the snai2 promoter/enhancer sequence was subcloned into the IS-eGFP transgenic vector (a gift from Dr. Robert Grainger)22,23. Primers used in cloning and subcloning are listed in Table S1. Constructs for preparing the in situ hybridization probes for snai2 and sox9 were obtained previously30, and the construct for expressing EnR-LefΔN-GR755A was generated in a previous study45. The mouse anti-Snai2 (DSHB 62.1E6, 1:50) and rabbit anti-GFP (Life Technologies A11122, 1:200), as well as Alexa Fluor 594 AffiniPure donkey anti-mouse and Alexa Fluor 488 AffiniPure donkey anti-rabbit antibodies (Jackson Immuno Research Laboratories 715-585-150 and 711-545-152, 1:500 for both), were used for immunohistochemistry.
Animals and transgenesis
Wild-type X. tropicalis adults (male and female) were purchased from NASCO, and the Wnt reporter line was generated in a previous study34. ISceI-mediated transgenesis was carried out as described by Ogino et al. to generate the snai2:eGFP transgenic founders22. Briefly, the IS-snai2:eGFP plasmid was digested with the I-SceI enzyme, and the reaction mixture was injected into fertilized X. tropicalis eggs. Embryos with eGFP expression in the migrating CNC (see Fig. S1B for an example) were selected and raised to adulthood. These transgenic founders were crossed with wild-type X. tropicalis frogs to generate heterozygotes, which were further inbred to obtain homozygotes.
Embryo manipulation
Embryo were obtained by natural mating and cultured in 0.1x MBS to desired stages as described previously30. For in situ hybridization and immunohistochemistry, embryos were fixed at desired stages and processed as described63. MO 13-3, the antisense MO for ADAM13, was synthesized by Gene Tools, and the sequence was reported previously30. For MO injections, 8-cell stage snai2:eGFP embryos were injected in a single dorsal-animal blastomere with 1.5 ng MO 13-3 using a PLI-100A microinjector (Harvard Apparatus), and Alexa Fluor 555 dextran (Invitrogen) was co-injected as a lineage tracer. For Wnt inhibitor treatment, embryos were cultured in XAV939 or IWR1-endo (both were from Selleckchem) from stage ~28 to ~35. Snai2-eGFP or Wnt reporter transgenic tadpoles were washed three times and subsequently cultured in 0.1x MBS until stage ~44 or ~47 (Fig. 6); wild-type tadpoles were immediately fixed and processed for in situ hybridization for snai2 or sox9 (Fig. 7A,B). For inhibition of Wnt target gene expression, 2-cell stage wild-type embryos were injected in one blastomere with 50 pg mRNA encoding EnR-LefΔN-GR755A, and cultured to stage ~28, when 10 mM DEX (Sigma-Aldrich D4902) or DMSO was added. Embryos were further cultured to stage ~35, fixed, and processed for in situ hybridization for snai2 or sox9.
Imaging
Fluorescent and bright-field images were taken with a Zeiss Axiozoom.V16 epifluorescence microscope. Image acquisition and processing for whole-mount embryos were carried out using an AxioCam MRc Rev3 camera and the ZEN 2.0 software package. To prepare head cartilage sections, stage ~46 tadpoles (wild-type and snai2:eGFP) were fixed as described63, embedded in Optimum Cutting Temperature media, and immediately frozen on dry ice. Embedded tadpoles were stored at −80 °C until being sectioned. Tadpoles were sectioned as previously described with a Leica CM3050 S cryostat at −35 °C64. Air-dried sections were visualized using a Zeiss Axiozoom.V16 epifluorescence microscope.
Phenotype scoring and statistics
Injected embryos were scored by comparing the injected side with the uninjected side of the same embryos, and Wnt inhibitor-treated embryos were scored by comparing with the DMSO-treated controls. The percentage of normal and reduced phenotypes were calculated, and Chi-squared tests were performed to compare the phenotypes in different treatment groups.
Ethics statement
Methods involving live animals were carried out in accordance with the guidelines and regulations approved and enforced by the Institutional Animal Care and Use Committee at West Virginia University and the University of Delaware.
Supplementary information
Acknowledgements
We thank Drs. Robert Grainger, Takuya Nakayama and Anoop Shah from the University of Virginia for help with transgenesis. This work was funded by the U.S. NIH (R01GM114105, R03DE022813 and P20GM104316 to SW, and R01EY015279 to MKD) and March of Dimes Foundation (1-FY10-399 to SW). Additionally, part of the imaging work was supported by NIH grant 1S10RR027273-01. Research in the Vleminckx laboratory is supported by the Research Foundation – Flanders (FWO-Vlaanderen) (Grants G0A1515N and G029413N) and by the Concerted Research Actions from Ghent University (BOF15/GOA/011); further support was obtained from the Hercules Foundation, Flanders (Grant AUGE/11/14).
Author Contributions
J.L., M.P., M.K.D. and S.W. designed the experiments. J.L., M.P., C.M., R.L. and S.W. performed the experiments; H.T.T. and K.V. generated the transgenic Wnt reporter line and provided assistance on experiments using this transgenic line. J.L., M.P. and S.W. analyzed the data and wrote the manuscript; M.K.D. and K.V. provided comments and assistance on manuscript preparation.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiejing Li and Mark Perfetto contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47665-9.
References
- 1.Simões-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–57. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronner ME, Simões-Costa M. The Neural Crest Migrating into the Twenty-First Century. Curr Top Dev Biol. 2016;116:115–34. doi: 10.1016/bs.ctdb.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayor R, Theveneau E. The neural crest. Development. 2013;140:2247–51. doi: 10.1242/dev.091751. [DOI] [PubMed] [Google Scholar]
- 4.Barraud P, et al. Neural crest origin of olfactory ensheathing glia. Proc Natl Acad Sci USA. 2010;107:21040–5. doi: 10.1073/pnas.1012248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mongera A, et al. Genetic lineage labeling in zebrafish uncovers novel neural crest contributions to the head, including gill pillar cells. Development. 2013;140:916–25. doi: 10.1242/dev.091066. [DOI] [PubMed] [Google Scholar]
- 6.Trainor PA. Developmental Biology: We Are All Walking Mutants. Curr Top Dev Biol. 2016;117:523–38. doi: 10.1016/bs.ctdb.2015.11.029. [DOI] [PubMed] [Google Scholar]
- 7.Aoto K, et al. Mef2c-F10N enhancer driven β-galactosidase (LacZ) and Cre recombinase mice facilitate analyses of gene function and lineage fate in neural crest cells. Dev Biol. 2015;402:3–16. doi: 10.1016/j.ydbio.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barriga EH, Trainor PA, Bronner M, Mayor R. Animal models for studying neural crest development: is the mouse different? Development. 2015;142:1555–60. doi: 10.1242/dev.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carney TJ, et al. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–30. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- 10.Rodrigues FS, Doughton G, Yang B, Kelsh RN. A novel transgenic line using the Cre-lox system to allow permanent lineage-labeling of the zebrafish neural crest. Genesis. 2012;50:750–7. doi: 10.1002/dvg.22033. [DOI] [PubMed] [Google Scholar]
- 11.Jimenez L, et al. Phenotypic chemical screening using a zebrafish neural crest EMT reporter identifies retinoic acid as an inhibitor of epithelial morphogenesis. Dis Model Mech. 2016;9:389–400. doi: 10.1242/dmm.021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkobtawi, M. et al. Characterization of Pax3 and Sox10 transgenic Xenopus laevis embryos as tools to study neural crest development. Dev Biol (2018). [DOI] [PMC free article] [PubMed]
- 13.Ossipova O, Kerney R, Saint-Jeannet JP, Sokol SY. Regulation of neural crest development by the formin family protein Daam1. Genesis. 2018;56:e23108. doi: 10.1002/dvg.23108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J, Severson C, Yang J, Wedlich D, Klymkowsky MW. Snail2 controls mesodermal BMP/Wnt induction of neural crest. Development. 2011;138:3135–45. doi: 10.1242/dev.064394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–77. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Saint-Jeannet JP, Klein PS. Wnt-frizzled signaling in neural crest formation. Trends Neurosci. 2003;26:40–5. doi: 10.1016/S0166-2236(02)00011-5. [DOI] [PubMed] [Google Scholar]
- 17.Vallin J, et al. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J Biol Chem. 2001;276:30350–8. doi: 10.1074/jbc.M103167200. [DOI] [PubMed] [Google Scholar]
- 18.Carl TF, Dufton C, Hanken J, Klymkowsky MW. Inhibition of neural crest migration in Xenopus using antisense slug RNA. Dev Biol. 1999;213:101–15. doi: 10.1006/dbio.1999.9320. [DOI] [PubMed] [Google Scholar]
- 19.Huang C, Kratzer MC, Wedlich D, Kashef J. E-cadherin is required for cranial neural crest migration in Xenopus laevis. Dev Biol. 2016;411:159–71. doi: 10.1016/j.ydbio.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Owens ND, et al. Measuring Absolute RNA Copy Numbers at High Temporal Resolution Reveals Transcriptome Kinetics in Development. Cell Rep. 2016;14:632–47. doi: 10.1016/j.celrep.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Session AM, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino H, McConnell WB, Grainger RM. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat Protoc. 2006;1:1703–10. doi: 10.1038/nprot.2006.208. [DOI] [PubMed] [Google Scholar]
- 23.Ogino H, McConnell WB, Grainger RM. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech Dev. 2006;123:103–13. doi: 10.1016/j.mod.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Saxena A, Peng BN, Bronner ME. Sox10-dependent neural crest origin of olfactory microvillous neurons in zebrafish. Elife (Cambridge) 2013;2:e00336. doi: 10.7554/eLife.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki J, Yoshizaki K, Kobayashi T, Osumi N. Neural crest-derived horizontal basal cells as tissue stem cells in the adult olfactory epithelium. Neurosci Res. 2013;75:112–20. doi: 10.1016/j.neures.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Lee YH, et al. Early development of the thymus in Xenopus laevis. Dev Dyn. 2013;242:164–78. doi: 10.1002/dvdy.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–40. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- 28.Locascio A, Manzanares M, Blanco MJ, Nieto MA. Modularity and reshuffling of Snail and Slug expression during vertebrate evolution. Proc Natl Acad Sci USA. 2002;99:16841–6. doi: 10.1073/pnas.262525399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–85. doi: 10.1016/S0012-1606(98)80005-5. [DOI] [PubMed] [Google Scholar]
- 30.Wei S, et al. ADAM13 induces cranial neural crest by cleaving class B Ephrins and regulating Wnt signaling. Dev Cell. 2010;19:345–52. doi: 10.1016/j.devcel.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cousin H, Abbruzzese G, Kerdavid E, Gaultier A, Alfandari D. Translocation of the cytoplasmic domain of ADAM13 to the nucleus is essential for Calpain8-a expression and cranial neural crest cell migration. Dev Cell. 2011;20:256–63. doi: 10.1016/j.devcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Jiejing, Perfetto Mark, Neuner Russell, Bahudhanapati Harinath, Christian Laura, Mathavan Ketan, Bridges Lance C., Alfandari Dominique, Wei Shuo. XenopusADAM19 regulates Wnt signaling and neural crest specification by stabilizing ADAM13. Development. 2018;145(7):dev158154. doi: 10.1242/dev.158154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei S, et al. Roles of ADAM13-regulated Wnt activity in early Xenopus eye development. Dev Biol. 2012;363:147–54. doi: 10.1016/j.ydbio.2011.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran HT, Sekkali B, Van Imschoot G, Janssens S, Vleminckx K. Wnt/beta-catenin signaling is involved in the induction and maintenance of primitive hematopoiesis in the vertebrate embryo. Proc Natl Acad Sci USA. 2010;107:16160–5. doi: 10.1073/pnas.1007725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borday C, et al. An atlas of Wnt activity during embryogenesis in Xenopus tropicalis. PLoS One. 2018;13:e0193606. doi: 10.1371/journal.pone.0193606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhatt, S., Diaz, R. & Trainor, P. A. Signals and switches in Mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol5 (2013). [DOI] [PMC free article] [PubMed]
- 37.Kurosaka H, Trainor PA, Leroux-Berger M, Iulianella A. Cranial nerve development requires co-ordinated Shh and canonical Wnt signaling. PLoS One. 2015;10:e0120821. doi: 10.1371/journal.pone.0120821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, et al. Spatiotemporal dynamics of canonical Wnt signaling during embryonic eye development and posterior capsular opacification (PCO) Exp Eye Res. 2018;175:148–158. doi: 10.1016/j.exer.2018.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noelanders, R. & Vleminckx, K. How Wnt Signaling Builds the Brain: Bridging Development and Disease. Neuroscientist (2016). [DOI] [PubMed]
- 40.Huang SM, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–20. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 41.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–7. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C, Carl TF, Trudeau ED, Simmet T, Klymkowsky MW. An NF-kappaB and slug regulatory loop active in early vertebrate mesoderm. PLoS One. 2006;1:e106. doi: 10.1371/journal.pone.0000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci USA. 2003;100:9360–5. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YH, Saint-Jeannet JP. Sox9 function in craniofacial development and disease. Genesis. 2011;49:200–8. doi: 10.1002/dvg.20717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deroo T, Denayer T, Van Roy F, Vleminckx K. Global inhibition of Lef1/Tcf-dependent Wnt signaling at its nuclear end point abrogates development in transgenic Xenopus embryos. J Biol Chem. 2004;279:50670–5. doi: 10.1074/jbc.M408969200. [DOI] [PubMed] [Google Scholar]
- 46.Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–9. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 47.Murray SA, Gridley T. Snail family genes are required for left-right asymmetry determination, but not neural crest formation, in mice. Proc Natl Acad Sci USA. 2006;103:10300–4. doi: 10.1073/pnas.0602234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray SA, Oram KF, Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–97. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- 49.Taneyhill LA, Bronner-Fraser M. Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Mol Biol Cell. 2005;16:5283–93. doi: 10.1091/mbc.e05-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu ZQ, et al. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci USA. 2012;109:16654–9. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakai D, Suzuki T, Osumi N, Wakamatsu Y. Cooperative action of Sox9, Snail2 and PKA signaling in early neural crest development. Development. 2006;133:1323–33. doi: 10.1242/dev.02297. [DOI] [PubMed] [Google Scholar]
- 52.LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- 53.Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69:3715–37. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maj E, et al. Controlled levels of canonical Wnt signaling are required for neural crest migration. Dev Biol. 2016;417:77–90. doi: 10.1016/j.ydbio.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 55.Dorsky RI, Moon RT, Raible DW. Control of neural crest cell fate by the Wnt signalling pathway. Nature. 1998;396:370–3. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- 56.Lee HY, et al. Instructive role of Wnt/beta-catenin in sensory fate specification in neural crest stem cells. Science. 2004;303:1020–3. doi: 10.1126/science.1091611. [DOI] [PubMed] [Google Scholar]
- 57.Hari L, et al. Lineage-specific requirements of beta-catenin in neural crest development. J Cell Biol. 2002;159:867–80. doi: 10.1083/jcb.200209039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yano F, et al. The canonical Wnt signaling pathway promotes chondrocyte differentiation in a Sox9-dependent manner. Biochem Biophys Res Commun. 2005;333:1300–8. doi: 10.1016/j.bbrc.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 59.Alexander C, Piloto S, Le Pabic P, Schilling TF. Wnt signaling interacts with bmp and edn1 to regulate dorsal-ventral patterning and growth of the craniofacial skeleton. PLoS Genet. 2014;10:e1004479. doi: 10.1371/journal.pgen.1004479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitlock KE, Smith KM, Kim H, Harden MV. A role for foxd3 and sox10 in the differentiation of gonadotropin-releasing hormone (GnRH) cells in the zebrafish Danio rerio. Development. 2005;132:5491–502. doi: 10.1242/dev.02158. [DOI] [PubMed] [Google Scholar]
- 61.Aguillon, R. et al. Cell-type heterogeneity in the early zebrafish olfactory epithelium is generated from progenitors within preplacodal ectoderm. Elife7 (2018). [DOI] [PMC free article] [PubMed]
- 62.Hansen A, Reiss JO, Gentry CL, Burd GD. Ultrastructure of the olfactory organ in the clawed frog, Xenopus laevis, during larval development and metamorphosis. J Comp Neurol. 1998;398:273–88. doi: 10.1002/(SICI)1096-9861(19980824)398:2<273::AID-CNE8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 63.Sive, H.L., Grainger, R.M. & Harland, R.M. Early Development of Xenopus Laevis. A Laboratory Manual. Cold Spring Harbor Laboratory Press (2000).
- 64.Zhang, S., Li, J., Lea, R. & Amaya, E. Assessing Primary Neurogenesis in Xenopus Embryos Using Immunostaining. J Vis Exp, e53949 (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).