Abstract
Germline pathogenic variants in BRCA1/2 account for one-third of familial breast cancers. The majority of BRCA1 function requires heterodimerization with BARD1. In contrast to BRCA1, BARD1 is a low-penetrance gene with an unclear clinical relevance, partly because of limited functional evidence. Using patient-derived lymphoblastoid cells, we functionally characterized two pathogenic variants (c.1833dupT, c.2099delG) and three variants of uncertain significance (VUSs) (c.73G>C, c.1217G>A, c.1918C>A). Three of these patients had breast cancers, whereas the remaining had colorectal cancers (n = 3). Both patients with pathogenic variants (c.1833dupT, c.2099delG) developed breast cancers with aggressive disease phenotypes such as triple-negative breast cancer and high cancer grades. As BARD1 encompasses multiple functional domains, including those of apoptosis and homologous recombination repair, we hypothesized that the function being impaired would correspond with the domain where the variant was located. Variants c.1918C>A, c.1833dupT, c.1217G>A, and c.2099delG, located within and proximal to apoptotic domains of ankyrin and BRCT, were associated with impaired apoptosis. Conversely, apoptosis function was preserved in c.73G>C, which was distant from the ankyrin domain. All variants displayed normal BRCA1 heterodimerization and RAD51 colocalization, consistent with their location being distal to BRCA1—and RAD51-binding domains. In view of deficient apoptosis, VUSs (c.1217G>A and c.1918C>A) may be pathogenic or likely pathogenic variants. In summary, functional analysis of BARD1 VUSs requires a combination of assays and, more importantly, the use of appropriate functional assays with consideration to the variant's location.
Keywords: neoplasm of the breast
INTRODUCTION
Approximately 10% of breast cancers are associated with a heritable component, of which one-third are attributable to germline pathogenic variants in BRCA1 and BRCA2 (Economopoulou et al. 2015). Beyond BRCA1/2, other breast cancer susceptibility genes include TP53, PTEN, CDH1, and PALB2. The PALB2 protein binds to BRCA2 and facilitates its nuclear localization and subsequent DNA repair function (Zhang et al. 2009). Similar to BRCA2, BRCA1 also has a binding partner—BARD1—that shares a high degree of structural and functional homology; both proteins contain a RING domain, a nuclear export signal, and two tandem BRCA1 carboxy-terminal (BRCT) domains and are involved in the homologous recombination repair pathway (Irminger-Finger and Jefford 2006; Stewart et al. 2018). Heterodimerization of BARD1–BRCA1 via the RING domain is crucial in the homologous recombination repair and transcriptional regulation functions of BRCA1 (Irminger-Finger and Jefford 2006). Studies in mice showed that BARD1-deficient mice displayed a similar phenotype as BRCA1-deficient mice (McCarthy et al. 2003). Independent of BRCA1, BARD1 is involved in stabilizing p53 tumor suppressor via its ANK and BRCT domains (Feki et al. 2005; Irminger-Finger et al. 2016).
Clinically, both BARD1 and BRCA1 germline pathogenic variants are enriched in more aggressive breast cancer phenotypes, such as triple-negative breast cancers (TNBCs), which are associated with higher rates of recurrence, progression, and mortality (Atchley et al. 2008; Maxwell et al. 2014; Couch et al. 2015; Buys et al. 2017). On a molecular level, breast cancers occurring in individuals with BARD1 germline pathogenic variants were found to share a similar somatic gene expression profile to those with BRCA1 pathogenic variants; for example, a breast cancer patient with a germline BARD1 deletion and tumor loss of heterozygosity showed a basal-like gene expression profile similar to those seen in cancers associated with BRCA1 germline pathogenic variants (Armes et al. 1998; Atchley et al. 2008; Sabatier et al. 2010).
Despite these similarities, the clinical significance of BARD1 remains uncertain. Unlike BRCA1, BARD1 germline pathogenic variants are found in <1% of breast cancer patients (Couch et al. 2015). As a result, there is insufficient epidemiologic evidence on the role of BARD1 as a breast cancer susceptibility gene. Moreover, the evidence can sometimes be controversial, further impeding our understanding of the clinical relevance of BARD1. For example, one of the most extensively studied BARD1 germline variants, c.1670G>C, was shown to be associated with a two- to fourfold increased breast cancer risk in some studies (Karppinen et al. 2004, 2006; De Brakeleer et al. 2010; Ratajska et al. 2012) but not in others (Jakubowska et al. 2008; Klonowska et al. 2015). Given that BARD1 is a low-penetrance gene, its pathogenic variants likely increased the cancer risk in concert with other low- to moderate-penetrance gene alleles. The interaction among the various genetic risk loci might have led to the varying penetrance estimations observed in the various studies. Overall, existing epidemiologic data are scarce and inconsistent. Functional studies can help circumvent this by reclassifying germline variants—hence, increasing the number of pathogenic variants available for inclusion in epidemiologic studies (Sauer and Andrulis 2005; Thouvenot et al. 2016).
Currently, BARD1 pathogenic variants found in breast cancer patients are largely truncating mutations assumed to encode for nonfunctional proteins (Ghimenti et al. 2002; Wu et al. 2006; De Brakeleer et al. 2010; Stewart et al. 2018). Little is known about the functional impact of the majority of BARD1 germline variants, many of which are missense. Most of these are classified as either variants with unknown clinical significance (VUSs) or benign variants based on in silico analyses with limited functional evidence (Ghimenti et al. 2002; Ishitobi et al. 2003; Karppinen et al. 2004; Irminger-Finger et al. 2016). The abundance of VUSs should be reevaluated to facilitate the implementation of epidemiologic studies. Functional studies aimed at evaluating VUSs should assess the various domains, such as those of DNA damage repair and apoptosis. Using clinical germline variants, we sought to demonstrate a systematic method of functionally evaluating BARD1 VUSs.
RESULTS
Clinical Presentation and Family History
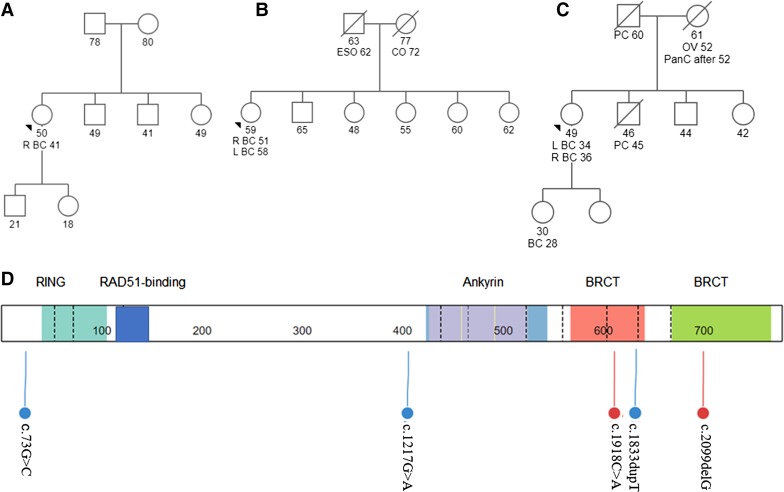
We identified six cases of BARD1 germline variants (pathogenic variants and VUSs). Three cases were found in patients with breast cancers, whereas the remaining three were in colorectal cancers (Table 1). Of the three patients with breast cancer, patient S531 was diagnosed with the aggressive TNBC phenotype at age 34 and has a strong family history of cancer, including a daughter diagnosed with breast cancer at age 28 (Fig. 1C). This patient was previously identified to carry a frameshift BARD1 variant c.2099delG, which results in the loss of the apoptotic domains. Interestingly, all three patients with colorectal cancers presented with colorectal cancers before the age of 50. Patient S490 presented at age 24 with stage IIIB disease and passed away a few years later, whereas patient S493 was diagnosed with stage IVA disease at age 41. Similarly, patient L75 presented with early-onset colorectal cancer with distant metastasis (Table 1). All three patients had missense VUSs, located in the apoptotic domains (c.1217G>A, c.1918C>A). No pathogenic variants were detected in the breast cancer (for S531) and colorectal cancer susceptibility genes (for L75, S490 and S493) (Supplemental Materials). Last, patient S69 was diagnosed with unilateral ER+/PR+/HER2− stage IIIA breast cancer with neither recurrence nor family history of cancers (Fig. 1A). She carried a missense variant in the nonfunctional region upstream of the RING domain (Fig. 1D).
Table 1.
Clinicopathological characteristics of the cases identified with BARD1 germline mutations
| Variant | Minor allele frequency | Patient demographics | Primary cancer (age at diagnosis) | Stage | Histological features | Other cancers (age at diagnosis) | Other variants | Cancer in first-degree relative (age at diagnosis) |
|---|---|---|---|---|---|---|---|---|
| S69 BARD1 c.73G>C p.Ala25Pro | None | Chinese female | Right breast cancer (41 yr) | IIIA | IDC grade 3 ER+/PR+/HER2− |
None | None | None |
| S519 BARD1 c.1833dupT p.Asp612Terfs | None | Chinese female | Right breast cancer (51 yr) | IIB | DCIS high grade ER+/PR−/HER2 unknown |
Contralateral breast cancer (58 yr) | None | Father: esophageal cancer (62 yr) Mother: colon cancer (72 yr) |
| S531 BARD1 c.2099delG p.Gly700Alafs | None | Mexican female | Left breast cancer (34 yr) | IIA | Medullary carcinoma ER−/PR−/HER2− |
Contralateral breast cancer (36 yr) | None | Father: prostate cancer (60 yr) Mother: ovarian cancer (52 yr) and pancreatic cancer (unknown) Brother: prostate cancer (45 yr) Daughter: breast cancer (28 yr) |
| L75 BARD1 c.1217G>A p.Arg406Gln | 0.00004124 | Chinese female | Descending colon cancer (37 yr) | IVB | Adenocarcinoma grade 2 | None | None | None |
| S490 BARD1 c.1217G>A p.Arg406Gln | 0.00004124 | Chinese female | Transverse colon cancer (24 yr) | IIIB | Adenocarcinoma grade 2 | None | None | None |
| S493 BARD1 c.1918C>A p.Leu640Ile | None | Malay male | Rectal cancer (41 yr) | IVA | Adenocarcinoma grade 2 | None | None | None |
Figure 1.
(A–C) Pedigrees of the cases identified with BARD1 germline mutation: (A) S69 BARD1 c.73G>C, (B) S519 BARD1 c.1833dupT, and (C) S531 BARD1 c.2099delG. (BC) Breast cancer, (CO) colon cancer, (ESO) esophageal cancer, (PanC) pancreatic cancer, (PC) prostate cancer. (D) Distribution of BARD1 germline mutation. Functional domains include the following: RING (residues 26–119), RAD51-binding (residues 123–162), ANK (residues 425–555), and BRCT (residues 566–777). The lolliplots were drawn using St. Jude PeCan Data Portal.
To establish the presence of clinical and tumor phenotype of BARD1 germline variants, we referred to the Cancer Genome Atlas (TCGA) data set. Given that the majority of BARD1 germline variants were reported among breast cancer patients, we examined the TCGA breast cancer subset for presence of BARD1 germline variants. Of the 1036 breast cancer patients, only 33 patients carried BARD1 germline variants (Supplemental Table S1). Of these, only two patients had pathogenic variants (c.729T>A, c.1690C>T). The remaining patients carried benign variants (c.1972C>T, c.2212A>G) and VUSs (c.581G>A, c.620A>G, c.1016G>A, c.1738G>A, c.2191C>G, c.2282G>A) (Supplemental Table S1). Both patients with the truncating pathogenic variants (c.729T>A, c.1690C>T) had infiltrating duct carcinoma, similar to the majority of the breast cancer patients.
We proceeded to investigate for a distinct mutational spectrum among tumors associated with BARD1 germline variants, based on the initial findings of a case report; Sabatier et al. (2010) found several features of BRCA-ness in a non-BCRA-related breast cancer containing heterozygous BARD1 germline variants with tumor loss of heterozygosity. Using the TCGA BRCA data set, we did not find any unique mutational spectrum among tumors associated with BARD1 germline variants (Supplemental Fig. S4). None of the tumors carried signature 3, which was associated with BRCA1 and BRCA2 variants (Polak et al. 2017).
Genomic Analyses
Overall, there were five unique germline variants in the six patients (Table 2); one VUS, BARD1 c.1217G>A, was seen in both patient S490 with stage IIIB colorectal cancer and patient L75 with metastatic colorectal cancer. Of all the variants, only BARD1 c.1217G>A was reported in the literature (Sauer and Andrulis 2005). Two were pathogenic/likely pathogenic frameshift variants (BARD1 c.1833dupT and BARD1 c.2099delG), and the remaining three were missense VUSs (BARD1 c.73G>C, BARD1 c.1217G>A, BARD1 c.1918C>A). The majority of these germline variants involved the ANK and BRCT domains (Fig. 1D). Only one, BARD1 c.73G>C, was located at the 5′ terminus proximal to the RING domain. Both pathogenic/likely pathogenic variants were located in the BRCT domain, consistent with the distribution of germline pathogenic variants reported in the literature (Fig. 1; Supplemental Materials).
Table 2.
Variant table
| Gene | Variant | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect (substitution, deletion, etc.) | dbSNP/dbVar ID | Allele registry ID | ClinVar variation ID | Genotype (heterozygous/homozygous) |
|---|---|---|---|---|---|---|---|---|---|---|
| BARD1 | c.73G>C p.Ala25Pro | 2 | NM_000465 | NP_000456 | Missense | Substitution | rL751646468 | CA333878 | 185,506 | Heterozygous |
| BARD1 | c.1833dupT p.Asp612Terfs | 2 | NM_000465 | NP_000456 | Nonsense | Duplication | rs1553614963 | CA658796163 | 530,030 | Heterozygous |
| BARD1 | c.2099delG p.Gly700Alafs | 2 | NM_000465 | NP_000456 | Nonsense | Deletion | rs1553612184 | CA658796159 | 530,114 | Heterozygous |
| BARD1 | c.1217G>A p.Arg406Gln | 2 | NM_000465 | NP_000456 | Missense | Substitution | rs587780014 | CA287510 | 127,713 | Heterozygous |
| BARD1 | c.1918C>A p.Leu640Ile | 2 | NM_000465 | NP_000456 | Missense | Substitution | rs1553612535 | CA350451296 | 530,056 | Heterozygous |
| BARD1 | c.1670G>C p.Cys557Ser | 2 | NM_000465 | NP_000456 | Missense | Substitution | rs28997576 | CA250552 | 8045 | Heterozygous |
Functional Analyses
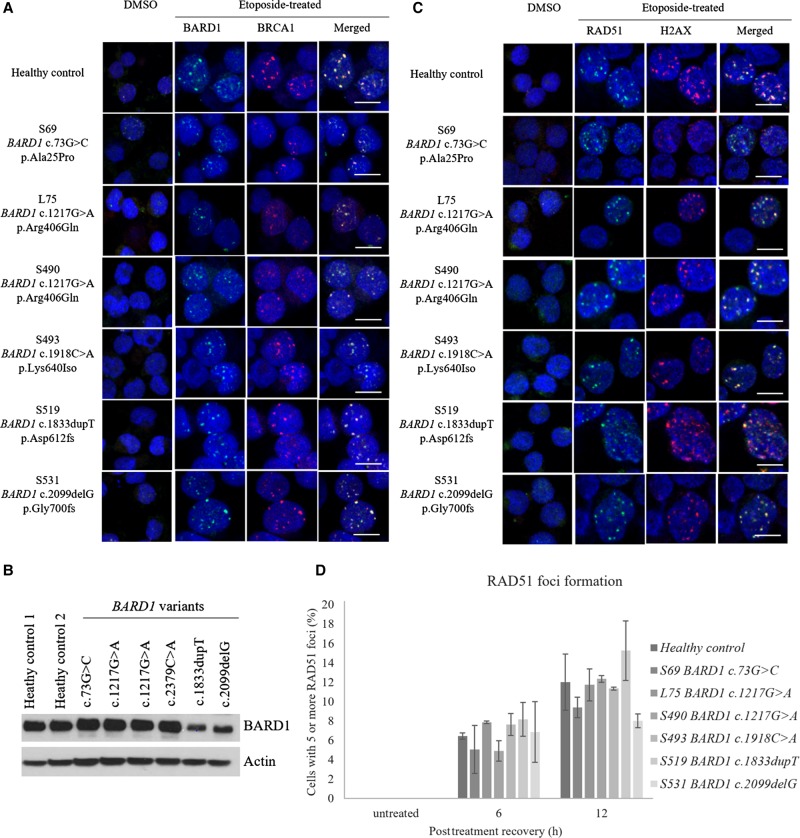
We evaluated each variant for the functions encoded by the various domains: RING domain for BARD1–BRCA1 colocalization, RAD51-binding domain for foci formation, and the ANK, BRCT domains for apoptosis. To induce double-stranded DNA breaks and subsequently apoptosis, cells were treated with etoposide. Colocalization of BARD1 and BRCA1 foci was observed at 24 h post–etoposide treatment (Fig. 2A). For the RAD51 foci assays, double-stranded DNA breaks were induced from the etoposide treatment, as evident from the rise in ɣ-H2AX foci (Fig. 2C). This was accompanied with a rise in RAD51 foci as homologous recombination repair was initiated (Fig. 2C,D). All the variants displayed normal BARD1–BRCA1 colocalization and RAD51 foci formation, consistent with the absence of mutations at the RING and RAD51-binding domains, respectively (Fig. 1D).
Figure 2.
(A) BARD1–BRCA1 colocalization was observed for all the BARD1 variants. BARD1 and BRCA1 foci were stained green and red, respectively. (B) Basal levels of BARD1 expression were lower in BARD1 c.1833dupT and BARD1 c.2099delG as compared to healthy controls. (C,D) RAD51 foci (green) were colocalized at the sites of DNA breaks as represented by ɣ-H2AX foci (red). Scale bar, 10 µm. No statistically significant difference in RAD51 localization was noted between the BARD1 variants and the healthy controls. An independent t-test was used to compare the results between the variants and healthy controls.
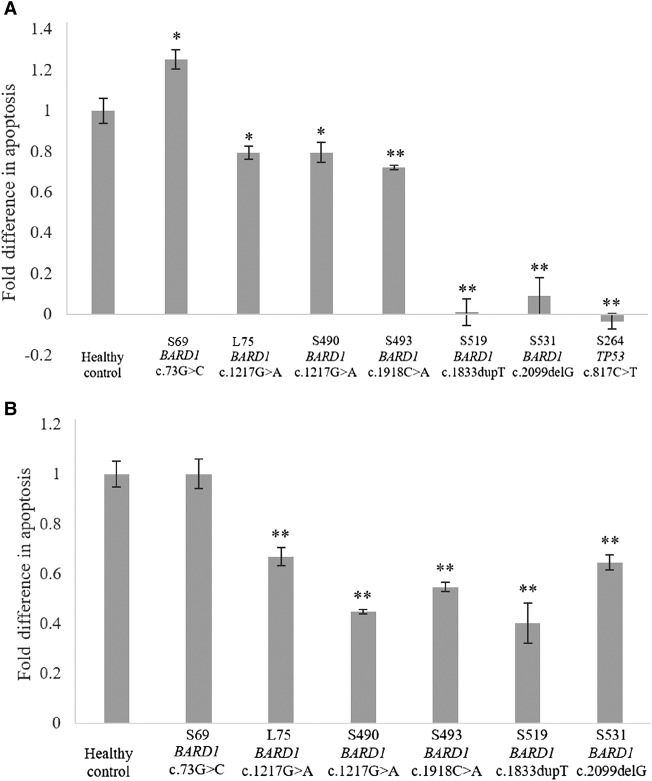
To investigate the variants’ impact on apoptosis, we measured the extent of DNA fragmentation and membrane phosphatidylserine translocation via TUNEL and Annexin V assays, respectively. Compared to the healthy controls, all the VUSs, except BARD1 c.73G>C, displayed a lower extent of apoptosis (Fig. 3A,B). This observed trend was consistent with the mutation sites of the VUSs in which all but BARD1 c.73G>C involved the ANK and BRCT domains. In addition, BARD1 c.1833dupT and BARD1 c.2099delG showed a greater extent of impaired apoptosis (Fig. 3A). Interestingly, patients with these variants also showed reduced BARD1 expression (Fig. 2B). To determine whether the reduced expression was the cause of the impaired apoptosis would require further studies. In view of the above findings, BARD1 c.1918C>A and BARD1 c.1217G>A might be pathogenic, whereas BARD1 c.73G>C is likely a benign variant (Table 3).
Figure 3.
(A) TUNEL and (B) Annexin V assays. Percentage rise in apoptosis for the variants was normalized to that of the healthy controls. When compared to the healthy control, posttreatment apoptosis was lower for all the BARD1 variants, except BARD1 c.73G>C. (*) P < 0.05, (**) P < 0.005. Cells containing TP53 c.817C>T were included as positive control of impaired apoptosis. An independent t-test was used to compare the results between the variants and healthy controls.
Table 3.
Compiled in silico prediction and functional results of the variants
| Variant | In silico predictiona | BARD1–BRCA1 colocalizationb | RAD51 foci formationb | TUNEL assayc | Annexin V assay | Overall functional status |
|---|---|---|---|---|---|---|
| S69 BARD1 c.73G>C p.Ala25Pro | Likely tolerated | Normal | Normal | Normal | Normal | Intact |
| L75 BARD1 c.1217G>A p.Arg406Gln | Likely tolerated | Normal | Normal | Impaired (0.8) | Impaired (0.7) | Possibly impaired apoptosis |
| S490 BARD1 c.1217G>A p.Arg406Gln | Likely tolerated | Normal | Normal | Impaired (0.8) | Impaired (0.4) | Possibly impaired apoptosis |
| S493 BARD1 c.1918C>A p.Leu640Ile | Conflicting | Normal | Normal | Impaired (0.7) | Impaired (0.5) | Possibly impaired apoptosis |
| S519 BARD1 c.1833dupT p.Asp612Terfs | Not applicable | Normal | Normal | Impaired (<0.2) | Impaired (0.4) | Impaired apoptosis |
| S531 BARD1 c.2099delG p.Gly700Alafs | Not applicable | Normal | Normal | Impaired (<0.2) | Impaired (0.7) | Impaired apoptosis |
aIn silico prediction results were based on PolyPhen-2, Align GVD, and SIFT prediction tools.
bFunction was considered as intact if not statistically different from healthy controls.
cRise in apoptosis for variants was normalized to that of healthy controls, in parentheses. Values were rounded to the nearest decimal.
DISCUSSION
Determining the pathogencity of a germline variant is critical for the management of patients and their families, and depends largely on both epidemiologic and functional evidence. Given the rarity of BARD1 germline variants, epidemiologic studies that require large samples of carriers may not be feasible (Lee et al. 2015). Hence, functional studies are crucial to the curation of BARD1 germline variants. Despite BARD1's indispensable role as the binding partner of BRCA1, most BARD1 VUSs (90%) and all of the BARD1 germline pathogenic variants have never been functionally assessed (Landrum et al. 2018). Here, we performed functional characterization of two pathogenic/likely pathogenic variants and three VUSs. The impaired apoptosis associated with variants c.1217G>A and c.1918C>A may provide evidence toward a likely pathogenic or pathogenic classification. However, there remains insufficient evidence for variant reclassification according to American College of Clinical Genetics (ACMG) criteria (Richards et al. 2015). Additional evidence, such as epidemiologic research, is still required.
Clinically, carriers of these pathogenic/likely pathogenic variants displayed several features suggestive of a more aggressive tumor phenotype, such as having early metastatic diseases, multiple tumors, and frequent tumor recurrence. Other studies have also suggested distinct and aggressive tumor phenotypes (i.e., TNBCs) associated with BARD1 germline pathogenic variants; empirically, a patient with a nonsense germline variant (c.1921C>T) was found to have multiple bilateral breast cancers (Atchley et al. 2008; Sabatier et al. 2010; Maxwell et al. 2014; Couch et al. 2015; Buys et al. 2017; Gass et al. 2017). However, with BARD1 being a low-penetrance gene, these phenotypes are unlikely caused solely by the respective BARD1 pathogenic variants (DeLeonardis et al. 2017). Low- to moderate-penetrance genes have been demonstrated in model-fitting studies, to act multiplicatively in raising cancer risk (Vahteristo et al. 2005). Studies on larger cohorts of patients with pathogenic BARD1 variants will be needed to further elucidate these clinicopathological characteristics and any potential modifying factors that may explain the variable cancer risks observed in previous BARD1 studies (Karppinen et al. 2004, 2006; Jakubowska et al. 2008; De Brakeleer et al. 2010; Ratajska et al. 2012; Klonowska et al. 2015).
More importantly, we showed that the type of aberrant protein function correlated with the domain location of the variant. Variants that displayed impaired apoptosis (c.1918C>A, c.1833dupT, c.1217G>A, and c.2099delG) were located within and proximal to the apoptotic domains of ankyrin and BRCT. On the other hand, none of these variants displayed abnormal BRCA1 heterodimerization or RAD51 colocalization given that they were located distal to the BRCA1- and RAD51-binding domains. Therefore, it is important to use the appropriate functional assay relevant to the site of mutation.
Assessing BARD1 functional status will require a composite of different assays. A recent functional analysis of 29 BARD1 missense variants evaluated for functional impairment using the homology-directed DNA repair (HDR) assay (Lee et al. 2015). The HDR assay measures the signal of green fluorescent protein in mutant cells containing double-stranded DNA breaks from the expression of endonuclease I-SceI. For many homologous recombination genes such as BRCA2, the HDR assay serves as a relatively high-throughput method of analyzing VUSs (Farrugia et al. 2008). In this study, 13 of the 29 BARD1 variants evaluated in the functional analysis were located in the ANK and BRCT domains and showed an intact homologous recombination function (Lee et al. 2015). The use of the Annexin V assay or other apoptotic assays might have provided further evidence of their functional impact. Similarly, in another study on BARD1 germline variants, two variants were found to be functionally intact using the apoptotic assays (Sauer and Andrulis 2005). Given that these variants were located in the BRCA1- and RAD51-binding domains (Sauer and Andrulis 2005), the addition of the HDR assay would have complemented the apoptosis findings.
The two BARD1 pathogenic variants included in our study, c.1833dupT and c.2099delG, were located within the BRCT domains. Unsurprisingly, they displayed impaired apoptosis but normal DNA damage repair. An indiscriminate use of the DNA damage repair assay alone would have otherwise missed the impaired apoptosis of these clinically pathogenic variants and would have misguided variant classification. Another of our variants, BARD1 c.1217C>A, was previously classified as benign based on its preserved homologous recombination function. Although we also observed intact DNA damage repair from our RAD51 assay, our additional apoptosis studies revealed impaired apoptosis in this variant (∼20%–60% lower than healthy controls). This may be because of the proximity of the variant to the ANK domain; structural analysis of the mutant protein would be useful to reevaluate the functional status of this variant.
In summary, we illustrated a systematic way of evaluating VUSs located in the DNA repair, apoptosis, and BRCA1-binding domains. However, a potential limitation of our assays involved the exclusion of more recent gene functions such as the repair of stalled replication forks and transcriptional regulation of estrogen metabolizing genes that are located in the BRCA1-binding and BRCT domains, respectively (Billing et al. 2018; Stewart et al. 2018). Ideally, functional assays should incorporate all the newly discovered gene functions. This may not be feasible as functional studies are costly and time-consuming. Nonetheless, reliable functional evidence can be efficiently obtained with the use of the appropriate assay, as emphasized in our study. Moreover, our choice of functional assays already covered both the BRCA1-binding and BRCT domains.
Ultimately, the overall evidence for variant classification pivots on the genetic and epidemiologic analyses (MacArthur et al. 2014). Large cohort studies on BARD1 pathogenic variants are still needed to assess the clinical relevance of this gene, as well as to elucidate potential genetic risk modifiers causing the observed variability of penetrance between and within families. To facilitate this, functional evaluation of the VUSs should be focused and specific to the variant's location.
METHODS
This study was approved by the SingHealth Centralized Institutional Review Board (IRB 2010/426/B) with signed informed consent from the participants.
Study Design
Patients referred to the Cancer Genetics Service at the National Cancer Centre Singapore between November 2014 and March 2018 were reviewed retrospectively for BARD1 germline pathogenic variants and VUSs. These variants were curated by a clinical laboratory accredited by the College of American Pathologists and Clinical Laboratory Improvement Amendments, in accordance with the criteria by the ACMG. Variant c.1918C>A was curated separately by us with reference to the ACMG guidelines as the carrier was not tested by clinical laboratories (Supplemental Table S2). Only patients who had provided research consent and those with previously banked lymphoblastoid cells were included. Clinicopathological data for age, sex, personal and cancers in first-degree relatives, tumor histology, American Joint Committee on Cancer (AJCC) staging, site, and immunohistochemistry (IHC) staining of estrogen, progesterone, and erbB-2 receptors were retrieved from electronic medical records. Pedigrees for each patient were constructed and drawn using Progeny Pedigree Drawing Software (Progeny Software LLC). To compare the overall distribution of BARD1 germline variants with that observed in our study, we performed a literature search using ClinVar.
Cell Culture and Treatment
Patient-derived lymphoblastoid cells were treated with 10 µM etoposide for 1 h to induce double-stranded DNA breaks, subsequently recovered by incubation in fresh medium. Vehicle control containing an equivalent volume of DMSO was used. Recovery durations of 12 h and 24 h were used for the assay on RAD51 foci formation and BARD1–BRCA1 colocalization, respectively. Cells were then harvested for immunofluorescence studies.
Immunofluorescence (IF) Studies of BARD1–BRCA1 Colocalization and RAD51 Foci Formation
Cells were fixed for 15 min using 4% paraformaldehyde, were permeabilized for 5 min using 0.3% Triton-X 100, and were blocked for 30 min with 10% goat serum in phosphate-buffered saline before incubation for 30 min at 37°C with primary antibodies. Slides were then stained with secondary antibodies for 1 h at room temperature before being mounted with Prolong Gold antifade reagent with DAPI (Life Technologies, ThermoFisher Scientific Corporation). The images were acquired by confocal microscope (Zeiss LSM800). Cells containing five or more foci were scored positive for RAD51 foci formation. At least 200 cells were counted for each slide, and duplicates were performed for each experiment.
Western Blot Analysis of BARD1 Basal Expression
Cell pellets were lysed in RIPA buffer supplemented with protease and phosphatase inhibitor cocktail (ThermoFisher Scientific Corporation), and protein extracts were quantified using the Pierce BCA protein assay kit (ThermoFisher Scientific Corporation). Proteins were then electrophoresed on sodium dodecyl sulfate polyacrylamide gel and transferred to polyvinylidene difluoride membrane (Milipore, Sigma-Aldrich). Membranes were blocked, and then incubated overnight with primary antibody followed by secondary antibodies. Immunoreactivity was detected with enhanced chemiluminescent HRP substrate (Advansta).
Apoptosis Studies
TUNEL assay
Cells were pretreated with 10 µM etoposide or DMSO in 5% RPMI for 24 h. Subsequently, cell apoptosis was measured with a microplate reader–based TiterTACS in situ apoptosis detection kit (R&D systems; 4822-96-K) as described by the manufacturer. Percentage rise in apoptosis was derived from the difference in absorbance between the etoposide-treated and DMSO-treated wells. Minimum of duplicates were performed for each experiment.
Annexin V assay
Cells were pretreated with 10 µM etoposide or DMSO in 5% RPMI for 6 h before incubation in fresh medium for up to 24 h. Subsequently, the cells were harvested and stained with Annexin V and propidium iodide according to manufacturer's protocol (BD Biosciences; 556547). Cells in early apoptosis are Annexin V positive and PI negative, whereas cells in late apoptosis or necrosis are both Annexin V and PI positive. For our study, we only considered apoptotic cells with Annexin V positive and PI negative. Cells with double staining were excluded, given that some of these cells might have died via necrosis. A minimum of triplicates was performed for each experiment.
ADDITIONAL INFORMATION
Data Deposition and Access
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The variants have been deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000920886, SCV000920887, SCV000920888, SCV000920889, and SCV000920890.
Ethics Statement
This study was approved by the Singhealth Centralized Institutional Review Board (IRB 2010/426/B) with signed informed consent from the participants.
Acknowledgments
We would like to thank the patients and research participants for their contribution to the study. We would like to acknowledge St. Jude PeCan Data Portal for the web application in generating Figure 1D and Supplemental Figures 1 and 2.
Author Contributions
M.R.T., S.T.C., S.H.C., C.E.L., N.D.B.I., and E.C. made substantial contributions to the design, administration, and conceptualization of the study and the interpretation of data and critically revised and drafted the manuscript. J.N. provided funding, acquisition of data, conception and design of the study, manuscript review, and editing. All authors approved the submission of the final manuscript and agreed to be accountable for all aspects of the work.
Funding
We thank our sources of support: National Medical Research Council (CSA) (NMRC/CSA-INV/0017/2017) and Singhealth Foundation Research Grant (SHF/PRISM002/2015) to J.N. and SingHealth (SMSTDA-Project FY2018) to M.R.T.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Armes JE, Egan M, Southey MC, Dite GS, McCredie MRE, Giles GG, Hopper JL, Venter DJ. 1998. The histologic phenotypes of breast carcinoma occurring before age 40 years in women with and without BRCA1 or BRCA2 germline mutations. Cancer 83: 2335–2345. [DOI] [PubMed] [Google Scholar]
- Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, Hortobagyi GN, Arun BK. 2008. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol 26: 4282–4288. 10.1200/JCO.2008.16.6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billing D, Horiguchi M, Wu-Baer F, Taglialatela A, Leuzzi G, Nanez SA, Jiang W, Zha S, Szabolcs M, Lin CS, et al. 2018. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol Cell 72: 127–139.e128. 10.1016/j.molcel.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buys SS, Sandbach JF, Gammon A, Patel G, Kidd J, Brown KL, Sharma L, Saam J, Lancaster J, Daly MB. 2017. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 123: 1721–1730. 10.1002/cncr.30498 [DOI] [PubMed] [Google Scholar]
- Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P, Olson JE, Godwin AK, Pankratz VS, Olswold C. 2015. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 33: 304 10.1200/JCO.2014.57.1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Brakeleer S, De Grève J, Loris R, Janin N, Lissens W, Sermijn E, Teugels E. 2010. Cancer predisposing missense and protein truncating BARD1 mutations in non–BRCA1 or BRCA2 breast cancer families. Hum Mutat 31: E1175–E1185. 10.1002/humu.21200 [DOI] [PubMed] [Google Scholar]
- DeLeonardis K, Sedgwick K, Voznesensky O, Matloff E, Hofstatter E, Balk S, Tung N. 2017. Challenges in interpreting germline mutations in BARD1 and ATM in breast and ovarian cancer patients. Breast J 23: 461–464. 10.1111/tbj.12764 [DOI] [PubMed] [Google Scholar]
- Economopoulou P, Dimitriadis G, Psyrri A. 2015. Beyond BRCA: new hereditary breast cancer susceptibility genes. Cancer Treat Rev 41: 1–8. 10.1016/j.ctrv.2014.10.008 [DOI] [PubMed] [Google Scholar]
- Farrugia DJ, Agarwal MK, Pankratz VS, Deffenbaugh AM, Pruss D, Frye C, Wadum L, Johnson K, Mentlick J, Tavtigian SV, et al. 2008. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res 68: 3523–3531. 10.1158/0008-5472.CAN-07-1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feki A, Jefford CE, Berardi P, Wu JY, Cartier L, Krause K-H, Irminger-Finger I. 2005. BARD1 induces apoptosis by catalysing phosphorylation of p53 by DNA-damage response kinase. Oncogene 24: 3726 10.1038/sj.onc.1208491 [DOI] [PubMed] [Google Scholar]
- Gass J, Tatro M, Blackburn P, Hines S, Atwal PS. 2017. BARD1 nonsense variant c. 1921C>T in a patient with recurrent breast cancer. Clin Case Rep 5: 104–107. 10.1002/ccr3.793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghimenti C, Sensi E, Presciuttini S, Brunetti IM, Conte P, Bevilacqua G, Caligo MA. 2002. Germline mutations of the BRCA1-associated ring domain (BARD1) gene in breast and breast/ovarian families negative for BRCA1 and BRCA2 alterations. Genes Chromosomes Cancer 33: 235–242. 10.1002/gcc.1223 [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I, Jefford CE. 2006. Is there more to BARD1 than BRCA1? Nat Rev Cancer 6: 382 10.1038/nrc1878 [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I, Ratajska M, Pilyugin M. 2016. New concepts on BARD1: regulator of BRCA pathways and beyond. Int J Biochem Cell Biol 72: 1–17. 10.1016/j.biocel.2015.12.008 [DOI] [PubMed] [Google Scholar]
- Ishitobi M, Miyoshi Y, Hasegawa S, Egawa C, Tamaki Y, Monden M, Noguchi S. 2003. Mutational analysis of BARD1 in familial breast cancer patients in Japan. Cancer Lett 200: 1–7. 10.1016/S0304-3835(03)00387-2 [DOI] [PubMed] [Google Scholar]
- Jakubowska A, Cybulski C, Szymańska A, Huzarski T, Byrski T, Gronwald J, Dębniak T, Górski B, Kowalska E, Narod SA, et al. 2008. BARD1 and breast cancer in Poland. Breast Cancer Res Treat 107: 119–122. 10.1007/s10549-007-9537-4 [DOI] [PubMed] [Google Scholar]
- Karppinen S-M, Heikkinen K, Rapakko K, Winqvist R. 2004. Mutation screening of the BARD1 gene: evidence for involvement of the Cys557Ser allele in hereditary susceptibility to breast cancer. J Med Genet 41: e114–e114. 10.1136/jmg.2004.020669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karppinen SM, Barkardottir RB, Backenhorn K, Sydenham T, Syrjäkoski K, Schleutker J, Ikonen T, Pylkäs K, Rapakko K, Erkko H, et al. 2006. Nordic collaborative study of the BARD1 Cys557Ser allele in 3956 patients with cancer: enrichment in familial BRCA1 and BRCA2 mutation-negative breast cancer but not in other malignancies. J Med Genet 43: 856 10.1136/jmg.2006.041731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonowska K, Ratajska M, Czubak K, Kuzniacka A, Brozek I, Koczkowska M, Sniadecki M, Debniak J, Wydra D, Balut M, et al. 2015. Analysis of large mutations in BARD1 in patients with breast and/or ovarian cancer: the Polish population as an example. Sci Rep 5: 10424 10.1038/srep10424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. 2018. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46: D1062–D1067. 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Banerjee T, Gillespie J, Ceravolo A, Parvinsmith MR, Starita LM, Fields S, Toland AE, Parvin JD. 2015. Functional analysis of BARD1 missense variants in homology-directed repair of DNA double strand breaks. Hum Mutat 36: 1205–1214. 10.1002/humu.22902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D, Manolio T, Dimmock D, Rehm H, Shendure J, Abecasis G, Adams D, Altman R, Antonarakis S, Ashley E. 2014. Guidelines for investigating causality of sequence variants in human disease. Nature 508: 469 10.1038/nature13127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KN, Wubbenhorst B, D'Andrea K, Garman B, Long JM, Powers J, Rathbun K, Stopfer JE, Zhu J, Bradbury AR, et al. 2014. Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genet Med 17: 630 10.1038/gim.2014.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EE, Celebi JT, Baer R, Ludwig T. 2003. Loss of Bard1, the heterodimeric partner of the BRCA1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol Cell Biol 23: 5056–5063. 10.1128/MCB.23.14.5056-5063.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, Rosebrock D, Livitz D, Kübler K, Mouw KW, et al. 2017. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 49: 1476 10.1038/ng.3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajska M, Antoszewska E, Piskorz A, Brozek I, Borg Å, Kusmierek H, Biernat W, Limon J. 2012. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res Treat 131: 89–97. 10.1007/s10549-011-1403-8 [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody W, Hegde M, Lyon E, Spector E. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier R, Adélaïde J, Finetti P, Ferrari A, Huiart L, Sobol H, Chaffanet M, Birnbaum D, Bertucci F. 2010. BARD1 homozygous deletion, a possible alternative to BRCA1 mutation in basal breast cancer. Genes Chromosomes Cancer 49: 1143–1151. 10.1002/gcc.20822 [DOI] [PubMed] [Google Scholar]
- Sauer MK, Andrulis IL. 2005. Identification and characterization of missense alterations in the BRCA1 associated RING domain (BARD1) gene in breast and ovarian cancer. J Med Genet 42: 633–638. 10.1136/jmg.2004.030049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MD, Zelin E, Dhall A, Walsh T, Upadhyay E, Corn JE, Chatterjee C, King MC, Klevit RE. 2018. BARD1 is necessary for ubiquitylation of nucleosomal histone H2A and for transcriptional regulation of estrogen metabolism genes. Proc Natl Acad Sci 115: 1316–1321. 10.1073/pnas.1715467115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouvenot P, Yamin BB, Fourrière L, Lescure A, Boudier T, Del Nery E, Chauchereau A, Goldgar DE, Houdayer C, Stoppa-Lyonnet D. 2016. Functional assessment of genetic variants with outcomes adapted to clinical decision-making. PLoS Genet 12: e1006096 10.1371/journal.pgen.1006096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahteristo P, Syrjäkoski K, Heikkinen T, Eerola H, Aittomäki K, von Smitten K, Holli K, Blomqvist C, Kallioniemi O-P, Nevanlinna H. 2005. BARD1 variants Cys557Ser and Val507Met in breast cancer predisposition. Eur J Hum Genet 14: 167 10.1038/sj.ejhg.5201542 [DOI] [PubMed] [Google Scholar]
- Wu JY, Vlastos AT, Pelte MF, Caligo MA, Bianco A, Krause KH, Laurent GJ, Irminger-Finger I. 2006. Aberrant expression of BARD1 in breast and ovarian cancers with poor prognosis. Int J Cancer 118: 1215–1226. 10.1002/ijc.21428 [DOI] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. 2009. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 19: 524–529. 10.1016/j.cub.2009.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The variants have been deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000920886, SCV000920887, SCV000920888, SCV000920889, and SCV000920890.