Abstract
Cholangiocarcinoma is a highly aggressive and lethal malignancy, with limited treatment options available. Recently, FGFR inhibitors have been developed and utilized in FGFR-mutant cholangiocarcinoma; however, resistance often develops and the genomic determinants of resistance are not fully characterized. We completed whole-exome sequencing (WES) of 11 unique tumor samples obtained from a rapid research autopsy on a patient with FGFR-fusion-positive cholangiocarcinoma who initially responded to the pan-FGFR inhibitor, INCB054828. In vitro studies were carried out to characterize the novel FGFR alteration and secondary FGFR2 mutation identified. Multisite WES and analysis of tumor heterogeneity through subclonal inference identified four genetically distinct cancer cell populations, two of which were only observed after treatment. Additionally, WES revealed an FGFR2 N549H mutation hypothesized to confer resistance to the FGFR inhibitor INCB054828 in a single tumor sample. This hypothesis was corroborated with in vitro cell-based studies in which cells expressing FGFR2–CLIP1 fusion were sensitive to INCB054828 (IC50 value of 10.16 nM), whereas cells with the addition of the N549H mutation were resistant to INCB054828 (IC50 value of 1527.57 nM). Furthermore, the FGFR2 N549H secondary mutation displayed cross-resistance to other selective FGFR inhibitors, but remained sensitive to the nonselective inhibitor, ponatinib. Rapid research autopsy has the potential to provide unprecedented insights into the clonal evolution of cancer throughout the course of the disease. In this study, we demonstrate the emergence of a drug resistance mutation and characterize the evolution of tumor subclones within a cholangiocarcinoma disease course.
Keywords: biliary tract neoplasm
INTRODUCTION
Cholangiocarcinoma is an aggressive and deadly rare cancer arising from bile duct epithelial cells with a 5-yr overall survival rate of <2% for advanced stage disease (PDQ Adult Treatment Editorial Board 2002; Razumilava and Gores 2014). Most patients with cholangiocarcinoma present with metastatic unresectable cancer, thus precluding curative therapy (Valle et al. 2010; Razumilava and Gores 2014). Given its poor prognosis and limited treatment options beyond first-line chemotherapy, development and optimization of novel therapies for cholangiocarcinoma are urgently needed.
The fibroblast growth factor receptor (FGFR) signaling pathway is aberrantly activated in ∼20% of cases of intrahepatic cholangiocarcinoma through various genomic alterations including point mutations, copy-number amplifications, and gene fusions (Roychowdhury et al. 2011; Wu et al. 2013). Extending beyond cholangiocarcinoma, alterations in the FGFR signaling pathway have been reported in non-small-cell lung carcinoma, endometrial cancer, and urothelial cancer (Roychowdhury et al. 2011; Wu et al. 2013). Currently, several tyrosine kinase inhibitors, covalent and noncovalent, nonselective and selective FGFR inhibitors are being assessed clinically in patients with metastatic cancer and have shown early responses in those patients with metastatic FGFR-mutant cancers (Gozgit et al. 2012; André et al. 2013; Angevin et al. 2013; Tabernero et al. 2015; Paik et al. 2017; Javle et al. 2018). Although genomic alterations in FGFR correlated with initial clinical responses to FGFR inhibitors, multiple secondary mutations in FGFR and other cellular signaling pathways have been identified in patients after treatment with FGFR inhibitors. Thus, elucidating the various acquired mechanisms of drug resistance to FGFR inhibitors will be critical for the development of new therapies to overcome resistance and improve the outcome of patients with FGFR-mutant cancers.
Tumor heterogeneity has been shown to negatively impact therapeutic response and contribute to treatment resistance in cancer patients, and thus it remains a major impediment to cancer treatment (Dexter and Leith 1986; Heppner and Miller 1989; Bedard et al. 2013; Fisher et al. 2013; Burrell and Swanton 2014). Both genetic and epigenetic mechanisms within the tumor itself as well as changes in the tumor microenvironment can drive the development of tumor heterogeneity (Heng et al. 2009; Junttila and de Sauvage 2013; Meacham and Morrison 2013). Genomic characterization of primary and recurrent/metastatic tumors from the same patient has further demonstrated spatial and temporal intrapatient tumor heterogeneity (ITH) (Bedard et al. 2013). Recent studies have evaluated ITH and clonal evolution through next-generation sequencing (NGS) methods, demonstrating the critical role of these processes in recurrence and development of therapeutic resistance in urothelial carcinoma, renal cell carcinoma, and acute myeloid leukemia (Ding et al. 2012; Gerlinger et al. 2012, 2014; Faltas et al. 2016). Studies like these, however, are limited in cholangiocarcinoma.
To date, the genomic landscape of cholangiocarcinoma has been largely characterized through tumor biopsies and surgical specimens and, therefore, may not accurately reflect the complex and heterogeneous nature of metastatic and drug-resistant disease (Zou et al. 2014; Ruzzenente et al. 2016; Farshidfar et al. 2017; Jusakul et al. 2017). Recently, Goyal et al. (2017) evaluated three patients with FGFR-fusion-positive cholangiocarcinoma who received the FGFR inhibitor BGJ398. Targeted gene panel sequencing using the commercial Guardant360 assay revealed an FGFR2 V564F gatekeeper mutation in plasma circulating tumor DNA (ctDNA) of all three patients and several additional FGFR2 mutations in two of the patients. One patient consented to rapid research autopsy, and this enabled the procurement of multiple metastatic tumors for genomic profiling with the FoundationOne assay (315-gene panel) to study acquired drug resistance to the drug BGJ398. This study successfully demonstrated the role of acquired mutations in resistance to BGJ398. It also demonstrated heterogeneity at time of autopsy, because eight of 12 tumor samples assessed lacked a secondary mutation in FGFR2. However, there are more than 10 FGFR inhibitors in active drug development in clinical trials, and mechanisms of resistance for each of these drugs remain a significant gap in knowledge. Prior research on acquired resistance mutations in KIT, ABL1, and ALK oncogenes with their respective kinase inhibitors demonstrates that cross-resistance and sensitivity for secondary mutations varies widely, and therefore understanding resistance profiles for other FGFR inhibitors will be essential. Further, evaluating additional patients receiving other FGFR inhibitors with an expanded scope of whole exome (more than 20,000 genes) will be critical to characterizing clonal heterogeneity and evolution with FGFR inhibitors.
In the current work, we present a patient with metastatic cholangiocarcinoma harboring a novel FGFR2–CLIP1 gene fusion who demonstrated a partial response followed by disease progression while on treatment with the FGFR-selective kinase inhibitor, INCB054828. Through rapid research autopsy of this patient and whole-exome sequencing (WES) of his metastatic cancer, we identified four unique tumor subclones and elucidated their evolution from the normal ancestral cell. Furthermore, we identified a posttreatment secondary kinase mutation in FGFR2 present in a single metastatic tumor sample and characterized its impact on sensitivity to a variety of FGFR inhibitors in vitro. The results of our in vitro drug sensitivity studies suggest that this mutation conferred resistance to INCB054828 in this patient and thus may have potential as a clinically useful biomarker of resistance. Importantly, characterizing tumor heterogeneity and the ability to detect clonal evolution in patients will facilitate approaches to prevent or overcome treatment resistance and disease recurrence.
RESULTS
Clinical Course
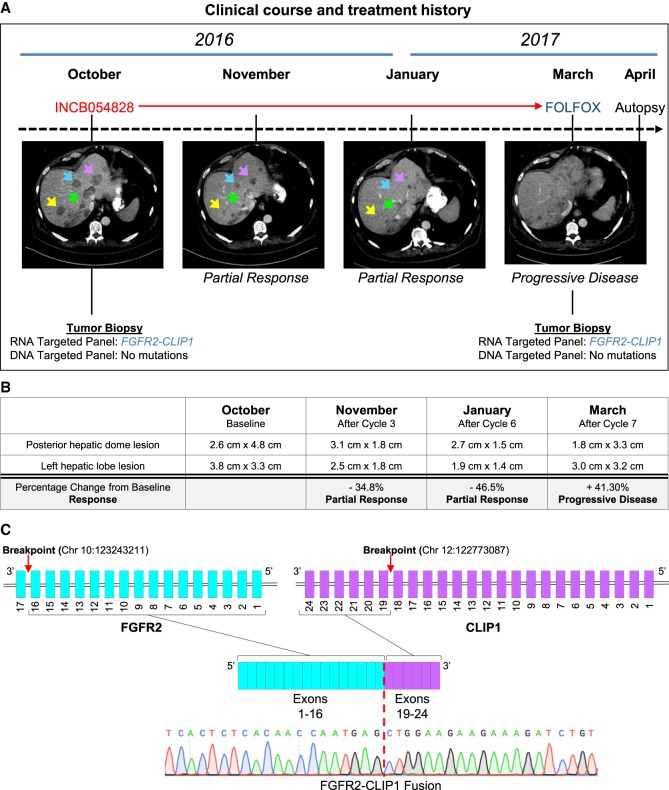
A 59-yr-old male presented clinically with abdominal pain and fullness in the fall of 2015. Abdominal CT and MRI scans revealed two small but suspicious-appearing lesions in the liver. He underwent biopsy of one liver lesion, and pathology demonstrated poorly differentiated adenocarcinoma with focal neuroendocrine differentiation (CK7+, CDX2+, synaptophysin/chromogranin+, CK20−, TTF1−, napsin−) consistent with pancreatic or biliary origin. A PET-CT scan showed localized cancer in the right hepatic lobe, and the patient subsequently underwent surgical resection with clear margins and no lymph node involvement. Surgical pathology confirmed intrahepatic cholangiocarcinoma, which was staged as T2aN0. The patient received no adjuvant therapy postsurgery. Five months later, in April 2016, surveillance MRI showed the emergence of new hepatic tumors, prompting palliative treatment with gemcitabine and cisplatin (Gem/Cis). Gemcitabine (1000 mg/m2) and cisplatin (25/m2) were given on day 1 and day 8 of a 21-d cycle. In June 2016, after two cycles of chemotherapy, CT scans revealed numerous hypodense lesions consistent with worsening of hepatic metastatic disease, and Gem/Cis was stopped. At this time, the patient underwent a repeat tumor biopsy and RNA profiling of his cancer using an NGS assay, OSU-SpARKFuse (Reeser et al. 2017), which revealed a novel gene fusion involving FGFR2 (exons 1–16) and CLIP1 (exons 19–24) (Fig. 1A; Table 1). The presence of the fusion was confirmed by reverse transcription PCR and Sanger sequencing with primers designed to flank the breakpoint (Fig. 1C; Supplemental Fig. S1A). CLIP1 is a CAP-Gly domain-containing linker protein 1 that has been shown to regulate the microtubule cytoskeleton. Based on the presence of this novel FGFR2–CLIP1 fusion in his cancer, at the beginning of October, the patient enrolled in a Phase I/II clinical trial (NCT02393248) evaluating the safety and tolerability of an oral pan-FGFR inhibitor, INCB054828. He received 13.5 mg once daily for days 1–14 per 21-d cycle. Disease assessment after cycles 3 (November) and 6 (January) showed robust partial response by RECIST criteria, consistent with this novel FGFR2 fusion being a driver of his metastatic cancer (Fig. 1A). As part of the study, two target lesions (posterior hepatic dome lesion and left hepatic lobe lesion) were tracked throughout the treatment course and had a 34.8% and 46.5% reduction from baseline after cycles 3 and 6, respectively (Fig. 1B). Prior to starting cycle 8, he was admitted to the hospital with significant weight loss and elevated liver function tests (LFTs), suggesting disease progression. After a total of 5 mo (7 cycles) on INCB054828, CT scans showed a 41.3% increase in size of the two target lesions confirming progressive disease (Fig. 1A,B). At this time, he underwent a repeat postprogression tumor biopsy that confirmed the continued presence of the FGFR2–CLIP1 fusion (Fig. 1A). One month after receiving the last dose of INCB045828, second-line chemotherapy (FOLFOX) was initiated. He received a single dose of oxaliplatin (190 mg) and fluorouracil (3975 mg). However, he passed away 11 d after receiving this single dose of FOLFOX as a result of liver failure. Prior to passing, he consented to our body donation study for patients with advanced cancer.
Figure 1.
Clinical description of a cholangiocarcinoma patient harboring an FGFR2–CLIP1 fusion. (A) A patient with metastatic cholangiocarcinoma underwent a liver biopsy and sequencing revealed an FGFR2–CLIP1 gene fusion. He received gemcitabine/cisplatin but after two cycles of therapy had disease progression. He was enrolled on INCB54828 and had a profound radiographic response. After 5.5 mo on therapy, he developed progression and passed away shortly thereafter from his disease. A repeat tumor biopsy at the time of progression did not reveal any secondary mutations. (B) Table summarizing two target lesions (posterior hepatic dome lesion and left hepatic lobe lesion) that were tracked throughout the treatment course and had a 34.8% and 46.5% reduction from baseline after cycles 3 and 6, respectively. (C) Schematic of the FGFR2–CLIP1 fusion involving exons 1–16 of FGFR2 and exons 19–24 of CLIP1. Chromatogram traces from Sanger sequencing of the tumor biopsy confirmed the presence of the fusion. Red dashed line indicates breakpoint within the sequence.
Table 1.
Variant table
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect (substitution, deletion, etc.) | dbSNP/dbVar ID | Genotype (heterozygous/homozygous) | ClinVar ID |
|---|---|---|---|---|---|---|---|---|
| FGFR2 and CLIP1 | RNA Chr 10:123243211 and Chr 12:122773087 DNA Chr 10:123241591 and Chr 12:122782142 |
N/A | N/A | Chromosomal rearrangement | Likely pathogenic | N/A | N/A | N/A |
| FGFR2 | Chr 10:123258036 | NM_000141.4, c.1645A>C | N549H | Single-nucleotide variant | Likely pathogenic | rs1057519045 | N/A | SUB5631497 |
Research Autopsy Reveals Clonal Heterogeneity in Cholangiocarcinoma
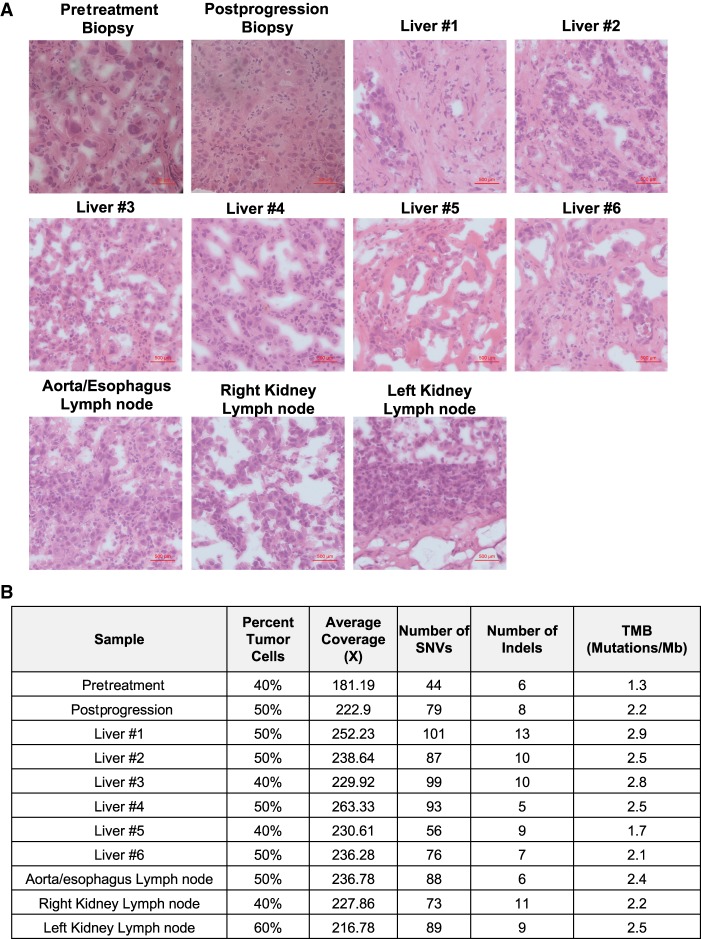
Upon death of this patient, a research autopsy was performed 8 hours postmortem. Gross examination revealed metastatic tumors involving the liver, omentum, and abdominal and retroperitoneal lymph nodes. Twenty-four liver tumor samples and five separate lymph nodes were procured at the time of autopsy. Although we attempted to sample distinct liver tumors, the patient's liver was predominately cancerous with limited grossly normal liver tissue present (Supplemental Fig. S1B). Samples used for subsequent analysis had at least 40% tumor content as determined by a board-certified pathologist (Fig. 2A,B). In total, a normal blood control and 11 tumor samples (one pretreatment tumor biopsy, one postprogression tumor biopsy, and nine autopsy tumor samples) were chosen for further analysis (Fig. 2B). Sanger sequencing confirmed that the FGFR2–CLIP1 fusion was present in each tumor sample (data not shown). DNA from these tumors were subjected to WES, yielding 231× average target coverage (Fig. 2B) and revealed a total of 979 somatic variants across all tumors (292 unique somatic variants) (Samorodnitsky et al. 2015a). Two hundred and forty-two of these mutations were unique to the postprogression and autopsy samples (Supplemental File S6). The tumor mutational burden (TMB) of samples ranged from 1.3 mutations/Mb in the pretreatment biopsy to 2.9 mutations/Mb in liver sample #1 (Fig. 2B), consistent with previous studies indicating low TMB in cholangiocarcinoma (Nakamura et al. 2015; Chalmers et al. 2017). All tumor samples were determined to be microsatellite stable (MSS) through analysis of 2539 loci by MANTIS (Kautto et al. 2017). Mutational signatures 16 and 19 were common across tumor samples. Signature 16 has been found in liver cancer and signature 19 has been found in pilocytic astrocytoma, however, their etiologies are unknown (Forbes et al. 2017).
Figure 2.
Tumor samples procured at research autopsy. (A) Hematoxylin and eosin (H&E) stains of representative slides taken from each tumor sample demonstrating abundant malignant cells. (B) Summary of estimated tumor content and WES metrics within each sample. (SNV) Single-nucleotide variant, (TMB) tumor mutational burden.
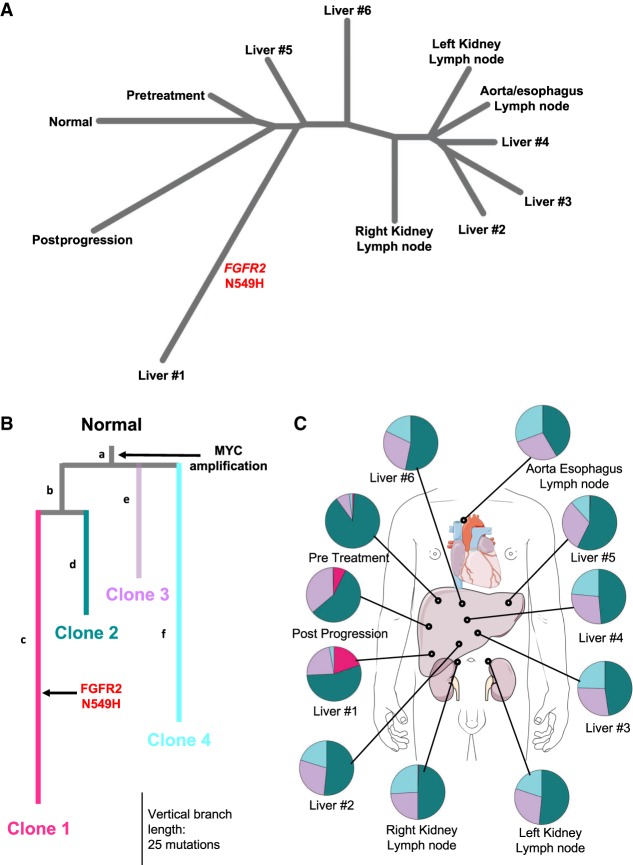
The somatic single-nucleotide variants (SNVs) called in each tumor sample were subsequently used to build a phylogenetic tree of tumor samples via the neighbor-joining (NJ) method (Fig. 3A; Saitou and Nei 1987). As expected, the pretreatment sample branched most closely to the normal cells; the two samples are separated by a relatively short genetic distance of 33.2 indicating a high degree of genetic similarity. The postprogression sample had the next closest genetic similarity to the normal, with a genetic distance of 37.6. The liver #1 sample was the most genetically unique tumor sample with a genetic distance of 100.1 from the normal. Liver samples #2, #3, and #4 were clustered with the aorta/esophagus lymph node and left kidney lymph node.
Figure 3.
Analysis of tumor heterogeneity. (A) Neighbor-joining tree over sets of somatic SNVs in each of 11 tumor samples, with normal defined as the empty set. (B,C) Subclonal inference from Canopy. Colors in B correspond to subclones in C. Letters identify branches of the tree. (B) Phylogenetic tree assessment with Canopy revealed four major clonal populations of cells. Each subclone is characterized by a group of mutations. MYC gain was truncal to all subclones. Clone 1 (pink) contained a unique FGFR2 N549H point mutation. Vertical distance corresponds to increased number of somatic mutations (SNVs and indels). (C) Prevalence of four tumor subclones within tumor samples.
We next utilized Canopy (Jiang et al. 2016) to computationally identify and characterize tumor subclones using both synonymous and nonsynonymous somatic SNVs, CNVs, and indels (Fig. 3B). This analysis revealed four tumor subclones across the 11 samples, with each subclone characterized by a unique group of genomic alterations (Fig. 3B; Supplemental Fig. S2; Supplemental Files S5, S7, S8). A four-clone model was selected because models with additional subclones yielded only marginal increases in BIC (Supplemental Fig. S2). Clones 2 (teal) and 3 (purple) were shared among all samples (Fig. 3C). Clone 4 (cyan) was seen in all except one autopsy sample (liver #1) and was not present in the pretreatment or posttreatment samples (Fig. 3C). Eighty-nine percent of the tumor cells in the pretreatment sample were estimated to be from clone 2 versus ∼40%–60% of the other samples (Fig. 3C). Clone 1 (pink) was primarily found in liver #1 (20%) and at low frequency in the posttreatment sample (7%) (Fig. 3C). This is consistent with the NJ tree, as the relatively large number of mutations unique to clone 1 accounts for the distance of liver #1 and the postprogression samples from all other samples.
Of the 292 distinct mutations (SNVs and indels) identified among these samples, only seven were truncal (i.e., common to all four subclones). Most notable among the truncal events (branch a) was a 21.9-Mb gain in Chromosome 8q (Chr 8: 124448804–146364022), containing MYC among other genes. Although Canopy did not identify nontruncal mutations shared by clones 3 and 4, post-hoc assignment was permitted to assign mutations to a hypothetical unique common ancestor. No such mutations were assigned, suggesting that clones 3 and 4 diverged relatively early in the tumor's evolution. Of clinical interest, WES revealed an FGFR2 kinase domain mutation, FGFR2 N549H in a single liver tumor, liver #1 (Fig. 3A). The FGFR2 N549H mutation occurs in the kinase hinge and has been shown to disengage the molecular breaker resulting in ligand-independent constitutive activation of the FGFR2 kinase (Chen et al. 2007). The FGFR2 N549H mutation was assigned uniquely to clone 1, which was the most genetically distinct subclone compared to the patient's normal blood DNA (Fig. 3B). Although clone 1 was predicted to be present at low frequency in the postprogression sample, FGFR2 N549H was not detected in this sample. ddPCR of all samples confirmed that the FGFR2 N549H mutation was unique to liver #1 (Supplemental Table S1). Of the 111 mutations unique to clone 1, this mutation was estimated to be the 63rd to occur. This led us to hypothesize that the N549H FGFR2 kinase domain mutation may have been partially responsible for driving resistance to INCB054828 in this patient, occurring along an existing clonal lineage. Driver mutation prediction with CHASM (Carter et al. 2009) predicted only FGFR2 N549H to be a statistically likely driver (defined as FDR-corrected P ≤ 0.05) (Supplemental File S9).
In Vitro Characterization of Acquired Mutations in the FGFR2–CLIP1 Fusion and Resistance to the FGFR Inhibitor INCB054828
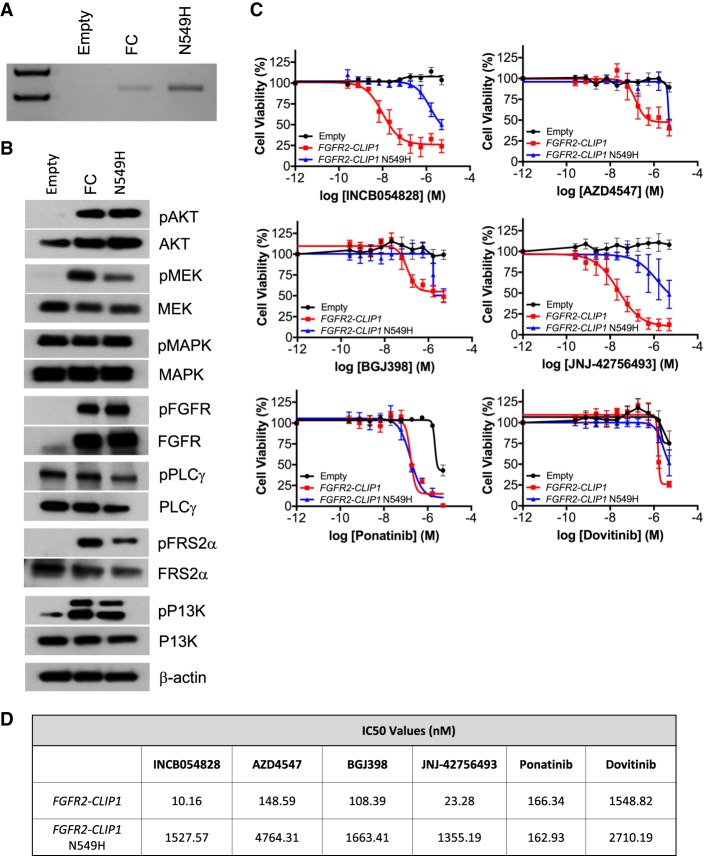
To confirm our clinical findings that the FGFR2–CLIP1 fusion is exquisitely sensitive to INCB054828 and explore the hypothesis that the FGFR2 N549H mutation confers resistance to INCB054828, we generated NIH3T3 cells that express either a control (Empty) vector, FGFR2–CLIP1 fusion (FC), or FGFR2–CLIP1 fusion with the N549H secondary mutation (N549H) and confirmed expression by RT-PCR (Fig. 4A) and Sanger sequencing (data not shown). Western blot analyses of FGFR2–CLIP1 fusion expression cells demonstrated increases in PI3K/AKT, MAPK/MEK, and FGFR2 signaling pathways with or without N549H (Fig. 4B).
Figure 4.
The FGFR2–CLIP1 is sensitive to FGFR inhibitors, whereas the N549H kinase domain mutation confers resistance. (A) RT-PCR confirmed the presence of the FGFR-CLIP1 fusion in the NIH3T3 FGFR2–CLIP1 (FC) cells and NIH3T3 FGFR2–CLIP1 N549H (N549H) cells. The fusion was not detected in the control vector (Empty) transduced cells. (B) Total cell lysates from NIH3T3 Empty, FC, and N549H cells were prepared and subjected to western blot analysis with antibodies against pAKT, AKT, pMEK, MEK, pMAPK, MAPK, pFGFR, FGFR, pPLCy, PLC, pFRS2, FRS2, pPI3K, PI3K, and β-actin. (C) IC50 curves of NIH3T3 Empty, FC, and N549H cells treated with FGFR inhibitors. Data from four replicate experiments are shown. Inhibitors include INCB054828, AZD4547, BGJ398 and JNJ42756493, ponatinib, and dovitinib. (D) IC50 values are reported for each inhibitor from the curves seen in C.
To evaluate the in vitro sensitivity of cells with the FGFR2–CLIP1 fusion and cells with the FGFR2–CLIP1 N549H to the FGFR inhibitor INCB054828, we treated NIH3T3 Empty, FGFR2–CLIP1, and FGFR2–CLIP1 N549H cells with increasing doses of INCB054828 or vehicle control (DMSO) ranging from 1.0 nM to 5000 nM and assessed cell viability after 72 h. Treatment of NIH3T3 FGFR2–CLIP1 (FC) cells with INCB054828 demonstrated substantial and reproducible inhibition of cell viability with an IC50 value of 10.16 nM (Fig. 4C,D). Consistent with our hypothesis, the FGFR2–CLIP1 N549H (N549H) cells were resistant to INCB054828 with an IC50 value of 1527.57 nM (Fig. 4C,D). Empty vector control cells (Empty) were not sensitive to INCB054828, which is expected as these cells do not express endogenous FGF ligands or FGF receptors (Fig. 4C). Thus, these data help explain this patient's clinical course with his initial FGFR2–CLIP1 fusion expressing tumor responding to INCB054828 followed by acquisition of resistance via the N549H mutation.
We subsequently extended these in vitro studies to include additional FGFR inhibitors that are currently being evaluated clinically in patients with metastatic cancer and have shown early responses in patients with FGFR-mutant cancers. AZD4547, BGJ398, and JNJ-42756493 are selective FGFR inhibitors, whereas ponatinib and dovitinib are nonspecific tyrosine kinase inhibitors that target BCR-ABL, VEGFR, PDGFR, SRC, RET, KIT, and FLT1 in addition to FGFR. Our results demonstrated that FGFR2–CLIP1 cells were sensitive to AZD4547, BGJ398, JNJ-42756493, and ponatinib with IC50 values of 148.59 nM, 108.39 nM, 23.28 nM, and 166.34 nM, respectively (Fig. 4C,D). The FGFR2–CLIP1 N549H cells were less sensitive to BGJ398, AZD4547, and JNJ-42756493, as demonstrated by higher IC50 values than the fusion alone (Fig. 4C,D). Interestingly, FGFR2–CLIP1 N549H cells demonstrated a similar sensitivity to ponatinib as cells with the fusion alone (Fig. 4C,D). Dovitinib was largely ineffective against FGFR2–CLIP1 without or with the secondary mutation (Fig. 4C,D). Empty vector control cells (Empty) were not sensitive to AZD-4547, BGJ398, or JNJ-42756493 (Fig. 4C,D). However, at the highest dose (5 µM) of ponatinib and dovitinib, the control cells (Empty) demonstrated decreased cell viability, which was not surprising as ponatinib and dovitinib are nonspecific inhibitors of FGFR (Fig. 4C,D). Taken together, these data demonstrate that the FGFR2–CLIP1 fusion confers sensitivity to some, but not all, FGFR inhibitors. The FGFR2 N549H secondary mutation confers resistance to most FGFR inhibitors, but ponatinib could be used to overcome this acquired drug resistance.
DISCUSSION
Tumor heterogeneity has been shown to have a critical role in response to therapy, development of resistance, and clinical outcome in patients with cancer (Rottenberg et al. 2012; Choi et al. 2017; Joung et al. 2017). Rapid research autopsy has emerged as a powerful strategy to study tumor heterogeneity, as it enables essentially unlimited sampling of all sites of metastatic disease throughout the body that would otherwise not be feasible through surgical resections or tumor biopsies (Krook et al. 2019). A number of recent studies have utilized rapid research autopsy to characterize tumor heterogeneity, clonal evolution, and mechanisms of acquired therapeutic resistance in breast, urothelial, pancreatic, and colorectal cancer. For instance, Saito et al. utilized research autopsy in breast cancer to assess trastuzumab resistance in primary versus metastatic sites (Saito et al. 2015). Faltas et al. (2016) performed rapid autopsy of two patients to construct phylogenetic trees of urothelial carcinoma. Here, we present our findings from rapid research autopsy of a patient with metastatic cholangiocarcinoma. This is the first study to evaluate clonal heterogeneity based on exome sequencing in cholangiocarcinoma, as well as the first description of acquired resistance to INCB54828, an oral FGFR inhibitor.
Most previous and current autopsy studies utilize methods such as clonal ordering (Merlo et al. 2006) and NJ (Saitou and Nei 1987) to identify and quantify relationships between different tumor regions and/or sites of metastatic cancer. In this study, we utilized the NJ method to generate a tumor-centric tree to assess similarities and differences among the pretreatment biopsy, postprogression biopsy, and nine unique tumors collected at the time of autopsy. The NJ analysis showed that the liver #1 sample with its unique FGFR2 N549H point mutation is an outlier versus the other liver samples from autopsy. NJ and related phylogenetic methods are powerful tools to assess high-level relatedness among tumors and identify exceptional tumors; however, they cannot capture the clonal heterogeneity present within discrete tumor masses or cross-seeding between sites.
Analysis of clonal evolution continues to develop as technical and computational challenges and the limited availability of large-scale autopsy data are overcome. In addition to NJ, we performed subclonal inference using Canopy (Jiang et al. 2016), which revealed four genetically distinct tumor subclones. Of these subclones, three were dominant across all 11 samples, with each subclone characterized by a specific set of mutations (Fig. 2B,C). The FGFR2 N549H mutation in clone 1 was unique to a single liver sample despite our in vitro data confirming its role as a resistance mutation. This pattern of site-unique FGFR2 resistance mutations was previously observed by Goyal et al. (2017) in which only four of 12 distinct metastatic autopsy samples were found to have acquired secondary mutations in FGFR2 serving to bypass the FGFR inhibitor effect. Each of these samples harbored unique FGFR mutations (K641R and N549H) with only one sample having two FGFR mutations (E565A and K641R). Meanwhile, the remaining eight sites were wild type (wt) for FGFR2. The observations seen by Goyal et al. along with our work presented here suggest that multiple independent drug resistance mechanisms, including FGFR-independent mechanisms, are likely contributing to tumor progression. Interestingly, in our model, clone 4 was specific to tumor samples collected at the time of autopsy, suggesting that either the biopsies missed a population of cells or that this subclone developed after the posttreatment biopsy. FOLFOX was administered after the collection of the posttreatment biopsy, but as this patient only received one dose of FOLFOX before passing away soon afterward, we do not believe that this single dose substantially affected the heterogeneity present at the time of autopsy. These findings provide evidence for the presumed notion that tumor biopsies do not accurately reflect the full complexity and heterogeneous nature of the disease. Clones 2–4 were seen in all autopsy samples at similar proportions. As the liver tumors were largely confluent at the time of autopsy (Supplemental Fig. S1B), multiple samples may have come from the same tumor. Another possibility is metastatic cross-seeding, as was observed by Savas et al. (2016) using their tool superFREQ (Flensburg et al. 2018) for subclonal analysis of four metastatic breast cancer cases, and by Brady et al. (2019) in pediatric osteosarcoma. Co-metastasis of multiple subclonal populations can also explain this distribution, as has been demonstrated to occur in breast ductal carcinoma (Casasent et al. 2018). Clone 2 was substantially reduced in the posttreatment and autopsy samples versus the pretreatment biopsy. One potential explanation is that clone 2 was more sensitive to INCB054828 than clones 1, 3, and 4. In clone 1, the decreased sensitivity is likely due to the FGFR2 N549H point mutation, evolving from a common lineage as clone 2. Although resistance mechanisms for clones 3 and 4 could not be determined through WES, and driver prediction did not indicate any other likely driver mutations, we hypothesize that there can be multiple independent drug resistance adaptations within a single patient. Previous studies have demonstrated that in addition to secondary kinase domain mutations, activation in the Akt, MAPK, and PTEN pathways can mediate resistance to FGFR inhibition (Datta et al. 2017; Goyal et al. 2017; Malchers et al. 2017). Although there was no evidence for PTEN mutations in this patient, transcriptome sequencing would be needed to assess the activation of other pathways. Studies are ongoing in our laboratory to identify FGFR-independent mechanisms of resistance and to define their contributions clinically, including RNA sequencing.
WES of this patient and derivation of a phylogenetic tree suggests that ancestral genotypes can persist throughout the disease course, despite the evolution of highly derived subclones. We note that only four SNVs, three indels, and one copy-number gain were detected in the trunk of this patient's phylogenetic tree (branch a), indicating that development of FGFR2-fusion-positive cholangiocarcinoma may only require a small number of other initiating events (Fig. 3B). For instance, clone 4 was only detected in autopsy samples, yet evolved from a distant ancestor to clones 1, 2, and 3. Clone 1 did not directly evolve from clone 2, but rather it shares a common ancestor with clone 2, which must have been extant before treatment (for clone 2 to be found in the pretreatment sample). Such persistent ancestral cells may serve as an “uncommitted” tumor reserve capable of developing new adaptations throughout the disease course. Subclonal analysis, such as in this study, permits the characterization of cancer as a dynamic process of multiple evolving and diverging cellular populations rather than a singular entity in a patient. This view of cancer permits somatic variants, a staple of cancer genomics, to be viewed in a new context. However, phylogeny inference from short-read bulk sequencing has several inherent limitations, most notably that phylogenetic solutions consistent with variant fractions and CNVs are frequently nonunique (Pradhan and El-Kebir 2018). Emerging long-read and single-cell sequencing technologies will permit more certain and accurate modeling of phylogeny by directly assessing the phasing of subclonal mutations.
Lessons learned from studying molecular mechanisms of resistance to ABL, EGFR, ALK, KIT, and RAF inhibitors in human cancers have highlighted the need for next-generation kinase inhibitors that are effective against acquired secondary resistance mutations (Demetri 2011; Roychowdhury and Talpaz 2011; Gainor and Shaw 2013; Lito et al. 2013; Van Allen et al. 2014; Hrustanovic et al. 2015). For example, Friboulet et al. demonstrated that crizotinib-induced resistance mutations in ALK-fusion-positive non-small-cell lung cancer (NSCLC) can be overcome by treatment with ceritinib (Friboulet et al. 2014). Furthermore, mutant-selective allosteric inhibitors have shown promise in overcoming the secondary EGFR resistance mutation T790M in NSCLC following EGFR-directed therapy (Jia et al. 2016). Thus, these studies may inform strategies to overcome secondary resistance mutations to FGFR-targeted therapies as several preclinical studies have demonstrated the emergence of a mutation at the gatekeeper residue or other residues within the ATP-binding pocket as well as other mutations in FGFR1–3 (Chell et al. 2013). Unfortunately, several potent and selective ATP-competitive small molecule FGFR inhibitors currently in clinical trials, including INCB054828, BGJ398, AZD4547, and LY2874455, share structural similarities and are ineffective in overcoming the gatekeeper mutations (Chae et al. 2017). Although not considered a gatekeeper mutation, the FGFR2 N549H mutation is in the vicinity of the ATP binding pocket. Notably, our in vitro findings provide further support for the cross-resistance of multiple FGFR inhibitors, as cells harboring the secondary FGFR mutation N549H were resistant to INCB054828, AZD4547, BGJ398, JNJ-42756493, and dovitinib. Because of this, there has been interest in the use of structure-based drug design to develop a class of next-generation inhibitors that would overcome resistance mutations located in the FGFR2 ATP-binding pocket (Tan et al. 2014). Interestingly, we demonstrated that FGFR2 N549H retained sensitivity to ponatinib. The clinical use of ponatinib in this context is supported by pharmacokinetic data in patients demonstrating a steady state ponatinib plasma concentration of 145 nM attained 4–8 h after receiving the maximum approved dose of 45 mg (Cortes et al. 2012). Unfortunately, there are serious adverse cardiovascular events associated with ponatinib, which are often dose-limiting (Gagnieu et al. 2013). Thus, the development of next-generation FGFR inhibitors has the potential to dramatically impact the clinical care of patients receiving FGFR-targeted therapies.
In summary, this work suggests that clonal heterogeneity contributes to acquired clinical resistance to the novel FGFR inhibitor, INCB054828, in cholangiocarcinoma. Although limited to a single patient, this is the first study, to our knowledge, to define a mechanism of acquired resistance to INCB054282 through a secondary mutation to the FGFR inhibitor, INCB054828. Through rapid research autopsy and WES, we determine the presence of four tumor subclones and elucidate their evolution in metastatic tissues over time in a patient with FGFR2-fusion-positive cholangiocarcinoma. Furthermore, we identified a posttreatment secondary kinase mutation in FGFR2, present in a single metastatic tumor sample demonstrating the significance of intertumor heterogeneity within the same patient. We characterized the impact of the N549H mutation on sensitivity to different FGFR inhibitors in vitro. The results of our in vitro drug sensitivity studies suggest that this mutation conferred resistance to INCB054828 in this patient and thus may have potential as a clinically useful biomarker of resistance. Overall, our findings suggest that secondary FGFR mutations are drivers of acquired clinical resistance. Understanding these mechanisms of resistance along with FGFR kinase domain-independent mechanisms of resistance will facilitate approaches to prevent or overcome treatment resistance and disease recurrence and guide clinical strategies for these patients.
METHODS
Research Autopsy and Patient Samples
The patient consented to an IRB-approved study for high-throughput sequencing of tumor and normal specimens (OSU-13053, NCT02090530) at the James Cancer Hospital and The Ohio State University. OSU-SpARKFuse, a targeted RNA-based NGS assay to detect gene fusions, and a targeted DNA sequencing assay to detect single-nucleotide variations were performed on tumor biopsy specimens as previously described (Reeser et al. 2017). The patient also consented to a body donation study. Upon death of this patient, next of kin informed the research team, who arranged for transportation to the OSU Regional Autopsy Center, and the autopsy was performed 8-h postmortem. Guided by radiographic scans, all visible malignant as well as adjacent normal tissues were collected and frozen in optimal cutting temperature (OCT) compound. Following the autopsy, the patient was returned to the funeral home.
Whole-Exome Sequencing
Genomic DNA was extracted from frozen tumor biopsy samples and tumors collected during autopsy using the QIAamp DNA Mini Kit. The QIAamp DNA Mini Blood Kit was used to extract genomic DNA from blood. WES was performed as described below. Briefly, the KAPA Hyper Prep Kit (Roche) was used for library preparation, and libraries were enriched using the xGEN Exome Research Panel v1.0 from Integrated DNA Technologies. 2 × 150-bp paired-end sequencing was performed on an Illumina HiSeq4000 at The Genomics Services Laboratory at Nationwide Children's Hospital (Columbus, Ohio).
Histology
Freshly collected tumor biopsy and autopsy samples were immediately embedded and frozen in OCT compound (Fisher). Frozen sections were cut from tumor biopsy and autopsy samples at 5 µm on a Leica Cryostat CM1950 for H&E staining. A board-certified pathologist reviewed representative slides for each tumor block for estimated tumor content.
Bioinformatics Analysis
All bioinformatics analyses were performed using the Oakley supercomputer at the Ohio Supercomputer Center (“Oakley supercomputer” 2012). Alignment of WES data to the human genome version hg19 was performed with Burrows–Wheeler Aligner (bwa) (Li and Durbin 2009) version 0.7.14. Duplicate reads were removed using Picard (“Picard Tools - By Broad Institute”) version 2.3.0. Picard and GATK (McKenna et al. 2010) version 3.5 were used to perform quality recalibration and local realignment around indels. SNV and indel calling were performed with VarScan2 (Koboldt et al. 2012) version 2.3.9 and bam-readcount (Larson and Abbott 2016) as previously described (Chen et al. 2019; Supplemental File S1). SNVs and indels were annotated using ANNOVAR (Wang et al. 2010) (revision #11f4bb, 2016-02-01). Putative driver mutation analysis was performed with CRAVAT (Douville et al. 2013) version 5.2.4, using the CHASM (Carter et al. 2009), algorithm version 3.1. TMB was computed as the sum of all called somatic SNVs and indels within the capture region in each sample, divided by the size of the capture region (∼38.9 Mb). Microsatellite instability (MSI) testing was performed with MANTIS (Kautto et al. 2017) using the recommended threshold of 0.4 to call MSI and a set of 2539 whole-exome microsatellite loci (Bonneville et al. 2017). Mutational signatures were called with deconstructSigs (Rosenthal et al. 2016) version 1.8.0 using the COSMIC Mutational Signatures set (Forbes et al. 2017), exome2genome trinucleotide frequency correction, and otherwise default settings (Supplemental File S2).
CNV calling was performed using FALCON (Chen et al. 2015) version 0.2, utilizing germline tumor and normal variants (Supplemental File S3). The QC procedure provided with Canopy (Jiang et al. 2016) was used to reduce false-positive segmentations, with default length and ΔCN settings. For each sample, rdep (read depth ratio) was the ratio of aligned reads in tumor versus normal. FALCON was initially run with threshold 0.3. Resulting CNVs were manually curated to identify genomic regions with major copy number >2 or minor copy number <0.5 in at least one sample. For each curated region, a common pair of breakpoints was estimated across all tumors, and FALCON was rerun with threshold 0.2 and set to the nearest SNPs to each breakpoint in the chromosome (Supplemental File S4). Matrices WM, Wm, εM, and εm (used for Canopy input) were obtained from FALCON output, and matrix Y was determined by calculating the overlap of mutations used for tree building with curated CNV regions.
To generate a sample-based phylogenetic tree, a distance matrix was first computed as follows:
where is the distance matrix, N is the number of samples, Si is the set of somatic SNVs called in sample i ∈ 1,…,N, and Δ is the set symmetric difference. The set of somatic SNVs in normal, corresponding to i = N + 1, is the empty set (by definition); therefore, the distance between normal and any tumor collapses to the number of SNVs in that tumor. The tree was generated over D via NJ (Saitou and Nei 1987) with RapidNJ (Simonsen et al. 2008) version 2.3.2 and visualized using Interactive Tree of Life (iTOL) (Letunic and Bork 2016) version 4.2.3. Normal is regarded as the root of the tree. Subclonal-based phylogenetic analysis with Canopy and maximum likelihood-based post-hoc assignment of somatic SNVs and indels to the resulting tree was performed as previously described (Chen et al. 2019). Canopy computes a Bayesian information criterion (BIC; Schwarz 1978) score for each potential number of subclones, which was used to determine the number of subclones that best represents the data. Note that Canopy estimates its normal cell fractions based on variant fractions and CNV data, and reported purities can differ from pathologist estimates, likely because different sections of the tumor blocks were sequenced than were reviewed by the pathologist (Supplemental File S5).
Mutations within each branch of the tree were temporally ordered using a Bradley–Terry model (Bradley and Terry 1952). For any tree edge z ∈ Z, we have a set of mutations Vz. For convenience, define VAFi(v) as the VAF of mutation v in sample i ∈ 1,…,N. Given variants v1, v2 ∈ Vz, we compute the score of v1 versus v2 in i as follows:
We now define , the wins matrix, over all samples:
We utilized the R package BradleyTerryScalable (Kaye and Firth 2017) version 0.1.0 with a = 1.1, which implements the maximum a priori estimate of Caron and Doucet (Caron and Doucet 2012). Results are returned as abilities, such that
where π1 and π2 are the abilities of mutations v1 and v2. Note that ability scores denote confidence in ordering, not the time intervals between acquisition of mutations. This analysis was performed independently for each edge of the clonal phylogeny tree, utilizing both mutations supplied to Canopy and those retroactively assigned to the tree.
Droplet Digital PCR and Analysis
Isolated genomic DNA was amplified using a custom-designed probe for the FGFR2 N549H point mutation (PrimePCR ddPCR Mutation Assay, Bio-Rad) and the ddPCR Supermix for Probes (Bio-Rad). The reaction mixture consisted of 250 ng of DNA template (8 µL), 10 µL of ddPCR Supermix for Probes (Bio-Rad), and 2 µL of the primer/probe mixture. Droplets were generated using the QX200 Droplet Generator (Bio-Rad) and then transferred to a 96-well plate (Eppendorf) for PCR amplification with the following conditions: 5 min at 95°C, 40 cycles of 94°C for 30 sec, 55°C for 1 min, followed by 98°C for 10 min (ramp rate 2°C/sec). Droplets were analyzed with the QX200 Droplet Reader (Bio-Rad) for fluorescent measurement of FAM and HEX probes. Gating was performed based on positive and negative controls, and mutant populations were identified. All reactions were run in duplicate. The ddPCR data were analyzed with QuantaSoft analysis software (Bio-Rad) to obtain fractional abundance of the mutant DNA alleles in the wt/normal background.
cDNA Plasmid Generation, Lentivirus Production, and Transduction
The FGFR2–CLIP1 fusion was produced and cloned into the pLVX-IRES-Puro vector (Clontech) by GenScript (Supplemental Fig. S1). Using site-directed mutagenesis, the FGFR2 N549H mutation was introduced into the fusion by GenScript. NIH3T3 cells were stably transduced with either empty, FGFR2–CLIP1 or FGFR2–CLIP1 N549H lentiviral vectors. Cells were selected in puromycin (1 µg/ml; Sigma) for 72 h prior to their use in downstream experiments.
RNA Isolation, RT-PCR, and Sanger Sequencing
RNA was isolated from cell lines, and cDNA was synthesized using the Quick-RNA MiniPrep Kit (Zymo) and the iScript cDNA Synthesis Kit (Bio-Rad), respectively. cDNA was subsequently PCR amplified with FGFR2–CLIP1 and FGFR2 N549H fusion specific primers (IDT). Primer sequences are listed in Table 2. The PureLink Quick PCR Purification Kit (Invitrogen) was used to purify amplified PCR product and samples were then Sanger sequenced (The Ohio State University Comprehensive Cancer Center Genomics Shared Resource, Columbus, OH).
Table 2.
Primer sequences
| Target | Forward | Reverse | Product size (bp) |
|---|---|---|---|
| FGFR2–CLIP1 | 5′-CAGAGACCAACGTTCAAGCA-3′ | 5′-CGGCATCCTTTTCTGTGAGT-3′ | 214 |
| N549H | 5′-GTGGCCGTGAAGATGTTGAA-3′ | 5′-AGGTATTCTCGGAGGTTGCC-3′ | 188 |
Primer sequences used for PCR and Sanger sequencing to confirm the presence of either the fusion or the mutation.
Cell Culture
NIH3T3 and HEK293T cell lines were purchased from American Type Culture Collection (ATCC) and cultured in a humidified incubator at 37°C and 5% CO2. Cells were cultured according to the ATCC-recommended protocols. All cell lines were routinely subjected to short tandem repeat profiling to confirm identities and mycoplasma testing using the e-Myco plus Mycoplasma PCR Detection Kit (Bulldog Bio).
Western Blotting
Western blot assays were carried out using established protocols and probed with the following antibodies: phospho-Akt (Ser473) 1:1000 (Cell Signaling 9271), Total Akt 1:1000 (Cell Signaling 9272), phospho-MEK1/2 1:5000 (Cell Signaling 9154), Total MEK1/2 1:5000 (Cell Signaling 9122), p44/42 MAPK (Erk1/2) 1:5000 (Cell Signaling 9101), Total MAPK 1:5000 (Cell Signaling 9102), phospho-FGF Receptor (Tyr653/654) 1:500 (Cell Signaling 3471), FGF Receptor 2 (D4L2V) 1:500 (Cell Signaling 23328), phospho-PLCγ1 (Tyr783) 1:1000 (Cell Signaling 14008), PLCγ1 (D9H10) 1:1000 (Cell Signaling 5690), phospho-FRS2-α (Tyr196) 1:1000 (Cell Signaling 3864), FRS2 1:1000 (abcam 10425), phospho-PI3 Kinase p85 (Tyr458)/p55 (Tyr199) 1:1000 (Cell Signaling 4228), PI3 Kinase p85 (19H8) 1:000 (Cell Signaling 4257), β-actin 1:10000 (Cell Signaling 4967).
Drug Sensitivity Assays
NIH3T3 Empty, FGFR2–CLIP1, FGFR2–CLIP1 N549H cells were plated at a density of 10,000 cells per well in 96-well plates. Cells were treated for 72 h with either INCB054828 (Incyte), BGJ398 (Cayman Chemical), JNJ-42756493 (Cayman Chemical), AZD-4547 (Cayman Chemical), ponatinib (Cayman Chemical), or dovitinib (Cayman Chemical) ranging from 0.01 to 5000 nM. Quantification of viable cells was assessed using an MTS/PMS colorimetric assay. IC50 values were calculated in Prism (GraphPad) using a four-parameter dose–response model.
ADDITIONAL INFORMATION
Data Deposition and Access
Data used for the analyses presented in the manuscript have been submitted to dbGaP (https://ncbi.nlm.nih.gov/gap) under the project accession number phs001830.v1.p1. The FGFR2-CLIP1 fusion gene variant and the secondary FGFR2 mutation identified in the patient have also been deposited to ClinVar (https://ncbi.nlm.nih.gov/clinvar/) under the accession numbers SCV000927106 and SCV000914229.1.
Ethics Statement
The patient consented to an IRB-approved study for high-throughput sequencing of tumor and normal specimens (OSU-13053, NCT02090530) at the James Cancer Hospital and The Ohio State University. OSU-SpARKFuse, a targeted RNA-based next-generation sequencing assay to detect gene fusions, and a targeted DNA sequencing assay to detect single-nucleotide variations were performed on tumor biopsy specimens as previously described (Samorodnitsky et al. 2015b; Reeser et al. 2017). The patient also consented to a body donation study.
Acknowledgments
We thank Jenny Badillo for her administrative support. We also thank current and past members of the Roychowdhury Precision Cancer Medicine Team, The Ohio State Comprehensive Cancer Center, and James Cancer Hospital and Pelotonia for its community support. We most importantly thank this patient and his family. We thank the Ohio Supercomputer Center for computing resources.
Author Contributions
M.A.K. and K.R.B. were involved in the generation of in vitro data. C.T. assisted with ddPCR experiments and analyses. A.M.S., T.D., and D.M.M. processed and prepared samples for next-generation sequencing. Computational analyses and interpretation were performed by R.B., A.P., E.S., and J.M. L.Y. supervised statistical analyses. P.A. supervised the rapid research autopsy. M.A.K., R.B., H.-Z.C., J.W.R., and M.R.W. completed dissection, collection, and processing of autopsy tumor samples. A.G.F. analyzed tumor samples for quality and tumor cell content. H.-Z.C., J.W.R., M.R.W., and S.R. received informed consent for the body donation and autopsy study. K.D. performed tumor biopsies, and S.R. treated this patient. M.A.K., R.B., and S.R. wrote, reviewed, and revised the manuscript. All authors have read and approved the final manuscript.
Funding
S.R. has received support from an American Cancer Society grant MRSG-12-194-01-TBG, the Prostate Cancer Foundation, NCI UH2CA202971 (OSU-SpARKFuse), NCI UH2CA216432 (MSIDx), American Lung Association, and Pelotonia. M.A.K. was in part supported by a T32 Oncology Training Grant (5T32CA009338) and Award Number Grant TL1TR002735 from the National Center for Advancing Translational Sciences. R.B. was in part supported by a T32 T32GM068412 and a Pelotonia graduate student fellowship. H.-Z.C. was supported by a Pelotonia postdoctoral fellowship and an ASCO Conquer Cancer Foundation Young Investigator Award. M.R.W. was supported by the Helene Fuld Health Trust Nursing Scholarship. Incyte Corporation provided INCB054828 for in vitro characterization but no funding was provided.
Competing Interest Statement
S.R. participated in Advisory Boards for Incyte Corporation (2017), AbbVie, Inc. (2017), and QED Therapeutics (2018). S.R. received honoraria from IDT Integrated DNA Technologies (2017) and Illumina (2018). Incyte Corporation provided INCB054828 for in vitro characterization but no funding was provided.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- André F, Bachelot T, Campone M, Dalenc F, Perez-Garcia JM, Hurvitz SA, Turner N, Rugo H, Smith JW, Deudon S, et al. 2013. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res 19: 3693–3702. 10.1158/1078-0432.CCR-13-0190 [DOI] [PubMed] [Google Scholar]
- Angevin E, Lopez-Martin JA, Lin CC, Gschwend JE, Harzstark A, Castellano D, Soria JC, Sen P, Chang J, Shi M, et al. 2013. Phase I study of dovitinib (TKI258), an oral FGFR, VEGFR, and PDGFR inhibitor, in advanced or metastatic renal cell carcinoma. Clin Cancer Res 19: 1257–1268. 10.1158/1078-0432.CCR-12-2885 [DOI] [PubMed] [Google Scholar]
- Bedard PL, Hansen AR, Ratain MJ, Siu L. 2013. Tumour heterogeneity in the clinic. Nature 501: 355–364. 10.1038/nature12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, Reeser JW, Yu L, Roychowdhury S. 2017. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol 1: 1–15. 10.1200/PO.17.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RA, Terry ME. 1952. Rank analysis of incomplete block designs: I. The method of paired comparisons. Biometrika 39: 324–345. 10.1093/biomet/39.3-4.324 [DOI] [Google Scholar]
- Brady SW, Ma X, Bahrami A, Satas G, Wu G, Newman S, Rusch M, Putnam DK, Mulder HL, Yergeau DA, et al. 2019. The clonal evolution of metastatic osteosarcoma as shaped by cisplatin treatment. Mol Cancer Res 17: 895–906. 10.1158/1541-7786.MCR-18-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, Swanton C. 2014. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol 8: 1095–1111. 10.1016/j.molonc.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron F, Doucet A. 2012. Efficient Bayesian inference for generalized Bradley–Terry models. J Comput Graph Stat 21: 174–196. 10.1080/10618600.2012.638220 [DOI] [Google Scholar]
- Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R. 2009. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res 69: 6660–6667. 10.1158/0008-5472.CAN-09-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasent AK, Schalck A, Gao R, Sei E, Long A, Pangburn W, Casasent T, Meric-Bernstam F, Edgerton ME, Navin NE. 2018. Multiclonal invasion in breast tumors identified by topographic single cell sequencing. Cell 172: 205–217.e12. 10.1016/j.cell.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae YK, Ranganath K, Hammerman PS, Vaklavas C, Mohindra N, Kalyan A, Matsangou M, Costa R, Carneiro B, Villaflor VM, et al. 2017. Inhibition of the fibroblast growth factor receptor (FGFR) pathway: the current landscape and barriers to clinical application. Oncotarget 8: 16052–16074. 10.18632/oncotarget.14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, et al. 2017. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9: 34 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chell V, Balmanno K, Little AS, Wilson M, Andrews S, Blockley L, Hampson M, Gavine PR, Cook SJ. 2013. Tumour cell responses to new fibroblast growth factor receptor tyrosine kinase inhibitors and identification of a gatekeeper mutation in FGFR3 as a mechanism of acquired resistance. Oncogene 32: 3059–3070. 10.1038/onc.2012.319 [DOI] [PubMed] [Google Scholar]
- Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, Miller WT, Mohammadi M. 2007. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell 27: 717–730. 10.1016/j.molcel.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bell JM, Zavala NA, Ji HP, Zhang NR. 2015. Allele-specific copy number profiling by next-generation DNA sequencing. Nucleic Acids Res 43: e23 10.1093/nar/gku1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Bonneville R, Yu L, Wing MR, Reeser JW, Krook MA, Miya J, Samorodnitsky E, Smith A, Martin D, et al. 2019. Genomic characterization of metastatic ultra-hypermutated interdigitating dendritic cell sarcoma through rapid research autopsy. Oncotarget 10: 277–288. 10.18632/oncotarget.26352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park S, Yoon Y, Ahn J. 2017. Improved prediction of breast cancer outcome by identifying heterogeneous biomarkers. Bioinformatics 33: 3619–3626. 10.1093/bioinformatics/btx487 [DOI] [PubMed] [Google Scholar]
- Cortes JE, Kantarjian H, Shah NP, Bixby D, Mauro MJ, Flinn I, O'Hare T, Hu S, Narasimhan NI, Rivera VM, et al. 2012. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med 367: 2075–2088. 10.1056/NEJMoa1205127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta J, Damodaran S, Parks H, Ocrainiciuc C, Miya J, Yu L, Gardner EP, Samorodnitsky E, Wing MR, Bhatt D, et al. 2017. Akt activation mediates acquired resistance to fibroblast growth factor receptor inhibitor BGJ398. Mol Cancer Ther 16: 614–624. 10.1158/1535-7163.MCT-15-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetri GD. 2011. Differential properties of current tyrosine kinase inhibitors in gastrointestinal stromal tumors. Semin Oncol 38: S10–S19. 10.1053/j.seminoncol.2011.01.018 [DOI] [PubMed] [Google Scholar]
- Dexter DL, Leith JT. 1986. Tumor heterogeneity and drug resistance. J Clin Oncol 4: 244–257. 10.1200/JCO.1986.4.2.244 [DOI] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. 10.1038/nature10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville C, Carter H, Kim R, Niknafs N, Diekhans M, Stenson PD, Cooper DN, Ryan M, Karchin R. 2013. CRAVAT: cancer-related analysis of variants toolkit. Bioinformatics 29: 647–648. 10.1093/bioinformatics/btt017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faltas BM, Prandi D, Tagawa ST, Molina AM, Nanus DM, Sternberg C, Rosenberg J, Mosquera JM, Robinson B, Elemento O, et al. 2016a. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat Genet 48: 1490–1499. 10.1038/ng.3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J, et al. 2017. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep 19: 2878–2880. 10.1016/j.celrep.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R, Pusztai L, Swanton C. 2013. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer 108: 479–485. 10.1038/bjc.2012.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flensburg C, Sargeant T, Oshlack A, Majewski I. 2018. SuperFreq: integrated mutation detection and clonal tracking in cancer. bioRxiv. 10.1101/380097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. 2017. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45: D777–D783. 10.1093/nar/gkw1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, Michellys PY, Awad MM, Yanagitani N, Kim S, et al. 2014. The ALK inhibitor ceritinib overcomes crizotinib resistance in non–small cell lung cancer. Cancer Discov 4: 662–673. 10.1158/2159-8290.CD-13-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnieu M-C, Heiblig M, Blond E, Legros L, Guillermin Y, Morisset S, Barraco F, Etienne M, Le Borgne O, Ruby J, et al. 2013. Cardio-vascular events occurring on ponatinib in chronic phase chronic myeloid leukemia patients, preliminary analysis of a multicenter cohort. Blood 122: 4020. [Google Scholar]
- Gainor JF, Shaw AT. 2013. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. J Clin Oncol 31: 3987–3996. 10.1200/JCO.2012.45.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, et al. 2014. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 46: 225–233. 10.1038/ng.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal L, Saha SK, Liu LY, Siravegna G, Leshchiner I, Ahronian LG, Lennerz JK, Vu P, Deshpande V, Kambadakone A, et al. 2017a. Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov 7: 252–263. 10.1158/2159-8290.CD-16-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozgit JM, Wong MJ, Moran L, Wardwell S, Mohemmad QK, Narasimhan NI, Shakespeare WC, Wang F, Clackson T, Rivera VM. 2012. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther 11: 690–699. 10.1158/1535-7163.MCT-11-0450 [DOI] [PubMed] [Google Scholar]
- Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu G, Ye CJ. 2009. Genetic and epigenetic heterogeneity in cancer: a genome-centric perspective. J Cell Physiol 220: 538–547. 10.1002/jcp.21799 [DOI] [PubMed] [Google Scholar]
- Heppner GH, Miller BE. 1989. Therapeutic implications of tumor heterogeneity. Semin Oncol 16: 91–105. [PubMed] [Google Scholar]
- Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, Okimoto RA, Lin L, Neel DS, Sabnis A, et al. 2015. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med 21: 1038–1047. 10.1038/nm.3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javle M, Lowery M, Shroff RT, Weiss KH, Springfeld C, Borad MJ, Ramanathan RK, Goyal L, Sadeghi S, Macarulla T, et al. 2018. Phase II study of BGJ398 in patients with FGFR-altered advanced cholangiocarcinoma. J Clin Oncol 36: 276–282. 10.1200/JCO.2017.75.5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Yun CH, Park E, Ercan D, Manuia M, Juarez J, Xu C, Rhee K, Chen T, Zhang H, et al. 2016. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 534: 129–132. 10.1038/nature17960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Qiu Y, Minn AJ, Zhang NR. 2016. Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing. Proc Natl Acad Sci 113: E5528–E5537. 10.1073/pnas.1522203113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JG, Oh BY, Hong HK, Al-Khalidi H, Al-Alem F, Lee HO, Bae JS, Kim J, Cha HU, Alotaibi M, et al. 2017. Tumor heterogeneity predicts metastatic potential in colorectal cancer. Clin Cancer Res 23: 7209–7216. 10.1158/1078-0432.CCR-17-0306 [DOI] [PubMed] [Google Scholar]
- Junttila MR, de Sauvage FJ. 2013. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501: 346–354. 10.1038/nature12626 [DOI] [PubMed] [Google Scholar]
- Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, et al. 2017. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov 7: 1116–1135. 10.1158/2159-8290.CD-17-0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautto EA, Bonneville R, Miya J, Yu L, Krook MA, Reeser JW, Roychowdhury S. 2017. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget 8: 7452–7463. 10.18632/oncotarget.13918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye E, Firth D. 2017. BradleyTerryScalable: fits the Bradley-Terry model to potentially large and sparse networks of comparison data (Version 0.1.0). Retrieved from https://CRAN.R-project.org/package=BradleyTerryScalable
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22: 568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook MA, Chen HZ, Bonneville R, Allenby P, Roychowdhury S. 2019. Rapid research autopsy: piecing the puzzle of tumor heterogeneity. Trends Cancer 5: 1–5. 10.1016/j.trecan.2018.11.004 [DOI] [PubMed] [Google Scholar]
- Larson D, Abbott T. 2016. bam-readcount. Retrieved from https://github.com/genome/bam-readcount
- Letunic I, Bork P. 2016. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Rosen N, Solit DB. 2013. Tumor adaptation and resistance to RAF inhibitors. Nat Med 19: 1401–1409. 10.1038/nm.3392 [DOI] [PubMed] [Google Scholar]
- Malchers F, Ercanoglu M, Schütte D, Castiglione R, Tischler V, Michels S, Dahmen I, Brägelmann J, Menon R, Heuckmann JM, et al. 2017. Mechanisms of primary drug resistance in FGFR1-amplified lung cancer. Clin Cancer Res 23: 5527–5536. 10.1158/1078-0432.CCR-17-0478 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. 2013. Tumour heterogeneity and cancer cell plasticity. Nature 501: 328–337. 10.1038/nature12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LMF, Pepper JW, Reid BJ, Maley CC. 2006. Cancer as an evolutionary and ecological process. Nat Rev Cancer 6: 924 10.1038/nrc2013 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, et al. 2015. Genomic spectra of biliary tract cancer. Nat Genet 47: 1003–1010. 10.1038/ng.3375 [DOI] [PubMed] [Google Scholar]
- Ohio Supercomputer Center. 2012. Oakley Supercomputer. http://osc.edu/ark:19495/hpc0cvqn
- Paik PK, Shen R, Berger MF, Ferry D, Soria JC, Mathewson A, Rooney C, Smith NR, Cullberg M, Kilgour E, et al. 2017. A phase Ib open-label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers. Clin Cancer Res 23: 5366–5373. 10.1158/1078-0432.CCR-17-0645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- PDQ Adult Treatment Editorial Board. 2002. Bile Duct Cancer (Cholangiocarcinoma) Treatment (PDQ®): Health Professional Version. In PDQ Cancer Information Summaries. National Cancer Institute (US), Bethesda, MD. [PubMed] [Google Scholar]
- Picard Tools—By Broad Institute. Retrieved from http://broadinstitute.github.io/picard/
- Pradhan D, El-Kebir M. 2018. On the non-uniqueness of solutions to the perfect phylogeny mixture problem. Paper presented at the Comparative Genomics, Cham. [Google Scholar]
- Razumilava N, Gores GJ. 2014. Cholangiocarcinoma. Lancet 383: 2168–2179. 10.1016/S0140-6736(13)61903-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeser JW, Martin D, Miya J, Kautto EA, Lyon E, Zhu E, Wing MR, Smith A, Reeder M, Samorodnitsky E, et al. 2017. Validation of a targeted RNA sequencing assay for kinase fusion detection in solid tumors. J Mol Diagn 19: 682–696. 10.1016/j.jmoldx.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. 2016. deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17: 31 10.1186/s13059-016-0893-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S, Vollebergh MA, de Hoon B, de Ronde J, Schouten PC, Kersbergen A, Zander SA, Pajic M, Jaspers JE, Jonkers M, et al. 2012. Impact of intertumoral heterogeneity on predicting chemotherapy response of BRCA1-deficient mammary tumors. Cancer Res 72: 2350 10.1158/0008-5472.CAN-11-4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S, Talpaz M. 2011. Managing resistance in chronic myeloid leukemia. Blood Rev 25: 279–290. 10.1016/j.blre.2011.09.001 [DOI] [PubMed] [Google Scholar]
- Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA, Quist MJ, et al. 2011. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med 3: 111ra121 10.1126/scitranslmed.3003161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzenente A, Fassan M, Conci S, Simbolo M, Lawlor RT, Pedrazzani C, Capelli P, D'Onofrio M, Iacono C, Scarpa A, et al. 2016. Cholangiocarcinoma heterogeneity revealed by multigene mutational profiling: clinical and prognostic relevance in surgically resected patients. Ann Surg Oncol 23: 1699–1707. 10.1245/s10434-015-5046-6 [DOI] [PubMed] [Google Scholar]
- Saito T, Kondo C, Shitara K, Ito Y, Saito N, Ikehara Y, Yatabe Y, Yamamichi K, Tanaka H, Nakanishi H. 2015. Comparison of intratumoral heterogeneity of HER2 expression between primary tumor and multiple organ metastases in gastric cancer: clinicopathological study of three autopsy cases and one resected case. Pathol Int 65: 309–317. 10.1111/pin.12290 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Samorodnitsky E, Jewell BM, Hagopian R, Miya J, Wing MR, Lyon E, Damodaran S, Bhatt D, Reeser JW, Datta J, et al. 2015a. Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum Mutat 36: 903–914. 10.1002/humu.22825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samorodnitsky E, Datta J, Jewell BM, Hagopian R, Miya J, Wing MR, Damodaran S, Lippus JM, Reeser JW, Bhatt D, et al. 2015b. Comparison of custom capture for targeted next-generation DNA sequencing. J Mol Diagn 17: 64–75. 10.1016/j.jmoldx.2014.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas P, Teo ZL, Lefevre C, Flensburg C, Caramia F, Alsop K, Mansour M, Francis PA, Thorne HA, Silva MJ, et al. 2016. The subclonal architecture of metastatic breast cancer: results from a prospective community-based rapid autopsy program “CASCADE”. PLoS Med 13: e1002204 10.1371/journal.pmed.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G. 1978. Estimating the dimension of a model. Ann Statist 6: 461–464. 10.1214/aos/1176344136 [DOI] [Google Scholar]
- Simonsen M, Mailund T, Pedersen CNS. 2008. Rapid neighbour-joining. Paper presented at the Algorithms in Bioinformatics, Berlin, Heidelberg. [Google Scholar]
- Tabernero J, Bahleda R, Dienstmann R, Infante JR, Mita A, Italiano A, Calvo E, Moreno V, Adamo B, Gazzah A, et al. 2015. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol 33: 3401–3408. 10.1200/JCO.2014.60.7341 [DOI] [PubMed] [Google Scholar]
- Tan L, Wang J, Tanizaki J, Huang Z, Aref AR, Rusan M, Zhu SJ, Zhang Y, Ercan D, Liao RG, et al. 2014. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci 111: E4869–E4877. 10.1073/pnas.1403438111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, Madhusudan S, Iveson T, Hughes S, Pereira SP, et al. 2010. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 362: 1273–1281. 10.1056/NEJMoa0908721 [DOI] [PubMed] [Google Scholar]
- Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV, et al. 2014. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 4: 94–109. 10.1158/2159-8290.CD-13-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, et al. 2013. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 3: 636–647. 10.1158/2159-8290.CD-13-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, Zhao X, Li Y, Li Q, Wang H, et al. 2014. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun 5: 5696 10.1038/ncomms6696 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used for the analyses presented in the manuscript have been submitted to dbGaP (https://ncbi.nlm.nih.gov/gap) under the project accession number phs001830.v1.p1. The FGFR2-CLIP1 fusion gene variant and the secondary FGFR2 mutation identified in the patient have also been deposited to ClinVar (https://ncbi.nlm.nih.gov/clinvar/) under the accession numbers SCV000927106 and SCV000914229.1.