Abstract
Heterozygous deleterious variants in PHIP have been associated with behavioral problems, intellectual disability/developmental delay, obesity/overweight, and dysmorphic features (BIDOD syndrome). We report an additional 10 individuals with pleckstrin homology domain-interacting protein (PHIP)-predicted deleterious variants (four frameshift, three missense, two nonsense, and one splice site; six of which are confirmed de novo). The mutation spectrum is diverse, and there is no clustering of mutations across the protein. The clinical phenotype of these individuals is consistent with previous reports and includes behavioral problems, intellectual disability, developmental delay, hypotonia, and dysmorphic features. The additional individuals we report have a lower frequency of obesity than previous reports and a higher frequency of gastrointestinal problems, social deficits, and behavioral challenges. Characterizing additional individuals with diverse mutations longitudinally will provide better natural history data to assist with medical management and educational and behavioral support.
Keywords: 2-3 toe cutaneous syndactyly, abdominal obesity, aggressive behavior, almond-shaped palpebral fissure, amblyopia, anteverted nares, attention deficit hyperactivity disorder, autism, blurred vision, chronic constipation, chronic fatigue, clinodactyly of the 5th finger, gastroesophageal reflux, generalized neonatal hypotonia, high forehead, intellectual disability, mild, mild global developmental delay, synophrys, thickened helices, thin upper lip vermilion
INTRODUCTION
PHIP (MIM# 612870) is located on 6q14.1 and encodes Pleckstrin homology domain-interacting protein (PHIP), a substrate receptor in a ubiquitin ligase pathway involved in DNA repair, genomic integrity, and cell proliferation and survival (Farhang-Fallah et al. 2000; Kato et al. 2000; Petroski and Deshaies 2005; Lee and Zhou 2007; Podcheko et al. 2007; Webster et al. 2016; Jansen et al. 2018). A de novo likely pathogenic variant in PHIP was first reported in a child as part of an exome sequencing study of 100 individuals with severe intellectual disability (IQ < 50) (de Ligt et al. 2012). Since then, 24 additional individuals with loss-of-function (eight nonsense, seven frameshift, three splice site, one translocation, and one gene deletion) or missense variants (n = 4) have been reported. The majority of these variants are de novo (n = 14), eight are of unknown inheritance, and one variant is inherited from an affected father to two siblings (Webster et al. 2016; Jansen et al. 2018). Individuals with PHIP variants have a common phenotype of behavioral problems, intellectual disability/developmental delay, obesity/overweight, and dysmorphic features (Webster et al. 2016; Jansen et al. 2018). Eight additional individuals were reported by Wang et al. (2016) in a Chinese autism study and included one de novo likely pathogenic frameshift variant and seven inherited missense variants, but without additional phenotypic or familial data to assess whether missense variants segregated with a neurobehavioral phenotype in the family to support pathogenicity. Here, we expand our previous report (Webster et al. 2016) and describe 10 additional individuals with heterozygous predicted deleterious PHIP variants with symptoms consistent with previous reports of behavioral problems and/or mental health diagnoses, intellectual disability/developmental delay, obesity/overweight, and dysmorphic features. Additionally, we report a high frequency of hypotonia and constipation.
RESULTS
Molecular Findings
Six of the 10 variants are predicted to result in loss of function via nonsense mediated decay (four frameshift and two nonsense), one is predicted to cause abnormal splicing per Human Splicing Finder (Desmet et al. 2009), and three are missense variants, each with high CADD v1.3 (Combined Annotation Dependent Depletion) scores (Table 1; Fig. 1; Rentzsch et al. 2019). Six of the 10 variants are de novo. Two were of uncertain inheritance because of lack of one parental sample, although the variants are not observed in the one available parent or unaffected sibling. One individual had results for neither parent, and one frameshift variant was inherited from a father whose clinical information was unavailable. Kinship analysis was performed for all tested samples, and biological parents were confirmed. None of the variants is present in population databases such as gnomAD and TOPMed. Previous genetic tests including fragile-X repeat analysis and chromosomal microarray were negative (Supplemental Note).
Table 1.
Genomic findings of heterozygous PHIP variants
| Subject ID | Chromosome | HGVS DNA reference (NM_017934.6) | HGVS protein reference (NP_060404.4) | Variant type (prediction score)a | ACMG/AMP-2015 classification (criteria Met) | dbSNP/dbVar ID | Parent of origin |
|---|---|---|---|---|---|---|---|
| 1 | Chr 6:79713538 (hg19) Chr6:79003821 (hg38) | c.1562A > G | p.(Lys521Arg) | Missense (CADD v1.3: 24) | Likely pathogenic (PS2, PM2, PP3) | Not available | De novo |
| 2 | Chr 6:79752620 (hg19) Chr6:79042903 (hg38) | c.540_541insA | p.(Gly181ArgfsTer12) | Frameshift | Likely pathogenic (PVS1, PM2) | Not available | Paternal |
| 3 | Chr 6:79735299 (hg19) Chr 6:790255582(hg38) | c.860C > A | p.(Ser287Tyr) | Missense (CADD v1.3: 24.8) | Likely pathogenic (PM2, PS2) | rs1085307845 | De novo |
| 4 | Chr 6:79671468 (hg19) Chr 6:78961751 (hg38) | c.3595delG | p.(Val1199Ter) | Nonsense | Pathogenic (PVS1, PS2, PM2) | Not available | De novo |
| 5 | Chr 6:79692625 (hg19) Chr 6:78982908 (hg38) | c.2744_2747del | p.(Lys915SerfsTer15) | Frameshift | Pathogenic (PVS1, PS2, PM2) | Not available | De novo |
| 6 | Chr 6:79679596 (hg19) Chr 6:78969879 (hg38) | c.3161delT | p.(Leu1054Ter) | Nonsense | Pathogenic (PVS1, PS2, PM2) | Not available | De novo |
| 7 | Chr 6:79752561 (hg19) Chr 6:79042844 (hg38) | c.598_599delACinsT | p.(Thr200LeufsTer8) | Frameshift | Likely pathogenic (PVS1, PM2) | rs1554210073 | Unknown (not paternal) |
| 8 | Chr 6:79668186-79668189 (hg19) Chr 6:78958469-78958472 (hg38) | c.3782 + 3_3782 + 6delAAGT | IVS32 + 3_IVS32 + 6delAAGT | Splice site (HSF 3.1: most probably affecting splicing) | Variant of unknown significance (PM2) | rs1131691771 | Unknown (not maternal) |
| 9 | Chr 6:79655778 (hg19) Chr 6:78946061 (hg38) | c.4570delT | p.(Ser1524LeufsTer22) | Frameshift | Likely pathogenic (PVS1, PM2) | Not available | Unknown |
| 10 | Chr 6:79735796 (hg19) Chr6:79026079 (hg38) | c.686C > T | p.(Ser229Leu) | Missense (CADD v1.3: 32) | Likely pathogenic (PM2, PS2) | Not available | De novo |
(HSF) Human Splicing Finder.
aCADD scores were only available for missense variants.
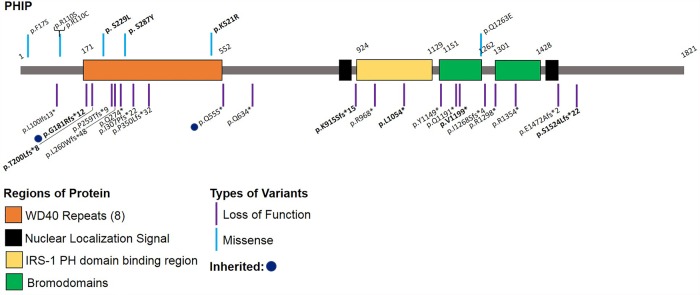
Figure 1.
Schematic 2D representation of the PHIP and its functional domains. Locations of likely pathogenic/pathogenic variants from previously reported individuals and the new variants reported herein indicated in bold. Schematic does not include splice site (three) or translocation (one) variants. (Adapted from Webster et al. 2016.)
Clinical Features
The 10 individuals with PHIP variants we describe include five females and five males, ranging from 16 mo to 15 yr of age, with an average age of 9 yr and 2 mo (Table 2). One individual was born prematurely, and five reported a variety of minor prenatal issues. Feeding difficulties (7/10), hypotonia (4/10), and neonatal jaundice (3/10) were the most commonly reported neonatal issues.
Table 2.
Clinical features of the new (n = 10) and previously reported cases (n = 24)
| New individuals included in this report | Average for new individuals in this report | Summary of previously reported cases | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individual ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | n = 10 | n = 25 |
| Age | 57/12 yr | 14/12 yr | 15 yr | 710/12 yr | 9 yr | 1210/12 yr | 94/12 yr | 12 yr | 153/12 yr | 35/12 yr | 92/12 yr (16 mo-15 yr) | 18 yr (5–52 yr) |
| Sex | F | M | F | F | F | M | F | M | M | M | F:M = 5:5 | F:M = 14:11 |
| Prenatal issues | None | IUGR Placental insufficiency Maternal hypertension |
Nuchal and arachnoid cysts (resolved in utero) LGA |
Maternal diabetes LGA |
None | None | None | LGA macrocephaly |
None | Unilateral ureterocele | 50% | 37.5% |
| Neonatal issues | Feeding difficulty | Feeding difficulty GERD PPHN of the newborn |
Feeding difficulty Jaundice |
Hypotonia Lethargy Jaundice |
None | Hypotonia | Feeding difficulty Jaundice |
Feeding difficulty Respiratory distress (required resuscitation) |
Feeding difficulty Hypotonia |
Feeding difficulty Hypotonia Congenital muscular torticollis |
Feeding difficulty (70%) Hypotonia (40%) |
Feeding difficulty (33%) |
| Developmental delay | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 100% | 92% |
| Age at sitting | 9 mo | Not achieved yet | 13 mo | 12 mo | 8 mo | 18 mo | 6 mo | 6.5 mo | 9 mo | 20 mo | 11.3 mo (n = 9) | Not available for all patients |
| Age at walking | 12 mo | Not achieved yet | 19 mo | 24 mo | 18 mo | 24 mo | 29 mo | 28 mo | 24 mo | 36 mo | 23.8 mo (n = 9) | Not available for all patients |
| Age at talking | 36 mo | 9 mo | Normal range | 36 mo | 9 mo | Normal range | 48 mo | Delayed | 42 mo | Does not speak in sentences yet | 30 mo (n = 6) | Not available for all patients |
| Intellectual disability | Yes, mild & hyperlexia | Yes | Yes, mild | Yes, mild | Yes, mild | Yes, mild | Yes, mild | Yes, mild | Yes, mild | Yes | 100% | 92% |
| Full scale IQ | n/a | n/a | 73 | 68 | n/a | 60 | 50-60 | n/a | n/a | n/a | n/a | |
| Vineland Overall Age (%) | 16% | 3% | 19% | 1% | 8% | 5% | 2% | 5% | 1% | n/a | 6.7% | n/a |
| Communication (%) | 42% | 9% | 23% | 3% | 8% | 14% | 2% | 4% | 1% | 11.8% | ||
| Daily living skills (%) | 16% | 2% | 50% | 1% | 19% | 4% | 4% | 16% | 1% | 12.6% | ||
| Socialization (%) | 2% | 12% | 5% | 1% | 5% | 5% | 2% | 5% | 1% | 4.2% | ||
| Motor(%) | 50% | 1% | - | 2% | - | - | - | - | - | 18% | ||
| Behavioral problems | ASD Attention problems SPD |
None | ASD features ADHD GAD Tantrums Impulse problems SPD Tics Trichotillomania |
ASD features Social anxiety Tantrums Impulse problems |
ADHD GAD Tantrums Impulse problems SPD features |
ASD ADD Tantrums Anxious in new situations |
ASD ADHD GAD PTSD Tantrums Impulse problems SPD features Tics |
ADD GAD/depression Tantrums Impulse problems SPD features |
ASD ADHD OCD Phobias Tantrums SPD features Enuresis nocturna |
None | 88% | 84% |
| Sleep problems | Yes | No | Yes | No | No | No | No | No | Yes | No | 30% | 24% |
| Dysmorphic features | High forehead Anteverted nares Long and smooth philtrum |
High forehead Almond shaped eyes Long and smooth philtrum Thin lips |
High forehead Synophrys Hypertelorism Almond shaped eyes Thick helices and earlobes Anteverted nares Thin lips Clinodactyly of 5th finger CAL |
High forehead Hypertelorism Almond shaped eyes Thick helices and earlobes Short nose and broad nasal tip Thin lips Clinodactyly Syndactyly of 2-3 toes CAL Hemangioma |
High forehead Hypertelorism Thick helices and earlobes Thin lips Clinodactyly of 5th finger Syndactyly of 2-3 toes |
High forehead Synophrys Almond shaped eyes Large ears Anteverted nares Short nose and broad nasal tip Long and smooth philtrum |
High forehead Large ears Thin lips Clinodactyly of 5th finger |
Large ears Clinodactyly of 5th finger CAL |
Almons shaped eyes Large ears Thin lips CAL |
Low-set posteriorly rotated ears with thick helices Arched eyebrows Synophrys Full and long eyelashes Epicanthus Anteverted nares Long philtrum Tongue-tie Transverse palmar crease Sacro-coccygeal dimple without medullar abnormality on MRI |
Facial features (100%) Other (60%) |
Facial features (96%) Other (79.2%) |
| Neurologic problems | None | Hypotonia | Hypotonia Abnormal gait |
Hypotonia Focal impaired awareness Difficulty walking Dysautonomia Hand tremors |
Truncal hypotonia Extremity hypertonia |
None | Truncal hypotonia Hypertonia of hands |
Hypotonia A single seizure event at 18 mo old Hand tremors |
Hypotonia Suspected seizure activity (generalized tonic-clonic) Chorea of limbs Migraine |
Hypotonia Febrile seizures Reduced white matter volume Cerebellar hypoplasia |
Hypotonia (80%) Seizures (20%) |
Hypotonia (32%) |
| Musculoskeletal problems | None | None | Easily fatigued Hypermobile joints | Easily fatigued Hypermobile joints |
Easily fatigued | Easily fatigued Pes planus |
Easily fatigued Pes planus |
Easily fatigued Pes planus |
Easily fatigued Hypermobile joints |
Unilateral genu recurvatum | Fatigue (70%) | Fatigue (40%) |
| Gastrointestinal problems | None | GERD (severe) | Constipation | Constipation (severe) GERD |
Constipation GERD |
Constipation | Constipation (resolved) | Constipation | Diarrhea Constipation GERD |
None | Constipation (70%) GERD (40%) |
Constipation(8%) GERD (4%) |
| Endocrine problems | None | Subclinical hypothyroidism | Borderline overweight (BMI = 23.9, z-score = 1) Delayed bone age Delayed menarche |
Obesity (BMI = 30.6, z-score = 2.71) | None | Unilateral cryptorchidism | Recent rapid weight gain | None | Overweight (BMI = 23.9, z-score = 1.09) | None | Obesity/
overweight (30%) Other (30%) |
Obesity (76%) Other (16.7%) |
| Eye problems | None | Pale optic nerve | Convergence problems | Amblyopia Myopia |
Convergence problems | Amblyopia Myopia |
Myopia Astigmatism Alternating exotropia/amblyopia |
None | Severe visual impairment Unilateral legally blind Amblyopia |
Myopia Strabismus |
80% | 72% |
| Infectious problems | None | None | None | Otitis media Upper respiratory |
None | None | UTI | Otitis media Upper respiratory |
Otitis media Upper respiratory TI |
UTI | 50% | 24% |
(F) Female, (M) male, (n/a) not available, (IUGR) intrauterine growth restriction, (PPHN) persistent pulmonary hypertension in the neonate, (GERD) gastroesophageal reflux disease, (LD) learning disability, (ASD) autism spectrum disorder, (SPD) sensory processing disorder, (ADHD/ADD) attention deficit (hyperactivity) disorder, (GAD) generalized anxiety disorder, (PTSD) posttraumatic stress disorder, (BMI) body mass index, (OM) otitis media, (UTI) urinary tract infection, (CAL) café au lait macule.
All individuals had delayed developmental milestones, and the majority were diagnosed with intellectual disability and/or learning disability (Table 2; Supplemental Note; Supplemental Table S1). The average ages for major developmental milestones were 11.3 mo for sitting (n = 9), 23.8 mo for walking (n = 9), and 30 mo for talking (n = 6). All individuals have intellectual disability (ID) or developmental delay (10/10), which is generally mild (IQ: 60–70) and in several cases was associated with higher verbal than performance IQ.
On the Vineland-II assessment of adaptive behaviors (n = 9), age-adjusted percentiles varied in communication (1%–42%; average:11.8%, median: 8%) and daily living skills (1%–50%; average: 12.6%, median 4%). Three children were in an appropriate age range for motor domain assessment, although there was a significance range of scores (1%–50%; average: 17.6%, median: 2%). However, all individuals had low socialization scores (1%–12%; average: 4.2%, median: 5%) and low overall composite scores (1%–19%; average: 6.7%, median 5%). Most individuals had behavioral diagnoses and/or significant behavioral challenges (8/9). The most common behavioral issues were attention-deficit/hyperactivity disorder (ADHD), attention deficit disorder (ADD) or attention span problems (7/9), anxiety and/or depression (7/9), aggression or tantrums (7/9), autism or features of autism spectrum disorder (ASD) (6/9), and impulse control problems (5/9). Sensory processing disorder (SPD) or similar symptoms were also common (7/9) and overlapped with autism or ASD features in 5/7 cases. Additionally, trichotillomania, emotional lability, fear of new situations, posttraumatic stress disorder (PTSD), and obsessive–compulsive disorder (OCD) were each noted in a single individual, and two individuals had a tic disorder. Sleep issues were noted in three individuals. Movement disorders included hand tremors in two individuals and chorea in another. Movement issues have worsened with age for two individuals.
Seizure or suspected seizure activity were observed in four individuals. One individual has complex partial seizures (focal impaired awareness), diagnosed at age 6 yr with a frequency of two to three episodes per day prior to medication. She has abnormal EEG (background slowing) and suspected absence seizures. One individual has suspected generalized tonic clonic seizure activity with approximately three seizures per year since age 11; however, 30-min EEGs were normal, and he has not been treated with medication. One individual had five episodes of febrile seizures before age 4. Another individual had a seizure at 18 mo old, but had normal EEGs and no further seizure activity.
All individuals have some dysmorphic features (10/10). The most common facial features were thick helices and earlobes/larger ears (8/10), high forehead (7/10), thin lips (6/10), almond shaped eyes (5/10), large ears (4/10), anteverted nares (4/10), and long and smooth philtrum (4/10). Hypertelorism (3/10), synophrys (3/10), short nose and broad nasal tip (2/10), and epicanthal folds (1/10) were less commonly noted. Other features included clinodactyly of the fifth finger (5/10), café-au-lait spots (4/10), and two-three toe syndactyly (2/10) (Table 2; Supplemental Table S1).
Most individuals have less severe medical issues compared to their neurodevelopmental and behavioral manifestations. Despite the previously reported association with obesity, only two individuals in this series were obese or overweight. Notably, one individual was borderline normal/overweight and another was within normal range but had recent rapid weight gain.
The majority of individuals (9/10) were hypotonic and/or fatigued easily. Some individuals were able to participate in age-appropriate motor activities (i.e., outdoor games and sports) with limited restriction, whereas others had more severe motor impairment.
Gastrointestinal problems were also commonly reported, primarily chronic constipation (7/10) and gastroesophageal reflux disease (GERD) (4/10). In some cases, these gastrointestinal issues were severe with four individuals requiring daily laxatives and one requiring a cecostomy tube. One individual had such severe reflux and feeding difficulties from birth that he still primarily feeds through a NG tube at 16 mo old.
Additionally, ophthalmologic problems were common (8/10); most commonly amblyopia (4/10) and also myopia, astigmatism, and convergence disorder. Strabismus was reported in a single individual. Several individuals (5/10) had recurrent infections including upper respiratory infections, otitis media, and urinary tract infections, although none were diagnosed with an immunodeficiency, and there were no atypical infections. Hypermobile joints and pes planus were each separately observed in 3/10 individuals. One of them is severely affected with chronic joint pain, and clinical diagnosis of hypermobile Ehlers–Danlos syndrome was made although no variants have been identified in genes known to be associated with Ehlers–Danlos syndrome. Endocrine problems other than obesity/overweight were infrequent (3/10) and included subclinical hypothyroidism, delayed menarche, and delayed bone age (2 yr). One individual had unilateral crypthorchidism.
DISCUSSION
We report 10 new individuals with PHIP variants with a clinical phenotype consistent with previously reported PHIP-related phenotype. The neurodevelopmental and behavioral phenotype of our newly reported individuals is similar to that in previous reports and represents the major challenge for these individuals (Webster et al. 2016; Jansen et al. 2018). Most individuals have borderline intellectual disability and fluent speech. We identified specific deficits in social skills as well as a high frequency of ADHD or attention problems, anxiety, depression, sensory processing disorder, aggressive tantrums, autism or ASD features, and impulse control problems. Many individuals had multiple behavioral issues.
Individuals in our cohort also share the common dysmorphic features with previously reported individuals. Other common medical problems associated with PHIP variants are ophthalmologic problems, hypotonia, easy fatiguability, gastrointestinal problems including constipation and reflux, hypermobile joints, recurrent minor infections, sleeping problems, and neonatal feeding difficulty. In many cases, these symptoms significantly impact daily life and require medical interventions (Fig. 2; Supplemental Table S1).
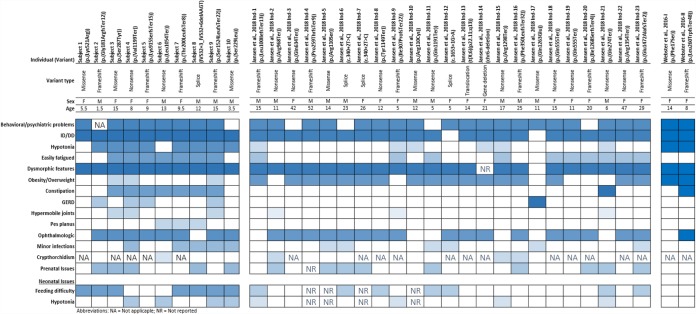
Figure 2.
Heatmap of most commonly seen clinical findings in at least one study among all reported individuals by this study, Webster et al. (2016), and Jansen et al. (2018). To emphasize the most commonly observed clinical findings within the cohorts and differences/similarities on the frequencies of some clinical findings between two cohorts, we created the heatmap of the findings by comparing the observed ratio of findings within each group for this study and Jansen et al. (2018). For Webster et al. (2016) individuals, a binary comparison is used because of the small number of individuals. Further clinical details are provided in Supplemental Table S1.
The two most notable differences between our cohort and previously reported individuals were the presence of significant gastrointestinal issues and relatively lower frequency of obesity in our series (Table 2; Fig. 2). In our cases, constipation (7/10), gastrointestinal reflux (4/10), and infant feeding problems (7/10) were common and, in some cases, severe. These symptoms were infrequently reported in previously reported cases: constipation (2/25), gastrointestinal reflux (1/25), and infant feeding problems (6/25). This difference could reflect absence of gastrointestinal data for other cohorts or a range of symptoms and symptom severity. Additionally, in previously reported cases, 75% were obese or overweight, whereas only three individuals (30%) in our cohort were obese or overweight. However, the average age of all previously reported obese/overweight individuals was 20.8 ± 13.5 (median = 15 yr) years, whereas individuals in this report had a mean age of 9.2 ± 4.7 (median = 9.25 yr) years. Weight gain seems to increase with puberty, and obesity tends to be abdominal. This could provide an opportunity to intervene before weight gain becomes significant through diet, exercise, and possibly medication.
PHIP was previously reported to encode multiple alternative isoforms (Podcheko et al. 2007; Webster et al. 2016; Jansen et al. 2018) probably because of different nomenclature used by different groups: NDRP by Kato et al. (2000), PHIP short isoform by Farhang-Fallah et al. (2000), and long isoform (PHIP1) by Podcheko et al. (2007). The long isoform (1821-amino-acid-long) isolated by Podcheko et al. 2007 seems to be the only protein product of PHIP. There are no recurrent variants or mutational hotspots in the protein. So far there is no phenotypic difference between missense and loss-of-function variants. Furthermore, although dysmorphic facial features are commonly reported across all individuals, there were no facial features specific to this condition that were recognizable.
PHIP is also known as DCAF14 (DDB1- and CUL4-associated factor 14) and is thought to assist in the binding of the ubiquitin complex to the target protein as a substrate receptor for the ubiquitin ligase complex that regulates cellular processes such as cell proliferation and survival, genomic integrity, and DNA repair (Petroski and Deshaies 2005; Lee and Zhou 2007; Webster et al. 2016). The PHIP protein has several known functional domains including a WD40 repeat domain, a pleckstrin homology domain binding region, nuclear localization signals, and bromodomains (Fig. 1; Farhang-Fallah et al. 2000; Kato et al. 2000; Podcheko et al. 2007; Jang et al. 2018). The PHIP WD40 region is likely involved in substrate binding and a recent study demonstrates that loss of the WD40 region prevents CUL4 recruitment to chromatin in the early stages of the cell cycle (Jang et al. 2018). They also showed that depletion of PHIP caused accumulation of some of the CUL4/DDB1 ubiquitin ligase products, including CDT1, p21, and SET8, on chromatin and results in abnormal cell cycle progression, genomic instability, and rereplication of genetic information (Jang et al. 2018). In addition, loss-of-function variants in CUL4B, a gene encoding a protein in the same functional pathway, are associated with an overlapping phenotype of intellectual disability, behavioral problems, and obesity (Tarpey et al. 2007). The three missense variants in our cohort all fall in the WD40 domain that is expected to be intolerant to variation (Gussow et al. 2016). Moreover, gnomAD constraint metrics for missense (Z-score = 5.14) and loss-of-function (pLI = 1, o/e = 0.6) variants are in favor of intolerance to variation (Karczewski et al. 2019 ). In mice, Phip is ubiquitously expressed and is expressed at high levels in skeletal muscle, brain, and pancreatic islet cells, consistent with the most common symptoms of PHIP-related phenotype such as hypotonia, neurobehavioral issues, and obesity (Podcheko et al. 2007).
The OMIM entry for PHIP-related phenotype (MIM# 617991) lists the acronym DIDOD (developmental delay, intellectual disability, obesity, and dysmorphic features). As behavioral problems are also among the most prominent and challenging problems in individuals with pathogenic PHIP variants, we propose to add “behavioral problems”; hence, the acronym BIDOD (behavioral problems, intellectual disability/developmental delay, obesity/overweight, and dysmorphic features) syndrome for PHIP-related phenotype.
In summary, the additional individuals we report are consistent with the phenotypic spectrum of previously reported individuals with PHIP-related BIDOD syndrome with the exception of higher frequency of gastrointestinal problems and an age-dependent relatively lower frequency of obesity/overweight during early childhood. Because the PHIP-related BIDOD syndrome is a relatively newly identified condition, more longitudinal phenotypic data are needed to further clarify the natural history and identify the most effective methods for symptom management.
METHODS
This study was approved by the Columbia University Institutional Review Board (Protocol No: IRB-AAAJ8651), and signed informed consents were obtained from participants. Participants who were clinically diagnosed contacted us to participate in our natural history study of individuals with predicted pathogenic/likely pathogenic PHIP variants. All participants had clinical exome sequencing or clinical panel gene testing because of behavioral problems and/or intellectual disability/developmental delay. None of the 10 individuals included has been previously reported in the literature. Clinical genetic test reports were reviewed, and all PHIP variants were classified according to the ACMG 2015 variant classification guidelines (Table 1; Richards et al. 2015). Medical records were reviewed, primary care takers were interviewed by telephone about the proband's medical history when possible, and adaptive development was assessed in nine individuals using the Vineland-II Adaptive Behavior Scales. Facial features were summarized after reviewing photographs, but only two families gave consent for publication of photos (Table 2; Fig. 3; Supplemental Note).
Figure 3.
Facial photos of the individuals 10 (left) and 4 (right) at 3 and 7.5 yr old, respectively.
ADDITIONAL INFORMATION
Data Deposition and Access
All reported variants, along with associated human phenotype ontology terms, are submitted to ClinVar (https://ncbi.nlm.nih.gov/clinvar/) under accession numbers SCV000899256.1 (p.Ser28Tyr), SCV000899257.1 (p.Thr200LeufsTer8), SCV000899258.1 (IVS32 + 3_IVS32 + 6delAAGT), SCV000899259.1 (p.Val1199Ter), SCV000899260.1 (p.Gly181ArgfsTer12), SCV000899261.1 (p.Ser1524LeufsTer22), SCV000899262.1 (p.Lys915SerfsTer15), SCV000899263.1 (p.Lys521Arg), SCV000899264.1 (p.Leu1054Ter), and SCV000899265.1 (p.Ser229Leu).
Ethics Statement
This study was approved by the Institutional Review Board of Columbia University under the protocol number IRB-AAAJ8651. Written consent was obtained from all families.
Acknowledgments
We thank the patients and their families for their participation and contribution of data. We thank the Center for Rare Childhood Disorders (C4RCD) Research Group for their support.
Author Contributions
K.E.C. collected and analyzed the data and drafted and critically reviewed the manuscript. V.O., A.W., and J.J. analyzed the data and critically reviewed the manuscript. E.H.G., K.R., J.M.H., A.V., and M.-N.B.D. provided the clinical data and critically reviewed the manuscript. W.K.C. conceived of the study, provided clinical data, analyzed the data, and drafted and critically reviewed the manuscript.
Funding
This work was supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) T35 training grant (5T35DK093430) to K.E.C. This work was supported from grants to W.K.C. from the JPB Foundation and SFARI.
Competing Interest Statement
Jane Juusola is an employee of GeneDx. Other authors declare no conflict of interest related to the presented study.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, et al. 2012. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 367: 1921–1929. 10.1056/NEJMoa1206524 [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Beroud C. 2009. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37: e67 10.1093/nar/gkp215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhang-Fallah J, Yin X, Trentin G, Cheng AM, Rozakis-Adcock M. 2000. Cloning and characterization of PHIP, a novel insulin receptor substrate-1 pleckstrin homology domain interacting protein. J Biol Chem 275: 40492–40497. 10.1074/jbc.C000611200 [DOI] [PubMed] [Google Scholar]
- Gussow AB, Petrovski S, Wang Q, Allen AS, Goldstein DB. 2016. The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol 17: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SM, Zhang Y, Utani K, Fu H, Redon CE, Marks AB, Smith OK, Redmond CJ, Baris AM, Tulchinsky DA, et al. 2018. The replication initiation determinant protein (RepID) modulates replication by recruiting CUL4 to chromatin. Nat Commun 9: 2782 10.1038/s41467-018-05177-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S, Hoischen A, Coe BP, Carvill GL, Van Esch H, Bosch DGM, Andersen UA, Baker C, Bauters M, Bernier RA, et al. 2018. A genotype-first approach identifies an intellectual disability-overweight syndrome caused by PHIP haploinsufficiency. Eur J Hum Genet 26: 54–63. 10.1038/s41431-017-0039-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP. 2019. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 10.1101/531210 [DOI] [Google Scholar]
- Kato H, Chen S, Kiyama H, Ikeda K, Kimura N, Nakashima K, Taga T. 2000. Identification of a novel WD repeat-containing gene predominantly expressed in developing and regenerating neurons. J Biochem 128: 923–932. 10.1093/oxfordjournals.jbchem.a022843 [DOI] [PubMed] [Google Scholar]
- Lee J, Zhou P. 2007. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell 26: 775–780. 10.1016/j.molcel.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20. 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Podcheko A, Northcott P, Bikopoulos G, Lee A, Bommareddi SR, Kushner JA, Farhang-Fallah J, Rozakis-Adcock M. 2007. Identification of a WD40 repeat-containing isoform of PHIP as a novel regulator of β-cell growth and survival. Mol Cell Biol 27: 6484–6496. 10.1128/MCB.02409-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. 2019. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47: D886–D894. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Raymond FL, O'Meara S, Edkins S, Teague J, Butler A, Dicks E, Stevens C, Tofts C, Avis T, et al. 2007. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet 80: 345–352. 10.1086/511134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Guo H, Xiong B, Stessman HA, Wu H, Coe BP, Turner TN, Liu Y, Zhao W, Hoekzema K, et al. 2016. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun 7: 13316 10.1038/ncomms13316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster E, Cho MT, Alexander N, Desai S, Naidu S, Bekheirnia MR, Lewis A, Retterer K, Juusola J, Chung WK. 2016. De novo PHIP-predicted deleterious variants are associated with developmental delay, intellectual disability, obesity, and dysmorphic features. Cold Spring Harb Mol Case Stud 2: a001172 10.1101/mcs.a001172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All reported variants, along with associated human phenotype ontology terms, are submitted to ClinVar (https://ncbi.nlm.nih.gov/clinvar/) under accession numbers SCV000899256.1 (p.Ser28Tyr), SCV000899257.1 (p.Thr200LeufsTer8), SCV000899258.1 (IVS32 + 3_IVS32 + 6delAAGT), SCV000899259.1 (p.Val1199Ter), SCV000899260.1 (p.Gly181ArgfsTer12), SCV000899261.1 (p.Ser1524LeufsTer22), SCV000899262.1 (p.Lys915SerfsTer15), SCV000899263.1 (p.Lys521Arg), SCV000899264.1 (p.Leu1054Ter), and SCV000899265.1 (p.Ser229Leu).