Abstract
Lipoprotein (a) [Lp(a)] is a genetically determined risk factor of coronary artery disease (CAD). Previous genome-wide association studies (GWASs), which were mostly carried out in Caucasians, have identified many Lp(a)-associated SNPs. Here, we performed a GWAS on Lp(a) levels and further explored the relationships between Lp(a)-associated SNPs and CAD severity in 1,403 Han Chinese subjects. We observed that elevated Lp(a) levels were significantly associated with the increased synergy between percutaneous coronary intervention with TAXUS and cardiac surgery (SYNTAX) score and the counts of heavily calcified lesions and long-range lesions (LRLs; P < 0.05), which are defined as lesions spanning >20 mm. Moreover, we identified four independent SNPs, namely, rs7770628, rs73596816, and rs6926458 in LPA, and rs144217738 in SLC22A2, that were significantly associated with Lp(a) levels. We also found that rs7770628 was associated with high SYNTAX scores [odds ratio (OR) (95% CI): 1.37 (1.05–1.80), P = 0.0213, false discovery rate (FDR) = 0.0852], and that rs7770628 and rs73596816 were associated with high risk of harboring LRLs [OR (95% CI): 1.53 (1.17–2.01), P = 0.0018, FDR = 0.0072 and 1.72 (1.19–2.49), P = 0.0040, FDR = 0.0080, respectively]. Our study was a large-scale GWAS to identify Lp(a)-associated variants in the Han Chinese population. Our findings highlight the importance and potential of Lp(a) intervention and expand our understanding of CAD prevention and treatment.
Keywords: single nucleotide polymorphism, SYNTAX score, genetics, lipid and lipoprotein metabolism, metabolic disease
Lipoprotein (a) [Lp(a)] is a type of complex particle that is linked to an apolipoprotein molecule by a disulfide bond and majorly consists of cholesterol-rich LDL (1, 2). High Lp(a) levels have been considered as an independent risk factor for atherosclerosis and coronary artery disease (CAD) for decades (3–6). The risk of myocardial infarction increases by 1.2 times with the doubling of serum Lp(a) levels (7). The prognostic utility of Lp(a) for CAD is attracting considerable attention, and remarkable effort has been devoted to the exploration of therapy targets (8, 9). Lp(a) level is undisputedly a genetically determined trait. A twin population study observed that more than 90% of the variation in Lp(a) levels is heritable (10). Large-scale genome-wide association studies (GWASs) (10, 11) and candidate gene studies (12) have identified sets of SNPs that are significantly associated with serum Lp(a) concentration. Several Lp(a)-associated SNPs are related to CAD risks or poor outcomes in patients with CAD. For example, rs3798220, one of the most-cited variants, accounts for a large extent of Lp(a) variation and increases the risk of CAD by 92% in the European population (12). However, studies on the Asian population have failed to replicate the association of rs3798220 with increased Lp(a) levels and the risk of cardiovascular events (13, 14). Racial differences should not be ignored, and GWASs on Asian populations are crucial to identify Lp(a)-associated variants and CAD development.
Therefore, our study aimed to identify Lp(a)-associated variants and evaluate their association with CAD severity. We recruited 1,403 Han Chinese patients who were diagnosed with CAD and who were undergoing percutaneous coronary intervention (PCI). We confirmed the association between increased Lp(a) levels and the CAD severity, which was evaluated through coronary angiography (CAG). Moreover, we identified four independent SNPs (i.e., rs7770628, rs73596816, and rs6926458 in LPA, and rs144217738 in SLC22A2) that were associated with Lp(a) variation by performing a GWAS. These SNPs were also independent of the KIV-2 copy number variation (CNV). Furthermore, we found that rs7770628 and rs73596816 are associated with CAD severity. This association indicates that rs7770628 and rs73596816 have potential roles in CAD progression.
MATERIALS AND METHODS
Study population
A total of 3,099 patients with CAD who were undergoing atorvastatin or rosuvastatin treatment at Guangdong General Hospital over the period of January 2010 to December 2013 were sequentially and prospectively enrolled. We included only those receiving PCI therapy in this study. The exclusion criteria were as follows: 1) 18 years < age > 80 years; 2) renal dysfunction [defined as serum creatinine (CREA) concentration greater than two times the normal upper limit (230 μmol/l) or a history of renal transplantation or dialysis]; 3) hepatic dysfunction [defined as serum alanine aminotransferase or aspartate aminotransferase (AST) transaminase concentration greater than two times the normal upper limit (80 U/l) or cirrhosis diagnosis]; 4) pregnant or lactating; 5) diagnosed with advanced cancer or undergoing hemodialysis; 6) history of thyroid disease and related medication; and 7) unavailable blood sample and CAG images. Baseline information, including demographics, medical history, biochemical measurements, and medication, was obtained from the hospital information database. The corrected LDL cholesterol (LDLCc) value was calculated by subtracting 30% of the Lp(a) mass from circulating LDL cholesterol (LDLC). This study was approved by the Medical Ethical Review Committee of Guangdong General Hospital (GDREC2010137 and GDREC2017071H) and conducted according to the Declaration of Helsinki. Informed consent (20170211) was obtained from all individual participants included in the study.
CAD diagnosis and severity evaluation via CAG
CAG was performed using the standard technique, and CAG images were acquired with from Syngo Dynamics cardiovascular imaging software (Siemens Medical Solutions USA, Inc., Malvern, PA). All angiograms were assessed by two expert cardiologists who were blinded to the genotype data. The SYNTAX score was used to determine the complexity and severity of CAD (15). The SYNTAX score characterizes the anatomy of coronary vasculature with respect to the number, location, and length of lesions, occurrence of total occlusions, bridging collaterals, bi/trifurcations, aorto-ostial lesions, tortuosity, calcification, thrombus, and diffusion of disease/small vessels. The SYNTAX score of each patient was calculated by using the online SYNTAX score calculator version 2.11.
We included two sub-items of the SYNTAX score [i.e., counts of heavily calcified lesions (HCLs) and long-range lesions (LRLs)] into the subsequent analyses. A scored lesion was defined as a ≥50% diameter stenosis in vessels ≥1.5 mm. A LRL was defined as a lesion spanning >20 mm, and a HCL was defined as a persisting opacification visible in more than one projection of the complete lumen of the coronary artery.
Lp(a) concentration determination
Fasting blood Lp(a) concentration was determined on the second day of each patient’s admission by biochemical methods. In brief, Lp(a) was measured in plasma samples by using sandwich ELISAs [Lp(a) ELISA kit; Yaji Biosystems, Shanghai, China) and a SYNCHRON LX20 UniCel DxC800 analyzer (Beckman Coulter Inc.). The Lp(a) concentrations of 1,123 subjects were measured.
SNPs and KIV-2 CNV genotyping
DNA was extracted with an established genomic DNA kit (TIANGEN; OSR-M102). Each subject was genotyped using a Global Screening Array bead chip from Illumina. Genotyping procedures followed the standard manual protocol for an Illumina Infinium HTS assay, and Genome Studio software and the calling algorithm from Illumina were used for normalized intensity data analyses. A total of 700,078 SNPs were genotyped for each individual.
Real-time fluorescence quantitative PCR was performed to detect the relative KIV repeat number by using the Bio-Rad CFX96 system. The synthesis of TaqMan® probe and primers for LPA KIV-2 exon2 and the detection experiment were formed by following established methods (7, 16). ΔCT values were obtained by subtracting the CT value of the single-copy gene from the CT value of KIV-2 and were used to represent KIV-2 CNV. Details are presented in the supplemental material.
Data analyses
Genotype imputation and quality control.
We applied a series of criteria for quality control (QC) and excluded unqualified samples and SNPs prior to imputation. Briefly, we excluded: 1) individuals with a missing rate of >5%; 2) SNPs with a missing rate of >5%; 3) SNPs with a P-value of Hardy-Weinberg disequilibrium test <1e-6; and 4) minor allele frequency (MAF) of <5%. Imputation was then carried out using IMPUTEv2 software (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html), and haplotype data of Chinese Han subsets in the 1000 Human Genomes Project phase III were used as reference. After imputation, post-QC procedures were applied to exclude those SNPs with: 1) low imputation quality of information <0.6; 2) a missing rate of >5%; 3) a P-value of Hardy-Weinberg disequilibrium test <1e-6; and 4) MAF of <5%.
GWAS analysis on Lp(a) levels.
Prior to the GWAS, a Shapiro-Wilk test was used to test the normality of Lp(a) concentrations, and an inverse-normal transformation was performed to normalize the highly skewed data. Each QCed SNP with a LP(a) level was subjected to an association test assuming a linear model under the additive mode using PLINK v1.07 software (http://zzz.bwh.harvard.edu/plink/). Sex and age were included for adjustment.
As the LPA gene is the most important determinant of Lp(a) levels, the association between Lp(a) level and other SNPs could be weakened and missed. We performed a further GWAS on Lp(a) by including Lp(a)-associated SNPs in the LPA gene into the original models, so as to identify additional variants for Lp(a) levels.
Independence of Lp(a)-associated SNPs.
The linkage disequilibrium (LD) between SNP pairs was accessed from the genotype data of the Han Chinese subsets (CHB and CHS subsets) in the 1000 Human Genomes Project. Conditional analyses were sequentially performed to evaluate the independence of each genome-wide significant SNP by including other SNPs in the original model. To evaluate the independence of Lp(a)-associated SNPs from KIV-2 CNV, we included ΔCT into the original linear model in genome-wide association analyses.
Association among Lp(a), LP(a)-associated variants, and CAD severity.
Linear and logistic regression analyses were used to assess the association among Lp(a), LP(a)-associated variants, and CAD severity. Confounding factors (i.e., sex, age, AST, CREA, hypertension and diabetes statuses, and lipid traits including LDLCc, HDL cholesterol, triglyceride, and total cholesterol) were included in the multivariate regression models for correction.
Statistical analyses were performed using R (version 3.4.3, http://www.R-project.org/).
RESULTS
Patient characteristics
We included 1,403 patients with CAD who were undergoing PCI therapy in this study after screening on the basis of inclusion and exclusion criteria. The subject screening procedure is shown in supplemental Fig. S1. The average Lp(a) level of the subjects was 30.27 ± 31.97 mg/dl. The average SYNTAX score of the subjects was 16.70 ± 10.93. The detailed demographic characteristics of the study population were summarized and are presented in Table 1. We compared the two subgroups of statin users to evaluate their homogeneity. The details are presented in the supplemental material and supplemental Table S1.
TABLE 1.
Baseline characteristics of the overall study population
| Characteristics | Value [N (%) or Mean ± SD] |
| Age | 62.73 ± 10.06 |
| Sex (male) | 1,119 (79.8) |
| BMI, kg/m2 | 24.27 ± 4.46 |
| Arrhythmia | 106 (7.6) |
| Diabetes | 385 (27.4) |
| Heart failure | 120 (8.6) |
| Hypertension | 816 (58.2) |
| Hyperlipidemia | 174 (12.4) |
| ALT, U/l | 27.58 ± 13.48 |
| AST, U/l | 26.72 ± 10.88 |
| CK, U/l | 86.16 ± 25.34 |
| CREA, umol/l | 91.96 ± 68.08 |
| CKMB, U/l | 7.38 ± 5.48 |
| TC, mmol/l | 4.36 ± 1.24 |
| LDLC, mmol/l | 2.65 ± 1.01 |
| LDLCc, mg/dl | 93.51 ± 38.67 |
| HDLC, mmol/l | 0.97 ± 0.25 |
| TG, mmol/l | 1.61 ± 1.14 |
| GLUC, mmol/l | 6.73 ± 2.69 |
| Lp(a), mg/dl | 30.27 ± 31.97 |
| ApoA, g/l | 1.05 ± 0.27 |
| BNP, pg/ml | 827.50 ± 1948.94 |
| β-blocker use | 1,254 (89.4) |
| ACEI use | 880 (62.7) |
| CCB use | 376 (26.8) |
| PPI use | 679 (48.4) |
| SYNTAX score | 16.70 ± 10.93 |
ACEI, angiotensin-converting enzyme inhibitor; ALT, alanine aminotransferase; BNP, brain natriuretic peptide; CCB, calcium channel blocker; CHOL, cholesterol; CK, creatine kinase; CKMB, creatine kinase MB; GLUC, glucose; HDLC, HDL cholesterol; PPI, proton pump inhibitor; SYNTAX score, synergy between PCI with TAXUS and cardiac surgery score; TRIG, triglyceride.
Elevated Lp(a) levels associated with CAD severity
We initially included the SYNTAX score as the major and composite index for CAD severity and two sub-items of the SYNTAX score, namely, HCL and LRL, as the minor indexes to evaluate the association between Lp(a) levels and CAD severity. Univariate and multivariate linear regression analyses, including confounding factors, were performed in succession. As expected, elevated Lp(a) levels were significantly associated with a high SYNTAX score (adjusted estimates: 1.23 ± 0.32, P = 1.28E-04) and increased LRL counts (adjusted estimates: 0.08 ± 0.02, P = 2.66E-04). However, no significant association was observed between Lp(a) levels and increased HCL counts.
We further grouped the cohort on the basis of the median value of the SYNTAX score and the presence of LRL and HCL and applied logistic regression analysis to analyze the association between Lp(a) levels and CAD severity. Elevated Lp(a) levels were significantly associated with the risk of having a high SYNTAX score and developing LRLs and HCLs (P < 0.05).
Details are presented in Table 2. In conclusion, elevated Lp(a) levels showed a significant association with high CAD severity in Han Chinese patients undergoing PCI.
TABLE 2.
Association between Lp(a) levels and CAD severity
| Severity of CAD | Univariate Analysis | Adjusted Analysis | Severity of CAD | Univariate Analysis | Adjusted Analysis | ||||
| Estimates ± SE | P | Estimates ± SE | P | OR (95% CI) | P | OR (95% CI) | P | ||
| SYNTAX score | 1.23 ± 0.32 | 1.28E-04 | 1.25 ± 0.32 | 9.10E-05 | High SYNTAX score | 1.27 (1.12–1.43) | 1.28E-04 | 1.29 (1.14–1.47) | 4.36E-05 |
| Counts of HCL | 0.02 ± 0.01 | 0.1180 | With HCL | 1.25 (1.01–1.56) | 0.0431 | 1.35 (1.02–1.80) | 0.0350 | ||
| Counts of LRL | 0.08 ± 0.02 | 2.66E-04 | 0.08 ± 0.02 | 0.0007 | With LRL | 1.23 (1.09–1.39) | 0.0010 | 1.22 (1.07–1.38) | 0.0021 |
Variables with P < 0.1 were entered into the multivariable model. Sex, age, AST, CREA, LDLCc, HDL cholesterol, triglyceride, total cholesterol, and hypertension and diabetes statuses were taken as covariates in adjusted analysis.
Identification of Lp(a)-associated variant via GWAS
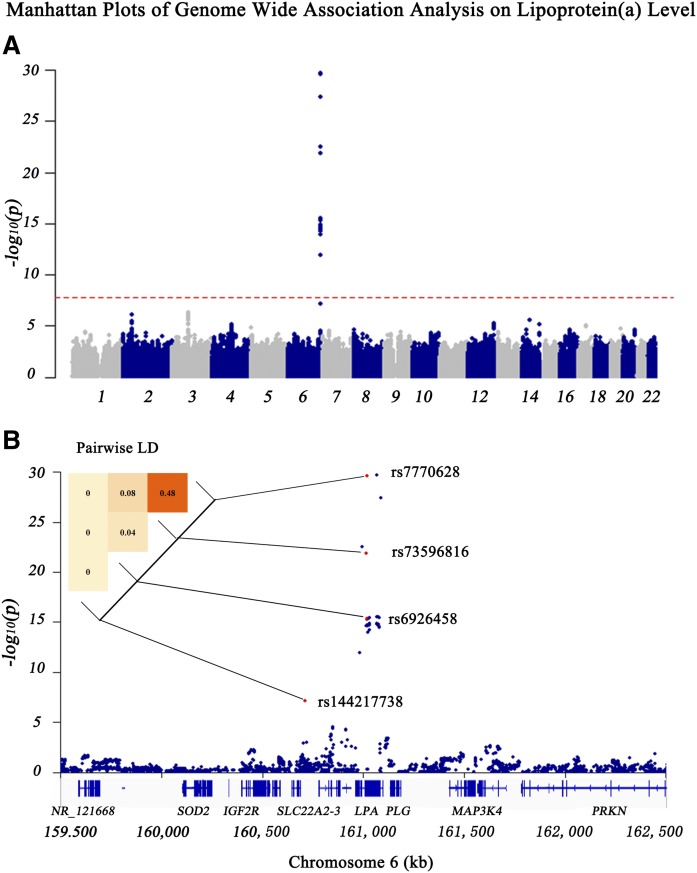
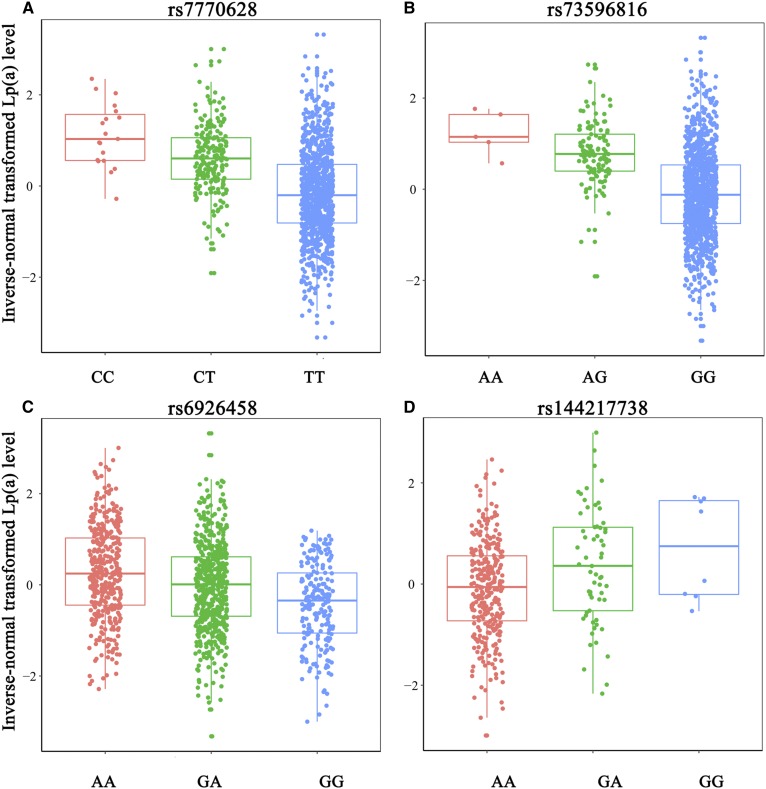
A total of 3,448,668 common SNPs were available for GWAS after imputation and QC procedures. The details of genotypic data processing are presented in supplemental Fig. S2. We performed a GWAS on each SNP against the inverse-normal transformed Lp(a) concentration to identify variants that affected Lp(a) levels. The threshold of genome-wide significance was defined as P < 0.05/3,448,668 = 1.45E-08. After correction for age and sex, 60 SNPs in the LPA gene reached the threshold (Fig. 1A, B). Conditional analyses were performed to candidate SNPs, and three SNPs in the LPA gene, namely, rs7770628, rs73596816, and rs6926458, showed their independence from each other. As shown in Table 3, rs7770628 (T > C) and rs73596816 (G > A) were related to increased Lp(a) levels [β ± SE = 0.73 ± 0.06, P = 2.01E-30 (Fig. 2A) and β ± SE = 0.86 ± 0.09, P = 1.26E-22 (Fig. 2B), respectively]. Moreover, rs6926458 (A > G) were related to reductions in Lp(a) levels [β ± SE = 0.34 ± 0.04, P = 4.59E-16 (Fig. 2C)].
Fig. 1.
Manhattan plots of genome-wide association analysis on Lp(a) level. A: Global Manhattan plot shows the association of each SNP in autosome with Lp(a). The red dashed line indicates the threshold of genome-wide significance. B: Local Manhattan plot is an amplified view of the global Manhattan plot showing the association of SNPs in a region centered by the lead SNP with Lp(a). The red dashed line indicates the threshold of genome-wide significance. The heat map in the top left corner of the plot shows the pairwise LD of the genome-wide significant SNPs (the darker the block, the more correlated the two SNPs). Numbers in the blocks show the r2 of SNP pairs. Gene regions are shown in the bottom of the plot.
TABLE 3.
GWAS-identified Lp(a)-associated SNPs
| Locus | SNP | MAF | EA/OAb | Number | Original Modelsa | Multi-SNP Modelsb | ||
| Beta ± SE | P | Beta ± SE | P | |||||
| LPA | rs7770628 | 0.11 | C/T | 1,147 | 0.73 ± 0.06 | 2.29E-30 | 0.43 ± 0.09 | 6.08E-06 |
| LPA | rs73596816 | 0.06 | A/G | 1,147 | 0.86 ± 0.09 | 1.27E-22 | 0.36 ± 0.12 | 1.75E-06 |
| LPA | rs6926458 | 0.33 | G/A | 1,147 | −0.34 ± 0.04 | 4.59E-16 | −0.21 ± 0.04 | 4.10E-03 |
| SLC22A2 | rs144217738 | 0.12 | G/A | 1,136 | 0.34 ± 0.06 | 6.90E-08 | 0.36 ± 0.06 | 6.06E-09 |
EA/OA, effect allele/other allele.
Sex and age were included into the models for adjustment.
Multi-SNP models included all four SNPs together to observe their independence from each other.
Fig. 2.
Box plots for Lp(a)-associated SNPs against Lp(a) levels. Four plots show the distribution of each genotype of Lp(a) level among four SNPs: rs7770628 (A); rs73596816 (B); rs6926458 (C); and rs144217738 (D).
Then, we performed further analyses by including three independent Lp(a)-associated SNPs in the LPA gene as covariates to identify other potential variants. Notably, we identified a SNP in SLC22A2, namely, rs144217738, which was nominally associated with Lp(a) levels (P = 6.90E-08) in the original model, but showed a genome-wide significant association [β ± SE = 0.36 ± 0.06, P = 6.06E-09 (supplemental Fig. S3, Fig. 2D)] when including three LPA SNPs as covariates in a multi-SNP model (Table 3). And four SNPs together accounted for 16.2% of the Lp(a) variation in this Chinese cohort. LD analyses were conducted on four SNPs and the r2 of two SNPs exceeding 0.5 in LD analysis was considered in LD (Fig. 1B). A full list of the 61 SNPs is shown in supplemental Table S4.
Independence of Lp(a)-associated SNPs from KIV-2 CNV
We randomly selected 645 subjects and used quantitative (q)PCR to quantify the copy number of KIV-2 in the LPA gene to evaluate independence of the Lp(a)-associated SNP from KIV-2 CNV. The mean ± SD of the ΔCT values was 1.79 ± 0.84 among all subjects. We first used linear regression analysis to evaluate the correlation between Lp(a) levels and KIV-2 CNV. As shown in supplemental Fig. S4, an increase in KIV-2 CNV was associated with a decrease in Lp(a) levels (univariate estimate: −0.14 ± 0.05, P = 0.0017; adjusted estimate with age and sex: −0.15 ± 0.05, P = 0.0014). This result was consistent with previously reported results. We included the ΔCT values in the original models and found that four identified SNPs were independent of KIV-2 CNV, given that they remained significantly associated with Lp(a) level (P < 0.0125 = 0.05/4). This result indicates that the effect of the four SNPs on Lp(a) is independent of that of KIV-2 CNV. Details are shown in supplemental Table S3.
Comparison of Lp(a) SNPs identified in previous studies
By referring to the published GWAS catalog and the Ensembl database, we found that rs7770628, rs73596816, and rs6926458 have been identified in previous GWASs or candidate gene studies, but have not been identified in the Chinese population. Furthermore, rs7770628 and rs73596816 have been reported to be associated with CAD risk. However, no study on Lp(a) has reported on rs144217738. We used the Linkage Disequilibrium Calculator in the Ensembl Project to identify the proxy SNP of rs144217738 and to search for rs144217738 in databases (e.g., Clinvar, GWAS catalog, and Ensembl) to verify its novelty.
We also explored the replication of some previously reported SNPs available in our data. For example, studies on Caucasian populations have reported that rs3798220 and rs7412 in the LPA gene are related to Lp(a) levels. However, we found that rs3798220 and rs7412 were nominally associated with Lp(a) levels (MAF = 0.09 and P = 0.0039 for rs3798220 and MAF = 0.06 and P = 0.0183 for rs7412). Although other published SNPs (e.g., rs10455872 and rs143431368) were also included in our genotyping panel, these SNPs were all rare in the Chinese population (both with MAF <0.01 in our cohort and 1000 Human Genomes Project subjects). A full list of previously reported SNPs associated with Lp(a) levels is shown in supplemental Table S5.
Association between Lp(a)-associated variants and CAD severity
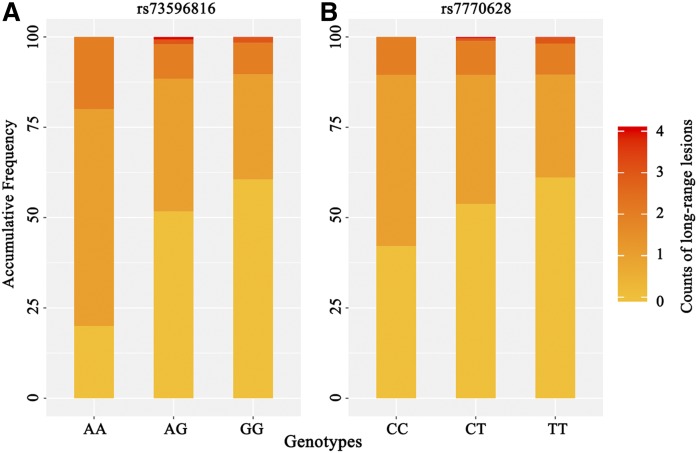
We conducted linear regression analyses on four SNPs and the indexes of CAD severity described above to explore whether the Lp(a)-associated variants could account for the difference in CAD severity among individuals. Although no significance was observed in analysis with the SYNTAX score and HCL counts (P > 0.05), we observed that the increase in the minor allele dose of rs73596816 was associated with increments in LRL counts (adjusted estimates: 0.19 ± 0.07, P = 0.0044, false discovery rate (FDR) = 0.0176; Fig. 3A). Furthermore, rs7770628 showed a nominally significant association with the LRL counts (adjusted estimates: 0.10 ± 0.05, P = 0.0348, FDR = 0.0696; Fig. 3B). Supplemental Table S2 provides the details.
Fig. 3.
Bar plots of two SNPs associated with count of LRLs. Each bar shows the proportion of harboring different counts of LRLs in each genotype (the darker the color of the block, the more LRLs it has). Both plots indicate that individuals carrying alleles with minor frequency are likely to be harboring more LRLs. Minor frequency alleles are A and C for rs73596816 (A) and rs7770628 (B), respectively.
Given the complexity of CAD etiology, the effects of the variants may be inadequately reflected in a moderate sample size and could not fit well in a linear relationship with CAD severity. We further divided the cohort into groups on the basis of the median value of the SYNTAX score [<15 (N = 649) and ≥15 (N = 725)], the presence of LRLs [without LRLs (N = 817) and with LRLs (N = 557)], and presence of HCLs [without HCLs (N = 1,274) and with HCLs (N = 100)]. Moreover, we used the logistic regression model to analyze the inter-group differences of Lp(a)-associated SNPs. Notably, an increase in minor allele dose of rs7770628 was associated with an increase in the risk of a high SYNTAX score [adjusted odds ratio (OR) (95% CI): 1.37 (1.05–1.80), P = 0.0213, FDR = 0.0852]. Carrying the minor alleles of rs7770628 and rs73596816 was associated with a high risk of harboring LRLs [adjusted OR (95% CI): 1.53 (1.17–2.01), P = 0.0018, FDR = 0.0072 and 1.72 (1.19–2.49), P = 0.0040, FDR = 0.0080, respectively]. No significant association with heavy calcification was found. Table 4 presents the details of these results. Moreover, when the Lp(a) level was included in the models, the relationship between rs7770628 and rs73596816 with CAD severity became insignificant.
TABLE 4.
Association of Lp(a)-associated SNPs with CAD severity
| Univariate Analysis | Adjusted Analysis | ||||||
| Item | EA/OA | Effect Allele Frequency | OR (95% CI) | P | OR (95% CI) | P | |
| SYNTAX score | <15 (N = 650) | ≥15 (N = 725) | |||||
| rs7770628 | C/T | 9.8% | 12.3% | 1.28 (1.01–1.63) | 0.0441 | 1.37 (1.05–1.80) | 0.0213 |
| rs73596816 | A/G | 5.2% | 6.1% | 1.18 (0.86–1.64) | 0.3130 | — | — |
| rs6926458 | G/A | 42.2% | 41.2% | 0.96 (0.98–1.36) | 0.5839 | — | — |
| rs144217738 | G/A | 10.9% | 12.5% | 1.17 (0.92–1.50) | 0.1963 | — | — |
| LRLs | Genotype | No LRL (N = 818) | With LRL (N = 557) | OR (95% CI) | P | OR (95% CI) | P |
| rs7770628 | C/T | 9.8% | 13.0% | 1.38 (1.09–1.38) | 0.0080 | 1.53 (1.17–2.01) | 0.0018 |
| rs73596816 | A/G | 4.8% | 7.1% | 1.52 (1.10–2.10) | 0.0110 | 1.72 (1.19–2.49) | 0.0040 |
| rs6926458 | G/A | 42.9% | 40.0% | 0.89 (0.76–1.04) | 0.1366 | — | — |
| rs144217738 | G/A | 11.0% | 12.9% | 1.18 (0.92–1.51) | 0.1786 | — | — |
| HCLs | Genotype | No HCL (N = 1275) | With HCL (N = 100) | OR (95% CI) | P | OR (95% CI) | P |
| rs7770628 | C/T | 13.5% | 10.9% | 1.27 (0.82–1.91) | 0.2610 | — | — |
| rs73596816 | A/G | 6.5% | 5.7% | 1.16 (0.62–2.00) | 0.6200 | — | — |
| rs6926458 | G/A | 42.5% | 41.6% | 1.04 (0.78–1.38) | 0.8070 | — | — |
| rs144217738 | G/A | 13.8% | 11.6% | 1.22 (0.78–1.83) | 0.3556 | — | — |
EA/OA, effect allele/other allele.
In summary, we tested the association of Lp(a) SNPs with CAD severity, and found that the minor frequency allele carriers of rs7770628 and rs73596816 were at a high risk of developing serious lesions in CAD.
DISCUSSION
Potential of Lp(a) as an indicator of CAD severity
In this study, we recruited 1,403 Han Chinese patients who were diagnosed with CAD and were undergoing PCI to identify Lp(a)-associated variants and evaluate their association with CAD severity. We observed that high Lp(a) levels were significantly associated with increased CAD severity, which is represented by a high SYNTAX score and the presence of LRLs and HCLs. This result is consistent with a recently reported finding demonstrating that elevated Lp(a) levels were associated with CAD severity (evaluated by Gensini score) in patients with type 2 diabetes mellitus (17). Moreover, the weaker association between Lp(a) and HCL than that between Lp(a) the other indexes indicates that Lp(a) may mainly affect CAD severity on the basis of lesion extent, but not through calcification. High SYNTAX scores, which indicate an extensive lesion, could predict high risks of poor outcomes during long-term follow-up (18–20). Hence, CAD severity, which is evaluated by the SYNTAX score, can reflect the prognosis of patients with CAD. For decades, researchers have continuously explored the clinical application of Lp(a) in CAD. The addition of Lp(a) to the Framingham risk score helped in the reclassification of patients with the risk of cardiovascular incidence (21). These findings support the conclusion that Lp(a) is a risk factor for CAD and is involved in atherosclerosis development (3, 4, 22). Therefore, we hypothesized that an elevated Lp(a) level could induce exacerbated lesions (e.g., accompanied by LRLs and HCLs) and might lead to poor outcomes in patients with CAD. This hypothesis requires further comprehensive and large-scale studies for verification.
Review of the mechanism of Lp(a) in CAD progression
The underlying mechanism of Lp(a) in CAD progression remains unclear. Previous works have reported that oxidized phospholipids carried by Lp(a) could mediate inflammatory activity by increasing in the arterial wall and enhancing the inflammatory response of monocytes (23). Furthermore, oxidized phospholipids promote proinflammatory reactions in the arterial wall and result in smooth muscle cell proliferation (24). The apo(a), another component of Lp(a), contains 10 types of kringle domains that are homologous to kringle IV, a plasminogen. The kringle domains in apo(a) could abolish the ligand interactions between kringles and lysine-containing substrates to decrease plasmin synthesis and inhibit fibrinolysis (25). These processes, which are mediated by Lp(a), are likely involved in CAD development. However, additional evidence for the underlying pathogenic roles of Lp(a) are still required.
Genetic regulation of plasma Lp(a) levels
The plasma Lp(a) level is a genetically influenced phenotype that is mainly controlled by the LPA locus and varies greatly because of the variants (26). Lp(a) levels vary from less than 0.1 mg/dl to more than 200 mg/dl among individuals. The Mendelian randomization approach and GWASs revealed that variants, mostly referring to the CNV of KIV-2 repeats and SNPs in the LPA gene (1), were associated with the Lp(a) levels. The size of the KIV-2 CNV is related to apo(a) size positively and inversely to the Lp(a) levels (26, 27) and apo(a) isoform size (28). The independence of Lp(a)-associated SNPs and apo(a) isoform size is essential to identify their independent effect for Lp(a) levels. Detection of the apo(a) isoforms’ size is not possible, resulting in a limitation of this study. However, published studies (7, 16) stated that the copy number of KIV-2 repeats in the LPA gene could represent the apo(a) isoforms’ size. Therefore, we applied qPCR to relatively quantify the copy number of KIV-2 in the LPA gene in reference to established methods to generate the ΔCT value and represent the apo(a) isoforms’ size.
Numerous SNPs have been identified and replicated in large-scale GWASs in Caucasian populations. However, few GWASs on Lp(a) in Asian populations, especially in Chinese populations, exist. A large-scale study estimated that rs3798220 and rs10455872 together account for 36% of the total variations in the Lp(a) level (12). This result has been replicated by other studies on Caucasian populations (29, 30). However, our previous study and other studies in the Asian population have failed to replicate the association between rs3798220 and Lp(a) variation (13, 14), whereas rs10455872 is extremely rare in the East Asian population (MAF = 0.005). Therefore, one of the possible reasons for lack of these associations in our study is the limited power generated from a minor sample size. This is a limitation of the present study and more studies on the Asian population are still necessary. Another reason was inferred that those Lp(a)-associated SNPs failed to be replicated did not independently affect Lp(a) levels but were in LD with the KIV-2 CNV among Europeans (13). Moreover, the size of the KIV-2 CNV varied considerably among races (31). The lack of associations between these two SNPs and Lp(a) was also observed in a study in an African-descent population (32). This study (32) identified another SNP, namely rs9457951, as potentially responsible for Lp(a) variation and independent of the KIV repeat copy number in African-descent populations. However, this SNP is rare in the non-African population according to data from the NCBI dbSNP database. A splice variant in LPA was identified in Finns, which was considered a population-specific locus associated with reduced Lp(a) levels (33). Therefore, the identification of variations associated with Lp(a) is essential.
Review of four Lp(a)-associated SNPs identified in our study
We conducted this GWAS and identified variants affecting Lp(a) levels in Chinese Han subjects. As previously described, we determined four independent SNPs significantly associated with Lp(a) levels in Chinese Han patients with CAD. Among the four SNPs, rs7770628, rs73596816, and rs6926458 in LPA were identified to be related to the Lp(a) level in other studies (10, 11), whereas rs144217738 in SLC22A2 was first discovered to be associated with the Lp(a) level in our study. The MAF of rs7770628 (C allele) was 40–50% in European populations but approximately 10% in Asian populations (recorded in the NCBI dbSNP database). Previous studies have reported variants in SLC22A family member 1-3 associated with Lp(a) levels, which supports our finding. Besides, a haplotype in the SLC22A3-LPAL2-LPA gene cluster was reported to be associated with Lp(a) level and CAD risk in Europeans (34); however, studies in Asians (35) and Hispanics (36) failed to replicate the association of haplotype and Lp(a) level or CAD risk. Therefore, the rs144217738 found in our study needs further research for confirmation. The difference of Lp(a)-associated variants among races and the underlying regulation mechanism in Lp(a) level remains unclear and requires further studies.
Genetic effects of LPA loci in CAD development
Lp(a) is one of the strongest genetic risk factors for CAD. Thus, the ability of the genetic variants associated with Lp(a) variation to account for the variation in risk of CAD among individuals must be evaluated, and new ideas for CAD prevention and treatment must be proposed. We found that rs7770628 and rs73596816 were associated with CAD severity. The presence of the copy of the minor allele (C allele) of rs7770628 increased the risks of having high SYNTAX scores by 37% in patients with CAD. Moreover, carrying the minor alleles of rs7770628 and rs73596816 may increase the risk of harboring LRLs by 53% and 72%, respectively. These two SNPs were reported to be associated with CAD risk (10). We considered these SNPs to be playing an important role in atherosclerosis development and CAD progression, especially affecting the lesion extent in patients with CAD.
Currently, numerous findings for Lp(a)-associated SNPs related to CAD risk or clinical endpoints are available. rs10455872 and rs3798220 are associated with reduced copy number of KIV-2 repeats and the risks of CAD and clinical endpoints (12, 33, 37–39). A genotype score including these two SNPs is predictive of the risk of developing CAD; nevertheless, this association is abolished when Lp(a) levels are adjusted (12). Moreover, rs10455872 was reported to be associated with coronary lesions and CAD severity (40). These two SNPs have been included in the multiloci risk score, which is a good predictor of CAD outcomes (41, 42). These results highlighted the hereditary susceptibility of CAD associated with sequence variants and their clinical application.
CONCLUSIONS
We performed a large-scale GWAS and identified four SNPs associated with Lp(a) levels in 1,403 Han Chinese patients with CAD. We evaluated the relationships among Lp(a) level, Lp(a)-associated SNPs, and CAD severity. High Lp(a) levels are associated with the increased risk of developing serious lesions in CAD. We replicated three SNPs in LPA, namely, rs7770628, rs73596816, and rs6926458, and identified one novel SNP in SLC22A2 that is rs144217738. These SNPs are associated with Lp(a) level in the Chinese Han population, which are independent of the KIV-2 repeats. Moreover, rs7770628 and rs73596816 are associated with the increased risk of developing serious lesions, especially LRLs, in CAD. These findings highlight the importance of Lp(a) intervention and deepen our understanding of CAD prevention and treatment and the importance of race variations in precision medical treatment.
Supplementary Material
Footnotes
Abbreviations:
- AST
- aspartate aminotransferase
- CAD
- coronary artery disease
- CAG
- coronary angiography
- CNV
- copy number variation
- CREA
- creatinine
- FDR
- false discovery rate
- GWAS
- genome-wide association study
- HCL
- heavily calcified lesion
- LD
- linkage disequilibrium
- LDLC
- LDL cholesterol
- LDLCc
- corrected LDL cholesterol
- Lp(a)
- lipoprotein (a)
- LRL
- long-range lesion
- MAF
- minor allele frequency
- OR
- odds ratio
- PCI
- percutaneous coronary intervention
- QC
- quality control
This study was supported by Ministry of Science and Technology of the People’s Republic of China Grant 2017YFC0909301, National Natural Science Foundation of China Grants 81872934 and 81673514, and Key Research and Development Program of Guangdong Province Grant 2019B020229003.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Schmidt K., Noureen A., Kronenberg F., and Utermann G.. 2016. Structure, function, and genetics of lipoprotein (a). J. Lipid Res. 57: 1339–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis K. L., Boffa M. B., Sahebkar A., Koschinsky M. L., and Watts G. F.. 2017. The renaissance of lipoprotein(a): Brave new world for preventive cardiology? Prog. Lipid Res. 68: 57–82. [DOI] [PubMed] [Google Scholar]
- 3.Berg K., Dahlen G., and Borresen A. L.. 1979. Lp(a) phenotypes, other lipoprotein parameters, and a family history of coronary heart disease in middle-aged males. Clin. Genet. 16: 347–352. [DOI] [PubMed] [Google Scholar]
- 4.Emerging Risk Factors Collaboration, Erqou S., Kaptoge S., Perry P. L., Di Angelantonio E., Thompson A., White I. R., Marcovina S. M., Collins R., Thompson S. G., et al. 2009. Lipoprotein(a) concentration and the risk of coronary heart disease, stroke, and nonvascular mortality. JAMA. 302: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foscolou A., Georgousopoulou E., Magriplis E., Naumovski N., Rallidis L., Matalas A. L., Chrysohoou C., Tousoulis D., Pitsavos C., and Panagiotakos D.. 2018. The mediating role of Mediterranean diet on the association between Lp(a) levels and cardiovascular disease risk: a 10-year follow-up of the ATTICA study. Clin. Biochem. 60: 33–37. [DOI] [PubMed] [Google Scholar]
- 6.Schatz U., Tselmin S., Muller G., Julius U., Hohenstein B., Fischer S., and Bornstein S. R.. 2017. Most significant reduction of cardiovascular events in patients undergoing lipoproteinapheresis due to raised Lp(a) levels - a multicenter observational study. Atheroscler. Suppl. 30: 246–252. [DOI] [PubMed] [Google Scholar]
- 7.Kamstrup P. R., Tybjaerg-Hansen A., Steffensen R., and Nordestgaard B. G.. 2009. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 301: 2331–2339. [DOI] [PubMed] [Google Scholar]
- 8.Ellis K. L., and Watts G. F.. 2018. Is lipoprotein(a) ready for prime-time use in the clinic? Cardiol. Clin. 36: 287–298. [DOI] [PubMed] [Google Scholar]
- 9.Watts G. F., and Boffa M. B.. 2018. Lipoprotein(a): lodestar for future clinical trials. Lancet. 392: 1281–1282. [DOI] [PubMed] [Google Scholar]
- 10.Mack S., Coassin S., Rueedi R., Yousri N. A., Seppala I., Gieger C., Schonherr S., Forer L., Erhart G., Marques-Vidal P., et al. 2017. A genome-wide association meta-analysis on lipoprotein (a) concentrations adjusted for apolipoprotein (a) isoforms. J. Lipid Res. 58: 1834–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melzer D., Perry J. R., Hernandez D., Corsi A. M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J. R., Paolisso G., et al. 2008. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet. 4: e1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke R., Peden J. F., Hopewell J. C., Kyriakou T., Goel A., Heath S. C., Parish S., Barlera S., Franzosi M. G., Rust S., et al. 2009. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 361: 2518–2528. [DOI] [PubMed] [Google Scholar]
- 13.Khalifa M., Noureen A., Ertelthalner K., Bandegi A. R., Delport R., Firdaus W. J., Geethanjali F. S., Luthra K., Makemaharn O., Pang R. W., et al. 2015. Lack of association of rs3798220 with small apolipoprotein(a) isoforms and high lipoprotein(a) levels in East and Southeast Asians. Atherosclerosis. 242: 521–528. [DOI] [PubMed] [Google Scholar]
- 14.Li Z. G., Li G., Zhou Y. L., Chen Z. J., Yang J. Q., Zhang Y., Sun S., and Zhong S. L.. 2013. Lack of association between lipoprotein(a) genetic variants and subsequent cardiovascular events in Chinese Han patients with coronary artery disease after percutaneous coronary intervention. Lipids Health Dis. 12: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sianos G., Morel M. A., Kappetein A. P., Morice M. C., Colombo A., Dawkins K., van den Brand M., Van Dyck N., Russell M. E., Mohr F. W., et al. 2005. The SYNTAX score: an angiographic tool grading the complexity of coronary artery disease. EuroIntervention. 1: 219–227. [PubMed] [Google Scholar]
- 16.Lanktree M. B., Rajakumar C., Brunt J. H., Koschinsky M. L., Connelly P. W., and Hegele R. A.. 2009. Determination of lipoprotein(a) kringle repeat number from genomic DNA: copy number variation genotyping using qPCR. J. Lipid Res. 50: 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H. W., Zhao X., Guo Y. L., Gao Y., Zhu C. G., Wu N. Q., and Li J. J.. 2018. Elevated lipoprotein (a) levels are associated with the presence and severity of coronary artery disease in patients with type 2 diabetes mellitus. Nutr. Metab. Cardiovasc. Dis. 28: 980–986. [DOI] [PubMed] [Google Scholar]
- 18.Ikeno F., Brooks M. M., Nakagawa K., Kim M. K., Kaneda H., Mitsutake Y., Vlachos H. A., Schwartz L., Frye R. L., Kelsey S. F., et al. ; BARI-2D Study Group . 2017. SYNTAX score and long-term outcomes: the BARI-2D trial. J. Am. Coll. Cardiol. 69: 395–403. [DOI] [PubMed] [Google Scholar]
- 19.Head S. J., Milojevic M., Daemen J., Ahn J. M., Boersma E., Christiansen E. H., Domanski M. J., Farkouh M. E., Flather M., Fuster V., et al. 2018. Mortality after coronary artery bypass grafting versus percutaneous coronary intervention with stenting for coronary artery disease: a pooled analysis of individual patient data. Lancet. 391: 939–948. [DOI] [PubMed] [Google Scholar]
- 20.Witberg G., Regev E., Chen S., Assali A., Barbash I. M., Planer D., Vaknin-Assa H., Guetta V., Vukasinovic V., Orvin K., et al. 2017. The prognostic effects of coronary disease severity and completeness of revascularization on mortality in patients undergoing transcatheter aortic valve replacement. JACC Cardiovasc. Interv. 10: 1428–1435. [DOI] [PubMed] [Google Scholar]
- 21.Willeit P., Kiechl S., Kronenberg F., Witztum J. L., Santer P., Mayr M., Xu Q., Mayr A., Willeit J., and Tsimikas S.. 2014. Discrimination and net reclassification of cardiovascular risk with lipoprotein(a): prospective 15-year outcomes in the Bruneck Study. J. Am. Coll. Cardiol. 64: 851–860. [DOI] [PubMed] [Google Scholar]
- 22.Kamstrup P. R., Tybjaerg-Hansen A., and Nordestgaard B. G.. 2012. Genetic evidence that lipoprotein(a) associates with atherosclerotic stenosis rather than venous thrombosis. Arterioscler. Thromb. Vasc. Biol. 32: 1732–1741. [DOI] [PubMed] [Google Scholar]
- 23.van der Valk F. M., Bekkering S., Kroon J., Yeang C., Van den Bossche J., van Buul J. D., Ravandi A., Nederveen A. J., Verberne H. J., Scipione C., et al. 2016. Oxidized phospholipids on lipoprotein(a) elicit arterial wall inflammation and an inflammatory monocyte response in humans. Circulation. 134: 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichikawa T., Unoki H., Sun H., Shimoyamada H., Marcovina S., Shikama H., Watanabe T., and Fan J.. 2002. Lipoprotein(a) promotes smooth muscle cell proliferation and dedifferentiation in atherosclerotic lesions of human apo(a) transgenic rabbits. Am. J. Pathol. 160: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boffa M. B., and Koschinsky M. L.. 2016. Lipoprotein(a): truly a direct prothrombotic factor in cardiovascular disease? J. Lipid Res. 57: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kronenberg F., and Utermann G.. 2013. Lipoprotein(a): resurrected by genetics. J. Intern. Med. 273: 6–30. [DOI] [PubMed] [Google Scholar]
- 27.Nikkola E., Ko A., Alvarez M., Cantor R. M., Garske K., Kim E., Gee S., Rodriguez A., Muxel R., Matikainen N., et al. 2017. Family-specific aggregation of lipid GWAS variants confers the susceptibility to familial hypercholesterolemia in a large Austrian family. Atherosclerosis. 264: 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rader D. J., Cain W., Ikewaki K., Talley G., Zech L. A., Usher D., and Brewer H. B.. 1994. The inverse association of plasma lipoprotein(a) concentrations with apolipoprotein(a) isoform size is not due to differences in Lp(a) catabolism but to differences in production rate. J. Clin. Invest. 93: 2758–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S. R., Prasad A., Choi Y. S., Xing C., Clopton P., Witztum J. L., and Tsimikas S.. 2017. LPA gene, ethnicity, and cardiovascular events. Circulation. 135: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chasman D. I., Shiffman D., Zee R. Y., Louie J. Z., Luke M. M., Rowland C. M., Catanese J. J., Buring J. E., Devlin J. J., and Ridker P. M.. 2009. Polymorphism in the apolipoprotein(a) gene, plasma lipoprotein(a), cardiovascular disease, and low-dose aspirin therapy. Atherosclerosis. 203: 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaw A., Boerwinkle E., Cohen J. C., and Hobbs H. H.. 1994. Comparative analysis of the apo(a) gene, apo(a) glycoprotein, and plasma concentrations of Lp(a) in three ethnic groups. Evidence for no common “null” allele at the apo(a) locus. J. Clin. Invest. 93: 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deo R. C., Wilson J. G., Xing C., Lawson K., Kao W. H., Reich D., Tandon A., Akylbekova E., Patterson N., Mosley T. H. Jr., et al. 2011. Single-nucleotide polymorphisms in LPA explain most of the ancestry-specific variation in Lp(a) levels in African Americans. PLoS One. 6: e14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim E. T., Wurtz P., Havulinna A. S., Palta P., Tukiainen T., Rehnstrom K., Esko T., Magi R., Inouye M., Lappalainen T., et al. ; Sequencing Initiative Suomi (SISu) Project . 2014. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 10: e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trégouët D. A., König I. R., Erdmann J., Munteanu A., Braund P. S., Hall A. S., Grosshennig A., Linsel-Nitschke P., Perret C., DeSuremain M., et al. 2009. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat. Genet. 41: 283–285. [DOI] [PubMed] [Google Scholar]
- 35.Lv X., Zhang Y., Rao S., Liu F., Zuo X., Su D., Wang M., Xia M., Guo H., Feng D., et al. 2012. Lack of association between four SNPs in the SLC22A3-LPAL2-LPA gene cluster and coronary artery disease in a Chinese Han population: a case control study. Lipids Health Dis. 11: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi L., Ma J., Qi Q., Hartiala J., Allayee H., and Campos H.. 2011. Genetic risk score and risk of myocardial infarction in Hispanics. Circulation. 123: 374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schunkert H., Konig I. R., Kathiresan S., Reilly M. P., Assimes T. L., Holm H., Preuss M., Stewart A. F., Barbalic M., Gieger C., et al. 2011. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch W., Mueller J. C., Schrempf M., Wolferstetter H., Kirchhofer J., Schomig A., and Kastrati A.. 2013. Two rare variants explain association with acute myocardial infarction in an extended genomic region including the apolipoprotein(A) gene. Ann. Hum. Genet. 77: 47–55. [DOI] [PubMed] [Google Scholar]
- 39.Qi Q., Workalemahu T., Zhang C., Hu F. B., and Qi L.. 2012. Genetic variants, plasma lipoprotein(a) levels, and risk of cardiovascular morbidity and mortality among two prospective cohorts of type 2 diabetes. Eur. Heart J. 33: 325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos P. C., Bueno C. T., Lemos P. A., Krieger J. E., and Pereira A. C.. 2014. LPA rs10455872 polymorphism is associated with coronary lesions in Brazilian patients submitted to coronary angiography. Lipids Health Dis. 13: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mega J. L., Stitziel N. O., Smith J. G., Chasman D. I., Caulfield M., Devlin J. J., Nordio F., Hyde C., Cannon C. P., Sacks F., et al. 2015. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 385: 2264–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khera A. V., Emdin C. A., Drake I., Natarajan P., Bick A. G., Cook N. R., Chasman D. I., Baber U., Mehran R., Rader D. J., et al. 2016. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. N. Engl. J. Med. 375: 2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.