In this study, Wang et al. address the differences between fetal and adult hematopoiesis. By single-cell RNA-seq analysis, the authors show that RNA-binding protein Lin28b alone is insufficient but together with the cytoplasmic RNA-binding protein Igf2bp3 reprograms adult blood cell progenitors to a fetal-like state. Taken together, these observations indicate that Lin28b–Igf2bp3 are developmental regulators that mediate the fetal–adult hematopoietic switch.
Keywords: regenerative medicine, cellular engineering, hematopoietic stem and progenitor cells, posttranscriptional regulation, ribonucleoprotein complexes, RNA-binding proteins, PAR-CLIP, cell fate specification, B-cell development, fetal hematopoiesis, gene regulatory networks, gene expression programs, single-cell RNA sequencing
Abstract
Fetal hematopoietic stem and progenitor cells (HSPCs) hold promise to cure a wide array of hematological diseases, and we previously found a role for the RNA-binding protein (RBP) Lin28b in respecifying adult HSPCs to resemble their fetal counterparts. Here we show by single-cell RNA sequencing that Lin28b alone was insufficient for complete reprogramming of gene expression from the adult toward the fetal pattern. Using proteomics and in situ analyses, we found that Lin28b (and its closely related paralog, Lin28a) directly interacted with Igf2bp3, another RBP, and their enforced co-expression in adult HSPCs reactivated fetal-like B-cell development in vivo more efficiently than either factor alone. In B-cell progenitors, Lin28b and Igf2bp3 jointly stabilized thousands of mRNAs by binding at the same sites, including those of the B-cell regulators Pax5 and Arid3a as well as Igf2bp3 mRNA itself, forming an autoregulatory loop. Our results suggest that Lin28b and Igf2bp3 are at the center of a gene regulatory network that mediates the fetal–adult hematopoietic switch. A method to efficiently generate induced fetal-like hematopoietic stem cells (ifHSCs) will facilitate basic studies of their biology and possibly pave a path toward their clinical application.
During mammalian development, the primary site of definitive hematopoiesis shifts from the fetal liver (FL) to the bone marrow (BM) around the time of birth (Kikuchi and Kondo 2006). The genetic program underlying this developmental transition as well as fetal hematopoiesis is poorly understood. The evolutionary conservation of these two distinct waves of hematopoiesis suggests that it may be important for optimal development of the immune system in mice and humans and should be considered to improve hematopoietic regeneration in the clinic. In current clinical practice, hematopoietic stem cell (HSC) transplantations use healthy adult donors as the source of BM-derived stem cells, consequently bypassing the fetal wave of hematopoiesis and regenerating only the adult layers of hematopoiesis. As a result, the reconstituted immune system in such patients may be missing subsets of cells that normally develop early during life. The recent report that innate lymphoid cells (ILCs) are not reconstituted in severe combined immunodeficiency (SCID) patients treated by nonmyeloablative BM HSC transplantation (Vély et al. 2016) provides an example suggesting that current methods may not be ideal. In the age of precision medicine, primary immunodeficiencies should be diagnosed prior to birth, and the transplantation of FL HSCs in utero may represent an ideal strategy for generating an intact immune system in these patients that would in theory cure them upon birth before major symptoms arise. Such an approach succeeded in sheep and monkeys (Flake et al. 1986; Harrison et al. 1989). However, acquiring human FL HSCs is challenging due to restrictions on obtaining healthy embryos from the second or third trimester (Panikkar et al. 2012). To address this unmet need, we propose a more efficient strategy to reprogram adult HSCs into their fetal-like counterparts, perhaps obviating the need for human FL donors. Specifically, we can induce such a reprogramming more efficiently using two defined factors, Lin28b and Igf2bp3, compared with either alone.
We discovered previously that expression of the cytoplasmic RNA-binding protein (RBP) Lin28b is associated with fetal but not adult hematopoiesis and that it is sufficient for orchestrating the genetic program that specifies fetal hematopoiesis (Yuan et al. 2012). In support of this idea, enforced expression of either Lin28 protein in hematopoietic stem and progenitor cells (HSPCs) from adult BM is sufficient to reactivate fetal-like lymphocyte development. A robust signature of fetal B-cell development is the generation of CD5+ innate-like B cells (referred to as B-1a). In contrast, the BM of adults mainly generate conventional B cells (referred to as B-2) (Yuan et al. 2012; Zhou et al. 2015; Kristiansen et al. 2016). Furthermore, the transduction of LIN28B is also sufficient to induce expression of fetal hemoglobin in adult human erythroblasts (Lee et al. 2013). While it appears that Lin28b is capable of autonomously specifying HSPC fate, the efficiency is not known. Here we took advantage of new RNA sequencing (RNA-seq) technology to assess the extent of transcriptome reprogramming at the single-cell level. Furthermore, molecular mechanisms of its posttranscriptional regulatory activity in hematopoietic cells have not been systematically defined.
Lin28 was first discovered as one of the heterochronic genes regulating Caenorhabditis elegans larval development (Ambros and Horvitz 1984; Moss et al. 1997), and mammals encode two paralogs: Lin28a and Lin28b (we refer to both paralogs together as “Lin28” here unless one of them is specified). However, only Lin28b is expressed in fetal HSPCs (Yuan et al. 2012). The cold-shock domain (CSD) and two zinc fingers (ZnFs) of Lin28 together mediate RNA binding with high affinity and distinct sequence specificity (Nam et al. 2011; Graf et al. 2013). It is well understood that Lin28 posttranscriptionally inhibits the maturation of the microRNA let-7 family (Heo et al. 2008; Newman et al. 2008; Rybak et al. 2008; Viswanathan et al. 2008). Nevertheless, this is unlikely to be its only function, considering that Lin28 proteins have been shown to bind thousands of transcripts and possibly affect their abundance and/or translation (Polesskaya et al. 2007; Xu and Huang 2009; Xu et al. 2009; Cho et al. 2012; Wilbert et al. 2012; Graf et al. 2013; Hafner et al. 2013). However, the Lin28-induced effects reported thus far tended to be marginal. Furthermore, the previously determined mRNA targets of Lin28b do not explain the mechanisms that promote fetal hematopoiesis. We reasoned that its key substrates and/or interacting partners could be specific to cellular context and thus searched for an experimentally tractable system to investigate Lin28b's mechanisms of action in HSPCs. Here we uncover gene regulatory networks (GRNs) connected to Lin28b to elucidate its role in (re)programming hematopoietic cell fate. As a result, we discovered Igf2bp3 to be a novel partner of Lin28b and provide a comprehensive blueprint of the genetic targets downstream from these two RBPs.
Results
A model system to expand the Lin28b GRN
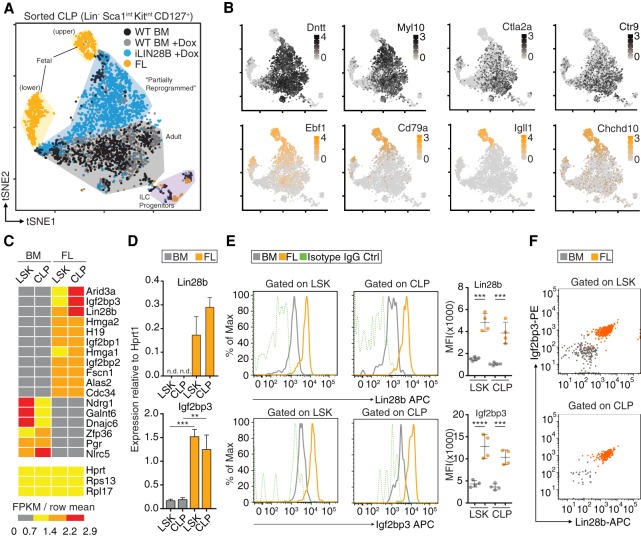
As an in vivo model system to reproducibly generate induced fetal-like HSCs (ifHSCs), we used a mouse engineered to express in a doxycycline (Dox)-inducible manner LIN28B tagged at the N terminus with the Flag epitope (Zhu et al. 2011), referred to here as the iLIN28B mouse (Supplemental Fig. S1A,B). We validated in this system that transgenic Flag-LIN28B protein is expressed in nearly 100% of HSPCs (Supplemental Fig. S1A). We showed previously that ectopic expression of either LIN28A or LIN28B phenotypically confers fetal-like properties to adult HSPCs (Yuan et al. 2012), but its effect on the transcriptome has not been characterized at the single-cell level. To address this, we performed single-cell RNA-seq (scRNA-seq) of common lymphoid progenitor (CLP) cells sorted from mouse FLs, adult BM of iLIN28B mice, and control mice either treated or untreated with Dox (Fig. 1A; Supplemental Table 1). We chose to analyze CLPs because we were particularly interested in how LIN28B might influence lymphoid lineage commitment. t-SNE (t-distributed stochastic neighbor embedding) analysis using the Seurat computational pipeline (Butler et al. 2018) revealed that FL CLPs consisted of two clusters of cells harboring distinct transcriptomes. One of them (the upper cluster) was characterized by the expression of the cell lineage determining transcription factor Ebf1 that is essential for B-cell development and has known function in FL CLPs (Fig. 1B; Lin and Grosschedl 1995; Zandi et al. 2008; Vilagos et al. 2012). In addition, Ebf1's direct target genes, including Cd79a, Igll1, and Chchd10 (Mansson et al. 2012), are also expressed, suggesting that it is functionally active (Fig. 1B). Intracellular fluorescence-activated cell sorting (icFACS) analysis confirmed that a fraction of FL CLPs are Ebf1+ (Supplemental Fig. S1C), consistent with the scRNA-seq result. While iLIN28B CLPs also up-regulate Ebf1 protein compared with adult BM CLPs, the levels are lower than in Ebf1+ FL CLPs (Supplemental Fig. S1C). A similar picture emerged for Hmga2 (Copley et al. 2013), a DNA-binding protein known to be expressed in FL HSPCs but not adult (Supplemental Fig. S1C). On the other hand, adult BM CLPs (±Dox) clustered separately from their FL counterparts and expressed, as expected, adult-specific markers (Fig. 1B), exemplified by Dntt (Benedict et al. 2000) and Myl10 (Oltz et al. 1992). icFACS of terminal deoxynucleotidyl transferase (TdT), the protein encoded by Dntt, confirmed that it is expressed in adult BM but not FL CLPs or progenitor B (pro-B) cells (Supplemental Fig. S1C); in both iLIN28B CLPs and pro-B cells, TdT is partially down-regulated but not as much as in FL. Of main interest in the t-SNE plot, CLPs from iLIN28B mice were in between FL and BM CLPs, suggesting that their transcriptomes underwent LIN28B-induced reprogramming toward FL CLPs (Fig. 1A). To our surprise, almost none of the iLIN28B CLPs fully adopt the FL phenotype. Consequently, we hypothesized that LIN28B may require additional factors in order to efficiently specify the fetal HSPC fate.
Figure 1.
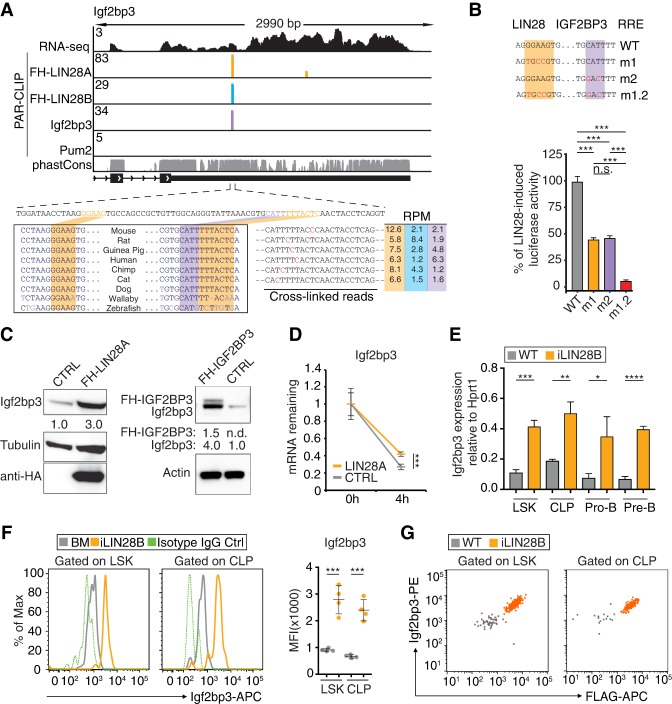
Expanding the Lin28b GRN. (A) The composite t-SNE plot depicts 3832 individual CLP cells color-coded by sample and comprising the following: 858 from adult BM of wild-type (WT) mice (black), 1083 from adult BM of WT mice treated with Dox (gray), 1238 from adult BM of iLIN28B mice treated with Dox (blue), and 653 from FLs of WT mice (orange). A few contaminating cells have been labeled as presumptive ILC progenitors based on gene expression. (B) The t-SNE plots are the same as in A, but color intensities represent log-normalized mRNA levels of the indicated marker genes for adult BM CLPs (gray; top row) and FL CLPs (orange; bottom row) in individual cells. (C) The heat map shows expression patterns of select transcripts from low-input RNA-seq of lineage-negative (Lin−) Sca1+ c-kit+ (LSK) and CLP cells sorted from FL or BM. Three housekeeping transcripts are included as controls. For each transcript, the ratios of FKPM (fragments per kilobase of transcript per million mapped reads) in sample over the row mean are plotted according to color scale. (D) The RT-qPCR analyses quantify Lin28b and Igf2bp3 mRNA expression normalized to Hprt1 in LSK and CLP cells sorted from FL or BM. (n.d.) Not detectable; (**) P ≤ 0.01; (***) P ≤ 0.001, two-tailed t-test. Error bars represent standard deviation of three biological replicates. (E) The histograms show Lin28b and Igf2bp3 expression in LSK and CLP cells from BM (gray) and FL (orange) based on FACS. Isotype IgG controls are also shown (green, dashed). The geometric means of fluorescence intensities (MFI) from biological replicates are plotted. (***) P ≤ 0.001; (****) P ≤ 0.0001, two-tailed t-test. Error bars represent standard deviation of four to five biological replicates. (F) The dot plots quantify Lin28b (X-axis) and Igf2bp3 (Y-axis) protein coexpression in LSK and CLP cells from BM and FL based on FACS. Results are representative of two independent experiments. Dots represent individual cells color-coded by sample.
Lin28b and Igf2bp3 are coexpressed in FL CLPs
To identify candidate Lin28 partners, we searched for transcripts in HSPCs with an expression pattern similar to Lin28b. We could not use our scRNA-seq data for this purpose, since we did not reach the sequencing saturation to detect many transcripts of interest (Supplemental Table 1; Ziegenhain et al. 2017). Thus, we performed bulk RNA-seq of two subpopulations from mouse FL and adult BM: lineage-negative (Lin−) Sca1+ c-kit+ (LSK) cells enriched for HSCs and CLP (Supplemental Table 2). Several factors were coexpressed with Lin28b, most strikingly Igf2bp1-3 (Fig. 1C; Supplemental Fig. S1D), another cytoplasmic RBP family that shares an oncofetal expression pattern with Lin28 (Boyerinas et al. 2008; Bell et al. 2013). We confirmed using RT-qPCR that Lin28b and Igf2bp1-3 were expressed in HSCs, CLPs, pro-B cells, and precursor B (pre-B) cells isolated from FL but not in the corresponding adult BM subsets (Fig. 1D; Supplemental Fig. S1E–H). Furthermore, this mRNA profile translated to elevated Igf2bp3 protein levels in HSPCs from FL compared with BM (Fig. 1E). To confirm at the single-cell level that these two RBPs are coexpressed within the same cell, we performed icFACS (Fig. 1F; Supplemental Fig. S1G). Indeed, the protein expression levels of Lin28b and Igf2bp3 correlated in FL HSPCs. Thus, we hypothesized that Igf2bp3 and Lin28b might collaborate to posttranscriptionally regulate the transcriptome of FL HSPCs.
Since our scRNA-seq results revealed that Lin28b may be involved in B lineage priming during fetal hematopoiesis, we adopted a B-cell-centric focus for the remainder of this study and chose as a model system 220-8, an immortalized mouse pro-B-cell line that provided us sufficient material for our studies (Alt et al. 1981, 1984; Muljo and Schlissel 2003). 220-8 cells were derived from adult BM and do not express either of the two Lin28 paralogs; therefore, we used retroviral vectors encoding human LIN28A or LIN28B tagged at the N terminus with the Flag-HA tandem epitopes (referred to here as 220-8-FH-LIN28A and -FH-LIN28B, respectively) to enforce stable expression and facilitate affinity purification. We determined that FH-LIN28A and FH-LIN28B were expressed at ∼264,000 and ∼66,000 copies per cell, respectively (Supplemental Fig. S2A,B), and were functional, considering that they suppressed mature let-7 microRNA (miRNA) expression (Supplemental Fig. S2C). For most of our in vitro experiments, we used the 220-8-FH-LIN28A cell line whose FH-LIN28A expression level approached endogenous LIN28B in the K562 erythroleukemia cell line, ∼650,000 copies per cell (Hafner et al. 2013).
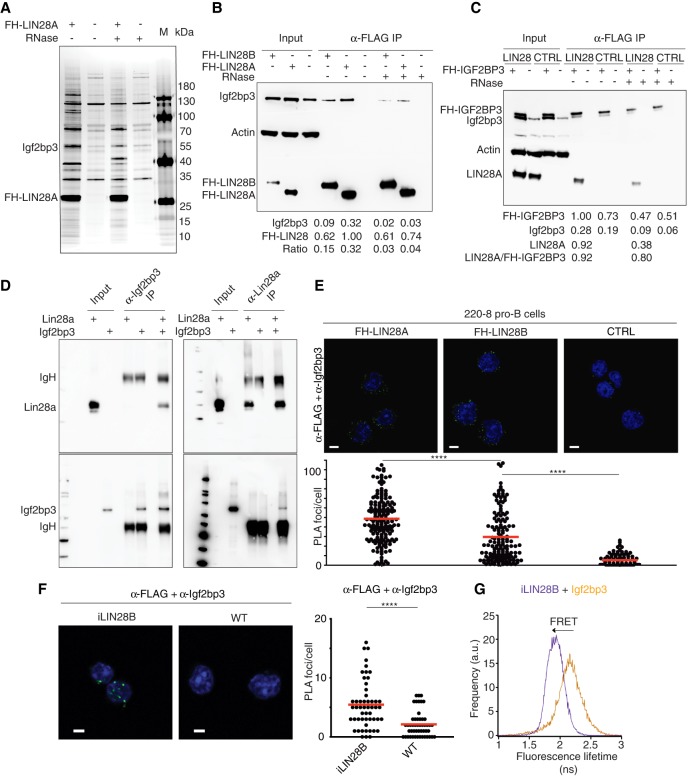
To identify proteins interacting with LIN28, we used mass spectrometry to analyze anti-Flag immunoprecipitates from lysates of 220-8-FH-LIN28A compared with untransduced cells (Fig. 2A; Supplemental Table 3). Interestingly, Igf2bp3 was one of the strongest LIN28-interacting proteins. This suggested that these two RBPs not only were coexpressed in fetal HSPCs but also formed a physical complex. We validated these immunoprecipitation results by Western blotting and found that the LIN28–Igf2bp3 interaction was weakened but persisted after treatment with RNase (Fig. 2B,C), suggesting that LIN28 and Igf2bp3 form a ribonucleoprotein (RNP) complex. Reciprocal immunoprecipitation of FH-IGF2BP3 confirmed its interaction with LIN28A (Fig. 2C). In addition, FH-IGF2BP3 pulled down endogenous Igf2bp3 (Fig. 2C), consistent with the ability of the Igf2bp family to form homodimers and heterodimers (Nielsen et al. 2004). Finally, both recombinant Lin28a and Igf2bp3 proteins purified from Escherichia coli also interacted, suggesting that formation of this complex did not require additional mammalian proteins or RNA (Fig. 2D). Taken together, our results affirmed our earlier hypothesis that Igf2bp3 collaborates with Lin28 proteins.
Figure 2.
Igf2bp3 and Lin28b proteins form an RNP complex. (A) Flag immunoprecipitation (Flag-IP) of lysates treated with or without RNase A from 220-8 pro-B cells transduced with FH-LIN28A (lanes 1,3) or mock-transduced 220-8 cells (lanes 2,4) and marker (M) fractionated by SDS-PAGE and silver-stained. Samples from the same immunoprecipitations were analyzed by mass spectrometry (Supplemental Table 3). (B) Flag-IP of lysates treated with or without RNase A from 220-8 cells either mock-transduced or transduced with FH-LIN28A or FH-LIN28B subjected to Western blotting and probed with anti-Igf2bp3, anti-Actin, and anti-HA antibodies. Background-corrected intensities for the indicated bands are listed below in arbitrary units along with ratios (Igf2bp3/FH-LIN28). Western blotting of corresponding amounts of input material from the same lysates serves as comparison. (C) Western blotting of input and Flag-IP material from lysates from 220-8 cells transduced first with either empty vector (MSCVpuro) or untagged LIN28A and then with either empty vector (MSCVneo) or FH-IGF2BP3 and probed with anti-Igf2bp3, anti-Actin, and anti-LIN28A antibodies. (D) Recombinant Lin28a and Igf2bp3 proteins purified from E. coli were coincubated in vitro, immunoprecipitated, and probed on a Western blot with the indicated antibodies. (E) The in situ proximity ligation assay (PLA) was performed on 220-8 cells transduced with either empty vector (WT), FH-LIN28A, or FH-LIN28B to detect Igf2bp3 proteins and Flag epitopes that are within ∼40 nm of each other. (Top row) Representative confocal microscopic images show fluorescent foci (green) that result from PLA. Nuclei were stained with DAPI (blue). Scale bar, 5 µm. (Bottom row) The number of fluorescent foci per cell is plotted for each sample. At least 90 cells from five random fields were counted per sample. (****) P ≤ 0.0001, Mann-Whitney U-test. (F) PLA was performed on adult BM pro-B cells from iLIN28B mice or WT littermate controls using anti-Flag and anti-Igf2bp3 primary antibodies similar to E. (****) P ≤ 0.0001, Mann-Whitney U-test. (G) Fluorescence lifetime in nanoseconds is plotted in a histogram in the presence of the donor only (anti-Igf2bp3 only; orange) or in the presence of a Förster resonance energy transfer (FRET) partner (anti-Igf2bp3 and anti-Flag costaining; purple) in adult BM pro-B cells from an iLIN28B mouse.
Igf2bp3 and LIN28B interact in situ
Next, we validated the interaction between Igf2bp3 and Lin28 proteins in situ. By confocal microscopy, we confirmed that FH-LIN28A and Igf2bp3 can be stained in cells using antibodies and localize primarily to the cytoplasm (Supplemental Fig. S2D). Proximity ligation assays (PLAs) (Söderberg et al. 2006) in 220-8 pro-B cells further revealed that Igf2bp3 colocalized with FH-LIN28A or FH-LIN28B within ∼40 nm of each other (Fig. 2E; Supplemental Fig. S2E–G; Supplemental Movie 1). In contrast, FH-LIN28A was not in proximity with two other cytoplasmic RBPs that served as specificity controls: Pumilio RNA-binding family member 2 (Pum2) and trinucleotide repeat containing 6A (Tnrc6a; also known as GW182) (Supplemental Fig. S2F,G). We further confirmed in primary pro-B cells from iLIN28B mice that endogenous Igf2bp3 colocalized with transgenic Flag-LIN28B protein (Fig. 2F). Finally, fluorescence lifetime imaging microscopy to measure Förster resonance energy transfer (FLIM-FRET), another nanoscale single-cell technique (Becker 2012), confirmed that LIN28 proteins heterodimerized with Igf2bp3 in 220-8 cells (Supplemental Fig. S2H,I) as well as in primary pro-B cells purified from iLIN28B mice (Fig. 2G).
Lin28b and Igf2bp3 share numerous overlapping binding sites on mRNA targets
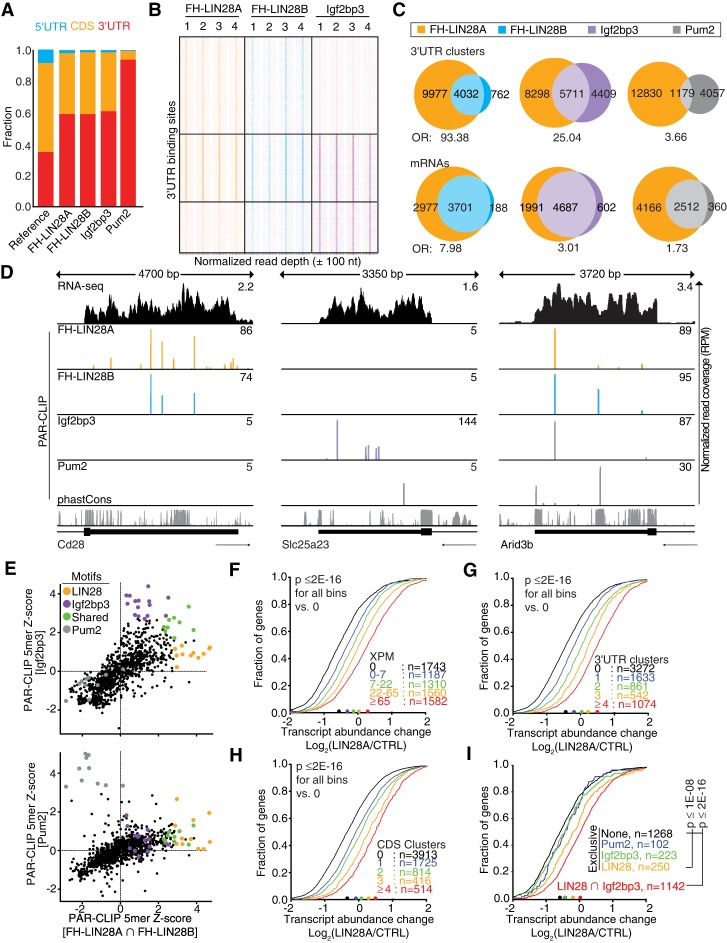
Based on their physical nanoscale proximity in the cytoplasm, we reasoned that Igf2bp3 and Lin28b work together to coregulate shared mRNA targets within the same GRN. Therefore, we mapped Lin28- and Igf2bp3-binding sites transcriptome-wide at nucleotide resolution by photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) (Hafner et al. 2010). Following metabolic labeling with 4-thiouridine (4SU) and UV cross-linking, we isolated RNA covalently linked to endogenous Igf2bp3 from 220-8 cells as well as FH-LIN28A or FH-LIN28B from stable 220-8 cell lines expressing the respective transgenes (Supplemental Fig. S3A). The RNA recovered from four biological replicates of each were converted into cDNA and deep-sequenced (Supplemental Table 4). Sequence reads uniquely aligning to the mouse genome were grouped into clusters by PARalyzer (Corcoran et al. 2011) to identify those enriched for T-to-C mutations, a hallmark of 4SU-mediated cross-linking. As a specificity control, we performed PAR-CLIP for Pum2, a cytoplasmic RBP with a well-defined RNA recognition element (RRE) in 3′ untranslated regions (UTRs) of mRNAs (Wang et al. 2002; Gerber et al. 2006; Hafner et al. 2010).
Most of the binding sites for all four RBPs distributed to exonic mRNA sequences, particularly the 3′ UTR (Fig. 3A; Supplemental Table 5), consistent with their predominantly cytoplasmic localization (Supplemental Fig. S2D). While the FH-LIN28A- and FH-LIN28B-binding profiles, as expected, were similar to each other (Hafner et al. 2013), Igf2bp3 unexpectedly bound a group of sites in common with LIN28 in addition to a distinct set of exclusive targets (Fig. 3B–D). In total, we identified 23,630 and 8099 binding sites for FH-LIN28A and FH-LIN28B, respectively, of which 59% were located in the 3′ UTR and largely overlapped (odds ratio [OR] = 93.38) (Fig. 3C). Surprisingly, 56% of the 10,120 Igf2bp3 3′ UTR-binding sites and 58% of the 6233 Igf2bp3-coding sequence (CDS)-binding sites also overlapped with FH-LIN28A sites (OR = 25.0 and 36.6, respectively) (Fig. 3B,C; Supplemental Fig. S3B). For comparison, although 87% of Pum2 target mRNAs were also bound by LIN28 proteins, only 23% of Pum2 and FH-LIN28A 3′ UTR-binding sites overlapped (OR = 3.66) (Fig. 3C). These results are in agreement with the direct interaction between Lin28b and Igf2bp3 described above (Fig. 2) and provided further evidence for a possible coregulation of mRNAs by these two RBP families.
Figure 3.
Lin28- and Igf2bp3-binding sites overlap extensively across the transcriptome. (A) Distributions of PAR-CLIP binding sites for FH-LIN28A/B, Igf2bp3, and Pum2 across mRNA exonic regions (5′ UTR, CDS, and 3′ UTR) are plotted. The reference bar shows the estimated fraction of each region within the transcriptome. (B) The heat map compares PAR-CLIP sequence read coverage centered (±100 nucleotides [nt]) around 3′ UTR-binding sites of FH-LIN28A (orange), FH-LIN28B (cyan), and Igf2bp3 (purple). Each column represents data from an independent PAR-CLIP experiment (replicates 1–4), and each row represents a specific 3′ UTR segment. Boxes separate distinctive target categories: LIN28-preferred (8352 rows), LIN28–Igf2bp3 shared (5711 rows), and Igf2bp3-preferred (4439 rows). The color intensities vary with PAR-CLIP read depth. (C, top panel) Venn diagrams show FH-LIN28A PAR-CLIP 3′ UTR-binding sites (clusters) overlapping by at least 1 nt with FH-LIN28B, Igf2bp3, and Pum2, respectively. (Bottom panel) Venn diagrams show mRNAs cotargeted by FH-LIN28A and FH-LIN28B, Igf2bp3, or Pum2, respectively. The OR of each overlap is indicated. (D) The genome browser tracks show the last exon for three exemplary mRNAs with LIN28A/B-exclusive (Cd28; left), Igf2bp3-exclusive (Slc25a23; middle), or shared binding sites (Arid3b; right). Binding sites from Pum2 PAR-CLIP are shown as a specificity control. The tracks showing PAR-CLIP sequence read coverage are colored by RBP. (Orange) LIN28A; (cyan) LIN28B; (purple) Igf2bp3; (gray) Pum2. RNA-seq coverage in 220-8 cells and the phastCons evolutionary conservation score for 21 mammalian genomes (Euarchontoglire clade) are shown in the top (black) and bottom (gray) tracks, respectively. (E) The dot plots depict Z-scores for the occurrence of all 1024 possible 5-mers in PAR-CLIP binding sites shared by LIN28A and LIN28B (FH-LIN28A ∩ FH-LIN28B) compared with Igf2bp3-binding sites (top panel) or Pum2-binding sites (bottom panel). The color code indicates 5-mers enriched in Igf2bp3 (purple), LIN28A/B (orange), both (green), or Pum2 PAR-CLIP experiments (gray). The sequences of top-scoring 5-mers are in Supplemental Table 6. (F–I) Cumulative distribution of log-transformed fold changes of mRNA expression comparing LIN28A transduced and empty vector transduced 220-8 cells determined by RNA-seq. FH-LIN28A target mRNAs were binned based on the number of cross-linked reads normalized per million (XPM) (F) or PAR-CLIP binding sites in the 3′ UTR (G) or CDS (H). (I) mRNAs were binned according to PAR-CLIP interactions. (Black) None; (blue line) only Pum2 sites; (green line) only Igf2bp3 sites; (orange line) only LIN28 sites; (red line) shared LIN28 and Igf2bp3 sites. The statistical significance of the shift compared with unbound targets was determined using a two-sided Kolmogorov-Smirnov (KS) test. Bin sizes are indicated.
Next, we aimed to identify the RREs that LIN28 and Igf2bp3 recognized in 220-8 cells and calculated enrichment (Z-score) of all possible pentamer (5-mer) motifs in our LIN28 and Igf2bp3 PAR-CLIP data sets (Fig. 3E; Supplemental Fig. S3C–F; Supplemental Table 6). Consistent with previous reports (Wilbert et al. 2012; Graf et al. 2013; Hafner et al. 2013), we found A/U-rich as well as GGAG-containing 5-mers among the top-scoring LIN28 RREs, corresponding with recognition by the CSDs and ZnF RNA-binding domains, respectively (Supplemental Fig. S3C; Supplemental Table 6; Nam et al. 2011; Graf et al. 2013). While T-to-C mutations indicative of cross-linking enriched within or close to the A/U-rich motifs (Supplemental Fig. S3D), they were found <5 bases away from the G-rich RREs, indicating that the ZnF domains likely are not compatible with UV cross-linking and explaining why the G-rich RREs were recovered in only a minority of binding sites in previous CLIP experiments (Cho et al. 2012; Wilbert et al. 2012; Graf et al. 2013; Hafner et al. 2013). Finally, the top-scoring motifs CAUCA and ACACA for Igf2bp3 and UGUANAUA for Pum2 were consistent with previous reports (Fig. 3E; Supplemental Fig. S3E,F; Supplemental Table 6; Hafner et al. 2010; Patel et al. 2012). Our data indicate that while pro-B cells possibly represent a valuable model system to study the gene regulatory impact of the LIN28 and Igf2bp3 collaboration, the molecular basis for their recruitment to RNA targets does not differ from previously studied cell types.
LIN28 and Igf2bp3 cooperatively stabilize mRNA targets independent of let-7
To quantify the impact of LIN28 binding on its targets, we expanded our analysis transcriptome-wide using RNA-seq by comparing mRNA levels in 220-8 cells transduced with empty vector or LIN28A. LIN28 transduction resulted in a significant increase in expression of its target mRNAs (Supplemental Table 7). This increase was dependent on the number of binding sites or the strength of FH-LIN28A or FH-LIN28B binding, as approximated by the number of cross-linked reads per target mRNA (XPM) (Fig. 3F–H; Supplemental Fig. S3G). The median mRNA abundance for the top FH-LIN28A targets (>65 XPM; n = 1582) increased by 1.8-fold versus only 1.2-fold increase for weakly bound targets (less than seven XPM; n = 1187) (Fig. 3F). The effect on target mRNA abundance was irrespective of the location of binding sites within the transcript, as LIN28 occupancy in either the CDS or 3′ UTR promoted expression (Fig. 3G,H). Indirect mRNA abundance increase by derepression of the 1074 predicted (Agarwal et al. 2015) miRNA let-7 targets may explain these effects for only <10% of LIN28 target mRNAs (Supplemental Fig. S3H,I). While we do not exclude the possibility that let-7 may play a complementary role in this GRN, in summary, our data suggest that let-7 down-regulation does not completely account for the mechanism of action of LIN28B in our cell lines.
We next investigated the extent of the collaboration between LIN28 and Igf2bp3 and whether cotargeted transcripts were more likely to be up-regulated by LIN28. We binned mRNA targets based on whether they were exclusively targeted by LIN28 proteins or Igf2bp3 or contained overlapping Igf2bp3- and LIN28-binding sites and quantified transcript abundance changes upon LIN28A transduction (Fig. 3I; Supplemental Table 7). Igf2bp3-exclusive targets were not significantly affected, similar to Pum2-exclusive targets. We found a modest 35% median increase of exclusive LIN28 targets. In contrast, the shared targets were up-regulated 1.6 times more efficiently (57% median increase) (Fig. 3I). In summary, our analyses are consistent with the notion that the Igf2bp3 and LIN28 families collaborate on a transcriptome-wide scale. The significant increase in target mRNA levels that correlate with LIN28 binding, coupled with the substantial overlap between Igf2bp3- and LIN28-binding sites when compared with LIN28 effects in other cell lines (Wilbert et al. 2012; Graf et al. 2013; Hafner et al. 2013), made the 220-8 cell line a suitable model system to identify coregulated targets with phenotypic impact.
Pax5 is a shared target of Lin28b and Igf2bp3 that is let-7-independent
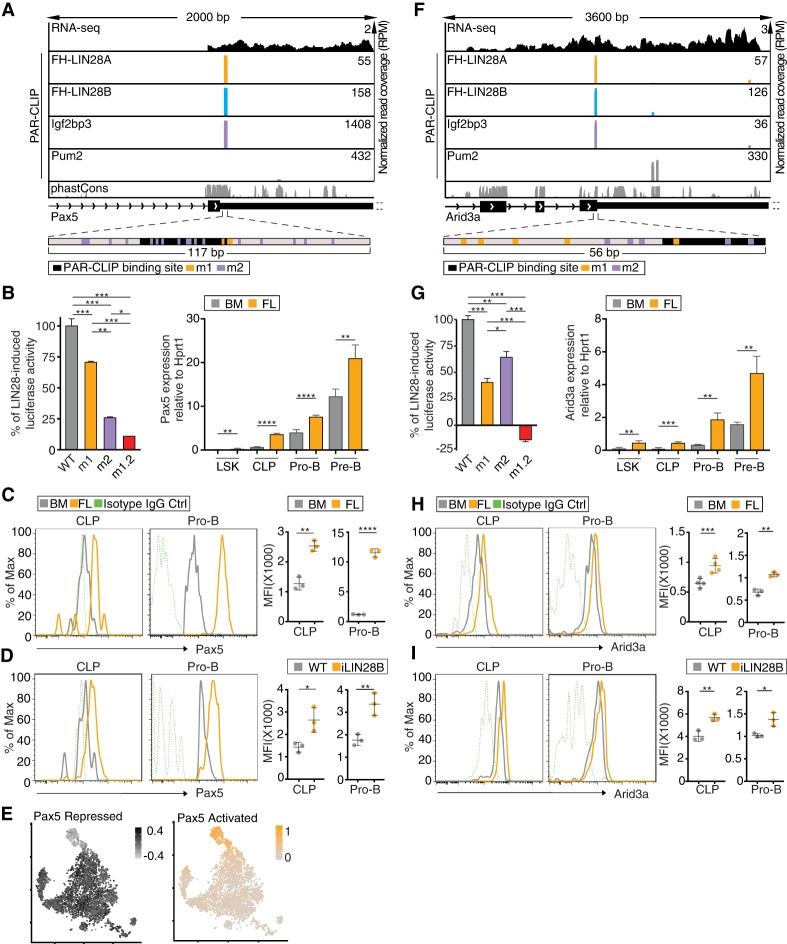
When we inspected the top 2500 shared PAR-CLIP targets, we found “DNA templated transcription” as the most significantly enriched gene ontology (GO) term (Supplemental Fig. S4A), and, among those, Pax5, a B-cell lineage-determining transcription factor, emerged near the top of the list. In previous CLIP experiments, Pax5 mRNA was not identified as a target of either LIN28 or IGF2BP3 because it is specifically expressed in B cells. Pax5 mRNA harbors a prominent binding site shared by LIN28 and Igf2bp3 in its 3′ UTR near the stop codon (Fig. 4A) and is not predicted to be a let-7 target (Agarwal et al. 2015). Thus, this 3′ UTR represented an ideal natural substrate to dissect the direct posttranscriptional regulatory effects of LIN28 and Igf2bp3. A 117-nucleotide (nt) fragment containing the recovered PAR-CLIP sites was fused to Renilla luciferase (Supplemental Fig. S4B) and transfected as a reporter plasmid into stable cell lines inducibly expressing FH-LIN28A. Overexpression of LIN28A increased the reporter gene expression by 33% (Supplemental Table 8), while mutation of the G-rich LIN28 RRE or all potential Igf2bp3 RREs dampened this increase by ∼30% and 75% (m1 and m2, respectively) (Fig. 4B), and double mutation nearly abolished the LIN28A-induced reporter activity (m1.2) (Fig. 4B). The Pax5 mRNA contains several additional shared, albeit weaker, binding sites further downstream in the 3′ UTR (Supplemental Fig. S5A). We also tested luciferase activity for three of these additional sites and observed a similar pattern of reduction in induced reporter activity upon mutation of one or both of the LIN28 or Igf2bp3 RREs (Supplemental Fig. S5B). These data suggest that cobinding by Igf2bp3 is necessary for full LIN28-dependent stimulation of reporter gene expression for multiple fragments of the Pax5 3′ UTR.
Figure 4.
Pax5 and Arid3a mRNAs are shared targets of LIN28 and Igf2pb3. (A,F) Genome browser tracks of binding sites in the Pax5 3′ UTR (A) and Arid3a CDS (F) are similar to Figure 3D. The browser windows are zoomed in on the primary shared binding site, and the 3′ UTRs of both transcripts extend further than depicted. The panels below the genome browser tracks depict the fragment of Pax5 3′ UTR or Arid3a CDS used in the dual-luciferase reporter constructs (shown in B,G). Locations of the binding site and mutations are shown by black (binding site), orange (m1), or purple (m2) bars. (B,G, left panel) Dual-luciferase reporter assays were performed using a sequence from a fragment of Pax5 3′ UTR (B) or Arid3a CDS (G) fused to Renilla luciferase (Rluc) and transfected into stable HEK293 cells inducibly expressing FH-LIN28A. WT indicates a fragment of the Pax5 3′ UTR or Arid3a CDS, m1 includes mutations in the LIN28 RRE, m2 includes mutations in the Igf2bp3 RRE, and m1.2 includes mutations in both. The Rluc signal was measured with or without FH-LIN28A transgene expression (±Dox) and was normalized to the signal from firefly luciferase (Fluc) encoded on the same reporter plasmid. The difference in normalized Rluc signal for WT with and without Dox was set to 100%, and the induced activity for m1, m2, and m1.2 was plotted relative to the WT baseline. Sequences are in Supplemental Table 10. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001, Tukey's HSD after adjustment for batch variation by two-way ANOVA. Bars represent mean ± standard error of mean (SEM) of three independent repeats. (Right panel) RT-qPCR analyses quantify Pax5 and Arid3a mRNA expression normalized to Hprt1 in LSK, CLP, pro-B, and pre-B cells sorted from FLs or BM. (**) P ≤ 0.01; (***) P ≤ 0.001; (****) P ≤ 0.0001, two-tailed t-test. Error bars represent standard deviation of three biological replicates. (C,H) Histograms depict Pax5 (C) and Arid3a (H) expression in CLP and pro-B cells from BM (gray) and FL (orange) based on FACS. Isotype IgG controls are also shown (green, dashed). The MFIs from biological replicates are plotted. (**) P ≤ 0.01; (***) P ≤ 0.001; (****) P ≤ 0.0001, two-tailed t-test. Error bars represent standard deviation of three to four biological replicates. (D,I) Histograms depict Pax5 (D) and Arid3a (I) expression in CLP and pro-B cells from BM of iLIN28B mice (orange) and WT littermate controls fed with Dox (gray). Isotype IgG controls are also shown (green, dashed). The MFIs from biological replicates are plotted. (*) P ≤ 0.05; (**) P ≤ 0.01, two-tailed t-test. Error bars represent standard deviation of three biological replicates. (E) tSNE plots show CLP cells colored by gene expression module scores of Pax5-activated targets (orange) and Pax5-repressed targets (black). Cells are colored according to scale, with darker-colored cells denoting higher aggregated expression of the respective sets of targets.
Next, we examined the pattern of Pax5 expression in vivo. In HSPCs sorted from FLs that have high Lin28b and Igf2bp3 expression, we surprisingly found a greater than threefold higher expression of Pax5 mRNA compared with their adult BM counterparts (Fig. 4B, right panel). Pax5 mRNA expression in iLIN28B BM HSPCs also increased approximately twofold compared with wild-type (WT) littermate controls (Supplemental Fig. S6A). This Pax5 mRNA increase in vivo was correlated with a greater than twofold increase in Pax5 protein content in CLP and pro-B cells from FL versus BM (Fig. 4C) as well as from iLIN28B versus WT mice (Fig. 4D). As evidence that this elevated Pax5 expression in FL CLPs resulted in transcriptional effects, we found a signature of its activity in our scRNA-seq data. Although Pax5 mRNA failed to be detected, we surmised that it should correlate with expression of its target genes, which include Ebf1, Cd79a, and Igll1 (McManus et al. 2011). Thus, the overall expression of a gene set of repressed or activated Pax5 targets (McManus et al. 2011) tended to be low or high, respectively, in the upper cluster of FL CLPs (Fig. 4E). However, we noticed that there was no clear sign of Pax5 activity in the partially reprogrammed iLIN28B CLPs (Fig. 4E), suggesting that Pax5 levels may still be limiting in iLIN28B HSPCs. Therefore, we asked whether additional Pax5 provided by lentiviral transduction could augment LIN28B-mediated reprogramming. Unexpectedly, Pax5 supplementation further increased the frequency of LSK+CD127+ HSPCs (Supplemental Fig. S6C), previously described by Dorshkind and colleagues (Montecino-Rodriguez et al. 2016) to be enriched in FL. Pax5 also increased the frequency of mature B-1a cells in the peritoneal cavity (Supplemental Fig. S6D). These results support the notion that Pax5 induction by iLIN28B alone is suboptimal. Thus, Pax5 exemplifies a heretofore unappreciated shared target of Lin28b and Igf2bp3 that is likely to play a role in fetal B-cell development.
Arid3a is a shared target of Lin28b, Igf2bp3, and let-7
Arid3a and Arid3b are predicted and validated targets of let-7 (Agarwal et al. 2015; Liao et al. 2016). Since we noticed that both Arid3a and Arid3b are also shared targets of LIN28 and Igf2bp3 (Figs. 3D [right panel], 4F), we constructed luciferase reporters to test the possibility that these two RBPs regulate Arid3a expression in addition to the previously reported role of let-7 (Supplemental Fig. S4B; Zhou et al. 2015). Overexpression of LIN28A increased Arid3a reporter gene expression, while mutation of the LIN28 or Igf2bp3 RREs dampened this increase, and double mutation completely abolished the induction of reporter by LIN28 (Fig. 4G, left panel). These data indicate that cobinding by Igf2bp3 in cis is necessary for full LIN28-dependent stimulation of reporter gene expression. As expected, in vivo, Arid3a mRNA and protein levels are higher in FL versus BM HSPCs (Fig. 4G [right panel], H) and are induced in iLIN28B mice (Supplemental Fig. S6B; Fig. 4I).
Next, we asked whether, like Pax5, Arid3a levels may still be limiting in iLIN28B HSPCs. Indeed, lentiviral Arid3a supplementation further increased LSK+CD127+ HSPCs and mature B-1a cells (Supplemental Fig. S6E,F), suggesting that enhancement of Arid3a expression by iLIN28B alone is suboptimal, and hypothesized that Igf2bp3 is necessary for full induction. Thus, both Arid3a and Pax5 transcripts are shared targets of LIN28B and Igf2bp3 and may in part explain the profound effects of this pair of RBPs on hematopoiesis.
LIN28 and Igf2bp3 levels are interdependent and regulated by a feed-forward loop
Beyond the widespread binding of LIN28 and Igf2bp3 to the same sites (Fig. 3B,C), we also observed autoregulation and cross-regulation between LIN28 and Igf2bp3. As reported previously, both LIN28A and LIN28B bound their own mRNAs, resulting in increased protein levels (Wilbert et al. 2012; Hafner et al. 2013). Furthermore, LIN28 and Igf2bp3 bound a deeply conserved site in the Igf2bp3 mRNA 3′ UTR (Fig. 5A), and we used reporter assays to separate direct LIN28-mediated effects from the previously reported indirect regulation via suppression of let-7 (Boyerinas et al. 2008; Zhu et al. 2011). We fused 131 nt of the Igf2bp3 3′ UTR containing the LIN28- and Igf2bp3-binding sites to Renilla luciferase (Supplemental Fig. S4B) and transfected the resulting plasmid into stable cell lines inducibly expressing FH-LIN28A. FH-LIN28A induction led to an ∼60% increase in reporter gene expression (Supplemental Table 8), and point mutations of the G-rich RRE (m1) (Fig. 5B) significantly dampened the effect of LIN28 on the reporter. Mutation of the Igf2bp3 RRE also reduced reporter expression (m2) (Fig. 5B), while the mutation of both RREs reduced it even further (m1.2) (Fig. 5B), suggesting that LIN28 and Igf2bp3 binding in cis results in cooperative regulation. Indeed, we found that in mouse 220-8 cell lines, enforced expression of either LIN28 or IGF2BP3 resulted in up-regulation of endogenous Igf2bp3 mRNA and protein (Fig. 5C). Furthermore, the half-life of endogenous Igf2bp3 mRNA increased upon transduction of LIN28A (Fig. 5D). In vivo, endogenous Igf2bp3 mRNA and protein levels also increased in iLIN28B HSPCs (Fig. 5E,F). Furthermore, Igf2bp3 levels correlated with transgenic LIN28B expression levels (Fig. 5G). In summary, these findings indicate that LIN28 and Igf2bp3 levels are autoregulated as part of a feed-forward positive feedback loop.
Figure 5.
Igf2bp3 and Lin28 proteins regulate Igf2bp3 expression in vitro and in vivo. (A) The genome browser tracks of the Igf2bp3 3′ UTR are formatted as in Figure 3D. (Bottom left panel) The alignment shows sequence conservation across the indicated vertebrates. The predicted LIN28 (orange) and Igf2bp3 (purple) RREs are highlighted. (Bottom right panel) Representative examples of common sequence reads from FH-LIN28A (orange), FH-LIN28B (cyan), and Igf2bp3 (purple) PAR-CLIP experiments are shown, with T-to-C mutations highlighted in red along with the sequence coverage in reads per million (RPM) color-coded by RBP. (B, top panel) Sequences of the WT RRE for the LIN28 ZnF domain and IGF2BP3 are shown. Mutations are indicated in red. (Bottom panel) Dual-luciferase reporter assay testing a fragment of the Igf2bp3 3′ UTR was performed as described in Figure 4C. (***) P ≤ 0.001, Tukey's HSD after adjustment for batch variation by two-way ANOVA. Bars represent mean ± SEM of three independent repeats. (C, left panel) Western blot of lysates from 220-8 cells transduced with empty vector (CTRL) or FH-LIN28A probed with anti-Igf2bp3, anti-Tubulin, and anti-HA antibodies. Quantitation of background-corrected intensities of the Igf2bp3 band normalized to tubulin is shown below the top panel. (Right panel) Western blo† of lysates from 220-8 cells transduced with empty vector (CTRL) or FH-IGF2BP3 probed with anti-Igf2bp3 and anti-Actin antibodies. Quantitation of background-corrected intensities of exogenous and endogenous Igf2bp3 bands normalized to actin is shown below the top panel. (D) RT-qPCR analyses quantify the fraction of Igf2bp3 mRNA remaining, normalized to U6 in 220-8 cell lines transduced with empty vector (CTRL) or untagged LIN28A after actinomycin D treatment for 4 h. (***) P ≤ 0.001, two-tailed t-test. Error bars represent standard deviation of three biological replicates. (E) RT-qPCR analyses quantify Igf2bp3 mRNA expression, normalized to Hprt1 in LSK, CLP, pro-B, and pre-B cells sorted from BM of iLIN28B mice and WT littermate controls fed with Dox. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001; (****) P ≤ 0.0001, two-tailed t-test. Error bars represent standard deviation of three biological replicates. (F) Histograms show Igf2bp3 expression in LSK and CLP cells from BM of iLIN28B mice (orange) and WT littermate controls fed with Dox (gray). Isotype IgG controls are also shown (green, dashed). The MFIs from biological replicates are plotted. (***) P ≤ 0.001, two-tailed t-test. Error bars represent standard deviation of four biological replicates. (G) The dot plots quantify transgenic Flag-LIN28B (X-axis) and endogenous Igf2bp3 (Y-axis) expression levels in LSK and CLP cells from BM of iLIN28B mice (orange) and WT littermate controls fed with Dox (gray). Results are representative of three biological replicates. Dots represent individual cells color-coded by sample.
Lin28b and Igf2bp3 cooperate in vivo
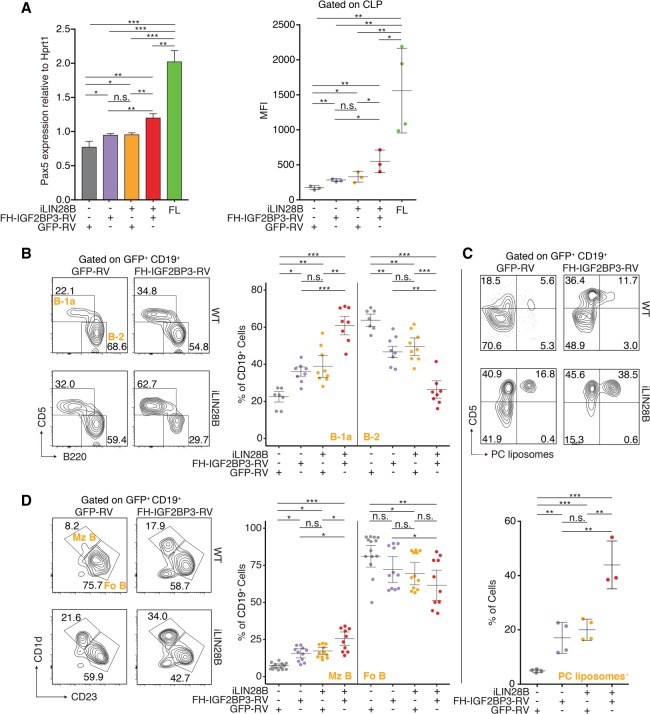
We found previously that LIN28B was sufficient to reprogram adult BM HSPCs to reactivate their potential for fetal-like B lymphopoiesis (Yuan et al. 2012) but hypothesized that additional factors exist to enhance LIN28B-induced hematopoietic reprogramming. Our extensive body of data nominated Igf2bp3 as a prime candidate. Therefore, we isolated HSPCs from WT and iLIN28B mouse BM and transduced them with empty vector (GFP-RV) or FH-IGF2BP3-RV. We then transplanted the transduced HSPCs into Rag1-deficient recipient mice. After 4 wk, we sorted GFP+ CLPs from the recipient mice and performed RT-qPCR to quantify Pax5 mRNA and icFACS to quantify Pax5 protein levels. We saw that LIN28B and IGF2BP3 cooperated in increasing Pax5 levels at both mRNA and protein levels but were not able to reach FL levels (Fig. 6A). We also assessed B-1a and marginal zone B (Mz B) cell development postreconstitution following transplantation into Rag1-deficient mice. Transduction of FH-IGF2BP3 alone increased the frequency of LSK+CD127+ HSPCs (Supplemental Fig. S6G), mature B-1a cells (Fig. 6B,C), and Mz B cells (Fig. 6D) to an extent comparable with LIN28B induction. Enforced expression of both factors, LIN28B and FH-IGF2BP3, resulted in an approximately threefold increase of LSK+CD127+ HSPCs (Supplemental Fig. S6G) and mature B-1a cell frequency (Fig. 6B,C) and a doubling of Mz B cell frequency (Fig. 6D) over WT controls, which constitutes a greater effect than either factor alone. A fraction of the induced B-1a cells bind phosphocholine (PC) via their membrane-bound immunoglobulin M (IgM), a signature function of native B-1a cells (Fig. 6C). In this regard, enforced expression of LIN28B and IGF2BP3 resulted in a greater percentage of PC binding among the induced B-1a cells compared with either factor alone (Fig. 6C). Overall, this expansion in B-1a and Mz B cells mirrored a depletion of B-2 cells, indicating that B lymphopoiesis in these adult mice shifted to a fetal program in a similar but enhanced manner, as we reported previously using LIN28 alone (Yuan et al. 2012). Taken together, our in vivo data further support the idea that LIN28B and IGF2BP3 collaborate, most likely by joint binding of target mRNAs, resulting in their increased levels (Fig. 7A,B).
Figure 6.
LIN28B and IGF2BP3 cooperation in adult BM HSPCs enhances fetal-like B-cell development. (A) Adult BM HSPCs from WT and iLIN28B mice were transduced with empty vector (GFP-RV) or FH-IGF2BP3-RV and transplanted into Rag1−/− recipient mice. GFP+ CLPs were sorted from the recipient mice after 4 wk. (Left panel) RT-qPCR was used to quantify Pax5 mRNA expression. (Right panel) icFACS was used to quantify Pax5 protein levels. The Pax5 mRNA and protein levels in FL CLP are also shown as positive controls. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001, two-tailed t-test. Error bars represent standard deviation of three to four biological replicates. (B) Adult BM HSPCs from WT and iLIN28B mice were transduced with empty vector (GFP-RV) or FH-IGF2BP3-RV and transplanted into Rag1−/− recipient mice. (Left panel) The dot plots represent FACS analyses of B-1a and B-2 cells in the peritoneal cavity of BM chimeric mice 4–6 wk after transplantation gated on transduced B cells (GFP+ CD19+). (Right panel) Frequencies of B-1a and B-2 cells from three independent rounds of transplantation are plotted. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001, Tukey's HSD after adjustment for batch variation by two-way ANOVA. Error bars represent the mean ± SEM. (C, top panel) The dot plots represent FACS analyses of B-1a cells as described in A and quantify the percentage of cells that recognize PC. (Bottom panel) Frequencies of B-1a cells that recognize PC are plotted. (**) P ≤ 0.01; (***) P ≤ 0.001, two-tailed t-test. Error bars represent standard deviation of three to four biological replicates. (D, left panel) The dot plots represent FACS analyses of Mz B (B220+ CD1d+ CD23−ve) and follicular B-2 (Fo B; B220+ CD1d−ve CD23+) cells among transduced splenic B cells in BM chimeras. (Right panel) Frequencies of Mz B and follicular B-2 cells from three independent rounds of transplantation are plotted. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001, Tukey's HSD after adjustment for batch variation by two-way ANOVA. Error bars represent the mean ± SEM.
Figure 7.
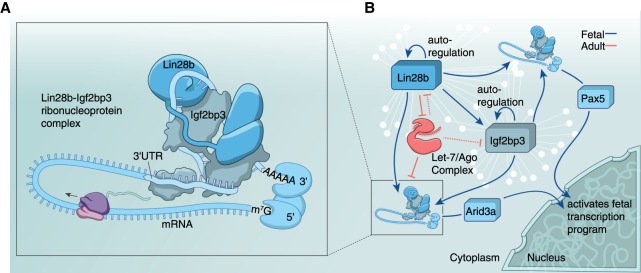
Lin28b and Igf2bp3 collaborate to activate the fetal transcription program by cobinding cognate mRNAs. (A) Graphical model (not drawn to scale) depicts how Lin28b and Igf2bp3 might collaborate by binding to the 3′ UTR of a cognate mRNA. Note that the stoichiometry of proteins within this RNP complex is not known. (B) A schematic of the molecular circuit in fetal HSPCs regulated by Lin28b and Igf2bp3 (blue represents fetal association) is based on the presented results suggesting that miRNA let-7 repression by Lin28b is only one aspect of this GRN. In adult HSPCs, let-7 (red represents adult association) is predicted to shut down this fetal network. White lines and nodes in the background represent the GRN comprised of thousands of transcripts bound by Lin28b and Igf2bp3. Pax5 and Arid3a are examples of such coregulated targets, which are transcription factors that transmit a molecular cascade from the cytoplasm into the nucleus to mediate a program of gene transcription.
Discussion
In summary, Lin28b can perform at least two biochemically distinct functions. By inhibiting let-7 biogenesis, it can indirectly cause derepression of a relatively small suite of mRNAs that includes Lin28b, Igf2bp3, and Arid3a, which are also predicted targets of let-7 (Agarwal et al. 2015). However, in an in vivo model system, while transgenic LIN28B protein levels in HSPCs from iLIN28B mice were comparable with the 220-8-FH-LIN28B cell line, let-7g levels decreased only modestly in primary CLP and pro-B cells (Supplemental Fig. S1A,B). This raises the question of whether in vivo effects of LIN28B are only dependent on repression of let-7. It has been well-documented that LIN28 proteins directly bind mRNA (Cho et al. 2012; Wilbert et al. 2012; Graf et al. 2013; Hafner et al. 2013); however, in all of these studies, the impact of LIN28 on the transcriptome remained limited, and thus no clear biological function for these LIN28-binding events emerged. In our study, we had the opportunity to functionally validate our targets in vivo. Possibly, in previous studies, the direct posttranscriptional function of LIN28 was studied in the wrong cellular context with missing crucial targets and/or cofactors. It is even possible that in a transformed cellular context, LIN28 and IGF2BP proteins oppose each other (Chatterji et al. 2018). Recently, Huang et al., (2018) reported that IGF2BP3 recognizes N6-methyladenosine (m6A) within the consensus GG(m6A)C motif and mediates promotion of mRNA stability and translation. This is consistent with our finding that shared LIN28–Igf2bp3 targets are stabilized. We report Pax5 mRNA to be one example that is independent of let-7 regulation, which also illustrates how Lin28b and Igf2bp3 cooperate to provide an interface between posttranscriptional and transcriptional regulatory networks to help orchestrate developmental programs (Fig. 7A,B). Interestingly, it was noted previously that in the absence of Pax5, the B-cell developmental block is earlier in the FL than in adult BM (Nutt et al. 1997), which might be due to the earlier onset of Pax5 expression that we observed in the FL (Fig. 4C,D). The specification of B-cell fates by transcription factors is well characterized, but the role of posttranscriptional regulation must be integrated with existing models of GRNs to provide a more realistic and predictive framework.
Our study demonstrated how RBPs can act as molecular switches to control cell fate specification, which is a broadly applicable principle. For instance, LIN28 can serve as one of four factors used to generate human induced pluripotent stem cells (Yu et al. 2007). However, the role of LIN28 in this somatic-to-embryonic stem cell reprogramming is not completely understood. Our work predicts that IGF2BP3 or a family member will partner with LIN28 in this instance also. It will be interesting to determine which mRNA targets are shared by IGF2BP3 and LIN28 in embryonic stem cells to mediate pluripotency. Previously, we found that LIN28B can induce fetal hemoglobin expression in adult human erythroblasts (Lee et al. 2013). Again, we predict that IGF2BP3 or a family member plays an important role in this instance too. Indeed, a role for a paralog, IGF2BP1, in activating fetal hemoglobin was reported recently (de Vasconcellos et al. 2017). The ability to induce fetal hemoglobin in adult erythroblasts is of medical interest, since it can ameliorate symptoms in β-thalassemia and sickle cell disease. Finally, one major impediment to performing in utero HSC transplantation is a source of healthy human FL donors. The ability to efficiently generate ifHSCs from adult BM could eventually bypass the need for human embryos and brings us one step closer to curing congenital hematological diseases in neonates before life-threatening consequences have had a chance to manifest. Furthermore, we predict that in utero hematopoietic reconstitution will better mimic natural ontogeny.
Materials and methods
Experimental procedures
Mice
All mice were maintained under specific pathogen-free conditions and treated in accordance with National Institute of Allergy and Infectious Diseases and Animal Care and Use Committee guidelines. C57BL/6N mice were purchased from Taconic. Embryonic day 16.5 (E16.5) fetuses were isolated from timed pregnant C57BL/6N mice. The Dox-inducible iLIN28B mice (Jackson Laboratory, stock no. 023911) (Zhu et al. 2011) were backcrossed to C57BL/6N for six to eight generations. To induce expression of iLIN28B, 4-wk-old mice were fed ad libitum sterile food pellets containing 200 mg of Dox per kilogram of diet (Bio Serv) for 4 wk. Littermates bearing rtTA*M2 but not tetO-LIN28B were also treated with Dox and used as WT controls alongside iLIN28B in experiments.
Cell sorting and scRNA-seq
FLs were harvested from E16.5 embryos and mechanically disrupted to obtain single-cell suspension. For BM cell isolation, femurs and tibias were cut from both ends, and the BM was flushed into a 15-mL centrifuge tube with 5% FCS (HyClone) in PBS followed by centrifugation at 300g for 10 min at 4°C. Reticulocytes were lysed using 1 mL of red blood cell lysis buffer (155 mM NH4Cl, 12 mM NaHCO3, 0.1 mM EDTA) for 5 min at room temperature and neutralized with RPMI 1640 cell culture medium (with 10% FCS). After centrifugation at 300g for 5 min at 4°C, the supernatant was discarded, and cells were resuspended in RPMI 1640 cell culture medium (with 10% FCS). All cells were filtered through a 70-µm nylon mesh. To enrich progenitor cells, cells expressing lineage markers were depleted. Cells are magnetically labeled with an antibody cocktail of biotinylated antibodies against so-called lineage markers for 10 min at 4°C followed by labeling with antibiotin MicroBeads for 15 min at 4°C. Cell suspension was loaded into a MACS mass spectrometry column (Miltenyi Biotec) after washing with 1 mL of PBS twice, and unlabeled cells passing through the column were collected (Lin− fraction). Lineage-depleted cells were stained for 30 min at 4°C with fluorochrome-conjugated monoclonal antibodies (Supplemental Table 9) at 1:100 dilution. Cell sorting was performed on a FACSAria III (BD Biosciences). LSK and CLP (Lin−Sca-1intc-Kitint CD127+) populations were harvested for molecular analyses.
Following FACS, suspensions of single cells were processed using the Chromium single-cell 3′ v1 chemistry multiplex kit (10X Genomics) following the manufacturer's instructions. Libraries were pooled in equal molar ratio, denatured, diluted to 10 pM, and sequenced using Illumina HiSeq2500 rapid mode twice at 50 cycles each (HiSeq Rapid PE cluster kit V2 and HiSeq Rapid SBS kit V2) to result in the following read lengths: 98 base pairs (bp) for read 1, 14 bp for i7 index, 8 bp for i5 index, and 10 bp for read 2.
Antibodies
For the antibodies used, see Supplemental Table 9.
FACS
Cells were stained for 30 min on ice with fluorochrome-conjugated antibodies (Supplemental Table 9). For FL staining, anti-CD11b/Mac-1 (M1/70) was omitted from the Lin cocktail (Morrison et al. 1995). For intracellular staining following surface staining, 1 mL of freshly prepared fixation/permeabilization (eBioscience) working solution was added to each sample and incubated for 60 min at 4°C in the dark. Cells were washed twice with 1 mL of permeabilization buffer. Antibodies to intracellular markers were diluted 1:100 in permeabilization buffer, and cells were incubated for 30 min at 4°C in the dark followed by two washes with 1 mL of permeabilization buffer. Analyses used either LSR II or FACSAria instruments (BD Biosciences). Data were collected for 0.2 × 106 to 3 × 106 cells and analyzed using FlowJo software (TreeStar).
RNA isolation
RNA was isolated from FACS-sorted cells using the NucleoSpin RNA XS kit (Takara Bio) per the manufacturer's instructions. RNA from 220-8 cells was isolated using the TRIzol reagent per the manufacturer's instructions (Thermo Fisher Scientific). RNA was quantitated using the Qubit RNA HS assay kit (Thermo Fisher Scientific).
Low-input RNA-seq
Low-input RNA-seq library preparation was performed per the manufacturer's instructions using the SMARTer stranded total RNA-seq Pico input mammalian kit (Takara Bio). cDNA libraries were sequenced twice for 75 cycles with index on a NextSeq500 instrument (Illumina).
DNA oligonucleotides
For the DNA oligonucleotides used, see Supplemental Tables 10 and 11.
RT-qPCR
First-strand cDNA was synthesized from 1000 ng of total RNA using the SuperScript III first strand synthesis supermix (Thermo Fisher Scientific) with oligo(dT) primers. Predesigned dual-labeled qPCR probes and primers for mRNA quantification were ordered from Integrated DNA Technologies (see Supplemental Table 11 for sequences). Real-time PCR was performed using the KAPA probe fast 2× master mix (KAPA biosystems) in a 7900HT Fast real-time PCR instrument (Applied Biosystems). TaqMan miRNA assays for let-7 family members and U6 snRNA (Rnu6) were purchased from Applied Biosystems and used according to the manufacturer's instructions. CT values from triplicate samples were averaged, and relative expression was calculated using the 2−ΔΔCT method.
Cell lines
The 220-8 mouse pro-B-cell line is a v-Abl-transformed clone derived from adult BALB/c BM with nonproductive VDJH rearrangements on both alleles (Alt et al. 1981). They were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum (HyClone), 2 mM L-glutamine, 100 µg/mL penicillin, 100 µg/mL streptomycin, and 50 µM 2-mercaptoethanol at 37°C in an atmosphere of 5% CO2.
Dox-inducible HEK293 cells expressing FH-LIN28A or FH-LIN28B were described previously (Hafner et al. 2013) and cultured in DMEM high glucose containing 10% FCS, 2 mM glutamine, 15 µg/mL blasticidin, and 100 µg/mL hygromycin. FH-LIN28A or FH-LIN28B expression was induced by addition of 1 µg/mL Dox to the cell medium for at least 16 h prior to harvesting.
Plasmids
FH-LIN28A and FH-LIN28B were amplified by PCR from cDNA clones provided by Markus Hafner (Hafner et al. 2013) followed by restriction digest with BglII and XhoI and ligation into pMSCVpuro (Clontech). FH-IGF2BP3 was PCR-amplified from cDNA clones provided by Markus Hafner (Hafner et al. 2010) and subcloned into pMSCVneo (Clontech) and pMSCV-IRES-GFP provided by Ken Murphy (Ranganath et al. 1998). Plasmid inserts were verified by sequencing (Macrogen).
Mouse mLin28a (mLin28a) plasmid (pET21b; Novagen) was constructed by Andrew Avery and Kannan Natarajan (Laboratory of Immunology, National Institutes of Health). The plasmid would translate to a mLin28a protein with a C-terminal glycine linker, TEV cleavage site, and hexahistidine tag. mIgf2bp3-GST plasmid was obtained from GeneCopoeia. Both plasmids were confirmed by sequencing (Macrogen).
To construct the psiCheck-2 (Promega) target luciferase reporter plasmids, we cloned a synthetic 117-bp fragment from the 3′ UTR of Pax5, a 56-bp fragment from Arid3a CDS, and a 131-bp fragment from Igf2bp3 3′ UTR, respectively. The fragments were inserted downstream from the Renilla luciferase gene using the XhoI and NotI restriction sites of the psiCheck-2 vector. Variants of the Pax5, Arid3a, and Igf2bp3 3′ UTR fragments with point mutations in the LIN28 and Igf2bp3 RREs were also synthesized and cloned into the psiCheck-2 vector using the above strategy (see Supplemental Table 10 for sequences).
Recombinant Lin28 and Igf2bp3 protein expression and purification
The mLin28a and Igf2bp3 plasmids were transformed into E. coli strain BL21cells (New England Biolabs) and cultured in LB (VWR) with 50 µg/mL ampicillin (BioGold) with shaking at 200 rpm at 37°C. Overnight cultures were split 1:5 (v/v) with fresh LB and grown at 200 rpm and 37°C until an O.D of 0.6–0.8. Recombinant protein expression was induced with 300 µM IPTG (BioGold) at 200 rpm for 4 h at 37°C. The cultures were harvested by centrifugation at 3000g for 20 min at 4°C, and the pellets were stored at −80°C.
Cell pellets containing overexpressed mLin28a were thawed at room temperature and resuspended in 30 mL of BugBuster (EMD Millipore) supplemented with Benzonase nuclease (EMD Millipore), rLysozyme solution (EMD Millipore), 0.5 mM PMSF (Sigma Aldrich), and cOmplete Mini EDTA-free protease inhibitor (Roche). The cell suspension was rocked slowly for 30 min at room temperature and centrifuged at 15,000g for 20 min at 4°C, and the supernatant was applied to a column (1.5 I.D. × 20 cm) containing 5 mL of HisPur Ni-NTA resin (Thermo Fisher Scientific) pre-equilibrated with cold running buffer (RB: 50 mM Tris at pH 8.0, 1 mM DTT, 100 µM ZnCl2). The Ni-NTA resin with the bound mLin28a was washed with five column volumes (CVs) of cold RB followed by five CVs of cold RB supplemented with 90 mM imidazole (Sigma Aldrich). The mLin28a was eluted from the Ni-NTA resin with cold RB containing 200 mM imidazole, and fractions of mLin28a were pooled. The mLin28a was further enriched with anion exchange (Hi-Trap Mono Q, GE Healthcare) and size exclusion (Sephadex-75, GE Healthcare) chromatography. mLin28a was concentrated with an Amicon Ultra 15 centrifugal filter with 10 K molecular weight cutoff (MWCO) (Millipore-Amicon) and the protein levels were determined with the Bio-Rad protein stain (Bio-Rad) compared with BSA standards (Sigma Aldrich). The purified mLin28a samples were aliquoted into 100-µL samples and stored at −80°C. Each step of the purification was monitored with SDS-PAGE, and protein enrichment was confirmed with Western blot analysis.
Cell pellets containing overexpressed mIgf2bp3-GST were thawed at 4°C and resuspended in 20 mL of cold RB2 (50 mM Tris at pH 8.0, 50 mM NaCl, 0.5 mM PMSF, complete EDTA-free protease inhibitor). This suspension was sonicated (Ultrasonics) for 6 min (5 sec on/5 sec off cycle, 0.25-in probe, 60% amplitude) at 4°C. During sonication, the sample tube was suspended in an ice slurry, with the lysate resuspended between every pulse. The disrupted cells were centrifuged at 15000g for 20 min at 4°C, and a clear supernatant was slow-tumbled with 3 mL of glutathione agarose resin (BPBio) pre-equilibrated with cold RB2 for 4 h at 4°C. The glutathione agarose resin with the bound mIgf2bp3-GST was spun down at 1000 rpm for 2 min at 4°C, and the supernatant was discarded. The resin with bound resin was resuspended in 50 mL of cold RB2 and spun down at 1000 rpm for 2 min at 4°C, and the supernatant was discarded. This was repeated one more time, and the washed resin with bound GST-tagged proteins were resuspend in 20 mL of cold RB2 and transferred to a column (1.5 I.D. × 20 cm). The resin with bound protein was washed with five CVs of cold RB2 followed by five CVs of cold RB2 supplemented with 0.5 mM reduced L-glutathione (BPBio). The bound protein was eluted from the column with cold RB2 containing 20 mM L-glutathione, and fractions containing mIgf2bp3-GST proteins were pooled. The buffer was exchanged (to 50 mM Tris at pH 8.0), and the mIgf2bp3-GST proteins were concentrated with Amicon Ultra 15 centrifugal filter with a 30 K MWCO (Millipore-Amicon). Protein amounts were determined using Bradford assay (Bio-Rad) using BSA as the standard (Sigma Aldrich). The purity of each sample was verified with SDS-PAGE and confirmed with Western blot analysis.
Binding studies of Lin28a and Igf2bp3-GST
Purified proteins (mLin28a or mIgf2bp3-GST) were slowly thawed at 4°C just prior to use. The binding reactions (50 µL) used 2 µg of mLin28a and/or 2 µg of the mIgf2bp-GST proteins added to RIPA buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 2 mM EDTA, 0.5% NP40). The reaction was initiated by transferring the reaction mixture for 1 h to 37°C (mixing every 10 min). The 50-µL reaction mixture was transferred to a tube containing 1 mL of RIPA buffer with 50 µL of Protein A Dynabeads (Thermo Fisher) and 2 µg of immobilized antibodies (Supplemental Table 9). The mixture was slowly tumbled for 1 h at 4°C and cleared as before, careful not to disturb the Dynabead pellet. After the last clearing, the tubes were centrifuged at 1000 rpm for 30 sec at 4°C and placed on the magnet, and the remaining supernatant was removed. Fifty microliters of 1× SDS PAGE (Invitrogen) loading buffer with DTT was added to the Dynabead pellet, which was then denatured for 3 min at 95°C for SDS-PAGE or stored at −80°C.
Retrovirus and lentivirus production
To make retroviral particles, a 10-cm culture dish of 80% confluent Platinum-E retroviral packaging cells (Cell Biolabs) was transfected in OptiMEM (Life Technologies) using 10 µg of the MSCV-based plasmid, 25 µL of P3000 (Life Technologies), and 25 µL of Lipofectamine 3000 (Life Technologies). After 6 h, culture medium was changed to DMEM (Life Technologies) with 10% FBS (Hyclone). After 48 h, viral supernatants were harvested and centrifuged at 1500 rpm for 5 min at 4°C to pellet cell debris. The supernatant was filtered through a 0.45-µm low-protein-binding membrane (Millipore) and used immediately or stored at −80°C until use.
To make lentiviral particles, pLenti-GFP, pLenti-Pax5, and pLenti-Arid3a (see “Plasmids”) were cotransfected with packaging plasmids pMD2.G and psPAX2 (Addgene plasmids 12259 and 12260) as described previously (Shalem et al. 2014). Briefly, a 10-cm culture dish of 80% confluent HEK293T cells was transfected in OptiMEM (Life Technologies) using 10 µg of the pLenti plasmid, 5 µg of pMD2.G, 7.5 µg of psPAX2, 25 µL of P3000 (Life Technologies), and 25 µL of Lipofectamine 3000 (Life Technologies). Lentiviruses were harvested as above.
Mass spectrometry
Approximately 5 × 107 FH-LIN28A and Puro (empty vector) 220-8 cells were washed three times in ice-cold PBS and lysed in 1× NP40 lysis buffer (50 mM HEPES at pH 7.5, 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5% [v/v] NP40, 0.5 mM DTT, complete EDTA-free protease inhibitor cocktail [Roche]) for 10 min on ice. Cell lysate was cleared by centrifugation at 13,000g for 15 min at 4°C. The protein content of the supernatants was determined by the Qubit protein assay kit. Lysates were subjected to immunoprecipitation by incubation for 1 h at 4°C using anti-Flag M2 magnetic beads (Sigma). Bead suspensions were incubated for 15 min at 22°C with and without RNase A/T1 mix (final concentration 100 U/µL RNaseT1/A) and subsequently cooled for 5 mi on icen. The immunoprecipitates were washed four times with immunoprecipitation wash buffer (50 mM HEPES-KOH at pH 7.5, 300 mM KCl, 0.05% [v/v] NP40, 0.5 mM DTT, complete EDTA-free protease), and FH-LIN28A RNP complexes were eluted with 3xFlag peptide.
Proteomic analysis was performed by Poochon Scientific. Briefly, equal amounts of immunoprecipitated proteins (45 µL) from each sample were loaded into wells of a 4%–12% NuPAGE gel. After electrophoresis, the gel was stained with SimplyBlue. The protein bands were excised and combined into two gel fractions for trypsin digestion. The gel samples were treated with DTT for reduction and then iodoacetamide for alkylation and further digested by trypsin in 25 mM NH4HCO3 solution. The tryptic peptides mixtures were cleaned using a C18 Zip-Tip and reconstituted in 20 µL of 0.1% formic acid before high-performance liquid chromatography and high-resolution tandem mass spectrometry (LC/MS/MS) analysis.
The LC/MS/MS analysis was performed using a Thermo Scientific Q-Exactive hybrid Quadrupole Orbitrap mass spectrometer and a Thermo Dionex UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). Peptides from trypsin digestion were loaded onto a peptide trap cartridge at a flow rate of 5 µL/min. The trapped peptides were eluted onto a reversed-phase 25-cm C18 PicoFrit column (New Objective) using a linear gradient of acetonitrile (3%–36%) in 0.1% formic acid. The elution duration was 120 min at a flow rate of 0.3 µL/min. Eluted peptides from the PicoFrit column were ionized and sprayed into the mass spectrometer using a Nanospray Flex Ion Source ES071 (Thermo Fisher Scientific) with the following settings: spray voltage, 1.6 kV, capillary temperature, 250°C.
The Q-Exactive instrument was operated in the data-dependent mode to automatically switch between full-scan mass spectrometry and MS/MS acquisition. Survey full-scan mass spectrometry spectra (m/z 300−1800) were acquired in the Orbitrap with 70,000 resolutions (m/z 200) after accumulation of ions to a 1 × 106 target value based on predictive automatic gain control (AGC). Dynamic exclusion was set to 20 sec. The 12 most intense multiply charged ions (z ≥ 2) were sequentially isolated and fragmented in the octopole collision cell by higher-energy collisional dissociation (HCD) using normalized HCD collision energy 25% with an AGC target 1 × 105 and a maximal injection time of 100 msec at 17,500 resolutions.
Raw data files obtained by LC/MS/MS were searched against the mouse protein sequence database using the Proteome Discoverer 1.4 software (Thermo) based on the Sequest and percolator algorithms. The false positive discovery rates (FDR) was set on 1%. We used peptide spectrum match counts (PSM#) to compare the protein relative abundance among samples.
Immunoprecipitation and Western analyses
Immunoprecipitations from ∼2 × 106 220-8 cells were performed as described above in “Mass Spectrometry.” Samples were denatured and subjected to electrophoresis in 4%–12% NuPAGE Bis-Tris gels (Life Technologies) and transferred to nitrocellulose membranes (Life Technologies). Blots were then incubated with primary antibodies for 2 h at 25°C and HRP-conjugated secondary antibody for 1 h at 25°C. Blots were developed with ECL Western blotting substrate (Thermo Fisher Scientific), and images were acquired using LAS-4000 (Fuji Film). Flag-BAP recombinant protein (Sigma) was diluted in PBS with 2% BSA to concentrations of 0.1–10 ng/µL for use as a standard and compared with lysate from the indicated number of 220-8 cells by Western blot using Flag M2 mouse monoclonal antibody (Sigma) at 1:2000 dilution. Blots were quantified with National Institutes of Health ImageJ (https://imagej.nih.gov/ij), and background was subtracted by rolling ball method with a radius of 150 pixels. When possible, signal from the protein band of interest were normalized to β-actin or tubulin that served as loading control.
Immunofluorescence (IF)
220-8 cells (5 × 105) were plated on each 12-mm poly-L-lysine-coated coverslip and allowed to settle for 30 min in a humidified chamber at 37°C. Cells were fixed with 4% paraformaldehyde (PFA) for 10 min at room tempterature and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. After three washes with 0.5% Tween-20 for 3 min each time, cells were blocked with 5% BSA/PBS for 30 min at 37°C and incubated with primary antibody (Supplemental Table 9) in 5% BSA/PBS overnight at 4°C. After primary antibody incubation, cells were washed five times with with 0.5% Tween-20 for 3 min each time and incubated with Alexa488- or Alexa555-conjugated secondary antibodies (Supplemental Table 9) in 0.5% Tween-20 for 1 h at room temperature. Cells were then washed five times with 0.5% Tween-20 for 3 min each time and mounted in mounting medium with DAPI (Sigma) for subsequent imaging.
Confocal images were acquired by Leica SP5 X-WLL with a 63× oil immersion lens. Image acquisition and processing software were Leica application suite and Imaris 8.3.1 (BitPlane).
PLAs
220-8 cells (3 × 106) were collected in a 5-mL polystyrene round-bottomed tube (Falcon). Cells were fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. After washing three times with 0.5% Tween-20 for 3 min each time, cells were blocked with 5% BSA/PBS for 30 min at 37°C and incubated with primary antibody (Supplemental Table 9) in 5% BSA/PBS overnight at 4°C. Primary antibodies were used at the same concentrations as for IF. After primary antibody incubation, PLA was performed according to the protocol of Duolink using PLA technology (Sigma). PLA signal was amplified and detected using Green detection reagent (Sigma). Samples were divided either to be mounted with mounting medium with DAPI (Sigma) on 12-mm poly-L-lysine-coated coverslips for confocal microscopy or left in tubes to be analyzed by flow cytometry on LSR-II with a 480-nm/100-mW laser and a 525/50-nm filter. Confocal images were acquired by Leica SP5 X-WLL with a 63× oil immersion lens. Image acquisition and processing software were Leica application suite and Imaris 8.3.1 (BitPlane). Flow cytometry data were analyzed by FlowJo 10.2 (FlowJo).
To perform PLAs in primary mouse cells, fresh BM cells were isolated from either iLIN28B or WT mice (see “FACS” for sorting strategy). PLAs, including signal detection, image acquisition, and analysis, were performed as mentioned above in 1.5-mL centrifugation tubes.
FLIM-FRET
FLIM-FRET assays were performed and analyzed as described previously (Ganesan et al. 2008). 220-8 cells (5 × 105) were plated on 12-mm poly-L-lysine-coated coverslips and allowed to settle for 30 min in a humidified chamber at 37°C. Samples were divided into four replicates for the Flag/Igf2bp3 pair and two replicates for the Flag/Pum2 pair. For the Flag/Igf2bp3 pairs, two of the replicates were incubated with FRET pair secondary antibodies, and two replicates were incubated with only donor secondary antibodies as donor-alone control. For the Flag/Pum2 pair, one replicate was incubated with FRET pair secondary antibodies, and the other was incubated with donor-only secondary antibody as donor-alone control. Cells were fixed with 4% PFA for 10 min at room temberature and permeabilized with 0.1% Triton X-100 for 10 min at room tempterature. After washing three times with 0.5% Tween-20 for 5 min each time, cells were blocked with 5% BSA/PBS for 30 min at 37°C and incubated with primary antibodies in 5% BSA/PBS overnight at 4°C. For the Flag/Igf2bp3 pair, each replicate was incubated with primary antibody pair anti-Flag (1:100; Sigma) and anti-Igf2bp3 (1:800; Millipore). For the Flag/Pum2 pair, each replicate was incubated with primary antibody pair anti-Flag (1:100; Sigma) and anti-Pum2 (1:200; Sigma). After primary antibody incubation, cells were washed three times with 0.5% Tween-20 for 3 min each time. For the Flag/Igf2bp3 pair, two replicates were incubated with either goat antimouse Alexa-488 (1:500; Life Technologies) and goat antirabbit Alexa-555 (1:1000; Life Technologies) or goat antimouse Alexa-555 (1:500; Life Technologies) and goat antirabbit Alexa-488 (1:1000; Life Technologies). Another two replicates (donor-alone control) were incubated with either goat antimouse Alexa-488 (1:500; Life Technologies) or goat antirabbit Alexa-488 (1:1000; Life Technologies). For the Flag/Pum2 pair, one replicate was incubated with goat antimouse Alexa-488 (1:500) and goat antirabbit Alexa-555 (1:1000), and the other replicate (donor-alone control) was incubated with goat antimouse Alexa-488 (1:500). All secondary antibody pairs were incubated for 1 h at room temperature. After secondary antibody incubation, samples were washed three times with 0.5% Tween-20 for 5 min each time. Coverslips were mounted in ProLong Diamond antifade mountant (Invitrogen).
To perform FLIM-FRET assays in primary mouse cells, fresh BM cells were isolated from either iLIN28B or WT mice (see “FACS” for sorting strategy). Sorted pro-B cells were prepared for primary antibody incubation in 1.5-mL centrifugation tubes as mentioned above followed by incubation with anti-Igf2bp3 (1:800) in 5% BSA/PBS overnight at 4°C. The next day, cells were washed twice with 0.5% Tween-20 for 5 min each time and divided into two replicates. One replicate was incubated with anti-Flag-Cy3 (1:100; Sigma, A9594) overnight at 4°C. The other replicate (donor-alone control) was incubated with goat antirabbit Alexa-488 (1:1000) for 1 h at room temperature, washed three times with 0.5% Tween-20 for 5 min each time, and mounted on a 5-mm poly-L-lysine-coated coverslip in ProLong Diamond antifade mountant. After overnight incubation with anti-Flag-Cy3 (1:100), cells were washed three times with 0.5% Tween-20 for 5 min each time and incubated with goat antirabbit Alexa-488 (1:1000) for 1 h at room temperature. After secondary antibody incubation, cells were washed three times with 0.5% Tween-20 for 5 min each time and mounted on a 5-mm poly-L-lysine-coated coverslip in ProLong Diamond antifade mountant.
For each sample, images of at least 10–20 cells were acquired on a Leica DMI 6000 SP5 inverted confocal microscope with a 63× oil immersion objective of NA 1.4 (Leica Microsystems). Alexa 488 excitation at 900 nm was achieved with a femtosecond mode-locked (80 MHz repetition rate) Mai-Tai HP pulsed multiphoton laser (Spectra Physics). Fluorescence was collected using a HPM100 hybrid detector R3809U-50 (Becker & Hickl; Hamamatsu Photonics) through a band-pass GFP filter at ET 525/50 (Chroma Technology Corp.). Fluorescence decays were resolved by time-correlated single-photon counting using a SPC150 acquisition board (Becker & Hickl). Images were acquired in 256 × 256 pixel format, collecting at least 1000 photons per pixel over 2–5 min. Fluorescence transients were analyzed and acquired with SPCImage software (Becker & Hickl), and results were exported and analyzed per single lifetime decay using an in-house-developed ImageJ protocol (National Institutes of Health). Histograms for the weighted mean average of the fluorescent lifetimes were derived from ImageJ and plotted as frequency distributions normalized and integrated for area under the curve using Igor-Pro (Wave Metrics Inc). The weighted mean lifetime was extracted from histograms of individual cells in ImageJ and converted to FRET efficiency relative to the donor-alone control according to the equation FRET efficiency (percentage) = 1 − {Tdonor/TDonorAcceptor [1-Td/(tDA) × 100]} and thus was used to allow statistical analysis.
PAR-CLIP
PAR-CLIP was performed as described previously (Hafner et al. 2010; Danan et al. 2016). Briefly, for each PAR-CLIP experiment, 400 × 106 to 600 × 106 cells were cultured in medium containing 100 µM 4SU for 16 h. Cells were washed and resuspended in 15 mL of PBS and irradiated in 15-cm plates with 1.5 mJ/cm2 312-nm wavelength UV. Cells were lysed in the equivalent of three cell pellet volumes and cleared by centrifugation at 13,000g. Cell lysates were treated with 1 U/µL RNase T1 and then immunoprecipitated using anti-Flag magnetic beads (M2, Sigma). For anti-Igf2bp3 (Millipore) and anti-Pum2 (Bethyl Labs), Protein G Dynabeads (Invitrogen) were used. The beads were taken up in one original bead suspension volume (10 µL of bead suspension per milliliter of lysate) and incubated in 15-mL centrifugation tubes on a rotating wheel for 1 h at 4°C. Beads were washed three times in 1 mL of immunoprecipitation wash buffer (50 mM HEPES-KOH at pH 7.5, 300 mM KCl, 0.05% [v/v] NP40, 0.5 mM DTT, complete EDTA-free protease inhibitor cocktail [Roche]) and resuspended in 1 vol of immunoprecipitation wash buffer. The RNA residing in the immunoprecipitate was further trimmed with 100 U/µL RNase T1. The beads were then washed three times in 1 mL of high-salt lysis buffer (50 mM HEPES-KOH at pH 7.5, 500 mM KCl, 0.05% [v/v] NP40, 0.5 mM DTT, complete EDTA-free protease inhibitor cocktail [Roche]) and resuspended in one bead volume of dephosphorylation buffer. RNA was dephosphorylated and radioactively labeled with [γ-32P] ATP. The protein–RNA complexes were separated by SDS-PAGE and transferred to nitrocellulose, and RNA–protein complexes were visualized by autoradiography. The radioactive bands were recovered, and the protein–RNA complex was digested with 1.2 mg/mL proteinase K (Roche) in proteinase K buffer (50 mM Tris-HCl at pH 7.5, 75 mM NaCl, 6.25 mM EDTA, 1% SDS) for 2 h at 37°C. Next, the RNA was isolated by acidic phenol/chloroform extraction and ethanol precipitation. The recovered RNA was converted into a cDNA library using a standard small RNA library protocol (Hafner et al. 2008) and sequenced on an Illumina HiSeq2000 or 2500 instrument.
Computational analyses
Single-cell mRNA-seq data demultiplexing and preprocessing
Sequenced scRNA-seq libraries were processed by the Cell Ranger version 2.0.2 pipeline using default parameters unless specified otherwise below (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger). Raw BCL files were demultiplexed and converted to FASTQ reads by cellranger mkfastq using bcl2fastq version 2.17. The genome reference mm10 (GRCm38) was set up by Cell Ranger mkref. The transcriptome reference Ensembl GRCm38.84 was set up by Cell Ranger mkgtf with the –attribute option to filter for protein_coding, large intergenic noncoding RNA (lincRNA), and antisense transcripts. Alignment, barcode processing, and quantification were performed using Cell Ranger count using STAR version 2.5.1b for alignment (Dobin et al. 2013) with all default parameters aside from the –expect-cells option to estimate expected cell recovery for each sample (Supplemental Table 1). Briefly, only reads with valid sample indexes, cell barcodes, and unique molecular identifiers (UMIs) that were confidently mapped with matching strand to annotated transcripts were counted. Reads were considered confidently mapped if they overlapped an exonic locus by at least 50%. Quantification results were converted from matrix market exchange format to a single tab-delimited table of UMI counts for each gene and each cell using in-house scripts. This gene–cell count table was read into the analysis package Seurat for downstream analyses.
For further details on the read alignment, barcode processing, and UMI assignment, see the Cell Ranger algorithms overview at https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/algorithms/overview. For cell recovery and gene detection metrics after processing by Cell Ranger, see Supplemental Table 1.
Single-cell mRNA-seq data downstream analysis
The gene–cell count table generated from Cell Ranger was loaded into R as a Seurat object for further analysis (R version 3.4.2 “Short Summer,” Seurat version 2.1.0) (Satija et al. 2015). The Seurat functions used and notable parameters are described below. Genes that were expressed in fewer than four cells were discarded. Cells with (1) >7% mitochondrial genes present, (2) <1500 total genes expressed, or (3) >3500 total genes expressed were discarded (function: FilterCells). After these filters, 3832 cells remained, with 14,251 genes detected overall. The remaining data were normalized by the “LogNormalize” method using the default scale factor of 10,000 followed by log transformation (function: NormalizeData). To find variable genes, each gene was assigned to one of 20 bins based on its average expression, and a dispersion z-score was calculated and normalized to the median absolute deviation within its assigned bin. Genes with 0.0125 < average expression <3.0 and dispersion z-score >0.5 were considered variable genes (function: FindVariableGenes). Cell cycle phases (S, G2M, or G1) were assigned to each cell by scoring for previously described markers (function: CellCycleScoring) (Kowalczyk et al. 2015). Variability due to cell cycle biases were regressed out prior to further dimensional reduction and clustering (function: ScaleData).
Linear dimensional reduction by PCA was performed using 1146 variable genes to calculate the first 20 PCs (function: RunPCA). Cells were clustered according to these 20 PCs with a resolution setting of 1.2 (function: FindClusters). Nonlinear dimensional reduction by tSNE was performed based on the same PCs using the fast Barnes-hut implementation and perplexity set to 45 (function: RunTSNE). The unsupervised tSNE clusters were plotted and cells were colored and manually grouped by their original identity (function: TSNEPlot). Differentially expressed markers for each cluster (unsupervised and manually grouped) were determined by Wilcoxon rank sum test, requiring a minimum fraction of detection of 0.25 and a minimum difference in average expression of 0.25 (log-scale) between the given cluster and all other clusters (functions: FindAllMarkers and FindMarkers). Gene module enrichment scores for Pax5-activated and Pax5-repressed targets were calculated using function AddModuleScore with default parameters and, as input, the respective pro-B-cell target lists (Revilla-I-Domingo et al. 2012).
Low-input RNA-seq analyses (FL/BM LSK and CLP)
Reads were aligned to the mm10 (GRCm38) mouse reference genome using STAR version 2.5.2 with default mapping parameters (Dobin et al. 2013). Expression values were normalized to fragments per kilobase of transcript per million mapped reads (FPKM) by RSEM versio 1.2.30 (Li and Dewey 2011) against the Ensembl GRCm38.p5 annotation. The expression heat map was generated by the R package pheatmap version 1.0.8 (https://CRAN.R-project.org/package=pheatmap).
Regular input RNA-seq analyses (220-8 control and hLIN28A)
Regular input RNA-seq analyses for 220-8 control and hLIN28A were performed as described previously (Escobar et al. 2014). Reads were aligned to the mm10 (GRCm38) genome by Bowtie version 2.2.5 and TopHat version 2.0.14 with default mapping parameters (Trapnell and Salzberg 2009; Langmead and Salzberg 2012). Expression values (FPKM) and fold changes were calculated by Cufflinks version 2.2.1 against the GRCm38.p4 annotation.
PAR-CLIP analyses
The PAR-CLIP sequencing data were processed by PARpipe, which handles the library preprocessing, alignment, clustering, annotation, and meta-analysis (https://ohlerlab.mdc-berlin.de/software/PARpipe_119). Adapters were trimmed using Cutadapt 1.12 (Martin 2011), and reads were aligned to the mm10 (GRCm38) genome using Bowtie version 1.1.1, allowing for up to one mismatch (Langmead et al. 2009). Aligned reads were sorted, converted to BAM files, and indexed with Samtools version 0.1.19 (Li et al. 2009). PARalyzer version 1.5 (Corcoran et al. 2011) was used to process reads into groups and clusters via filters for (1) minimum read length, (2) minimum read count, (3) minimum unique T-to-C locations, and (4) minimum T-to-C conversions. Reads, groups, and clusters were annotated to the features in the Gencode vM9 annotation file for reference chromosomes, scaffolds, assembly patches, and alternative loci.
For each RBP, we produced a high-confidence set of binding sites that overlapped in at least three of four replicates. We discarded sites that were annotated to mitochondrial transcripts or to transcripts that were <500 bp in length. Furthermore, sites that overlapped across all RBPs over a Pum2 CDS cluster or over a Pum2 site lacking TGTAN were discarded as background. In-house scripts were used to count and visualize the proportions of binding to 3′ UTRs, 5′ UTRs, and CDSs.
5-mer Z-scoring
Exclusive sets of reads for LIN28 (FH-LIN28A ∩ FH-LIN28B), Igf2bp3, and Pum2 were isolated using a series of commands in BEDTools version 2.25 (Quinlan and Hall 2010). First, the collapsed PARalyzer-used reads for each RBP were expanded and converted to the BED file format using an in-house script. We added ±25 bp to the intervals of these reads in order to account for potential enrichment or depletion of 5-mers upstream of and downstream from the target sites. The BEDTools utility “intersect with option -v” was used to subtract reads of a given RBP that overlapped with those of the other RBPs. For LIN28, the FH-LIN28A and FH-LIN28B reads were intersected and pooled together prior to subtraction against Igf2bp3 and Pum2. The BEDTools utility getfasta was used to recover the genomic nucleotide sequences corresponding to the remaining intervals in these BED files. To produce background, we used an in-house script to scramble the target read sequences while preserving GC content. We then counted the 5-mers for target and for background, respectively, and calculated Z-score enrichment by proportion Z = (X − µ)/σ. X is the proportion count/total counts for a given 5-mer, and µ and σ are the average and standard deviation, respectively, of 5-mer proportions in background. The Z-scores for LIN28 versus Igf2bp3 and LIN28 versus Pum2 were plotted, and the top-scoring motifs were highlighted manually.
Genome browser track generation and visualization
RNA-seq alignment files were filtered for uniquely mapped reads, normalized to reads per million (RPM), and then converted to the bigwig file format. PAR-CLIP alignment files of PARalyzer-used reads were expanded to reflect true coverage (originally collapsed in PARpipe preprocessing). All track processing was performed using Samtools version 0.1.19 (Li et al. 2009), Bedtools 2.25.0 (Quinlan and Hall 2010), and the University of California at Santa Cruz bedGraphToBigWig (Kent et al. 2010) utility. The phastCons conservation track mm10.60way.phastCons60wayEuarchontoGlire.bw was downloaded from http://hgdownload.cse.ucsc.edu/goldenpath/mm10/phastCons60way (Miller et al. 2007). All tracks were visualized in Integrative Genomics Viewer version 2.3 (Robinson et al. 2011). Editing and recoloring were performed with Inkscape and Adobe Illustrator software.
Cumulative distribution plots
Cumulative distribution plots were generated in R version 3.2.3 using the built-in functions plot(), lines(), ecdf(), and points(). Significance of distribution shifts against background were tested by two-sided Kolmogorov-Smirnov test ks.test().
T-to-C enrichment profiles
Genomic positions and frequency of the T-to-C mutations were identified from a BAM file containing PARalyzer-used reads generated by the PARpipe analysis pipeline (https://ohlerlab.mdc-berlin.de/software/PARpipe_119) using a custom script (JAVA). Coordinates of enriched 5-mer sequence elements for the examined RBPs (LIN28A, LIN28B, Pum2, and Igf2bp3) within the RBP sites as found by PARalyzer (Corcoran et al. 2011) were identified. The frequency of T-to-C mismatches in each base position between 10 nt upstream of and 10 nt downstream from the genomic position of the 5-mer motifs was summed up and plotted using R.
Calculation of cluster overlap significance (Venn diagrams)
P-values were calculated using Bedtools version 2.25.0 (Quinlan and Hall 2010) using procedure fisher. This procedure implements Fisher's exact test to calculate the probability that the two interval sets overlap using a heuristic that estimates the number of possible intervals. This estimate is dependent on the total size of genomic locations where the intervals can be located. We therefore calculated the possible target size for each of the pairwise comparisons. Total potential target size was calculated as a sum of 3′ UTR sizes for targeted genes (those with at least one 3′ UTR cluster; Mouse Ensembl release 86). Analysis was restricted to 3′ UTR clusters. Similar analysis was performed for CDS regions for Supplemental Figure S3.
Heat map of spatial enrichment
Spatial enrichment of raw reads from 12 independent PAR-CLIP experiments around detected PAR-CLIP clusters was plotted using ngs.plot version 2.61 (Shen et al. 2014). LIN28A and Igf2bp3 clusters were first ordered by their classification (LIN28-exclusive, LIN28 and Igf2bp3 shared clusters, and then Igf2bp3-exclusive clusters) and then by chromosomal position. Analysis was restricted to 3′ UTR clusters only. Read density (all reads were truncated to 20 nt) was then plotted in a ±100-nt window relative to cluster centers.
K-mer analysis
The positions of all 1024 5-mers within 108 nt from PAR-CLIP cluster centers were determined using Bedtools version 2.25.0 (Quinlan and Hall 2010) and MEME suite version 4.11.2 (Bailey et al. 2009). The absolute positions of the pentamers in the genome were converted into distances to PAR-CLIP cluster centers on corresponding transcripts retaining strandness. The obtained tables of k-mer positions were further processed using R version 3.2. To extract information regarding pentamers that are most differentially enriched, we calculated a positional K-mer imbalance score:
where count1 and count2 are K-mer counts at a given position in data sets 1 and 2, respectively. K-mer imbalance is distributed between −1 (k-mer detected only in data set 2) and +1 (K-mer is detected in data set 1 only). For plotting, we further calculated position-specific k-mer Z-scores by normalizing K-merimbalance by the standard deviation of imbalance over our ±108-nt window:
All pentamers were then ranked by average k-mer z-scores in the central 10-nt windows, and the 20 top pentamers most strongly enriched in LIN28A, LIN28B, and Igf2bp3 data sets were plotted. For the comparison, we used only those exclusive LIN28A or LIN28B clusters that did not overlap Igf2bp3 clusters and Igf2bp3 clusters that did not overlap LIN28A clusters.
GO term enrichment analysis
For GO term enrichment analysis, we used DAVID version 6.8 (https://david.ncifcrf.gov) with default settings (Huang et al. 2009).
Data accessiblility
The NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) accession numbers are GSE98466 for the RNA-seq and PAR-CLIP data and GSE108129 for the scRNA-seq data.
Supplementary Material
Acknowledgments
We thank George Daley (Harvard) for iLIN28B mice; Fred Alt (Harvard) for 220-8 cells; Ke Wang (National Institute of Allergy and Infectious Diseases [NIAID]) for cell sorting; Thomas Tuschl (Rockefeller University), Pavel Morozov (Rockefeller University), Gokhul Kilaru (NIAID), Dwayne McCully (National Institute of Arthritis and Musculoskeletal and Skin Diseases [NIAMS]), and Neelanjan Mukherjee (University of Colorado at Denver) for help with the PAR-CLIP computational analysis pipeline; Gustavo Gutierrez-Cruz and Stefania Dell'Orso (NIAMS) for sequencing; Patrick Burr (NIAID) for some RNA-seq; Jens Durruthy (10X Genomics) for expert advice on scRNA-seq; Hong-Wei Sun (NIAMS) for expert advice on inferential statistics; Ryan Kissinger (NIAID) for graphic art in Figure 7, A and B; NIAID Office of Cyber Infrastructure and Computational Biology for high-performance computing; Avinash Bhandoola (National Cancer Institute [NCI]), John O'Shea (NIAMS), Shalini Oberdoerffer (NCI), Philipp Oberdoerffer (NCI), Astrid Haase (National Institute of Diabetes and Digestive and Kidney Diseases), and Ron Germain (NIAID) for comments and suggestions; and Lonelyss Charles (National Insitutes of Health Library Editing Service) for reviewing the manuscript. Our work was funded by the National Institutes of Health Intramural Research Program of the National Institute of Allergy and Infectious Diseases and National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Author contributions: C.K., M.H., and S.A.M. conceived the study. S.W., B.C., Y.S., P.K., M.W., A.F., P.T.S., X.L., R.L., S.G., C.K., M.H., and S.A.M. performed the methodology. B.C., Y.S., P.K., A.F., S.G., and M.H. were responsible for the software used. S.W. and Y.S. performed the validation. S.W., B.C., Y.S., P.K., A.F., S.G., and M.H. performed the formal analysis. S.W., Y.S., X.W., and R.L. performed the investigations. B.C. curated the sequencing data. C.K., M.H., and S.A.M. wrote the original draft of the manuscript. All authors reviewed and edited the manuscript. S.W., B.C., Y.S., P.K., A.F., and S.G. performed visualization. M.H. and S.A.M. acquired funding and resources and managed and supervised the project.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.325100.119.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4: 1–38. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. 1981. Organization and reorganization of immunoglobulin genes in A-MuLV-transformed cells: rearrangement of heavy but not light chain genes. Cell 27: 381–390. 10.1016/0092-8674(81)90421-9 [DOI] [PubMed] [Google Scholar]
- Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. 1984. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J 3: 1209–1219. 10.1002/j.1460-2075.1984.tb01955.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226: 409–416. 10.1126/science.6494891 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker W. 2012. Fluorescence lifetime imaging - techniques and applications. J Microsc 247: 119–136. 10.1111/j.1365-2818.2012.03618.x [DOI] [PubMed] [Google Scholar]
- Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. 2013. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci 70: 2657–2675. 10.1007/s00018-012-1186-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict CL, Gilfillan S, Thai TH, Kearney JF. 2000. Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev 175: 150–157. 10.1111/j.1600-065X.2000.imr017518.x [DOI] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. 2008. Identification of let-7-regulated oncofetal genes. Cancer Res 68: 2587–2591. 10.1158/0008-5472.CAN-08-0264 [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji P, Hamilton KE, Liang S, Andres SF, Wijeratne HRS, Mizuno R, Simon LA, Hicks PD, Foley SW, Pitarresi JR, et al. 2018. The LIN28B–IMP1 post-transcriptional regulon has opposing effects on oncogenic signaling in the intestine. Genes Dev 32: 1020–1034. 10.1101/gad.314369.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. 2012. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 151: 765–777. 10.1016/j.cell.2012.10.019 [DOI] [PubMed] [Google Scholar]
- Copley MR, Babovic S, Benz C, Knapp DJHF, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K, et al. 2013. The Lin28b–let-7–Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol 15: 916–925. 10.1038/ncb2783 [DOI] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. 2011. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol 12: R79 10.1186/gb-2011-12-8-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danan C, Manickavel S, Hafner M. 2016. PAR-CLIP: a method for transcriptome-wide identification of RNA binding protein interaction sites. Methods Mol Biol 1358: 153–173. 10.1007/978-1-4939-3067-8_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vasconcellos JF, Tumburu L, Byrnes C, Lee YT, Xu PC, Li M, Rabel A, Clarke BA, Guydosh NR, Proia RL, et al. 2017. IGF2BP1 overexpression causes fetal-like hemoglobin expression patterns in cultured human adult erythroblasts. Proc Natl Acad Sci 114: E5664–E5672. 10.1073/pnas.1609552114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar TM, Kanellopoulou C, Kugler DG, Kilaru G, Nguyen CK, Nagarajan V, Bhairavabhotla RK, Northrup D, Zahr R, Burr P, et al. 2014. MiR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity 40: 865–879. 10.1016/j.immuni.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flake AW, Harrison MR, Adzick NS, Zanjani ED. 1986. Transplantation of fetal hematopoietic stem cells in utero: the creation of hematopoietic chimeras. Science 233: 776–778. 10.1126/science.2874611 [DOI] [PubMed] [Google Scholar]
- Ganesan S, Rohde G, Eckermann K, Sroka K, Schaefer MKE, Dohm CP, Kermer P, Haase G, Wouters F, Bähr M, et al. 2008. Mutant SOD1 detoxification mechanisms in intact single cells. Cell Death Differ 15: 312–321. 10.1038/sj.cdd.4402262 [DOI] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci 103: 4487–4492. 10.1073/pnas.0509260103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf R, Munschauer M, Mastrobuoni G, Mayr F, Heinemann U, Kempa S, Rajewsky N, Landthaler M. 2013. Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation. RNA Biol 10: 1146–1159. 10.4161/rna.25194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12. 10.1016/j.ymeth.2007.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Max KEA, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, Tuschl T. 2013. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA 19: 613–626. 10.1261/rna.036491.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MR, Crombleholme TM, Tarantal AF, Nathan Slotnick R, Golbus MS, Zanjani ED. 1989. In-utero transplantation of fetal liver haemopoietic stem cells in monkeys. Lancet 334: 1425–1427. 10.1016/S0140-6736(89)92036-9 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284. 10.1016/j.molcel.2008.09.014 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. 2018. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20: 285–295. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26: 2204–2207. 10.1093/bioinformatics/btq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Kondo M. 2006. Developmental switch of mouse hematopoietic stem cells from fetal to adult type occurs in bone marrow after birth. Proc Natl Acad Sci 103: 17852–17857. 10.1073/pnas.0603368103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, Regev A. 2015. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res 25: 1860–1872. 10.1101/gr.192237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen TA, Jaensson Gyllenbäck E, Zriwil A, Björklund T, Daniel JA, Sitnicka E, Soneji S, Bryder D, Yuan J. 2016. Cellular barcoding links B-1a B cell potential to a fetal hematopoietic stem cell state at the single-cell level. Immunity 45: 346–357. 10.1016/j.immuni.2016.07.014 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, De Vasconcellos JF, Yuan J, Byrnes C, Noh SJ, Meier ER, Kim KS, Rabel A, Kaushal M, Muljo SA, et al. 2013. LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood 122: 1034–1041. 10.1182/blood-2012-12-472308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao TT, Hsu WH, Ho CH, Hwang WL, Lan HY, Lo T, Chang CC, Tai SK, Yang MH. 2016. Let-7 modulates chromatin configuration and target gene repression through regulation of the ARID3B complex. Cell Rep 14: 520–533. 10.1016/j.celrep.2015.12.064 [DOI] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376: 263–267. 10.1038/376263a0 [DOI] [PubMed] [Google Scholar]
- Mansson R, Welinder E, Ahsberg J, Lin YC, Benner C, Glass CK, Lucas JS, Sigvardsson M, Murre C. 2012. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc Natl Acad Sci 109: 21028–21033. 10.1073/pnas.1211427109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17: 10 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, Jaritz M, Tagoh H, Busslinger M. 2011. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J 30: 2388–2404. 10.1038/emboj.2011.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, Burhans R, King DC, Baertsch R, Blankenberg D, et al. 2007. 28-Way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res 17: 1797–1808. 10.1101/gr.6761107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecino-Rodriguez E, Fice M, Casero D, Berent-Maoz B, Barber CL, Dorshkind K. 2016. Distinct genetic networks orchestrate the emergence of specific waves of fetal and adult B-1 and B-2 development. Immunity 45: 527–539. 10.1016/j.immuni.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. 1995. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci 92: 10302–10306. 10.1073/pnas.92.22.10302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss E, Lee R, Ambros V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646. 10.1016/S0092-8674(00)81906-6 [DOI] [PubMed] [Google Scholar]
- Muljo SA, Schlissel MS. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol 4: 31–37. 10.1038/ni870 [DOI] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. 2011. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147: 1080–1091. 10.1016/j.cell.2011.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. 2008. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549. 10.1261/rna.1155108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Kristensen MA, Willemoës M, Nielsen FC, Christiansen J. 2004. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Res 32: 4368–4376. 10.1093/nar/gkh754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Urbanek P, Rolink A, Busslinger M. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev 11: 476–491. 10.1101/gad.11.4.476 [DOI] [PubMed] [Google Scholar]
- Oltz EM, Yancopoulos GD, Morrow MA, Rolink A, Lee G, Wong F, Kaplan K, Gillis S, Melchers F, Alt FW. 1992. A novel regulatory myosin light chain gene distinguishes pre-B cell subsets and is IL-7 inducible. EMBO J 11: 2759–2767. 10.1002/j.1460-2075.1992.tb05341.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikkar B, Smith N, Brown P. 2012. Reflexive research ethics in fetal tissue xenotransplantation research. Account Res 19: 344–369. 10.1080/08989621.2012.728910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, et al. 2012. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev 26: 43–53. 10.1101/gad.177428.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. 2007. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev 21: 1125–1138. 10.1101/gad.415007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath S, Ouyang W, Bhattacharya D, Sha WC, Grupe A, Peltz G, Murphy KM. 1998. GATA-3-dependent enhancer activity in IL-4 gene regulation. J Immunol 161: 3822–3826. [PubMed] [Google Scholar]
- Revilla-I-Domingo R, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, Smeenk L, Trupke J, Sommer A, Jaritz M, et al. 2012. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J 31: 3130–3146. 10.1038/emboj.2012.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. 2008. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10: 987–993. 10.1038/ncb1759 [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33: 495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana EN, Hartenian E, Zhang F. 2014. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343: 84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. 2014. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15: 284 10.1186/1471-2164-15-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3: 995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Salzberg SL. 2009. How to map billions of short reads onto genomes. Nat Biotechnol 27: 455–457. 10.1038/nbt0509-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vély F, Barlogis V, Vallentin B, Neven B, Perchet T, Petit M, Yessaad N, Touzot F. 2016. Evidence of innate lymphoid cell redundancy in humans. Nat Immunol 17: 1291–1299. 10.1038/ni.3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilagos B, Hoffmann M, Souabni A, Sun Q, Werner B, Medvedovic J, Bilic I, Minnich M, Axelsson E, Jaritz M, et al. 2012. Essential role of EBF1 in the generation and function of distinct mature B cell types. J Exp Med 209: 775–792. 10.1084/jem.20112422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100. 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TMT. 2002. Modular recognition of RNA by a human Pumilio-homology domain. Cell 110: 501–512. 10.1016/S0092-8674(02)00873-5 [DOI] [PubMed] [Google Scholar]
- Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, Yan BY, Nathanson JL, Hutt KR, Lovci MT, et al. 2012. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell 48: 195–206. 10.1016/j.molcel.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Huang Y. 2009. Histone H2a mRNA interacts with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res 37: 4256–4263. 10.1093/nar/gkp372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Zhang K, Huang Y. 2009. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA 15: 357–361. 10.1261/rna.1368009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920. 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. 2012. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science 335: 1195–1200. 10.1126/science.1216557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M. 2008. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol 181: 3364–3372. 10.4049/jimmunol.181.5.3364 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Li Y-S, Bandi S, Tang L, Shinton SA, Hayakawa K, Hardy RR. 2015. Lin28b promotes fetal B lymphopoiesis through the transcription factor Arid3a. J Exp Med 212: 569–580. 10.1084/jem.20141510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Ng SC, Segr AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. 2011. The Lin28/let-7 axis regulates glucose metabolism. Cell 147: 81–94. 10.1016/j.cell.2011.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. 2017. Comparative analysis of single-cell RNA sequencing methods. Mol Cell 65: 631–643.e4. 10.1016/j.molcel.2017.01.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.