In this study, Brennan et al. identify protein complex stoichiometry imbalances as a major cause of protein aggregation in aneuploid cells. They propose that proteotoxic stress is a universal feature of aneuploid cells and show that degradation and aggregation of excess polypeptides function as a form of dosage compensation.
Keywords: aneuploidy, protein aggregation, protein homeostasis
Abstract
Aneuploidy, a condition characterized by chromosome gains and losses, causes reduced fitness and numerous cellular stresses, including increased protein aggregation. Here, we identify protein complex stoichiometry imbalances as a major cause of protein aggregation in aneuploid cells. Subunits of protein complexes encoded on excess chromosomes aggregate in aneuploid cells, which is suppressed when expression of other subunits is coordinately altered. We further show that excess subunits are either degraded or aggregate and that protein aggregation is nearly as effective as protein degradation at lowering levels of excess proteins. Our study explains why proteotoxic stress is a universal feature of the aneuploid state and reveals protein aggregation as a form of dosage compensation to cope with disproportionate expression of protein complex subunits.
Eukaryotes have a problem: Subunits of protein complexes are not encoded in operons. Although eukaryotes have evolved to coordinate expression of subunits of the same complex (Li et al. 2014; Taggart and Li 2018), changes in gene dosage of a subset of subunits of a protein complex, transient gene copy number imbalances during DNA replication, or fluctuations in gene expression can disrupt this coordinate expression, leading to the production of complex subunits that lack their binding partners. These orphan subunits have the potential to misfold and cause proteotoxic stress. How eukaryotic cells deal with stoichiometric imbalances is a fundamental yet largely unexplored question.
Aneuploidy represents an especially dramatic case of gene dosage alteration because changes in autosome copy number generally lead to a corresponding change in the levels of RNAs and proteins produced by genes located on aneuploid chromosomes (Pavelka et al. 2010; Torres et al. 2010; Dephoure et al. 2014). Not surprisingly, these dramatic alterations in cellular protein composition significantly impact cellular physiology, causing cell proliferation defects, metabolic alterations, and oxidative stress (for review, see Santaguida and Amon 2015). Protein homeostasis defects are especially prevalent in aneuploid cells. In budding yeast, many different aneuploidies harbor more protein aggregates, display decreased chaperone activity, and exhibit sensitivity to conditions that interfere with proteasomal degradation (Torres et al. 2007; Oromendia et al. 2012). In mammals, aneuploidy also disrupts protein homeostasis, leading to altered autophagy, abnormal protein folding, and accumulation of protein aggregates (Tang et al. 2011; Stingele et al. 2012; Donnelly and Storchová 2014; Santaguida et al. 2015). Why protein aggregation is so widespread in aneuploid cell lines was not understood.
We previously generated a series of haploid yeast strains, each harboring an extra copy of one of the 16 yeast chromosomes (referred to here as disomes) (Torres et al. 2007). Analysis of the localization of Hsp104, a disaggregase that associates with protein aggregates (Liu et al. 2010), showed that gain of any of the 16 yeast chromosomes causes an increase in the number of cells harboring Hsp104 foci, providing evidence for increased protein aggregation (Oromendia et al. 2012). Here we determine the molecular basis of this phenotype. We found that subunits of protein complexes encoded on excess chromosomes aggregate in aneuploid cells. Our analyses also provide insights into how cells respond to protein stoichiometry imbalances. Protein aggregation can quantitatively deplete excess protein from the cytosol. We conclude that protein aggregation commonly performs functional dosage compensation.
Results
Identification of proteins that aggregate in aneuploid yeast cells
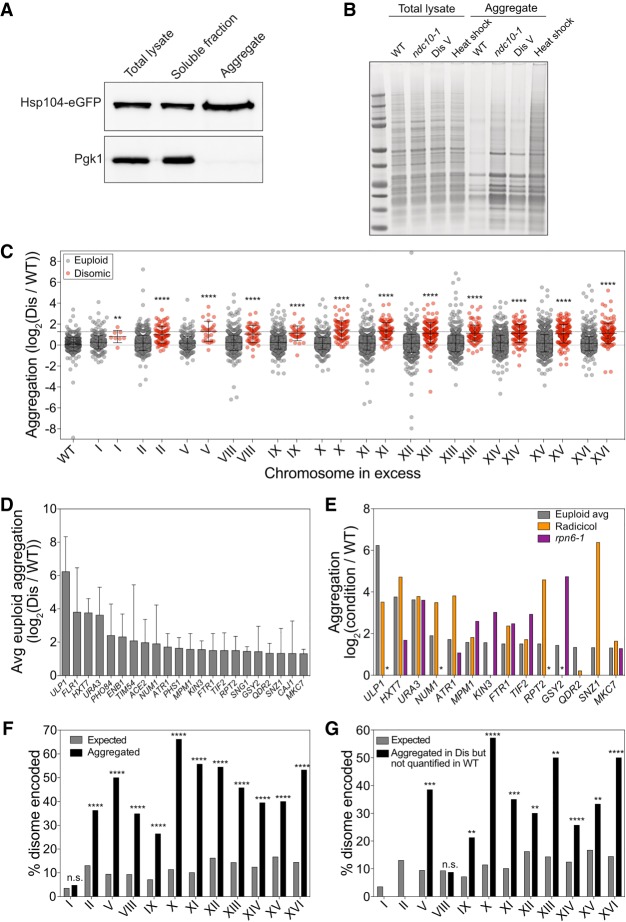
Previous studies of proteotoxic stress in aneuploidy had shown that cells with defined chromosome gains or randomly generated unknown karyotypes harbored increased levels of protein aggregates (Oromendia et al. 2012; Stingele et al. 2012; Santaguida et al. 2015). To gain insight into why protein aggregation is so common in aneuploid cells, we determined the composition of protein aggregates in disomic yeast strains. We isolated protein aggregates from disomic yeasts strains by a stringent differential centrifugation method (Koplin et al. 2010). Hsp104 was enriched in aggregate fractions (Fig. 1A); however, we note that aggregates isolated in this manner may not contain all Hsp104-decorated aggregates and also may contain aggregates not recognized by Hsp104. Analysis of protein aggregates by SDS-PAGE revealed that protein aggregation is increased in aneuploid cells. Cells that missegregate chromosomes at a high frequency because they carry a temperature-sensitive allele in the kinetochore component encoding gene NDC10 harbored high levels of protein aggregates (Fig. 1B). Increased amounts of aggregated proteins were also observed in haploid cells disomic for chromosome V (Fig. 1B).
Figure 1.
Identification of proteins that aggregate in aneuploid yeast cells. (A) Total lysate, aggregates, and soluble fractions obtained from exponentially growing cells expressing Hsp104-eGFP (A31392) were analyzed for Hsp104 and Pgk1 abundance. (B) Protein aggregates and total lysates were prepared from euploid cells (wild type [WT], A35797), ndc10-1 cells (ndc10-1, A13413) grown for 4 h at 30°C, disome V cells (disome [Dis] V, A28265), and euploid cells (A2587) after an 8-min heat shock at 42°C (heat shock). Total lysates and aggregate fractions were subjected to SDS-PAGE and stained with Coomassie. (C) WT and disome cells were grown to exponential phase in synthetic complete (SC) medium containing heavy lysine and light lysine, respectively. Aggregated proteins are separated into two dot plots, with red dots indicating proteins encoded on the duplicated chromosome and gray dots indicating proteins encoded on euploid chromosomes. The first column represents aggregates purified from a mixed sample of heavy lysine-labeled WT and light lysine-labeled WT. Lines represent mean and standard deviation (SD). The top dashed line at log2 1.27 shows the cutoff used to define aggregating proteins. (**) P < 0.01; (****) P < 0.0001, Mann-Whitney test. (D) The average (Avg) aggregate enrichment of proteins encoded on euploid chromosomes that were identified in aggregates of at least three out of 12 disomes. Only proteins with an average enrichment of ≥log2 1.27 as measured in C are shown. Error bars indicate SD. (E) The enrichment of proteins from D was compared with their enrichment in aggregates purified from cells treated with radicicol (orange) or cells harboring the rpn6-1 allele (purple) from Supplemental Figure S4. An asterisk indicates proteins that were not quantified in either the radicicol or rpn6-1 experiments because they did not pass the detection threshold in aggregates purified from the reference strain but were readily detected in aggregates isolated from radicicol-treated or rpn6-1 cells. (F) The percentage of proteins encoded by the duplicated chromosome that were enriched at a level greater than the aggregation threshold of log2 1.27 (black bars) and the percentage of proteins encoded by the duplicated chromosome as a fraction of the whole proteome (gray bars). (n.s.) Not significant; (****) P < 0.0001, cumulative distribution function for a hypergeometric distribution. (G) The percentage of proteins encoded by the duplicated chromosome that were not quantified by stable isotope labeling by amino acids in cell culture (SILAC) mass spectrometry (MS) because the heavy-labeled (WT) peptides did not pass the detection threshold (black bars) and the percentage of proteins encoded by the duplicated chromosome as a fraction of the whole proteome (gray bars). (n.s.) Not significant; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001, cumulative distribution function for a hypergeometric distribution.
Having established that aneuploidy causes an increase in protein aggregates that can be isolated by differential centrifugation, we used stable isotope labeling by amino acids in cell culture (SILAC) mass spectrometry (MS) to identify proteins that preferentially aggregate in 12 different disomic yeast strains (Fig. 1C; Supplemental Fig. S1A; Supplemental Data S1; Ong et al. 2002; Shevchenko et al. 2006). Reproducibility was high between individual experiments: 70% of proteins were identified in repeats of individual experiments (Supplemental Fig. S1B,C). Although biological replicates were well correlated, the mean of the SILAC ratios for all proteins combined in aggregates varied between replicates of the same disome (e.g., for disome II, the means were 0.59, 0.69, and 0.30). To account for this variability and to be able to conduct analyses on the aggregate data set as a whole, we mean-centered all experiments such that the mean relative enrichment was equal across experiments (Fig. 1C). Each experiment was mean-centered to 0 by subtracting the mean of all SILAC ratios in that experiment from all data points. To return the normalized values to a baseline that more closely resembles the increase in protein aggregation in disomic strains observed in the raw data, a constant (log2 0.27) was added to all normalized data points. This constant is the mean log2 ratio of all euploid-encoded proteins in the data set prior to normalization. Of note, we also identified proteins that were enriched in aggregates isolated from euploid strains compared with disome strains. However, in triplicate experiments for disome II, only four proteins (1.4%) were enriched more than twofold in aggregates from euploid cells, and their enrichment across replicate experiments was highly variable (Supplemental Fig. S1D,E).
Which proteins aggregate in disomic yeast strains? The similar banding patterns of wild-type (WT) and aneuploid aggregates on SDS-PAGE gels (Fig. 1B) indicated that aggregates were composed of the same proteins but that they aggregate more in aneuploid strains than in euploid strains. Comparison of the banding pattern of protein aggregates on SDS-PAGE with the banding pattern of purified ribosomes further suggested that protein aggregates of both euploid and disomic yeast strains were enriched for ribosomes (Supplemental Fig. S2A). To estimate the contribution of ribosomes to protein aggregates in disomic yeast strains, we first determined the abundance of proteins in aggregates in each strain relative to its euploid reference by summing the raw total intensity of all heavy-labeled peptides and all light-labeled peptides and then calculating a ratio of the two (Supplemental Fig. S2B). Nine out of 12 disomic strains contained more aggregated protein than euploid controls by this estimate. We then calculated the signal of each ribosomal protein as a percentage of the total signal for all aggregated proteins and determined that 75% of aggregated proteins were ribosomal proteins. Interestingly, the disomic strains with fewer ribosomes aggregating were the same strains that showed lower levels of total aggregate burden (Supplemental Fig. S2B,C), confirming that ribosomes make up the majority of aggregating proteins in disomic yeast strains. Two lines of evidence indicate that it is assembled ribosomes rather than individual subunits that accumulate in aggregates. First, almost all excess ribosomal subunits are quantitatively degraded in disomic yeast strains (Dephoure et al. 2014). Second, the Coomassie staining pattern of protein aggregates on SDS-PAGE resembles the pattern of purified intact ribosomes (Supplemental Fig. S2A). We conclude that ribosomes are abundant in aggregates purified by our method in both euploid and aneuploid yeast strains but aggregate more in disomes.
To determine which proteins other than ribosomes are found in aggregates purified by our differential centrifugation method, we assessed the presence of known phase-separated structures such as P bodies, stress granules, or nucleoli in purified aggregates (Banani et al. 2017) using the annotations by Jain et al. (2016). P-body proteins were significantly underrepresented in aggregates compared with total lysates (Supplemental Fig. S2D). We also identified fewer stress granule proteins and nucleolar proteins in aggregates compared with total lysates; however, this difference was not significant (Supplemental Fig. S2D). We conclude that components of phase-separated structures are not significantly enriched in our aggregate preparations.
Next, we conducted an unbiased gene ontology (GO) analysis (Supplemental Fig. S2E) to determine which proteins were in fact enriched in aggregates isolated from aneuploid strains. Macromolecular complex was the GO term most significantly associated with proteins in aggregates, suggesting that complex subunits may be predisposed to aggregation. Ribosomes were the second most significant GO term, confirming our observation that the organelle is enriched in our aggregate preparation. The term “nonmembrane-bound organelle” (fourth most significant GO term) includes ribosomes, which likely drives the significance of this GO term. The third most significant GO term was “cell,” which we interpret to mean that all GO terms with P-values larger than this generic GO term, which includes the GO terms mitochondrion, nucleolus, and membrane, are not likely to be meaningful. This conclusion is supported by our observation that proteins known to be components of the nucleolus are not enriched in aggregates isolated from disomic yeast strains.
Next, we examined the physical and chemical properties of aggregating proteins. For this analysis, we compared aggregating proteins with the whole yeast proteome and proteins identified in lysates from which the aggregates were purified. Proteins in both aggregates and total lysates had more disordered regions than the whole proteome (Supplemental Fig. S3A). Hydrophobicity did not affect aggregation propensity (Supplemental Fig. S3B). Amino acid composition appeared to affect protein aggregation in aneuploid yeast strains. Aggregated proteins were more basic and contained slightly more aromatic residues than proteins identified in lysates (Supplemental Fig. S3C–E). Whether these features influence aggregation propensity remains to be determined. However, based on our knowledge of the effects of aneuploidy on cell physiology and the fact that “macromolecular complex” was the most significant GO term describing proteins that aggregate in disomic yeast strains, we can envision two classes of proteins that aggregate in disomic yeast strains: Class 1 is proteins that rely extensively on protein folding pathways to achieve their native conformation. Such proteins could aggregate because protein quality control is compromised in aneuploid yeast strains (Torres et al. 2010; Oromendia et al. 2012). Aggregation of these proteins ought to be independent of the identity of the aneuploid chromosomes and should occur in multiple different disomic yeast strains. Class 2 is proteins encoded by the duplicated chromosome. Such proteins could aggregate because they are in excess. We first focused on proteins in class 1.
Proteins that aggregate in multiple disomes also aggregate in cells with compromised protein quality control
Aneuploid cells experience proteotoxic stress (Tang et al. 2011; Oromendia et al. 2012; Stingele et al. 2012; Santaguida et al. 2015). Proteins that rely extensively on protein folding pathways to achieve their native conformation could thus aggregate in aneuploid cells more than in euploid cells. To identify proteins that exhibit an increased aggregation due to the aneuploid state, we identified proteins that (1) were encoded on euploid chromosomes, (2) aggregated in at least three different disomic yeast strains, and (3) had an average enrichment of at least 2.4-fold (log2 1.27) in disomic aggregates. We arrived at this cutoff for enrichment of proteins in disome aggregates using a false discovery rate (FDR) of 5% (see the Materials and Methods). This analysis identified 22 proteins (Fig. 1D; Supplemental Data S2).
If proteins aggregate in multiple different disomic strains because proteostasis is compromised, they should also aggregate in euploid cells in which protein quality control pathways are inhibited. To test this, we analyzed protein aggregates of cells harboring a temperature-sensitive mutation in the proteasome subunit encoding gene RPN6 (Isono et al. 2005) and of cells treated with the Hsp90 inhibitor radicicol (Supplemental Fig. S4; Supplemental Data S2). For the 22 proteins identified as aggregating in multiple disomic strains, we obtained quantitative information for 14 in aggregates of either rpn6-1 cells, radicicol-treated cells, or both. Of these, 13 (94%) were enriched 2.4-fold (log2 1.27) in aggregates of rpn6-1 cells, radicicol-treated cells, or both (P < 0.001, hypergeometric cumulative distribution function) (Fig. 1E). We conclude that proteostasis deficiency causes protein aggregation in aneuploid cells regardless of whether gene copy number is altered.
Duplicated proteins are highly enriched in aneuploid aggregates
We next examined proteins encoded on disomic chromosomes. These proteins were indeed significantly enriched in aggregates isolated from their respective disomic strains (Fig. 1C, red dots). To determine the number of proteins that aggregate because they are produced from two rather than one gene copy, we used a cutoff of 2.4-fold (log2 1.27; based on a 5% FDR, described above) to define aggregate enrichment. We identified 437 proteins that were enriched in aggregates in a disome-specific manner (Fig. 1F; Supplemental Data S1). For example, chromosome II encodes for 13% of the yeast proteome when duplicated, yet 36% of proteins that aggregate in strains disomic for chromosome II are encoded on chromosome II. Similarly dramatic results are observed in all disomic strains (Fig. 1F). We further note that this enrichment of disome-encoded proteins in aggregates underestimates proteins that aggregate because they are produced in excess. Proteins that fall below an established signal to noise ratio in either the heavy or light channel cannot be included because a SILAC ratio cannot be calculated. To mitigate this limitation, we examined proteins that did not pass the signal to noise threshold in one channel but had a signal to noise ratio of at least twice the threshold in the other channel (see the Materials and Methods). This identified an additional 320 proteins that were quantified only in disome aggregates compared with just 72 that were identified only in aggregates of the euploid control strain (Supplemental Data S1). Importantly, 92 of the proteins quantified only in aggregates of disomic strains were encoded on the disomic chromosome, as opposed to just two that were quantified only in aggregates of euploid strains (Fig. 1G). We conclude that at least 529 proteins (437 proteins identified as enriched in disomes compared with WT aggregates + 92 identified only in disome aggregates) aggregate when their gene copy number is increased by twofold.
Increasing gene copy number by one causes protein aggregation in human cells
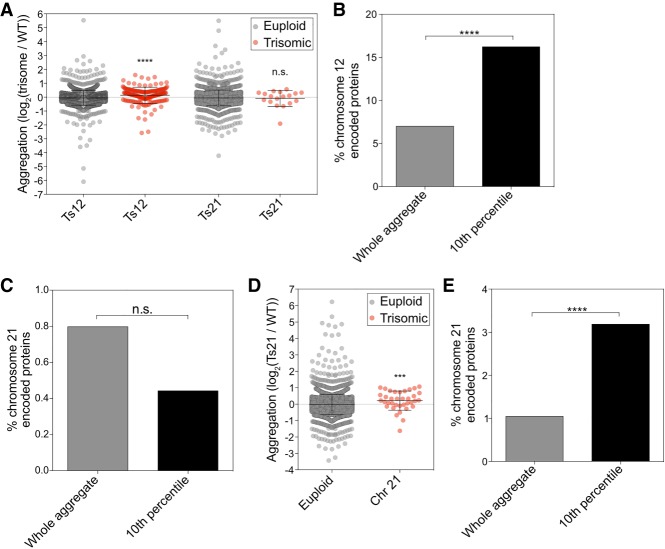
Is protein aggregation a feature of aneuploidy that is conserved across eukaryotes? To address this question, we analyzed protein aggregation in near-diploid human RPE-1 cells and two derivatives that were trisomic for chromosome 12 or 21 (Stingele et al. 2012). As in yeast, proteins most enriched in aggregates were encoded on the trisomic chromosome. Within the 10th percentile of proteins most enriched in trisomy 12 aggregates, 16.2% of proteins were encoded by chromosome 12 (Fig. 2A,B; Supplemental Data S3). This enrichment was highly significant (Fisher's exact test, P < 0.0001). In contrast, proteins encoded by other chromosomes had no significant enrichment, with the exception of proteins encoded by chromosome 4, which were slightly depleted (Fisher's exact test, P = 0.0297). We conclude that chromosome 12-encoded proteins are enriched in aggregates of trisomy 12 cells.
Figure 2.
Aggregate analysis in trisomic human cells. (A) RPE-1 cells and RPE-1 cells trisomic for either chromosome 12 or chromosome 21 were cultured in medium containing heavy lysine or light lysine, respectively, for 10 generations. Within each experiment, euploid-encoded proteins (gray dots) were plotted separately from trisome-encoded proteins (red dots). (****) P < 0.0001; (n.s.) not significant, Mann-Whitney test. (B,C) Enrichment of chromosome 12-encoded (B) and chromosome 21-encoded (C) proteins in total aggregates (gray bars) and among the top 10% most highly enriched aggregated proteins (black bars). (n.s.) Not significant; (****) P < 0.0001, cumulative distribution function for a hypergeometric distribution. (D) Euploid and trisomy 21 cells were treated with 25 nM chloroquine and 1 µM MG-132, and aggregates were plotted as in A. (***) P < 0.001, Mann-Whitney test. (E) Enrichment of chromosome 21-encoded proteins in total aggregates (gray bar) and among the top 10% most highly enriched aggregated proteins (black bar). (****) P < 0.0001, cumulative distribution function for a hypergeometric distribution. (Ts) Trisome; (Chr) chromosome.
Enrichment of chromosome 21-encoded proteins was not evident in aggregates purified from trisomy 21 cell lines (Fig. 2A,C; Supplemental Data S3), most likely due to the fact that chromosome 21 is the gene-poorest chromosome in humans. To increase protein aggregation in trisomy 21 cells, we prevented protein degradation by inhibiting the proteasome and lysosomal degradation with MG-132 and chloroquine, respectively. In this experiment, we observed that within the 10th percentile of proteins most enriched in trisomy 21 aggregates, 3.2% were encoded by chromosome 21. Overall, proteins encoded on chromosome 21 represent only 1% of proteins in aggregates (Fig. 2D,E; Supplemental Data S3). This enrichment was highly significant (Fisher's exact test, P = 0.0005). In contrast, proteins encoded on other chromosomes did not show significant enrichment. Among the chromosome 21-encoded proteins enriched in aggregates were two proteins known to contribute to disease phenotypes in trisomy 21: DOPEY2, a protein implicated in causing mental retardation (Rachidi et al. 2005), and APP, the precursor of the Alzheimer's disease-associated amyloid-β. We conclude that aggregation of proteins encoded on excess chromosomes is also a feature of aneuploidy in humans. We note that the enrichment of proteins encoded on excess chromosomes is less apparent in trisomic human cells than in disomic yeast cells. This is to be expected. Gain of a copy of a chromosome in a diploid cell causes fewer protein imbalances than gain of a chromosome in a haploid cell.
Stoichiometric imbalance of protein complexes can cause protein aggregation
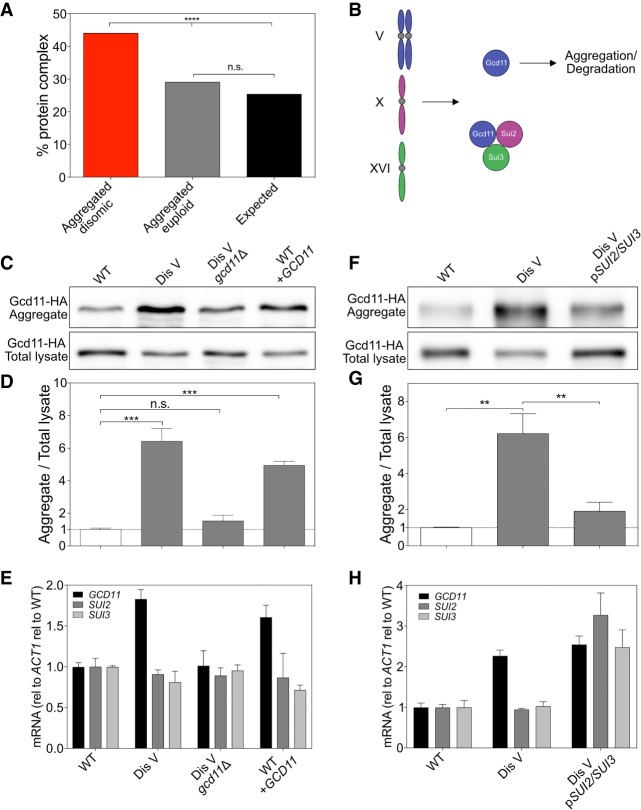
We next examined the mechanism by which altering gene copy number causes protein aggregation in aneuploid cells. Protein complex analysis using the annotation by Pu et al. (2009) showed that 44.2% of the 529 proteins that aggregated in disomic yeast strains when encoded on an excess chromosome were subunits of protein complexes (Fig. 3A). By comparison, only 29.2% of proteins encoded by euploid chromosomes that were enriched in aggregates were subunits of protein complexes (Fig. 3A).
Figure 3.
Stoichiometric imbalances of protein complex subunits cause protein aggregation. (A) The percentage of proteins encoded by duplicated chromosomes enriched by log2 ≥1.27 in aggregates (red), the percentage of proteins encoded by euploid chromosomes enriched by log2 ≥1.27 in aggregates (gray), and the percentage of proteins in the genome (black) that are annotated to form protein complexes by Pu et al. (2009) were calculated. (****) P < 0.0001; (n.s.) not significant, cumulative distribution function for a binomial distribution. (B) Diagram of eIF2 complex stoichiometry in disome V cells. (C) Gcd11-HA in aggregates and total lysates in WT (A40189), disome V (A40190), disome V GCD11-HA/gcd11Δ (A40191), and WT URA3::GCD11-HA (WT + GCD11; A40192) cells. Only one of the two GCD11 genes in disome V cells was tagged with HA to ensure that protein levels are comparable between strains. (D) Quantification of Western blots in C. n = 3; SD. (***) P < 0.001, t-test. (E) Cells were grown as in C, and mRNA levels for eIF2 subunits were determined. Values were normalized to ACT1 and then to WT expression levels. n = 3; SD. (F–H) Western blot, relative aggregation quantification, and mRNA expression for WT (A40193), disome V (A40194), and disome V pSUI2/SUI3 (A40195) cells as described in C–E. n = 3; SD. (**) P < 0.01, t-test. (Dis) Disome; (rel) relative.
We hypothesized that the enrichment of duplicated protein complex subunits in disome aggregates was due to protein complex subunits requiring binding to other subunits to acquire their native state. This hypothesis predicts that when gene copy number of the other complex subunits is altered in accordance with expression of the subunit produced in excess, aggregation should be prevented. We tested this prediction by studying the eIF2 complex, which is required for translation initiation and is composed of three subunits. The eIF2 γ subunit Gcd11 is encoded on chromosome V (Fig. 3B). Gcd11 was found in aggregates isolated from yeast strains disomic for this chromosome but returned to euploid levels in aggregates from disome V strains in which one copy of GCD11 was deleted (Fig. 3C–E). In fact, a single extra copy of GCD11 in an otherwise euploid strain was sufficient to cause Gcd11 aggregation (Fig. 3C–E). Thus, increased gene dosage of GCD11 is necessary and sufficient to cause aggregation of the protein. It is worth noting that Gcd11 aggregated less when expressed in excess by itself than when overexpressed due to an extra copy of chromosome V. This observation suggests that proteotoxic stress caused by disomy of chromosome V exaggerates aggregation of Gcd11.
To test whether aggregation of Gcd11 in disome V strains can be prevented by doubling the gene copy number of the eIF2 α subunit encoding gene SUI2 and the eIF2 β subunit encoding gene SUI3, we introduced a centromeric plasmid carrying both genes under their native promoters into disome V strains. Expression of SUI2 and SUI3 dramatically reduced Gcd11 aggregation in disome V cells (Fig. 3F–H). We note that expression of SUI2 and SUI3 also increased Gcd11 levels in lysates (Fig. 3F). A pool of Gcd11 is degraded when expressed in excess (Dephoure et al. 2014). Increased expression of SUI2 and SUI3 likely also protects Gcd11 from degradation. We conclude that stoichiometric imbalance of protein complexes can cause protein aggregation in aneuploid cells.
Excess proteins either aggregate or are degraded
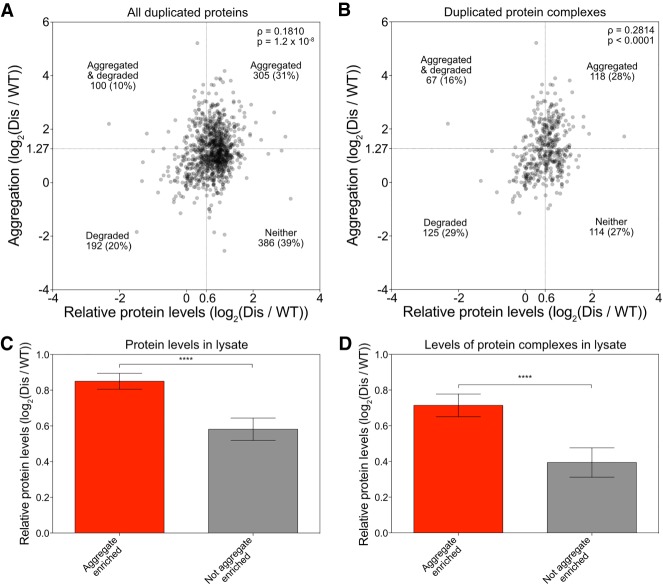
Previous studies showed that many proteins that function in protein complexes are degraded when in excess in aneuploid cells, restoring their levels to euploid or near-euploid levels (Dephoure et al. 2014; Ori et al. 2016; Gonçalves et al. 2017). We observed that many proteins aggregate when in excess. An important question regarding these two observations is whether excess subunits are down-regulated by both mechanisms or are neutralized by one or the other. To distinguish between these possibilities, we analyzed protein degradation and aggregation propensity for all disome-encoded proteins identified in our protein aggregate analysis and the protein degradation analysis of Dephoure et al. (2014). Note that both data sets were created with the same strains. Analysis of the relative abundance of proteins encoded on disomic chromosomes confirmed that the two data sets were indeed similar to each other despite slight differences in growth conditions (Supplemental Fig. S5).
We first asked whether relative abundance of a protein in total lysates correlates with protein aggregation; in other words: Is a protein more likely to aggregate when its levels in total lysate are higher than in a euploid strain? We indeed observed a weak but significant correlation (Fig. 4A). This observation indicates that proteins that are dosage-compensated by protein degradation are less likely to be found in aggregates. Because excess cytoplasmic ribosomal subunits are almost exclusively degraded (Dephoure et al. 2014), we also asked whether the correlation was driven by ribosomal proteins. Removing cytoplasmic ribosomal proteins from the data set weakened the correlation between abundance of proteins in extracts and their propensity to aggregate, but it was still significant (Supplemental Fig. S6A).
Figure 4.
Excess proteins are either aggregated or degraded. (A,B) Correlation between enrichment in protein aggregates (measured in Fig. 1C) and relative protein levels (measured in disome lysates by Dephoure et al. 2014) was determined for all proteins encoded by the duplicated chromosome quantified in both data sets (A) and duplicated subunits of protein complexes (B). Spearman correlation of 0.1810 (P = 1.2 × 10−8) in A and 0.2814 (P < 0.0001) in B. Dashed lines indicate thresholds for proteins that are considered aggregated (Y-axes) or degraded (X-axes). The number of proteins that fall into each quadrant is indicated. (C,D) All duplicated proteins (C) and duplicated complex subunits (D) were separated into two bins: (1) aggregated proteins (red bars), which were defined as proteins with an enrichment of at least log2 1.27 in disomic aggregates, and (2) nonaggregated proteins (gray bars), which were defined as proteins with an enrichment of log2 ≤0.727. Average relative levels in disome lysates as measured by Dephoure et al. (2014) are plotted. Error bars represent 95% confidence intervals. (****) P < 0.0001, Mann-Whitney test. (Dis) Disome.
To further assess whether protein aggregation and degradation are mutually exclusive, we analyzed the data using cutoffs previously set to define a protein to aggregate or be degraded when in excess. Dephoure et al. (2014) defined a disome-encoded protein as degraded when its abundance in extracts relative to a euploid reference was 1.52 (log2 0.6) instead of the expected 2 (log2 1). We defined any disome-encoded protein as aggregated when it was found enriched 2.4-fold (log2 1.27) in aggregates (Fig. 1). Based on these criteria, only 100 out of 983 (10%) disomic proteins were considered to both aggregate and be degraded when in excess. This is evident by the fact that the top left quadrant in the graph in Figure 4A is underpopulated (Fisher's exact test, P = 0.0045). This mutually exclusive behavior of disome-encoded proteins was not driven by ribosomal proteins because we observed the same underpopulation of the top left quadrant when we removed ribosomal proteins (Supplemental Fig. S6A). When we restricted our analysis to subunits of protein complexes, only 67 out of 424 (16%) proteins were both enriched in disome aggregates and degraded when in excess (Fisher's exact test, P = 0.0012) (Fig. 4B), although we note that this observed lack of overlap between aggregation and degradation was, to a significant extent, driven by ribosomal proteins (Supplemental Fig. S6B).
Another way to assess whether disome-encoded proteins aggregate or are degraded but not both is to ask whether disome-encoded proteins that are highly enriched in aggregates are present in total lysates at the twofold level expected for disome-encoded proteins or whether their levels are lower. We found that disome-encoded proteins that are enriched in aggregates are present at levels close to the expected level of twofold, indicating that they are not degraded (Fig. 4C). In contrast, levels of proteins that were detected in aggregates but not enriched in aggregates were lower in total lysates (Fig. 4C). Removal of ribosomal subunits from the analysis did not alter this conclusion (Supplemental Fig. S6C). The results were even more striking when we focused our analysis on disome-encoded proteins that are part of protein complexes (Fig. 4D; Supplemental Fig. S3D). We conclude that, when present in excess, proteins have a preferred fate of either degradation or aggregation.
Protein complex subunits that aggregate when in excess have lower turnover rates than degraded subunits
What determines whether excess protein complex subunits aggregate or are degraded? Computational analyses revealed that both classes of subunits harbor large protein–protein interfaces within their complexes compared with proteins that neither aggregate nor are degraded (Fig. 5A; Supplemental Data S4; Marsh et al. 2013). This is consistent with earlier work in mammalian cells demonstrating that protein complex subunits with larger interfaces tend to be unstable and rapidly degraded when not bound to their partners (McShane et al. 2016). If overexpressed relative to other components of the complex, these large interfaces could facilitate inappropriate protein–protein interactions, leading to aggregation instead of degradation (Levy et al. 2012).
Figure 5.
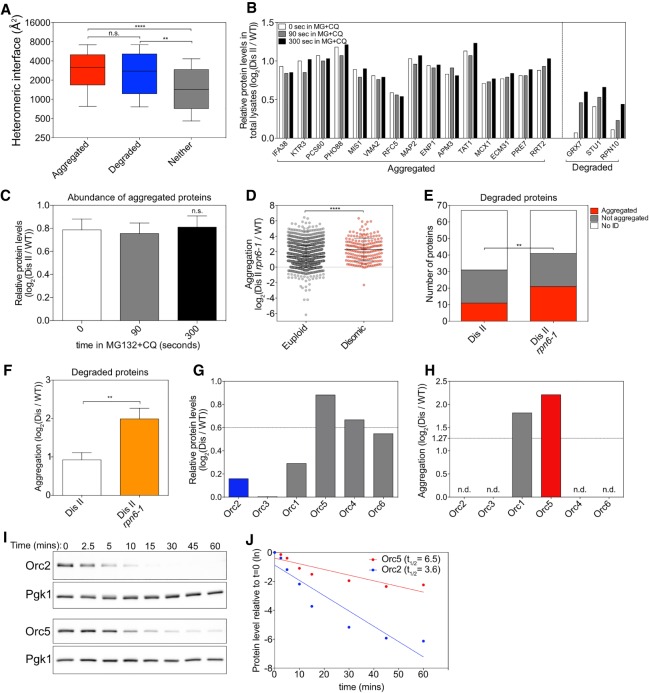
Protein half-life determines whether a protein aggregates or is degraded. (A) Proteins encoded on duplicated chromosomes were separated into three categories: aggregated, degraded, and neither. Aggregated proteins (red) and degraded proteins (blue) were defined as in Figure 4A. Proteins that were identified in our analysis and that by Dephoure et al. (2014) but did not pass the threshold for aggregation or degradation were considered neither aggregated nor degraded (gray). The heteromeric interface sizes of the proteins in each category are plotted as box plots, with whiskers representing the 10th–90th percentile. (****) P < 0.0001; (**) P < 0.01; (n.s.) not significant, Mann-Whitney test. (B) The change in levels of proteins encoded on chromosome II in disome II in total lysates of cells treated with 100 µM MG-132 (MG) and 10 mM chloroquine (CQ) relative to WT (data from Dephoure et al. 2014). Examples of aggregating proteins as determined in Figure 1C and of degraded proteins as determined by Dephoure et al. (2014) are shown. White bars indicate relative levels immediately before the addition of MG-132 and chloroquine, and gray bars and black bars indicate relative levels 90 and 300 sec thereafter, respectively. (C) Mean levels of all aggregating proteins as measured in B at each time point. Error bars indicate SEM. (n.s.) Not significant, Wilcoxon matched-pairs signed rank test. (D) Disome II rpn6-1 (A40196) or WT (A23504) cells were grown to exponential phase at 30°C in SC medium containing light lysine and heavy lysine, respectively, and aggregating proteins were identified. Lines indicate mean and SD. (****) P < 0.0001, Mann-Whitney test. (E) Proteins considered degraded when duplicated by Dephoure et al. (2014) were examined in aggregates purified from disome II cells (shown in Fig. 1C) and disome II rpn6-1 cells (shown in D). A protein was considered to aggregate when it was enriched by more than log2 1.27 in aggregates (red) and not aggregated when enriched by less than log2 1.27 (gray). “No ID” indicates proteins that were not identified in aggregates (white). (**) P < 0.01, cumulative distribution function for a binomial distribution. (F) Degree of aggregation was determined for all proteins in E in disome II aggregates and disome II rpn6-1 aggregates. Bars represent SD. (**) P < 0.01, Mann-Whitney test. (G,H) Relative protein levels as determined by Dephoure et al. (2014) (G) and relative aggregation as measured in Figure 1C (H) for origin recognition complex (ORC) subunits when encoded by disomic chromosomes. (n.d.) Not detected in aggregates. (I,J) Cells were grown to exponential phase at 30°C in YEP medium containing 2% raffinose. Expression of HA-tagged ORC2 (A40197) and ORC5 (A40198) was induced with 2% galactose for 20 min. Next, protein synthesis was halted by the addition of 2% glucose and 0.5 mg/mL cycloheximide (t = 0). Protein levels were determined. Pgk1 was used as a loading control (I). Protein levels were quantified relative to the loading control and normalized to the 0-min time point (J). (Dis) Disome; (ln) natural logarithm.
Given that both excess protein complex subunits that are degraded and those that aggregate harbor large protein-binding interfaces, we hypothesized that their fate could be determined by differences in recognition by the ubiquitin/proteasome or lysosomal degradation systems. Previous work showed that degradation of proteins that are encoded on a disomic chromosome can be prevented by treating disomic cells with the proteasome inhibitor MG-132 and the lysosomal-degradation inhibitor chloroquine (Fig. 5B, right part of the graph; Dephoure et al. 2014). Importantly, we found that preventing protein degradation did not affect overall levels of individual proteins that aggregate (Fig. 5B, left part of the graph) and average levels of all proteins that aggregate (Fig. 5C; Supplemental Data S4). Therefore, proteins found in aggregates generally have low turnover rates.
To further assess whether protein stability determines whether a protein aggregates, we analyzed the effects of proteasome inhibition on the fate of proteins that are normally degraded when in excess. Partial inhibition of the proteasome using a temperature-sensitive rpn6-1 mutant grown at the semipermissive temperature (30°C) caused a general increase in protein aggregation for all proteins in disome II cells (Fig. 5D; Supplemental Data S4). Sixty-seven disomic proteins are normally degraded in cells disomic for chromosome II (Dephoure et al. 2014). We identified 31 of these in our analysis. Eleven out of 31 (35.5%) aggregated in disome II cells with a functional proteasome. In disome II rpn6-1 cells, we identified 41 of these 67 proteins. Twenty-one (51.2%) of these were enriched in aggregates (Fig. 5E). We then asked whether the 67 proteins that were typically degraded when duplicated in disome II cells were enriched in aggregates when proteasome function is compromised. In cells with normal proteasome function, the 67 proteins had a mean enrichment of 1.9 (log2 0.92) in disome II aggregates. In disome II rpn6-1 cells, their mean enrichment increased to 4.0 (log2 2.0), indicating that many of these usually degraded proteins are now deposited in protein aggregates (Fig. 5F).
We next determined the effects of protein stability on the choice between degradation and aggregation by analyzing the origin recognition complex (ORC), in which some subunits are degraded when in excess while others are not (Dephoure et al. 2014). The ORC is a six-subunit complex essential for the initiation of DNA replication (Bell et al. 1993). All six subunits are encoded on different chromosomes in yeast.
Orc2 levels are mostly attenuated (reduced to 1.1 [log2 0.16] relative to WT) by degradation when the ORC2 gene is present in two copies instead of one (Fig. 5G; Dephoure et al. 2014; ). In contrast, Orc5 is not attenuated (present at 1.8 [log2 0.88] relative to WT) by degradation but is highly enriched in aggregates when encoded by two gene copies (Fig. 5G,H). To mimic stoichiometric imbalance, we transiently overexpressed ORC2 and ORC5 individually from the galactose-inducible GAL1-10 promoter. Half-life measurements showed that overproduced Orc5 was twice as stable as overproduced Orc2 (Fig. 5I,J). We conclude that the stability of a protein can determine whether it aggregates or is degraded when produced in excess. Our data further indicate that surprisingly subtle half-life differences can determine a protein's dosage compensation fate. What minimal half-life is required for a protein to be eliminated by degradation when in excess remains to be determined.
Dosage compensation by protein aggregation
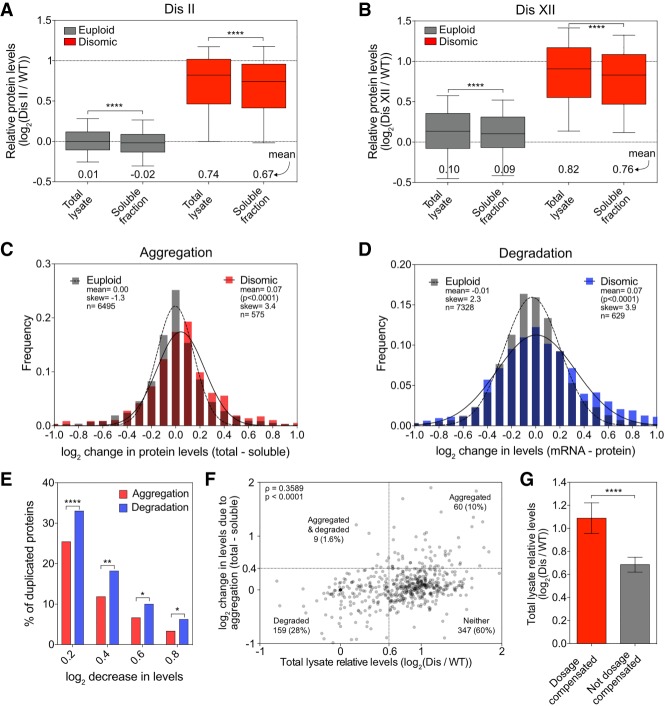
Degradation of excess subunits of protein complexes serves as a form of dosage compensation (Dephoure et al. 2014). Can protein aggregation serve the same purpose by sequestering excess subunits in an inaccessible aggregate compartment? To address this question, we quantified the relative abundance of proteins that remained in the soluble fraction following aggregate isolation in yeast strains disomic for chromosome II or XII and compared it with their abundance in extracts prior to aggregate removal (referred to here as total lysate) (Supplemental Fig. S1A). We observed a small but nevertheless significant difference in relative abundance of proteins encoded on euploid chromosomes between soluble fractions and total lysates (Fig. 6A,B; Supplemental Data S5), which is consistent with the observation that proteins encoded on euploid chromosomes are also found enriched in aggregates isolated from aneuploid cells (Fig. 1C). However, depletion of proteins encoded on disomic chromosomes from the soluble fraction was much more dramatic. Their mean relative abundance in the soluble fraction was noticeably decreased to 1.59-fold (log2 0.67) compared with 1.66-fold (log2 0.74) in total lysates for disome II and 1.69-fold (log2 0.76) compared with 1.77-fold (log2 0.82) for disome XII (Fig. 6A,B; Supplemental Data S5).
Figure 6.
Dosage compensation by protein aggregation. (A,B) Total lysate and soluble fractions, obtained as described in Figure 1A, were analyzed for disome II (A) and disome XII (B). Proteins encoded by euploid chromosomes (gray) and proteins encoded by the duplicated chromosome (red). Data are represented as box plots, with whiskers extending to 10th and 90th percentiles. Means are indicated below. The top dashed line represents the theoretical mean for proteins encoded by the duplicated chromosome. (****) P < 0.0001, Mann-Whitney test. (C) Changes in protein levels due to aggregation for proteins quantified in A and B were calculated for every protein by subtracting its log2 ratio in the soluble fraction from its log2 ratio in the total lysate. Disome II and disome XII data were pooled and separated into two subsets: euploid chromosome-encoded proteins (gray) and disomic chromosome-encoded proteins (red). Frequency distributions of each subset were then generated using a bin size of 0.1. Distributions were fit to Gaussian curves for euploid (dashed line) and disome (solid line). P-value shows that mean change of disome-encoded proteins is significantly different from the mean change of euploid-encoded proteins (Mann-Whitney test). (D) Changes in protein levels due to degradation for disome II and disome XII were calculated by subtracting the relative protein level from the relative mRNA level for each gene as measured by Dephoure et al. (2014). Frequency distributions were generated, and curve fitting was performed on the pooled data as in C. P-value shows that mean change of disome-encoded proteins is significantly different from the mean change of euploid-encoded proteins (Mann-Whitney test). (E) The percentage of proteins encoded by the duplicated chromosome that decreased in levels by at least a log2 of 0.2, 0.4, 0.6, and 0.8 due to aggregation (red bars) was calculated using the data described in C, and the percentage of those that decreased due to degradation (blue bars) was calculated using the data in D. (****) P < 0.0001; (**) P < 0.01; (*) P < 0.05, χ2 test. (F) The correlation between protein levels in total lysate and reduction in levels due to aggregation was determined for proteins encoded on duplicated chromosomes from the pooled data set of disome II (A) and disome XII (B). Spearman correlation of 0.3589 (P < 0.0001). Dashed lines indicate thresholds for proteins being considered dosage-compensated by aggregation (Y-axes) or degradation (X-axes). The number of proteins that fall into each quadrant is indicated (note that 19 data points fell outside the range of the axes but were included in the calculations). (G) Proteins encoded on duplicated chromosomes were separated into two categories: Dosage-compensated proteins (red) were defined as their levels being reduced by at least log2 0.4 in the soluble fraction relative to the total lysate. “Not dosage-compensated” proteins (gray) were defined as their levels being reduced by less than log2 0.4. Mean levels in total lysates are plotted; error bars indicate 95% confidence intervals. (****) P < 0.0001, Mann-Whitney test. (Dis) Disome.
The decrease of disome-encoded proteins in the soluble fraction could be due to many proteins experiencing small amounts of aggregation or a few proteins aggregating to a large degree. To distinguish between these possibilities, we first calculated the change in levels for each protein by subtracting protein levels in the soluble fraction from those in the total lysate. We then pooled the data from the disome II and disome XII analysis and generated two bins: proteins that were encoded on disomic chromosomes and proteins encoded on euploid chromosomes. As expected, the changes in levels of the 6495 proteins encoded by euploid chromosomes were evenly distributed (Fig. 6C; gray bars); changes in levels of the 575 proteins encoded by disomic chromosomes were not. Both the mean and skew significantly deviated from the expected distribution (Fig. 6C). Of the 575 disome-encoded proteins, 147 (26%) were depleted by 1.15-fold (log2 0.2) in the soluble fraction compared with total lysate (Fig. 6E), and 12% were depleted by 1.32-fold (log2 0.4). Full attenuation, defined as a 1.74-fold (log2 0.8) decrease, occurred for 3% of proteins (Fig. 6E). This indicates that reduction in protein levels for duplicated proteins in the soluble fraction is largely due to many proteins decreasing by a small degree. However, full attenuation by aggregation also occurs.
How does dosage compensation by aggregation compare with dosage compensation by protein degradation? Previous studies showed that 21% of proteins decrease by 1.52-fold (log2 0.6) due to either down-regulation of gene expression or protein degradation (Dephoure et al. 2014). To assess what fraction of the proteome is attenuated solely by protein degradation, we subtracted protein levels from RNA levels in the data set published by Dephoure et al. (2014). We then pooled data for disome II and disome XII, allowing us to examine 7328 measurements for proteins encoded on euploid chromosomes and 629 measurements for disome II- and disome XII-encoded proteins. As expected, the changes in levels of the 7328 proteins encoded by euploid chromosomes were evenly distributed (Fig. 6D, gray bars). The 629 proteins encoded by disomic chromosomes behaved differently. Both the mean and skew were larger than observed in the expected distribution (Fig. 6D). Of the 629 disome-encoded proteins, 209 (33.2%) were depleted by 1.15-fold (log2 0.2) relative to RNA levels (Fig. 6E), and 18.4% were depleted by 1.32-fold (log2 0.4). Full attenuation (1.74-fold [log2 0.8]) occurred for 6.4% of proteins (Fig. 6E). We conclude that small degrees of attenuation of disome-encoded proteins occur by both aggregation and degradation. Reduction to levels approaching those seen in euploid cells occurs predominantly through protein degradation.
Finally, we asked whether dosage compensation of a particular protein was mediated by both protein degradation and aggregation or one or the other in our disome II and XII data set. First, we determined whether a correlation existed between the relative abundance of a disome-encoded protein in the total lysate and the degree of dosage compensation by aggregation. This was the case, indicating that proteins with higher relative expression levels are more likely to be dosage-compensated by aggregation (Fig. 6F). To further assess whether protein aggregation and degradation are mutually exclusive, we analyzed the data using cutoffs defined previously (Dephoure et al. 2014). Disome-encoded proteins present in the total lysate at levels below 1.52 (log2 0.6; instead of the expected 2 [log2 1]) were considered dosage-compensated by degradation. Disome-encoded proteins that were depleted in the soluble fraction by at least 1.52 (log2 0.6) compared with the total lysate were considered dosage-compensated by aggregation. Based on these criteria, only nine out of 575 (1.6%) disomic proteins were dosage-compensated by both aggregation and degradation, as evident by the fact that the top left quadrant in the graph in Figure 6F is underpopulated (Fisher's exact test, P = 0.0011). We also asked whether disome-encoded proteins that are dosage-compensated by aggregation are present in total lysates at the twofold level expected for disome-encoded proteins. This was the case (Fig. 6G), demonstrating that they are not degraded. In contrast, levels of proteins considered not dosage-compensated by aggregation were lower in total extracts. Our data indicate that excess proteins are dosage-compensated by aggregation or degradation but rarely by both.
Discussion
Our analysis of protein aggregation in disomic yeast strains provided insight into why protein aggregation is a universal feature of the aneuploid state and revealed protein aggregation as a means of dosage compensation. This dosage compensation not only is relevant in aneuploid cells but could very well contribute to stoichiometry control in euploid cells that encounter stoichiometric imbalances due to transient imbalances caused by variability in gene expression. Remarkably, aggregation is nearly as effective as protein degradation at lowering levels of excess proteins. Whether protein aggregation serves a cytoprotective role in situations where high levels of unassembled complex subunits are present in cells (i.e., in aneuploid cells) is an important question that remains to be answered.
Which proteins aggregate in aneuploid cells?
Our analysis of protein aggregation in a series of disomic yeast strains provided insight into why protein aggregation is a universal feature of the aneuploid state. We identified proteins encoded on the disomic chromosomes as well as proteins encoded on euploid chromosomes to be enriched in aggregates.
Among the proteins found in aggregates of multiple different aneuploid strains, ribosomes were the most prominent. Ribosomes comprise ∼75% of aggregates isolated from both euploid cells and aneuploid cells but aggregate more readily in aneuploid cells, as judged by the fact that they harbor more aggregates. Why ribosomes are enriched in aggregates of disomic yeast strains is not clear. Perhaps a higher fraction of ribosomes is defective in disomic yeast strains, causing them to aggregate.
Ribosomes were not the only proteins commonly found in aggregates of aneuploid yeast strains. Interestingly, the proteins that aggregated in multiple different aneuploid cell lines also aggregated when protein quality control pathways were impaired. This observation raises the interesting possibility that proteins that are especially dependent on protein quality control pathways to maintain their solubility are “canary in the coal mine” proteins of the state of cellular protein quality control. It will be interesting to determine which properties make the canary proteins so sensitive to perturbations in protein homeostasis.
We also analyzed proteins that aggregate because their encoding genes were duplicated due to disomy. These proteins were strongly enriched for subunits of protein complexes, leading us to hypothesize that subunits of heteromeric protein complexes present at levels twice that of their binding partners are prone to aggregation. We went on to demonstrate that this was true in the case of the eIF2 complex. Our analysis of human trisomic cell lines further revealed that aggregation of proteins that are in excess is surprisingly sensitive to alterations in gene expression. Changing gene expression by as little as 50% causes aggregation of many proteins.
Cellular response to excess subunits of protein complexes
Our findings have important implications for euploid cells. Although expression of subunits of the same complex is tightly coordinated in eukaryotes (Li et al. 2014; Taggart and Li 2018), it is likely that even euploid cells encounter transient stoichiometric imbalances. Variability in expression of individual subunits can lead to stoichiometric imbalances, where individual subunits lack binding partners or where protein complexes are only partially assembled.
Our study shows that 61.5% of proteins either aggregate or are degraded when produced in excess. Among proteins that function in complexes, 73% of proteins either aggregate or are degraded. Interestingly, the 27% that are neither aggregated nor degraded have significantly smaller heteromeric protein-binding interfaces than protein complex subunits that aggregate or are degraded. We propose that excess proteins that do not aggregate or get degraded are simply less aggregation-prone and lack the signals that target them for degradation. Whether there are features that distinguish proteins that are degraded from proteins that are aggregated remains to be determined.
Our data also indicate that aggregation and degradation of proteins encoded on disomic chromosomes is largely mutually exclusive. Intuitively, this makes sense: If excess proteins are degraded, there is no excess protein to aggregate. What determines whether excess proteins are degraded or aggregate is their half-life when unbound. This is what we observed for Orc2 and Orc5. Overexpressed Orc2 has a shorter overall half-life, presumably because excess Orc2 is degraded. Orc5 has a longer overall half-life when overexpressed because excess Orc5 aggregates. What was surprising was that a change in protein half-life of twofold appeared to determine whether an overexpressed protein aggregates or is eliminated by degradation. What determines the half-life of a protein and what minimal half-life is required to be eliminated by degradation when in excess remains to be determined.
Dosage compensation by protein aggregation
Our results indicate that protein aggregation is nearly as effective as protein degradation at lowering levels of excess proteins. Using stringent cutoffs to designate a protein being dosage-compensated, we found that 12% of disome-encoded proteins were depleted from the soluble fraction by 1.32-fold due to aggregation; depletion by protein degradation occurred for 18.4% of proteins.
We hypothesize that protein aggregation could serve a cytoprotective function that shields aneuploid and euploid cells from toxic activities of excess protein complex subunits. Testing this hypothesis requires the development of methods that prevent protein aggregation in aneuploid cells, a task that has proven difficult. Protein aggregation as a cytoprotective mechanism has been proposed during cellular and organismal aging (David et al. 2010; Walther et al. 2015), suggesting that it could also serve this function in aneuploid cells. However, it is worth noting that the aggregation that occurs during aging does not function to normalize stoichiometric imbalances but rather to protect cells from highly abundant proteins that exceed their solubility during aging. Protein aggregation has also been found to protect cells during stress by sequestering misfolded proteins and targeting them to specific subcellular compartments (Escusa-Toret et al. 2013; Miller et al. 2015). Aggregation of disease-inducing proteins such as amyloid β, which is associated with Alzheimers disease, has also been proposed to protect neurons from toxic oligomers (Caughey and Lansbury 2003). Our observation that widespread aggregation serves the purpose of protecting cells from gene dosage excess raises the intriguing possibility that aggregation may be one mechanism that allows cancer cells to tolerate aneuploidy. Analysis of the degree of protein aggregation in cancer cell lines that are highly aneuploid and thus experience large-scale stoichiometric imbalances yet have high proliferative potential could shed light on this question.
Materials and methods
Yeast strains, plasmids, and growth conditions
All yeast strains are derivatives of W303 and are described in Supplemental Table S1. Primers are listed in Supplemental Table S2, and plasmids are listed in Supplemental Table S3. Yeast strains were generated and manipulated as described previously (Guthrie and Fink 1991). Cells were grown at 30°C in YEP supplemented with 2% raffinose (YEP-R), 2% raffinose + 2% galactose (YEP-RG), or 2% glucose (YEP-D) or in synthetic complete medium supplemented with 2% glucose (SC-D). For SILAC experiments, cells were grown overnight in SC-D medium containing 100 µg/mL heavy (13C6 15N2 [K8]; Cambridge Isotope Labs) or light (K4) (Sigma-Aldrich) L-lysine. Cells were then diluted to OD600 = 0.05 to 0.1 and grown to OD600 = 0.4–1.0 before harvesting lysates. Strains harboring temperature-sensitive mutations in NDC10 or RPN6 were grown overnight at room temperature and grown to exponential phase at room temperature before shifting to the semipermissive temperature of 30°C. ndc10-1 strains were grown for 4 h and rpn6-1 strains were grown for 6 h at 30°C before harvesting for aggregate purification. For radicicol treatments, cells harboring a deletion in the gene encoding the drug transporter Pdr5 were grown to exponential phase in SC medium at 30°C, and radicicol (Cayman Chemical Company) or DMSO was added to the culture at a final concentration of 70 µM. Cells were grown in the presence of radicicol or DMSO for 30 min at 30°C before harvesting for aggregate purification.
Disomes used in this study are derivatives of those published in Torres et al. (2007). Gene deletions, fusion proteins, and promoter swaps were generated using PCR-based methods (Longtine et al. 1998) in a WT W303 yeast strain. Disomes carrying gene manipulations were constructed by crosses. Karyotypes of key disomic strains were verified by whole-genome sequencing.
GCD11-HA and GCD11Δ were generated by PCR-based methods (Longtine et al. 1998). GCD11-HA with its native promoter and terminator was cloned into p24 by Gibson cloning (Gibson et al. 2009) and integrated at ura3 by NcoI digestion. The pSUI2/SUI3 CEN plasmid was constructed by Gibson cloning (Gibson et al. 2009) SUI2 and SUI3 with their native promoters and terminators into p158.
Cell culture and SILAC labeling of RPE-1 cells
RPE-1 WT (control) cells were grown in heavy (K8) SILAC DMEM (DMEM with high glucose minus glutamine, lysine, and arginine) supplemented with 10% dialyzed FBS (Gibco), 50 µg/mL K8 lysine (Cambridge Isotope Labs), 85 µg/mL arginine (Sigma-Aldrich), and 2 mM glutamine/1 mM sodium pyruvate (Thermo Fisher). RPE-1 cells trisomic for chromosome 12 or 21 were grown in light (K0) SILAC DMEM (same formulation as above, with 50 µg/mL K0 lysine; Sigma-Aldrich). Cells were cultured for eight generations prior to testing K8 labeling efficiency by MS and then expanded for harvest at passage 10. Prior to harvest, cells were treated with 25 µM chloroquine for 16 h followed by addition of 1 µM MG-132 (Calbiochem) for 6 h to induce protein aggregation. Cells were harvested by trypsinization, neutralized in SILAC DMEM, and washed with PBS; an equal number of heavy and light cells were mixed (WT + Ts12; WT + Ts21), treated with sodium azide, and snap-frozen for aggregate purification.
Aggregate purification and analysis
Aggregate purification was carried out based on methods published by Koplin et al. (2010) with some modifications to the cell lysis procedure. Cells were grown to exponential phase in SC medium under the conditions indicated above. Sodium azide was added to a final concentration of 50 mM in the cultures before pelleting cells. Ten OD600 units of cell pellets were washed with 50 mM sodium azide and snap-frozen in liquid nitrogen. Frozen pellets were then resuspended in 1 mL of buffer containing 1 M sorbitol, 100 mM sodium citrate, 60 mM EDTA, 10 mM DTT (pH 7.0; SCED), 1 mM PMSF, and 2× protease inhibitor tablets (Roche). Cells were pelleted, resuspended in 1 mL of SCED buffer containing 1 mg/mL 20T zymolyase (MP Biomedicals), and rotated gently for 8 min at room temperature. Digested cells were pelleted by spinning at 250g for 5 min at 4°C. Pellets were washed gently in ice-cold SCED buffer containing 1 mM PMSF and 2× protease inhibitors. Cells were lysed by resuspending in 1.5 mL of ice-cold lysis buffer containing 20 mM NaPi (pH 6.8), 10 mM DTT, 1 mM EDTA, 0.1% Tween 20 (Sigma-Aldrich), 1 mM PMSF, and 5× protease inhibitors and then sonicated for eight pulses at output level 4 and 50% duty cycle. Lysates were cleared by spinning at 850g for 2 min at 4°C, yielding the total lysate fraction. Protein concentrations of the cleared lysate were determined by Bradford (Bio-Rad). For quantitative Western blotting and Coomassie staining, samples were diluted to the same protein concentration. A small aliquot of the total lysate fraction was taken for analysis, and aggregates were pelleted from the remainder of the total lysate by spinning the extract at 16,000g for 20 min at 4°C. The supernatant was removed and reserved as the soluble fraction. Pellets were then washed twice by resuspending in buffer containing 20 mM NaPi (pH 6.8), 2% NP40, 1 mM PMSF, and 2× protease inhibitors and then sonicated for six pulses at output level 4 and 50% duty cycle. A final wash was carried out in buffer containing 20 mM NaPi (pH 6.8), 1 mM PMSF, and 2× protease inhibitors, and then pellets were sonicated for four pulses at output level 2 and 60% duty cycle, yielding the aggregate fraction. All centrifugations during the wash steps were at 16,000g for 20 min at 4°C.
For Western blot analysis and Coomassie staining analysis, total lysate fractions were boiled in SDS loading buffer. Aggregate fractions were resuspended in buffer containing 200 mM NaPi (pH 6.8), 5% SDS, 8 M Urea, 10 mM DTT, and 0.01% bromophenol blue (HU buffer) and then vortexed at 1500 rpm for 5 min at 60°C. For Coomassie staining, SDS-PAGE gels were stained with Imperial protein stain (Thermo Scientific) per the manufacturer's instructions. For Western blotting, proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and detected by the following antibodies: Hsp104-eGFP was detected using a mouse anti-GFP antibody (JL-8 epitope; Clontech) at a dilution of 1:1000. Pgk1 was detected using a mouse anti-Pgk1 antibody (Invitrogen) at a 1:10,000 dilution. Gcd11-HA was detected using a mouse anti-HA antibody (HA.11 epitope; BioLegend). Quantification was performed using Fiji software (Schindelin et al. 2012).
Ribosome purification
Cells were grown to exponential phase, pelleted by centrifugation, washed with water, and then snap-frozen in liquid nitrogen. Cells were incubated in lysis buffer (20 mM HEPES at pH 7.4, 100 mM potassium acetate, 2 mM magnesium acetate, 3 mM DTT, protease inhibitor tablets, 1 mg/mL zymolyase) for 5 min at 4°C and then lysed by French press (Sim-Aminco). The lysate was cleared by centrifuging at 19,000 rpm (Beckman Coulter, JA25.50 rotor) for 20 min at 4°C. The cleared lysates were applied to a 30% sucrose gradient containing 20 mM HEPES (pH 7.4), 500 mM potassium acetate, 2 mM magnesium acetate, and 3 mM DTT. Ribosomes were pelleted by centrifugation at 50,000 rpm (Beckman Coulter, type 70 Ti rotor) for 4 h at 4°C. The pellet was dissolved in lysis buffer and analyzed by SDS-PAGE.
Quantitative proteomics of yeast aggregates
Five OD600 units of heavy-labeled cells were mixed with five OD600 units of light-labeled cells, and aggregates were harvested as described above. Purified SILAC-labeled aggregates were boiled in SDS lysis buffer (50 mM Tris at pH 7.5, 2 mM EDTA, 2% SDS) and separated on 4%–12% Tris-glycine by SDS-PAGE gels (Invitrogen). Proteins were visualized by Coomassie staining (45% methanol, 5% glacial acetic acid, 3 g/L Coomassie brilliant blue G-250) and subjected to gel band cutting (Shevchenko et al. 2006), destaining, and in-gel Lys-C (Wako Chemicals) digest at a 50:1 protein to protease ratio to liberate peptides. Following digest, the buffer-containing peptides and gel slices were acidified using 50% acetonitrile/5% formic acid, and recovered peptides were vacuum-centrifuged to near dryness. Each gel fraction was desalted via StageTip, dried via vacuum centrifugation, and reconstituted in 5% acetonitrile and 5% formic acid for liquid chromatography-tandem MS (LC-MS/MS) processing.
Quantitative proteomics of RPE-1 cell aggregates
RPE-1 cells were cultured and treated as described above. Aggregate purification was carried out as described for yeast cells with the following modifications. Cells were collected, treated with sodium azide, and snap-frozen. Frozen cell pellets were resuspended in cold lysis buffer (described above) and sonicated. Samples were analyzed using the same mass spectrometric method and data processing workflow as yeast aggregates (see below).
LC-MS
MS data were collected using a Q Exactive mass spectrometer (Thermo Fisher) coupled with a Famos autosampler (LC Packings) and an Accela600 LC pump (Thermo Fisher). Peptides were separated on an ∼18-cm column with 100 µm inner diameter packed with Accucore150 resin (2.6 μm, 150 Å; Thermo Fisher Scientific); ∼1 µg of peptides was loaded onto the column for each analysis. Peptides were separated with a 2-h gradient of 5%–26% acetonitrile in 0.125% formic acid with a flow rate of ∼300 nL/min. For the MS1 scan, resolution was set to 70,000 with an automatic gain control (AGC) target of 1 × 106 and a maximum injection time of 250 msec. We selected the top 20 precursors for HCD MS2 analysis with the following parameters: resolution, 17,500; AGC, 1 × 105; maximum injection time, 90 msec; isolation window, 2 Th; normalized collision energy (NCE), 25; and centroid spectrum data type. In addition, unassigned and singly charged species were excluded from MS2 analysis, and dynamic exclusion was set to automatic.
For data analysis, mass spectra were processed using a Sequest-based in-house software pipeline. MS spectra were converted to mzXML using a modified version of ReAdW.exe. MS2 spectra were searched with a database, including all predictive ORFs for entries from the yeast SGD (http://www.yeastgenome.org/download-data; downloaded March 12, 2014). For human aggregate samples, database searching included all entries from the human UniProt database (August 10, 2011). Both yeast and human databases were concatenated with a reverse database composed of all protein sequences in reverse order. Searches were performed using a 50-ppm precursor ion tolerance. Product ion tolerance was set to 0.03 Th. Oxidation of methionine residues (+15.995 Da) and heavy lysine (K8) incorporation (+8.0142) were set as a variable modification. Peptide spectral matches (PSMs) were altered to a 1% FDR (Elias and Gygi 2007, 2010). PSM filtering was performed using a linear discriminant analysis, as described previously (Huttlin et al. 2010), while considering the following parameters: XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy. PSMs were identified, quantified, collapsed to a 1% FDR, and then further collapsed to a final protein-level FDR of 1%. Furthermore, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides. Four gel band regions were cut per sample (individual lanes) of the SDS-PAGE gel; these four regions were processed separately, and each is an independent MS sample (to decrease the complexity of peptides in each run). Protein assembly was used to group the four runs of a given sample together for batched analysis and calculation of log2(heavy/light) ratios. The resulting data were filtered for SILAC-quantified proteins based on the summed signal to noise for heavy and light peptides; contaminant peptides identified in the search were removed at this step. An intensity cutoff of 10 was applied for summed heavy plus light channels, and each individual channel had an intensity cutoff of 5 (to avoid identifying proteins with an intensity of 0 in either the light or heavy channel; i.e., proteins found in only one condition or the other). A SILAC-labeled total lysate reference sample (to allow for normalization of SILAC mixing) was obtained as described above. Proteins were pelleted by trichloroacetic acid (TCA) precipitation, dried via vacuum centrifugation, and digested with Lys-C. Resulting peptides were desalted using StageTips, and MS data collection and analysis were performed as described above for aggregate samples.
SILAC MS of total lysate and soluble fractions
Total lysate samples were prepared, and soluble fractions (supernatant after aggregate isolation) were collected as described above. Approximately 800 µg of protein per sample was pelleted via TCA precipitation and dried via vacuum centrifugation. Samples were reduced and alkylated followed by digestion with Lys-C. Resulting peptides were desalted using StageTips, and samples were dried via vacuum centrifugation.
Each sample was fractionated using offline basic pH reversed-phase chromatography (BPRP HPLC) (Wang et al. 2011). We used an Agilent 1200 pump equipped with a degasser and a photodiode array (PDA) detector (set at 220- and 280-nm wavelength) from Thermo Fisher Scientific. Peptides were subjected to a 50-min linear gradient from 5% to 35% acetonitrile in 10 mM ammonium bicarbonate (pH 8) at a flow rate of 0.6 mL/min over an Agilent 300Extend C18 column (3.5-μm particles, 4.6 mm ID, and 220 mm in length). The peptide mixture was fractionated into a total of 96 fractions, which were consolidated into 24, from which 12 nonadjacent samples were analyzed (Paulo et al. 2016). Samples were subsequently acidified with 1% formic acid and vacuum-centrifuged to near dryness. Each consolidated fraction was desalted via StageTip, dried again via vacuum centrifugation, and reconstituted in 5% acetonitrile and 5% formic acid for LC-MS/MS processing. Samples were analyzed using a Q Exactive mass spectrometer using the same instrument method and data processing as described above for yeast aggregate samples.
Real-time RT-qPCR
Cells were grown to exponential phase in SC medium containing 2% glucose, as for aggregate purification. Five OD600 units of culture were pelleted by centrifugation at 3000 rpm for 2 min at 4°C. The pellet was resuspended in 1 mL of cold SC medium, transferred to a 2-mL microfuge tube, and pelleted by centrifugation at 3000 rpm for 2 min at 4°C. The pellet was flash-frozen in liquid nitrogen and stored at −80°C. To extract total RNA, ∼200 μL of glass beads, 400 μL of TES buffer (10 mM Tris at pH 7.5, 10 mM EDTA, 0.5% SDS), and 400 µL of acid phenol:chloroform (pH 4.5) were added to the cell pellet, and the tubes were vortexed for 30 min at 65°C. The phases were separated by centrifugation, and the top phase was transferred to a new tube containing 1 mL of 120 mM sodium acetate in ethanol to precipitate RNA at 4°C. Precipitates were collected by centrifugation and resuspended in 100 μL of DEPC-treated water. Total RNA was further purified using the RNeasy minikit (Qiagen), including DNase treatment, according to the manufacturer's instructions. cDNA was synthesized from 750 ng of total RNA using the SuperScript III first strand synthesis supermix kit (Invitrogen) with random hexamer primers according to the manufacturer's instructions. Real-time qPCR reactions were run using the SYBR Premix Ex Taq Perfect real-time kit (TaKaRa Bio) and a Roche LightCycler 480 (Roche) according to the manufacturer's instructions. qPCR primers are listed in Supplemental Table S2.
Turnover measurements for aggregated proteins
Protein turnover for aggregated proteins in disome II cells was determined by examining protein levels relative to WT before treatment with MG-132 and choloroquine and 90 and 300 sec thereafter. Data were generated by Dephoure et al. (2014).
Orc protein half-life measurements
Cells were grown to exponential phase at 30°C in YEP medium containing 2% raffinose. Expression of HA-tagged ORC2 and ORC5 was induced by diluting cultures into YEP medium containing 2% raffinose and 2% galactose for 20 min. The 0-min time point was taken, and protein synthesis was halted by the addition of 2% glucose and 0.5 mg/mL cycloheximide (Sigma-Aldrich). Time points were taken at the indicated times after the addition of glucose and cycloheximide. For each time point, ∼0.5 OD600 units of culture were pelleted by centrifugation at 3000 rpm for 2 min at 4°C. Cells were incubated at 4°C in 5% TCA for at least 10 min. Cell pellets were washed once with acetone and dried. Cells were lysed with glass beads in 100 μL of lysis buffer (50 mM Tris-Cl at pH 7.5, 1 mM EDTA, 2.75 mM dithiothreitol [DTT], complete protease inhibitor cocktail [Roche]) with a bead beater. Samples were boiled in 1× SDS loading buffer. Following SDS-PAGE and transfer of proteins to a nitrocellulose membrane, proteins were detected with the following antibodies. Pgk1 was detected using a mouse anti-Pgk1 antibody (Invitrogen) at a 1:10,000 dilution. Orc2-HA and Orc5-HA were detected using a mouse anti-HA antibody (HA.11 epitope; BioLegend). Quantification was performed using Fiji software (Schindelin et al. 2012). Protein levels were calculated by subtracting a background measurement for each band, dividing the intensity of the Orc band by the intensity of the Pgk1 band, and normalizing all time points to the 0-min time point. Half-life calculations were made by fitting curves to an exponential decay function in Prism (Graphpad).
SILAC MS data normalization
To account for imperfect mixing of heavy- and light-labeled cells, log2 ratios of proteins identified in aggregates were normalized by subtracting the average log2 ratio of all euploid-encoded proteins in the total lysate obtained before pelleting aggregates for each experiment.
To control for nonbiological batch effects between disomic cell lines, each experiment was mean-centered to 0 by subtracting the mean of all SILAC ratios in that experiment from all data points. To return the normalized values to a baseline that more closely resembles the increase in protein aggregation in disomic strains observed in the raw data, a constant (log2 0.27) was added to all normalized data points. This constant is the mean log2 ratio of all euploid-encoded proteins in the data set prior to normalization.
Cutoff determination for aggregation
To decide what level of enrichment in disome aggregates constituted calling a protein aggregated, we used a 5% FDR obtained from the analysis of aggregates obtained from a WT(heavy)–WT(light) SILAC MS analysis. In this analysis, only 5% of identified proteins were enriched by greater than log2 1.27 in WT(heavy) aggregates.
Signal to noise quantification of relative aggregate burden
To account for the fact that some proteins are more abundant in aggregates than others, aggregate burden for each disome relative to WT was calculated by comparing the signal to noise ratio of all heavy-labeled proteins with the signal to noise ratio of all light-labeled proteins in protein aggregates. The sum of all light-labeled signal to noise ratio was divided by the sum of all heavy-labeled signal to noise ratio. This ratio was then normalized by dividing the ratio of light signal to noise ratio to the heavy signal to noise ratio for the total lysate.
GO analysis
GO component analysis was performed using the Saccharomyces Genome Database (SGD) GO term finder (accessed October 12, 2018).
Aggregate property calculations
Disorder predictions for protein sequences were calculated using IUPred on the “long” setting. For each sequence, per-residue disorder scores were averaged across the full length of the protein. Grand average hydropathy (GRAVY) scores, isoelectric points, aliphatic indices, and aromaticity scores were calculated using the YeastMine tool on SGD (accessed October 31, 2018).
Proteins not quantified due to low signal in the WT channel
In our MS analysis pipeline, peptides for which either individual analysis channel had an intensity of <5 were discarded to avoid identifying proteins with an intensity of 0 in either the light or heavy channel; i.e., proteins found in only one condition or the other because a SILAC ratio could not be calculated. We examined proteins that failed to pass the signal to noise threshold in one channel but had a signal to noise ratio of at least twice the threshold in the other channel. Using this method, we found 320 proteins that were quantified only in the disome channel compared with 72 quantified in only the WT channel (Supplemental Data S1). Importantly, 92 of the proteins quantified only in the disome channel were encoded by the duplicated chromosome as opposed to just two duplicated gene-encoded proteins quantified in only the WT channel. It is possible that proteins that are identified exclusively in the disome channel never aggregate in WT cells. It is also possible that with increased coverage, these proteins would pass the signal threshold in both channels. To distinguish between these possibilities, we took advantage of a difference in coverage between two of our replicate experiments for disome II. In the low-coverage experiment (366 proteins quantified), nine proteins were not quantified due to low signal in the WT channel. In the high-coverage experiment (847 proteins quantified), we identified six of these proteins, and all six passed the detection threshold in both channels, allowing us to calculate a SILAC ratio. Four of these six proteins were considered aggregating using 2.4-fold (log2 of 1.27) as a cutoff. We conclude that proteins that cannot be quantified by SILAC MS due to low signal in WT cells are also likely enriched in aneuploid aggregates.
Heteromeric interface size determination
We searched for subunits of heteromeric protein complexes in the Protein Data Bank (on February 2, 2017) with >90% sequence identity to Saccharomyces cerevisiae genes. We selected a single heteromeric structure for each gene. For genes with multiple structures available, we selected the structure with the greatest number of unique subunits in the complex followed by the greatest number of atoms present in the polypeptide chain for ties. The total amount of heteromeric interface formed by each subunit was calculated using AREAIMOL (Winn et al. 2011).
Statistical analysis
The statistical tests used are indicated in the figure legends, Materials and Methods, and/or Results. Values of n, the definition of center, error bars (e.g., standard deviation and confidence intervals), and significance levels are reported in the figures and/or figure legends. All box plots represent median and interquartile range, with whiskers indicating the 10th–90th percentile. All of the indicated statistical tests were performed using Matlab or Prism.
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al. 2016) partner repository in seven parts with the following data set identifiers: PXD011874, PXD011875, PXD011876, PXD011877, PXD011878, PXD011915, and PXD011916.
Supplementary Material
Acknowledgments
We thank Arzu Sandikci for providing purified ribosomes; Steve Bell for assistance with ORC experiments; and David Pincus, Luke Berchowitz, and members of the Amon laboratory for suggestions and critical reading of this manuscript. This work was supported by National Institutes of Health grants CA206157 and GM118066 to A.A., who is an investigator of the Howard Hughes Medical Institute and the Paul F. Glenn Center for Biology of Aging Research at Massachusetts Institute of Technology. J.A.M. is supported by a Medical Research Council Career Development Award (MR/M02122X/1). S.S. was supported by the American Italian Cancer Foundation (AICF), a Fellowship in Cancer Research from Marie Curie Actions and the Italian Association for Cancer Research (AIRC), and a Koch Institute Quinquennial Cancer Research Fellowship.
Author contributions: C.M.B., L.P.V., J.N.W., S.S., J.A.P., Z.S., J.W.H, J.A.M., and A.A. designed the experiments. C.M.B., L.P.V., J.N.W., S.S., J.A.P., J.A.M., and A.A. performed experiments and analyzed results. C.M.B., L.P.V., J.N.W., J.A.M., and A.A. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.327494.119.
References
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Kobayashi R, Stillman B. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262: 1844–1849. 10.1126/science.8266072 [DOI] [PubMed] [Google Scholar]
- Caughey B, Lansbury PT. 2003. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci 26: 267–298. 10.1146/annurev.neuro.26.010302.081142 [DOI] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. 2010. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol 8: e1000450 10.1371/journal.pbio.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O'Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. 2014. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife 3: e03023 10.7554/eLife.03023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Storchová Z. 2014. Dynamic karyotype, dynamic proteome: buffering the effects of aneuploidy. Biochim Biophys Acta 1843: 473–481. 10.1016/j.bbamcr.2013.11.017 [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214. 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. 2010. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol 604: 55–71. 10.1007/978-1-60761-444-9_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa-Toret S, Vonk WIM, Frydman J. 2013. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol 15: 1231–1243. 10.1038/ncb2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gonçalves E, Fragoulis A, Garcia-Alonso L, Cramer T, Saez-Rodriguez J, Beltrao P. 2017. Widespread post-transcriptional attenuation of genomic copy-number variation in cancer. Cell Syst 5: 386–398.e4. 10.1016/j.cels.2017.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. 1991. Methods in enzymology: molecular biology of Saccharomyces cerevisiae. Academic Press, San Diego. [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189. 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Saito N, Kamata N, Saeki Y, Toh-E A. 2005. Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J Biol Chem 280: 6537–6547. 10.1074/jbc.M409364200 [DOI] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. 2016. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. 2010. A dual function for chaperones SSB–RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol 189: 57–68. 10.1083/jcb.200910074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy ED, De S, Teichmann SA. 2012. Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc Natl Acad Sci 109: 20461–20466. 10.1073/pnas.1209312109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157: 624–635. 10.1016/j.cell.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero A, Hao X, Öling D, Grantham J, Nyström T. 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140: 257–267. 10.1016/j.cell.2009.12.031 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961.<953::AID-YEA293>3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- Marsh JA, Hernández H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA. 2013. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell 153: 461–470. 10.1016/j.cell.2013.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane E, Sin C, Zauber H, Wells JN, Donnelly N, Wang X, Hou J, Chen W, Storchová Z, Marsh JA, et al. 2016. Kinetic analysis of protein stability reveals age-dependent degradation. Cell 167: 803–815.e21. 10.1016/j.cell.2016.09.015 [DOI] [PubMed] [Google Scholar]
- Miller SBM, Mogk A, Bukau B. 2015. Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J Mol Biol 427: 1564–1574. 10.1016/j.jmb.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386. 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- Ori A, Iskar M, Buczak K, Kastritis P, Parca L, Andrés-Pons A, Singer S, Bork P, Beck M. 2016. Spatiotemporal variation of mammalian protein complex stoichiometries. Genome Biol 17: 47 10.1186/s13059-016-0912-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. 2012. Aneuploidy causes proteotoxic stress in yeast. Genes Dev 26: 2696–2708. 10.1101/gad.207407.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, O'Connell JD, Everley RA, O'Brien J, Gygi MA, Gygi SP. 2016. Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. J Proteomics 148: 85–93. 10.1016/j.jprot.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. 10.1038/nature09529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu S, Wong J, Turner B, Cho E, Wodak SJ. 2009. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res 37: 825–831. 10.1093/nar/gkn1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi M, Lopes C, Costantine M, Delabar J-M. 2005. C21orf5, a new member of Dopey family involved in morphogenesis, could participate in neurological alterations and mental retardation in Down syndrome. DNA Res 12: 203–210. 10.1093/dnares/dsi004 [DOI] [PubMed] [Google Scholar]
- Santaguida S, Amon A. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 16: 473–485. 10.1038/nrm4025 [DOI] [PubMed] [Google Scholar]
- Santaguida S, Vasile E, White E, Amon A. 2015. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev 29: 2010–2021. 10.1101/gad.269118.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. 2012. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol 8: 608 10.1038/msb.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart JC, Li G-W. 2018. Production of protein-complex components is stoichiometric and lacks general feedback regulation in eukaryotes. Cell Syst 7: 580–589.e4. 10.1016/j.cels.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-C, Williams BR, Siegel JJ, Amon A. 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512. 10.1016/j.cell.2011.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. 10.1126/science.1142210 [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83. 10.1016/j.cell.2010.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–D456. 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P, Morimoto RI, Dobson CM, Vendruscolo M, Mann M, et al. 2015. Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161: 919–932. 10.1016/j.cell.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, et al. 2011. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11: 2019–2026. 10.1002/pmic.201000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al. 2016) partner repository in seven parts with the following data set identifiers: PXD011874, PXD011875, PXD011876, PXD011877, PXD011878, PXD011915, and PXD011916.