In this review, Core et al. discuss the recent advances in our understanding of the early steps in Pol II transcription, highlighting the events and factors involved in the establishment and release of paused Pol II. They also discuss a number of unanswered questions about the regulation and function of Pol II pausing.
Keywords: gene expression, gene regulation, Pol II pausing, transcription regulation
Abstract
Precise spatio–temporal control of gene activity is essential for organismal development, growth, and survival in a changing environment. Decisive steps in gene regulation involve the pausing of RNA polymerase II (Pol II) in early elongation, and the controlled release of paused polymerase into productive RNA synthesis. Here we describe the factors that enable pausing and the events that trigger Pol II release into the gene. We also discuss open questions in the field concerning the stability of paused Pol II, nucleosomes as obstacles to elongation, and potential roles of pausing in defining the precision and dynamics of gene expression.
Cell type- and condition-specific patterns of gene expression involve tight control of multiple steps in the transcription cycle. Early regulatory events govern the association of DNA-binding transcription factors (TFs) with their target motifs and the recruitment of RNA polymerase II (Pol II) and general TFs (GTFs) to the promoter region of a gene. These events enable transcription initiation and the synthesis of a short, nascent RNA. However, Pol II then pauses promoter-proximally, awaiting further signals before entering the gene body. The fate of the paused early elongation complex is absolutely decisive for gene output. If paused Pol II successfully transitions into productive elongation, a functional full-length mRNA can be made. However, a failure in the maturation of paused Pol II toward productive RNA synthesis short circuits the process of gene expression. Thus, understanding the factors that govern stable Pol II pausing and release are of paramount importance toward an appreciation of gene regulation in human health and disease.
The phenomenon of promoter-proximal Pol II pausing was described in the mid 1980s by John Lis (Gilmour and Lis 1986; Rougvie and Lis 1988) during study of the Drosophila heat-shock protein (hsp) genes. Pol II was found to be recruited to hsp promoters prior to heat stress and to begin RNA synthesis. However, Pol II halted within the initially transcribed region, remaining tightly associated with the short, nascent RNA. Importantly, this paused Pol II could be released into productive elongation under conditions that triggered gene activity. These findings from the heat-shock system in Drosophila came together with discoveries from a number of mammalian systems, including the well-studied mammalian β-globin locus, at which engaged Pol II was found to be enriched in the first half of the gene (Gariglio et al. 1981). Similar contemporaneous reports of regulation during early elongation of the HIV LTR, c-myc, and Fos genes emerged (Kao et al. 1987; Krumm et al. 1992; Strobl and Eick 1992; Plet et al. 1995). Together, these studies signified that early elongation is a key checkpoint in gene expression. However, despite >30 yr of research, the mechanisms underlying the regulation of early elongation remain an area of vigorous study and even controversy to this day. In particular, active debates remain about how much of the paused elongation complex proceeds into productive RNA synthesis versus undergoing promoter-proximal termination (Henriques et al. 2013; Jonkers et al. 2014; Chen et al. 2015; Krebs et al. 2017; Nilson et al. 2017; Shao and Zeitlinger 2017; Erickson et al. 2018).
An obstacle to dissecting the mechanisms of Pol II pausing is the lack of an in vitro system that accurately recapitulates the position, duration, and regulation of pausing as it occurs in cells. Although several promising biochemical systems have been developed to study early elongation (Adelman et al. 2005; Cheng and Price 2007; Luse et al. 2011; Li and Gilmour 2015; Vos et al. 2018b), a majority of these require reduced concentrations or omission of specific ribonucleotide triphosphate (NTP) substrates to artificially stall Pol II in the promoter-proximal region. Although a great deal has been learned from these systems, our inability to generate paused Pol II in vitro at physiological NTP concentrations indicates that we still lack some key factor or feature in these cell-free experiments. Therefore, the revolution in our understanding of pausing came with the development of genomic techniques for monitoring Pol II distribution and dynamics in metazoan cells. Indeed, early mapping of Pol II using chromatin immunoprecipitation (ChIP)–chip revealed peaks of Pol II near many promoters (Kim et al. 2005; Guenther et al. 2007), indicating that Pol II goes through rate-limiting steps near the transcription start site (TSS) before being released into the gene. Further work revealed that the majority of active genes showed promoter-proximal enrichment of the Pol II signal and that these polymerases were often engaged in early elongation, suggesting that promoter-proximal pausing was a widespread phenomenon (Muse et al. 2007; Zeitlinger et al. 2007). Upon the development of global run-on sequencing (GRO-seq), it became clear that the peaks of Pol II near promoters represented elongation complexes that were paused but competent to resume transcription elongation (Core et al. 2008). In fact, subsequent work has demonstrated that the vast majority of all Pol II detected near promoters in ChIP or permanganate footprinting assays (Table 1) represents polymerase that is actively engaged in elongation and capable of resuming synthesis in vitro (Core et al. 2012; Li et al. 2013; Pugh and Venters 2016).
Table 1.
Assays for the detection of elongation complexes and paused Pol II
Thanks to the growing repertoire of approaches to study transcription elongation in vivo (Table 1), the field now appreciates that pausing of Pol II in early elongation is an obligate part of the transcription cycle; a step that is universally experienced by Pol II at all genes. In particular, potent small molecule inhibitors that block pause release were demonstrated to broadly trap Pol II at promoters and abrogate nearly all RNA synthesis in Drosophila and mammalian cells (Chao and Price 2001; Ni et al. 2008; Rahl et al. 2010; Henriques et al. 2013; Jonkers et al. 2014). The efficiency of pause release is thus recognized as a central determinant of gene expression levels, raising great interest in the mechanisms that drive this process. Moreover, pausing has been shown recently to be a universal hallmark of Pol II transcription, with Pol II transcribed enhancers, upstream antisense RNAs, and long noncoding RNAs exhibiting pausing that appears identical to that described at promoters (Bunch et al. 2014; Core et al. 2014; Andersson et al. 2015; Scruggs et al. 2015; Bunch et al. 2016; Henriques et al. 2018; Rennie et al. 2018; Tome et al. 2018). Thus, a better understanding of pausing is also necessary toward developing models for the regulated expression and functions of noncoding RNA species.
In this review, we present recent advances in our understanding of the early steps in Pol II transcription, highlighting the events and factors involved in the establishment and release of paused Pol II. We describe current models for pausing control and will discuss a number of unanswered questions about the regulation and function of Pol II pausing.
Pausing takes center stage in gene regulation
The transcription cycle involves a number of sequential steps, many of which are regulated in response to cellular signals or cues (Fig. 1). DNA sequences around promoters must be rendered accessible to the transcription machinery, which often involves cooperation between TFs (Fig. 1A), coactivators such as the histone acetyltransferase p300/CBP and/or chromatin remodeling machines such as Swi/Snf (e.g. Chrom. mod) (Fig. 1A, green). Together, these factors stimulate binding by the GTFs (Fig. 1B, brown) and association of the Mediator complex (Fig. 1B, blue), which facilitate recruitment of Pol II (Fig. 1B, red) to form the preinitiation complex (PIC). Within the PIC, Pol II and the GTF TFIID make extensive contacts with DNA that extend ∼40 bp on either side of the TSS (Nogales et al. 2017). These interactions include binding to core promoter motifs associated with the TSS itself as well as those upstream of and downstream from the TSS (described in detail below).
Figure 1.
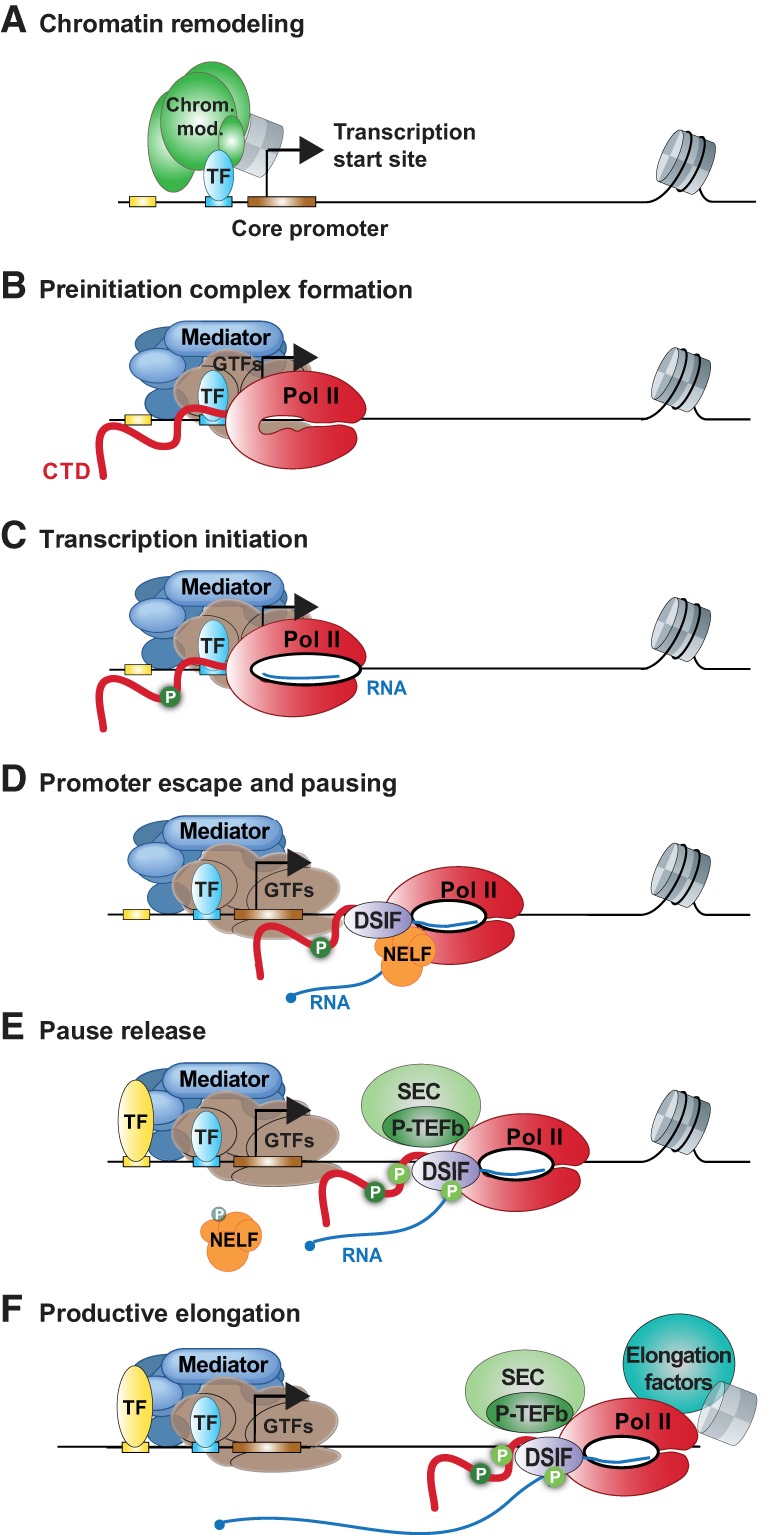
Outline of early steps in the transcription cycle. Promoter DNA is shown with the TSS depicted by an arrow. TF-binding motifs and core promoter sequences are designated, with the average position of the first nucleosome shown, ∼140 bp downstream from the TSS. Distinct protein factors and complexes are involved in each step as labeled in the figure. Phosphorylation of DSIF (dichloro-1-β-D-ribofuranosylbenzimidazole [DRB] sensitivity-inducing factor), NELF and the Pol II CTD is shown as a green “P.” Active positive transcription elongation factor b (P-TEFb) is typically found within the superelongation complex (SEC) as shown. Elongation factors that facilitate productive elongation through chromatin include Spt6, FACT, and polymerase-associated factor 1 (PAF1).
The C-terminal domain (CTD) of the Pol II Rpb1 subunit (Fig. 1, shown as red tail on Pol II) is comprised of numerous repeats of the consensus sequence Y1S2P3T4S5P6S7. The Pol II CTD is largely unphosphorylated during initiation, allowing stabilizing interactions of the unmodified CTD with the Mediator complex (Wong et al. 2014; Sato et al. 2016; Tsai et al. 2017). However, across the transcription cycle, the CTD repeats are posttranslationally modified at distinct sites, which stimulates stage-appropriate interactions with transcription initiation, elongation, and RNA processing factors (Buratowski 2009).
The association of the multimeric TFIIH complex with the growing transcriptional assembly allows the TFIIH-associated “helicase” to translocate DNA toward Pol II. This drives opening of the DNA strands to form a transcription bubble (Fig 1C) and allows entry of the template DNA into the Pol II active site (Fishburn et al. 2016; Schilbach et al. 2017; Dienemann et al. 2019). The polymerase is thus able to initiate RNA synthesis, catalyzing the formation of a nascent RNA that is complementary to template DNA sequences. As synthesis begins, the RNA remains associated with the template DNA to form an RNA–DNA hybrid within the Pol II active site (Fig. 1C, RNA, blue). The RNA–DNA hybrid imparts considerable stability to the elongation complex, and, as it grows, the polymerase becomes increasingly committed to the DNA template (Luse 2013). During productive elongation, an RNA–DNA hybrid of 8–9 nt is maintained: with each nucleotide of RNA added to the growing 3′ end of the transcript, the transcription bubble moves forward to expose the next template DNA base. Simultaneously, 1 bp of RNA–DNA is disrupted at the 5′ end of the hybrid to allow RNA to exit the polymerase and template and nontemplate DNA to reanneal. However, during early elongation, tight contacts with promoter DNA and GTFs often persist, preventing the polymerase from moving forward along DNA as the RNA chain is extended and causing “scrunching” of an extended transcription bubble within Pol II (Fig. 1C; Fazal et al. 2015). This conformation is energetically unfavorable, and, after >10 nt of RNA has been formed, the upstream region of the extended bubble collapses (Holstege et al. 1997; Hieb et al. 2006; Luse 2013), allowing template and nontemplate DNA to reassociate. The energy gained through this process has been proposed to propel the polymerase forward, facilitating the disruption of upstream contacts with DNA and the general transcription machinery and enabling a step called promoter escape.
Concomitant with initiation, the TFIIH-associated kinase Cdk7 phosphorylates the CTD on serine residues at positions 5 and 7 of the repeat (Fig 1C, dark-green P) (Buratowski 2009; Glover-Cutter et al. 2009; Kim et al. 2009; Eick and Geyer 2013; Rodriguez-Molina et al. 2016). The role of Ser7 phosphorylation (Ser7-P) at protein-coding genes is still enigmatic, but biochemical evidence indicates that this modification helps prime the CTD for subsequent phosphorylation events (Czudnochowski et al. 2012). Ser5 phosphorylation (Ser5-P), however, has clear roles in the early stages of mRNA transcription. Ser5-P on the CTD disrupts interactions with Mediator (Wong et al. 2014), facilitating promoter escape. Furthermore, the Ser5-P CTD is recognized by the enzymes that cap the nascent RNA 5′ end to protect it against degradation (Ho and Shuman 1999; Rodriguez et al. 2000; Fabrega et al. 2003). Indeed, RNA capping commences as soon at the nascent transcript emerges from the RNA exit channel, at ∼19–22 nt in length (Rasmussen and Lis 1993,1995; Chiu et al. 2002; Tome et al. 2018).
The emerging nascent RNA also serves as a recognition site for factors such as Spt5 that associate selectively with the Pol II elongation complex (Fig. 1D; Missra and Gilmour 2010). Spt5 and its binding partner Spt4 form a heterodimer called DSIF (dichloro-1-β-D-ribofuranosylbenzimidazole [DRB] sensitivity-inducing factor) (discussed in detail below) that influences several steps in transcription elongation. Spt5 docks on Pol II near the RNA exit channel (Bernecky et al. 2017; Vos et al. 2018a,b), where it facilitates capping of the nascent RNA (Pei and Shuman 2002). The negative elongation factor (NELF) complex then recognizes the Pol II–Spt5 interface and associates with the elongation complex as it transcribes through the promoter-proximal region (Fig. 1D). Notably, early elongation is slower and more prone to pausing than synthesis further downstream (Luse 2013; Jonkers et al. 2014), and Pol II typically pauses within the region 25–50 nt downstream from the TSS. As described in detail below, pausing can be influenced by the DNA and RNA sequences within the Pol II active site (Nechaev et al. 2010) and the adoption of a nonproductive conformation of the RNA–DNA hybrid (Vos et al. 2018a,b). The paused state is stabilized by both Spt5 and NELF (Yamaguchi et al. 1999), with NELF in particular preventing reactivation of the Pol II catalytic site (Adelman and Henriques 2018; Vos et al. 2018a,b).
Release of paused Pol II into productive RNA synthesis is triggered by the activity of the kinase, positive transcription elongation factor b (P-TEFb) (Fig. 1E). P-TEFb is recruited to promoters through interactions with TFs, Mediator, and coactivators (Peterlin and Price 2006; Takahashi et al. 2011). Critically, a majority (>75%) of enzymatically active P-TEFb is typically found within a larger superelongation complex (SEC) (Luo et al. 2012) that encompasses a broad array of factors that aid in recruitment of P-TEFb to active genes and stimulation of RNA synthesis (He et al. 2010; Lin et al. 2010; Sobhian et al. 2010; Li and Gilmour 2013) P-TEFb is known to phosphorylate many proteins, including Spt5, NELF, and the Pol II CTD; however, it is the phosphorylation of Spt5 that is most directly linked to pause release (Fig. 1E, light-green P; Guo et al. 2000; Yamada et al. 2006; Cheng and Price 2007). Mechanistically, the phosphorylation of Spt5 appears to trigger dissociation of NELF from Pol II, enabling Pol II reactivation and resumption of elongation (Wu et al. 2003; Yamada et al. 2006; Cheng and Price 2007).
Due to the cell's ability to tune the duration of promoter-proximal pausing through transacting factors DSIF, NELF, and P-TEFb, this step has been termed “regulated pausing” (Weber et al. 2014; Chiu et al. 2018). In contrast, pausing due solely to cis-acting nucleic acid sequence or universal structural barriers such as nucleosomes has been called “ubiquitous” or “intrinsic pausing” (Landick 2006; Teves et al. 2014; Weber et al. 2014). Given the tight wrapping of DNA around the histone proteins, nucleosomes are inherent barriers to Pol II elongation that must be contended with across the full length of the gene. Indeed, to facilitate passage through such ubiquitous obstacles, eukaryotic cells have evolved a number of specialized elongation factors, including polymerase-associated factor 1 (PAF1), FACT, and Spt6. Following pause release, the polymerase moves downstream into the gene body and soon encounters the first nucleosome, typically centered around +140 bp downstream from the TSS (Mavrich et al. 2008; Tolstorukov et al. 2009; Weber et al. 2014). During this transition, Pol II assembles the repertoire of transcription elongation factors that facilitate transcription through nucleosomes, thereby preventing long-lived pausing at nucleosomal boundaries (Fig. 1F, elongation factors; Kwak et al. 2013; Weber et al. 2014).
Recent structural studies have illuminated how the recruitment of elongation factors can be triggered by pause release (Vos et al. 2018a,b). First, the dissociation of NELF during pause release exposes a binding site for PAF1 on the elongation complex (Shi et al. 1996, Vos et al. 2018a). Accordingly, PAF1 associates with Pol II just as it encounters the first nucleosome on downstream DNA (Adelman et al. 2006; Van Oss et al. 2016). Second, the Spt6 protein and linker region leading to the Pol II CTD are phosphorylated by P-TEFb, stimulating interactions between Spt6 and elongating Pol II (Vos et al. 2018a). The FACT complex also associates with the elongation complex at this stage (Belotserkovskaya et al. 2004) through interactions with Pol II and histones. Spt6 and FACT both have histone chaperone activity allowing them to facilitate the disassembly and reassembly of nucleosomes as Pol II passes through. Thus, the binding of elongation factors facilitates Pol II movement through nucleosomes and maintains chromatin organization across the gene body (Wada et al. 2000; Kaplan et al. 2003; Ardehali et al. 2009).
Many factors govern the establishment and release of paused Pol II
As described above, a number of factors converge to establish paused Pol II and mediate its release. Below, we outline the current state of knowledge for each of the factors that affect regulated pausing. We emphasize that the level of paused Pol II should be considered an equilibrium between the rate of Pol II recruitment, initiation, and entry into the paused state and the rate of pause release into either productive elongation or termination (Fig. 2). Critically, paused Pol II will not accumulate near promoters if recruitment or initiation rates are slow, nor if pause release or termination rates are high. Thus, the fact that a majority of genes show accumulation of paused Pol II is strong evidence that recruitment of Pol II is typically much faster than pause release or termination. We therefore focus below on the factors that influence the rate limiting steps of Pol II pausing and its escape downstream into productive RNA synthesis.
Figure 2.
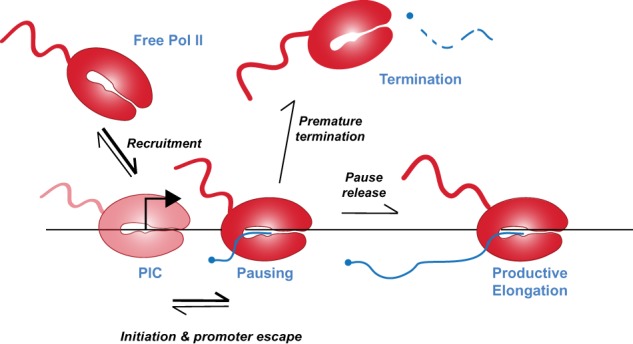
Rate constants favor the establishment of paused Pol II. The steps leading toward productive elongation and the relative rates of transitions between each step are depicted, with thicker arrows indicating faster rates. Note that Pol II in the PIC and paused forms are not coincident on the same DNA due to steric hindrance. The accumulation of Pol II in early elongation at most promoters indicates that the relative rates toward establishing paused Pol II (recruitment, initiation, and promoter escape) are typically faster than rates of Pol II release from pausing into productive elongation or premature termination. Upon termination, Pol II releases nascent RNA, which is rapidly degraded, and the polymerase is recycled into the pool of free Pol II.
DNA or RNA sequence
Sequence elements in the DNA and RNA can modulate pausing by directing the binding of trans-acting factors or directly altering the efficiency of catalysis by RNA polymerases. Binding sites for TFs (Fig. 3A) such as Drosophila GAGA factor (GAF) serve to recruit GAF and associated chromatin remodeling machines (e.g., NURF) (Tsukiyama et al. 1994; Fuda et al. 2015), thereby opening the chromatin in the vicinity of promoters. This exposes core promoter sequences that are instrumental for directing the position, level, and orientation of transcription initiation by recruiting GTFs. Drosophila DNA elements enriched in core promoters that promote PIC assembly include the TATA-box, Initiator (Inr), MTE, and the downstream promoter element (DPE) motifs (Fig. 3A; Vo Ngoc et al. 2017). Although similar sequences have been noted in mammals, the consensus motifs are considerably more degenerate and have lower information content (Ohler 2006).
Figure 3.
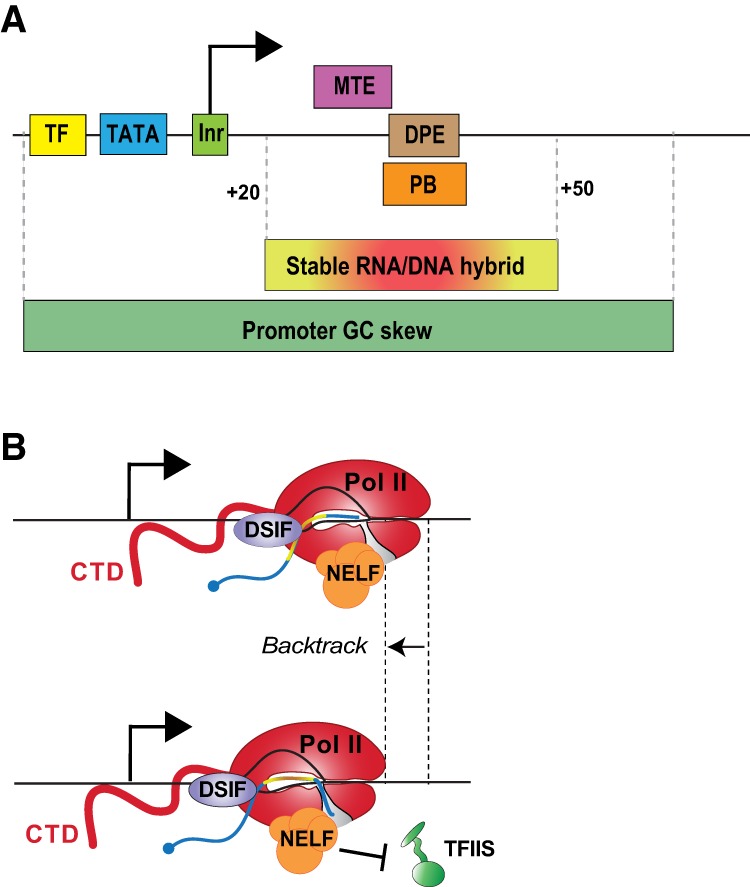
Core promoter elements and sequence features that lead to efficient promoter-proximal pausing. (A) TFs bind their sites and, with the help of the core promoter elements, recruit the transcription machinery to the promoter. DNA elements enriched in core promoters that promote PIC assembly include the TATA-box (−28 to −35 with respect to the TSS), Initiator (−4 to +2), MTE (+18 to +29), and DPE (+28 to +32) motifs. Drosophila promoters enriched in paused Pol II display a variant of the DPE called the pause button (PB, +25 to +35). Many of these core promoter elements play a dual role in initiation and pausing. The region around the pause site (+20 to +50) often contains sequences that produce locally strong RNA/DNA hybrids (red–yellow box) that can induce pausing and polymerase backtracking. A high GC skew in promoters and enrichment of G bases in the nontemplate strand of pause sites, especially in mammals, can lead to efficient pausing by creating strong RNA/DNA hybrids or possibly DNA and RNA structures that favor pausing. (B) Depiction of how strong RNA/DNA hybrids and pausing factors lead to pausing and backtracking of Pol II. After Pol II transcribes past a sequence with a strong RNA/DNA hybrid (red-yellow line) relative to the downstream sequence, the polymerase can pause. During pausing, the polymerase often backtracks such that the stronger more stable RNA/DNA hybrid is held within the Pol II active site. This movement causes the 3′ end of the RNA to be extruded though the funnel (gray), preventing further transcription elongation. NELF (orange) occludes the binding site of TFIIS (green) on the outer surface of Pol II, preventing TFIIS from reaching into the funnel to stimulate cleavage of the backtracked RNA and realignment of the RNA 3′ end with the polymerase active site.
Notably, many motifs important for stable promoter opening and PIC formation positively correlate with the level of paused Pol II (Hendrix et al. 2008; Gilchrist et al. 2010; Kwak et al. 2013). For instance, combinations of GAF, Inr, and DPE motifs reveal an additive effect with the presence of each element and levels of pausing observed in Drosophila (Hendrix et al. 2008). In addition, the agreement of promoter sequences to the consensus for these motifs also correlates positively with pausing levels (Kwak et al. 2013). In Drosophila, the pause button motif (Fig. 3A, PB) is highly enriched at paused genes and is similar to and overlaps with the position of the consensus DPE (Hendrix et al. 2008). Intriguingly, prominent pausing of Pol II often occurs at sequences with combinations of pause button, GAF, and/or Inr elements (Hendrix et al. 2008), suggesting that these motifs can work cooperatively with one another to recruit and retain paused Pol II.
The position of core promoter elements relative to the TSS is also suggested to affect pausing location and intensity. For instance, pausing is at a maximum at Drosophila promoters when the TATA, Inr, MTE, or pause button are at their consensus position as compared with when the element is shifted up- or downstream (Kwak et al. 2013). Also, the sites of pausing can track with the position of downstream elements within a limited range (Kwak et al. 2013). Thus, a “tethering” model has emerged wherein consensus promoter elements create stable PICs and early elongation complexes. In this model, multiple strong interactions with factors at the promoter stimulate recruitment of Pol II but also impede promoter escape, as such interactions need to be broken as Pol II transitions toward elongation. Indeed, TFIID interacts with both the Inr centered at the TSS and the DPE located at the site of pausing, putting TFIID in position to simultaneously interact with GTFs at the promoter and the early elongation complex (Sypes and Gilmour 1994; Burke and Kadonaga 1996). In addition, strong core promoter motifs can retain a scaffold of PIC components following promoter escape, allowing them to recruit new Pol II very efficiently. This would ensure that additional Pol II is available to quickly replace polymerases released from pausing, maintaining high occupancy of the pause site.
DNA elements can also directly alter polymerase elongation rate and processivity as a result of the extensive interactions between the polymerase and nucleic acids. Sequence-dependent pausing of elongation occurs in all organisms from bacteria to humans, and recent genomic examination of pausing at high resolution has revealed multiple conserved aspects of DNA sequence that contribute to this process (Larson et al. 2014). A common observation is that the RNA/DNA hybrid that forms within the active site of paused Pol II typically has a high G/C content flanked by A/T-rich regions (Chan and Landick 1993; Nechaev et al. 2010; Gressel et al. 2017; Sheridan et al. 2019) The stability of G/C-rich RNA/DNA hybrids within the active site presents an obstacle to forward translocation of the polymerase. When forward movement becomes energetically unfavorable (Fig. 3B), the polymerase becomes susceptible to backtracking upstream along the template, which dislodges the 3′ end of the nascent RNA from the catalytic site. Such a conformation is clearly refractory to RNA synthesis and can lead to transcriptional arrest (Kireeva and Kashlev 2009). There is strong evidence that Pol II backtracks during early elongation and that backtracking contributes to the longevity of the paused state (Adelman et al. 2005; Nechaev et al. 2010; Weber et al. 2014; Sheridan et al. 2019). Moreover, this influence of RNA/DNA hybrid stability is observed at mammalian CpG island promoters that display a clear GC skew that correlates well with pausing (Core et al. 2008; Kellner et al. 2015).
Nascent RNA secondary structures that influence the entry into or exit from a paused state are well documented in prokaryotes (Hein et al. 2014); however, eukaryotic examples of this are rare. Transcription of the human immunodeficiency virus (HIV) in humans is a notable exception, and HIV was one of the first models of promoter-proximal pausing and premature transcription termination (Feinberg et al. 1991; Kessler and Mathews 1991, 1992). The HIV virus integrates into the host genome, where viral gene expression is limited by regulation of early elongation. The initially transcribed region of HIV forms an RNA stem–loop structure termed the “transactivation response” or TAR element. NELF stabilizes pausing at the HIV promoter (Zhang et al. 2007), potentially through interactions with TAR RNA (Palangat et al. 1998; Fujinaga et al. 2004). Full-length HIV RNA synthesis requires expression of a viral transcriptional activator, Tat. The Tat protein can interact with P-TEFb and TAR, enabling recruitment of P-TEFb to the HIV promoter. This stimulates pause release through dissociation of NELF and phosphorylation of Spt5 (Fujinaga et al. 2004). Although there is no evidence for formation of RNA structures similar to TAR at cellular genes, the RNA-binding domain within NELF-E was shown recently to bind a short “K-turn” structure (Pagano et al. 2014) that is modestly enriched near Drosophila promoters. Thus, whereas RNA binding by NELF does not appear to be broadly necessary for pausing (Missra and Gilmour 2010; Gressel et al. 2017; Vos et al. 2018b), it may stabilize NELF interactions with elongating Pol II at select genes.
Intriguingly, secondary structure predictions suggest that the nontemplate DNA strand just upstream of paused Pol II has an increased likelihood to form structures such as G quadruplexes (Szlachta et al. 2018). Structure formation by the nontemplate DNA strand could be envisioned to deter its reannealing with the template strand after transcription, potentially enabling the formation of R loops between nascent RNA and the DNA template. Given the recent interest in R-loop formation near promoters (Chédin 2016; Chen et al. 2017) and the impact of R loops on genome stability (Santos-Pereira and Aguilera 2015), this will be an interesting subject for future study. Finally, we note that while sequence elements in the DNA or RNA present inherent obstacles to a transcribing polymerase, they alone do not dictate the longevity of promoter-proximal pausing. Instead, sequence features that slow elongation by Pol II are envisioned to create a window of opportunity for the association and action of protein factors such as those described below.
Spt5 and the DSIF complex
Spt5 is a well-studied and essential elongation factor that is necessary for both coding and noncoding RNA transcription in higher eukaryotes (Henriques et al. 2018). Spt5 homologs exist in bacteria and archaea, and the binding surfaces for these proteins on RNA polymerases are highly conserved (Hartzog and Fu 2013; Schulz et al. 2016; Bernecky et al. 2017). Notably, eukaryotic Spt5 binds Pol II as a heterodimer with the Spt4 protein to form a complex called DSIF (Wada et al. 1998a; Ivanov et al. 2000; Yamaguchi et al. 2013). The name DSIF originated from in vitro transcription assays studying early elongation, in which the presence of Spt5/Spt4 was essential for transcription to be sensitive to P-TEFb kinase inhibitors such as DRB (Wada et al. 1998a). Accordingly, as described in detail in the section on P-TEFb, Spt5 is the critical target phosphorylated by P-TEFb to mediate pause release (Wada et al. 1998b).
Notably, the region in Spt5 that is phosphorylated by P-TEFb contains a series of amino acid repeats that resemble the Pol II CTD (Yamada et al. 2006), suggesting that phosphorylation of Spt5, like that of the Pol II CTD, helps coordinate interactions across each stage of the transcription cycle. However, despite decades of research, many important questions remain concerning Spt5 activity and domain structure, and this is an area of active research (Shetty et al. 2017). In particular, the defects caused by Spt5 mutations that impact cell viability or organismal development are poorly understood. Interestingly, several mutations that lead to developmental defects are predicted to destabilize Spt5 structure, and careful examination of Spt5 levels in missense mutations reveals that many are protein hypomorphs (Cooper et al. 2005; Jennings 2013; Bernecky et al. 2017). Thus, reduced cellular levels of Spt5 appear sufficient to cause cell type- or condition-specific defects.
Features of the Spt5–Pol II interface enforce specific recognition of elongation complexes by Spt5. Regions of Pol II bound by Spt5 are occluded during transcription initiation by GTFs, such as TFIIE (Larochelle et al. 2012; Schulz et al. 2016). The release of initiation factors during promoter escape thus exposes these interaction surfaces to enable Spt5 binding to Pol II. Furthermore, stable association of Spt5 with polymerase requires nascent RNA emerging from the elongation complex (Missra and Gilmour 2010). This requirement ensures that Spt5 association occurs shortly after promoter escape, as Pol II forms an RNA of 18–20 nt in length (Missra and Gilmour 2010). Recently, RNA binding has been mapped to the KOW5 motif of Spt5, which was shown to be critical for both high-affinity binding of Spt5 to the elongation complex and promoter-proximal pausing (Qiu and Gilmour 2017). Once bound, Spt5 stimulates rapid and efficient capping of RNA through direct associations with the capping enzyme (Wen and Shatkin 1999; Pei and Shuman 2002).
Purified DSIF/Spt5 and its bacterial homolog NusG are stimulatory to transcription elongation, serving to suppress pausing during RNA synthesis. Spt5 and NusG bind near the clamp domain that engages downstream DNA where they appear to hold the clamp in a closed position to maintain stabilizing contacts with DNA during translocation (Weixlbaumer et al. 2013; Bernecky et al. 2017). This favorable stabilizing conformation of Spt5 on Pol II is evident in both the active and paused elongation complexes, implying that Spt5 consistently serves this positive role (Bernecky et al. 2017; Vos et al. 2018a,b). Furthermore, Spt5 is stimulatory to elongation in vitro in the absence of NELF, particularly at low NTP concentrations (Wada et al. 1998a; Cheng and Price 2007). Thus, the conserved activity of DSIF/Spt5 is that of a positive elongation factor (Hartzog and Fu 2013).
The pause-enhancing activity of Spt5 activity in early Pol II elongation results from Spt5-mediated recruitment of NELF to the Pol II elongation complex (Cheng and Price 2007). Accordingly, phosphorylation of Spt5 stimulates productive elongation at least in part because it triggers the dissociation of NELF from the elongation complex. This transition thus allows Spt5 to resume a positive role in elongation. Following pause release, Spt5 remains associated with Pol II across the gene body, where it serves as a recruitment platform for a number of factors that stimulate Pol II activity and RNA processing.
TFIIS
Classic in vitro studies demonstrated that when Pol II encounters an obstacle that blocks forward movement, such as a DNA-bound protein, nucleosome, or certain DNA sequences, the polymerase becomes prone to sliding backward along the DNA (Fig. 3B; Izban and Luse 1992; Reines 1992; Bondarenko et al. 2006). This backtracking causes transcriptional arrest, since the 3′ end of the RNA becomes disengaged from the Pol II active site, preventing additional nucleotide incorporation. The protein factor TFIIS rescues Pol II from the backtracked state. Like Spt5, TFIIS activity and mechanism is highly conserved from man to eubacteria (Kireeva and Kashlev 2009).
Mechanistically, TFIIS appears to recognize the conformation of arrested Pol II and bind near the funnel through which incoming NTPs are thought to reach the active site. TFIIS then extends a finger-like domain into the funnel, toward the Pol II active site, to stimulate endonucleolytic cleavage of the nascent RNA. This process releases a short RNA and creates a new RNA 3′ end that is properly aligned with the enzymatic active site. Recent structural studies have elucidated the molecular details of this reaction (Kettenberger et al. 2003; Cheung and Cramer 2011) and revealed the proximity of TFIIS and backtracked RNA in the funnel. Importantly, these findings illuminate that extensive backtracking of RNA into the funnel would create a steric clash between RNA and TFIIS, explaining why backtracking of more than ∼5 nt is rare and much less efficiently restarted by TFIIS.
The importance of TFIIS to maintaining the activity of promoter-proximally paused Pol II was elucidated using in vitro transcription assays of the Drosophila hsp70 promoter (Adelman et al. 2005). These experiments demonstrated that Pol II could not readily elongate through the defined sites of pausing in the absence of TFIIS and suggested a propensity for promoter-proximally paused Pol II to undergo backtracking and arrest. Accordingly, depletion of TFIIS from Drosophila cells resulted in a slower, weaker activation of the heat-shock genes upon thermal stress. Thus, TFIIS-mediated cleavage was proposed to be critical for efficient resumption of elongation by paused Pol II. The scope of TFIIS activity was emphasized by genomic analyses of the short RNAs associated with paused Pol II in Drosophila using Start-seq (start site-associated RNA sequencing) (Table 1). Mapping of RNA 3′ ends at single-nucleotide resolution in control cells versus those depleted of TFIIS, revealed that paused Pol II often backtracks by 2–5 nt and requires TFIIS to regain elongation-competence (Nechaev et al. 2010). Recent work in human cells confirmed these findings and highlighted that Pol II is particularly susceptible to backtracking during early elongation, where TFIIS is critical for efficient pause release during responses to both heat stress and hypoxia (Sheridan et al. 2019).
NELF
The four-subunit NELF complex (NELF-A, NELF-B, NELF-C/D isoforms of the same gene, and NELF-E) binds the Pol II–Spt5 interface during early elongation (Yamaguchi et al. 2002). In addition to requiring Spt5 for tight association with Pol II, NELF subunits display a number of RNA-binding motifs, suggesting that RNA recognition could be involved in NELF association with the elongation complex (Fujinaga et al. 2004; Pagano et al. 2014; Vos et al. 2016, 2018b). NELF homologs have not been identified in lower organisms (e.g., bacteria, yeast, worms, or plants), suggesting that NELF plays a specialized function in gene regulation in animals (Narita et al. 2003; Yamaguchi et al. 2013). Interestingly, the organisms that exhibit stable pausing of Pol II are those that that exhibit a NELF complex, implying a functional connection. A phenomenon similar to pausing has been noted at a subset of genes in Saccharomyces pombe, which lacks NELF subunits (Booth et al. 2018). However, the slowed promoter-proximal elongation observed in S. pombe is less widespread and pausing is much shorter-lived than in higher organisms (Booth et al. 2018). These findings support biochemical studies of NELF, which indicate that that NELF is not required to initiate Pol II pausing but instead plays a role in stabilizing and extending the lifetime of a paused complex (Cheng and Price 2007; Henriques et al. 2013; Vos et al. 2018b).
Interestingly, the loss of NELF reduces the accumulation of paused Pol II at many promoters, without causing an increase of polymerase within gene bodies (Gilchrist et al. 2008; Gilchrist et al. 2010; Core et al. 2012; Williams et al. 2015). Thus, Pol II that fails to undergo stable pausing appears susceptible to termination of transcription. Indeed, measurements of the stability of paused Pol II confirmed that promoter-proximal Pol II turnover was much more rapid in cells lacking NELF (Henriques et al. 2013; Shao and Zeitlinger 2017). Furthermore, the destabilization of Pol II pausing also reduces the local retention of a scaffold of GTFs that form during initiation (Gilchrist et al. 2010). Thus, diminished pause stability can inhibit the process of new initiation (Boettiger and Levine 2009; Gilchrist et al. 2010; Lagha et al. 2013). Importantly, the loss of Pol II and GTFs from the promoter region allows for the encroachment of nucleosomes over the TSS, further blunting gene activity (Gilchrist et al. 2010; Gilchrist et al. 2012).
Recent structural studies shed light on the mechanism through which NELF stabilizes pausing of Pol II: NELF interacts broadly with the binding surface occupied by TFIIS (Fig. 3B). Thus, association of NELF with Pol II blocks recognition by TFIIS, thereby inhibiting rescue from backtracking and arrest (Palangat et al. 2005; Vos et al. 2018b). In addition, NELF binding is thought to stabilize the distortion of the Pol II active site that is characteristic of pausing, further deterring RNA elongation. As anticipated, escape of Pol II from pausing into productive RNA synthesis involves release of NELF from the Pol II complex (Wu et al. 2003). The mechanistic basis for this process clearly involves the P-TEFb-mediated phosphorylation of Spt5 but may include phosphorylation of NELF as well (Fujinaga et al. 2004).
P-TEFb
The kinase activity of P-TEFb is critical for release of paused Pol II into productive elongation, both in vivo and in vitro. P-TEFb, comprised predominantly of Cyclin T1 and Cdk9, was first discovered by David Price (Marshall and Price 1995; Price 2000) as the activity in nuclear extracts required to overcome a block to early elongation in vitro. This powerful biochemical system was then used by the Price and Handa laboratories (Wada et al. 1998a; Yamaguchi et al. 1999; Cheng and Price 2007) to uncover the mechanisms at play in establishing the early elongation block, leading to the discoveries of both DSIF and NELF complexes. Importantly, depletion of either DSIF or NELF from nuclear extracts greatly reduced pausing in early elongation and rendered transcription independent of P-TEFb activity (Wada et al. 1998a; Yamaguchi et al. 1999; Cheng and Price 2007). These biochemical assays thus revealed that P-TEFb is needed specifically to relieve DSIF/NELF-mediated inhibition of early elongation.
To identify the functional target of P-TEFb in pause release, a partially purified system was implemented in which individual components of the elongation complex were prephosphorylated by P-TEFb prior to addition to an in vitro transcription reaction. Critically, these experiments showed that addition of prephosphorylated Spt5 to transcription assays prevented NELF from eliciting negative effects on Pol II elongation, dramatically reduced promoter-proximal pausing, and made elongation insensitive to P-TEFb inhibition (Cheng and Price 2007). Furthermore, specific mutations in Spt5 were described that interfered with its pausing function, and in vitro transcription with these mutants did not require P-TEFb activity (Guo et al. 2000). These findings were confirmed in vivo, when mutants in Spt5 that prevent P-TEFb-mediated phosphorylation were shown to prevent Pol II release into gene bodies, even under conditions where P-TEFb is fully functional in phosphorylation of the Pol II CTD (Yamada et al. 2006). Thus, Spt5 emerges as the critical target of P-TEFb activity for pause release, both in vitro and in vivo.
Nonetheless, P-TEFb phosphorylates a large number of factors in vivo, and there is great interest in defining all potentially relevant targets of P-TEFb activity. However, this has been complicated by the sheer number of proteins phosphorylated by P-TEFb in metazoan cells, with recent work suggesting over 100 putative P-TEFb-phosphorylated proteins (Sanso et al. 2016). Interestingly, P-TEFb targets include factors involved in transcription elongation, chromatin modifications, and RNA processing, suggesting that P-TEFb activity broadly coordinates pause release with transcription through chromatin and production of a mature mRNA. Furthermore, P-TEFb can phosphorylate the Pol II CTD at Ser2 residues (Ser2-P), which is considered a mark of active, productive elongation. Biochemical studies reveal that P-TEFb activity toward Ser2 of the CTD is only efficient if the CTD is “primed” by prior phosphorylation of Ser7 (Czudnochowski et al. 2012). Since Ser7-P is deposited by the TFIIH-associated kinase Cdk7, this suggests that the Ser2-P modification can only occur after successful transcription initiation has taken place. Thus, the enzymatic specificity of P-TEFb appears to be tuned to enable CTD Ser2 phosphorylation only at the appropriate stage of the transcription cycle (Buratowski 2009). Nonetheless, P-TEFb does not carry out the bulk of Pol II CTD Ser2-P; instead the kinases Cdk12 and Cdk13 contribute the majority of this modification (Bartkowiak et al. 2010; Blazek et al. 2011). In particular, Cdk12 associates with Pol II as it elongates downstream into the gene body, and Ser2-P levels increase markedly across gene bodies, with a peak at the 3′ ends (Mayer et al. 2010). Accordingly, binding partners of Pol II that recognize Ser2-P CTD are enriched >2 kb into the gene and encompass a number of chromatin-modifying and RNA processing complexes that help Pol II achieve maximum elongation rates (Bartkowiak et al. 2010; Jonkers et al. 2014).
The availability of P-TEFb kinase inhibitors such as DRB and the more specific molecule flavopiridol (FP) have enabled a detailed study of transcription in the absence of P-TEFb activity in living cells. Foundational experiments have documented that blocking P-TEFb activity traps Pol II in early elongation at nearly all mRNA promoters (Chao and Price 2001; Ni et al. 2008; Rahl et al. 2010; Henriques et al. 2013; Jonkers et al. 2014). Importantly, this failure of Pol II to escape the promoter-proximal region is evident within several minutes of FP treatment, bolstering conclusions that this is a direct effect of P-TEFb inhibition. Such experiments, however, must be performed and interpreted with caution: FP is a noncovalent inhibitor with off target effects on other kinases at high doses. Therefore care must be taken to determine the optimal concentration and time of treatment for FP. To date, short 5- to 10-min time courses of FP treatment with 200–500 nM concentration, coupled with nascent RNA sequencing (RNA-seq) strategies, have proven most useful for defining the immediate consequences of P-TEFb inhibition on Pol II elongation (Henriques et al. 2013; Jonkers et al. 2014). Notably, increasingly specific and covalent Cdk9 inhibitors continue to be developed (e.g. NVP-2) (Gressel et al. 2017; Olson et al. 2018), and recent experiments with these molecules broadly confirm the requirement for P-TEFb activity for productive Pol II elongation.
Although P-TEFb is active as a dimeric Cyclin/Cyclin-dependent Kinase pair, it is typically found as part of larger complexes. When associated with the 7SK noncoding RNA, MeCPE, LARP7, and HEXIM proteins, P-TEFb activity is inhibited (Zhou et al. 2012). Activation of P-TEFb therefore requires its regulated dissociation from the inhibitory 7SK complex. Several proteins are known to liberate P-TEFb from 7SK complexes, including the HIV Tat protein and the bromodomain-containing protein Brd4 (Barboric et al. 2007; Schroder et al. 2012). Many cellular stresses and signaling pathways liberate P-TEFb from 7SK, enabling large-scale activation of gene expression (Quaresma et al. 2016). Although much of the 7SK complex is thought to be sequestered away from chromatin, there is evidence that this inactive P-TEFb complex can be recruited to select loci, where P-TEFb can be released from 7SK and HEXIM1, to activate the kinase activity locally (D'Orso and Frankel 2010; Faust et al. 2018).
The most active form of P-TEFb is associated with the SEC (He et al. 2010; Lin et al. 2010; Sobhian et al. 2010). The SEC contains a number of factors that can stimulate catalysis by Pol II, such as ELL (Shilatifard et al. 1996), as well as a number of proteins that can facilitate SEC/P-TEFb recruitment to target genes. For example, components of the SEC interact with Mediator, allowing for recruitment of P-TEFb and pause release at Meditator-occupied loci (Takahashi et al. 2011). In addition, P-TEFb is thought to interact with a number of TFs and RNA-binding proteins that help guide it to specific gene regions (e.g., NF-kB DDX21, SRSF2) (Barrandon et al. 2007; Van Herreweghe et al. 2007; Ji et al. 2013; Calo et al. 2015). Artificial recruitment experiments using the Drosophila hsp70 promoter demonstrated that recruitment of P-TEFb to the unstimulated promoter was sufficient to increase gene activity (Lis et al. 2000). Accordingly, there is strong evidence that the level of P-TEFb brought to a promoter can dictate the rate of pause release and thus gene expression. Therefore, the more rapidly a gene recruits P-TEFb, the shorter-lived pausing will be at the promoter and the more efficiently Pol II will be released into productive RNA synthesis.
Brd4 and bromodomain and extraterminal domain (BET) proteins
The BET proteins have two tandem bromodomains that can recognize acetylated lysine residues and an extraterminal domain that interacts with regulators and effector proteins such as JMJD6 (Konuma et al. 2017; Xu and Vakoc 2017). BET family bromodomains associate with acetylated histones and TFs, which directs these proteins toward active chromatin regions such as promoters and enhancers. The BET protein Brd4 has received significant attention for its role in gene activation, since its extended CTD can bind P-TEFb (Bisgrove et al. 2007). Moreover, interactions with Brd4 dissociate P-TEFb from the inactivating 7SK complex, increasing the cellular pool of “free” and active P-TEFb (Schroder et al. 2012). In addition, association of Brd4 with P-TEFb increases kinase activity, suggesting an allosteric stimulatory effect of Brd4-P-TEFb interactions (Schroder et al. 2012; Itzen et al. 2014). Accordingly, RNAi-mediated depletion of Brd4 was found to reduce levels of P-TEFb on chromatin, phosphorylation of the Pol II CTD, and gene activity (Jang et al. 2005; Yang et al. 2005).
These findings suggested a model wherein Brd4 would release P-TEFb from 7SK, recruit P-TEFb to selected loci through Brd4 acetyl-lysine binding, and stimulate P-TEFb kinase activity for targeted gene activation. The implication of this model is that manipulation of Brd4 could be a powerful means to control gene output. The fortuitous development of potent small molecule inhibitors of BET–bromodomain binding to acetyl lysine enabled testing of this model (e.g., JQ1, I-BET) (Filippakopoulos et al. 2010; Dawson et al. 2011; Xu and Vakoc 2017). These BET-inhibitor compounds had strong effects on Brd4 association with chromatin and gene expression, demonstrating that Brd4 stimulates both mRNA and noncoding RNA transcription (Filippakopoulos et al. 2010; Dawson et al. 2011; Kanno et al. 2014; Xu and Vakoc 2017). However, it remained unclear which function of Brd4 was critical for the stimulation of gene activity, particularly because Brd4 depletion or inhibition required hours or days to achieve full effect, raising the possibility of indirect consequences. In particular, it was noted in gene expression analysis that BET-inhibition caused significant up-regulation of HEXIM1, a central component of the inactive 7SK complex, which would cause cellular sequestration of P-TEFb (Anand et al. 2013). Thus, clarifying the direct role of Brd4 would require faster-acting inhibition strategies.
Recent work using an optimized chemical degrader of Brd4 called dBET6 has begun to tease apart the many roles of Brd4. Importantly, rapid, nearly complete loss of Brd4 phenocopies a loss of P-TEFb, with a broad collapse of gene expression across the genome (Winter et al. 2017). Surprisingly, this occurs without a detectable effect of dBET6 on P-TEFb binding to chromatin, demonstrating that Brd4 is not critical for targeting of P-TEFb to gene or enhancer regions. Thus, these studies suggest that a key role of Brd4 could be in allosteric activation of P-TEFb, to enable chromatin bound Cdk9 to act efficiently on the transcription machinery (Schroder et al. 2012; Itzen et al. 2014). Clearly, this is an exciting area that invites additional research.
Pausing as a regulatory strategy
Pausing of Pol II occurs at genes across the spectrum of expression levels, from the most active genes, to those showing very little activity. However, paused Pol II is very infrequently found at genes that are not expressed (<1% of paused genes are deemed inactive by nascent RNA-seq techniques), suggesting that pause release is an inherently leaky process. Thus, we suggest that pausing should be considered a means to tune gene expression levels, rather than as an “on–off switch.” In cases where gene transcription is observed to be low, pausing can be viewed as maintaining the gene's potential to be reactivated at a later time. In light of this, it is worth noting that pausing appears to be enriched at genes where small changes in expression are highly relevant to the underlying biology, such as signaling molecules, kinases, receptors, and many TFs (Adelman et al. 2009; Gilchrist et al. 2012).
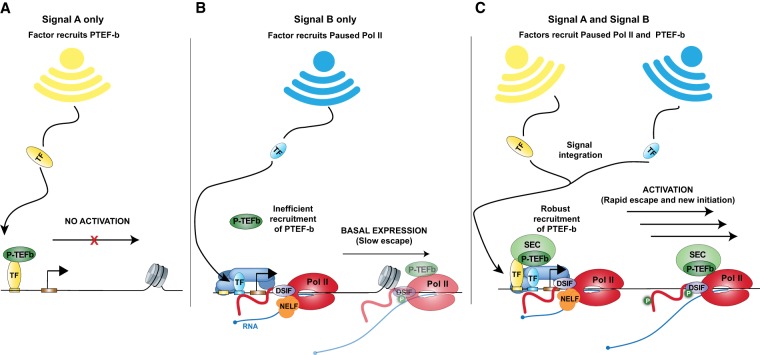
Integration of signals
One benefit of separate regulation of transcription initiation and elongation is that signaling pathways can affect one or both steps, thus allowing integration of opposing or reinforcing signals (Fig. 4). The outcome is dynamic and has precise control of gene expression programs over both short and long periods. For example, some TFs can recruit Pol II and establish pausing (Fig. 4A), while others can trigger pause release (Fig. 4B). Combinations of TFs can thus work cooperatively to tune the levels of Pol II engaged at a gene promoter and the rate of release into productive elongation (Fig. 4C). The Drosophila GAF and heat shock factor (HSF) are a notable duo of TFs that work together in this way to potentiate and activate promoters, respectively (Duarte et al. 2016). Examples of signaling systems where initiation and pausing are differentially modulated to induce a rapid response include immune responses (Adelman et al. 2009; Gilchrist et al. 2012), hormone signaling (Hah et al. 2011; Gupte et al. 2013), or early development (Boettiger and Levine 2009; Marks et al. 2012; Lagha et al. 2013; Saunders et al. 2013; Williams et al. 2015). Critically, any significant increase in initiation frequency would also require a decrease in pausing duration, since the occupancy of the promoter-proximal region by paused Pol II sterically blocks initiation of additional polymerases (Gressel et al. 2017; Shao and Zeitlinger 2017).
Figure 4.
Pausing can serve as a mechanism to integrate multiple signaling events. TFs can stimulate initiation, elongation, or both. (A) When a signal responsive activator only stimulates elongation, for example, by recruiting P-TEFb, activation does not occur since polymerases were not initiated at the promoter prior to the TF binding. (B) If a signaling pathway and TF stimulates initiation only, then polymerases will pause proximally. Low levels of pause escape, possibly stimulated by Mediator-dependent P-TEFb recruitment, establishes a basal level of transcription. (C) When both signals are active, high levels of initiation and pause escape lead to robust transcriptional activation. By altering the level of input from either signaling pathway, the cell can fine tune the transcription levels of genes.
Pausing has been associated with dynamic control of gene expression across developmental time and in processes involving multiple cell generations. For example, genes that are active but not paused during Drosophila early embryonic development tend to be completely repressed in later stages, whereas genes that are active and paused early in development display oscillating patterns of gene expression at later points of development (Saunders et al. 2013). The corresponding hypothesis is that pausing factors maintain a paused state over time, allowing varying levels of gene activation in response to other signals later in development. Pausing also enables synchrony in a short time scale across cell populations that is critical for coordinating rapid tissue development in early Drosophila development (Boettiger and Levine 2009; Lagha et al. 2013). Also, loss of NELF in mouse embryonic stem cells or the pause-inducing function of Spt5 in zebrafish perturb differentiation (Guo et al. 2000; Amleh et al. 2009; Williams et al. 2015). Together, these observations suggest that potentiation and plasticity of gene expression afforded by pausing is critical during development.
A pertinent goal moving forward in regard to signal-dependent regulation is to determine which TFs—and hence which signals—regulate pause potentiation versus escape into productive RNA synthesis. In this regard, a pause-inducing factor could facilitate any event that leads to the establishment of paused Pol II, such as Pol II recruitment, initiation, or the association of NELF and Spt5. On the other hand, recruitment of active P-TEFb to promoters is the rate-limiting step and hence most critical component of pause escape. Thus, it is not surprising that cells use a multipronged approach to recruit P-TEFb during both basal and activated transcription (as described above). Uncovering the direct mechanisms that lead from signaling events to selective recruitment of P-TEFb to promoters remains a topic of intense investigation.
Pol II as a chromatin remodeling enzyme
RNA polymerase is the cell's most powerful chromatin remodeler and can dramatically impact promoter chromatin architecture. As Pol II translocates along DNA, the torsional stress creates positive supercoils ahead of the polymerase that destabilize nucleosomes (Teves et al. 2014). In addition, Pol II directly or indirectly associates with an entourage of chromatin modifiers, remodelers, and chaperones that alter the chromatin architecture both ahead of and in the wake of transcribing polymerases. In the case of pausing, the high occupancy of Pol II creates a strong barrier to the local assembly of nucleosomes (Gilchrist et al. 2008; Gilchrist et al. 2010; Gilchrist et al. 2012; Teves et al. 2014). In fact, at mammalian promoters that are divergent, the entire distance between divergently paused Pol II is open and accessible for TFs to bind (Core et al. 2014; Scruggs et al. 2015; Chen et al. 2016). Consistently, ablation of NELF results in decreased pausing, increased nucleosome occupancy at many promoters, and decreased transcription of many genes. Promoters with sequences that are highly amenable to bending around histone proteins are especially susceptible to repression after NELF removal, suggesting that paused Pol II can successfully compete with encroaching nucleosomes to maintain an active chromatin architecture (Gilchrist et al. 2010). This same arrangement and sensitivity to reduction of pausing is observed at transcribed enhancers that are often divergent (Henriques et al. 2018). The emerging model is that promoter proximal pausing on either side of promoter or enhancer regions likely serves as bookends to keep repressive chromatin forces from occluding regulatory sequences such as TF motifs.
Pausing as a modulator of DNA repair, cell cycle progression, and cell identity
Transcription-coupled nucleotide excision repair (TC-NER) is responsible for recognizing and repairing genotoxic stresses such as UV exposure. This response is generally rapid and is important for attenuation of acute and long-term effects of genotoxic stress. In this process, Pol II stalls at a DNA lesion and triggers recruitment of specialized NER proteins. Lavigne et al. (2017) reported recently that UV-induced stress results in a uniform release of paused Pol II from promoters that coincides with increased TC-NER within active genes. This wave of elongating polymerases is thought to facilitate scanning and repair of UV-induced regions to protect the transcribed genome (Lavigne et al. 2017). A plausible mechanism for UV-induced pause escape comes from evidence that UV damage releases P-TEFb from the repressive 7SK snRNP, increasing the pool of active P-TEFb (Nguyen et al. 2001; Byers et al. 2005). Thus, paused Pol II can act as a reservoir of polymerases that can be rapidly released to detect lesions and ultimately protect actively expressed genes from potentially harmful mutations.
Storage of Pol II at promoters may also help in maintenance of cell type-specific gene expression programs during different phases of the cell cycle or through multiple cell divisions. Recent work has shown that Pol II occupancy at promoters increases as active transcription decreases when mouse myoblasts transition to quiescence (Gala et al. 2018). When released from quiescence, pausing levels decrease to normal as transcription resumes. This is a prime example of dynamic regulation afforded by pausing, and the potentiation of later activation may help maintain the correct transcription program once cell growth cues are received. For instance, without pausing during quiescence, the chromatin might adopt a closed or repressive architecture that would inhibit binding of TFs needed for the return to growth. This could cause deleterious alterations in the transcriptional program needed for exit from quiescence.
Similarly, preferential reactivation of genes upon exit from mitosis may involve “bookmarking” of specific genes by the presence of paused Pol II, select TFs and histone modifications (Michelotti et al. 1997; Kadauke and Blobel 2013). The antirepressive and kinetic advantage afforded by bookmarking is hypothesized to be important for cell identity (Kadauke et al. 2012; Teves et al. 2016, 2018; Liu et al. 2017). Although recent work has shown that low levels of transcription are maintained for most genes in mitotic cells (Palozola et al. 2017), a small fraction of genes have higher transcription in mitosis than asynchronous cells. The emerging question is how are certain genes maintained in a lowly active or preferentially active state during mitosis? It is noteworthy that the characteristics associated with bookmarked promoters or enhancers—namely, the persistence of an open chromatin environment among otherwise high nucleosome occupancy—is also a hallmark of paused promoters as described above. However, a clear link between pausing and mitotic bookmarking has yet to be uncovered.
What causes the polymerase to pause promoter-proximally?
Analyses of transcription by RNA polymerases from bacteria to humans have demonstrated that early elongation complexes are not as stable or processive as complexes found downstream within gene bodies (Roberts 1988; Kireeva and Kashlev 2009; Luse 2013). Indeed, even in the absence of chromatin, RNA elongation is considerably slower without the full repertoire of elongation-stimulatory factors, rendering the polymerase highly sensitive to sequence-dependent pausing (Roberts 1988; Kireeva and Kashlev 2009; Luse 2013). Single-nucleotide resolution analyses have demonstrated that the position of pausing is highly focused between +25 and +35 nt downstream from the TSS in Drosophila and mammalian species (Fig. 5A; Nechaev et al. 2010; Kwak et al. 2013; Williams et al. 2015). However, the exact sites of pausing can vary from gene to gene depending on the promoter and initially transcribed sequence, and pausing can occur up to 50 nt downstream. As described above, the sequence features that contribute to pausing are multipartite and interconnected, making them difficult to study in isolation or to predict computationally. Indeed, sequences within the RNA–DNA hybrid, the duplex DNA immediately downstream from the polymerase, and the collection of nucleic acids held within the polymerase have all been implicated in determining the stability of the elongation complex and the efficiency of NTP incorporation. Therefore, models for promoter-proximal pausing in all organisms include a role for DNA/RNA sequence in determining both the position and duration of pausing (Roberts 1988; Chan and Landick 1993; Nechaev et al. 2010; Kwak et al. 2013; Hein et al. 2014). Moreover, the sequence surrounding the pause site directly influences the propensity of the elongation complex to backtrack (Fig. 3), contributing to NELF and TFIIS-mediated regulation.
Figure 5.
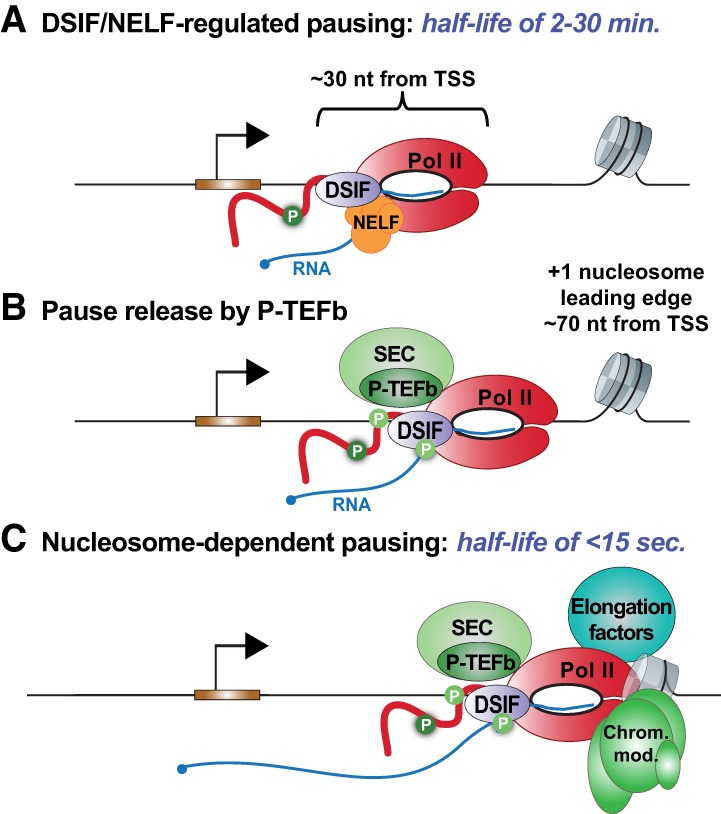
Distinct locations and mechanisms underlie regulated Pol II pausing and nucleosome-dependent pausing. (A) Pausing regulated by DSIF and NELF occurs with a predominant peak located 30-nt downstream from the TSS (range = 25–50 nt). This pausing can be long-lived and is clearly modulated to tune gene expression. Release of DSIF/NELF-mediated pausing is triggered by recruitment of P-TEFb to the gene promoter. (B) Upon pause release, Pol II soon encounters the first downstream nucleosome, called the +1 nucleosome. (C) The presence of a nucleosome can transiently halt RNA synthesis and induce Pol II arrest. Association of elongation factors and chromatin modifying complexes greatly facilitate Pol II movement through nucleosomes in vivo, making this pause short lived.
It is tempting to speculate that DSIF/NELF-regulated Pol II pausing might be affected by the first nucleosome encountered. However, careful analyses of the positions and stability of paused Pol II argue against such a model. Elegant single-nucleotide mapping of Pol II with respect to nucleosomes indicates that regulated pausing occurs well before the first nucleosome is encountered (Kwak et al. 2013; Li and Gilmour 2013; Weber et al. 2014). Moreover, genes with the highest levels of promoter-proximally paused Pol II exhibit very low occupancy of downstream nucleosomes, implying no direct involvement of nucleosomes in regulated pausing (Gilchrist et al. 2010; Li and Gilmour 2013).
Structural studies of paused RNA polymerase complexes from bacteria and mammals provide important insights into pausing by revealing a substantial deformation of the polymerase active site that precludes further nucleotide addition. In particular, recent structures of mammalian Pol II reveal that the RNA–DNA hybrid adopts a “tilted” conformation in the paused complex, wherein the RNA has translocated out of the active site, but the DNA has not (Vos et al. 2018b). This leaves the catalytic site without an available, unpaired DNA base to template new NTP incorporation. Such a distortion of the active site conformation would block additional RNA synthesis, while allowing the polymerase to remain stably associated with the nascent RNA. Interaction with the NELF complex constrains paused Pol II in this inactive conformation, structurally revealing how NELF stabilizes Pol II pausing. Moreover, NELF occupies and blocks a binding surface recognized by TFIIS, thereby preventing TFIIS-mediated Pol II reactivation. Upon P-TEFb driven pause release and dissociation of NELF, the DNA–RNA hybrid adopts an active conformation that is compatible with RNA extension; however, the molecular details of this transition have yet to be elucidated.
Following pause release by P-TEFb, downstream nucleosomes can represent additional obstacles to productive elongation (Fig. 5B). Indeed, Pol II pauses briefly as it enters the first nucleosome, as contacts between DNA and histones are broken (Fig. 5C; Kwak et al. 2013; Weber et al. 2014). Interestingly, this type of short-lived pausing is more prominent at constitutively active genes that exhibit lower levels of DSIF/NELF-regulated pausing (Li and Gilmour 2013). By comparison, the nucleosomes downstream from highly paused promoters show lower stability and higher turnover, suggesting that the establishment of paused Pol II destabilizes nucleosome structure and/or cause partial histone loss (Teves et al. 2014; Mieczkowski et al. 2016; Voong et al. 2016). Moreover, chromatin remodelers could be recruited to paused Pol II along with elongation factors (e.g., Spt6, FACT, and PAF1) to lower the nucleosomal barrier to productive elongation upon pause release (Brown et al. 1996; Teves et al. 2014). Although there is no current evidence that intrinsic, nucleosome-dependent pausing is regulated during gene activation, the presence of histone variants such as H2A.Z within the initial nucleosomes reduces the appearance of pausing in this region (Weber et al. 2014). H2A.Z-containing nucleosomes can recruit chromatin remodelers and are often heavily acetylated, suggesting several mechanisms that could be used to diminish the extent to which nucleosomes are obstacles to elongation.
Interestingly, Pol II slowed by the first several nucleosomes becomes susceptible to termination (Chiu et al. 2018). These data raise the intriguing possibility that proper maturation of the transcription complex during release from regulated pausing is important to prepare Pol II to efficiently transcribe through nucleosomes. In support of this idea, P-TEFb activity stimulates the association of Spt6, PAF1, and components of the SEC with Pol II during the transition to productive elongation. A prediction of this model is that Pol II, which elongates downstream in the absence of the appropriate factors, should be particularly prone to termination at nucleosomal boundaries. Consistent with this, during activation of the Hsp70 gene in the absence of P-TEFb, Pol II elongates through the typical sites of pausing but appears to stall downstream during passage through the first several nucleosomes (Ni et al. 2008). In future work, it will be interesting to further test this model and identify the factors involved in modulating nucleosome disassembly and transcription termination.
How stable is paused Pol II?
The accumulation of Pol II observed just downstream from most metazoan promoters clearly indicates that the release from pausing is a kinetic bottleneck in the transcription cycle, such that pause release or premature termination occurs on a time scale that is much slower than the processes of transcription initiation and promoter escape. However, since initiation and establishment of the paused state are generally accepted to take only a few seconds (Darzacq et al. 2007; Steurer et al. 2018) the relative genomic distribution of Pol II in ChIP-seq (ChIP combined with high-throughput sequencing) or PRO-seq (precision run-on sequencing) experiments cannot alone distinguish whether Pol II pauses for 1 min, 10 min, or 1 hr prior to release. Therefore, additional strategies are required to more directly measure the duration of Pol II pausing in living cells. Much of this work has used the drug triptolide (Trp) to inhibit transcription initiation through blocking the activity of the TFIIH helicase (Titov et al. 2011). In the absence of new initiation, one can measure the occupancy of engaged Pol II in the promoter region over time, to calculate the decay of the pre-existing paused complex. The earliest of these studies interrogated the half-life of promoter Pol II using total Pol II ChIP-qPCR and quantification of short TSS-associated RNAs (start-RNAs) held within paused Pol II and reported strong agreement between these measures (Henriques et al. 2013). These findings revealed that short nascent RNAs released by paused Pol II are almost immediately degraded by the exosome and demonstrated that either RNA- or ChIP-based methods could be used to define the stability of paused polymerase (Henriques et al. 2013). Critically, even though this study only evaluated pausing at a dozen genes, there was a clear diversity in stability of Pol II across these loci: Whereas the average half-life was on the order of 5–10 min, examples of genes with much shorter—and much longer—durations were described.
A number of subsequent studies have made use of Trp to monitor pausing duration in a variety of cell types and conditions, using a broad repertoire of techniques to measure paused Pol II levels (Buckley et al. 2014; Jonkers et al. 2014; Chen et al. 2015; Krebs et al. 2017; Nilson et al. 2017; Shao and Zeitlinger 2017; Erickson et al. 2018). Despite these differences, all such studies have shown that Pol II displays highly variable, gene-specific profiles of stability. Pol II signal was dramatically reduced at some genes by the earliest time point tested (often ∼2 min), whereas other genes displayed stably paused Pol II after 20 min or even 1 hr (Henriques et al. 2013; Buckley et al. 2014; Jonkers et al. 2014; Chen et al. 2015; Krebs et al. 2017; Nilson et al. 2017; Shao and Zeitlinger 2017; Erickson et al. 2018; Steurer et al. 2018). As a result of this striking variability among genes and the different strategies, numbers of genes, and thresholds used for data analysis in the individual studies, reported average half-lives of paused Pol II span from ∼2 to 30 min. As anticipated, all of these estimates for the duration of pausing are >10-times longer than the typical duration of TF-binding events or transcription initiation. Thus, all measurements of the lifetime of paused Pol II strongly support pausing as being a central rate-limiting step. However, the variability of reported average pausing durations has led to something of a controversy in the field (Price 2018). In this regard, we emphasize that the critical relevant aspect of these studies of paused Pol II stability is not the average value obtained but instead is in the incredible diversity of behaviors uncovered. In short, the very fact that paused Pol II is highly unstable at some genes and remarkably stable at others means that characterizing the “average” gene is less relevant than defining the mechanisms that distinguish a promoter with rapid Pol II turnover from one with very stable pausing.
Intriguingly, genomic analysis of Pol II stability using Start-seq across a time course of triptolide treatment in Drosophila (Krebs et al. 2017) revealed that pausing of Pol II at mRNA promoters is significantly longer-lived than pausing at enhancers (Henriques et al. 2018). In this analysis, nearly half of enhancers showed pausing half-lives of <2.5 min, whereas only 17% of protein-coding genes displayed such Pol II rapid turnover. This finding has both technical and mechanistic implications. Technically, it places limits on the ability to measure Pol II half-lives in vivo using imaging strategies that cannot distinguish between promoter-associated versus enhancer-associated Pol II (Darzacq et al. 2007; Steurer et al. 2018), since such studies will blur together the distinct Pol II turnover rates at enhancers and promoters. Moreover, given that enhancers and other transcribed noncoding RNA loci are far more numerous than mRNA genes, the majority of Pol II detected will not represent Pol II at protein coding genes. Thus dynamic measurements obtained using these strategies should be interpreted with caution. Mechanistically, these findings raise central questions about how the stability of early elongation complexes is determined and what factors or features underlie the striking difference in Pol II status at protein-coding versus regulatory loci. Biochemically, elongation complexes comprised of Pol II, the template DNA, and the nascent RNA have extraordinary stability (Kireeva et al. 2000). Engaged Pol II complexes are retained on chromatin following stringent washing conditions that disrupt a majority of other protein–protein and protein–DNA interactions (Gariglio et al. 1974; Wuarin and Schibler 1994), and paused Pol II can be subjected to RNase and DNase treatment without the polymerase disengaging the template or the nascent RNA that it protects (Coppola and Luse 1984; Cai and Luse 1987). Remarkably, Pol II within RNase-treated samples or samples stored in liquid nitrogen for ∼30 yr even retain their ability to resume transcription in run-on reactions (Jackson et al. 1998; Core et al. 2008; Chu et al. 2018). This extreme stability has been observed in the absence of auxiliary factors, indicating that it is an inherent property of elongating Pol II. Therefore, we propose that the lack of stable pausing at enhancers results from a termination factor that actively dismantles the engaged transcription complex. Although such termination factors have been described in bacterial and yeast systems (e.g., Rho factor in Escherichia coli; NNS pathway in Saccharomyces cerevisiae) (Porrua and Libri 2015; Ray-Soni et al. 2016), the mechanisms underlying premature termination in metazoa remain to be elucidated.
Is pausing near promoters mechanistically similar to pausing within gene bodies?
Following release from promoter-proximal pausing, Pol II is known to pause transiently or slow elongation rate within genes, including near splice junctions and 3′ ends (Brodsky et al. 2005; Alexander et al. 2010). However, there are several salient differences between regulated pausing of Pol II in early elongation and pausing observed elsewhere along the gene body, including the duration of pausing and the evidence for regulation of the paused state. As described above, pausing near promoters is stable and can persist for many minutes; in contrast, pausing near splice junctions is very short lived (≪15 sec) (Singh and Padgett 2009; Danko et al. 2013; Kwak et al. 2013; Jonkers et al. 2014). This efficiency of productive elongation stems in part from the entourage of elongation factors associated with Pol II in gene bodes, which render Pol II refractory to intrinsic pausing caused by sequence context or nucleosomal obstacles (Teves et al. 2014; Jonkers and Lis 2015). Accordingly, recent studies of TFIIS activity demonstrated that Pol II backtracking and arrest is highly focused near promoters, with little evidence of backtracking or arrest at splice junctions (Weber et al. 2014; Sheridan et al. 2019). Moreover, the NELF complex, which is critical to promoter-proximal pausing, does not play a role in pausing within gene bodies, and the detergent Sarkosyl, which is essential for paused Pol II to resume elongation and “run-on” in vitro, is not needed for Pol II within genes to restart RNA synthesis (Core et al. 2012). Thus, pausing in early elongation exhibits features and sensitivities not observed for productive elongation within genes, supporting distinct mechanisms of pausing and its regulation.
Conclusions: pausing as the gateway to productive mRNA synthesis
Accumulation of Pol II near most promoters demonstrates that relative rates of termination and pause release are much slower than rates of recruitment and initiation. Regulatory processes typically target rate-limiting steps, and, consistently, the release of paused Pol II has emerged as a central point of gene control. Currently, the field is striving to go beyond relative rates for each step in the transcription cycle, to measure directly the rate constants at individual genes. In these studies, it will be most helpful to study dynamic systems, where one can observe the kinetics of each step as genes are activated or repressed. Furthermore, we must account for the full range of rates for the establishment and release of paused Pol II across all promoters and use the resulting classifications to generate new hypotheses regarding what sequence elements or protein factors influence the ultimate fate of early transcribing Pol II. Although it remains unclear how frequently termination of promoter Pol II occurs in vivo, there is growing evidence for rapid Pol II turnover at certain metazoan genes and enhancers, suggesting an underappreciated layer of transcriptional control. In light of the new questions put forth, known or undescribed protein factors and DNA/RNA structures can now be examined with respect to their contribution to pause polymerase fate and turnover. Thus, much remains to be discovered about the critical step of Pol II pausing and how the transition to productive elongation is orchestrated in metazoan cells.
Acknowledgments
We thank our colleagues for their helpful advice and discussions during the writing of this review (in alphabetical order): Jay Bradner, Steve Buratowski, Patrick Cramer, Steve Henikoff, Mike Levine, John Lis, Ali Shilatifard, and Eric Wagner. This work was supported by in part by the National Institutes of Health Common Fund 4D Nucleome Program Transformative Collaborative Project Award (TCPA-2017-ADELMAN) to K.A., and National Institutes of Health award R35-GM128857 to L.C.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.325142.119.
References
- Adelman K, Henriques T. 2018. Transcriptional speed bumps revealed in high resolution. Nature 560: 560–561. 10.1038/d41586-018-05971-8 [DOI] [PubMed] [Google Scholar]
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell 17: 103–112. 10.1016/j.molcel.2004.11.028 [DOI] [PubMed] [Google Scholar]
- Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT. 2006. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol 26: 250–260. 10.1128/MCB.26.1.250-260.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. 2009. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci 106: 18207–18212. 10.1073/pnas.0910177106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD, Innocente SA, Barrass JD, Beggs JD. 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593. 10.1016/j.molcel.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amleh A, Nair SJ, Sun J, Sutherland A, Hasty P, Li R. 2009. Mouse cofactor of BRCA1 (Cobra1) is required for early embryogenesis. PLoS One 4: e5034 10.1371/journal.pone.0005034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, et al. 2013. BET bromodomains mediate transcriptional pause release in heart failure. Cell 154: 569–582. 10.1016/j.cell.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Sandelin A, Danko CG. 2015. A unified architecture of transcriptional regulatory elements. Trends Genet 31: 426–433. 10.1016/j.tig.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. 2009. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J 28: 1067–1077. 10.1038/emboj.2009.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Matija Peterlin B, Zhou Q. 2007. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res 35: 2003–2012. 10.1093/nar/gkm063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. 2007. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol 27: 6996–7006. 10.1128/MCB.00975-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. 2010. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24: 2303–2316. 10.1101/gad.1968210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Saunders A, Lis JT, Reinberg D. 2004. Transcription through chromatin: understanding a complex FACT. Biochim Biophys Acta 1677: 87–99. 10.1016/j.bbaexp.2003.09.017 [DOI] [PubMed] [Google Scholar]
- Bernecky C, Plitzko JM, Cramer P. 2017. Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat Struct Mol Biol 24: 809–815. 10.1038/nsmb.3465 [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci 104: 13690–13695. 10.1073/pnas.0705053104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM. 2011. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 25: 2158–2172. 10.1101/gad.16962311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. 2009. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325: 471–473. 10.1126/science.1173976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko VA, Steele LM, Újvári A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. 2006. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell 24: 469–479. 10.1016/j.molcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Booth GT, Parua PK, Sanso M, Fisher RP, Lis JT. 2018. Cdk9 regulates a promoter-proximal checkpoint to modulate RNA polymerase II elongation rate in fission yeast. Nat Commun 9: 543 10.1038/s41467-018-03006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA. 2005. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol 6: R64 10.1186/gb-2005-6-8-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev 10: 1479–1490. 10.1101/gad.10.12.1479 [DOI] [PubMed] [Google Scholar]
- Buckley MS, Kwak H, Zipfel WR, Lis JT. 2014. Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights into its regulation. Genes Dev 28: 14–19. 10.1101/gad.231886.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch H, Zheng X, Burkholder A, Dillon ST, Motola S, Birrane G, Ebmeier CC, Levine S, Fargo D, Hu G, et al. 2014. TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat Struct Mol Biol 21: 876–883. 10.1038/nsmb.2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch H, Lawney BP, Burkholder A, Ma D, Zheng X, Motola S, Fargo DC, Levine SS, Wang YE, Hu G. 2016. RNA polymerase II promoter-proximal pausing in mammalian long non-coding genes. Genomics 108: 64–77. 10.1016/j.ygeno.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546. 10.1016/j.molcel.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev 10: 711–724. 10.1101/gad.10.6.711 [DOI] [PubMed] [Google Scholar]
- Byers SA, Price JP, Cooper JJ, Li Q, Price DH. 2005. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem 280: 16360–16367. 10.1074/jbc.M500424200 [DOI] [PubMed] [Google Scholar]
- Cai H, Luse DS. 1987. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation, and elongation complexes. J Biol Chem 262: 298–304. [PubMed] [Google Scholar]
- Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J. 2015. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518: 249–253. 10.1038/nature13923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CL, Landick R. 1993. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol 233: 25–42. 10.1006/jmbi.1993.1482 [DOI] [PubMed] [Google Scholar]
- Chao SH, Price DH. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276: 31793–31799. 10.1074/jbc.M102306200 [DOI] [PubMed] [Google Scholar]
- Chédin F. 2016. Nascent connections: R-loops and chromatin patterning. Trends Genet 32: 828–838. 10.1016/j.tig.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Gao X, Shilatifard A. 2015. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev 29: 39–47. 10.1101/gad.246173.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pai AA, Herudek J, Lubas M, Meola N, Jarvelin AI, Andersson R, Pelechano V, Steinmetz LM, Jensen TH, et al. 2016. Principles for RNA metabolism and alternative transcription initiation within closely spaced promoters. Nat Genet 48: 984–994. 10.1038/ng.3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen JY, Zhang X, Gu Y, Xiao R, Shao C, Tang P, Qian H, Luo D, Li H, et al. 2017. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol Cell 68: 745–757.e5. 10.1016/j.molcel.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Price DH. 2007. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem 282: 21901–21912. 10.1074/jbc.M702936200 [DOI] [PubMed] [Google Scholar]
- Cheung AC, Cramer P. 2011. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471: 249–253. 10.1038/nature09785 [DOI] [PubMed] [Google Scholar]
- Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. 2002. Tat stimulates cotranscriptional capping of HIV mRNA. Mol Cell 10: 585–597. 10.1016/S1097-2765(02)00630-5 [DOI] [PubMed] [Google Scholar]
- Chiu AC, Suzuki HI, Wu X, Mahat DB, Kriz AJ, Sharp PA. 2018. Transcriptional pause sites delineate stable nucleosome-associated premature polyadenylation suppressed by U1 snRNP. Mol Cell 69: 648–663.e7. 10.1016/j.molcel.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu T, Rice EJ, Booth GT, Salamanca HH, Wang Z, Core LJ, Longo SL, Corona RJ, Chin LS, Lis JT, et al. 2018. Chromatin run-on and sequencing maps the transcriptional regulatory landscape of glioblastoma multiforme. Nat Genet 50: 1553–1564. 10.1038/s41588-018-0244-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373. 10.1038/nature09652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Armstrong J, Moens CB. 2005. Zebrafish foggy/spt 5 is required for migration of facial branchiomotor neurons but not for their survival. Dev Dyn 234: 651–658. 10.1002/dvdy.20584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola JA, Luse DS. 1984. Purification and characterization of ternary complexes containing accurately initiated RNA polymerase II and less than 20 nucleotides of RNA. J Mol Biol 178: 415–437. 10.1016/0022-2836(84)90151-7 [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall J, Lis J. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848. 10.1126/science.1162228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. 2012. Defining the status of RNA polymerase at promoters. Cell Rep 2: 1025–1035. 10.1016/j.celrep.2012.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. 2014. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet 46: 1311–1320. 10.1038/ng.3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czudnochowski N, Bosken CA, Geyer M. 2012. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun 3: 842 10.1038/ncomms1846 [DOI] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. 2013. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 50: 212–222. 10.1016/j.molcel.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. 2007. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14: 796–806. 10.1038/nsmb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533. 10.1038/nature10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienemann C, Schwalb B, Schilbach S, Cramer P. 2019. Promoter distortion and opening in the RNA polymerase II cleft. Mol Cell 73: 97–106.e4. 10.1016/j.molcel.2018.10.014 [DOI] [PubMed] [Google Scholar]
- D'Orso I, Frankel AD. 2010. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol 17: 815–821. 10.1038/nsmb.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte FM, Fuda NJ, Mahat DB, Core LJ, Guertin MJ, Lis JT. 2016. Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev 30: 1731–1746. 10.1101/gad.284430.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D, Geyer M. 2013. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 113: 8456–8490. 10.1021/cr400071f [DOI] [PubMed] [Google Scholar]
- Erickson B, Sheridan RM, Cortazar M, Bentley DL. 2018. Dynamic turnover of paused Pol II complexes at human promoters. Genes Dev 32: 1215–1225. 10.1101/gad.316810.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrega C, Shen V, Shuman S, Lima CD. 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell 11: 1549–1561. 10.1016/S1097-2765(03)00187-4 [DOI] [PubMed] [Google Scholar]
- Faust TB, Li Y, Bacon CW, Jang GM, Weiss A, Jayaraman B, Newton BW, Krogan NJ, D'Orso I, Frankel AD. 2018. The HIV-1 Tat protein recruits a ubiquitin ligase to reorganize the 7SK snRNP for transcriptional activation. Elife 7: e31879 10.7554/eLife.31879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal FM, Meng CA, Murakami K, Kornberg RD, Block SM. 2015. Real-time observation of the initiation of RNA polymerase II transcription. Nature 525: 274–277. 10.1038/nature14882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg MB, Baltimore D, Frankel AD. 1991. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci 88: 4045–4049. 10.1073/pnas.88.9.4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J, Galburt E, Hahn S. 2016. Transcription start site scanning and the requirement for ATP during transcription initiation by RNA polymerase II. J Biol Chem 291: 13040–13047. 10.1074/jbc.M116.724583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Guertin MJ, Sharma S, Danko CG, Martins AL, Siepel A, Lis JT. 2015. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet 11: e1005108 10.1371/journal.pgen.1005108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24: 787–795. 10.1128/MCB.24.2.787-795.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gala HP, Saha D, Venugopal N, Aloysius A, Dhawan J. 2018. RNA polymerase II pausing regulates a quiescence-dependent transcriptional program, priming cells for cell cycle reentry. bioRxiv. 10.1101/250910 [DOI] [Google Scholar]
- Gariglio P, Buss J, Green MH. 1974. Sarkosyl activation of RNA polymerase activity in mitotic mouse cells. FEBS Lett 44: 330–333. 10.1016/0014-5793(74)81170-1 [DOI] [PubMed] [Google Scholar]
- Gariglio P, Bellard M, Chambon P. 1981. Clustering of RNA polymerase B molecules in the 5′ moiety of the adult β-globin gene of hen erythrocytes. Nucleic Acids Res 9: 2589–2598. 10.1093/nar/9.11.2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev 22: 1921–1933. 10.1101/gad.1643208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551. 10.1016/j.cell.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. 2012. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev 26: 933–944. 10.1101/gad.187781.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. 1986. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol 6: 3984–3989. 10.1128/MCB.6.11.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. 2009. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol 29: 5455–5464. 10.1128/MCB.00637-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressel S, Schwalb B, Decker TM, Qin W, Leonhardt H, Eick D, Cramer P. 2017. CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife 6: 29736 10.7554/eLife.29736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88. 10.1016/j.cell.2007.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Yamaguchi Y, Schilbach S, Wada T, Lee J, Goddard A, French D, Handa H, Rosenthal A. 2000. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature 408: 366–369. 10.1038/35042590 [DOI] [PubMed] [Google Scholar]
- Gupte R, Muse GW, Chinenov Y, Adelman K, Rogatsky I. 2013. Glucocorticoid receptor represses proinflammatory genes at distinct steps of the transcription cycle. Proc Natl Acad Sci 110: 14616–14621. 10.1073/pnas.1309898110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. 2011. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145: 622–634. 10.1016/j.cell.2011.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Fu J. 2013. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta 1829: 105–115. 10.1016/j.bbagrm.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38: 428–438. 10.1016/j.molcel.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Johnston J, Zeitlinger J. 2015. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol 33: 395–401. 10.1038/nbt.3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein PP, Kolb KE, Windgassen T, Bellecourt MJ, Darst SA, Mooney RA, Landick R. 2014. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat Struct Mol Biol 21: 794–802. 10.1038/nsmb.2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. 2008. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci 105: 7762–7767. 10.1073/pnas.0802406105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. 2013. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell 52: 517–528. 10.1016/j.molcel.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, Adelman K. 2018. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev 32: 26–41. 10.1101/gad.309351.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieb AR, Baran S, Goodrich JA, Kugel JF. 2006. An 8 nt RNA triggers a rate-limiting shift of RNA polymerase II complexes into elongation. EMBO J 25: 3100–3109. 10.1038/sj.emboj.7601197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3: 405–411. 10.1016/S1097-2765(00)80468-2 [DOI] [PubMed] [Google Scholar]
- Holstege FC, Fiedler U, Timmers HT. 1997. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J 16: 7468–7480. 10.1093/emboj/16.24.7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen F, Greifenberg AK, Bösken CA, Geyer M. 2014. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res 42: 7577–7590. 10.1093/nar/gku449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol 20: 2970–2983. 10.1128/MCB.20.9.2970-2983.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS. 1992. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′–5′ direction in the presence of elongation factor SII. Genes Dev 6: 1342–1356. 10.1101/gad.6.7.1342 [DOI] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EM, Cook PR. 1998. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell 9: 1523–1536. 10.1091/mbc.9.6.1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19: 523–534. 10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Jennings BH. 2013. Pausing for thought: disrupting the early transcription elongation checkpoint leads to developmental defects and tumourigenesis. Bioessays 35: 553–560. 10.1002/bies.201200179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. 2013. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153: 855–868. 10.1016/j.cell.2013.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502. 10.1126/science.1141319 [DOI] [PubMed] [Google Scholar]
- Jonkers I, Lis JT. 2015. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16: 167–177. 10.1038/nrm3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. 2014. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife 3: e02407 10.7554/eLife.02407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Blobel GA. 2013. Mitotic bookmarking by transcription factors. Epigenetics Chromatin 6: 6 10.1186/1756-8935-6-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. 2012. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150: 725–737. 10.1016/j.cell.2012.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR, Vahedi G, Heightman TD, Garcia BA, Reinberg D, et al. 2014. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 21: 1047–1057. 10.1038/nsmb.2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330: 489–493. 10.1038/330489a0 [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099. 10.1126/science.1087374 [DOI] [PubMed] [Google Scholar]
- Kellner WA, Bell JS, Vertino PM. 2015. GC skew defines distinct RNA polymerase pause sites in CpG island promoters. Genome Res 25: 1600–1609. 10.1101/gr.189068.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M, Mathews MB. 1991. Tat transactivation of the human immunodeficiency virus type 1 promoter is influenced by basal promoter activity and the simian virus 40 origin of DNA replication. Proc Natl Acad Sci 88: 10018–10022. 10.1073/pnas.88.22.10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M, Mathews MB. 1992. Premature termination and processing of human immunodeficiency virus type 1-promoted transcripts. J Virol 66: 4488–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. 2003. Architecture of the RNA polymerase II–TFIIS complex and implications for mRNA cleavage. Cell 114: 347–357. 10.1016/S0092-8674(03)00598-1 [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. 2005. A high-resolution map of active promoters in the human genome. Nature 436: 876–880. 10.1038/nature03877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Suh H, Cho EJ, Buratowski S. 2009. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J Biol Chem 284: 26421–26426. 10.1074/jbc.M109.028993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Kashlev M. 2009. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci 106: 8900–8905. 10.1073/pnas.0900407106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Komissarova N, Waugh DS, Kashlev M. 2000. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem 275: 6530–6536. 10.1074/jbc.275.9.6530 [DOI] [PubMed] [Google Scholar]
- Konuma T, Yu D, Zhao C, Ju Y, Sharma R, Ren C, Zhang Q, Zhou MM, Zeng L. 2017. Structural mechanism of the oxygenase JMJD6 recognition by the extraterminal (ET) domain of BRD4. Sci Rep 7: 16272 10.1038/s41598-017-16588-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Imanci D, Hoerner L, Gaidatzis D, Burger L, Schübeler D. 2017. Genome-wide single-molecule footprinting reveals high RNA polymerase II turnover at paused promoters. Mol Cell 67: 411–422.e4. 10.1016/j.molcel.2017.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M. 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter–proximal region. Genes Dev 6: 2201–2213. 10.1101/gad.6.11.2201 [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953. 10.1126/science.1229386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, et al. 2013. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153: 976–987. 10.1016/j.cell.2013.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R. 2006. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans 34: 1062–1066. 10.1042/BST0341062 [DOI] [PubMed] [Google Scholar]
- Larochelle S, Amat R, Glover-Cutter K, Sansó M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. 2012. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 19: 1108–1115. 10.1038/nsmb.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. 2014. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344: 1042–1047. 10.1126/science.1251871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne MD, Konstantopoulos D, Ntakou-Zamplara KZ, Liakos A, Fousteri M. 2017. Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat Commun 8: 2076 10.1038/s41467-017-02145-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gilmour DS. 2013. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J 32: 1829–1841. 10.1038/emboj.2013.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gilmour DS. 2015. Reconstitution of factor-dependent, promoter proximal pausing in Drosophila nuclear extracts. Methods Mol Biol 1276: 133–152. 10.1007/978-1-4939-2392-2_7 [DOI] [PubMed] [Google Scholar]
- Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS. 2013. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol Cell 50: 711–722. 10.1016/j.molcel.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37: 429–437. 10.1016/j.molcel.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev 14: 792–803. [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Pelham-Webb B, Di Giammartino DC, Li J, Kim D, Kita K, Saiz N, Garg V, Doane A, Giannakakou P, et al. 2017. Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep 19: 1283–1293. 10.1016/j.celrep.2017.04.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, Marshall S, Florens L, Washburn MP, Shilatifard A. 2012. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol Cell Biol 32: 2608–2617. 10.1128/MCB.00182-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luse DS. 2013. Promoter clearance by RNA polymerase II. Biochim Biophys Acta 1829: 63–68. 10.1016/j.bbagrm.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luse DS, Spangler LC, Újvári A. 2011. Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. J Biol Chem 286: 6040–6048. 10.1074/jbc.M110.174722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, Lis JT. 2016. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat Protoc 11: 1455–1476. 10.1038/nprot.2016.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. 2012. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149: 590–604. 10.1016/j.cell.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 270: 12335–12338. 10.1074/jbc.270.21.12335 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. 2008. Nucleosome organization in the Drosophila genome. Nature 453: 358–362. 10.1038/nature06929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278. 10.1038/nsmb.1903 [DOI] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. 2015. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161: 541–554. 10.1016/j.cell.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D. 1997. Marking of active genes on mitotic chromosomes. Nature 388: 895–899. 10.1038/42282 [DOI] [PubMed] [Google Scholar]
- Mieczkowski J, Cook A, Bowman SK, Mueller B, Alver BH, Kundu S, Deaton AM, Urban JA, Larschan E, Park PJ, et al. 2016. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat Commun 7: 11485 10.1038/ncomms11485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A, Gilmour DS. 2010. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci 107: 11301–11306. 10.1073/pnas.1000681107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511. 10.1038/ng.2007.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, et al. 2003. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol 23: 1863–1873. 10.1128/MCB.23.6.1863-1873.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338. 10.1126/science.1181421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414: 322–325. 10.1038/35104581 [DOI] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. 2008. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol 28: 1161–1170. 10.1128/MCB.01859-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson KA, Lawson CK, Mullen NJ, Ball CB, Spector BM, Meier JL, Price DH. 2017. Oxidative stress rapidly stabilizes promoter-proximal paused Pol II across the human genome. Nucleic Acids Res 45: 11088–11105. 10.1093/nar/gkx724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Patel AB, Louder RK. 2017. Towards a mechanistic understanding of core promoter recognition from cryo-EM studies of human TFIID. Curr Opin Struct Biol 47: 60–66. 10.1016/j.sbi.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Grosso ARF, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ. 2015. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161: 526–540. 10.1016/j.cell.2015.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U. 2006. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res 34: 5943–5950. 10.1093/nar/gkl608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, Zhang T, Kwiatkowski N, Boukhali M, Green JL, et al. 2018. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol 14: 163–170. 10.1038/nchembio.2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Kwak H, Waters CT, Sprouse RO, White BS, Ozer A, Szeto K, Shalloway D, Craighead HG, Lis JT. 2014. Defining NELF-E RNA binding in HIV-1 and promoter-proximal pause regions. PLoS Genet 10: e1004090 10.1371/journal.pgen.1004090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palangat M, Meier TI, Keene RG, Landick R. 1998. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol Cell 1: 1033–1042. 10.1016/S1097-2765(00)80103-3 [DOI] [PubMed] [Google Scholar]
- Palangat M, Renner DB, Price DH, Landick R. 2005. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc Natl Acad Sci 102: 15036–15041. 10.1073/pnas.0409405102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palozola KC, Donahue G, Liu H, Grant GR, Becker JS, Cote A, Yu H, Raj A, Zaret KS. 2017. Mitotic transcription and waves of gene reactivation during mitotic exit. Science 358: 119–122. 10.1126/science.aal4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Shuman S. 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem 277: 19639–19648. 10.1074/jbc.M200015200 [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305. 10.1016/j.molcel.2006.06.014 [DOI] [PubMed] [Google Scholar]
- Plet A, Eick D, Blanchard JM. 1995. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene 10: 319–328. [PubMed] [Google Scholar]
- Porrua O, Libri D. 2015. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol 16: 190–202. 10.1038/nrm3943 [DOI] [PubMed] [Google Scholar]
- Price DH. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol 20: 2629–2634. 10.1128/MCB.20.8.2629-2634.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH. 2018. Transient pausing by RNA polymerase II. Proc Natl Acad Sci 115: 4810–4812. 10.1073/pnas.1805129115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh BF, Venters BJ. 2016. Genomic organization of human transcription initiation complexes. PLoS One 11: e0149339 10.1371/journal.pone.0149339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Gilmour DS. 2017. Identification of regions in the Spt5 subunit of DRB sensitivity-inducing factor (DSIF) that are involved in promoter-proximal pausing. J Biol Chem 292: 5555–5570. 10.1074/jbc.M116.760751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaresma AJC, Bugai A, Barboric M. 2016. Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res 44: 7527–7539. 10.1093/nar/gkw585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445. 10.1016/j.cell.2010.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci 90: 7923–7927. 10.1073/pnas.90.17.7923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. 1995. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J Mol Biol 252: 522–535. 10.1006/jmbi.1995.0517 [DOI] [PubMed] [Google Scholar]
- Ray-Soni A, Bellecourt MJ, Landick R. 2016. Mechanisms of bacterial transcription termination: all good things must end. Annu Rev Biochem 85: 319–347. 10.1146/annurev-biochem-060815-014844 [DOI] [PubMed] [Google Scholar]
- Reines D. 1992. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem 267: 3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Rennie S, Dalby M, Lloret-Llinares M, Bakoulis S, Dalager Vaagenso C, Heick Jensen T, Andersson R. 2018. Transcription start site analysis reveals widespread divergent transcription in D. melanogaster and core promoter-encoded enhancer activities. Nucleic Acids Res 46: 5455–5469. 10.1093/nar/gky244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein–DNA interactions detected at single-nucleotide resolution. Cell 147: 1408–1419. 10.1016/j.cell.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JW. 1988. Phage λ and the regulation of transcription termination. Cell 52: 5–6. 10.1016/0092-8674(88)90523-5 [DOI] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol 20: 104–112. 10.1128/MCB.20.1.104-112.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Molina JB, Tseng SC, Simonett SP, Taunton J, Ansari AZ. 2016. Engineered covalent inactivation of TFIIH-kinase reveals an elongation checkpoint and results in widespread mRNA stabilization. Mol Cell 63: 433–444. 10.1016/j.molcel.2016.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi MJ, Lai WKM, Pugh BF. 2018. Simplified ChIP-exo assays. Nat Commun 9: 2842 10.1038/s41467-018-05265-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804. 10.1016/S0092-8674(88)91087-2 [DOI] [PubMed] [Google Scholar]
- Sanso M, Levin RS, Lipp JJ, Wang VY, Greifenberg AK, Quezada EM, Ali A, Ghosh A, Larochelle S, Rana TM, et al. 2016. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev 30: 117–131. 10.1101/gad.269589.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira JM, Aguilera A. 2015. R loops: new modulators of genome dynamics and function. Nat Rev Genet 16: 583–597. 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Tsai KL, Yu X, Sardiu M, Saraf A, Washburn MP, Florens L, Asturias FJ, Conaway RC, et al. 2016. Role for the MED21-MED7 hinge in assembly of the mediator-RNA polymerase II holoenzyme. J Biol Chem 291: 26886–26898. 10.1074/jbc.M116.756098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Sutcliffe C, Lis JT, Ashe HL. 2013. Extensive polymerase pausing during Drosophila axis patterning enables high-level and pliable transcription. Genes Dev 27: 1146–1158. 10.1101/gad.215459.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilbach S, Hantsche M, Tegunov D, Dienemann C, Wigge C, Urlaub H, Cramer P. 2017. Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551: 204–209. 10.1038/nature24282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, Lau J, Bisgrove D, Schnolzer M, Verdin E, et al. 2012. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem 287: 1090–1099. 10.1074/jbc.M111.282855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S, Gietl A, Smollett K, Tinnefeld P, Werner F, Grohmann D. 2016. TFE and Spt4/5 open and close the RNA polymerase clamp during the transcription cycle. Proc Natl Acad Sci 113: E1816–25. 10.1073/pnas.1515817113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scruggs BS, Gilchrist DA, Nechaev S, Muse GW, Burkholder A, Fargo DC, Adelman K. 2015. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol Cell 58: 1101–1112. 10.1016/j.molcel.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Zeitlinger J. 2017. Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet 49: 1045–1051. 10.1038/ng.3867 [DOI] [PubMed] [Google Scholar]
- Sheridan RM, Fong N, D'Alessandro A, Bentley DL. 2019. Widespread backtracking by RNA Pol II is a major effector of gene activation, 5′ pause release, termination, and transcription elongation rate. Mol Cell 73: 107–118.e4. 10.1016/j.molcel.2018.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A, Kallgren SP, Demel C, Maier KC, Spatt D, Alver BH, Cramer P, Park PJ, Winston F. 2017. Spt5 plays vital roles in the control of sense and antisense transcription elongation. Mol Cell 66: 77–88.e5. 10.1016/j.molcel.2017.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol 16: 669–676. 10.1128/MCB.16.2.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. 1996. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271: 1873–1876. 10.1126/science.271.5257.1873 [DOI] [PubMed] [Google Scholar]
- Singh J, Padgett RA. 2009. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16: 1128–1133. 10.1038/nsmb.1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 38: 439–451. 10.1016/j.molcel.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steurer B, Janssens RC, Geverts B, Geijer ME, Wienholz F, Theil AF, Chang J, Dealy S, Pothof J, van Cappellen WA, et al. 2018. Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II. Proc Natl Acad Sci 115: E4368–E4376. 10.1073/pnas.1717920115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl LJ, Eick D. 1992. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J 11: 3307–3314. 10.1002/j.1460-2075.1992.tb05409.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sypes MA, Gilmour DS. 1994. Protein/DNA crosslinking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res 22: 807–814. 10.1093/nar/22.5.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szlachta K, Thys RG, Atkin ND, Pierce LCT, Bekiranov S, Wang YH. 2018. Alternative DNA secondary structure formation affects RNA polymerase II promoter-proximal pausing in human. Genome Biol 19: 89 10.1186/s13059-018-1463-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. 2011. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146: 92–104. 10.1016/j.cell.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, Weber CM, Henikoff S. 2014. Transcribing through the nucleosome. Trends Biochem Sci 39: 577–586. 10.1016/j.tibs.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Teves SS, An L, Hansen AS, Xie L, Darzacq X, Tjian R. 2016. A dynamic mode of mitotic bookmarking by transcription factors. Elife 5: e22280 10.7554/eLife.22280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, An L, Bhargava-Shah A, Xie L, Darzacq X, Tjian R. 2018. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. Elife 7: e35621 10.7554/eLife.35621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. 2011. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol 7: 182–188. 10.1038/nchembio.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstorukov MY, Kharchenko PV, Goldman JA, Kingston RE, Park PJ. 2009. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res 19: 967–977. 10.1101/gr.084830.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tome JM, Tippens ND, Lis JT. 2018. Single-molecule nascent RNA sequencing identifies regulatory domain architecture at promoters and enhancers. Nat Genet 50: 1533–1541. 10.1038/s41588-018-0234-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Yu X, Gopalan S, Chao TC, Zhang Y, Florens L, Washburn MP, Murakami K, Conaway RC, Conaway JW, et al. 2017. Mediator structure and rearrangements required for holoenzyme formation. Nature 544: 196–201. 10.1038/nature21393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. 1994. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367: 525–532. 10.1038/367525a0 [DOI] [PubMed] [Google Scholar]
- Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. 2007. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J 26: 3570–3580. 10.1038/sj.emboj.7601783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oss SB, Shirra MK, Bataille AR, Wier AD, Yen K, Vinayachandran V, Byeon IL, Cucinotta CE, Heroux A, Jeon J, et al. 2016. The histone modification domain of Paf1 complex subunit Rtf1 directly stimulates H2B ubiquitylation through an interaction with Rad6. Mol Cell 64: 815–825. 10.1016/j.molcel.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo Ngoc L, Wang YL, Kassavetis GA, Kadonaga JT. 2017. The punctilious RNA polymerase II core promoter. Genes Dev 31: 1289–1301. 10.1101/gad.303149.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voong LN, Xi L, Sebeson AC, Xiong B, Wang JP, Wang X. 2016. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell 167: 1555–1570.e15. 10.1016/j.cell.2016.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Pollmann D, Caizzi L, Hofmann KB, Rombaut P, Zimniak T, Herzog F, Cramer P. 2016. Architecture and RNA binding of the human negative elongation factor. Elife 5: e14981 10.7554/eLife.14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, Cramer P. 2018a. Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature 560: 607–612. 10.1038/s41586-018-0440-4 [DOI] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Urlaub H, Cramer P. 2018b. Structure of paused transcription complex Pol II–DSIF–NELF. Nature 560: 601–606. 10.1038/s41586-018-0442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. 1998a. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12: 343–356. 10.1101/gad.12.3.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. 1998b. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J 17: 7395–7403. 10.1093/emboj/17.24.7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Orphanides G, Hasegawa J, Kim DK, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Oh S, Reinberg D, et al. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol Cell 5: 1067–1072. 10.1016/S1097-2765(00)80272-5 [DOI] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. 2014. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell 53: 819–830. 10.1016/j.molcel.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Leon K, Landick R, Darst SA. 2013. Structural basis of transcriptional pausing in bacteria. Cell 152: 431–441. 10.1016/j.cell.2012.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ. 1999. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev 13: 1774–1779. 10.1101/gad.13.14.1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, Adelman K. 2015. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell 58: 311–322. 10.1016/j.molcel.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, Reyes JM, di Iulio J, Souza A, Ott CJ, et al. 2017. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell 67: 5–18.e19. 10.1016/j.molcel.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Jin Y, Struhl K. 2014. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol Cell 54: 601–612. 10.1016/j.molcel.2014.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev 17: 1402–1414. 10.1101/gad.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. 1994. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol 14: 7219–7225. 10.1128/MCB.14.11.7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Vakoc CR. 2017. Targeting cancer cells with BET bromodomain inhibitors. Cold Spring Harb Perspect Med 7: a026674 10.1101/cshperspect.a026674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 21: 227–237. 10.1016/j.molcel.2005.11.024 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97: 41–51. 10.1016/S0092-8674(00)80713-8 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. 2002. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol 22: 2918–2927. 10.1128/MCB.22.9.2918-2927.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Shibata H, Handa H. 2013. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim Biophys Acta 1829: 98–104. 10.1016/j.bbagrm.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19: 535–545. 10.1016/j.molcel.2005.06.029 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516. 10.1038/ng.2007.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Klatt A, Gilmour DS, Henderson AJ. 2007. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J Biol Chem 282: 16981–16988. 10.1074/jbc.M610688200 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143. 10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]