Abstract
Reactivation of the cell cycle, including DNA replication, might play a major role in Alzheimer's disease (AD). A more than diploid DNA content in differentiated neurons might alternatively result from chromosome mis-segregation during mitosis in neuronal progenitor cells. It was our objective to distinguish between these two mechanisms for aneuploidy and to provide evidence for a functional cell cycle in AD. Using slide-based cytometry, chromogenic in situ hybridization, and PCR amplification of alu-repeats, we quantified the DNA amount of identified cortical neurons in normal human brain and AD and analyzed the link between a tetraploid DNA content and expression of the early mitotic marker cyclin B1. In the normal brain, the number of neurons with a more than diploid content amounts to ∼10%. Less than 1% of neurons contains a tetraploid DNA content. These neurons do not express cyclin B1, most likely representing constitutional tetraploidy. This population of cyclin B1-negative tetraploid neurons, at a reduced number, is also present in AD. In addition, a population of cyclin B1-positive tetraploid neurons of ∼2% of all neurons was observed in AD. Our results indicate that at least two different mechanisms need to be distinguished giving rise to a tetraploid DNA content in the adult brain. Constitutional aneuploidy in differentiated neurons might be more frequent than previously thought. It is, however, not elevated in AD. In addition, in AD some neurons have re-entered the cell cycle and entirely passed through a functional interphase with a complete DNA replication.
Keywords: cell cycle, cyclin B1, neurodegeneration, chromogenic in situ hybridization, CISH, slide-based cytometry, SBC, quantitative real-time PCR
Introduction
Neurodegeneration in Alzheimer's disease (AD) follows a certain hierarchy affecting some brain areas earlier than others (Braak and Braak, 1991). The pattern of disease progression, which might provide the key to understand AD pathomechanism, matches the regional degree of neuronal plasticity and inversely recapitulates ontogenetic and phylogenetic brain development (Arendt, 2001).
As a likely explanation for this selective neuronal vulnerability, it has been hypothesized that adult neurons with a high plastic potential rest in a labile state of differentiation, which is prone to a loss of differentiation control and a subsequent dedifferentiation that renders these neurons highly vulnerable for neurodegeneration (Arendt, 2001). This hypothesis is supported by observations on the expression of developmentally regulated genes in AD that correspond to a condition of dedifferentiation and links neurodegeneration to cell-cycle-related events (Heintz, 1993; Arendt et al., 2000). The re-expression of a multitude of cell-cycle regulators known to control the activation and progression of the cell cycle in dividing cells has been observed previously in degenerating neurons in AD (Smith and Lippa, 1995; Arendt et al., 1996, 1998; McShea et al., 1997; Nagy et al.,1997a,b; Vincent et al., 1997; Busser et al., 1998; Ding et al., 2000). Moreover, applying fluorescence in situ hybridization (FISH) technique to paraffin sections of the adult human CNS, Yang et al. (2001) could provide direct evidence for aneuploidy in AD, but not in normal brain. More recently, however, two groups (Rehen et al., 2005; Yurov et al., 2005) applying FISH to sorted isolated cellular nuclei reported a considerable degree of constitutional aneuploidy in the normal adult human brain, including triploidy and tetraploidy, corroborating cytophotometric observations made several decades ago (Brodskij and Kusc, 1962; Lapham and Johnstone, 1963; Mares et al., 1973). This kind of constitutional trisomy and tetrasomy might arise at least in part through chromosome mis-segregation during mitosis in neuronal progenitor cells and can persist into adulthood (A. H. Yang et al., 2003). Developmental deficiencies in chromosome segregation giving rise to aneuploid neuronal progenitors have also been suggested as a critical pathogenetic mechanism in AD (Potter, 1991). Alternatively, trisomic and tetrasomic cells could be the result of cell-cycle re-entry processes and subsequent partial or full DNA replication of mature neurons in the adult CNS, which is potentially associated with neuronal death (Herrup and Arendt, 2002).
To understand the pathogenetic pathways leading to neurodegeneration in AD and to design potential strategies of prevention that would require to target either the developing or the mature nervous system, it would be essential to determine whether the unscheduled cell-cycle events represent the failure of a cycling neuronal precursor to exit the cell cycle or whether a fully mature neuron has re-entered the cell cycle in response to a stimulus. Here, we developed a novel strategy of single-cell DNA quantification of identified neurons on brain slices, based on slide-based cytometry (SBC), PCR amplification of alu repeats, and chromogenic in situ hybridization (CISH), three independent methods with a high intermethod reliability, and applied this method to quantify the DNA amount of identified neurons in AD and normal adults.
To determine whether the more than diploid DNA content, seen both in control and AD, represents the failure of a cycling neuronal precursor to exit the cell cycle during brain development or whether a fully mature neuron has re-entered the cell cycle, we analyzed the link between DNA content and expression of cyclin B1, a marker of early mitosis and, thus, an indication that cells have entirely passed through S phase.
Materials and Methods
Case recruitment and tissue preparation.
Brains from 13 patients with AD (mean age ± SD, 76.3 ± 8.1 years) and 13 healthy controls dying without any history of neurological or psychiatric illness (71.7 ± 10.3 years) were used. Each case of AD met the National Institute of Neurologic and Communicative Disorders and Stroke and Alzheimer's Disease and Related Disorders Association criteria for definite diagnosis of AD. Severity of AD pathology was scored according to Braak and Braak (1991); six cases were classified as stage I-II (early stage of the disease), and seven cases as stage V-VI (advanced stage of the disease). Tissue blocks from the entorhinal (Brodmann area 28) and occipital (Brodmann area 17) cortices were fixed in 4% phosphate-buffered paraformaldehyde (4% PFA in PBS), pH 7.4, for 9 d and cryoprotected in 30% sucrose. Sections of 30 μm thickness were cut on a freezing microtome.
Immunohistochemistry.
For SBC, slices were processed using immunofluorescence staining as described previously (Mosch et al., 2006). Neurons were identified with the monoclonal mouse-anti-pan-neurofilament antibody (SMI 311, 1:750; Sternberger Monoclonals, Lutherville, MD). Cyclin B was detected with the polyclonal rabbit-anti-cyclin B1 (clone H-433, 1:500; Santa Cruz Biotechnology, Heidelberg, Germany) and the DNA was stained with propidium iodide (PI; Sigma, Taufkirchen, Germany). Slices for microdissection and chromogenic in situ hybridization were processed with an avidin-biotin system. Bound SMI 311 antibody was incubated with secondary biotin-conjugated goat-anti-mouse antibody (1:500; Dianova, Hamburg, Germany). Subsequently, sections were processed with Extravidin-peroxidase conjugate (1:500; Sigma) and 0.04% 3,3′-diaminobenzidine (DAB)/0.015% H2O2 (Sigma) as chromogen. Finally, sections were mounted onto slides, air dried for microdissection, or further processed using CISH.
Slide-based cytometry.
SBC was performed using a laser-scanning cytometer (CompuCyte, Cambridge, MA) and the appropriate software, WinCyte version 3.4. The conditions for SBC were optimized for the present application as described previously (Lenz et al., 2004; Mosch et al., 2006). Each fluorescent event was recorded with respect to size, perimeter, x–y position on the object slide, and maximum (maximum pixel) and overall integral fluorescence intensity. The relative DNA content of the cells was determined by the integral PI fluorescence values and these data were further analyzed using the cell-cycle software ModfitLT, version 2.0 (Verity Software House, Topsham, ME). The entire parahippocampal gyrus was scanned with 80,000–120,000 analyzed cells for each specimen. The determination of the numerical neuronal density was performed using the tool of WinCyte to record the x–y position of each fluorescence event. A square region with a side length of 1 mm was created and the number of neurons within this region randomly placed 10 times through the entire entorhinal cortex was determined. The relative number of cyclin B1-positive neurons also acquired by means of WinCyte.
Chromogenic in situ hybridization.
Hybridization was performed with a ZytoDotCEN 17 probe (ZytoVision, Bremerhaven, Germany), which targets α-satellite sequences of the centromere of chromosome 17. The digoxigenin-labeled probe was immunohistochemically visualized using peroxidase-conjugated Fab fragments of an anti-digoxigenin antibody from sheep (Roche Diagnostics, Mannheim, Germany) and nickelammoniumsulfate/DAB/0.015% H2O2 as chromogen. Fixed human lymphocytes dropped on object slides and HeLa cells cultured under standard conditions and grown on coverslips were used as controls. A total of 400–500 neuronal nuclei sampled throughout all cortical layers of the entorhinal cortex were analyzed for each case.
Microdissection and quantitative PCR.
Single neurons, identified by immunoreactivity for neurofilaments (SMI 311) were cut from brain slices with a laser microdissector (PALM MicroBeam; P.A.L.M. Microlaser Technologies, Bernried, Germany) and subsequently subjected to DNA quantification. DNA content of individual neurons was quantified through real-time PCR amplification of alu repeats (Walker et al., 2003), a class of short interspersed elements in the eukaryotic genome, which reach a copy number of ∼1 million in primates (Houck et al., 1979; Batzer and Deininger, 2002). Alu repeats were chosen because of their high copy number and low level of polymorphism compared with other short interspersed elements in the eukaryotic genome (Roy-Engel et al., 2001). The residual risk of an artificial influence by different copy numbers or single nucleotide polymorphisms in several individuals was avoided by the intraindividual comparison of two different brain areas of each patient. Alu oligonucleotide primers alu-for 5′-GTGGCTCACGCCTGTAATCCC-3′ and alu-rev 5′-ATCTCGGCTCACTGCAA-CCTC-3′, localized in conserved regions of the alu repeats, were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Real-time PCR quantification was accomplished in a Rotor-Gene 2000 (Corbett Research, Sydney, Australia). Data were analyzed by the Rotor-Gene 2000 software Rotorgene, version 4.6, and statistics were performed using PlotIT 3.2 (SPE Software, Ville de St-Georges, Quebec, Canada). Human lymphocytes treated identically to human brain tissue were used for control. A DNA amount of 2.07 ± 0.6 pg (mean ± SD) and 4.06 ± 0.5 pg was obtained for one single and two lymphocytes, respectively. For each case, at least 20 SMI 311-immunoreactive cells sampled from all layers of the entorhinal cortex were captured with the microdissector and processed for PCR.
Intermethod reliability of DNA quantification by SBC and quantitative PCR.
Intermethod reliability of all three methods for DNA quantification (SBC, CISH, and quantitative PCR) was evaluated by subsequent application of the methods to 48 identified neurons from one case of AD. The specimen was initially processed for immunofluorescence and analyzed by SBC with a laser-scanning cytometer. Subsequently, the slice was subjected to CISH, combined with conventional Nissl staining. Thereafter, identified cells were captured with laser microdissection and subjected to quantitative PCR. For each cell, the PI integral obtained by LSC was plotted against the number of spots and the amount of DNA calculated from the PCR (Fig. 1).
Figure 1.
Intermethod reliability of three independent methods for single-cell DNA quantification. A set of 48 identical microscopically identified neurons of the entorhinal cortex in a patient with early AD was evaluated through subsequent application of SBC, CISH, and PCR amplification of alu repeats. Tissue sections were first processed for SBC, followed by hybridization with the chromosome 17 probe. Subsequently, identified neurons were captured through laser microdissection and subjected to PCR amplification of alu repeats. A–C, Regression analyses reveal correlation coefficients according to Bravais-Pearson of r = 0.92 for the LSC data versus hybridization results (CISH; A), of r = 0.80 for LSC versus PCR amplification (B) and r = 0.78 for PCR amplification versus hybridization (C). All correlation coefficients are significant at p < 0.001. D, The combined set of data obtained with all three methods.
Results
In the present study, we analyzed the single-cell DNA content at the level of individual neurons in the human entorhinal cortex of normal adult human brain and in patients with AD. This investigation was based on a novel strategy of single-cell DNA quantification of identified neurons that combines SBC, CISH, and PCR amplification of alu repeats, three independent methods with a high intermethod reliability in conjunction with the analysis of expression of cyclin B1, an early mitotic marker.
Intermethod reliability for single-cell DNA quantification
Intermethod reliability of all three methods of single-cell DNA quantification was evaluated through subsequent application of SBC, CISH, and PCR amplification of alu repeats to 48 identical microscopically identified pyramidal neurons of the entorhinal cortex in a patient with early AD.
After processing for CISH, neurons contained either one, two, three, or four hybridization spots. When the PI integral was plotted against the number of hybridization spots, a highly significant correlation was obtained (Fig. 1 A). Similarly, the PI integral showed a highly significant correlation with the DNA content determined by PCR amplification of alu repeats (Fig. 1 B). The DNA amount determined by PCR amplification of alu repeats also correlated highly significantly with the number of hybridization spots (Fig. 1 C). Regression analysis between data obtained by SBC, PCR, and CISH indicates a high intermethod reliability between all three techniques (Fig. 1 A–D). Based on these correlations, the DNA amount (mean ± SD) of a single diploid neuron identified by two hybridization spots was determined as 2.52 ± 0.87 pg, whereas the DNA content of a neuron with four spots amounted to 5.94 ± 1.28 pg. The mean single-cell DNA content (±SD) of lymphocytes treated identically to brain tissue and used for control experiments was determined at 2.07 ± 0.6 pg. The DNA amount of neurons with either one or three hybridization spots was determined as 0.99 ± 0.21 and 4.12 ± 1.45 pg, respectively. Differences in the DNA content were statistically significant (Student's t test) for one-spot versus two-spot cells (p = 0.0000001), two-spot versus three-spot cells (p = 0.001), and three-spot versus four-spot cells (p = 0.023). The largest variance in the single neuron DNA content was observed for neurons with three spots, ranging from 2.1 to 7.2, thus spanning an interval of almost 4n (for comparison, one hybridization spot, 0.75–1.3 pg; two spots, 0.5–4.9 pg; four spots, 4.2–7.8 pg). Despite true aneuploidy, one needs to consider sectioning artifacts through the nucleus to explain this large variance. It can, therefore, not be excluded that a certain fraction of three-spot cells might in fact represent tetraploid cells and, similarly, one-spot cells might represent diploid neurons. Although we tried to minimize the likelihood for these artifacts by optimizing the section thickness (Lenz et al. 2004), we cannot rule out that our data represent some underestimation of the number of hyperploid neurons.
Quantification of neuronal loss and single-neuron DNA amount by slide-based cytometry
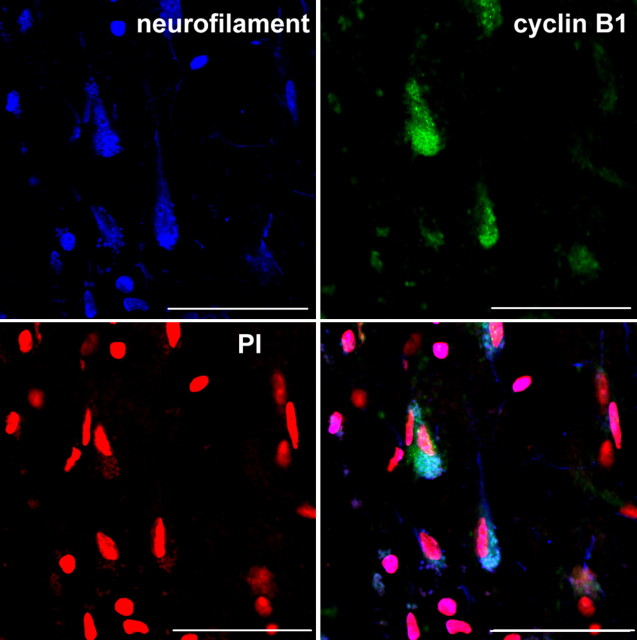
Neurons of the entorhinal cortex were identified by immunocytochemical detection with the pan-neuronal neurofilament marker SMI 311. Sections were counter reacted for the presence of cyclin B1, and nuclear DNA was stained by PI (Fig. 2). Cyclin B1 was localized preferentially in the neuronal cytoplasm and was found in all cases with AD. Controls could be separated into two different groups based on the expression of cyclin B1. In 7 of 13 controls, cyclin B1 was clearly detected in ∼30 to 80% of neurons, whereas the other six control cases showed only a marginal indication of cyclin B1 expression in <10% of neurons. In the additional analysis, both groups were analyzed separately, designated as control group I (marginal cyclin B1 expression) and control group II (strong cyclin B1 expression).
Figure 2.
Confocal image of a brain slice as used for SBC analysis. Immunohistochemical double staining for neurofilament (SMI 311, blue) and cyclin B1 (green) of pyramidal neurons in the human entorhinal cortex, counter reacted with PI (red). Scale bars, 200 μm.
Loss of neurons in the entorhinal cortex of AD brain was quantified by SBC. The numerical density of neurons showed a significant disease-related decline and, compared with control group I, decreased to 85% in group II, to 72% in early AD, and to 52% in advanced AD cases (Fig. 3 A). The frequency of neurons expressing cyclin B1 was increased in AD cases. Although the relative fraction of neurons expressing cyclin B1 was not different between control group II (42 ± 4%) and early AD (39 ± 3%), it was significantly increased in advanced AD stages (61 ± 4%) (Fig. 3 B). Analysis of numerical cell density in conjunction with cyclin B1 expression (supplemental Table 1, available at www.jneurosci.org as supplemental material) shows that neuron loss in AD affects both cyclin B1-positive and cyclin B1-negative neurons, albeit to a different extent. Whereas in early AD, cyclin B1-positive neurons are more severely reduced in number than cyclin B1-negative neurons, the opposite is true for advanced AD. This could reflect a progressive rate of conversion of neurons toward cyclin B1 expression and persistence at this stage for a longer period. Alternatively, a selective loss of cyclin B1-negative neurons cannot be ruled out.
Figure 3.
Quantification of neuronal loss and single-neuron DNA amount by SBC. A, Analysis of neuron number showing the decline with progression of the disease. B, Neuronal cyclin B1 expression is elevated in advanced AD stages. C–E, Single-cell DNA amount in neurons of the entorhinal cortex in controls and patients with AD. Shift of neurons from diploid toward triploid and tetraploid DNA amount in AD cases. Tetraploid neurons express cyclin B1 in AD, but not in control cases. F, Distribution of entorhinal cortical neurons with a tetraploid content of DNA according to the presence or absence of cyclin B1 expression. In both control groups, only cyclin B1-negative neurons were present, which are reduced in number in AD cases. In addition, a cyclin B1-positive cell population appears in AD and represents the majority of tetraploid cells under disease conditions. Statistics were performed for control versus AD groups with an additional analysis for early AD versus advanced AD in B. Shown are mean values ± SEM. **p ≤ 0.01; *p ≤ 0.05, Student's t test.
DNA analysis by SBC identified the majority of neurons in controls as diploid. In control groups I and II, respectively, 89 ± 4 and 88 ± 5% of neurons had a 2n DNA content, whereas 11 ± 4 and 12 ± 5% had a hyperploid DNA content between 2n (diploid) and 4n (tetraploid). (Here we use the term “hyperploid” to designate the group of cells with >2n and <4n DNA content.)
In both control groups, 0.4 ± 0.3% of neurons contained a tetrasomic DNA content (Fig. 3 C). In AD, a significant shift of neurons from the diploid to hyperploid and tetraploid DNA content was observed. Neurons with a diploid DNA content were reduced to 77 ± 2 and 70 ± 7% in early and advanced AD, respectively, whereas the fraction of neurons containing DNA between 2n and 4n were increased to 21 ± 2 and 29 ± 8%. The frequency of neurons with a tetrasomic content of DNA was increased in early AD by factor six (2.4 ± 0.7%) and more than doubled in advanced AD (1.1 ± 0.7%).
Analysis of the numerical cell density of neurons with a DNA amount of 2n, 2n–4n, or 4n (supplemental Table 2, available at www.jneurosci.org as supplemental material) indicated an increase in the number of neurons with >2n DNA from 38 and 35 neurons per square millimeter in control groups I and II to 57 and 53 in early and advanced AD, respectively. Against the background of a neuronal loss of ∼28 and 48% in early and advanced AD, respectively, compared with control group I, this reflects a remarkable increase that is most likely attributable to a disease-related reactivation of DNA replication. Alternatively, this shift could theoretically also arise from a selective loss of diploid neurons, although we did not obtain any indications for this.
To determine, whether the shift in the single neuron DNA content in AD reflects a constitutional hyperploidy or is the result of cell-cycle activation in mature neurons, we analyzed the link between the DNA content and the expression of cyclin B1 as an early mitotic marker (Figs. 3 D,E). Cyclin B1 expression occurs at the end of the S phase and reaches peak expression during G2 and M phase (Ito, 2000); the simultaneous analysis of single neuron DNA content and cyclin B1 expression might, therefore, allow to identify neurons that are in an active cycle and progressed beyond the S phase.
There were no significant differences between control and AD cases with respect to the presence of cyclin B1 expression in neurons with a DNA content of 2n or between 2n and 4n. Control brains and AD cases, however, clearly dissociated for neurons with a tetraploid DNA content. In controls, all neurons with a 4n content of DNA were negative for cyclin B1. This population of cyclin B1-negative neurons with a tetraploid content of DNA was also present in AD, although it was reduced in number compared with controls (Fig. 3 F). In addition, a population of cyclin B1-positive 4n neurons was observed in AD, representing the majority of neurons with a tetraploid content of DNA.
Chromogenic in situ hybridization
CISH with a chromosome 17 probe consistently revealed distinct hybridization spots in all cell types, including neurons, astrocytes, and microglial cells. In control hybridizations on lymphocytes, >99% of cells displayed two hybridization spots, which reveals a low rate of aneuploidy (Rehen et al., 2001). In cultured HeLa cells, cells with two (58% of cells), three (15%), or four spots (25%) were observed in agreement with previous studies (Hake et al., 2006), demonstrating the high proliferation rate of these cells (Figs. 4, 5).
Figure 4.
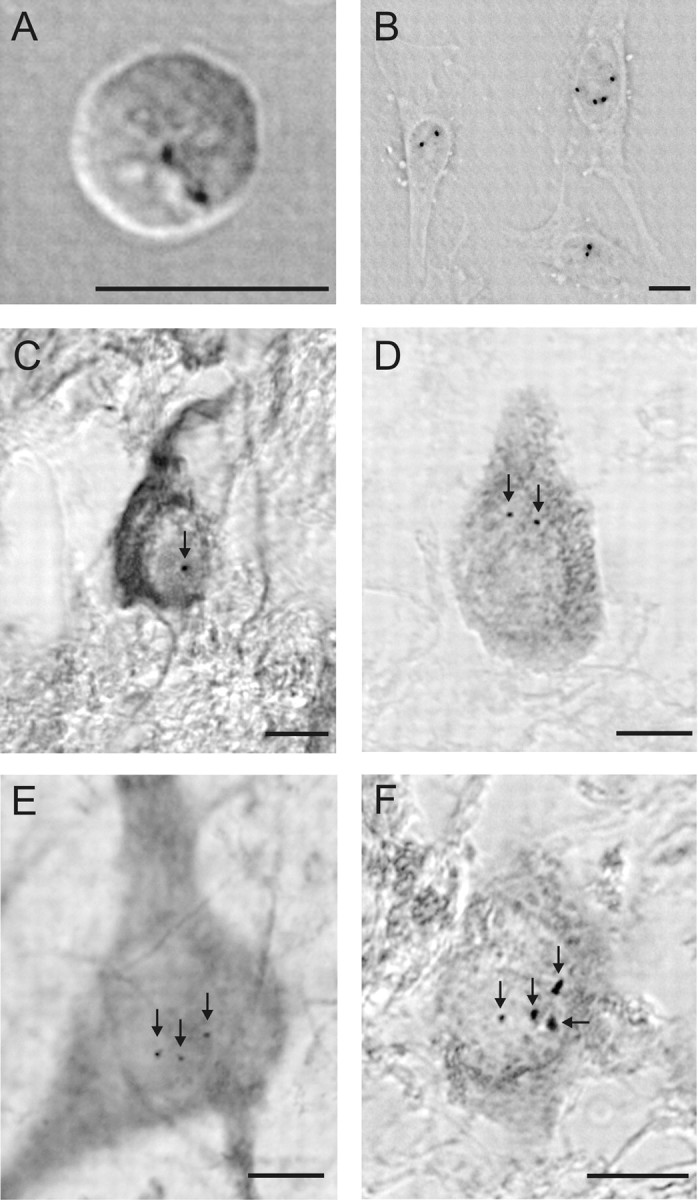
CISH signals with a chromosome 17 probe in lymphocytes, HeLa cells and neurons of the entorhinal cortex in one advanced case of AD. A, Two spots are present in 99% of all lymphocytes. B, Proliferating HeLa cells show either two or four spots, indicating a diploid and tetraploid DNA amount. C–F, Neurons in an advanced case of AD showing one, two, three, or four spots (arrows). Scale bars, 10 μm.
Figure 5.
Quantitation of CISH signals in neurons of the entorhinal cortex in both control and AD groups. AD cases compared with controls show the same percentage of cells without or with one spot, but a shift of neurons from two spots toward three and four spots. Statistics were performed for control versus AD groups. Mean values ± SEM are shown. ***p ≤ 0.001; **p ≤ 0.01, Student's t test. In validation experiments, the probe was applied to a proliferating culture of HeLa cells and to PFA-fixed lymphocytes pretreated identically to brain slices.
After hybridization of brain sections, two hybridization spots were obtained for the majority of neurons in the entorhinal cortex of both controls and AD patients (control group I, 72 ± 1%; control group II, 71 ± 3%; AD early, 64 ± 1%; AD advanced, 67 ± 2%) (Figs. 4, 5). In addition, neurons with none, one, or three spots were less frequently observed. The fraction of neurons without any hybridization signal or one spot was not different between controls and AD, and might result from sectioning artifacts partially cutting the nucleus. A small but constant number of neurons in both controls and AD displayed four hybridization spots.
The number of nuclei with three hybridization spots were identical in both control groups (group I, 6 ± 1%; group II, 6.9 ± 0.5%), but about doubled in AD patients (AD early, 12 ± 4%; AD advanced, 13 ± 1%). A small portion of 0.23 ± 0.2 and 0.3 ± 0.1% of neurons showed four hybridization spots in control groups I and II, respectively. The relative number of these neurons with four spots was increased in AD by factor four to five (early AD, 1.3 ± 0.5%; advanced AD, 1.5 ± 0.7%).
DNA quantification by PCR
Subsequently to CISH, the single-cell DNA content was further analyzed in the same cases by laser capture microdissection of neurons in the entorhinal cortex individually identified under the microscope and subsequent PCR amplification of alu repeats. The frequency distribution of single-cell DNA content obtained by this method is displayed in Figure 6 A. Comparing AD to controls, a shift toward higher size classes and differences in the shape of the distribution becomes apparent. The distributions of control groups have a single maximum at 2.5–3.5 pg per cell, which corresponds to the size for a 2n DNA content as determined in initial validation experiments (see above) (Fig. 1). In addition, AD groups displayed a second maximum in the size group of 6.5–7.5 pg per cell, most likely representing tetraploid neurons. This shift toward a higher single neuron DNA content in AD cases is similarly apparent on histograms obtained by SBC (Fig. 6 B).
Figure 6.
A, Frequency distribution of single-neuron DNA content determined by PCR amplification of alu repeats in both control and AD groups. Shift toward higher DNA content in AD cases compared with controls and appearance of a second peak displaying a doubled DNA content are shown (i.e., tetraploid neurons). B, PI integral histograms of single-neuron DNA content obtained by SBC. Each case is represented by a single graph. Similarly to the PCR data, a shift toward a higher DNA content is observed in AD.
To exclude pre-existing individual variations in neuronal ploidy as potential cause for AD-related shifts in the amount of neuronal DNA, we comparatively analyzed two brain regions with a comparable cellular composition but different vulnerability toward AD pathology. In contrast to the analysis described above, measurements were confined only to pyramidal neurons. Quantification of single-cell DNA by PCR amplification of alu repeats was performed in parallel on pyramidal neurons of the entorhinal cortex, affected early during the course of AD by neurofibrillary degeneration, and in the occipital cortex, which shows a pathological involvement only at the most advanced stages. By this kind of intraindividual control, potential interindividual differences with influence on the measurement such as resulting from a polymorphism of alu repeats were excluded. In control brains, there was no difference between the DNA content of pyramidal neurons in the entorhinal and occipital cortices (Fig. 7). In AD cases, however, the average DNA content of individual neurons in the entorhinal cortex was significantly elevated compared with the occipital cortex.
Figure 7.
PCR analysis of the DNA content in layer V pyramidal neurons of the entorhinal (EC) and occipital cortices (OC) in six control and six AD cases to exclude interindividual differences in alu repeats. Contrary to controls, the average DNA content of entorhinal neurons was elevated compared with occipital neurons in AD cases. Shown are mean values ± SEM of 10–15 cells per specimen. * p ≤ 0.05, Student's t test.
Discussion
In the present study, we determined the DNA content of cortical neurons in both controls and AD patients and the potential association with expression of cyclin B1 as an indicator of cell cycle activation and progression beyond S phase.
The results document a diploid DNA content in the majority of neurons. A constant number between 6 and 12% of neurons in normal adult human brain, however, contains DNA of >2n, and ∼0.25 to 0.5% of neurons even reaches a DNA content of 4n. This percentage of neurons with a tetraploid DNA content is somewhat smaller than the rate reported previously for tetraploid neurons in the adult human CNS of ∼1% determined by fluorescence in situ hybridization for the chromosome 21 in combination with flow cytometry of isolated nuclei (Rehen et al., 2005). As shown in the present study, these neurons do not express cyclin B1 and, thus, did not re-enter the cell cycle and most likely represent a constitutional tetraploidy derived from chromosome mis-segregation during mitosis in neuronal progenitor cells (A. H. Yang et al., 2003).
The percentage of hyperploid neurons in the normal adult brain in the present study reached a rate of up to 12%. Based on an FISH analysis with probes against six different chromosomes, a rate of hyperploidy (per individual chromosome) for adult neurons of the human brain between 0.1 and 0.8% was determined previously, which corresponds to a cumulative number of hyperploid nuclei for all chromosomes in the two-digit range indicating a large-scale genomic variation in differentiated neurons (Yurov et al., 2005). In line with these data are recent reports describing large-scale copy-number polymorphisms for certain genes among normal healthy human individuals (Iafrate et al., 2004; Sebat et al., 2004).
In AD, the relative number of neurons containing a DNA amount of >2n is increased, and in most advanced stages of the disease reaching about twice the frequency seen in controls. Although hyperploidy can vary within neuronal populations from single individuals (Rehen et al., 2005), this progressive shift from a diploid DNA content to a content of >2n between early to more advanced stages of the disease argues against an increase in constitutional hyperploidy to account for the increased neuronal amount of DNA in AD.
Contrary to controls, the majority of neurons in AD with a tetraploid content of DNA express cyclin B1. This association between an elevated content of DNA and expression of cyclin B1 in AD indicates that some neurons have reactivated their cell cycle and progressed toward the S phase and beyond. The majority of these neurons stay in the S phase with incomplete DNA replication, whereas only ∼1–2% contain a double DNA content, suggesting a complete DNA replication (Fig. 8).
Figure 8.
Synoptic overview on results of the current approach to identify cycling neurons in the adult human brain based on the simultaneous determination of single-neuron DNA content and expression of cyclin B1. Cyclin B1 expression occurs at the end of S phase and reaches peak expression during G2 and M phase. Neurons with a DNA amount of >2n that simultaneously express cyclin B1 are likely to have progressed in an active cycle beyond the S phase. In AD, ∼2% of neurons are tetraploid expressing cyclin B1, indicating complete replication, whereas ∼20% express cyclin B1 and contain DNA between 2n and 4n, indicating incomplete replication.
These results are in agreement with previous studies of Yang et al. (2001), who detected ∼3 to 4% of aneuploid hippocampal pyramidal cells in AD by applying FISH to paraffin-embedded sections. The authors further supposed that, because of a restricted hybridization efficiency, this result might even be an underestimation of the real number of aneuploid neurons. The present study indicates that “cycling neurons” (i.e., neurons that have a DNA content of >2n and at the same time express cyclin B1) are well in the range of ∼20%, which is much higher than thought previously. Reasons for an incomplete replication of DNA could be an accumulation of DNA damage and a decreased expression and activity of replication enzymes (Kuenzle, 1985; Mazzarello et al., 1992; Lovell et al., 2000; Nouspikel and Hanawalt, 2003). Developmental deficiencies in chromosome segregation giving rise to aneuploid neuronal progenitors have also been suggested as a critical pathogenetic mechanism in AD (Potter, 1991). This “chromosome malsegregation hypothesis” is supported by a large body of evidence arising from cytogenetic studies performed to assess structural and numerical chromosome aberrations in cultured peripheral cells of AD patients, as well as an increased risk to develop AD and show chromosomal malsegregation in lymphocytes in young mothers of adults with trisomy 21 (White et al., 1981; Moorhead and Heyman, 1983; Cherry et al., 1992; Percy et al., 1993; Schupf et al., 1994; Migliore et al., 1997, 1999, 2006). In particular, as shown in a series of elegant studies in the laboratory of H. Potter (Li et al., 1997; Geller and Potter, 1999; Boeras et al., 2007), mutations in the presenilin genes that cause AD also cause chromosome instability. Neuronal aneuploidy and polyploidy might contribute to inflammation, amyloidogenic processing of amyloid precursor protein, and apoptosis, which all are critical features of AD (Potter, 2005). It might, therefore, be tempting to assume that aneuploid neurons are highly prone to death and, thus, preferentially undergo cell death during the course of AD with only a small number (but still higher than controls) surviving.
As shown by the PCR analysis, the increase in the neuronal content of DNA is pronounced in the entorhinal cortex, a brain region affected by neurofibrillary degeneration already in early stages of AD, whereas the occipital cortex, known to be involved in AD pathology only in the most advanced stages of the disease, shows only marginal changes of neuronal DNA content. It might, thus, be suggested that the reactivation of the cell cycle and progression beyond the S phase might be directly related to cell death in AD, and does not occur to a major extent in brain regions that play minor roles in pathology. In the entorhinal cortex, we determined the extent of neuronal death in advanced stages of AD at ∼50% (Fig. 3 A). Assuming a constant rate of cell death over the duration of the disease, which might be estimated with at least ∼15 years (corresponding to 5478 d), we can theoretically calculate a rate of neuronal death of ∼0.01% or 1 in 10,000 per day. If all those 20% of neurons that are in an active cell cycle will eventually die, this process would take ∼2000 d (20% divided by 0.01%) or >5 years. This conclusion on cell death in AD as a very slow process is in agreement with previous studies suggesting that affected neurons can survive for years or even decades before they die Morsch et al., 1999; Yang et al., 2001; (Guillozet-Bongaarts et al., 2006).
Despite the expression of cyclin B1 at the G2–M phase transition in AD, there is an additional constitutive cyclin B1 expression that is independent of the cellular DNA content both in AD and a subgroup of controls. This expression of cyclin B1 in neurons of the normal adult human brain had similarly been observed in a number of previous studies (Vincent et al., 1997; Busser et al., 1998; Smith et al., 1999; Y. Yang et al., 2003). Referring to this, alternative cellular functions unrelated to cell cycle regulation in differentiated neurons are discussed, such as regulation of neuronal differentiation and plasticity, initiation of apoptosis, or phosphorylation of tau protein (Tamaru et al., 1993; Shi et al., 1994; Vincent et al., 1997; Schmetsdorf et al., 2005). Our data show that the relative number of cyclin B1-positive neurons increases only in more advanced cases of AD, but is similar in control and early AD cases. The transition from control to early AD, however, is accompanied by a loss of neurons of ∼15%. In a previous study, Y. Yang et al. (2003) documented a pronounced expression of cyclin B1 in cases with MCI which potentially progress toward AD (Y. Yang et al., 2003). It might thus be suggested that cyclin B1-expressing neurons are particularly vulnerable to cell death. The progressive shift from cyclin B1 negativity to positivity might be counterbalanced by an increasing rate of death of these neurons, masking their initial rise in early stages of the disease.
This is the first reported application of SBC and quantitative single-cell PCR to quantify the DNA amount of individually identified neurons in the adult human CNS, and we demonstrate a high reliability of both methods together with CISH. Our work extends previous studies analyzing the state of aneuploidy in the adult human CNS, applying FISH to sorted isolated nuclei (Rehen et al., 2005; Yurov et al., 2005) or paraffin-embedded tissue sections (Yang et al., 2001), and avoids potential artifacts associated with cell separation and hybridization (Yurov et al., 2005) or large-scale copy number polymorphisms (Sebat et al., 2004). The use of cytometry together with molecular biology methods is a promising approach to studying mechanisms at molecular and cellular levels (Darzynkiewicz et al., 2001). Such single-cell analysis with different methods reflects the concept of cytomics and will forward our understanding of the molecular architecture and functionality of neuronal systems (Valet et al., 2004; Kriete, 2005). As in our studies the DNA content of single neurons determined by SBC is far better correlated to CISH than is the qPCR measurement, we conclude that SBC is a very reliable technique for future studies on the single cell DNA content in tissue sections.
The present study indicates that the frequency of neurons in the adult human brain with a tetraploid level of DNA might be higher than previously thought. In the normal brain, tetraploid neurons are not in an active cycle and most likely are the result of chromosome mis-segregation during mitosis. In AD, some neurons can re-enter the cell cycle and entirely pass through a functional interphase with a complete DNA replication. Thus, at least two different mechanisms can be distinguished giving rise to a more than diploid content of DNA in the adult human brain.
Footnotes
This work was supported by the Bundesministerium für Bildung, Forschung und Technologie, Interdisziplinäres Zentrum für Klinische Forschung at the University of Leipzig (01KS9504, Project C1; MD/PhD grant to M.M.), the Research Training School Grant GRK 1097 “InterNeuro,” and by a grant from the Saxon Ministry of Science and the Fine Arts (Grant 11-899981016, Art.1 HWP to B.M.). We thank Dr. J. Bocsi (Cardiac Center Leipzig, Leipzig, Germany) for providing the cell cycle software, Prof. C. Hoang-Vu (Experimental and Oncological Surgery Research Group, University Department of General, Visceral, and Vascular Surgery, Martin Luther University, Halle, Germany) for providing the microdissector, H. Gruschka for assistance in preparing the brain sections, and G. Baumbach (Faculty of Biology, Pharmacy and Psychology, Leipzig, Germany) for preparing the lymphocytes.
References
- Arendt T. Alzheimer's disease as a disorder of mechanisms underlying structural brain self-organization. Neuroscience. 2001;102:723–765. doi: 10.1016/s0306-4522(00)00516-9. [DOI] [PubMed] [Google Scholar]
- Arendt T, Rodel L, Gartner U, Holzer M. Expression of the cyclin-dependent kinase inhibitor p16 in Alzheimer's disease. NeuroReport. 1996;7:3047–3049. doi: 10.1097/00001756-199611250-00050. [DOI] [PubMed] [Google Scholar]
- Arendt T, Holzer M, Gartner U. Neuronal expression of cycline dependent kinase inhibitors of the INK4 family in Alzheimer's disease. J Neural Transm. 1998;105:949–960. doi: 10.1007/s007020050104. [DOI] [PubMed] [Google Scholar]
- Arendt T, Holzer M, Stobe A, Gartner U, Luth HJ, Bruckner MK, Ueberham U. Activated mitogenic signaling induces a process of dedifferentiation in Alzheimer's disease that eventually results in cell death. Ann NY Acad Sci. 2000;920:249–255. doi: 10.1111/j.1749-6632.2000.tb06931.x. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Boeras DI, Granic A, Padmanabhan J, Crespo NC, Rojiani AM, Potter H. Alzheimer's presenilin 1 cause chromosome missegregation and aneuploidy. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2006.10.027. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol (Berl) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Brodskij VJ, Kusc AA. Izmenenie cisla polyploidnych kletok v postembryonalom razvitiji tkanej krysy. Dokl Akad Nauk SSSR Otd Biol. 1962;162:713–716. [Google Scholar]
- Busser J, Geldmacher DS, Herrup K. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer's disease brain. J Neurosci. 1998;18:2801–2807. doi: 10.1523/JNEUROSCI.18-08-02801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry LM, Funk J, Lesser JM, Lesam M. Gender differences and their interpretation of genetic instability in Alzheimer's disease. Mutat Res. 1992;275:57–76. doi: 10.1016/0921-8734(92)90009-e. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Smolewski P, Bedner E. Use of flow and laser scanning cytometry to study mechanisms regulating cell cycle and controlling cell death. Clin Lab Med. 2001;21:857–873. [PubMed] [Google Scholar]
- Ding XL, Husseman J, Tomashevski A, Nochlin D, Jin LW, Vincent I. The cell cycle Cdc25A tyrosine phosphatase is activated in degenerating postmitotic neurons in Alzheimer's disease. Am J Pathol. 2000;157:1983–1990. doi: 10.1016/S0002-9440(10)64837-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller LN, Potter H. Chromosome missegregation and trisomy 21 mosaicism in Alzheimer's disease. Neurobiol Dis. 1999;6:167–179. doi: 10.1006/nbdi.1999.0236. [DOI] [PubMed] [Google Scholar]
- Guillozet-Bongaarts AL, Cahill ME, Cryns VL, Reynolds MR, Berry RW, Binder LI. Pseudophosphorylation of tau at serine 422 inhibits caspase cleavage: in vitro evidence and implications for tangle formation in vivo . J Neurochem. 2006;97:1005–1014. doi: 10.1111/j.1471-4159.2006.03784.x. [DOI] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- Heintz N. Cell death and the cell cycle: a relationship between transformation and neurodegeneration? Trends Biochem Sci. 1993;18:157–159. doi: 10.1016/0968-0004(93)90103-t. [DOI] [PubMed] [Google Scholar]
- Herrup K, Arendt T. Re-expression of cell cycle proteins induces neuronal cell death during Alzheimer's disease. J Alzheimers Dis. 2002;4:243–247. doi: 10.3233/jad-2002-4315. [DOI] [PubMed] [Google Scholar]
- Houck CM, Rinehart FP, Schmid CW. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979;132:289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Ito M. Factors controlling cyclin B expression. Plant Mol Biol. 2000;43:677–690. doi: 10.1023/a:1006336005587. [DOI] [PubMed] [Google Scholar]
- Kriete A. Cytomics in the realm of systems biology. Cytometry A. 2005;68:19–20. doi: 10.1002/cyto.a.20184. [DOI] [PubMed] [Google Scholar]
- Kuenzle CC. Enzymology of DNA replication and repair in the brain. Brain Res. 1985;357:231–245. doi: 10.1016/0165-0173(85)90026-8. [DOI] [PubMed] [Google Scholar]
- Lapham LW, Johnstone MA. Cytologic and cytochemical studies of neuroglia. Arch Neurol. 1963;9:194–202. doi: 10.1001/archneur.1963.00460080104013. [DOI] [PubMed] [Google Scholar]
- Lenz D, Mosch B, Bocsi J, Arendt T, Tarnok A. Assessment of DNA replication in central nervous system by laser scanning cytometry. Proc SPIE. 2004;5322:146–156. [Google Scholar]
- Li J, Xu M, Zhou H, Ma J, Potter H. Alzheimer presenilins in the nuclear membrane, interphase kinetochores and centrosomes suggest a role in chromosome segregation. Cell. 1997;90:917–927. doi: 10.1016/s0092-8674(00)80356-6. [DOI] [PubMed] [Google Scholar]
- Lovell MA, Xie C, Markesbery WR. Decreased base excision repair and increased helicase activity in Alzheimer's disease brain. Brain Res. 2000;855:116–123. doi: 10.1016/s0006-8993(99)02335-5. [DOI] [PubMed] [Google Scholar]
- Mares V, Lodin Z, Sacha J. A cytochemical and autoradiographic study of nuclear DNA in mouse Purkinje cells. Brain Res. 1973;53:273–289. doi: 10.1016/0006-8993(73)90214-x. [DOI] [PubMed] [Google Scholar]
- Mazzarello P, Poloni M, Spadari S, Focher F. DNA repair mechanisms in neurological diseases: facts and hypotheses. J Neurol Sci. 1992;112:4–14. doi: 10.1016/0022-510x(92)90125-5. [DOI] [PubMed] [Google Scholar]
- McShea A, Harris PL, Webster KR, Wahl AF, Smith MA. Abnormal expression of the cell cycle regulators P16 and CDK4 in Alzheimer's disease. Am J Pathol. 1997;150:1933–1939. [PMC free article] [PubMed] [Google Scholar]
- Migliore L, Testa A, Scarpato R, Pavese N, Petrozzi L, Bonuccelli U. Spontaneous and induced aneuploidy in peripheral blood lymphocytes of patients with Alzheimer's disease. Hum Genet. 1997;101:299–305. doi: 10.1007/s004390050632. [DOI] [PubMed] [Google Scholar]
- Migliore L, Botto N, Scarpato R, Petrozzi L, Cipriani G, Bonuccelli U. Preferential occurrence of chromosome 21 malsegregation in peripheral blood lymphocytes of Alzheimer disease patients. Cytogenet Cell Genet. 1999;87:41–46. doi: 10.1159/000015389. [DOI] [PubMed] [Google Scholar]
- Migliore L, Boni G, Bernardini R, Trippi F, Colognato R, Fontana I, Coppedè F, Sbrana I. Susceptibility to chromosome malsegregation in lymphocytes of women who had a Down syndrome child in young age. Neurobiol Aging. 2006;27:710–716. doi: 10.1016/j.neurobiolaging.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Moorhead PS, Heyman A. Chromosome studies of patients with Alzheimer's disease. Am J Med Genet. 1983;14:545–556. doi: 10.1002/ajmg.1320140319. [DOI] [PubMed] [Google Scholar]
- Morsch R, Simon W, Coleman PD. Neurons may live for decades with neurofibrillary tangles. J Neuropathol Exp Neurol. 1999;58:188–197. doi: 10.1097/00005072-199902000-00008. [DOI] [PubMed] [Google Scholar]
- Mosch B, Mittag A, Lenz D, Arendt T, Tarnok A. Laser scanning cytometry in human brain slices. Cytometry A. 2006;69:135–138. doi: 10.1002/cyto.a.20228. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Esiri MM, Smith AD. Expression of cell division markers in the hippocampus in Alzheimer's disease and other neurodegenerative conditions. Acta Neuropathol (Berl) 1997a;93:294–300. doi: 10.1007/s004010050617. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Esiri MM, Cato AM, Smith AD. Cell cycle markers in the hippocampus in Alzheimer's disease. Acta Neuropathol (Berl) 1997b;94:6–15. doi: 10.1007/s004010050665. [DOI] [PubMed] [Google Scholar]
- Nouspikel T, Hanawalt PC. When parsimony backfires: neglecting DNA repair may doom neurons in Alzheimer's disease. Bioessays. 2003;25:168–173. doi: 10.1002/bies.10227. [DOI] [PubMed] [Google Scholar]
- Percy ME, Markovic VD, Dalton AJ, McLachlan DR, Berg JM, Rusk AC, Somerville MJ, Chodakowski B, Andrews DF. Age-associated chromosome 21 loss in Down syndrome: possible relevance to mosaicism and Alzheimer's disease. Am J Med Genet. 1993;45:584–588. doi: 10.1002/ajmg.1320450513. [DOI] [PubMed] [Google Scholar]
- Potter H. Review and hypothesis: Alzheimer disease and Down syndrome-chromosome 21 nondisjunction may underlie both disorders. Am J Med Genet. 1991;48:1192–1200. [PMC free article] [PubMed] [Google Scholar]
- Potter H. Cell cycle and chromosome segregation defects in Alzheimer's disease. In: Copani A, Nicoletti F, editors. Cell-cycle mechanisms and neuronal death. Georgetown, TX: Landes; 2005. pp. 55–78. [Google Scholar]
- Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH, Chun J. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci USA. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, Fontanoz M, Chun J. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel AM, Carroll ML, Vogel E, Garber RK, Nguyen SV, Salem AH, Batzer MA, Deininger PL. Alu insertion polymorphisms for the study of human genomic diversity. Genetics. 2001;159:279–290. doi: 10.1093/genetics/159.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmetsdorf S, Gartner U, Arendt T. Expression of cell cycle-related proteins in developing and adult mouse hippocampus. Int J Dev Neurosci. 2005;23:101–112. doi: 10.1016/j.ijdevneu.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Schupf N, Kapell D, Lee JH, Ottman R, Mayeux R. Increased risk of Alzheimer's disease in mothers of adults with Down's syndrome. Lancet. 1994;344:353–356. doi: 10.1016/s0140-6736(94)91398-6. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Shi L, Nishioka WK, Th'ng J, Bradbury EM, Litchfield DW, Greenberg AH. Premature p34cdc2 activation required for apoptosis. Science. 1994;263:1143–1145. doi: 10.1126/science.8108732. [DOI] [PubMed] [Google Scholar]
- Smith MZ, Nagy Z, Esiri MM. Cell cycle-related protein expression in vascular dementia and Alzheimer's disease. Neurosci Lett. 1999;271:45–48. doi: 10.1016/s0304-3940(99)00509-1. [DOI] [PubMed] [Google Scholar]
- Smith TW, Lippa CF. Ki-67 immunoreactivity in Alzheimer's disease and other neurodegenerative disorders. J Neuropathol Exp Neurol. 1995;54:297–303. doi: 10.1097/00005072-199505000-00002. [DOI] [PubMed] [Google Scholar]
- Tamaru T, Trigun SK, Okada M, Nakagawa H. Identification of cells expressing a D type G1 cyclin in matured brain: implication for its role in neuronal function. Neurosci Lett. 1993;153:169–172. doi: 10.1016/0304-3940(93)90314-b. [DOI] [PubMed] [Google Scholar]
- Valet G, Leary JF, Tarnok A. Cytomics-new technologies: towards a human cytome project. Cytometry A. 2004;59:167–171. doi: 10.1002/cyto.a.20047. [DOI] [PubMed] [Google Scholar]
- Vincent I, Jicha G, Rosado M, Dickson DW. Aberrant expression of mitotic cdc2/cyclin B1 kinase in degenerating neurons of Alzheimer's disease brain. J Neurosci. 1997;17:3588–3598. doi: 10.1523/JNEUROSCI.17-10-03588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA, Kilroy GE, Xing J, Shewale J, Sinha SK, Batzer MA. Human DNA quantitation using Alu element-based polymerase chain reaction. Anal Biochem. 2003;315:122–128. doi: 10.1016/s0003-2697(03)00081-2. [DOI] [PubMed] [Google Scholar]
- White BJ, Crandall C, Goldsmit J, Morrow CH, Alling DW, Carleton Gajdusek DC, Tjio JH. Cytogenetic studies of familial and sporadic Alzheimer disease. Am J Med Genet. 1981;10:77–89. doi: 10.1002/ajmg.1320100110. [DOI] [PubMed] [Google Scholar]
- Yang AH, Kaushal D, Rehen SK, Kriedt K, Kingsbury MA, McConnell MJ, Chun J. Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. J Neurosci. 2003;23:10454–10462. doi: 10.1523/JNEUROSCI.23-32-10454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer's disease. J Neurosci. 2001;21:2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Mufson EJ, Herrup K. Neuronal cell death is preceded by cell cycle events at all stages of Alzheimer's disease. J Neurosci. 2003;23:2557–2563. doi: 10.1523/JNEUROSCI.23-07-02557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Monakhov VV, Soloviev IV, Vostrikov VM, Vorsanova SG. The variation of aneuploidy frequency in the developing and adult human brain revealed by an interphase FISH study. J Histochem Cytochem. 2005;53:385–390. doi: 10.1369/jhc.4A6430.2005. [DOI] [PubMed] [Google Scholar]