Abstract
Disrupted-In-Schizophrenia 1 (DISC1) is a candidate gene for susceptibility to schizophrenia. DISC1 is reported to interact with NudE-like (NUDEL), which forms a complex with lissencephaly-1 (LIS1) and 14-3-3ε. 14-3-3ε is involved in the proper localization of NUDEL and LIS1 in axons. Although the functional significance of this complex in neuronal development has been reported, the transport mechanism of the complex into axons and their functions in axon formation remain essentially unknown. Here we report that Kinesin-1, a motor protein of anterograde axonal transport, was identified as a novel DISC1-interacting molecule. DISC1 directly interacted with kinesin heavy chain of Kinesin-1. Kinesin-1 interacted with the NUDEL/LIS1/14-3-3ε complex through DISC1, and these molecules localized mainly at cell bodies and partially in the distal part of the axons. DISC1 partially colocalized with Kinesin family member 5A, NUDEL, LIS1, and 14-3-3ε in the growth cones. The knockdown of DISC1 by RNA interference or the dominant-negative form of DISC1 inhibited the accumulation of NUDEL, LIS1, and 14-3-3ε at the axons and axon elongation. The knockdown or the dominant-negative form of Kinesin-1 inhibited the accumulation of DISC1 at the axons and axon elongation. Furthermore, the knockdown of NUDEL or LIS1 inhibited axon elongation. Together, these results indicate that DISC1 regulates the localization of NUDEL/LIS1/14-3-3ε complex into the axons as a cargo receptor for axon elongation.
Keywords: DISC1, Kinesin-1, axonal transport, axon elongation, schizophrenia, NUDEL
Introduction
Schizophrenia is a complex genetic disorder with fairly high heritability. Recently, several genes have been identified as candidate genes for susceptibility to schizophrenia, including neuregulin 1 (Stefansson et al., 2002), dysbindin (Straub et al., 2002), G72 (Chumakov et al., 2002), catechol-O-methyltransferase (Egan et al., 2001; Bilder et al., 2002; Shifman et al., 2002), and others (Craddock et al., 2005; Harrison and Weinberger, 2005). Although candidate genes for schizophrenia have been identified, the molecular mechanisms underlying the disease are essentially unknown.
Disrupted-In-Schizophrenia 1 (DISC1) is one of the most probable candidate genes for susceptibility to schizophrenia (Millar et al., 2000; Blackwood et al., 2001; Craddock et al., 2005). In a Scottish family, the chromosome translocation (1;11)(q42.1;q14.3) was associated with major psychiatric illnesses, with a predominance of schizophrenic symptomatology (Blackwood et al., 2001). This translocation interrupts the coding sequence of DISC1, leading to the reduction of DISC1 expression (Millar et al., 2005) or deletion of the C-terminal region. Haplotype transmission analysis using single-nucleotide polymorphisms (SNPs) from the 1q42 region indicated that the DISC1 gene plays an important role in the etiology of schizophrenia (Hennah et al., 2003; Hodgkinson et al., 2004; Callicott et al., 2005). DISC1 interacts with several proteins, including NudE-like (NUDEL) (Ozeki et al., 2003; Millar et al., 2003; Morris et al., 2003), fasciculation and elongation protein zeta 1 (FEZ1) (Miyoshi et al., 2003), lissencephaly-1 (LIS1) (Brandon et al., 2004), and phosphodiesterase 4B (PDE4B) (Millar et al., 2005). Among these proteins, the association with NUDEL and LIS1 supports the notion that DISC1 contributes to the neuronal development and morphology (Kholmanskikh et al., 2003).
LIS1 mutation leads to type 1 lissencephaly, which is characterized by profuse migration defects during brain development (Reiner et al., 1993; Hattori et al., 1994). LIS1-deficient cerebellar granule neurons show defects in neuronal migration and axon extension (Kholmanskikh et al., 2003). NUDEL is identified as a LIS1-binding protein (Niethammer et al., 2000; Sasaki et al., 2000). The NUDEL/LIS1 complex is initially distributed in the centrosome or microtubule organizing center (MTOC) and then transported into the axonal growth cones (Niethammer et al., 2000; Sasaki et al., 2000). It is also known that the NUDEL/LIS1 complex accumulates with plus-end tracking proteins, including cytoplasmic linker protein 170 (CLIP-170) and end-binding protein 1 (EB1) near the plus ends of microtubules, which terminate in growth cones (Coquelle et al., 2002).
14-3-3ε binds to NUDEL phosphorylated by cyclin-dependent kinase (cdk5) and maintains NUDEL phosphorylation (Toyo-oka et al., 2003). Mice deficient in 14-3-3ε show defects in brain development and neuronal migration (Toyo-oka et al., 2003), similar to mice deficient in LIS1 (Hirotsune et al., 1998; Gambello et al., 2003). Deficiency of 14-3-3ε causes mislocalization of the NUDEL/LIS1 complex from axons, suggesting that 14-3-3ε regulates the axonal targeting of the NUDEL/LIS1 complex by sustaining NUDEL phosphorylation (Toyo-oka et al., 2003). However, it remains unclear how 14-3-3ε or other component regulates the transport of the NUDEL/LIS1 complex into axons.
In this study, we identified Kinesin-1, a microtubule-dependent and plus-end directed motor, as a DISC1-interacting molecule. Our results show that DISC1 links Kinesin-1 to the NUDEL/LIS1/14-3-3ε complex, serves as the cargo receptor, and regulates the transport of the complex to axons, leading to axon elongation.
Materials and Methods
Materials and chemicals.
The fragment of DISC1–wild type (WT) (1–832) was inserted into pMAL-c2 (New England Laboratories, Beverly, MA), pEF–BOS–Myc, pEGFP–C-terminal 1 (C1), or pEGFP–N-terminal 3 (N3) (Clontech, Palo Alto, CA). The fragments of DISC1–N-terminal of break point (NB) (1–597) or DISC1–N-terminal half (NH) (1–361) were inserted into pEGFP–C1 or pEGFP–N3, respectively. The fragments of DISC1–C-terminal of break point (CB) (598–832) or DISC1–C-terminal half (CH) (362–832) were inserted into pEGFP–C1. The fragment of 14-3-3ε was inserted into pEF–BOS–glutathione S-transferase (GST). The fragment of Kinesin family member 5A (KIF5A)–Full (1–1027) was inserted into pGEX–4T-2 (Amersham Biosciences, Buckinghamshire, UK), pEF–BOS–hemagglutinin (HA), or pCAGGS—Myc. The fragments of KIF5A–Headless (HL) (402–1027), KIF5A–C1 (706–1027), or KIF5A–C-terminal 2 (C2) (830–1027) were inserted into pCAGGS–Myc. The RNA interference (RNAi)-resistant (RNAiR) NUDEL or LIS1 was created by three silence mutations into human NUDEL at nucleotides 265–298 (5′-CAGAGCTATAAGCAAGTGTCTGTGTTAGAAGATG-3′) and human LIS1 at nucleotides 1049–1083 (5′-GGAAGTTTATTTTGAGCTGCGCAGATGACAAGACC-3′), respectively (the underlining indicates the mutations).
Antibodies against 14-3-3ε (Transduction Laboratories, Lexington, KY), KIF5A, Tau-1, cytoplasmic dynein intermediate chain (DIC) (Chemicon, Temecula, CA), GFP (MBL, Nagoya, Japan), Rho GDP dissociation inhibitor (RhoGDI), cytoplasmic dynein heavy chain (CDHC) (Santa Cruz Biotechnology, Santa Cruz, CA), and β-tubulin (Sigma, St. Louis, MO) were purchased. Anti-LIS1 antibody was prepared as reported previously (Sapir et al., 1997). Four kinds of rabbit anti-DISC1 antibodies were prepared by Biologica (Nagoya, Japan) and purified by use of peptides (251–268, 571–586, 644–661, and 722–738 amino acids of rat DISC1) as antigens. Rabbit anti-kinesin light chain (KLC) antibody was raised against GST–KLC1. Because this antigen contains the conserved sequence of KLCs, this antibody recognizes KLC1 and KLC2 (Kimura et al., 2005).
Affinity column chromatography and mass spectral analyses.
Rat brain lysate was loaded onto beads coated with maltose-binding protein (MBP) or MBP–DISC1–WT. The proteins bound to the affinity columns were eluted by the addition of buffer containing 500 mm NaCl. The eluates were subjected to SDS-PAGE, and the bound proteins were evaluated by silver staining. We identified proteins by comparing the molecular weights determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and theoretical peptide masses from the proteins registered in the National Center for Biotechnology Information database.
In vitro binding assay.
GST or MBP fusion proteins were expressed in Escherichia coli BL21(DE3) and purified according to the instructions of the manufacturer. MBP or MBP–DISC1 (400 pmol) was mixed with glutathione–Sepharose 4B beads (Amersham Biosciences) coated with 100 pmol of GST alone, GST–KLC1, or GST–KIF5A in buffer (20 mm Tris-HCl, 1 mm EDTA, and 1 mm DTT). The bound MBP fusion proteins were coeluted with GST fusion proteins. Portions of the eluates were subjected to SDS-PAGE, followed by immunoblot analysis with anti-MBP antibody.
GST pull-down assay.
COS7 cells were transfected in various combinations (supplemental Fig. 1B,D, available at www.jneurosci.org as supplemental material) with Lipofectamine (Invitrogen, Carlsbad, CA) and cultured for 48 h. Cells were lysed with buffer [20 mm Tris-HCl, 1 mm EDTA, 150 mm NaCl, 1 mm DTT, 0.1% (w/v) Triton X-100, 10 μm p-amidinophenyl methanesulfonyl fluoride hydrochloride, and 10 μg/ml leupeptin]. The lysate was sonicated and then clarified by centrifugation at 12,000 × g for 10 min at 4°C. The soluble supernatant was incubated with glutathione–Sepharose 4B beads (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material) or glutathione–Sepharose 4B beads coated with 50 pmol of GST alone or GST–KIF5A (supplemental Fig. 1D, available at www.jneurosci.org as supplemental material). Portions of the eluates were subjected to SDS-PAGE, followed by immunoblot analyses with the indicated antibodies.
Coimmunoprecipitation assay.
COS7 cells were transfected in various combinations (see Fig. 2 B) (supplemental Fig. 1A,C, available at www.jneurosci.org as supplemental material) with Lipofectamine and cultured for 48 h. Preparations of the soluble supernatant were described above. The soluble supernatant was incubated with various antibodies and protein A Sepharose (Amersham Biosciences). The precipitated proteins were eluted with SDS sample buffer. Portions of the eluates were subjected to SDS-PAGE, followed by immunoblot analysis with the indicated antibodies.
Figure 2.

Complex formation with DISC1, Kinesin-1, and NUDEL in vitro and in vivo. A, An in vitro binding assay was performed using purified recombinant proteins. Beads immobilized with GST alone, GST–KLC1, or GST–KIF5A were incubated with MBP alone or MBP–DISC1. Bound proteins were analyzed by immunoblotting with antibodies against MBP and GST. The lower bands represent degradation products of MBP–DISC1. B, Lysates from transfected COS7 cells expressing NUDEL–GFP, Myc–DISC1, and HA–KIF5A were used in an immunoprecipitation assay with antibody against HA. The expression levels of NUDEL–GFP, Myc–DISC1, or HA–KIF5A were almost equal in each lane (data not shown). Bound proteins were analyzed by immunoblotting with antibodies against GFP, Myc, and HA. The lower bands represent degradation products of HA–KIF5A in immunoblotting with antibody against HA. C, D, Mapping of the regions in KIF5A or DISC1 required for binding to DISC1 (C) or KIF5A (D), respectively. E, F, Lysates from differentiated PC12 cells were used in an immunoprecipitation assay with antibodies (Ab) against DISC1 (E) or KIF5A (F). Bound proteins and inputs of assay were analyzed by immunoblotting with antibodies against the indicated proteins. Aliquots of original samples (10% Input) and eluates (30%) were subjected to SDS-PAGE.
PC12 cells were differentiated with nerve growth factor (NGF) for 48 h. Lysates from differentiated PC12 cells were used in an immunoprecipitation assay with antibodies against DISC1 (see Fig. 2 E) or KIF5A (see Fig. 2 F) as described above. The precipitated proteins were eluted with 500 mm NaCl to separate the fraction including the precipitated antibodies.
PC12 cells were transfected with scramble or rat DISC1-specific (rDISC1)–small interfering RNAs (siRNAs) and then differentiated with NGF for 48 h. Lysates from transfected PC12 cells were used in an immunoprecipitation assay with antibody against NUDEL (supplemental Fig. 3A, available at www.jneurosci.org as supplemental material).
Preparation and culture of rat hippocampal neurons.
Hippocampal neurons were prepared from embryonic day 18 rat embryos using papain (Inagaki et al., 2001). Neurons were seeded on poly-d-lysine- and laminin-coated coverslips and cultured in Neurobasal medium (Invitrogen) supplemented with B-27 supplement (Invitrogen) and 1 mm glutamine. To get the enlarged images of the axonal growth cones, neurons were seeded on poly-d-lysine-coated coverslips (see Fig. 3 F–K). These enlarged axonal growth cones are structurally similar to the smaller one and that on laminin (Dent and Gertler, 2003).
Figure 3.
Colocalization of DISC1 with Kinesin-1, NUDEL, LIS1, and 14-3-3ε. A–E, Rat primary hippocampal neurons showed staining of DISC1 and KIF5A (A), LIS1 (B), 14-3-3ε (C), or NUDEL-GFP (D), and NUDEL and LIS1 (E) at DIV3. Arrows indicate the distal parts of axons. Scale bars, 20 μm. F–K, Enlarged growth cones of rat primary hippocampal neurons show the staining of DISC1 and KIF5A (F), LIS1 (G), or 14-3-3ε (H), and NUDEL and LIS1 (I), and tubulin and DISC1 (J) or KIF5A (K). The peripheral regions of the growth cones are visualized by staining of F-actin with Alexa 647-conjugated phalloidin (blue).
Transfection of siRNAs.
A 21-oligonucleotide siRNA duplex was synthesized to target the rat DISC1–1 sequence 5′-GTGTTGGAGGGTCAGCTGATT-3′, the rat DISC1–2 sequence 5′-CGCGGAGAAGCAGAAGGTTTT-3′, the rat KLC1 sequence 5′-ATACGACGACGACATCTCT-3′, and the rat KLC2 sequence 5′-TCTGGTGATCCAGTATGCT-3′. A scramble sequence, 5′-CAGTCGCGTTTGCGACTGG-3′, was used as a negative control. For knockdown of DISC1 or KLCs, we used siRNAs of DISC1–1 and DISC1–2 or of KLC1 and KLC2, respectively. For knockdown of NUDEL, LIS1, or CDHC, we used pSilencer2.1–NUDEL or pSilencer2.1–LIS1 (Shu et al., 2004) or four kinds of CDHC–siRNAs (He et al., 2005), respectively. Transfection of siRNA into PC12 cells was performed with Lipofectamine 2000 (Invitrogen). The protein concentration of cell lysate was estimated by use of a BCA kit (Bio-Rad, Hercules, CA), and equal amounts of cell lysate were analyzed by immunoblotting with the indicated antibodies (see Figs. 4 A, 6 A). Transfection of siRNA (1.2 μg) and/or expression plasmid encoding various cDNA (1.0 μg) into hippocampal neurons was performed using a calcium phosphate method.
Figure 4.

Effect of DISC1 knockdown on localization of NUDEL, LIS1, and 14-3-3ε in the distal part of the axon. A, PC12 cells were transfected with scramble or rDISC1–siRNAs and then differentiated with NGF for 48 h. Lysates were analyzed by immunoblotting with antibodies against DISC1, NUDEL, LIS1, 14-3-3ε, and β-tubulin. B, Hippocampal neurons transfected with expression plasmid encoding EGFP–GST and scramble or rDISC1–siRNAs at DIV1 were detected by EGFP fluorescence and staining of Tau-1, which is a maker of axon at DIV3. Hippocampal neurons transfected with Cy3-labeled scramble or rDISC1–siRNAs were detected by Cy3 fluorescence and the staining of Tau-1. More than 90% of the cells transfected with EGFP–GST and rDISC1–siRNAs or Cy3–rDISC1–siRNAs showed the decreased immunoreactivity of DISC1. Axons of transfected neurons were scored according to axon length. C, Hippocampal neurons transfected with expression plasmid encoding EGFP–GST and scramble or rDISC1–siRNAs were detected by EGFP fluorescence and staining of 14-3-3ε. Hippocampal neurons transfected with Cy3-labeled scramble or rDISC1–siRNAs were detected by Cy3 fluorescence and staining of 14-3-3ε and F-actin. The accumulation of 14-3-3ε in the distal part of axons was determined, as described in Materials and Methods. B, C, Numbers of cells used for each calculation are >50, and the values shown are means ± SE of three independent experiments (Student's t test, **p < 0.01). D, E, G, Hippocampal neurons were transfected with rDISC1–siRNAs. The localization of LIS1, 14-3-3ε, or tubulin of DISC1-knockdown neurons is shown by staining of DISC1 and LIS1, 14-3-3ε, or tubulin. The distal parts of axons of the transfected cells are visualized by staining of F-actin with Alexa 647-conjugated phalloidin (blue). F, Hippocampal neurons transfected with rDISC1–siRNAs and expression plasmid encoding NUDEL–GFP are shown by GFP fluorescence and staining of DISC1 and F-actin (blue). D–G, Arrows indicate DISC1-knockdown neurons. Enlarged images of the axonal growth cones are shown. Scale bars, 20 μm.
Figure 6.
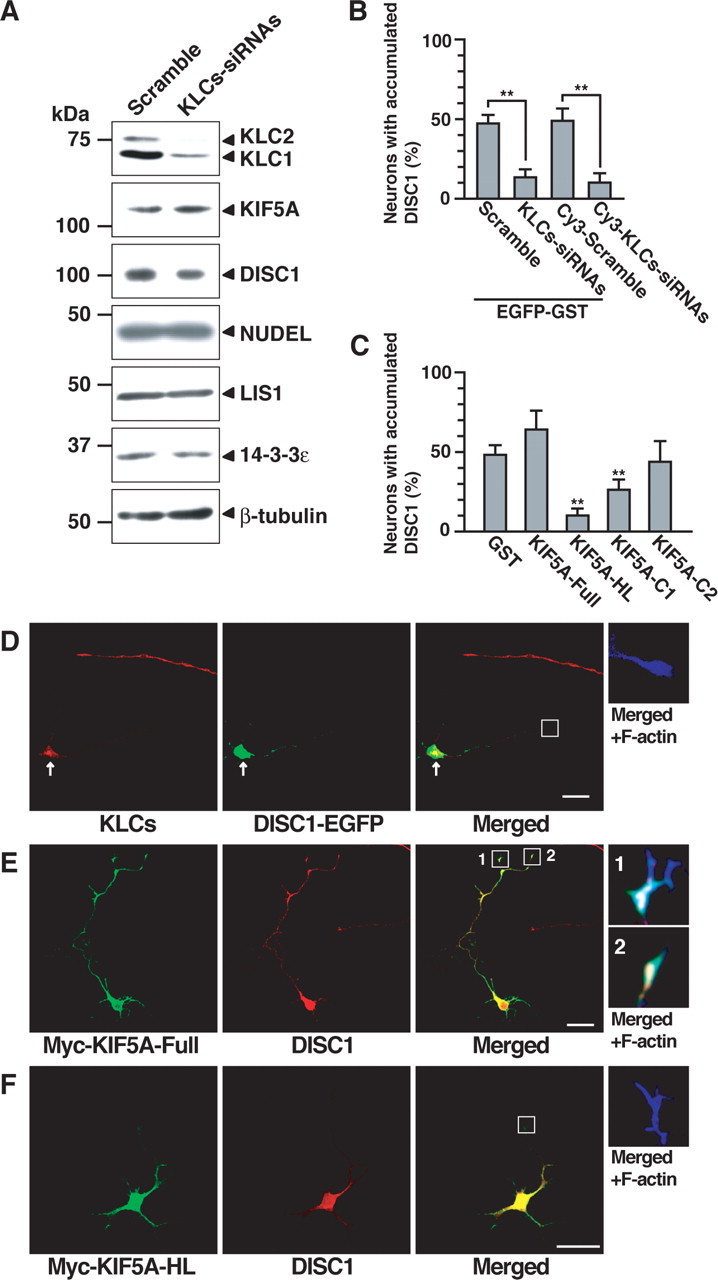
Effect of KLCs knockdown and dominant-negative form of KIF5A on accumulation of DISC1 in the distal part of the axon. A, PC12 cells were transfected with scramble or KLCs–siRNAs and then differentiated with NGF for 48 h. Lysates were analyzed by immunoblotting with antibodies against the indicated proteins. B, Hippocampal neurons transfected with expression plasmid encoding EGFP–GST and scramble or KLCs–siRNAs at DIV1 were detected by EGFP fluorescence and staining of DISC1 and F-actin at DIV3. Hippocampal neurons transfected with Cy3-labeled scramble or KLCs–siRNAs were detected by Cy3 fluorescence and staining of DISC1 and F-actin. Axons of transfected neurons were scored using a percentage of the numbers of accumulated DISC1 in the distal part of the axon. More than 90% of the cells transfected with EGFP–GST and KLCs–siRNAs or Cy3–KLCs–siRNAs showed the decreased immunoreactivity of KLCs. C, Neurons transfected with Myc–GST or Myc–KIF5A mutants were identified by Myc staining and were scored using a percentage of the numbers of accumulated DISC1 in the distal parts of axons. B, C, Numbers of cells used for each calculation are >50, and the values shown are means ±SE of three independent experiments (Student's t test, **p<0.01).D, Hippocampal neurons transfected with KLCs-siRNAs and expression plasmid encoding DISC1-EGFP are shown by EGFP fluorescence and staining of KLCs (red) and F-actin (blue). Arrows indicate KLCs-knockdown neurons. E, F, Hippocampal neurons transfected with expression plasmid encoding Myc-KIF5A-Full (E) or Myc-KIF5A-HL (F) are shown by staining of Myc (green), DISC1 (red), and F-actin (blue). D-F, Enlarged images of the axonal growth cones are shown. Scale bars, 20 μm.
Immunofluorescence analysis.
In the case of nonlabeled siRNAs, hippocampal neurons were cotransfected with nonlabeled siRNAs and EGFP or EGFP–GST, and EGFP-positive neurons were counted. In the case of cyanine 3 (Cy3)-labeled siRNAs, Cy3-positive neurons were counted. Axons were determined by Tau-1 staining and its morphology (Craig and Banker, 1994) (see Fig. 4 B, 5 D, 8 A,B) (supplemental Fig. 2, available at www.jneurosci.org as supplemental material).
Figure 5.

Effect of N-terminal fragment of DISC1 on accumulation of 14-3-3ε in the distal part of the axon. A–C, Hippocampal neurons transfected with expression plasmid encoding EGFP-tagged GST (A), DISC1–WT (B), or DISC1–NB (C) are shown by EGFP fluorescence and staining of 14-3-3ε (red) and F-actin (blue). Enlarged images of the axonal growth cones are shown. D, E, Hippocampal neurons transfected with expression plasmid encoding EGFP–GST or EGFP–DISC1 mutants. The EGFP-positive neurons were scored by axon length (D) and the numbers of accumulated 14-3-3ε in the distal part of axons at DIV3 (E). Numbers of cells used for each calculation are >50, and the values shown are means ± SE of three independent experiments (Student's t test, **p < 0.01). F, G, Hippocampal neurons transfected with rDISC1–siRNAs, and expression plasmid encoding EGFP–DISC1–WT (F) or EGFP–DISC1–NB (G) at DIV1 are shown by EGFP fluorescence and the staining of DISC1 (red) and 14-3-3ε (blue) at DIV3. Enlarged images of the axons are shown. Arrows indicate endogenous DISC1-knockdown neurons. Scale bars, 20 μm.
Figure 8.
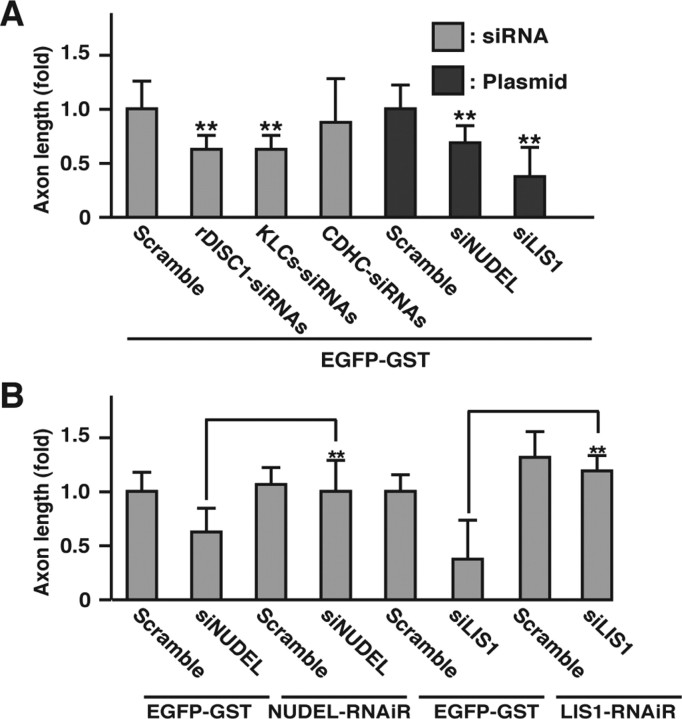
Inhibition of axon elongation by knockdown of NUDEL or LIS1 but not cytoplasmic dynein. A, Hippocampal neurons transfected with EGFP–GST and scramble, DISC1–siRNAs, KLCs–siRNAs, CDHC–siRNAs, or pSilencer encoding scramble, siNUDEL, or siLIS1 at DIV1 were detected by EGFP fluorescence and staining of Tau-1 at DIV3. Axons of transfected neurons were scored according to axon length. Numbers of cells used for each calculation are >50, and the values shown are means ± SE of triplicates (Student's t test, **p < 0.01). B, NUDEL- or LIS1-knockdown hippocampal neurons transfected with expression plasmid encoding EGFP–GST, EGFP–NUDEL–RNAiR, or EGFP–LIS1–RNAiR. The EGFP-positive neurons were scored by axon length. Numbers of cells used for each calculation are >50, and the values shown are means ± SE of three independent experiments (Student's t test, **p < 0.01).
The accumulations of 14-3-3ε (see Figs. 4 C, 5 E), DISC1 (Fig. 6 B,C), specifically Rac1-associated protein 1 (Sra-1) (supplemental Fig. 4E, available at www.jneurosci.org as supplemental material), or Numb (supplemental Fig. 4F, available at www.jneurosci.org as supplemental material) mean >150% increment of fluorescence intensity of 14-3-3ε, DISC1, Sra-1, or Numb in the central regions of growth cones compared with the average of those in the intermediate regions of axonal shafts between cell bodies and the distal parts of axons, as described previously (Kawano et al., 2005). The cell tracker 5-chloromethylfluorescein diacetate or Orange 5-(and-6)-(((4-chloromethhyl)benzoyl)amino)tetramethylrhodamine (Invitrogen), which is a cytosolic marker, was used to normalize, and the above analyses were performed in same neurons.
Hippocampal neurons were fixed with 3.7% formaldehyde in PBS for 10 min and treated with PBS that contained 0.05% Triton X-100 for 10 min. Neurons were incubated with primary antibodies overnight at 4°C, washed, and incubated for 1 h with second antibodies. Immunofluorescence analyses were examined with a laser scanning confocal microscope (model LSM510; Zeiss, Oberkochen, Germany).
Time-lapse observation.
Hippocampal neurons were seeded on a 3.5 cm glass-bottom dish and then transfected with expression plasmid encoding EGFP only, EGFP–GST, or NUDEL–GFP at day in vitro (DIV) 1 and with Cy3–scramble, Cy3–rDISC1–siRNAs, or Cy3–KLCs–siRNAs at DIV2. At DIV4, the areas of distal part of axons in Cy3- and EGFP-positive neurons were photobleached in various squares with 75 laser irradiations and then observed at 37°C for 4 min in a microincubator (Sankei, Tokyo, Japan) using a laser scanning confocal microscope equipped with Zeiss Axiovert 100 M (Plan Apochromat 63×/1.40 numerical aperture oil immersion objective) (see Fig. 7 A–E). The supplemental movie files (QuickTime Movie) were produced using Adobe ImageReady 2.0 (Adobe Systems, San Jose, CA).
Figure 7.

Effect of knockdown of DISC1 or KLCs on transport of NUDEL–GFP. A–D, Hippocampal neurons were transfected with expression plasmid encoding NUDEL–GFP at DIV1 and then transfected with Cy3–scramble (A, B) or Cy3–rDISC1–siRNAs (C, D) at DIV2. Under these conditions, NUDEL–GFP was localized in the distal part of the axon at DIV4. The areas of distal part of axons in Cy3- and GFP-positive neurons were photobleached in various squares and then observed at DIV4. The numbers of no more than 70% of recovered NUDEL–GFP fluorescence in the axonal growth cone within 4 min. E, Quantification of the recovery of the fluorescence of NUDEL–GFP at the photobleached area in the distal parts of axons of Cy3–scramble-, Cy3–rDISC1–siRNAs-, or Cy3–KLCs–siRNAs-transfected neurons. The growth cones with >70% fluorescent recovery in 4 min after photobleach were considered as “growth cones with recovered NUDEL–GFP.” Data are means ± SE of five independent experiments (total numbers of cells is 50). **p < 0.01, Student's t test.
Results
Identification of Kinesin-1 as a novel DISC1-interacting molecule
To detect DISC1-interacting molecules, cytosolic extract from rat brain was loaded onto an affinity column coated with maltose-binding protein (MBP) or MBP–DISC1. Several proteins with molecular mass of ∼180, 125, 120, 42, 40, 31, and 30 kDa were detected in the eluate from the MBP–DISC1 affinity column but not from the MBP affinity column (Fig. 1 A,B). To distinguish DISC1-interacting molecules from degradation products of bait, immunoblot analyses were performed using anti-MBP and anti-DISC1 antibodies. Molecular mass of ∼150 or 50 kDa was full-length or degradation product of MBP–DISC1, respectively. By mass spectral analyses, proteins with molecular mass of ∼180, 125, 120, 31, and 30 kDa were identified as rat ADP-ribosylation factor guanine nucleotide-exchange factor 2 (ARFGEF2), KIF5B, phosphofurin acidic cluster sorting protein 1 (PACS-1), 14-3-3ε, and receptor for activated C kinase (RACK1), respectively. The 42 and 40 kDa proteins were not identified. ARFGEF2 regulates the activity of ARF, which plays an important role in intracellular vesicular trafficking (Togawa et al., 1999). KIF5B belongs to the kinesin heavy chain family and acts as a motor molecule (Gudkov et al., 1994). Kinesin-1 is a tetramer of two kinesin heavy chains (KIF5A, KIF5B, or KIF5C) and two kinesin light chains (KLC1 or KLC2) (Brady, 1985; Vale et al., 1985) and acts as a molecular motor on microtubules that are essential for anterograde axonal transport (Miki et al., 2001; Kamal and Goldstein, 2002). PACS-1 regulates the localization of the trans-Golgi network (Wan et al., 1998). RACK1 is identified as an adaptor for PKC and is thought to function for several signaling enzymes, but its function is unclear in neuronal development (Schechtman and Mochly-Rosen, 2001).
Figure 1.
Identification of Kinesin-1 as a novel DISC1-interacting molecule. A, B, Cytosolic extract from rat brain were used in affinity-column chromatography with MBP–DISC1. Aliquots from eluates were resolved by SDS-PAGE using 6% gel (A) to high-molecular-weight proteins and 12% gel (B) to low-molecular-weight proteins, followed by silver staining. Arrows indicate the DISC1-interacting proteins. Bound proteins were analyzed by mass spectral analyses. Asterisks indicate full-length or degradation product of MBP–DISC1. C, The eluates of DISC1 affinity-column chromatography were analyzed by immunoblotting with antibodies against kinesin family, including KIF5B, KLCs, KIF5A, KIF3B, and KIF2. D, The eluates of DISC1 affinity-column chromatography were analyzed by immunoblotting with antibodies against the NUDEL/LIS1/14-3-3ε/cytoplasmic dynein complex, including 14-3-3ε, NUDEL, LIS1, DIC, and CDHC, and RhoGDI as a negative control. Aliquots of original samples (2% Input) and eluates (10%) were subjected to SDS-PAGE.
14-3-3ε binds to phosphorylated NUDEL and forms a complex with NUDEL/LIS1/cytoplasmic dynein (Toyo-oka et al., 2003). The band corresponding to NUDEL, which was reported as a DISC1-interacting molecule, was not detected by silver staining because it might be covered with several degradation products of MBP–DISC1. The immunoblot analyses revealed an interaction of DISC1 with KIF5A, KLC1, KLC2, NUDEL, LIS1, DIC, and CDHC but not with other kinesins, including KIF3B and KIF2, and RhoGDI, an abundant cytoplasmic protein (Fukumoto et al., 1990), as a negative control (Fig. 1 C,D). Because the results of affinity-column chromatography show that DISC1 binds to Kinesin-1 and 14-3-3ε, both of which appear to be involved in the DISC1 transport to axons, we focused on Kinesin-1 and the NUDEL/LIS1/14-3-3ε complex among DISC1-interacting molecules.
Interaction of DISC1 with Kinesin-1 and NUDEL in vitro
To examine whether DISC1 directly interacts with kinesin heavy chain or light chain, an in vitro binding assay was performed using purified recombinant proteins. We mainly used KIF5A in this study, because KIF5A and KIF5C are specifically expressed in brain, whereas KIF5B is ubiquitously expressed in various tissues (Niclas et al., 1994; Miki et al., 2001). Affinity beads coated with GST, GST–KLC1, or GST–KIF5A were mixed with MBP or MBP–DISC1. MBP–DISC1 bound directly to GST–KIF5A but not to GST alone or GST–KLC1 (Fig. 2 A). DISC1 also interacted with KIF5C (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material), indicating that DISC1 binds to Kinesin-1 through direct interaction with KIF5s.
To examine whether DISC1 forms a ternary complex with KIF5A and NUDEL, Myc–DISC1, HA–KIF5A, and NUDEL–GFP were transfected into COS7 cells. When HA–KIF5A was immunoprecipitated, NUDEL–GFP was coimmunoprecipitated only in the presence of Myc–DISC1 (Fig. 2 B), suggesting that KIF5A interacts with NUDEL through DISC1. DISC1 appears to form a more stable complex with KIF5A in the presence of NUDEL (Fig. 2 B). To examine whether DISC1 forms a complex with NUDEL and 14-3-3ε, Myc–DISC1, NUDEL–GFP, and GST–14-3-3ε were transfected into COS7 cells. When GST–14-3-3ε was precipitated, Myc–DISC1 and NUDEL–GFP were coprecipitated as well (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material).
We examined which domain of DISC1 interacts with KIF5A using deletion mutants of Myc–KIF5A. When DISC1–EGFP was immunoprecipitated from COS7 cells, Myc–KIF5A–Full, Myc–KIF5A–HL, or Myc–KIF5A–C1 was coimmunoprecipitated with DISC1–EGFP, whereas Myc–KIF5A–C2 was not (Fig. 2 C) (supplemental Fig. 1C, available at www.jneurosci.org as supplemental material). We next examined which domain of DISC1 interacts with KIF5A using deletion mutants of DISC1–EGFP in a GST pull-down assay. GST–KIF5A bound to Myc–DISC1–WT, Myc–DISC1–NB, and Myc–DISC1–NH but not to Myc–DISC1–CH and Myc–DISC1–CB (Fig. 2 D) (supplemental Fig. 1D, available at www.jneurosci.org as supplemental material). To narrow down the domain of DISC1 required for the binding with KIF5A, we performed in vitro binding assay. GST–KIF5A bound to MBP–DISC1 (341–400 amino acids) (data not shown). We also confirmed that DISC1–CB interacts with NUDEL (Fig. 2 D), as described previously (Morris et al., 2003; Ozeki et al., 2003). These results indicate that the N-terminal region of DISC1 (341–400 amino acids) interacts with the C-terminal region of the KIF5A, presumably through the region containing 706–829 amino acids of KIF5A.
Interaction of DISC1 with Kinesin-1, NUDEL, LIS1, and 14-3-3ε in vivo
To examine the physiological interaction of Kinesin-1, DISC1, NUDEL, LIS1, and 14-3-3ε, we used differentiated PC12 cells in biochemical experiments, because the DISC1 expression is increased after the differentiation by NGF (Ozeki et al., 2003). When DISC1 was immunoprecipitated from PC12 cells, KIF5A, NUDEL, LIS1, and 14-3-3ε were coimmunoprecipitated with DISC1 (Fig. 2 E). When KIF5A or NUDEL was immunoprecipitated, DISC1 was reciprocally coimmunoprecipitated with KIF5 or NUDEL, respectively (Fig. 2 F) (supplemental Fig. 3A, available at www.jneurosci.org as supplemental material). These results indicate that DISC1 forms a complex with Kinesin-1, NUDEL, LIS1, and 14-3-3ε under physiological conditions.
We examined whether DISC1 colocalized with Kinesin-1, NUDEL, LIS1, and 14-3-3ε using cultured rat hippocampal neurons, as a model system (Craig and Banker, 1994). DISC1 localized mainly at cell bodies and partially in the distal part of the axon in cultured rat hippocampal neurons at DIV3 (Fig. 3 A–D). DISC1 colocalized with KIF5A, LIS1, and 14-3-3ε in the distal part of the axon (Fig. 3 A–C). Because anti-DISC1 and anti-NUDEL antibodies were rabbit polyclonal ones, we could not perform double immunostaining with both antibodies. Instead, we showed colocalization of NUDEL–GFP and DISC1 (Fig. 3 D) and of endogenous NUDEL and LIS1 (Fig. 3 E) in the distal part of the axon.
The axonal growth cone contains both filopodia and lamellipodial, and its area is classified into the peripheral, transition, and central regions (Dent and Gertler, 2003). The peripheral region is composed of actin filaments, and the central region is composed of the microtubule terminal projecting from the axon shaft (Dent and Gertler, 2003). The high-magnification view in the growth cones of axons revealed that DISC1, KIF5A, LIS1, 14-3-3ε, and NUDEL mainly localized in the central region and diffusely localized in the peripheral and transition regions of the growth cone (Fig. 3 F–I). DISC1 partially colocalized with KIF5A, LIS1, and 14-3-3ε in the growth cones (Fig. 3 F–H). NUDEL partially colocalized with LIS1 in the growth cones (Fig. 3 I). DISC1 and KIF5A showed the similar punctate structures along microtubules in the growth cones (Fig. 3 J,K). Based on these results of the high-magnification view in the growth cones together with the finding that DISC1 forms a complex with KIF5A, DISC1 appears to form a complex with Kinesin-1 along microtubules.
Roles of DISC1 in the localization of NUDEL, LIS1, and 14-3-3ε in the distal part of axons
To examine the roles of DISC1 in the distribution of NUDEL, LIS1, and 14-3-3ε, we performed a knockdown experiment of DISC1 using RNAi. Because the expression levels of DISC1 in hippocampal neurons were low, PC12 cells were transfected with rDISC1–siRNAs for immunoblot analysis. The expression of DISC1 was knocked down by rDISC1–siRNAs, whereas the levels of NUDEL, LIS1, 14-3-3ε, and β-tubulin were not affected (Fig. 4 A). The expression levels of rat DISC1 were reduced to ∼10–20% by rDISC1–siRNAs compared with control scramble (Fig. 4 A).
We then examined the effect of DISC1 knockdown on neuronal morphology using hippocampal neurons transfected with rDISC1–siRNAs. To identify the transfected cells, EGFP–GST was cotransfected with rDISC1–siRNAs in rat hippocampal neurons at DIV1, and the cell morphologies were observed at DIV3. In rDISC1–siRNAs-transfected neurons, the immunoreactivity of DISC1 was markedly decreased and the length of the axons was reduced compared with those of scramble-transfected neurons (Fig. 4 B) (supplemental Fig. 2A,B, available at www.jneurosci.org as supplemental material), suggesting that DISC1 plays an essential role in axon elongation.
To examine the roles of DISC1 in the distribution of NUDEL, LIS1, and 14-3-3ε, the immunofluorescence analysis was performed using hippocampal neurons. The knockdown of DISC1 inhibited the accumulation of LIS1, 14-3-3ε, or NUDEL–GFP in the distal part of the axon but not in the cell body (Fig. 4 C–F). In contrast, in scramble-transfected neurons, LIS1, 14-3-3ε, or NUDEL–GFP localized in the distal part of the axons, as observed in nontransfected neurons (Fig. 3 B–D). Essentially similar results were obtained when Cy3-labeled scramble or rDISC1–siRNAs were used to identify the transfected cells instead of unlabeled siRNAs (Fig. 4 C).
To examine the possibility that the knockdown of DISC1 affects the major cytoskeletal architectures and thereby nonspecifically modulates the protein transport, we determined the localization of actin filaments, tubulin, Tau, and EB1, a plus end binding protein of microtubules (Mimori-Kiyosue et al., 2000). The knockdown of DISC1 did not affect the localization of actin filaments (Fig. 4 D–F), tubulin (Fig. 4 G), Tau (supplemental Fig. 2, available at www.jneurosci.org as supplemental material), and EB1 (supplemental Fig. 3B,C, available at www.jneurosci.org as supplemental material). We have shown previously that collapsin response mediator protein-2 (CRMP-2) links KLCs of Kinesin-1 to specifically Rac1-associated protein (Sra-1) or Numb and regulates their transport along microtubules to axons (Arimura and Kaibuchi, 2005; Kawano et al., 2005). The knockdown of CRMP-2 or KLCs inhibited the accumulation of Sra-1 or Numb in the distal part of axons (supplemental Fig. 4E,F, available at www.jneurosci.org as supplemental material) (Arimura and Kaibuchi, 2005; Kawano et al., 2005). The knockdown of DISC1 did not affect the localization of Sra-1 or Numb (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). These results suggest that DISC1 specifically regulated the localization of LIS1, 14-3-3ε, and NUDEL–GFP without affecting the major cytoskeletal architectures. However, we cannot completely neglect the possibility that axonal structures are changed by the DISC1 knockdown because DISC1 may transport cytoskeletal elements.
Based on these results in DISC1-knockdown neurons, we wondered whether DISC1 regulates the complex formation of NUDEL, LIS1, and 14-3-3ε. We investigated whether DISC1 affects the complex formation by an immunoprecipitation assay using scramble–siRNAs-transfected or rDISC1–siRNAs-transfected PC12 cells. When NUDEL was immunoprecipitated from scramble-transfected PC12 cells, DISC1, LIS1, and 14-3-3ε were coimmunoprecipitated with NUDEL (supplemental Fig. 3A, available at www.jneurosci.org as supplemental material). Similarly, LIS1 and 14-3-3ε were coimmunoprecipitated with NUDEL from DISC1-knockdown PC12 cells (supplemental Fig. 3A, available at www.jneurosci.org as supplemental material), indicating that the knockdown of DISC1 did not affect the formation of the NUDEL/LIS1/14-3-3ε complex.
Effect of N-terminal fragment of DISC1 on axon elongation
To examine the effect of the N-terminal fragment of DISC1 (DISC1–NB or DISC1–NH), which interacts with Kinesin-1 but not with the NUDEL/LIS1/14-3-3ε complex on axon elongation and the localization of the NUDEL complex in the axon, we expressed various EGFP-tagged DISC1 mutants (Fig. 5 A–E). DISC1–NB or DISC1–NH accumulated in the distal part of the axon and inhibited axon elongation (Fig. 5 D), as described previously (Ozeki et al., 2003). DISC1–NB or DISC1–NH inhibited the accumulation of 14-3-3ε (Fig. 5 C,E), LIS1, and NUDEL (supplemental Fig. 5, available at www.jneurosci.org as supplemental material) (data not shown) in the distal part of the axon, suggesting that DISC1–NB or DISC1–NH acts as a dominant-negative mutant for endogenous DISC1. We speculated that DISC1–CH and DISC1–CB mutants also act as the dominant-negative mutants for endogenous DISC1 because they interact with NUDEL but not with Kinesin-1. However, DISC1–CH and DISC1–CB did not affect neuronal morphologies or the distribution of 14-3-3ε (Fig. 5 D,E). Although we do not know the exact reason why DISC1–CH or DISC1–CB does not alter the distribution of NUDEL/LIS1/14-3-3ε, one possible explanation may be that DISC1–CH or DISC1–CB has a lower affinity for NUDEL/LIS1/14-3-3ε than wild-type DISC1 has. To confirm the specificity of DISC1 knockdown on neuronal morphology and the localization of NUDEL, LIS1, and 14-3-3ε, rescue experiments were performed using human DISC1 cDNA. The expression of human DISC1 cDNA was not inhibited by rDISC1–siRNAs (data not shown). The inhibition of the defective localization of 14-3-3ε was rescued by DISC1–WT but not by DISC1–NB, which lacks the NUDEL-binding region (Fig. 5 F,G), indicating that DISC1 is required for the proper localization of the NUDEL/LIS1/14-3-3ε complex, and DISC1–NB results in the loss-of-function mutant under DISC1-knockdown conditions.
Roles of Kinesin-1 in the localization of DISC1 in the distal part of axons
To examine the role of Kinesin-1 in the localization of DISC1 and the NUDEL/LIS1/14-3-3ε complex, we used the knockdown of KLCs instead of that of KIF5s. Because KIF5A, KIF5B, and KIF5C have functional redundancy, we tried triple knockdowns of KIF5s but have not succeeded to date. It has been reported that KLCs are essential for the proper function or localization of KIF5A and KIF5B using mice deficient in KLCs (Rahman et al., 1999), although it remains unclear how deficiency of KLCs inhibits KHCs function (Verhey et al., 1998; Adio et al., 2006). Thus, we performed the knockdown of KLCs by KLCs–siRNAs in PC12 cells (Kimura et al., 2005). The expression levels of rat KLC1 and KLC2 were reduced to ∼10–20% by KLCs–siRNAs compared with control scramble (Fig. 6 A). The levels of KIF5A, DISC1, NUDEL, LIS1, 14-3-3ε, and β-tubulin were not affected by the knockdown of KLCs (Fig. 6 A). In KLCs–siRNAs-transfected hippocampal neurons, the immunoreactivity of KLCs was clearly reduced (Fig. 6 D), and the length of the axons was reduced compared with those of scramble-transfected neurons (see Fig. 8 A), as reported previously (Kimura et al., 2005). In KLCs-knockdown neurons, endogenous DISC1 or DISC1–EGFP was not accumulated in the distal part of the axon but was mainly present within the cell body (Fig. 6 B,D), and the accumulation of NUDEL–GFP, LIS1, and 14-3-3ε was also reduced in the distal part of the axon (data not shown), whereas the accumulation of actin filaments was not affected (Fig. 6 D). Essentially similar results were obtained when Cy3-labeled scramble or KLCs–siRNAs were used to identify the transfected cells instead of unlabeled siRNAs (Fig. 6 B).
To confirm the specific effect of KIF5A on the localization of DISC1, we expressed KIF5A–HL, which is thought to impair the motor function of Kinesin-1 and acts as the dominant-negative form (Nakata and Hirokawa, 2003), inhibited the accumulation of DISC1, but not that of actin filaments, in the distal part of the axon (Fig. 6 C,E,F). KIF5A–C1 containing the DISC1-binding region also inhibited the accumulation of DISC1, whereas KIF5A–C2 lacking the DISC1-binding region did not (Fig. 6 C). These results indicate that Kinesin-1 is required for the proper localization of DISC1.
Regulation of the transport of NUDEL by DISC1 and Kinesin-1
To confirm the link between the DISC1- or KLCs-knockdown phenotype and the disrupted transport of NUDEL, LIS1, and 14-3-3ε, we monitored the transport of NUDEL by time-lapse imaging of NUDEL–GFP. Hippocampal neurons were transfected with NUDEL–GFP at DIV1 and then transfected with Cy3-labeled siRNAs at DIV2. Under these conditions, NUDEL–GFP was localized in the distal part of the axon at DIV4. In the neurons containing Cy3–siRNA in the cell body at DIV4, photobleaching of NUDEL–GFP was performed and followed by time-lapse analysis (Fig. 7 A–D). We counted the numbers of no more than 70% of recovered NUDEL–GFP fluorescence to the axonal growth cone within 4 min (Fig. 7 E). The control neurons transfected with Cy3–scramble showed a steady recovery of fluorescence in the photobleached area (Fig. 7 A,B,E) (supplemental Movie 1, available at www.jneurosci.org as supplemental material). In contrast, a delay in recovery was observed in DISC1- or KLC-knockdown neurons (Fig. 7 C–E) (supplemental Movies 2, 3, available at www.jneurosci.org as supplemental material). When the neurons were treated with nocodazole and placed on ice for 5 min, which destroys the microtubules (Galbraith et al., 1999), the movement of NUDEL–GFP was partially prevented, whereas that of EGFP alone or EGFP–GST (supplemental Movies 4, 5, available at www.jneurosci.org as supplemental material) was not impaired, and neither placing on ice for 5 min nor nocodazole treatment alone, which is insufficient to destroy the microtubules, prevented NUDEL–GFP movement (supplemental Movie 6, available at www.jneurosci.org as supplemental material) (Galbraith et al., 1999; Kimura et al., 2005). The movement of NUDEL–GFP was not completely prevented under these conditions, because a part of movement of NUDEL–GFP might be attributable to simple diffusion. These results indicate that most of the recovery of NUDEL–GFP is attributable to transport on the rail of microtubules, because the transport along microtubules is faster than the simple diffusion and so is inhibited by microtubule disruption (Galbraith et al., 1999) and by the knockdown of DISC1 or KLC.
Regulation of axon elongation by NUDEL and LIS1 but not by cytoplasmic dynein
Because the functions of NUDEL and LIS1 in axon elongation of hippocampal neurons remain essentially unknown, we examined the effect of NUDEL or LIS1 knockdown using the RNAi plasmids (Shu et al., 2004) on morphologies of hippocampal neurons. In NUDEL- and LIS1-knockdown neurons, the length of the axons was reduced compared with those of scramble-transfected neurons (Fig. 8 A), suggesting that NUDEL and LIS1 regulate axon elongation. To confirm these results, rescue experiments were performed using RNAiR mutants of NUDEL and LIS1 with three silence mutations. The results of immunoblot analyses revealed that the expressions of these mutants were not decreased by the cotransfection of each RNAi plasmids (data not shown). The inhibition of axon elongation was rescued by NUDEL–RNAiR or LIS1–RNAiR, respectively (Fig. 8 B).
We next examined the effect of cytoplasmic dynein knockdown on axon elongation. It has been reported that the knockdown of CDHC has no effect on the formation and growth of axons in cultured rat sympathetic neurons (He et al., 2005). Consistently, we found that the transfection of CDHC–siRNAs reduced the expression levels of CDHC (data not shown), and the suppression of CDHC expression did not affect the axon growth in hippocampal neurons (Fig. 8 A). Therefore, our results together with the previous observation indicate that cytoplasmic dynein is not essential for axon elongation.
Discussion
Regulation of the localization of NUDEL, LIS1, and 14-3-3ε by DISC1
The NUDEL/LIS1/14-3-3ε complex is transported to the vicinity of the centrosome or the MTOC and the distal parts of neurites (Leventer et al., 2001; Toyo-oka et al., 2003). Our results indicate that the NUDEL/LIS1/14-3-3ε complex is transported to the axon by Kinesin-1 through DISC1. Previously, it has been reported that DISC1 localizes in the centrosome (Morris et al., 2003; Ozeki et al., 2003). DISC1 appears to be essential for maintaining the NUDEL/LIS1/dynein complex at the centrosome and regulates neuronal migration (Kamiya et al., 2005), although the mode of actions of DISC1 on NUDEL/LIS1/dynein remains essentially unknown. We did not observe the effect of DISC1 on neuronal migration, because cultured hippocampal neurons did not migrate dynamically in vitro. We focused on the accumulation of DISC1 in the distal part of axons, so we could not see the accumulation of DISC1 at the centrosome in hippocampal neurons. NUDEL and LIS1 at the centrosome play a crucial role in nuclear migration through the action of dynein, whereas they promote axon elongation in the distal part of axon. Because both neuronal migration and axon elongation are critical steps in neuronal development, DISC1 plays a vital role by regulating the proper localization of the NUDEL and LIS1 at the centrosome and the distal part of the axon. Although the functions of the NUDEL/LIS1 complex at the distal part of the axon are not fully understood, LIS1-deficient cerebellar granule neurons show defects in axon extension (Kholmanskikh et al., 2003). We found that the knockdown of NUDEL or LIS1 inhibited axon elongation (Fig. 8 A), and NUDEL–RNAiR or LIS1–RNAiR rescued this inhibition (Fig. 8 B), indicating that NUDEL and LIS1 play essential roles in axon elongation in hippocampal neurons.
DISC1 is essential for maintaining cytoplasmic dynein at the centrosome (Kamiya et al., 2005), and Kinesin-1 is required for the polarized localization of dynein to microtubule plus ends in Aspergillus nidulans (Zhang et al., 2003). In contrast, we found that the localization of CDHC at axons did not apparently change in DISC1- or KLCs-knockdown neurons. The dynein activity is essential for neuronal migration (Shu et al., 2004; Tsai and Gleeson, 2005), whereas it is not necessary for axon elongation (Fig. 8 A). It has been reported that dynein activity is not critically required for the initial neuronal development, including elaboration of neuritis and axon specification (Chuang et al., 2005). Thus, NUDEL and LIS1 at growth cone appear to regulate axon elongation in a manner independent of cytoplasmic dynein. LIS1 directly interacts with CLIP-170, another member of +Tips (Coquelle et al., 2002), and appears to tether microtubule plus ends to the cortical actin meshwork, thereby regulating neuronal motility (Kholmanskikh et al., 2006). Thus, the mode of action of DISC1, NUDEL, and LIS1 at the growth cone is different from that in centrosome.
Dysfunction of the cargo receptor in neuronal development
The results of this study suggest that DISC1 serves as the cargo receptor of the NUDEL/LIS1/14-3-3ε complex and Kinesin-1. The KIF5A-binding region of DISC1 (341–400 amino acids) is different from the FEZ1-binding region of DISC1 (446–633 amino acids) (Miyoshi et al., 2003) or PDE4B-binding region of DISC1 (219–283 amino acids) (Millar et al., 2005). In the accompanying paper (Shinoda et al., 2006), we identified growth factor receptor-bound protein 2 (Grb2) as a DISC1-interacting protein using membrane extract of rat brain. Grb2 acts as an adaptor molecule that links receptor tyrosine kinases and the Ras–ERK pathway (Lowenstein et al., 1992; Takenawa et al., 1998; Buday, 1999). DISC1 also forms a ternary complex with Grb2 and Kinesin-1 and regulates the transport of Grb2 to the distal parts of axons in a Kinesin-1-dependent manner. We found that the transported Grb2 by DISC1 is involved in neurotrophin-induced axonal elongation. Thus, it can be possible that DISC1 links Kinesin-1 to FEZ1 or PDE4B. It is worthwhile examining whether DISC1 serve as the cargo receptor not only for the NUDEL complex and Grb2 but also for FEZ1 or PDE4B for the future studies.
We recently found that CRMP-2 links Kinesin-1 to tubulin heterodimers, Sra-1, and Numb and regulates their transport to the growing axon (Arimura and Kaibuchi, 2005; Kawano et al., 2005; Kimura et al., 2005). CRMP-2 binds to tubulin heterodimers to promote microtubule assembly, thereby enhancing axon elongation (Fukata et al., 2002). Based on these observations, we proposed that CRMP-2 is the cargo receptor of Kinesin-1 and regulates neuronal development. CRMP-2 was also independently identified as dihydropyrimidinase-related protein-2 (DRP-2) (Hamajima et al., 1996). DRP-2 is a candidate gene for susceptibility to schizophrenia (Johnston-Wilson et al., 2000; Nakata et al., 2003). The expression of DRP-2 is significantly decreased in the frontal cortex regions of schizophrenics (Johnston-Wilson et al., 2000). SNPs analyses have shown that the DRP-2 gene is associated with paranoid-type schizophrenia (Nakata et al., 2003). DISC1 participates in the transport of the NUDEL/LIS1/14-3-3ε complex and Grb2 through interaction with KHC of Kinesin-1, whereas CRMP-2 participates in the transport of tubulin heterodimers, Sra-1, and Numb through interaction with KLC of Kinesin-1 (Arimura and Kaibuchi, 2005; Kawano et al., 2005; Kimura et al., 2005). Although DISC1 and CRMP-2 have different partner proteins, they have similar modes of action that link motor proteins to transport proteins. Consistently, we found that the knockdown of CRMP-2 inhibits the accumulation of Sra-1 and Numb at the growth cones (Arimura and Kaibuchi, 2005; Kawano et al., 2005), whereas the knockdown of DISC1 dose not affect it (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). In addition, it has been reported that several diseases associate with impaired cargo transport (Vale, 2003; Hirokawa and Takemura, 2004). Thus, it is possible that dysfunction of the cargo receptors, such as DISC1 and CRMP-2/DRP-2, may impair neuronal development, leading to psychiatric disorders, such as schizophrenia.
Effect of DISC1 lacking the NUDEL-binding region on axon elongation
The pathogenesis of schizophrenia appears to be related to neuronal development based on reports of adverse events during the prenatal and perinatal periods that correlate with the presence of cognitive and behavioral signs during childhood and adolescence (Weinberger, 1987). Moreover, postmortem and neuroimaging studies of schizophrenics delineate changes in brain structure and volume that appear to arise from a reduction of neuritic processes, such as axons, dendrites, and synapses (Harrison, 1999; Shenton et al., 2001; Lewis and Levitt, 2002). Thus, the products of susceptibility genes for schizophrenia are thought to have important roles in brain development. In a Scottish family, the chromosome translocation interrupts the coding sequence of DISC1, leading to the reduction of DISC1 expression or the production of the disease form of DISC1. Millar et al. (2005) have claimed that they could not detect a putative C-terminal truncated DISC1 protein in the translocation (1;11) lymphoblastoid cell lines. The existence of the mutant DISC1 in the Scottish family is still controversial (Sawa and Snyder, 2005). In this study, we found that the knockdown of DISC1 or DISC1 lacking the NUDEL-binding region inhibited the accumulation of NUDEL, LIS1, and 14-3-3ε at the axons and reduced axonal growth. DISC1 lacking the NUDEL-binding region also did not interact with Grb2 (Shinoda et al., 2006). Placing these observations in the context of the pathogenesis of schizophrenia, translocation or SNPs of the DISC1 gene may cause dysfunctions of DISC1 that inhibit the transport of the NUDEL/LIS1 complex and Grb2 and then disrupt neuronal development, which may relate to abnormal synaptic activity or connectivity in schizophrenia (Harrison, 1999; Lewis and Levitt, 2002). Studies in mice are underway to expand our understanding of the roles of DISC1 and the DISC1-interacting molecules in the pathogenesis of schizophrenia.
Footnotes
This work was supported by Grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, Special Coordination Funds for Promoting Science and Technology, the Pharmaceuticals and Medical Devices Agency, Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists, the 21st Century Centre of Excellence Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and the Research Grant (15A-2) for Nervous and Mental Disorders from the Ministry of Health, Labour, and Welfare. We thank Dr. T. Nagase and Dr. L. Goldstein for kindly providing cDNAs, Dr. O. Reiner for kindly providing antibody, Dr. N. Ozaki for his critical and helpful discussions, Dr. A. Sawa for helpful discussion, Drs. N. Arimura, Y. Fukata, T. Watanabe, and S. Wang for providing materials, N. Hattori, S. Kozawa, M. Yamamoto, and Y. Yamashita for their technical assistance, and T. Ishii for secretarial assistance.
References
- Adio S, Reth J, Bathe F, Woehlke G. Regulation mechanisms of Kinesin-1. J Muscle Res Cell Motil. 2006;27:153–160. doi: 10.1007/s10974-005-9054-1. [DOI] [PubMed] [Google Scholar]
- Arimura N, Kaibuchi K. Key regulators in neuronal polarity. Neuron. 2005;48:881–884. doi: 10.1016/j.neuron.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Bilder RM, Volavka J, Czobor P, Malhotra AK, Kennedy JL, Ni X, Goldman RS, Hoptman MJ, Sheitman B, Lindenmayer JP, Citrome L, McEvoy JP, Kunz M, Chakos M, Cooper TB, Lieberman JA. Neurocognitive correlates of the COMT Val(158)Met polymorphism in chronic schizophrenia. Biol Psychiatry. 2002;52:701–707. doi: 10.1016/s0006-3223(02)01416-6. [DOI] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St. Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders–cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady ST. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran-Jimeniz B, Camargo LM, Oliver KR, Beher D, Shearman MS, Whiting PJ. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Buday L. Membrane-targeting of signalling molecules by SH2/SH3 domain-containing adaptor proteins. Biochim Biophys Acta. 1999;1422:187–204. doi: 10.1016/s0304-4157(99)00005-2. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, Verchinski BA, Meyer-Lindenberg A, Balkissoon R, Kolachana B, Goldberg TE, Weinberger DR. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci USA. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JZ, Yeh TY, Bollati F, Conde C, Canavosio F, Caceres A, Sung CH. The dynein light chain Tctex-1 has a dynein-independent role in actin remodeling during neurite outgrowth. Dev Cell. 2005;9:75–86. doi: 10.1016/j.devcel.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, Barry C, Tanaka H, La Rosa P, Puech A, Tahri N, Cohen-Akenine A, Delabrosse S, Lissarrague S, Picard FP, Maurice K, Essioux L, Millasseau P, Grel P, et al. Genetic and physiological data implicating the new human gene G72 and the gene for d-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA. 2002;99:13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle FM, Caspi M, Cordelieres FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N, De Mey JR, Reiner O. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol Cell Biol. 2002;22:3089–3102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, O'Donovan MC, Owen MJ. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet. 2005;42:193–204. doi: 10.1136/jmg.2005.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- Dent EW, Gertler FB. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, Ueda T, Kikuchi A, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- Galbraith JA, Reese TS, Schlief ML, Gallant PE. Slow transport of unpolymerized tubulin and polymerized neurofilament in the squid giant axon. Proc Natl Acad Sci USA. 1999;96:11589–11594. doi: 10.1073/pnas.96.20.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello MJ, Darling DL, Yingling J, Tanaka T, Gleeson JG, Wynshaw-Boris A. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudkov AV, Kazarov AR, Thimmapaya R, Axenovich SA, Mazo IA, Roninson IB. Cloning mammalian genes by expression selection of genetic suppressor elements: association of kinesin with drug resistance and cell immortalization. Proc Natl Acad Sci USA. 1994;91:3744–3748. doi: 10.1073/pnas.91.9.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamajima N, Matsuda K, Sakata S, Tamaki N, Sasaki M, Nonaka M. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene. 1996;180:157–163. doi: 10.1016/s0378-1119(96)00445-3. [DOI] [PubMed] [Google Scholar]
- Harrison PJ. The neuropathology of schizophrenia: A critical review of the data and their interpretation. Brain. 1999;122:593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- Hattori M, Adachi H, Tsujimoto M, Arai H, Inoue K. Miller-Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase. Nature. 1994;370:216–218. doi: 10.1038/370216a0. [DOI] [PubMed] [Google Scholar]
- He Y, Francis F, Myers KA, Yu W, Black MM, Baas PW. Role of cytoplasmic dynein in the axonal transport of microtubules and neurofilaments. J Cell Biol. 2005;168:697–703. doi: 10.1083/jcb.200407191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Varilo T, Kestila M, Paunio T, Arajarvi R, Haukka J, Parker A, Martin R, Levitzky S, Partonen T, Meyer J, Lonnqvist J, Peltonen L, Ekelund J. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003;12:3151–3159. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors in neuronal development, intracellular transport and diseases. Curr Opin Neurobiol. 2004;14:564–573. doi: 10.1016/j.conb.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Goldman D, Jaeger J, Persaud S, Kane JM, Lipsky RH, Malhotra AK. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J Hum Genet. 2004;75:862–872. doi: 10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N, Chihara K, Arimura N, Menager C, Kawano Y, Matsuo N, Nishimura T, Amano M, Kaibuchi K. CRMP-2 induces axons in cultured hippocampal neurons. Nat Neurosci. 2001;4:781–782. doi: 10.1038/90476. [DOI] [PubMed] [Google Scholar]
- Johnston-Wilson NL, Sims CD, Hofmann JP, Anderson L, Shore AD, Torrey EF, Yolken RH. Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. Mol Psychiatry. 2000;5:142–149. doi: 10.1038/sj.mp.4000696. [DOI] [PubMed] [Google Scholar]
- Kamal A, Goldstein LS. Principles of cargo attachment to cytoplasmic motor proteins. Curr Opin Cell Biol. 2002;14:63–68. doi: 10.1016/s0955-0674(01)00295-2. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Yoshimura T, Tsuboi D, Kawabata S, Kaneko-Kawano T, Shirataki H, Takenawa T, Kaibuchi K. CRMP-2 is involved in Kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol Cell Biol. 2005;25:9920–9935. doi: 10.1128/MCB.25.22.9920-9935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholmanskikh SS, Dobrin JS, Wynshaw-Boris A, Letourneau PC, Ross ME. Disregulated RhoGTPases and actin cytoskeleton contribute to the migration defect in Lis1-deficient neurons. J Neurosci. 2003;23:8673–8681. doi: 10.1523/JNEUROSCI.23-25-08673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholmanskikh SS, Koeller HB, Wynshaw-Boris A, Gomez T, Letourneau PC, Ross ME. Calcium-dependent interaction of Lis1 with IQGAP1 and Cdc42 promotes neuronal motility. Nat Neurosci. 2006;9:50–57. doi: 10.1038/nn1619. [DOI] [PubMed] [Google Scholar]
- Kimura T, Arimura N, Fukata Y, Watanabe H, Iwamatsu A, Kaibuchi K. Tubulin and CRMP-2 complex is transported via Kinesin-1. J Neurochem. 2005;93:1371–1382. doi: 10.1111/j.1471-4159.2005.03063.x. [DOI] [PubMed] [Google Scholar]
- Leventer RJ, Cardoso C, Ledbetter DH, Dobyns WB. LIS1: from cortical malformation to essential protein of cellular dynamics. Trends Neurosci. 2001;24:489–492. doi: 10.1016/s0166-2236(00)01887-7. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to Ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci USA. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Millar JK, Christie S, Porteous DJ. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun. 2003;311:1019–1025. doi: 10.1016/j.bbrc.2003.10.101. [DOI] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, Thomson PA, Hill EV, Brandon NJ, Rain JC, Camargo LM, Whiting PJ, Houslay MD, Blackwood DH, Muir WJ, Porteous DJ. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T, Kuroda S, Katayama T, Tohyama M. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 2003;8:685–694. doi: 10.1038/sj.mp.4001352. [DOI] [PubMed] [Google Scholar]
- Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J Cell Biol. 2003;162:1045–1055. doi: 10.1083/jcb.200302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Ujike H, Sakai A, Takaki M, Imamura T, Tanaka Y, Kuroda S. The human dihydropyrimidinase-related protein 2 gene on chromosome 8p21 is associated with paranoid-type schizophrenia. Biol Psychiatry. 2003;53:571–576. doi: 10.1016/s0006-3223(02)01729-8. [DOI] [PubMed] [Google Scholar]
- Niclas J, Navone F, Hom-Booher N, Vale RD. Cloning and localization of a conventional kinesin motor expressed exclusively in neurons. Neuron. 1994;12:1059–1072. doi: 10.1016/0896-6273(94)90314-x. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH, Ross CA, Sawa A. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci USA. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Kamal A, Roberts EA, Goldstein LS. Defective kinesin heavy chain behavior in mouse kinesin light chain mutants. J Cell Biol. 1999;146:1277–1288. doi: 10.1083/jcb.146.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Sapir T, Elbaum M, Reiner O. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 1997;16:6977–6984. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Sawa A, Snyder SH. Genetics. Two genes link two distinct psychoses. Science. 2005;310:1128–1129. doi: 10.1126/science.1121114. [DOI] [PubMed] [Google Scholar]
- Schechtman D, Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- Shenton ME, Dickey CC, Frumin M, McCarley RW. A review of MRI findings in schizophrenia. Schizophr Res. 2001;49:1–52. doi: 10.1016/s0920-9964(01)00163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Bronstein M, Sternfeld M, Pisante-Shalom A, Lev-Lehman E, Weizman A, Reznik I, Spivak B, Grisaru N, Karp L, Schiffer R, Kotler M, Strous RD, Swartz-Vanetik M, Knobler HY, Shinar E, Beckmann JS, Yakir B, Risch N, Zak NB, et al. A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet. 2002;71:1296–1302. doi: 10.1086/344514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda T, Taya S, Tsuboi D, Hikita T, Matsuzawa R, Kuroda S, Iwamatsu A, Kaibuchi K. DISC1 regulates neurotrophin-induced axon elongation via interaction with Grb2. J Neurosci. 2006;26:4–14. doi: 10.1523/JNEUROSCI.3825-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O'Neill FA, Walsh D, Kendler KS. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Miki H, Matuoka K. Signaling through Grb2/Ash-control of the Ras pathway and cytoskeleton. Curr Top Microbiol Immunol. 1998;228:325–342. doi: 10.1007/978-3-642-80481-6_12. [DOI] [PubMed] [Google Scholar]
- Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, Hirotsune S, Wynshaw-Boris A. 14-3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Lizotte DL, Abramson T, Barenboim L, Schnapp BJ, Rapoport TA. Light chain-dependent regulation of Kinesin's interaction with microtubules. J Cell Biol. 1998;143:1053–1066. doi: 10.1083/jcb.143.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li S, Fischer R, Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell. 2003;14:1479–1488. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]