Abstract
Fragile X syndrome, as well as other forms of mental retardation and autism, is associated with altered dendritic spine number and structure. Fragile X syndrome is caused by loss-of-function mutations in Fragile X mental retardation protein (FMRP), an RNA-binding protein that regulates protein synthesis in vivo. It is unknown whether FMRP plays a direct, cell-autonomous role in regulation of synapse number, function, or maturation. Here, we report that acute postsynaptic expression of FMRP in Fmr1 knock-out (KO) neurons results in a decrease in the number of functional and structural synapses without an effect on their synaptic strength or maturational state. Similarly, neurons endogenously expressing FMRP (wild-type) have fewer synapses than neighboring Fmr1 KO neurons. An intact K homology domain 2 (KH2) RNA-binding domain and dephosphorylation of FMRP at S500 were required for the effects of FMRP on synapse number, indicating that FMRP interaction with RNA and translating polyribosomes leads to synapse loss.
Keywords: Fragile X syndrome, FMRP, hippocampus, synapse, pruning, translation
Introduction
Altered dendritic spine number and structure is associated with mental retardation and autism (Kaufmann and Moser, 2000; Bagni and Greenough, 2005; Pickett and London, 2005), suggesting that deficits in synaptic connectivity and function contributes to the etiology of these prevalent diseases. Fragile X syndrome (FXS) is the most common inherited form of mental retardation and is caused by loss of function mutations in the Fragile X mental retardation gene (FMR1). FXS affects ∼1/4000 males and 1/8000 females and is characterized by several physical, behavioral, and cognitive abnormalities, including mild to severe mental retardation, hyperactivity, and autism (O'Donnell and Warren, 2002). Cortical neurons in patients with FXS and Fmr1 knock-out (KO) mice have increased dendritic spine density, as well as longer spines, reminiscent of immature filopodia (Irwin et al., 2001; Nimchinsky et al., 2001; Bagni and Greenough, 2005; Antar et al., 2006; Grossman et al., 2006a,b). From these findings, it has been suggested that the protein product of Fmr1, Fragile X mental retardation protein (FMRP), facilitates synapse elimination or pruning and regulates synapse maturation (Weiler and Greenough, 1999; Bagni and Greenough, 2005; Antar et al., 2006). FMRP is an RNA-binding protein and functions to regulate mRNA translation in neurons and synapses (Feng, 2002; O'Donnell and Warren, 2002; Weiler et al., 2004; Bagni and Greenough, 2005; Grossman et al., 2006a). Although the lifelong loss of FMRP in humans and mice results in altered dendritic spines, it is unknown whether FMRP directly regulates synapse number, function, or maturation in a cell autonomous manner or whether these changes can be reversed with postnatal expression of FMRP. Furthermore, no role for FMRP regulated translation in mammalian synapse development has been established.
Determining the function of FMRP in neurons is critical to understanding deficits in neuronal function that result from its absence, as in FXS. In the present study, we investigate the role of FMRP in synaptic function and synapse maturation in vitro by acutely expressing FMRP in Fmr1 KO neurons and directly comparing them to neighboring Fmr1 KO neurons using both electrophysiological and immunocytochemical techniques. We find that acute postsynaptic expression of FMRP in Fmr1 KO neurons in culture reduces the number of functional and structural synaptic connections without altering the strength or maturational state of the remaining synapses. As observed with acute exogenous expression of FMRP, wild-type (WT) neurons have fewer synapses than their Fmr1 KO neighbors in dissociated cocultures. Mutations of FMRP in the K homology domain 2 (KH2) RNA-binding region or that mimic phosphorylated FMRP are unable to affect synapse number or function. These results support the idea that FMRP directly regulates synapse number postnatally through postsynaptic interactions with RNA and regulation of translation.
Materials and Methods
Hippocampal slice cultures and FMRP transfection.
Organotypic hippocampal slice cultures were prepared from postnatal day 6 (P6) WT or Fmr1 KO mice bred from the congenic C57BL/6 mouse strain (Jackson Laboratories, Bar Harbor, ME) using previously published protocols (Stoppini et al., 1991). Biolistic transfection and gold bullet preparation were performed with the Helios Gene Gun system (Bio-Rad, Hercules, CA) according to the manufacturer's protocols (McAllister, 2004). All FMRP-green fluorescent protein (GFP) constructs were obtained from Dr. Jennifer Darnell at Rockefeller University and are driven by the human FMR1 promoter. Construction of the GFP-tagged FMRP (wtFMRP-GFP) as well as I304N FMRP and arginine/glycine-rich box (RGG) deletion (ΔRGG) FMRP have been described previously (Darnell et al., 2005b). The S500A and S500D mutations were introduced into wtFMRP-GFP by QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) of the KpnI-fragment, which was subcloned into pBluescript for mutagenesis and then replaced in the WT construct.
Dissociated culture and immunocytochemistry.
Dissociated CA3–CA1 hippocampal cultures (dentate gyrus was cutoff) were prepared from P0–P1 WT and Fmr1 KO mice using modified, previously published protocols (Brewer et al., 1993). Neurons were plated at a density of 250 neurons/mm2 on poly-d-lysine/laminin or matrigel coated coverslips. At 7 days in vitro (DIV), cultures were transfected with either calcium phosphate or Lipofectamine 2000 (Invitrogen, Eugene, OR). At 12 DIV, cells were fixed in 4% paraformaldehyde (PFA)/4% sucrose for 15 min at 37°C, blocked in PBS/10% goat serum and labeled with 1° antibody against the extracellular N terminus of glutamate receptor 1 (GluR1; 1:50; Calbiochem, La Jolla, CA). For the postsynaptic marker 95 kDa postsynaptic density protein (PSD-95) and synapsin, neurons were permeabilized with 0.2% Triton-X for 1 h before treatment with 1° antibodies to PSD-95 (1:800; Affinity Bioreagents, Golden, CO), synapsin (1:1,000; provided by Dr. Thomas Sudhof, University of Texas Southwestern Medical Center), or 2F5 monoclonal FMRP antibody (1:400) (Gabel et al., 2004) (provided by Dr. Justin Fallon, Brown University, Providence, RI, and Dr. Jennifer Darnell, Rockefeller University, New York, NY) for 1 h and AlexaFluor-conjugated 2° antibodies (1:300, 1 h; Invitrogen). Hippocampal slice cultures were fixed in 4% PFA (4°C, overnight) permeabilized with 0.7% Triton-X (4°C, overnight). The slices were treated with 2F5 FMRP antibody (1:400) and AlexaFluor 2° antibody (1:300 both overnight at 4°C).
Fluorescence was detected using a Nikon (Tokyo, Japan) TE2000 inverted microscope equipped with a cooled CCD camera (dissociated neuron culture) or a Zeiss (Oberkochen, Germany) LSM 510 Meta confocal microscope (slice sections). Images were analyzed and quantitated using MetaMorph software (Universal Imaging, Downingtown, PA). Healthy neurons are first identified by their smooth soma and multiple processes under differential interference contrast (DIC) microscopy. For synaptic staining, immunoreactive puncta are defined as discrete points along a dendrite (within 50 μm from the soma) with fluorescence intensity at least twice the background staining of a region adjacent to the dendrite. Significant differences between Fmr1 KO neurons and WT or FMRP-transfected neurons were determined with an unpaired t test. For all group data, averages +SEM are plotted and n (number of cells) is on each bar (*p < 0.05; **p < 0.01; ***p < 0.001).
Electrophysiology.
Simultaneous whole-cell recordings were obtained from CA1 pyramidal neurons in slice cultures visualized using infrared-DIC and GFP fluorescence to identify transfected and nontransfected neurons (Gibson et al., 2006). Recordings were made at 30°C in a submersion chamber perfused at 3 ml/min with artificial CSF (ACSF) containing the following (in mm): 119 NaCl, 2.5 KCl, 26 NaHCO3, 1 NaH2PO4, 11 d-Glucose, 3 CaCl2, 2 MgCl2, 0.1 picrotoxin, 0.002 2-chloro-adenosine, 0.1% DMSO, pH 7.28, 320 mOsm and saturated with 95% O2/5%CO2. For all intracellular recordings, the neuron was clamped at −60 mV through whole-cell recording pipettes (∼3–7 MΩ) filled with an intracellular solution containing the following (in mm): 2.5 BAPTA, 125 Cs-Meth, 6 CsCl, 3 NaCl, 10 HEPES, 10 sucrose, 2 QX-314, 10 tetraethylammonium-Cl, 4 ATP-Mg, 0.4 GTP-Na, 14 phosphocreatine-Tris, pH 7.2, 285 mOsm. For isolated NMDA receptor (NMDAR) measurements, the ASCF was supplemented with 20 μm DNQX and 20 μm glycine and the neuron was clamped at +50 mV. For mEPSC measurements, the ACSF was supplemented with 1 μm TTX. Synaptic responses were evoked by single bipolar electrode placed in stratum radiatum of area CA1 (along the Schaeffer collaterals) 50–200 μm from the recorded neurons with monophasic current pulses (5–40 μA, 200 μs). For minimal stimulation experiments, a glass theta-tube electrode was filled with ACSF and positioned in the stratum radiatum along the Schaeffer collaterals ∼20–50 μm from the recorded neurons. Once a synaptic response was obtained, the stimulation intensity was gradually decreased until synaptic failures and synaptic successes could be clearly distinguished in both neurons (typically 0.5–5 μA). Capacitance, series resistance, and input resistance were measured in voltage clamp with a 400 ms, −10 mV step from a −60 mV holding potential (filtered at 30 kHz, sampled at 50 kHz). The capacitance was calculated by first obtaining the decay time constant of a current transient induced by a voltage step (the faster time constant of a double-exponential decay fitted to the first 20 ms) and then dividing this by the series resistance. Cells were only used for analysis if the series resistance was <30 MΩ and was stable throughout the experiment. Input resistance ranged from 75 to 350 MΩ. Data were not corrected for junction potential.
Synaptic potentials were filtered at 2 kHz, acquired and digitized at 10 kHz on a personal computer using custom software (Labview; National Instruments, Austin, TX). Time constants (τ) of the decay of isolated NMDAR EPSCs were determined by fitting the decay of the maximum amplitude of NMDAR EPSC with a double exponential in LabView using the Levenberg-Marquardt algorithm to determine the least-squares set of coefficients that best fit the set of input data points (X,Y) as expressed by a nonlinear function y = f(x,a), where a is the set of coefficients. Miniature EPSCs (mEPSCs) were detected off-line using an automatic detection program (MiniAnalysis; Synaptosoft, Decatur, GA) with a detection threshold set at a value greater than at least 2 SD of the noise values, followed by a subsequent round of visual confirmation. The detection threshold remained constant for the duration of each experiment. Failures and successes were defined as responses with amplitudes less than or >10 pA, respectively, followed by a subsequent round of visual confirmation for each qualification without knowledge of the transfection state of the neuron. The percentage of silent synapses calculated as 1 − ln(failure rate at −60 mV)/ln(failure rate at +50), as described previously (Liao et al., 1995). Significant differences between transfected and nontransfected neurons were determined using a paired t test.
Results
Acute expression of FMRP reduces evoked excitatory synaptic transmission
To determine whether FMRP acutely regulates evoked excitatory synaptic transmission, we acutely expressed a GFP-tagged human FMRP (wtFMRP-GFP) in organotypic hippocampal slice cultures prepared from P6 Fmr1 KO mice. Neurons were biolistically transfected at 3 DIV with wtFMRP-GFP driven via the endogenous human FMR1 promoter to avoid strong overexpression. wtFMRP-GFP displays similar polyribosome association and localization as endogenous FMRP, suggesting that addition of the N-terminal GFP does not affect the protein's function (Antar et al., 2004, 2006; Darnell et al., 2005b). Consistent with the pattern of endogenous FMRP expression, wtFMRP-GFP was punctate throughout the soma and dendritic arborization (Fig. 1A) (Antar et al., 2004). To quantify the levels of wtFMRP-GFP expression after transfection, immunocytochemistry for FMRP in untransfected wild-type and transfected Fmr1 KO slice cultures was performed (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). Somatic wtFMRP-GFP expression in transfected Fmr1 KO neurons was ∼180% of that observed in WT neurons (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material). Although transfected FMRP levels are greater than endogenous FMRP levels in unstimulated WT neurons, these levels are within the physiological range of FMRP induced by experience or neuronal activity (Weiler et al., 1997; Todd and Mack, 2000; Hou et al., 2006).
Figure 1.
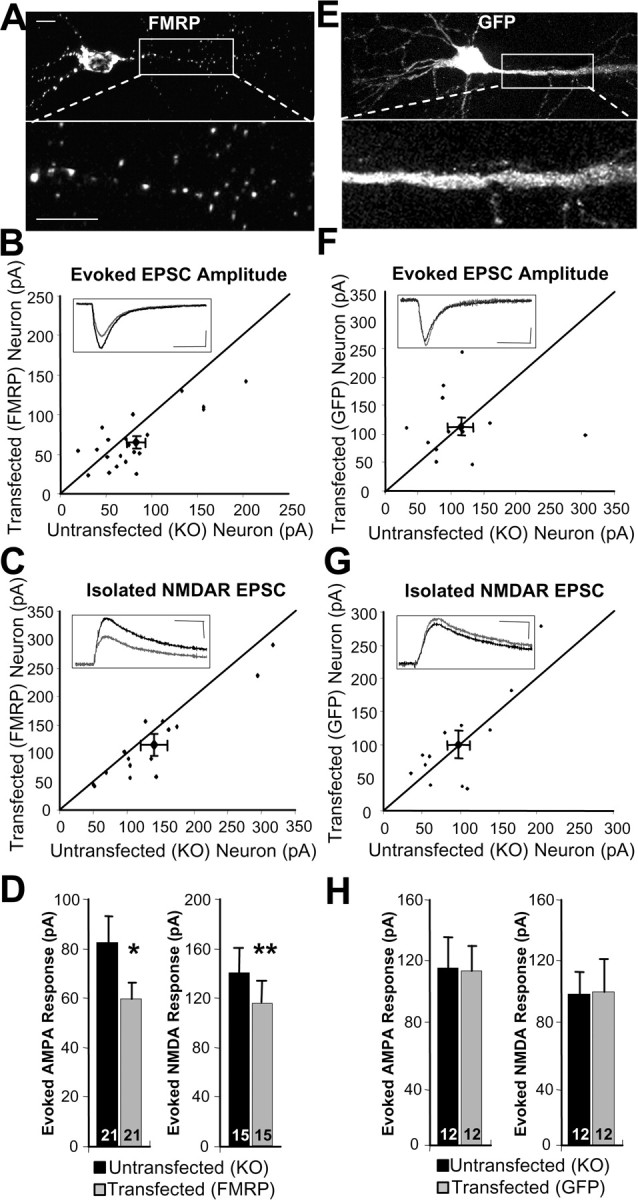
Acute postsynaptic FMRP expression reduces evoked AMPAR and NMDAR mediated EPSCs. A, Expression pattern of FMRP-GFP fusion protein in a CA1 pyramidal neuron in hippocampal slice culture. Scale bars, 10 μm. B, C, Dot plot of the peak amplitude of the evoked AMPAR- (B) or NMDAR-mediated EPSCs (C) of paired recordings from an untransfected Fmr1 KO versus a neighboring FMRP transfected neuron. In this and all figures, the diagonal line represents the values where the EPSC amplitudes from transfected and untransfected cells are equal. The large diamond represents mean ± SEM. Inset, Average of 25 consecutive traces from a representative experiment. Black trace is the untransfected neuron, gray trace is the transfected neuron. Scale bar is 20 pA and 20 ms. Stimulation artifact has been digitally removed for clarity. D, Average AMPAR and NMDAR EPSC amplitude from untransfected KO and FMRP transfected cells. E, As in A, but image shows GFP expression pattern. F, G, As in B and C, but dot plots represent peak evoked AMPAR-mediated (F) and NMDAR-mediated (G) EPSCs from neighboring untransfected and GFP-expressing Fmr1 KO neurons. H, Average AMPAR and NMDAR EPSC amplitude from untransfected KO and GFP transfected cells. In this and all figures, averages are plotted + SEM and n is plotted on each bar. *p < 0.05; **p < 0.01.
Three to 7 d posttransfection (equivalent postnatal day 12–16), simultaneous whole-cell patch-clamp recordings were obtained from untransfected Fmr1 KO and neighboring transfected wtFMRP-GFP-expressing CA1 pyramidal neurons (hereafter referred to as “neuron pairs”). AMPA receptor (AMPAR)-mediated EPSCs were evoked with extracellular stimulation of Schaffer collateral axons and were measured in the neuron pairs held at −60 mV. We observed an ∼20% reduction in the amplitude of EPSCs in neurons transfected with wtFMRP-GFP (Fig. 1B,D). No significant difference was observed between transfected and untransfected neurons in resting membrane potential, input resistance, or whole-cell capacitance (supplemental Table 1, available at www.jneurosci.org as supplemental material), indicating that overall neuronal health, subthreshold membrane conductances, and size were unaffected by transfection of wtFMRP-GFP.
Although AMPA and NMDA receptors typically colocalize at excitatory synapses, they can be differentially regulated (Petralia et al., 1999; Groc and Choquet, 2006). To determine whether wtFMRP-GFP expression affects NMDAR-mediated EPSCs, we measured the peak amplitude of isolated NMDAR-mediated EPSCs in neuron pairs, by voltage clamping neurons at +50 mV in the presence of the AMPAR antagonist, DNQX. Like AMPAR-mediated EPSCs, isolated NMDAR-mediated EPSCs were reduced by ∼20% after FMRP expression (Fig. 1C,D). Although the amplitude of NMDAR-mediated EPSCs was reduced in wtFMRP-GFP-expressing neurons, there was no change in the time constants of decay of the isolated NMDAR-mediated currents (supplemental Table 1, available at www.jneurosci.org as supplemental material). Because the decay rate of NMDAR-mediated currents declines over the course of synapse maturation and is regulated by the NMDAR subunit composition (Cull-Candy et al., 2001), these results indicate that wtFMRP-GFP does not alter NMDAR subunit composition or functional synapse maturation. Transfection of GFP alone in Fmr1 KO slice cultures had no effect on AMPAR- or NMDAR-mediated EPSCs (Fig. 1E–H). These data indicate that acute, postnatal expression of FMRP reduces evoked synaptic transmission.
FMRP reduces the frequency of miniature EPSCs
An FMRP-induced decrease in evoked EPSCs may occur through a decrease in synapse number, strength of individual synapses, or presynaptic release probability. To begin to differentiate between these possibilities, we measured the effects of FMRP on spontaneous (action-potential independent) mEPSCs in neuron pairs in the presence of TTX (1 μm) (Fig. 2A). mEPSC amplitude is indicative of the strength of individual functional synapses, whereas mEPSC frequency is dependent on synapse number as well as presynaptic release probability. The frequency of mEPSCs was reduced by ∼35% in wtFMRP-GFP-expressing cells as compared with untransfected Fmr1 KO neurons, whereas the amplitude of mEPSCs was unchanged (Fig. 2B–D). Neither the frequency nor the amplitude of mEPSCs was altered after GFP expression (Fig. 2E–H). Therefore, postsynaptic FMRP expression reduces synaptic transmission not by reducing the strength of individual synapses, but by either decreasing presynaptic glutamate release probability or the number of synapses.
Figure 2.

Postsynaptic FMRP reduces the frequency, but not amplitude of miniature EPSCs. A, Representative traces of mEPSCs simultaneously recorded from an untransfected Fmr1 KO and a neighboring FMRP transfected neuron. Calibration: 10 pA, 500 ms. B, C, Dot plot representation of the frequency (B) and amplitude (C) of mEPSCs in paired recordings. Inset, Cumulative probability distributions for both untransfected neurons (solid black line) and transfected, FMRP-expressing neurons (dotted line). Each point on the curve represents the average mEPSC frequency and amplitude from an individual neuron. The x-axis is the mEPSC frequency (B) or amplitude (C). The y-axis is the cumulative probability. D, Average mEPSC frequency and amplitude in untransfected KO and FMRP transfected cells. E, Representative traces from untransfected and neighboring GFP-expressing Fmr1 KO neurons. Calibration: 20 pA, 500 ms. F, G, Dot plots of mEPSC frequency (F) and amplitude (G) from neighboring untransfected and GFP-expressing Fmr1 KO CA1 pyramidal neurons. Inset, Cumulative probability distribution for both untransfected neurons (solid black line) and transfected, GFP-expressing neurons (dotted line). H, Average mEPSC frequency and amplitude in untransfected KO and GFP transfected cells. **p < 0.01.
Acute expression of FMRP increases synaptic failures, but does not affect short-term plasticity or the percentage of silent synapses
To determine effects of postsynaptic wtFMRP-GFP expression on glutamate release probability, we measured paired-pulse facilitation of EPSCs after rapid, successive stimuli (Fig. 3A) in neuron pairs. Paired-pulse facilitation is inversely proportional to presynaptic release probability and provides a relative indicator of this measure (Manabe et al., 1993). Although wtFMRP-GFP expression reduced the amplitude of the first EPSC (Fig. 3C,D, S1), similar to our previous results (Fig. 1B), there was no effect on paired-pulse facilitation at a range of interstimulus intervals (Fig. 3B). These results indicate that postsynaptic expression of FMRP does not affect the presynaptic release probabilities of axons providing input to that neuron.
Figure 3.

Postsynaptic FMRP increases synaptic failures, but does not affect paired-pulse facilitation or silent synapses. A, Representative experiment. Raw, Average of 25 consecutive traces simultaneously recorded from an untransfected Fmr1 KO and a neighboring FMRP transfected neuron. Calibration: 20 pA, 20 ms. Scaled, Traces from Raw have been scaled so that the S1 EPSC amplitude of the transfected neuron is equal to that of the untransfected neuron. B, Paired-pulse facilitation values (mean ± SEM) at a range of interstimulus intervals is not different between FMRP transfected and untransfected KO neurons as determined with a repeated measures ANOVA (F(1,30) = 0.642; p = 0.42). C, Dot plot representation of the peak amplitude of the first response (S1) in paired recordings from an untransfected Fmr1 KO neuron and FMRP-transfected neuron (n = 18). D, Average EPSC amplitude for S1. E, Representative minimal stimulation experiment. Evoked responses were obtained in simultaneous recordings from an untransfected Fmr1 KO neuron and FMRP transfected neuron using minimal stimulation. Shown are 25 consecutive traces separated into failures and successes for an FMRP-transfected and untransfected neuron held at −60 mV (left) and +50 mV (right) and an average of the successes. Calibration: 20 pA, 10 ms. F, Average percentage of synaptic failures at −60 mV and +50 mV, the peak amplitude of synaptic successes at −60 mV (synaptic potency), and the percentage of silent synapses in FMRP transfected and untransfected neurons. *p < 0.05; **p < 0.01.
One explanation that can account for the observed reduction in evoked synaptic responses and mEPSC frequency in FMRP expressing neurons, without a change in mEPSC amplitude or presynaptic release probability, is a reduction in the number of functional synaptic connections. To examine this idea, we used a “minimal stimulation” protocol to stimulate a small number of axons, allowing us to compare the relative number of functional synaptic connections one axon makes onto both an Fmr1 KO untransfected and wtFMRP-GFP-expressing neuron. Because of the probabilistic nature of glutamate release, we observed both failures and successes of synaptic transmission (Fig. 3E). Simultaneous voltage-clamp recordings of neuron pairs revealed a ∼60% increase in synaptic failures in neurons expressing wtFMRP-GFP (Fig. 3F). Furthermore, the amplitude of successes, or synaptic potency, was reduced by ∼35% in neurons expressing wtFMRP-GFP (Fig. 3F). Because mEPSC amplitude is unchanged (Fig. 2C,D), the decrease in synaptic potency is likely caused by a decrease in the number of synaptic contacts per axon. It is important to note that the failure rate is also representative of the presynaptic release probability of a synaptic bouton. Therefore, it is possible that the changes we observe in failure rate with wtFMRP-GFP expression could be presynaptic in nature. However, based on the findings that FMRP does not alter paired-pulse facilitation, we hypothesize that FMRP results in a decrease in functional synapse number.
A hallmark of excitatory synapse maturation is the insertion of postsynaptic of AMPARs, which occurs after NMDARs (Wu et al., 1996; Petralia et al., 1999). As a result, there is a developmental decline in the number of “silent synapses,” synapses that contain NMDARs, but not AMPARs (Durand et al., 1996). To determine whether FMRP regulated functional synapse maturation, we measured silent synapses in simultaneous recordings of neuron pairs. Synaptic failures and successes in response to minimal stimulation of presynaptic axons were measured at holding potentials of −60 mV and +50 mV. Expression of wtFMRP-GFP increased synaptic failures measured at both −60 mV and +50 mV, but did not affect the percentage of silent synapses (Fig. 3F). Thus, FMRP expression appears to reduce the number of synaptic connections without altering the function or maturational state of the remaining synapses.
Exogenous and endogenous postnatal expression of FMRP reduces structural synapse number
To examine whether acute FMRP expression resulted in a decrease in the number of structural synapses, we acutely expressed wtFMRP-GFP into dissociated hippocampal cultures prepared from Fmr1 KO mice. After transfection of wtFMRP-GFP, neurons were stained for the AMPAR subunit GluR1 (surface), the postsynaptic marker PSD-95, or the presynaptic marker synapsin. Synapsin puncta were quantified at synapses on dendrites expressing or not expressing FMRP. wtFMRP-GFP expression reduced the number of synapses as measured with all three synaptic markers by 30–40% as compared with neighboring untransfected KO neurons (Fig. 4A–C). wt-FMRP did not affect the fluorescent intensity of remaining synaptic puncta, but did reduce PSD-95 puncta size (supplemental Table 2, available at www.jneurosci.org as supplemental material). As a control for transfection and GFP expression, exogenous expression of two different FMRP-GFP mutants with single site mutations had no effect on synapse number (Figs. 4A–C, 6M,N). These results, together with our functional synapse measurements in slice culture, indicate that acute postsynaptic FMRP expression reduces the number of structural and functional synapses.
Figure 4.
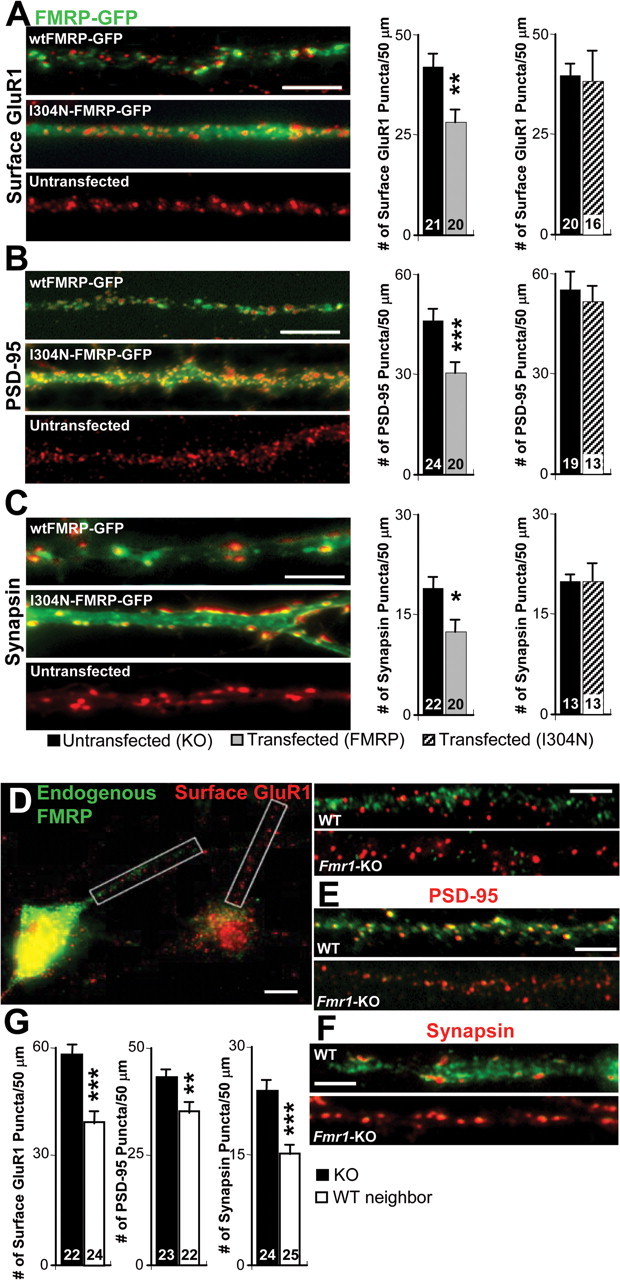
Acute exogenous and endogenous FMRP reduce the number of synapses as measured with immunocytochemical markers. A, Left, Representative dendrites of Fmr1 KO dissociated hippocampal cultures transfected with either wtFMRP-GFP or I304N-FMRP-GFP (green) and labeled for surface GluR1 (red). Right, Average number of surface GluR1 puncta in neurons transfected with either wtFMRP-GFP or I304N-FMRP-GFP and neighboring untransfected KO neurons. B, Left, As in A, except red is PSD-95 immunofluorescence. Right, Average number of PSD-95 puncta. C, Left, As in A, except red is synapsin immunofluorescence. Right, Average number of synapsin puncta. D, Left, Representative image of side-by-side wild-type and Fmr1 KO neurons in culture. Green is endogenous FMRP immunofluorescence. Red is surface GluR1 immunofluorescence. Right, Dendrites of the wild-type and Fmr1 KO neuron. E, As in A, except red is PSD-95 immunofluorescence. F, As A, except red is synapsin immunofluorescence. G, Average number of surface GluR1, PSD-95, and synapsin puncta in wild-type neurons and neighboring Fmr1 KO neurons. *p < 0.05; **p < 0.01; ***p < 0.001. Scale bars: A–C, 10 μm; D, left, 10 μm; D, right, 5 μm; E, F, 10 μm
Figure 6.

Dephosphorylated FMRP reduces synapse function and number. A, Expression pattern of S500A-FMRP-GFP in CA1 pyramidal neuron in hippocampal slice cultures. B, Dot plot of the peak amplitude of the evoked EPSC of paired recordings from untransfected Fmr1 KO and neighboring S500A-FMRP transfected neuron. C, Representative traces of mEPSCs simultaneously recorded from untransfected Fmr1 KO and S500A-FMRP transfected neuron. D, E, Dot plot of the frequency (D) and amplitude (E) of mEPSCs from untransfected Fmr1 KO versus neighboring S500A-FMRP-transfected neurons. Insets, Cumulative probability distribution for both untransfected neurons (solid black line) and transfected S500A-FMRP-expressing neurons (dotted line). F, Group data (mean + SEM) of B, D, and E. G–L, As in A–F, except the transfected neurons express S500D-FMRP-GFP. M, Left, Dendrites of Fmr1 KO dissociated hippocampal cultures transfected with either S500A-FMRP-GFP or S500D-FMRP-GFP (green) and immunofluorescence for PSD-95 (red). Right, Average number of PSD-95 puncta in neurons transfected with either S500A-FMRP-GFP or S500D-FMRP-GFP and neighboring untransfected neurons. N, As in A, except red is synapsin immunofluorescence. On the right is the average number of synapsin puncta. *p < 0.05; **p < 0.01. Scale bars: A, G, N, 10 μm; M, 5 μm. Calibrations: B, inset, 50 pA, 20 ms; C, I, 20 pA, 500 ms; H, inset, 25 pA, 20 ms
In the above experiments, we have used exogenous, acute expression of FMRP to evaluate its effects on synapses. To determine whether WT neurons expressed fewer synapses than Fmr1 KO neurons, we prepared dissociated cocultures containing both WT and Fmr1 KO neurons. This allowed us to make side-by-side comparisons of synapse number on FMRP-expressing (WT) and KO neurons on the same coverslip as performed with wtFMRP-GFP transfected neurons (Fig. 4D). Immunocytochemistry for both FMRP and synaptic markers was performed on these cocultures at 11–12 DIV to identify synapses on WT and Fmr1 KO neurons. As observed with acute FMRP expression, WT neurons displayed 20–35% fewer synapses as measured with all synaptic markers compared with neighboring Fmr1 KO neurons (Fig. 4D–G). WT neurons also displayed smaller PSD-95 puncta similar to wtFMRP-GFP transfected neurons (supplemental Table 2, available at www.jneurosci.org as supplemental material). Importantly, the number of synaptic puncta in WT neurons is not different from Fmr1 KO neurons transfected with wtFMRP-GFP, indicating that the increased synapse number in Fmr1 KO neurons can be rescued by acute, postnatal, exogenous FMRP expression (supplemental Table 2, available at www.jneurosci.org as supplemental material). This data further indicates that decreases in synapse number are not merely a result of acute, exogenous FMRP expression, but reflect the function of endogenous FMRP.
Synaptic function in WT versus Fmr1 KO mice
Our findings of reduced synapse number and function after FMRP expression suggest that WT neurons may have reduced synaptic function compared with Fmr1 KO neurons. We measured evoked AMPAR-mediated EPSCs and mEPSCs in single neurons of WT organotypic slice cultures and compared these values with those obtained from untransfected Fmr1 KO neurons in previous experiments. We failed to detect any differences in synaptic function between WT and Fmr1 KO slice cultures (AMPAR-mediated EPSCs: WT, 109.6 ± 10.3 pA, n = 72; Fmr1 KO, 100.4 ± 5.4 pA, n = 126; p = 0.4; mEPSC frequency: WT, 1.89 ± 0.23 Hz, n = 34; Fmr1 KO, 1.60 ± 0.14 Hz, n = 95; p = 0.3; mEPSC amplitude: WT, 28.2 ± 0.8 pA, n = 34; Fmr1 KO, 25.9 ± 0.6 pA, n = 95; p = 0.1). However, there is a high level of variability in evoked EPSCs and mEPSCs between slice cultures, even between slices prepared from the same animal. Specifically, the mathematical variance and SD were greater (∼40%) between neurons on different slices than between neurons within the same slice. These data suggest that side-by-side comparisons of FMRP expressing and Fmr1 KO neurons are required to detect the 20–35% differences in synaptic function which we observe after acute FMRP transfection. In support of this view, when we minimize variability by coculturing wild-type and Fmr1 KO neurons, we observe a difference in the number of synapses between neighboring neurons (Fig. 4D–G).
The KH2 RNA-binding domain, but not the RGG box is required for FMRP regulation of synapse number and function
FMRP is an RNA-binding protein that binds to as many as 400 brain mRNAs and associates with large polyribosome complexes in brain and synaptoneurosomes (Feng et al., 1997a; O'Donnell and Warren, 2002; Khandjian et al., 2004; Stefani et al., 2004). FMRP interacts with RNA through several RNA-binding motifs, including two heterogeneous nuclear ribonucleoprotein (hnRNP)-KH domains (KH1 and KH2) and an arginine/glycine-rich RNA-binding motif (RGG box) (Darnell et al., 2001, 2005a; Schaeffer et al., 2001). The KH2 domain associates with a specific RNA structure termed a “kissing complex” (Darnell et al., 2005a). A single point mutation in the KH2 domain of FMRP (an Ile to Asn switch at residue 304, originally found in a patient with a particularly severe form of FXS) abolishes FMRP interactions with kissing complex RNAs, but also reduces dimerization with other FMRP molecules and polyribosome association (Feng et al., 1997b; Laggerbauer et al., 2001; Darnell et al., 2005a,b). To test whether the FMRP-dependent reduction in synaptic function required interaction with kissing complex RNAs or polyribosomes, we expressed the I304N-FMRP-GFP construct in Fmr1 KO slice cultures and examined synaptic function. Although the total level of expression of this construct was not different, the expression pattern of I304N-FMRP-GFP in dendrites was more diffuse than wtFMRP-GFP (Figs. 4A–C, 5A, supplemental Table 1, available at www.jneurosci.org as supplemental material). The diffuse expression pattern of I304N-FMRP-GFP in neurons is consistent with previous reports that I304N-FMRP associates with a smaller messenger RNP and is diffusely distributed in cultured cell lines (Feng et al., 1997b; Schrier et al., 2004; Darnell et al., 2005b). Transfection of I304N-FMRP-GFP into Fmr1 KO neurons had no effect on evoked synaptic transmission, mEPSC frequency, or amplitude (Fig. 5B–F). Similarly, I304N-FMRP-GFP expressed in Fmr1 KO dissociated hippocampal cultures did not affect the number, size, or intensity of synapse marker puncta (Fig. 4A–C, supplemental Table 2, available at www.jneurosci.org as supplemental material). These results indicate that postsynaptic interactions of FMRP with polyribosomes or kissing complex mRNAs are required for regulation of synapse number.
Figure 5.
An intact KH2 RNA-binding domain of FMRP, but not an RGG box, is required to reduce synapse function. A, Expression pattern of I304N-FMRP-GFP fusion protein in a CA1 pyramidal neuron in slice culture. B, Dot plot of the peak amplitude of the evoked AMPAR-EPSCs from paired recordings of untransfected Fmr1 KO versus I304N-FMRP transfected neurons. Inset, Representative evoked EPSCs from one neuron pair. C, Representative traces of mEPSCs simultaneously recorded from an untransfected Fmr1 KO and I304N-FMRP transfected neuron. D, E, Dot plot representation of the frequency (D) and amplitude (E) of mEPSCs in Fmr1 KO versus neighboring I304N-FMRP transfected neurons. Inset, Cumulative probability distribution of mEPSC frequency and amplitude for both untransfected neurons (solid black line) and transfected I304N-FMRP-expressing neurons (dotted line). F, Group data (mean + SEM) of B, D, and E. G–L, As in A–F, except the transfected neurons express ΔRGG-FMRP-GFP. *p < 0.05. Scale bars: A, G, 10 μm. Calibrations: B, H, insets, 20 pA, 20 ms; C, I, 10 pA, 500 ms.
The RNA-binding domain of FMRP with the highest reported affinity for RNA is the RGG-box, which associates with a tertiary RNA structure termed a “G-quartet” (Darnell et al., 2001; Schaeffer et al., 2001). To examine whether association of FMRP with G-quartet containing mRNAs is important for the regulation of synapse function by FMRP, a mutant construct was used in which the entire RGG-box was deleted (ΔRGG-FMRP-GFP) (Fig. 5G–L) (Darnell et al., 2005b). When this construct was introduced into Fmr1 KO neurons, the expression level and pattern was similar to wtFMRP-GFP (Fig. 5G, supplemental Table 1, available at www.jneurosci.org as supplemental material). Expression of ΔRGG-FMRP-GFP reduced both the amplitude of evoked AMPAR-mediated responses and the frequency of mEPSCs without altering the amplitude of mEPSCs (Fig. 5H–L), implying that, like wild-type FMRP, this mutant construct reduced the number of functional synaptic connections. These results suggest that postsynaptic FMRP interactions with G-quartet containing RNAs are not required to regulate synapse number.
A nonphosphorylatable form of FMRP at S500 mimics wild-type FMRP and induces synapse loss
Our data suggest that acute expression of FMRP in Fmr1 KO neurons decreases synapse number through the ability of FMRP to regulate translation, as expression of the I304N-FMRP-GFP construct, which is unable to regulate translation, has no effect on synapse number. In mammalian neurons, FMRP is associated with translating polyribosomes and is required for glutamate stimulated translation at synapses, suggesting FMRP functions to allow or stimulate translation (Todd et al., 2003; Stefani et al., 2004; Weiler et al., 2004; Hou et al., 2006). However, in vitro FMRP suppresses translation of its target mRNAs (Laggerbauer et al., 2001; Li et al., 2001; Sung et al., 2003), and the brains of Fmr1 KO mice have elevated protein synthesis rates in vivo, and increased levels of specific proteins (Sung et al., 2003; Zalfa et al., 2003; Qin et al., 2005; Hou et al., 2006). These results are more consistent with a role for FMRP as a translational suppressor. Furthermore, translational suppression of target mRNAs by the Drosophila homolog of FMRP, dFXR, is implicated in regulation of axonal and dendritic development (Zhang et al., 2001; Lee et al., 2003). Previously, it has been proposed that phosphorylation of FMRP on a conserved serine residue (Ser500 in human FMRP) switches FMRP from a translational activator to a suppressor (Ceman et al., 2003). An FMRP mutant, S500A-FMRP, which cannot be phosphorylated (mimicking the dephosphorylated state), is largely associated with actively translating polyribosomes, whereas a phosphorylation site mimic of FMRP, S500D-FMRP, is more strongly associated with stalled polyribosomes. To determine whether FMRP association with stalled or translating polysomes resulted in synapse loss, we transfected either of the phosphorylation site FMRP mutants (S500A-FMRP-GFP or S500D-FMRP-GFP) into Fmr1 KO slice cultures. Both S500A-FMRP-GFP and S500D-FMRP-GFP displayed a similar expression level and punctate expression pattern to wtFMRP-GFP (Fig. 6A,G, supplemental Table 1, available at www.jneurosci.org as supplemental material). S500A-FMRP-GFP expression resulted in a 35–40% decrease in both evoked AMPAR EPSCs and mEPSC frequency, similar to wtFMRP-GFP, with no change in mEPSC amplitude (Fig. 6B–F) or paired-pulse facilitation (supplemental Table 1, available at www.jneurosci.org as supplemental material). Consistent with its effects on synapse function in slice cultures, transfection of S500A-FMRP-GFP into dissociated Fmr1 KO neuron cultures reduced synapse number by 25–30% as measured with immunocytochemistry for presynaptic and postsynaptic markers, similar to wtFMRP-GFP (Fig. 6M,N). In contrast, S500D-FMRP-GFP had no effect on any measure of synaptic function or number (Fig. 6G–N, supplemental Table 1, available at www.jneurosci.org as supplemental material). Therefore, S500A-FMRP-GFP mimics wtFMRP-GFP with regard to synapse loss and suggests that at least some of wtFMRP-GFP exists in a dephosphorylated state which leads to a reduction in synapse number. Furthermore, these data imply that FMRP association with translating polyribosomes leads to synapse loss.
Discussion
FMRP negatively regulates synapse number
Changes in dendritic spine number and shape have been observed in FXS patients and Fmr1 KO mice, both in vivo and in cultures (Braun and Segal, 2000; Irwin et al., 2001; Nimchinsky et al., 2001; Bagni and Greenough, 2005; Antar et al., 2006; Grossman et al., 2006a,b). How these morphological changes relate to functional synapses and whether FMRP plays a direct role in determining synapse number or function was unknown. Here, we demonstrate that acute expression of FMRP in Fmr1-KO neurons at near-endogenous levels reduces synapse number without changing the function or maturational state of the remaining synapses. Furthermore, Fmr1-KO neurons have more synapses compared with their wild-type neighbors in dissociated culture and acute postnatal expression of FMRP is sufficient to reverse this overabundance. Our results are supported by observations of increased dendritic spine number on cortical neurons of FXS patients and Fmr1 KO mice that have developed in vivo (Irwin et al., 2001; Nimchinsky et al., 2001; Bagni and Greenough, 2005; Grossman et al., 2006b). Our results extend on these structural studies and find that postsynaptic FMRP directly regulates functional synapse number.
Does FMRP regulate synapse formation, maturation, or pruning?
The abundance of filopodial-like spines in FXS patients and Fmr1 KO mice have prompted the idea that synapses in Fmr1 KO are more immature (Braun and Segal, 2000; Irwin et al., 2001, 2002; Nimchinsky et al., 2001; Antar et al., 2006; Grossman et al., 2006b). Acute FMRP expression did not effect short-term plasticity, mEPSC amplitude, the decay of NMDAR EPSCs, or percentage of silent synapses (Figs. 1–3), indicating that FMRP is not regulating presynaptic release probability, the strength of individual synapses, or their functional maturation. FMRP may play a role in the earliest stages of synapse development that occur before glutamate receptor insertion, making it less likely that a synapse is created (Waites et al., 2005). Alternatively, FMRP may facilitate elimination or pruning synapses after they are formed. Drosophila FMRP (dFXR) reduces synaptic bouton number at the neuromuscular junction as well as dendritic arborization in the CNS (Zhang et al., 2001; Pan et al., 2004), which could also be attributable to either inhibition of growth or pruning. In support of a role for FMRP in dendritic pruning, spiny stellate cells in the barrel cortex of Fmr1 KO mice fail to prune their dendrites from the barrel septa (Galvez and Greenough, 2005). More experiments are needed to determine the roles for FMRP in synapse formation and elimination.
Here, we observed that acute postsynaptic FMRP expression resulted in a reduction in synapse function and number compared with neighboring untransfected Fmr1 KO neurons (Figs. 1–4). Similarly, fewer synapses were observed in WT neurons in dissociated neuron cultures compared with their Fmr1 KO neighbors, indicating that our results with acute expression of FMRP reflect endogenous FMRP function (Fig. 4). Because the effects of acute FMRP expression on synapse function are moderate (20–35%), the large variability across slice culture preparations may make it difficult to detect differences in evoked EPSCs and mEPSCs between WT and Fmr1 KO cultures. It may be necessary to make comparisons between neighboring neurons in the same culture. Alternatively, or in addition, there may be intercellular interactions between FMRP-expressing and Fmr1 KO neurons, which result in Fmr1 KO neurons having more synapses. This would be relevant to females with FXS who have a mosaic expression of FMRP.
FMRP regulates synapse number through postsynaptic KH2 domain interactions
Previous work has focused on two RNA tertiary structures that are important for FMRP-RNA interactions. The RGG box of FMRP binds with high specificity and affinity to mRNAs containing a G-quartet structure and is implicated in their translational suppression (Brown et al., 2001; Darnell et al., 2001; Schaeffer et al., 2001), whereas the KH domains KH1 and KH2 interact with kissing complex RNA structures (Darnell et al., 2005a). FMRP with a deleted RGG box (ΔRGG-FMRP) functioned just as well as wtFMRP in reduction of synapse number (Fig. 5). This result suggests that interactions of FMRP with postsynaptic G-quartet-containing mRNAs such as microtubule-associated protein 1b (MAP1b), or the PSD-95-associated protein SAPAP4 are not required for FMRP regulation of synapse number in mammals (Brown et al., 2001; Darnell et al., 2001). In contrast to the RGG box deletion, a single point mutation in the KH2 domain (I304N FMRP) abolished the effects of FMRP on synapse number. I304N FMRP, which was first discovered in a patient with a particularly severe form of FXS (De Boulle et al., 1993), fails to dimerize and associate with large translating polyribosomes (Feng et al., 1997b; Laggerbauer et al., 2001; Darnell et al., 2005b). I304N FMRP abolishes binding to kissing complex RNAs and no longer acts as a translational suppressor (Laggerbauer et al., 2001; Darnell et al., 2005a). Thus, it is likely that FMRP regulation of translation, either through interaction with kissing-complex-containing mRNAs or polyribosomes, is critically important for the ability of FMRP to reduce synapse number. In addition, whereas wild-type FMRP and all other mutant constructs of FMRP displayed a punctate expression pattern, the I304N-FMRP mutant construct was much more diffuse (Figs. 4, 5), as observed in cultured cell lines (Schrier et al., 2004; Darnell et al., 2005b). These puncta observed with endogenous or wtFMRP-GFP may be polyribosomes or RNA granules because they contain RNA and move via microtubules in dendrites (Antar et al., 2004, 2005; Kanai et al., 2004). This result indicates that FMRP association with polyribosomes and/or RNA granules is required to induce synapse loss and implicates translational regulation of FMRP target mRNAs in this process.
FMRP mutant that associates with translating polyribosomes leads to synapse loss
FMRP has been implicated in both translational suppression and activation. Such dual translational regulation by RNA-binding proteins is important to mediate specific and localized protein expression (Huang and Richter, 2004; Kindler et al., 2005; Wells, 2006). Phosphorylation of RNA-binding proteins such as cytoplasmic polyadenylation-binding protein (CPEB) and zip code-binding protein (ZBP) is a mechanism by which the switch from repression to activation (or derepression) occurs (Huang and Richter, 2004; Huttelmaier et al., 2005; Wells, 2006). Like CPEB and ZBP, phosphorylation of FMRP on a conserved serine (406 in Drosophila, 499 in mouse, and 500 in human) has been suggested to be a mechanism to regulate translational suppression and activation by FMRP. In vitro phosphorylation of Drosophila FMR affected binding to poly U, but not poly G RNA (Siomi et al., 2002). However, mimicking phosphorylation of murine FMRP at Ser499 with Asp does not affect binding to known target mRNAs, but rather increases association of FMRP with stalled polysomes in vivo. Substitution of Ala for Ser, thereby mimicking the dephosphorylated form, recruited FMRP to translating polysomes (Ceman et al., 2003). From these findings, it was suggested that in the phosphorylated state, FMRP binds mRNAs that are initiated but not actively elongating (stalled polysomes); FMRP dephosphorylation results in a conformational change to facilitate elongation (Ceman et al., 2003). A dephosphomimic of FMRP (S500A) reduces synapse function and number in the same manner as wild-type FMRP, whereas the phosphomimic of FMRP (S500D) had no effect (Fig. 6). These results suggest that the ability of FMRP to regulate synapse number requires association with translating polyribosomes, whereas suppression of protein synthesis prevents synaptic regulation. Although we cannot rule out the possibility that the S500 mutation affects other aspects of FMRP function, we propose a model in which FMRP dephosphorylation and stimulation of translation of mRNA target(s) leads to synapse loss. This idea is in contrast to conclusions in the dFXR-null fly that translational suppression of MAP1b or Rac1 is thought to lead to a reduction in presynaptic and postsynaptic structures, respectively (Zhang et al., 2001; Lee et al., 2003). However, it is likely in both species that FMRP-mediated translational suppression and activation are important for proper synapse development. Determining how FMRP phosphorylation is regulated and, in turn, how FMRP phosphorylation regulates translation in neurons and dendrites will provide more detailed insight into how FMRP controls synapse number.
Translational regulation by FMRP is also implicated in acute long-term synaptic depression (LTD) in mature neurons (>P21) (Huber et al., 2002; Koekkoek et al., 2005; Hou et al., 2006; Nosyreva and Huber, 2006). Therefore, FMRP may regulate translation of the same or different proteins to coordinate rapid changes in function with long-term changes in synapse structure. Because FMRP binds to as many as 4% of brain mRNAs, the challenge now lies in determining which mRNAs of postsynaptically expressed proteins are regulated by FMRP and lead to LTD and synapse loss (Sung et al., 2000; Brown et al., 2001; Miyashiro et al., 2003). We would anticipate that these mRNA targets are dendritically expressed and require an intact KH2 domain of FMRP for translational regulation.
Footnotes
This work was supported by National Institutes of Health Grants 1F31NS050992 (B.E.P.) and NS045711 (K.M.H.), and a McKnight Foundation and FRAXA Research Foundation grant to K.M.H. K.M.H. is a Southwestern Medical Foundation endowed scholar in biomedical research. We thank Drs. Robert and Jennifer Darnell for the FMRP-GFP plasmids and discussions, Drs. Justin Fallon and Jennifer Darnell for 2F5 FMRP antibody, Maggie Waung for assistance with the dissociated neuron cultures, Dr. Jay Gibson for assistance with dual patch clamping, and members of the Huber lab and Dr. Ege Kavalali for discussions and comments on this manuscript.
References
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and Fmr1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci. 2006 doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Braun K, Segal M. FMRP involvement in formation of synapses among cultured hippocampal neurons. Cereb Cortex. 2000;10:1045–1052. doi: 10.1093/cercor/10.10.1045. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005a;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Mostovetsky O, Darnell RB. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005b;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Durand GM, Kovalchuk Y, Konnerth A. Long-term potentiation and functional synapse induction in developing hippocampus. Nature. 1996;381:71–75. doi: 10.1038/381071a0. [DOI] [PubMed] [Google Scholar]
- Feng Y. Fragile X mental retardation: misregulation of protein synthesis in the developing brain? Microsc Res Tech. 2002;57:145–147. doi: 10.1002/jemt.10063. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997a;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997b;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Gabel LA, Won S, Kawai H, McKinney M, Tartakoff AM, Fallon JR. Visual experience regulates transient expression and dendritic localization of fragile X mental retardation protein. J Neurosci. 2004;24:10579–10583. doi: 10.1523/JNEUROSCI.2185-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez R, Greenough WT. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am J Med Genet A. 2005;135:155–160. doi: 10.1002/ajmg.a.30709. [DOI] [PubMed] [Google Scholar]
- Gibson JR, Bartley AF, Huber KM. Role for the subthreshold currents ILeak and IH in the homeostatic control of excitability in neocortical somatostatin-positive inhibitory neurons. J Neurophysiol. 2006;96:420–432. doi: 10.1152/jn.01203.2005. [DOI] [PubMed] [Google Scholar]
- Groc L, Choquet D. AMPA and NMDA glutamate receptor trafficking: multiple roads for reaching and leaving the synapse. Cell Tissue Res. 2006;326:423–438. doi: 10.1007/s00441-006-0254-9. [DOI] [PubMed] [Google Scholar]
- Grossman AW, Aldridge GM, Weiler IJ, Greenough WT. Local protein synthesis and spine morphogenesis: fragile X syndrome and beyond. J Neurosci. 2006a;26:7151–7155. doi: 10.1523/JNEUROSCI.1790-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AW, Elisseou NM, McKinney BC, Greenough WT. Hippocampal pyramidal cells in adult Fmr1 knockout mice exhibit an immature-appearing profile of dendritic spines. Brain Res. 2006b;1084:158–164. doi: 10.1016/j.brainres.2006.02.044. [DOI] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Huang YS, Richter JD. Regulation of local mRNA translation. Curr Opin Cell Biol. 2004;16:308–313. doi: 10.1016/j.ceb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile-X mental retardation. Proc Natl Acad Sci USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, Swain RA, Weiler IJ, Greenough WT. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Idupulapati M, Gilbert ME, Harris JB, Chakravarti AB, Rogers EJ, Crisostomo RA, Larsen BP, Mehta A, Alcantara CJ, Patel B, Swain RA, Weiler IJ, Oostra BA, Greenough WT. Dendritic spine and dendritic field characteristics of layer V pyramidal neurons in the visual cortex of fragile-X knockout mice. Am J Med Genet. 2002;111:140–146. doi: 10.1002/ajmg.10500. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Huot ME, Tremblay S, Davidovic L, Mazroui R, Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc Natl Acad Sci USA. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koekkoek SK, Yamaguchi K, Milojkovic BA, Dortland BR, Ruigrok TJ, Maex R, De Graaf W, Smit AE, VanderWerf F, Bakker CE, Willemsen R, Ikeda T, Kakizawa S, Onodera K, Nelson DL, Mientjes E, Joosten M, De Schutter E, Oostra BA, Ito M, et al. Deletion of FMR1 in Purkinje cells enhances parallel fiber LTD, enlarges spines, and attenuates cerebellar eyelid conditioning in Fragile X syndrome. Neuron. 2005;47:339–352. doi: 10.1016/j.neuron.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel E, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Hessler NA, Malinow R. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature. 1995;375:400–404. doi: 10.1038/375400a0. [DOI] [PubMed] [Google Scholar]
- Manabe T, Wyllie DJ, Perkel DJ, Nicoll RA. Modulation of synaptic transmission and long-term potentiation: effects on paired pulse facilitation and EPSC variance in the CA1 region of the hippocampus. J Neurophysiol. 1993;70:1451–1459. doi: 10.1152/jn.1993.70.4.1451. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Biolistic transfection of cultured organotypic brain slices. Methods Mol Biol. 2004;245:197–206. doi: 10.1385/1-59259-649-5:197. [DOI] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosyreva ED, Huber KM. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J Neurophysiol. 2006;95:3291–3295. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Pan L, Zhang YQ, Woodruff E, Broadie K. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr Biol. 2004;14:1863–1870. doi: 10.1016/j.cub.2004.09.085. [DOI] [PubMed] [Google Scholar]
- Petralia RS, Esteban JA, Wang YX, Partridge JG, Zhao HM, Wenthold RJ, Malinow R. Selective acquisition of AMPA receptors over postnatal development suggests a molecular basis for silent synapses. Nat Neurosci. 1999;2:31–36. doi: 10.1038/4532. [DOI] [PubMed] [Google Scholar]
- Pickett J, London E. The neuropathology of autism: a review. J Neuropathol Exp Neurol. 2005;64:925–935. doi: 10.1097/01.jnen.0000186921.42592.6c. [DOI] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrier M, Severijnen LA, Reis S, Rife M, van't Padje S, van Cappellen G, Oostra BA, Willemsen R. Transport kinetics of FMRP containing the I304N mutation of severe fragile X syndrome in neurites of living rat PC12 cells. Exp Neurol. 2004;189:343–353. doi: 10.1016/j.expneurol.2004.05.039. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Higashijima K, Ishizuka A, Siomi H. Casein kinase II phosphorylates the fragile X mental retardation protein and modulates its biological properties. Mol Cell Biol. 2002;22:8438–8447. doi: 10.1128/MCB.22.24.8438-8447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Sung YJ, Conti J, Currie JR, Brown WT, Denman RB. RNAs that interact with the fragile X syndrome RNA binding protein FMRP. Biochem Biophys Res Commun. 2000;275:973–980. doi: 10.1006/bbrc.2000.3405. [DOI] [PubMed] [Google Scholar]
- Sung YJ, Dolzhanskaya N, Nolin SL, Brown T, Currie JR, Denman RB. The fragile X mental retardation protein FMRP binds elongation factor 1A mRNA and negatively regulates its translation in vivo. J Biol Chem. 2003;278:15669–15678. doi: 10.1074/jbc.M211117200. [DOI] [PubMed] [Google Scholar]
- Todd PK, Mack KJ. Sensory stimulation increases cortical expression of the fragile X mental retardation protein in vivo. Brain Res Mol Brain Res. 2000;80:17–25. doi: 10.1016/s0169-328x(00)00098-x. [DOI] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci USA. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites CL, Craig AM, Garner CC. Mechanisms of vertebrate synaptogenesis. Annu Rev Neurosci. 2005;28:251–274. doi: 10.1146/annurev.neuro.27.070203.144336. [DOI] [PubMed] [Google Scholar]
- Weiler IJ, Greenough WT. Synaptic synthesis of the Fragile X protein: possible involvement in synapse maturation and elimination. Am J Med Genet. 1999;83:248–252. doi: 10.1002/(sici)1096-8628(19990402)83:4<248::aid-ajmg3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler IJ, Spangler CC, Klintsova AY, Grossman AW, Kim SH, Bertaina-Anglade V, Khaliq H, de Vries FE, Lambers FA, Hatia F, Base CK, Greenough WT. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc Natl Acad Sci USA. 2004;101:17504–17509. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG. RNA-binding proteins: a lesson in repression. J Neurosci. 2006;26:7135–7138. doi: 10.1523/JNEUROSCI.1795-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Malinow R, Cline HT. Maturation of a central glutamatergic synapse. Science. 1996;274:972–976. doi: 10.1126/science.274.5289.972. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]



