Abstract
Silent information regulator-2 (SIR2) proteins regulate lifespan of diverse organisms, but their distribution and roles in the CNS remain unclear. Here, we show that sirtuin 2 (SIRT2), a mammalian SIR2 homolog, is an oligodendroglial cytoplasmic protein and localized to the outer and juxtanodal loops in the myelin sheath. Among cytoplasmic proteins of OLN-93 oligodendrocytes, α-tubulin was the main substrate of SIRT2 deacetylase. In cultured primary oligodendrocyte precursors (OLPs), SIRT2 emergence accompanied elevated α-tubulin acetylation and OLP differentiation into the prematurity stage. Small interfering RNA knockdown of SIRT2 increased the α-tubulin acetylation, myelin basic protein expression, and cell arbor complexity of OLPs. SIRT2 overexpression had the opposite effects, and counteracted the cell arborization-promoting effect of overexpressed juxtanodin. SIRT2 mutation concomitantly reduced its deacetylase activity and its impeding effect on OLP arborization. These results demonstrated a counterbalancing role of SIRT2 against a facilitatory effect of tubulin acetylation on oligodendroglial differentiation. Selective SIRT2 availability to oligodendroglia may have important implications for myelinogenesis, myelin–axon interaction, and brain aging.
Keywords: sirtuin 2, tubulin acetylation/deacetylation, oligodendrocyte differentiation, myelination, RNA interference, cytoskeleton
Introduction
Yeast silent information regulator 2 (SIR2) protein regulates transcription silencing at ribosomal DNA, HM mating type loci, and telomeres (Shore et al., 1984; Schnell and Rine, 1986; Smith and Boeke, 1997; Imai et al., 2000). It suppresses the generation of toxic extrachromosomal DNA circles, and mediates the lifespan extension by calorie restriction (Sinclair and Guarente, 1997; Lin et al., 2002). In Caenorhabditis elegans, acquisition of an additional copy of Sir2.1 gene similarly results in long lifespan beyond the wild-type range (Tissenbaum and Guarente, 2001). In mammals, SIR2 is represented by seven homologs (named sirtuins), among which sirtuin 1 (SIRT1) has been shown to be related with calorie restriction-induced antiaging effects in rodents and in cultured human cells (Cohen et al., 2004; Picard et al., 2004). Apart from histones, studies have identified nonhistone substrates and corresponding functions of sirtuins. SIRT1 deacetylates p53 tumor suppressor protein to promote cell survival (Luo et al., 2001; Vaziri et al., 2001). It may also bind other transcription factors such as Ku70, NF-κB (nuclear factor-κB), FOXO (Forkhead box class O), and HIC1 (hypermethylated in cancer 1) to repress expression of other genes or itself (Cohen et al., 2004; Motta et al., 2004; Yeung et al., 2004; Chen et al., 2005). Furthermore, SIRT1 is capable to repress terminal differentiation of myocytes (Fulco et al., 2003). SIRT2 has been shown to deacetylate α-tubulin and control mitotic exit of cell cycles (Dryden et al., 2003; North et al., 2003).
Several lines of evidences have suggested the importance of sirtuins in the CNS. Studies have implicated histone deacetylases (HDACs), to which SIR2 belongs (Gray and Ekstrom, 2001), in regulating neural/glial development and axonal degeneration. Inhibition of class I and class II HDACs not only induced adult neural progenitors to differentiate toward neurons as opposed to a fate toward glia (Hsieh et al., 2004), but also delayed oligodendrocyte (OL) lineage progression (Marin-Husstege et al., 2002; Shen et al., 2005). As in the other systems, HDACs may also participate in the formation of transcriptional complexes to regulate neuronal or glial differentiation (Ballas et al., 2001). Moreover, SIRT1 seems involved in the delayed Wallerian degeneration in the peripheral nervous system of C57BL/Wlds mouse (Araki et al., 2004; Wang et al., 2005). More recently, proteomic studies have revealed the presences of SIRT2 in myelin sheath (Vanrobaeys et al., 2005; Roth et al., 2006). However, the exact distribution and functional roles of SIR2 in the CNS are still mostly unexplored.
In the present study, we aimed at mapping SIRT2 expression, deciphering its possible roles, and elucidating the molecular mechanisms underlying its effects in the rat CNS. Our results indicated that SIRT2 was an oligodendroglial protein. It impeded oligodendroglial differentiation through its tubulin deacetylation activity and might function to prevent overdifferentiation or early aging of the cells. Preliminary results of the present study have been reported previously in abstract form (Cao et al., 2004).
Materials and Methods
Cloning, in vitro expression, and small interfering RNA knockdown of rat SIRT2.
For details, refer to the study by Zhang et al. (2005). Briefly, cDNA clones from a rat brain cDNA library (Invitrogen, Carlsbad, CA) were sequenced and analyzed by Blast searches (Altschul et al., 1997). Digoxigenin-labeled cRNA probes were synthesized by in vitro transcription against 274 unannotated cDNA sequences, and used for in situ hybridization histochemistry (ISH) on CNS histological sections. Sirt2 was identified for additional characterization because of the predominant expression of the mRNA in neuroglial cells.
The nucleotides 625–1053 of rat Sirt2 open reading frame (ORF), encoding a C-terminal SIRT2 fragment (SIRT2c) was amplified by PCR and subcloned into pET41a (Novagen, Madison, WI) for in vitro expression. The GST-SIRT2c fusion protein was glutathione S-transferase (GST) affinity purified and used to immunize adult New Zealand White rabbits (n = 2) for the generation of anti-SIRT2 polyclonal antibody. For expression in mammalian cells, ORF sequence of Sirt2 cDNA was amplified by PCR and cloned into pXJ40 (Manser et al., 1997) or pEGFP-C1 (Clontech, Palo Alto, CA), creating pXJ-Sirt2 and pEGFP-Sirt2, respectively. Point mutations at SIRT2 amino acid residue 131 (N131A), 133 (D133A), 150 (H150Y), were generated with Genetailor site-directed mutagenesis kit (Invitrogen).
Three small interfering RNA (siRNA) duplexes against rat SIRT2 (NM_001008368) at nucleotides 518–540, 1390–1412, and 1927–1949 were commercially synthesized (1stBase, Singapore). Transfection reagents were used for introducing the mammalian cell expression plasmids and siRNAs into cultured oligodendrocytes (see below). siRNAs for HDAC6 (nucleotides 128–150; accession number XM_228753) and juxtanodin (JN) (nucleotides 3235–3257; accession number DQ119821), both of which showed no expression in oligodendrocyte precursors (OLPs) of the present differentiation stages, were used as controls.
Antibodies.
For purifying the rabbit anti-SIRT2 polyclonal antibody, the GST-SIRT2c recombinant protein was firstly resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (PerkinElmer, Boston, MA). Then, the membrane slices that contained the recombinant protein were excised, minced, and used for affinity purification of anti-SIRT2 antibody.
Anti-acetylated α-tubulin 6-11B-1 [immunocytochemistry (ICC), 1:1000; immunoblotting (IB), 1:2000], anti-α-tubulin B-5-1-2 (ICC, 1:1000; IB, 1:2000), anti-FLAG (ICC, 1:1000; IB, 1:2000), anti-β-actin (IB, 1:5000), anti-pan-sodium channel (NavP) (ICC, 1:200), and anti-neurofilament 200 (NF200) (ICC, 1:1000) were all purchased from Sigma (St. Louis, MO). Anti-pan-actin C4 (IB, 1:2000), anti-myelin basic protein (MBP) (ICC, 1:200), and anti-glial fibrillary acidic protein (GFAP) (ICC, 1:200) were obtained from Chemicon (Temecula, CA). Other primary antibodies included anti-3′-cyclic nucleotide-3′-phosphodiesterase (CNP) (ICC, 1:200; IB, 1:400; US Biological, Swampscott, MA), anti-OX42 (ICC, 1:50; Harlan Sera-Lab, Sussex, UK), anti-potassium channel Kv1.2 (ICC, 1:300; Upstate Biotech, Charlottesville, VA), anti-enhanced green fluorescent protein (EGFP) (ICC, 1:500; IB, 1:1000; Invitrogen), anti-lysine-14-acetylated histone H3 (IB, 1:1000; Upstate Biotech), anti-lysine-16-acetylated histone H4 (IB, 1:1000; Serotec, Oxford, UK), and anti-acetylated lysine antibody (IB, 1:250; Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies comprised goat anti-rabbit and/or -mouse IgG conjugated to alkaline phosphatase (Chemicon) or biotin (Vector Laboratories, Burlingame, CA), and Alexa Fluor 488- or 568-conjugated goat anti-rabbit or -mouse IgG or IgM antibodies (Invitrogen).
Experimental animals.
All animal used in this study were approved by the Institutional Animal Care and Use Committee at the National University of Singapore. For antibody generation, two New Zealand White rabbits were injected with antigen every 2 weeks (described above). Ten days after the sixth injection, blood was withdrawn from the rabbits under anesthesia (ketamine, 35 mg, and xylazine, 5 mg/kg of body weight). Killing was then performed by intracardiac injection of Nembutal (100 mg/kg). For histology, adult Wistar rats were anesthetized and killed with Nembutal (100 mg/kg of body weight), and subsequently perfused with saline and 3% paraformaldehyde (or 4% for immunoelectron microscopy). Brain sections of the CNS were then prepared on a cryostat (for light microscopy) or vibratome (for immunoelectron microscopy). For Western blots or primary cell culture, the animals were anesthetized and killed with Nembutal (100 mg/kg of body weight), and their CNS or cerebra were dissected for extracting total protein or primary OLP cells.
Western blotting, immunoprecipitation, and in vitro tubulin deacetylase assay.
For Western blotting, samples were resolved by SDS-PAGE and transferred to PVDF membranes (PerkinElmer). After being blocked by nonfat milk (Bio-Rad, Hercules, CA), membranes were probed with designated primary antibodies at room temperature overnight. The membrane was then washed and incubated with alkaline phosphatase-conjugated secondary antibody. Immunodetection was performed using CDP-Star chemiluminescence reagent (Roche, Basel, Switzerland). For immunoprecipitation, 2 mg of transfected OLN-93 cell lysate, generated using M-PER Mammalian Protein Extraction Reagent (Pierce, Rockford, IL) supplemented with protease inhibitor mixture (Pierce), was first precleared with 30 μl of protein A-agarose (Amersham Biosciences, Piscataway, NJ) at 4°C for 3 h. The supernatant after that was collected and incubated sequentially with 6 μg of anti-EGFP for 8 h at 4°C and 50 μl of protein A-agarose for 4 h at 4°C. The immunoprecipitates were finally collected by centrifugation. For in vitro α-tubulin deacetylase assay, refer to the study by North et al. (2003). Briefly, we resuspended the immunoprecipitate as described above in 150 μl of deacetylase buffer (50 mm Tris-HCl, pH 9.0, 4 mm MgCl2, and 0.2 mm DTT) and divided it into portions of 50 μl each to which 40 μg of OLN-93 cell lysate was added together with nicotinamide adenosine dinucleotide (NAD) and/or nicotinamide (NAM). After incubation for 2 h at room temperature, reactions were terminated by adding SDS-PAGE sample buffer.
Cell cultures and siRNA or plasmid transfection.
Isolation and culture of primary OLPs followed previous protocols (McCarthy and de Vellis, 1980; Armstrong, 1998). Briefly, cerebra of 1- to 2-d-old Wistar rats were dissected and minced, and the dissociated cell suspension was cultured for ∼10 d in DMEM (Sigma) plus 10% FCS, 1 mm sodium pyruvate, and 25 μg/ml gentamicin before immature OLPs were isolated by shaking and selective detachment. The OLPs were then maintained proliferating for 1–3 d in DMEM plus sodium pyruvate (1 mm), gentamicin (25 μg/ml), insulin (5 μg/ml), transferrin (5 μg/ml), selenium (30 nm), glutamine (2 mm), T4 (thyroxine) (0.4 μg/ml), T3 (triiodothyronine) (0.3 μg/ml), FCS (0.5%), PDGF (platelet-derived growth factor) (10 ng/ml; Sigma), and bFGF (basic fibroblast growth factor) (10 ng/ml; Invitrogen). One to 6 d before transfection or fixation for immunocytochemistry, the mitogens were withdrawn from the medium to allow the cells to differentiate on poly-d-lysine (10 μg/ml; Sigma) coated Thermanox pretreated coverslips (Nunc, Roskilde, Denmark) in the culture plates. OLN-93 cells were grown in DMEM supplemented with 10% FCS.
The primary OLPs and OLN-93 cells were transfected using Effectene Transfection Reagent (Qiagen, Hilden, Germany) or Lipofectamine RNAiMax (Invitrogen) as instructed by the manufacturers. NAM or NAD (Sigma) was added at the indicated concentrations. Forty-eight hours after transfection, cells were harvested and lysed for Western blotting by M-PER Mammalian Protein Extraction Reagent (Pierce), or fixed with 3% paraformaldehyde for immunocytochemistry.
Immunocytochemistry, double labeling, and immunoelectron microscopy.
For details, refer to the studies by Liang et al. (2000) and Zhang et al. (2005). Briefly, for in situ hybridization histochemistry, in vitro-transcribed digoxigenin-labeled cRNA probe was generated using the same Sirt2 cDNA (as template) as for antibody development (nucleotides 625–1053 of the open reading frame). It was hybridized to histological sections of rat CNS at 0.6 μg/ml concentration for overnight at 62°C. The hybridization signals were finally detected by alkaline phosphatase-conjugated anti-digoxigenin antibody and the NBT (nitroblue tetrazolium)-BCIP (5-bromo-4-chloro-3′-indolylphosphate) reaction. For immunofluorescence double/triple labeling, histological sections of the rat CNS or fixed cells were washed with PBS and preincubated with PBS-T-NGS (0.1 m PBS, 0.3% Triton X-100, and 6% normal goat serum) to block nonspecific binding. Primary antibodies were then added for overnight incubation at room temperature. Secondary antibodies conjugated with either Alexa Fluor 488 or 568 were used for detecting the primary antibodies. For immunoperoxidase (avidin–biotin peroxidase method) and immunoelectron microscopy, refer to protocols described previously (Liang et al., 2000; Zhang et al., 2005).
Data analyses.
All data were verified by at least two repeats of the experiments. Immunofluorescence microscopic results were analyzed using a laser-scanning confocal microscope (Olympus Fluoview FV1000, Olympus, Tokyo, Japan). Western films were scanned by GS-710 Calibrated Imaging Densitometer (Bio-Rad), and the band density was calculated by multiplying the mean OD (after subtracting background) with the band area (in square millimeters) using Quantity-One (version 3.1; Bio-Rad) software. Relative expression abundance was calculated by dividing the ODs of SIRT2 or CNP bands by corresponding β-actin bands of the same developmental stage.
For complexity analyses, OLPs or OLN93 cells were classified as simple if they only had one to three primary processes, as intermediate if they had over three straight radial primary branches with small secondary processes, as complex if many tertiary and additional branches emerged, and as highly complex if the distal processes became wooly or fused to form lamellopodia around the cell bodies (see examples in Results) (Marin-Husstege et al., 2002). For semiquantitative analyses of SIRT2, CNP, or acetylated α-tubulin (AcTub) levels in differentiating OLPs (see Fig. 6), “−” denotes undetectable signals (for SIRT2 or CNP) and “BL” denotes basic-level expression (for AcTub); “+” indicates clear expression (for SIRT2 or CNP) or elevation (for AcTub) of signals mainly in the cell bodies; “++” refers to expression (for SIRT2 or CNP) or increase (for AcTub) of signals in both cell bodies and processes; “+++” stands for high levels of expression throughout the cell.
Figure 6.
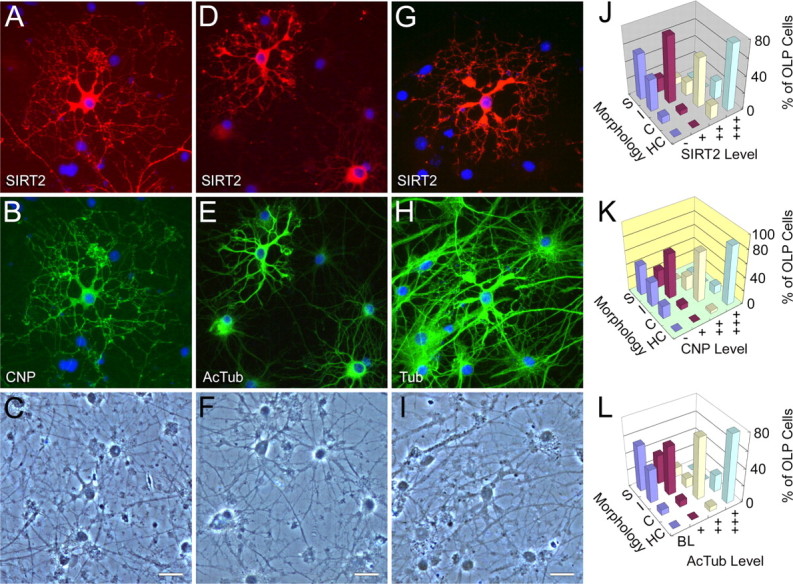
SIRT2, tubulin acetylation, CNP, and oligodendrocyte differentiation. A–I, Double-immunofluorescence photomicrographs showing close correlation of SIRT2 expression (A, D) with CNP (B) and levels of α-tubulin acetylation (E) in cultured OLPs. Levels of total α-tubulin (H) were, however, not closely associated with SIRT2 expression (G). Morphologies of the immunonegative cells could also be appreciated in the respective bottom panels of phase-contrast photomicrographs (C, F, I). Note the increased α-tubulin acetylation and emergence/increases of SIRT2 and CNP immunoreactivities in the prematurity-stage oligodendrocytes, compared with the earlier-stage cells that showed no SIRT2 or CNP signals, and low levels of α-tubulin acetylation. Tub, α-Tubulin; AcTub, acetylated α-tubulin. Scale bars, 20 μm. J–L, Three-dimensional bar charts showing the differentiation progression of the arborization of OLP in relation to the emergence/increase of cellular expression levels of SIRT2, CNP and acetylated α-tubulin. S, I, C, and HC, Simple, intermediate, complex, and highly complex cellular arborization, respectively. BL, −, +, ++, and +++ are used to designate the levels of expression (see Materials and Methods). “% of OLP cells” on the z-axis represents, in each of the four expression levels, the percentage of cells in each complexity category relative to the total.
Results
Molecular features and developmental expression profile of SIRT2 in rat CNS
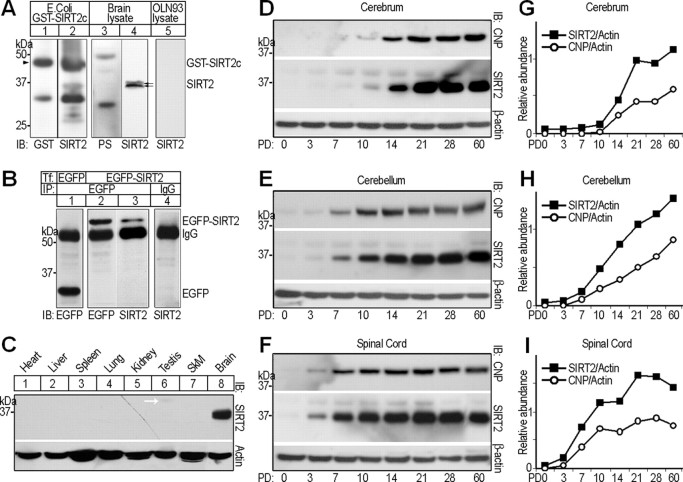
The rat Sirt2 mRNA contained an open reading frame encoding a 351 aa protein. On Western blots, an in-house produced, affinity-purified anti-SIRT2 antibody detected a doublet band of ∼37 kDa in adult rat brain protein samples (Fig. 1A, lane 4), whereas preimmunization serum could not reveal immunoreactivity at this position (Fig. 1A, lane 3). The antibody also bound the recombinant GST-SIRT2c fusion protein, so did a commercial GST antibody (Fig. 1A, lanes 1 and 2). To further determine the specificity of the antibody, we inserted Sirt2 open reading frame into pEGFP-C1 mammalian expression vector (Clontech). OLN-93 (a rat oligodendrocyte cell line) (Richter-Landsberg and Heinrich, 1996) cells transfected with the pEGFP-Sirt2 were lysed, immunoprecipitated using anti-EGFP antibody, and then subjected to immunoblots. The band for the recombinant EGFP-SIRT2 emerged at ∼66 kDa, as detected by either anti-SIRT2 or anti-EGFP antibody (Fig. 1B, lanes 2 and 3). In the lysate of OLN-93 transfected with empty pEGFP-C1 vector, EGFP was precipitated and migrated to ∼29 kDa (Fig. 1B, lane 1). In contrast, irrelevant IgG (anti-FLAG M2; Sigma) cannot precipitate the recombinant protein (Fig. 1B). Together, the antibody proved adequately specific for additional experiments.
Figure 1.
Molecular features of rat SIRT2. A, Western blots, showing specificity tests of rabbit polyclonal anti-SIRT2 antibody. Labels above and below blots indicate the protein sample origins and antibodies used for immunoblotting detection (IB), respectively. GST-SIRT2c, Recombinant GST-SIRT2 C terminus (209–351) protein expressed in Escherichia coli. PS, Preimmunization serum. The double arrows indicate the SIRT2 doublet detected by rabbit anti-SIRT2. The arrowhead points to the position of GST-SIRT2c. B, OLN-93 cells were transiently transfected (Tf) with either pEGFP-C1 vector (lane 1) or pEGFP-Sirt2 (lanes 2–4). Forty-eight hours after transfection, lysates of the cells were subjected to immunoprecipitation (IP) using anti-EGFP or irrelevant IgG, followed by IB with antibodies indicated below. C, Multitissue Western blot probed with rabbit polyclonal anti-SIRT2 antibody. The bottom panel shows the pan-actin immunoreactivity as loading control. SkM, Skeleton muscle. D–I, Western blot analyses comparing the expression profiles of SIRT2 and CNP in postnatal day 0, 3, 7, 10, 14, 21, 28, or 60 rat cerebrum, cerebellum, and spinal cord, respectively. Each lane was loaded with 20 μg of soluble tissue lysate. Signals for β-actin served as loading controls. Quantitative analyses of expression levels of SIRT2 and CNP relative to β-actin in the different CNS regions are shown on the right (G–I). PD, Postnatal day.
On multitissue Western blots probed with the above anti-SIRT2 antibody, the 37 kDa SIRT2 doublet band was detected in brain protein sample, but not in that of the heart, liver, spleen, lung, kidney, testis, or skeletal muscle, although a weak 41 kDa band (white arrow), which we presumed to be a SIRT2 splicing variant, was seen in testis protein lysate (Fig. 1C).
To investigate the developmental expression of SIRT2, protein samples of the cerebra, cerebella, and cervical spinal cords of rats aged 0, 3, 7, 10, 14, 21, 28, and 60 postnatal days were subjected to IB. In the cerebrum, the 37 kDa SIRT2 was marginal at postnatal day 7 (PD7), clearly visible at PD10, increased sharply at PD14, and approximated adult level at PD21 (Fig. 1D). In both cervical spinal cord and cerebellum, the signal appeared at PD3, but higher abundance and earlier approximation to adult level (PD10) was detected in spinal cord (Fig. 1E,F). In comparison, immunoblotting signals for the 42 kDa CNP emerged at approximately the same time as SIRT2 in all the three CNS regions, and cofluctuated with those of SIRT2 along the various postnatal ages (Fig. 1D–I).
SIRT2 is a cytoplasmic protein of oligodendroglia and myelin
As detected by ISH using a digoxigenin-labeled cRNA probe, Sirt2 mRNA-positive signals were seen throughout most CNS regions. Positive cell bodies in the gray matter were typically dispersed, round-shaped, and of relatively small sizes, whereas those in the white matter were more variable in morphology (Fig. 2A–D). Some proximal processes extending from the positive cell bodies, especially in such CNS areas as the cerebellar and cerebral cortices, also contained Sirt2 mRNA (Fig. 2A,B).
Figure 2.
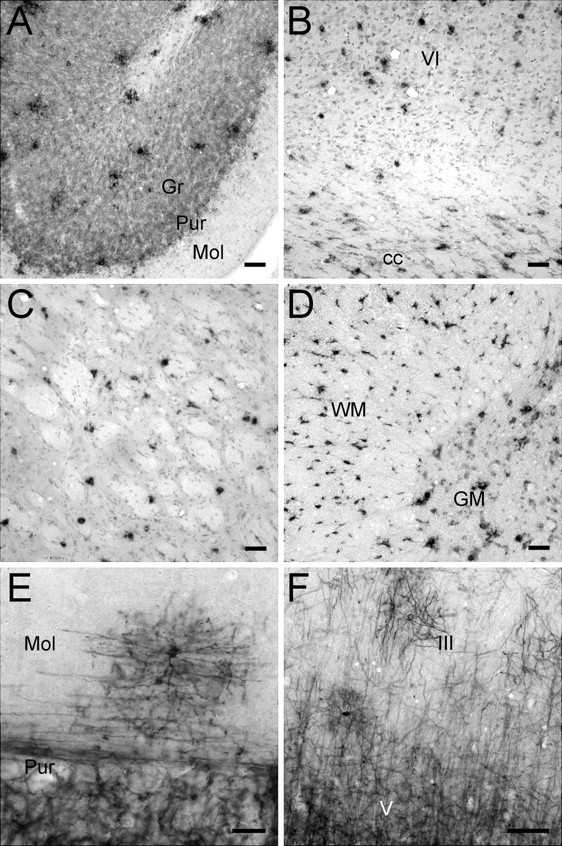
Distribution of SIRT2 mRNA and protein in rat CNS. A–F, In situ hybridization histochemistry (A–D) and immunohistochemistry (E, F) results showing the distribution of SIRT2-expressing cells in the cerebellum (A, E), cerebral cortex (B, F), caudate–putamen (C), and cervical spinal cord (D). III, V, and VI, Layers III, V, and VI of cerebral cortex; cc, corpus callosum; Gr, granule cell layer of cerebellar cortex; Mol, molecular layer of cerebellar cortex; Pur, Purkinje cell layer of cerebellar cortex; GM, gray matter of spinal cord; WM, white matter of spinal cord. Scale bars: A–D, 50 μm; E, F, 20 μm.
ICC with the in-house anti-SIRT2 antibody revealed mostly cellular processes and structures along axons or axon bundles throughout the CNS. Relatively few cell bodies of relatively small sizes showed SIRT2 immunoreactivity (Fig. 2E,F). In the cerebral cortex, SIRT2 immunoreactivity increased gradually from superficial to deep layers and to the subcortical white matter (Fig. 2F). In the cerebellar cortex, SIRT2 immunoreactivity was abundant in the granule and Purkinje cell layers. The molecular layer, however, was mostly devoid of SIRT2 signals with the exception of a few scattered cells and related processes near the Purkinje cell layer (Fig. 2E).
Double immunofluorescence established that SIRT2 expression in the CNS was primarily confined to oligodendrocytes. Colocalization between SIRT2 and CNP, a well characterized oligodendrocyte marker in the CNS, was seen in all CNS regions examined (Fig. 3A–D). Figure 3A shows the cerebellar cortex where SIRT2 immunoreactive signals overlapped those of CNP in the oligodendroglial processes, along myelinated axons in the Purkinje cell and granule cell layers, and in the subcortical white matter. The CNP-positive perikarya of oligodendrocytes, however, were frequently devoid of detectable SIRT2 (Fig. 3A). Similar colocalization of SIRT2 and CNP in oligodendroglial processes/along myelinated axons, and absence of SIRT2 in the oligodendrocytic cell bodies were found in, among others, the caudate nucleus and in the gray and white matters of cervical spinal cord (Fig. 3B–D). This phenomenon may suggest the main functional site of SIRT2 in oligodendroglial processes rather than in cell bodies. Double labeling against SIRT2 and MBPs, another type of marker proteins for oligodendroglia and myelin sheath, confirmed the above findings (Fig. 3I,J). SIRT2 was not detected in astrocytes marked by GFAP, in microglia marked by OX42, nor in neurons marked by NF200 (Fig. 3E–H).
Figure 3.
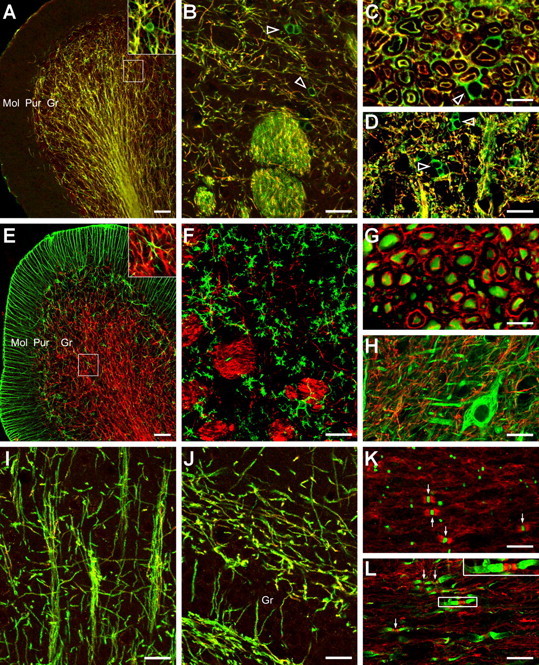
SIRT2 is an oligodendroglial protein. A–H, Double-immunofluorescence labeling of SIRT2 (red in all panels) plus CNP (A–D), GFAP (E), OX42 (F), neurofilament (G, H), or MBP (I, J) in the rat cerebellar cortex (A, E), caudate–putamen (B, F), cerebral cortex (I; layers III and IV), dentate gyrus (J), cross (C, G) or longitudinal sections (D, H) of cervical spinal white (C, G) or gray (D, H) matter. The inset in A or E is the observation of the marked area at a higher magnification. In A–D, note that, although it overlapped CNP (green) in most oligodendrocytic processes, SIRT2 (red) was undetected or only faintly positive in most oligodendroglial cell bodies that showed strong CNP staining (open arrowheads). K, L, On longitudinal sections of cervical spinal white matter, double-immunofluorescence labeling (K) of SIRT2 (red) together with pan-sodium channel (NavP, green), or triple labeling (L) of SIRT2 (red) together with NavP (green) and potassium channel Kv1.2 (green) showing SIRT2 distribution in the juxtanodal and paranodal domains of myelin sheath. The arrows point to examples of the NavP clusters at the nodes of Ranvier. The inset in L shows an example from the marked area at a higher magnification. Note the SIRT2-positive bands flanking the node of Ranvier and in between the strongly Kv1.2-positive juxtaparanodes. Gr, Granule cell layer of cerebellar cortex (A, E) or the dentate gyrus (J). Mol, Molecular layer of cerebellar cortex; Pur, Purkinje cell layer of cerebellar cortex. Scale bars: B, D, F, H–J, 20 μm; A, E, 50 μm; C, G, K, L, 10 μm.
Under high magnification, in certain axon bundles near the pia surface of cross-sectioned cervical spinal white matter, SIRT2-positive profiles, similar to those of CNP labeling, were observed in concentric double rings around the NF200-positive neuronal axons. The inner ring often seemed in contact with the centrally positioned axons, and was separated from the outer ring by a SIRT2-negative zone. Careful comparison with SIRT2 immunoelectron microscopic data (see below) suggested that the concentric rings most possibly corresponded to the cytoplasm-containing innermost and outermost layers of myelin sheaths, respectively (Fig. 3C,G). In most other axon bundles of cervical spinal cord, SIRT2 signals could be seen around the NF200-positive axons but mostly did not directly contact or overlap the latter (Fig. 3C,G).
On longitudinal sections of cervical spinal white matter, as revealed by multiple immunofluorescence, SIRT2 was frequently found enriched in the juxtanodal and paranodal regions in between the sodium channel-clustered node of Ranvier and the potassium channel Kv1.2-enriched juxtaparanodal domain (Fig. 3K,L).
Under electron microscope, SIRT2 immunoreactivity was mainly found in the cytoplasm-containing noncompact portions of myelin sheaths such as the abaxonal (outer) layer (Fig. 4A,D), the juxtanodal/paranodal terminal loops (Fig. 4B,C,E,F), and in processes of oligodendroglia. Electron-dense immunoreactive products were also occasionally found in the perikaryal cytoplasm, but only rarely in the marginal heterochromatin areas of oligodendrocytic nucleus (Fig. 4A). No SIRT2 was detected in the compact laminas of myelin sheath, and no association of the signals with cytoplasmic organelles could be unequivocally established.
Figure 4.
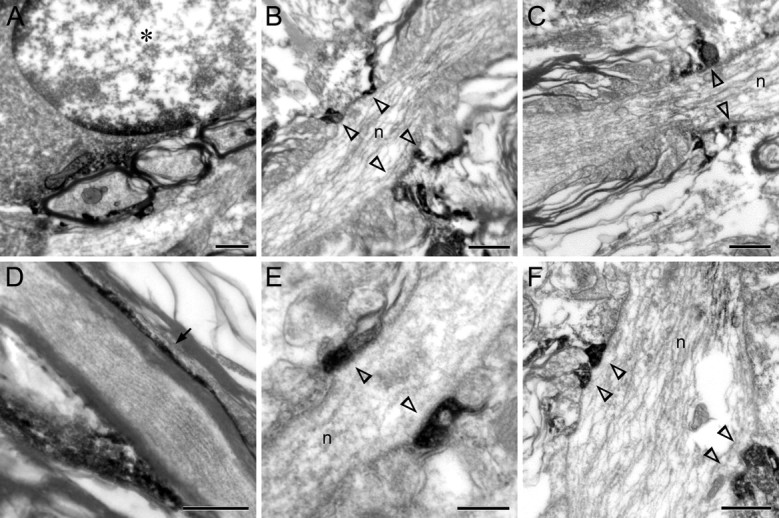
SIRT2 localization in oligodendrocytes/myelin sheaths under electron microscope. A, Immunoelectron micrograph of cervical spinal cord showing occasional SIRT2 immunoreactivity in the perikaryal cytoplasm and in marginal heterochromatin clumps of nucleus of oligodendrocyte (denoted by an asterisk). B–F, Immunoelectron micrographs of cervical spinal cord (B, C, F) or corpus callosum (D, E) showing SIRT2 immunoreactivity in the cytoplasm-containing outer layer of myelin sheaths (D, arrow) or the juxtanodal loops of myelin sheaths (B, C, E, F, open arrowheads) on the sides of the nodes of Ranvier (marked by “n” in the middle of the axons). Scale bars: A–D, F, 0.5 μm; E, 0.2 μm.
α-Tubulin was the main cytoplasmic substrate of NAD-dependent SIRT2 deacetylase
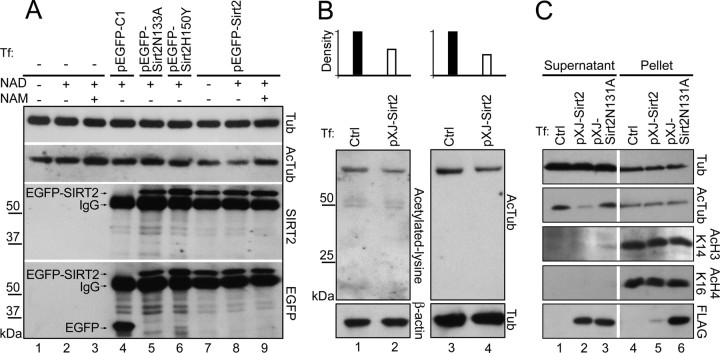
Western blot analysis showed no endogenous SIRT2 expression in OLN-93 rat oligodendroglia cell line (Figs. 1A, lane 5; 5A, lanes 1–3). To identify candidate SIRT2 substrates in oligodendrocyte cytoplasm, the lysate supernatant (containing mainly cytoplasmic proteins) of pXJ-Sirt2 transfected OLN-93 cells was subjected to Western blots probed with an anti-acetylated-lysine antibody (AKL5C1) (Matsuyama et al., 2002) or a mouse monoclonal antibody specific to lysine 40-acetylated α-tubulin (6-11B-1) (Piperno et al., 1987). The former antibody revealed a predominant positive band at 52 kDa. This signal was presumed to be acetylated α-tubulin because (1) anti-acetylated α-tubulin detected a single band at exactly the same position, (2) in comparison with normal OLN-93 cell lysate, the lysate with overexpressed FLAG-SIRT2 showed significantly decreased levels of both the acetylated lysine (48% reduction compared with control) and acetylated α-tubulin (57% reduction compared with control) immunoreactivities. Levels of total α-tubulin were not affected by FLAG-SIRT2 overexpression (Fig. 5B). The anti-acetylated lysine antibody also stained two other bands in OLN-93 immunoblots, but in comparison with acetylated α-tubulin these signals were much weaker, and seemingly not much influenced by overexpression of FLAG-SIRT2 (Fig. 5B).
Figure 5.
Cytoplasmic α-tubulin is the substrate of SIRT2 in OLN93 cells. A, OLN-93 cells were transiently transfected (Tf) as indicated, lysed, and immunoprecipitated with anti-EGFP. The immunoprecipitates were then added to OLN-93 cell lysate (as the source for acetylated α-tubulin) together with various combinations of NAD (1 mm) or NAM (5 mm), and incubated for 2 h at room temperature. Samples after that were subjected to Western blot analyses (5 μg each lane). B, C, OLN-93 cells were transiently transfected with pXJ-Sirt2, lysed, and centrifuged; the supernatant (B, C) and pellet (C) of the cell lysate were subjected to Western blots. Crtl, Normal OLN-93 cell lysate. The bar charts on the top of B represent the densitometric analysis of the bands in lanes 1–4. The band densities of acetylated-lysine in lane 2 and acetylated-α-tubulin in lane 4 (both of which transfected with pXJ-Sirt2) are expressed as percentages of the control groups in lanes 1 and 3, respectively.
In addition to using α-tubulin as its substrate, SIRT2 also deacetylates histones in vitro and in vivo (North et al., 2003; Vaquero et al., 2006). We next tested whether histone acetylation levels in the transfected cells were altered. In addition to the supernatant portion mentioned above, the pellet of the cell lysate in which most nuclear proteins existed was also subjected to Western blots. As shown in Figure 5C, almost all the recombinant FLAG-SIRT2 was found in the supernatant portion. In the presence of FLAG-SIRT2 rather than FLAG-SIRT2N131A, α-tubulin was obviously deacetylated in the cytoplasm. Conversely, the acetylation levels of the histone H3 lysine 14 and histone H4 lysine 16 (North et al., 2003; Vaquero et al., 2006) were not significantly changed (Fig. 5C). These results suggested that the main substrate for SIRT2 was α-tubulin in the cytoplasm of OLN-93 cells.
To further determine the α-tubulin deacetylase activity of rat SIRT2, EGFP-SIRT2 or its point-mutated forms were overexpressed in OLN-93 cells, immunoprecipitated by EGFP antibody, and used as enzymes to treat untransfected normal OLN-93 cell lysate as a substrate. Western blots were finally used to semiquantitatively assess the tubulin deacetylase activity of the immunoprecipitates. The results showed that, in the presence of NAD (1 mm), the addition of immunoprecipitated EGFP-SIRT2, rather than EGFP, caused a significant decrease of acetylated α-tubulin in the cell lysate (Fig. 5A, lane 8). The EGFP-SIRT2 did not influence the level of total α-tubulin, and its tubulin deacetylase activity was abolished when NAD was removed or sufficient NAM (5 mm) was added into the system (Fig. 5A, lanes 7 and 9). Point-mutated SIRT2 s, in which aspartic acid-133 of SIRT2 was replaced by alanine (SIRT2D133A) or histidine-150 by tyrosine (SIRT2H150Y) (Finnin et al., 2001), showed significantly diminished deacetylase activity (Fig. 5A, lanes 5 and 6). SIRT2N131A (replacement of asparagine-131 by alanine) exhibited similar reduction of deacetylase activity (data not shown). These results indicated that SIRT2 could deacetylate α-tubulin in an NAD-dependent manner.
Association among SIRT2 expression, tubulin acetylation levels, and oligodendrocyte maturation in culture
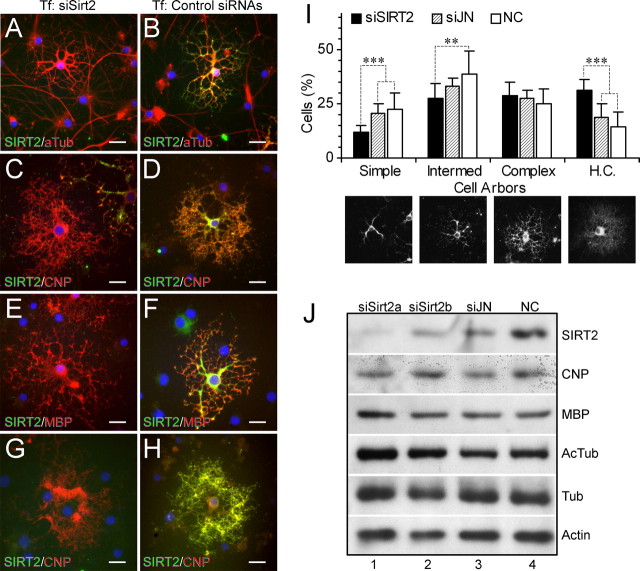
We next investigated the relationships between SIRT2 expression, tubulin acetylation, and development of oligodendroglia in cultured primary OLPs extracted from postnatal day 1–2 rat cerebra. After 3–4 d of differentiation, many of the OLPs started to express SIRT2. Surprisingly, abundance of endogenous SIRT2 expression was positively correlated with α-tubulin acetylation levels in the same cells. Subcellular distribution of acetylated α-tubulin and SIRT2 also mostly overlapped. More importantly, the process arbors of SIRT2-positive and high-AcTub primary OLPs were clearly more complex in morphology than those of the surrounding SIRT2-negative and base-level-AcTub cells. Primary and secondary processes of the former were often tortuous along the way and enlarged at the distal end from where many finer processes originated. In contrast, processes of the SIRT2-negative OLPs often radiated out straight from the cell bodies, and gave rise to relatively fewer additional branches (Fig. 6D–F). Levels of total α-tubulin, however, were not closely correlated with SIRT2 expression (Fig. 6G–I).
Additional double immunofluorescent stainings proved that the concurrent SIRT2 emergence, α-tubulin acetylation increase, and typical morphological changes of cultured primary OLPs marked the entry of oligodendrocyte differentiation into the prematurity stage (Barry et al., 1996; Armstrong, 1998; Baumann and Pham-Dinh, 2001; Woodruff et al., 2001). All cells positive for SIRT2 became immunoreactive for the oligodendroglial functional protein CNP as well, whereas those lacking SIRT2 remained negative for CNP. The two enzymes showed overall covariation in levels, and colocalization at subcellular domains (Fig. 6A–C). Semiquantitative analyses indicated close correlations among levels of SIRT2, AcTub, and CNP and progression of OLP arbor differentiation (Fig. 6J–L).
The raised α-tubulin acetylation levels despite the appearance of SIRT2 tubulin deacetylase activities in the prematurity-stage oligodendrocytes seemed paradoxical. We hypothesized that the heightened α-tubulin acetylation (possibly because of activities of certain acetyltransferases) and CNP activity promoted OLP differentiation. The concurrent SIRT2 increase functioned as a counterbalance to prevent uncontrolled tubulin acetylation, to offset overdifferentiation, and to avoid premature myelination in the CNS. To test this hypothesis, the following overexpression and siRNA knockdown experiments were performed.
SIRT2 transfection lowered α-tubulin acetylation levels and inhibited OLP differentiation
Two days after Sirt2 transfection, primary OLP cells (in their third or fourth day of differentiation) showed retarded morphological differentiation (Fig. 7A,C,F,G) and marked decreases in α-tubulin acetylation levels (Fig. 7A). Many of the transfected cells expressed considerable levels of intrinsic CNP, but failed to assume the typical morphology of the normally CNP-positive prematurity-stage oligodendrocytes (Fig. 6F). Process arbors of these cells still appeared relatively immature, much like those of earlier-stage cells (Fig. 7A,C,F,G). In contrast, morphologies of cells expressing EGFP only or FLAG-juxtanodin 141 (FLAG-JN141) (Zhang et al., 2005) appeared to be highly complicated, similar to untransfected neighboring cells (Fig. 7D,E,H). Overall, at the end of the 3–4 d differentiation, the OLP cells untransfected or transfected with control plasmids exhibited mostly intermediate or complex morphology, whereas the majority of the OLPs transfected by Sirt2 were simple or intermediate in cell arbor complexity (for criteria, see Materials and Methods). In comparison to the JN141 transfected controls, for example, the percentage of simple cells increased by threefold in the pXJ-Sirt2 transfected group, whereas those with complex morphology dropped from 53.6 to 34.9% (Fig. 7I) (average of three experiments; n > 300 cells for all groups).
Figure 7.
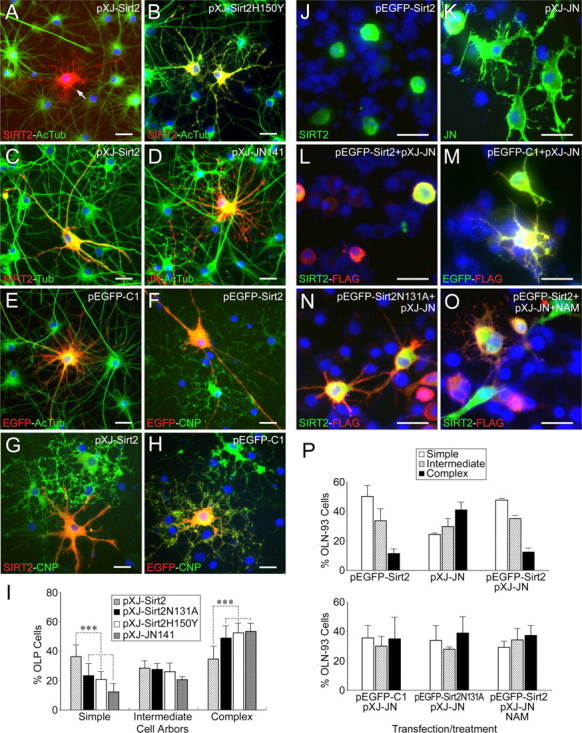
Inhibitory effects of SIRT2 overexpression on oligodendrocyte differentiation. A–H, Sirt2-transfected OLP cells showed decreased α-tubulin acetylation levels (A, arrow), and simpler morphology compared with neighboring untransfected cells (A, C, F, G), with cells transfected with SIRT2H150Y, the partially inactive form of SIRT2 deacetylase (B), and with pEGFP-C1 or JN141-transfected controls (E, H, D). Total α-tubulin levels in the Sirt2-transfected cells were not significantly affected (C). For the visualization of the much higher intensity of overexpressed SIRT2 signals in the transfected cells, normal endogenous SIRT2 signals in the neighboring cells, if any, were underexposed and therefore could not be optimally appreciated in some of the panels (A, B, C, G). I, Statistical analyses of morphological maturation of primary OLPs transfected with pXJ-Sirt2, pXJ-Sirt2N131A, pXJ-Sirt2H150Y, and pXJ-JN141 plasmids. Data are from three independent experiments (n > 300 transfected cells for each transfection group). ***p < 0.001 (Student's t tests). Error bars indicate SD. J–O, pEGFP-Sirt2 transfected OLN93 cells showed much fewer cell processes (J), in comparison with cells transfected by pXJ-JN alone (K) or pXJ-JN plus pEGFP-C1 (M). When cotransfected with pXJ-JN, pEGFP-Sirt2 counterregulated the arborization-promoting effect of the former (L), whereas the inhibitory effect of pEGFP-Sirt2N131A was much less obvious (N). Addition of nicotinamide (5 mm) to the culture medium partly abolished the arborization-inhibiting effect of SIRT2 (O). Cells in all groups except those shown in O were transfected in the presence of NAD. P, Statistical analyses showing the counteractive effect of overexpressed SIRT2 on JN overexpression-promoted OLN-93 cellular arborization. OLN-93 cells were transfected or treated as indicated in J–O. Data are from two independent experiments (n > 300 transfected cells for each transfection group). Plasmids/drugs for transfection/treatment of cultures are indicated on the top right of A–H and J–O, and antibodies for immunofluorescence on the bottom left. Refer to Figure 6 for abbreviations. Scale bars, 20 μm.
To test whether the deacetylase activity of SIRT2 is essential for its inhibitory effect on OL arborization, SIRT2H150Y (which mostly lost deacetylase activity) (Fig. 5A) (Finnin et al., 2001) was used to transfect OLP cells. The results showed that the deacetylase activity loss of SIRT2H150Y almost completely abolished the inhibitory effect of wild-type SIRT2 on OL arbor complexity (Fig. 7B,I). Transfection of another deacetylase-inactive mutant of SIRT2, SIRT2N131A, gave rise to similar results (Fig. 7I).
To further determine the effects of SIRT2, we cotransfected OLN-93 cells with both pEGFP-Sirt2 and pXJ-juxtanodin. Consistent with our previous study (Zhang et al., 2005), cells overexpressing FLAG-tagged JN alone showed marked increases in the process arbor complexity (43.1% complex; 25.6% simple) (Fig. 7K,P). In contrast, cells overexpressing both FLAG-JN and EGFP-SIRT2 showed remarkably less developed branches or only rounded-up cell bodies (13.1% complex; 50.0% simple) (Fig. 7L,P), much similar to cells overexpressing only SIRT2 (Fig. 7J,P). The simultaneous overexpression of SIRT2N131A or EGFP together with JN did not significantly counteract the arborization-promoting effect of the latter (Fig. 7M,N,P). Addition of 5 mm nicotinamide to the JN-SIRT2 cotransfected culture partly blocked the arborization-inhibiting effect of SIRT2 (Fig. 7O,P).
Overall, the simplification of cell morphology and the decrease of α-tubulin acetylation in the SIRT2-transfected primary OLP cells contrasted strongly with OLPs expressing endogenous SIRT2. Together with data from the cotransfection experiments, these results supported the notion that SIRT2 decelerated oligodendroglial maturation through its tubulin deacetylase activity.
SIRT2 siRNA knockdown reduced tubulin deacetylation and accelerated OLP differentiation
To verify the above SIRT2 effects from a different angle, we used siRNAs to interfere with intrinsic SIRT2 expression in cultured primary OLPs starting at 2–4 d into their differentiation. Transfections by siRNAs for JN or HDAC6, both of which showed no detectable expression within the OLP differentiation time window under the present study (data not shown), served as controls.
All three Sirt2 siRNA duplexes depleted or lowered SIRT2 immunoreactivity in a majority of the cultured OLPs. The knockdown effect of the siRNA targeting Sirt2 nucleotides 518–540 (NM_001008368) was nearly complete and lasted for at least 8 d. When examined 3–4 d into the knockdown, OLPs showed no obvious changes in the levels and distribution of CNP or MBPs, nor did the cells display marked alteration in their process arbors (Fig. 8A–F). After 7–8 d of nullified/reduced Sirt2 expression by the siRNAs, however, significantly more OLPs in the experimental cultures adopted complex or highly complex morphologies, compared with JN/HDAC6 siRNA-treated or with siRNA-omitted controls. Percentage of OLPs with simple morphologies, indicative of less differentiation, accordingly decreased in the experimental group (Fig. 8I). Accompanying the morphologically enhanced differentiation in the experimental group were the decreased deacetylation of tubulin and increased expression of an immature form of MBP at ∼50 kDa (Ursell et al., 1995). Immunoreactivities for CNP, total tubulin, or β-actin in the OLPs, as detected by Western blotting analyses, were not significantly altered by SIRT2 siRNAs (Fig. 8J). JN or HDAC6 siRNAs transfection gave rise to no significant influence on differentiation or gene expression of OLPs at the present developmental stage.
Figure 8.
Prolonged SIRT2 knockdown promoted OLP differentiation. A–F, siRNA knockdown of endogenous SIRT2 for 3 d (days 2–5 into differentiation) did not significantly alter the morphological complexity of OLPs or distribution of acetylated α-tubulin (A), CNP (C), and MBPs (E) compared with cells transfected with JN or HDAC6 siRNAs (B, D, F). OLPs within 10 d into differentiation had no JN or HDAC6 immunoreactivity (data not shown). Thus, the latter two siRNAs served here as negative controls. G–J, siRNA knockdown of endogenous SIRT2 for 6 or 8 d (days 2–8 or 2–10 into differentiation) promoted the differentiation of OLPs as evidenced by increased morphological complexity (I) and upregulation of acetylated α-tubulin and MBP expression (J) in comparison with untransfected or JN/HDAC6 siRNA-treated controls. No difference was observed in the expression of CNP, total α-tubulin and β-actin between the SIRT2 knockdown and the control groups (J). Examples of cell morphology and CNP expression in the 8 d SIRT2 knockdown and untransfected control are shown in G and H, respectively. I, Error bars indicate SD. Grayscale pictures under the graph represent typical examples for the four morphological hierarchies. HC, Highly complex; NC, untransfected negative control; siJN, JN siRNA 3235–3257; siSIRT2a, Sirt2 siRNA 518–540; siSIRT2b, Sirt2 siRNA 1390–1412. ***p < 0.001; **p < 0.01, Student's t tests. Refer to Figures 6 and 7 for additional abbreviations. Plasmids/siRNAs for transfection are indicated on the top of the columns, and antibodies for immunofluorescence on the bottom left of individual panels. Scale bars, 20 μm.
Discussion
The present article presents our findings of SIRT2 as an oligodendroglia-specific protein that exerted its negative control on oligodendroglial differentiation or aging through deacetylating microtubule cytoskeleton. These results revealed the connection between the highly conserved lifespan-extending SIR2 gene family and oligodendroglial development or CNS myelination, and will hopefully contribute to the understanding of brain aging and related demyelinating or neurodegenerative diseases.
Rat SIRT2 as a specific marker for oligodendrocytes
By means of ISH, immunohistochemistry, and double labeling, the present study identified SIRT2 as a member of the protein family that was preferentially expressed by oligodendrocytes. This result agrees with previous proteomic studies that have identified SIRT2 in the myelin sheath (Vanrobaeys et al., 2005; Roth et al., 2006). In comparison with various markers of developing and mature oligodendrocytes/myelin sheath, SIRT2 expression colocalized with that of CNP except in oligodendrocytic cell bodies where little SIRT2 was found. The two also covaried along the progression of oligodendrocyte development in vivo and in vitro. In this connection, it is noteworthy that CNP, like SIRT2, is also a tubulin-associated oligodendroglial protein in the CNS (Bifulco et al., 2002; Lee et al., 2005).
Unlike the absence of SIRT2 in the majority of oligodendroglial perikarya in the adult CNS, clear SIRT2 signals were observed in the cell bodies of cultured primary OLPs. This dissimilarity in subcellular distribution of the protein most likely reflected differences between mature oligodendrocytes in the adult CNS and oligodendrocyte precursors in the culture.
SIRT2 as a differentiation inhibitor of oligodendroglia
Among others, cytoskeleton-associated oligodendroglial proteins such as CNP, juxtanodin, and mayven, have been recently reported to regulate oligodendrocytic process outgrowth, gene expression, and/or myelin–axon interaction (Jiang et al., 2005; Lee et al., 2005; Zhang et al., 2005). The present results showed that SIRT2 is yet another member of the protein family, but functioned as a “braking” mechanism in oligodendroglial development. Overexpression of SIRT2 significantly inhibited the oligodendroglial arborization and tubulin acetylation, indicating retardation of the course of specialization of the cells, whereas siRNA knockdown of endogenous SIRT2 had the opposite effects. This inhibitory role of SIRT2 is further supported by our finding that overexpression of the protein counteracted the facilitatory effects of JN on arborization of OLN-93 oligodendrocyte cell line. Interestingly, SIRT1 has also been implicated in repressing myocyte differentiation (Fulco et al., 2003).
Activity blockade of HDAC activities by trichostatin A (TSA) and valproic acid delays oligodendroglial differentiation (Marin-Husstege et al., 2002; Liu et al., 2003; Hsieh et al., 2004; Shen et al., 2005). This was confirmed by our experiments (data not shown). Because SIRT2 is not inhibited by TSA (North et al., 2003; the present study), these results clearly indicate the involvement of other HDACs in regulating oligodendroglial differentiation. Interestingly, HDAC6, another known tubulin deacetylase, was coimmunoprecipitated with SIRT2 when overexpressed together with SIRT2 (Hubbert et al., 2002; North et al., 2003).
Apart from regulating differentiation, the abundant SIRT2 immunoreactivity in adult oligodendrocytes (the present study) entails functional roles of the protein in the mature CNS. Under electron microscope, SIRT2 was mainly found in the noncompact outer and terminal loops of myelin sheath. This subcellular positioning and the tubulin deacetylation activity seemingly pointed to possible roles of the protein in subcytoarchitecture, molecular trafficking, or myelin–axon signaling of oligodendroglia. Other cytoskeleton-related oligodendroglial proteins like CNP and juxtanodin have also been implicated in similar functions (Bifulco et al., 2002; Lappe-Siefke et al., 2003; Zhang et al., 2005).
SIRT2, tubulin deacetylation, and oligodendroglial differentiation
The present results demonstrated functional roles of tubulin acetylation in maturation of oligodendroglia, as well as SIRT2-mediated tubulin deacetylation in decelerating the differentiation/aging of the cell. Our data indicated that SIRT2 action in oligodendrocytes was most likely mediated through tubulin deacetylation. First, as revealed by our in vivo and in culture results (Figs. 3, 4), SIRT2 was primarily an oligodendroglial cytoplasmic protein mainly localized to the cellular processes and noncompact cytoplasm-containing parts of myelin sheath, in agreement with previous data (Perrod et al., 2001; North et al., 2003; Vanrobaeys et al., 2005). Second, SIRT2 was colocalized with acetylated α-tubulin in cultured primary OLPs. Third, SIRT2 mutants (SIRT2N131A and SIRT2H150Y) simultaneously diminished its tubulin deacetylase activity and its inhibition of OLP morphological differentiation, and siRNA knockdown of SIRT2 gave rise to similar effects. Finally, comparison of Western blotting results from the anti-acetylated lysine, anti-acetylated α-tubulin, and anti-acetylated histones H3/H4 antibodies identified α-tubulin as the main substrate for SIRT2 among OLN-93 proteins (Fig. 5B,C). In comparison with histones, tubulin has been demonstrated as a preferred substrate of SIRT2 (North et al., 2003). Our in vitro deacetylation test also indicated NAD-dependent tubulin deacetylase activity of SIRT2. Hence tubulin deacetylation through SIRT2 probably represented yet another important mechanism in the complex cytoskeletal control of oligodendrogenesis and myelinogenesis, in parallel to those mediated through other cytoskeleton-related proteins such as MAP2 (microtubule-associated protein 2), Tau, CNP, and juxtanodin (LoPresti and Konat, 2001; Richter-Landsberg, 2001; Lee et al., 2005; Zhang et al., 2005).
It remains unclear how tubulin deacetylation by SIRT2 was translated into altered cytoplasmic extensions and delayed differentiation of oligodendrocytes. A plausible hypothesis might be that deacetylation lowered microtubule stability, reduced tubulin polymerization, increased depolymerization, or altered the binding of tubulin to associated proteins, as previously suggested in other cell types (Piperno et al., 1987; Robson and Burgoyne, 1989; Hubbert et al., 2002).
SIRT2 has also been shown to interact with histones H3 and H4 (North et al., 2003; Vaquero et al., 2006) and transcription factor HoxA10 (Bae et al., 2004). Our electron microscopic observation also found SIRT2, albeit at lower levels than in the cytoplasm, in the nuclei of a limited number of oligodendrocytes. It thus cannot be entirely ruled out that SIRT2 may use other cytoplasmic or nuclear proteins to exert its influence in oligodendroglia/myelin sheaths. It awaits future investigations to determine the circumstances/factors that govern the substrate selection (tubulin vs histones, for example) and subcellular distribution (cytoplasmic vs nuclear) of SIRT2. Interestingly, histone H4 has been shown a preferred substrate of SIRT2 during mitosis of other cell types (Vaquero et al., 2006).
SIRT2, brain aging, and neurodegeneration?
The SIR2 protein family extends lifespan or slows down aging in various species/cell types ranging from yeast, Drosophila, and C. elegans to mouse and rat (Blander and Guarente, 2004). In view of the antagonistic effects of SIRT2 on oligodendrocyte differentiation, the presence of the protein in the adult CNS could presumably help prevent overdifferentiation or premature senescence of oligodendrocytes or myelin sheaths. Alternatively, this negative influence of SIRT2 on oligodendroglial differentiation may help preserve an adequate pool of immature oligodendrocytes for remyelination or CNS self-repair when needed. Indirectly, the SIRT2 effects on oligodendroglia and myelin could also presumably influence neuronal survival or aging. It is well documented that structural/functional abnormality of oligodendroglia or myelin sheath may cause axon degeneration or neuronal death (Bifulco et al., 2002). Therefore, it could be intriguing to investigate CNS aging and neurodegeneration in relation to SIRT2 and to glia–neuron interaction.
Footnotes
This work was supported by Biomedical Research Council Grants BMRC/06/1/21/19/460, BMRC/04/1/21/19/305, and BMRC/01/1/21/19/179 (F.L.) and National Medical Research Council of Singapore Grant NMRC/0946/2005 (F.L.). We thank Dr. Zhicheng Xiao and Dr. Qidong Hu (Institute of Molecular and Cell Biology, Singapore) for help in primary oligodendrocyte culture and generous provision of OLN-93 cell line. We also are grateful to Dr. Zhiwei Feng (National Neuroscience Institute, Singapore) and Dr. Boon Chuan Low (National University of Singapore, Singapore) for supplying plasmid DNAs.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Armstrong RC. Isolation and characterization of immature oligodendrocyte lineage cells. Methods. 1998;16:282–292. doi: 10.1006/meth.1998.0685. [DOI] [PubMed] [Google Scholar]
- Bae NS, Swanson MJ, Vassilev A, Howard BH. Human histone deacetylase SIRT2 interacts with the homeobox transcription factor HOXA10. J Biochem (Tokyo) 2004;135:695–700. doi: 10.1093/jb/mvh084. [DOI] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Barry C, Pearson C, Barbarese E. Morphological organization of oligodendrocyte processes during development in culture and in vivo. Dev Neurosci. 1996;18:233–242. doi: 10.1159/000111413. [DOI] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Bifulco M, Laezza C, Stingo S, Wolff J. 2′,3′-Cyclic nucleotide 3′-phosphodiesterase: a membrane-bound, microtubule-associated protein and membrane anchor for tubulin. Proc Natl Acad Sci USA. 2002;99:1807–1812. doi: 10.1073/pnas.042678799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Cao Q, Zhang B, Wong PHT, Ling EA, Liang F. Cloning and neuroglial expression of rat sirtuin2. Soc Neurosci Abstr. 2004;30:405. 12. [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jiang S, Avraham HK, Park SY, Kim TA, Bu X, Seng S, Avraham S. Process elongation of oligodendrocytes is promoted by the Kelch-related actin-binding protein Mayven. J Neurochem. 2005;92:1191–1203. doi: 10.1111/j.1471-4159.2004.02946.x. [DOI] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Lee J, Gravel M, Zhang R, Thibault P, Braun PE. Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J Cell Biol. 2005;170:661–673. doi: 10.1083/jcb.200411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Hatanaka Y, Saito H, Yamamori T, Hashikawa T. Differential expression of gamma-aminobutyric acid type B receptor-1a and -1b mRNA variants in GABA and non-GABAergic neurons of the rat brain. J Comp Neurol. 2000;416:475–495. [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Liu A, Muggironi M, Marin-Husstege M, Casaccia-Bonnefil P. Oligodendrocyte process outgrowth in vitro is modulated by epigenetic regulation of cytoskeletal severing proteins. Glia. 2003;44:264–274. doi: 10.1002/glia.10290. [DOI] [PubMed] [Google Scholar]
- LoPresti P, Konat GW. Hydrogen peroxide induces transient dephosphorylation of tau protein in cultured rat oligodendrocytes. Neurosci Lett. 2001;311:142–144. doi: 10.1016/s0304-3940(01)02137-1. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Manser E, Huang HY, Loo TH, Chen XQ, Dong JM, Leung T, Lim L. Expression of constitutively active alpha-PAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, Horinouchi S, Yoshida M. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Perrod S, Cockell MM, Laroche T, Renauld H, Ducrest AL, Bonnard C, Gasser SM. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J. 2001;20:197–209. doi: 10.1093/emboj/20.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Landsberg C. Organization and functional roles of the cytoskeleton in oligodendrocytes. Microsc Res Tech. 2001;52:628–636. doi: 10.1002/jemt.1047. [DOI] [PubMed] [Google Scholar]
- Richter-Landsberg C, Heinrich M. OLN-93: a new permanent oligodendroglia cell line derived from primary rat brain glial cultures. J Neurosci Res. 1996;45:161–173. doi: 10.1002/(SICI)1097-4547(19960715)45:2<161::AID-JNR8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Robson SJ, Burgoyne RD. Differential localisation of tyrosinated, detyrosinated, and acetylated alpha-tubulins in neurites and growth cones of dorsal root ganglion neurons. Cell Motil Cytoskeleton. 1989;12:273–282. doi: 10.1002/cm.970120408. [DOI] [PubMed] [Google Scholar]
- Roth AD, Ivanova A, Colman DR. New observations on the compact myelin proteome. Neuron Glia Biol. 2006;2:15–21. doi: 10.1017/S1740925X06000068. [DOI] [PubMed] [Google Scholar]
- Schnell R, Rine J. A position effect on the expression of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:494–501. doi: 10.1128/mcb.6.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984;3:2817–2823. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Ursell MR, McLaurin J, Wood DD, Ackerley CA, Moscarello MA. Localization and partial characterization of a 60 kDa citrulline-containing transport form of myelin basic protein from MO3–13 cells and human white matter. J Neurosci Res. 1995;42:41–53. doi: 10.1002/jnr.490420106. [DOI] [PubMed] [Google Scholar]
- Vanrobaeys F, Van CR, Dhondt G, Devreese B, Van BJ. Profiling of myelin proteins by 2D-gel electrophoresis and multidimensional liquid chromatography coupled to MALDI TOF-TOF mass spectrometry. J Proteome Res. 2005;4:2283–2293. doi: 10.1021/pr050205c. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng EE, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhai Q, Chen Y, Lin E, Gu W, McBurney MW, He Z. A local mechanism mediates NAD-dependent protection of axon degeneration. J Cell Biol. 2005;170:349–355. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff RH, Tekki-Kessaris N, Stiles CD, Rowitch DH, Richardson WD. Oligodendrocyte development in the spinal cord and telencephalon: common themes and new perspectives. Int J Dev Neurosci. 2001;19:379–385. doi: 10.1016/s0736-5748(00)00083-6. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Cao Q, Guo A, Chu H, Chan YG, Buschdorf JP, Low BC, Ling EA, Liang F. Juxtanodin: an oligodendroglial protein that promotes cellular arborization and 2′,3′-cyclic nucleotide-3′-phosphodiesterase trafficking. Proc Natl Acad Sci USA. 2005;102:11527–11532. doi: 10.1073/pnas.0500952102. [DOI] [PMC free article] [PubMed] [Google Scholar]