Abstract
Copy number variants, such as duplications and hemizygous deletions at chromosomal loci of up to a few million base pairs, are highly associated with psychiatric disorders. Hemizygous deletions at human chromosome 22q11.2 were found to be associated with elevated instances of schizophrenia and autism spectrum disorder in 1992 and 2002, respectively. Following these discoveries, many mouse models have been developed and tested to analyze the effects of gene dose alterations in small chromosomal segments and single genes of 22q11.2. Despite several limitations to modeling mental illness in mice, mouse models have identified several genes on 22q11.2—Tbx1, Dgcr8, Comt, Sept5, and Prodh—that contribute to dimensions of autism spectrum disorder and schizophrenia, including working memory, social communication and interaction, and sensorimotor gating. Mouse studies have identified that heterozygous deletion of Tbx1 results in defective social communication during the neonatal period and social interaction deficits during adolescence/adulthood. Overexpression of Tbx1 or Comt in adult neural progenitor cells in the hippocampus delays the developmental maturation of working memory capacity. Collectively, mouse models of variants of these 4 genes have revealed several potential neuronal mechanisms underlying various aspects of psychiatric disorders, including adult neurogenesis, microRNA processing, catecholamine metabolism, and synaptic transmission. The validity of the mouse data would be ultimately tested when therapies or drugs based on such potential mechanisms are applied to humans.
Keywords: 22q11.2, ASD, CNV, intellectual disability, mouse model, schizophrenia
Significance Statement.
The biological mechanisms of psychiatric disorders are still poorly understood, thereby hindering the development of mechanism-based novel therapies. Genetic variants, collectively termed copy number variants (CNVs), are robustly and reproducibly associated with many psychiatric disorders. For example, hemizygous deletions at the human chromosome 22q11.2 are associated with elevated rates of schizophrenia, autism, and intellectual disability; duplication at this locus is associated with autism and intellectual disability. Research in this area started to delve into the mechanistic bases of these CNVs. Studies using mouse models have identified several genes encoded on chromosome 22q11.2 (Tbx1, Dgcr8, Comt, Sept5, and Prodh) that directly contribute to certain aspects of autism and schizophrenia. The neuronal processes affected by these individual genes include adult neurogenesis, microRNA processing, catecholamine metabolism, and synaptic transmission. Understanding these potential mechanisms could help develop new therapies or drugs to better treat patients with psychiatric disorders.
Introduction
Since the 1950s, psychopharmacology has drastically transformed the field of psychiatry. This transformation was not guided by a rational approach based on mechanistic knowledge; instead, many drugs were serendipitously identified to be beneficial for treating mental illness. Chlorpromazine was initially used as an anti-histamine drug to assuage low blood pressure and rapid heart rate during surgical shock; however, its antipsychotic effects were first recognized by Henri Laborit in 1952 and it was widely used for the treatment of schizophrenia by Pierre Deniker, Jean Delay, J. M. Harl, and Heinz Lehmann. Imipramine was initially developed by the pharmaceutical company Geigy as a chlorpromazine-like compound to combat psychosis of schizophrenia. Although imipramine did not achieve this intended effect, its beneficial effects on depression were recognized by Roland Kuhn in 1956. John Cade discovered the beneficial effects of lithium in patients with mania in 1949; this discovery was forgotten but was later recognized thanks to the work of Morgens Schou. The serendipitous discoveries of these drugs freed many patients from semi-permanent confinement in mental hospitals, which were often located in alienating remote locations, or from prolonged psychoanalysis treatment with little tangible improvement. Thus, these drugs and their sister compounds, which were subsequently developed, became a solid starting point to gain knowledge of their sites of actions, which provided valuable insights into the mechanisms underlying psychiatric disorders.
Nearly half a century later, we are witnessing yet another transformation in the field of psychiatry. Rapid advances in the field of genetics have identified cases of duplication and hemizygous deletions of chromosomal loci, collectively termed copy number variants (CNVs), which show a robust and reproducible association with a wide range of psychiatric disorders. CNVs provide new opportunities to explore the mechanisms underlying psychiatric disorders such as schizophrenia, autism spectrum disorder (ASD), bipolar disorder, intellectual disability (ID), and attention-deficit/hyperactivity disorder (ADHD).
CNVs
CNVs are deletions and duplications of large stretches of chromosomes, which can extend up to a few million bases in length. In 2007 and 2008, a number of studies reported the association of many CNVs with schizophrenia and autism. Each CNV was found to have unprecedented levels of association with mental illness (Sebat et al., 2007; Szatmari et al., 2007; Ullmann et al., 2007; Brunetti-Pierri et al., 2008; Christian et al., 2008; Marshall et al., 2008; Mefford et al., 2008; Sharp et al., 2008; Stefansson et al., 2008; Walsh et al., 2008; Xu et al., 2008). While several syndromic cases of developmental disorders had been known to be associated with copy number variations (Lee and Lupski, 2006), these studies presented a novel view that CNVs have a much broader contribution to nonsyndromic, as well as syndromic, cases of many psychiatric disorders.
Three features distinguish CNVs from widely studied common genetic variants such as single nucleotide polymorphisms (SNPs). First, CNVs are rare variants; each CNV is found in less than 1% of patients with schizophrenia or ASD (Kirov et al., 2014; Rees et al., 2014b). Second, unlike common variants (e.g., SNPs) that individually raise the risk for mental illness only slightly, the rates of psychiatric disorders among carriers of each CNV are extraordinarily high. The odds ratios for schizophrenia reach 67.7 with 22q11.2 deletions, 20.6 with 16p11.2 distal deletions, and higher than 10 with 2q16.3 deletions, 15q13.3 deletions, 7q11.23 duplications, 9p24.3 deletions/duplications, and 8q22.2 deletions (Marshall et al., 2017). Similarly, the odds ratios are high for ASD and ID with these and other CNVs (Cooper et al., 2011; Girirajan et al., 2011; Malhotra and Sebat, 2012). Thirdly, each CNV is associated, to varying degrees, with multiple disorders such as schizophrenia, ASD, ID, bipolar disorder, depression, and ADHD (Elia et al., 2011; Malhotra and Sebat, 2012; Kirov et al., 2014). The pleiotropic actions of CNVs do not conform to the existing clinical classification of psychiatric disorders, which is based on symptomatic clustering. However, the CNV effects are hardly surprising considering that the clinical diagnosis of one psychiatric disorder is often followed by another, even among idiopathic cases (Plana-Ripoll et al., 2019).
Despite promising leads, the precise mechanistic links between CNVs and their symptomatic manifestations still remain poorly understood. In humans, analysis has been largely correlative. Due to technical limitations, studies have been unable to establish the causative effects of CNV-associated alterations in gene expression patterns and regional activity patterns in the brain with mental illness.
One way of viewing association is that all the genes encoded in a CNV contribute in some way to any given clinically defined psychiatric disorder. This could occur through the collective actions of all genes, particularly in the cases of large-sized CNVs. Consistent with this possibility, the severity of the phenotype increases with the size of the CNV and each of large de novo CNVs does not contain ASD-associated genes identified by exome sequencing (Sanders et al., 2015). An alternative view is that a single driver gene within a CNV is critical. This notion is supported by evidence that each of small de novo CNVs contained a single ASD-associated gene identified by exome sequencing, and no cases were found in which many such genes were encoded in a single small de novo CNV (Sanders et al., 2015). However, exome sequencing might not be ideal to identify driver genes if their variants detected by exome sequencing simply do not exist or do not frequently occur. Moreover, hemizygous deletion or duplication might produce more severe effects than some variants that exome sequencing identify. There are cases where a single gene can recapitulate the effects of a large deletion (e.g., 17p11.2) (Lee and Lupski, 2006). Thus, it is still possible that a larger size CNV increases the chances of affecting a few risk genes with large effects, and a small CNV contains more than one contributory gene.
Analyses of biological pathways have elegantly demonstrated select pathways in CNV cases in humans. However, some technical limitations need to be considered. First, some CNV-encoded genes are not listed in data sets for pathway analyses because their functions are not well characterized or they contribute to peripheral phenotypes and used as negative controls. Second, some analyses are done using all CNVs as a set. Such grouping is ideal to identify contributory genes and biological pathways common to all CNVs but not those unique to each CNV.
A third possibility is that more than one, but not all, gene in each CNV, regardless of its size, is contributory to psychiatric disorders. Consistent with this hypothesis, many genes known to be involved in diverse functions, such as synaptic functions and activity-regulated cytoskeleton-associated proteins, are represented in CNVs (Walsh et al., 2008; Pinto et al., 2014; Chang et al., 2015; Sanders et al., 2015; Marshall et al., 2017). Data coming from mouse models lend further support for this hypothesis that several, but not all, genes even in a large CNV are contributory and the degree of contribution varies among such genes (see Figure 1B–C) (Hiroi et al., 2012, 2013, 2018; Hiroi and Nishi, 2016; Nishi and Hiroi, 2016; Zinkstok et al., 2019).
Figure 1.
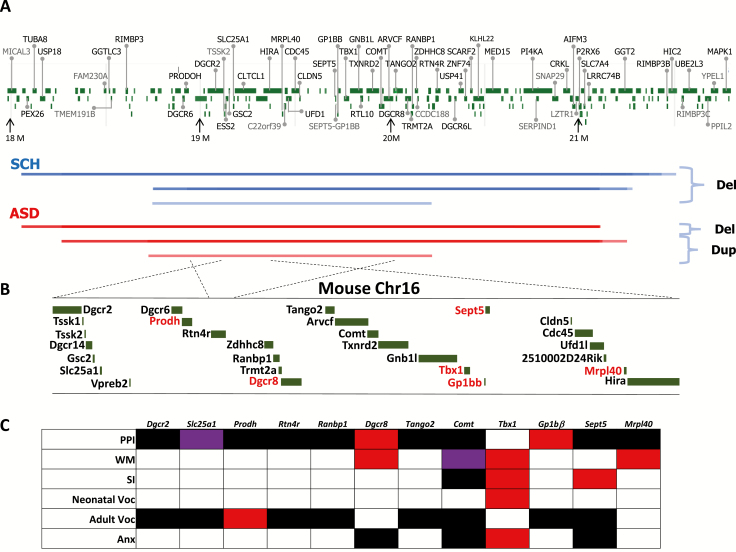
22q11.2 CNVs and mouse models. (A) Human 22q11.2 hemizygous deletions and duplications associated with schizophrenia and autism spectrum disorder (ASD). The names of protein-coding genes only are provided for clarity. Color density increases with segment recurrence. Data were tallied from studies that examined schizophrenia and ASD patients with 22q11.2 CNV (Szatmari et al., 2007; Weksberg et al., 2007; Marshall et al., 2008, 2017; Kirov et al., 2009; Pinto et al., 2010; Girirajan et al., 2011; Sanders et al., 2011, 2015; Ahn et al., 2014; Szatkiewicz et al., 2014; Li et al., 2016; Rees et al., 2016; Kushima et al., 2018). The starting and ending addresses of deletions and duplications are based on the addresses indicated in their publications, but all addresses were converted into those of GRCh38.p12. (B) Mouse chromosome 16, a homolog of the minimal nested segment of human 22q11.2 CNV. (C) Mouse models of single gene deletions. Only those genes in which deletions were examined in congenic (10 or higher backcrosses) or co-isogenic mouse models were studied because biased genetic background in noncongenic mice results in interpretative difficulty (Hiroi, 2018). Red indicates phenotypes consistent with 22q11.2 hemizygous deletions in humans. Black indicates no detectable effect. Purple indicates phenotypes opposite to those of human 22q11.2 deletion carriers. Anx, anxiety-related behaviors; PPI, prepulse inhibition; Pup, neonatal vocalization; SI, social interaction or sociability; Voc Adult, adult vocalization; WM, working memory. Neonatal vocalization was recorded on and before neonatal day 12 under maternal separation, which reflects ASD-related pup social communication with a mother (Takahashi et al., 2016; Esposito et al., 2017; Kikusui and Hiroi, 2017; ÓBroin, 2018). Adult vocalization was recorded at 9 weeks of age in a view jar, arena, click box, geotaxis grid, and tube and collectively analyzed. Adult vocalization could reflect an adverse reaction to stress or expressions of anxiety. As no second mouse was present in any of the experimental settings, it does not reflect interactive social behavior. Please note that the data presented here are based on information available as of April 2, 2019. We set a lenient significance level at P < .05. Readers are advised to check the site for updates at http://www.mousephenotype.org/. Prodh (Paterlini et al., 2005; Koscielny et al., 2014); Dgcr2 (Koscielny et al., 2014); Slc25a1 (Koscielny et al., 2014); Mrpl40 (Devaraju et al., 2017); Sept5 (Harper et al., 2012; Hiroi et al., 2012; Koscielny et al., 2014); Gp1bβ (Koscielny et al., 2014); Tbx1 (Hiramoto et al., 2011; Takahashi et al., 2016); Comt (Papaleo et al., 2008; O’Tuathaigh et al., 2010, 2012; Koscielny et al., 2014); Tango2 (Koscielny et al., 2014); Dgcr8 (Fénelon et al., 2013; Ouchi et al., 2013; Chun et al., 2017); Ranbp1 (Koscielny et al., 2014); Rtn4r (Koscielny et al., 2014).
22q11.2 CNV
Unlike most CNVs that were more recently discovered in 2007 and 2008, the association between several CNVs and syndromic developmental disorders was known much earlier (Lee and Lupski, 2006). Robert Shprintzen and his colleagues at the Albert Einstein College of Medicine and the Montefiore Hospital were following up on patients exhibiting a cluster of physical symptoms including cleft palate, congenital heart defects, characteristic facial features, and learning disability (Shprintzen et al., 1978). They were found to have high rates of ID and delays in cognitive and language development (Golding-Kushner et al., 1985). In 1992, Shprintzen and colleagues reported that some of those patients also suffered from schizophrenia (Shprintzen et al., 1992). In the same year, several research teams, including Shprintzen’s, reported that the patients exhibiting the above-mentioned physical abnormalities carried deletions at chromosome 22q11.2 (Driscoll et al., 1992; Scambler et al., 1992). Together, these data established both an association between schizophrenia/learning disability and the physical symptoms, and an association between the physical symptoms and 22q11.2 hemizygous deletions. Since then, the direct association of 22q11.2 deletions with schizophrenia, learning disability, ID, anxiety, and ADHD has been consistently replicated by his group and others (Goldberg et al., 1993; Chow et al., 1994; Karayiorgou et al., 1995; Lindsay et al., 1995; McDonald-McGinn et al., 1997; Murphy et al., 1999; Sugama et al., 1999).
Although early estimates were confounded by small sample sizes and ascertainment biases, current estimates, which are based on large sample sizes, indicate that 22q11.2 deletions occur at the rate of approximately 0.3% in general schizophrenia samples (Kirov et al., 2014; Rees et al., 2014b; Marshall et al., 2017). Conversely, among carriers of 22q11.2 deletions, 30% of the adults develop schizophrenia (Schneider et al., 2014), 13% to 27% are diagnosed with ASD depending on their age, 16% to 36% are diagnosed with ADHD, and 24% to 36% are diagnosed with anxiety (Niklasson et al., 2002; Fine et al., 2005; Antshel et al., 2007; Kates et al., 2007; Schneider et al., 2014; Wenger et al., 2016; Hoeffding et al., 2017).
One important detail for consideration is that the cohorts of patients with schizophrenia used in large-scale studies and meta-analyses do not systematically include or exclude patients with ID diagnoses. Some cohorts were formed using inpatient samples that included patients with low IQ, and other cohorts may have excluded patients with ID. The exclusion of the ID comorbidity from schizophrenia samples is expected to underestimate the actual rate of such CNVs in schizophrenia, as almost half of the patients with 22q11.2 deletion are diagnosed with ID.
Duplication of 22q11.2 was also identified by a team of researchers at the Albert Einstein College of Medicine (Edelmann et al., 1999). Many cases of 22q11.2 duplication were shown to exhibit epilepsy, ID, ADHD, and ASD (Ensenauer et al., 2003; Hassed et al., 2004; Portnoï et al., 2005, 2009; Yobb et al., 2005; de La Rochebrochard et al., 2006; Alberti et al., 2007; Engels et al., 2007; Mukaddes and Herguner, 2007; Torres-Juan et al., 2007; Descartes et al., 2008; Ramelli et al., 2008; Wentzel et al., 2008; Yu et al., 2008; Lo-Castro et al., 2009; Soysal et al., 2011; Wenger et al., 2016) at rates higher than those of noncarriers (Hoeffding et al., 2017; Olsen et al., 2018). Schizophrenia samples have shown lower rates of 22q11.2 duplication compared with controls; therefore, this variant is considered to be a protective factor (Rees et al., 2014a). However, a recent study with a larger sample size did not find this effect to be statistically significant after genome-wide correction (Marshall et al., 2017). Additional cases of schizophrenia, psychosis, paranoia, hallucination, suicidal ideation, and mood disorders among 22q11.2 duplication carriers have also been reported (van Amelsvoort et al., 2016; Olsen et al., 2018; Woestelandt, 2018). Again, interpretation of these results should also consider how the samples were collected, as most duplication carriers have cognitive deficits and many are diagnosed with ID.
There are some individual variations in the locations and sizes of 22q11.2 duplication and hemizygous deletions. The majority of 22q11.2 deletions are approximately 3.0 Mb in size, but there are nested cases of 2.0-Mb or 1.5-Mb deletions and duplications; nested 1.5-Mb deletions help narrow down critical regions within the larger deletions (see Figure 1A).
Because the smallest nested deletion is still 1.5 Mb long, it is not possible to analyze how each 22q11.2 gene contributes to the symptoms of psychiatric disorders in humans. While there are individual cases of much smaller deletions (Girirajan et al., 2011), they are too few to statistically determine the definitive role of the encoded genes in mental illness. Researchers have attempted to correlate SNPs of some individual 22q11.2-encoded genes with psychiatric conditions in humans, but the results have been mixed and inconsistent, perhaps due to the weak effects of SNPs compared with deletions or duplications.
Predictive Power of Dimensions
The presence of a CNV by itself is a reliable predictive risk factor for the onset of psychiatric disorders. However, penetrance of CNVs is incomplete, and additional predictors that signal the impending onset of psychiatric disorders are needed. The underlying impetus to identify such predictors is that they might serve not only as a warning signal but also as a good starting point for early intervention. Moreover, identifying the mechanisms underlying those predictors may allow preventive therapeutic options to be used long before the onset of psychiatric disorders.
There are some known factors that predict the future onset of psychiatric disorders (Ozonoff et al., 2014). Typically, ASD is clinically diagnosed in children when they are 2 to 3 years of age. However, certain prognostic features, such as reduced eye contact (Jones and Klin, 2013) and atypical preverbal vocalizations (Esposito et al., 2017), could signal the future onset of ASD at least in a subpopulation (Zwaigenbaum et al., 2013; Estes et al., 2015; Ozonoff et al., 2018). Atypical cries do not optimally facilitate bonding or reciprocity between babies and mothers because the emotional state of such atypical cries is not easily understood (Esposito and Venuti, 2010) or is negatively perceived (Esposito et al., 2013) by mothers. Delayed motor and language development (Ozonoff et al., 2010, 2014) are also noted among incipient ASD babies; however, delayed motor development might not be specific to ASD, as it is also seen in children with other developmental issues (Iverson et al., 2019).
Individuals who develop schizophrenia after adolescence show atypical developmental trajectories in some domains before symptoms reach diagnostic thresholds. School performance, behavioral development, and various cognitive capacities, particularly complex cognition and social cognition, start to precipitously lag behind their peers 10 years before the onset of psychosis (van Oel et al., 2002; Ullman et al., 2012; Gur et al., 2014a). Children and adolescents who later develop schizophrenia lag in working memory, attention, and processing speed (David et al., 1997; Davidson et al., 1999; Fuller et al., 2002; Woodberry et al., 2008; Reichenberg et al., 2010; Bora et al., 2014; Meier et al., 2014; Bora and Pantelis, 2015; Seidman et al., 2016). Cognitive deficits in individuals with schizophrenia have been shown to affect a wide range of capacities, including attention, working memory, executive function, episodic memory, semantic memory, visual memory, verbal ability and learning, spatial memory and reasoning, face memory, emotion differentiation, verbal reasoning, list memory, processing speed, and fluency (Saykin et al., 1991; Yung and McGorry, 1996; Fioravanti et al., 2005; Piskulic et al., 2007; Goldenberg et al., 2012; Schaefer et al., 2013; Vangkilde et al., 2016).
Evidence from 22q11.2 deletion cases is consistent with this line of evidence obtained from idiopathic cases. The most profoundly impaired capacities are facial memory, identification of emotions from facial expressions, emotion intensity differentiation, and age differentiation of faces; other capacities are also impaired, including executive functions, verbal and spatial memory, verbal and nonverbal reasoning, and spatial reasoning. A delay in the developmental maturation of most of these features can be seen as early as 8 years of age among 22q11.2 deletion carriers compared with individuals with developmental delays or typically developing individuals (Gur et al., 2014b). In particular, some carriers of 22q11.2 deletions show a decline in IQ scores from childhood to adulthood, and the degree of this decline predicts the subsequent onset of psychosis (Gothelf et al., 2005, 2013; Vorstman et al., 2015).
Modeling CNVs
Model organisms complement human studies because specific genes or small chromosomal segments within a CNV can be isolated and the actual functional impact of their dose alterations on central nervous system functions and behaviors can be evaluated, which is not possible in humans. Moreover, the neuronal and molecular alterations caused by deletion or overexpression of each gene can be evaluated in model organisms. Worms, flies, fish, and mice have been used as models to identify the causal roles of genes in the manifestation of behavioral and neural phenotypes (Guna et al., 2015). However, there are some limitations. First, homologs of many human 22q11.2 genes are not present in lower species. Many human 22q11.2 protein-coding gene and microRNA homologs are not present in flies and worms (Guna et al., 2015). Second, relating genes to behaviors that are relevant to human psychiatric conditions is challenging in these organisms. The behavioral repertoire of worms, flies, and fish is limited, and higher-order cognitive functions that are affected in individuals with schizophrenia and ASD cannot be easily modeled. Third, there are obvious differences in the structure of the central nervous system between humans and flies, worms, and fish. Fourth, there are significant differences in the developmental stages between humans and those species.
Compared with those lower species, the murine and nonhuman primate models have fewer flaws. Many mouse models with large and small segmental deletions and duplication have been generated and tested. However, correlating behaviors to human psychiatric disorders is still challenging. There is no Diagnostic and Statistical Manual of Mental Disorders for mice or even nonhuman primates. There is no reliable or objective way to model hallucinations, delusion, and speech in mice or nonhuman primates. The anatomical basis for even more elemental cognitive functions (e.g., cognitive flexibility) might be different between humans and such model organisms; the degree of brain development is quantitatively and qualitatively different between humans and model organisms such as rodents and nonhuman primates (Cáceres et al., 2003; Semple et al., 2013; Rilling, 2014). Fundamental, motivational behaviors that are thought to occur across species show differences between species. Social behaviors in different species are based on different sensory cues—for example, smell and auditory cues in rodents vs visual and auditory cues in humans. Even nonhuman primates cannot completely overcome such limitations.
Nonetheless, some aspects or dimensions that are not necessarily specific to a disorder can be examined in both rodents and humans (Morris and Cuthbert, 2012). Mice exhibit certain behavioral dimensions that are thought to be relevant to human psychiatric conditions and are thus ideal for determining the genetic basis for those behaviors (Hiroi et al., 2012, 2013; Hiroi and Nishi, 2016; Nishi and Hiroi, 2016; Hiroi, 2018). For example, cognitive decline, including working memory deficits, is seen in patients with schizophrenia, ADHD, and ASD. Impaired social behaviors are seen, in various forms, in patients with ASD, schizophrenia, depression, and ADHD. Prepulse inhibition (PPI) is disrupted in patients with schizophrenia, obsessive compulsive disorder, and ADHD (Geyer, 2006). Examining such effects in mice may identify potential substrates involved in the causal relationship between CNVs and some aspects of psychiatric disorders (Hiroi and Nishi, 2016; Nishi and Hiroi, 2016; Hiroi, 2018).
Don’t Blame Behavior, Blame Experimental Designs
Variable or absent behavioral phenotypes have led some researchers to claim that mouse phenotypes, especially behavior, are inherently variable, and therefore neuronal and anatomical phenotypes should be used for analyzing the impact of genetic variants in mouse models. This misperception has arisen partly due to a lack of attention to the genetic background. Low reproducibility is not unique to behavioral phenotypes—anatomical, synaptic, molecular, and gene expression phenotypes are equally variable and not reproducible or interpretable in mutant models whose genetic background is not controlled.
When genetic background is systematically biased near the gene of interest in wild-type and mutant littermates, different mouse cohorts show varied degrees of such biases; so do independently developed mutant mouse lines of the gene. Such mutant and wild-type mice littermates do not share equally shuffled alleles near the deleted gene; more alleles of embryonic stem cells (many are of a 129 inbred mouse line) and of the breeder mice (often C57BL/6J), respectively, are present in mutant and wild-type littermates near the target gene because of lower recombination rates between that gene and nearby alleles compared with distant alleles (Hiroi, 2018). In such mice or noncongenic mice, any phenotypic difference in behavior, anatomy, synaptic, and neurochemical functions or gene expression between wild-type and mutant mice cannot be attributed to the target gene (Gerlai, 2001; Wolfer et al., 2002; Crusio, 2004; Zoghbi and Warren, 2010; Hiroi, 2018).
This point has been elegantly proven by a series of studies that have shown some reproducible phenotypic differences in gene expression patterns that can be truly attributed to the target gene when the genetic background is made increasingly homogeneous between mutant and wild-type littermates (Valor and Grant, 2007; Yang et al., 2007; Ricard et al., 2010; O’Leary and Osborne, 2011). Confounding gene expression alterations resulting from allelic differences between noncongenic mutant and wild-type mice are likely to impact other phenotypes, either independently or interactively with the target genes, as different inbred mouse lines have different baselines for the shape and volume of various anatomical structures (Wahlsten et al., 2003; Chen et al., 2006; Routh et al., 2009), the number of neurons (Schwegler et al., 1996a, 1996b; Kempermann and Gage, 2002; Yilmazer-Hanke et al., 2003; Routh et al., 2009), the shape and number of dendritic spines (Restivo et al., 2006), the degree of synaptic plasticity (Nguyen et al., 2000a, 2000b; Moore et al., 2011), and behavioral features including PPI (Crawley et al., 1997), working memory (Crawley et al., 1997), social behaviors (Crawley et al., 1997; Faure et al., 2017), and neonatal vocalization (Scattoni et al., 2008, 2011; Faure et al., 2017). In fact, a behavioral phenotypic difference appears or disappears when the genetic background is altered by the type of inbred mice used (Suzuki et al., 2009b; Hiroi et al., 2012).
One obvious solution to this problem is to make the genetic background homogeneous, as much as practically possible, by backcrossing a mutant to one inbred mouse line for over 10 generations (i.e., congenic mouse). Many well-designed studies have painstakingly developed congenic mutant mice. Although it is not possible to make the allelic differences between congenic mutant and wild-type littermates completely homogenous (Bolivar et al., 2001; Flaherty and Bolivar, 2007; Hiroi, 2018), it still considerably reduces interpretative uncertainty. Another more fundamental approach is to generate a mutant mouse with embryonic stem cells derived from C57BL/6N mice and breed with C57BL/6N mice (co-isogenic mouse). The International Mouse Phenotyping Consortium (http://www.mousephenotype.org/) routinely generates and tests many co-isogenic mouse models for PPI and adult vocalization (Koscielny et al., 2014). The use of co-isogenic mutant mice completely eliminates the confounding effects of systematic bias in allelic distributions near the target gene. Moreover, standardized behavioral assays are applied by the International Mouse Phenotyping Consortium, further increasing the level of reproducibility and interpretability.
Unfortunately, many studies have failed to reproduce original published results of noncongenic mutant mice. Some studies have indicated in vague terms that their mice are of “pure” C57BL/6J background or of “a C57BL/6J background.” Often the cited references therein indicate that the original mice are noncongenic mice. Yet the authors do not elaborate on what is meant by “pure” and how “a C57BL/6J background” was achieved; technically, a completely homogenous genetic background—if that is meant by “pure”—is not possible when starting with noncongenic mice (Bolivar et al., 2001; Flaherty and Bolivar, 2007). Even when the data are reproducible in noncongenic mice, they might simply reflect the hard-to-break allelic differences near the target gene rather than the actual effects of target genes. Therefore, we will limit our discussion of mouse models to congenic or co-isogenic mice.
In Search of Individual 22q11.2 Driver Genes
Mouse models were developed to recapitulate the effects of human 22q11.2 duplications and deletions, small chromosomal segments, or individual genes on psychiatric disorders. We reported the first segmental duplication model of a 200-kb human chromosomal segment, which included human SEPT5, GP1BB, TBX1, and GNB1L (Hiroi et al., 2005). This co-isogenic model was hyperactive, showed progressive exacerbation of hyperactivity in a stressful open field, and was unable to engage in reciprocal social interaction or building a nest. Satisfying predictive validity, 3 weeks of treatment with the antipsychotic drug clozapine attenuated the exacerbated, compulsive, and repetitive hyperactivity in these mice.
Our subsequent analysis focused on an adjacent approximately 190-kb human chromosomal segment, including TXNRD2, COMT, and ARVCF (Suzuki et al., 2009a). Congenic mice that overexpressed this segment were selectively impaired in the developmental maturation of working memory capacity from adolescence to adulthood; they were indistinguishable from wild-type littermates in acoustic PPI, reciprocal social interaction, or behaviors related to fear, anxiety, and mood. COMT, among the 3 encoded genes, is likely to be a contributory gene, as individuals with a high-activity allele of COMT show a blunted expansion of working memory capacity from adolescence to adulthood in humans (Dumontheil et al., 2011).
These 2 mouse models demonstrated that social/repetitive behaviors and developmental maturation of working memory are selectively impacted by high gene doses of the 200-kb and 190-kb chromosomal segments, respectively.
There are also mouse models of large hemizygous deletions of 22q11.2. A congenic Dgcr2-Ufd1l deletion model (Chun et al., 2014) and a co-isogenic Dgcr2-Hira deletion model (Didriksen et al., 2017) consistently showed defective acoustic PPI (see Figure 1B for gene locations on the murine chromosome). A congenic Dgcr2-Hira deletion model (Sigurdsson et al., 2010) and co-isogenic Dgcr2-Hira deletion model (Nilsson et al., 2016) exhibited deficits in the acquisition phase of working memory.
Known biology guided early studies of single 22q11.2 driver genes, and genes of unknown functions were not examined. For example, early studies focused on the genes involved in synaptic transmission and metabolism of glutamate and dopamine because those neurotransmitters had been implicated in several psychiatric disorders. However, some results from those studies have been irreproducible partly because most studies used noncongenic mice. Moreover, known biology was also limited at that time. Since then, studies using congenic and co-isogenic mice have provided a wealth of reliable data, which have drastically revised our understanding of the contributions of individual 22q11.2-encoded genes to dimensional behaviors of psychiatric disorders, and new potential genes and neurobiological bases have emerged (Hiroi et al., 2013; Hiroi and Nishi, 2016; Nishi and Hiroi, 2016; Hiroi, 2018; Zinkstok et al., 2019).
The current mouse data do not support the view that all 22q11.2-encoded genes equally contribute to each dimension of mental illness (see Figure 1C). For example, there are 22q11.2 genes that globally affect many dimensions of psychiatric disorders (e.g., Tbx1). Other genes are essential for a select set of behavioral dimensions (e.g., Sept5, Dgcr8, Comt, and Prodh). Deletions of many other 22q11.2 genes have not had an effect on the behavioral dimensions tested so far; however, a more extensive characterization of behavioral dimensions may reveal specific aspects relevant to mental illness that may be regulated by those genes (Hiroi et al., 2012, 2013, 2018; Hiroi and Nishi, 2016; Nishi and Hiroi, 2016).
Tbx1
Tbx1 encodes a transcription factor and is 1 of the 4 protein-coding genes encoded in the 200-kb segment that we identified (Hiroi et al., 2005). When compared with wild-type littermates, congenic Tbx1 heterozygous mice scored lower in reciprocal social interaction and a working memory index and higher in anxiety-related behaviors. They also exhibited higher levels of an approach response to a nonsocial object at 2 months of age (Hiramoto et al., 2011). While cases of human Tbx1 mutations exist and are associated with ASD (Gong et al., 2001; Paylor et al., 2006; Ogata et al., 2014), they carry additional mutations and other variants (Ogata et al., 2014); therefore, a causal role of TBX1 in ASD could not be definitively established. Our mouse studies complemented this limitation of the human studies.
Atypical behaviors in Tbx1 heterozygous mice start to appear as early as on postnatal day 8. The sequence of vocal calls is altered in Tbx1 heterozygous pups, which lowers the incentive for mothers to approach them (Takahashi et al., 2016). In these mice, the atypical call sequence at the age of postnatal day 8 serves as a predictor for atypical social and cognitive dimensions at adolescence. This is consistent with the observations made in babies incipient for idiopathic ASD—those that were eventually diagnosed with ASD had emitted atypical cries (Esposito et al., 2017). As atypical cries among incipient ASD babies are often prognostic of the future outcome, such neonatal behavioral dimensions can be utilized to understand the mechanistic basis of the developmental trajectory of ASD (Esposito et al., 2017; Kikusui and Hiroi, 2017; ÓBroin, 2018).
Adult neurogenesis has emerged as a potential mechanistic basis for low working memory capacity. Tbx1 protein expression is enriched in adult neural progenitor cells in the adolescent mouse brain (Hiramoto et al., 2011). When this gene is overexpressed in adult neural progenitor cells in the hippocampus, the mice exhibit a blunting of working memory capacity at 2 months of age; however, the same treatment has no detectable effect at 1 month of age (Boku et al., 2018). These observations suggest that Tbx1 might impact the developmental maturation of working memory capacity via its effects on adult neurogenesis in 22q11.2 duplication cases. This might model the adult onset of working memory deficits seen in adult ASD patients, but it is unlikely that human TBX1 plays a role in the early onset of cognitive impairments in patients with ID.
Dgcr8
Dgcr8 is another 22q11.2 gene that impacts more than one known dimension. Heterozygous mice are impaired in acoustic PPI, working memory, and spatial memory; however, anxiety-related behaviors are not affected. Dgcr8 impact on social interaction has not been examined (Fénelon et al., 2013; Ouchi et al., 2013; Chun et al., 2017). Potential mechanisms through which Dgcr8 deficiency alters behavioral dimensions include adult neurogenesis (Ouchi et al., 2013), disturbances in microRNA expression (Fénelon et al., 2013; Chun et al., 2017), short-term synaptic plasticity in the cortex (Fénelon et al., 2013), and synaptic transmission of the thalamocortical projection (Chun et al., 2017).
Comt
Although Comt is one of the 22q11.2 genes that is hemizygous in 22q11.2 deletion cases, heterozygous or homozygous deletion of this gene does not recapitulate the impacts of the whole genome deletion, suggesting that a low dose of Comt does not, by itself, contribute to the dimensions of 22q11.2 deletion cases. Homozygous deletion of Comt also has no effect on PPI (O’Tuathaigh et al., 2012; Koscielny et al., 2014). Neither homozygous nor heterozygous deletion of Comt affects sociability (O’Tuathaigh et al., 2010, 2012). Heterozygous or homozygous deletion of Comt has no detrimental effect on working memory (Papaleo et al., 2008; O’Tuathaigh et al., 2010).
Our recent work implicates adult neurogenesis as a potential mechanism through which a high gene dose of COMT blunts the developmental maturation of working memory capacity at adolescence (Suzuki et al., 2009a; Boku et al., 2018). This mechanism could be targeted for cognitive improvement in the duplication cases of 22q11.2. The methylating action of COMT on catecholamines and other catechol-carrying molecules might underlie this action of COMT overexpression on working memory.
Sept5
Congenic Sept5 homozygous mice have lower basal levels of social interaction compared with wild-type littermates (Harper et al., 2012; Hiroi et al., 2012). This observation is consistent with one reported case of SEPT5 and GP1BB deletion with developmental delays (Bartsch et al., 2011). Sept5 inhibits excess release of dopamine and glutamate and contributes to the formation of axonal and dendritic arborization (Beites et al., 1999, 2001; Dong et al., 2003; Yang et al., 2010). However, social interaction is enhanced when Sept5 is overexpressed in the dorsal hippocampus and amygdala in mice (Harper et al., 2012); this suggests that high doses of this gene, at least in these 2 structures, do not faithfully recapitulate 22q11.2 hemizygous deletion and duplication cases.
Genes encoding synaptic functions and activity-regulated cytoskeleton-associated proteins are considered to be functional categories for schizophrenia-associated CNVs (Marshall et al., 2017). SEPT5 is implicated in cytoskeletal organization and synaptic transmission (Beites et al., 1999; Yang et al., 2010) and is a component of the biological pathways of synaptic genes implicated in schizophrenia with CNVs (Marshall et al., 2017) and in 22q11.2 Gene Interaction Network and 22q11.2-linked pathway interactions (Bassett et al., 2017) in humans. However, homozygous or heterozygous deletion of Sept5 in mice does not impair PPI, a sensorimotor gating dimension of schizophrenia (Koscielny et al., 2014). Similarly, proteome analyses in humans with 22q11.2 hemizygosity identified SLC25A1 and its family members as critical for synaptic morphology and plasticity and mitochondrial functions (Gokhale et al., 2019). However, deletions of these genes do not impair PPI in mice (Figure 1) (Koscielny et al., 2014). These observations in humans and mice may not be conflicting because PPI is just one dimensional aspect of schizophrenia and is not a proxy for clinically defined schizophrenia. Sept5 and SLC25A1 deficiency might have a rather selective effect on some unidentified behavioral dimensions in patients with schizophrenia.
When the genetic background of Sept5 homozygous mice was experimentally altered using different inbred mouse lines, the degree of social interaction deficits changed (Suzuki et al., 2009b; Harper et al., 2012; Hiroi et al., 2012). Because of wide variations between individual SNPs in the genome, epistatic interactions between a CNV and SNPs outside or inside a CNV might phenotypically manifest differently among carriers. This may explain the diverse clinical diagnoses of the 22q11.2 CNV.
Prodh
Deletion of Prodh alone does not cause PPI deficits in a congenic genetic background (Paterlini et al., 2005). Further, a co-isogenic Prodh mouse is normal in PPI (Koscielny et al., 2014). Male co-isogenic Prodh homozygous mice exhibit atypical adult vocalization; female homozygous mice are normal (Koscielny et al., 2014). Because adult vocalization was measured in many experimental settings such as open field, transparent tube, and cylinder and collectively analyzed in this assay, it is not clear whether such atypical vocalizations in males were an adverse reaction to stress or expressions of anxiety; it is not an expression of interactive social behavior, as no second mouse was present in any of the experimental settings.
Almost all genes encoded in the 22q11.2 CNV have some neuronal and molecular functions; however, mouse data indicate that not all encoded genes contribute to dimensions of psychiatric disorders. From a phenotypic perspective (see Figure 1C), genes that have been found to reduce PPI include Gp1bβ and Dgcr8; PPI is not affected by the deletion of Prodh, Dgcr2, Mrpl40, Sept5, Comt, Tango2, Ranbp1, and Rtn4r. Working memory is negatively affected by the deletion of Mpril40, Tbx1, and Dgcr8; Comt deletion improves working memory. Sociability or reciprocal social interaction is reduced by the deletion of Sept5 and Tbx1, but not by Comt deletion. Neonatal vocalization under a maternal separation test is negatively impacted by the heterozygous deletion of Tbx1. Adult vocalization under stress- or anxiety-provoking settings is reduced by the deletion of Prodh; the deletions of Dgcr2, Slc25a1, Sept5, Gp1bβ, Comt, Tango2, Ranbp1, and Rtn4r have no obvious effect. Anxiety-related behaviors are induced by the deletion of Tbx1 but not by the deletion of Sept5, Comt, or Dgcr8.
There are 22q11.2 genes that induce robust anatomical abnormalities (e.g., Dgcr2 and Ranbp1) (Paronett et al., 2015; Molinard-Chenu and Dayer, 2018) without affecting PPI or adult vocalization (Koscielny et al., 2014). It is possible that the behavioral dimensions affected by such anatomical alterations have not yet been identified and examined. As more phenotypes are characterized, a more complete picture is likely to emerge. Nonetheless, neuronal, molecular, or gene expression phenotypes in the mouse brain should have an effect on behavioral dimensions before they can be safely considered as factors that contribute to the development of human psychiatric disorders (Hiroi et al., 2013; Hiroi and Nishi, 2016; Nishi and Hiroi, 2016; Hiroi, 2018).
Conclusions
CNVs have provided much hope for a deeper understanding of the mechanistic basis of mental illness. The pleiotropic actions of a CNV on many psychiatric disorders might challenge and revise the current paradigms of distinct psychiatric disorders. We are, however, still constrained by our limited technical capability to fully tap the potential of CNVs to improve our understanding of mental illness and develop better therapeutic options. Nonetheless, some hypotheses have emerged from mouse models of CNVs. An in-depth characterization of CNV-encoded genes in brain biology and their behavioral manifestation in mouse models is a prerequisite to reconstruct the precise functional gene-behavior relationship and to develop mechanism-based therapy for the various dimensional aspects of mental illness.
Acknowledgments
We thank Drs George Kirov, Jonathan Sebat, Santhosh Girirajan, Søren Brunak, Ruairidh King, and Kenny Ye for their comments. Research reported in this publication was partly supported by the National Institute of Mental Health of the National Institutes of Health under Award Numbers R01MH099660, R01DC015776, R21HD053114, and U54HD090260 to N.H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or any other individual.
Statement of Interest
None.
References
- Ahn K, Gotay N, Andersen TM, Anvari AA, Gochman P, Lee Y, Sanders S, Guha S, Darvasi A, Glessner JT, Hakonarson H, Lencz T, State MW, Shugart YY, Rapoport JL (2014) High rate of disease-related copy number variations in childhood onset schizophrenia. Mol Psychiatry 19:568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti A, Romano C, Falco M, Calì F, Schinocca P, Galesi O, Spalletta A, Di Benedetto D, Fichera M (2007) 1.5 mb de novo 22q11.21 microduplication in a patient with cognitive deficits and dysmorphic facial features. Clin Genet 71:177–182. [DOI] [PubMed] [Google Scholar]
- Amelsvoort TV, Denayer A, Boermans J, Swillen A (2016) Psychotic disorder associated with 22q11.2 duplication syndrome. Psychiatry Res 236:206–207. [DOI] [PubMed] [Google Scholar]
- Antshel KM, Aneja A, Strunge L, Peebles J, Fremont WP, Stallone K, Abdulsabur N, Higgins AM, Shprintzen RJ, Kates WR (2007) Autistic spectrum disorders in velo-cardio facial syndrome (22q11.2 deletion). J Autism Dev Disord 37:1776–1786. [DOI] [PubMed] [Google Scholar]
- Bartsch I, Sandrock K, Lanza F, Nurden P, Hainmann I, Pavlova A, Greinacher A, Tacke U, Barth M, Busse A, Oldenburg J, Bommer M, Strahm B, Superti-Furga A, Zieger B (2011) Deletion of human GP1BB and SEPT5 is associated with bernard-soulier syndrome, platelet secretion defect, polymicrogyria, and developmental delay. Thromb Haemost 106:475–483. [DOI] [PubMed] [Google Scholar]
- Bassett AS, et al. ; International 22q11.2DS Brain and Behavior Consortium (2017) Rare genome-wide copy number variation and expression of schizophrenia in 22q11.2 deletion syndrome. Am J Psychiatry 174:1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beites CL, Xie H, Bowser R, Trimble WS (1999) The septin cdcrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci 2:434–439. [DOI] [PubMed] [Google Scholar]
- Beites CL, Peng XR, Trimble WS (2001) Expression and analysis of properties of septin cdcrel-1 in exocytosis. Methods Enzymol 329:499–510. [DOI] [PubMed] [Google Scholar]
- Boku S, Izumi T, Abe S, Takahashi T, Nishi A, Nomaru H, Naka Y, Kang G, Nagashima M, Hishimoto A, Enomoto S, Duran-Torres G, Tanigaki K, Zhang J, Ye K, Kato S, Männistö PT, Kobayashi K, Hiroi N (2018) Copy number elevation of 22q11.2 genes arrests the developmental maturation of working memory capacity and adult hippocampal neurogenesis. Mol Psychiatry 23:985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar VJ, Cook MN, Flaherty L (2001) Mapping of quantitative trait loci with knockout/congenic strains. Genome Res 11:1549–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora E, Lin A, Wood SJ, Yung AR, McGorry PD, Pantelis C (2014) Cognitive deficits in youth with familial and clinical high risk to psychosis: a systematic review and meta-analysis. Acta Psychiatr Scand 130:1–15. [DOI] [PubMed] [Google Scholar]
- Bora E, Pantelis C (2015) Meta-analysis of cognitive impairment in first-episode bipolar disorder: comparison with first-episode schizophrenia and healthy controls. Schizophr Bull 41:1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti-Pierri N, et al. (2008) Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet 40:1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C (2003) Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci U S A 100:13030–13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D (2015) Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci 18:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Kovacevic N, Lobaugh NJ, Sled JG, Henkelman RM, Henderson JT (2006) Neuroanatomical differences between mouse strains as shown by high-resolution 3D MRI. Neuroimage 29:99–105. [DOI] [PubMed] [Google Scholar]
- Chow EW, Bassett AS, Weksberg R (1994) Velo-cardio-facial syndrome and psychotic disorders: implications for psychiatric genetics. Am J Med Genet 54:107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, Karamohamed S, Badner JA, Matsui S, Conroy J, McQuaid D, Gergel J, Hatchwell E, Gilliam TC, Gershon ES, Nowak NJ, Dobyns WB, Cook EH Jr (2008) Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry 63:1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Westmoreland JJ, Bayazitov IT, Eddins D, Pani AK, Smeyne RJ, Yu J, Blundon JA, Zakharenko SS (2014) Specific disruption of thalamic inputs to the auditory cortex in schizophrenia models. Science 344:1178–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Du F, Westmoreland JJ, Han SB, Wang YD, Eddins D, Bayazitov IT, Devaraju P, Yu J, Mellado Lagarde MM, Anderson K, Zakharenko SS (2017) Thalamic mir-338-3p mediates auditory thalamocortical disruption and its late onset in models of 22q11.2 microdeletion. Nat Med 23:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, et al. (2011) A copy number variation morbidity map of developmental delay. Nat Genet 43:838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN, Belknap JK, Collins A, Crabbe JC, Frankel W, Henderson N, Hitzemann RJ, Maxson SC, Miner LL, Silva AJ, Wehner JM, Wynshaw-Boris A, Paylor R (1997) Behavioral phenotypes of inbred mouse strains: implications and recommendations for molecular studies. Psychopharmacology (Berl) 132:107–124. [DOI] [PubMed] [Google Scholar]
- Crusio WE. (2004) Flanking gene and genetic background problems in genetically manipulated mice. Biol Psychiatry 56:381–385. [DOI] [PubMed] [Google Scholar]
- David AS, Malmberg A, Brandt L, Allebeck P, Lewis G (1997) IQ and risk for schizophrenia: a population-based cohort study. Psychol Med 27:1311–1323. [DOI] [PubMed] [Google Scholar]
- Davidson M, Reichenberg A, Rabinowitz J, Weiser M, Kaplan Z, Mark M (1999) Behavioral and intellectual markers for schizophrenia in apparently healthy male adolescents. Am J Psychiatry 156:1328–1335. [DOI] [PubMed] [Google Scholar]
- de La Rochebrochard C, Joly-Helas G, Goldenberg A, Durand I, Laquerriere A, Ickowicz V, Saugier-Veber P, Eurin D, Moirot H, Diguet A, de KF, Tiercin C, Mace B, Marpeau L, Frebourg T (2006) The intrafamilial variability of the 22q11.2 microduplication encompasses a spectrum from minor cognitive deficits to severe congenital anomalies. Am J Med Genet A 140:1608–1613. [DOI] [PubMed] [Google Scholar]
- Descartes M, Franklin J, Diaz de Ståhl T, Piotrowski A, Bruder CE, Dumanski JP, Carroll AJ, Mikhail FM (2008) Distal 22q11.2 microduplication encompassing the BCR gene. Am J Med Genet A 146A:3075–3081. [DOI] [PubMed] [Google Scholar]
- Devaraju P, Yu J, Eddins D, Mellado-Lagarde MM, Earls LR, Westmoreland JJ, Quarato G, Green DR, Zakharenko SS (2017) Haploinsufficiency of the 22q11.2 microdeletion gene mrpl40 disrupts short-term synaptic plasticity and working memory through dysregulation of mitochondrial calcium. Mol Psychiatry 22:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didriksen M, Fejgin K, Nilsson SR, Birknow MR, Grayton HM, Larsen PH, Lauridsen JB, Nielsen V, Celada P, Santana N, Kallunki P, Christensen KV, Werge TM, Stensbol TB, Egebjerg J, Gastambide F, Artigas F, Bastlund JF, Nielsen J (2017) Persistent gating deficit and increased sensitivity to NMDA receptor antagonism after puberty in a new mouse model of the human 22q11.2 microdeletion syndrome: a study in male mice. J Psychiatry Neurosci 41:150381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Ferger B, Paterna JC, Vogel D, Furler S, Osinde M, Feldon J, Bueler H (2003) Dopamine-dependent neurodegeneration in rats induced by viral vector-mediated overexpression of the parkin target protein, CDCrel-1. Proc Natl Acad Sci U S A 100:12438–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DA, Spinner NB, Budarf ML, McDonald-McGinn DM, Zackai EH, Goldberg RB, Shprintzen RJ, Saal HM, Zonana J, Jones MC (1992) Deletions and microdeletions of 22q11.2 in velo-cardio-facial syndrome. Am J Med Genet 44:261–268. [DOI] [PubMed] [Google Scholar]
- Dumontheil I, Roggeman C, Ziermans T, Peyrard-Janvid M, Matsson H, Kere J, Klingberg T (2011) Influence of the COMT genotype on working memory and brain activity changes during development. Biol Psychiatry 70:222–229. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE (1999) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167. [DOI] [PubMed] [Google Scholar]
- Elia J, et al. (2011) Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet 44:78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels H, Brockschmidt A, Hoischen A, Landwehr C, Bosse K, Walldorf C, Toedt G, Radlwimmer B, Propping P, Lichter P, Weber RG (2007) DNA microarray analysis identifies candidate regions and genes in unexplained mental retardation. Neurology 68:743–750. [DOI] [PubMed] [Google Scholar]
- Ensenauer RE, Adeyinka A, Flynn HC, Michels VV, Lindor NM, Dawson DB, Thorland EC, Lorentz CP, Goldstein JL, McDonald MT, Smith WE, Simon-Fayard E, Alexander AA, Kulharya AS, Ketterling RP, Clark RD, Jalal SM (2003) Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am J Hum Genet 73:1027–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Hiroi N, Scattoni ML (2017) Cry, baby, cry: expression of distress as a biomarker and modulator in autism spectrum disorder. Int J Neuropsychopharmacol 20:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Nakazawa J, Venuti P, Bornstein MH (2013) Componential deconstruction of infant distress vocalizations via tree-based models: a study of cry in autism spectrum disorder and typical development. Res Dev Disabil 34:2717–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Venuti P (2010) Understanding early communication signals in autism: A study of the perception of infants’ cry. J Intellect Disabil Res 54:216–223. [DOI] [PubMed] [Google Scholar]
- Estes A, Zwaigenbaum L, Gu H, St John T, Paterson S, Elison JT, Hazlett H, Botteron K, Dager SR, Schultz RT, Kostopoulos P, Evans A, Dawson G, Eliason J, Alvarez S, Piven J; IBIS network (2015) Behavioral, cognitive, and adaptive development in infants with autism spectrum disorder in the first 2 years of life. J Neurodev Disord 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure A, Pittaras E, Nosjean A, Chabout J, Cressant A, Granon S (2017) Social behaviors and acoustic vocalizations in different strains of mice. Behav Brain Res 320:383–390. [DOI] [PubMed] [Google Scholar]
- Fénelon K, Xu B, Lai CS, Mukai J, Markx S, Stark KL, Hsu PK, Gan WB, Fischbach GD, MacDermott AB, Karayiorgou M, Gogos JA (2013) The pattern of cortical dysfunction in a mouse model of a schizophrenia-related microdeletion. J Neurosci 33:14825–14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine SE, Weissman A, Gerdes M, Pinto-Martin J, Zackai EH, McDonald-McGinn DM, Emanuel BS (2005) Autism spectrum disorders and symptoms in children with molecularly confirmed 22q11.2 deletion syndrome. J Autism Dev Disord 35:461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravanti M, Carlone O, Vitale B, Cinti ME, Clare L (2005) A meta-analysis of cognitive deficits in adults with a diagnosis of schizophrenia. Neuropsychol Rev 15:73–95. [DOI] [PubMed] [Google Scholar]
- Flaherty L, Bolivar V (2007) Congenic and consomic strains. In: Neurobehavioral genetics, 2nd edition (Jones BC, Mormede P, eds), pp115–127. New York: Taylor & Francis. [Google Scholar]
- Fuller R, Nopoulos P, Arndt S, O’Leary D, Ho BC, Andreasen NC (2002) Longitudinal assessment of premorbid cognitive functioning in patients with schizophrenia through examination of standardized scholastic test performance. Am J Psychiatry 159:1183–1189. [DOI] [PubMed] [Google Scholar]
- Gerlai R. (2001) Gene targeting: technical confounds and potential solutions in behavioral brain research. Behav Brain Res 125:13–21. [DOI] [PubMed] [Google Scholar]
- Geyer MA. (2006) The family of sensorimotor gating disorders: comorbidities or diagnostic overlaps? Neurotox Res 10:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, Shafer N, Bernier R, Ferrero GB, Silengo M, Warren ST, Moreno CS, Fichera M, Romano C, Raskind WH, Eichler EE (2011) Relative burden of large cnvs on a range of neurodevelopmental phenotypes. PLoS Genet 7:e1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A, et al. (2019) Systems analysis of the 22q11.2 microdeletion syndrome converges on a mitochondrial interactome necessary for synapse function and behavior. J Neurosci 39:3561–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R, Motzkin B, Marion R, Scambler PJ, Shprintzen RJ (1993) Velo-cardio-facial syndrome: a review of 120 patients. Am J Med Genet 45:313–319. [DOI] [PubMed] [Google Scholar]
- Goldenberg PC, Calkins ME, Richard J, McDonald-McGinn D, Zackai E, Mitra N, Emanuel B, Devoto M, Borgmann-Winter K, Kohler C, Conroy CG, Gur RC, Gur RE (2012) Computerized neurocognitive profile in young people with 22q11.2 deletion syndrome compared to youths with schizophrenia and at-risk for psychosis. Am J Med Genet B Neuropsychiatr Genet 159B:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding-Kushner KJ, Weller G, Shprintzen RJ (1985) Velo-cardio-facial syndrome: language and psychological profiles. J Craniofac Genet Dev Biol 5:259–266. [PubMed] [Google Scholar]
- Gong W, Gottlieb S, Collins J, Blescia A, Dietz H, Goldmuntz E, McDonald-McGinn DM, Zackai EH, Emanuel BS, Driscoll DA, Budarf ML (2001) Mutation analysis of TBX1 in non-deleted patients with features of DGS/VCFS or isolated cardiovascular defects. J Med Genet 38:E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothelf D, Eliez S, Thompson T, Hinard C, Penniman L, Feinstein C, Kwon H, Jin S, Jo B, Antonarakis SE, Morris MA, Reiss AL (2005) COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat Neurosci 8:1500–1502. [DOI] [PubMed] [Google Scholar]
- Gothelf D, Schneider M, Green T, Debbané M, Frisch A, Glaser B, Zilkha H, Schaer M, Weizman A, Eliez S (2013) Risk factors and the evolution of psychosis in 22q11.2 deletion syndrome: a longitudinal 2-site study. J Am Acad Child Adolesc Psychiatry 52:1192–1203.e3. [DOI] [PubMed] [Google Scholar]
- Guna A, Butcher NJ, Bassett AS (2015) Comparative mapping of the 22q11.2 deletion region and the potential of simple model organisms. J Neurodev Disord 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RC, Calkins ME, Satterthwaite TD, Ruparel K, Bilker WB, Moore TM, Savitt AP, Hakonarson H, Gur RE (2014a) Neurocognitive growth charting in psychosis spectrum youths. JAMA Psychiatry 71:366–374. [DOI] [PubMed] [Google Scholar]
- Gur RE, Yi JJ, McDonald-McGinn DM, Tang SX, Calkins ME, Whinna D, Souders MC, Savitt A, Zackai EH, Moberg PJ, Emanuel BS, Gur RC (2014b) Neurocognitive development in 22q11.2 deletion syndrome: comparison with youth having developmental delay and medical comorbidities. Mol Psychiatry 19:1205–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper KM, Hiramoto T, Tanigaki K, Kang G, Suzuki G, Trimble W, Hiroi N (2012) Alterations of social interaction through genetic and environmental manipulation of the 22q11.2 gene sept5 in the mouse brain. Hum Mol Genet 21:3489–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassed SJ, Hopcus-Niccum D, Zhang L, Li S, Mulvihill JJ (2004) A new genomic duplication syndrome complementary to the velocardiofacial (22q11 deletion) syndrome. Clin Genet 65:400–404. [DOI] [PubMed] [Google Scholar]
- Hiramoto T, Kang G, Suzuki G, Satoh Y, Kucherlapati R, Watanabe Y, Hiroi N (2011) Tbx1: identification of a 22q11.2 gene as a risk factor for autism spectrum disorder in a mouse model. Hum Mol Genet 20:4775–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N. (2018) Critical reappraisal of mechanistic links of copy number variants to dimensional constructs of neuropsychiatric disorders in mouse models. Psychiatry Clin Neurosci 72:301–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N, Hiramoto T, Harper KM, Suzuki G, Boku S (2012) Mouse models of 22q11.2-associated autism spectrum disorder. Autism Open Access Suppl 1:001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N, Nishi A (2016) Dimensional deconstruction and reconstruction of CNV-associated neuropsychiatric disorders. In: Modeling the psychopathological dimensions of schizophrenia: from molecules to behavior (Pletnikov MV, Waddington JL, eds), pp 285–302. Amsterdam: Academic Press, Elsevier. [Google Scholar]
- Hiroi N, Takahashi T, Hishimoto A, Izumi T, Boku S, Hiramoto T (2013) Copy number variation at 22q11.2: from rare variants to common mechanisms of developmental neuropsychiatric disorders. Mol Psychiatry 18:1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N, Zhu H, Lee M, Funke B, Arai M, Itokawa M, Kucherlapati R, Morrow B, Sawamura T, Agatsuma S (2005) A 200-kb region of human chromosome 22q11.2 confers antipsychotic-responsive behavioral abnormalities in mice. Proc Natl Acad Sci U S A 102:19132–19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffding LK, Trabjerg BB, Olsen L, Mazin W, Sparsø T, Vangkilde A, Mortensen PB, Pedersen CB, Werge T (2017) Risk of psychiatric disorders among individuals with the 22q11.2 deletion or duplication: a Danish nationwide, register-based study. JAMA Psychiatry 74:282–290. [DOI] [PubMed] [Google Scholar]
- Iverson JM, et al. (2019) Early motor abilities in infants at heightened versus low risk for ASD: a baby siblings research consortium (BSRC) study. J Abnorm Psychol 128:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W, Klin A (2013) Attention to eyes is present but in decline in 2-6-month-old infants later diagnosed with autism. Nature. 504:427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK (1995) Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A 92:7612–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kates WR, Antshel KM, Fremont WP, Shprintzen RJ, Strunge LA, Burnette CP, Higgins AM (2007) Comparing phenotypes in patients with idiopathic autism to patients with velocardiofacial syndrome (22q11 DS) with and without autism. Am J Med Genet A 143A:2642–2650. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Gage FH (2002) Genetic influence on phenotypic differentiation in adult hippocampal neurogenesis. Brain Res Dev Brain Res 134:1–12. [DOI] [PubMed] [Google Scholar]
- Kikusui T, Hiroi N (2017) A self-generated environmental factor as a potential contributor to atypical early social communication in autism. Neuropsychopharmacology 42:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O’Donovan MC; International Schizophrenia Consortium; Wellcome Trust Case Control Consortium (2009) Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet 18:1497–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Rees E, Walters JT, Escott-Price V, Georgieva L, Richards AL, Chambert KD, Davies G, Legge SE, Moran JL, McCarroll SA, O’Donovan MC, Owen MJ (2014) The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry 75:378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscielny G, et al. (2014) The international mouse phenotyping consortium web portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res 42:D802–D809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushima I, et al. (2018) Comparative analyses of copy-number variation in autism spectrum disorder and schizophrenia reveal etiological overlap and biological insights. Cell Rep 24:2838–2856. [DOI] [PubMed] [Google Scholar]
- Lee JA, Lupski JR (2006) Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron 52:103–121. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. (2016) Genome-wide analysis of the role of copy number variation in schizophrenia risk in chinese. Biol Psychiatry 80:331–337. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Morris MA, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, Shprintzen R, Antonarakis SE, Baldini A, Pulver AE (1995) Schizophrenia and chromosomal deletions within 22q11.2. Am J Hum Genet 56:1502–1503. [PMC free article] [PubMed] [Google Scholar]
- Lo-Castro A, Galasso C, Cerminara C, El-Malhany N, Benedetti S, Nardone AM, Curatolo P (2009) Association of syndromic mental retardation and autism with 22q11.2 duplication. Neuropediatrics 40:137–140. [DOI] [PubMed] [Google Scholar]
- Malhotra D, Sebat J (2012) Cnvs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148:1223–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, et al. (2008) Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 82:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, et al. ; Psychosis Endophenotypes International Consortium; CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium (2017) Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, et al. (1997) The 22q11.2 deletion: screening, diagnostic workup, and outcome of results; report on 181 patients. Genet Test 1:99–108. [DOI] [PubMed] [Google Scholar]
- Mefford HC, et al. (2008) Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med 359:1685–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier MH, Caspi A, Reichenberg A, Keefe RS, Fisher HL, Harrington H, Houts R, Poulton R, Moffitt TE (2014) Neuropsychological decline in schizophrenia from the premorbid to the postonset period: evidence from a population-representative longitudinal study. Am J Psychiatry 171:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinard-Chenu A, Dayer A (2018) The candidate schizophrenia risk gene DGCR2 regulates early steps of corticogenesis. Biol Psychiatry 83:692–706. [DOI] [PubMed] [Google Scholar]
- Moore SJ, Throesch BT, Murphy GG (2011) Of mice and intrinsic excitability: genetic background affects the size of the postburst afterhyperpolarization in CA1 pyramidal neurons. J Neurophysiol 106:1570–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SE, Cuthbert BN (2012) Research domain criteria: cognitive systems, neural circuits, and dimensions of behavior. Dialogues Clin Neurosci 14:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaddes NM, Herguner S (2007) Autistic disorder and 22q11.2 duplication. World J Biol Psychiatry 8:127–130. [DOI] [PubMed] [Google Scholar]
- Murphy KC, Jones LA, Owen MJ (1999) High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry 56:940–945. [DOI] [PubMed] [Google Scholar]
- Nguyen PV, Abel T, Kandel ER, Bourtchouladze R (2000a) Strain-dependent differences in LTP and hippocampus-dependent memory in inbred mice. Learn Mem 7:170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PV, Duffy SN, Young JZ (2000b) Differential maintenance and frequency-dependent tuning of LTP at hippocampal synapses of specific strains of inbred mice. J Neurophysiol 84:2484–2493. [DOI] [PubMed] [Google Scholar]
- Niklasson L, Rasmussen P, Oskarsdóttir S, Gillberg C (2002) Chromosome 22q11 deletion syndrome (CATCH 22): neuropsychiatric and neuropsychological aspects. Dev Med Child Neurol 44:44–50. [DOI] [PubMed] [Google Scholar]
- Nilsson SR, Fejgin K, Gastambide F, Vogt MA, Kent BA, Nielsen V, Nielsen J, Gass P, Robbins TW, Saksida LM, Stensbøl TB, Tricklebank MD, Didriksen M, Bussey TJ (2016) Assessing the cognitive translational potential of a mouse model of the 22q11.2 microdeletion syndrome. Cereb Cortex 26:3991–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi A, Hiroi N (2016) Genetic mechanisms emerging from mouse models of CNV-associated neuropsychiatric disorders. In: The neurobiology of schizophrenia (Abel T, Nickl-Jockschat T, eds), pp 397–417. New York: Academic Press/Elsevier. [Google Scholar]
- ÓBroin P, Becket MV, Takahashi T, Izumi T, Ye K, Kang K, Pouso P, Topolski M, Pena JL, Hiroi N (2018) Computational analysis of neonatal mouse ultrasonic vocalization. Current Protocols in Mouse Biology 8:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Niihori T, Tanaka N, Kawai M, Nagashima T, Funayama R, Nakayama K, Nakashima S, Kato F, Fukami M, Aoki Y, Matsubara Y (2014) TBX1 mutation identified by exome sequencing in a Japanese family with 22q11.2 deletion syndrome-like craniofacial features and hypocalcemia. PLoS One 9:e91598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary J, Osborne LR (2011) Global analysis of gene expression in the developing brain of gtf2ird1 knockout mice. PLoS One 6:e23868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L, et al. (2018) Prevalence of rearrangements in the 22q11.2 region and population-based risk of neuropsychiatric and developmental disorders in a Danish population: a case-cohort study. Lancet Psychiatry 5:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Tuathaigh CM, Hryniewiecka M, Behan A, Tighe O, Coughlan C, Desbonnet L, Cannon M, Karayiorgou M, Gogos JA, Cotter DR, Waddington JL (2010) Chronic adolescent exposure to δ-9-tetrahydrocannabinol in COMT mutant mice: impact on psychosis-related and other phenotypes. Neuropsychopharmacology 35:2262–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Tuathaigh CM, Clarke G, Walsh J, Desbonnet L, Petit E, O’Leary C, Tighe O, Clarke N, Karayiorgou M, Gogos JA, Dinan TG, Cryan JF, Waddington JL (2012) Genetic vs. pharmacological inactivation of COMT influences cannabinoid-induced expression of schizophrenia-related phenotypes. Int J Neuropsychopharmacol 15:1331–1342. [DOI] [PubMed] [Google Scholar]
- Ouchi Y, Banno Y, Shimizu Y, Ando S, Hasegawa H, Adachi K, Iwamoto T (2013) Reduced adult hippocampal neurogenesis and working memory deficits in the dgcr8-deficient mouse model of 22q11.2 deletion-associated schizophrenia can be rescued by IGF2. J Neurosci 33:9408–9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, Iosif AM, Baguio F, Cook IC, Hill MM, Hutman T, Rogers SJ, Rozga A, Sangha S, Sigman M, Steinfeld MB, Young GS (2010) A prospective study of the emergence of early behavioral signs of autism. J Am Ac Child Ado Psy 49:256–266. [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, Young GS, Belding A, Hill M, Hill A, Hutman T, Johnson S, Miller M, Rogers SJ, Schwichtenberg AJ, Steinfeld M, Iosif AM (2014) The broader autism phenotype in infancy: when does it emerge? J Am Acad Child Adolesc Psychiatry 53:398–407.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, Young GS, Brian J, Charman T, Shephard E, Solish A, Zwaigenbaum L (2018) Diagnosis of autism spectrum disorder after age 5 in children evaluated longitudinally since infancy. J Am Acad Child Adolesc Psychiatry 57:849–857.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaleo F, Crawley JN, Song J, Lipska BK, Pickel J, Weinberger DR, Chen J (2008) Genetic dissection of the role of catechol-O-methyltransferase in cognition and stress reactivity in mice. J Neurosci 28:8709–8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronett EM, Meechan DW, Karpinski BA, LaMantia AS, Maynard TM (2015) Ranbp1, deleted in digeorge/22q11.2 deletion syndrome, is a microcephaly gene that selectively disrupts layer 2/3 cortical projection neuron generation. Cereb Cortex 25:3977–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterlini M, Zakharenko SS, Lai WS, Qin J, Zhang H, Mukai J, Westphal KG, Olivier B, Sulzer D, Pavlidis P, Siegelbaum SA, Karayiorgou M, Gogos JA (2005) Transcriptional and behavioral interaction between 22q11.2 orthologs modulates schizophrenia-related phenotypes in mice. Nat Neurosci 8:1586–1594. [DOI] [PubMed] [Google Scholar]
- Paylor R, Glaser B, Mupo A, Ataliotis P, Spencer C, Sobotka A, Sparks C, Choi CH, Oghalai J, Curran S, Murphy KC, Monks S, Williams N, O’Donovan MC, Owen MJ, Scambler PJ, Lindsay E (2006) Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci U S A 103:7729–7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, et al. (2010) Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, et al. (2014) Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 94:677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskulic D, Olver JS, Norman TR, Maruff P (2007) Behavioural studies of spatial working memory dysfunction in schizophrenia: a quantitative literature review. Psychiatry Res 150:111–121. [DOI] [PubMed] [Google Scholar]
- Plana-Ripoll O, et al. (2019) Exploring comorbidity within mental disorders among a Danish national population. JAMA Psychiatry. doi: 10.1001/jamapsychiatry.2018.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoï MF. (2009) Microduplication 22q11.2: a new chromosomal syndrome. Eur J Med Genet 52:88–93. [DOI] [PubMed] [Google Scholar]
- Portnoï MF, Lebas F, Gruchy N, Ardalan A, Biran-Mucignat V, Malan V, Finkel L, Roger G, Ducrocq S, Gold F, Taillemite JL, Marlin S (2005) 22q11.2 duplication syndrome: two new familial cases with some overlapping features with digeorge/velocardiofacial syndromes. Am J Med Genet A 137:47–51. [DOI] [PubMed] [Google Scholar]
- Ramelli GP, Silacci C, Ferrarini A, Cattaneo C, Visconti P, Pescia G (2008) Microduplication 22q11.2 in a child with autism spectrum disorder: clinical and genetic study. Dev Med Child Neurol 50:953–955. [DOI] [PubMed] [Google Scholar]
- Rees E, et al. ; Wellcome Trust Case Control Consortium (2014a) Evidence that duplications of 22q11.2 protect against schizophrenia. Mol Psychiatry 19:37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E, Kendall K, Pardiñas AF, Legge SE, Pocklington A, Escott-Price V, MacCabe JH, Collier DA, Holmans P, O’Donovan MC, Owen MJ, Walters JTR, Kirov G (2016) Analysis of intellectual disability copy number variants for association with schizophrenia. JAMA Psychiatry 73:963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, Mahoney-Davies G, Legge SE, Moran JL, McCarroll SA, O’Donovan MC, Owen MJ, Kirov G (2014b) Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry 204:108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenberg A, Caspi A, Harrington H, Houts R, Keefe RS, Murray RM, Poulton R, Moffitt TE (2010) Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: a 30-year study. Am J Psychiatry 167:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restivo L, Roman FS, Ammassari-Teule M, Marchetti E (2006) Simultaneous olfactory discrimination elicits a strain-specific increase in dendritic spines in the hippocampus of inbred mice. Hippocampus 16:472–479. [DOI] [PubMed] [Google Scholar]
- Ricard G, Molina J, Chrast J, Gu W, Gheldof N, Pradervand S, Schütz F, Young JI, Lupski JR, Reymond A, Walz K (2010) Phenotypic consequences of copy number variation: insights from smith-magenis and potocki-lupski syndrome mouse models. PLoS Biol 8:e1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilling JK. (2014) Comparative primate neuroimaging: insights into human brain evolution. Trends Cogn Sci 18:46–55. [DOI] [PubMed] [Google Scholar]
- Routh BN, Johnston D, Harris K, Chitwood RA (2009) Anatomical and electrophysiological comparison of CA1 pyramidal neurons of the rat and mouse. J Neurophysiol 102:2288–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, et al. (2011) Multiple recurrent de novo cnvs, including duplications of the 7q11.23 Williams Syndrome region, are strongly associated with autism. Neuron 70:863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, et al. ; Autism Sequencing Consortium (2015) Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87:1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Gur RC, Gur RE, Mozley PD, Mozley LH, Resnick SM, Kester DB, Stafiniak P (1991) Neuropsychological function in schizophrenia. Selective impairment in memory and learning. Arch Gen Psychiatry 48:618–624. [DOI] [PubMed] [Google Scholar]
- Scambler PJ, Kelly D, Lindsay E, Williamson R, Goldberg R, Shprintzen R, Wilson DI, Goodship JA, Cross IE, Burn J (1992) Velo-cardio-facial syndrome associated with chromosome 22 deletions encompassing the digeorge locus. Lancet 339:1138–1139. [DOI] [PubMed] [Google Scholar]
- Scattoni ML, Gandhy SU, Ricceri L, Crawley JN (2008) Unusual repertoire of vocalizations in the BTBR T+tf/J mouse model of autism. PLoS One 3:e3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, Ricceri L, Crawley JN (2011) Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav 10:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J, Giangrande E, Weinberger DR, Dickinson D (2013) The global cognitive impairment in schizophrenia: consistent over decades and around the world. Schizophr Res 150:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, et al. ; International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome (2014) Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the international consortium on brain and behavior in 22q11.2 deletion syndrome. Am J Psychiatry 171:627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegler H, Boldyreva M, Linke R, Wu J, Zilles K, Crusio WE (1996a) Genetic variation in the morphology of the septo-hippocampal cholinergic and gabaergic systems in mice: II. Morpho-behavioral correlations. Hippocampus 6:535–545. [DOI] [PubMed] [Google Scholar]
- Schwegler H, Boldyreva M, Pyrlik-Göhlmann M, Linke R, Wu J, Zilles K (1996b) Genetic variation in the morphology of the septo-hippocampal cholinergic and gabaergic system in mice. I. Cholinergic and gabaergic markers. Hippocampus 6:136–148. [DOI] [PubMed] [Google Scholar]
- Sebat J, et al. (2007) Strong association of de novo copy number mutations with autism. Science 316:445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman LJ, Shapiro DI, Stone WS, Woodberry KA, Ronzio A, Cornblatt BA, Addington J, Bearden CE, Cadenhead KS, Cannon TD, Mathalon DH, McGlashan TH, Perkins DO, Tsuang MT, Walker EF, Woods SW (2016) Association of neurocognition with transition to psychosis: baseline functioning in the second phase of the North American Prodrome Longitudinal Study. JAMA Psychiatry 73:1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple BD, Blomgren K, Gimlin K, Ferriero DM, Noble-Haeusslein LJ (2013) Brain development in rodents and humans: identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol 106–107:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, et al. (2008) A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet 40:322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Lewin ML, Sidoti EJ, Berkman MD, Argamaso RV, Young D (1978) A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: velo-cardio-facial syndrome. Cleft Palate J 15:56–62. [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg R, Golding-Kushner KJ, Marion RW (1992) Late-onset psychosis in the velo-cardio-facial syndrome. Am J Med Genet 42:141–142. [DOI] [PubMed] [Google Scholar]
- Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA, Gordon JA (2010) Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature 464:763–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soysal Y, Vermeesch J, Davani NA, Şensoy N, Hekimler K, İmirzalıoğlu N (2011) Molecular characterization of microduplication 22q11.2 in a girl with hypernasal speech. Genet Mol Res 10:2148–2154. [DOI] [PubMed] [Google Scholar]
- Stefansson H, et al. (2008) Large recurrent microdeletions associated with schizophrenia. Nature 455:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugama S, Namihira T, Matsuoka R, Taira N, Eto Y, Maekawa K (1999) Psychiatric inpatients and chromosome deletions within 22q11.2. J Neurol Neurosurg Psychiatry 67:803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Harper KM, Hiramoto T, Funke B, Lee M, Kang G, Buell M, Geyer MA, Kucherlapati R, Morrow B, Männistö PT, Agatsuma S, Hiroi N (2009a) Over-expression of a human chromosome 22q11.2 segment including TXNRD2, COMT and ARVCF developmentally affects incentive learning and working memory in mice. Hum Mol Genet 18:3914–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Harper KM, Hiramoto T, Sawamura T, Lee M, Kang G, Tanigaki K, Buell M, Geyer MA, Trimble WS, Agatsuma S, Hiroi N (2009b) Sept5 deficiency exerts pleiotropic influence on affective behaviors and cognitive functions in mice. Hum Mol Genet 18:1652–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatkiewicz JP, et al. (2014) Copy number variation in schizophrenia in Sweden. Mol Psychiatry 19:762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, et al. (2007) Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39:319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Okabe S, Broin PÓ, Nishi A, Ye K, Beckert MV, Izumi T, Machida A, Kang G, Abe S, Pena JL, Golden A, Kikusui T, Hiroi N (2016) Structure and function of neonatal social communication in a genetic mouse model of autism. Mol Psychiatry 21:1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Juan L, Rosell J, Morla M, Vidal-Pou C, García-Algas F, de la Fuente MA, Juan M, Tubau A, Bachiller D, Bernues M, Perez-Granero A, Govea N, Busquets X, Heine-Suñer D (2007) Mutations in TBX1 genocopy the 22q11.2 deletion and duplication syndromes: a new susceptibility factor for mental retardation. Eur J Hum Genet 15:658–663. [DOI] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos AC, Banna L, Brereton AV, Hill A, Bisgaard AM, Müller I, Hultschig C, Erdogan F, Wieczorek G, Ropers HH (2007) Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat 28:674–682. [DOI] [PubMed] [Google Scholar]
- Ullman VZ, Levine SZ, Reichenberg A, Rabinowitz J (2012) Real-world premorbid functioning in schizophrenia and affective disorders during the early teenage years: a population-based study of school grades and teacher ratings. Schizophr Res 136:13–18. [DOI] [PubMed] [Google Scholar]
- Valor LM, Grant SG (2007) Clustered gene expression changes flank targeted gene loci in knockout mice. PLoS One 2:e1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oel CJ, Sitskoorn MM, Cremer MP, Kahn RS (2002) School performance as a premorbid marker for schizophrenia: a twin study. Schizophr Bull 28:401–414. [DOI] [PubMed] [Google Scholar]
- Vangkilde A, Jepsen JR, Schmock H, Olesen C, Arnarsdóttir S, Baaré WF, Plessen KJ, Didriksen M, Siebner HR, Werge T, Olsen L (2016) Associations between social cognition, skills, and function and subclinical negative and positive symptoms in 22q11.2 deletion syndrome. J Neurodev Disord 8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorstman JA, et al. ; International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome (2015) Cognitive decline preceding the onset of psychosis in patients with 22q11.2 deletion syndrome. JAMA Psychiatry 72:377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlsten D, Metten P, Crabbe JC (2003) Survey of 21 inbred mouse strains in two laboratories reveals that BTBR T/+ tf/tf has severely reduced hippocampal commissure and absent corpus callosum. Brain Res 971:47–54. [DOI] [PubMed] [Google Scholar]
- Walsh T, et al. (2008) Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320:539–543. [DOI] [PubMed] [Google Scholar]
- Weksberg R, Stachon AC, Squire JA, Moldovan L, Bayani J, Meyn S, Chow E, Bassett AS (2007) Molecular characterization of deletion breakpoints in adults with 22q11 deletion syndrome. Hum Genet 120:837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger TL, Miller JS, DePolo LM, de Marchena AB, Clements CC, Emanuel BS, Zackai EH, McDonald-McGinn DM, Schultz RT (2016) 22q11.2 duplication syndrome: elevated rate of autism spectrum disorder and need for medical screening. Mol Autism 7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzel C, Fernström M, Ohrner Y, Annerén G, Thuresson AC (2008) Clinical variability of the 22q11.2 duplication syndrome. Eur J Med Genet 51:501–510. [DOI] [PubMed] [Google Scholar]
- Woestelandt L, Novo A, Philippe A, Guyaux N, Rio M, Romano S, Robel L (2018) PDD-NOS, psychotic features and executive function deficits in a boy with proximal 22q11.2 microduplication: evolution of the psychiatric symptom profile from childhood to adolescence. Eur J Med Genet 61:280–283. [DOI] [PubMed] [Google Scholar]
- Wolfer DP, Crusio WE, Lipp HP (2002) Knockout mice: simple solutions to the problems of genetic background and flanking genes. Trends Neurosci 25:336–340. [DOI] [PubMed] [Google Scholar]
- Woodberry KA, Giuliano AJ, Seidman LJ (2008) Premorbid IQ in schizophrenia: a meta-analytic review. Am J Psychiatry 165:579–587. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M (2008) Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet 40:880–885. [DOI] [PubMed] [Google Scholar]
- Yang S, Farias M, Kapfhamer D, Tobias J, Grant G, Abel T, Bućan M (2007) Biochemical, molecular and behavioral phenotypes of rab3a mutations in the mouse. Genes Brain Behav 6:77–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YM, Fedchyshyn MJ, Grande G, Aitoubah J, Tsang CW, Xie H, Ackerley CA, Trimble WS, Wang LY (2010) Septins regulate developmental switching from microdomain to nanodomain coupling of ca(2+) influx to neurotransmitter release at a central synapse. Neuron 67:100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmazer-Hanke DM, Roskoden T, Zilles K, Schwegler H (2003) Anxiety-related behavior and densities of glutamate, GABAA, acetylcholine and serotonin receptors in the amygdala of seven inbred mouse strains. Behav Brain Res 145:145–159. [DOI] [PubMed] [Google Scholar]
- Yobb TM, Somerville MJ, Willatt L, Firth HV, Harrison K, MacKenzie J, Gallo N, Morrow BE, Shaffer LG, Babcock M, Chernos J, Bernier F, Sprysak K, Christiansen J, Haase S, Elyas B, Lilley M, Bamforth S, McDermid HE (2005) Microduplication and triplication of 22q11.2: a highly variable syndrome. Am J Hum Genet 76:865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Cox K, Friend K, Smith S, Buchheim R, Bain S, Liebelt J, Thompson E, Bratkovic D (2008) Familial 22q11.2 duplication: a three-generation family with a 3-mb duplication and a familial 1.5-mb duplication. Clin Genet 73:160–164. [DOI] [PubMed] [Google Scholar]
- Yung AR, McGorry PD (1996) The initial prodrome in psychosis: descriptive and qualitative aspects. Aust N Z J Psychiatry 30:587–599. [DOI] [PubMed] [Google Scholar]
- Zinkstok J, Boot E, Bassett AS, Hiroi N, Butcher NJ, Vingerhoets C, Vorstman JAS, van Amelsvoort TAMJ (2019) The 22q11.2 deletion syndrome from a neurobiological perspective. Lancet Psychiatry. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Warren ST (2010) Neurogenetics: advancing the “next-generation” of brain research. Neuron 68:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaigenbaum L, Bryson S, Garon N (2013) Early identification of autism spectrum disorders. Behav Brain Res 251:133–146. [DOI] [PubMed] [Google Scholar]