Abstract
DNA topoisomerase I is an essential nuclear enzyme involved in resolving the torsional stress associated with DNA replication, transcription, and chromatin condensation. Here we report the discovery of a seizure-suppressor mutant, top1JS, which suppresses seizures in a Drosophila model of human epilepsy. A P-element mutagenesis screen using easily shocked seizure-sensitive mutant as a genetic background identified top1JS, which plays a novel role in regulating nervous system excitability. Plasmid rescue, excision, complementation, and sequencing analyses verified that top1JS results from a P-element insertion in the 5′ untranslated region. Quantitative reverse transcription analysis on wild-type and mutant fly heads showed that the top1JS mutation causes reduced transcription level in the CNS, suggesting a partial loss-of-function mutation. Electrophysiological experiments revealed normal seizure thresholds in top1JS mutants, which are different from other seizure suppressors identified previously, suggesting a novel mechanism underlying seizure suppression by top1JS. The pharmacological camptothecin feeding experiment and cell death analysis suggested that the seizure suppression by top1JS may occur via increased neuronal apoptosis. Furthermore, overexpression of the DIAP1 (Drosophila inhibitor of apoptosis 1) gene rescues top1JS suppression, providing additional support for a neural apoptosis suppression mechanism. The top1JS mutation is the first viable partial loss-of-function mutation identified in higher eukaryotes, and the results presented here point to a novel function for topo I in construction and/or maintenance of circuits required for seizure propagation in vivo.
Keywords: topoisomerase I, seizure, suppression, camptothecin, epilepsy, Drosophila
Introduction
Eukaryotic DNA topoisomerases are ubiquitous and essential nuclear enzymes involved in various DNA transactions (Wang, 1996, 2002; Champoux, 2001). DNA topoisomerases are classified as type I and type II; they differ structurally and mechanistically (Champoux, 2001). Eukaryotic type I topoisomerase (topo IB) plays important roles in DNA replication, RNA transcription, DNA recombination, chromosome condensation, and the maintenance of genomic stability (Champoux, 2001; Wang, 2002). Immunohistochemical analysis in selected mouse brain regions reveals that a significant level of topo I activity is present in the brain, and the level of topo I activity varies among different brain regions (Plaschkes et al., 2005). The cerebellum, visual cortex, and the striatum exhibit higher topo I activity than the hippocampus and hypothalamus. Dual-label immunofluorescence analysis reveals that topo I-expressing neurons are both inhibitory and noninhibitory neurons (Plaschkes et al., 2005).
Mammalian DNA topoisomerases are the targets of several anticancer drugs used today in the clinic, including the topo I inhibitor camptothecin (CPT) and its derivatives (Wang, 1994; Wang et al., 1997; Pommier, 1998; Pommier et al., 1999; Li and Liu, 2000). Several in vitro studies have shown that CPT induces apoptosis in cultured neurons (Morris and Geller, 1996; Park et al., 1997). Although apoptotic cell death occurs physiologically during development, neuronal apoptosis is also associated with various neurodegenerative disorders and conditions of neural trauma including Alzheimer's disease, Parkinson's disease, Huntington's disease, amyotrophic lateral sclerosis, AIDS dementia, epilepsy, retinitis pigmentosa, cerebellar degeneration, and ischemic brain injury and stroke (Thompson, 1995). It is not known how these anticancer agents signal a postmitotic neuron to execute its apoptotic pathway.
The work presented here describes a mutant screen that allows us to identify novel loss-of-function seizure suppressors by P-element mutagenesis. Seizure-suppressor genes are identified by mutations that revert the seizure-sensitive phenotype in a Drosophila model of epilepsy (Kuebler et al., 2001). Seizure-suppressor mutants were isolated by mobilizing a P element in an easily shocked (eas) mutant background and looking for second-site mutations that reduce seizure susceptibility. Of multiple seizure-suppressor genes identified, we focus on top1JS, a new allele of the gene top1 that has not been previously implicated in seizure susceptibility. The top1 gene has been primarily associated with growth and development in Drosophila (Lee et al., 1993). Mutants of top1 exhibit abnormal proliferation and defective nuclear morphology in ovary cells: there are extranumeral germ-line cells in individual egg chambers, and the follicle cells are underreplicated (Zhang et al., 2000). Topo I is maternally stored in embryos, and embryos from mutant mothers frequently show disrupted nuclear divisions with defects in chromosome condensation and segregation. Topo I localizes to the nuclei during interphase and prophase but disperses into the cytoplasm at metaphase. The cytological and genetic analyses of the top1 mutants suggest that topo I plays critical roles in many developmental stages active in cell proliferation.
We report a surprising new role for topo I in the nervous system as a seizure suppressor. The top1 mutation top1JS isolated in this study acts as a general seizure suppressor, reducing the seizure susceptibility of eas, slamdance (sda), and bang-senseless (bss) seizure-sensitive flies. Plasmid rescue, P-element excision, complementation, and sequence analyses verified that top1JS is a new allele of the top1 gene resulting from a P-element insertion in the 5′ untranslated region (UTR). Reverse transcription (RT) analysis on wild-type and mutant fly heads showed that the top1JS mutation causes reduced transcription level in the CNS, suggesting a partial loss-of-function mutation. Electrophysiological experiments revealed normal seizure thresholds in the top1JS mutation that are different from other seizure suppressors identified previously, suggesting a novel mechanism underlying seizure suppression. Pharmacological experiment and cell death analysis suggested that the seizure suppression by top1JS may occur via the increased neuronal apoptosis. Overexpression of the Drosophila inhibitor of apoptosis 1 (DIAP1) gene rescues top1JS suppression, providing additional support for a neural apoptosis suppression mechanism. Finally, we discussed the potential practical implications based on genetic and drug-feeding experiments.
Materials and Methods
Fly stocks.
Drosophila strains were maintained on standard cornmeal–molasses agar medium at room temperature (22–25°C). The eas gene is located at 14B on the cytological map and encodes an ethanolamine kinase (Pavlidis et al., 1994). The eas allele used in this study is easPC80, which is caused by a 2 bp deletion that introduces a frame shift; the resulting truncated protein lacks a kinase domain and abolishes all enzymatic activity (Pavlidis et al., 1994). The bang-sensitive (BS) phenotype of eas flies is completely penetrant; thus, in this study, we use eas BS mutation as a genetic background to screen its suppressors by P-element hybrid dysgenesis. The sda gene is located at 97D on the third chromosome and encodes an aminopeptidase N. The allele used in this study is sda iso7.8, which is caused by a 2-bp insertion in the 5′ untranslated region; phenotypes appear to be a result of enzyme underproduction (Zhang et al., 2002). The single P-element stock used in this screen is y w P{lacw}3–76a, which contains a single P{lacw} insertion at the cytological position of 18C-D. The topo I alleles used in this study are as follows: top1G0229, top1G0134, top1G0201, top177, and top1112. All alleles are lethal and behave genetically as nulls, indicating that the top1 gene is essential for viability. To figure out the exact insertion sites of top1G0229, top1G0134, and top1G0201 alleles, plasmid rescue was performed on these three alleles. Results showed that top1G0229 allele is caused by a P{lacw} insertion in the first intron of top1 at the scaffold position 257493/257494; the top1G0134 allele is caused by a P{lacw} insertion in the second exon of top1 at the scaffold position 257709/257710; and the top1G0229 allele is caused by a P{lacw} insertion in the fourth intron of top1 at the scaffold position 260024/260025 (Fig. 1). In addition, one previously studied allele, top1112, is attributable to a deletion in the 3′ untranslated region of top1 (Lee et al., 1993; Zhang et al., 1996). The deficiency stock used in this work is Df(1)ED7294, which has a deletion in the region between 13B1 and 13C3. The top1 rescue construct used here is P{top1 ± 10}, which contains a 10 kb genomic SpeI–BamHI fragment. This 10 kb genomic fragment includes the entire top1 transcribed region and 1.3 kb of upstream and 0.5 kb of downstream sequences; it contains no complete gene other than the 8.5 kb top1 gene (Lee et al., 1993) (http://flybase.bio.indiana.edu). All above fly stocks were obtained from Bloomington Stock Center at Indiana University. The transgenic upstream activator sequence (UAS)–DIAP1 line was obtained from Dr. John Nambu (University of Massachusetts, Amherst, Amherst, MA).
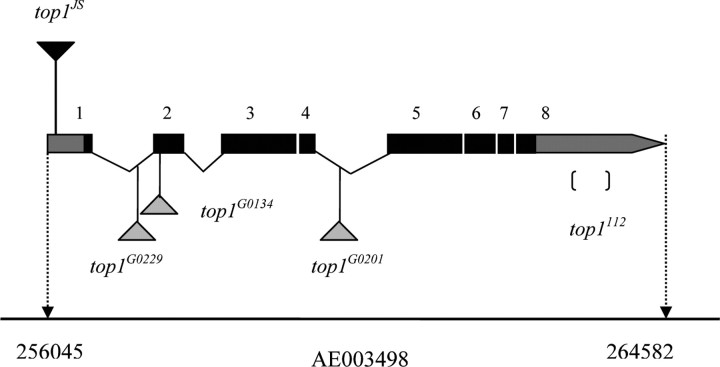
Figure 1.
The top1 locus at 13B6 on chromosome X, scaffold AE003498. The top locus spans 8.5 kb and encodes three different transcripts: top1-RA (5325 nt), top1-RB (2379 nt), and top1-RC (4940 nt). For simplicity, only the transcript corresponding to top1-RA is shown. The exons are represented as numbered boxes. The gray areas correspond to untranslated regions (including exon 2–7, part of exon 1, and part of exon 8), whereas the black areas correspond to protein-coding regions. Alleles of top1G0229, top1G0134, and top1G0201 are P-element insertion lines. The top1112 allele has a deletion in 3′ untranslated region. The P element responsible for the top1JS mutation is inserted in the 5′ untranslated region, between nucleotides 256075 and 256076 of genomic scaffold AE003498, corresponding to 31 and 32 of the top1 mRNA variant A and C, which is 257 bp away from the translation start site.
P-element mutagenesis screen scheme and behavioral tests.
Female y w eas P[lacw] flies were crossed to males w; Sp/CyO, δ 2, 3 Dr/TM3 Sb, and δ 2, 3 male progeny were crossed to virgin C(1)DX females. The male flies were tested for BS behavior: BS flies were discarded, but exceptional bang-resistant (BR) flies were kept for additional analysis. By using this P-element mutagenesis screening scheme, X chromosomal recessive and dominant suppressors and autosomal-dominant suppressors can be isolated. Of 26,833 male flies screened, 34 suppressors were isolated from behavior testing. Seizure suppression varied from 90% (strongest) to 10% (weakest); top1JS suppressed eas by ∼63%.
BS behavioral tests were performed on flies 2–3 d after eclosion. Flies were anesthetized with CO2 before collection and tested the following day to eliminate any CO2 effects on behavior. For testing, 15–20 flies were placed in a food vial and stimulated with a VWR Vortex mixer (VWR International, West Chester, PA) at maximum speed for 10 s. BS paralytic mutants undergo seizures characterized by brief hyperactivity (2 s) and temporary paralysis (20–300 s). Recovery times were calculated by measuring the time from the end of the vortex until the flies resumed an upright standing position.
Electrophysiology.
All flies were used for electrophysiology 2–3 d after eclosion. Flies were mounted for electrophysiology without anesthesia by capturing and immobilizing them with a vacuum line. Immobilized flies were then attached to a pin glued to the dorsal thorax with Super Glue gel (Scotch; 3M, St. Paul, MN). Two uninsulated tungsten electrodes were inserted into the brain for stimulation. The ground electrode was inserted into the abdomen. Recording electrodes were uninsulated tungsten electrodes inserted into the dorsal longitudinal muscles (DLMs) or tergotrochanter muscles identified by their thoracic insertion sites (Tanouye and Wyman, 1980). Two types of electrical stimulation were used: single-pulse stimulation and high-frequency (HF) stimulation. Single-pulse stimuli (0.2 ms duration, 0.5 Hz) were delivered to the brain to drive the giant fiber (GF). During the course of each experiment, the GF was stimulated continuously with single-pulse stimuli to assess GF circuit function. HF stimulation (0.5 ms pulses at 200 Hz for 300 ms) was used to elicit seizures. Seizure-like activity in Drosophila is observed as uncontrolled, high-frequency (>100 Hz) neuronal firing, most conveniently examined in motoneurons. Seizures are extensive: >30 motoneurons innervate at least seven different thoracic muscle groups participate in seizures (Kuebler and Tanouye, 2000). To determine seizure threshold, HF stimuli were initially given to flies at predicted intensities, depending on their genotypes. If the stimulus fails to elicit a seizure, the intensity was subsequently increased at 1 V increments until a seizure was induced. The threshold was determined for an individual fly as the lowest intensity at which seizures occurred. The fly was allowed to rest 15 min after each HF stimulation.
Molecular mapping of top1JS.
The combination of plasmid rescue and sequencing approaches was used to determine the exact P-element insertion site of top1JS. The plasmid rescue was performed as described previously with some modifications (Wilson et al., 1989). Drosophila genomic DNA was isolated as described on the Berkeley Drosophila Genome Project website (http://www.fruitfly.org/about/methods/inverse.pcr.html). Genomic DNA (2–5 μg) was digested with EcoRI, and the fragments were self-ligated with T4 DNA ligase. Ligated products were transformed into DH5α electrocompetent cells, and the transformants were selected on ampicillin plates (100 mg/ml). Plasmid DNA from positive clones was isolated and digested with EcoRI and BamHI to find different digestion patterns. Plasmid DNA was sequenced and aligned to the genome database [National Center for Biotechnology Information (NCBI) blast program]. The approximate insertion site was determined using a primer near the end of the P-element sequence (CGACGGGACCACCTTATGTTATTTCATCATG). The exact insertion site was then determined by sequencing back toward the P element using a primer specific for the flanking genomic region of top1 (TCCCTGCTTCACA-CCATCCTTC).
RT-PCR analysis.
Adult wild-type Canton Special (CS) and top1JS fly heads were decapitated using a razor, and fly heads were collected in 1.5 ml centrifuge tubes. Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA), and 2.5 μg was reverse transcribed using Sprint PowerScript PrePrimed Single Shot (random hexamer primers; Clontech, Mountain View, CA). The primer pair used to amplify top1 transcripts generated a 499 bp product. Their sequences are as follows: forward, 5′-AGTCGCGACAAGGATCGACACA-3′ and reverse, 5′CGGCAACGTTTCCATTGCCAT-3′. The internal control was fly actin gene CG7478-PA (Act79B) amplified with the primer pair to amplify the actin gene: forward, 5′-ATCCAGGTATCGCTGACCGTATGC-3′ and reverse, 5′-AAAGAAGGCGTTGCCGCTCGTTTC-3′. This primer pair generated a 435 bp gene product. The cDNA products from reverse transcription were serially diluted for sensitivity. PCRs were performed with TaqDNA polymerase (Qiagen, Valencia, CA) in a reaction volume of 50 μl. The PCR was performed in a thermal cycler with the following cycling parameters: 2 min at 94°C and then 28 cycles consisting of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C; after completion of the last cycle, a final incubation at 72°C for 7 min was performed. The PCRs were analyzed on a 1.5% agarose gel. The RT-PCR products were quantified using ImageJ 1.36 (http://rsb.info.nih.gov/ij). The relative top1 transcript levels in the test and control lanes were determined by normalizing the signal intensity of the top1 band to that of the corresponding Act79B band.
Drug feeding.
The topoisomerase I inhibitor CPT (C9911; Sigma, St. Louis, MO) was used. CPT feeding solutions contained 5% sucrose to encourage flies to ingest drug and were mixed with formula 4–24 Blue Drosophila medium (Carolina Biological Supply, Burlington, NC) in a 1:1 ratio of dry food to drug solution to create the desired feeding medium. For example, to make 4 μm CPT feeding medium, a 1:1 blend of 8 μm CPT solution with dry food would be made. Progeny were reared in the CPT-containing food and tested at 2–3 d after eclosion. Different BS mutants tolerated drug differently: pupal lethality was observed at different CPT concentrations for different BS mutants; eas mutants could tolerate higher concentration CPT than sda and bss mutants.
Apoptosis analysis.
Heads from 2-d-old wild-type and top1JS mutant flies were collected and frozen in OCT embedding medium (Tissue Tek; Miles, Elkhart, IN). Cryostat sections (10 μm) were collected on positively charged slides (Fisherbrand Superfrost Plus; Fisher Scientific, Houston, TX) and fixed in 2% glutaraldehyde in PBS (freshly prepared) for 20 min at room temperature. Slides were then washed with PBS and incubated in freshly prepared permeabilization solution (0.1% Triton X-100, 0.1% sodium citrate) for 2 min on ice. Terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) assay was performed using In Situ Cell Death Detection Kit, Fluorescein (Roche Diagnostics, Indianapolis, IN) according to manufacturer's directions. Slides was washed with PBS to stop the reaction and then imaged by confocal microscope.
For agarose gel electrophoresis, genomic DNA samples from wild-type, eas top1JS double-mutant, and top1JS mutant flies were electrophoretically separated on 1.5% agarose gel containing ethidium bromide (0.5 μg/ml). DNA was visualized by UV transillumination.
Results
Isolation and molecular basis of the top1JS mutation
In a screen for new seizure suppressors, the top1JS mutation was found to reduce the behavioral paralysis of eas flies after a mechanical shock by ∼63% and decrease the seizure susceptibility to an electrical shock by three to four times. The top1JS mutation was mapped by both molecular and genetic methods. Molecularly, plasmid rescue was used to isolate genomic DNA flanking the insertion site of the P element responsible for top1JS. The flanking DNA was then sequenced and compared with the information available in the Drosophila genome database using the NCBI blast program. This comparison identified the location of the top1JS insertion within the top1 gene in the 13B6 region of the X chromosome (Fig. 1). By sequencing the flanking genomic DNA back toward the P element, the exact location of the insertion was identified. The P element responsible for the top1JS mutation is inserted in the 5′ untranslated region between nucleotides 256075 and 256076 of the genomic scaffold AE003498, corresponding to 31 and 32 of the top1 mRNA variants A and C; 257 bp away from the translation start site.
The top1JS mutation raises seizure thresholds and shortens the recovery time of eas flies
Drosophila BS paralytics form a class of behavioral mutants with enhanced seizure-sensitive phenotypes. The class includes the following: bss, bas (bang-sensitive), eas (ethanolamine kinase) (Pavlidis et al., 1994), tko (technical knockout) (mitochondrial ribosomal protein) (Royden et al., 1987), kdn (knockdown) (citrate synthase) (Fergestad et al., 2006), and sda (aminopeptidase) (Zhang et al., 2002). All of these BS mutants have been shown to have low seizure thresholds compared with wild-type flies in electrophysiological experiments (Kuebler and Tanouye, 2000; Kuebler et al., 2001). Among BS mutations, eas is considered intermediate in terms of both the degree to which it reduces seizure threshold and the ease with which it can be suppressed by second-site modifiers (Kuebler and Tanouye, 2000; Kuebler et al., 2001). We examined the seizure thresholds of suppressors in an eas background to see whether double mutants have higher seizure thresholds.
To further investigate a role for top1 in seizure suppression, we examined additional top1 mutants in an eas background. All of the top1 alleles studied are X-chromosomal lethals necessitating examination in heteroallelic combinations with top1JS. Interestingly, top1JS complemented the lethality of all lethal top1 alleles. However, some, but not all, lethal alleles acted to suppress seizures (Fig. 2A,B). Thus, top1G0229 acts to suppress seizures as indicated by the genotype: top1G0229 eas/top1JS eas that showed 63% suppression of the behavioral paralytic phenotype compared with their top1G0229 eas/+ eas controls. In addition, electrophysiology showed an increased seizure threshold (8.2 ± 1.9 V) compared with their top1G0229 eas/+ eas controls (3.4 ± 0.2 V). Formally, top1G0229 in heteroallelic combination with top1JS appears to be a recessive seizure suppressor of eas. Qualitatively similar results were observed with other mutations in heteroallelic combination with top1JS: top1112 (34% BS suppression, 6.3 ± 2.5 V seizure threshold), Df(1)ED7294 (68% BS suppression, 10.4 ± 2.7 V seizure threshold). In contrast, several top1 lethal mutations show no evidence of seizure suppressor function: they were found to complement seizure suppression by top1JS (Fig. 2A,B). For example, top1G0201 does not appear to be a seizure suppressor, as indicated by 0% suppression seen in flies of the genotype top1G0201 eas/top1JS eas identical to their control genotype top1G0201 eas/+ eas. The two genotypes also show similar seizure thresholds: 3.5 ± 0.2 V for the experimental compared with 3.3 ± 0.1 V for the control. Qualitatively similar results were seen with top1G0134 in heteroallelic combination with top1JS (5% BS suppression, 4.2 ± 1.2 V seizure threshold).
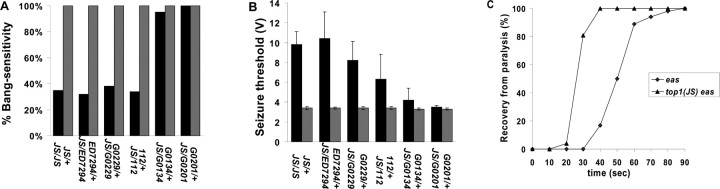
Figure 2.
Behavioral bang sensitivity and seizure threshold in heteroallelic combination of top1JS and various other top1 mutants in an eas mutant background. The data here reveal that combinations of top1JS allele with different top1 alleles display different suppression efficiency in the eas mutant background. A, Behavioral bang sensitivity in various top1 allelic combinations. B, Seizure thresholds in various top1 allelic combinations. Seizure thresholds are consistent with behavioral testing. Black bars represent the behavioral bang sensitivity and seizure thresholds in different top1 allelic combinations with top1JS in an eas background. Gray bars represent heterozygous top1 alleles in an eas background; they act as controls in this assay. C, Comparison of recovery time in eas and top1JS eas double mutants. Shown is the percentage of flies recovered with time and a cumulative measure that includes the initial behavior seizure, the paralysis period, and the recovery seizure, which are not indicated separately. The number of flies standing at intervals after the shock was recorded until the entire population had recovered. For each genotype, n ≥ 80 flies tested for behavior; n ≥ 8 flies tested for electrophysiology; n ≥ 30 flies recorded for recovery time. For behavioral bang sensitivity and seizure threshold, results from the allelic combinations of top1JS/top1JS, top1JS/Df (1)ED7294, top1JS/top1G0229, and top1JS/top1112 are statistically different from their corresponding controls (p < 0.05), whereas results from the allelic combinations of top1JS/top1G0134 and top1JS/top1G0201 are not statistically different from their corresponding controls (p > 0.05).
In some top1JS eas flies, paralysis is completely eliminated; others become paralyzed but recover more quickly, a phenomenon previously seen in BS mutants treated with human anticonvulsant drugs (Reynolds et al., 2004; Tan et al., 2004; Song and Tanouye, 2006), as well as double-mutant combinations of BS mutants with their corresponding seizure-suppressor mutants (Song and Tanouye, 2006). Normally, eas mutants require 44 ± 11 s to recover from paralysis when flies are 2 to 3 d old; it only takes same-age top1JS eas flies 27 ± 4 s to recover (Fig. 2C). Recovery time is another criterion to judge the seizure severity in BS mutants and double mutants of BS mutants with seizure-suppressor mutants.
Seizure suppression by top1JS mutation is reverted by introducing wild-type top1
To further investigate whether the suppression by top1JS is caused by reduced expression of top1, we crossed top1JS eas double mutants to flies carrying a genomic duplication of the top1 gene. The top1 rescue construct used here is P {top1 ± 10}, which contains a 10 kb genomic SpeI–BamHI fragment. This 10 kb genomic fragment includes the entire top1 transcribed region, 1.3 kb of upstream and 0.5 kb of downstream sequences; it contains no complete gene other than top1 (Lee et al., 1993) (http://flybase.bio.indiana.edu). We found that the addition of an extra copy of top1+ product successfully restored the behavioral bang-sensitive phenotype of top1JS eas double mutants. The percentage of bang sensitivity increased from 36% in top1JS eas double-mutant flies to 100% in top1JS eas; P {top1 ± 10}/+ flies. In electrophysiology experiments, the top1JS eas double mutants have seizure thresholds at 9.1 ± 1.1 V, and the addition of an extra copy of top1+ decreased the seizure threshold of top1JS eas to 3.4 ± 0.2 V, which is very similar to the seizure threshold of the eas mutant (Fig. 3). The restoration of seizure sensitivity in both behavioral testing and electrophysiological experiments by adding top1+ is another piece of evidence that top1JS reduces top1 expression, leading to seizure suppression.
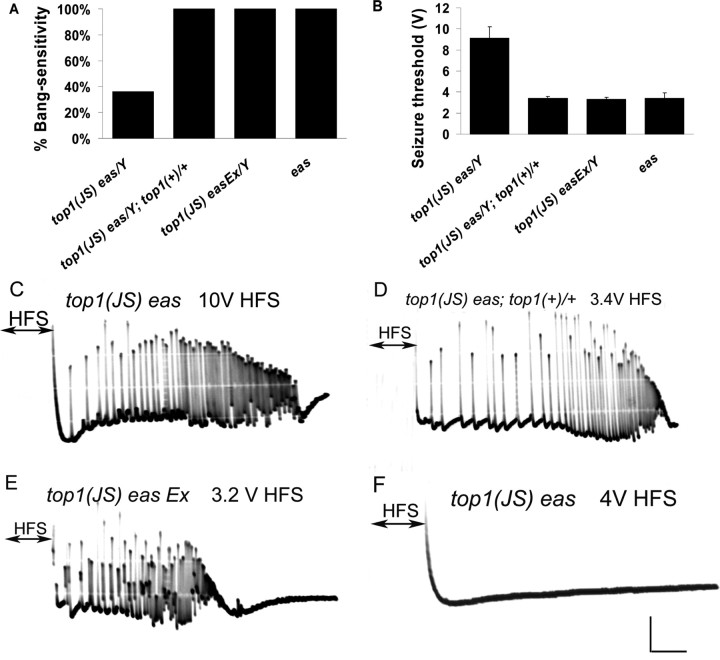
Figure 3.
Behavioral and electrophysiological evidences that top1JS causes seizure suppression. A, Behavioral evidence that seizure suppression is caused by top1JS. The top1 rescue construct (top1+) used here is P {top1 ± 10}, which contains the entire top1 transcribed region and no other complete genes. In the presence of top1+, top1JS eas double mutants show 100% bang sensitivity, which is the same as eas mutants (100% BS; p < 0.01), suggesting the rescue of the seizure suppression phenotype. The top1JS easEx is a precise excision line in which the P element is precisely removed, and the behavioral phenotypes of those flies get reverted to 100% bang sensitivity (p < 0.01). B, Electrophysiological evidence that seizure suppression is caused by top1JS. Seizure thresholds are consistent with the behavioral phenotypes: both rescue and excision lines have decreased seizure thresholds, similar to the eas mutant (p < 0.01). C, A representative seizure in a top1JS eas fly recorded from the DLM after a high-frequency brain stimulus (HFS) of 10 V. The DLM shows aberrant high-frequency firing during the seizure phase. D, A seizure is elicited in a top1JS eas; top1(+)/+ fly after a 3.4 V HFS. E, A seizure is elicited in a top1JS easEx fly after a 3.2 V HFS. F, Failure to elicit seizure in a top1JS eas double-mutant fly after a 3.4 V HFS. Calibration: 10 mV, 200 ms. For each genotype, n ≥ 80 flies tested for behavior; n ≥ 8 flies tested for electrophysiology.
Precise excision of the P element in the double-mutant top1JS eas restores seizure susceptibility similar to the eas mutant
Excision experiments were performed by reintroducing the transposase Delta 2,3 into the top1JS eas double mutants. Among eight putative precise excision lines, one precisely excised line was verified according to genomic PCR and sequencing assays. Behavioral and electrophysiological experiments showed that this precise excision line restores the behavioral bang-sensitive phenotype to 100% and the seizure threshold to 3.3 ± 0.2 V (Fig. 3). The other seven putative precise excision lines turned out to contain small insertions varying from several base pairs to ∼200 bp. Interestingly, despite these small insertions, flies showed bang-sensitive behavioral phenotypes and low seizure threshold, similar to the precise excision lines. Surprisingly, we did not find any imprecise excision lines that deleted top1. Our expectation was that top1 deletions would be bang resistant (viable) or lethal; this would be somewhat similar to Df(1)ED7294, which deletes a region including the top1 gene.
The top1JS mutation reveals seizure threshold similar to the wild type
When analyzed outside the BS background by electrophysiology, top1JS flies exhibit normal seizure thresholds like those of wild-type flies (Fig. 4A): the wild-type female fly has a seizure threshold of 41.5 ± 5.6V, whereas the top1JS mutant female has a seizure threshold of 36.7 ± 2.7V. In addition, the heteroallelic combinations top1JS/top1G0229, top1JS/top1112, top1JS/top177, top1JS/top1G0134, and top1JS/top1G0201 have seizure thresholds similar to wild type (Fig. 4A). To further study whether alterations in individual neuron excitability lead to seizure suppression, GF thresholds were measured in the top1 mutants. Results in Figure 4B showed that stimulus voltages required for activation of the GF in top1JS mutants and were indistinguishable from wild-type flies. This suggests that altered single neuron excitability is probably not an explanation for seizure suppression by the top1 mutants.
Figure 4.
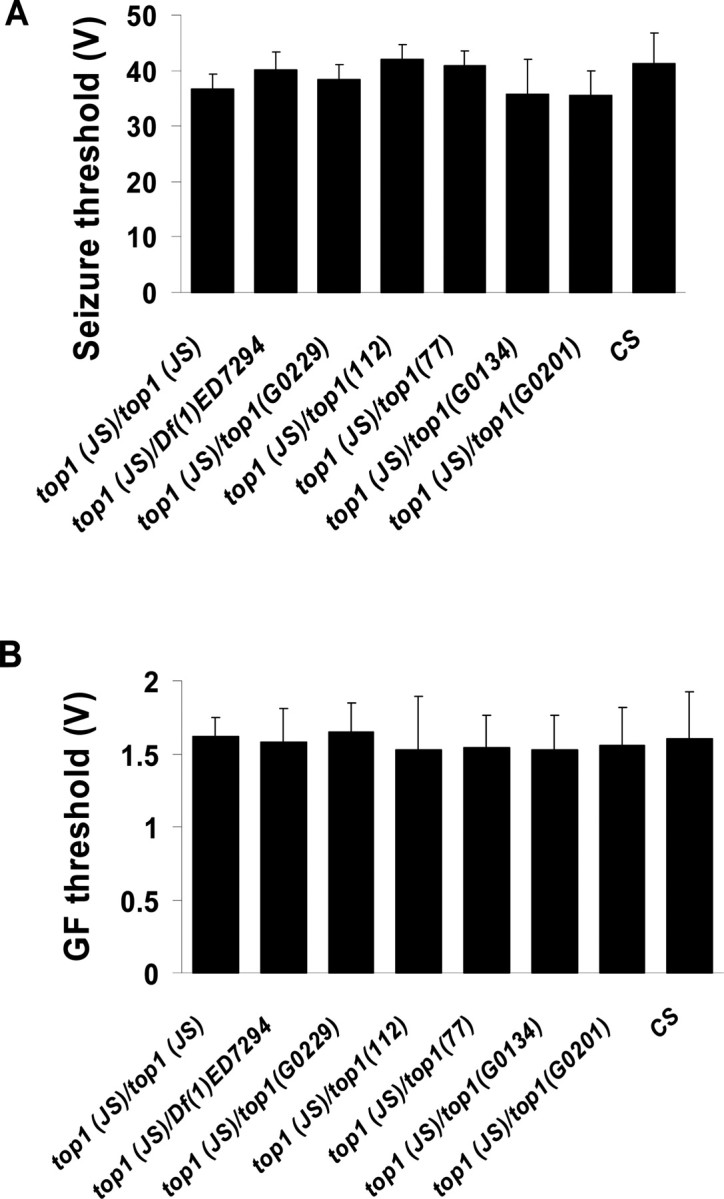
Seizure thresholds and GF thresholds of top1JS and its various heteroallelic combinations. A, Seizure thresholds of top1JS mutation and its various heteroallelic combinations. B, GF thresholds of top1JS and its various heteroallelic combinations. GF thresholds were measured by determining the minimum voltage required to elicit a stable, short-latency DLM response (∼1.4 ms) after 0.5 ms test pulses delivered at 1 Hz. For each genotype, n ≥ 8 flies tested. For both seizure threshold and GF threshold, results of different allelic combinations do not show significant difference compared with wild-type CS flies (p > 0.05).
All previously characterized seizure suppressors identified by reverse genetics exhibited elevated seizure thresholds that are significantly higher than those of wild type. Thus, the seizure-suppressor mutants shakB2 (gap junction) and para (voltage-gated Na+ α-subunit) exhibit seizure thresholds of 80.63 ± 8.71 and 65.0 ± 7.2 V at 300 ms HF stimuli, respectively; the seizure suppressor mutants mlenapts (double-strand RNA helicase), ShKS133 (voltage-gated K+ α-subunit), and mei-P26EG (RBCC-NHL protein) exhibit seizure thresholds of 72.2 ± 7.3, 83.8 ± 12.8, and 91.4 ± 5.1 V at 400 ms HS stimuli, respectively (Kuebler et al., 2001; Glasscock et al., 2005; Song and Tanouye, 2006). The ectopic neural expression of esg (escargot) (transcription factor of snail family) raised the seizure threshold to 55.5 ± 13.2 V (Hekmat-Scafe et al., 2005). The elevated seizure threshold was thought to relate reduced seizure susceptibility (Kuebler et al., 2001). The normal seizure threshold and GF threshold displayed in top1JS mutation may indicate a different mechanism of seizure suppression than other mutants above. A more likely possibility is that a positive feedback loop, which recruits multiple neurons simultaneously is disrupted so that the seizure would be suppressed or shortened. Another possibility is that suppression is a result of synaptic inhibition: the recruitment of inhibitory circuits, instead of excitatory (seizure) circuits, could account for the suppression and shortening of seizures we see near the suppression threshold.
The top1JS mutant acts as a general seizure suppressor
Additional genetic experiments revealed that the top1JS mutation interacts with other BS mutations, suggesting that it functions as a general seizure suppressor (Fig. 5). In general, bss is regarded as the most difficult BS mutant to suppress, followed by eas, and then sda. Mutants of bss have a seizure threshold at 3.2 ± 0.3 V, whereas sda flies have seizure thresholds ∼6 V, higher than those of both eas and bss. As shown in Figure 5A, the bang-sensitive phenotype of sda was strongly suppressed by the top1JS mutation, resulting in 63% suppression for top1JS; sda double mutants. Control flies of both sda and top1JS/+; sda/sda showed 100 and 95% bang sensitivity, respectively. In electrophysiology experiments, top1JS elevated sda seizure threshold to 14.8 ± 4.9 V, much higher than that of the sda mutant itself (Fig. 5). The behavioral and electrophysiological data indicated that top1JS acts as a general seizure suppressor and does not interact solely with the eas mutation. To further investigate a role for top1 in seizure suppression, we examined additional top1 mutants in a sda background. As for eas, some, but not all, top1 alleles acted to suppress sda seizures (Fig. 5). Thus, top1112 acts to suppress sda seizures as indicated by the genotype: top1JS/top1112; sda that showed 61% suppression of the behavioral paralytic phenotype compared with their control genotypes. In addition, electrophysiology showed an increased seizure threshold (14.2 ± 1.8 V), compared with their control genotypes (∼6 V). Finally, top1112 in heteroallelic combination with top1JS appears to be a recessive seizure suppressor of sda. Several top1 lethal mutations show no evidence of seizure suppression function: they were found to complement seizure suppression by top1JS. That is, top1G0229, top1G0201, and top1G0134 all do not suppress sda seizures in heteroallelic combination with top1JS. For top1G0201 and top1G0134, these findings are similar to eas; these top1 mutations did not act as seizure suppressors. In contrast, the findings with top1G0229 are especially interesting because it is a potent suppressor of eas seizures (Fig. 2) but not sda seizures.
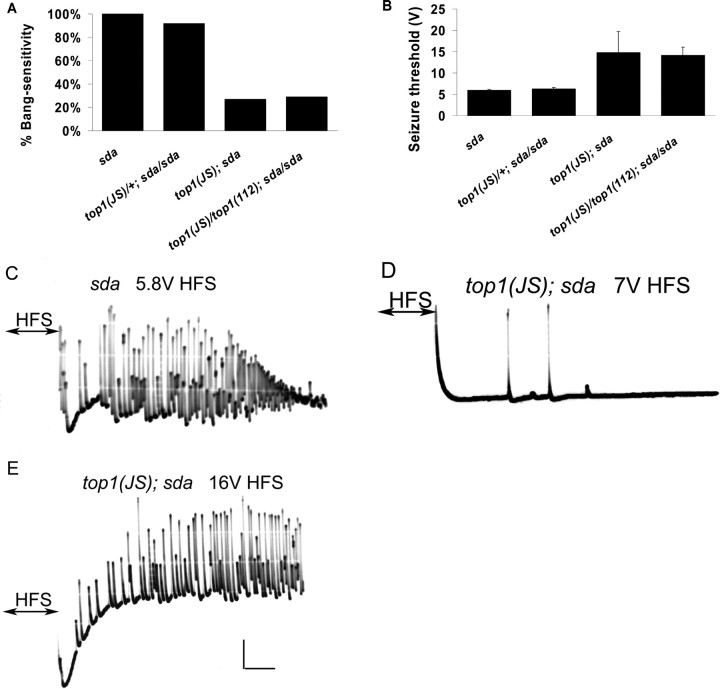
Figure 5.
The top1 mutation suppresses seizures in sda flies. A, Behavioral suppression of seizures in sda flies by top1JS and top1JS/top1112 heteroallelic combination. The top1JS mutation and top1JS/top1112 heteroallelic combination reduce bang-sensitive paralysis in sda flies to 27 and 29%, respectively (p < 0.01). Suppression of the sda BS behavior by top1JS is semidominant because top1JS heterozygotes are 92% bang sensitive. B, Electrophysiological suppression of seizures in sda flies by top1JS and top1JS/top1112 heteroallelic combination. Seizure thresholds are elevated in flies of genotypes top1JS; sda and top1JS/top1112; sda (p < 0.01). All sda genotypes were examined in females. C, A representative seizure in an sda fly recorded from the DLM after a high-frequency brain stimulus (HFS) of 5.8 V. D, failure to elicit seizure in a top1JS fly after a 7 V HFS. The trace shown here is absent of seizure activity, and only a few DLM responses can be seen after administration of a 7 V HFS. E, A seizure is elicited in a top1JS; sda double mutant after a 16 V HFS. Calibration: 10 mV, 200 ms. For each genotype, n ≥ 80 flies tested for behavior; n ≥ 8 flies tested for electrophysiology.
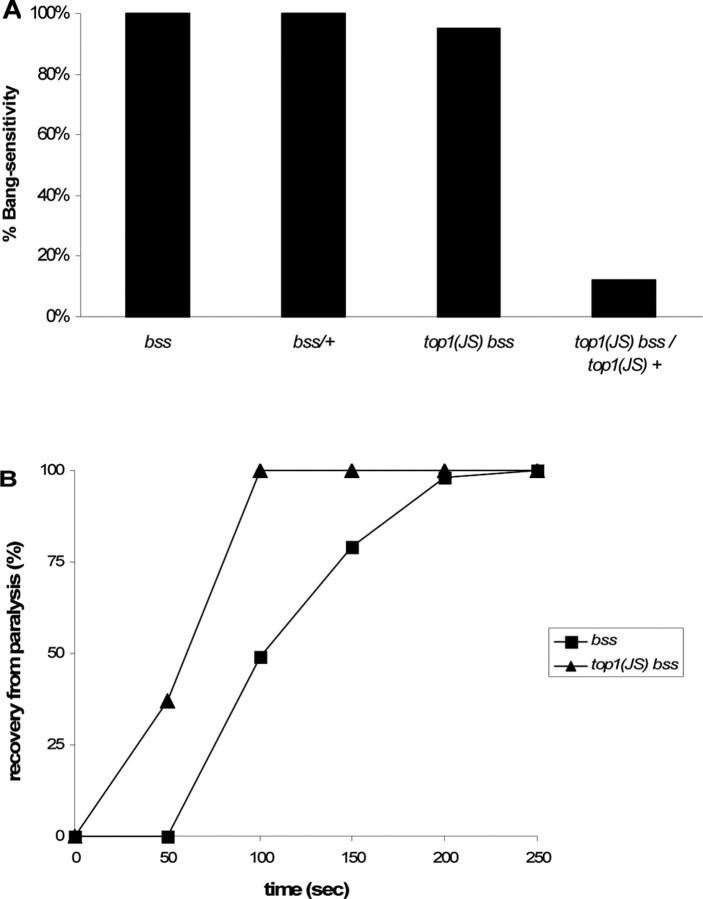
The bang-sensitive behavioral phenotype of bss was not suppressed by top1JS. Thus, top1JS bss double mutants showed 100% bang sensitivity. However, top1JS bss does appear to change some bss phenotypes, indicating that it may act to reduce the severity of bss seizures. This is most apparent in a reduction of paralytic behavioral recovery time. Thus, recovery time of the bss mutant is 120 s (1–2 d); recovery time is shortened to 77 s in the bss double mutants (Fig. 6). It has been shown previously that a reduction in recovery time, mainly a reduction in tonic–clonic spontaneous activity in bss mutants, is the first indication of seizure suppression (Kuebler et al., 2001; Song and Tanouye, 2006). In addition, the top1JS mutation suppresses the behavioral bang sensitivity of bss/+ heterozygotes. The bss mutant behaves as a dominant mutation in behavior, that is, bss/+ heterozygotes show 100% bang sensitivity. In contrast, top1JS bss/top1JS + flies only show 12% bang sensitivity (Fig. 6), suggesting suppression of the BS phenotype of bss/+ by the top1JS mutation.
Figure 6.
Seizure suppression in bss flies. A, Behavioral bang sensitivity is reduced in bss/+ heterozygous flies in the presence of top1JS. Note that bss/+ flies show 100% bang sensitivity; however, when it is placed in a top1JS homozygous background, behavioral bang sensitivity is reduced to 12% (p < 0.01). B, The recovery time is shortened in double-mutant top1JS bss flies. The bss mutant has an average paralysis time of 120 s, whereas the double-mutant top1JS bss shorten the paralysis time to 77 s. Shown is the percentage of flies recovered with time and a cumulative measure that includes the initial behavior seizure, the paralysis period, and the recovery seizure, which are not indicated separately. The number of flies standing at intervals after the shock was recorded until the entire population had recovered. For behavioral bang-sensitivity test, n ≥ 50 flies tested; for the recovery time, n ≥ 30 flies recorded.
The top1JS mutant has a reduced topoisomerase I mRNA transcription level in the CNS
Reverse-transcription PCR showed that top1 transcripts were reduced in the heads of top1JS mutants. Wild-type flies displayed a strong expression level of top1 transcripts, whereas the top1JS mutant displayed a weak expression level of top1 transcripts (Fig. 7). There are three top1 transcripts in Drosophila referred to as top1-RA (5325 nt), top1-RB (2379 nt), and top1-RC (4940 nt). Because of the presence of large overlapping regions among the three transcripts, this assay could not differentiate the transcription level of individual mRNA transcript. Shown in Figure 7 is the mRNA transcription level of top1: the mutant transcription level is ∼12.5-fold less than wild type. This transcription profile indicated that top1JS is a partial loss-of-function mutation caused by the disruption of 5′ UTR, and seizure suppression by the top1JS mutation is caused by reduced level of top1 transcripts.
Figure 7.
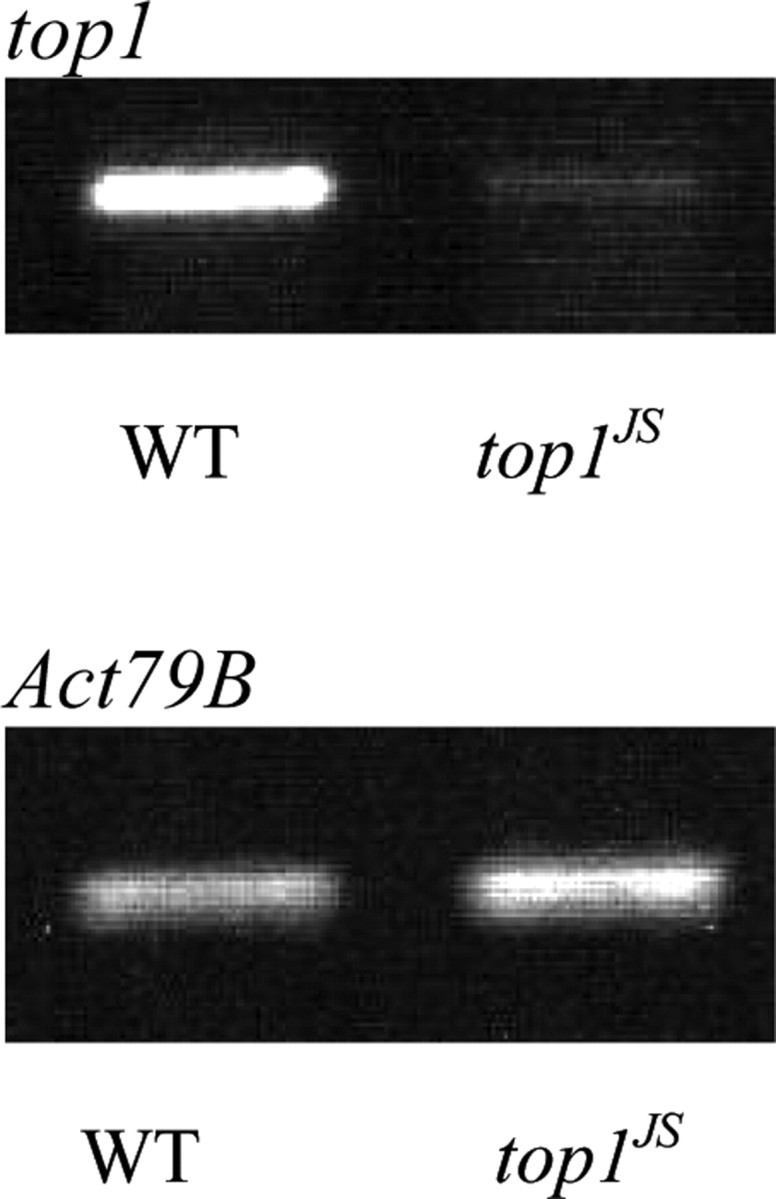
RT-PCR reveals a reduction in the top1 mRNA transcription level. Shown are the results of an RT-PCR analysis of RNA prepared from the heads of wild-type (WT) flies and top1JS mutant flies. Primers used in this assay amplified part of the first exon of the top1 gene. The internal control used in this assay is fly actin gene CG7478-PA (Act79B). The heads of top1JS mutants display a significant reduction (∼12.5-fold) in the level of top1 transcripts. Calibration: 10 mV, 200 ms.
Camptothecin reduces recovery time in BS mutants
We examined effects of the topo I inhibitor camptothecin (CPT) in three BS mutants: eas, sda, and bss. These mutants were raised in various concentrations of the drug, and their bang sensitivity was examined. In our studies, we did not see the elimination of the BS phenotype in any of our mutants treated with CPT. However, we did observe a reduction in the paralytic recovery time of all three BS mutants (Table 1). Treatment of bss with 1 μm CPT shortened the recovery time to 55 ± 9 s (n = 65) compared with the untreated (118 ± 43 s; n = 80) recovery time. There was also a reduction in the eas recovery time, albeit, more modest than seen for bss. Treatment of eas with 4 μm CPT shortened the recovery time to 30 ± 5 s (n = 62) compared with the untreated (44 ± 11 s; n = 80) recovery time. We also observed a small but statistically significant reduction in the sda recovery time. Treatment of sda with 1 μm CPT shortened the recovery time to 21 ± 3 s (n = 72) compared with the untreated (25 ± 6 s; n = 80) recovery time. These observations are generally consistent with those described previously using anti-epileptic agents fed to BS flies. Paralytic recovery time is reduced by phenytoin, gabapentin, KBr, and carbenoxolone: anti-epileptic drugs with different targets (Reynolds et al., 2004; Tan et al., 2004; Song and Tanouye, 2006). Some anti-epileptic drugs (carbamageprine, ethosuximide, and vigabactrin) have no effect on BS flies (Reynolds et al., 2004). Anti-epileptic drugs generally do not eliminate the BS phenotype of mutants (Reynolds et al., 2004).
Table 1.
Effects of camptothecin on the recovery time of various BS mutants
| Genotype | Recovery time (s) |
p value | |||
|---|---|---|---|---|---|
| Control (−CPT) | n | Test (+CPT) | n | ||
| bss | 118 ± 43 | 80 | 55 ± 9 | 65 | <0.01 |
| eas | 44 ± 11 | 80 | 30 ± 5 | 62 | <0.01 |
| sda | 25 ± 6 | 80 | 21 ± 3 | 72 | <0.05 |
Adult flies were fed on the CPT-containing medium, and their progeny were tested at 1–3 d after eclosion for the recovery time. Control flies were fed CPT-free medium. Results are statistically significant for all three mutants (p < 0.05).
The top1JS mutant has increased cell death
Seizure suppression by the top1JS mutant might be caused by neuronal cell death. This is consistent with increased neuronal cell death observed with treatment of the topoisomerase I inhibitor camptothecin (Morris et al., 1996). TUNEL assay showed increased apoptosis in top1JS mutant brain compared with that observed in wild-type flies (Fig. 8A,B). In addition, gel electrophoresis of top1JS mutant and top1JS eas double-mutant DNA showed considerable fragmentation compared with wild-type (Fig. 8C). The indication is that the top1JS mutation causes considerable cell death, much of it in the nervous system.
Figure 8.
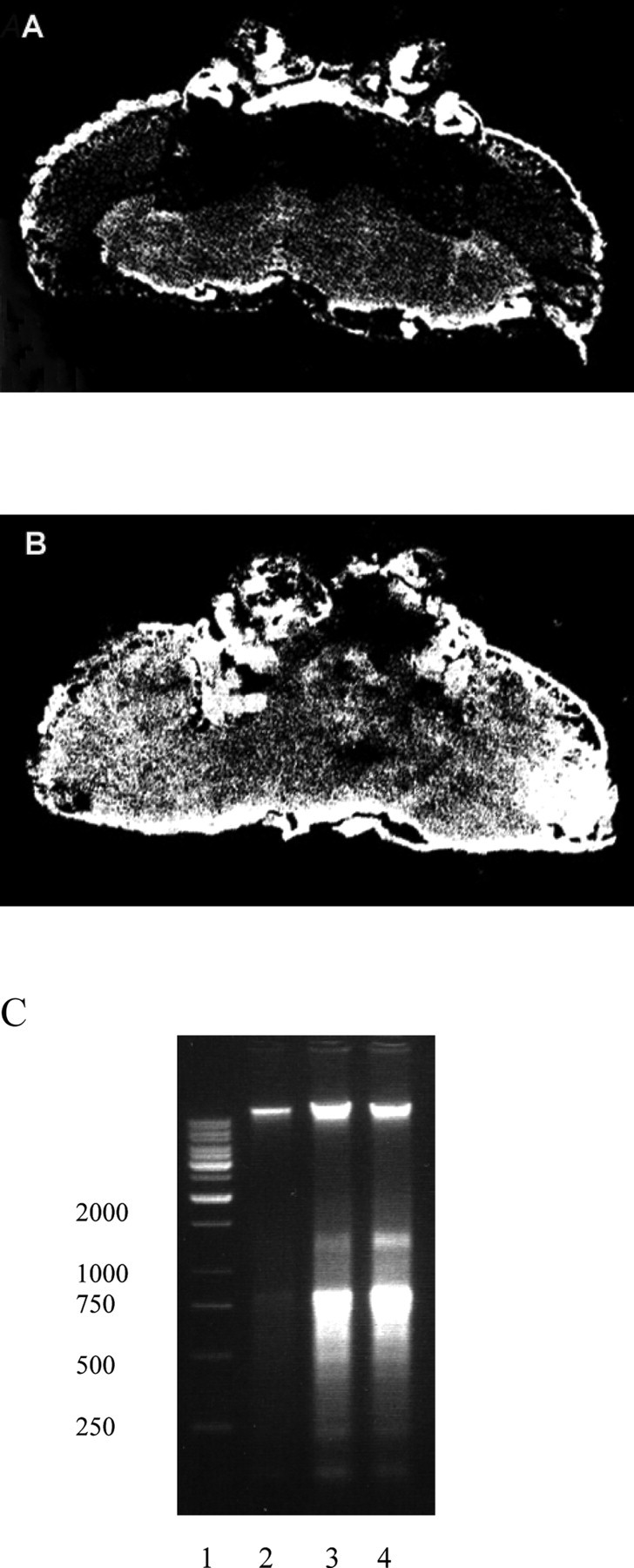
The top1JS mutation causes apoptosis. A, B, TUNEL assay. Representative sections from adult brain of wild-type (A) and top1JS mutant (B) flies. White indicates TUNEL signal. Weak diffuse TUNEL is observed in wild type. In mutant, TUNEL signal is more widespread and intense. C, DNA analysis by 1.5% agarose gel electrophoresis of genomic DNA. Lane 1, DNA ladder, 1 kb (Fermentas, Hanover, MD). Lane 2, Genomic DNA from wild-type flies. Lane 3, DNA from top1JS eas double-mutant flies. Lane 4, DNA from top1JS mutant flies. Fragmentation is observed in mutant DNA.
Overexpression of DIAP1 in the nervous system restores seizure sensitivity in top1JS eas
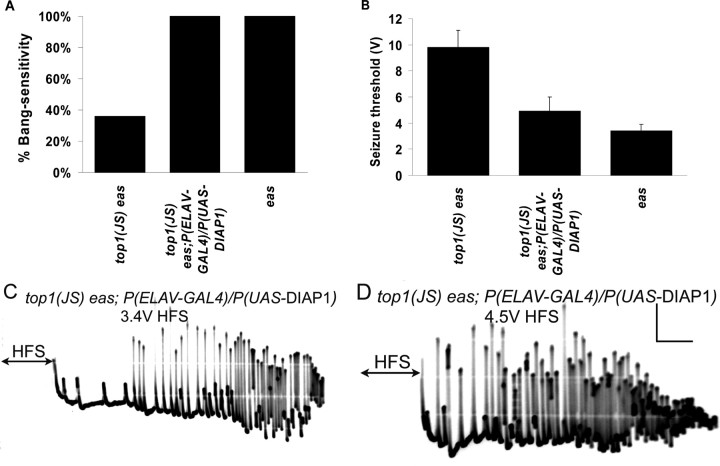
DIAP1, which is encoded by the th (thread) gene, is an antiapoptotic protein. DIAP1 contains two BIR (baculoviral inhibitor of apoptosis repeats) domains that are required for its antiapoptotic function. The GAL4/UAS expression system was used: the embryonic lethal abnormal vision (ELAV)–GAL4 construct leads to neuronal expression of GAL4 transcriptional activator, which drives expression of UAS–DIAP1 in all neurons. Overexpression of DIAP1 in the nervous system should suppress neuronal apoptosis, leading to the rescue of any neuronal apoptosis phenotypes caused by the top1JS mutation. This rescue could be reflected by restored seizure sensitivity in top1JS eas double mutants. Indeed, flies with the genotype top1JS eas; P(ELAV-GAL4)/P(UAS-DIAP1) revealed substantial seizure sensitivity (Fig. 9): the behavioral bang sensitivity was restored to 100%, similar to the eas single mutant (Fig. 9A); the electrophysiological seizure threshold was decreased to 4.9 ± 1.1 V, significantly lower than the 9.8 ± 1.3 V in top1JS eas double mutants (Fig. 9B–D). This result suggests that the seizure suppression caused by the top1JS mutant is a result of increased neural apoptosis, pointing to a novel function for topo I in construction and/or maintenance of a circuit required for seizure propagation in vivo.
Figure 9.
Overexpression of DIAP1 in the nervous system restores seizure sensitivity of top1JS eas double mutants. A, Behavioral bang sensitivity is restored in top1JS eas; P(ELAV-GAL4)/P(UAS-DIAP1) flies (p < 0.01, compared with top1JS eas). B, The seizure threshold is decreased in top1JS eas; P(ELAV-GAL4)/P(UAS-DIAP1) flies compared with top1JS eas (p < 0.01), suggesting increased seizure sensitivity. C, A seizure is triggered in a top1JS eas; P(ELAV-GAL4)/P(UAS-DIAP1) fly at a 3.4 V HFS. D, A seizure is triggered in a top1JS eas; P(ELAV-GAL4)/P(UAS-DIAP1) fly at a 4.5 V HFS. Calibration: 10 mV, 200 ms. For each genotype, n ≥ 30 flies tested for behavior; n ≥ 6 flies tested for electrophysiology.
Discussion
Discovery of a novel seizure-suppressor mutation: top1JS
The identification of top1 as a seizure suppressor is surprising because the biological functions with which it has been associated are not obviously electrical excitability functions. The top1 gene appears to play an important role in the nervous system as a key regulator of seizure susceptibility. The top1JS mutation shows the ability to suppress seizures in multiple BS strains, including eas, sda, and bss mutants. These BS mutants are well characterized on a behavioral and electrophysiological level, and their seizure activity shows numerous similarities with seizure activity in humans, making them a useful tool for identifying new seizure suppressors (Benzer, 1971; Kuebler and Tanouye, 2000). Two of these BS mutants examined (eas and sda) encode very different products: eas encodes ethanolamine kinase involved in synthesis of the phosphatidyl ethanolamine in neuronal membrane, and sda encodes an aminopeptidase N (Pavlidis et al., 1994; Zhang et al., 2002). The gene product of bss is still unknown but thought to encode another very different product (M. Tanouye, unpublished results). Therefore, these three BS mutations may increase seizure susceptibility of flies by different mechanisms. The top1JS mutation can suppress seizures in eas and sda flies to a similar extent, with the suppression slightly better in top1JS; sda double mutants than in top1JS eas double mutants, whereas the suppression of behavioral bang sensitivity in bss flies is not very efficient. However, the recovery time after paralysis is drastically shortened, and the tonic–clonic spontaneous seizures during paralysis are drastically reduced in double-mutant top1JS bss flies. In addition, top1JS suppresses seizures in bss/+ heterozygotes to 12% behaviorally (100% in bss/+ flies). This is consistent with previous observations on general seizure suppressors that bss is the strongest of the three mutations, eas is the intermediate, and sda is the weakest (in terms of reduction of seizure thresholds and the ease with which it can be suppressed by secondary mutations that reduce nervous system excitability).
The partial loss-of-function top1JS mutations that act to suppress seizures cause no other obvious phenotypes whether in a wild-type or a seizure-sensitive mutant background. Thus, top1JS mutant flies show no obvious nervous system excitability defects: they are not temperature paralytic (hypoexcitability), and they do not exhibit leg-shaking under ether anesthesia (hyperexcitability). In addition, the mutant flies show no notable defects in other specific behaviors: they eat, jump, fly, groom, court, and mate normally. The top1 alleles characterized previously all exhibit homozygous lethal phenotype, suggesting an essential role of the top1 gene in Drosophila development. The top1JS mutation is the first homozygous viable Drosophila partial loss-of-function top1 mutation identified thus far. Molecular experiments verified that the mutation has a reduced mRNA transcription level compared with wild-type flies.
There appears to be two salient phenotypes associated with top1 mutations: (1) recessive lethal, suggesting a vital function, and (2) a recessive seizure suppression function. A parsimonious model for the results from different top1 alleles might have top1JS as the weakest allele because it is the only viable one. Furthermore, the lethal alleles would be expected to have stronger phenotypes indicative of greater loss-of-function. This does not appear to be the case: top1 gene function seems more complicated based on several observations involving allelic specificity. Although viable, top1JS appears to be the strongest seizure suppressor: greater seizure suppression is seen for top1JS homozygotes than for any of the heteroallelic combinations. Surprisingly, in heteroallelic combinations, some top1 lethal alleles (top1G0134, top1G0201) complement suppression of eas seizures. Another interesting observation from examination of different top1 alleles comes from results with top1G0229. In heteroallelic combination with top1JS, this allele suppresses eas seizures, but not sda seizures. The surprise is that several previous studies have shown that sda is generally easier to suppress than eas. The explanation for these top1 allelic complexities is not entirely clear at present. The P-element insertion sites differ for the different top1 alleles, and part of the explanation may come from differential exon usage in different top1 isoforms.
Possible mechanism of seizure suppression by top1JS
The mechanism by which topo I influences seizure susceptibility is not clear. Mouse brain shows substantial levels of topo I activity varying among different brain regions (Plaschkes et al., 2005). Immunohistochemistry shows that the highest topo I levels are observed in inhibitory neurons. In vitro studies show that topo I disruption leads to neuronal cell death, evidenced by accompanying DNA fragmentation, chromatin condensation, cell blebbing, cytoplasmic shrinkage, fragmentation of neurites, and other characteristics of apoptotic cell death (Morris and Geller, 1996). RNA and protein inhibitors prevent apoptosis attributable to topo I disruption. This indicates that topo I-dependent apoptosis requires active transcription and translation of downstream proteins involved in apoptosis and other topo I-interacting proteins. One possible mechanism is adapted from a recently proposed model of topo I action in DNA relaxation and damage control (Leppard and Champoux, 2005). Transcription in neurons generates supercoiled DNA that must be continuously relaxed to sustain high levels of RNA synthesis. Binding of topo I and DNA forms the cleavable complex leading to relaxation. A reduced level of topo I attributable to mutation or CPT treatment leads to partial inhibition of transcription. A series of events including relocalization of topo I from the nucleolus to the nucleoplasm leads to downstream events: the synthesis of proteins involved in the apoptosis. Electrophysiological evidence provided in this study is consistent with the above explanation. In addition, apoptosis analysis showed that top1JS caused considerable amounts of cell death. Furthermore, overexpression of DIAP1 in the nervous system resulted in the restoration of seizure sensitivity in top1JS eas double mutants, suggesting increased apoptosis in the top1JS mutant is the cause of seizure suppression.
The top1JS mutation has a normal seizure threshold compared with wild-type flies when analyzed outside a BS background. A model that accounts for several major features of Drosophila seizure susceptibility and its modification by genetic mutations was proposed (Kuebler et al., 2001). This model proposed that the number of input neurons makes a difference in seizure thresholds among wild-type, BS mutant, seizure suppressor mutant, and double mutants of BS mutant with suppressor mutant. In a normal wild-type fly, stimulation of two input neurons (each input neuron represents a population of neurons with similar properties or an extensive neural circuit) with a high-frequency electrical stimulus triggers a seizure, and synaptic potentials from these two neurons summate in the Sei (seizure) neuron temporally and spatially. Seizure is triggered through the seizure output neural circuit once the threshold is reached. In a BS mutant, it is easier to trigger a seizure, and stimulation of a single neuron is sufficient. A low-voltage high-frequency stimulus is sufficient to trigger a seizure. In a seizure suppressor mutant, it is difficult to trigger a seizure and necessary to trigger three input neurons. Therefore, in this case, a high-voltage high-frequency stimulus is required. In a double mutant, the seizure threshold is elevated and higher than a BS mutant, so only two neurons are stimulated to trigger a seizure, and they are summated temporally and spatially. Based on this model, neuronal cell death caused by the top1JS mutation decreases the number of neurons being stimulated during the high-frequency stimulation. Therefore, a normal seizure threshold was seen in the top1JS mutant. The neuronal cell death proposal can also explain the discrepancy seen in electrophysiology and behavior. The top1JS mutation can suppress bss/+ flies to 12% bang sensitivity behaviorally (100% bang sensitivity in bss/+ flies); however, the seizure threshold remains unchanged compared with that of bss/+ flies.
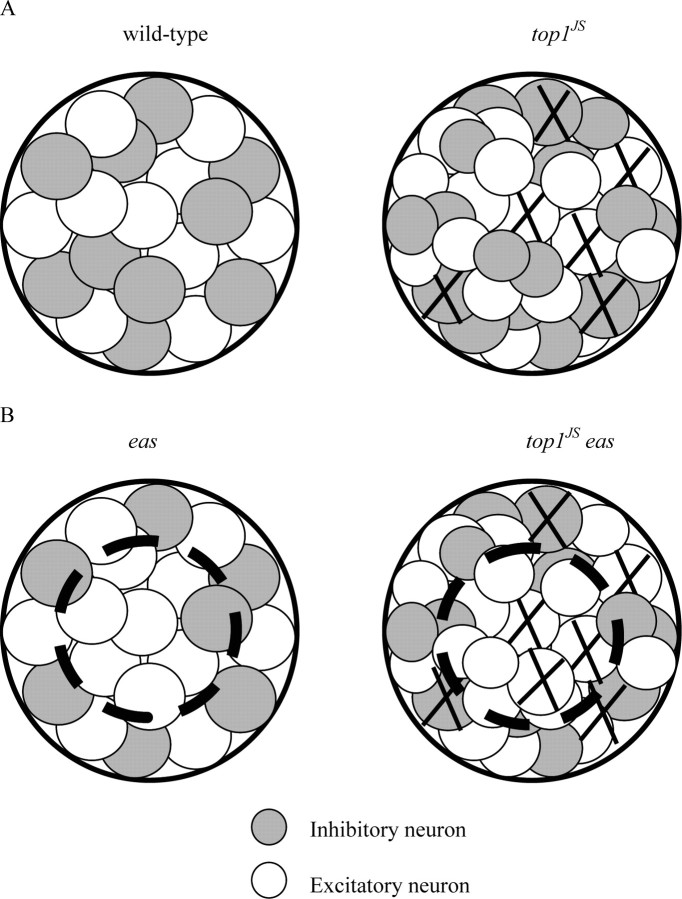
The above model seems to present us with a paradox. Seizure threshold of the top1JS mutant is similar to the wild type because of increased neuronal apoptosis; therefore, seizure threshold of double-mutant top1JS eas should be lower than the eas mutant itself also because of increased neuronal apoptosis. However, in contrast, seizure threshold of the top1JS eas double mutant is actually higher than the eas single mutant, as seen in the previous electrophysiological assay. One possible explanation for this paradox could be that different genetic backgrounds generate different ratios of excitatory neurons to inhibitory neurons in the Sei neuron pool (Fig. 10). In the wild-type and top1JS mutant flies, excitatory and inhibitory neurons are balanced in the Sei neuron pool, although the top1JS Sei neuron pool contains more neurons than the wild-type because of its seizure-resistant property. When neuronal apoptosis occurs, excitatory and inhibitory neurons are killed at the same level; therefore, the total number of neurons is similar in the wild-type and the top1JS mutant, leading to similarities in seizure threshold (Fig. 10A). In the double-mutant top1JS eas and eas mutant flies, excitatory neurons are more prevalent than inhibitory neurons in the Sei neuron pool because of the seizure-sensitive property of eas mutant background. Therefore, when neuronal apoptosis occurs, more excitatory neurons are killed than inhibitory neurons in the double-mutant top1JS eas. In this case, to reach the seizure threshold, more (excitatory/inhibitory) neurons are required to be recruited to trigger a seizure, which results in the increased threshold in top1JS compared with the eas mutant (Fig. 10B).
Figure 10.
Sei neuron pools in different genetic backgrounds. A, The Sei neuron pool in wild type and the top1JS mutant. Excitatory and inhibitory neurons are balanced in the Sei neuron pool, although the top1JS Sei neuron pool contains more neurons than the wild-type because of its seizure-resistant property. B, The Sei neuron pool in the eas mutant and top1JS eas double mutant. Excitatory neurons are more prevalent than inhibitory neurons in the Sei neuron pool because of the seizure-sensitive property of eas mutant background. The dot-circled area represents the neurons involved in seizure generation, because lower seizure thresholds are required in eas and top1JS eas than the wild type and top1JS.
Practical implications from genetic and CPT-feeding analyses
Epilepsy is the most common chronic neurological disorder, affecting ∼2 million people in the United States population (McNamara, 1999; Shneker and Fountain, 2003). However, only approximately two-thirds of patients experience symptomatic relief with antiepileptic drugs currently on the market, and many of these responders experience periodic breakthrough seizures and toxic nervous system side effects. The present work is an example of how Drosophila genetics can lead to identification of new genes and new drug targets relevant to human disease. The identification of topo I as a seizure suppressor is surprising because DNA topoisomerases have not previously been associated with seizure or seizure control. For top1JS mutants, there are no apparent defects in nervous system functions or other specific behaviors implying that limited disruption of topo I can be well tolerated. However, top1 is an essential gene and severe top1 mutations are lethal, implying that top1 function must be adjusted to an ideal range: one that suppresses seizures but is not otherwise deleterious. CPT acts as an inhibitor of topo I in mammals. CPTs have been described as one of the most promising anticancer drugs of the 21st century. Irinotecan (CPT-11) and topotecan are water-soluble derivatives approved by the Food and Drug Administration for treatment of colorectal and ovarian cancer. CPT has not previously been associated with the treatment of epilepsy, but potential application in the clinic is suggested by this work. CPT did not affect BS paralysis in flies but did affect recovery time. Although not as impressive as top1JS mutation in terms of seizure suppression, CPT treatment does not cause detectable neuronal apoptosis in brain sections of viable long-term feeding progeny (data not shown). The assumption is that CPT treatment may have weaker effects than the genetic mutation, which is consistent with the weaker behavioral seizure suppression by CPT. Moreover, CPT results are comparable with others observed with known human anti-epileptic drugs. A preliminary comparison of feeding experiments suggests that CPT may be a less effective anti-epileptic agent compared with phenytoin, but may be better than valproate, KBr, and carbenoxolone (Kuebler and Tanouye, 2002; Reynolds et al., 2004; Tan et al., 2004; Song and Tanouye, 2006).
Footnotes
This work was supported by a National Institutes of Health research grant and an Epilepsy Foundation grant to M.T. We thank the Bloomington Drosophila Stock Center and Dr. John Nambu (University of Massachusetts, Amherst, Amherst, MA) for Drosophila stocks used in this study. We also thank Kevin Qian for help with statistical analysis on CPT feeding experiments, Zejuan Sheng for confocal microscopy, and Drs. Hekmat-Scafe and Glasscock for discussions throughout the project.
References
- Benzer S. From the gene to behavior. J Am Med Assoc. 1971;218:1015–1022. [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Fergestad T, Bostwick B, Ganetzky B. Metabolic disruption in Drosophila bang-sensitive seizure mutants. Genetics. 2006;173:1357–1364. doi: 10.1534/genetics.106.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasscock E, Singhania A, Tanouye MA. The mei-P26 gene encodes a RING finger B-box coiled-coil-NHL protein that regulates seizure susceptibility in Drosophila. Genetics. 2005;170:1677–1689. doi: 10.1534/genetics.105.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Dang KN, Tanouye MA. Seizure suppression by gain-of-function escargots mutations. Genetics. 2005;169:1477–1493. doi: 10.1534/genetics.104.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuebler D, Tanouye MA. Modifications of seizure susceptibility in Drosophila. J Neurophysiol. 2000;83:998–1009. doi: 10.1152/jn.2000.83.2.998. [DOI] [PubMed] [Google Scholar]
- Kuebler D, Tanouye M. Anticonvulsant valproate reduces seizure-susceptibility in mutant Drosophila. Brain Res. 2002;958:36–42. doi: 10.1016/s0006-8993(02)03431-5. [DOI] [PubMed] [Google Scholar]
- Kuebler D, Zhang H, Ren X, Tanouye MA. Genetic suppression of seizure susceptibility in Drosophila. J Neurophysiol. 2001;86:1211–1225. doi: 10.1152/jn.2001.86.3.1211. [DOI] [PubMed] [Google Scholar]
- Lee MP, Brown SD, Chen A, Hsieh T-S. DNA topoisomerase I is essential in Drosophila melanogaster. Proc Natl Acad Sci USA. 1993;90:6656–6660. doi: 10.1073/pnas.90.14.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppard J, Champoux JJ. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- Li T, Liu L. Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol. 2000;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- McNamara JO. Emerging insights into the genesis of epilepsy. Nature. 1999;399(Suppl):A15–A22. doi: 10.1038/399a015. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Geller HM. Induction of neuronal apoptosis by camptothecin, an inhibitor of DNA topoisomerase I: evidence for cell cycle-independent toxicity. J Cell Biol. 1996;134:757–770. doi: 10.1083/jcb.134.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Morris EJ, Greene LA, Geller HM. G1/S cell cycle blockers and inhibitors of cyclin dependent kinases suppress camptothecin-induced neuronal apoptosis. J Neurosci. 1997;17:1256–1270. doi: 10.1523/JNEUROSCI.17-04-01256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Ramaswami M, Tanouye MA. The Drosophila easily shocked gene: a mutation in a phospholipid synthetic pathway causes seizure, neuronal failure, and paralysis. Cell. 1994;79:23–33. doi: 10.1016/0092-8674(94)90397-2. [DOI] [PubMed] [Google Scholar]
- Plaschkes I, Silverman FW, Priel E. DNA topoisomerase I in the mouse central nervous system: age and sex dependence. J Comp Neurol. 2005;493:357–369. doi: 10.1002/cne.20793. [DOI] [PubMed] [Google Scholar]
- Pommier Y. Diversity of DNA topoisomerases I and inhibitors. Biochimie. 1998;80:255–270. doi: 10.1016/s0300-9084(98)80008-4. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Pourquier P, Urasaki Y, Wu J, Laco GS. Topoisomerase I inhibitors: selectivity and cellular resistance. Drug Resist Updat. 1999;2:307–318. doi: 10.1054/drup.1999.0102. [DOI] [PubMed] [Google Scholar]
- Reynolds ER, Stauffer EA, Feeney L, Rojahn E, Jacobs B, McKeever C. Treatment with the antiepileptic drugs phenytoin and gabapentin ameliorates seizure and paralysis of Drosophila bang-sensitive mutants. J Neurobiol. 2004;58:503–513. doi: 10.1002/neu.10297. [DOI] [PubMed] [Google Scholar]
- Royden C, Pirrotta V, Jan L. The tko locus, site of a behavioral mutation in D. melanogaster, codes for a protein homologous to prokaryotic ribosomal protein S12. Cell. 1987;51:165–173. doi: 10.1016/0092-8674(87)90144-9. [DOI] [PubMed] [Google Scholar]
- Shneker BF, Fountain NB. Epilepsy. Dis Mon. 2003;49:426–478. doi: 10.1016/s0011-5029(03)00065-8. [DOI] [PubMed] [Google Scholar]
- Song J, Tanouye MA. Seizure suppression by shakB2, a gap junction mutation in Drosophila. J Neurophysiol. 2006;95:627–635. doi: 10.1152/jn.01059.2004. [DOI] [PubMed] [Google Scholar]
- Tan JS, Lin F, Tanouye MA. Potassium bromide, an anticonvulsant, is effective at alleviating seizures in the Drosophila bang-sensitive mutant bang senseless. Brain Res. 2004;1020:45–52. doi: 10.1016/j.brainres.2004.05.111. [DOI] [PubMed] [Google Scholar]
- Tanouye MA, Wyman RJ. Motor outputs of the giant nerve fiber in Drosophila. J Neurophysiol. 1980;44:405–421. doi: 10.1152/jn.1980.44.2.405. [DOI] [PubMed] [Google Scholar]
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Wang H, Morris Natschke S, Lee K. Recent advances in discovery and development of topoisomerase inhibitors as antitumor agents. Med Res Rev. 1997;17:367–425. doi: 10.1002/(sici)1098-1128(199707)17:4<367::aid-med3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases as targets of therapeutics: an overview. Adv Pharmacol. 1994;29:1–9. doi: 10.1016/s1054-3589(08)60537-2. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wilson C, Pearson R, Bellen H, O'Kane C, Grossniklaus U, et al. P-element mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- Zhang CX, Lee MP, Chen AD, Brown SD, Hsieh T. Isolation and characterization of a Drosophila gene essential for early embryonic development and formation of cortical cleavage furrows. J Cell Biol. 1996;134:923–934. doi: 10.1083/jcb.134.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CX, Chen AD, Gettel NJ, Hsieh T-S. Essential functions of DNA topoisomerase I in Drosophila melanogaster. Dev Biol. 2000;222:27–40. doi: 10.1006/dbio.2000.9704. [DOI] [PubMed] [Google Scholar]
- Zhang H, Tan J, Reynolds E, Kuebler D, Faulhaber S, Tanouye M. The Drosophila slamdance gene: a mutation in an aminopeptidase can cause seizure, paralysis and neuronal failure. Genetics. 2002;162:1283–1299. doi: 10.1093/genetics/162.3.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]