Abstract
Although visual pigments play key structural and functional roles in photoreceptors, the relationship between the properties of mammalian cone pigments and those of mammalian cones is not well understood. We generated transgenic mice with rods expressing mouse short-wave cone opsin (S-opsin) to test whether cone pigment can substitute for the structural and functional roles of rhodopsin and to investigate how the biophysical and signaling properties of the short-wave cone pigment (S-pigment) contribute to the specialized function of cones. The transgenic S-opsin was targeted to rod outer segments, and formed a pigment with peak absorption at 360 nm. Expression of S-opsin in rods lacking rhodopsin (rho−/−) promoted outer segment growth and cell survival and restored their ability to respond to light while shifting their action spectrum to 355 nm. Using the spectral separation between S-pigment and rhodopsin, we found that the two pigments produced similar photoresponses. Dark noise did not increase in transgenic rods, indicating that thermal activation of S-pigment might not contribute to the low sensitivity of mouse S-cones. Using rod arrestin knock-out animals (arr1−/−), we found that the physiologically active (meta II) state of S-pigment decays 40 times faster than that of rhodopsin. Interestingly, rod arrestin was efficient in deactivating S-pigment in rods, but its deletion did not have any obvious effect on dim-flash response shutoff in cones. Furthermore, transgenic cone arrestin was not able to rescue the slow shutoff of S-pigment dim-flash response in arr1−/− rods. Thus, the connection between rod/cone arrestins and S-pigment shutoff remains unclear.
Keywords: opsins, arrestin, phototransduction, visual pigment, photoreceptor, retina
Introduction
Cone photoreceptors and their visual pigments, especially in mammals, have proven challenging for study compared with their rod counterparts. In most mammalian species, cones make up <5% of the photoreceptors (Carter-Dawson and LaVail, 1979). In addition to low abundance, their diversity also complicates the functional characterization of cones. Short-, middle-, and long-wave-sensitive cones are found in trichromats such as humans and Old World primates, whereas all other mammalian dichromats use middle- and short-wave-sensitive cones. The separation of the short-wave from the middle/long-wave pigment occurred >500 million years ago (Mollon, 1989), indicating the evolutionary significance of short-wave cones. Yet, very little is known about the properties of mammalian short-wave pigments.
In salamander, short-wave cones (S-cones) are more sensitive than red cones (L-cones) and are comparable in sensitivity to rods (Perry and McNaughton, 1991). This is consistent with the lower dark noise in salamander S-cones compared with L-cones (Rieke and Baylor, 2000) produced by thermal activation of the pigment (Kefalov et al., 2003). In contrast, in monkey, S-cones are comparable in sensitivity to L-cones and significantly less sensitive than rods (Schnapf et al., 1990). Whereas monkey L-cones also exhibit a high level of dark noise, consistent with a high rate of thermal activation of their pigment, the dark noise in mammalian S-cones has not been measured and the rate of thermal activation of mammalian short-wave pigment (S-pigment) is not known. The response properties of mouse S-pigment have been investigated in wild-type mouse cones (Nikonov et al., 2006) and in the photoreceptors of nrl knock-out mice (nrl−/−), which display cone-like properties (Nikonov et al., 2005). In both of these cases, however, S-pigment is coexpressed with middle-wave pigment (M-pigment) in the same cell, and in different proportions across cells, making the data analysis difficult. As a result, little is known about the biochemical and biophysical properties of S-pigment and how these attributes contribute to shaping the responses from S-cones.
Studies of transgenic photoreceptors expressing cone opsin provide an effective alternative approach by which to examine some of the biophysical properties of cone visual pigments and how they may affect the functional properties of cones. We describe a study of the S-pigment by expressing it as a transgene in mouse rods. By crossing these mice with an existing rhodopsin knock-out mouse line (Lem et al., 1999), we also produced rods that expressed S-pigment alone. With these and related mouse lines, we were able to investigate the targeting of S-pigment to the rod outer segment, the effect of S-pigment on the long-term viability of rod cells, the ability of S-pigment to activate rod transducin, and its inactivation by rhodopsin kinase and rod- or cone-arrestin. We also measured the lifetime of S-pigment meta-II state in situ and investigated the possible contribution of thermal activation of S-pigment to the low sensitivity of mouse S-cones.
Materials and Methods
Generation of S-opsin and S-opsin-1D4 transgenic mice.
The use of mice in these experiments was in accordance with the guidelines established by the National Institutes of Health and the Society for Neuroscience Policy on the Use of Animals in Neuroscience Research. The 1.2 kb S-opsin cDNA coding sequence was synthesized by reverse transcription-PCR with primer Sopsin_F1 (CCGCTCGAGGGTGATAGCAGAAGAATCGTC) and Sopsin_R1 (GAAGATCTCACCAGAGTGCCACCAC) using mRNA prepared from mouse retinas. The PCR product coding for S-opsin was cloned into pBluescript KS vector with EcoR V site (pBlopsin) and sequenced. To make the S-opsin-1D4 construct, the DNA sequence coding for rhodopsin 1D4 epitope (ETSQVAPA) was inserted immediately before the S-opsin stop codon by two-step PCR. All of the following cloning steps were performed on the pBluescript KS vector using a three-piece ligation involving three restriction sites. The S-opsin coding sequence was ligated with a 0.6 kb mp1 fragment for polyadenylation site at 3` end (XhoI, BglII, and BamHI site; pBlopsin-mp1) and a 4.4 kb fragment of the rhodopsin promoter region at 5′ end (KpnI, XhoI, and XbaI sites; pRhMBO) (Lem et al., 1991). The pRhoMBO plasmid was purified by CsCl2 gradient and digested with KpnI and XbaI to yield the 6.2 kb insert fragment, which was then purified by QIAEXII gel extraction kit (Qiagen, Hilden, Germany) and Elutip-d column (Schleicher and Schuell BioScience, Keene, NH).
The 6.2 kb insert fragment was microinjected into zygotes from C57BI/6J and DBA/2J F1 strains according to standard procedures. Founders and transgene-positive mice were identified by PCR of DNA obtained from tail biopsies. S-opsin or S-opsin-1D4 mice were subsequently crossed with rhodopsin−/− (rho−/−) (Lem et al., 1999) and/or arr1−/− mice (Xu et al., 1997), as well as a transgenic line of mice that expressed cone arrestin in place of rod arrestin in rods (Chan et al., 2007).
Characterization of S-opsin and S-opsin-1D4 transgenic mice using immunohistochemistry, morphologic, and Western blot analyses.
Mice were killed by CO2 inhalation followed by cervical dislocation. The superior pole was marked by cauterization before enucleation. Eyecups were immersed in fixative (4.0% paraformaldehyde, 0.5% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.2) for 1 h at 4°C, washed extensively, and cryoprotected with 30% sucrose in 0.1 m cacodylate buffer, pH 7.2, overnight. The eyecups were embedded in OCT (Tissue-Tek) and immersed in liquid nitrogen. Frozen sections (10 μm) were obtained using a cryostat (Leica, Nussloch, Germany). Sections were incubated with a 1:100 dilution of antibody made against S-opsin N-terminal sequence H2N-SGEDDFYLFQNISSV-COOH (MBO_N) (QCB; Biosource, Camarillo, CA) for 1 h and a 1:500 dilution of goat anti rabbit IgG conjugated with FITC or Texas Red (Vector Laboratories, Burlingame, CA). Prepared sections were viewed using confocal microscopy (LSM510; Zeiss, Oberkochen, Germany).
For light-microscopic analysis of retinal morphology, eyecups were fixed overnight in 1/2 Karnovsky buffer (2.5% glutaraldehyde, 2% paraformaldehyde in 0.1 m cacodylate buffer, pH 7.2), washed, and embedded in epoxy resin as described previously (Concepcion et al., 2002). The epon-embedded eyes were sectioned into 1 μm thickness and stained with Richardson's stain (0.5% methylene blue, 0.5% Borax, and 0.5% Azure II).
For Western blot analysis, retinas from 1-month-old mice were dissected under infrared light in cold PBS. Each retina was homogenized in 100 μl of buffer [80 mm Tris-HCl, 4 mm MgCl2, 1 mm CaCl2, and protease inhibitor mixture (Boehringer Mannheim, Indianapolis, IN), pH 8.0]. Twenty units of DNase I (Boehringer Mannheim) were then added, and the mixture was incubated at 22°C for 30 min. An equal volume of SDS-loading buffer was added to each sample and the indicated amounts were separated on a 12% Tris-glycine polyacrylamide gel (Invitrogen, Eugene, OR). The proteins were transferred onto a nitrocellulose membrane and incubated with mAb 1D4 (1:10000), mAb 4D2 (1:10000), or MBO_N (1:1000) for 1 h and then with goat anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase for 30 min. The ECL system (GE Healthcare Bio-Sciences, Piscataway, NJ) was used for immunodetection.
Absorbance measurement of purified S-opsin-1D4.
All of the following steps were performed under infrared light. S-opsin-1D4rho−/− mice were dark-adapted overnight. Their retinas were dissected out and incubated for 4 h at room temperature in solubilization buffer (20 mm HEPES, 150 mm NaCl, 3 mm MgCl2, and 1% dodecyl maltoside, pH 7.4) containing protease inhibitor mixture (1 tablet per 10 ml of buffer; Roche, Indianapolis, IN). The samples were centrifuged at 15,000 × g for 20 min. The supernatant was incubated with 1D4-Sepharose equilibrated with solubilization buffer (50 μl of 1D4-Sepharose per retina) at 4°C overnight. The S-opsin-1D4-bound 1D4-Sepharose was washed twice with buffer (5 mm HEPES, 0.02% dodecyl maltoside, pH 7.4). The bound S-opsin-1D4 pigment was eluted by incubating the Sepharose beads with 100 μm 1D4 peptide (Biomer Technology, Hayward, CA) in elution buffer (5 mm MES, 0.02% dodecyl maltoside, pH 6.0) for 2 h. A spectrometric scan of the supernatant was taken in a Beckman (Fullerton, CA) spectrophotometer.
Single-cell recordings.
Mice were housed in a 12 h light/dark cycle and were dark-adapted overnight before the experiment. Tissue preparation and recording techniques followed procedures described previously (Burns et al., 2002). Briefly, an animal was killed by CO2 asphyxiation followed by cervical dislocation and the eyes were removed under dim red light. All further manipulations were performed under infrared light. The eyes were hemisected and the retinas removed from the pigment epithelium and chopped into small pieces with a razor blade. Small pieces of the retina were placed in the experimental chamber on the stage of an inverted microscope and perfused with bicarbonate-buffered solution [containing (in mm) 112.5 NaCl, 3.6 KCl, 2.4 MgCl2, 1.2 CaCl2, 10 HEPES, pH 7.4, 20 NaHCO3, 3 Na succinate, 0.5 Na glutamate, 0.02 EDTA, and 10 glucose]. The solution was bubbled with 95% O2/5% CO2, and warmed to 36–38°C in a flow heater (Matthews, 1999) before it entered the experimental chamber. Membrane current from rods were recorded with a suction electrode from individual outer segments projecting from a piece of retina. Cone recordings were done by drawing a portion of the inner segment of a single cone from a piece of retina in the suction electrode using rod transducin knock-out animals (Nikonov et al., 2005). The recording electrode was filled with (in mm) 140 NaCl, 3.6 KCl, 2.4 MgCl2, 1.2 CaCl2, 3 HEPES, pH 7.4, 0.02 EDTA, and 10 glucose. Twenty millisecond flashes were delivered from a calibrated light source via computer-controlled shutters. Flashes of visible light (≥400 nm) were delivered from an optical bench with a tungsten filament light source. Flashes of UV light (≤400 nm) were delivered by fiber optics from a xenon arc lamp. Light intensity and wavelength were changed by using calibrated neutral-density and interference filters. The current was amplified, low-pass filtered at 30 Hz, digitized at 1 kHz, and stored on a computer for subsequent analysis. Photosensitivity was calculated from the dim-flash response as the ratio between response amplitude and the number of photons delivered by the flash (pA photons−1/μm−2). The amplitude of the single-photon response was calculated from a series of 30 identical dim-flash responses as the response ensemble variance-to-mean ratio. Integration time was calculated as the integral of the dim-flash response with the transient-peak amplitude normalized to unity.
Results
Expression of S-opsin in rod photoreceptors
The mouse rod opsin promoter was used to express the mouse S-opsin (which in mouse absorbs in the UV part of the light spectrum) in rods. An additional construct was made to express S-opsin-1D4, which contains the C-terminal eight amino acids of the rhodopsin sequence, ETSQVAPA, appended to the C terminus of S-opsin (Fig. 1A). The last five residues (QVAPA) are known to be necessary and sufficient for directing rhodopsin trafficking from the inner segment, where nascent proteins are synthesized, to the outer segment, the site of phototransduction (Deretic et al., 1998; Tam et al., 2000; Concepcion et al., 2002). It is also the epitope recognized by a mouse rhodopsin monoclonal antibody, 1D4 (Hodges et al., 1988), which allowed us to quantify the expression of S-opsin-1D4 in relation to that of endogenous rhodopsin. In this way, we could compare the abilities of the rod- and cone-targeting sequences to direct the proper localization of the S-opsin to the rod outer segment.
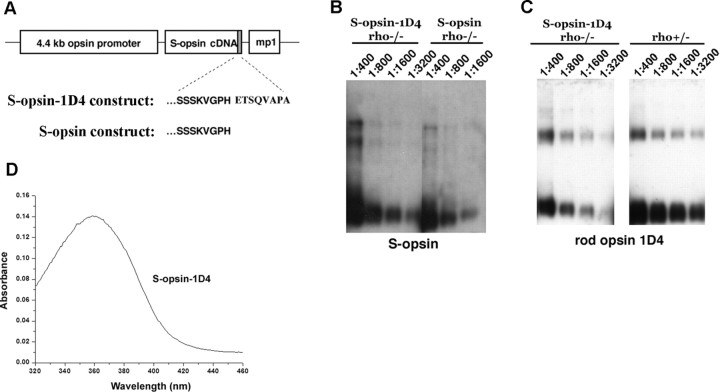
Figure 1.
Expression of S-opsin in transgenic mouse rods. A, The mouse rod opsin (Lem et al., 1991) promoter was used to direct expression of mouse S-opsin in rods. One construct contained the full coding sequence of S-opsin, whereas S-opsin-1D4 contained the terminal eight amino acid residues from rhodopsin (ETSQVAPA) appended to the C terminus of S-opsin. B, Western blot of serially diluted retinal extracts from age-matched S-opsin-1D4rho−/− and S-opsinrho−/− mice probed with an antibody against S-opsin. The ratios indicate the fraction of a retina loaded per lane. C, Western blot of serially diluted retinal extracts from age-matched S-opsin-1D4rho−/− and rho+/− mice probed with the monoclonal antibody 1D4, which recognizes the terminal eight amino acid residues on rhodopsin. The presence of 1D4 on S-opsin-1D4 allows for direct comparison of expression levels between S-opsin and rhodopsin in rho+/− retinas. Ratios indicate the fraction of retina loaded per lane. D, Spectrometric measurement of purified S-opsin-1D4 shows peak absorbance at 360 nm.
The expression levels of S-opsin and S-opsin-1D4 in the rhodopsin knock-out (rho−/−) background appeared to be similar, judged by Western blotting of a serial dilution of retinal homogenates with an antibody that recognizes the N-terminal region of S-opsin (Fig. 1B). At the same time, the expression level of S-opsin-1D4 in the rho−/− background was assessed in relation to that of rhodopsin in rho+/− mice by using the 1D4 antibody, which should recognize rhodopsin and S-opsin-1D4 equally (Fig. 1C). The comparison between S-opsin-1D4rho−/− and rho+/− samples showed S-opsin-1D4 in rho−/− mice to be expressed at ≤1/8 of rhodopsin in rho+/− retinas. The expression level of S-opsin in rho−/− retina, in turn, was ∼30-fold more than that of endogenous S-opsin expressed in wild-type retina (data not shown). The outer segments made by these S-opsin-1D4rho−/− rods were shortened and similar in length to those of cones (Fig. 2, compare A, E). Thus, perhaps serendipitously, the expression of S-opsin in rho−/− rods approximated its expression in cones, given that the ratio of rods to cones in the murine retina is 33:1 (Carter-Dawson and LaVail, 1979). The absorption spectrum of purified S-opsin-1D4 pigment from S-opsin-1D4rho−/− mouse retinas had a peak at 360 nm (Fig. 1D), consistent with the previously measured maximal absorption at 359 nm (Yokoyama et al., 1998). Thus, the expressed S-opsin appeared to be correctly folded and able to bind to 11-cis-retinal to form cone pigment in the rod outer segment.
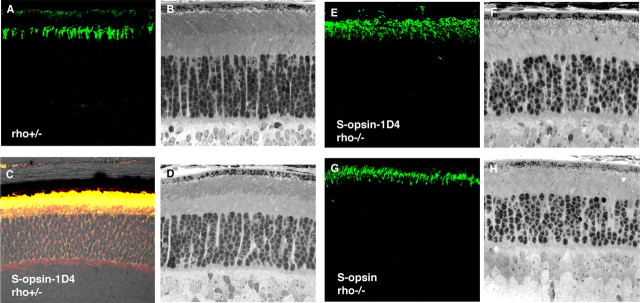
Figure 2.
Localization of S-opsin and retinal morphology in 3-week-old mice. A, C, E, G, Localization of S-opsin (green) visualized by immunofluorescence microscopy of frozen retinal sections. B, D, F, H, Retinal morphology of the corresponding genotype. A, S-opsin reactivity in endogenous cones of the control rho+/− retina. B, Transgenic S-opsin-1D4rho+/− labeled with S-opsin (green) and rhodopsin (red) show colocalization (yellow). Retinal morphology is mostly normal in these genotypes (B, D). S-opsin-1D4 and S-opsin are correctly localized to the outer segments when they are expressed in the rho−/− background (E, G). Disorganized outer segment structures are seen in these mice (F, H).
In 3-week-old mice, the location of S-opsin in the retina was examined using immunofluorescence. In control rho+/− retina sections, endogenous S-opsin reactivity (Fig. 2A, green) was more concentrated in a subset of cones at the inferior pole of the retina (supplemental Fig. 1, available at www.jneurosci.org as supplemental material, for the superior–inferior distribution of S-opsin along the vertical meridian of the retina). In S-opsin-1D4rho+/− retina, S-opsin-1D4 (red) was correctly targeted to the rod outer segment along with rhodopsin (green) (Fig. 2C, merged to give yellow, showing colocalization). The same was found in S-opsin-1D4rho−/− retina (Fig. 2E, green), indicating that, rather than simply cotrafficking with rhodopsin (Concepcion et al., 2002), S-opsin-1D4 had an intrinsic ability to traffic to the rod outer segment. Interestingly, S-opsin behaved the same as S-opsin-1D4 in the rho−/− background (Fig. 2G), indicating a conservation of the C-terminal sequence between rhodopsin and S-opsin for polarized transport to the outer segment. In parallel with the above observations, bright-field microscopy indicated that the presence of S-opsin-1D4 in rho+/− rods had no deleterious effect on the latter's survival or their outer-segment morphology (Fig. 2, compare B, D), and likewise for S-opsin (data not shown). At the same time, the presence of S-opsin in rho−/− rods promoted the formation of rod outer segments (Fig. 2H), which were otherwise absent in rho−/− rods (Humphries et al., 1997; Lem et al., 1999). The rod-outer-segment structure, however, appeared disorganized and, as mentioned above, was shorter than that in rho+/− retinas (Fig. 2, compare B, H) (for cell structure under EM, see supplemental Fig. 2, available at www.jneurosci.org as supplemental material). The same was true for S-opsin-1D4 (Fig. 2F).
At 8 weeks of age, the morphology of retinas expressing S-opsin remained mostly the same as that at 3 weeks. Specifically, in the rho+/− background, S-opsin had no long-term deleterious effect on rod cells (Fig. 3, compare A, B). Interestingly, the amount of rhodopsin was approximately twofold lower in S-opsinrho+/− and S-opsin-1D4rho+/− when compared with transgene-negative rho+/− littermates (Fig. 3E). The twofold rhodopsin decrease was confirmed using differential absorbance measurements: the amount of rhodopsin in S-opsinrho+/− was 0.06 ± 0.01 nmol per retina (mean ± SD; n = 3) compared with 0.11 ± 0.1 nmol per retina (n = 3) in transgene-negative rho+/− littermates. Similar downregulation of rhodopsin was observed for S-opsin-1D4rho+/− retinas. Perhaps this downregulation of rhodopsin occurred because the multiple copies of the transgene bearing the rhodopsin promoter titrated away the transcription factors for driving endogenous rhodopsin. In the rho−/− background, S-opsin allowed the rods to retain some outer-segment structure (Fig. 3D), and significantly slowed the time course of rod-cell death found in rho−/− retina (Fig. 3C) (Humphries et al., 1997; Lem et al., 1999). Thus, expression of cone pigment in rods was able to at least partially reverse the effects of rhodopsin deletion on the structure of their outer segments and their survival. In addition, as described below, the expression of cone pigment in rho−/− rods restored their ability to respond to light.
Figure 3.
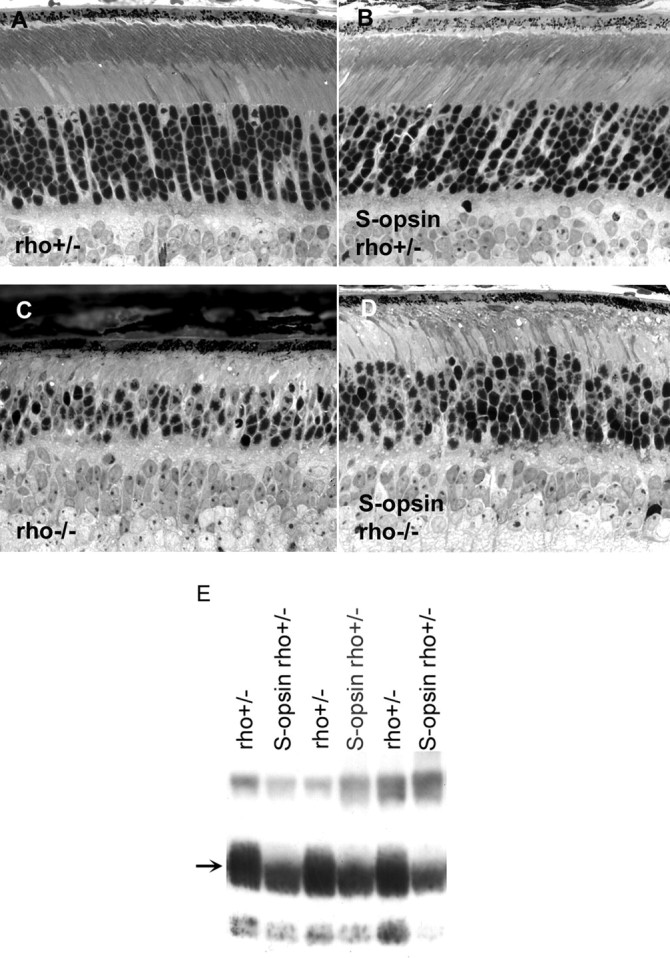
Analysis of retinal morphology and rhodopsin content in 2-month-old mice. A, Retinal morphology is largely normal in rho+/− mice. B, Retinal morphology is unchanged by the expression of S-opsin in this genetic background. C, The outer segment does not form in the absence of rhodopsin, and the number of rods decreases as a function of age. D, The presence of S-opsin delays retinal degeneration in rho−/− mice (compare C, D). E, Rhodopsin content is lowered when S-opsin is expressed; transgene-negative rho+/− littermates show higher rhodopsin content. Individual retinas were homogenized, and an equal fraction (1/3200) from each retina was loaded per lane. The proteins were blotted onto nitrocellulose and probed with 4D2, a monoclonal antibody against rhodopsin (arrow).
Expressed S-opsin is functional in rod cells
Individual rods from 4- to 12-week-old mice were studied by suction-electrode recording. Although either rho+/− rods or rho+/+ rods could be chosen as control, we used rho+/− rods simply because there was less rhodopsin in them, so the situation was slightly closer to the transgenic lines. Interestingly, the dark currents of rho+/−, S-opsinrho+/−, S-opsin-1D4rho+/−, S-opsinrho−/−, and S-opsin-1D4rho−/− rods were all comparable (Table 1), even though S-opsinrho−/− and S-opsin-1D4rho−/− rods had disorganized and significantly shortened outer segments (Fig. 3D). Possibly, the rods selected for recording had more “normal” outer segments than average.
Table 1.
Parameters for the flash responses of control and transgenic mouse rods
| rho+/− (n = 24) | S-opsinrho−/−(n = 20) | S-opsin-1D4rho−/−(n = 20) | S-opsinrho+/− (n = 13) | S-opsin-1D4rho+/−(n = 13) | |
|---|---|---|---|---|---|
| Dark current, Id (pA) | 19.1 ± 1.1 | 16.6 ± 1.8 | 15.6 ± 1.2 | 15.7 ± 0.9 | 19.1 ± 1.5 |
| Sensitivity at λmax′ S (pA photons−1/μm2) | 1.3 ± 0.2 × 10 − 1 (500 nm) | 7.4 ± 1.6 × 10 − 3 (360 nm) | 5.0 ± 0.5 × 10 − 3 (360 nm) | 0.7 ± 0.1 × 10 − 1 (500 nm) | 1.1 ± 0.1 × 10 − 1 (500 nm) |
| Single-photon response, a (pA) | 0.56 ± 0.04 | 0.30 ± 0.03 | 0.21 ± 0.01 | 0.43 ± 0.07 (500 nm) | 0.72 ± 0.07 (500 nm) |
| 0.42 ± 0.06 (360 nm) | 0.77 ± 0.06 (360 nm) | ||||
| Time to peak (ms) | 158 ± 5 | 153 ± 8 | 155 ± 9 | 122 ± 5 (500 nm) | 137 ± 2 (500 nm) |
| 122 ± 7 (360 nm) | 137 ± 2 (360 nm) | ||||
| Integration time (ms) | 254 ± 16 | 221 ± 16 | 204 ± 17 | 200 ± 12 (500 nm) | 224 ± 18 (500 nm) |
| 190 ± 14 (360 nm) | 216 ± 16 (360 nm) | ||||
| Normalized sensitivity at λmax, S/I d (photons−1/μm2) | 6.8 ± 0.7 × 10 − 3 (500 nm) | 5.1 ± 1.0 × 10 − 4 (360 nm) | 3.5 ± 0.4 × 10 − 4 (360 nm) | 4.2 ± 0.3 × 10 − 3 (500 nm) | 5.7 ± 0.6 × 10 − 3 (500 nm) |
| Dark current variance (pA2) | 0.11 ± 0.01 (n = 14) | 0.10 ± 0.01 (n = 14) | 0.08 ± 0.01 (n = 13) | 0.10 ± 0.01 (n = 11) | 0.11 ± 0.01 (n = 14) |
Values given are mean ± SEM.
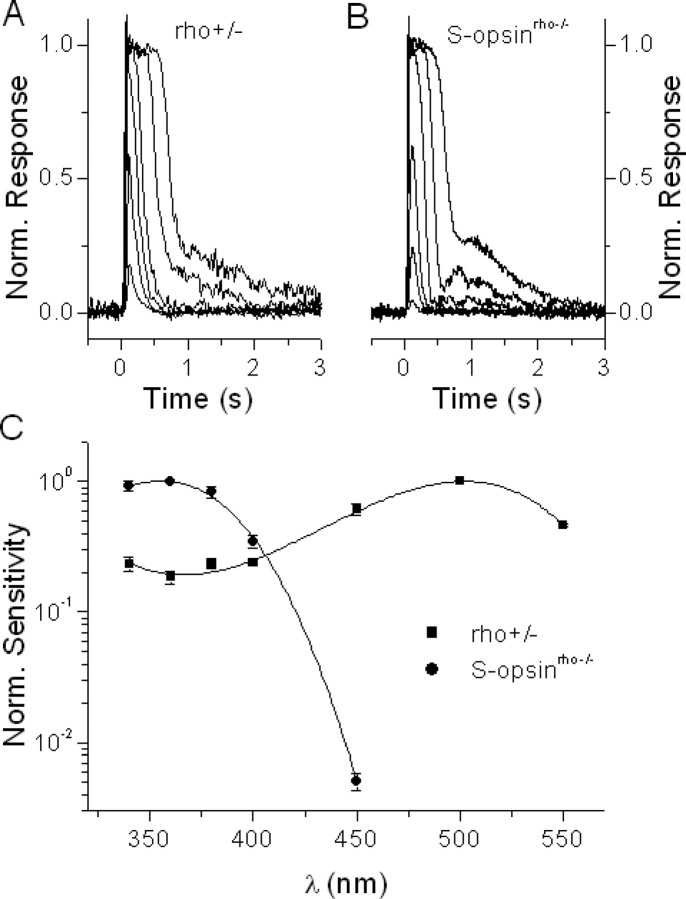
The light responses from rho+/− rods and S-opsinrho−/− rods had very similar waveforms (Fig. 4A,B). However, the spectral sensitivity of S-opsinrho−/− rods peaked at 355 nm, versus 500 nm for that of rho+/− cells (Fig. 4C). The same was found for S-opsin-1D4rho−/− rods (data not shown). The good match between this action-spectrum λmax and the absorption-spectrum λmax of S-pigment (360 nm) (Fig. 1D) confirmed that the photoresponses in these rods were indeed generated by the S-pigment. Thus, S-pigment was able to activate the rod phototransduction cascade in the absence of the native pigment, rhodopsin.
Figure 4.
Transgenic S-opsin expressed in rods produces rod-like photoresponses while inducing S-cone-like spectral sensitivity. A, Family of rhodopsin flash responses from control rho+/− rods. B, Family of S-pigment flash responses from transgenic S-opsinrho−/− rods. Flashes for both figures were delivered at t = 0 s and flash intensity was increased in 0.6 log units. C, Average spectral sensitivities of control rho+/− rods (squares, normalized at 500 nm) and of transgenic S-opsinrho−/− rods (circles, normalized at 360 nm). Error bars indicate SEM. Solid lines represent a third-order polynomial fit to each spectrum.
We compared the peak sensitivity at 360 nm of S-opsinrho−/− and S-opsin-1D4rho−/− rods with the peak sensitivity at 500 nm of control rho+/− cells. After normalizing each cell's sensitivity against its dark current (Table 1, S/Id), we found that the average sensitivities of S-opsinrho−/− and S-opsin-1D4rho−/− rods were 7.5 and 5.1%, respectively, that of rho+/− rods. The rather similar sensitivities of the two transgenic lines were consistent with the comparable expression levels of S-opsin and S-opsin-1D4 in these lines as measured by Western blot (Fig. 1B). The significantly lower sensitivity of rods expressing S-pigment vs rhodopsin could be attributable to differences in the efficiency of activation of the rod phototransduction cascade by the two pigments, or it could reflect a lower S-pigment expression level as suggested by the Western blot data (Fig. 1C).
To resolve this issue, we first compared the amplitude and kinetics of single-photon responses produced by S-pigment in rho−/− rods and by rhodopsin in rho+/− rods. Transgenic and control rods were stimulated with a series of 30 identical dim flashes at wavelengths of 360 and 500 nm, respectively. For each cell, the amplitude of the single-photon response was calculated from the response ensemble variance-to-mean ratio. The time-to-peak and the integration time (time integral of the normalized response) were measured from the corresponding mean response. Overall, the single-photon responses of S-opsinrho−/− and S-opsin-1D4rho−/− rods had somewhat smaller amplitudes, slightly shorter integration times, but similar times-to-peak compared with rho+/− rods (Table 1). The interpretation of these small differences, however, is complicated by the degeneration of S-opsinrho−/− and S-opsin-1D4rho−/− rods. For example, changes in pigment content and outer-segment morphology can affect the properties of the photoresponse (Calvert et al., 2001). For these reasons, we did not pursue these small differences further.
Rods coexpressing S-pigment and rhodopsin were better suited for comparison of the photoresponses by the two pigments as they had intact cellular morphology and the two pigments functioned in identical conditions. S-opsinrho+/− and S-opsin-1D4rho+/− rods showed a higher sensitivity in the UV region than rho+/− controls (Fig. 5A), reflecting the contribution of S-pigment to their responses at these wavelengths. The peak sensitivities of these transgenic rods at 500 nm were slightly lower than that of rho+/− rods (Table 1). This was most likely because of the lower expression of rhodopsin in the presence of the transgene (Fig. 3E). The action spectrum of S-opsinrho+/− rods could be fitted by a linear combination of the action spectra of rho+/− and S-opsinrho−/− rods (from Fig. 4C) in an 88%:12% ratio (Fig. 5B). A corresponding fit for the action spectrum of the S-opsin-1D4rho+/− rods could be done by an 86%:14% ratio of rho+/− and S-opsin-1D4rho−/− action spectra (Fig. 5C). These ratios are similar to the 86%:14% ratio of rhodopsin and S-opsin in these rods derived by mass spectrometry (Shi et al., 2005). The comparable contribution of S-pigment to the overall pigment content and to the action spectrum indicates that S-pigment and rhodopsin trigger single-photon responses with similar amplitudes in a given cell.
Figure 5.
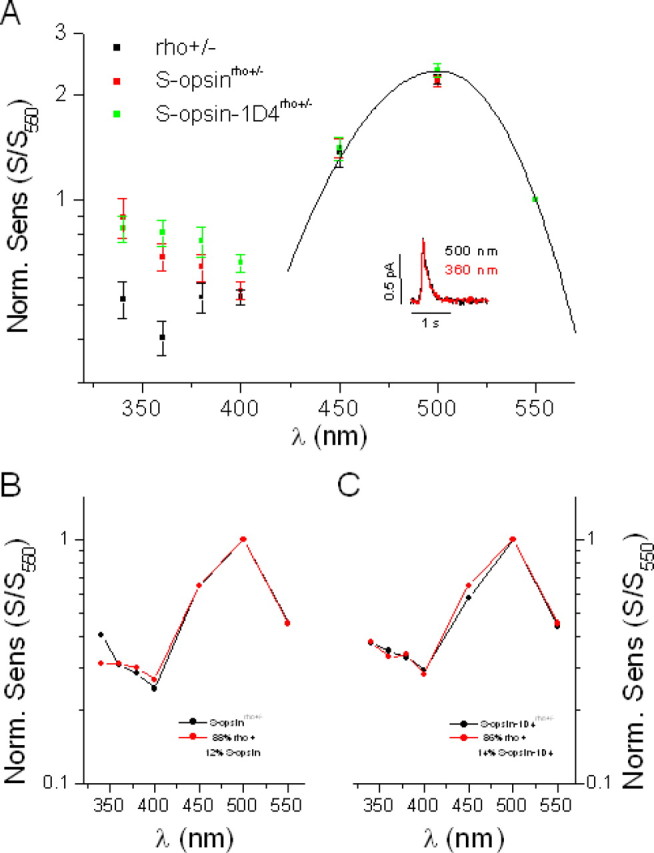
Transgenic expression of S-opsin in rho+/− rods enhances their sensitivity to UV light without affecting their response properties. A, Comparison of spectral sensitivities of control rho+/− rods (black), transgenic S-opsinrho+/− rods (red), and transgenic S-opsin-1D4rho+/− rods (green). Spectral sensitivities are shown normalized at 550 nm, where contribution of S-pigment to the overall action spectrum in the transgenic cells is negligible (Fig. 4C). This point is also supported by the good fit at long wavelengths of both control and transgenic action spectra to the rhodopsin A1 template with λmax = 500 nm (solid line). The inset shows the similar amplitudes and kinetics of the single photon responses from one S-opsinrho−/− rod at 500 nm (black) and 360 nm (red). B, The action spectrum of S-opsinrho+/− rods (black) can be fit by a combination of the action spectra of rho+/− rods (Fig. 5A) and S-opsinrho−/− rods (Fig. 4C) in an 88%:12% ratio (red). C, Similar fit for the S-opsin-1D4rho+/− rod action spectrum (black) with an 86%:14% ratio of rho+/− and S-opsin-1D4rho+/− rod spectra (red). Error bars indicate SEM.
To directly compare the single-photon responses generated by S-pigment and rhodopsin in an S-opsinrho+/− rod, we stimulated the cell with a series of 30 identical dim flashes at 360 nm and at 500 nm. The amplitude of the single-photon response at each wavelength was calculated from the response ensemble variance-to-mean ratio. At 500 nm, the response was generated exclusively by rhodopsin because S-pigment absorption at this wavelength was negligible (Figs. 1D, 4C). At 360 nm, in contrast, the absorption of rhodopsin was only 0.17 of its peak absorption at 500 nm whereas S-pigment absorption was at its maximum (Fig. 4C). Together with the 12% S-opsin contribution to the action spectrum of S-opsinrho+/− rods, the overall contribution of S-pigment to the photoresponse at 360 nm can be calculated as follows:
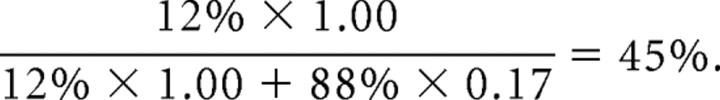 |
The same calculation yields 49% for the fraction of the overall response at 360 nm produced by S-pigment-1D4. Thus, in cells coexpressing S-pigment and rhodopsin, the two pigments had comparable contributions to the overall response at 360 nm. If S-pigment were to produce a single-photon response significantly different from that by rhodopsin, the overall single-photon response at 360 nm would also be expected to differ from that at 500 nm. However, the single-photon responses at 360 and 500 nm had identical amplitudes, whether in S-opsinrho+/− or in S-opsin-1D4rho+/− rods (Fig. 5A, inset, Table 1). The kinetics of the responses elicited at these two wavelengths were also very similar (Fig. 5A, inset, Table 1). Thus, when coexpressed in the same rod, S-pigment and rhodopsin generated single-photon responses with very similar amplitudes and kinetics. In other words, S-pigment and rhodopsin activated the rod phototransduction cascade with similar efficiencies, and were inactivated comparably by the rod shutoff mechanisms.
A previous study showed that transgenic expression of L-pigment in Xenopus rods results in a significant increase in their dark noise (Kefalov et al., 2003). This effect is caused by the 10,000-fold higher rate of spontaneous thermal activation of L-pigment compared with rhodopsin in these cells. To estimate the spontaneous thermal activity of S-pigment expressed in mouse rods, we measured the level of noise in darkness in S-opsinrho+/−, S-opsin-1D4rho+/− rods, and rho+/− rods. Surprisingly, we found no difference in the current variance between them (Table 1), indicating that, in contrast to the case of L-pigment in Xenopus rods, there was no significant increase in the dark noise produced by the expression of S-pigment in mouse rods.
Measurement of meta II decay rate of S-pigment in situ
Meta-II rhodopsin is partially inactivated by phosphorylation by rhodopsin kinase, followed by the binding of arrestin to the phosphorylated rhodopsin for complete inactivation (for review, see Makino et al., 2003). In arr1−/− rods, the response displays a biphasic shutoff, with a fast initial inactivation caused by phosphorylation followed by a slow tail that represents the decay of meta II rhodopsin (Xu et al., 1997). From rho+/−arr1−/− rods, we measured the average time constant of rhodopsin meta II decay to be 49.5 ± 4.3 s (n = 7) (Fig. 6A), consistent with previous work (Xu et al., 1997). By breeding S-opsinrho−/− mice with arr1−/− mice, we obtained S-opsinrho−/−arr1−/− animals which allowed us to measure the decay rate of S-pigment meta II. The average time constant of this decay (Fig. 6B) was only 1.4 ± 0.1 s (n = 20); similarly, from S-opsin-1D4rho−/−arr1−/− rods, it was 1.2 ± 0.1 s (n = 19). Thus, the meta II decay of S-pigment measured in intact rods, was ∼40 times faster than that of rhodopsin. Indeed, the decay of photoactivated S-pigment was so fast that these rods recovered within seconds even from saturating flashes (Fig. 6C). Despite this fast decay of S-pigment meta II, response shutoff was slower in S-opsinrho−/− lacking arrestin (Fig. 6B and 6C) compared with control S-opsinrho−/− rods with arrestin (Figs. 4B, 5A, inset). This indicates that rod arrestin does contribute to the inactivation of transgenic S-opsin meta II in rod cells (Fig. 7A).
Figure 6.
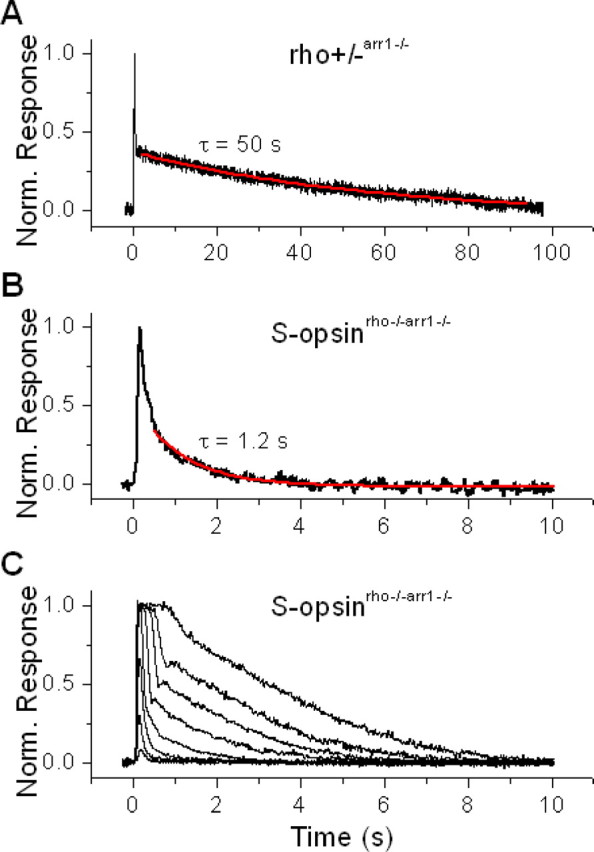
The physiologically active (meta II) state of S-pigment decays significantly faster than that of rhodopsin. A, Fitting the slow recovery phase of normalized dim-flash response from rho+/− arr1−/− rod with a single exponential decay function gave time constant of rhodopsin meta II of 50 s. B, Similar fit of the dim-flash response from S-opsinrho−/−arr1−/− rod gave an S-pigment meta II time constant of 1.2 s. C, In contrast to the case of rhodopsin, S-pigment meta II decay is sufficiently fast to allow collection of a complete family of flash responses, from threshold to saturation.
Figure 7.
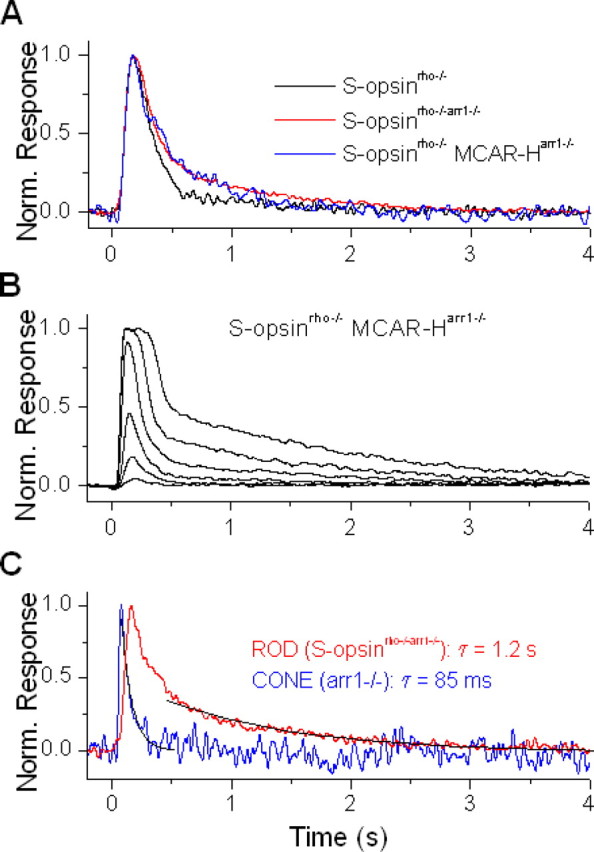
Cone arrestin is less efficient than rod arrestin at inactivating transgenic S-pigment, expressed in rods. A, Normalized dim-flash responses from transgenic S-pigment in control rod (S-opsinrho−/−, black), rod lacking rod arrestin (S-opsinrho−/−arr1−/−, red), and rod lacking rod arrestin but expressing cone arrestin (S-opsinrho−/− MCAR-Harr1−/−, blue). Deletion of rod arrestin results in slower recovery of the dim-flash response, which is not reversed by the transgenic expression of cone arrestin. B, Family of flash responses from S-opsinrho−/− MCAR-Harr1−/− rod. Cone arrestin partially reverses the slow decay of the responses to brighter flashes (compare with Fig. 6C). C, Comparison of normalized dim-flash responses from S-opsinrho−/−arr1−/− rod (red) and arr1−/− cone (blue), with recovery fit by a single exponential decay function (black) in each case. The cone-response recovery is significantly faster than the decay of S-opsin meta II, which is rate limiting for the recovery of the rod.
Efficiency of cone arrestin in deactivation of S-pigment
We showed above that, in the rod environment, rod arrestin contributes to the inactivation of transgenic S-pigment. Mouse cones and the cone-like photoreceptors of nrl−/− mice are known to express rod arrestin (Mears et al., 2001; Zhu et al., 2005) and a separate cone-specific arrestin [mouse cone arrestin (MCAR)] (Craft et al., 1994; Zhu et al., 2002). To find out the contribution of cone arrestin to the inactivation of cone pigment, we reconstituted these two proteins in transgenic mouse rods. We examined rods from S-opsinrho−/−MCARarr1−/− mice, obtained by crossing S-opsinrho−/−arr1−/− mice with transgenic mice that expressed cone arrestin in place of rod arrestin in their rods (Chan et al., 2007). We used two transgenic cone arrestin lines: MCAR-L and MCAR-H, which express fourfold and 45-fold, respectively, more cone arrestin in rods than the endogenous level of cone arrestin expressed in cones (Chan et al., 2007). Surprisingly, even for the high-expression line, the dim-flash response decay of S-opsinrho−/−MCARarr1−/− rods was not any faster than that of S-opsinrho−/−arr1−/− rods, and both were slower than the decay of S-opsinrho−/− rods (Fig. 7A). In other words, whereas rod arrestin was effective in inactivating S-pigment, cone arrestin was apparently not. More quantitatively, we used the integration time of the dim-flash response as a measure of S-pigment deactivation. The average integration times of dim-flash responses for S-opsinrho−/−arr1−/− and S-opsin-1D4rho−/−arr1−/− rods were 413 ± 38 ms (n = 18) and 544 ± 40 ms (n = 13), respectively, significantly higher than those of the corresponding rods still with rod arrestin (221 ± 16 ms and 204 ± 17 ms, respectively) (Table 1). The dim-flash integration time for S-opsinrho−/−MCARarr1−/− rods was 410 ± 86 ms (n = 7) for the MCAR-H line and 516 ± 63 ms (n = 9) for the MCAR-L line, no faster than that in the absence of arrestin at all. With brighter flashes, S-opsinrho−/−MCARarr1−/− rods did show a discernibly faster response recovery than S-opsinrho−/−arr1−/− rods (compare Figs. 6C, 7B). Thus, cone arrestin had some ability to inactivate phosphorylated S-pigment meta II [S*–P (phosphorylated activated S-opsin)], although weakly. Notably, cone arrestin is also not very effective in inactivating rhodopsin when expressed as a transgene in arr1−/− rods (Chan et al., 2007).
How about the role of rod and cone arrestins in the cone environment? We have not studied the effect of deleting cone arrestin in cones for lack of the required mouse line, but we did examine the dim-flash responses of M-pigment in cones lacking rod arrestin. The average inactivation time constant of arr1−/− cones was 73 ± 17 ms (n = 10), surprisingly similar to that of wild-type cones reported previously (Nikonov et al., 2006), and suggesting that rod arrestin is either not required or nonrate-limiting for dim-flash response shutoff in cones, at least in the presence of cone arrestin (see also Lyubarsky et al., 2000). However, as described previously, in the absence of rod arrestin in S-opsinrho−/−arr1−/− rods the response triggered by S-pigment clearly decayed much more slowly and with kinetics determined by S-pigment meta II decay (Fig. 7C). In summary, although in transgenic rods S-pigment activity can be affected strongly by rod arrestin and only weakly by cone arrestin, the exact role of S-pigment shutoff by arrestins in the recovery of the light response in native cones remains unclear.
Discussion
Rods lacking rhodopsin do not develop outer segments and are not able to respond to light (Humphries et al., 1997; Lem et al., 1999). We report in this study that the expression of transgenic S-opsin was able to promote outer-segment growth and survival of these cells. The ability of S-opsin to target correctly to the rod outer segment without endogenous rhodopsin implicates commonalities in the minimal trafficking domain at the C terminus of these visual pigments. The expression of S-opsin also restored the light response of rho−/− rods, with a λmax at 355 nm corresponding to that of S-pigment.
We observed in a previous study that, at 10% expression level, a rhodopsin-phosphorylation-site mutant, S338A, was able to restore outer-segment formation and a retinal morphology that resembled wild-type mice (Concepcion et al., 2002). However, expression of comparable levels of S-opsin or S-opsin-1D4 was not able to substitute for rhodopsin to the similar degree as S338A in promoting rod survival or outer-segment formation. These results point toward additional differences in structural elements required for outer-segment formation in rods and cones.
The coexpression of S-pigment and rhodopsin in transgenic rho+/− rods allowed a comparison of the signaling properties of the two pigments side-by-side in the same cell. We found that S-pigment and rhodopsin produce responses of similar amplitude, speed, and waveform. Thus, the two pigments activate rod transducin with the same efficiency, and both are inactivated by rhodopsin kinase and rod arrestin equally well. These results on S-pigment support and extend our previous finding that, when coexpressed in transgenic Xenopus rods, L-pigment and rhodopsin produce identical photoresponses (Kefalov et al., 2003). Hence, the gain and kinetics of the response are determined not by the visual pigment, but by the downstream phototransduction proteins (Makino et al., 2003; Krispel et al., 2006). The slightly faster response kinetics of S-opsinrho+/− and S-opsin-1D4rho+/− rods may be caused by a decrease in the overall pigment content in these rods compared with rho+/− rods (Fig. 3E), because the response is speeded when the amount of rhodopsin is decreased (Calvert et al., 2001; Jastrzebska et al., 2006). Potentially, the same reason may explain the faster response recovery in S-opsinrho−/− and S-opsin-1D4rho−/− rods.
The finding that S-pigment and rhodopsin produce responses of similar amplitudes has allowed us to derive the S-opsin expression level from the action spectra of transgenic cells. From the spectral sensitivity of transgenic rho+/− rods, we arrived at an expression level of 12–14% (12% for S-opsin and 14% for S-opsin-1D4) with respect to the endogenous rhodopsin in these cells. This estimate is remarkably similar to the estimates by mass spectrometry (14%) from retinas of the same genotype. Incidentally, using the same approach, the average expression of S-pigment in rho−/− rods was also 13% that of rhodopsin in transgenic rho+/− cells (15 and 10.2% for S-opsinrho−/− and S-opsin-1D4rho−/− rods, respectively). These numbers are derived from the 7.5 and 5.1% sensitivities of these cells vs control rho+/− rods and the twofold lower rhodopsin levels in transgenic rho+/− rods vs control rho+/− rods. Thus, the expression of S-opsin in all transgenic rods was ∼13% that of rhodopsin in transgenic rods. The low level of transgenic S-opsin expression despite its being driven by the rhodopsin promoter, capable of high levels of expression of transgenic rhodopsin (Olsson et al., 1992; Mendez et al., 2000), may reflect unknown control elements or a less stable S-opsin in the rod environment. Previously, we found that the expression of L-opsin in Xenopus rods was also low (Kefalov et al., 2003).
L-pigment undergoes spontaneous (thermal) activation in amphibian photoreceptors (Rieke and Baylor, 2000; Sampath and Baylor, 2002) with a rate constant ∼10,000-fold as high as that of rhodopsin (Kefalov et al., 2003). This high spontaneous activity is one of the mechanisms contributing to the low sensitivity of amphibian L-cones compared with rods. What about the S-pigment? A mouse rho+/− rod, with 50% of wild-type rhodopsin, contains 3.5 × 107 rhodopsin molecules (Lyubarsky et al., 2004), and this number decreases by roughly another 50%, to 1.75 × 107 when transgenic S-opsin is expressed (see Results). In such a cell, an average expression of 13% S-pigment thus gives 2.3 × 106 S-opsin molecules. The molecular rate of thermal activation of L-pigment at 37°C has been estimated to be 5.6 × 10−6 s−1 (Kefalov et al., 2003). If L- and S- pigments had similar molecular rates of thermal activation, one would expect 2.3 × 106 × 5.6 × 10−6 s−1 = 13 thermal activations per second. Based on background-light experiments on wild-type rods (data not shown), this level of constitutive activity should produce a more than twofold increase in dark noise, which we did not observe in S-opsinrho+/− rods (Table 1). There are at least two possibilities. First, as in amphibians, mouse S-pigment could be much more stable than L-pigment (Rieke and Baylor, 2000). Second, the difference in chromophore in the transgenic cone pigment, A1 in mouse rods, and A2 in amphibian rods could affect significantly the thermal rates, as reported for rod pigment (Donner et al., 1990). Ultimately, this issue can be resolved by studying transgenic mice with rods expressing A1 L-cone pigment, which would allow direct comparison with the previous Xenopus study. These experiments are under way. In any case, the lack of increase in dark noise in transgenic S-opsin mouse rods indicates that the rate of thermal activation of A1 S-pigment in mouse is significantly lower than that of A2 L-pigment in amphibians. Concomitant with the low thermal activity of amphibian S-pigment (Rieke and Baylor, 2000), salamander S-cones are as sensitive to light as the corresponding rods (Perry and McNaughton, 1991; Ma et al., 2001), an observation consistent with spontaneous pigment activity playing a significant role in setting cone sensitivity under dark-adapted conditions. However, mammalian S-cones have a considerably lower sensitivity to light than mammalian rods (Nikonov et al., 2006), despite the apparently low thermal activity of mouse S-pigment found here. Thus, at least for mammalian S-cones, there seems to be other mechanisms downstream of the pigment that confer low sensitivity (Tachibanaki et al., 2001).
The rho−/− rods expressing S-opsin and lacking arrestin have allowed us to measure the lifetime of the meta II of S-pigment. We found that it decays ∼40 times faster than rhodopsin meta II, with a time constant of ca. 1.3 s. This value, derived in situ at 37°C, should be more precise than the previous in vitro biochemical estimate of <30 s at 22°C for mouse S-pigment (Kusnetzow et al., 2004) or 12 min at −10°C for chicken S-pigment (Imai et al., 1997). Although the meta II lifetime normally does not dominate the decay of the cone light response, the amount of cone pigment activated by intense light may exceed the cell capacity for phosphorylation such that the rapid decay of meta II may become important for response decline. The short lifetime of S-pigment meta II may also explain the previous finding by others that, in the absence of pigment phosphorylation, the decline of the response triggered by M-pigment is slowed dramatically, but not of that triggered by the S-pigment (Nikonov et a., 2005). Presumably, M-pigment meta II decay is substantially slower than S-pigment meta II decay. Indeed, from the data of Nikonov et al. (2005), we measured an S-pigment response decline half-time of 0.7 s in nrl−/−grk1−/− cones, which, for a single-exponential decay, corresponds to a time constant of 1.0 s, similar to the 1.3 s recovery of S-pigment responses in our transgenic rods.
Finally, we found that cone arrestin when expressed in rods does not efficiently inactivate rhodopsin or the transgenic S-pigment. This is in contrast to the high efficiency with which the native rod arrestin inactivated both pigments in rods. At the same time, consistent with previous ERG studies (Lyubarsky et al., 2000), we found that the deletion of rod arrestin (normally also expressed in mouse cones) does not affect the shutoff of M-pigment dim-flash responses in individual cones. How can these discrepancies be reconciled? One noted difference between the rod and cone environments is the faster and wider dynamic changes in calcium concentration within cones after light exposure (Sampath et al., 1999). Calcium modulates multiple steps in phototransduction including pigment phosphorylation, synthesis of cGMP by guanylate cyclase, and the affinity for cGMP of the cGMP-gated channel (Nakatani et al., 2002). Each of these feedbacks speeds response recovery. It is thought that these calcium feedbacks are stronger/faster in cones (for review, see Korenbrot and Rebrik, 2002). Possibly, as a result, the actions of the arrestins on pigment deactivation are partially masked. Unfortunately, because of the copresence of rod and cone arrestins in cones, our findings currently are unable to provide an unambiguous conclusion about their roles in the cone response.
Footnotes
This work was supported by National Institutes of Health Grants EY12155 (J.C.) and EY06837 (K.-W.Y.), a Karl Kirchgessner Foundation Vision Research grant (V.J.K.), and the Arnold and Mabel Beckman Macular Research Center (J.C.). V.J.K. is a recipient of a Career Development Award from Research to Prevent Blindness. We thank Dr. Paul Hargrave for generous gifts of the rhodopsin antibody R2-12N and Drs. Rosalie Crouch, Carter Cornwall, and Petri Ala-Laurila for their comments on this manuscript.
References
- Burns ME, Mendez A, Chen J, Baylor DA. Dynamics of cyclic GMP synthesis in retinal rods. Neuron. 2002;36:81–91. doi: 10.1016/s0896-6273(02)00911-x. [DOI] [PubMed] [Google Scholar]
- Calvert PD, Govardovskii VI, Krasnoperova N, Anderson RE, Lem J, Makino CL. Membrane protein diffusion sets the speed of rod phototransduction. Nature. 2001;411:90–94. doi: 10.1038/35075083. [DOI] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J Comp Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- Chan S, Rubin WW, Mendez A, Liu X, Song X, Hanson SM, Craft CM, Gurevich VV, Burns ME, Chen J. Functional comparisons of visual arrestins in rod photoreceptors of transgenic mice. Invest Ophthalmol Vis Sci. 2007;48:1968–1975. doi: 10.1167/iovs.06-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion F, Mendez A, Chen J. The carboxyl-terminal domain is essential for rhodopsin transport in rod photoreceptors. Vision Res. 2002;42:417–426. doi: 10.1016/s0042-6989(01)00195-x. [DOI] [PubMed] [Google Scholar]
- Craft CM, Whitmore DH, Wiechmann AF. Cone arrestin identified by targeting expression of a functional family. J Biol Chem. 1994;269:4613–4619. [PubMed] [Google Scholar]
- Deretic D, Schmerl S, Hargrave PA, Arendt A, McDowell JH. Regulation of sorting and post-Golgi trafficking of rhodopsin by its C-terminal sequence QVS(A)PA. Proc Natl Acad Sci USA. 1998;95:10620–10625. doi: 10.1073/pnas.95.18.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner K, Firsov ML, Govardovskii VI. The frequency of isomerization-like “dark” events in rhodopsin and porphyropsin rods of the bull-frog retina. J Physiol (Lond) 1990;428:673–692. doi: 10.1113/jphysiol.1990.sp018234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges RS, Heaton RJ, Parker JM, Molday L, Molday RS. Antigen-antibody interaction. Synthetic peptides define linear antigenic determinants recognized by monoclonal antibodies directed to the cytoplasmic carboxyl terminus of rhodopsin. J Biol Chem. 1988;263:11768–11775. [PubMed] [Google Scholar]
- Humphries MM, Rancourt D, Farrar GJ, Kenna P, Hazel M, Bush RA, Sieving PA, Sheils DM, McNally N, Creighton P, Erven A, Boros A, Gulya K, Capecchi MR, Humphries P. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat Genet. 1997;15:216–219. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- Imai H, Terakita A, Tachibanaki S, Imamoto Y, Yoshizawa T, Shichida Y. Photochemical and biochemical properties of chicken blue-sensitive cone visual pigment. Biochemistry. 1997;36:12773–12779. doi: 10.1021/bi970809x. [DOI] [PubMed] [Google Scholar]
- Jastrzebska B, Fotiadis D, Jang GF, Stenkamp RE, Engel A, Palczewski K. Functional and structural characterization of rhodopsin oligomers. J Biol Chem. 2006;281:11917–11922. doi: 10.1074/jbc.M600422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefalov V, Fu Y, Marsh-Armstrong N, Yau KW. Role of visual pigment properties in rod and cone phototransduction. Nature. 2003;425:526–531. doi: 10.1038/nature01992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenbrot JI, Rebrik TI. Tuning outer segment Ca2+ homeostasis to phototransduction in rods and cones. Adv Exp Med Biol. 2002;514:179–203. doi: 10.1007/978-1-4615-0121-3_11. [DOI] [PubMed] [Google Scholar]
- Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME. RGS expression rate-limits recovery of rod photoresponses. Neuron. 2006;51:409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Kusnetzow AK, Dukkipati A, Babu KR, Ramos L, Knox BE, Birge RR. Vertebrate ultraviolet visual pigments: protonation of the retinylidene Schiff base and a counterion switch during photoactivation. Proc Natl Acad Sci USA. 2004;101:941–946. doi: 10.1073/pnas.0305206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lem J, Applebury ML, Falk JD, Flannery JG, Simon MI. Tissue-specific and developmental regulation of rod opsin chimeric genes in transgenic mice. Neuron. 1991;6:201–210. doi: 10.1016/0896-6273(91)90356-5. [DOI] [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolo M, Makino CL, Sidman RL. Morphological, physiological, and biochemical changes in rhodopsin knock-out mice. Proc Natl Acad Sci USA. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubarsky AL, Chen C, Simon MI, Pugh EN., Jr Mice lacking G-protein receptor kinase 1 have profoundly slowed recovery of cone-driven retinal responses. J Neurosci. 2000;20:2209–2217. doi: 10.1523/JNEUROSCI.20-06-02209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubarsky AL, Daniele LL, Pugh EN., Jr From candelas to photoisomerizations in the mouse eye by rhodopsin bleaching in situ and the light-rearing dependence of the major components of the mouse ERG. Vision Res. 2004;44:3235–3251. doi: 10.1016/j.visres.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Ma J, Znoiko S, Othersen KL, Ryan JC, Das J, Isayama T, Kono M, Oprian DD, Corson DW, Cornwall MC, Cameron DA, Harosi FI, Makino CL, Crouch RK. A visual pigment expressed in both rod and cone photoreceptors. Neuron. 2001;32:451–461. doi: 10.1016/s0896-6273(01)00482-2. [DOI] [PubMed] [Google Scholar]
- Makino CL, Wen XH, Lem J. Piecing together the timetable for visual transduction with transgenic animals. Curr Opin Neurobiol. 2003;13:404–412. doi: 10.1016/s0959-4388(03)00091-6. [DOI] [PubMed] [Google Scholar]
- Matthews HR. A compact modular flow heater for the superfusion of mammalian cells. J Physiol (Lond) 1999;518:13P. [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Mendez A, Burns ME, Roca A, Lem J, Wu LW, Simon MI, Baylor DA, Chen J. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28:153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Mollon JD. “Tho' she kneel'd in that place where they grew...” The uses origins of primate colour vision. J Exp Biol. 1989;146:21–38. doi: 10.1242/jeb.146.1.21. [DOI] [PubMed] [Google Scholar]
- Nakatani K, Chen C, Yau KW, Koutalos Y. Calcium and phototransduction. Adv Exp Med Biol. 2002;514:1–20. doi: 10.1007/978-1-4615-0121-3_1. [DOI] [PubMed] [Google Scholar]
- Nikonov SS, Daniele LL, Zhu X, Craft CM, Swaroop A, Pugh EN., Jr Photoreceptors of Nrl−/− mice coexpress functional S- and M-cone opsins having distinct inactivation mechanisms. J Gen Physiol. 2005;125:287–304. doi: 10.1085/jgp.200409208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonov SS, Kholodenko R, Lem J, Pugh EN., Jr Physiological features of the S- and M-cone photoreceptors of wild-type mice from single-cell recordings. J Gen Physiol. 2006;127:359–374. doi: 10.1085/jgp.200609490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson JE, Gordon JW, Pawlyk BS, Roof D, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- Perry RJ, McNaughton PA. Response properties of cones from the retina of the tiger salamander. J Physiol (Lond) 1991;433:561–587. doi: 10.1113/jphysiol.1991.sp018444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieke F, Baylor DA. Origin and functional impact of dark noise in retinal cones. Neuron. 2000;26:181–186. doi: 10.1016/s0896-6273(00)81148-4. [DOI] [PubMed] [Google Scholar]
- Sampath AP, Baylor DA. Molecular mechanism of spontaneous pigment activation in retinal cones. Biophys J. 2002;83:184–193. doi: 10.1016/S0006-3495(02)75160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath AP, Matthews HR, Cornwall MC, Bandarchi J, Fain GL. Light-dependent changes in outer segment free-Ca2+ concentration in salamander cone photoreceptors. J Gen Physiol. 1999;113:267–277. doi: 10.1085/jgp.113.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapf JL, Nunn BJ, Meister M, Baylor DA. Visual transduction in cones of the monkey Macaca fascicularis. J Physiol (Lond) 1990;427:681–713. doi: 10.1113/jphysiol.1990.sp018193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi GW, Chen J, Concepcion F, Motamedchaboki K, Marjoram P, Langen R. Light causes phosphorylation of non-activated visual pigments in intact mouse rod photoreceptor cells. J Biol Chem. 2005;280:41184–41191. doi: 10.1074/jbc.M506935200. [DOI] [PubMed] [Google Scholar]
- Tachibanaki S, Tsushima S, Kawamura S. Low amplification and fast visual pigment phosphorylation as mechanisms characterizing cone photoresponses. Proc Natl Acad Sci USA. 2001;98:14044–14049. doi: 10.1073/pnas.241396898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Moritz OL, Hurd LB, Papermaster DS. Identification of an outer segment targeting signal in the COOH terminus of rhodopsin using transgenic Xenopus laevis. J Cell Biol. 2000;151:1369–1380. doi: 10.1083/jcb.151.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Dodd RL, Makino CL, Simon MI, Baylor DA, Chen J. Prolonged photoresponses in transgenic mouse rods lacking arrestin. Nature. 1997;389:505–509. doi: 10.1038/39068. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB, Kawamura S. Regeneration of ultraviolet pigments of vertebrates. FEBS Lett. 1998;423:155–158. doi: 10.1016/s0014-5793(98)00086-6. [DOI] [PubMed] [Google Scholar]
- Zhu X, Ma B, Babu S, Murage J, Knox BE, Craft CM. Mouse cone arrestin gene characterization: promoter targets expression to cone photoreceptors. FEBS Lett. 2002;524:116–122. doi: 10.1016/s0014-5793(02)03014-4. [DOI] [PubMed] [Google Scholar]
- Zhu X, Wu K, Rife L, Brown B, Craft CM. Rod arrestin expression and function in cone photoreceptors. Invest Ophthalmol Vis Sci. 2005;46:1179. [Google Scholar]