Abstract
The repressor element 1 (RE1) silencing transcription factor (REST) helps preserve the identity of nervous tissue by silencing neuronal genes in non-neural tissues. Moreover, in an epithelial model of tumorigenesis, loss of REST function is associated with loss of adhesion, suggesting the aberrant expression of REST-controlled genes encoding this property. To date, no adhesion molecules under REST control have been identified. Here, we used serial analysis of chromatin occupancy to perform genome-wide identification of REST-occupied target sequences (RE1 sites) in a kidney cell line. We discovered novel REST-binding motifs and found that the number of RE1 sites far exceeded previous estimates. A large family of targets encoding adhesion proteins was identified, as were genes encoding signature proteins of neuroendocrine tumors. Unexpectedly, genes considered exclusively non-neuronal also contained an RE1 motif and were expressed in neurons. This supports the model that REST binding is a critical determinant of neuronal phenotype.
Keywords: REST, transcription, serial analysis of chromatin occupancy, binding motif, synaptic transmission, neuroendocrine tumors
Introduction
The repressor element 1 (RE1) silencing transcription factor [REST (also called NRSF)] restricts expression of some genes to neurons by repression through a common genetic element, RE1 (also called NRSE). Within the developing nervous system, REST transiently represses the synaptotagmin 4, brain-derived neurotrophic factor, and calbindin genes in dividing progenitors, and its loss at terminal differentiation allows their expression in mature neurons (Ballas et al., 2005). In contrast, outside of the nervous system, REST normally mediates long-term silencing of these genes (Lunyak et al., 2002; Roopra et al., 2004; Ballas et al., 2005). The extent to which these findings apply to other neuronal genes is not known. Progress in addressing this question has been hampered by the lack of a well defined consensus REST DNA sequence motif, which is unusually large, and a reliable methodology for interrogating functional binding in vivo. A recent in silico study of REST-binding sites, however, has estimated that only 11% of neuronal-specific genes contained a REST-binding site, suggesting that the types of genes regulated by REST might represent a relatively small subset of the neuronal gene repertoire (Mortazavi et al., 2006).
Several human pathologies, including certain epithelial and neuroendocrine cancers, as well as paraneoplastic diseases (Coulson, 2005), are characterized by aberrant expression of neuronal genes, some of which contain a putative REST-binding or RE1 site (Westbrook et al., 2005). The mechanism by which these RE1-containing neuronal genes are induced is not known. In general, the contribution of REST repression to expression levels of its target genes has been poorly studied. Ectopic overexpression of REST in the rat neuronal PC12 cell line blocks induction of sodium channel mRNA in response to differentiation by NGF, but basal levels of sodium channel gene expression were only affected mildly (Ballas et al., 2001). During mouse and chick development, only one or two neuronal genes were derepressed outside of nervous tissues after gene deletion or expression of a dominant-negative form of REST (DN REST) (Chen et al., 1998). In contrast, expression of a recombinant REST-VP16 fusion protein, which has the repressor domains of REST replaced with the activation domain of the adenovirus activator, VP16, is sufficient to convert a muscle cell line into one that exhibits a morphology characteristic of neurons (Watanabe et al., 2004). Finally, one neuronal gene is reported as repressed in vivo by REST loss of function, which is more consistent with REST having an activator function (Kallunki et al., 1998).
Here, we sought to address, in a global and unbiased manner, the constellation of genes regulated by REST in vivo, and the impact of REST loss of function on expression of a subset of these genes by using a method, serial analysis of chromatin occupancy (SACO), that we used previously to identify the transcriptome regulated by the transcriptional activator cAMP response element-binding protein (CREB) (Impey et al., 2004). Four new findings resulted from our studies. First, sequence motif analysis of the SACO-delineated REST-binding regions revealed that the canonical RE1 sequence is actually a subset of a much larger, more complex motif, suggesting that previous studies may have significantly underestimated the number of functional REST-binding sites. Second, and unexpectedly, unlike most vertebrate transcription factor binding motifs, the majority of RE1 sites appear to be occupied in cells that express REST. Third, the discovery of novel RE1 motifs allowed the identification of neuronal regulated genes that had not been detected previously. Fourth, not all REST target genes were derepressed by loss of functional REST, suggesting a complex mechanism for normal neuronal gene repression.
Materials and Methods
Cell culture.
The murine kidney cell line (TCMK1) was obtained from American Type Culture Collection (Manassas, VA) and maintained in MEM with 10% FBS, 1% sodium pyruvate, 1% nonessential amino acids, and 1.5 mg/ml sodium bicarbonate. Primary mouse embryonic fibroblasts (MEFs) were derived from embryonic day 14.5 (E14.5) embryos and maintained in DMEM containing 10% FBS, 2 mm l-glutamine, and 1% nonessential amino acids. For the trichostatin A (TSA) experiment, MEFs were treated for 16 h with 300 nm TSA (Sigma, St. Louis, MO). Primary mouse cortical neurons were isolated from E15.5 embryos and maintained in Neurobasal medium with 2% B27 and 500 mm l-glutamine. The cortical neurons were treated with 5 mm cytosine arabinoside (Sigma) after 3 d in vitro to inhibit the growth of dividing cells. All cultures were maintained at 37°C in an atmosphere of 5% CO2.
Chromatin immunoprecipitation analyses.
Cross-linked chromatin for the chromatin immunoprecipitation (ChIP) analysis (Ballas et al., 2001) was immunoprecipitated using polyclonal antibodies directed against either the N terminus of REST (REST-N) (Chong et al., 1995) or the C terminus of REST (REST-C) (Ballas et al., 2005). After the reversal of cross-links, the ChIP DNA was purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA), and DNA samples were analyzed by 40–50 cycles of PCR or 40 cycles of real-time PCR (primer sequences are available upon request).
SACO library construction and data analysis.
A REST SACO library was constructed from TCMK1 cells according to Impey et al. (2004). Briefly, ChIP DNA from 5 × 107 TCMK1 cells was blunt ended with the DNA Terminator End Repair kit (Lucigen, Middleton, WI), extracted with phenol/chloroform, and ethanol (EtOH) precipitated. The DNA was ligated to adapters, purified (QIAquick PCR purification kit; Qiagen), resuspended in 50 μl of 10 mm Tris, pH 8, and 1 μl was amplified by PCR in a 100 μl reaction containing 100 pmol of primer (5′bio-CCGGTCTACTGAATTCCGAAC), 2 μl of Platinum Taq polymerase (Invitrogen, San Diego, CA), 2 mm MgCl2, and 0.75 mm dNTPs (Roche Diagnostics, Indianapolis, IN) for one cycle at 95°C for 50 s; 25 cycles at 94°C for 15 s, 55°C for 30 s, and 68°C for 4 min; followed by one cycle at 68°C for 7 min. The PCRs were purified (QIAquick PCR purification kit; Qiagen), and 10 μg of DNA was digested with 6 μl (60 U) of NlaIII (New England Biolabs, Ipswich, MA) in a 200 μl reaction at 37°C for 2 h. The digests were extracted with phenol/chloroform, precipitated with EtOH, and resuspended in 24 μl of DEPC water. To create ditags, 12 μl of this DNA was ligated to 36 pmol of LongSAGE adapter A with 2 μl of 10× T4 ligase buffer. The remaining 12 μl was ligated to 36 pmol of LongSAGE adapter B. Both reactions were heated to 50°C for 2 min before adding 1 μl of T4 ligase and incubating at 16°C for 12 h. The ligations were purified (QIAquick PCR purification kit; Qiagen), eluted with 200 μl of DEPC water, and added to streptavidin-coated magnetic beads (Dynal M280; Invitrogen) in 200 μl of 2× BW buffer (10 mm Tris, pH 8, 2 m NaCl, 0.4 mg/ml BSA, and 1 mm EDTA). After 1 h of incubation at 25°C on a rotator, the beads were washed six times for 5 min with 1× BW buffer and three times with 10 mm Tris, pH 8. To release the ditags from the cassettes, the ligation products were digested with MmeI (New England Biolabs) as follows: the beads were washed twice with 200 μl of 1× NEB buffer 4/1× S-adenosyl methionine (SAM; 1 μl of 32 nm SAM in 800 μl of 1× NEB buffer 4) and then resuspended in 70 μl of 10 mm Tris, pH 8. The samples were incubated in 10 μl of 1× buffer 4, 10 μl of 10× SAM (1 μl of 32 mm SAM in 79 μl of DEPC water), and 10 μl of MmeI for 2.5 h at 37°C. The supernatants were pooled in 100 μl of 10 mm Tris, pH 8, extracted with phenol/chloroform, and precipitated with EtOH. Sample was resuspended in 10 mm Tris, pH 8, and concatemerized with T4 DNA ligase for 12 h at 25°C. The sample was subjected to LongSAGE as described by Saha et al. (2002) with a few modifications: concatemers were isolated using the agarose gel method, NEB T4 DNA ligase (2000 U/μl), and GlycoBlue (Ambion, Austin, TX) were used instead of T4 DNA ligase and glycogen. Sequencing of the REST-SACO library was performed by the Cold Spring Harbor Laboratory Genome Research Center.
Bioinformatics.
The concatemer sequences from the library were extracted from chromatograms using “phred” (Ewing and Green, 1998). A custom Perl script was used to separate ditags at all CATGs, and the resulting SACO GSTs were mapped to genomic CATG sites using a C program. A set of programs for analyzing SACO data is available at http://genome.bnl.gov/SACO/. Data from the REST SACO library are available at http://genome.bnl.gov/REST/. Additional details on binding site information (generation of search matrices and score cutoffs) are in the supplemental material (available at www.jneurosci.org).
Gel shift assays.
Nuclear extracts were prepared from NIH3T3 cells as described by Wadman et al. (1997), and gel shift reactions were performed essentially as described by Wood et al. (2003). The nuclear extracts were preincubated on ice, with or without competitor DNA, for 30 min in 19 μl of solution containing 20 mm HEPES, pH 7.9, 100 mm KCl, 5 mm MgCl2, 8% (v/v) glycerol, and 1 μg of salmon sperm DNA. Radioactive probe, 0.8 nm at ∼20,000–25,000 cpm, was added to each reaction and incubated at room temperature for 30 min. For supershift experiments, 1 μl of REST-N serum (Chong et al., 1995) was added to the reactions and incubated for an additional 30 min at room temperature. The reactions were run on 6% native polyacrylamide/1× TGE (20 mm Tris, 384 mm glycine, and 2 mm EDTA) at 150 V for 90 min at 4°C. For the competition assays, 10 nm, 100 nm, and 1 μm nonradioactive probes were used. Radiolabeled DNA probes were end labeled using T4 kinase (New England Biolabs) and [32P]dATP. The following oligonucleotides were PAGE purified and annealed in buffer A (100 mm KCl and 30 mm HEPES): expanded RE1, 5′-CAGGAACTTCAGCACCCGCGGGGCGGACAGCGCCTACCGCA-3′ and 5′-TGCGGTAGGCGTCTGTCCGCCCCGCGGGTGCTGAAGTTCCTG-3′; compressed RE1, 5′-GGGAGATTTCAGGACGCGGACAGCGCCCTGAATT-3′ and 5′-AATTCAGGGCGCTGTCCGCGTCCTGAAATCTCCC-3′.
Adenoviral vectors and transduction.
Adenovirus-bearing cDNAs for either DN REST [as described by Chong et al. (1995)] or green fluorescent protein (GFP) were introduced into MEFs by calcium phosphate precipitation (Fasbender et al., 1998).
RNA extraction, reverse transcription-PCR, and real-time PCR.
Total RNA from primary cortical neurons and MEFs transduced with adenovirus was extracted using the RNeasy kit (Qiagen) and treated with DNase (DNA-free kit; Ambion). Mouse thymus RNA was purchased from Ambion. Reverse transcription was performed using Superscript III and Oligo (dT)12–18 primers (Invitrogen). Quantitative PCR was performed in an Applied Biosystems (Foster City, CA) PRISM 7900HT Fast Real-Time PCR system with SYBR green PCR master mix. Primers were designed using Primer3 software (Massachusetts Institute of Technology, Cambridge, MA; http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). For analysis of cDNA levels, primers were designed around exon/intron boundaries and, for ChIP confirmation, primers were designed around each putative REST-binding site. All PCRs were run under the same cycling conditions (95°C/10 min; 95°C/15 s and 57°C/1 min for 40 cycles). The relative abundance of each cDNA for the DN REST experiments was determined by using a standard curve generated from 10-fold serial dilutions of mouse whole-brain cDNA and normalized to Gapdh cDNA levels. To analyze changes in gene expression, the ratio of the means between control and experimental samples from three independent experiments was calculated, and error bars represent the SD of these means. Significance of changes was determined using Student's t test. To compare the relative expression levels of T-cell-expressed genes in thymus and primary cortical neurons, standard curves were first generated from 10-fold serial dilutions of thymus cDNA to ensure that the primers were amplifying efficiently across a broad range of cDNA concentrations. The average cycle threshold (Ct) value for each gene from three independent samples was then compared between thymus and primary cortical neurons. A Ct value of 30 was chosen to represent genes expressed at low levels. For REST ChIP confirmation, the relative occupancy of putative binding sites from two independent immunoprecipitation reactions was determined using a standard curve generated from 10-fold serial dilutions of input DNA, and this value was expressed as a percentage of input DNA. As a negative control for the ChIP experiments, primers were designed around a region of the REST gene that lacked a REST-binding site. All primer sequences are available upon request.
Luciferase assay.
The thymidine kinase (TK) promoter from pRL-TK Renilla luciferase expression vector (Promega, Madison, WI) was ligated to the BglII/HindIII sites of pGL3-basic (Promega) to generate pGL3-TK. RE1 sites and their flanking sequences were cloned into pCR4-TOPO using the TA TOPO cloning kit (Invitrogen). The inserts from pCR4-TOPO were excised with SpeI/BglII and cloned into the NheI/BglII sites of pGL3-TK to create the TK-RE1 and TK-expanded RE1 reporter constructs. RE1 sites were deleted or mutated using QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). All constructs were verified by sequence analysis. For the assay, TCMK1 cells (1 × 105 cells per well on 12-well dishes) were transfected with 250 ng of luciferase reporter vector along with 20 ng of pRL-TK and 250 ng of an expression vector for DN REST (Chong et al., 1995) or an empty vector (pcDNA; Invitrogen). Forty-eight hours after transfection, the ratio of firefly versus Renilla luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega) following the manufacturer's protocol. The mean changes in luciferase activity were measured across three independent samples for reporters cotransfected with DN REST or pcDNA. Error bars represent the SDs of these means, and significance was determined using the Student's t test.
Results
Genome-wide mapping of REST-binding sites by SACO
Using a polyclonal antibody (REST-N) that recognizes the N-terminal domain of REST (Chong et al., 1995), we used SACO to identify REST-binding sites in the TCMK1 mouse kidney cell line. The REST-SACO library contained 41,257 genomic signature tags (GSTs), of which 29,807 were analyzed further (sup-plemental Fig. 1, available at www.jneurosci.org as supplemental material) (library information is available at http://genome.bnl.gov/REST/).
Figure 1.
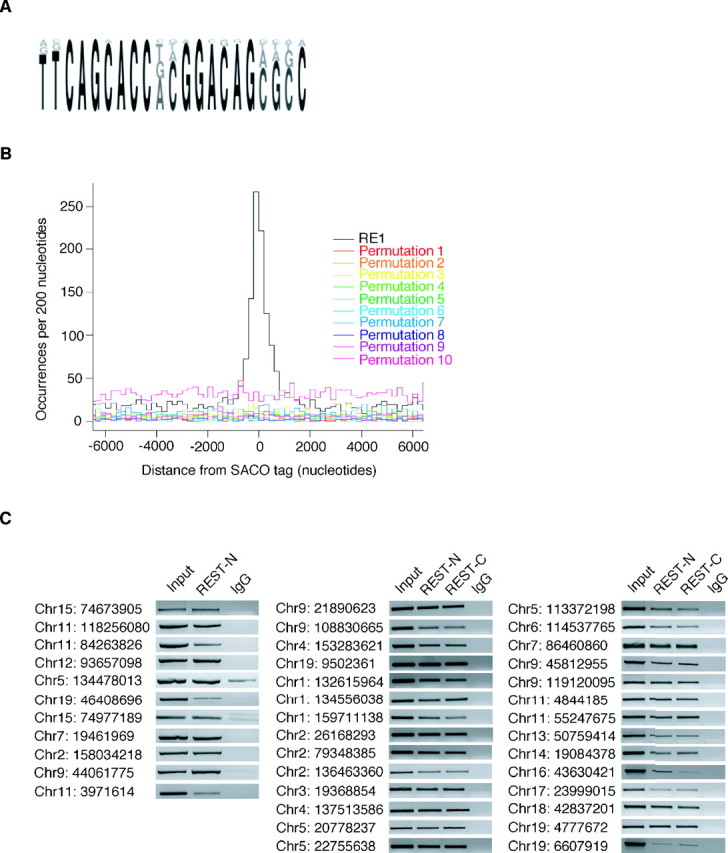
REST occupies SACO-identified RE1 sites in vivo. A, A sequence logo representing the PWM used to identify RE1 sites in the genome. B, Histograms of GST-RE1 offsets showing that the sequences surrounding the SACO sites are enriched only for RE1s and not for permutations of the RE1. C, ChIP showing REST binding to SACO-identified RE1 sites in TCMK1 kidney cells. The chromosome (Chr) number and position of each site is indicated on the left (University of California, Santa Cruz, Mm5 build). Immunoprecipitations were performed with antibodies directed against either REST-N or REST-C or with a rabbit IgG antibody. Sonicated genomic DNA served as a positive control for each RE1 site (Input).
To determine whether the genomic locations identified by SACO were associated with a canonical REST-binding site (RE1), we generated a position weight matrix (PWM) based on the sequences of 20 RE1 sites that bound REST in functional assays and four RE1-like sites that did not (Schoenherr et al., 1996) (Fig. 1A) (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). In comparing the genomic locations of the RE1 sites and REST GSTs, we found that 893 RE1 sites were within 3 kb of a GST, the distance representing the upper limit of our ChIP fragment size. As a control, 10 permutations of the RE1 site were generated by randomly scrambling the nucleotide positions constituting the 21 bp sequence. When the genomic locations of these 10 permutations were compared with those of the REST GSTs, no colocalization was observed (Fig. 1B). Furthermore, loci associated with 42,000 randomly chosen GSTs were not enriched for RE1 motifs (data not shown).
REST occupancy at genomic loci identified by SACO
We used ChIP analysis in TCMK1 kidney cells to validate REST occupancy of the identified RE1 sites that colocalized with REST GSTs (Fig. 1C) (supplemental Table 1, available at www.jneurosci.org as supplemental material). Thirty-nine RE1 sites were interrogated for REST binding using both the antibody used to construct the library (REST-N) and a second antibody that recognized a different portion of REST (REST-C) (Ballas et al., 2005). All 39 sites tested showed REST occupancy with both antibodies; no enrichment was seen in the IgG control. As an additional control, we performed PCR using primers that amplified a region within the REST gene that is 5 kb from the closest RE1 site (data not shown). No significant enrichment was seen for this region, indicating that REST bound specifically to the SACO-identified RE1 sites.
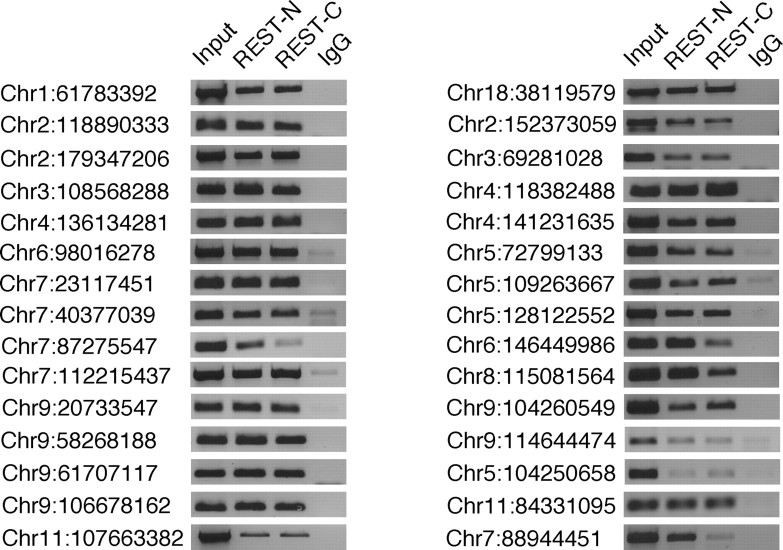
A question that arose from our initial analysis of SACO-identified RE1 sites was whether REST occupied a majority or only a subset of RE1 sites in non-neuronal cells. In an attempt to answer this, we analyzed 15 RE1 sites that were identified by SACO (Fig. 2, left) (supplemental Table 2, available at www.jneurosci.org as supplemental material) and 15 non-SACO sites that were chosen randomly from the set of genomic PWM-identified RE1 s (Fig. 2, right) (supplemental Table 2, available at www.jneurosci.org as supplemental material). REST occupied both sets of RE1 sites in TCMK1 kidney cells.
Figure 2.
REST occupies the majority of RE1 sites in kidney cells. Chromatin immunoprecipitations were performed on randomly chosen RE1 sites that were identified either by SACO (left) or bioinformatically (right) in the mouse kidney cell line, TCMK1. Immunoprecipitations were performed with antibodies directed against either REST-N or REST-C or with a rabbit IgG antibody. Sonicated DNA was used as the positive control for each RE1 site (Input). The chromosome (Chr) number and position of each RE1 site are indicated on the left (University of California, Santa Cruz, Mm5 build).
The RE1 sites identified by SACO were not restricted to the promoter regions of putative target genes [University of California, Santa Cruz (Santa Cruz, CA) database, http://genome.ucsc.edu, Mm5]. For the target genes Celsr3, Syt7, Neurod1, and Htr6, the RE1 site was located within the promoter region, intron, exon, and downstream of the transcriptional stop site, respectively (Fig. 3A). This distribution was representative of the 1606 putative REST target genes that were within 50 kb of a SACO-identified RE1 site. Four hundred thirty-four (27%) genes had RE1 sites 10–50 kb upstream of the 5′ transcript start site, 131 (8%) were within 10 kb from the transcript start, 310 (19%) were located within an intron, 71 (4%) were within an exon, 154 (10%) were within 10 kb downstream of a transcriptional stop site, and 506 (32%) were 10–50 kb downstream from the transcriptional stop site (Fig. 3B).
Figure 3.

REST-binding sites are not restricted to the promoter regions of target genes. A, Diagrams showing REST GSTs colocalized with RE1 sites for four distinct transcripts. The position of each RE1 site (star) is shown relative to the SACO GSTs and the transcriptional start sites (arrows). Introns are represented by solid lines, exons are represented by black boxes, and intergenic regions are represented by dotted lines (University of California, Santa Cruz, Mm5 build). Chr, Chromosome. B, Locations of the 893 SACO-identified RE1 sites within 50 kb of putative target genes.
Characterization of putative REST target genes
To identify potential REST target genes, we further characterized the 893 SACO-identified RE1 sites using information from the University of California, Santa Cruz database (Mm5). For this analysis, we focused on genes within 50 kb of an RE1 site. Examination of these genes showed that they encoded proteins involved in all facets of neural development and function (Fig. 4A). Although some of these targets had been identified previously, many were new. Strikingly, REST targets represented several multigene families encoding cadherins, olfactory receptors, synaptotagmins, solute carriers, and voltage-dependent calcium and potassium channels. In addition to coding genes, RE1 sites also colocalized with microRNA transcripts, several of which have not been identified previously (Fig. 4B).
Figure 4.
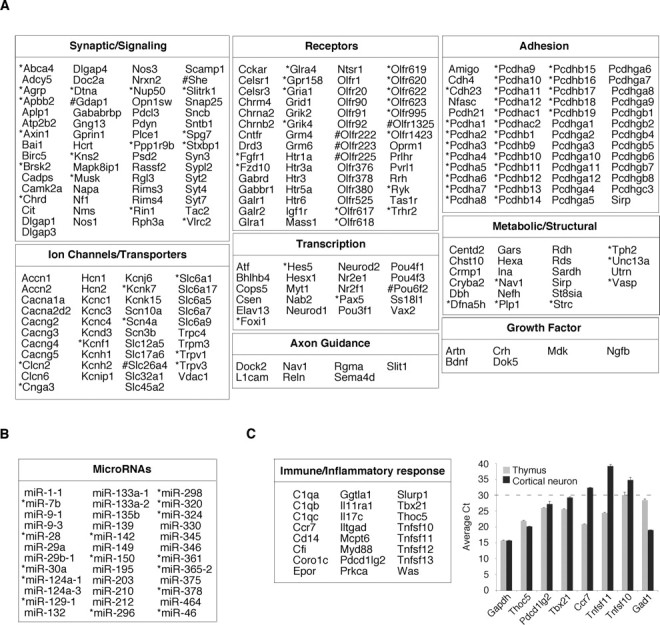
Categorization of putative target genes at SACO-identified REST-binding sites. A, Chart showing genes within 50 kb of SACO-identified RE1 sites, expanded RE1 sites (*), and compressed RE1 sites (#) involved in nervous system development or function. B, Table showing microRNA transcripts within 50 kb of a REST-binding site. C, Left, Table of genes involved in the immune/inflammatory response. C, Right, Quantitative RT-PCR analysis of six T-cell-expressed genes in mature cortical neurons (black bars) and thymus (gray bars). The average Ct for each gene (n = 3) is on the y-axis, and the dotted line demarcates the boundary between genes with moderate-to-high expression levels (Ct < 30) and those that are expressed at low levels (Ct > 30). For comparison, the relative expression of Gapdh, a ubiquitously expressed gene, and Gad1, a neuronal gene, are also shown.
Our analysis also identified genes whose function and expression were not restricted to the nervous system (supplemental Fig. 3, available at www.jneurosci.org as supplemental material), including those encoding proteins involved in immune and inflammatory responses (Fig. 4C, left). Like neuronal-specific genes, some of these immune targets also represented gene families, such as complement and the tumor necrosis factor superfamily (Tnfsf). To test whether these T-cell-expressed genes were also expressed in the nervous system, we used real-time reverse transcription (RT)-PCR to measure their relative expression levels in thymus and mature cortical neurons. A Ct value between 30 and 40 was taken to indicate low-to-negligible levels of transcript. Three genes, Thoc5, Pdcd1lg2, and Tbx21, were also expressed in neurons, exhibiting Ct values between 20 and 30 (Fig. 4C, right), and Thoc5 was expressed at the same level in neurons and thymus. As a control, we examined expression of the neuronal-specific Gad1 gene in both thymus and mature cortical neurons. As predicted, it had a Ct value of ∼30 in thymus, consistent with an extremely low level of expression in non-neuronal tissues. Thus, our SACO analysis suggests that some REST-modulated genes not previously considered to be neuronal are nonetheless expressed in the nervous system.
Contribution of REST to basal expression levels of target genes
To examine the functional effects of REST binding, we virally transduced primary MEFs with DN REST and analyzed the expression of 44 SACO-identified target genes (Fig. 5). In addition to DN REST, shRNAs targeting REST were also used to analyze the contribution of REST to gene expression of select genes, and the results were similar to those presented here (data not shown). From the DN REST analysis, three classes of genes emerged. In one class, represented by 19 genes, expression of DN REST resulted in increased transcript levels ranging from 1.5- to nearly 50-fold over levels in control infected cells (Fig. 5A, left).
Figure 5.
REST regulation of its target gene repertoire is more complex than simple repression. A, Left, Real-time RT-PCR analysis of REST target genes that were derepressed in the presence of DN REST. Fold changes in MEFs virally transduced with cDNA for DN REST (Ad-dnREST) are shown relative to transductions with GFP cDNA (Ad-GFP). Levels of cDNA were measured 48 h after transduction and normalized to endogenous GAPDH. A, Middle, Relative luciferase activity was lower in TCMK1 cells for constructs bearing RE1 sites from the derepressed class of genes (TK-RE1) compared with constructs with the RE1 sites deleted (TK-ΔRE1). A, Right, Derepression of luciferase reporter genes was seen only in TCMK1 cells cotransfected with DN REST and constructs bearing the RE1 site. Fold changes in luciferase activity are shown relative to cells expressing a control construct (pcDNA). B, Left, Real-time RT-PCR analysis of REST target genes that were unchanged in the presence of DN REST. B, Middle, Relative luciferase activity was lower in TCMK1 cells for constructs bearing RE1 sites from the unchanged class of genes (TK-RE1) than for constructs with the RE1 sites deleted (TK-ΔRE1). B, Right, Derepression of luciferase reporter genes was seen only in TCMK1 cells cotransfected with DN REST and constructs bearing the RE1 site. C, Left, Real-time RT-PCR analysis of REST target genes that were repressed in the presence of DN REST. C, Middle, Relative luciferase activity was lower in TCMK1 cells for constructs bearing RE1 sites from the repressed class of genes (TK-RE1) than for constructs with the RE1 sites deleted (TK-ΔRE1). C, Right, Derepression of luciferase reporter genes was seen only in TCMK1 cells cotransfected with DN REST and constructs bearing the RE1 site. D, Real-time RT-PCR analysis of REST target genes in MEFs virally transduced with either dominant-negative REST (Ad-dnREST) or GFP (Ad-GFP). Changes in mRNA levels were measured 48 h after transduction of either untreated MEFs or MEFs that were treated for 16 h with the HDAC inhibitor TSA (300 nm). All fold changes are shown relative to MEFs transduced with Ad-GFP, and the expression of each gene was normalized to endogenous GAPDH levels. Error bars represent the SD from three independent experiments. *p ≤ 0.05; **p ≤ 0.005; Student's t test.
Transcript levels of CoREST, a gene whose transcription is not regulated by REST, were unchanged in these cells. To confirm that the derepression required the RE1 site, we constructed luciferase reporter genes containing the RE1 site and flanking sequences of the Glra1 and Syt7 genes upstream of the minimal TK promoter and introduced them into the TCMK1 kidney cells (Fig. 5A, middle). TCMK1 cells were used for these studies because of their high transfection efficiency compared with MEFs. The RE1 site for the Gad1 gene, a gene previously shown to be repressed by REST (Lunyak et al., 2002), served as a control. As expected, recombinant luciferase activity was lower for all three reporter constructs containing an RE1 site, consistent with REST repressor function. This repression was mediated by REST, because cotransfection of each reporter construct together with the DN REST vector resulted in a threefold or greater increase in luciferase activity over control transfected cells (Fig. 5A, right).
In contrast with the above class, we identified 20 genes whose endogenous expression levels were not changed after expression of DN REST (Fig. 5B, left). We analyzed the RE1 sites in four of these genes (Cacna1 h, Nfasc, Reln, and Crh) by luciferase reporter analysis as above. All of the reporter genes were repressed by the presence in cis of the RE1 sequence (Fig. 5B, middle), and simultaneous expression of DN REST resulted in derepression (Fig. 5B, right).
Five genes (Ihh, Mdk, Cutl2, Epha8, and Syt2), representing the third class of REST targets, showed a significant decrease, rather than increase, in their transcriptional activity after expression of DN REST (Fig. 5C, left). This suggests that REST could also act as an activator for these genes. We directly investigated this idea by constructing a luciferase reporter vector containing the RE1 site for the Syt2 gene. Surprisingly, as in the other two gene classes, luciferase activity was lower with this RE1 sequence in cis (Fig. 5C, middle), and the reporter's expression increased after coexpression with DN REST (Fig. 5C, right). These results indicated that REST still functioned as a repressor at this promoter and suggested that some of the activator functions ascribed previously to REST are indirect.
A recent report suggests that repressors other than REST are responsible for preventing the expression of certain neuronal genes (Kim et al., 2006). Therefore, we examined, for six genes, whether additional repressor mechanisms might explain the inability of these genes to be derepressed by DN REST. MEFs were treated with the more global inhibitor of histone deacetylase (HDAC) activity, TSA, and expression of the six test genes was reexamined by RT-PCR (Fig. 5D). Three genes, Prg4, Nfasc, and Reln, were derepressed in the presence of TSA alone or with TSA and DN REST, but not by DN REST alone, consistent with the presence of an additional repressor or more generalized repression mediated by chromatin inaccessibility. In contrast, the remaining three genes, Crh, Cacna1 h, and Drd3, were not affected by treatment with TSA, suggesting that an HDAC-independent mechanism, or lack of an activator, prevented expression.
Discovery of novel REST-binding motifs
Not all of the REST-SACO GSTs localized with readily identifiable RE1 sites, suggesting that REST may bind to a different DNA element either alone or in conjunction with another DNA binding protein. To attempt to address this question, we asked whether any specific DNA elements were highly represented within 2 kb loci containing one or more GST but no canonical RE1 site within 3 kb. Every combination of an 8 nt sequence (65,536 in total) was used to search for enrichment. The two 8-mer sequences corresponding to the 5′ or 3′ end of the canonical RE1 site were identified. Surprisingly, the 5′ and 3′ sites were found to colocalize and define a novel expanded RE1 site containing insertions of 3–9 nt between the 11th and 12th nucleotide of the previously defined canonical consensus site (Fig. 6A). New PWMs were derived from the dataset of 21-mer RE1s by allowing for an insertion of 3–9 nt. Using these parameters, we were able to identify another 4631 novel RE1 sites in the genome, 394 of which were within 3 kb of a SACO GST (information on all sites is available at http://genome.bnl.gov/REST/). ChIP analysis confirmed that in vivo REST occupied several of these expanded RE1 motifs in TCMK1 kidney cells (Fig. 6B) (supplemental Table 3, available at www.jneurosci.org as supplemental material). Interestingly, in vivo REST binding to the RE1 motif containing a 4 nt insertion was weaker than to the others we tested, perhaps reflecting the possibility that this is not a preferred motif (data not shown).
Figure 6.
Discovery of expanded RE1 sites. A, Sequence logo representation of the PWM used to identify expanded RE1 sites in the genome. NNN represents 3–9 bp that comprise the insertion. B, Quantitative PCR of chromatin immunoprecipitations in TCMK1 kidney cells showing REST binding to expanded RE1 sites. Fold changes in DNA immunoprecipitated with antibodies directed against REST-N or REST-C or with a control rabbit IgG antibody are shown relative to sonicated genomic DNA (Input). The chromosome (Chr) number and position of the expanded RE1 sites are indicated (University of California, Santa Cruz, Mm5 build), as are the number of nucleotides that comprise each insertion (Split3 indicates that 3 bp make up the insertion). Error bars represent the SD from three independent experiments.
In addition to the expanded RE1 site, a compressed form of the RE1 site was also discovered (Fig. 7A, left). Because the nucleotides located at positions 10 and 11 of the canonical site are the least conserved within and among RE1s in mammalian species, we asked whether removing these two nucleotides and replacing them with either 0 or 1 random nucleotide would yield functional REST-binding sites. Indeed, those sites with 1 nt inserted between the ninth and 12th position of the canonical site were found to bind REST by ChIP analysis in TCMK1 kidney cells (Fig. 7A, right) (supplemental Table 4, available at www.jneurosci.org as supplemental material). We identified 186 of these sites, 27 of which were selected for by SACO. Furthermore, gel shift assays performed on NIH3T3 nuclear extracts using radiolabeled oligonucleotide probes encompassing either the expanded or compressed RE1 for the Scg3 and Slc26a4 genes, respectively, showed binding activity that was specific for REST (Fig. 7B).
Figure 7.
Discovery of compressed RE1 sites. A, Left, Sequence logo representation of the PWM used to identify compressed RE1 sites in the genome. A, Right, ChIP analysis showing REST binding to SACO-identified compressed RE1 sites in TCMK1 kidney cells. The chromosome (Chr) number and position of each site is indicated on the left (University of California, Santa Cruz, Mm5 build). Immunoprecipitations were performed with antibodies directed against either REST-N or REST-C or with a rabbit IgG antibody. Sonicated genomic DNA served as a positive control for each RE1 site (Input). B, Electromobility shift assays showing that NIH3T3 nuclear extracts contain binding activities for the expanded and compressed RE1 motifs. Left, Radiolabeled probes containing the expanded RE1 motif from the Scg3 gene (lanes 1–3) and the compressed RE1 motif from the Slc26a4 gene (lanes 4–6) were shifted (arrow) in the presence of nuclear extract (lanes 2, 5), whereas no shift was seen in the absence of extract (lanes 1, 4). The addition of REST-N antibody resulted in a supershift of the protein/DNA-bound complexes (lanes 3, 6). Right, Nonlabeled competitor probes (10 nm, 100 nm, and 1 μm) effectively competed for binding to NIH3T3 extract in the presence of radiolabeled oligonucleotide probes for both the expanded RE1 (lanes 1–5) and compressed RE1 (lanes 6–10). C, ChIP analyses showing REST binding to SACO-identified genomic locations not associated with RE1 sites. The chromosome number and primer locations used to amplify each SACO locus are indicated on the left (University of California, Santa Cruz, Mm5 build).
Even with the novel sites, there were still GSTs that did not associate with any recognizable REST-binding site. To test whether REST occupied these “orphan” loci, we performed ChIP analysis on twenty 2 kb loci that contained at least two distinct GSTs but no obvious REST-binding site within 3 kb. REST occupied 10 of these sites in TCMK1 kidney cells (Fig. 7C). These results strongly suggest that in these instances, REST interaction is complex and may involve association with another DNA binding factor or binding through a DNA element unrelated to the known RE1.
Functional analysis of the expanded RE1 motif
To examine the function of REST at the expanded (3–9 nt insertion) RE1 sites, we performed luciferase reporter analyses (Fig. 8A). For the expanded RE1 sites containing 3, 5, 6, 7, and 9 nt, relative luciferase activities were lower in reporter genes with these sites, consistent with REST-mediated repression. With one exception, cotransfection of cells with DN REST and these reporter constructs caused a threefold or greater increase in luciferase activity. The construct containing an expanded RE1 site with an 8 nt insertion was not derepressed by DN REST, suggesting that an additional repressor element resides in its flanking sequence.
Figure 8.
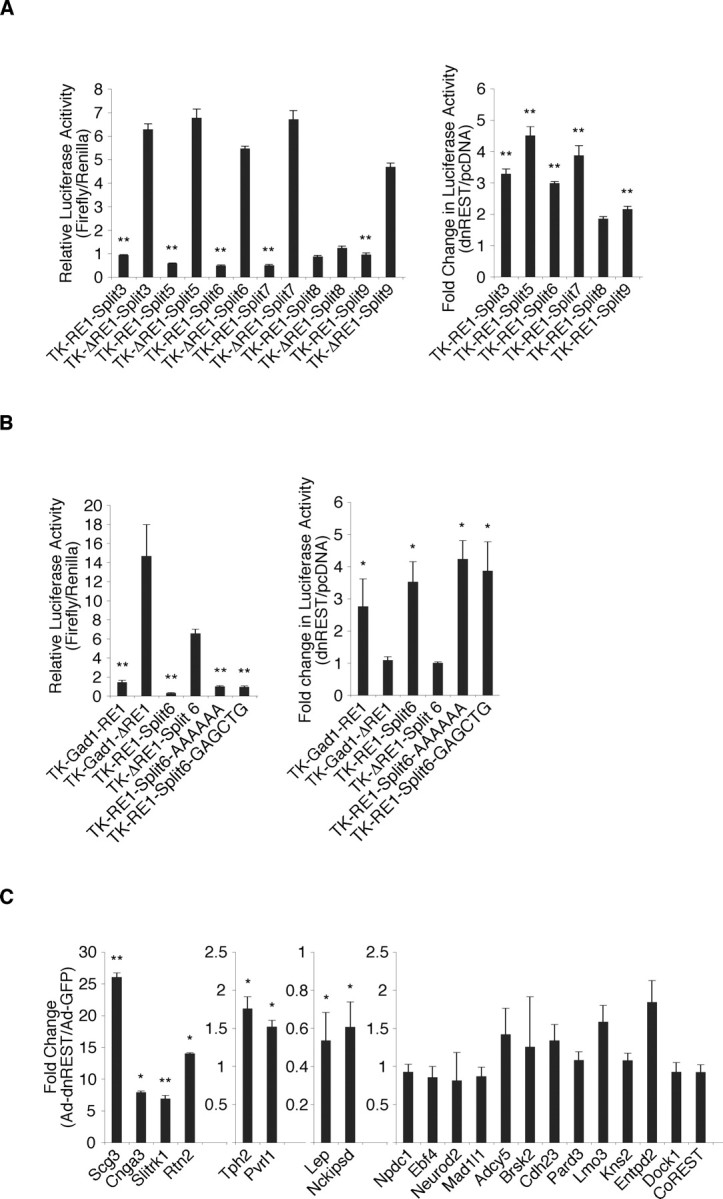
REST regulates genes with the expanded motif. A, REST is a repressor at the expanded RE1 sites. Left, Relative luciferase activity is lower in TCMK1 kidney cells 48 h after transfection with constructs bearing expanded RE1 sites (TK-RE1-splitN) compared with constructs lacking them (TK-ΔRE1-splitN). Right, Coexpression of reporter constructs with DN REST resulted in derepression of expanded RE1-containing constructs (TK-RE1-splitN). Fold changes are relative to cells expressing a control construct (pcDNA). B, The inserted nucleotides of the expanded RE1 site do not contribute to REST function. Left, Relative luciferase activity is lower in TCMK1 kidney cells 48 h after transfection with constructs bearing the Gad1-RE1 (positive control), RE1-split6 site, and RE1-split6 sites whose insertions were swapped with 6 A nucleotides or a random sequence (GAGCTG) compared with constructs lacking the sites (TK-Gad1-ΔRE1 and TK-ΔRE1-split6). Right, Coexpression of reporter constructs with DN REST resulted in derepression for constructs containing the GAD1 RE1 and the intact and mutated RE1-split6 sites. Fold changes in luciferase activity in cells expressing DN REST are shown relative to cells expressing a control construct (pcDNA). C, Real-time RT-PCR analysis of MEFs virally transduced with DN REST (Ad-dnREST) resulted in derepression, repression, or no change in cDNA levels of REST target genes. Levels of each cDNA are shown relative to MEFs transduced with a control virus (Ad-GFP) and were normalized to endogenous GAPDH. Error bars represent the SD from three independent experiments. *p ≤ 0.05; **p ≤ 0.005; Student's t test.
To determine whether the sequence of the inserted nucleotides was important for repressor function, we performed an additional luciferase reporter analysis. The 6 nt that separated the 5′ and 3′ ends of the REST-binding site were mutated to six adenines or an irrelevant 6-mer (GAGCTG). Mutating the sequences had no effect on repressor activity (Fig. 8B, left) or the response of the luciferase reporters to DN REST (Fig. 8B, right), suggesting that specific insert sequences are not required for REST binding.
Annotation of genes associated with the expanded RE1 motif showed that they represented functional classes similar to those associated with the canonical RE1 site (Fig. 4A) (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). As before, REST regulation of these genes fell into three different functional categories as determined by our DN REST analysis. Of the 20 genes examined, six (Scg3, Cnga3, Slitrk1, Rtn2, Tph2, and Pvrl1) were derepressed from 1.5- to nearly 30-fold relative to controls, two (Lep and Nckipsd) were repressed, and the remaining 12 genes were unchanged (Fig. 8C). Thus, REST regulation of genes containing the expanded RE1 motif, like the traditional motif, is complex and likely to involve additional factors.
Discussion
REST target genes were identified originally by the presence of a similar putative repressor-binding motif in genes encoding two neuronal-specific proteins, the voltage-dependent sodium channel and the synaptic protein, SCG10. Promoter fragments of these genes bound the REST repressor in vitro, and reporter genes containing the promoter were derepressed after loss of REST function in transient transfection analyses of non-neuronal cells (Chong et al., 1995; Schoenherr and Anderson, 1995; Schoenherr et al., 1996). These first studies suggested that REST-repressed genes were required for synaptic function, but whether other aspects of the neuronal phenotype were under REST control was not explored. Since that time, it has been suggested, primarily based on bioinformatics approaches, that many other synaptic proteins, as well as proteins involved in neurosecretion, are under REST control (Lunyak et al., 2002; Bruce et al., 2004; Sun et al., 2005; Johnson et al., 2006; Mortazavi et al., 2006; Wu and Xie, 2006). Only a subset of the REST-binding sites, however, has been validated by ChIP, leaving open the question of the functional relevance of these sites. Because of its potentially unique function in regulating protein networks specifying the neuronal phenotype, we sought to identify REST-occupied genes on a more global scale and to determine the contribution of REST to their regulation.
We chose SACO methodology, as opposed to Chip-on-chip, because it avoids certain biases in the experimental design. First, because SACO is a sequence-based approach, it does not require hybridization analysis to identify the occupied loci, so each functional binding site in the genome is interrogated without bias. Second, because SACO does not depend on promoter arrays, REST-binding sites located at large distances from promoters can still be identified. Third, unlike the case of tiled arrays that contain gaps caused by repetitive sequences, all REST-binding sites should be available to the SACO analysis. In addition to CREB, sequence-based approaches similar to SACO have been used to identify the genetic networks regulated by other transcriptional activators, including E2F4, p53, Oct4, Nanog, and c-myc, as well as networks associated with histones modified by acetylation (Chen and Sadowski, 2005; Kim et al., 2005; Loh et al., 2006; Wei et al., 2006; Zeller et al., 2006). With respect to REST, whereas previous in silico and sequence-based approaches have identified genes associated with a consensus 21 bp REST-binding site, using SACO, we first found, unexpectedly, that the REST-binding site was both compressed and expanded compared with this original motif. Interestingly, close inspection of the original 21 bp RE1 motif indicated that nucleotides 10 and 11 were two of the least-highly conserved nucleotides across mammalian species. This is precisely where the additional nucleotides were either inserted (for the expanded sites) or removed (for the compressed sites). Thus, we propose that the consensus RE1 motif used to generate our original matrix, and the previous matrices of others, is only a subclass of the authentic REST-binding site.
The new expanded and compressed RE1 motifs increased the number of potential REST-binding sites in the mouse genome from 4991, based on our original RE1 motif, to 9808. This number is greater than the number of all SACO-identified REST-binding sites (1314), likely because we were unable to sequence the SACO library to saturation because of contamination of the initial TCMK1 cell line with human cells. Nonetheless, ChIP analysis of a large set of genes, identified through bioinformatics as containing REST-binding sites, indicated that the majority of sites were occupied in these cells. This finding deviates from the results of another study in U373 glioma cells showing that neuronal genes containing REST-binding sites were occupied, whereas others were not (Bruce et al., 2004). It is possible that the level of REST expression in the kidney cell line was higher than in the U373 cells, thereby permitting detection of REST binding to lower-affinity sites. Consistent with this idea, ectopic expression of REST resulted in occupancy of the previously unoccupied sites.
Of the 1314 SACO-identified RE1 sites, 27 represented the compressed class, 893 were the 21 bp consensus RE1s, and 394 contained inserts of 3–9 nt. It is possible that the overrepresentation of the consensus RE1 motif represents evolution to a higher-affinity binding site, but this has not been tested experimentally. Ontological analysis of genes associated with the new RE1 motifs revealed genes that had not been identified previously, including some encoding proteins involved in lipid metabolism and cell cycle control. Of particular interest, however, was the finding that REST potentially linked, at the transcriptional level, gene products dedicated to a critical neuronal function, specifically, neurotransmission. This process lies at the very heart of nervous system function and requires coordination of protein functions in both the presynaptic and postsynaptic membranes. Our SACO analysis suggests that REST regulates gene products critical to synaptic transmission on both sides of the process. For example, genes encoding synaptotagmins and RIMs, which mediate vesicle release in the presynaptic cell, contain RE1 motifs occupied by REST. Similarly, genes required for the postsynaptic response, encoding neurotransmitter receptors and voltage-dependent potassium and calcium channels, contain functional REST-binding motifs. If RE1-containing genes that were not identified by SACO are also considered, the list of gene products dedicated to neurotransmission is even larger, encompassing synaptophysin, neurexins, complexin on the presynaptic side, and many more neurotransmitter receptors on the postsynaptic side. Thus, the primary protein complexes that distinguish neurons from all other cell types, and that are critical for mediating neuronal plasticity, are linked at the transcriptional level by REST.
Our studies suggest that this REST gene network may ultimately provide a molecular signature of cancers that acquire a neuroendocrine phenotype. A subset of the events underlying neurotransmission is dedicated to neurosecretion, and many of the genes involved in the latter contain REST-binding sites, as indicated by our results and those of Sun et al. (2005). Because cells in certain prostate cancers begin to express a neuroendocrine phenotype, we asked whether this phenotype might be associated with dysregulation of genes under REST control. Indeed, in a mouse model of metastatic neuroendocrine cancer in prostate tissue (Hu et al., 2002), in which 32 mRNAs, representing nine different aspects of the neuroendocrine phenotype, were upregulated, 26 of 32 contained a REST-binding site motif. Four of the six genes that did not contain an RE1 motif, however, have family members with the motif, so erroneous assignment of the highly homologous genes cannot be formally excluded. The increased expression of neuroendocrine genes in the mouse prostate cancer model is consistent with previous studies showing that certain colon and lung cancers are associated with mutations in REST that inhibit repression (Coulson et al., 2000; Hu et al., 2002; Gurrola-Diaz et al., 2003: Neumann et al., 2004; Westbrook et al., 2005). In a mouse model of colorectal cancer, in which mutated REST was identified as an underlying cause of loss of adhesiveness, no neuronal target genes encoding adhesion molecules were identified (Westbrook et al., 2005). Interestingly, SACO analysis identified a large family of adhesion proteins, protocadherins, which have been associated with human prostate and colon cancers (Okazaki et al., 2002; Yang et al., 2005; Terry et al., 2006). It will be interesting to determine whether they are also dysregulated in the colorectal mouse models.
An unexpected finding from our SACO analysis was that individual members of large multigene families were associated with RE1 motifs. For example, there were 10 individual RE1 motifs associated with 23 olfactory receptor genes just on mouse chromosome 2 alone. The finding of individual RE1 motifs on related, presumably duplicated genes extends to many other gene families as well, including those encoding calcium and potassium voltage-dependent ion channels, transient receptor potential channels, solute ion exchangers, and complement proteins. What these families may have in common, in addition to the individual RE1 motifs, is the selective expression of a family member in a particular cell type. For the olfactory receptors, for example, an individual vertebrate sensory neuron expresses only one or two members of the gene family because of an enhancer that selects them, specifically, from the entire genetic repertoire (Lomvardas et al., 2006). Activating specific genes that are otherwise repressed by REST is an economical mechanism that might be evolutionarily favored. In contrast to this situation, other genes share repression mediated from a single RE1 site (Lunyak et al., 2002). This spreading mechanism may have evolved to repress a large set of disparate genes in situ, for example in non-neuronal cells, which either never had RE1 sites or lost their own RE1 sites during evolution. In this scenario, the RE1 motif may be providing a boundary for the silenced chromatin.
Our functional studies indicate that the contribution of REST repression to basal expression levels is context dependent. Several studies have suggested a role for REST in regulating genes important for function of non-neuronal cells including smooth muscle (Cheong et al., 2005) and heart (Schoenherr et al., 1996; Kuwahara et al., 2001, 2003; Ogawa et al., 2002). Additionally, one in silico study reported no significant enrichment (11%) of RE1 sites in neuronal-specific genes (Mortazavi et al., 2006). In agreement with this, in our analysis of the RE1-associated genes with identifiers in the gene ontology database (Amigo), only ∼18% were classified as being neuronal. Based on our RT-PCR analysis of immune genes containing RE1 sites, however, which indicated that they were expressed in neurons and T-cells, it appears that REST regulates genes encoding proteins involved in both neuronal and non-neuronal functions. If REST sites are always occupied, as our data suggests, it will be important to determine how REST repression in non-neuronal cells is prevented to allow expression of these genes.
Footnotes
This work was supported by a grant from the National Institutes of Health to G.M., and G.M. is an Investigator of the Howard Hughes Medical Institute. J.J.D. and S.R.M. were supported by the United States Department of Energy Prime Contract DE-AC02-98CH10886 with Brookhaven National Laboratory. We thank Dr. Nurit Ballas (Stony Brook University) for her advice and encouragement throughout the course of this study.
References
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm PB, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel GM. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sadowski I. Identification of the mismatch repair genes PMS2 and MLH1 as p53 target genes by using serial analysis of binding elements. Proc Natl Acad Sci USA. 2005;102:4813–4818. doi: 10.1073/pnas.0407069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Cheong A, Bingham AJ, Li J, Kumar B, Sukumar P, Munsch C, Buckley NJ, Neylon CB, Porter KE, Beech DJ, Wood IC. Downregulated REST transcription factor is a switch enabling critical potassium channel expression and cell proliferation. Mol Cell. 2005;20:45–52. doi: 10.1016/j.molcel.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Coulson JM, Edgson JL, Woll PJ, Quinn JP. A splice variant of neuron-restrictive silencer factor repressor is expressed in small cell lung cancer: a potential role in derepression of neuroendocrine genes and a useful clinical marker. Cancer Res. 2000;60:1840–1844. [PubMed] [Google Scholar]
- Coulson SM. Transcriptional regulation: cancer, neurons and the REST. Curr Biol. 2005;15:R665–R668. doi: 10.1016/j.cub.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Gurrola-Diaz C, Lacroix J, Dihlmann S, Becker C-M, von Knebel Doeberitz M. Reduced expression of the neuron restrictive silencer factor permits transcription of glycine receptor α1 subunit in small-cell lung cancer cells. Oncogene. 2003;22:5636–5645. doi: 10.1038/sj.onc.1206790. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Fasbender A, Lee JH, Walters RW, Moninger TO, Zabner J, Welsh MJ. Incorporation of adenovirus in calcium phosphate precipitates enhances gene transfer to airway epithelia in vitro and in vivo. J Clin Invest. 1998;102:184–193. doi: 10.1172/JCI2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Ippolito JE, Garabedian EM, Humphrey PA, Gordon JI. Molecular characterization of a metastatic neuroendocrine cell cancer arising in the prostates of transgenic mice. J Biol Chem. 2002;277:44462–44474. doi: 10.1074/jbc.M205784200. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeny S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Johnson R, Gamblin RJ, Ooi L, Bruce AW, Donaldson IJ, Westhead DR, Wood IC, Jackson RM, Buckley NJ. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 2006;34:3862–3877. doi: 10.1093/nar/gkl525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki P, Edelman GM, Jones FS. The neural restrictive silencer element can act as both a repressor and enhancer of L1 cell adhesion molecule gene expression during postnatal development. Proc Natl Acad Sci USA. 1998;95:3233–3238. doi: 10.1073/pnas.95.6.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bhinge AA, Morgan XC, Iyer VR. Mapping DNA-protein interactions in large genomes by sequence tag analysis of genomic enrichment. Nat Methods. 2005;2:47–53. doi: 10.1038/nmeth726. [DOI] [PubMed] [Google Scholar]
- Kim M-Y, Jeong BC, Lee JH, Kee HJ, Kook H, Kim NS, Kim YH, Kim J-K, Ahn KY, Kim KK. A repressor complex, AP4 transcription factor and geminin, negatively regulates expression of target genes in nonneuronal cells. Proc Natl Acad Sci USA. 2006;103:13074–13079. doi: 10.1073/pnas.0601915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara K, Saito Y, Ogawa E, Takahashi N, Nakagawa Y, Naruse Y, Harada M, Hamanaka I, Izumi T, Miyamoto Y, Kishimoto I, Kawakami R, Nakanishi M, Mori N, Nakao K. The neuron-restrictive silencer element-neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol Cell Biol. 2001;21:2085–2097. doi: 10.1128/MCB.21.6.2085-2097.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara K, Saito Y, Takano M, Arai Y, Yasuno S, Nakagawa Y, Takahashi N, Adachi Y, Takemura G, Horie M, Miyamoto Y, Morisaki T, Kuratomi S, Noma A, Fujiwara H, Yoshimasa Y, Kinoshita H, Kawakami R, Kishimoto I, Nakanishi M, et al. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 2003;22:6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y-H, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interaction and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze S-H, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Leeper-Thompson EC, Garcia ST, Myers RM, Wold B. Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann SB, Seitz R, Gorzella A, Heister A, von Knebel Doeberitz M, Becker C-M. Relaxatin of glycine receptor and onconeural gene transcription control of NRSF deficient small cell lung cancer cell lines. Mol Brain Res. 2004;120:173–181. doi: 10.1016/j.molbrainres.2003.10.021. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Saito Y, Kuwahara K, Harada M, Miyamoto Y, Hamanaka I, Kajiyama N, Takahashi N, Izumi T, Kawakami R, Kishimoto I, Naruse Y, Mori N, Nakao K. Fibronectin signaling stimulates BNP gene transcription by inhibiting neuron-restrictive silencer element-dependent repression. Cardiovasc Res. 2002;53:451–459. doi: 10.1016/s0008-6363(01)00492-8. [DOI] [PubMed] [Google Scholar]
- Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation. Carcinogenesis. 2002;23:1139–1148. doi: 10.1093/carcin/23.7.1139. [DOI] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Saha S, Sparks AB, Rago C, Akmaev B, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE. Using the transcriptome to annotate the genome. Nat Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YM, Greenway DJ, Johnson R, Street M, Belyaev ND, Deuchars J, Bee T, Wilde S, Buckley NJ. Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Mol Biol Cell. 2005;16:5630–5638. doi: 10.1091/mbc.E05-07-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry S, Queires L, Gil-Diez-de-Medina S, Chen MW, de la Taille A, Allory Y, Tran PL, Abbou CC, Buttyan R, Vacherot F. Protocadherin-PC promotes androgen-independent prostate cancer cell growth. Prostate. 2006;66:1100–1113. doi: 10.1002/pros.20446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Gratz GG, Agulnick AD, Heiner W, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Kameoka S, Gopalakrishnan V, Aldape KD, Pan ZZ, Lang FF, Majumder S. Conversion of myoblasts to physiologically active neuronal phenotype. Genes Dev. 2004;18:889–900. doi: 10.1101/gad.1179004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C-L, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Westbrook TF, Martin ES, Schlabach RR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, DePinho RA, Chin L, Elledge SJ. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Wood IC, Belyaev ND, Bruce AW, Jones C, Mistry M, Roopra A, Buckley NJ. Interaction of the repressor element 1-silencing transcription factor (REST) with target genes. J Mol Biol. 2003;334:863–874. doi: 10.1016/j.jmb.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chen MW, Terry S, Vacherot F, Chopin DK, Bemis DL, Kitajewski J, Benson MC, Guo Y, Buttyan R. A human- and male-specific protocadherin that acts through the wnt signaling pathway to induce neuroendocrine transdifferentiation of prostate cancer cells. Cancer Res. 2005;65:5263–5271. doi: 10.1158/0008-5472.CAN-05-0162. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Zhao XD, Lee CWH, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu YT, Weng Z, Kuznetsov VA, Sung W-K, Ruan Y, Dang CV, Wei C-L. Global mapping of c-myc binding sites and target gene network in human B cells. Proc Natl Acad Sci USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]