Abstract
We investigated the ubiquitin-like modification of GABAA receptor-associated protein (GABARAP) and its function. A fusion protein of GABARAP with v5 in the N terminus and myc in the C terminus was expressed in rat cultured hippocampal neurons and PC12 cells. Western blotting with antibodies to v5 and myc revealed that the C terminus of GABARAP was cleaved off. Cleavage was blocked by mutating the C-terminal Gly116 to Ala, suggesting that G116 is required for the processing. Unlike ubiquitin, GABARAP was not incorporated covalently into higher-molecular-weight protein complexes. Nor was GABARAP degraded by ubiquitinylation through the proteasome, although GABARAP formed noncovalent dimers. Immunofluorescent confocal microscopy demonstrated that recombinantly expressed GABARAP was diffusely localized in PC12 cells. However, prevention of C-terminal processing in the mutant GABARAPG116A resulted in redistribution to the Golgi. In neurons, punctate cytoplasmic staining of GABARAP was seen in soma and processes, whereas GABARAPG116A was limited to soma. Compared with wild-type GABARAP, the colocalization and interaction of GABARAPG116A with GABAA receptors were significantly reduced, resulting in a reduction in expression of receptors in the plasma membrane. When α1β2γ2S-containing GABAA receptors were expressed in oocytes, the increased surface expression of GABAA receptors, as shown by increased GABA currents and surface-accessible GABAA receptor subunit polypeptides resulting from GABARAP coexpression, was prevented by mutation G116A. In addition, the distribution of NSF (N-ethylmaleimide-sensitive factor) was affected in GABARAPG116A-expressing neurons. These results suggest that glycine 116 is required for C-terminal processing of GABARAP and that processing is essential for the localization of GABARAP and its functions as a trafficking protein.
Keywords: GABAA receptors, GABARAP, trafficking, ubiquitinylation-like modification, NSF, inhibitory neurotransmission
Introduction
GABAA receptors are ligand-gated ion channels that mediate rapid inhibitory synaptic transmission in the CNS and serve as the target for many important neuroactive drugs (Macdonald and Olsen, 1994; Wallner et al., 2003). However, the molecular mechanisms regulating GABAA receptor trafficking and function are poorly understood. GABAA receptor-associated protein (GABARAP), a 117 aa protein, was first isolated by yeast two-hybrid screening using the intracellular domain of the γ2 subunit of the GABAA receptor as bait (Wang et al., 1999). In QT-6 cells, GABARAP clustered recombinant GABAA receptors, changed channel kinetics, and altered the dose–response curve for GABA (Chen et al., 2000). Furthermore, the single channel conductance was significantly higher in L929 cells coexpressing α1β2γ2S subunits with GABARAP than without (Everitt et al., 2004), showing high conductance states similar to those observed in native receptors (Fatima-Shad and Barry, 1992; Curmi et al., 1993; Birnir et al., 1994, 2001). Recently, we and others reported that overexpression of GABARAP increased the cell-surface number of GABAA receptors (Leil et al., 2004; Boileau et al., 2005; Chen et al., 2005). Immunostaining microscopy showed that GABARAP colocalized with intracellular vesicular GABAA receptors and to some extent with plasma membrane GABAA receptors but not with synaptically localized GABAA receptors and gephyrin (Kneussel et al., 2000; Kittler et al., 2001; Leil et al., 2004). Moreover, GABARAP has been shown to interact with soluble and polymerized tubulin (Wang and Olsen, 2000) and with the ATPase N-ethylmaleimide-sensitive factor (NSF), a chaperone that activates soluble NSF attachment protein receptor (SNARE) proteins in membrane fusion events (Kittler et al., 2001; Leil et al., 2004). This evidence implies that GABARAP might be an important factor regulating the intracellular trafficking of GABAA receptors, despite the fact that loss of GABARAP in knock-out mice does not eliminate synaptic targeting of GABAA receptors in neurons (O'Sullivan et al., 2005).
Nuclear magnetic resonance (NMR) spectroscopy and x-ray crystallography demonstrated that one surface of GABARAP containing a ubiquitin-fold that is conserved in the family of GABARAP (Coyle et al., 2002; Knight et al., 2002; Stangler et al., 2002). Sequence analysis showed that G116 is conserved in all proteins of the GABARAP family: GEC1, GATE-16, LC-3, and yeast Apg8. The homologs of GABARAP can be modified in the C terminus, in a manner resembling ubiquitin (Ichimura et al., 2000; Tanida et al., 2003; Yamazaki-Sato et al., 2003). Ubiquitin, as well as several families of nonubiquitin proteins, such as SUMO (Muller et al., 2001), show an E1–E3 cascade of C-terminal modifications leading to covalent transfer to other proteins (Hochstrasser, 2000). Likewise, the GABARAP family modification first requires a proteolytic cleavage to expose glycine (amino acid 116 in GABARAP), additional activation by E1- and E2-like enzymes, and finally conjugation to other biomolecules, in this case phospholipids. In yeast homolog Apg8, the activated C terminus is attached to a chemically identified lipid phosphatidyl ethanolamine (Ichimura et al., 2000), apparently to promote membrane attachment (Kabeya et al., 2004). This study demonstrated that GABARAP might also conjugate to phospholipids, possibly via the same mechanism. This modification is critical for GABARAP to control intracellular GABAA receptor trafficking.
Materials and Methods
Vector construction.
v5, GABARAP, and GABARAPG116A were amplified by PCR from pcDNA3.1/v5 and pGEX/GABARAP, respectively. The mutation glycine 116 to alanine (GABARAPG116A) was mutagenized using mutant primer in the C terminus. The PCR products were cloned into pCR2.1 according to the manufacturer's instruction. After sequencing, the inserts containing v5, GABARAP, or GABARAPG116A were subcloned into the mammalian expression vector pcDNA 3.1 containing myc as the C-terminal tag. For experiments using Xenopus laevis oocytes, the GABARAP and GABARAPG116A were subcloned into pBluescript II SK vector. The Adeno-X Expression System (Clontech, Mountain View, CA) was used to construct the Ad expression vectors. The v5-GABARAP-myc or v5-GABARAP-myc cDNAs were inserted into pShuttle vector. The expression cassette from the pShuttle was excised and inserted into the Adeno-X viral DNA. Recombinant Adeno-X viral DNA was purified and analyzed by restriction mapping to confirm that it contained the v5-GABARAP-myc or v5-GABARAPG116A-myc. The recombinant adenoviral DNA was digested with PacI and transfected into human embryonic kidney 293 (HEK293) cells by Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Lysates were used to reinfect HEK293 cells for large-scale production.
Cell culture and transfection.
Rat adrenal pheochromocytoma cells (PC12) were maintained in DMEM (Invitrogen), supplemented with 10% horse serum and 5% heat-inactivated fetal bovine serum equilibrated with 5% CO2/95% air at 37°C. PC12 cells with 80% confluence were transfected with pcDNA3.1/v5-GABARAP-myc-his or pcDNA3.1/v5-GABARAP116A-myc-his by Lipofectamine 2000.
Primary neuron culture and infection.
Low-density hippocampal neurons from embryonic day 18 Sprague Dawley rat were prepared by papain-dissociation (Worthington Biochemical, Lakewood, NJ) and cultured in neurobasal medium with B27 serum-free supplement and penicillin and streptomycin (Invitrogen). Briefly, embryos were removed on embryonic day 18 from maternal rats anesthetized with halothane and killed by decapitation. Hippocampi were dissected and placed in Ca2+- and Mg2+-free HEPES-buffered HBSS, pH 7.45. Tissues were dissociated by papain digestion followed by trituration through a 10 ml pipette and papain inhibitor treatment. Cells were pelleted and resuspended in B27 serum-free supplement with penicillin and streptomycin (100 U/ml and 100 μg/ml, respectively). Dissociated cells were then plated at a density of 50,000 cells per well onto 12 mm round coverslips that had been precoated with poly-d-lysine (0.05 mg/ml) and washed with H2O. The neurons were grown in a humidified atmosphere of 5% CO2/95% air at 37°C and fed every 7 d by exchange of 50% of the medium. Seven days after culturing in vitro, cells were infected with adenovirus expressing v5-GABARAP-myc-his or v5-GABARAP116A-myc-his.
Immunostaining and confocal microscopy.
Forty-eight hours after transfection or 4 d after the infection of the virus, the cells on glass coverslips coated with poly-d-lysine were fixed with 3.75% paraformaldehyde in PBS for 15 min at 4°C. For the surface-expressed receptor, cells were washed with PBS and incubated with primary antibody against the N terminus of γ2 (1:100) in 1% BSA for 1 h followed by a 1 h incubation of secondary antibody conjugated with fluorescent dye. For the intracellular proteins, cells were permeabilized with 0.02% Triton X-100 for 5 min followed by the primary-secondary antibody incubation (anti-γ2, 1:100; anti-v5, 1:200; anti-NSF, 1:200; secondary antibody, 1:400). For the dual-fluorescence staining with germ agglutinin and 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI), cells were incubated for another 15 min at room temperature. After mounting the coverslip on glass slides, cells were analyzed by Leica TCS SP MP inverted confocal microscopy. Z-stacks of the images were collected with an optical thickness of 1 μm and analyzed by MetaVue (Universal Imaging, Downingtown, PA) and ImagePro Plus software (Media Cybernetics, Silver Spring, MD).
Coimmunoprecipitation and Western blot.
Forty-eight hours after the transfection or 4 d after infection, whole-cell protein was lysed by radioimmunoprecipitation assay (RIPA) buffer [150 mm NaCl, 1% NP-40, 0.5% sodium deoxycholate, 50 mm Tris and proteinase inhibitor mixture (1 mm phenylmethylsulfonyl fluoride, 20 μg/ml aprotinin, 20 μg/ml leupeptin)]. After incubation on ice for 0.5 h, cell suspensions were centrifuged at 15,000 × g for 15 min at 4°C. The immunocomplexes were recovered with the aid of Protein G agarose (Sigma, St. Louis, MO). Proteins in cell lysates and the immunocomplexes were loaded by SDS-PAGE and transferred to polyvinylidene difluoride membrane by a semidry method. The membrane was then incubated in PBS buffer and 0.05% Tween with 5% nonfat milk and primary antibody at room temperature for 1 h. After incubation with secondary antibody for 1 h, the membrane was visualized by enhanced chemiluminescence (GE Healthcare, Piscataway, NJ). The blot was scanned and analyzed with Quantity One software (Bio-Rad, Hercules, CA).
Xenopus laevis oocyte expression system.
Capped mRNA (cRNA) was synthesized by in vitro transcription from Apa I-linearized cDNA constructs using the mMessage Machine kit (Ambion, Austin, TX) as described previously (Chang et al., 2003; Chen et al., 2005). Oocytes were injected with a total volume of 50 nl of cRNA mixed in a ratio of 1:1:2:4 (α1:β2:γ2: H2O, GABARAP or mutant GABARAP). Oocytes were maintained in six-well plates at 17–19°C in SOS solution (in mm: 100 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, and 5 HEPES), supplemented with 50 μg/ml gentamycin and 100 μg/ml streptomycin and penicillin, and were used for electrophysiological experiments 3–5 d after injection.
Two-electrode voltage-clamp analysis.
Oocytes under a two-electrode voltage clamp (holding potential −70 mV) were gravity perfused continuously with ND96 recording solution containing (in mm) 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, and 5 HEPES, pH 7.4, at a rate of ∼5 ml/min. In general, drugs and reagents were dissolved in ND96. A standard two-electrode voltage-clamp recording was performed using an Axoclamp-2A amplifier (Molecular Devices, Union City, CA) interfaced to a computer with a DigiData 1322-A device (Molecular Devices). Electrodes were filled with 3 m KCl and had a resistance of 0.5–1.5 MΩ. Data acquisition and analyses were performed using pClamp 8.2 (Molecular Devices) and Prism software (GraphPad Software, San Diego, CA).
Cell-surface biotinylation assay.
Surface-expressed proteins were biotinylated by the membrane impermeable EZ-link sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate (sulfo-NHS-SS-biotin; Pierce, Rockford, IL). Four days after injection, oocytes (10 oocytes per group) were washed with ND96 solution three times, incubated with 1 mg/ml sulfo-NHS-SS-biotin at room temperature for 30 min, and then washed with 25 mm Tris, pH 8.0, followed by two washes with PBS. Oocytes were homogenized in 200 μl of RIPA buffer (150 mm NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mm Tris, pH 7.5). The yolk and cellular debris were removed after three centrifugations at 3600 × g for 10 min. A 20 μl aliquot was taken and mixed with 2× SDS loading buffer to detect total proteins. The remaining supernatant was incubated with streptavidin beads at 4°C overnight. The eluted proteins and whole proteins were detected by Western blot using anti-γ2.
Results
GABARAP shows C-terminal cleavage prevented by mutation of glycine 116 to alanine
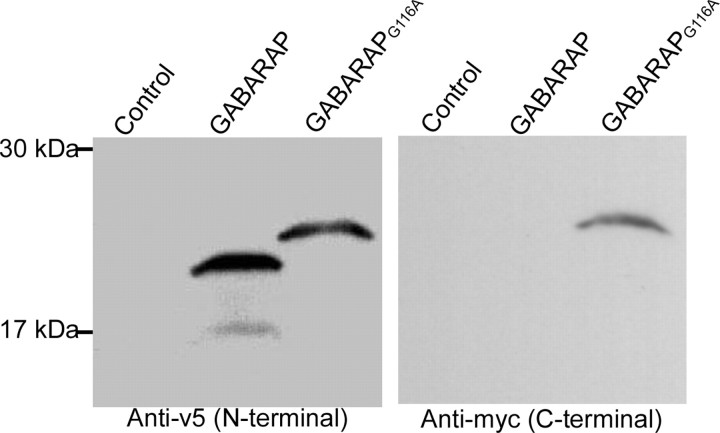
To examine the role of Gly116 in the C-terminal modification of GABARAP, a mammalian expression vector containing either GABARAP or mutant GABARAPG116A tagged by v5 in the N terminus and myc in the C terminus, was transfected into PC12 cells. Forty-eight hours after the transfection, whole-cell protein was extracted and loaded on SDS-PAGE gels followed by immunostaining with anti-v5 and anti-myc antibody. In wild-type GABARAP-transfected cells (Fig. 1), a 22 kDa band was detected by the anti-v5 antibody. However, there was no band visible with anti-myc staining, indicating that the C-terminal myc was completely cleaved off from GABARAP. In addition, other lower-molecular-weight (Mr) bands were also visible with anti-v5 staining, suggesting additional modification of GABARAP similar to Apg8, possibly a lipid adduct conjugation. In mutant GABARAPG116A-transfected cells, a larger band (24 kDa) is seen, which is stained by both anti-v5 and anti-myc staining, indicating that the G116A mutation was not modified on the C terminus, and mutation of Gly116 to Ala prevents C-terminal cleavage of GABARAP.
Figure 1.
Mutation of Gly116 of GABARAP abolishes C-terminal processing. Whole-cell proteins from PC12 cells transfected with v5-GABARAP-myc or v5-GABARAPG116A-myc were extracted and detected by anti-v5 and anti-myc in a Western blot. A 22 kDa band was detected only by anti-v5 in wild-type GABARAP-treated neurons (lane 2 in each gel). In addition, another two smaller-Mr bands were also visible, indicating some additional modification, such as lipidation. In mutant GABARAPG116A-treated cells, a 24 kDa band was found with both anti-v5 and anti-myc staining (lane 3 in each gel), indicating that the C-terminal tag was cleaved off in wild-type GABARAP but not in the mutant.
GABARAP itself does not undergo ubiquitinylation or transfer to other proteins
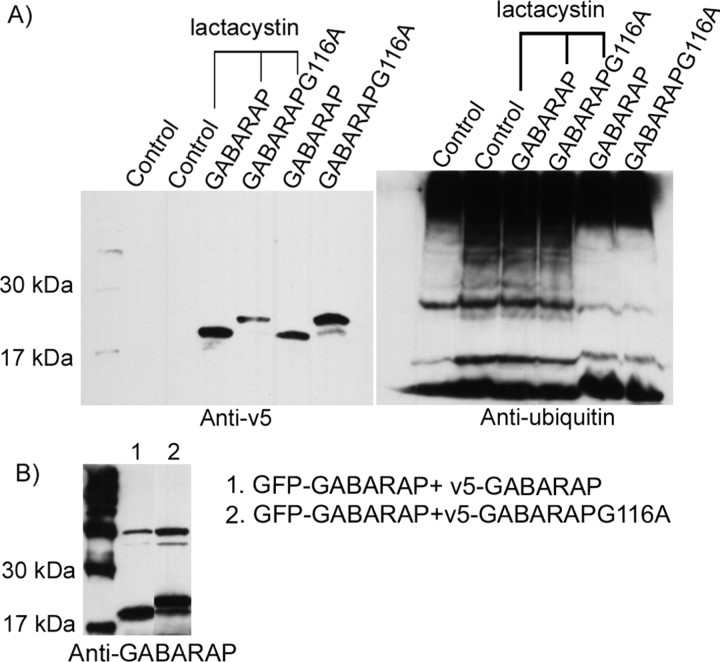
The C-terminal modification of GABARAP is similar to the initial steps in ubiquitinylation. To determine whether GABARAP itself undergoes ubiquitinylation, neurons infected with GABARAP or GABARAPG116A were treated with 5 μm lactacystin, a proteasome inhibitor, for 12 h. Western blots with anti-v5 antibody from three independent experiments did not reveal any change in the molecular weight or quantity of either wild-type or mutant GABARAP after lactacystin treatment. In addition, densitometry analysis showed no difference between wild-type GABARAP and mutant GABARAPG116A before or after lactacystin treatment (Fig. 2A, left). This suggests that GABARAP is not covalently attached to other proteins, and it is not degraded through the proteasome. The same blot was stripped to remove anti-v5 and stained by anti-ubiquitin. Anti-ubiquitin recognized ubiquitin (8.5 kDa), its polymers (17 and 25 kDa), and ubiquitinylated proteins (higher-Mr proteins) in the groups without lactacystin treatment. After lactacystin treatment, the quantity of ubiquitin and polyubiquitinylated proteins were increased (Fig. 2A, right). A band indicating ubiquitin coupling to GABARAP was not detected, suggesting that GABARAP does not undergo the ubiquitinylation process.
Figure 2.
A, GABARAP is not degraded by ubiquitinylation through the proteasome. Four days after infection of v5-GABARAP-myc or v5-GABARAPG116A-myc, hippocampal neurons were treated with 5 μm lactacystin for 12 h. Western blot with anti-v5 demonstrated that, unlike ubiquitin, which showed accumulated Mr bands after lactacystin treatment, neither GABARAP nor GABARAPG116A showed any alteration in molecular weight or quantity after lactacystin treatment. B, GABARAP formed dimers or higher-order polymers in PC12 cells. v5-GABARAP-myc or v5-GABARAPG116A-myc was cotransfected with GFP–GABARAP into PC12 cells for 48 h. Anti-v5-protein G-agarose immunoprecipitated the whole-cell proteins followed by anti-GABARAP staining in a Western blot. A 40 kDa band, corresponding to GFP–GABARAP, was detected along with a 22 or 24 kDa band equivalent to v5-GABARAP-myc and v5-GABARAPG116A-myc, respectively, indicating the formation of homodimers or polymers.
GABARAP forms noncovalent dimers or higher-order polymers
Previous studies have suggested that GABARAP may be capable of dimerization. To identify whether GABARAP forms dimers in cells, green fluorescent protein (GFP)-GABARAP and v5-GABARAP or v5-GABARAPG116A were cotransfected into PC12 cells. Forty-eight hours after transfection, whole-cell proteins were extracted in RIPA buffer and recovered by anti-v5 and Protein G agarose. The immunocomplexes were loaded on SDS-PAGE gels followed by anti-GABARAP staining. v5-GABARAP or v5-GABARAPG116A was detected at 22 and 24 kDa, respectively. An additional 40 kDa band corresponding to the Mr of GFP–GABARAP was also seen in both groups, indicating that GFP–GABARAP was coimmunoprecipitated with both wild-type GABARAP and GABARAPG116A. Anti-GFP staining confirmed that the 40 kDa band contained GFP-fusion protein (Fig. 2B).
Disruption of the modification by mutation of Gly116 to Ala changes the subcellular localization of GABARAP in PC12 cells
To address the effect of C-terminal modification on the subcellular localization of GABARAP, we examined the distribution of GABARAP and GABARAPG116A by staining with anti-v5 and anti-myc 48 h after transfection in PC12 cells (Fig. 3A). Whereas v5-immunoreactivity was evident in both wild-type GABARAP and GABARAPG116A-positive cells, myc immunoreactivity was detected only in GABARAPG116A-positive cells, further confirming our Western blot results that the C terminus of GABARAP is posttranslationally cleaved off. GABARAP immunoreactivity was detected ubiquitously in PC12 cells; however, GABARAPG116A immunoreactivity was localized peripherally near the cytoplasmic membrane. To further characterize the subcellular localization of GABARAPG116A, we performed triple-labeling experiments with markers for the Golgi apparatus [wheat germ agglutinin (WGA)] and the nucleus (DAPI). Compared with wild-type GABARAP that was found in all parts of the cell, distinct from and within the Golgi and nucleus, GABARAPG116A only colocalized with the Golgi marker WGA, not DAPI. In addition, the distribution of the Golgi apparatus was changed by the mutation (Fig. 3B), suggesting that GABARAP is important for maintaining the normal distribution and function of the Golgi apparatus. It also implies that mutation of Gly116 prevents trafficking of GABARAP through the Golgi.
Figure 3.
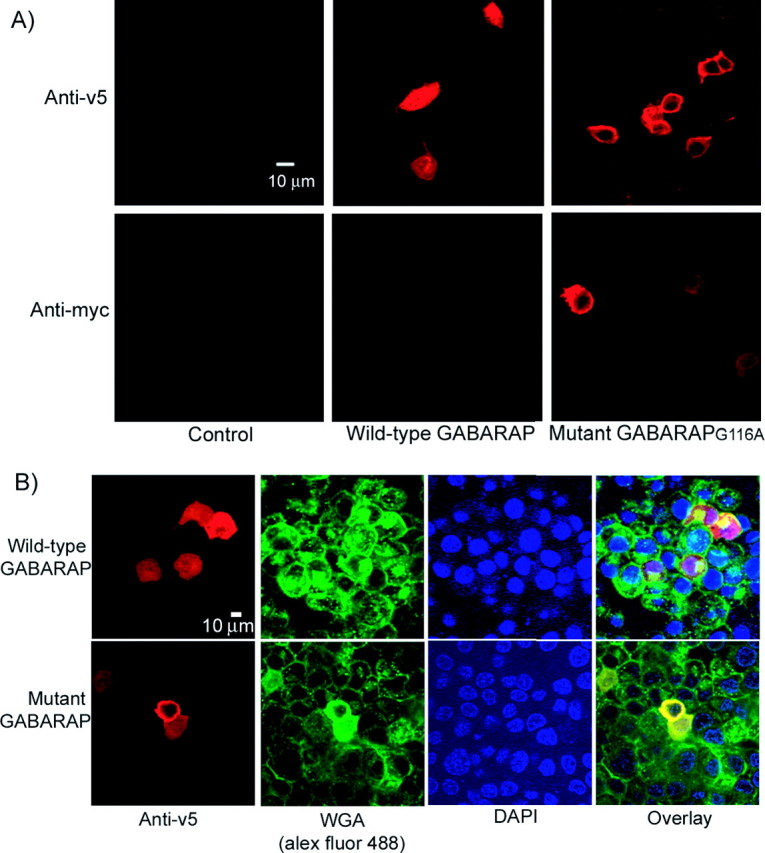
Disruption of the modification by mutation of Gly116 to Ala changes the subcellular localization of GABARAP in PC12 cells. A, Three days after transfection, PC12 cells were fixed, permeabilized, and then stained with anti-v5 and anti-myc, respectively. Mutant GABARAPG116A was detected by both anti-v5 and anti-myc immunostaining, whereas wild-type GABARAP could only be detected with the anti-v5 antibody. Wild-type GABARAP was ubiquitously located in the cytoplasm; however, mutant GABARAPG116A was limited in the peripheral part of the cells. B, Triple-immunofluorescent staining was performed in permeabilized PC12 cells with anti-v5 (red), WGA Alexa Fluor 488 (the marker of Golgi; green), and DAPI (the marker of nuclei; blue). The merged image showed that wild-type GABARAP localized ubiquitously in the cell, whereas mutant GABARAPG116A colocalized primarily with WGA.
Mutation of Gly116 to Ala changes the subcellular localization of GABARAP and GABAA receptors and reduces the interaction between GABAA receptors and GABARAP in hippocampal neurons
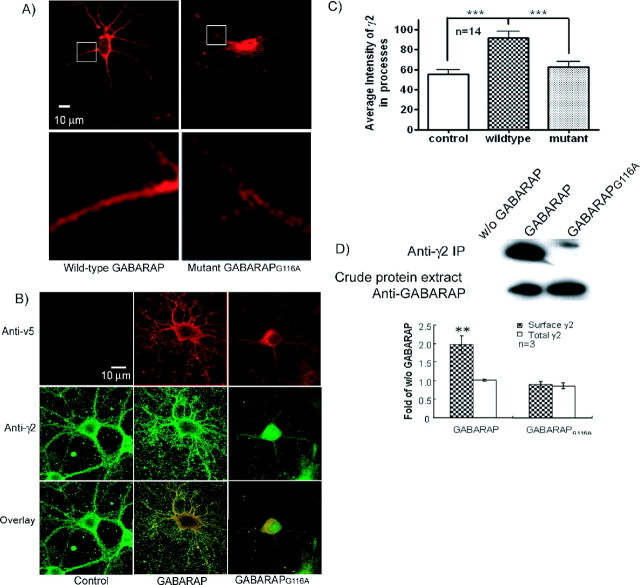
To investigate the effect of the Gly116 mutation of GABARAP on native GABAA receptors, an adenoviral vector containing either GABARAP or GABARAPG116A was used to infect low-density cultured hippocampal neurons (7 d after in vitro culture). In agreement with the findings reported in PC12 cells, Western blotting of extracts from these neurons showed that C-terminal processing only occurs in wild-type GABARAP and was abolished in GABARAPG116A (data not shown). Anti-v5 staining of infected permeabilized neurons revealed punctate staining of GABARAP in the cell soma and processes (Fig. 4A, left). In contrast, GABARAPG116A was localized primarily to the soma, a pattern similar to that observed in PC12 cells (Fig. 4A, right). Image analysis of the neuronal processes indicated that the intensity in the neuronal processes of wild-type GABARAP (95.58 ± 8.92; n = 16) is significantly higher (p < 0.01) than that of mutant GABARAPG116A (62.4 ± 5.57; n = 14), whereas the intensity of the v5 immunoreactivity of whole cells has no difference between these two groups.
Figure 4.
Mutation of Gly116 to Ala changes the subcellular localization of GABARAP and GABAA receptors and reduces the interaction between GABAA receptor and GABARAP in hippocampal neurons. A, Low-density cultures of rat hippocampal neurons were infected by adenoviral v5-GABARAP-myc and v5-GABARAPG116A-myc. Four days after infection, neurons (11 d) were stained with anti-v5 to show the distribution of GABARAP. Wild-type GABARAP localized ubiquitously throughout neurons, including both cell body and processes; however, most of the mutant GABARAPG116A localized merely in the cell body. Image analysis of neuronal processes demonstrated that the intensity of GABARAP in neuronal processes is higher than that of mutant GABARAPG116A. B, Confocal images of permeabilized primary cultured hippocampal neurons showed colocalization of GABARAP (red) and γ2 subunits (green). GABARAPG116A has reduced colocalization with GABAA receptors compared with wild-type GABARAP as indicated by Pearson correlation (p < 0.001). C, Overexpression of GABARAP increased the intensity of γ2 subunit in neuronal processes compared with the noninfected neurons. Mutant GABARAPG116A showed no effect on the γ2 subunits. D, Mutation of Gly116 decreased the interaction of GABARAP with the γ2 subunit of the receptor. Top, Whole-cell proteins extracted 4 d after infection with GABARAP or GABARAPG116A were immunoprecipitated by polyclonal anti-γ2 and protein G-agarose followed by Western blot using anti-v5. Anti-γ2 pulled down more wild-type GABARAP than GABARAPG116A. Bottom, Western blot with anti-v5 showed the same level of protein expression of wild-type GABARAP and GABARAPG116A. Error bars represent SEM.
The effect of the Gly116 mutation of GABARAP on its colocalization with native GABAA receptors was examined by immunostaining with anti-v5 and anti-γ2 antibodies in permeabilized cultured hippocampal neurons (Fig. 4B). In GABARAP-infected neurons, GABAA receptors were located in both cell body and processes, with the punctate staining located mainly in the processes. Compared with wild-type GABARAP-positive neurons, more GABAA receptors were localized in the cell body rather than processes in mutant GABARAPG116A-infected neurons. Image analysis indicated that overexpression of wild-type GABARAP increased the intensity of GABAA receptors in the neuronal processes compared with noninfected neurons (p < 0.001). The mutant GABARAPG116A did not demonstrate this enhancement effect. The total cell γ2 content shows no difference among these three groups (Fig. 4C). GABARAP was colocalized highly with γ2-containing GABAA receptors, chiefly in the base and branches of processes, but a greater fraction of the GABAA receptors were located in the soma with mutant GABARAPG116A. Pearson's correlation indicated that wild-type GABARAP colocalized more extensively with GABAA receptors than did the mutant GABARAPG116A (0.714 ± 0.0156 and 0.334 ± 0.0255, respectively; p < 0.001; n = 36–42).
To determine whether mutation of Gly116 directly affects the interaction of GABARAP with GABAA receptors, coimmunoprecipitation using anti-γ2 antibody was used. Both wild-type and mutant GABARAP were pulled down along with the γ2 subunit, although the quantity of GABARAPG116A pulled down was considerably lower, indicating that the degree of interaction between γ2 subunits and GABARAPG116A in cells was weaker than that between γ2 subunits and wild-type GABARAP (Fig. 4D). This result is consistent with the confocal analysis, which showed that mutation of GABARAP Gly116 reduced colocalization with the γ2 subunit-containing GABAA receptors.
Mutation in GABARAPG116A abolishes the GABARAP enhancement of cell-surface GABAA receptors
Intact, fixed hippocampal neurons transfected with wild-type or mutant GABARAP were stained with anti-γ2 (nonpermeabilized) and anti-v5 antibody (after permeabilization). GABAA receptors containing γ2 subunits were found in clusters at the plasma membrane, a fraction of which were colocalized with wild-type GABARAP (Fig. 5A). GABARAPG116A displayed dramatically reduced colocalization with surface clusters of these GABAA receptors (Fig. 5A). The surface expression of the γ2 subunit was significantly increased by overexpression of GABARAP in the hippocampal neurons (p < 0.05) but not by overexpression of GABARAPG116A (Fig. 5B). This suggests that GABARAP, but not GABARAPG116A, is capable of trafficking GABAA receptors to the plasma membrane, as we reported previously for wild-type GABARAP (Leil et al., 2004).
Figure 5.
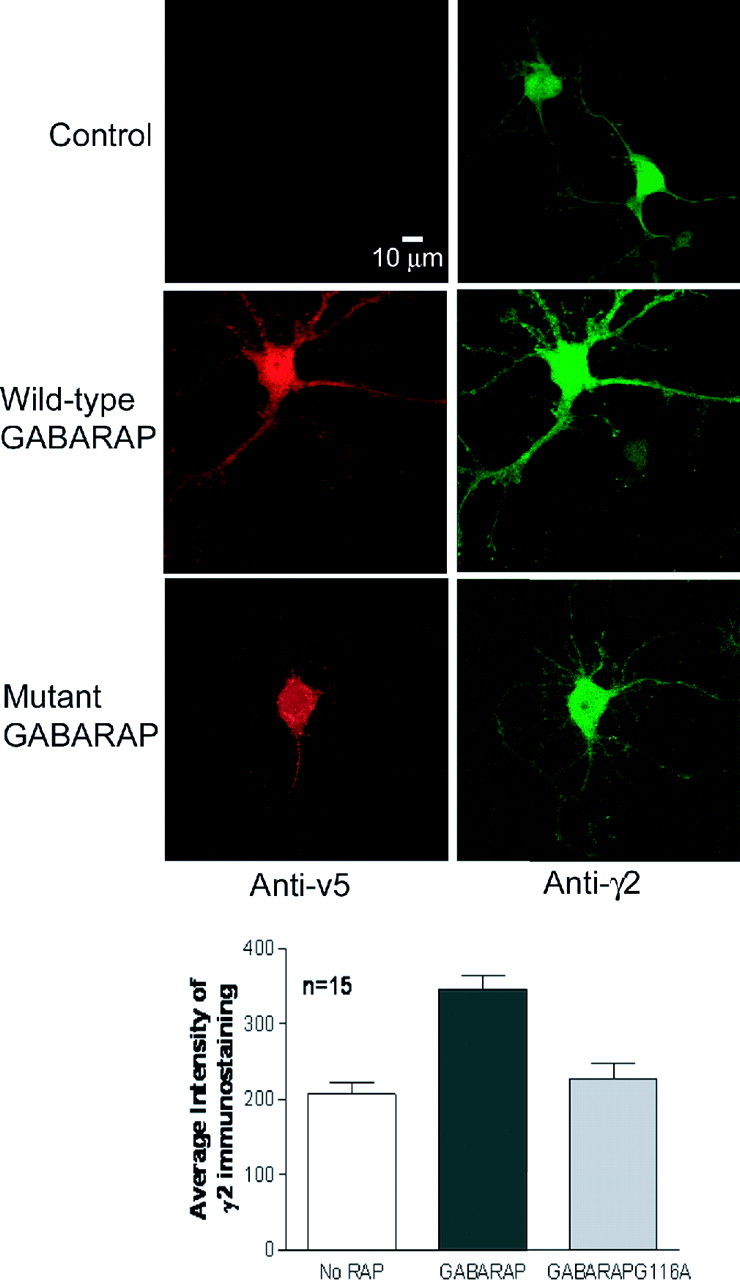
Mutation in GABARAPG116A abolished the GABARAP enhancement of cell-surface GABAA receptors. A, Confocal image of surface-expressed γ2 subunits (green; nonpermeabilized) in cultured hippocampal neurons overexpressing wild-type GABARAP or GABARAPG116A (red anti-v5 staining; permeabilized). B, Image analysis demonstrated that neurons transfected with wild-type GABARAP show significantly increased levels of γ2 immunostaining at the cell surface (p < 0.05), whereas those transfected with GABARAPG116A do not. Error bars represent SEM.
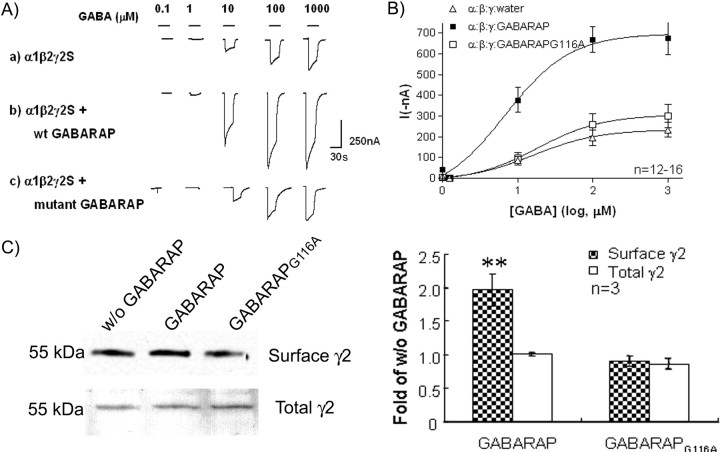
Mutation in GABARAPG116A blocks stimulation of GABA receptor channel expression in X. laevis oocytes
We have reported previously that GABARAP increased both GABA currents and cell-surface content of GABAA receptor polypeptides in X. laevis oocytes (Chen et al., 2005). The effects of the Gly116 mutation of GABARAP were studied in this system. As shown in Figure 6, A and B, in agreement with previous results, coexpression of GABARAP increased the GABA currents by twofold to threefold compared with currents obtained from oocytes in which GABARAP was not coexpressed. The maximum currents (in nanoamperes) in the GABARAP-expressing group were 697.8 ± 84.3 (n = 16). However, coexpression of mutant GABARAPG116A and GABAA subunits resulted in GABA-evoked currents of ∼306.5 ± 67.8 nA (n = 12) that were similar to those derived from cells expressing GABAA receptor subunits alone but significantly less than currents obtained from cells coexpressing wild-type GABARAP. The concentration–response curve for GABA revealed that the change was mainly caused by a decrease in the maximum response, because there is little change in the EC50. Measurement of surface GABAA receptors by cell-surface biotinylation demonstrated that wild-type GABARAP was able to increase the level of the γ2 subunit at the cell surface, whereas GABARAPG116A was not (Fig. 6C). This suggests that C-terminal processing of GABARAP is required for trafficking of GABAA receptors.
Figure 6.
Mutation in GABARAPG116A blocks stimulation of GABAA receptor channel surface expression. A, B, Two-electrode voltage clamp recording showed that GABARAPG116A eliminated the enhancement effect on GABA current induced in oocytes. A, Example of GABA-induced currents in oocytes expressing α1β2γ2S subunits with water, GABARAP, or GABARAPG116A in a ratio of 1:1:2:4. B, GABA dose–response curve. In Xenopus oocytes expressing α1β2γ2S subunits, GABARAP increased the maximal evoked GABA currents (without affecting EC50), whereas GABARAPG116A did not. The maximum chloride currents elicited by 0.1–1000 μm GABA in oocytes coexpressed with α1β2γ2S GABAA subunits and GABARAPG116A were not statistically different (p < 0.001) from currents obtained from oocytes expressing α1β2γ2S subunits alone. C, Surface biotinylation assay demonstrated that GABARAP enhanced the cell-surface expression of GABAA receptor in oocytes. GABARAPG116A blocked the enhancement (as shown in the top lanes). However, GABARAP has no effect on the biosynthesis of GABAA receptor γ2 subunit. Error bars represent SEM.
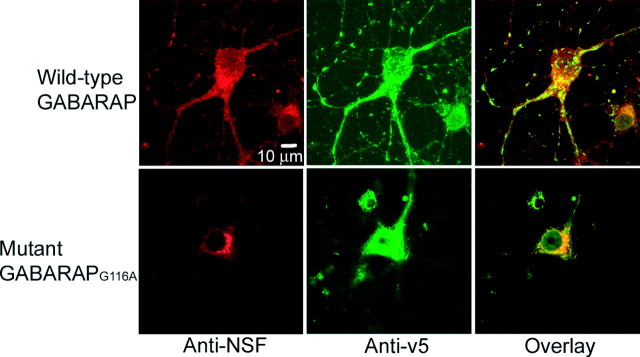
Mutation of Gly116 of GABARAP disrupts subcellular localization of NSF
To examine the effects of mutant GABARAPG116A on NSF, permeabilized cultured hippocampal neurons were labeled with polyclonal goat anti-v5 and monoclonal anti-NSF followed by secondary antibody incubation. In three experiments, NSF was evident in both the cell body and processes of GABARAP-infected neurons and showed a high degree of colocalization with GABARAP. This is illustrated for one cell in Figure 7. However, in neurons that were infected with GABARAPG116A (n = 7), NSF was found to be redistributed mainly to the cell body, as was GABARAP, with the two proteins still colocalized. Image analysis by Pearson's correlation indicated no significant difference between GABARAP versus GABARAPG116A colocalization with NSF.
Figure 7.
Mutation of Gly116 of GABARAP disrupts subcellular localization of NSF. Permeabilized cultured hippocampal neurons (11 d) were stained by anti-v5 (green) and anti-NSF (red) 4 d after infection. In GABARAPG116A-positive neurons, the distribution of NSF was disrupted.
Discussion
The presence of the major ubiquitin fold in GABARAP suggests that it may undergo processing in a manner similar to ubiquitin. Our results verify that GABARAP does undergo C-terminal processing, which is blocked by mutation of glycine 116 to alanine (Fig. 1). These results are in agreement with those found in starved HEK cells in which GABARAP, GEC1, GATE-16, and LC-3 were modified in the C terminus (Tanida et al., 2003). However, unlike ubiquitinylation, in which ubiquitin is covalently attached to other proteins, GABARAP does not appear to be incorporated covalently into higher-Mr protein complexes either singly or in multiple copies (Fig. 2A). GABARAP itself is not degraded via ubiquitin-mediated proteolysis because inhibition of proteasome activity did not result in accumulation of high-Mr GABARAP protein complexes. GABARAP does form a noncovalent dimer (Nymann-Andersen et al., 2002) and the C-terminal mutation did not interfere with the dimerization (Fig. 2B). However, we could not exclude the possibility that GABARAP might also form noncovalent polymers.
GABARAP is related in both amino acid sequence and three-dimensional structure to homologous proteins from yeast (Apg8 or AUT7P) and mammals [GEC1, GATE-16, and light chain 3 (LC3) of microtubule-associated protein]. This posttranslational processing of the C terminus is conserved in all of the proteins of the GABARAP family (Ichimura et al., 2000; Kabeya et al., 2000; Tanida et al., 2003). Yeast Apg8 was the first of the GABARAP family shown to undergo this modification. During the modification, the last amino acid in the C terminus is first cleaved by the activating cysteine protease Apg4, leaving glycine116 exposed (Ichimura et al., 2000). This penultimate glycine, which is conserved in all proteins in this family, is activated by Apg7 and transferred to Apg3, which are E1 and E2 enzymes, respectively (Komatsu et al., 2001; Yamazaki-Sato et al., 2003). Finally, it is conjugated to the membrane lipid phosphatidyl ethanolamine (PE) (Ichimura et al., 2000), catalyzed by an as yet unidentified E3. The lipid PE can also be removed by the priming protease Apg4, releasing the C-terminal glycine that is then exposed for a new processing cycle (Hemelaar et al., 2003). The human homologs of Apg4, Apg7, and Apg3 were recently cloned and their enzymatic activities on GABARAP and its homologs were verified (Tanida et al., 2001, 2002, 2004; Hemelaar et al., 2003; Scherz-Shouval et al., 2003; Yamazaki-Sato et al., 2003). Yeast Apg8 was shown to give two bands by SDS-PAGE, the unmodified protein with apparent Mr of 14 kDa and the lipid adduct at slightly lower apparent Mr (12 kDa). This latter band was more enriched in the membrane fraction of cell homogenates. A similar lower-Mr band (Fig. 1, left, lane 2) with hydrophobic properties, presumably a lipid adduct, was also observed for the GABARAP family (Tanida et al., 2003; Kabeya et al., 2004). However, no E3 enzyme for this modification has been identified. E3 enzymes are substrate specific and thus assessment of specific product formation is required for identification of the specific E3.
Conjugation of the yeast homolog of GABARAP, Apg8, with PE results in its tight association with the autophagosome vesicle membrane (Ichimura et al., 2000). Many cytosolic proteins involved in receptor trafficking or docking associate with membranes via lipophilic substitutents. For example, membrane localization and clustering of glutamate receptors and other ion channels by the synaptic PDZ [postsynaptic density-95 (PSD-95)/Discs large/zona occludens-1]-containing protein PSD-95 depends on palmitoylation (El-Husseini et al., 2000). Palmitoylation of ABP (AMPA receptor-binding protein) targets it to the membrane and allows it to associate with GluR2 (glutamate receptor 2) at the plasma membrane in spines of cultured hippocampal neurons (DeSouza et al., 2002). Lipidation of PICK1 (protein interacting with C kinase 1) allows it to traffic AMPA receptors to synapses (Jin et al., 2006). Subcellular fractionation studies showed that GABARAP is associated with a membrane compartment in HEK cells (Tanida et al., 2003), suggesting that GABARAP might conjugate with a lipid to increase its affinity for membranes. Like the lipid adduct of yeast Apg8, which runs at slightly lower apparent Mr (Ichimura et al., 2000; Kabeya et al., 2004), it is possible that the smaller protein observed in our Western blots probed with anti-v5 is the lipidated form of GABARAP (Fig. 1). Disruption of the C-terminal processing by mutation of glycine 116 blocked GABARAP lipidation, thereby preventing GABARAP from being transported out of the trans-Golgi (Fig. 2B,C).
GABARAP was shown previously to increase the cell-surface expression of GABAA receptors (Leil et al., 2004; Boileau et al., 2005; Chen et al., 2005). Applying GABARAP IgG from Stiff-Person Syndrome patients decreased the surface GABAA receptors in cultured hippocampal neurons (Raju et al., 2006). Here, we provide additional evidence that GABARAP enhances the surface expression of GABAA receptors, suggesting that GABARAP facilitates the intracellular trafficking of GABAA receptors. GABARAP has been shown to interact with some trafficking factors, such as microtubules, NSF, and transferrin receptors. In addition, GATE-16 has been shown to enhance the ATPase activity of NSF and is believed to be a component of the intra-Golgi transport machinery (Legesse-Miller et al., 1998; Sagiv et al., 2000; Muller et al., 2002). In this study, blocking the processing of GABARAP by G116A interferes with its intracellular trafficking and results in its subcellular redistribution. At the same time, the subcellular localization of GABAA receptors was also changed, further supporting the conclusion that GABARAP participates in intracellular trafficking of GABAA receptors.
Control of neurotransmitter receptor expression at the postsynaptic membrane is of critical importance for functional neurotransmission. Sorting, targeting, clustering, and degradation of receptors as dynamic processes play a key role in the construction and functional maintenance of synapses. It is well established that the sorting of newly synthesized membrane proteins to distinct domains in polarized cells appears to begin in the Golgi/trans-Golgi network (TGN), where proteins can segregate and exit in separate transport vesicles (Keller et al., 2001). GABARAPG116A showed a reduced intracellular colocalization pattern with GABAA receptors (Fig. 4). Immunoprecipitation results indicated that anti-γ2 pulls down less mutant GABARAPG116A than wild-type GABARAP, confirming that the interaction between GABARAPG116A and γ2-containing GABAA receptors in cells was decreased. The unmatched subcellular distribution of GABAA receptor and GABARAPG116A suggests that there are some other factors regulating the intracellular trafficking of GABAA receptors. Recent studies using the yeast two-hybrid system identified some new GABAA receptor-associated proteins (Chen and Olsen, 2007), such as the GABARAP homolog GEC1 (Mansuy et al., 2004), Golgi-specific DHHC zinc finger protein (GODZ) (Keller et al., 2004), Plic-1 (Bedford et al., 2001), and HAP (Huntingtin-associated protein) (Kittler et al., 2004). These proteins can also traffic GABAA receptors, through binding other subunits of the pentamer, out of the TGN to a final destination. Because there is no apparent change in synaptic expression of GABAA receptors in cortex of GABARAP knock-out mice (O'Sullivan et al., 2005), this also suggests that other GABAA receptor-associated proteins might substitute for, or supplement, the function of GABARAP. Coexpression of mutant GABARAPG116A in both neurons and oocytes expressing α1β2γ2S subunits showed less cell-surface-expressed GABAA receptor. This loss of receptor surface expression may have been attributable to the abolition of the interaction between GABARAPG116A and intracellular transportation tubulovesicular membranes containing GABAA receptors, resulting in a reduction in the intracellular trafficking of GABAA receptor to the cell surface. In addition to the enhancement of the surface expression of the GABAA receptors by GABARAP in oocytes (Fig. 6), GABARAP may also change the kinetics as reported by Chen et al. (2000) and Everitt et al. (2004). However, oocytes studied with the two-electrode voltage clamp are not technically appropriate for kinetics studies. Oocytes may lack clustering factors, such as gephyrin, needed to demonstrate the kinetics changes as observed in QT-6 cells and L929 cells.
Another possible conclusion is that GABARAP affects the distribution of GABAA receptor through NSF. In the study of colocalization between GABARAP and NSF, we observed that the subcellular localization of NSF was disturbed by the mutant GABARAPG116A. NSF has been shown to participate in the fusion of transport vesicles with the membrane of the acceptor or target compartment. In this fusion process, v-SNARE, a specific protein expressed in the intracellular vesicle, binds to a receptor on the target membrane (t-SNARE). NSF is recruited to this complex via soluble NSF attachment proteins (SNAPs) that can bind directly to the SNARE proteins and stimulate the ATPase activity of NSF. In consequence, the SNARE complexes dissociate after membrane fusion, allowing the individual SNARE proteins to be recycled for subsequent rounds of fusion (Rothman, 1994). By controlling the localization of NSF, the modification of GABARAP might also affect the function of NSF. NSF functions as the controller of the fusion of post-Golgi transport vesicles or fusion of intracellular vesicles with plasma membranes. In other words, NSF and GABARAP together may control either the intracellular trafficking and/or the membrane insertion of GABAA receptors.
In conclusion, we provide evidence that GABARAP is associated with the intracellular tubulovesicular transport pathway through its lipid-modified C terminus. At the same time, it also interacts with γ subunits of GABAA receptors located in vesicles and transported along microtubules to cytoplasmic membranes. The mutant GABARAPG116A cannot associate with trafficking vesicles in the TGN because of the lack of a hydrophobic C-tail, thereby losing its ability to traffic GABAA receptors. As a consequence, the number of GABAA receptors reaching specific target destinations at the plasma membrane was reduced in GABARAPG116A-expressing neurons.
Footnotes
This work was supported by funds provided by the state of California for medical research on alcohol and substance abuse (to R.W.O.). We thank Dr. Xin Liu for assisting the image analysis and Dr. Andrea Everitt for discussing this manuscript.
References
- Bedford FK, Kittler JT, Muller E, Thomas P, Uren JM, Merlo D, Wisden W, Triller A, Smart TG, Moss SJ. GABA(A) receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat Neurosci. 2001;4:908–916. doi: 10.1038/nn0901-908. [DOI] [PubMed] [Google Scholar]
- Birnir B, Everitt AB, Gage PW. Characteristics of GABAA channels in rat dentate gyrus. J Membr Biol. 1994;142:93–102. doi: 10.1007/BF00233386. [DOI] [PubMed] [Google Scholar]
- Birnir B, Eghbali M, Cox GB, Gage PW. GABA concentration sets the conductance of delayed GABAA channels in outside-out patches from rat hippocampal neurons. J Membr Biol. 2001;181:171–183. doi: 10.1007/s00232-001-0021-5. [DOI] [PubMed] [Google Scholar]
- Boileau AJ, Pearce RA, Czajkowski C. Tandem subunits effectively constrain GABAA receptor stoichiometry and recapitulate receptor kinetics but are insensitive to GABAA receptor-associated protein. J Neurosci. 2005;25:11219–11230. doi: 10.1523/JNEUROSCI.3751-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Olcese R, Olsen RW. A single M1 residue in the beta2 subunit alters channel gating of GABAA receptor in anesthetic modulation and direct activation. J Biol Chem. 2003;278:42821–42828. doi: 10.1074/jbc.M306978200. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang H, Vicini S, Olsen RW. The gamma-aminobutyric acid type A (GABAA) receptor-associated protein (GABARAP) promotes GABAA receptor clustering and modulates the channel kinetics. Proc Natl Acad Sci USA. 2000;97:11557–11562. doi: 10.1073/pnas.190133497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZW, Olsen RW. GABAA receptor associated proteins: a key factor regulating GABAA receptor function. J Neurochem. 2007;100:279–294. doi: 10.1111/j.1471-4159.2006.04206.x. [DOI] [PubMed] [Google Scholar]
- Chen ZW, Chang CS, Leil TA, Olcese R, Olsen RW. GABAA receptor-associated protein regulates GABAA receptor cell-surface number in Xenopus laevis oocytes. Mol Pharmacol. 2005;68:152–159. doi: 10.1124/mol.104.009878. [DOI] [PubMed] [Google Scholar]
- Coyle JE, Qamar S, Rajashankar KR, Nikolov DB. Structure of GABARAP in two conformations: implications for GABA(A) receptor localization and tubulin binding. Neuron. 2002;33:63–74. doi: 10.1016/s0896-6273(01)00558-x. [DOI] [PubMed] [Google Scholar]
- Curmi JP, Premkumar LS, Birnir B, Gage PW. The influence of membrane potential on chloride channels activated by GABA in rat cultured hippocampal neurons. J Membr Biol. 1993;136:273–280. doi: 10.1007/BF00233666. [DOI] [PubMed] [Google Scholar]
- DeSouza S, Fu J, States BA, Ziff EB. Differential palmitoylation directs the AMPA receptor-binding protein ABP to spines or to intracellular clusters. J Neurosci. 2002;22:3493–3503. doi: 10.1523/JNEUROSCI.22-09-03493.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Craven SE, Chetkovich DM, Firestein BL, Schnell E, Aoki C, Bredt DS. Dual palmitoylation of PSD-95 mediates its vesiculotubular sorting, postsynaptic targeting, and ion channel clustering. J Cell Biol. 2000;148:159–172. doi: 10.1083/jcb.148.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt AB, Luu T, Cromer B, Tierney ML, Birnir B, Olsen RW, Gage PW. Conductance of recombinant GABA (A) channels is increased in cells co-expressing GABA(A) receptor-associated protein. J Biol Chem. 2004;279:21701–21706. doi: 10.1074/jbc.M312806200. [DOI] [PubMed] [Google Scholar]
- Fatima-Shad K, Barry PH. A patch-clamp study of GABA- and strychnine-sensitive glycine-activated currents in post-natal tissue-cultured hippocampal neurons. Proc Biol Sci. 1992;250:99–105. doi: 10.1098/rspb.1992.0136. [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Biochemistry. All in the ubiquitin family. Science. 2000;289:563–564. doi: 10.1126/science.289.5479.563. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Jin W, Ge WP, Xu J, Cao M, Peng L, Yung W, Liao D, Duan S, Zhang M, Xia J. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J Neurosci. 2006;26:2380–2390. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoe-Pognetto M, Luscher B. The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J Neurosci. 2004;24:5881–5891. doi: 10.1523/JNEUROSCI.1037-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P, Toomre D, Diaz E, White J, Simons K. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat Cell Biol. 2001;3:140–149. doi: 10.1038/35055042. [DOI] [PubMed] [Google Scholar]
- Kittler JT, Rostaing P, Schiavo G, Fritschy JM, Olsen R, Triller A, Moss SJ. The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABA(A) receptors. Mol Cell Neurosci. 2001;18:13–25. doi: 10.1006/mcne.2001.1005. [DOI] [PubMed] [Google Scholar]
- Kittler JT, Thomas P, Tretter V, Bogdanov YD, Haucke V, Smart TG, Moss SJ. Huntingtin-associated protein 1 regulates inhibitory synaptic transmission by modulating gamma-aminobutyric acid type A receptor membrane trafficking. Proc Natl Acad Sci USA. 2004;101:12736–12741. doi: 10.1073/pnas.0401860101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneussel M, Haverkamp S, Fuhrmann JC, Wang H, Wassle H, Olsen RW, Betz H. The gamma-aminobutyric acid type A receptor (GABAAR)-associated protein GABARAP interacts with gephyrin but is not involved in receptor anchoring at the synapse. Proc Natl Acad Sci USA. 2000;97:8594–8599. doi: 10.1073/pnas.97.15.8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight D, Harris R, McAlister MS, Phelan JP, Geddes S, Moss SJ, Driscoll PC, Keep NH. The X-ray crystal structure and putative ligand-derived peptide binding properties of gamma-aminobutyric acid receptor type A receptor-associated protein. J Biol Chem. 2002;277:5556–5561. doi: 10.1074/jbc.M109753200. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Tanida I, Ueno T, Ohsumi M, Ohsumi Y, Kominami E. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1–E2 complex formation. J Biol Chem. 2001;276:9846–9854. doi: 10.1074/jbc.M007737200. [DOI] [PubMed] [Google Scholar]
- Legesse-Miller A, Sagiv Y, Porat A, Elazar Z. Isolation and characterization of a novel low molecular weight protein involved in intra-Golgi traffic. J Biol Chem. 1998;273:3105–3109. doi: 10.1074/jbc.273.5.3105. [DOI] [PubMed] [Google Scholar]
- Leil TA, Chen ZW, Chang CS, Olsen RW. GABAA receptor-associated protein traffics GABAA receptors to the plasma membrane in neurons. J Neurosci. 2004;24:11429–11438. doi: 10.1523/JNEUROSCI.3355-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald RL, Olsen RW. GABAA receptor channels. Annu Rev Neurosci. 1994;17:569–602. doi: 10.1146/annurev.ne.17.030194.003033. [DOI] [PubMed] [Google Scholar]
- Mansuy V, Boireau W, Fraichard A, Schlick JL, Jouvenot M, Delage-Mourroux R. GEC1, a protein related to GABARAP, interacts with tubulin and GABA(A) receptor. Biochem Biophys Res Commun. 2004;325:639–648. doi: 10.1016/j.bbrc.2004.10.072. [DOI] [PubMed] [Google Scholar]
- Muller JM, Shorter J, Newman R, Deinhardt K, Sagiv Y, Elazar Z, Warren G, Shima DT. Sequential SNARE disassembly and GATE-16-GOS-28 complex assembly mediated by distinct NSF activities drives Golgi membrane fusion. J Cell Biol. 2002;157:1161–1173. doi: 10.1083/jcb.200202082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- Nymann-Andersen J, Wang H, Olsen RW. Biochemical identification of the binding domain in the GABA(A) receptor-associated protein (GABARAP) mediating dimer formation. Neuropharmacology. 2002;43:476–481. doi: 10.1016/s0028-3908(02)00165-x. [DOI] [PubMed] [Google Scholar]
- O'Sullivan GA, Kneussel M, Elazar Z, Betz H. GABARAP is not essential for GABA receptor targeting to the synapse. Eur J Neurosci. 2005;22:2644–2648. doi: 10.1111/j.1460-9568.2005.04448.x. [DOI] [PubMed] [Google Scholar]
- Raju R, Rakocevic G, Chen Z, Hoehn G, Semino-Mora C, Shi W, Olsen R, Dalakas MC. Autoimmunity to GABAA-receptor-associated protein in stiff-person syndrome. Brain. 2006;129:3270–3276. doi: 10.1093/brain/awl245. [DOI] [PubMed] [Google Scholar]
- Rothman JE. Intracellular membrane fusion. Adv Second Messenger Phosphoprotein Res. 1994;29:81–96. doi: 10.1016/s1040-7952(06)80008-x. [DOI] [PubMed] [Google Scholar]
- Sagiv Y, Legesse-Miller A, Porat A, Elazar Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 2000;19:1494–1504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Sagiv Y, Shorer H, Elazar Z. The COOH terminus of GATE-16, an intra-Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J Biol Chem. 2003;278:14053–14058. doi: 10.1074/jbc.M212108200. [DOI] [PubMed] [Google Scholar]
- Stangler T, Mayr LM, Willbold D. Solution structure of human GABA(A) receptor-associated protein GABARAP: implications for biological function and its regulation. J Biol Chem. 2002;277:13363–13366. doi: 10.1074/jbc.C200050200. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Tanida I, Komatsu M, Ueno T, Kominami E. GATE-16 and GABARAP are authentic modifiers mediated by Apg7 and Apg3. Biochem Biophys Res Commun. 2003;300:637–644. doi: 10.1016/s0006-291x(02)02907-8. [DOI] [PubMed] [Google Scholar]
- Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- Wallner M, Hanchar HJ, Olsen RW. Ethanol enhances alpha 4 beta 3 delta and alpha 6 beta 3 delta gamma-aminobutyric acid type A receptors at low concentrations known to affect humans. Proc Natl Acad Sci USA. 2003;100:15218–15223. doi: 10.1073/pnas.2435171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Olsen RW. Binding of the GABA(A) receptor-associated protein (GABARAP) to microtubules and microfilaments suggests involvement of the cytoskeleton in GABARAPGABA(A) receptor interaction. J Neurochem. 2000;75:644–655. doi: 10.1046/j.1471-4159.2000.0750644.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW. GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature. 1999;397:69–72. doi: 10.1038/16264. [DOI] [PubMed] [Google Scholar]
- Yamazaki-Sato H, Tanida I, Ueno T, Kominami E. The carboxyl terminal 17 amino acids within Apg7 are essential for Apg8 lipidation, but not for Apg12 conjugation. FEBS Lett. 2003;551:71–77. doi: 10.1016/s0014-5793(03)00899-8. [DOI] [PubMed] [Google Scholar]