Abstract
Neurofibromatosis type 1 (NF1) is a dominant genetic disorder that causes tumors of the peripheral nervous system. In addition, >40% of afflicted children have learning difficulties. The NF1 protein contains a highly conserved GTPase-activating protein domain that inhibits Ras activity, and the C-terminal region regulates cAMP levels via G-protein-dependent activation of adenylyl cyclase. Behavioral analysis indicates that learning is disrupted in both Drosophila and mouse NF1 models. Our previous work has shown that defective cAMP signaling leads to the learning phenotype in Drosophila Nf1 mutants. In the present report, our experiments showed that in addition to learning, long-term memory was also abolished in Nf1 mutants. However, altered NF1-regulated Ras activity is responsible for this defect rather than altered cAMP levels. Furthermore, by expressing clinically relevant human NF1 mutations and deletions in Drosophila Nf1-null mutants, we demonstrated that the GAP-related domain of NF1 was necessary and sufficient for long-term memory, whereas the C-terminal domain of NF1 was essential for immediate memory. Thus, we show that two separate functional domains of the same protein can participate independently in the formation of two distinct memory components.
Keywords: neurofibromatosis type 1, long-term memory, learning, Drosophila, cognitive disorder, human disease
Introduction
Neurofibromatosis type 1 (NF1) is one of the most common neurogenetic disorders with a prevalence of 1 in 3500 (Stephens et al., 1987). NF1 is predominantly identified by neurofibromas, benign tumors of the peripheral nervous system, as well as malignant peripheral nerve sheath tumors (Stephens et al., 1987). Learning disabilities are commonly observed in 30–60% of afflicted children (North, 2000). The NF1 protein has a central GAP-related domain (GRD) that accelerates inactivation of Ras (Ballester et al., 1990). Although no direct correlation has been established between specific mutations and phenotypes, a missense mutation that abolishes the Ras-GAP function of NF1 was found in human patients with multiple symptoms including learning disability and mental retardation, suggesting that loss of the GAP function of NF1 may underlie cognitive dysfunction (Klose et al., 1998). In addition to regulating Ras activity, NF1 has been shown to regulate cAMP levels in both Drosophila and mouse models (Guo et al., 1997, 2000; The et al., 1997; Tong et al., 2002; Dasgupta et al., 2003; Hannan et al., 2006). Interestingly, although no specific region of the protein has been associated with any NF1 disease phenotypes (Fahsold et al., 2000; Messiaen et al., 2000; Mattocks et al., 2004), our recent report demonstrated that the GRD is sufficient for mediating Ras-dependent regulation of signal transduction pathways, whereas the C-terminal region is required for G-protein-dependent adenylyl cyclase (AC) activation (Hannan et al., 2006).
In Drosophila, Nf1-null mutants exhibit compromised learning, or immediate memory, in the Pavlovian olfactory conditioning paradigm. This behavioral phenotype is attributed to disruption in the rutabaga-encoded adenylyl cyclase pathway (Guo et al., 2000). In the Morris water maze, Nf1 +/− mice exhibit a spatial learning defect that is resulting from increased Ras activity (Costa et al., 2001, 2002; Li et al., 2005). Such discrepancy is likely caused by the vast temporal difference between the two training paradigms. It only takes minutes to train and test flies (Tully and Quinn, 1985), whereas for mice, it takes two training sessions per day and 6 d to complete the training (Morris, 1984). In addition, injection of a protein synthesis inhibitor to the lateral ventricle of the mice significantly reduces their performance in the water maze (Meiri and Rosenblum, 1998). This suggests that the behavioral phenotype exhibited by the Nf1 +/− mice may actually be a form of long-lasting memory that requires repetitive training sessions and is dependent on protein synthesis. In this report, we demonstrated that Nf1 mutant flies also exhibit abolished long-term memory (LTM). Expressing the highly conserved human NF1 (hNF1) protein in Nf1-null mutant flies, including variants containing clinically relevant missense mutations as well as large deletions, allowed us to identify the structural and/or functional requisites for these behaviors. Our analyses revealed that the GRD is required for LTM, whereas sequences in the C-terminal region regulate immediate memory.
Materials and Methods
Fly stocks.
Flies were raised at room temperature (22 to 24°C) on standard cornmeal medium. The Nf1 mutants, Nf1P1 and Nf1P2, together with the parental K33 line were obtained from A. Bernards (Massachusetts General Hospital, Boston, MA). The Gal4 driver line, elav-Gal4;Nf1P1 (Williams et al., 2001), was obtained from A. Sehgal (University of Pennsylvania, Philadelphia, PA). Construction of UAS-hNF1 transgenes and generation of transgenic fly lines carrying normal hNF1 and human NF1 point mutants and deletion mutants were described previously (Hannan et al., 2006). Transcription of UAS-hNF1 transgenes in flies was controlled using a nervous system specific X chromosome line, elav-Gal4 (see above). The crossing schemes designed to generate progeny carrying one copy of the transgene and one copy of the Gal4 driver in the Nf1 mutant background are outlined (see Fig. 2 A).
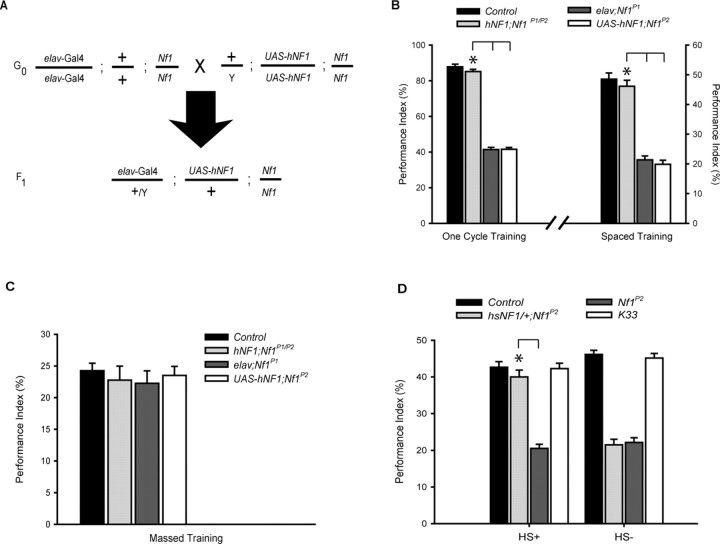
Figure 2.
Rescue of learning and LTM defects by expressing hNF1 as well as heat shock NF1 (hsNF1) transgene in Nf1-null mutants. A, Crosses performed to generate F1 progeny expressing UAS-hNF1 constructs under the control of the pan-neuronal elav-Gal4 driver. B, Rescue of learning and LTM by expressing hNF1 transgene in Nf1-null mutants. Transgenic flies expressing hNF1 pan-neuronally (elav/+/Y;UAS-hNF1/+;Nf1P1/P2) exhibit significant increases (*p < 0.001) in both learning (left) and LTM (right) from parental lines (elav;Nf1P1 and UAS_hNF1;Nf1P2). The wild-type control is 2202u, an isogenic line from which transgenic parental lines were generated (Hannan et al., 2006). C, Normal ARM performance in all transgenic lines. None of the transgenes show any nonspecific effect on ARM (n = 4 PIs per group). D, Acute expression of NF1 rescues LTM. Heat shock-induced expression of NF1 (hsNF1/+;Nf1P2) before spaced training significantly rescues (*p < 0.001) the LTM defect found in Nf1 mutants when compared with both 2202u (control) and K33 wild-type flies. This indicates the importance of NF1 in LTM formation. HS+, raised at 18°C and shifted to 30°C for 30 min 2 h before training; HS−, no heat shock treatment. PI scores are expressed as mean ± SEM, n = 8 unless otherwise indicated.
One-cycle training.
Flies were trained and tested with the classical (Pavlovian) conditioning protocol of Tully and Quinn (1985). Briefly, ∼100 flies were trapped in a training chamber that is lined with an electrifiable copper grid. Two odors were then delivered to the flies sequentially through air current, with the first odor (conditioned stimulus; CS+) delivery paired with electric shock (unconditioned stimulus), but no shock was received with the delivery of the second odor (CS−). Each odor was delivered in an interval of 1 min, with 45 s of fresh air after the delivery of each odor. This procedure constituted one training cycle. To test for learning, flies were transferred to a choice point where the two odors were presented to them by two converging air currents. Flies were given 120 s to choose between the two arms of the T-maze from which the odors were delivered. At the end of this period, flies were trapped inside individual arms, anesthetized, and counted. To eliminate odor bias, the concentrations of the two odors, which are aversive to untrained flies, were calibrated such that untrained flies distributed themselves 50:50 in the T-maze.
Performance index.
Two groups of flies were always trained and tested in one complete experiment; for one group, methylcyclohexanol (MCH) was CS+ and benzaldehyde (BA) was CS−, whereas for the second group BA was CS+ and MCH was CS−. The “probability correct” of each reciprocal group was calculated as the number of flies avoiding CS+ minus those avoiding CS− divided by the total number of flies in the T-maze arms. The resulting two probability corrects are then averaged and normalized to become one performance index (PI), which can range from 0 (a 50:50 distribution reflecting no learning) to 100 (all flies learned to avoid shock-paired odor). All statistical analyses in this study were performed using the paired Student's t test.
Long-term memory.
This training paradigm is in accordance to a previous report (Yin et al., 1994). Extended training procedures were performed with an automated training system in which fresh air was bubbled at 750 ml/min through one of the three channels in a “bubbler manifold” (custom built by General Valve, Fairfield, NJ). One channel was for “fresh” air, a second was for BA, and the third was for MCH. Each channel contained two vials, one with 10 ml of distilled water and the other with either pure heavy mineral oil (Fisher Scientific, Houston, TX) alone or with a particular dilution of BA or MCH (Fluka, Neu-Ulm, Germany). Switching of bubbler channels and of a relay to deliver electric shock pulses to the flies was computer controlled (system custom designed by Island Motion, Tappan, NY). During massed training, flies received 10 training cycles (as above) delivered one right after the other. For spaced training, flies received 10 training cycles with a 15 min rest interval between each cycle. To assay memory retention, flies were tapped gently from the training chamber into their usual food vials and stored at 18°C for the duration of 24 h. Flies were then transferred to the choice point of the T-maze where the usual 2 min test trial was performed.
Cycloheximide feeding and heat shock treatment.
The cycloheximide (CXM) feeding regimen was as reported previously (Yin et al., 1994). Briefly, groups of ∼100 flies were placed in feeding tubes that contained one Whatman (Maidstone, UK) filter paper strip soaked with 125 μl of solution mixture. Solution mixture contained 35 mm (CXM+) in 4% sucrose or 4% sucrose (CXM−) and was fed to the flies at 25°C for 12–15 h before training and again at 18°C during the 24 h retention period. Flies were allowed to clean themselves in standard food vials 30 min before training.
The heat shock protocol is similar to that reported previously (Guo et al., 2000). Heterozygous transgenic flies (hsNF1/+;Nf1P2) were used to avoid potential recessive effects of the insertion on behavior. Flies were raised at 18°C and moved to 30°C for 30 min. After a 2 h resting period at room temperature (20–24°C), flies were subjected to spaced training and tested 24 h later at 25°C.
Olfactory acuity and shock reactivity.
Odor-avoidance responses to BA or MCH were quantified with the method of Boynton and Tully (1992). Briefly, groups of ∼100 untrained flies received a 2 min test trial in the T-maze. Different groups were given a choice between either BA or MCH versus fresh room air. PIs were calculated as above. Shock-avoidance responses to 60 V were quantified with the method of Dura et al. (1993). Briefly, groups of ∼100 untrained flies received a 2 min test trial in the T-maze. Each arm of the T-maze contained an electric shock grid, and different groups of flies were given a choice between shock versus no shock. PIs were calculated as above. Both olfactory acuity and shock reactivity were normal for all genotypes (Table 1).
Table 1.
Performance indexes for shock reactivity and olfactory avoidance
| Genotypes | Shock Reactivity (60 V) | Odor avoidance |
|
|---|---|---|---|
| BA | MCH | ||
| 2202u | 85 ± 3 | 79 ± 3 | 82 ± 3 |
| K33 | 78 ± 3 | 79 ± 3 | 77 ± 4 |
| elav/+/Y;UAS-hNF1/+;Nf1 P1/P2 | 83 ± 2 | 80 ± 2 | 80 ± 3 |
| Nf1 P1 | 79 ± 2 | 76 ± 2 | 76 ± 3 |
| Nf1 P2 | 79 ± 3 | 77 ± 2 | 77 ± 3 |
| elav/+/Y;UAS-GRD1;Nf1 P1/P2 | 83 ± 4 | 79 ± 3 | 83 ± 2 |
| elav/+/Y;UAS-GRD2;Nf1 P1/P2 | 81 ± 2 | 79 ± 2 | 82 ± 3 |
| elav/+/Y;UAS-GRDdel;Nf1 P1/P2 | 80 ± 3 | 79 ± 3 | 78 ± 2 |
| elav/+/Y;UAS-R1276P;Nf1 P1/P2 | 80 ± 2 | 80 ± 3 | 77 ± 2 |
| elav/+/Y;UAS-R1391S;Nf1 P1/P2 | 81 ± 2 | 80 ± 3 | 81 ± 2 |
| elav/+/Y;UAS-K1423E;Nf1 P1/P2 | 82 ± 2 | 81 ± 3 | 80 ± 4 |
| elav/+/Y;UAS-Nterm;Nf1 P1/P2 | 81 ± 3 | 79 ± 3 | 80 ± 3 |
| elav/+/Y;UAS-Cterm;Nf1 P1/P2 | 79 ± 2 | 80 ± 3 | 78 ± 4 |
All wild-type, transgenic, and mutants flies used in this study have normal responses to aversive odors and electric shocks. All scores are expressed as mean PI ± SEM. For all shock reactivity and odor avoidance assays, n = 4. No statistical difference at the level of α = 0.05 is detected for sensorimotor activities and odor avoidance.
Results
Expression of human NF1 transgene in Nf1-null mutants can rescue immediate memory and LTM defects
To dissect the long-term memory phenotype of Nf1 mutants, we subjected flies to massed (10 cycles with no rest interval) or spaced (10 training cycles with 15 min rest intervals) training protocols before we tested for their memory 24 h later (see Materials and Methods). At the time of testing, spaced-trained flies exhibit two memory components, anesthesia-resistant memory (ARM) and LTM. LTM is protein-synthesis dependent whereas ARM is not. However, flies that received massed training will only exhibit ARM (Tully et al., 1994; Dubnau and Tully, 1998). In our analyses, mutants that are defective in LTM but exhibit normal ARM performance will be categorized as LTM mutants. All flies in this study are able to detect odors and shock (see Table 1).
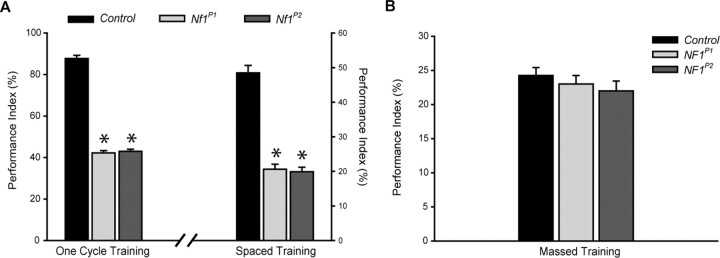
Two Nf1-null mutants were used in this study, Nf1P1 and Nf1P2, neither of which showed any detectable NF1 protein expression and both of which are defective in olfactory associative learning (The et al., 1997; Guo et al., 2000). K33 flies, the parental line of the Nf1 mutants, were used as a wild-type control. We first confirmed the Nf1 mutant learning phenotype by testing them immediately after one cycle of training (see Materials and Methods). Consistent with our previous report (Guo et al., 2000), these mutants exhibit significantly lower learning performance when compared with wild-type control flies (Fig. 1 A). Both Nf1-null mutants also display compromised 24 h memory after spaced training compared with the parental line (Fig. 1 A), whereas they exhibit normal ARM, measured 24 h after massed training (Fig. 1 B). This indicates that NF1 is specifically affecting the LTM component of 24 h memory, in addition to its effect on learning.
Figure 1.
Learning and LTM defects, but normal ARM in Nf1-null mutants. A, Learning and LTM defects in Nf1 mutants. Compared with the K33 (control) parental group, the Nf1P1 and Nf1P2 mutants display significantly lower performance (*p < 0.001) in learning (one cycle training) and in LTM (spaced training; for details, refer to Materials and Methods). B, Normal ARM performance in Nf1 mutants. Memory performance was tested 24 h after massed training. Nf1 mutants perform similar to the parental K33 (control) flies after massed training. This indicates that the 24 h memory defect observed in these mutants is in fact an LTM defect because ARM is normal. PI scores are expressed as mean ± SEM, n = 8.
The Drosophila NF1 protein has 60% identity with the human NF1 ortholog (The et al., 1997), and previous experiments show that the human protein can function in place of the fly protein to rescue body size and stimulation of AC activity (Tong et al., 2002; Hannan et al., 2006). Amino acid residues that are normally conserved between the two species are found mutated in NF1 patient samples, suggesting their potential functional significance in the fly (Hannan et al., 2006). We hypothesized that the human protein would be able to rescue the behavioral phenotypes encountered in Nf1 mutants. To examine whether hNF1 can rescue fly Nf1 mutant behavioral phenotypes, we expressed the hNF1 protein in the null mutant background using the elav-GAL4 driver, which has a pan-neuronal expression pattern (for crossing scheme, see Fig. 2 A). The transgenic parental lines, elav;Nf1P1 and UAS-hNF1;Nf1P2, were generated using an isogenic line 2202u, which displays similar LTM performance to K33 (Fig. 2 D). The 2202u line is used as the wild-type control in Figure 2 and Figure 4. When compared with the parental control lines (elav;Nf1P1 and UAS-hNF1;Nf1P2), the expression of hNF1 in the hNF1;Nf1P1/P2 progeny (elav/+/Y;UAS-hNF1/+;Nf1P1/P2) significantly rescued both learning and LTM to wild-type level (Fig. 2 B) and also retained normal level of ARM (Fig. 2 C). Thus, human NF1 is also conserved for behavioral function with the Drosophila ortholog. The rescue of LTM by hNF1 suggests that NF1 is essential for the formation of LTM, in addition to its established role in learning.
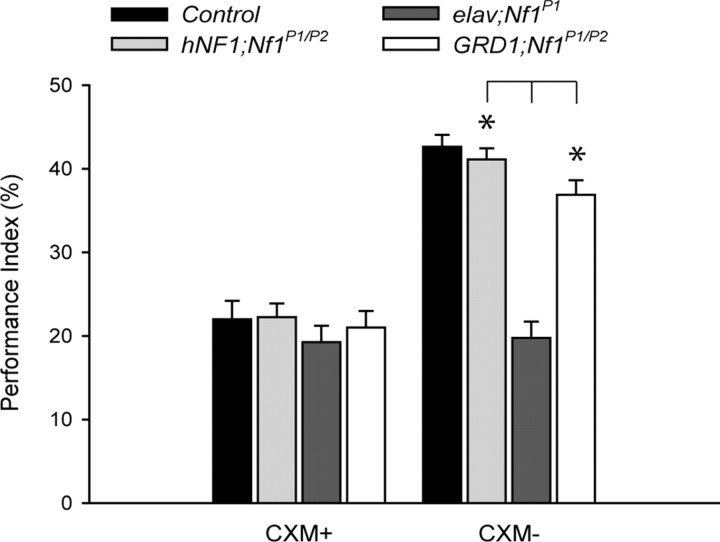
Figure 4.
CXM abolished LTM performance in wild-type and hNF1 transgenic flies. CXM, a protein synthesis inhibitor, was fed to the wild-type, hNF1, and GRD1 flies before spaced training and again during the 24 h retention period (see Materials and Methods). For the CXM-treated group (CXM+), LTM for wild-type control, hNF1 and GRD1 flies were reduced to Nf1P1 mutant levels. However, when hNF1 and GRD1 flies were treated with vehicle, they still displayed significant rescue compared with the elav;Nf1P1 parental control (right; *p < 0.001). These results indicate that NF1 is indeed important for the formation of protein synthesis-dependent memory and that the GRD fragment specifically rescues LTM after spaced training. PI scores are expressed as mean ± SEM, n = 8; p < 0.001.
To rule out any developmental abnormalities that may contribute to the LTM defect observed in Nf1 mutants, we used a heat shock promoter to induce expression of the Drosophila Nf1 gene in the Nf1P2 mutant background by temperature shifting before training (see Materials and Methods). According to our previous study, this heat shock-induced expression was enough to rescue the learning phenotype (Guo et al., 2000). Acute expression of the Nf1 gene before spaced training significantly rescued the LTM defect in the Nf1P2 mutant background (Fig. 2 D). These results indicate that NF1 is required acutely for the formation of LTM.
The GRD region of NF1 is required for its function in LTM
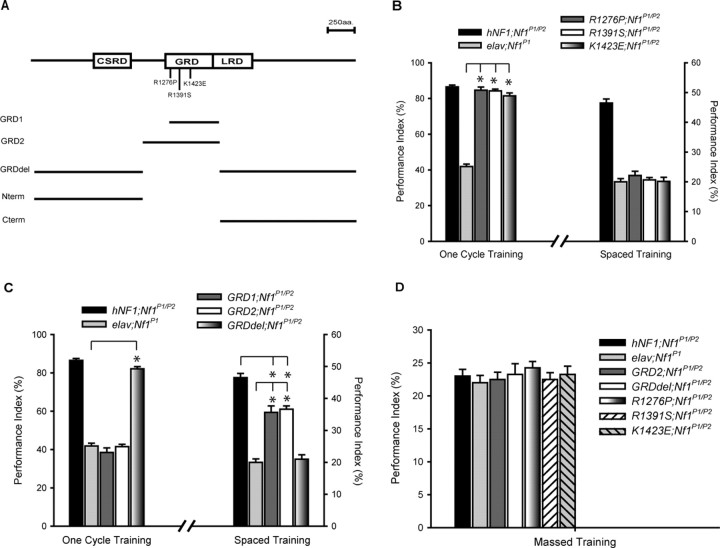
To gain insights into the underlying mechanisms of the LTM phenotype, we examined various point mutations observed in NF1 patients that selectively disrupt NF1-regulated signal transduction pathways (Fig. 3 A). Two of the clinically identified hNF1 mutations, R1391S and K1423E, exhibit greatly reduced affinity for Ras (Gutmann et al., 1993; Poullet et al., 1994), whereas R1276P has >8000-fold reduced GAP activity compared with wild-type NF1 protein (Klose et al., 1998). Flies expressing any of the three hNF1 point mutations (elav/+/Y;UAS-R1276P/+;Nf1P1/P2; elav/+/Y;UAS-R1391S/+;Nf1P1/P2; elav/+/Y;UAS-K1423E/+;Nf1P1/P2) display normal learning (Fig. 3 B) and ARM (Fig. 3 D) but defective LTM performance (Fig. 3 B). This suggests that the GAP activity of NF1 as well as its interaction with the Ras protein is extremely important for NF1-dependent LTM.
Figure 3.
The GRD domain and GAP activity are necessary and sufficient for LTM formation, whereas NF1 without the GRD domain rescues learning. A, Positions of three hNF1 missense mutations and size of five hNF1 deletion constructs that have been expressed in Drosophila Nf1-null mutants. Refer to Results for a detailed description of these mutants. CSRD, Cys-Ser rich domain; LRD, Leu-rich domain; GRD1 and GRD2, GRD fragments of different sizes; GRDdel, NF1 protein with the GRD domain deleted; Nterm, N-terminal fragment. B, GRD point mutations restore learning to wild-type level but fail to rescue LTM. The three GRD point mutations are able to significantly rescue (*p < 0.001) the learning defect in the Nf1 mutant (elav;Nf1P1) to the same extent as the full-length human NF1 transgene. However, the three point mutations are not able to rescue the LTM defect of Nf1 mutants (right). C, Rescue of LTM but not learning by GRD fragments. Flies expressing GRDdel significantly rescue (*p < 0.001) learning to the wild-type level, whereas flies expressing the GRD fragments, GRD1 and GRD2, do not rescue learning (left). Mutant flies expressing both GRD fragments exhibit partial yet significant rescue (*p < 0.001) of LTM compared with the Nf1 mutant (right). When compared with flies expressing full-length hNF1 transgene, mutants expressing the GRD fragments are significantly lower in LTM performance (*p < 0.001), indicating only partial rescue of LTM. In contrast, mutants expressing the GRD-deleted protein show no rescue of LTM (right). D, Normal ARM performance in wild-type and mutant transgenic lines. None of the transgenes shows any nonspecific effect on ARM (n = 4 PIs per group), indicating that NF1 is only involved in LTM. PI scores are expressed as mean ± SEM, n = 8 unless otherwise indicated.
To further evaluate the importance of the GRD for the NF1 behavioral phenotypes, we generated transgenic flies expressing hNF1 protein fragments of different sizes that contain the GRD, named GRD1 (elav/+/Y;UAS-GRD1;Nf1P1/P2) and GRD2 (elav/+/Y;UAS-GRD2;Nf1P1/P2), as well as an hNF1 protein that has a deletion of the GRD, named GRDdel (elav/+/Y;UAS-GRDdel;Nf1P1/P2) (Fig. 3 A). Expression of GRDdel rescues learning of Nf1 mutant flies to wild-type level, whereas both GRD1 and GRD2 flies (Fig. 3 C) show similar learning performance to Nf1-null mutants (Fig. 1 A), suggesting that the GRD is not important for NF1-dependent learning. We also tested GRDdel, GRD1, and GRD2 flies for 24 h memory after spaced training. LTM is rescued, although partially, in flies expressing GRD1 and GRD2 but not GRDdel (Fig. 3 C), whereas ARM is not affected by any of the fragments tested (Fig. 3 D). This verifies that the GRD fragment is indeed functional for behavioral rescue and also illustrates that the GRD is an essential region for NF1-dependent LTM.
To verify that rescue of 24 h memory after spaced training by hNF1 or the GRD fragment is indeed rescue of the LTM defect, we fed flies with a protein synthesis inhibitor, CXM, before and after spaced training (see Materials and Methods). As mentioned above, LTM is protein synthesis-dependent whereas ARM is not. Therefore, LTM should be sensitive to CXM treatment, as shown previously (Tully et al., 1994; Yin et al., 1994). Twenty-four hour memory performance was compromised when wild-type control flies, and flies expressing hNF1 (hNF1;Nf1P1/P2) or GRD fragment alone (GRD1;Nf1P1/P2), were subjected to CXM treatment (Fig. 4). This indicates that NF1, and especially the GRD, is indeed required for protein synthesis-dependent memory, LTM.
The C-terminal region of NF1 is essential for learning
Because expression of the GRDdel fragment rescues learning as shown above (Fig. 3 C), we hypothesized that regions important for NF1-dependent learning lie outside of the GRD. Two different truncated hNF1 transgenes were used to test this hypothesis; the N-terminal (elav/+/Y;UAS-Nterm;Nf1P1/P2) construct contains regions upstream of the GRD, whereas the C-terminal (Cterm; elav/+/Y;UAS-Cterm;Nf1P1/P2) construct contains regions downstream of the GRD (Fig. 3 A). Biochemical assays indicate that Cterm is functional for NF1-dependent neuropeptide and neurotransmitter stimulation of AC activity (Hannan et al., 2006). The Cterm fragment also rescues the cAMP-dependent Nf1 mutant body size defect, whereas the N-terminal region and the GRD do not rescue body size (Hannan et al., 2006). Neither transgene was able to rescue the LTM defect in the null mutant background (Fig. 5), consistent with the absence of the GRD region in these constructs. The Cterm fragment, however, rescues learning significantly (Fig. 5), suggesting that elements within this region are crucial for NF1 to mediate learning. Together, these data indicate that the different structural/functional relationships revealed by biochemical assays in our previous study (Hannan et al., 2006) also have a correspondingly distinct effect on the role of NF1 in different phases of learning and memory.
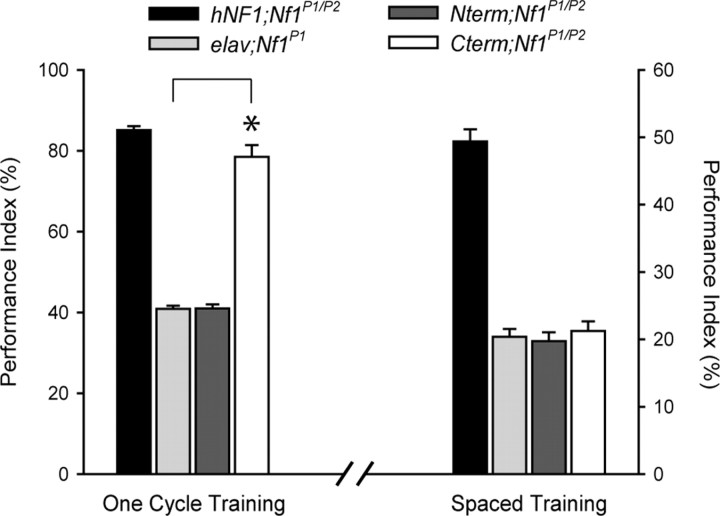
Figure 5.
Rescue of learning by Cterm fragment. Flies expressing Cterm (see Fig. 3 A) exhibit complete rescue of learning compared with the wild-type transgene (*p < 0.001), whereas the N-terminal fragment (Nterm) (see Fig. 3 A) has no effect on the learning score (left). Both fragments are unable to restore LTM performance (right). These data indicate region-specific functionality of the NF1 protein for distinct memory phases (i.e., the GRD is required for LTM) and the C-terminal is essential for learning. PI scores are expressed as mean ± SEM, n = 8.
Discussion
In this study, we have dissected the functional significance of two NF1 protein regions using the Pavlovian olfactory conditioning paradigm in Drosophila. The C-terminal region contains sequences that are essential for immediate memory, whereas the GRD is required for LTM formation. These two regions also possess distinct biochemical properties by which they individually mediate different signaling pathways (Hannan et al., 2006). These unique properties of NF1 suggest that different signal transduction pathways contribute to distinct phases of memory.
The Morris water maze, for testing spatial learning performance in mice, requires the subject to find a platform submerged under water by using spatial cues in the environment. This task requires two training sessions per day and, in the case of Nf1 +/− mice, 6 d to complete the training regimen (Silva et al., 1997). The amount of time for this task is significantly longer than the 4 min required for training flies in the Pavlovian olfactory learning task (Tully and Quinn, 1985; Guo et al., 2000). In fact, the water maze paradigm is strikingly similar to the spaced training we used for LTM induction in flies, both of which have repetitive training as well as resting components. This similarity is indeed valid because both paradigms have been shown to produce protein synthesis-dependent memory (Tully et al., 1994; Meiri and Rosenblum, 1998). This study resolves the discrepancy of different pathways underlying the behavioral phenotypes exhibited by these two NF1 animal model systems. Our results indicate that different phases of memory were examined in previous reports.
According to earlier findings, the GRD deletion and point mutants used in this study are also defective in mediating growth factor-stimulated Ras-dependent AC activity (Hannan et al., 2006). The three GRD point mutants have been shown to be essential for the affinity of NF1 for Ras as well as GAP activity (Gutmann et al., 1993; Poullet et al., 1994; Klose et al., 1998). In the mammalian system, growth factor receptors have been demonstrated to be an essential component for the maintenance of long-term potentiation, an electrophysiological phenomenon suggested to be the underlying mechanism for learning and memory (Bramham and Messaoudi, 2005). Ras signaling has also been shown to play an important role in synaptic plasticity as well as learning and memory (Brambilla et al., 1997; Atkins et al., 1998). The epidermal growth factor receptor (EGFR) was shown to be important for Ras-mediated AC stimulation in our previous study (Hannan et al., 2006). The effects of the GRD point mutants on LTM suggest that the EGFR and Ras pathway may be an important mechanism for LTM in flies, as illustrated in our working model (Fig. 6). Additional experiments assaying the LTM performance of Ras and EGFR mutants will be needed to confirm this hypothesis.
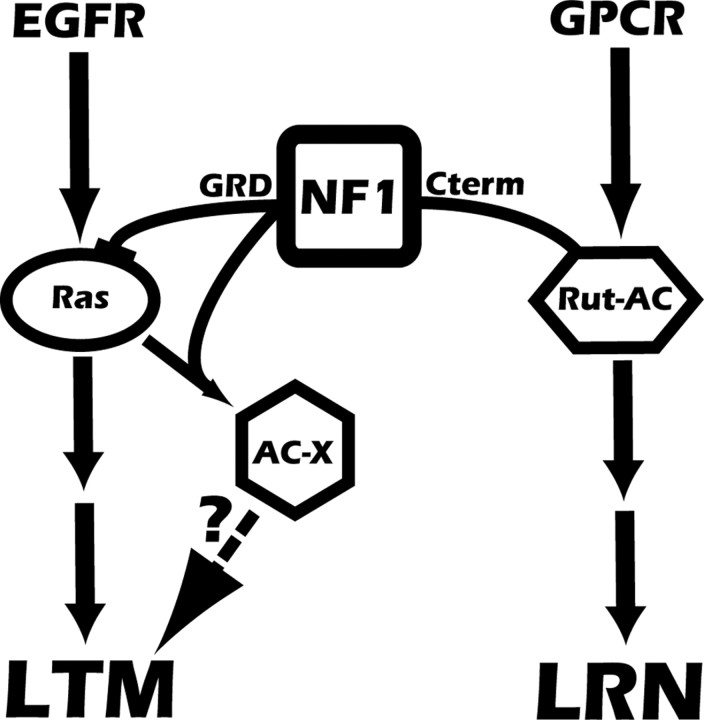
Figure 6.
Working model for regulation of distinct memory processes by different domains of NF1. In this model, two different signaling pathways underlie distinct phases of memory formation, which are both mediated by NF1. The GRD domain, with its GAP activity and interaction with the Ras protein, is necessary and sufficient for mediating EGFR signaling (Hannan et al., 2006) as well as LTM (Fig. 3 C). Thus, EGFR may be an essential signaling mechanism to mediate LTM in flies. Also shown is the synergistic stimulation of an unknown AC (AC-X) by NF1 and Ras proteins (Hannan et al., 2006). This AC-X may be the downstream target of Ras and NF1 governing LTM formation. The C-terminal of the NF1 protein has been shown to mediate G-protein signaling (Hannan et al., 2006) and is essential to regulate learning or immediate memory (Fig. 5). Therefore, signaling molecules, such as serotonin and histamine, whose downstream signaling pathways are mediated by NF1 (Hannan et al., 2006), may be important for learning or immediate memory. GPCR, G-protein coupled receptor; Rut-AC, rutabaga-encoded adenylyl cyclase; LRN, learning.
Combining our present behavioral data together with the former biochemical analysis (Hannan et al., 2006), we proposed a working model as shown in Figure 6. Two independent pathways are mediated by different regions of the NF1 protein. The C-terminal region controls the G-protein-dependent AC pathway, which can be stimulated by neurotransmitters such as serotonin and histamine (Hannan et al., 2006). This NF1-cAMP pathway is important for learning (Fig. 5) (Guo et al., 2000). The GRD region regulates Ras activity, which can be stimulated by growth factors, such as epidermal growth factor, to induce cAMP production (Hannan et al., 2006). This NF1-Ras pathway is essential for LTM formation. This requires normal GAP activity of NF1-GRD and interaction with the Ras protein (Fig. 3 B). Although our data showed that fragments containing the GRD can only partially rescue the LTM defect, this may be because of insufficient conformational support of the GRD fragments to fully restore wild-type function. The fact that deletion of the GRD from the NF1 protein (Fig. 3 C) eliminates the ability to rescue the LTM defect suggests the importance of the GRD in the role of NF1 in regulating LTM formation.
This report is the first step in gaining insight into the nature of the cognitive defects found in NF1 patients using the Drosophila model system. Interestingly, the NF1 protein presents a unique case of having distinct regions governing two independent steps of an important cognitive process. These NF1 protein regions that are involved in different phases of learning and memory contain different types of post-translational modification sites, such as phosphorylation sites for protein kinase A and protein kinase C (Mangoura et al., 2006), and binding sites for proteins such as syndecan (Hsueh et al., 2001). It will be interesting to investigate the role that these sites play in governing the behavioral outputs assayed in this report to find out the exact mechanisms and pathways that govern the distinct behaviors of learning and memory.
Footnotes
This work was supported by grants from the National Institutes of Health (2R01 NS34779-06) and DART Neuroscience to Y.Z. and the United States Army Neurofibromatosis Research Program Grants DAMD17-99-1-9500 (to F.H. and Y.Z.), W81XWH-04-1-025 (to F.H.), and W81XWH-05-1-0142 (to Y.Z.). We thank Andre Bernards for providing Nf1 mutant flies and human NF1 clones.
References
- Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- Ballester R, Marchuk D, Boguski M, Saulino A, Letcher R, Wigler M, Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990;63:851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Boynton S, Tully T. latheo, a new gene involved in associative learning and memory in Drosophila melanogaster, identified from P element mutagenesis. Genetics. 1992;131:655–672. doi: 10.1093/genetics/131.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla R, Gnesutta N, Minichiello L, White G, Roylance AJ, Herron CE, Ramsey M, Wolfer DP, Cestari V, Rossi-Arnaud C, Grant SG, Chapman PF, Lipp HP, Sturani E, Klein R. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Messaoudi E. BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog Neurobiol. 2005;76:99–125. doi: 10.1016/j.pneurobio.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Costa RM, Yang T, Huynh DP, Pulst SM, Viskochil DH, Silva AJ, Brannan CI. Learning deficits, but normal development and tumor predisposition, in mice lacking exon 23a of Nf1. Nat Genet. 2001;27:399–405. doi: 10.1038/86898. [DOI] [PubMed] [Google Scholar]
- Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- Dasgupta B, Dugan LL, Gutmann DH. The neurofibromatosis 1 gene product neurofibromin regulates pituitary adenylate cyclase-activating polypeptide-mediated signaling in astrocytes. J Neurosci. 2003;23:8949–8954. doi: 10.1523/JNEUROSCI.23-26-08949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, Tully T. Gene discovery in Drosophila: new insights for learning and memory. Annu Rev Neurosci. 1998;21:407–444. doi: 10.1146/annurev.neuro.21.1.407. [DOI] [PubMed] [Google Scholar]
- Dura JM, Preat T, Tully T. Identification of linotte, a new gene affecting learning and memory in Drosophila melanogaster . J Neurogenet. 1993;9:1–14. doi: 10.3109/01677069309167272. [DOI] [PubMed] [Google Scholar]
- Fahsold R, Hoffmeyer S, Mischung C, Gille C, Ehlers C, Kucukceylan N, Abdel-Nour M, Gewies A, Peters H, Kaufmann D, Buske A, Tinschert S, Nurnberg P. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am J Hum Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HF, The I, Hannan F, Bernards A, Zhong Y. Requirement of Drosophila NF1 for activation of adenylyl cyclase by PACAP38-like neuropeptides. Science. 1997;276:795–798. doi: 10.1126/science.276.5313.795. [DOI] [PubMed] [Google Scholar]
- Guo HF, Tong J, Hannan F, Luo L, Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila . Nature. 2000;403:895–898. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Boguski M, Marchuk D, Wigler M, Collins FS, Ballester R. Analysis of the neurofibromatosis type 1 (NF1) GAP-related domain by site-directed mutagenesis. Oncogene. 1993;8:761–769. [PubMed] [Google Scholar]
- Hannan F, Ho I, Tong JJ, Zhu Y, Nurnberg P, Zhong Y. Effect of neurofibromatosis type I mutations on a novel pathway for adenylyl cyclase activation requiring neurofibromin and Ras. Hum Mol Genet. 2006;15:1087–1098. doi: 10.1093/hmg/ddl023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Roberts AM, Volta M, Sheng M, Roberts RG. Bipartite interaction between neurofibromatosis type I protein (neurofibromin) and syndecan transmembrane heparan sulfate proteoglycans. J Neurosci. 2001;21:3764–3770. doi: 10.1523/JNEUROSCI.21-11-03764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose A, Ahmadian MR, Schuelke M, Scheffzek K, Hoffmeyer S, Gewies A, Schmitz F, Kaufmann D, Peters H, Wittinghofer A, Nurnberg P. Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum Mol Genet. 1998;7:1261–1268. doi: 10.1093/hmg/7.8.1261. [DOI] [PubMed] [Google Scholar]
- Li W, Cui Y, Kushner SA, Brown RA, Jentsch JD, Frankland PW, Cannon TD, Silva AJ. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15:1961–1967. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Mangoura D, Sun Y, Li C, Singh D, Gutmann DH, Flores A, Ahmed M, Vallianatos G. Phosphorylation of neurofibromin by PKC is a possible molecular switch in EGF receptor signaling in neural cells. Oncogene. 2006;25:735–745. doi: 10.1038/sj.onc.1209113. [DOI] [PubMed] [Google Scholar]
- Mattocks C, Baralle D, Tarpey P, ffrench-Constant C, Bobrow M, Whittaker J. Automated comparative sequence analysis identifies mutations in 89% of NF1 patients and confirms a mutation cluster in exons 11–17 distinct from the GAP related domain. J Med Genet. 2004;41:e48. doi: 10.1136/jmg.2003.011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri N, Rosenblum K. Lateral ventricle injection of the protein synthesis inhibitor anisomycin impairs long-term memory in a spatial memory task. Brain Res. 1998;789:48–55. doi: 10.1016/s0006-8993(97)01528-x. [DOI] [PubMed] [Google Scholar]
- Messiaen LM, Callens T, Mortier G, Beysen D, Vandenbroucke I, Van Roy N, Speleman F, Paepe AD. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Morris R. Developments of a water-maze procedure for studying spatial learning in the rat. J Neurosci Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- North K. Neurofibromatosis type 1. Am J Med Genet. 2000;97:119–127. doi: 10.1002/1096-8628(200022)97:2<119::aid-ajmg3>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Poullet P, Lin B, Esson K, Tamanoi F. Functional significance of lysine 1423 of neurofibromin and characterization of a second site suppressor which rescues mutations at this residue and suppresses RAS2Val-19-activated phenotypes. Mol Cell Biol. 1994;14:815–821. doi: 10.1128/mcb.14.1.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Frankland PW, Marowitz Z, Friedman E, Laszlo GS, Cioffi D, Jacks T, Bourtchuladze R. A mouse model for the learning and memory deficits associated with neurofibromatosis type I. Nat Genet. 1997;15:281–284. doi: 10.1038/ng0397-281. [DOI] [PubMed] [Google Scholar]
- Stephens K, Riccardi VM, Rising M, Ng S, Green P, Collins FS, Rediker KS, Powers JA, Parker C, Donis-Keller H. Linkage studies with chromosome 17 DNA markers in 45 neurofibromatosis 1 families. Genomics. 1987;1:353–357. doi: 10.1016/0888-7543(87)90037-1. [DOI] [PubMed] [Google Scholar]
- The I, Hannigan GE, Cowley GS, Reginald S, Zhong Y, Gusella JF, Hariharan IK, Bernards A. Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science. 1997;276:791–794. doi: 10.1126/science.276.5313.791. [DOI] [PubMed] [Google Scholar]
- Tong J, Hannan F, Zhu Y, Bernards A, Zhong Y. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat Neurosci. 2002;5:95–96. doi: 10.1038/nn792. [DOI] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster . J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila . Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–2256. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila . Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]